Abstract
High-risk human papillomaviruses (HPVs) are associated with carcinomas of the cervix and other genital tumors. The HPV oncoprotein E6 is essential for oncogenic transformation. We identify here hADA3, human homologue of the yeast transcriptional coactivator yADA3, as a novel E6-interacting protein and a target of E6-induced degradation. hADA3 binds selectively to the high-risk HPV E6 proteins and only to immortalization-competent E6 mutants. hADA3 functions as a coactivator for p53-mediated transactivation by stabilizing p53 protein. Notably, three immortalizing E6 mutants that do not induce direct p53 degradation but do interact with hADA3 induced the abrogation of p53-mediated transactivation and G1 cell cycle arrest after DNA damage, comparable to wild-type E6. These findings reveal a novel strategy of HPV E6-induced loss of p53 function that is independent of direct p53 degradation. Given the likely role of the evolutionarily conserved hADA3 in multiple coactivator complexes, inactivation of its function may allow E6 to perturb numerous cellular pathways during HPV oncogenesis.
Studies with viral oncogenes that induce dominant cellular transformation have led to the identification and elucidation of a number of biochemical pathways that are perturbed in human cancer (1, 56, 82). One group of tumor viruses directly implicated in the pathogenesis of human cancer is the “high-risk” subgroup of human papillomaviruses (HPVs), exemplified by HPV type 16 (HPV16), which are thought to be causally linked to the development of >90% cases of cervical cancer (81, 82). In vitro studies have defined two HPV oncogenes, E6 and E7, that are nearly always expressed in HPV-associated carcinomas and cell lines derived from them (5, 63).
The ability of HPV E6 and/or E7 oncogenes to induce the immortalization of human epithelial cells in vitro has provided an invaluable tool to elucidate the mechanisms by which these oncogenes function (6, 30, 50, 73). E6 and E7 are thought to alter the cell growth control mechanisms by inactivating key cellular regulatory proteins (as reviewed in references 81 and 82). For example, E7 protein binds to and inactivates the function of the retinoblastoma tumor suppressor protein (14, 51) by targeting it for degradation via the ubiquitin-proteasome pathway (8, 25). Similarly, E6 interacts with the p53 tumor suppressor protein through the ubiquitin ligase E6AP and targets it for degradation (6, 61, 73, 74), leading to loss of p53-mediated cell cycle arrest and apoptotic responses to DNA damage (29, 44).
While E6-induced loss of p53 function represents a critical element of E6-induced cellular transformation, recent studies have implicated a number of additional cellular targets of E6. Indeed, E6 is now known to directly interact with the calcium-binding protein ERC55 (E6BP, E6-binding protein) (10), focal adhesion component paxillin (69), tumor suppressor protein hDlg (the human homologue of the Drosophila disks large protein) (37, 42), a protein that is required for both the initiation and elongation steps of chromosomal DNA replication Mcm7 (i.e., minichromosome maintenance protein 7) (39, 40), transcriptional factor interferon regulatory factor-3 (IRF-3) (59), transcriptional factor c-myc (28), the proapoptotic protein Bak (Bcl-2 homologous antagonist/killer) (67, 68), the Rap-GTPase activating protein E6TP1 (E6 targeted protein 1) (19), the transcriptional coactivator CBP/p300 (55, 80), the tyrosine kinase Tyk2 (protein-tyrosine kinase 2) (45), the tumor suppressor protein hScrib (the human homologue of the Drosophila Scribble [Vartul] tumor suppressor protein) (52), the PKC-related Rho-regulated serine-threonine kinase PKN (a novel protein kinase with a catalytic domain homologous to that of the protein kinase C) (20), multi-PDZ-domain protein 1 (MUPP1) (43), membrane-associated guanylate kinase protein (MAGI-1) (24), and G-protein pathway suppressor 2 (Gps2) (13). In addition to these direct E6 targets, E6 is now known to rapidly induce an increase in the cellular telomerase activity, and E6-immortalized cells exhibit increased telomerase activity (21, 38, 58, 66). Thus, it is likely that the dominant transforming ability of E6 and other viral oncogenes reflects their ability to concurrently alter multiple cellular controls that ensure the untransformed state of a cell. Identification and characterization of E6 targets is therefore likely to contribute to our understanding of cellular tumor suppressor pathways.
In this study, we have used the yeast two-hybrid interaction system to identify a novel E6 target, hADA3. hADA3 is the human homologue of the yeast transcriptional coactivator yADA3 (alteration or deficiency in activation) (31, 60). Genetic studies in yeast have identified ADA3 as a critical component of coactivator complexes that link transcriptional activators, bound to specific promoters, to histone acetylation and basal transcriptional machinery (as reviewed in reference 7). In yeast, ADA3 and its associated adaptor ADA2 are part of biochemical complexes that recruit GCN5 (i.e., general control nonrepressed 5), a histone acetyltransferase (HAT), to promoters (15, 32, 60, 71).
We demonstrate here that hADA3 binds to the high-risk (cancer-associated) but not the low-risk (benign lesion-associated) HPV E6 proteins and to immortalization-competent but not the immortalization-defective HPV16 E6 mutants. Additionally, E6 induces a dramatic decrease in the levels of hADA3 protein. We demonstrate that hADA3 directly interacts with the p53 protein in vitro and in vivo. Furthermore, overexpression of hADA3 markedly enhances the p53-mediated sequence-specific transactivation of p53 target genes. Most significantly, the E6 mutants that are incapable of interacting with and targeting p53 for degradation inhibit the p53-mediated transactivation and G1 cell cycle arrest. These results demonstrate that E6 inactivates the function of an evolutionarily conserved transcriptional coactivator and provide a second mechanism of E6-induced inhibition of p53 function independent of E6-induced p53 degradation. Given the strong likelihood that human ADA3, similar to its yeast counterpart, will function as a coactivator in multiple mammalian transcriptional pathways, the inactivation of ADA3 function by the high-risk HPV E6, and possibly by other viral oncoproteins, may represent a crucial part of the viral oncogenic strategy.
MATERIALS AND METHODS
Yeast two-hybrid constructs and screening.
A Matchmaker two-hybrid “prey” cDNA library in pGAD10 vector was constructed from mRNA derived from a normal mammary epithelial cell line 76N (19, 20) by using a mixture of oligo(dT) and random hexanucleotide primers (custom made through Clontech, Palo Alto, Calif.). The library contained 1.5 × 106 primary recombinants with an average insert size of 1.5 kb. The HPV16 E6 sequences encoding residues 2 to 158 were inserted into pGBT9 vector for use as a bait (19). Yeast two-hybrid screening was performed as described previously (19, 20). HPV16 E6-interacting proteins scoring positive by growth on Trp−, Leu−, and His− selection medium, as well as expression of β-galactosidase activity in comparison with a negative control bait pLam 5′ (human lamin-Gal4 DNA-binding domain hybrid; Clontech), were processed further.
Cells and media.
Saos-2 and U2OS osteosarcoma cell lines were grown in α-MEM (Life Technologies, Grand Island, N.Y.) supplemented with 10% fetal calf serum (HyClone, Logan, Utah). 293T cells were grown in Dulbecco modified Eagle medium (Life Technologies, Grand Island, N.Y.) supplemented with 10% fetal calf serum. Breast epithelial cell strain 76N and its immortal derivatives (76-E6, 76-F2V, 76-8S9A10T, and 76-Y54H) were grown in DFCI-1 medium as described previously (6, 8, 12, 47).
Plasmid constructs.
Full-length hADA3 and hADA2 were amplified from normal mammary gland cDNA (Clontech) by using the Advantage cDNA amplification kit and the following primers: ADA3-sense (ATGCGGCCGCCATGAGTGAGTTGAAAGACTGCCCC), ADA3-antisense (ATTCTAGACTACCCATCCAGCAGCTTCAGGA), ADA2-sense (ATGCGGCCGCATGGCCCTTTTAGAAGCTGTGAT), and ADA2-antisense (ATGGATCCTTAGCCTTTAGTGATGTATCCTTCT). The PCR incorporated an N-terminal FLAG tag in ADA3, as well as in ADA2. The resultant PCR products were cloned in pCR3.1, a mammalian expression vector (Invitrogen), and their sequences were verified. A pCR3.1 construct encoding the untagged or FLAG-tagged wild-type human p53 was derived by subcloning the cDNA insert from pCMV-p53 (obtained from Bert Vogelstein, Johns Hopkins Medical Center) into pCR3.1 vector. GST-hADA3(75-432) was constructed by subcloning the corresponding sequence from the original pGAD10 clone into pGEX-4T1 vector (Roche). GST-hADA2 and GST-hADA3 constructs in pGEX-4T1 encoding the complete polypeptides fused to glutathione S-transferase (GST), respectively, were derived by subcloning these inserts from pCR3.1 constructs or by using the PCR (for GST-p53). Histidine-tagged hADA3 was constructed by cloning the PCR-derived cDNA fragment in pEF-His vector (Invitrogen). GST-E6AP (amino acids [aa] 37 to 865) was obtained from Peter Howley (Harvard Medical School) (33, 34). HPV16, HPV18, HPV11, HPV6 E6, the HPV16 E6 mutants in pSG5 vector used for in vitro translation, and GST-NES1 have been described previously (19, 46). pEF-16E6-MYC, encoding Myc-tagged E6, was kindly provided by M. Ishibashi (Aichi Cancer Center, Aichi, Japan) (37). GST-p300 (aa 300 to 528) was obtained from S. Grossman (Dana-Farber Cancer Institute, Harvard Medical School) (27). The p53-responsive luciferase reporters incorporating the promoter elements of Bax, cyclin G, Gadd45, mdm2, and IGFBP-3 were obtained from Carol Prives (Columbia University, New York, N.Y.) and pG13 and MG15 reporters were provided by Wafik El-Diery (University of Pennsylvania) originally obtained from Bert Vogelstein (Johns Hopkins School of Medicine). The pEGFP-N1 construct was purchased from Clontech.
