Abstract
A cDNA library enriched with Myc-responsive cDNAs but depleted of myc cDNAs was used in a functional screen for growth enhancement in c-myc-null cells. A cDNA clone for mitochondrial serine hydroxymethyltransferase (mSHMT) that was capable of partial complementation of the growth defects of c-myc-null cells was identified. Expression analysis and chromatin immunoprecipitation demonstrated that mSHMT is a direct Myc target gene. Furthermore, a separate gene encoding the cytoplasmic isoform of the same enzyme is also a direct target of Myc regulation. SHMT enzymes are the major source of the one-carbon unit required for folate metabolism and for the biosynthesis of nucleotides and amino acids. Our data establish a novel functional link between Myc and the regulation of cellular metabolism.
The importance of myc in oncogenesis has been recognized for over 20 years, starting with the isolation of v-myc oncogenes and the identification of c-myc as a site for retroviral insertion and chromosomal rearrangements. However, it has become increasingly clear that myc also plays a direct role in the control of normal cell proliferation, since the loss of myc expression is detrimental for normal animal development (9) and causes the arrest of growth of primary mouse fibroblasts (10). A somatic knockout of the c-myc genes in immortalized rat fibroblasts results in a remarkably stable prolongation of the cell cycle to nearly three times the length of that in the parental cells (25). Thus, an investigation of myc function is central to gaining a more general understanding of the regulation of cellular growth and the disruptions of growth networks that contribute to cancer.
Myc is a transcriptional factor that forms a heterodimer with Max through its helix-loop-helix-leucine zipper domain (15). The Myc/Max heterodimer binds to target genes at a consensus CACGTG motif and activates transcription at least in part by modifying the local chromatin through histone acetylation (4, 11, 26). Myc presumably controls cellular growth through the regulation of a set of target genes, although the identities of the sets of genes that control individual cellular phenotypes remain largely unknown (8). A large number of genes demonstrating a Myc-dependent pattern of expression have been identified recently through the use of differential screening and microarrays (3, 7, 16, 28, 35), but the direct participation of Myc in most gene regulation has not been verified. The most compelling evidence for direct Myc-dependent regulation is the ability to cross-link Myc to a specific chromosomal consensus binding site. Bona fide Myc target genes identified to date are involved in the control of cell proliferation (genes for cdk4, cyclin D2, Id2, Cul1, etc.) (5, 18, 23, 30) and in certain steps in the biosynthesis of nucleotides (CAD) and polyamines (ODC) (1, 27). The findings for CAD and ODC imply that in addition to controlling the cell cycle, Myc also regulates the growth of the cell. In support of this idea, the loss of myc gene function in Drosophila melanogaster results in decreased body sizes of adult flies, and overexpression of Myc results in an increased cell size without an increase in cell number (20). Furthermore, c-myc-null fibroblasts have reduced rates of protein synthesis (25), and overexpression of Myc stimulates expression of ribosomal proteins (3, 16). However, despite a growing number of Myc-responsive genes, the activities of Myc, especially the control of cell proliferation, have not been attributed in full to the transcriptional regulation of a particular gene or group of genes.
In the present paper, we report the isolation of a novel Myc target gene through functional screening of a myc-depleted cDNA library in c-myc-null rat fibroblasts. The gene encodes mitochondrial serine hydroxymethyltransferase (mSHMT), whose overexpression partially rescues the slow growth of c-myc-null fibroblasts. We demonstrate that the gene encoding cSHMT, the cytoplasmic isoform of mSHMT, is also a Myc-responsive gene, emphasizing the importance of the SHMT enzyme pathway in Myc-induced phenotypes. Reactions catalyzed by the mSHMT and cSHMT enzymes provide a major source of the one-carbon unit required for a variety of metabolic reactions in the cell. Our data from functional screening identify new Myc target genes involved in a central metabolic process in the cell.
MATERIALS AND METHODS
Cell culture, Myc-ER system, transfection, and retroviral infection.
Rat-1 fibroblasts, rat c-myc-null fibroblasts (HO15.19 cells), HO15.19 cells expressing a cdk4-cyclin D1 fusion protein (DK cells), HO15.19 cells expressing the c-Myc-estrogen receptor (ER) chimeric protein, retroviral producer PhoeNX cells, and HEK 293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Raji cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum. Retroviral infection of HO15.19 and DK cells was performed according to published protocols (31) by using PhoeNX cells. PhoeNX cells were transfected via the calcium phosphate method. To obtain a transduced cell population, LXSH-based retroviruses, HO15.19 cells, and DK cells were selected for resistance to hygromycin (150 μg/ml) for 7 days. HO15.9 cells expressing cMyc-ER chimeric protein were passaged several times under subconfluent conditions (<50% confluence) to obtain cells in an exponential phase of growth. 4-Hydroxytamoxifen (4-OH; Sigma) was added directly to the medium at a final concentration of 200 nM. RNA was collected at 2, 4, 8, and 16 h after the addition of 4-OH.
Library preparation and CFSE selection.
