Abstract
The p53 tumor suppressor regulates the cellular response to genetic damage through its function as a sequence-specific transcription factor. Among the most well-characterized transcriptional targets of p53 is the mdm2 oncogene. Activation of mdm2 is critical in the p53 pathway because the mdm2 protein marks p53 for proteosome-mediated degradation, thereby providing a negative-feedback loop. Here we show that the ATM-related TRRAP protein functionally cooperates with p53 to activate mdm2 transcription. TRRAP is a component of several multiprotein acetyltransferase complexes implicated in both transcriptional regulation and DNA repair. In support of a role for these complexes in mdm2 expression, we show that transactivation of the mdm2 gene is augmented by pharmacological inhibition of cellular deacetylases. In vitro analysis demonstrates that p53 directly binds to a TRRAP domain previously shown to be an activator docking site. Furthermore, transfection of cells with antisense TRRAP blocks p53-dependent transcription of mdm2. Finally, using chromatin immunoprecipitation, we demonstrate direct p53-dependent recruitment of TRRAP to the mdm2 promoter, followed by increased histone acetylation. These findings suggest a model in which p53 directly recruits a TRRAP/acetyltransferase complex to the mdm2 gene to activate transcription. In addition, this study defines a novel biochemical mechanism utilized by the p53 tumor suppressor to regulate gene expression.
The gene encoding p53 is the most frequently mutated locus in human cancer (reviewed in reference 64). p53 is a tumor suppressor which induces either cell cycle arrest or apoptosis in response to DNA damage. These effects rely on the ability of p53 to function as a sequence-specific transcription factor. Following DNA damage, multiple signaling pathways result in the stabilization of the normally short-lived p53 protein. Stabilized p53 then activates or represses the transcription of a number of downstream target genes, many of which play critical roles in cell cycle arrest or apoptosis. The biochemical mechanism by which p53 represses certain target loci was recently described in studies showing that p53 recruits histone deacetylase (HDAC) complexes (49). In contrast, the precise biochemical mechanism by which p53 activates transcription is still being elucidated. Interactions between p53 and the TATA-binding protein (TBP) and between p53 and several of the TBP-associated factors have been reported (11, 17, 37, 39, 42, 59-61). These interactions are likely to play an important role in the loading of the RNA polymerase holoenzyme onto target gene promoters. They do not, however, explain how p53 resolves the inhibitory effects of the chromatin structure at target genes prior to polymerase loading. It has recently been learned that sequence-specific transcription factors such as p53 overcome the repressive effects of chromatin structure via the recruitment of multiprotein enzyme complexes (21, 54). In fact, DNA footprinting studies of p53 have demonstrated that the region protected at p53 binding sites is much larger than expected (71), suggesting that p53 recruits a large multiprotein complex to target genes. These complexes fall into two major groups, those that directly modify nucleosomal histones and those that reposition nucleosomes on DNA in an ATP-dependent manner. Among the best-characterized chromatin-modifying complexes are histone acetyltransferases (HATs). The enzymatic subunits of these complexes add acetyl groups to conserved lysines in the amino-terminal tails of histones (10). This modification either relaxes the interaction between the histones and DNA or triggers the recruitment of other proteins involved in transcriptional regulation. Recently, an in vitro study demonstrated that recruitment of the p300 acetyltransferase to the p21 gene by p53 results in acetylation of promoter histones and increased transcription (16). The in vivo interactions between p53 and chromatin-modifying complexes have not been as thoroughly examined.
The ATM-related TRRAP protein is the only subunit shared by the mammalian HAT complexes, PCAF/SAGA, TFTC, STAGA, and NuA4/Tip60 (8, 57, 63). Furthermore, TRRAP appears to be the subunit directly bound by sequence-specific activators (9, 53). TRRAP was originally identified based on its interaction with the c-Myc and E2F1 oncoproteins (34, 45, 46). A variety of evidence indicates that TRRAP is likely to be recruited by other sequence-specific transcription factors as well. Furthermore, TRRAP/HAT complexes TFTC, STAGA, and NuA4/Tip60 have recently been implicated in DNA repair (7, 25, 44). Based on the observations that the previously identified ATM-related proteins function upstream of p53 (30, 33, 69, 72) and that some p53 target genes are regulated through changes in histone acetylation (27, 49, 55), we undertook the present study to determine whether p53 and TRRAP might functionally interact.
The mdm2 oncogene was first identified as an amplified locus in transformed murine fibroblasts (12). Subsequently mdm2 overexpression has been documented in a number of human cancers as well (47, 52). Induction of mdm2 transcription is largely controlled by p53 through a pair of tandem binding sites in intron 1 (3, 28). The binding of p53 to these sites is cooperative and activates a cryptic promoter. Basal transcription of mdm2 is p53 independent and regulated by a classical promoter located upstream of exon 1 (3, 28, 70).
mdm2 encodes a ubiquitin ligase that has p53 as one of its major substrates (23, 24, 31). This establishes a negative-feedback loop in which the product of a p53-activated gene catalyzes the degradation of p53. Because the mdm2 gene is among the most thoroughly characterized transcriptional targets of p53 and because of its role in human cancer, we utilized it as a target to assess whether p53 and TRRAP cooperate in transcriptional regulation. We demonstrate here that expression of TRRAP is sufficient to activate transcription of the mdm2 promoter, but not other promoters, in p53-expressing cells. Transactivation of mdm2 by TRRAP depends on both the expression of p53 and on the presence of the p53 binding sites in the mdm2 promoter. Furthermore, we demonstrate that TRRAP and p53 interact in vitro and that TRRAP is directly recruited by p53 to the endogenous mdm2 promoter in vivo. These observations suggest that, like other members of the ATM family of proteins (30, 33, 69, 72), TRRAP functionally interacts with the p53 tumor suppressor. The recruitment of TRRAP/HAT complexes to the mdm2 promoter by p53 results in increased histone acetylation and provides a mechanistic symmetry to previous studies showing that recruitment of deacetylase complexes by p53 mediates transcriptional repression (49). This is analogous to what has been documented for the transcription factors of the Myc, E2F, and nuclear hormone receptor families (14, 18, 34, 40, 46). Furthermore, this study documents a novel biochemical mechanism (i.e., TRRAP/acetyltransferase targeting) which is critical to p53 function.
MATERIALS AND METHODS
Cell lines, transfection, reporter assays, and adenovirus infection.
Human embryonic kidney cell line 293 and human lung cancer cell line H1299 were obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle medium (GIBCO) with 10% fetal calf serum (HyClone). Transient transfections of 293 cells were performed by using calcium phosphate with modifications described previously (45). H1299 cells were transiently transfected with Lipofectamine 2000 (Life Technologies, Inc.) according to the manufacturer's instructions. For studies involving sodium butyrate (Sigma), cells were treated immediately following transfection with the HDAC inhibitor for 16 h at a final concentration of 100 μM or 1 mM. Luciferase reporter assays were performed in accordance with the manufacturer's recommendation (Promega). High-titer p53-expressing adenovirus stocks were obtained from M. Herlyn and used to infect H1299 cells at a multiplicity of infection of 20. Infections proceeded for 4 h in serum-free media, at which time normal medium was added. Cells were harvested 24 h postinfection.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed essentially as described previously (40) with commercially available reagents (Upstate Biotechnology). Briefly, cells were treated for 10 min at room temperature with 1% formaldehyde to cross-link nuclear proteins to chromatin. Cross-linking was stopped by the addition of 125 mM glycine (pH 8.0). Cells were then lysed in sodium dodecyl sulfate (SDS)-containing lysis buffer, and chromatin was sheared by sonication. Immunoprecipitations were performed overnight at 4°C with rotation. Precipitates were captured onto protein A/G beads, washed, and eluted. Cross-linking was reversed by incubation at 65°C for 16 h, and DNA was purified by proteinase K treatment and phenol-chloroform extraction. PCR was performed using the following primers, which flank the p53 response element in the human mdm2 gene: 5′ primer, 5′-TGGGCAGGTTGACTCAGCTTTTCCTC-3′; 3′ primer, 5′-TTCCGAAGCTGGAATCTGTGACCTGC-3′. For ChIP studies of human cells transfected with the murine mdm2 luciferase plasmid, the 5′ primer was located upstream of the p53 binding sites and the 3′ primer was located within the luciferase gene to avoid coamplification of the endogenous mdm2 locus. The primer sequences were as follows: 5′ primer, 5′-GTTATTTAAACGCTGCCCCGTTTCCG-3′; 3′ primer, 5′-TCGAAGTATTCCGCGTACGTGATGTTC-3′.
