Abstract
Previous studies have established an important role of histone acetylation in transcriptional control by nuclear hormone receptors. With chromatin immunoprecipitation assays, we have now investigated whether histone methylation and phosphorylation are also involved in transcriptional regulation by thyroid hormone receptor (TR). We found that repression by unliganded TR is associated with a substantial increase in methylation of H3 lysine 9 (H3-K9) and a decrease in methylation of H3 lysine 4 (H3-K4), methylation of H3 arginine 17 (H3-R17), and a dual modification of phosphorylation of H3 serine 10 and acetylation of lysine 14 (pS10/acK14). On the other hand, transcriptional activation by liganded TR is coupled with a substantial decrease in both H3-K4 and H3-K9 methylation and a robust increase in H3-R17 methylation and the dual modification of pS10/acK14. Trichostatin A treatment results in not only histone hyperacetylation but also an increase in methylation of H3-K4, increase in dual modification of pS10/acK14, and reduction in methylation of H3-K9, revealing an extensive interplay between histone acetylation, methylation, and phosphorylation. In an effort to understand the underlying mechanism for an increase in H3-K9 methylation during repression by unliganded TR, we demonstrated that TR interacts in vitro with an H3-K9-specific histone methyltransferase (HMT), SUV39H1. Functional analysis indicates that SUV39H1 can facilitate repression by unliganded TR and in so doing requires its HMT activity. Together, our data uncover a novel role of H3-K9 methylation in repression by unliganded TR and provide strong evidence for the involvement of multiple distinct histone covalent modifications (acetylation, methylation, and phosphorylation) in transcriptional control by nuclear hormone receptors.
In eukaryotic cells, genomic DNA is complexed with histone and nonhistone proteins to form chromatin. The N-terminal tails of histone proteins are known to undergo a number of posttranslational covalent modifications, including acetylation, methylation, phosphorylation, ubiquitination, and ADP-ribosylation (55). Such posttranslational modifications have long been thought to play important roles in chromatin structure and function (50, 54). The identification and characterization of a large number of histone acetyltransferases and histone deacetylases (HDACs) over the last several years have firmly established the functional importance of histone acetylation in chromatin structure and function (13, 40, 46, 60). First reported in 1964, histones have long been known to undergo methylation (33). Early studies have identified methylation in multiple lysine residues, including lysines 4, 9, 27, and 36 of H3 and lysine 20 in H4. In addition, histones can be methylated on arginine residues. The identification and characterization of several histone methyltransferases (HMTs) (8, 36, 39, 44, 47, 52, 53) have provided new insights into the role of histone methylation in chromatin function and gene transcription.
Human (SUV39H1) and fission yeast (Clr4) homologs of Drosophila Su(var)3-9 have recently been shown to encode HMTs that selectively methylate histone H3 at K9 (39). The HMT activity of these proteins is conferred through their highly conserved SET domain (39), and recently, this modification has been shown to generate a binding site for heterochromatin protein 1 (HP1) (2, 22), providing a strong link between H3-K9 methylation, gene silencing, and heterochromatin formation. H3-K9 methylation also inhibits the phosphorylation of serine 10 in H3, a modification that has been shown to be involved in gene activation and chromosome condensation/segregation (11, 28, 29). Importantly, H3-K9 methylation has also been found to occur outside of heterochromatin, as recent studies showed that the retinoblastoma protein interacts with and targets SUV39H1 for methylation and repression of the cyclin E promoter (35, 49). Besides SUV39 family proteins, one additional HMT, G9a, has been reported to methylate K9 as well as K27 of H3 (47).
HMTs that specifically methylate H3-K4 have also been identified recently (6, 36, 52). One H3-K4 HMT, Set9, has been shown to potentiate transcription activation through its ability to methylate H3-K4. Methylation of H3-K4 antagonizes methylation in H3-K9, facilitates acetylation by p300, and precludes binding of the NURD complex to H3 N-terminal tails, supporting a role for H3-K4 methylation in transcription activation (36, 52). However, work with Saccharomyces cerevisiae indicates that H3-K4 methylation may be involved in transcription repression (6). In S. cerevisiae, Set1 is likely to be the major if not the only enzyme responsible for H3-K4 methylation. Genetic experiments indicate that Set1 is required for ribosomal DNA (rDNA) silencing and silencing at telomeres and mating type loci (HML) (6). Thus, the effect of H3-K4 methylation on transcription seems to be context dependent.
Outside of lysine methylation of histones, two arginine-specific HMTs, PRMT1 and CARM1, have been reported to function as coactivators for nuclear hormone receptors (NR) (3, 8, 30, 47, 53). PRMT1 methylates H4 arginine 3 (H4-R3) (53), and CARM1 methylates H3 arginine 17 (H3-R17) as well as arginines 3 and 26 (41). Notably, methylation of H4-R3 facilitates subsequent acetylation by p300, providing a molecular explanation for the observed coactivator activity of PRMT1. Collectively, such diversity in histone N-terminal tail modifications and their reported effects in combination has led to the proposal of a “histone code” hypothesis, which is defined by the ability of distinct or similar histone modifications to act in combination to bring about distinct biological events (19, 43).
Nuclear hormone receptors are a large group of structurally related transcription factors whose transcriptional activities are generally regulated by lipophilic ligands (31). As members of the NR superfamily, the thyroid hormone receptor (TR) and retinoic acid receptor (RAR) have the capacity to alternately repress and activate transcription depending on the absence or presence of their cognate ligands (12, 18, 32, 38, 48). Studies over the last several years indicate that TR and RAR make use of distinct cofactors and chromatin modification to accomplish their dual functions in transcription (48, 56). While activation by liganded receptors is associated with the recruitment of coactivators and targeted acetylation of chromatin (9, 42), repression by unliganded receptors is mediated through interaction with the corepressors SMRT and N-CoR and recruitment of HDAC activities (10, 14, 16, 24, 25). Although HMTs such as PRMT1 and CARM1 have been shown to facilitate transcriptional activation by NRs (3, 8, 53), it has not yet been reported whether histone methylation on lysines is also involved in transcriptional control by NRs.
In this study, we investigated the involvement of multiple histone modifications in transcription regulation by TR with chromatin immunoprecipitation (ChIP) assays. We demonstrate that transcriptional regulation by TR involves, in addition to the expected changes in acetylation, methylation of H3-K4, H3-K9, and H3-R17 and phosphorylation of H3-S10. Among these modifications, H3-K9 methylation increases significantly during repression by unliganded TR and is reduced during activation by liganded TR. We show that TR interacts with an H3-K9-specific HMT, SUV39H1, and that SUV39H1 facilitates repression by unliganded TR in an HMT activity-dependent manner. Together, our data uncover a novel role of H3-K9 methylation in repression by unliganded TR and provide strong evidence for the involvement of multiple distinct types of histone covalent modifications (acetylation, methylation, and phosphorylation) in transcriptional control by nuclear receptors.
