Abstract
Telomere maintenance is required for chromosome stability, and telomeres are typically replicated by the action of the reverse transcriptase telomerase. In both tumor and yeast cells that lack telomerase, telomeres are maintained by an alternative recombination mechanism. Genetic studies have led to the identification of DNA polymerases, cell cycle checkpoint proteins, and telomere binding proteins involved in the telomerase pathway. However, how these proteins affect telomere-telomere recombination has not been identified to date. Using an assay to trace the in vivo recombinational products throughout the course of survivor development, we show here that three major replicative polymerases, α, δ, and ɛ, play roles in telomere-telomere recombination and that each causes different effects and phenotypes when they as well as the telomerase are defective. Polymerase δ appears to be the main activity for telomere extension, since neither type I nor type II survivors arising via telomere-telomere recombination were seen in its absence. The frequency of type I versus type II is altered in the polymerase α and ɛ mutants relative to the wild type. Each prefers to develop a particular type of survivor. Moreover, type II recombination is mediated by the cell cycle checkpoint proteins Tel1 and Mec1, and telomere-telomere recombination is regulated by telomere binding protein Cdc13 and the Ku complex. Together, our results suggest that coordination between DNA replication machinery, DNA damage signaling, DNA recombination machinery, and the telomere protein-DNA complex allows telomere recombination to repair telomeric ends in the absence of telomerase.
Telomeres are dynamic DNA-protein complexes that protect the ends of linear chromosomes, preventing detrimental chromosome rearrangements and defending against genomic instability and the associated risk of cancer (33, 44, 53). Telomeric DNA is synthesized by the enzyme telomerase (37, 45, 50). In certain human cells, telomerase activity is absent, and telomeres are gradually shortened with successive cell divisions due to incomplete replication, which eventually causes replicative senescence. Once telomeres become sufficiently short, they are thought to lose the ability to protect the ends of the chromosomes from being recognized as broken ends, which would be subject to active recombinational repair. Continuous telomere shortening in human fibroblasts leads to chromosome fusions, crisis, and apoptosis (2). Very few human cells can bypass the crisis either through telomerase reactivation or through an alternative recombination pathway for telomere lengthening (ALT) (6, 15, 41).
Telomeric DNA in the yeast Saccharomyces cerevisiae consists of ∼350 ± 75 bp of TG1-3/C1-3A DNA. Internal to the TG1-3/C1-3A tracts are middle repetitive DNA elements, called X and Y′ (33, 53). The telomeric TG1-3/C1-3A DNA forms a complex nonnucleosomal chromatin structure, called the telosome (52). The major component in yeast telosomes is a double-stranded, sequence-specific DNA-binding protein-Rap1p complex including Rif1p and Rif2p. The copy number of the Rap1p complex negatively regulates telomere length (31, 51). Telomeric end-binding proteins Cdc13p and the Ku complex also play important roles in the protection of the chromosomal ends and the recruitment of telomerase to the telomeres (4, 10, 21, 36, 39). In telomerase-proficient cdc13-1 cells, telomeres harbor long single-stranded tails at the semipermissive temperature (18). Reduction of telomere length has also been observed in Ku mutants (3). Additionally, telomere maintenance is under checkpoint control. Mutations in S. cerevisiae Tel1 and Mec1, homologs of the human checkpoint gene ATM, lead to shortened telomeres (22, 42, 54). This suggests that proteins involved in DNA damage checkpoints also play a role in maintaining telomere length.
Even in organisms that normally rely on telomerase, telomerase-independent mechanisms of telomere maintenance exist. Although most cells in S. cerevisiae (30, 46), Kluyveromyces lactis (32), and Schizosaccharomyces pombe (34) that lack the gene for a telomerase component die, survivors arise relatively frequently in all three organisms. In both S. cerevisiae and K. lactis, generation of survivors requires RAD52-dependent recombination. In S. cerevisiae the majority of cells that survive in the absence of telomerase activity have multiple tandem copies of the subtelomeric Y′ element and very short terminal tracts of TG1-3/C1-3A DNA (30, 48) (type I survivors). In a minor fraction (∼10%) of the survivors (type II), the lengths of the telomere sequence are increased by various amounts, to 10 kb or longer (48). The appearance of the type II survivor depends on the presence of Rad50p, Rad59p, and Sgs1p (5, 11, 12, 24, 49), and it is magnified by deficiencies in mismatch repair (43). The structure of type II telomeres in Saccharomyces resembles that of 10 to 15% of human cell lines and tumors that maintain telomeric DNA by the ALT pathway (6, 15, 41).
DNA polymerases are involved in genome replication, recombination, and repair. Genetic studies of S. cerevisiae show that polymerases α, δ, and ɛ share the duty of replicating the cellular genome and that all three polymerases are essential for growth (reviewed in reference 25). Eukaryotic polymerase α forms a complex with primase. During initiation of DNA replication, the primase synthesizes a short RNA primer on the leading strand at the replication origin and the primer for the Okazaki fragments on the lagging strand. The primer is transferred to polymerase α directly and further extended by polymerase α (17). Polymerase δ, along with its accessory factors PCNA and RFC, is then recruited to the initial extension and replaces the primase-polymerase α complex for a highly processive synthesis (25). Polymerase ɛ has also been proposed to work in cooperation with polymerase δ during leading- and lagging-strand synthesis, but its precise role remains to be determined (25).
All forms of DNA replication are executed by the actions of primers, templates, and DNA polymerases. In a telomerase-proficient strain, the leading strand is synthesized by the addition of a G-rich strand, using the reverse transcriptase telomerase (29) and the RNA template (46). The lagging strand is concurrently elongated by the major replicative DNA polymerases α and δ using the newly formed G strand as the template (14), and these polymerases may be recruited to telomeres through Cdc13p (16, 40). A mutation in polymerase α causes a telomerase-mediated increase in telomere length (1). Mutations in polymerases δ and ɛ show relatively minor or irreproducible variations in average telomere length (1, 14).
Although the DNA polymerases, cell cycle checkpoint proteins, and telomere binding proteins employed in the telomerase pathway have been experimentally determined, the roles of these proteins in the telomere-telomere recombination pathway remain obscure. Here, we show that all three replicative polymerase-defective, telomerase-minus mutants display distinct telomere phenotypes which reflect the in vivo functions of these polymerases in telomere-telomere recombination. Polymerase δ is essential for telomere lengthening in both type I and type II recombination, whereas polymerase α and ɛ mutants each preferably employ a certain type of recombination. Type II recombination is mediated by the cell cycle checkpoint proteins Tel1 and Mec1. Moreover, the telomere binding proteins Cdc13 and the Ku complex both exhibit abilities for the generation and maintenance of recombination events, respectively.