In all cases, samples were removed from PCR mixtures at 20, 25, 30, 35, and 40 cycles and products were resolved on 1.5% agarose gels and visualized by ethidium bromide staining to ensure that amplification was maintained in the linear range.
Immunoprecipitation and Western blotting.
Immunoprecipitations and Western blotting conditions have been described previously (45). Briefly, cells were harvested, washed twice in cold phosphate-buffered saline, and lysed in a solution containing 0.5% NP-40, 150 mM sodium chloride, 20 mM sodium phosphate, 5 mM EDTA, and 10% glycerol with protease inhibitors. Lysates were cleared of debris by centrifugation and then either resolved directly by SDS-polyacrylamide gel electrophoresis (PAGE) or incubated overnight at 4°C with immunoprecipitating antibodies. Precipitates were captured onto protein A or G beads by 4 h of incubation at 4°C, washed four times in lysis buffer, and boiled in SDS-PAGE sample buffer prior to electrophoresis. Antibodies to TRRAP, MAX, and p53 (Santa Cruz Biotechnology), as well as mdm2 (Oncogene Research Products), FLAG (Sigma), acetylated histone H3 and H4 (Upstate Biotechnology Inc.), and acetylated lysine (Cell Signaling Technology) were obtained from commercial sources. Autoradiography was performed using the ECL system (Amersham).
GST pull-downs.
Production and purification of glutathione S-transferase (GST)-TRRAP fusion proteins were performed as described previously (53). Affinity purification of baculovirus-expressed FLAG-tagged p53 has also been described (38). Binding reaction mixtures included 5 μg of individual purified GST-TRRAP proteins on glutathione-Sepharose 4b beads (Amersham Pharmacia) and 1 μl of purified p53 in a total volume of 1 ml. Reaction mixtures also included 250 mM NaCl, 0.5% NP-40, 25 mM HEPES, 10% glycerol, and a protease inhibitor cocktail (Roche Diagnostics). After a 16-h incubation at 4°C, beads were washed three times with binding buffer. Bound proteins were recovered from the beads by boiling the beads in SDS-PAGE sample buffer and then were resolved by SDS-PAGE. Gels were blotted and probed for p53 by using monoclonal antibody D01.
Plasmids and site-directed mutagenesis.
Cytomegalovirus (CMV)-driven TRRAP expression vectors have been described previously (53). The murine mdm2 promoter-luciferase reporter was obtained from D. George and contains the 840-bp XhoI fragment from the murine mdm2 locus. The human mdm2 promoter-luciferase reporter was provided by M. Oren and contains the 350-bp HincII-XhoI fragment of the human gene. The mutant version of this construct contains the 300-bp PstI-XhoI fragment of the same gene. Both constructs have been described in detail previously (73). All other reporter constructs have also been described previously (1, 5, 19, 26, 41, 66). CMV-driven p53 expression vectors were obtained from A. Levine and R. Hay (56). Acetylation site mutations were introduced by site-directed mutagenesis according to the manufacturer's instructions (Stratagene).
RESULTS
The ATM-related TRRAP protein activates mdm2 transcription.
We originally identified the ATM-related TRRAP protein based on its physical and functional interaction with the oncogenic transcription factors c-Myc and E2F1 (45). Subsequently, our work and that of several other groups established that TRRAP is a subunit of several distinct acetyltransferase complexes (8, 43, 57, 63). Because these complexes are more abundant than c-Myc and E2F1 and because they predate these transcription factors in evolution, it appears likely that the TRRAP complexes function as coactivators for other transcription factors as well. A common feature of c-Myc and E2F1 is that their function is inhibited by deacetylase complexes (22, 32, 40, 74). The deacetylases presumably inhibit c-Myc and E2F1 activity by reversing the effects of TRRAP/HAT recruitment. As for c-Myc and E2F1, p53 target gene transcription has recently been shown to be repressed by HDAC recruitment (49). We therefore hypothesized that TRRAP might functionally interact with p53 to activate transcription. To test this hypothesis, we assayed the effect of TRRAP overexpression on p53-mediated transcription of mdm2. Because TRRAP forms the physical link between sequence-specific transcription factors such as c-Myc, E2F1, and p53 and the enzymatic subunits of the acetyltransferase complexes (9, 53), we reasoned that overexpression might augment recruitment of the TRRAP complexes. Furthermore, our studies demonstrate that TRRAP is the limiting subunit in the HAT complexes (our unpublished results). As such, TRRAP overexpression is predicted to lead to the de novo assembly of additional HAT complexes, resulting in the availability of more complexes for recruitment by sequence-specific transcription factors such as c-Myc, E2F1, and possibly p53.
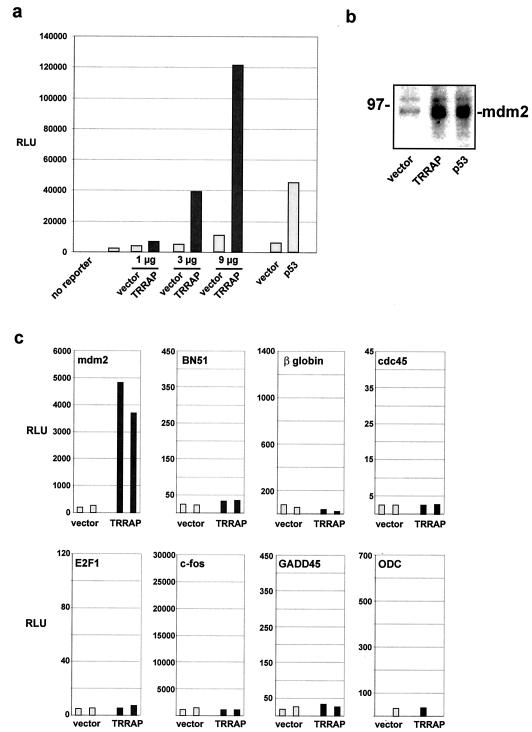
To assess whether the TRRAP acetyltransferase complexes play a role in mdm2 transcription, a luciferase reporter driven by the murine mdm2 regulatory region was transiently transfected into 293 human embryonic kidney cells along with an expression vector encoding full-length TRRAP. The luciferase reporter contains 840-bp of the murine mdm2 gene, including the p53 response elements located in intron 1. As shown in Fig. 1a, overexpression of TRRAP activates the mdm2 promoter in a dose-dependent manner. Over the course of several similar experiments, expression of TRRAP resulted in a 10- to 50-fold induction of the cotransfected mdm2 promoter-luciferase reporter. As expected, p53 expression also resulted in robust induction of the mdm2 promoter. We next assessed whether the effect of TRRAP on the mdm2 promoter-reporter construct was evident at the endogenous mdm2 gene. Again, 293 cells were transfected with expression vectors for either TRRAP or p53. Immunoblotting of lysates from transfected cells showed that mdm2 levels were increased by either TRRAP or p53 expression, relative to levels in vector-transfected controls (Fig. 1b). To determine whether TRRAP overexpression has a global role in augmenting transcription, the effect of TRRAP overexpression on the promoters from a variety of different types of cellular genes was tested. This study included promoters from other oncogenes, cell type-specific promoters, and promoters displaying a wide range of basal activities. The results from a representative sample of these other promoters are shown in Fig. 1c. Cotransfection of the TRRAP expression vector with these reporters into 293 cells reveals that TRRAP does not globally activate transcription, and its effects here are largely specific to the mdm2 promoter.
FIG. 1.