MATERIALS AND METHODS
Plasmid constructs.
The 4xUAS.TRβA reporter was described previously (24). The 4xUAS.TK-CAT reporter was kindly provided by Katia Georgopoulos (Harvard Medical School). The preparation of single-stranded DNA of reporters was done as described previously (59). The construct for in vitro synthesis of mRNA encoding GAL4-TR (GAL4 DNA-binding domain [DBD], amino acids 1 to 147) fused to the ligand-binding domain of Xenopus TRβA (amino acids 86 to 369) has been described (24). To generate a construct for in vitro synthesis of mRNA encoding human HMT SUV39H1, the full-length SUV39H1 was inserted in frame into a modified pSP64(polyA) (Promega) vector containing an N-terminal Flag tag (F-SUV39H1). The HMT-defective mutant SUV39H1m (H324K) was generated by PCR-based mutagenesis, and the mutation was verified by DNA sequencing.
Microinjection of Xenopus oocytes and subsequent analyses of protein expression, chromatin structure, and transcription.
Preparation and microinjection of mRNAs and reporter DNA into stage VI Xenopus oocytes were done as described previously (59). All mRNAs for microinjection were synthesized in vitro with SP6 mMessage mMachine kits from Ambion. Unless indicated in the figure legends, mRNA was injected at a concentration of 100 ng/μl (18.4 nl/oocyte), and single-stranded reporter DNA at a concentration of 50 ng/μl (18.4 nl/oocyte) according to the experimental scheme indicated in each figure. The injected oocytes were incubated at 18°C overnight and processed for transcriptional analysis by primer extension and assayed for chromatin structure by partial micrococcal nuclease digestion as described previously (59). The internal control for transcription is the primer extension product of the Xenopus storage histone H4 mRNA (58). Western blotting analyses for expression of F-SUV39H1 and F-SUV39H1m were carried out with a Flag tag-specific antibody (M2) purchased from Sigma, whereas Western analysis of GAL4-TR expression was performed with a GAL4 DBD-specific antibody purchased from Santa Cruz Biotechnology (sc-510).
ChIP assays.
The ChIP assays for histone methylation and acetylation were performed essentially as described previously (24). In brief, after overnight incubation, the groups of injected oocytes were treated with 1% formaldehyde for 10 min. The oocytes were washed twice with phosphate-buffered saline (PBS) solution and incubated with 100 mM Tris (pH 9.4)-10 mM dithiothreitol (DTT) at 30°C for 15 min. The oocytes were then rinsed once and resuspended in homogenization buffer (50 μl/oocyte) (20 mM Tris [pH 7.6], 60 mM KCl, and 1 mM DTT) and homogenized by pipetting. The homogenates were sonicated in a cold room to break chromatin into average 300- to 500-bp fragments. After centrifugation with a benchtop microcentrifuge (15 min at 13,000 rpm) to remove insoluble materials, the extracts were diluted onefold with ChIP I buffer (0.1% sodium deoxycholate, 1% Triton X-100, 2 mM EDTA, 50 mM HEPES [pH 7.6], 150 mM NaCl, 1 mM DTT, and 0.4 mM phenylmethylsulfonyl fluoride [PMSF]) and used for ChIP assays (each reaction with 100 μl of extract and 1 μl of individual antibody) as described previously (42).
The antibodies against acetylated H3 and H4, methylated H4-R3 and H3-R17, and H3-S10/acK14 were purchased from Upstate Biotechnology. The antibodies against methylated H3-K4 and H3-K9 were described previously (6, 39). The PCR A reaction (5′ primer, TGCCAGGGCCTATTTTGAATC, and 3′ primer, AGAGCCTGAGTGAAGCCCATAAG) generated a 100-bp PCR product, whereas the PCR B reaction (5′ primer, CAGTGCTGCAATGATACCGCGAGA, and 3′ primer, GAGGCGGATAAAGTTGCAGGACCA) generated a 115-bp fragment. The final PCRs were carried out with inclusion of 1 μCi of [32P]dCTP in each PCR, and the PCR product was visualized by autoradiography after fractionation on a 6% native polyacrylamide gel. The quantification of ChIP results was carried out with a PhosphorImager and by setting the value in control samples to 1.
HMT activity assays.
The HMT activity assays with calf thymus core histones as substrates were performed primarily as described previously (45). Briefly, when HeLa or Xenopus oocyte extracts were used, 5 μg of HeLa or Xenopus oocyte extract was incubated in 25 μl of reaction mix with 2.5 μg of calf thymus core histones (Roche Molecular Biochemicals) along with 0.5 μCi of S-adenosyl-l-[methyl-3H]methionine ([3H]SAM) in HMT buffer (50 mM Tris [pH 8.0], 1 mM PMSF, and 0.5 mM DTT) at 30°C for 1 h. The core histones were fractionated by sodium dodecyl sulfate-15% polyacrylamide gel electrophoresis (SDS-15% PAGE) followed by Coomassie staining and fluorography.
To assay for HMT activity in corepressor complexes of Sin3, Mi-2/NURD, SMRT, and N-CoR, the corresponding complexes were immunoprecipitated from 500 μg of HeLa nuclear extracts by incubating in 200 μl of immunoprecipitation buffer (25 mM Tris [pH 8.0], 150 mM NaCl, 1 mM DTT, 0.1% NP-40, and 1 mM PMSF) with antibodies (5 μl) specific for Sin3A (AK-11; Santa Cruz Biotechnology), CHD4 (kindly provided by Weidong Wang, National Institute on Aging), SMRT (raised against SMRT amino acids 1165 to 1363), and N-CoR (raised against N-CoR amino acids 1 to 373) (25), respectively. After 1 h of incubation in a cold room with rotation, 10 μl of 50% protein A-Sepharose beads was added to each tube and further incubated for 2 h. The beads were then collected by centrifugation and washed five times with the immunoprecipitation buffer and a final wash with HMT buffer. The beads from each immunoprecipitation were divided into two halves; one was used for the HMT activity assay as described above and the other was used for the HDAC activity assay as described previously (25). In the experiment shown in Fig. 4B, the HMT assays were carried out as described previously except that unlabeled SAM substrate (100 μM final concentration) was used. H3-K9 methylation was subsequently detected by Western blotting analysis with the antibody against H3-K9.
FIG. 4.
TSA does not appear to directly affect histone methylation. (A) HMT activity in Xenopus oocytes was detected by an in vitro methylation assay with core histones as the substrate and [3H]SAM as the donor. The methylation of core histones was revealed by fluorogram after separation on an SDS-15% PAGE gel. (B) Ability of TSA to inhibit specifically the HMT activity toward H3-K9 in Xenopus oocyte extracts. The experiments were performed essentially as for panel A except unlabeled SAM was used. After separation by SDS-15% PAGE, proteins were transferred to a nitrocellulose membrane, and H3-K9 methylation was detected by Western analysis with the methylated H3-K9-specific antibody (αmK9-H3).
