Abstract
The desmoglein 1 (Dsg1) and desmocollin 1 (Dsc1) isoforms of the desmosomal cadherins are expressed in the suprabasal layers of epidermis, whereas Dsg3 and Dsc3 are more strongly expressed basally. This differential expression may have a function in epidermal morphogenesis and/or may regulate the proliferation and differentiation of keratinocytes. To test this hypothesis, we changed the expression pattern by overexpressing human Dsg3 under the control of the keratin 1 (K1) promoter in the suprabasal epidermis of transgenic mice. From around 12 weeks of age, the mice exhibited flaking of the skin accompanied by epidermal pustules and thinning of the hair. Histological analysis of affected areas revealed acanthosis, hypergranulosis, hyperkeratosis, localized parakeratosis, and abnormal hair follicles. This phenotype has some features in common with human ichthyosiform diseases. Electron microscopy revealed a mild epidermal spongiosis. Suprabasally, desmosomes showed incorporation of the exogenous protein by immunogold labeling but were normal in structure. The epidermis was hyperproliferative, and differentiation was abnormal, demonstrated by expression of K14 in the suprabasal layer, restriction of K1, and strong induction of K6 and K16. The changes resembled those found in previous studies in which growth factors, cytokines, and integrins had been overexpressed in epidermis. Thus our data strongly support the view that Dsg3 contributes to the regulation of epidermal differentiation. Our results contrast markedly with those recently obtained by expressing Dsg3 in epidermis under the involucrin promoter. Possible reasons for this difference are considered in this paper.
Desmosomes are complex intercellular junctions that link the keratin filaments of adjacent cells, providing mechanical strength to epithelial tissues such as the epidermis. The desmosomal cadherins desmoglein (Dsg) and desmocollin (Dsc) are calcium-dependent adhesion molecules that exist as three isoforms, each the product of a different gene. Dsg2 and Dsc2 are expressed in all desmosome-containing tissues, whereas Dsg1, Dsc1, Dsg3, and Dsc3 are restricted to stratified epithelia. In the epidermis, a stratified epithelium, isoforms 1 and 3 show a reciprocal expression pattern (4, 25, 28, 31, 44). Dsg3 has a strong basal distribution associated with proliferating cells, and Dsg1 has a suprabasal expression in the terminally differentiating layers (3, 4, 39). Thus, there is an increase in the ratio of Dsg1 to Dsg3 from the basal to the apical surface of the epidermis. It is likely that individual desmosomes in the cell layers where Dsg1 and Dsg3 expression overlap contain both isoforms, as has been demonstrated for desmocollin isoforms (31). This differential expression of desmosomal glycoproteins within the epidermis has been suggested to form adhesive gradients that may provide positional information to keratinocytes and thereby participate in the regulation of the proliferation and differentiation of the tissue. It was demonstrated in another system, the mammary gland, that desmosomal adhesion does indeed play an important role in epithelial morphogenesis (38).
The importance of Dsg3 function in the maintenance of normal cell-to-cell adhesion in the epidermis has been demonstrated in studies of the human disease pemphigus vulgaris (PV) and of Dsg3-knockout and transgenic mice. In PV, autoantibodies to Dsg3 bind to the extracellular region of the desmosomes, resulting in loss of adhesion (acantholysis) immediately above the basal layer (29). The resulting epidermal blistering may be fatal if inadequately treated. Targeted disruption of Dsg3 in mice produces a phenotype resembling PV, with loss of keratinocyte adhesion, leading to epidermal and oral mucosal blistering, together with hair loss and runting (26, 27). Transgenic mice expressing a mutant, truncated form of Dsg3 under the control of the keratin 14 (K14) promoter also have epidermal abnormalities, including flaking skin and blackening of the tail tip, which were associated with a reduction in the size and number of desmosomes (1).
One approach to investigate the role of desmosomal cadherins and other proteins in epidermis is to misexpress them with differentiation-specific promoters. Thus, Henkler et al. expressed Dsc1 in the basal layer of mouse epidermis by using the K14 promoter but found no effect on epidermal differentiation (21). In contrast, Elias et al. expressed Dsg3 in the upper epidermis under the control of the involucrin promoter and found dramatic effects on the structure and function of the stratum corneum (15). The epidermis resembled oral mucosa and showed greatly increased transepidermal water loss (TEWL), which was lethal.
We expressed Dsg3 in the upper epidermis under the control of the K1 promoter. Our results showed substantial perturbation of epidermal proliferation and differentiation producing a condition with some features of chronic dermatitis and ichthyosis. Interestingly, this phenotype contrasts markedly with that obtained by Elias et al. (15) and shows more resemblance to phenotypes produced by overexpression of various cytokines and integrins.
MATERIALS AND METHODS
Construction of the expression vector and generation of transgenic mice.
Two overlapping cDNA clones of human DSG3 (hDSG3), E12 and E33, were kindly supplied by John Stanley (2). The full-length cDNA of 3.3 kb was assembled from the two clones by utilizing a unique XbaI site in the overlap region. The human K1 targeting vector was kindly supplied by Dennis Roop (37, 18). In order to aid later transgene excision, the K1 promoter was cut with EcoRI and cloned into the EcoRI site of pBluescript (Clontech), which had been previously modified by the removal of five enzyme sites, SmaI, BamHI, SpeI, XbaI, and NotI. The full-length hDSG3 cDNA was then cloned into the NotI site of the new K1 targeting vector (Fig. 1A). The K1-hDSG3 transgene was excised from the plasmid sequences of the targeting vector by cutting with BssHII. The transgene DNA was recovered by using the QIAEX II kit (Qiagen) and diluted in microinjection buffer (5 mM Tris, 0.1 mM EDTA [pH 7.4]) to a concentration of 5 ng/μl. Microinjection of C57BL/6 × CBA embryos was carried out in M2 medium, and embryos were cultured overnight to the two-cell stage in M16 medium and implanted into pseudopregnant recipient females (22). These experiments were carried out under Home Office license no. 40/2179.