In vitro binding assays.
The indicated 35S-labeled proteins were generated from their pCR3.1 expression constructs by using a wheat germ lysate-based coupled in vitro transcription-translation system (TNT Wheat Germ Lysate System; Promega) in the presence of [35S]cysteine or [35S]methionine as described previously (19). Aliquots of 35S-labeled proteins were incubated with 1 μg of GST or appropriate GST fusion proteins noncovalently bound to glutathione beads in 300 μl of lysis buffer (100 mM Tris, pH 8.0; 100 mM NaCl; 0.5% Nonidet P-40) for 2 h at 4°C, and bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by fluorography.
In vivo association between hADA3 to 16E6.
293T cells were plated at 106 per 100-mm dish and transfected with 0.5 μg of pCR3.1 construct, encoding the FLAG-tagged hADA3, and 2.5 μg of pEF-myc-16E6, either alone or in combination, by the calcium phosphate method (6). After 48 h, cell lysates were prepared in lysis buffer (100 mM Tris, pH 8.0; 100 mM NaCl; 0.5% NP-40; 1 mM phenylmethylsulfonyl fluoride). Lysates were precleared twice with protein G-agarose, and equal aliquots were used for immunoprecipitation with anti-FLAG antibody (M2; Sigma, St. Louis, Mo.), followed by immunoblotting with anti-myc (9E10) or anti-FLAG monoclonal antibodies. Enhanced chemiluminescence (ECL) was used for detection (Amersham, Piscataway, N.J.).
E6-induced degradation of hADA3.
A total of 5 × 105 293T cells per 100-mm dish were transfected with 0.5 μg of pCR3.1-FLAG-hADA3, with or without the indicated amounts of pEF-myc-tagged HPV16 E6, by the calcium phosphate method. Five hundred nanograms of green fluorescent protein (GFP) DNA was cotransfected to assess the transfection efficiency. The total amount of DNA was held constant by adding vector DNA. Cell lysates were prepared after 48 h in 1× sample buffer (50 mM Tris, pH 8.0; 2% SDS; 10% glycerol), and 30-μg aliquots of lysate protein were immunoblotted with anti-FLAG antibody to detect ADA3 or with anti-myc antibody to detect the E6 protein. Parallel plates were used to isolate total RNA for Northern blotting of the introduced hADA3 and GFP (as a control) by using standard procedures (19).
Pulse-chase analysis.
A total of 5 × 105 293T cells per 100-mm dish were cotransfected with 0.5 μg of hADA3 and either 2 μg of vector or HPV16 E6. After 48 h, cells were metabolically pulse-labeled with 300 μCi of [35S]methionine or [35S]cysteine (Express35S35S; NEN) for 30 min and chased for the indicated time periods. Equal amounts of radiolabeled lysates (based on counts) were immunoprecipitated with anti-hADA3 serum (generated against a peptide representing ADA3 residues 418 to 432 [Animal Pharm Services, Inc.]) and analyzed by SDS-PAGE, followed by fluorography.
In vitro E6-induced hADA3 degradation.
p53, hADA3, HPV16 E6, and 6 E6 were translated in vitro in the presence of [35S]cysteine (E6 proteins) or [35S]methionine (hADA3 and p53) by using a rabbit reticulocyte lysate-based coupled transcription-translation system (TNT Rabbit Reticulocyte Lysate System; Promega) as suggested by the supplier. To assess degradation, 5-μl aliquots of in vitro-translated 35S-labeled hADA3 or p53 were incubated with 5 μl of HPV16 E6, 6 E6, or water-primed (control) lysate. After 15 h at 30°C, the degradation reactions were terminated by adding sample buffer, and the proteins were resolved by SDS-PAGE and visualized by fluorography.
In vivo association between p53 and hADA3.
A total of 5 × 105 U2OS cell, expressing the endogenous wild-type p53 (2) were transfected with 2.5 μg of pCR3.1-hADA3 or vector, by using the Fugene reagent, according to the supplier's instructions (Roche). After 48 h cell lysates were prepared, and 1-mg aliquots of cell lysate protein were precleared with protein G-agarose and subjected to immunoprecipitation with an anti-p53 antibody (ab-1; Oncogene Research). Immunoblotting for ADA3 was performed with a rabbit anti-ADA3 antiserum, followed by ECL detection.
To assess the association of p53 and hADA3 in 293T cells, 1 μg of FLAG-p53 was transfected with or without 8 μg of His-tagged hADA3 by the calcium phosphate method. At 48 h posttransfection, cells were lysed in binding buffer (50 mM Tris, pH 8.0; 100 mM NaCl; 0.5% Nonidet P-40), and 400 μg of protein was incubated with His-Bind resin (Novagen) for 5 min at 4°C. Beads were washed five times with binding buffer, and bound proteins were eluted by boiling in sample buffer, followed by SDS-PAGE and immunoblotting with anti-FLAG antibody to detect p53. The membrane was reprobed with anti-His antibody (Santa Cruz) to assess the expression of hADA3.
p53-dependent transactivation of luciferase reporters.
A total of 5 × 105 Saos-2 cells (p53-negative osteosarcoma cell line) per 100-mm dish were transfected with various expression plasmids indicated in the figure legends, together with the indicated luciferase reporters. Each dish also received 20 ng of simian virus 40-Renilla luciferase reporter (pRL-SV40) to correct for differences in transfection. Total DNA was kept constant by adding the vector DNA. Luciferase activity was measured 24 h later by using a dual-luciferase kit (Promega).
Analysis of cell cycle arrest after DNA damage.
A total of 5 × 105 cells per 100-mm-diameter dish of normal mammary epithelial cell strain 76N, E6-immortalized 76N (76-E6), E6-F2V mutant-immortalized 76N (76-F2V), E6-8S9A10T mutant-immortalized 76N (E6-8S9A10T), and E6-Y54H mutant-immortalized 76N (76-Y54H) were cultured in DFCI-1 medium for 48 h. Cells were then treated with 0.5 μg of adriamycin/ml for 24 h to induce DNA damage. Cells were trypsinized, fixed in 70% ethanol, treated with RNase A (50 μg/ml for 30 min at 37°C), and stained with propidium iodide (50 μg/ml for 30 min). The cell cycle distribution was quantified by fluorescence-activated cell sorting (Coulter, Miami, Fla.).
RESULTS
Identification of hADA3 as a novel HPV16 E6-interacting protein.
As we have reported previously, screening of a mammary epithelial cell yeast two-hybrid library with HPV16 E6 as a bait identified 28 specific E6-interacting clones (out of 3.96 × 106 screened) (19, 20). Homology analysis by a gapped BLAST search of the NCBI database showed one clone (clone 36) to correspond to residues 75 to 432 of the 432-aa human ADA3 (53). Human ADA3 is evolutionarily conserved, with significant homology with yeast and Drosophila ADA3 (53).
Expression pattern of hADA3.
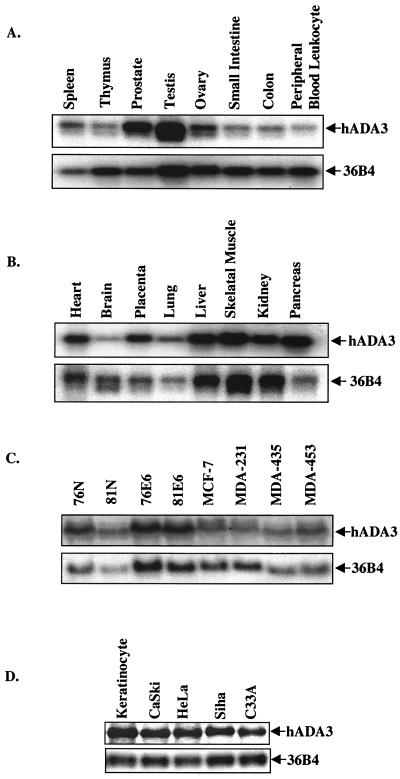
To verify that hADA3 corresponds to an expressed human gene and to assess its expression, we performed Northern blotting of total RNA from normal, immortal, and tumor cell lines, as well as in multiple human tissues by using multiple tissue RNA blots (Clontech). One major transcript of approximately 1.3 kb was observed in all tissues and in vitro established cell lines tested, although the levels varied in different tissues (Fig. 1). These results indicated that the novel E6 target hADA3 indeed represented the product of a transcribed human gene.