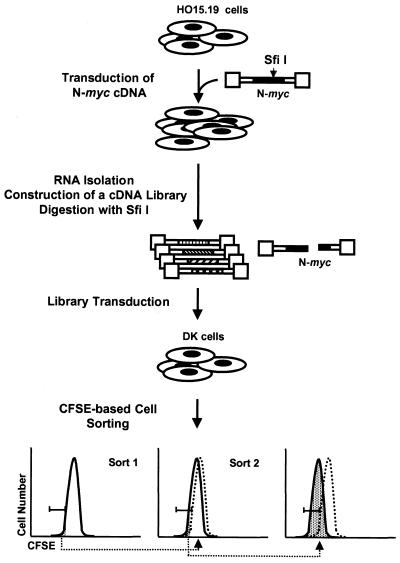
HO15.19 cells were transduced with pLXSH vector carrying mouse N-myc cDNA, which contains a unique SfiI site. Digestion with SfiI disrupts the N-myc coding region, rendering it functionally inactive. After the cells were infected and selected for hygromycin, total cellular RNA was isolated with TRIzol reagent (GIBCO/BRL) and poly(A) RNA was isolated with oligo(dT) cellulose (Ambion). cDNA was prepared with a SuperScript Choice System for cDNA Synthesis kit (GIBCO/BRL) and cloned into the modified pREBNA vector (32). The modified pREBNA vector contains bacterial ori sequences downstream of the polylinker and Lox sequences inserted into the NheI site in the 5′ and 3′ long terminal repeats. The library was digested extensively with SfiI, followed by transfection into packaging cells and subsequent infection into DK cells according to standard protocols. After infection, cells were stained with 10 μM carboxyfluorescein diacetate succinilmidyl ester (CFSE; Molecular Probes) in suspension in growth medium (3 × 106 cells/ml) for 5 min at 37°C, washed three times with 10 ml of ice-cold medium, and plated on 150-mm-diameter tissue culture dishes (106 cells/dish). CFSE is a vital fluorescent dye that irreversibly binds to cellular macromolecules. After 5 days, cells were collected and analyzed by flow cytometry, and a population of sorted cells (usually the 2 to 5% of cells with the lowest content of CFSE) was replated. After the cell population on the plates was expanded, cells were subjected to the second round of CFSE staining and sorting. To rescue retroviral vectors integrated into the cellular genome, we utilized a strategy proposed earlier (17). Briefly, 105 cells obtained after the second sort were infected with a retrovirus encoding the Cre protein, which binds to Lox sites in long terminal repeats of the integrated pREBNA vector and excises a circular double-stranded DNA containing one chimeric LTR, a cDNA insert, and bacterial ori sequences. At 36 to 48 h postinfection, cells were lysed in buffer P1 from a QIAGEN plasmid minikit, followed by treatment with solutions P2 and P3, according to the kit's instructions. Plasmid DNA was cloned through standard transformation of competent Escherichia coli bacteria.
Chromatin immunoprecipitation (ChIP).
HEK 293 or Raji cells were cross-linked by adding formaldehyde (final concentration, 1%; Fisher Scientific) directly to the cells on a rocking tissue culture dish for 10 min at room temperature. Cross-linking was terminated by the addition of glycine to a final concentration of 125 mM. Fixed cells were washed with ice-cold phospate-buffered saline containing 0.5 mM phenylmethylsulfonyl fluoride (PMSF) and then scraped in swelling buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid), pH 8.0], 85 mM KCl, 0.5% NP-40, 0.5 mM PMSF, and 100 ng (each) of leupeptin and aprotinin per ml}. Cells were incubated on ice for 20 min, followed by Dounce homogenization. Nuclei were harvested by centrifugation (4,500 × g) and then resuspended in sonication buffer (0.1% sodium dodecyl sulfate, F buffer [39a], 0.5 mM PMSF, 100 ng [each] of leupeptin and aprotinin per ml) and sonicated on ice to obtain 1,000- to 2,000-bp-long DNA fragments. After sonication, lysates were cleared by microcentrifugation at 14,000 rpm. Lysates were normalized according to measurements of their optical densities at 260 nanometers. Each lysate was diluted six times with dilution buffer (F buffer, 0.5 mM PMSF, 100 ng [each] of leupeptin and aprotinin per ml) and normalized by dilution in IP buffer (0.015% sodium dodecyl sulfate, F buffer, 0.5 mM PMSF, and 100 ng [each] of leupeptin and aprotinin per ml). The normalized chromatin lysates were precleared overnight with preblocked protein A/G beads. Protein A or G beads were preblocked by incubation for 3 h at 4°C in IP buffer containing 1 μg of salmon sperm DNA per ml and 1 μg of bovine serum albumin per ml. Precleared lysates were incubated at 4°C overnight with 1.5 μg of anti-Myc (N262; Santa Cruz) or 5 μl of anti-Gal4 (sc-429; Santa Cruz). Beads were washed and eluted, followed by reversal of the cross-linking (22) and DNA isolation performed with a QIAGEN PCR purification kit. Eluted material was subjected to quantitative PCR amplification. PCR was performed with the following sets of primers: human mSHMT promoter-specific primers SC-25 (CGAGTTGCGATGCT-GTACTTCTCT) and SC-26 (GCTCGGTTGCATCATCTGCA), human cSHMT promoter-specific primers cprSHT5 (AGCGACAGGCTTAAGTGAGG) and cprSHT3 (GTGCCACCAGTCCCAGAC), human TERT-specific primers SC-9 (AGTGGATTCGCGGGCACAGA) and SC-10 (AGCACCTCGCGGTAGTGGCT), human chromosome 22-specific primers Chr22a (TTACAGGTAAGCACCTCCATGACC) and Chr22b (GCAAAAGCTACCATTTAGGAACCC), rat mSHMT-specific primers RmSHMTs (CTGGATGACCAGTGGAAAGG) and RmSHMT (GACCCTTGCGTGATGAAAGT), rat nucleolin-specific primers NUCLa (CTGGGAGGGCGATGTAGAGT) and NUCLb (GGAAGGGGGTTATCTCGAAG), and rat glucokinase-specific primers GLUa (TGCCCGATTTTCATCTTCTT) and GLUb (CCAAGGACTTCCGCACTAAC).