Expression of the TRRAP cofactor stimulates transcription of the mdm2 promoter. (a) 293 cells were transiently transfected with a reporter plasmid in which the 840 bp flanking the murine mdm2 intron 1 promoter drives expression of the luciferase gene. In addition, cells were cotransfected with a CMV-driven expression vector encoding acetyltransferase complex subunit TRRAP (dark bars). Control transfections included the empty CMV expression vector (light bars). Where indicated, cells were transfected with a CMV-driven p53 expression vector or its empty vector control. Cells were harvested 24 h posttransfection, and luciferase activity was determined and expressed as relative light units (RLU). Transfections included 300 ng of mdm2 promoter-driven reporter plasmid and the indicated amounts of TRRAP expression vector. In addition, a parallel transfection included 5 μg of p53 expression vector. (b) 293 cells were transfected with expression vectors for either TRRAP (15 μg) or p53 (4 μg) as indicated. Cells were harvested as in panel a, and lysates were resolved by SDS-PAGE, blotted, and probed for mdm2. Migration of a size marker is indicated at left in kilodaltons. (c) Luciferase reporters driven by the promoters of the indicated cellular genes (500 ng) were cotransfected with 4 μg of the CMV-driven TRRAP expression vector (dark bars) or its empty vector control (light bars). Paired bars represent data from duplicate transfections.
Activation of mdm2 transcription by TRRAP requires p53.
293 cells express the adenovirus E1B oncoprotein, which partially inhibits p53 function (35, 62, 68). Because the studies shown in Fig. 1 were conducted with 293 cells, further study was needed to determine whether p53 function is essential for the effect of TRRAP on mdm2 expression. To address this, 293 cells were transfected with a human mdm2 promoter construct which was either wild type or had its p53-responsive element deleted (Fig. 2a) (73). As for the mouse mdm2 promoter, overexpression of TRRAP resulted in a dose-dependent activation of wild-type mdm2 promoter activity. As expected, expression of p53 also dramatically activated mdm2 transcription. When assayed with the mdm2 promoter lacking a functional p53 response element, transactivation by TRRAP was absent. p53 activation of the mutant mdm2 promoter was also inhibited. These data confirm that the effect of TRRAP on mdm2 transcription depends on functional p53 binding sites within the promoter.
FIG. 2.
Activation of mdm2 transcription by TRRAP requires p53. (a) A luciferase reporter (500 ng) driven by a fragment of the human mdm2 promoter encompassing the intron 1 p53 response element was transfected into 293 cells (left). Parallel transfections were performed with an isogenic reporter in which the p53 response element was deleted by a 5′ truncation (right). Transfections also included the TRRAP expression vector or its empty vector control as indicated. Transfections were performed in duplicate and included increasing amounts of plasmid (1, 3, and 7 μg). Positive-control transfections included the mdm2 reporter and a p53 expression vector (5 μg). Luciferase activity was assayed and expressed as relative light units (RLU). (b) p53-negative cell line H1299 was transfected as indicated. Cells were transfected with the murine mdm2 promoter-luciferase reporter (300 ng) used in Fig. 1a and CMV-driven expression vectors for either TRRAP (3 μg) or p53 (1 μg). Control transfections included appropriate amounts of empty CMV vector. Duplicate transfections were performed, and cells were harvested 24 h posttransfection.
p53 is a member of a family of transcription factors which all share DNA binding specificity (29). We therefore set out to determine whether mdm2 induction by TRRAP requires p53 itself or another family member. H1299 cells, human lung cancer cells which completely lack p53 expression (13), were chosen as a genetic background for these studies. Using these cells, we addressed whether coexpression of TRRAP and p53 has a synergistic effect on the mdm2 promoter. For the purpose of this experiment, a suboptimal level of p53 was expressed in order that any synergy with TRRAP not be masked by the effect of p53 alone. In contrast to the results for 293 cells, TRRAP overexpression in H1299 cells failed to activate mdm2 transcription (Fig. 2b). Similarly, expression of very low levels of p53 failed to activate mdm2 transcription. Remarkably, coexpression of TRRAP and p53 acted synergistically to activate mdm2 transcription by approximately 35-fold. Considered together, these results indicate that the effect of TRRAP on mdm2 transcription is strictly dependent on p53.
Blocking TRRAP function with antisense treatment blocks mdm2 transactivation by p53.
As a definitive test of whether TRRAP plays a role in the p53-mediated activation of mdm2 transcription, we set up an assay in which TRRAP expression was blocked by antisense treatment. We have previously shown that transfection of a TRRAP antisense vector in which the first 1.5 kb of the 13-kb human TRRAP cDNA is expressed in antisense orientation results in a dramatic and specific decrease in TRRAP protein levels in 293 cells (45). In Fig. 3, we tested the ability of this reagent to block p53's effect on mdm2 transcription in 293 cells. Remarkably, expression of antisense TRRAP blocked p53 function in a dose-dependent manner. In parallel transfections, the empty vector into which the antisense cDNA was cloned had no effect on p53 activity. Furthermore, this effect of antisense TRRAP is not due to a global squelching of transcription because several other promoters have been similarly assayed and found to be completely unaffected by treatment with antisense TRRAP (data not shown). These data clearly indicate that TRRAP function is required for the transcriptional activation of the mdm2 oncogene by p53.
FIG. 3.
Expression of antisense TRRAP blocks p53-mediated transcription of mdm2. 293 cells were transiently transfected with the murine mdm2 promoter-luciferase reporter described for Fig. 1 (500 ng). Transfections also included 1 μg of a CMV-driven expression vector for p53 or an empty vector control as indicated. The p53 expression vector was cotransfected with increasing amounts of a CMV-driven expression vector for antisense TRRAP or the empty vector control (1 and 3 μg), as indicated. At 24 h posttransfection, cells were lysed and luciferase activity was determined.
The ATM homology domains of TRRAP are required for mdm2 transactivation.
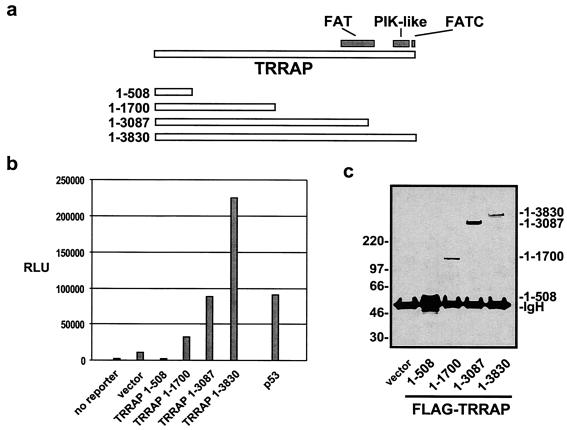
TRRAP is a member of the ATM family (45). These large nuclear proteins share three highly conserved domains, which have been termed FAT, PI3K-like, and FATC (Fig. 4a) (6). Because the other mammalian ATM family proteins interact with p53 either genetically or biochemically (30, 33, 69, 72), we examined whether the cooperation between TRRAP and p53 was dependent on TRRAP's FAT, PI3K-like, and FATC domains. A series of FLAG-tagged TRRAP deletion mutants was expressed in 293 cells and then individually assayed for their abilities to activate the mdm2 promoter (Fig. 4b). As seen previously, expression of full-length TRRAP (amino acids 1 to 3830) resulted in a 20-fold increase in mdm2 transcription. Deletion of the carboxy-terminal PI3K-like and FATC domains (in construct 1-3087) resulted in a loss of 60% of the transactivation potential. Deletion of the final ATM family homology domain (FAT) from TRRAP (in construct 1-1700) resulted in an 85% decrease in mdm2 transactivation. Western blot analysis for the common FLAG epitope shows that full-length and truncated TRRAP proteins are expressed at comparable levels (Fig. 4c). These observations demonstrate that the domains shared by TRRAP and the other ATM family proteins are critical for TRRAP's ability to regulate the p53-dependent transcription of mdm2.
FIG. 4.