FIG. 1.
ADA3 mRNA expression in human tissues and cultured cells. Blots containing 2 μg of poly(A)+ mRNA per lane (Tissue-Blot [Clontech]) (A and B) or 20 μg of total RNA (C and D) from normal (76N and 81N), immortal (76E6 and 81E6), and tumor (MCF-7, MDA-MB-231, MDA-MB-435, and MDA-MB-453) mammary epithelial cells (C) or from normal keratinocytes and cervical carcinoma cell lines (HeLa, CasKi, Siha, and C33A) (D) were probed with a 32P-labeled 1.3-kb hADA3 cDNA probe, followed by autoradiography. Hybridization with the 36B4 probe was used as a loading control. The arrows point to the major 1.3-kb transcript, which is seen at various levels in all cell lines and tissues.
High-risk but not low-risk E6 proteins bind hADA3 protein in vitro.
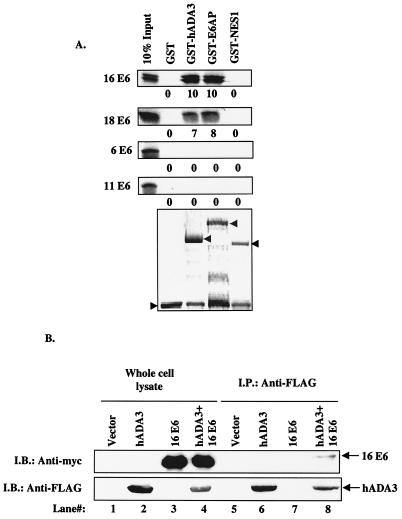
To further confirm the interaction between E6 proteins and hADA3 identified by the yeast two-hybrid approach, we assessed the binding of in vitro-translated 35S-labeled HPV16, HPV18, HPV6, or HPV11 E6 polypeptides to GST-ADA3 (hADA3 aa 75 to 432 fused to the C terminus of GST). As expected, substantial binding of HPV16 and HPV18 “high-risk” E6 proteins to GST-E6AP (aa 37 to 865, used as a positive control) (34), but little binding of HPV6 or HPV11 “low-risk” E6 proteins, was observed. GST alone and GST-NES1 (46), used as negative controls, showed no binding. Importantly, similar to GST-E6AP, GST-hADA3 showed substantial binding to high-risk HPV (HPV16 and HPV18) E6 proteins, whereas little binding was seen to low-risk HPV (6 and 11) E6 proteins (Fig. 2A, upper panel). Coomassie blue staining of the gel confirmed the presence of respective GST fusion proteins in binding reactions (Fig. 2A, lower panel). Thus, similar to E6AP, hADA3 selectively interacts with high-risk HPV E6 proteins.
FIG. 2.
In vitro and in vivo association between HPV E6 proteins and hADA3. (A) In vitro binding of high-risk (HPV16 and HPV18) and low-risk (HPV6 and HPV11) E6 proteins to hADA3. 35S-labeled E6 proteins were generated by in vitro translation with wheat germ lysate, and aliquots were allowed to bind to GST, GST-hADA3 (aa 75 to 432), GST-E6AP (as a positive control), or GST-NES1 (as a negative control) coated on glutathione-Sepharose beads. Bound E6 proteins were resolved by SDS-PAGE and visualized by fluorography. The input lane contains 10% of the aliquots used in the binding reactions. The numbers under each lane indicate the percent binding compared to the input, as determined by densitometry. The bottom panel shows Coomassie blue staining of the GST fusion proteins used in binding reactions. (B) In vivo association of hADA3 with HPV16 E6. A total of 106 293T cells per 100-mm dish were transfected with 0.5 μg of pCR3.1-FLAG-hADA3 or 2.5 μg of pEF-myc-HPV16 E6 (16 E6) alone or in combination, as indicated. After 48 h, the cell lysates were subjected to anti-FLAG immunoprecipitation, followed by either anti-myc (upper panel, lanes 5 to 8) or anti-FLAG (lower panel, lanes 5 to 8) immunoblotting (I.B.). Whole-cell lysates (10% input, lanes 1 to 4) were directly blotted with anti-myc antibody (upper panel) or anti-FLAG antibody (lower panel) to assess the expression of HPV16 E6 and hADA3, respectively.
HPV16 E6 protein binds to hADA3 protein in vivo.
To assess whether E6-hADA3 interaction can take place in mammalian cells in vivo, 293T cells were transfected with plasmids encoding FLAG-tagged hADA3 and myc-tagged HPV16 E6. The hADA3 protein was immunoprecipitated with an anti-FLAG antibody, and coimmunoprecipitated E6 was detected by anti-myc immunoblotting. As expected, no E6 protein was detected in the immunoprecipitates of cells transfected with vector alone or hADA3 or E6 constructs transfected individually (Fig. 2B, upper panel, lanes 5, 6, and 7). In contrast, E6 protein was clearly detected in anti-FLAG immunoprecipitates of cells cotransfected with hADA3 and E6 (Fig. 2B, upper panel, lane 8). Anti-FLAG (Fig. 2B, lower panel, lanes 1 to 4) and anti-myc immunoblotting of whole-cell lysates (upper panel, lanes 1 to 4) indicated the expression of E6 and hADA3 proteins in the appropriate lanes. Similar results were observed upon transfection of E6 and hADA3 constructs in Cos-7 cells (data not shown). These results clearly demonstrate that E6 and hADA3 associate in vivo.
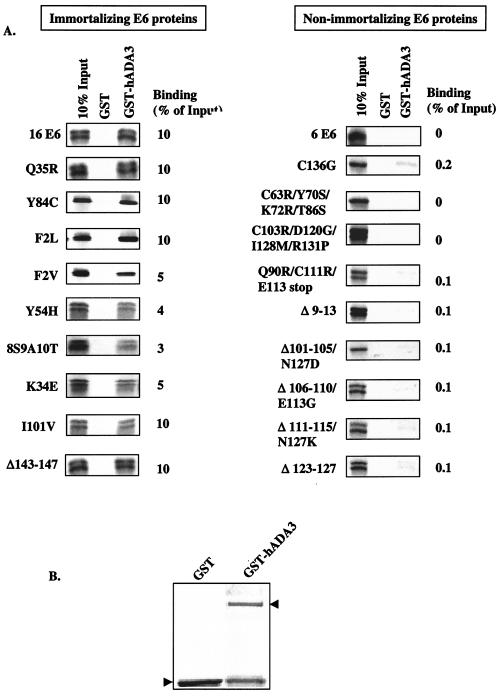
Immortalizing but not the nonimmortalizing E6 mutants bind hADA3.
We previously characterized a large panel of HPV16 E6 mutants, and the binding of these mutants to E6AP, E6BP, p53, and E6TP1 has been well defined (12, 21, 47). We utilized these mutants to determine whether a correlation existed between their ability to bind to ADA3 and their known behavior as immortalizing or nonimmortalizing proteins in mammary epithelial cells (12, 21, 47). Binding was assessed in an in vitro assay by using GST-hADA3 fusion protein and in vitro-translated [35S]cysteine-labeled E6 proteins (19, 20). These results demonstrated that, similar to wild-type E6, those E6 mutants that are known to induce the immortalization of mammary epithelial cells (MEGs), were able to bind to hADA3 (Fig. 3A, left panel). In contrast, none of the E6 mutants that are deficient in mammary epithelial cell immortalization showed binding to hADA3 over background (Fig. 3A, right panel). Figure 3B shows the presence of GST fusion proteins in the binding reactions, as assessed by Coomassie blue staining. These results show that binding to hADA3 strongly correlates with the ability of HPV16 E6 to immortalize epithelial cells.
FIG. 3.
In vitro binding of immortalizing (left panel) and nonimmortalizing (right panel) HPV16 E6 mutants to hADA3. (A) Equal aliquots of in vitro-translated 35S-labeled E6 proteins were allowed to bind to GST or GST-hADA3 (aa 75 to 432), and bound E6 proteins were then resolved by SDS-PAGE and visualized by fluorography. The input lane contains 10% of the aliquots used in the binding reactions. The numbers next to each lane indicate the percent binding compared to the input. Δ, Deletion. The immortalizing versus nonimmortalizing designation of each E6 mutant is derived from a mammary epithelial cell immortalization assay reported previously (12, 47). All mutant proteins are derivative of HPV16 E6. (B) The bottom panel shows Coomassie blue staining of the GST fusion proteins used in the binding reactions.
Loss of hADA3 protein upon coexpression of HPV16 E6.