RESULTS
Methylation of H3-K9 increased during repression by unliganded TR and decreased during activation by liganded TR.
We have shown previously that expression of TR and its heterodimer partner retinoid X receptor in Xenopus oocytes represses transcription in the absence of T3 and activates transcription in the presence of T3 from a Xenopus TRβA promoter-based reporter assembled into chromatin via a replication-coupled pathway (57, 59). We have demonstrated more recently by chromatin immunoprecipitation (ChIP) assays that repression by unliganded TR/retinoid X receptor or a fusion of the GAL4 DBD (1 to 147) and TR ligand-binding domain (83 to 309) (referred to as GAL4-TR hereafter) is correlated with the recruitment of corepressor SMRT and N-CoR complexes and deacetylation of histones over the promoter region (24).
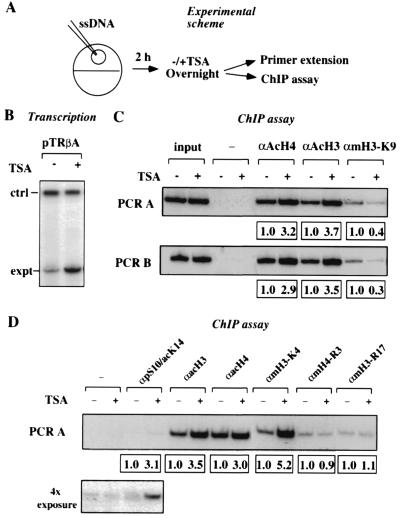
Given the recent evidence for functional significance of histone methylation in transcriptional control and the proposed histone code hypothesis (19, 43), we sought to investigate the involvement of histone methylation in transcriptional regulation by TR. Considering the potential role of H3-K9 methylation in transcriptional repression, we first tested whether repression by unliganded TR could also utilize K9 methylation in H3. To this end, we used an experimental scheme (Fig. 1A) to express GAL4-TR in Xenopus oocytes by injection of its mRNA synthesized in vitro into the cytoplasm. We then assembled a modified pTRβA reporter containing four copies of the GAL4 binding site (4xUAS.TRβA) into chromatin via a replication-coupled chromatin assembly pathway by injection of its single-stranded DNA into the nuclei of oocytes (1). The injected DNA was shown to assemble into chromatin by partial micrococcal nuclease digestion followed by Southern hybridization (Fig. 1B). As expected, primer extension analysis revealed that GAL4-TR repressed transcription in the absence of T3 and activated transcription in the presence of T3 from the injected reporter (Fig. 1D). The internal control represents the primer extension product from the endogenous storage histone H4 mRNA and served as a loading control.
FIG. 1.
Changes in H3-K9 methylation during repression and activation by unliganded and liganded TR. (A) Diagram showing the experimental scheme. (B) Diagram illustrating the structure of the 4xUAS.TRβA reporter. (C) Partial micrococcal nuclease digestion (MNase) assay demonstrated the assembly of injected DNA into chromatin. Two hours after DNA injection, oocytes were homogenized and aliquoted into three tubes. Chromatin was then partially digested with a decreased concentration of MNase. The DNA was recovered by phenol-chloroform extraction after treatment with proteinase K and detected by Southern analysis with 32P-labeled 4xUAS.TRβA reporter. The positions of DNA fragments corresponding to mono-, di-, and trinucleosome are indicated. (D) Expression of the GAL4-TR fusion protein repressed transcription in the absence of T3 and activated transcription in the presence of T3 from the 4xUAS.TRβA reporter. The control (ctrl) represents the primer extension product from the endogenous storage histone H4 mRNA. Also shown is the expression of GAL4-TR by Western blotting analysis with a GAL4 DBD-specific antibody (sc-510; Santa Cruz Biotechnology). (E) ChIP assays revealed the involvement of H3-K9 methylation in transcriptional regulation by TR. The levels of H3-K9 methylation (boxed values below the gels) were determined by ChIP assays with a K9-methylated-H3-specific antibody. The acetylation of H3 was assayed with an antibody specific for acetylated H3 (06-599; Upstate Biotechnology). The control (−) ChIP assay was performed with addition of protein A-Sepharose but without specific antibody. As illustrated in panel B, the PCR A reaction allows analysis of the promoter vicinity, whereas PCR B allows analysis of a region approximately 2 kb downstream of the transcriptional start site. Note that changes in H3-K9 methylation and acetylation were significant in the PCR A region but not in the PCR B region. The data were quantified with a phosphoimager, and the results were based on at least two independent experiments.
We next used ChIP assays to determine whether the level of H3-K9 methylation changed under different conditions with an antibody specific for H3 methylated at K9 (anti-mH3-K9). This antibody has been well demonstrated to be specific for K9-methylated H3 in both Western blotting analysis and ChIP assays (22, 27, 37, 39). In parallel, we also analyzed the level of H3 acetylation by ChIP assays with an acetylated-H3-specific antibody. While control ChIP assays (protein A-Sepharose beads only) gave virtually no background, a low level of signal was detected over both the promoter vicinity (PCR A) and a distal region about 2 kb downstream of the transcriptional start site (PCR B) with the anti-mH3-K9 antibody (Fig. 1E). This result indicates that some H3 proteins in our chromatin templates were methylated at K9. Notably, the level of K9 methylation increased significantly (4.8-fold) in the promoter vicinity region (PCR A, compare lanes 8 and 7) when transcription was repressed by unliganded TR. In contrast, the level of K9 methylation decreased during activation by liganded TR (PCR A, compare lanes 9 and 7).
Since no significant difference in K9 methylation was observed at the distal region (PCR B), the increased K9 methylation during repression by unliganded TR appears to be a localized event targeted to the promoter vicinity. ChIP assays with anti-acetylated H3 antibody revealed a decrease in acetylation during repression by unliganded TR (PCR A, compare lane 11 with 10). Although not obvious in this experiment, a slight increase in acetylation of H3 was usually observed during activation in the promoter region. Collectively, these results showed that K9 methylation increased in the state of repression by unliganded TR and decreased in the state of activation by liganded TR.
Involvement of other histone modifications in transcriptional regulation by TR.
We next used ChIP assays to assess the possible involvement of other histone modifications in both repression by unliganded TR and activation by liganded TR. A representative result is shown in Fig. 2. Consistent with the earlier result that K4 in H3 is a major site of histone methylation in ciliates, yeast cells, and human HeLa cells (45), H3-K4 methylation was readily detected in the control by ChIP assays with a methylated H3-K4-specific antibody (αmH3-K4). Interestingly, in comparison to the control, the level of H3-K4 methylation decreased to about 50% in the presence of unliganded TR and further decreased to about 30% in the presence of liganded TR. In multiple experiments, we have not observed any significant change in methylation of H4-R3 under either repression or activation conditions. However, a robust increase (4.7-fold) in methylation of H3-R17 was observed during transcriptional activation by liganded TR. In addition, a decrease in methylation of H3-R17 during repression by unliganded TR was also observed. The increase in H3-R17 methylation during activation by liganded TR is consistent with the earlier finding that CARM1, the HMT that methylates H3-R17, is a coactivator for nuclear hormone receptors (3, 7). We suggest that the increase in methylation of H3-R17 is a result of specific targeting of a Xenopus CARM1-like protein by liganded TR.