To normalize samples by the amount of nonspecific DNA, we amplified regions in the promoters of the rat PCNA gene (PCNa [CGAAGCACCCAGGTAAGTGT] and PCNb [ATCGTATCCGTGGTTTGAGC]) and in the third intron of the human β-globin gene (SC-46 [ATCTTCC-TCCCACAGCTCCT] and SC-47 [TTTGCAGCCTCACCTTCTTT]). One of the primers in each pair was end labeled with [γ-32P]ATP. Amplification was performed in a model T3 thermocycler (Biometra) for 31 cycles at 94°C for 45 s, 60°C for 45 s, and 72°C for 60 s, followed by a final extension step at 72°C for 5 min. The optimal number of cycles for exponential amplification was determined by kinetic analysis. PCR products were resolved on a 4 or 6% polyacrylamide gel. PCR products were quantitated and normalized with the Molecular Dynamics PhoshphorImaging system.
RESULTS
Functional screening for Myc-responsive genes.
The creation of c-myc-null cells through targeted knockout of both somatic genes in a rat fibroblast cell line has contributed significantly to an understanding of Myc function (6, 25, 44). These cells offer an opportunity to assess Myc-dependent phenotypes without manipulation of the cellular environment and to assay the contribution of individual potential target genes to the growth control. Attempts to complement c-myc-null cells with cDNA expression libraries resulted in the finding that only c- and N-myc cDNAs would suffice, implying a unique function for Myc in cell proliferation (2, 29). These data also suggested that Myc fulfills its function through the regulation of more than one downstream effector. Thus, in the course of the selection, c-myc-null cells expressing c- or N-myc cDNA will most likely outgrow cells in which expression of a cDNA of a Myc target gene only partially complements the cellular growth defects. Since at least one of the myc family genes is expressed in every cell type, all previously available cDNA libraries invariably contain myc cDNAs. To overcome this obstacle, we prepared cDNA from N-myc-reconstituted c-myc-null cells and depleted it from the full-length N-myc cDNA (Fig. 1 and Materials and Methods), thus creating a library enriched with cDNAs of N-Myc target genes yet lacking functional N-myc cDNA.
FIG. 1.
Outline of the experimental strategy. The dotted lines indicate the CFSE profiles of initial populations before the first cell sorting. The shaded area below the peak designates cells taken for the next step of analysis. Note the gradual decrease in the intensity of CFSE staining of cells after each sorting. See details in the text and Materials and Methods.
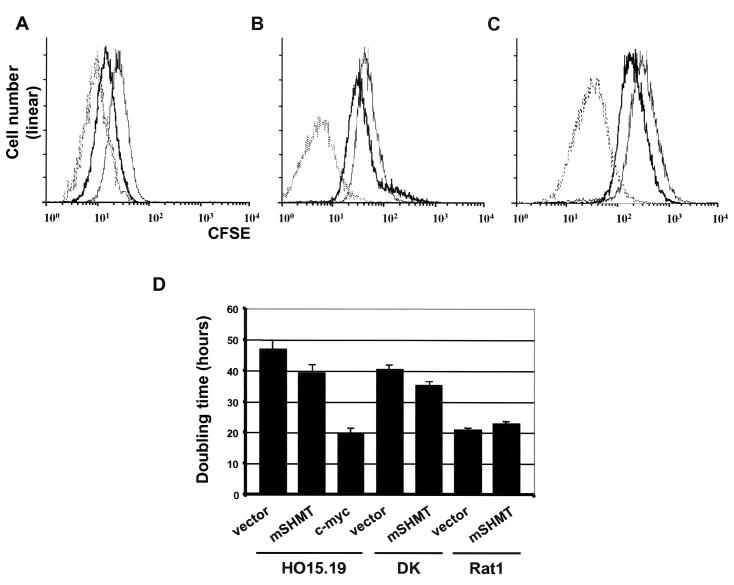
The Myc-depleted cDNA library was introduced into DK cells, a c-myc-null cell line which overexpresses a cdk4-cyclin D1 fusion protein (34) that partially complements their growth defects (Fig. 2D). To select for the faster-growing cells, we utilized a CFSE-fluorescence-activated cell sorting (FACS) methodology described earlier (29). Briefly, cells are pulse-labeled with the irreversibly binding vital fluorescent dye CFSE and propagated for 5 days in culture to allow the dye to be diluted through cell division. Cells are then subjected to FACS based on the intensity of staining. In 5 days, fast-growing cells will retain smaller amounts of CFSE than slow-growing cells, since the number of cell divisions corresponds to a CFSE dilution factor. After two rounds of sorting, a population of faster-growing cells was obtained, although the rate of their growth was lower than that of Myc-reconstituted c-myc-null cells (Fig. 2A). Retroviral inserts were rescued from the faster-growing population (Materials and Methods) and subjected to sequence analysis. Isolated cDNAs with intact coding regions included proteosome component C3, Fos-related antigen 1 (Fra1), ribosomal protein S28, vacuolar H+ ATPase subunit E, ribosomal protein S20, and mSHMT. We also isolated several truncated cDNAs for a2(I)collagen.
FIG. 2.