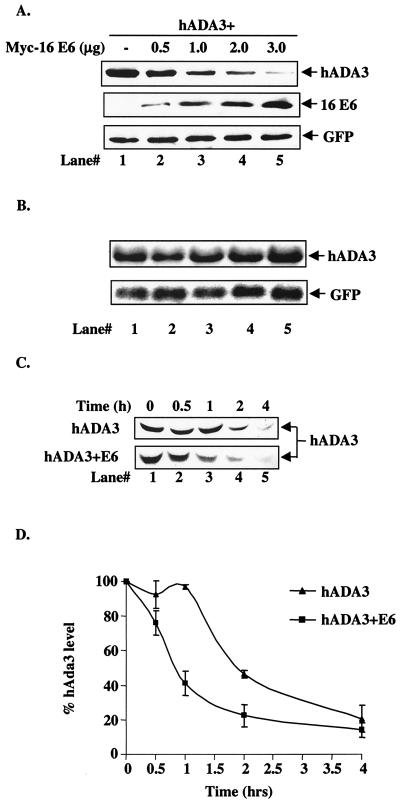
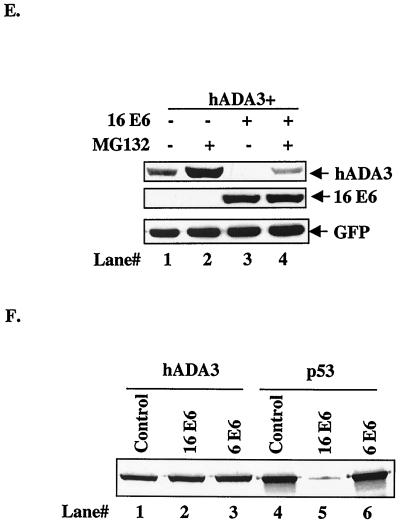
Among the previously identified E6-binding proteins, p53 (via E6AP), hMCM7, Bak, E6TP1, Vartul, hDlg, and Gps2 (13, 19, 33, 34, 39, 40, 49, 52, 61, 68) are known to be targeted for ubiquitin-dependent proteasome-mediated degradation, whereas ERC55, paxillin, IRF-3, p300, and PKN are not destabilized (10, 20, 55, 59, 69, 80). To begin to assess whether E6 may target hADA3 for degradation, FLAG-tagged hADA3 in pCR3.1 vector and increasing amounts of myc-tagged HPV16 E6 in pEF vector were transiently expressed in 293T cells. Due to the unavailability of blotting anti-E6 antibodies, a myc-tagged E6 was used. Cell lysates after transfection were subjected to immunoblotting with anti-FLAG to detect hADA3 and anti-myc to detect myc-tagged E6. Significantly, coexpression of HPV16 E6 protein dose dependently reduced the level of hADA3 protein (Fig. 4A, lanes 1 to 5). Parallel plates of transfected cells from the experiment shown in Fig. 4A were used to isolate total RNA and subjected to Northern blotting with 1.3-kb hADA3 cDNA as a probe. These analyses showed the hADA3 mRNA levels to be invariant upon cotransfection with E6 (Fig. 4B), a finding consistent with E6-induced loss of hADA3 protein via degradation. The levels of GFP mRNA and protein, used as a transfection efficiency control, showed no change (Fig. 4A and B). Due to the unavailability of antibodies that recognize endogenous hADA3 protein, we transfected 293T cells with hADA3 alone or together with HPV16 E6 to determine hADA3 protein stability. Transfected cells were pulse-labeled with [35 S]methionine and [35S]cysteine, followed by a chase for various time periods. Anti-ADA3 immunoprecipitates of these lysates were subjected to fluorography. As seen in Fig. 4C (a representative autoradiogram) and D (data plotted from three independent experiments), hADA3 turnover was substantially faster, compared to controls, in cells cotransfected with E6. Furthermore, treatment of transfected cells with proteasome inhibitor MG132 (8, 25) resulted in substantial inhibition of E6-induced loss of hADA3 protein (Fig. 4E, lane 4, versus lane 3). Taken together, these findings strongly support the conclusion that in vivo interaction with E6 leads to hADA3 degradation. Notably, the presence of 16 E6 did not enhance the degradation of hADA3 in vitro under conditions in which p53 was clearly degraded (Fig. 4F). These results suggest that certain factors that are only present in human cells but not in the rabbit reticulocyte lysates are required for E6-induced degradation of hADA3.
FIG. 4.
Reduction of hADA3 protein levels upon coexpression of HPV16 E6. (A) A total of 5 × 105 293T cells per 100-mm dish were transfected with 0.5 μg each of pCR3.1-FLAG-hADA3 either alone or together with increasing amounts of pEF-myc-HPV16 E6, as indicated. Five hundred nanograms of GFP DNA was included to assess the transfection efficiency. Total amount of DNA was kept constant by adding vector DNA. Cell lysates were prepared at 48 h posttransfection, and 30-μg aliquots of lysate protein were subjected to anti-FLAG immunoblotting (upper panel), followed by anti-myc (middle panel) and anti-GFP (lower panel) reblotting. (B) Duplicate plates of the transfections shown in panel A were used to isolate total cellular RNA, and 20-μg aliquots were used for Northern blotting with a 32P-labeled 1.3-kb hADA3 cDNA probe, followed by autoradiography (upper panel). A GFP cDNA probe was included in the hybridization mix as a control (lower panel). (C) Pulse-chase analysis of hADA3. 293T cells were transfected with 0.5 μg of pCR3.1-hADA3 alone or together with 2 μg of vector or pCR3.1-HPV16 E6. After 48 h, cells were metabolically pulse-labeled with [35S]methionine plus [35S]cysteine for 30 min and chased for the indicated time periods (shown in hours). Equal amounts of radiolabeled lysates (based on the counts per minute) were immunoprecipitated with anti-hADA3 antiserum and resolved by SDS-PAGE, followed by fluorography. A representative experiment is shown. (D) The amount of hADA3 was calculated by densitometry, and the percentage of hADA3 left was plotted against time indicated. The mean ± the standard deviation (SD) of three independent experiments is shown. (E) 293T cells were transfected with 0.5 μg of pCR3.1-FLAG-hADA3 alone or with 2.5 μg of pEF-myc-16 E6. At 20 h after transfection, the cells were either mock treated or treated with MG132 (50 μM) for 4 h. Immunoblotting was performed as for panel A. (F) Lack of in vitro degradation of hADA3 by E6. The 35S-labeled hADA3, p53, HPV6 E6, and HPV16 E6 proteins were generated by in vitro translation in rabbit reticulocyte lysates in the presence of [35S]cysteine (E6) or [35S]methionine (ADA3 and p53). Aliquots of 35S-labeled hADA3 (lanes 1 to 3) or p53 (lanes 4 to 6) were incubated with water-primed lysate (control; lanes 1 and 4) or HPV16 E6 (lanes 2 and 5) or HPV6 E6 (lanes 3 and 6) proteins for 15 h at 30°C. The hADA3 or p53 remaining at the end of the incubation period was resolved by SDS-PAGE and visualized by fluorography.
hADA3 interacts with p53 protein in vitro.
In view of the selective interaction of immortalizing HPV E6 proteins with hADA3 and E6-induced hADA3 degradation, it became evident that E6 may inhibit hADA3 function as part of its oncogenic process. However, the function(s) of human ADA3 has not been described, although its ability to assemble into ADA-like multiprotein complexes in mammalian cells (53) suggested that it could function as a transcriptional coactivator, as does its yeast homologue (7). Of direct relevance to E6-mediated epithelial cell immortalization, which involves a critical role for inactivation of p53 function (12), we investigated the possibility that hADA3 may function as a p53 coactivator and that its interaction with E6 may provide an additional mechanism for E6-induced loss of p53 function. This idea was consistent with previous observations that the human p53 transactivation domain fused to the Gal4 DNA-binding domain was transcriptionally competent in yeast and that this transactivation function required yADA3 (9).
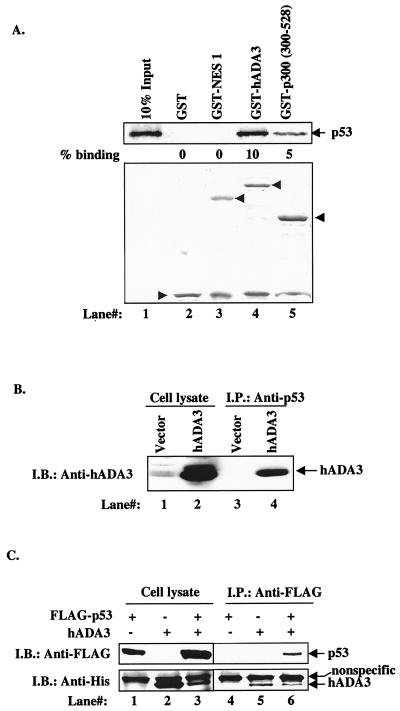
As a first step, we sought to determine whether hADA3 might directly interact with human p53 in a way analogous to the physical interaction of yADA3 with transcriptional activators for which it functions as a coactivator (9). Therefore, we assessed the binding of in vitro-translated 35S-labeled p53 to GST-hADA3. As expected, p53 showed substantial binding to GST-p300 (aa 300 to 528, used as a positive control). GST alone or GST-NES1, used as negative controls, showed no binding. Importantly, p53 showed clear binding to GST-hADA3, similar to its binding to GST-p300 (Fig. 5A). Thus, p53 and hADA3 directly interact with each other.