FIG. 2.
Involvement of multiple histone modifications in transcriptional regulation by TR. The experiments were performed as in Fig. 1 except ChIP assays were carried out with specific antibodies as indicated. A fivefold longer exposure is shown for αS10/acK14. Note that changes in methylation of H3-K4, H3-R17, and dual modification S10/acK14 were significant in the PCR A region but not in the PCR B region.
Finally, as phosphorylation of H3-S10 has been linked directly to acetylation of H3-K14 and the transcriptional activation process, we also examined the levels of this dual modification (pS10/acK14) under both repression and activation conditions. Although the signals in ChIP assays were weak, this dual modification showed a clear increase during activation (4.3-fold) and decrease in repression (0.3-fold). This result reveals a direct correlation of the dual modification in S10 and K14 with transcription activation and is consistent with the proposed role of phosphorylation of S10 in stimulation of acetylation and subsequent transcriptional activation (11, 28). Similar to H3-K9 methylation, all these observed changes in histone modifications appear to be targeted to the promoter vicinity, as in each case no significant change was detected in the distal region (PCR B). Collectively, our results reveal that, in addition to acetylation and deacetylation, histone methylation and phosphorylation are also involved in transcriptional regulation by TR. Furthermore, our results indicate that distinct but complex patterns of histone modifications are associated with repression by unliganded TR and activation by liganded TR.
Functional interplay between histone acetylation and other modifications.
The above results revealed the involvement of multiple histone modifications in transcriptional regulation by TR. One possibility is that each of the observed modifications reflects the recruitment of a specific enzyme (such as HMTs) by either unliganded or liganded TR. Alternatively, it is also possible that some of the modifications may be dependent on or the consequence of other histone modifications. For instance, it has been shown that methylation of H3-K9 interferes with phosphorylation of S10 and that methylation of H3-K4 facilitates subsequent histone acetylation by p300 (36, 39, 52).
Since repression by unliganded TR is associated with histone deacetylation as a result of specific targeting of the HDAC3-containing SMRT/N-CoR complexes (24) and activation by liganded TR is associated with recruitment of histone acetyltransferases such as CBP/p300 (9, 42), we next sought to determine the effect of histone acetylation on other histone modifications. To do so, we took advantage of our previous observation that treatment with trichostatin A (TSA), an HDAC-specific inhibitor, can induce transcription activation and histone hyperacetylation from repressive chromatin (24). As illustrated in Fig. 3A, groups of oocytes were injected with single-stranded DNA of the 4xUAS.pTRβA reporter to assemble DNA into repressive chromatin, treated or not with TSA (0.3 μM) overnight, and then assayed for transcription by primer extension and histone modifications by ChIP assays. The results in Fig. 3B showed that TSA treatment enhanced transcription from the injected reporter. In agreement with its ability to inhibit HDAC activity, TSA treatment increased the levels of acetylation of both H3 and H4 (Fig. 3C). Note that the effect of TSA on chromatin acetylation was global, being detected in both the promoter vicinity (PCR A) and the distal region (PCR B) in the pTRβA reporter and all other regions that we analyzed (data not shown).
FIG. 3.
TSA treatment reveals extensive interplay between histone acetylation, methylation, and phosphorylation. (A) Diagram illustrating the experimental scheme. Note that TSA treatment was performed 2 h after DNA injection to allow complete assembly of injected DNA into chromatin. (B) TSA treatment resulted in transcription activation. The expected primer extension product from the TRβA promoter is marked with an asterisk. The control (ctrl) was the primer extension product from endogenous storage histone H4 mRNA. (C) ChIP assays reveal the relationship of anticorrelation between histone acetylation and H3-K9 methylation. The levels of H3-K9 methylation and acetylation of H3 and H4 were analyzed on both the promoter vicinity (PCR A) and the distal region (PCR B). (D) TSA treatment resulted in a drastic increase in H3-K4 methylation and in dual modification of S10 phosphorylation and K14 acetylation. A fourfold-longer exposure is also shown for pS10/acK14. Note that TSA treatment did not result in any significant change in H3-R17 and H4-R3 methylation.
In contrast to the increase in histone acetylation, there was a concomitant decrease in the level of H3-K9 methylation after TSA treatment in all regions tested (Fig. 3C). Assuming that TSA has no direct inhibitory effect on the HMT(s) that methylates H3-K9 (see below), this result indicates that the level of histone acetylation has a significant influence on the level of K9 methylation. Thus, not only can H3-K9 methylation interfere with S10 phosphorylation and subsequent acetylation (39); our result indicates that histone acetylation can also antagonize H3-K9 methylation.
We next extended our study to examine the effect of TSA treatment on the dual modification pS10/acK14 and methylation of H3-K4, H4-R3, and H3-R17. As shown in Fig. 3D, TSA treatment led to an increase (3.1-fold) in the dual modification pS10/acK14. This result is consistent with the correlation of this modification with transcription activation. Interestingly, TSA treatment also induced a strong increase in H3-K4 methylation (5.2-fold). Thus, although no increase in H3-K4 methylation was observed during activation by TR, H3-K4 methylation was induced by histone hyperacetylation. On the other hand, TSA treatment did not result in any significant change in methylation of H4-R3 and H3-R17, suggesting that the methylation of these sites is not directly influenced by levels of histone acetylation. In summary, our results show that histone hyperacetylation antagonizes H3-K9 methylation and facilitates H3-K4 methylation and S10 phosphorylation, thus revealing extensive interplay between histone acetylation and other modifications.
TSA does not appear to inhibit H3-K9 methylation directly.
One possible conclusion from the above results is that TSA directly inhibits (H3-K9) or promotes (H3-K4) HMT activity, providing an alternative explanation for the observed decrease in H3-K9 methylation and increase in H3-K4 methylation after TSA treatment. To test this possibility, we examined whether TSA could inhibit histone methylation in vitro by using Xenopus oocyte extracts. We first demonstrated that Xenopus oocyte extract contains HMT activities that can methylate both core histone H3 and H4 (Fig. 4A). To look specifically at the effect on H3-K9 methylation, we then repeated the experiment in Fig. 4A but replaced the [3H]SAM with unlabeled SAM in methylation reactions. Methylated histones were then fractionated by SDS-15% PAGE, transferred to nitrocellulose, and examined for H3-K9 methylation by Western blotting with the anti-mH3-K9 antibody.