MATERIALS AND METHODS
Strain preparation.
The yeast strains carrying tlc1::LEU2 and rad52::HIS3 plus pRAD52 were described previously (48). STY95 (YPH501 tlc1/TLC1) was transformed with pSD218 (a gift of D. Gottschling) (14) cut with PflMI to make STY539. A 5-fluorouracil-resistant colony purified from these strains was used to generate several potential clones of STY540 (YPH501 tlc1/TLC1 cdc17-2/CDC17), which were then sporulated and screened for a segregant unable to grow at 37°C. The yeast strains STY549 (tlc1/TLC1 cdc2-2/CDC2), STY535 (tlc1/TLC1 pol2-3/POL2 plus YCppol2-18), STY736 (YPH501 tlc1/TLC1 mec1::LEU2::hisG/MEC1 sml1::LEU2::hisG/SML1 tel1::MET17/TEL1 met17::LEU2::hisG/MET17), STY518 (YPH501 tlc1/TLC1 cdc13-1/CDC13), and STY524 (YPH501 tlc1/TLC1 hdf1::HIS3/HDF1) were made by mating STY106 (YPH α tlc1) or STY107 (YPH atlc1) plus pTLC1 (48) with UCC5898 (a gift of D. Gottschling), YHA300 plus YCppol2-18 (a gift of A. Sugino), STY584 (YPH amec1::LEU2::hisG sml1::LEU2::hisG tel1::MET17 met17::LEU2::hisG) (a gift of Y. Tsukamoto), STY515 (YPH acdc13-1) (a gift of A. Taggart), and STY502 (YPH α hdf1::HIS3) (a gift of W. Tham) plus pRS316HDF1 (a gift of Y. Tsukamoto), respectively. STY600 (tlc1/TLC1 cdc2-2/CDC2 rad52/RAD52 plus pRAD52) was made by mating a tlc1 cdc2-2 spore from STY549 with a YPH rad52::HIS3 haploid plus pRAD52 (48). These heterozygous diploids were then sporulated and screened for segregants that were unable to grow at 37°C or for the HIS3 and LEU2 markers from their original deletions by replica plating to selective medium and by Southern blot analysis.
Sample preparation and Southern blot analysis.
Spore cells were serially diluted into or restreaked onto YEPD medium. Liquid cultures were generated by inoculating spore colonies from the tetrad plate into 10 ml of liquid YEPD medium. Cultures were diluted repeatedly 1:10,000 or 3:10,000 into fresh medium at 48 or 72 h. A solid-plate study was performed by repeatedly streaking spore colonies from the tetrad plates on YEPD plates. Optimal semipermissive temperatures were determined and used: 27°C for the cdc13-1 mutant and 30°C for polymerase mutants (1) (data not shown). Southern blotting with a telomeric C1-3A probe was carried out as previously described (48). When DNA was examined from individual colonies, the colony was expanded in 2 ml of liquid medium to obtain enough DNA for Southern analysis. DNA was digested with XhoI or a mixture of AluI, HaeIII, HinfI, and MspI, which cut AGCT, GGCC, GANTC, and CCGG sequences, respectively. Data shown here are representative of two or more experiments from independent spores.
Senescence and recovery growth curve analysis.
Two independent spores of each genotype were used to inoculate cultures. Cultures were inoculated directly from the tetrad master plate into liquid YEPD medium on day 0 and allowed to grow overnight on a 27°C or 30°C roller drum. Then, 24 h later, the cell concentration in each culture was determined by hemacytometer, and each culture was diluted to a concentration of 2 × 105 cells/ml in fresh medium. This procedure was repeated for 10 days. The concentrations of the nine cultures for each strain on each day were then averaged, and the results of the senescence and recovery rate were graphed.
tlc1 hdf1 survival assay.
STY524 (YPH501 tlc1/TLC1 hdf1::HIS3/HDF1)/pRS316HDF1 (plasmid bearing the HDF1 gene and the URA3 marker) was sporulated and screened for Ura+ tlc1 and tlc1 hdf1 segregants. Four independent type I and type II survivors were isolated by solid YEPD plate restreaking and Southern blot analysis in each tlc1 and tlc1 hdf1 strain carrying pRS316HDF1. Cells from the plates lacking uracil were used to inoculate YEPD medium, grown for 24 h, and plated on both 5-fluorouracil and YEPD plates. The ratio of survivor maintenance was calculated by dividing the cell numbers on 5-fluorouracil plates by those on YEPD plates.
RESULTS
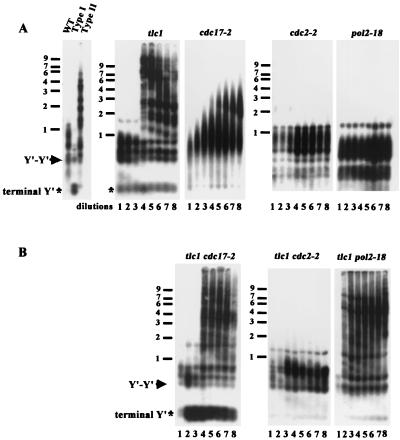
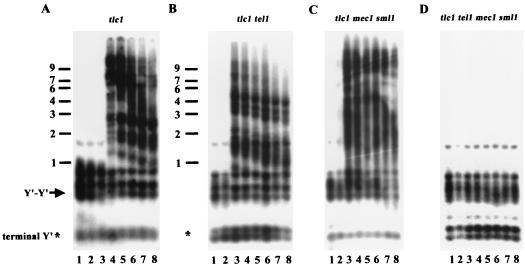
In the tlc1 strain that lacks telomerase activity, yeast cells gradually lose viability after 40 to 60 generations of propagation (46). However, a subset of cells in the culture eventually bypass the senescence and survive. Two classes of survivors, type I and type II, have been identified and can be distinguished by two different Southern blot analyses. In the first Southern blot analysis, the genomic DNA is digested with a mixture of four restriction enzymes to very small fragments. The telomere sequences, which are not cut by these enzymes, remain relatively large. Using this technique, DNA from wild-type YPH499 cells yields C1-3A-hybridizing fragments of ∼400 ± 75 bp from Y′ telomeres, ∼500-bp fragments from tandem Y′ DNA, and fragments of up to 1.1 kb from X telomeres (48) (Fig. 1, wild type). DNA from type I survivors yields fragments of less than 300 bp from Y′ telomeres (asterisk, terminal Y′, Fig. 1A), in addition to the ∼500-bp fragment from tandem Y′ elements (arrow, Fig. 1A). In contrast, type II telomeric DNA digested with these enzymes yields many differently sized fragments of up to 10 kb or even larger (type II, Fig. 1A).