FIG. 5.
In vitro and in vivo interaction between hADA3 and p53. (A) In vitro binding between hADA3 and p53. Binding reaction mixtures included equal aliquots of [35S]methionine-labeled p53 generated by in vitro translation (as in Fig. 4F) and 1 μg of GST, GST-hADA3, GST-p300 (as a positive control), or GST-NES1 (as a negative control). Bound p53 protein was resolved by SDS-PAGE and detected by fluorography. The numbers under each lane indicate the percent binding as based on input control. The bottom panel shows Coomassie blue staining of the GST fusion proteins used in the binding reactions. (B) In vivo association of endogenous p53 with transfected hADA3. A total of 5 × 105 U2OS cells were transfected with 2.5 μg of pCR3.1 vector or pCR3.1-hADA3 by using the Fugene reagent. After 48 h, 1-mg aliquots of cell lysate proteins were subjected to anti-p53 immunoprecipitation (I.P.) and immunoblotted (I.B.) with an anti-hADA3 serum (lanes 3 and 4). Whole-cell lysates (50 μg) were directly blotted with anti-hADA3 serum (lanes 1 and 2) to assess the expression of hADA3. (C) In vivo association between transfected p53 and hADA3. A total of 106 293T cells were transfected with 1 μg of FLAG-p53 and 8.0 μg of His-tagged hADA3 alone or in combination. At 40 h after transfection, the cells were lysed in binding buffer, and 400-μg aliquots of lysates were allowed to bind with His-Bind resin (Novagen). The bead-bound proteins were subjected to SDS-PAGE, followed by anti-FLAG immunoblotting to detect transfected p53 (upper panel, lanes 4 to 6). The membrane was reprobed with an anti-His antibody to assess the levels of hADA3 (lower panel, lanes 4 to 6). The cell lysate (lanes 1 to 3) represents 40-μg aliquots of lysate protein.
p53 protein associates with hADA3 protein in vivo.
Given the direct in vitro binding between hADA3 and p53 (Fig. 5A), we examined whether these proteins associate in vivo by using two different cell types. First, p53-positive U2OS osteosarcoma cells (2) were transfected with pCR3.1 vector or pCR3.1-hADA3, and cell lysates were subjected to anti-p53 immunoprecipitations, followed by immunoblotting with a rabbit anti-ADA3 antiserum to detect p53-associated hADA3. hADA3 was clearly coimmunoprecipitated with endogenous p53 in hADA3-transfected U2OS cells (Fig. 5B, lane 4). In another experiment, we cotransfected hADA3 in pEF-His vector and FLAG-tagged p53 in pCR3.1 vector in 293T cells, affinity isolated hADA3 on His-Bind resin, and assessed the presence of associated FLAG-p53 by using anti-FLAG immunoblotting. As with endogenous p53 (above), a clear association between exogenously expressed p53 and ADA3 was seen (Fig. 5C). Immunoblotting of whole-cell lysates showed the expected expression of proteins. These results clearly demonstrate that hADA3 and p53 associate in vivo.
hADA3 enhances the p53-dependent transactivation of a p53-consensus site containing reporters.
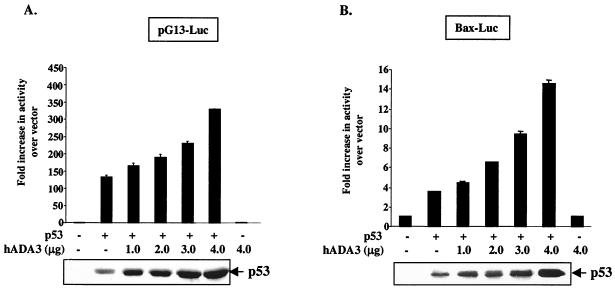
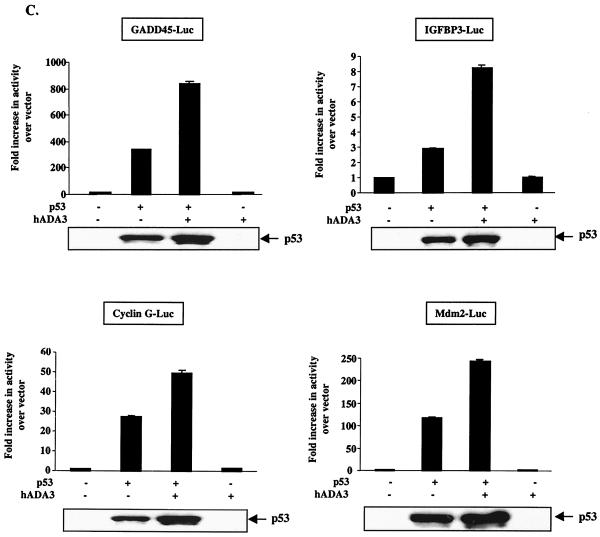
Given the direct interaction of hADA3 with p53 and its assembly into chromatin-associated complexes bound to p53 target promoters, we sought to determine whether hADA3 indeed functions as a coactivator for p53. For this purpose, p53-negative Saos-2 cells were cotransfected with a fixed amount of pCR3.1 constructs of p53 and increasing amounts of hADA3, together with either pG13-luciferase (a p53-responsive reporter containing 13 repeats of a consensus p53-binding site upstream of firefly luciferase) (36) or Bax-luciferase (the promoter of p53-responsive gene cloned upstream of luciferase) (18). Luciferase activity was quantified in cell lysates 24 h posttransfection. Consistent with previously published results (36), little luciferase activity was observed in vector-transfected cells, with about a 100-fold increase in pG13-luciferase activity (Fig. 6A) and a 4-fold increase in Bax-luciferase activity (Fig. 6B) upon cotransfection with p53. Significantly, cotransfection of hADA3 dose dependently increased the activity of both reporters (Fig. 6A and B). No activity was observed when hADA3 was transfected without p53, demonstrating that the hADA3-induced increase in transactivation was p53 dependent (Fig. 6). Notably, Western blot analysis of cell lysates used in luciferase assay showed an hADA3 dose-dependent increase in p53 protein levels. These data suggest that, similar to other coactivators such as p300 (26, 27), hADA3 coactivator function may also involve stabilization of p53 protein. No transactivation of MG15-luciferase (containing a mutated p53-binding consensus and used as a negative control) (36) was seen upon transfection with p53 or p53 together with hADA3 (data not shown). We further confirmed the ability of hADA3 to enhance the p53-mediated transactivation by using luciferase reporters linked to promoters of several p53 target genes such as GADD45, IGFBP3, cyclin G, and mdm2 (57). In each case, hADA3 enhanced the p53-mediated transactivation of the reporters, whereas no transactivation was observed when hADA3 alone was transfected (Fig. 6C). Similar to Fig. 6A and B, cotransfection of hADA3 with p53 increased the levels of p53 protein (bottom panel). Taken together, these results clearly demonstrate that hADA3 functions as a coactivator for p53-dependent transactivation in mammalian cells.
FIG. 6.
Enhancement of p53-dependent transactivation by hADA3. A total of 5 × 105 Saos-2 cells were transfected with 25 ng (A) or 50 ng (B and C) of pCR3.1-p53 and pCR3.1-hADA3 (4 μg [C] or as indicated [A and B]), together with 250 ng (A) or 1 μg (B and C) of the indicated promoter-luciferase reporters. The total DNA amount for each transfection was kept constant by adding vector DNA. After 24 h of transfection, cells were lysed, and the luciferase activity was measured. Transactivation results were calculated as fold activation over vector. A total of 20 ng of pRL-SV40 was concurrently transfected and used to correct for differences in transfection. Data indicate mean ± SD of triplicates of a representative experiment. Experiments were repeated at least three times. A “−” indicates the vector DNA. The lysates of cells used in luciferase assays were analyzed by immunoblotting with an anti-p53 antibody (DO1) to assess the expression of p53 (lower panels).
HPV16 E6 inhibits the coactivator function of hADA3.