The results indicated that addition of TSA (1 μM) had no effect on H3-K9 methylation (Fig. 4B). Indeed, we did not observe any inhibition of H3-K9 methylation in this in vitro assay with concentrations of TSA ranging from 0.5 to 20 μM that were sufficient to inhibit HDAC activity. We thus conclude that TSA has no direct effect on H3-K9 methylation and the effect of TSA on H3-K9 methylation in Xenopus oocytes is not due to direct inhibition of H3-K9 HMT activity by TSA.
Corepressor complexes such as Sin3, Mi-2/NURD, SMRT, and N-CoR contain no detectable HMT activity.
Strong evidence indicates that repression by unliganded TR requires histone deacetylation and is mediated through specific recruitment of two closely related corepressors, SMRT and N-CoR (10, 16, 20, 24). Both SMRT and N-CoR comprise a category of class I HDAC-containing complexes by association with HDAC3 (14, 25). Two other class I HDAC-containing complexes, the HDAC1/2-containing Sin3 and Mi-2/NURD complexes, also contribute to repression by unliganded TR (15, 34, 61), most likely through their effect on global deacetylation of chromatin (24). The result that H3-K9 methylation increases significantly during repression by unliganded TR raises the intriguing possibility that some of the HDAC-containing complexes involved in TR repression may contain H3-K9 HMT activity.
To test this idea, we immunoprecipitated each complex from HeLa nuclear extracts and tested HMT activity in vitro. As a positive control, HeLa nuclear extract methylated both core histones H3 and H4 in vitro in the presence of [3H]SAM under our assay conditions (see Materials and Methods). However, extensive effort failed to detect significant HMT activity in all four immunoprecipitated corepressor complexes (Fig. 5A, upper panel). On the other hand, all the immunoprecipitated complexes exhibited HDAC activity toward core histones labeled in vitro (Fig. 5B). Furthermore, highly purified Sin3 and Mi-2/NURD complexes also contain no HMT activity (Yi Zhang, personal communication). The absence of HMT activity in all four HDAC-containing corepressor complexes is also consistent with a recent report on the purification of HMTs from HeLa nuclear extracts (36). In this report, both H3-K9-specific HMTs (SUV39H1-like and G9a-like activity) were found to migrate as smaller complexes in comparison to the known gel filtration profiles of corepressors SMRT, N-CoR, Sin3A, and NURD complexes.
FIG. 5.
Corepressor complexes implicated in TR repression contain no detectable HMT activity. (A) The corepressor complexes Sin3, SMRT, N-CoR, and Mi-2/NURD were individually immunoprecipitated from HeLa nuclear extracts with complex-specific antibodies as indicated and tested for HMT activity with core histones as substrates. Note that no detectable HMT activity was found in each immunoprecipitated complex, although HeLa nuclear extract (lane 1) contained abundant HMT activity toward H3 and less for H4. Also shown is Coomassie staining of core histones. In contrast, all immunoprecipitated corepressor complexes exhibited HDAC activity, as revealed by an in vitro deacetylation assay (B). Lane 1, immunoprecipitation with rabbit immunoglobulin G as a control for the deacetylation assay.
SUV39H1 interacts with TR and facilitates repression by unliganded TR.
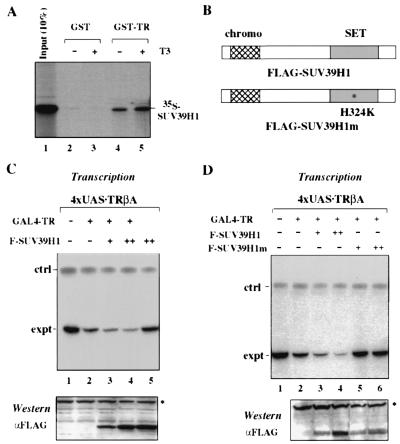
Our ChIP assay results reveal that repression by unliganded TR is associated not only with histone deacetylation but also with increased H3-K9 methylation. Given the result that corepressor complexes did not appear to contain HMT activity, we next tested whether TR could interact with SUV39H1, the HMT that preferentially methylates H3-K9 (39). With an in vitro pulldown assay, we found that in vitro-translated SUV39H1 bound to glutathione S-transferase (GST)-TR (Fig. 6A). Although the interaction between GST-TR and SUV39H1 was not influenced by the presence of T3, the interaction was dependent on the presence of TR, because no interaction was detected with control GST alone (Fig. 6A).
FIG. 6.
SUV39H1 interacts with TR and facilitates TR repression in an HMT activity-dependent manner. (A) Binding of SUV39H1 to GST-TR as shown by an in vitro pulldown assay. In vitro-translated, [35S]methionine-labeled SUV39H1 was tested for binding to GST or a GST-TR fusion protein. The binding of SUV39H1 to GST-TR was specific because no binding to GST was detected under the same conditions. Note that the binding of SUV39H1 to GST-TR was not affected by the presence of 1 μM T3. (B) Diagrams showing the structure of the wild-type and mutant SUV39H1s containing a Flag tag at the N terminus. The mutant carries a mutation of histidine 324 to lysine. (C) Expression of wild-type SUV39H1 in Xenopus oocytes facilitates repression by unliganded TR. Xenopus oocytes were injected with mRNA encoding GAL4-TR (10 ng/μl) alone or together with mRNA encoding wild-type SUV39H1 (50 ng/μl for lane 3 and 150 ng/μl for lanes 4 and 5) and double-stranded DNA from the 4xUSA.TRβA reporter. After overnight incubation, total RNA was recovered from the injected oocytes, and the level of transcription was determined by primer extension. Also shown in the bottom panel is the expression of Flag-SUV39H1 in each group of injected Xenopus oocytes as detected by Western analysis with a Flag-specific antibody. The asterisk indicates a nonspecific band detected in Western analysis which served as a loading control. (D) HMT activity is required for SUV39H1 to promote TR repression. The experiment was carried out as for panel C except mutant SUV39H1 was also included. Note that the mutant SUV39H1 completely lost the ability to enhance repression by unliganded TR (compare lanes 5 and 6 with 2 to 4).
The above result prompted us to test whether expression of SUV39H1 could facilitate repression by unliganded TR. For convenience in the repression assay, we assembled the 4xUAS.pTRβA reporter into chromatin by injecting DNA in the double-stranded form (which gives a relatively high basal level of transcription in comparison to chromatin assembled through injection of single-stranded DNA [59]). To observe the effect of SUV39H1 on TR repression, we also chose to inject a relatively low concentration of GAL4-TR mRNA (10 ng/μl), which by itself led to a moderate repression (Fig. 6C, compare lane 2 with 1). Importantly, expression of a Flag-tagged SUV39H1 (F-SUV39H1) with GAL4-TR enhanced repression in a dose-dependent manner (Fig. 6C and D, compare lanes 3 and 4 with 2), whereas the expression of F-SUV39H1 alone had no effect on transcription (Fig. 6C, lane 5). Furthermore, the expression of SUV39H1 had no effect on the activation by GAL4-TR in the presence of T3 (data not shown). The expression of F-SUV39H1 in oocytes was confirmed by Western analysis with a Flag tag-specific antibody (Fig. 6C, lower panel).