The ATM homology domains of TRRAP are required for optimal mdm2 transcription. (a) The TRRAP protein shares three carboxy-terminal domains of homology with the other ATM family proteins, FAT, PIK-like, and FATC. Carboxy-terminal truncation TRRAP mutants were created from a CMV expression vector. Numbers at left indicate the TRRAP amino acids encoded by each construct, with the 1-3830 construct representing full-length TRRAP. (b) Cotransfection of 293 cells with the mdm2 luciferase reporter (300 ng) and the TRRAP expression vectors (5 μg) diagrammed in panel a. A control transfection included the mdm2 reporter and a CMV-driven p53 vector (1 μg). Transfections were performed in duplicate, and bars represent averaged luciferase activity expressed as relative light units (RLU). (c) Western blot of protein expression levels for the TRRAP truncation mutants used in panel b. After transfection, 293 cells were lysed and immunoprecipitations were performed with anti-FLAG antibodies. Precipitates were resolved by SDS-4 to 12% PAGE. After being transferred, the blot was probed for the FLAG epitope common to all the TRRAP mutants. Migration of size standards is indicated at left in kilodaltons. The position of the heavy chain from the immunoprecipitating antibody is also indicated (IgH).
Inhibition of deacetylase activity activates mdm2 transcription.
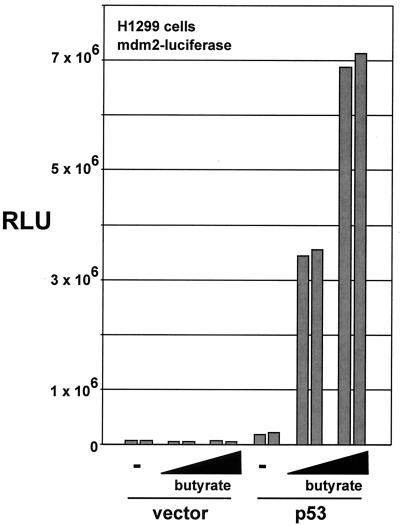
The ability of TRRAP to act synergistically with p53 in the activation of mdm2 transcription is predicted to depend on the function of the HAT complexes with which TRRAP associates. If recruitment of endogenous TRRAP/HAT complexes is critical for p53-dependent activation of mdm2 expression, then pharmacological inhibition of cellular HDACs might augment p53-mediated transcription of mdm2. To test this, 293 cells were transfected with the mdm2 promoter-luciferase reporter construct and either a p53 expression vector or the empty vector as a control (Fig. 5). Cells were then treated with the HDAC inhibitor sodium butyrate at two different concentrations, as indicated. In mock-treated cells, the suboptimal expression of p53 resulted in a modest increase in mdm2 promoter activity. The addition of sodium butyrate augmented this effect in a dose-dependent manner, confirming that increased levels of acetylation result in increased mdm2 transcription.
FIG. 5.
Inhibition of cellular deactylases augments mdm2 transcription. H1299 cells were cotransfected with the mdm2 luciferase reporter (500 ng) and a p53 expression vector (3 μg) or empty vector as indicated. Following transfection, cells were treated for 16 h with sodium butyrate (100 μM or 1 mM) or left untreated. Transfections were performed in duplicate as represented by each pair of bars. Cells were lysed 16 h posttransfection, and luciferase activity was determined.
Acetylation of p53 is not required for mdm2 transcription.
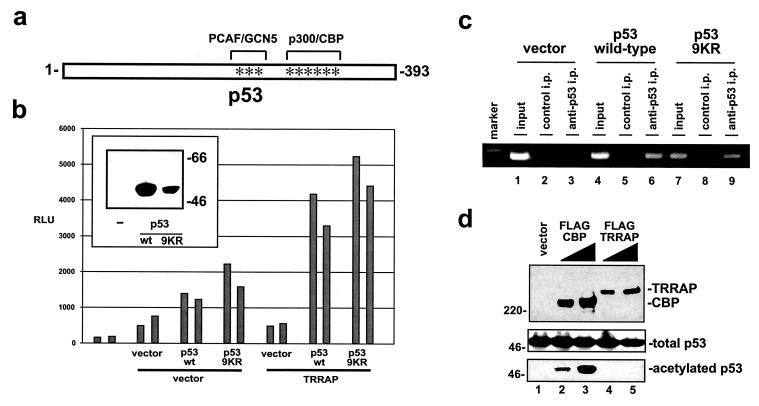
The p53 protein itself is acetylated by TRRAP-associated HATs PCAF and hGCN5 (36). The presence of TRRAP in HAT complexes and the observation that inhibiting HDACs augments p53's ability to transactivate the mdm2 promoter suggest that the acetylation of either mdm2-associated histones or of p53 itself might be responsible for the cooperation between p53 and TRRAP. To distinguish between these possibilities, p53-negative H1299 cells were transfected with expression vectors for TRRAP and either wild-type p53 or a p53 mutant in which nine lysines in the carboxy terminus representing potential acetylation sites were mutated to arginines (9KR mutant) (Fig. 6a). Arginine substitutions were selected because they conserve the positive charge of the lysines while removing the potential for acetylation (56). When assayed with the mdm2 promoter, the 9KR mutant exhibited an equal or greater transactivation potential than wild-type p53 (Fig. 6b). The synergy between TRRAP and p53 in H1299 cells observed in Fig. 2b was reassessed by using the 9KR mutant. As observed with wild-type p53, coexpression of TRRAP and the 9KR mutant resulted in synergistic activation of the mdm2 promoter. As assessed by Western blotting, the steady-state levels of wild-type p53 and 9KR mutants were comparable (Fig. 5b, inset). These data suggest that the acetylation of p53 itself is not absolutely essential for p53's ability to cooperate with the ATM-related TRRAP protein in transcriptional regulation of mdm2. This is consistent with a recent in vitro study of the cooperation between p53 and p300 in p21 expression, where p53 acetylation by p300 was also shown to be dispensable for transcription (16), and with other studies (56). It remained possible that the 9KR mutant was significantly different from wild-type p53 in its ability to cooperate with TRRAP but that this difference was masked by a compensatory change in DNA binding affinity. To assess the relative abilities of wild-type p53 and the 9KR mutant to bind the mdm2 promoter, we employed the ChIP technique. For these studies, H1299 cells were transfected with either the wild-type or 9KR p53 expression vectors in combination with the mdm2 luciferase reporter. After transfection, cells were treated with formaldehyde to cross-link nuclear proteins to the DNA sites they bind in vivo. After lysis, the chromatin was sheared and protein/DNA complexes were immunoprecipitated with antibodies directed against p53. After reversal of the cross-links and purification of the DNA from immunoprecipitates, PCR was used to determine the relative levels of binding of wild-type and 9KR p53 to the mdm2 promoter contained in the cotransfected reported plasmid. The PCR primers were designed to specifically amplify a region of the transfected mdm2 promoter which includes the p53 binding sites. By placing the downstream primer within the luciferase reporter gene, we were able to eliminate coamplification of the endogenous mdm2 gene. The PCR products were analyzed by agarose gel electrophoresis as shown in Fig. 6c. In the absence of p53 expression, no occupancy of the mdm2 promoter is evident (lane 3), while expression of either wild-type or 9KR versions of p53 resulted in similar levels of occupancy of these sites (compare lanes 6 and 9). Amplification of input chromatin indicated that each group had been transfected with similar levels of mdm2 promoter-luciferase plasmid (input, lanes 1, 4, and 7).
FIG. 6.