FIG. 1.
Replicative polymerases exhibit distinct phenotypes in telomere-telomere recombination. Liquid cultures were generated as described in Materials and Methods and diluted repeatedly 3:10,000 into fresh YEPD medium at 72 h at 30°C, the semipermissive temperature for these polymerase mutants. Genomic DNA from single mutants (A) and double mutants (B) from each dilution was digested with a combination of AluI, HaeIII, HinfI, and MspI, fractionated through 1% agarose, and transferred to a nylon filter. The filter was hybridized subsequently to a C1-3A probe. An equal amount of DNA was loaded in each lane. An asterisk marks the position of critically short telomeres, and the arrow indicates the TG1-3/C1-3A fragments between Y′-Y′ tandem repeats. WT, wild type. Size markers (kilobases) are shown on the left.
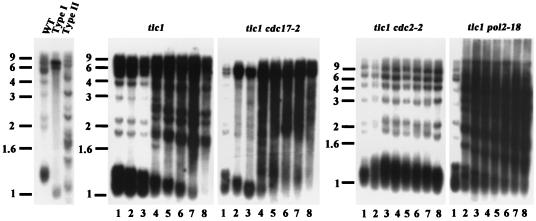
While the type II extension pattern is most easily distinguished by the four-base cutter assay, slight telomere lengthening and the type I telomere pattern are more easily visualized by XhoI digestion Southern blot analysis. XhoI cleaves 0.9 kb from the 3′ end of the Y′ element. In type I survivors, three major XhoI fragments (∼1.0, 6.7, and 5.2 kb) hybridized to the telomere probe. The ∼1.0-kb terminal fragment from Y′ telomeres consisted mainly of Y′ DNA plus a short stretch of TG1-3/C1-3A repeat. The strong hybridization at 6.7 and 5.2 kb contained the tandemly repeated Y′ long and Y′ short elements, respectively (Fig. 2) (48). In contrast, the type II survivors exhibited many XhoI fragments of different sizes.
FIG. 2.
Restriction analysis by XhoI digestion supports the involvement of replicative polymerases in telomere-telomere recombination. The experiment was conducted as described for Fig. 1, except that XhoI was used for restriction digestion. Size markers (kilobases) are shown on the left.
In a tlc1 mutant liquid culture, although ∼90% of the survivors are type I, they grow slowly and are quickly overtaken by the type II survivors, which have a growth rate similar to that of wild-type cells (48). This is also exacerbated by the ability to gradually convert from the type I to the type II pattern of telomere structure (48). In the liquid culture assay (49), when cultures starting from freshly dissected spores were repeatedly diluted 1:10,000 at 48-h intervals, dramatic telomere lengthening could be observed after several dilutions. In this study, we used this liquid culture system for all mutant strains and then conducted both the four-base cutter and XhoI Southern analyses. For each strain, we show data for either one or both of the Southern analyses.
Replicative polymerases exhibit distinct roles on telomere-telomere recombination.
We were interested in finding which DNA polymerases were employed for telomere-telomere recombination. To examine the role of replicative polymerases in telomere-telomere recombination in tlc1 strains, mutations of the three major replicative polymerases (8, 13, 35) were generated in these strains. To evaluate if polymerase α participates in telomere-telomere recombination in tlc1 cells, a temperature-sensitive mutation (cdc17-2) in the polymerization domain (8) was used. Tetrads from heterozygous tlc1/TLC1 cdc17-2/CDC17 diploid strains were dissected, and individual tlc1 and tlc1 cdc17-2 spore colonies were inoculated into liquid medium and cultured at a semipermissive temperature (30°C). When both cultures reached the stationary phase, DNA was prepared (dilution 1, Fig. 1A), and the culture was diluted into fresh medium. This DNA isolation and dilution protocol was repeated seven times (D2 to D8).
DNAs from the cultures were first analyzed by digestion with four-base cutters as described above, followed by Southern hybridization. In the tlc1 cultures, telomeres were suddenly and dramatically lengthened between D3 and D4 (Fig. 1A). At the same time, the growth rate of the cultures increased gradually to the wild-type level (data not shown). In cdc17-2 (polα) cells, gradual telomere lengthening was observed (Fig. 1A). However, a different result was seen in the tlc1 cdc17-2 mutant. At the D1 stage, the telomeres of tlc1 cdc17-2 mutant were 200 bp shorter than at the equivalent tlc1 D1 stage (Fig. 1B). This telomere pattern was more similar to the D3 of the tlc1 culture. The intensities of the Y′ telomeric fragment at ∼250 bp and the Y′-Y′ tandem repeat at ∼500 bp were increased at the D2 stage. This result indicated that telomere lengthening in cdc17-2 cells is telomerase dependent and type I-like Y′-Y′ recombination became a preferred pathway in the polymerase α-defective telomerase-minus strain.
Interestingly, this type I-like Y′-Y′ pattern was not stably maintained. While the main pattern was unchanged, the intensity of the Y′-Y′ tandem repeat fragment at the D3 stage of the tlc1 cdc17-2 mutant decreased. This indicated that most telomeres had not only remained short, but had also reduced the copy numbers of the Y′-Y′ tandem repeats at the D3 stage. Similar to what was observed in tlc1 cells, at the D4 stage the type II pattern was established suddenly, indicating that type II survivors started to take over the culture at this stage in tlc1 cdc17-2 (polα) cells.
We next determined whether two other major replicative polymerases, δ and ɛ, were involved in telomere-telomere recombination. The tlc1 cdc2-2 (polδ [13]) double mutant exhibited no long telomeric or subtelomeric lengthening throughout the eight dilutions, suggesting that both type I and type II lengthening was mainly dependent on polymerase δ for long-tract extension (Fig. 1B). In contrast, telomere shortening occurred much earlier, and the type II long telomere pattern was quickly generated at the D2 stage of the tlc1 pol2-18 cells (polɛ [35]) (Fig. 1B). This experiment suggested that type II-like telomere-telomere recombination is a preferred pathway to repair the telomeric damage in the tlc1 pol2-18 (polɛ) strains.