HMT activity is required for SUV39H1 to enhance TR repression.
With the observation that expression of SUV39H1 was able to enhance repression by unliganded TR, we next wished to determine whether this enhancement required the HMT activity of SUV39H1. To this end, we constructed a mutant SUV39H1 with histidine 324 (H) changed to lysine (K) (Fig. 6B). This mutation has been shown to completely inactivate the SUV39H1 HMT activity (39). When tested together with the wild-type SUV39H1, this mutant clearly lost its ability to enhance TR repression (Fig. 6D). Furthermore, expression of this mutant partially impaired the repression by unliganded TR in several independent experiments (Fig. 6D, compare lanes 5 and 6 with 2). Western analysis with a Flag-specific antibody confirmed that the functional difference between wild-type and mutant SUV39H1s was not due to their differential expression in Xenopus oocytes. Collectively, these results demonstrate that SUV39H1 facilitates repression by unliganded TR in an HMT activity-dependent manner.
DISCUSSION
In this study, we demonstrated that, in addition to histone acetylation and deacetylation, other histone modifications, including methylation and phosphorylation, are also involved in transcriptional control by TR.
Involvement of H3-K9 methylation in repression by unliganded TR.
In addition to the expected histone deacetylation, we demonstrated that repression by unliganded TR is accompanied by an increase in H3-K9 methylation (Fig. 1), a decrease in methylation of H3-K4 and H3-R17 and an decrease in phosphorylation of S10 (Fig. 2). While histone acetylation has been well documented in transcriptional regulation, recent studies have provided evidence for a diverse role of histone methylation in gene expression (8, 39, 53). Both lysine and arginine residues in the N-terminal tails of core histones can be methylated, and the effect of methylation on transcription appears to be site specific.
The identification of SUV39H1 and its yeast homologue Clr4 as the H3-K9-specific HMT provides the first direct connection between H3-K9 methylation and heterochromatin gene silencing. The role of SUV39H1 and H3-K9 methylation appears not to be restricted to heterochromatin gene silencing, as recent studies showed that the retinoblastoma protein interacts with SUV39H1 and targets it to the cyclin E promoter for H3-K9 methylation and transcription repression (35, 49). Our finding that H3-K9 methylation increases significantly during repression by unliganded TR is consistent with a role of H3-K9 methylation in transcription repression. In support of this, we demonstrated that SUV39H1 interacts with TR in vitro and facilitates TR repression in an HMT activity-dependent manner (Fig. 6). These results indicate that H3-K9 methylation is unlikely to be a simple consequence of TR repression but contributes to repression by unliganded TR.
As H3-K9 methylation has been shown to create a binding site for HP1 in the N-terminal tail of H3 (2, 22), one mechanism by which H3-K9 methylation facilitates TR repression is to recruit HP1. Future work will test whether HP1 is indeed recruited to the promoter region during repression by unliganded TR. Alternatively, H3-K9 methylation could enhance repression through its antagonistic effect on S10 phosphorylation and histone acetylation, both correlating with transcriptional activation by liganded TR. Consistent with this idea, we found that S10 phosphorylation decreases during repression by unliganded TR and increases during activation by liganded TR.
Given the conservation in the mechanisms of transcriptional regulation by the NR superfamily, our results suggest that the H3-K9 methylation is likely to extend far beyond heterochromatic gene silencing and be widely used for transcriptional repression by NRs and other transcription factors as well. In this regard, it is noteworthy that H3-K9 methylation appears nearly unchanged in bulk H3 prepared from primary mouse embryonic fibroblasts derived from Suv39 h1 and Suv39 h2 double-knockout mice, suggesting that H3-K9 methylation is widely present in euchromatin (39).
Correlation of H3-K4 methylation with transcription.
In contrast to the increase in H3-K9 methylation, we also observed a decrease in H3-K4 methylation during repression by unliganded TR. Recent studies with ChIP assays on fission yeast and the chicken β-globin locus indicate that H3-K9 methylation is restricted to the heterochromatic regions, whereas H3-K4 methylation is specific to the surrounding euchromatic regions (27, 37). In addition, immunofluorescence studies indicate that H3-K4 methylation is largely absent from the inactive X chromosome, which is enriched in H3-K9 methylation (5). Moreover, it has been shown recently that methylation of H3-K4 by Set9, also called Set7, antagonizes H3-K9 methylation and precludes the association of the NURD complex with the H3 tail (36, 52). Set9 enhanced transcriptional activation by GAL4-VP16, and, importantly, a mutation in Set9 that abolished its enzymatic activity also impaired the ability of Set9 to stimulate activation by GAL4-VP16 (36). Collectively, these results define H3-K4 methylation as a marker for euchromatin and suggest a role for H3-K4 methylation in transcription activation (45).
While the decrease in H3-K4 methylation during repression by unliganded TR is consistent with such an idea, it was surprising to find that H3-K4 methylation decreased further during activation by liganded TR (Fig. 2). Our results on H3-K4 methylation indicate a lack of direct correlation of this modification with transcription. Indeed, work with S. cerevisiae indicates that H3-K4 methylation may be involved in transcription repression as well. In S. cerevisiae, Set1 is likely to be the major if not the only enzyme responsible for H3-K4 methylation (6). Genetic experiments indicate that Set1 is required for rDNA silencing and silencing at telomeres and mating type loci (6). It is unknown currently how the same modification can be involved in such apparently opposite transcriptional events. However, according to the histone code hypothesis (19, 43), it is likely that the overall pattern of histone modifications, not a single modification, determines the outcome on chromatin structure and function. Thus, a single modification can have distinct effects on transcription depending on the combination with other histone modifications.
Involvement of methylation at H3-R17 and H4-R3 in transcriptional activation by liganded TR.
In addition to H3-K9 and K4 methylation, the level of H3-R17 methylation showed a reduction during repression by unliganded TR and an increase during activation by liganded TR (Fig. 2). The NR coactivator CARM1 has been shown to methylate R3, R17, and R26 in H3 (41) and facilitate NR activation in concert with SRC/p160 family coactivators (7, 8, 21). More recently, activation of the estrogen receptor-regulated pS2 gene was shown to be associated with a dramatic increase in H3-R17 methylation, which correlated with specific recruitment of CARM1 (3). Thus, the increased H3-R17 methylation that we observed is likely a result of specific recruitment of a Xenopus CARM1-like activity by liganded TR. Interestingly, although the H4-R3 methylation is largely carried out in mammalian cells by PRMT1, which is also a coactivator for NRs (53), we have not observed any change in H4-R3 methylation during repression by unliganded TR and activation by liganded TR. Whether this result reflects the absence of a PRMT1-like activity in Xenopus oocytes remains to be determined.