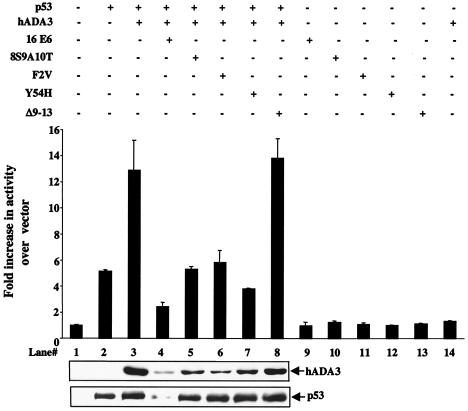
Given the ability of E6 to interact with and induce the degradation of hADA3, we examined whether E6 inhibits the coactivator function of hADA3. Since E6 is known to target p53 protein for ubiquitin-dependent proteasome-mediated degradation (48), wild-type E6 alone could not be employed in these experiments. However, we and others have previously characterized three E6 mutants—F2V, 8S9A10T, and Y54H—that do not induce degradation of p53 but do induce cellular immortalization (12, 17, 38, 47) (Fig. 7, lower panel). Our binding experiments (Fig. 3) indicated that these mutants were competent at binding to hADA3, thus providing reagents to assess E6-induced perturbation of ADA3 function in the context of p53 transactivation. Saos-2 cells were transfected with pCR3.1 constructs of p53, hADA3, wild-type E6, or E6 mutants individually or in combination, as indicated, together with a Bax-luciferase reporter (Fig. 7). As expected, hADA3 significantly enhanced the p53-dependent transactivation of Bax promoter (Fig. 7, lane 2 versus lane 3). Neither hADA3 nor E6 or its mutants induced the luciferase activity on their own (Fig. 7, lanes 9 to 14). Cotransfection of wild-type E6 markedly inhibited the hADA3/p53-mediated transactivation (Fig. 7, lane 4), as expected. Importantly, each of the E6 mutants that do not directly interact with p53 was able to induce a marked decrease in the level of luciferase activity induced by cotransfection of p53 and ADA3 (Fig. 7, lanes 5 to 7). Significantly, the Δ9-13 E6 mutant, which does not bind to hADA3, was incapable of inhibiting hADA3/p53-dependent transactivation (Fig. 7, lane 8). The ability of E6 and its mutants to abrogate hADA3 function was accompanied by the expected reduction in hADA3 levels (Fig. 7, bottom panel). These results clearly demonstrate that interaction of E6 with hADA3 abrogates its coactivator function.
FIG. 7.
Inhibition of hADA3 coactivator function by HPV16 E6 proteins. A total of 5 × 105 Saos-2 cells were cotransfected with 1 μg of Bax-luciferase reporter with or without 50 ng of pCR3.1-p53, 3 μg of pCR3.1-hADA3, and 2.5 μg of pCR3.1-16 E6 or its mutants, as indicated (+). The total DNA amount for each transfection was kept constant by adding vector DNA. After 24 h of transfection, the luciferase activity was measured as described in the legend for Fig. 6. For the expression of hADA3 and p53, the lysates described above were analyzed by Western blotting with anti-hADA3 serum (upper panel) or anti-p53 antibody (lower panel).
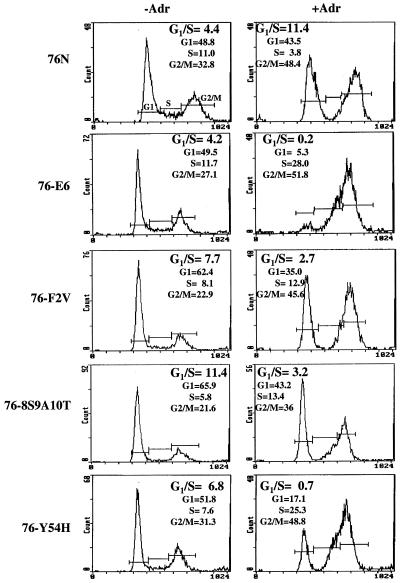
Based on the results presented above, we sought to determine whether the E6 mutants examined above compromised the p53 function. For this purpose, we assessed the ability of the DNA-damaging agent adriamycin to induce G1 cell cycle arrest in mammary epithelial cells stably expressing E6 mutants (8S9A10T, F2V, and Y54H) compared to wild-type E6-immortalized MECs and parental normal MECs (76N). As expected, adriamycin treatment of normal MECs led to G1 cell cycle arrest (an increase in the G1/S ratio from 4.4 to 11.4) (Fig. 8). Wild-type E6-immortal cells showed complete abrogation of G1 arrest (a clear decrease in G1/S ratio from 4.2 to 0.2). Importantly, cells immortalized with these three E6 mutants behaved similarly to wild-type E6-immortalized cells in that the G1 cell cycle arrest response was disrupted. Taken together, these results suggest that E6 can abrogate p53 function by a mechanism that does not involve direct E6-dependent p53 degradation.
FIG. 8.
Abrogation of adriamycin-induced G1 cell cycle arrest in cells expressing p53-nondegrading E6 mutants. A total of 5 × 105 cells of normal mammary epithelial cell strain (76N) or its derivatives immortalized with HPV16 E6 (76-E6), HPV16-F2V (76-F2V), HPV16-8S9A10T (76-8S9A10T), or HPV16-Y54H (76-Y54H) were cultured for 48 h and then treated with 0.5 μg of adriamycin/ml for 24 h. The cells were fixed, treated with RNase A, and stained with propidium iodide, and then their DNA content was quantified with a fluorescence-activated cell sorter. The G1, S, and G2/M phases of the cell cycle are indicated.
DISCUSSION
Increasing evidence supports the concept that many viral transforming proteins induce oncogenesis by interacting with and perturbing the function of key cellular proteins involved in the maintenance of normal cellular behavior. Importantly, many of the viral oncogene targets have been demonstrated to play key roles in human cancer, thus providing a clear rationale for searching for new targets of viral oncoproteins. As an example, the HPV oncoprotein E6 abolishes the function of p53 tumor suppressor protein, whose mutations or deletions constitute the single most frequent genetic lesion in human malignancies (as reviewed in references 44 and 57).
We have identified here human ADA3, a homologue of the yeast ADA3 transcriptional coactivator, as a novel HPV E6 target. We demonstrate that ADA3 functions as a coactivator for p53-dependent transcriptional activation, a conclusion consistent with findings reported while this study was in preparation (this report and reference 72). Importantly, we demonstrate that E6 abrogates the coactivator function of hADA3. Given the evolutionary conservation of ADA3 structure and function from yeast to humans and the likelihood that hADA3 will provide coactivator function for a number of transcriptional activators, we suggest that abrogation of hADA3 function by HPV E6 represents an important element of E6-induced oncogenic transformation. A number of observations discussed below support this view.
We demonstrate that the high-risk (cancer-associated) but not the low-risk (benign lesion-associated) HPV E6 proteins bind to hADA3. Furthermore, all of the immortalization-competent but none of the immortalization-defective HPV16 E6 mutants were able to interact with hADA3. This tight correlation between hADA3 binding and immortalizing ability of E6 mutants is remarkable. Notably, the immortalizing E6 mutants that bound to ADA3 included three examples (F2V, Y54H, and 8S9A10T) that are known to be defective in inducing the degradation of p53 (12, 17, 38, 47). Finally, recent studies have demonstrated that a prominent mechanism by which E6 abrogates the function of cellular regulatory proteins is by targeting them for ubiquitin-dependent degradation (19, 33, 34, 39, 49, 61, 68). As we and others have shown, degradation of these targets, such as p53, strongly correlates with the immortalizing ability of E6 (12, 17). We show here that interaction with E6 also targets hADA3 for degradation. The mechanism of hADA3 degradation appears to be distinct from that of E6-induced p53 degradation since E6 did not target hADA3 for degradation in vitro, whereas clear degradation of p53 was observed under these conditions. Interestingly, these results are reminiscent of another E6-binding protein Gps2, which also undergoes E6-induced degradation in vivo but not in vitro (13). Furthermore, we and others have reported earlier that HPV oncoprotein E7 induces the degradation of retinoblastoma protein in vivo; however, no degradation was observed under in vitro conditions (8, 25). It remains possible that in vitro degradation conditions vary for different proteins. Further studies should identify whether E6 partner E6AP or another ubiquitin ligase is involved in hADA3 degradation. Collectively, these results, together with the fact that hADA3 functions as a coactivator for the tumor suppressor protein p53, strongly support a likely role for E6-hADA3 interaction in E6-dependent oncogenic transformation.
Our findings have important implications for strategies used by viral oncogenes, such as E6, to abrogate p53 function. Previously, E6-induced p53 degradation has been well defined as an important mechanism to inactivate p53 function (12, 48). E6 has also been shown to interact with p300 and inhibit its coactivator function for p53 (55, 80). We show that E6 mutants, which are incapable of inducing p53 degradation, are nonetheless capable of inactivating hADA3 function. These mutants are also able to inhibit p53-dependent transactivation and disrupt p53-mediated G1 cell cycle arrest after DNA damage. Thus, our results identify a third potential mechanism by which E6 oncoprotein can inhibit p53 function.