Overexpression of mSHMT enhances proliferation of c-myc-null cells. (A) FACS profiles of DK cells (thin line), c-myc-null cells (HO15.19) infected with a c-myc-expressing vector (dotted line), and DK cells infected with a cDNA expression library (bold line) after two rounds of sorting. (B) FACS profiles of DK cells infected with either empty vector (thin line) or an mSHMT-expressing vector (bold line) after one round of sorting. c-myc-null cells (HO15.19) infected with a c-myc expression vector before the sorting are shown with a dotted line. (C) CFSE profile of c-myc-null cells (HO15.19) infected as described for panel B and selected for resistance to hygromycin (see Materials and Methods). (D) Doubling times of DK, c-myc-null, and Rat-1 cells expressing the constructs indicated below each bar.
mSHMT partially rescues the slow-growth phenotype of c-myc-null cells.
cDNAs identified in the course of the screening were individually transduced into DK cells in parallel with an empty vector as a control and subjected to the original selection. Cells with low levels of CFSE staining were collected with a cell sorter, and the growth rate of the sorted cells was determined by FACS analysis. Among all tested cDNAs, overexpression of only mSHMT resulted in a low but reproducible increase in the growth rate of DK cells compared to that of cells transduced with vector only (Fig. 2B). The gene encoding mSHMT and a different gene encoding its cytoplasmic isoform, cSHMT, are both nuclear genes. SHMT enzymes catalyze the conversion of serine and tetrahydrofolate (THF) into glycine and 5,10-methylene-THF. This reaction is a major source of one-carbon-unit substituted folate cofactors necessary for a variety of metabolic reactions, including thymidylate and de novo purine biosynthesis, biosynthesis of methionine and histidine, and the catabolism of methyl groups and formate (reviewed in reference 39). It has also been suggested that SHMT activity may regulate cell growth and proliferation (37, 38, 41).
We were interested in whether overexpression of mSHMT rescues the slow growth of the original c-myc-null cells (HO15.19). To this end, we transduced an mSHMT-expressing vector or an empty vector into c-myc-null cells or DK cells, followed by selection on hygromycin for 7 days. The growth rate of the resulting cell populations was determined by both CFSE staining and the direct determination of the cell doubling time (Fig. 2). Overexpression of mSHMT partially rescued the slow-growth phenotype of HO15.19 and DK cells, as visualized by the peak of staining with CFSE (Fig. 2C). The doubling time of the c-myc-null cells decreased from 47.1 ± 1.9 h to 39.0 ± 1.4 h for myc-null mSHMT-expressing cells (n = 4, P = 0.0173) and from 40.9 ± 1.8 h for DK cells to 35.5 ± 1.3 h for DK mSHMT-expressing cells (n = 3, P = 0.0333). It is noteworthy that overexpression of mSHMT in the parental Rat-1 cells did not enhance their growth rate (Fig. 2D).
mSHMT and cSHMT genes are direct targets of Myc.
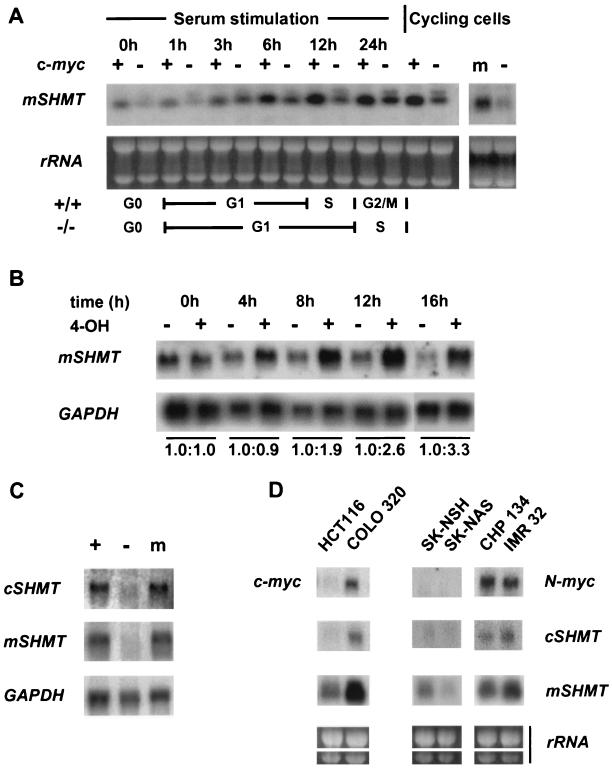
Our next goal was to determine if mSHMT is indeed a Myc target gene. We used Northern blot analysis of RNAs isolated from c-myc-null cells and parental Rat-1 cells (which possess wild-type c-myc genes). As shown in Fig. 3A, mSHMT expression is strongly induced by serum in Rat-1 cells whereas the induction is severely attenuated in c-myc-null cells. We also compared the profiles of mSHMT expression in logarithmically growing Rat-1 cells, c-myc-null cells, and c-myc-null cells reconstituted with a c-myc cDNA. The level of mSHMT RNA was enhanced in Myc-reconstituted c-myc-null cells compared to that of the original c-myc-null cells transduced with empty vector, implying that mSHMT is a downstream target of c-Myc. We also noted a second, slower-migrating mSHMT RNA in serum-stimulated myc-null cells that is under investigation. We note that mSHMT is still expressed basally in log-phase myc-null cells and is inducible to some extent by serum, indicating that other factors also regulate mSHMT expression.
FIG. 3.
Genes for mSHMT and cSHMT are Myc-responsive genes. (A) RNA isolated from serum-stimulated or logarithmically growing c-myc-null cells (lanes marked −), parental Rat-1 cells (lanes marked +), or c-myc-null cells expressing a c-myc cDNA (lane m) was probed in a Northern blot with an mSHMT-specific probe. Ethidium bromide-stained 28 and 18S rRNAs are shown in each lane as a loading control. The numbers above the blots indicate times after serum induction. (B) c-myc-null cells expressing c-Myc-ER were passaged several times under subconfluent conditions (<50% confluence) to obtain cells in an exponential phase of growth. 4-OH was added directly to the medium at a final concentration of 200 nM. RNA was collected following the addition of 4-OH at the time intervals shown above the blots. RNA was also collected from untreated cells at the same time intervals. RNA was probed sequentially by Northern blotting with mSHMT and GAPDH probes. A number below a blot indicates the fold induction of the mSHMT-specific message normalized to the amount of GAPDH in each lane at each time point. (C) RNA isolated from logarithmically growing c-myc-null cells (lane marked −), parental Rat-1 cells (lane marked +), or c-myc-null cells expressing a c-myc cDNA (lane m) was sequentially hybridized by Northern blotting with probes specific for mSHMT, cSHMT, and GAPDH. (D) RNA was isolated from logarithmically growing colon carcinoma cells (HCT116 and Colo320) or neuroblastoma cells (SK-NSH, SK-NAS, CHP-134, IMR32) and sequentially hybridized by Northern blotting with probes specific to the mSHMT, cSHMT, c-myc, or N-myc gene. Ethidium bromide-stained 28 and 18S rRNAs are shown in each lane as a loading control.