Correlation of S10 phosphorylation with activation by liganded TR.
In addition to methylation on various sites described above, the dual modification pS10/acK14 in H3 displays a decrease during repression and drastic increase during activation. While we have yet to examine S10 phosphorylation independent of K14 acetylation, the fact that no significant change in acetylation of H3 and H4 was observed during activation by liganded TR in comparison to the controls (Fig. 1) implies an increase in S10 phosphorylation during activation. In S. cerevisiae, S10 phosphorylation is coupled to K14 acetylation and is required for transcriptional activation by a subset of Gcn5-regulated genes (28). In vitro, S10 phosphorylation has been shown to promote subsequent histone acetylation by several histone acetyltransferases (11, 28). Thus, an increase in S10 phosphorylation during activation by liganded TR is consistent with a role of S10 phosphorylation in promoting transcription activation. So far, several S10-specific kinases have been identified, including the Ip11/Aurora family kinases and the kinase Snf1 (17, 29). It is unknown currently whether the enhanced S10 phosphorylation is a result of the direct or indirect recruitment of an S10-specific kinase by liganded TR.
Interplay between histone acetylation and other modifications.
By using the HDAC inhibitor TSA, we showed that TSA treatment not only induced histone hyperacetylation but also resulted in a reduction in H3-K9 methylation, increase in H3-K4 methylation and S10 phosphorylation, and no change in methylation of H3-R17 and H4-R3. As TSA had no effect on H3-K9 methylation, at least in vitro (Fig. 4), the reduction in H3-K9 methylation was unlikely a direct inhibition by TSA of H3-K9 methylation. Instead, this result most likely reflects a functional interplay between histone acetylation and H3-K9 methylation. Such an antagonistic outcome between H3-K9 methylation and acetylation is not surprising, given the exclusive nature of H3-K9 methylation and acetylation. Nevertheless, such an inverse correlation between H3-K9 methylation and acetylation further enforces the idea that H3-K9 methylation is directly correlated with transcription repression.
In addition, TSA treatment also led to a strong increase in H3-K4 methylation (Fig. 3). As with H3-K9 methylation, our preliminary results indicate that TSA has no effect on H3-K4 methylation in a Xenopus oocyte extract in vitro. We suggest that the effect of TSA on H3-K4 again is a direct or indirect result of histone hyperacetylation. As TSA treatment induced a strong increase in H3-K4 methylation, our result suggests that histone hyperacetylation is likely to facilitate subsequent methylation on H3-K4. Nevertheless, as the level of H3-K4 methylation was actually found to decrease during activation by liganded TR (Fig. 2), H3-K4 methylation is unlikely to be essential for the transcriptional activation process. As discussed above, the effect of H3-K4 methylation on transcription may well depend on its combination with other histone modifications.
Furthermore, TSA treatment also led to a strong increase in dual modification of S10 phosphorylation and K14 acetylation. Although S10 phosphorylation has been shown to facilitate subsequent histone acetylation both in vitro and in vivo, histone acetylation on K14 appears to have no effect on S10 phosphorylation (11, 28). How TSA treatment promotes S10 phosphorylation is thus not clear. One possibility is that, although acetylation at K14 alone has no effect on S10 phosphorylation, acetylation on multiple sites in H3 or other histones and/or methylation at H3-K4 may facilitate S10 phosphorylation. Alternatively, the increased S10 phosphorylation may not be a result linked directly to histone acetylation per se but a consequence of transcription activation. Taken together, our results provide strong evidence for extensive functional interplay between histone acetylation, methylation, and phosphorylation.
SUV39H1 facilitates TR repression in an HMT-dependent manner.
As repression by unliganded TR is believed to be mediated through corepressor complexes, the strong increase in H3-K9 methylation during repression by unliganded TR suggests that one or more of the HDAC-containing corepressor complexes involved in TR repression may potentially contain HMT activity responsible for H3-K9 methylation. It is thus rather disappointing that our extensive efforts failed to reveal HMT activity in several well-characterized corepressor complexes, including HDAC3-containing SMRT and N-CoR complexes and HDAC1/2-containing Sin3 and Mi-2/NURD complexes (14, 25, 51, 61-63). Despite ample HMT activities in HeLa nuclear extracts, neither immunoprecipitated corepressor complexes SMRT, N-CoR, Sin3, and Mi-2/NURD nor their highly purified fractions contained detectable HMT activity (Fig. 5). Instead, an interaction between TR and SUV39H1 was observed in an in vitro GST pulldown assay. Although this interaction was independent of hormone in our in vitro interaction assay and remains to be confirmed in vivo, the interaction is likely of functional significance, as expression of SUV39H1 in Xenopus oocytes facilitated repression by unliganded TR (Fig. 6). Importantly, the ability of SUV39H1 to enhance TR repression is directly correlated with its HMT activity, since an SUV39H1 mutant (H324K) defective in HMT activity was also impaired in enhancing repression by unliganded TR (Fig. 6).
While our results demonstrate a role of H3-K9 methylation in repression by unliganded TR, it is unknown whether recruitment of a Xenopus homolog of SUV39H1 is responsible for increased H3-K9 methylation during repression by unliganded TR. Given the strong antagonism between histone acetylation and H3-K9 methylation as revealed by TSA treatment, it is also possible that the increased H3-K9 methylation may not be entirely due to the specific recruitment of an HMT by unliganded TR. Instead, histone hypoacetylation as a result of specific recruitment of SMRT and N-CoR complexes could presumably generate a better substrate for H3-K9 methylation and thus lead to an increased methylation in K9.
Is histone methylation reversible?
As discussed above, we observed significant alterations in histone methylation after TSA treatment and during repression or activation by unliganded and liganded TR, respectively. Our results thus raise an intriguing question: whether histone methylation is reversible. On the basis of thermodynamic principles, methyl-lysine and methyl-arginine are believed to be stable. However, demethylation could theoretically be catalyzed by histone demethylases, although the existence of histone demethylases has yet to be demonstrated. The results that H3-K9 methylation increases during repression by unliganded TR and decreases during activation by liganded TR or after treatment with TSA suggest that site-specific histone methylation is reversible. The decrease in H3-K9 methylation is unlikely to be due to the effect of liganded TR or TSA on chromatin assembly, as chromatin assembly via the replication-coupled pathway is a fast event (about 30 min after injection) and was complete before addition of hormone or TSA (2 h after injection).
If histone demethylases do not exist, an alternative approach through proteolytic processing has been proposed for removal of methylated histone tails (19). However, if histone methylation is widely involved in regulation of gene expression, as shown here and proposed in the histone code hypothesis (19, 43), it will likely be reversible in order to meet the acute response in control of gene expression.