The type I extension pattern is more easily visualized by XhoI digestion and Southern blot analysis. In agreement with the four-base cutter results in Fig. 1, the XhoI digestion Southern blot analysis of the tlc1 cdc17-2 (polα) mutant showed that the Y′ amplification signal at 6.7 and 5.2 kb developed at the D1 stage and declined slightly at D3 (Fig. 2). After D3, although type II survivors took over the culture, type I-like Y′-Y′ amplification was preferred and persisted to D8. The results of the XhoI digestion patterns for the tlc1 cdc2-2 (polδ) and tlc1 pol2-18 (polɛ) mutants also reinforced those of the four-base cutter experiment. A slight lengthening of the telomere at the D2 and D3 stages and the rapid type II development at the D2 stage could be observed for the tlc1 cdc2-2 (polδ) and tlc1 pol2-18 (polɛ) mutants, respectively.
Accelerated senescence phenotype in telomerase-deficient and polymerase-defective mutants.
To further evaluate the influence of replicative polymerases on the senescence and recovery rate of the tlc1 mutant, cells harboring double mutations were tested at the semipermissive temperature in liquid cultures and on solid plates. Cultures inoculated from freshly dissected spores were grown to the stationary phase and then diluted to the same cell densities. Cell numbers were determined every 24 h under a microscope over a period of 10 days (Fig. 3A).
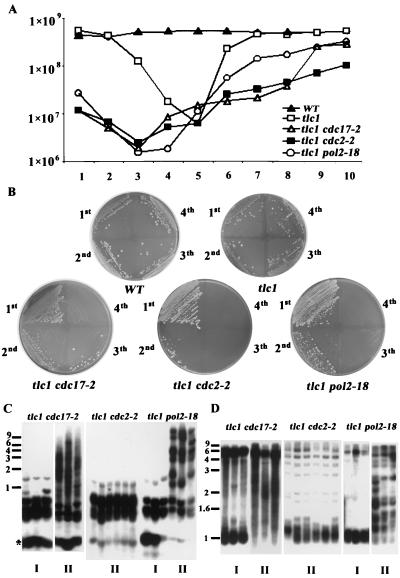
FIG. 3.
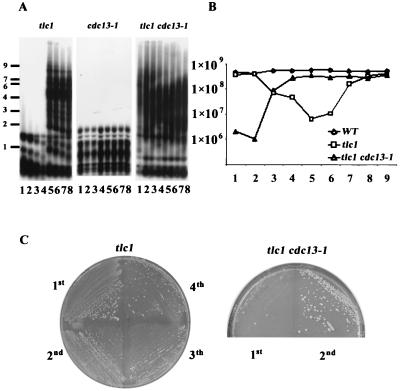
Accelerated senescent phenotype in telomerase-deficient and polymerase-defective mutants. (A) Cultures of wild-type (WT), tlc1, tlc1 cdc17-2, tlc1 cdc2-2, and tlc1 pol2-18 strains were inoculated from spores and grown to the stationary phase at 30°C. Fresh cultures were then inoculated to 2 × 105 cells/ml and grown for 24 h, and cell numbers from the overnight cultures were determined and graphed. (B) Each strain was repeatedly streaked on solid YEPD plates and grown for 3 days at 30°C. It was noticeable that all telomerase-minus, polymerase-defective spores senesced faster, and survivors showed up much more seldom than those of the tlc1 mutant. (C and D) Analysis of telomeric pattern of single survivors. Survivors were generated from the solid-plate streaking. DNAs from each strain were prepared, and Southern blot analysis was performed as described for Fig. 1 and 2 by either four-base cutter (C) or XhoI (D) digestion. Three type I and three type II survivors from the tlc1 cdc17-2 and tlc1 pol2-18 mutants and six survivors from the tlc1 cdc2-2 mutant are shown here. Size markers (kilobases) are shown at the left.
The tlc1 strain displayed the slowest growth rate at day 5. While the growth curve of all three single polymerase mutants showed no significant difference compared to that of the wild type (data not shown), all three double polymerase and telomerase mutants displayed the lowest growth rates at day 3, after which the cultures gradually returned to the wild-type growth rate. The phenotype of these double mutant strains was similar to the fast senescence and recovery phenotype of the tlc1 rad51 cells; survivors in this genetic background lose viability at the third dilution (28). The growth curve analysis was consistent with our four-base cutter Southern analysis, where all three polymerase-deficient telomerase-minus strains established telomere or subtelomere lengthening earlier than the tlc1 strain (Fig. 1B and 2).
In the polymerase α-deficient, telomerase-minus (tlc1 cdc17-2) strain, the growth curve exhibited two steps of recovery. The first recovery was observed between day 3 and day 5, and then a second recovery was observed from day 7. This result is consistent with the interpretation from the Southern analysis (Fig. 1 and 2) that the first recovery was due to the presence of the type I survivors, whereas the second and faster growth should be due to contributions from the type II survivors. In contrast, the double tlc1 rev3 mutant, which encodes a translesion repair DNA polymerase ζ, did not display any major difference in the growth and telomere phenotypes as observed in these replicative polymerase-deficient, telomerase-minus mutants (data not shown).
Distinct patterns of individual survivors from telomerase-deficient and polymerase-defective mutants.
Next, we were interested in evaluating the effect of replicative polymerases on the formation of individual type I and type II survivors. We recovered survivors in the form of single colonies by repeatedly streaking mutant spores on solid YEPD plates. All telomerase-minus, polymerase-defective spores senesced faster, and survivors showed much more difficulty than those of the tlc1 mutant (Fig. 3B). Moreover, survivors from the tlc1 cdc17-2 (polα) and tlc1 cdc2-2 (polδ) mutants were more difficult to obtain than those from the tlc1 pol2-18 (polɛ) mutant (data not shown). These results suggest that polymerases α and δ play more significant roles than polymerase ɛ in the telomere-telomere recombination pathway.
Genomic DNA from these survivors was analyzed by the four-base cutter (Fig. 3C) and the XhoI (Fig. 3D) Southern analyses. Six independent survivors from each mutant strain are shown in Fig. 3C and 3D. Although the characteristic type I and type II survivors could be recovered from the tlc1 cdc17-2 (polα) and tlc1 pol2-18 (polɛ) mutants, all the recovered tlc1 cdc2-2 (polδ) survivors (20 of 20 in Table 1) had barely elongated telomeres. These tlc1 cdc2-2 survivors had completely lost the ability to elongate long fragments, having neither Y′-Y′ nor TG1-3-TG1-3 lengthening. Instead they showed a unique phenotype of slight lengthening of the TG1-3 telomere of survivors (Fig. 3C). The lengths of these telomeres ranged from 300 to 600 bp, and these elongated telomeres were still subject to dynamic shortening and lengthening (data not shown).
TABLE 1.