Relatively little is known about hADA3 functions in mammalian systems except for its ability to function as a p53 coactivator (72). In yeast, ADA3 forms a complex with ADA2 and GCN5 HAT, which interacts with ADA2 (32). This trimolecular complex is crucial for a variety of transcriptional pathways in yeast. Importantly, the heterologous reconstitution analyses that identified the requirement of yeast ADA3 for p53 transactivation domain function in yeast also demonstrated a crucial role for ADA2 and GCN5 (9), indicating that this complex likely functions as a unit (65). Relatively little is known about the mammalian ADA complex. However, hADA3, when transfected in HeLa cells, was found to be part of a multiprotein complex that included hADA2, hGCN5, and the closely related gene product P/CAF, suggesting that mammalian cells likely assemble hADA3 into complexes that may be similar, if not identical, to those characterized at length in yeast (53). It is likely, however, that mammalian ADA complexes may be more complex. For example, P/CAF directly interacts with p300, another HAT protein that functions as a coactivator for p53 and a variety of other transactivators (62, 64, 77). Furthermore, hADA3 itself interacts with p300 (72), suggesting that hADA3 may participate in multiple complexes with distinct HAT proteins in mammalian cells. It will be of obvious importance to explore whether E6 targets the various hADA3-containing complexes equally or in a selective manner.
Genetic studies in yeast have also demonstrated a crucial role of ADA complex in the transactivation function of mammalian nuclear hormone receptors such as glucocorticoid receptor, retinoid receptors, estrogen receptor, and thyroid hormone receptor (3, 31, 70). If mammalian ADA complex is also found to participate in nuclear hormone function, as is likely, then E6-mediated loss of hADA3 function could perturb these key transactivation pathways involved in cell growth regulation and differentiation. Studies to address such a possibility are currently under way in our laboratory.
A key to coactivator function is the ability of these proteins to directly interact with transcriptional activators and facilitate their sequence-specific transcription by interacting with basal transcriptional machinery and by recruiting HAT machinery (16, 54, 75). We demonstrated, both in vitro and in vivo, that hADA3 interacts with p53 and that hADA3 functions as a coactivator of p53-mediated transactivation of a variety of target promoters, as well as a consensus p53-binding site linked to a reporter. Interestingly, we found that hADA3 dose dependently stabilized the p53 protein, suggesting that increase in p53 levels may contribute to hADA3 coactivator function. Interestingly, p300 has also been shown to stabilize p53 by interfering with mdm2 interaction with p53 (27, 35). Future studies should determine whether mdm2 plays any role in ADA3-mediated stabilization of p53. Since there are other coactivators for p53, and each coactivator can function in the context of multiple transactivators, the precise role of ADA3 versus other coactivators for p53-mediated physiological functions will require additional studies. Indeed, there are conflicting reports on how critical p300 and CBP coactivators are for p53 function, and knockout mice with individual deletions of these coactivators show distinct phenotypes (4, 26, 41, 78, 79). Notably, recent analyses showed that knockouts of P/CAF and hGCN5, which are highly related, also have vastly different phenotypes (76).
Our results, identifying the evolutionarily conserved hADA3 as a target of E6 protein, together with recent studies, suggest that inactivation of coactivator function may be a critical viral oncogenic strategy and that coactivators may contribute to cellular tumor suppressor mechanisms. For example, high-risk HPV E6 oncoproteins, E1A, and polyomavirus large T bind to and inactivate p300 function (11, 55, 77, 80). Recent studies have demonstrated that although mice lacking both copies of p300-related CBP coactivator are embryonically lethal, CBP+/− mice are cancer prone (41, 78). Notably, alterations or mutations of p300 have been observed in human leukemia and in colorectal, breast, and pancreatic cancers (22, 23). Further studies are warranted to directly define the role of hADA3 and other virally targeted coactivators in human oncogenesis.
In conclusion, we identified hADA3 as a novel cellular target of the high-risk HPV E6 oncoproteins and we demonstrated that E6 abrogates the coactivator function of hADA3. We identified p53 as a target of hADA3 coactivator function, leading us to conclude that abrogation of hADA3 function provides an additional strategy for HPV E6 to inactivate p53 function. Given the evolutionary conservation of hADA3, it is likely that viral oncoprotein-mediated inactivation of its function plays an important role in oncogenesis.
Acknowledgments
We thank Peter Howley (Harvard Medical School, Boston, Mass.) for E6AP plasmids, Elliot Androphy (University of Massachusetts Medical Scohol, Boston) for E6 mutants, Bert Vogelstein (Johns Hopkins School of Medicine, Baltimore, Md.) and Waffik El-Diery (University of Pennsylvania, Philadelphia, Pa.) for p53-binding promoter constructs, and Steve Grossman (Dana-Farber Cancer Institute, Boston, Mass.) for GST-p300.
This work was supported by NIH grants CA81076 and CA70195 to V.B., and CA87986, CA75075, and CA76118 to H.B. A.K., Y.Z., and G.M. are recipients of fellowships from the Massachusetts Department of Public Health.
A.K., Y.Z., and G.M. contributed equally to this study.
REFERENCES
- 1.Ali, S. H., and J. A. DeCaprio. 2001. Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin. Cancer Biol. 11:15-23. [DOI] [PubMed] [Google Scholar]
- 2.Allan, L. A., and M. Fried. 1999. p53-dependent apoptosis or growth arrest induced by different forms of radiation in U2OS cells: p21WAF1/CIP1 repression in UV-induced apoptosis. Oncogene 18:5403-5412. [DOI] [PubMed] [Google Scholar]
- 3.Anafi, M., Y. F. Yang, N. A. Barlev, M. V. Govindan, S. L. Berger, T. R. Butt, and P. G. Walfish. 2000. GCN5 and ADA adaptor proteins regulate triiodothyronine/GRIP1 and SRC-1 coactivator-dependent gene activation by the human thyroid hormone receptor. Mol. Endocrinol. 14:718-732. [DOI] [PubMed] [Google Scholar]
- 4.Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 5.Baker, C. C., W. C. Phelps, V. Lindgren, M. J. Braun, M. A. Gonda, and P. M. Howley. 1987. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol. 61:962-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Band, V., J. A. De Caprio, L. Delmolino, V. Kulesa, and R. Sager. 1991. Loss of p53 protein in human papillomavirus type 16 E6-immortalized human mammary epithelial cells. J. Virol. 65:6671-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, S. L. 1999. Gene activation by histone and factor acetyltransferases. Curr. Opin. Cell Biol. 11:336-341. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 9.Candau, R., D. M. Scolnick, P. Darpino, C. Y. Ying, T. D. Halazonetis, and S. L. Berger. 1997. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene 15:807-816. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J. J., C. E. Reid, V. Band, and E. J. Androphy. 1995. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science 269:529-531. [DOI] [PubMed] [Google Scholar]
- 11.Cho, S., Y. Tian, and T. L. Benjamin. 2001. Binding of p300/CBP co-activators by polyoma large T antigen. J. Biol. Chem. 276:33533-33539. [DOI] [PubMed] [Google Scholar]
- 12.Dalal, S., Q. Gao, E. J. Androphy, and V. Band. 1996. Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells. J. Virol. 70:683-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degenhardt, Y. Y., and S. Silverstein. 2001. Gps2, a protein partner for human papillomavirus E6 proteins. J. Virol. 75:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyson, N., P. M. Howley, K. Munger, and E. Harlow. 1989. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 243:934-937. [DOI] [PubMed] [Google Scholar]
- 15.Eberharter, A., D. E. Sterner, D. Schieltz, A. Hassan, J. R. Yates, S. L. Berger, and J. L. Workman. 1999. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6621-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards, D. P. 2000. The role of coactivators and corepressors in the biology and mechanism of action of steroid hormone receptors. J. Mammary Gland Biol. Neopl. 5:307-324. [DOI] [PubMed] [Google Scholar]
- 17.Foster, S. A., G. W. Demers, B. G. Etscheid, and D. A. Galloway. 1994. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J. Virol. 68:5698-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedlander, P., Y. Haupt, C. Prives, and M. Oren. 1996. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Mol. Cell. Biol. 16:4961-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, Q., S. Srinivasan, S. N. Boyer, D. E. Wazer, and V. Band. 1999. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol. Cell. Biol. 19:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, Q., A. Kumar, S. Srinivasan, L. Singh, H. Mukai, Y. Ono, D. E. Wazer, and V. Band. 2000. PKN binds and phosphorylates human papillomavirus E6 oncoprotein. J. Biol. Chem. 275:14824-14830. [DOI] [PubMed] [Google Scholar]
- 21.Gao, Q., L. Singh, A. Kumar, S. Srinivasan, D. E. Wazer, and V. Band. 2001. Human papillomavirus type 16 E6-induced degradation of E6TP1 correlates with its ability to immortalize human mammary epithelial cells. J. Virol. 75:4459-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gayther, S. A., S. J. Batley, L. Linger, A. Bannister, K. Thorpe, S. F. Chin, Y. Daigo, P. Russell, A. Wilson, H. M. Sowter, J. D. Delhanty, B. A. Ponder, T. Kouzarides, and C. Caldas. 2000. Mutations truncating the EP300 acetylase in human cancers. Nat. Genet. 24:300-303. [DOI] [PubMed] [Google Scholar]