Carboxy-terminal acetylation of p53 is not required for synergy with TRRAP. (a) The p53 protein contains multiple carboxy-terminal lysines which are targets for acetylation by HAT PCAF/hGCN5 or p300/CBP (asterisks). To eliminate potential effects of acetylation, these and adjacent lysines were mutated to arginine, resulting in the compound mutant designated 9KR. The lysines mutated represent amino acids 319, 320, 321, 370, 372, 373, 381, 382, and 386 in human p53. (b) p53-negative H1299 cells were cotransfected with the mdm2 luciferase reporter (300 ng) and expression vectors for TRRAP (5 μg) and either wild-type (wt) or mutant (9KR) p53 (1.5 μg) as indicated. Paired bars represent data from duplicate transfections, and control transfections included equivalent amounts of appropriate empty vectors. (Inset) Western blots of H1299 lysates from parallel transfections were probed for p53 to determine protein expression levels. Migration of size markers is indicated at right in kilodaltons. (c) ChIP analysis was performed on 293 cells after transfection with the murine mdm2 promoter-luciferase construct and expression vectors for wild-type or 9KR versions of p53, as indicated. DNA/protein complexes were immunoprecipitated with antibodies against p53 (lanes 3, 6, and 9) or control antibodies (lanes 2, 5, and 8). Precipitates were washed, cross-linking was reversed, and DNA was purified. PCR was then performed using primers that flank the p53 binding sites in the transfected mdm2 promoter. A parallel PCR was performed using purified DNA from each of the three transfection groups that was not subjected to immunoprecipitation (lanes 1, 4, and 7). Migration of a 564-bp size marker is shown for reference (marker). (d) H1299 cells were transfected with a p53 expression vector (5 μg) and two concentrations of vectors encoding FLAG epitope-tagged versions of either CBP (lanes 2 and 3) or TRRAP (lanes 4 and 5). A control transfection included the empty expression vector at the higher dose (lane 1). Cell lysates were produced and either resolved directly or subjected to immunoprecipitation with antisera directed against acetylated lysine residues. After Western blotting, lysates were probed for the FLAG epitope shared by CBP and TRRAP (top) or for p53 (middle). Antiacetyllysine immunoprecipitates were also probed for p53 (bottom).
As a direct test of whether TRRAP expression results in increased p53 acetylation, we used antiacetyllysine antibodies to immunoprecipitate acetylated p53 from H1299 cells expressing TRRAP and p53 (Fig. 6d). As a positive control, cells were transfected with an expression vector for CBP, which is known to acetylate p53 under these in vivo conditions. Antiacetyllysine immunoprecipitates from these cells were probed for p53. Acetylation of p53 was detected in cells expressing CBP but not in cells expressing TRRAP (Fig. 6d, bottom). Levels of transfected p53 in all groups were comparable (Fig. 6d, middle), and TRRAP and CBP were expressed at similar levels as well (Fig. 6d, top). These data suggest that TRRAP cooperates with p53 not by inducing its acetylation but rather by mediating recruitment of acetyltransferase complexes to the mdm2 promoter.
p53 binds directly to TRRAP in vitro and in vivo.
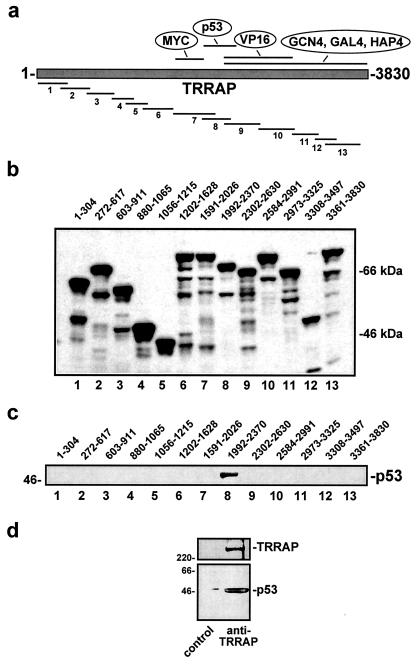
The simplest model that explains the data presented here is one in which TRRAP facilitates the recruitment of one or more of its associated HAT complexes to p53. p53 then targets the TRRAP/HAT complex to the mdm2 promoter to acetylate histones and enhance transcription. To test this, we produced 13 150- to 350-amino-acid fragments of TRRAP recombinantly as GST fusion proteins (Fig. 7a). After purification and normalization (Fig. 7b), these proteins were used as bait in in vitro pull-down reactions where purified, baculovirus-encoded p53 served as the prey. This analysis revealed that TRRAP amino acids 1992 to 2370 serve as the major p53 docking site (Fig. 7c). This region overlaps the docking domain on TRRAP recently described for acidic activators (9) and is directly adjacent to the TRRAP domain targeted by the Myc family of activators (53). Weak binding by p53 to a second region of TRRAP (amino acids 2973 to 3325) was also observed in some experiments (data not shown). These results support a model in which p53, like other activators, recruits acetyltransferase complexes by directly binding to their common subunit, TRRAP.
FIG. 7.
p53 directly binds to TRRAP in vitro and in vivo. (a) Fragments of TRRAP spanning the entire protein were expressed as GST fusions in Escherichia coli. The p53 binding domain defined in panel c is indicated. (b) Purified GST-TRRAP fusion proteins were resolved by SDS-PAGE, blotted, and probed for GST to document relative expression levels and confirm predicted molecular weights. The TRRAP amino acids encompassed in each fusion protein are indicated. (c) After purification, GST-TRRAP fusion proteins were used as bait in in vitro pull-down reactions. FLAG-tagged p53 expressed and affinity purified from baculovirus-infected cells served as the target protein in these reactions. After a washing, binding reaction mixtures were resolved by SDS-PAGE, blotted, and probed for p53. (d) Nondenaturing lysates of 293 cells were subjected to immunoprecipitation using antibodies directed against TRRAP or a control antibody. Precipitates were resolved, blotted, and probed for both TRRAP (top) and p53 (bottom). Migration of size markers is indicated at left in kilodaltons.
To examine whether an interaction between p53 and TRRAP could be observed in vivo, nondenaturing lysates from 293 cells were subjected to immunoprecipitation using antibodies directed against TRRAP. Precipitates were analyzed for TRRAP and for coprecipitation of p53 by Western blotting (Fig. 7d). As expected, precipitates contained significant TRRAP protein. In addition, p53 was also specifically present in TRRAP immunoprecipitations, suggesting that p53 recruits TRRAP in vivo.
p53 directly recruits TRRAP to the endogenous mdm2 promoter in vivo.
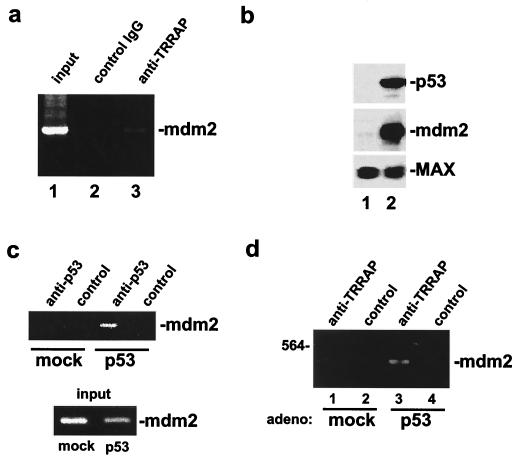
To assess whether TRRAP is physically recruited by p53 to the endogenous mdm2 promoter, we again employed the ChIP technique. For these studies, 293 cells were treated with formaldehyde. After lysis and shearing, protein/DNA complexes were immunoprecipitated with antibodies directed against TRRAP. PCR was then used to determine whether TRRAP is recruited to the mdm2 promoter. Primers were designed to specifically amplify a 397-bp region of the endogenous mdm2 promoter which includes the p53 binding sites. The results of the anti-TRRAP ChIP analysis demonstrate that TRRAP is indeed recruited to this region of the mdm2 promoter (Fig. 8a). No PCR product was observed in control immunoprecipitations performed with preimmune serum. To assess whether TRRAP recruitment to the mdm2 promoter is mediated by p53, we again utilized p53-negative cell line H1299. To quantitatively rescue p53 expression in these cells, we performed infections with a p53-expressing adenovirus. H1299 cells infected with a green fluorescent protein-expressing adenovirus served as a control. As a confirmation that p53 protein levels were rescued in H1299 cells by adenovirus infection, we performed Western analysis of p53- and mock-rescued cell lysates (Fig. 8b). The results of this analysis showed that the p53 protein was specifically expressed in p53 virus-infected cells. Furthermore, blotting for the mdm2 protein demonstrated that adenovirus-mediated rescue of p53 expression was sufficient to activate transcription of the endogenous mdm2 gene. Blots were probed for the constitutively expressed MAX protein as a control. To confirm that adenovirus rescue of p53 expression resulted in the binding of p53 to its sites in the endogenous mdm2 promoter, we performed ChIP analysis using antibodies directed against p53. As expected, rescue of p53 expression resulted in the binding of p53 to the mdm2 promoter (Fig. 8c). We next assessed whether TRRAP recruitment to the mdm2 promoter was p53 dependent. ChIP analysis using anti-TRRAP antibodies demonstrated that TRRAP was recruited to the endogenous mdm2 promoter only when p53 expression was rescued by adenovirus infection (Fig. 8d). These observations confirm that p53 is both necessary and sufficient to recruit the TRRAP cofactor to the endogenous mdm2 gene.