Distribution of survivor types
| Genotype | No. of survivors (% of total)
|
||
|---|---|---|---|
| Total studied | Type I | Type II | |
| tlc1 | 92 | 86 (93) | 6 (7) |
| tlc1 cdc17-2 | 62 | 24 (39) | 38 (61)a |
| tlc1 cdc2-2 | 20 | 0 (0) | 20 (100)b |
| tlc1 pol2-18 | 50 | 10 (20)c | 40 (80) |
| tlc1 tel1 | 86 | 86 (100) | 0 (0) |
| tlc1 mec1 sml1 | 86 | 86 (100) | 0 (0) |
| tlc1 tel1 mec1 sml1 | 86 | 86 (100) | 0 (0) |
| tlc1 cdc13-1 | 63 | 7 (11) | 56 (89) |
All type II survivors from the tlc1 cdc17-2 (polα) mutant had noticeable type I-like Y′-Y′ amplification.
All the recovered tlc1 cdc2-2 (polδ) survivors had barely elongated telomeres, which is an atypical type II pattern.
All type I survivors from the tlc1 pol2-18 (polɛ) mutant had much less Y′-Y′ amplification than the typical type I pattern of the tlc1 survivors.
These telomeres were long enough for the maintenance of viability of the tlc1 cdc2-2 survivors. Of the 61 tlc1 cdc17-2 (polα) survivors analyzed (Table 1), 24 (39%) showed the type I pattern and 38 (61%) showed the type II-like pattern. However, all 38 type II survivors from the tlc1 cdc17-2 mutant had noticeable Y′-Y′ amplification products (Fig. 3D and data not shown). This result indicated that Y′-Y′ amplification is a favored repair pathway in telomerase-minus, polymerase α-defective cells. In contrast, of the 50 tlc1 pol2-18 (polɛ) survivors that were assayed, 10 (20%) showed the type I-like pattern and 40 (80%) showed the type II pattern. However, all 10 type I survivors from the tlc1 pol2-18 mutant had much less Y′-Y′ amplification than the typical type I pattern of the tlc1 survivor (Fig. 3D and data not shown). This result supported the observation that polymerase ɛ has a more important role in type I than in type II recombination.
tlc1 cdc2-2 survivors occur through RAD52-dependent type II-like telomere-telomere recombination.
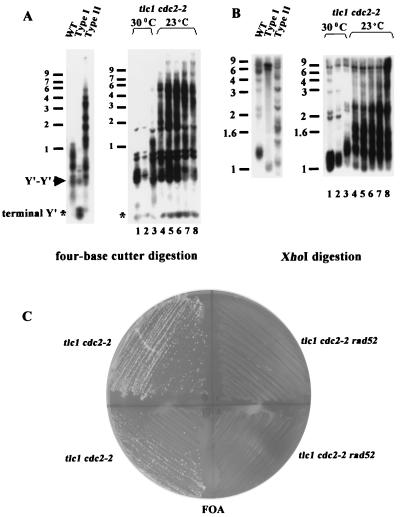
Because the pattern of tlc1 cdc2-2 (polδ) survivors was quite different from the typical patterns of type I and type II survivors, we were aware that these survivors might be derived from intragenic or extragenic suppressors or that a third undiscovered pathway for telomere maintenance might have been elicited. To clarify these possibilities, we first tested whether reintroduction of the wild-type activity of polymerase δ after the establishment of survivors would rescue the pattern of tlc1 cdc2-2 survivors. We found that the tlc1 cdc2-2 survivors quickly converted to the typical type II pattern once the survivors were cultured at 23°C in liquid culture (Fig. 4A and B). Given the facts above, we hypothesized that the slight lengthening of the telomere is generated by Rad52-dependent telomere-telomere recombination.
FIG. 4.
Maintenance of tlc1 cdc2-2 survivors requires Rad52p. (A and B) Cultures of the tlc1 cdc2-2 strain were inoculated from spore colonies and grown to the stationary phase at 30°C. Stationary-phase cultures were diluted repeatedly 3:10,000 into fresh medium at 72 h as described for Fig. 1. The cultures were grown at 30°C for the first two dilutions and switched to 23°C, the permissive temperature, for the remaining dilutions. DNA was prepared for Southern analysis as described for Fig. 1A and 2B. Size markers (kilobases) are shown on the left. (C) Spores of rad52 tlc1 cdc2-2 and tlc1 cdc2-2 strains carrying a RAD52 URA3 plasmid were isolated on complete medium lacking uracil. Survivors were generated by restreaking spore colonies on YEPD plates. Shown here are two tlc1 cdc2-2 (left) and two rad52 tlc1 cdc2-2 (right) survivors without the RAD52 plasmid on a 5-fluorouracil (FOA) plate at 30°C. The plate is the first restreak from survivors on the YEPD plates. WT, wild type.
To determine if Rad52p is required to maintain tlc1 cdc2-2 survivors, we isolated independent spores in tlc1 cdc2-2 rad52 and tlc1 cdc2-2 strains carrying a RAD52 URA3 plasmid. Survivors were generated by restreaking spores on YEPD plates. Cells that had lost the RAD52 plasmid were identified by their ability to grow on plates containing 5-fluorouracil. While tlc1 cdc2-2 survivors could grow on the 5-fluorouracil plate, rad52 tlc1 cdc2-2 survivors did not grow at all, even on the first restreak after loss of the RAD52 plasmid. This result indicated that deletion of RAD52 totally prevents the formation of tlc1 cdc2-2 survivors (Fig. 4C). This argued that the cdc2-2 (polδ) mutant at the semipermissive temperature is somewhat leaky and that the observed slight elongation represents a residual amount of true type II recombination. Altogether, these experiments clearly established that tlc1 cdc2-2 mutants survived through RAD52-dependent homologous recombination.
Type II telomere-telomere recombination is mediated by the TEL1 and MEC1 genes.
Checkpoint responses maintain the order and fidelity of the eukaryotic cell cycle, and defects in checkpoints contribute to genetic instability and cancer. The involvement of checkpoint proteins in telomere maintenance was implicit from the consequences of telomere shortening in tel1 and mec1 strains (22, 42). Further evidence of Tel1p's involvement in telomere maintenance came from the recent observation that its human homolog ATM directly phosphorylates Nbs1 (19, 54). Nbs1 forms a complex with hMre11 and Rad50 at double-strand breaks. Previously, several groups demonstrated that RAD50 is essential for type II survivor development (5, 11, 49). This led us to speculate that TEL1 and/or MEC1 also contributes to the regulation of type II telomere-telomere recombination.