To further prove that mSHMT regulation was Myc dependent, we employed a Myc-ER-inducible system (24). Myc-null cells were infected with a vector that expresses a chimeric protein consisting of c-Myc fused to the hormone-binding domain of the ER. Under normal conditions, the c-Myc-ER chimera is not transported to the nucleus and thus does not activate Myc-responsive genes. Upon addition of 4-OH, subcellular translocation regulates c-Myc-ER activity. We tested the expression level of mSHMT in c-myc-null Myc-ER-expressing cells treated or not treated with 4-OH. 4-OH induces a substantial up-regulation of mSHMT expression, whereas there was no induction of mSHMT in untreated cells (Fig. 3B) or in cells expressing the vector only with or without 4-OH (data not shown). Induction of Myc-ER by 4-OH in these cells fully restores their growth rate to wild-type levels (data not shown). mSHMT is also induced 2.4-fold by Myc-ER in the presence of cycloheximide (data not shown).
As mentioned above, there are two SHMTs (mitochondrial and cytoplasmic) that are major suppliers of one-carbon units for cellular biosynthesis. Since we demonstrated that mSHMT is a Myc target gene, we were interested in determining whether the expression of cSHMT is also Myc regulated. To this end, we hybridized RNA obtained from Rat-1 cells, c-myc-null cells, and Myc-reconstituted c-myc-null cells by employing Northern blotting with a probe specific to mouse cSHMT. As shown in Fig. 3C, the level of cSHMT mRNA is significantly higher in cycling Rat-1- or Myc-reconstituted c-myc-null cells than in cycling c-myc-null cells infected with empty vector. Expression of cSHMT was also induced in Myc-ER cells by treatment with 4-OH, suggesting that the cSHMT gene is also a Myc target gene (data not shown).
Since c-myc and N-myc genes are often amplified in tumors, we were interested in learning whether the level of mSHMT or cSHMT mRNA is also elevated in tumor cells with large amounts of the Myc protein. SHMT expression was analyzed in colon carcinoma cell lines with amplified (Colo320) or normal (HCT116) c-myc genes and in neuroblastoma lines with amplified (CHP134, IMR32) or normal (SK-NSH, SK-NAS) N-myc genes. As shown in Fig. 3D, expression of both SHMT genes was higher in tumor cell lines with amplified myc genes than in cells with lower Myc levels.
In vivo cross-linking of c-Myc to the SHMT promoters.
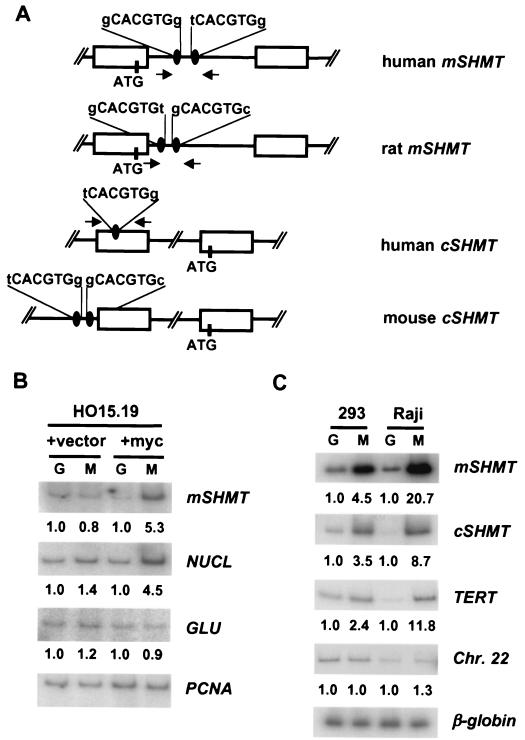
Sequence analysis of cytosolic SHMT and mSHMT genes in different species revealed that their promoters contained one or two Myc consensus binding sites (Fig. 4A). We wanted to determine if Myc actually binds to these sites in vivo. To this end, we obtained cross-linked chromatin from either myc-null cells and Myc-reconstituted null cells (Fig. 4B) or from human HEK293 cells and Raji cells, which have high levels of the c-Myc protein (Fig. 4C). The chromatin was immunoprecipitated with either anti-c-Myc antibodies or with control anti-Gal4 antibodies (ChIP). After reversion of the cross-linking, DNA was purified and subjected to PCR with primers flanking the region of the SHMT promoters encompassing the potential Myc binding sites (Fig. 4A). As a control for DNA quantitation and nonspecific background, we performed PCR on the same DNA samples with primers to chromosomal sites that are not expected to bind to Myc protein in vivo. In myc-null cells, there was no enrichment in the anti-Myc ChIP experiment for the Myc target gene nucleolin or for mSHMT, nor was there any enrichment for the nontarget gene products glucokinase (which contains a Myc/Max consensus site) and PCNA. In contrast, reconstitution of Myc expression in myc-null cells led to four- to fivefold enrichment of both mSHMT and nucleolin genomic DNA fragments in the anti-Myc ChIP but not in the control anti-Gal4 ChIP samples (Fig. 4B). No binding of Myc to glucokinase or PCNA was observed. In Raji and HEK293 cells, there was a substantial enrichment for mSHMT and cSHMT genomic DNA in the anti-Myc ChIP (3.5- to 20-fold) that paralleled the enrichment for the Myc target gene TERT (2.4- to 11-fold) (Fig. 4C). There was no enrichment of a nonfunctional Myc consensus sequence on chromosome 22 or of the nontarget β-globin gene. Equivalent background signals were obtained for all samples with the control β-globin primers. It is noteworthy that the highest signal-to-noise ratio (20-fold) was observed for the mSHMT gene in Raji anti-Myc ChIP samples, which have the highest Myc protein levels. Thus, ChIP demonstrates the direct binding of c-Myc to both the mSHMT and cSHMT promoters, consistent with the above data on Myc-dependent expression of these genes.