FIG. 8.
p53 mediates recruitment of TRRAP to the endogenous mdm2 promoter in vivo. (a) ChIP analysis was performed on 293 cells after formaldehyde cross-linking. DNA/protein complexes were immunoprecipitated with antibodies against TRRAP (lane 3) or control antibodies (lane 2). Precipitates were washed, cross-linking was reversed, and DNA was purified. PCR was then performed using primers which flank the p53 binding sites in the endogenous human mdm2 promoter. A parallel PCR was performed using purified DNA that was not subjected to immunoprecipitation (lane 1). The position of the 397-bp mdm2 promoter PCR product is indicated at the right. IgG, immunoglobulin G. (b) H1299 cells were rescued for p53 expression by infection with a p53-expressing adenovirus (lane 2). As a control, a mock rescue was performed by infection with a GFP-expressing adenovirus (lane 1). Whole-cell lysates from infected H1299 cells were resolved by SDS-PAGE and transferred to membranes. Blots were probed with antibodies against p53 or the mdm2 protein. As a loading control, blots were probed for the constitutively expressed MAX protein. (c) ChIP analysis was performed on H1299 cells in which p53 expression was rescued by adenovirus infection as in panel b. Again, mock-rescued cells served as a control. Immunoprecipitation of DNA/protein complexes was performed by using an anti-p53 antibody or a control antibody. As above, precipitated DNA was purified and subjected to PCR to amplify the mdm2 promoter region containing the p53 binding sites. DNA purified (but not immunoprecipitated) from mock- and p53-rescued cells (input; bottom) was subjected to PCR for the same region of the mdm2 promoter. (d) ChIP and PCR were performed as in panel a using antibodies against TRRAP or a control antibody. Analysis was performed on p53- (lanes 1 and 2) and mock-rescued cells (lanes 3 and 4). The position of the amplified mdm2 promoter fragment is indicated at right, and the migration of the 564-bp size marker is indicated at the left.
Recruitment of TRRAP to the mdm2 promoter is correlated with increased histone acetylation.
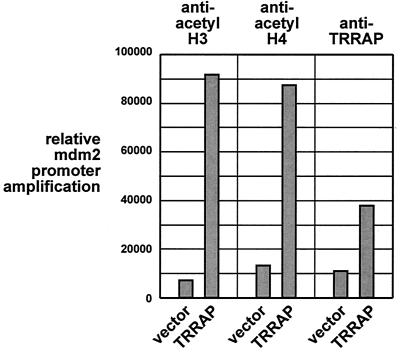
TRRAP is a component of several HAT complexes. We therefore examined whether TRRAP recruitment to the mdm2 promoter resulted in increased histone acetylation. Using antibodies directed against the acetylated forms of either histone H3 or H4, ChIP assays were performed on 293 cells transfected with the TRRAP expression vector and with the mdm2 promoter-luciferase plasmid. Primers flanking the p53 binding sites in this plasmid were used to amplify DNA associated with acetylated H3 or H4. As evident in Fig. 9, overexpression of TRRAP resulted in a significant increase in the level of TRRAP bound to the mdm2 promoter. Using antibodies directed against acetylated H3, we found a dramatic increase in H3 acetylation at the mdm2 promoter (right) following TRRAP recruitment. A similar increase in histone H4 acetylation was also observed (middle). These data suggest that TRRAP recruitment to the mdm2 promoter results in increased acetylation of both histone H3 and H4. Among the TRRAP/HAT complexes are those which independently catalyze both of these modifications.
FIG. 9.
TRRAP expression results in increased acetylation of histones H3 and H4 at the mdm2 promoter. 293 cells were transfected with the murine mdm2 promoter luciferase construct and a CMV-driven TRRAP expression vector (or empty vector), as indicated. ChIP analysis was then performed as for Fig. 6c using antibodies against TRRAP (right) or acetylated forms of histone H3 (left) or H4 (middle). The ChIP PCR products generated from the transfected mdm2 promoter were resolved by agarose gel electrophoresis, stained with ethidium bromide, quantitated, and normalized to input DNA. The resulting values were expressed graphically.
DISCUSSION
The gene encoding the p53 tumor suppressor is the most commonly mutated locus in human cancer (64). p53 functions as a sequence-specific transcription factor to regulate genes controlling growth arrest and apoptosis (15, 64). Until recently, little was known of the biochemical mechanism utilized by p53 to modulate transcription. Several studies have recently provided indications that the role of p53 in target gene expression may involve altering levels of histone acetylation. For example, Murphy et al. demonstrated that p53-mediated repression of the Map4 and stathmin genes relies on recruitment of HDAC complexes (49). Furthermore, a recent in vitro study showed that activation of at least one p53 target gene, that encoding p21, may result from recruitment of the HAT p300 to the p21 promoter (16). The present study was undertaken to define the biochemical mechanism utilized by p53 to activate transcription in vivo by examining well-characterized p53 target gene mdm2. We report here that p53-dependent expression of mdm2 involves recruitment of HAT complexes containing the ATM-related TRRAP cofactor. Specifically, we demonstrated that p53 and TRRAP act synergistically to activate mdm2 transcription and that both proteins are necessary for optimal mdm2 expression. Two potential models for the mechanism by which p53 and TRRAP cooperate were initially considered. In the first model, TRRAP simply functions as a transcriptional cofactor for p53 by mediating the recruitment of STAGA, TFTC, hSAGA/PCAF, or NuA4/Tip60 acetyltransferase complexes. These complexes in turn would catalyze the modification of histones associated with the mdm2 promoter and thereby facilitate transcription. The second model is one in which TRRAP overexpression results in the stabilization of the p53 protein via its acetylation or through some other posttranscriptional mechanism. The data presented here support the first model, as an acetylation-negative p53 mutant still cooperates with TRRAP, no TRRAP-induced acetylation of p53 was observed, and direct measurement of p53 protein levels revealed no stabilization following TRRAP overexpression (Fig. 6). It is worth noting that the acetylation-negative p53 mutant utilized here lacks the carboxy-terminal lysines which have been shown to be modified by the TRRAP-associated HAT PCAF and by p300/CBP (20, 36). Most importantly, we show that TRRAP and p53 interact directly in vitro and in vivo, that blocking TRRAP function with antisense treatment blocks p53-mediated transcription of mdm2, that TRRAP is directly recruited to the endogenous mdm2 promoter by p53, and that this recruitment correlates with increased histone acetylation.
In an elegant study of p53 acetylation, Barlev et al. recently reported that the transcriptional cofactors TRRAP and CBP are bound more tightly to acetylated than nonacetylated p53 (4). Our studies of the physical interaction between p53 and TRRAP were conducted using only nonacetylated p53, but the Barlev study suggests that the binding we observed would be increased severalfold by p53 acetylation. As in our study, Barlev et al. found no correlation between p53 DNA binding affinity and its acetylation status. Like us, they also found that TRRAP (or CBP) recruitment to a p53 target gene promoter correlated with increased levels of histone acetylation. Our failure to find a defect in mdm2 transcription when p53 acetylation sites were mutated differs from what Barlev et al. found at the p21 promoter. However, our study is consistent with several previous reports that found no transcriptional defect for acetylation-deficient p53 on the p21 promoter or on synthetic reporter genes (16, 50, 56). The p21 promoter contains a single, upstream p53 binding site, while there are two intronic binding sites for p53 in the mdm2 gene. The presence of only a single binding site in the p21 gene may make it more sensitive to the effects of increased cofactor recruitment following p53 acetylation, while the dual binding sites in mdm2 may make increased cofactor recruitment unnecessary for maximum transcription. Clearly the effect of p53 acetylation on the transcription of individual target genes requires further study.