To test this possibility, we isolated otherwise isogenic tlc1, tlc1 tel1, tlc1 mec1 sml1, and tlc1 tel1 mec1 sml1 spore products (the sml1 mutation suppresses the inviability caused by mec1 mutations) by dissecting heterozygous diploids and restreaking them multiple times on plates until survivors appeared. DNA was prepared from individual survivors, and telomere structure was determined by Southern hybridization. Significantly, all of the tlc1 tel1, tlc1 mec1 sml1, and tlc1 tel1 mec1 sml1 survivors established a type I pattern (86 of 86 survivors for all three mutants, Table 1). These data suggested that Tel1p and Mec1p mediate the generation of type II survivors.
It is possible that some type II events might occur in these strains but are too rare to be recovered by solid-plate restreaking. To confirm the role of Tel1p and Mec1p in the generation of type II survivors, we also examined the formation of survivors in the liquid assay, which favors isolation of type II survivors due to their faster growth rate (48). We inoculated tlc1, tlc1 tel1, tlc1 mec1 sml1, and tlc1 tel1 mec1 sml1 spore clones into liquid medium. The cultures were diluted 1:10,000 every 2 days for a total of eight dilutions. DNA was prepared prior to each dilution and analyzed by Southern hybridization. As shown in Fig. 5, while type II survivors took over the tlc1, tlc1 tel1, and tlc1 mec1 sml1 cultures (Fig. 5A to C) at, respectively, the fourth, third, and third dilutions, the type II pattern was not detected in the tlc1 tel1 mec1 sml1 quadruple mutant (Fig. 5D), even after prolonged subculturing. The earlier appearance of type II survivors in the tlc1 tel1 and tlc1 mec1 sml1 strains is consistent with the faster senescence of tlc1 tel1 and tlc1 mec1 sml1 cells on solid plates (data not shown). These results suggested that the generation of type II survivors is mediated by the combined actions of Tel1p and Mec1p. In the absence of either one of them, the cells still contained residual activity provided by the other for the generation of type II survivors.
FIG. 5.
TEL1 and MEC1 genes are required for type II telomere-telomere recombination. Freshly dissected tlc1 (A), tlc1 tel1 (B), tlc1 mec1 sml1 (C), and tlc1 tel1 mec1 sml1 (D) spores were inoculated into YEPD medium. At 48-h intervals, DNA was isolated, and the cultures were then diluted 1:10,000 into fresh medium. This process was performed eight times. DNA was prepared from each time point for each strain and analyzed as for Fig. 1. Size markers (kilobases) are shown at the left.
Telomeric single-stranded binding protein Cdc13 and end-binding protein Ku regulate recombination events.
Several reports provide evidence that the telomere binding proteins Cdc13 and Ku can regulate the telomerase pathway both positively and negatively (4, 10, 21, 37, 39). To determine whether these proteins could also be factors for telomere-telomere recombination, we first isolated otherwise isogenic tlc1 and tlc1 cdc13-1 spore clones. The liquid serial dilution and four-base cutter assays were conducted at the semipermissive temperature, as described above, and the DNA from each strain was examined by Southern hybridization (Fig. 6A). Compared to the tlc1 strain, which took five dilutions to establish, the type II pattern of the tlc1 cdc13-1 strain was established right from the beginning of the culture. In agreement with this finding, tlc1 cdc13-1 cells senesced much faster than the tlc1 strain in the liquid senescence and recovery assay. The tlc1 cdc13-1 strain dropped to its lowest growth rate at day 2 (Fig. 6B). Consistent with its rapid rate of senescence, the tlc1 cdc13-1 mutant grown on a solid streaking plate displayed a similar phenotype. Survivors appeared right at the first restreak of spore colonies (Fig. 6C).
FIG. 6.
Telomeric single-stranded binding protein Cdc13 influences survivor formation. (A) Liquid cultures and telomere Southern analysis were performed as described for Fig. 1 except that the cultures were grown at 27°C, the semipermissive temperature for the tlc1 cdc13-1 strain. Size markers (kilobases) are shown at the left. (B) The senescence and recovery rate of the wild-type (WT), tlc1, and tlc1 cdc13-1 strains were determined as described for Fig. 3A except that the cultures were grown at 27°C. (C) Growth of the tlc1 cdc13-1 strain on solid plates was examined by repeated streaking on YEPD plates and growth at 27°C.
The Ku complex has been shown to bind telomeres (21) and recruit telomerase (39). Therefore, we tested whether the Ku complex also participates in regulating the maintenance of survivors. Since the tlc1 hdf1 double mutation is lethal, to obtain survivors we generated a tlc1/TLC1 hdf1/HDF1 heterozygous diploid containing plasmid pRS316HDF1. Haploid tlc1 and tlc1 hdf1 spores containing the pRS316HDF1 plasmid were made by tetrad dissection, and type I and type II survivors were generated from these strains. To determine the influence of the HDF1-containing plasmid on the maintenance of tlc1 and tlc1 hdf1 survivors, we plated both type I and type II survivors on 5-fluorouracil plates to select for loss of the pRS316HDF1 plasmid. The survival rates for both type I (9.24%) and type II (42.4%) tlc1 hdf1 cells on 5-fluorouracil plates were significantly lower than those of the type I (93%) and type II (92%) tlc1 controls (Table 2). This experiment suggested that the Ku complex may be required for end protection and survivor maintenance in the telomerase-minus strain. Moreover, type I survivors (9.24%) relied more on Ku proteins for their viability than type II survivors (42.4%).
TABLE 2.
Influence of the Ku complex on maintenance of survivors
| Survivor type | Genotype | % 5-Fluorouracil-resistant cells |
|---|---|---|
| I | tlc1 hdf1 + pRS316HDF1 | 9.24 |
| tlc1 + pRS316HDF1 | 93 | |
| II | tlc1 hdf1 + pRS316HDF1 | 42.4 |
| tlc1 + pRS316HDF1 | 91.8 |
DISCUSSION
To better understand the effect of proteins involved in telomere-telomere recombination, we began to examine factors in various pathways more systematically. In this study, our liquid serial dilution and four-base cutter assays provided a way to detect the in vivo extension products of recombination by replicative polymerases. During DNA replication, the leading and lagging strands are synthesized concurrently. The three major polymerase mutants exhibit different telomere phenotypes in telomerase-deficient cells. All three replicative polymerases play roles in the telomere-telomere recombination pathway, and recombination events extend all the way to the ends of chromosomes, which may indicate that telomere-telomere recombination is a form of break-induced replication (11, 38, 49).