- 23.Giles, R. H., D. J. Peters, and M. H. Breuning,. 1998. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 14:178-183. [DOI] [PubMed] [Google Scholar]
- 24.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grossman, S. R. 2001. p300/CBP/p53 interaction and regulation of the p53 response. Eur. J. Biochem. 268:2773-2778. [DOI] [PubMed] [Google Scholar]
- 27.Grossman, S. R., M. Perez, A. L. Kung, M. Joseph, C. Mansur, Z. X. Xiao, S. Kumar, P. M. Howley, and D. M. Livingston. 1998. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell 2:405-415. [DOI] [PubMed] [Google Scholar]
- 28.Gross-Mesilaty, S., E. Reinstein, B. Bercovich, K. E. Tobias, A. L. Schwartz, C. Kahana, and A. Ciechanover. 1998. Basal and human papillomavirus E6 oncoprotein-induced degradation of Myc proteins by the ubiquitin pathway. Proc. Natl. Acad. Sci. USA 95:8058-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen, R., and M. Oren. 1997. p53; from inductive signal to cellular effect. Curr. Opin. Genet. Dev. 7:46-51. [DOI] [PubMed] [Google Scholar]
- 30.Hawley-Nelson, P., K. H. Vousden, N. L. Hubbert, D. R. Lowy, and J. T. Schiller. 1989. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 8:3905-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henriksson, A., T. Almlof, J. Ford, I. J. McEwan, J. A. Gustafsson, and A. P. Wright. 1997. Role of the Ada adaptor complex in gene activation by the glucocorticoid receptor. Mol. Cell. Biol. 17:3065-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horiuchi, J., N. Silverman, G. A. Marcus, and L. Guarente. 1995. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol. Cell. Biol. 15:1203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 13:775-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawai, H., L. Nie, D. Wiederschain, and Z. M. Yuan. 2001. Dual role of p300 in the regulation of p53 stability. J. Biol. Chem. 276:45928-45932. [DOI] [PubMed] [Google Scholar]
- 36.Kern, S. E., K. W. Kinzler, A. Bruskin, D. Jarosz, P. Friedman, C. Prives, and B. Vogelstein. 1991. Identification of p53 as a sequence-specific DNA-binding protein. Science 252:1708-1711. [DOI] [PubMed] [Google Scholar]
- 37.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:11612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klingelhutz, A. J., S. A. Foster, and J. K. McDougall. 1996. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature 380:79-82. [DOI] [PubMed] [Google Scholar]
- 39.Kuhne, C., and L. Banks. 1998. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J. Biol. Chem. 273:34302-34309. [DOI] [PubMed] [Google Scholar]
- 40.Kukimoto, I., S. Aihara, K. Yoshiike, and T. Kanda. 1998. Human papillomavirus oncoprotein E6 binds to the C-terminal region of human minichromosome maintenance 7 protein. Biochem. Biophys. Res. Commun. 249:258-262. [DOI] [PubMed] [Google Scholar]
- 41.Kung, A. L., V. I. Rebel, R. T. Bronson, L. E. Ch'ng, C. A. Sieff, D. M. Livingston, and T. P. Yao. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272-277. [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, S. S., R. S. Weiss, and R. T. Javier. 1997. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:6670-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 45.Li, S., S. Labrecque, M. C. Gauzzi, A. R. Cuddihy, A. H. Wong, S. Pellegrini, G. J. Matlashewski, and A. E. Koromilas. 1999. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene 18:5727-5737. [DOI] [PubMed] [Google Scholar]
- 46.Liu, X. L., D. E. Wazer, K. Watanabe, and V. Band. 1996. Identification of a novel serine protease-like gene, the expression of which is downregulated during breast cancer progression. Cancer Res. 56:3371-3379. [PubMed] [Google Scholar]
- 47.Liu, Y., J. J. Chen, Q. Gao, S. Dalal, Y. Hong, C. P. Mansur, V. Band, and E. J. Androphy. 1999. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J. Virol. 73:7297-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maki, C. G., J. M. Huibregtse, and P. M. Howley. 1996. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 56:2649-2654. [PubMed] [Google Scholar]
- 49.Mantovani, F., P. Massimi, P., and L. Banks. 2001. Proteasome-mediated regulation of the hDlg tumor suppressor protein. J. Cell Sci. 114:4285-4292. [DOI] [PubMed] [Google Scholar]
- 50.Munger, K., W. C. Phelps, V. Bubb, P. M. Howley, and R. Schlegel. 1989. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J. Virol. 63:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munger, K., B. A. Werness, N. Dyson, W. C. Phelps, E. Harlow, and P. M. Howley. 1989. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 8:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakagawa, S., and J. M. Huibregtse. 2000. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20:8244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 54.Orlando, V., and R. Paro. 1995. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr. Opin. Genet. Dev. 5:174-179. [DOI] [PubMed] [Google Scholar]
- 55.Patel, D., S. M. Huang, L. A. Baglia, and D. J. McCance. 1999. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 18:5061-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pipas, J. M., and A. J. Levine. 2001. Role of T antigen interactions with p53 in tumorigenesis. Semin. Cancer Biol. 11:23-30. [DOI] [PubMed] [Google Scholar]
- 57.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 58.Ratsch, S. B., Q. Gao, S. Srinivasan, D. E. Wazer, and V. Band. 2001. Multiple genetic changes are required for efficient immortalization of different subtypes of normal human mammary epithelial cells. Radiat. Res. 155:143-150. [DOI] [PubMed] [Google Scholar]
- 59.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saleh, A., V. Lang, R. Cook, and C. J. Brandl. 1997. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J. Biol. Chem. 272:5571-5578. [DOI] [PubMed] [Google Scholar]
- 61.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 62.Schiltz, R. L., and Y. Nakatani. 2000. The PCAF acetylase complex as a potential tumor suppressor. Biochim. Biophys. Acta 1470:M37-M53. [DOI] [PubMed] [Google Scholar]
- 63.Schwarz, E., U. K. Freese, L. Gissmann, W. Mayer, B. Roggenbuck, A. Stremlau, and H. zur Hausen. 1985. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314:111-114. [DOI] [PubMed] [Google Scholar]
- 64.Shikama, N., J. Lyon, and N. B. La Thangue. 1997. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 7:230-236. [DOI] [PubMed] [Google Scholar]
- 65.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoppler, H., D. P. Hartmann, L. Sherman, and R. Schlegel. 1997. The human papillomavirus type 16 E6 and E7 oncoproteins dissociate cellular telomerase activity from the maintenance of telomere length. J. Biol. Chem. 272:13332-13337. [DOI] [PubMed] [Google Scholar]
- 67.Thomas, M., and L. Banks. 1998. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene 17:2943-2954. [DOI] [PubMed] [Google Scholar]
- 68.Thomas, M., and L. Banks. 1999. Human papillomavirus (HPV) E6 interactions with Bak are conserved amongst E6 proteins from high and low risk HPV types. J. Gen. Virol. 80:1513-1517. [DOI] [PubMed] [Google Scholar]
- 69.Tong, X., and P. M. Howley. 1997. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 94:4412-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.vom Baur, E., M. Harbers, S. J. Um, A. Benecke, P. Chambon, and R. Losson. 1998. The yeast Ada complex mediates the ligand-dependent activation function AF-2 of retinoid X and estrogen receptors. Genes Dev. 12:1278-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang, L., L. Liu, and S. L. Berger. 1998. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 12:640-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang, T., T. Kobayashi, R. Takimoto, A. E. Denes, E. L. Snyder, W. S. el-Deiry, and R. K. Brachmann. 2001. hADA3 is required for p53 activity. EMBO J. 20:6404-6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wazer, D. E., X. L. Liu, Q. Chu, Q. Gao, and V. Band. 1995. Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc. Natl. Acad. Sci. USA 92:3687-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Werness, B. A., A. J. Levine, and P. M. Howley. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248:76-79. [DOI] [PubMed] [Google Scholar]
- 75.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 76.Yamauchi, T., J. Yamauchi, T. Kuwata, T. Tamura, T. Yamashita, N. Bae, H. Westphal, K. Ozato, and Y. Nakatani. 2000. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97:11303-11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 78.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 79.Yuan, Z. M., Y. Huang, T. Ishiko, S. Nakada, T. Utsugisawa, H. Shioya, Y. Utsugisawa, Y. Shi, R. Weichselbaum, and D. Kufe. 1999. Function for p300 and not CBP in the apoptotic response to DNA damage. Oncogene 18:5714-5717. [DOI] [PubMed] [Google Scholar]
- 80.Zimmermann, H., R. Degenkolbe, H. U. Bernard, and M. J. O'Connor. 1999. The human papillomavirus type 16 E6 oncoprotein can downregulate p53 activity by targeting the transcriptional coactivator CBP/p300. J. Virol. 73:6209-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.zur Hausen, H. 1999. Viruses in human cancers. Eur. J. Cancer 35:1878-1885. [DOI] [PubMed] [Google Scholar]
- 82.zur Hausen, H. 2000. Papillomaviruses causing cancer: evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 92:690-698. [DOI] [PubMed] [Google Scholar]