We have reported here a direct interaction between p53 and TRRAP. Another recently published study found a direct interaction between p53 and ADA3 (65). Like TRRAP, ADA3 is a component of the PCAF/SAGA, STAGA, and TFTC acetyltransferase complexes. Considered together with our results, these data suggest that multiple subunits of these complexes (e.g., TRRAP and ADA3) can make independent, direct contact with p53. This scenario is similar to what has recently been described for the SWI/SNF chromatin-remodeling complex. There, a single sequence-specific activator was shown to bind several SWI/SNF subunits independently (51). The ability to interact with multiple subunits in a single cofactor complex may serve to stabilize recruitment by sequence-specific activators such as p53.
TRRAP is the only subunit shared by mammalian HAT complexes PCAF/SAGA, NuA4/Tip60, STAGA, and TFTC. Among these, NuA4/Tip60, STAGA, and TFTC have been firmly implicated not just in transcription but also in the repair of DNA damage (7, 25, 44). This dual role in transcription and DNA repair is common to p53 and the TFTC and NuA4/Tip60 complexes. The remaining TRRAP complex, PCAF/SAGA, has been postulated to function with p53 (58), perhaps in a signaling capacity, to enhance its tumor suppressor effects. This hypothesis was based in part on the homology between TRRAP and the ATM family of proteins, all of which can transmit signals to p53 in the form of increased phosphorylation. Whether there is a physiological link between p53 and the DNA damage repair and apoptosis functions of the TRRAP acetyltransferase complexes is worthy of further investigation, particularly in light of the transcriptional link reported here.
Several issues related to p53 function are raised by this studies. Among the most important issues that remain to be addressed is how p53 mediates recruitment of the correct type of chromatin-modifying complex to each target gene. For example, how can p53 recruit HDACs to the Map4 and stathmin genes to repress transcription while recruiting p300 to the p21 gene and recruiting TRRAP complexes to the mdm2 gene to activate transcription? A second issue raised by these studies is whether p53 preferentially recruits any of the distinct TRRAP/HAT complexes to the mdm2 promoter. Our histone H3 and H4 acetylation data suggest that at least two distinct TRRAP complexes are recruited to the endogenous mdm2 promoter. Current efforts are aimed at identifying these complexes.
It is also of interest to determine the extent to which TRRAP plays a role in the regulation of other p53 target genes. Evidence for Gadd45 (Fig. 1) suggests that the synergistic transactivation of the mdm2 promoter by p53 and TRRAP is not common to all p53 targets. This may be due to different chromatin contexts in which distinct p53 target gene promoters reside. For example, nucleosomal histones at the mdm2 promoter may be in such a configuration that their repositioning is necessary before transcription can occur, and recruitment of the TRRAP-associated HATs may be critical for this process. Conversely, the positioning of nucleosomes on the promoters of other p53 target genes may not be inhibitory to transcription or may require modifications distinct from those catalyzed by the TRRAP/HAT complexes. Finally, the robust TRRAP responsiveness of the mdm2 promoter may result from the unique juxtaposition of two p53 binding sites in this gene (2, 67) or from their intronic location.
In general, human cancers accumulate mutations that impair the p53 pathway (64). These mutations can be within the p53 gene itself or in genes encoding other components of the p53 pathway. In tumors carrying wild-type p53, overexpression of the p53-degrading mdm2 protein is a common mechanism utilized to block p53 function (reviewed in reference 48). Our demonstration that the TRRAP/HAT complexes play a role in mdm2 expression may have important implications in this subset of human tumors.
This study documents a novel biochemical mechanism utilized by p53 to activate transcription. In addition, p53 (along with c-Myc and E2F1) represents the third transcriptional regulator critical in human cancer that utilizes the TRRAP acetyltransferase complexes to activate gene expression. Finally, the direct recruitment of TRRAP acetyltransferase complexes to the mdm2 promoter by p53 to activate transcription provides a mechanistic symmetry to previous studies showing that recruitment of HDAC complexes by p53 mediates transcriptional repression.
Acknowledgments
We thank Maureen Murphy, Donna George, Shelley Berger, Ramin Shiekhattar, Amanda Norvell, Paul Lieberman, and Ronen Marmorstein for helpful discussions regarding this work. We also thank K. Satyamoorthy, C. Berking, and M. Herlyn for advice regarding adenoviral infections. In addition, we thank Linda Penn, Arnold Levine, Donna George, Bruno Amati, John Cleveland, Michael Simonson, Joseph Nevins, Kiyoshi Ohtani, Moshe Oren, Bert Vogelstein, and Wafik El-Deiry for plasmids and adenoviral stocks.
S.B.M. is a Special Fellow of the Leukemia and Lymphoma Society of America. This work was supported by grants to S.B.M. from the NIH (CA090464), The V Foundation, and The Emerald Foundation.
REFERENCES
- 1.Arata, Y., M. Fujita, K. Ohtani, S. Kijima, and J. Y. Kato. 2000. Cdk2-dependent and -independent pathways in E2F-mediated S phase induction. J. Biol. Chem. 275:6337-6345. [DOI] [PubMed] [Google Scholar]
- 2.Barak, Y., E. Gottlieb, T. Juven-Gershon, and M. Oren. 1994. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 8:1739-1749. [DOI] [PubMed] [Google Scholar]
- 3.Barak, Y., T. Juven, R. Haffner, and M. Oren. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J. 12:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 5.Bello-Fernandez, C., G. Packham, and J. L. Cleveland. 1993. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 90:7804-7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosotti, R., A. Isacchi, and E. L. Sonnhammer. 2000. FAT: a novel domain in PIK-related kinases. Trends Biochem. Sci. 25:225-227. [DOI] [PubMed] [Google Scholar]
- 7.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brand, M., K. Yamamoto, A. Staub, and L. Tora. 1999. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J. Biol. Chem. 274:18285-18289. [DOI] [PubMed] [Google Scholar]
- 9.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 10.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 11.Buschmann, T., Y. Lin, N. Aithmitti, S. Y. Fuchs, H. Lu, L. Resnick-Silverman, J. J. Manfredi, Z. Ronai, and X. Wu. 2001. Stabilization and activation of p53 by the coactivator protein TAFII31. J. Biol. Chem. 276:13852-13857. [DOI] [PubMed] [Google Scholar]
- 12.Cahilly-Snyder, L., T. Yang-Feng, U. Francke, and D. L. George. 1987. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat. Cell Mol. Genet 13:235-244. [DOI] [PubMed] [Google Scholar]
- 13.Chen, J. Y., W. D. Funk, W. E. Wright, J. W. Shay, and J. D. Minna. 1993. Heterogeneity of transcriptional activity of mutant p53 proteins and p53 DNA target sequences. Oncogene 8:2159-2166. [PubMed] [Google Scholar]
- 14.Eisenman, R. N. 2001. Deconstructing myc. Genes Dev. 15:2023-2030. [DOI] [PubMed] [Google Scholar]
- 15.el-Deiry, W. S., S. E. Kern, J. A. Pietenpol, K. W. Kinzler, and B. Vogelstein. 1992. Definition of a consensus binding site for p53. Nat. Genet. 1:45-49. [DOI] [PubMed] [Google Scholar]
- 16.Espinosa, J. M., and B. M. Emerson. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 8:57-69. [DOI] [PubMed] [Google Scholar]
- 17.Farmer, G., J. Colgan, Y. Nakatani, J. L. Manley, and C. Prives. 1996. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol. Cell. Biol. 16:4295-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 19.Greasley, P. J., C. Bonnard, and B. Amati. 2000. Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res. 28:446-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 21.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 22.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89:341-347. [DOI] [PubMed] [Google Scholar]