Acknowledgments
We thank Katia Georgopoulos for providing the 4xUAS.TK-CAT reporter and Weidong Wang for antibody against CHD4. We also thank Rafael Herrera, Neil McKenna, and Fred Pereira for critical reading of and comments on the manuscript.
This work was supported by National Institutes of Health grants DK56324 and DK58679 to J. Wong and GM40922 to C. D. Allis.
REFERENCES
- 1.Almouzni, G., and A. P. Wolffe. 1993. Replication-coupled chromatin assembly is required for the repression of basal transcription in vivo. Genes Dev. 7:2033-2047. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, U. M., S. Daujat, S. J. Nielsen, K. Nightingale, and T. Kouzarides. 2002. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 3:39-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco, J. C., S. Minucci, J. Lu, X. J. Yang, K. K. Walker, H. Chen, R. M. Evans, Y. Nakatani, and K. Ozato. 1998. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 12:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30:73-76. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, D., S. M. Huang, and M. R. Stallcup. 2000. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J. Biol. Chem. 275:40810-40816. [DOI] [PubMed] [Google Scholar]
- 8.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 96:393-403. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454-457. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 12.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 13.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 14.Guenther, M. G., W. S. Lane, W. Fischle, E. Verdin, M. A. Lazar, and R. Shiekhattar. 2000. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 14:1048-1057. [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 16.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C. K. Glass, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 18.Hu, X., and M. A. Lazar. 2000. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol. Metab. 11:6-10. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Jepsen, K., O. Hermanson, T. M. Onami, A. S. Gleiberman, V. Lunyak, R. J. McEvilly, R. Kurokawa, V. Kumar, F. Liu, E. Seto, S. M. Hedrick, G. Mandel, C. K. Glass, D. W. Rose, and M. G. Rosenfeld. 2000. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 102:753-763. [DOI] [PubMed] [Google Scholar]
- 21.Koh, S. S., D. Chen, Y. H. Lee, and M. R. Stallcup. 2001. Synergistic enhancement of nuclear receptor function by p160 coactivators and two coactivators with protein methyltransferase activities. J. Biol. Chem. 276:1089-1098. [DOI] [PubMed] [Google Scholar]
- 22.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 23.Li, J., B. W. O'Malley, and J. Wong. 2000. p300 requires its histone acetyltransferase activity and SRC-1 interaction domain to facilitate thyroid hormone receptor activation in chromatin. Mol. Cell. Biol. 20:2031-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J., Q. Lin, W. Wang, P. Wade, and J. Wong. 2002. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes Dev. 16:687-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, J., J. Wang, Z. Nawaz, J. M. Liu, J. Qin, and J. Wong. 2000. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 19:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Q., A. Imhof, T. N. Collingwood, F. D. Urnov, and A. P. Wolffe. 1999. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 18:5634-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken β-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 28.Lo, W. S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu, C. D. Allis, R. Marmorstein, and S. L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 29.Lo, W. S., L. Duggan, N. C. Tolga, Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 30.Ma, H., C. T. Baumann, H. Li, B. D. Strahl, R. Rice, M. A. Jelinek, D. W. Aswad, C. D. Allis, G. L. Hager, and M. R. Stallcup. 2001. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr. Biol. 11:1981-1985. [DOI] [PubMed] [Google Scholar]
- 31.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 33.Murray, K. 1964. The occurrence of e-N-methyllysine in histone. Biochemistry 3:10-15. [DOI] [PubMed] [Google Scholar]
- 34.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89:373-380. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 36.Nishioka, K., S. Chuikov, K. Sarma, H. Erdjument-Bromage, C. D. Allis, P. Tempst, and D. Reinberg. 2002. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 16:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 38.Ordentlich, P., M. Downes, and R. M. Evans. 2001. Corepressors and nuclear hormone receptor function. Curr. Top. Microbiol. Immunol. 254:101-116. [DOI] [PubMed] [Google Scholar]
- 39.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 40.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 41.Schurter, B. T., S. S. Koh, D. Chen, G. J. Bunick, J. M. Harp, B. L. Hanson, A. Henschen-Edman, D. R. Mackay, M. R. Stallcup, and D. W. Aswad. 2001. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry 40:5747-5756. [DOI] [PubMed] [Google Scholar]
- 42.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 43.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 44.Strahl, B. D., P. A. Grant, S. D. Briggs, Z.-W. Sun, J. R. Bone, J. A. Caldwell, S. Mollah, R. G. Cook, J. Shabanowitz, D. F. Hunt, and C. D. Allis. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22:1298-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 47.Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276:25309-25317. [DOI] [PubMed] [Google Scholar]
- 48.Urnov, F. D., and A. P. Wolffe. 2001. A necessary good: nuclear hormone receptors and their chromatin templates. Mol. Endocrinol. 15:1-16. [DOI] [PubMed] [Google Scholar]
- 49.Vandel, L., E. Nicolas, O. Vaute, R. Ferreira, S. Ait-Si-Ali, and D. Trouche. 2001. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol. Cell. Biol. 21:6484-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Holde, K. E. 1988. Chromatin. Springer-Verlag, New York, N.Y.
- 51.Wade, P. A., P. L. Jones, D. Vermaak, and A. P. Wolffe. 1998. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr. Biol. 8:843-846. [DOI] [PubMed] [Google Scholar]
- 52.Wang, H., R. Cao, L. Xia, H. Erdjument-Bromage, C. Borchers, P. Tempst, and Y. Zhang. 2001. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 8:1207-1217. [DOI] [PubMed] [Google Scholar]
- 53.Wang, H., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 54.Wolffe, A. P. 1998. Chromatin: structure and function, p. 97-108. Academic Press, San Diego, Calif.
- 55.Wolffe, A. P., and J. J. Hayes. 1999. Chromatin disruption and modification. Nucleic Acids Res. 27:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolffe, A. P., J. Wong, and D. Pruss. 1997. Activators and repressors: making use of chromatin to regulate transcription. Genes Cells 2:291-302. [DOI] [PubMed] [Google Scholar]
- 57.Wong, C. W., and M. L. Privalsky. 1998. Transcriptional repression by the SMRT-mSin3 corepressor: multiple interactions, multiple mechanisms, and a potential role for TFIIB. Mol. Cell. Biol. 18:5500-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong, J., D. Patterton, A. Imhof, D. Guschin, Y. B. Shi, and A. P. Wolffe. 1998. Distinct requirements for chromatin assembly in transcriptional repression by thyroid hormone receptor and histone deacetylase. EMBO J. 17:520-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong, J., Y. B. Shi, and A. P. Wolffe. 1995. A role for nucleosome assembly in both silencing and activation of the Xenopus TRβA gene by the thyroid hormone receptor. Genes Dev. 9:2696-2711. [DOI] [PubMed] [Google Scholar]
- 60.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 61.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357-364. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, Y., G. LeRoy, H. P. Seelig, W. S. Lane, and D. Reinberg. 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95:279-289. [DOI] [PubMed] [Google Scholar]