FIG. 4.
c-Myc binds to the promoters of mSHMT and cSHMT genes in vivo. (A) Schematic presentation of human, mouse, and rat mSHMT and cSHMT promoters. Open boxes represent exons. Myc binding sites and ATG codons are shown. Arrows indicate the positions and direction of primers used in PCR. (B and C) HO15.19 cells infected with empty vector (+vector lanes), HO15.19 cells infected with c-myc-expression vector (+myc lanes), HEK 293 cells, or Raji cells were cross-linked and lysed, and chromatin was immunoprecipitated with c-Myc-specific (M lanes) or Gal4-specific (G lanes) antibodies, followed by the reversal of the cross-linking and DNA isolation. Isolated DNA was used in PCR with radiolabeled oligonucleotides flanking Myc binding sites in the promoters of the genes indicated to the right of the blots. For negative controls, two types of PCR were performed. The first one was carried out with oligonucleotides flanking CACGTG sites that do not interact with c-Myc in the promoter of the rat glucokinase gene (GLU) (11) (B) or somewhere on human chromosome 22 (Chr. 22) (4) (C). Another PCR was performed with oligonucleotides flanking a DNA region containing no CACGTG sites in the rat PCNA gene (B) or in the third intron of a silent human β-globin gene (C). Quantitation of the PCR products was performed with the Molecular Dynamics IhoshphorImaging system. A number below the a blot indicates the fold difference in the signal intensity, determined by dividing a given PCR signal by a corresponding PCNA-specific (B) or β-globin-specific (C) signal and by the ratio of these signals in the corresponding Gal4 lane.
DISCUSSION
Several approaches have been used to discover genes whose expression is modulated by Myc in normal or transformed cells. This study represents the first successful functional screen for cDNAs that can bypass Myc function. Two previous attempts to complement the growth defect in myc-null cells led to the repeated cloning of Myc itself: either c-Myc or N-Myc (2, 29). These studies emphasized the unique nature of the Myc pathway in cellular physiology but offered little insight into the functional downstream target genes. Here we introduce a novel functional screen for downstream effectors of Myc function by using a powerful sorting technique for cellular growth rate and a library depleted of myc cDNAs. We show that an important aspect of Myc function is to regulate cellular one-carbon metabolism through the transcriptional regulation of genes encoding both mSHMT and cSHMT. Neither of these genes has been previously identified as a Myc target by subtractive or differential hybridization. Both genes have functional Myc/Max binding sites near or within their TATA-less promoters, and both genes can be cross-linked to Myc in vivo. SHMT genes are Myc dependent in steady-state log-phase growth in rat fibroblasts and are induced by Myc-ER in 4-OH-regulated Myc-ER-reconstituted cells. These data establish that mSHMT and cSHMT are direct targets of Myc in human and rodent cells and that Myc is a rate-limiting factor for their expression.
SHMT catalyzes the conversion of serine and THF to glycine and methylene-THF (reviewed in reference 39). This reaction generates most of the one-carbon units for thymidylate, purine, and methionine synthesis. Two separate genes encode the mitochondrial (mSHMT) and cytoplasmic (cSHMT) forms of the enzyme (13, 14, 40). It remains unclear why a cell has both enzymes, since the mitochondrial form is the main source of one-carbon units for both mitochondrial and cytoplasmic folate metabolism and nucleoside biosynthesis (12). Interestingly, culturing of myc-null cells or myc-null cells overexpressing cyclin D1/cdk4 in high concentrations of nucleotides, formate, or methionine does not provide any growth enhancement (data not shown), indicating that it is not possible to complement this biochemical pathway by simple substrate replacement. This suggests that localized SHMT activity produces one-carbon-unit-substituted folate cofactors that may have a unique influence on cell growth and/or proliferation. In Saccharomyces cerevisiae, the two SHMT enzymes are not essential due to redundant biosynthetic pathways. However, strains defective in both enzymes exhibit severe growth defects under specific culture conditions (21). Furthermore, CHO cells lacking mSHMT are glycine auxotrophs (33). We speculate that mSHMT activity may become rate limiting for growth in c-myc-null cells in part because of other metabolic pathways that are simultaneously down-regulated in response to the Myc deficiency.