In fact, evidence is steadily accumulating that recombination is accomplished through the help of the replication machinery (23, 26, 27). The primase-polymerase α complex is required for the initial synthesis at leading and lagging strands during DNA replication. In the case of type II telomere-telomere recombination, the leading strand is synthesized from the 3′-hydroxyl group of the G strand. Since there is no autonomously replicating sequence in TG1-3/C1-3A repeats but autonomously replicating sequences are contained in the Y′ elements (9), defective polymerase α may cause more severe damage within Y′s at the leading strand during DNA replication. This stalled replication intermediate in Y′s may promote strand transfer to other Y′s by the break-induced replication pathway. Alternatively, given the potential secondary structure in G-rich single-stranded sequences that may form during type II lengthening, the priming step at the lagging strand for type II recombination may face stronger hindrance in the polymerase α-defective cells. This may explain why type I recombination is a preferred pathway in telomerase-minus, polymerase α-deficient cells.
Polymerases δ and ɛ have been proposed to synthesize both leading and lagging strands in a highly coordinated way during processive DNA replication (7, 25). Here we observed separate phenotypes for these two polymerases. In the telomerase-deficient, polymerase δ-defective strain, telomeres completely lost the ability to amplify long-tract telomeric DNA, so there was neither Y′-Y′ amplification nor TG1-3-TG1-3 recombination (Fig. 2). This experiment strongly supports the hypothesis that polymerase δ is the major polymerase for processive long-tract lengthening of both type I and type II telomere-telomere recombination. Without its function for long-tract elongation, the telomeres of survivors maintain short telomeres, in a RAD52-dependent manner, that are sufficiently long to sustain cell viability (Fig. 4). In contrast, in the telomerase-deficient polymerase ɛ-defective strain, the telomere can still be extremely elongated by the processive extension ability from polymerase δ. Polymerase ɛ has a much more important role in type I than type II recombination, which is reflected by the fact that in the absence of polymerase ɛ, it becomes more difficult to elongate multiple copies of the long Y′ elements immediately (Fig. 2 and 3D). Our telomere recombination study recapitulates but also conflicts with certain features from the study of the mating type switch at the MAT locus. Double-strand break repair at the MAT locus may be carried out by polymerases α, ɛ, and δ (23). We show here that polymerase δ is the major polymerase for long-tract extension of telomeres, whereas polymerase ɛ had more of an effect than polymerase δ in the gene conversion pathway during mating type switching (23). This discrepancy probably only reflects the difference in the mutant strains used in these two studies. The pol3-1 mutant used in the MAT locus study is defective in the 3′→5′ exonuclease/proofreading domain of polymerase δ, whereas the cdc2-2 mutant used in the telomere study is defective in the polymerase domain of polymerase δ (13). This exonuclease/proofreading pol3-1 mutation has been shown to have no significant effect on the telomere-telomere recombination pathway (43).
Several pathways other than the DNA recombination machinery also govern the process of telomere-telomere recombination. Here we show that type II telomere-telomere recombination is coordinately controlled by Tel1p and Mec1p (Fig. 5). Tel1p, Mec1p, and their human homolog ATM have been found to regulate DNA damage repair and telomere maintenance. Given the finding that ATM can phosphorylate Nbs1 when it responds to DNA damage (19, 54), Tel1p and Mec1p may provide a similar function to convert the Rad50-Mre11-Xrs2 complex through Xrs2p to an active form, which is necessary to generate a single-stranded overhang that turns the telomeric terminus into a suitable substrate for the process of type II telomere-telomere recombination (5, 11, 49). Furthermore, given the evidence that Tel1p and Mec1p may govern the action of Rad50-Mre11-Xrs2 at the telomere, it would be of interest to investigate, at internal chromosomal double-strand breaks, whether Rad50-Mre11-Xrs2 is regulated by the combined action of Tel1p and Mec1p.
In addition to DNA damage-signaling proteins, telomeric single-strand binding protein Cdc13 and end-binding protein Ku perform positive and negative roles not only in telomerase-mediated telomere maintenance (4, 10, 21, 37, 39), but also in telomere-telomere recombination. Recently, Grandin et al. (20) also showed that type II survivors arose exclusively from the telomerase-minus cdc13-1 strain. The cdc13-1 strain exhibits a single-stranded tail phenotype at its semipermissive temperature (18). The prompt development of type II survivors in the tlc1 cdc13-1 strain (Fig. 6A) may reflect that the single-stranded TG1-3 tail is highly recombigenic and may be the in vivo substrate during telomere-telomere recombination. Furthermore, the double-stranded end-binding complex Ku is also required for the telomere protection of survivors. Our results demonstrate that type II cells are more resistant to the loss of Ku function than type I cells (Table 2). They may also indicate that the topology and structure of the type II ends are different from those of the wild-type and type I survivors.
The structure of telomeres in Saccharomyces type II survivors is reminiscent of that in human cell lines and tumors that maintain telomeric DNA by the telomerase-independent ALT pathway. It seems that at least two steps are involved in type II and ALT survival. The first is the generation of the long extension on one (or a few) critically short telomere(s) in the population (47), which will give the dying clone a slight selective advantage. The second is the propagation of that structure through a RAD52-dependent process to the remaining telomeres in that cell and then into the population for the viable maintenance of cancerous cells. Different proteins, e.g., Rad50, Sgs1, Tel1, Mec1, Ku, Cdc13, and replicative polymerases, could affect either or both of these steps in a positive or negative way.
Acknowledgments
We are very grateful for the suggestions and other support of V. Zakian, J. Haber, and S. Brill. We also thank D. Gottschling, A. Sugino, Y. Tsukamoto, A. Taggart, and W. Tham for providing yeast strains and plasmids and J. Lin, C. Chen, M. Chen, and A. Gabriel for critical comments on the manuscript.
This work was supported by grants from the National Science Council of Taiwan and the National Taiwan University College of Medicine to S. Teng.