- 23.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 24.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 25.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, D. G., K. Ohtani, and J. R. Nevins. 1994. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 8:1514-1525. [DOI] [PubMed] [Google Scholar]
- 27.Juan, L. J., W. J. Shia, M. H. Chen, W. M. Yang, E. Seto, Y. S. Lin, and C. W. Wu. 2000. Histone deacetylases specifically down-regulate p53-dependent gene activation. J. Biol. Chem. 275:20436-20443. [DOI] [PubMed] [Google Scholar]
- 28.Juven, T., Y. Barak, A. Zauberman, D. L. George, and M. Oren. 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8:3411-3416. [PubMed] [Google Scholar]
- 29.Kaelin, W. G., Jr. 1999. The p53 gene family. Oncogene 18:7701-7705. [DOI] [PubMed] [Google Scholar]
- 30.Khanna, K. K., K. E. Keating, S. Kozlov, S. Scott, M. Gatei, K. Hobson, Y. Taya, B. Gabrielli, D. Chan, S. P. Lees-Miller, and M. F. Lavin. 1998. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat. Genet. 20:398-400. [DOI] [PubMed] [Google Scholar]
- 31.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 32.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate Mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 33.Lakin, N. D., B. C. Hann, and S. P. Jackson. 1999. The ataxia-telangiectasia related protein ATR mediates DNA-dependent phosphorylation of p53. Oncogene 18:3989-3995. [DOI] [PubMed] [Google Scholar]
- 34.Lang, S. E., S. B. McMahon, M. D. Cole, and P. Hearing. 2001. E2f transcriptional activation requires trrap and gcn5 cofactors. J. Biol. Chem. 276:32627-32634. [DOI] [PubMed] [Google Scholar]
- 35.Lechner, M. S., D. H. Mack, A. B. Finicle, T. Crook, K. H. Vousden, and L. A. Laimins. 1992. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J. 11:3045-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, X., C. W. Miller, P. H. Koeffler, and A. J. Berk. 1993. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol. Cell. Biol. 13:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, Y., A. L. Colosimo, X. J. Yang, and D. Liao. 2000. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol. Cell. Biol. 20:5540-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu, H., and A. J. Levine. 1995. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc. Natl. Acad. Sci. USA 92:5154-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo, R. X., A. A. Postigo, and D. C. Dean. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92:463-473. [DOI] [PubMed] [Google Scholar]
- 41.Marhin, W. W., S. Chen, L. M. Facchini, A. J. Fornace, Jr., and L. Z. Penn. 1997. Myc represses the growth arrest gene gadd45. Oncogene 14:2825-2834. [DOI] [PubMed] [Google Scholar]
- 42.Martin, D. W., R. M. Munoz, M. A. Subler, and S. Deb. 1993. p53 binds to the TATA-binding protein-TATA complex. J. Biol. Chem. 268:13062-13067. [PubMed] [Google Scholar]
- 43.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 273:23781-23785. [DOI] [PubMed] [Google Scholar]
- 44.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMahon, S. B., H. A. VanBuskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 46.McMahon, S. B., M. A. Wood, and M. D. Cole. 2000. The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20:556-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Momand, J., D. Jung, S. Wilczynski, and J. Niland. 1998. The MDM2 gene amplification database. Nucleic Acids Res. 26:3453-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Momand, J., H. H. Wu, and G. Dasgupta. 2000. MDM2—master regulator of the p53 tumor suppressor protein. Gene 242:15-29. [DOI] [PubMed] [Google Scholar]
- 49.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura, S., J. A. Roth, and T. Mukhopadhyay. 2000. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol. Cell. Biol. 20:9391-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oliner, J. D., K. W. Kinzler, P. S. Meltzer, D. L. George, and B. Vogelstein. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80-83. [DOI] [PubMed] [Google Scholar]
- 53.Park, J., S. Kunjibettu, S. B. McMahon, and M. D. Cole. 2001. The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 15:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 55.Richon, V. M., T. W. Sandhoff, R. A. Rifkind, and P. A. Marks. 2000. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl. Acad. Sci. USA 97:10014-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez, M. S., J. M. Desterro, S. Lain, D. P. Lane, and R. T. Hay. 2000. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol. 20:8458-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saleh, A., D. Schieltz, N. Ting, S. B. McMahon, D. W. Litchfield, J. R. Yates III, S. P. Lees-Miller, M. D. Cole, and C. J. Brandl. 1998. Tra1p is a component of the yeast ADA/SPT transcriptional regulatory complexes. J. Biol. Chem. 273:26559-26570. [DOI] [PubMed] [Google Scholar]
- 58.Schiltz, R. L., and Y. Nakatani. 2000. The PCAF acetylase complex as a potential tumor suppressor. Biochim. Biophys. Acta 1470:M37-M53. [DOI] [PubMed] [Google Scholar]
- 59.Seto, E., A. Usheva, G. P. Zambetti, J. Momand, N. Horikoshi, R. Weinmann, A. J. Levine, and T. Shenk. 1992. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA 89:12028-12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thut, C. J., J. L. Chen, R. Klemm, and R. Tjian. 1995. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science 267:100-104. [DOI] [PubMed] [Google Scholar]
- 61.Uesugi, M., and G. L. Verdine. 1999. The alpha-helical FXXPhiPhi motif in p53: TAF interaction and discrimination by MDM2. Proc. Natl. Acad. Sci. USA 96:14801-14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van den Heuvel, S. J., T. van Laar, W. M. Kast, C. J. Melief, A. Zantema, and A. J. van der Eb. 1990. Association between the cellular p53 and the adenovirus 5 E1B-55kd proteins reduces the oncogenicity of Ad-transformed cells. EMBO J. 9:2621-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vassilev, A., J. Yamauchi, T. Kotani, C. Prives, M. L. Avantaggiati, J. Qin, and Y. Nakatani. 1998. The 400 kDa subunit of the PCAF histone acetylase complex belongs to the ATM superfamily. Mol. Cell 2:869-875. [DOI] [PubMed] [Google Scholar]
- 64.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 65.Wang, T., T. Kobayashi, R. Takimoto, A. E. Denes, E. L. Snyder, W. S. el-Deiry, and R. K. Brachmann. 2001. hADA3 is required for p53 activity. EMBO J. 20:6404-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, Y., and M. S. Simonson. 1996. Voltage-insensitive Ca2+ channels and Ca2+/calmodulin-dependent protein kinases propagate signals from endothelin-1 receptors to the c-fos promoter. Mol. Cell. Biol. 16:5915-5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Waterman, J. L., J. L. Shenk, and T. D. Halazonetis. 1995. The dihedral symmetry of the p53 tetramerization domain mandates a conformational switch upon DNA binding. EMBO J. 14:512-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 69.Woo, R. A., K. G. McLure, S. P. Lees-Miller, D. E. Rancourt, and P. W. Lee. 1998. DNA-dependent protein kinase acts upstream of p53 in response to DNA damage. Nature 394:700-704. [DOI] [PubMed] [Google Scholar]
- 70.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126-1132. [DOI] [PubMed] [Google Scholar]
- 71.Xiao, G., D. White, and J. Bargonetti. 1998. p53 binds to a constitutively nucleosome free region of the mdm2 gene. Oncogene 16:1171-1181. [DOI] [PubMed] [Google Scholar]
- 72.Yarosh, D. B., P. D. Cruz, I. Dougherty, N. Bizios, J. Kibitel, K. Goodtzova, D. Both, S. Goldfarb, B. Green, and D. Brown. 2000. FRAP DNA-dependent protein kinase mediates a late signal transduced from ultraviolet-induced DNA damage. J. Investig. Dermatol. 114:1005-1010. [DOI] [PubMed] [Google Scholar]
- 73.Zauberman, A., D. Flusberg, Y. Haupt, Y. Barak, and M. Oren. 1995. A functional p53-responsive intronic promoter is contained within the human mdm2 gene. Nucleic Acids Res. 23:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]