The full extent of the metabolic pathways that are under Myc control will require further study. The SHMT genes join those for CAD, ODC, LDH-A, IRP2, and others as direct Myc targets involved in basic cellular metabolism (1, 27, 36, 43). Both the LDH-A and IRP2 genes are components of general cellular growth pathways, but they have not been shown to contribute directly to the growth rate, as we show here for mSHMT. The importance of Myc in the control of cellular growth is highlighted by studies of Drosophila, where Myc gain of function or loss of function controls cell size without any concomitant change in cell number (20). These observations link Myc expression to cellular growth and only indirectly to the cell cycle. Overexpression of Myc in B cells also leads to an increase in cell size (19, 34a). In contrast to these influences on cellular growth, several mammalian genes with direct roles in the control of the cell cycle have been shown to be Myc targets, including the genes for cyclin D2, cdk4, and cul1. cul1 has been reported to rescue the growth defect in primary myc-null fibroblasts (30), and cdk4 has a modest growth-enhancing effect in immortalized c-myc-null fibroblasts (18). Cyclin D2 is reported to be required for Myc-dependent cell proliferation (5), but ectopic expression of cyclin D2 does not stimulate the growth of c-myc-null cells (2). Finally, reduced expression of c-myc led to smaller embryos with fewer cells in most major organs, rather than smaller cells (42). Thus, it appears that in mammals, Myc controls both basic cellular growth pathways and progression through the cell cycle. The identification of mSHMT as a Myc target for cellular growth provides an important insight into the ability of Myc to regulate cellular metabolism. A screen for cDNAs that synergize with mSHMT to further enhance the growth rate of Myc-deficient cells may reveal additional rate-limiting components of the Myc pathway.
Acknowledgments
We thank Marcelo Wood for RNA samples and Julie Goodliffe for a critical reading of the manuscript and for helpful discussions.
This work was supported by grants from the NIH to M.D.C. and J.M.S.
REFERENCES
- 1.Bello-Fernandez, C., G. Packham, and J. L. Cleveland. 1993. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90:7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berns, K., C. Martins, J. H. Dannenberg, A. Berns, H. te Riele, and R. Bernards. 2000. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene 19:3330-3334. [DOI] [PubMed] [Google Scholar]
- 3.Boon, K., H. N. Caron, R. van Asperen, L. Valentijn, M. C. Hermus, P. van Sluis, I. Roobeek, I. Weis, P. A. Voute, M. Schwab, and R. Versteeg. 2001. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 20:1383-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard, C., O. Dittrich, A. Kiemaier, K. Dohmann, A. Menkel, M. Eilers, and B. Luscher. 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 15:2042-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchard, C., K. Thieke, A. Maier, R. Saffrich, J. Hanley-Hyde, W. Ansorge, S. Reed, P. Sicinski, J. Bartek, and M. Eilers. 1999. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 18:5321-5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush, A., M. Mateyak, K. Dugan, A. Obaya, S. Adachi, J. Sedivy, and M. D. Cole. 1998. c-myc null cells misregulate cad and gadd45 but not other proposed c-myc targets. Genes Dev. 12:3797-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coller, H. A., C. Grandori, P. Tamayo, T. Colbert, E. S. Lander, R. N. Eisenman, and T. R. Golub. 2000. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc. Natl. Acad. Sci. USA 97:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang, C. V. 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol. Cell. Biol. 19:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, A. C., M. Wims, G. D. Spotts, S. R. Hann, and A. Bradley. 1993. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7:671-682. [DOI] [PubMed] [Google Scholar]
- 10.de Alboran, I. M., R. C. O'Hagan, F. Gartner, B. Malynn, L. Davidson, R. Rickert, K. Rajewsky, R. A. DePinho, and F. W. Alt. 2001. Analysis of C-MYC function in normal cells via contitional gene-targeted mutation. Immunity 14:45-55. [DOI] [PubMed] [Google Scholar]
- 11.Frank, S. R., M. Schroeder, P. Fernandez, S. Taubert, and B. Amati. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15:2069-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, T. F., J. P. Rife, and V. Schirch. 2001. The role of serine hydroxymethyltransferase isozymes in one-carbon metabolism in MCF-7 cells as determined by 13C NMR. Arch. Biochem. Biophys. 393:42-50. [DOI] [PubMed] [Google Scholar]
- 13.Garrow, T. A., A. A. Brenner, V. M. Whitehead, X. N. Chen, R. G. Duncan, J. R. Korenberg, and B. Shane. 1993. Cloning of human cDNAs encoding mitochondrial and cytosolic serine hydroxymethyltransferases and chromosomal localization. J. Biol. Chem. 268:11910-11916. [PubMed] [Google Scholar]
- 14.Girgis, S., I. M. Nasrallah, J. R. Suh, E. Oppenheim, K. A. Zanetti, M. G. Mastri, and P. J. Stover. 1998. Molecular cloning, characterization and alternative splicing of the human cytoplasmic serine hydroxymethyltransferase gene. Gene 210:315-324. [DOI] [PubMed] [Google Scholar]
- 15.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16:653-699. [DOI] [PubMed] [Google Scholar]
- 16.Guo, Q. M., R. L. Malek, S. Kim, C. Chiao, M. He, M. Ruffy, K. Sanka, N. H. Lee, C. V. Dang, and E. T. Liu. 2000. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 60:5922-5928. [PubMed] [Google Scholar]
- 17.Hannon, G. J., P. Sun, A. Carnero, L. Y. Xie, R. Maestro, D. S. Conklin, and D. Beach. 1999. MaRX: an approach to genetics in mammalian cells. Science 283:1129-1130. [DOI] [PubMed] [Google Scholar]