REFERENCES
- 1.Adams, A. K., and C. Holm. 1996. Specific DNA replication mutations affect telomere length in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artandi, S. E., and R. A. DePinho. 2000. A critical role for telomeres in suppressing and facilitating carcinogenesis. Curr. Opin. Genet. Dev. 10:39-46. [DOI] [PubMed] [Google Scholar]
- 3.Boulton, S. J., and S. P. Jackson. 1996. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 24:4639-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent nonhomologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 17:1819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brad, J., R. A. Marciniak, M. Mcvey, S. A. Stewart, W. C. Hahn, and L. Guarente. 2001. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 20:905-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan, T. M., Englezou, A., Dalla-Pozza, L., Dunham, M. A., and Reddel, R. R. 1997. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3:1271-1274. [DOI] [PubMed] [Google Scholar]
- 7.Burgers, P. M. 1998. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma 107:218-227. [DOI] [PubMed] [Google Scholar]
- 8.Carson, M., and L. Hartwell. 1985. CDC17: an essential gene that prevents telomere elongation in yeast. Cell 42:249-257. [DOI] [PubMed] [Google Scholar]
- 9.Chan, C. S., and B. K. Tye. 1983. Organization of DNA sequences and replication origins at yeast telomeres. Cell 33:563-573. [DOI] [PubMed] [Google Scholar]
- 10.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Q., A. Ijpma, and C. W. Greider, 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen, H., and D. A. Sinclair. 2001. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl. Acad. Sci. USA 98:3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conrad, M., and C. S. Newlon. 1983. Saccharomyces cerevisiae cdc2 mutants fail to replicate approximately one-third of their nuclear genome. Mol. Cell. Biol. 3:1000-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diede, S. J., and D. E. Gottschling. 1999. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99:723-733. [DOI] [PubMed] [Google Scholar]
- 15.Dunham, M. A., A. A. Neumann, C. L. Fasching, and R. R. Reddel. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26:447-450. [DOI] [PubMed] [Google Scholar]
- 16.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 17.Frick, D. N., and C. C. Richardson. 2001. DNA primases. Annu. Rev. Biochem. 70:39-80. [DOI] [PubMed] [Google Scholar]
- 18.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatei, M., D. Young, K. M. Cerosaletti, A. Desai-Mehta, et al. 2000. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat. Genet. 25:115-119. [DOI] [PubMed] [Google Scholar]
- 20.Grandin, N., C. Damon, and M. Charbonneau. 2001. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 20:6127-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gravel, S., M. Larrivee, P. Labrecque, and R. Wellinger. 1998. Yeast Ku as a regulator of chromosomal DNA end structure. Science 280:741-744. [DOI] [PubMed] [Google Scholar]
- 22.Greenwell, P. W., S. L. Kronmal, S. E. Porter, J. Gassenhuber, B. Obermaier, and T. D. Petes. 1995. TEL1: a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82:823-829. [DOI] [PubMed] [Google Scholar]
- 23.Holmes, A. M., and J. E. Haber. 1999. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell 96:415-424. [DOI] [PubMed] [Google Scholar]
- 24.Huang, P., F. E. Pryde, D. Lester, R. L. Maddison, R. Borts, I. D. Hickson, and E. J. Louis. 2001. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 11:125-129. [DOI] [PubMed] [Google Scholar]
- 25.Hubscher, U., H. P. Nasheuer, and J. E. Syvaoja. 2000. Eukaryotic DNA polymerases, a growing family. Trends Biochem. Sci. 25:143-147. [DOI] [PubMed] [Google Scholar]
- 26.Kraus, E., W., Leung, and J. E. Haber. 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzminov, A. 2001. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. USA 98:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le, S., J. Moore, J. Haber, and C. Greider. 1999. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics 152:143-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lingner, J., T. R. Hughes, A. Shevchenko, M. Mann, V. Lundblad, and T. R. Cech. 1997. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276:561-567. [DOI] [PubMed] [Google Scholar]
- 30.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est-senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 31.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 32.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822-1834. [DOI] [PubMed] [Google Scholar]
- 33.McEachern, M. J., A. Krauskopf, and E. Blackburn. 2000. Telomeres and their control. Annu. Rev. Genet. 34:331-358. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 35.Navas, T. A., Z. Zhou, and S. J. Elledge. 1995. DNA polymerase å links the DNA replication machinery to the S phase checkpoint. Cell 80:29-39. [DOI] [PubMed] [Google Scholar]
- 36.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 37.Nugent, C. I., and V. Lundblad. 1998. The telomerase reverse transcriptase: components and regulation. Genes Dev. 12:1073-1085. [DOI] [PubMed] [Google Scholar]
- 38.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson, S. E., A. E. Stellwagen, S. J. Diede, M. S. Singer, et al. 2001. The function of a stem-loop in telomerase RNA is linked to the DNA repair protein Ku. Nat. Genet. 27:64-67. [DOI] [PubMed] [Google Scholar]
- 40.Qi, H., and V. A. Zakian. 2000. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 14:1777-1788. [PMC free article] [PubMed] [Google Scholar]
- 41.Reddel, R. R., T. M. Bryan, L. M. Colgin, K. T. Perrem, and T. R. Yeager. 2001. Alternative lengthening of telomeres in human cells. Radiat. Res. 155:194-200. [DOI] [PubMed] [Google Scholar]
- 42.Ritchie, K. B., J. C. Mallory, and T. D. Petes. 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6065-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rizki, A., and V. Lundblad. 2001. Defects in mismatch repair promote telomerase-independent proliferation. Nature 411:713-716. [DOI] [PubMed] [Google Scholar]
- 44.Shay, J. W., Y. Zou, E. Hiyama, and W. E. Wright. 2001. Telomerase and cancer. Hum. Mol. Genet. 10:677-685. [DOI] [PubMed] [Google Scholar]
- 45.Shore, D. 2001. Telomeric chromatin: replicating and wrapping up chromosome ends. Curr. Opin. Genet Dev. 11:189-198. [DOI] [PubMed] [Google Scholar]
- 46.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 47.Teng, S. C., C. Epstein, Y. L. Tsai, H. W. Cheng, H. Chen, and J. J. Lin. 2002. Induction of global stress response in Saccharomyces cerevisiae cells lacking telomerase. Biochem. Biophys. Res. Commun. 291:714-721. [DOI] [PubMed] [Google Scholar]
- 48.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teng, S. C., J. Chang, B. McCowan, and V. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 50.Weilbaecher, R. G., and V. Lundblad. 1999. Assembly and regulation of telomerase. Curr. Opin. Chem. Biol. 3:573-577. [DOI] [PubMed] [Google Scholar]
- 51.Wotton, D., and D. Shore. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748-760. [DOI] [PubMed] [Google Scholar]
- 52.Wright, J. H., D. E. Gottschling, and V. A. Zakian. 1992. Saccharomyces telomeres assume a nonnucleosomal chromatin structure. Genes Dev. 6:197-210. [DOI] [PubMed] [Google Scholar]
- 53.Zakian, V. A. 1996. Structure, function and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30:141-172. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, B. B., and S. J. Elledge. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433-439. [DOI] [PubMed] [Google Scholar]