- 18.Hermeking, H., C. Rago, M. Schuhmacher, Q. Li, J. F. Barett, A. J. Obaya, B. C. O'Connell, M. K. Mateyak, W. Tam, F. Kohlhuber, C. V. Dang, J. M. Sedivy, D. Eick, B. Vogelstein, and K. W. Kinzler. 2000. Identification of CDK4 as a target of c-MYC. Proc. Natl. Acad. Sci. USA 97:2229-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iritani, B. M., and R. N. Eisenman. 1999. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc. Natl. Acad. Sci. USA 96:13180-13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston, L. A., D. A. Prober, B. A. Edgar, R. N. Eisenman, and P. Gallant. 1999. Drosophila myc regulates cellular growth during development. Cell 98:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kastanos, E. K., Y. Y. Woldman, and D. R. Appling. 1997. Role of mitochondrial and cytoplasmic serine hydroxymethyltransferase isozymes in de novo purine synthesis in Saccharomyces cerevisiae. Biochemistry 36:14956-14964. [DOI] [PubMed] [Google Scholar]
- 22.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 23.Lasorella, A., M. Noseda, M. Beyna, Y. Yokota, and A. Iavarone. 2000. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407:592-598. [DOI] [PubMed] [Google Scholar]
- 24.Littlewood, T. D., D. C. Hancock, P. S. Danielian, M. G. Parker, and G. I. Evan. 1995. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 23:1686-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateyak, M. K., A. J. Obaya, S. Adachi, and J. M. Sedivy. 1997. Phenotypes of Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 8:1039-1048. [PubMed] [Google Scholar]
- 26.McMahon, S. B., M. A. Wood, and M. D. Cole. 2000. The c-Myc cofactor TRRAP recruits the histone acetylase hGCN5. Mol. Cell. Biol. 20:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miltenberger, R. J., K. A. Sukow, and P. J. Farnham. 1995. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol. Cell. Biol. 15:2527-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nesbit, C. E., J. M. Tersak, L. E. Grove, A. Drzal, H. Choi, and E. V. Prochownik. 2000. Genetic dissection of c-myc apoptotic pathways. Oncogene 19:3200-3212. [DOI] [PubMed] [Google Scholar]
- 29.Nikiforov, M. A., I. Kotenko, O. Petrenko, A. Beavis, L. Valenick, I. Lemischka, and M. D. Cole. 2000. Complementation of Myc-dependent cell proliferation by cDNA expression library screening. Oncogene 19:4828-4831. [DOI] [PubMed] [Google Scholar]
- 30.O'Hagan, R. C., M. Ohh, G. David, I. M. de Alboran, F. W. Alt, W. G. Kaelin, and R. A. DePinho. 2000. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 14:2185-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrenko, O., A. Beavis, M. Klaine, R. Kittappa, I. Godin, and I. R. Lemischka. 1999. The molecular characterization of the fetal stem cell marker AA4. Immunity 10:691-700. [DOI] [PubMed] [Google Scholar]
- 33.Pfendner, W., and L. I. Pizer. 1980. The metabolism of serine and glycine in mutant lines of Chinese hamster ovary cells. Arch. Biochem. Biophys. 200:503-512. [DOI] [PubMed] [Google Scholar]
- 34.Rao, R. N., N. B. Stamm, K. Otto, S. Kovacevic, S. A. Watkins, P. Rutherford, S. Lemke, K. Cocke, R. P. Beckmann, K. Houck, D. Johnson, and B. J. Skidmore. 1999. Conditional transformation of rat embryo fibroblast cells by a cyclin D1-cdk4 fusion gene. Oncogene 18:6343-6356. [DOI] [PubMed] [Google Scholar]
- 34a.Schuhmacher, M., M. S. Stage, A. Pajic, A. Polack, U. H. Weidle, G. W. Bornkann, D. Eick, and F. Kohlhuber. 1999. Control of cell growth by c-Myc in the absence of cell division. Curr. Biol. 9:1255-1258. [DOI] [PubMed] [Google Scholar]
- 35.Schuhmacher, M., F. Kohlhuber, M. Holzel, C. Kaiser, H. Burtscher, M. Jarsch, G. W. Bornkamm, G. Laux, A. Polack, U. H. Weidle, and D. Eick. 2001. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29:397-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim, H., C. Dolde, B. C. Lewis, C. S. Wu, G. Dang, R. A. Jungmann, R. Dalla-Favera, and C. V. Dang. 1997. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA 94:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snell, K. 1985. Enzymes of serine metabolism in normal and neoplastic rat tissues. Biochim. Biophys. Acta 843:276-281. [DOI] [PubMed] [Google Scholar]
- 38.Snell, K. 1980. Liver enzymes of serine metabolism during neonatal development of the rat. Biochem. J. 190:451-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snell, K., U. Baumann, P. C. Byrne, K. J. Chave, S. B. Renwick, P. G. Sanders, and S. K. Whitehouse. 2000. The genetic organization and protein crystallographic structure of human serine hydroxymethyltransferase. Adv. Enzyme Regul. 40:353-403. [DOI] [PubMed] [Google Scholar]
- 39a.Sommer, A., K. Bousset, E. Kremmer, M. Austin, and B. Lüscher. 1998. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J. Biol. Chem. 273:6632-6642. [DOI] [PubMed] [Google Scholar]
- 40.Stover, P. J., L. H. Chen, J. R. Suh, D. M. Stover, K. Keyomarsi, and B. Shane. 1997. Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J. Biol. Chem. 272:1842-1848. [DOI] [PubMed] [Google Scholar]
- 41.Thorndike, J., T. T. Pelliniemi, and W. S. Beck. 1979. Serine hydroxymethyltransferase activity and serine incorporation in leukocytes. Cancer Res. 39:3435-3440. [PubMed] [Google Scholar]
- 42.Trumpp, A., Y. Refaell, T. Oskarsson, S. Gasser, M. Murphy, G. R. Martin, and J. M. Bishop. 2001. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414:768-773. [DOI] [PubMed] [Google Scholar]
- 43.Wu, K. J., A. Polack, and R. Dalla-Favera. 1999. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-MYC. Science 283:676-679. [DOI] [PubMed] [Google Scholar]
- 44.Xiao, Q., G. Claassen, J. Shi, S. Adachi, J. Sedivy, and S. R. Hann. 1998. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 12:3803-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]