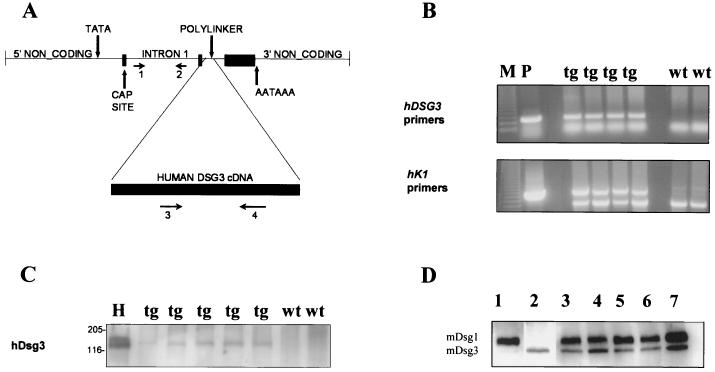
FIG. 1.
Detection of the K1-Dsg3 transgene in mouse epidermis. (A) K1-Dsg3 targeting construct based on that used in reference 18. PCR primers 1 through 4 (horizontal arrows) for identifying transgenic mice are shown. (B) Potential transgenic mice were screened by PCR using hDsg3-specific and human K1 vector-specific (hK1) primers (shown in panel A) on tail DNA. Diluted plasmid DNA (P), used for microinjection, served as a positive control. A 250-bp fragment was amplified by the hDsg3 primers in the transgenic but not the wild-type mice, and a 600-bp product was detected by the K1 vector-specific primers in the transgenic mice but not the wild-type mice, but a lower, nonspecific band was detected in all mice, serving as an internal positive control (M, 100-bp DNA ladder). tg, transgenic mice; wt = wild-type mice. (C) Western blot of epidermal lysates with the hDsg3-specific antibody; HaCaT cells (H) were a positive control. Molecular masses (in kilodaltons) are indicated to the left of the blot. (D) Western blot of epidermal cell lysates with mDsg3- and mDsg1-specific antibodies. Lane 1 is a sample of epidermal protein from a wild-type mouse blotted for mDsg1 only; lane 2 is a sample from the same mouse as used in lane 1 but blotted for mDsg3 only. Lanes 3 through 7 show samples from mice blotted for mDsg1 and mDsg3 simultaneously. Lanes 3, 5, and 7, tg mice; lanes 4 and 6, wild-type mice (the sample in lane 4 is from the same mouse as used in lanes 1 and 2).
Isolation of genomic DNA and PCR analysis.
Genomic DNA was isolated from tail samples obtained from weaned mice as follows. Tissue was digested in 0.4 mg of proteinase K/ml in cell lysis solution (Flowgen) overnight at 55°C and centrifuged briefly. After adding 200 μl of protein precipitation solution (Flowgen) to the supernatant and centrifuging at 13,000 × g for 5 min, DNA was precipitated from the supernatant with isopropanol. DNA was diluted in TE buffer. Transgene integration was determined by PCR on genomic DNA samples with hDSG3-specific primers and human K1 vector-specific primers (18) (Fig. 1A and B).
Histology.
Samples of epidermis taken from the back, belly, ear, tail, and footpad were fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS). Tissues were then washed in PBS and processed for paraffin embedding. Five-micrometer-thick sections were attached to poly-l-lysine-coated slides and stained with hematoxylin and eosin stain.
Source of antibodies.
The primary antibodies used in Western blotting and immunohistochemical analysis were as follows: a rabbit polyclonal antibody raised against the whole extracellular domain of hDsg3 (45); 11-5F, a mouse monoclonal antibody against bovine desmoplakin (33); rabbit anti-mouse Dsc3 (Ke Lui and Carolyn Byrne, personal communication); rabbit anti-murine Dsg1 (mDsg1) (15); rabbit anti-mDsg3 (15); mouse anti-plakoglobin (Transduction Laboratories); MK1, rabbit anti-mouse K1 (BAbCo); MK14, rabbit anti-mouse K14 (BAbCo); RPmK16, rabbit anti-mouse K16 (34); NCL-CK6, anti-mouse K6 (Novocastra Laboratories); rabbit anti-mouse loricrin (BAbCo); rabbit anti-mouse involucrin (BAbCo); rabbit anti-mouse filaggrin (20); and rat monoclonal antibody against Ki67 antigen (17).
Immunohistochemistry. (i) Immunoperoxidase staining.
Skin tissue was fixed in 4% paraformaldehyde in PBS and embedded in paraffin wax, and 5-μm-thick sections were attached to poly-l-lysine-coated slides. Sections were deparaffinized, rehydrated through a graded ethanol series, and washed in PBS. Antigen retrieval was performed by heating slides to 95°C for 4 to 15 min in 0.01 M citrate buffer (pH 6) in a microwave oven. The sections were then immunostained by the ABC peroxidase method (Vector) with diaminobenzidine (Sigma) as the enzyme substrate and hematoxylin as a counterstain.
(ii) Immunofluorescence.
Frozen sections were fixed in ice-cold acetone-methanol (1:1, vol/vol) for 20 min, followed by a 15-min wash in PBS. The sections were blocked in 10% normal goat serum for 1 h, followed by incubation with the PV serum (kindly provided by T. Hashimoto) for 1 h at room temperature. After washing in PBS, the fluoroscein isothiocyanate-labeled secondary antibody was added to the sections for 30 min.
Toluidine blue staining.
Excised whole skin was fixed in 4% paraformaldehyde and dehydrated into 100% ethanol. Tissue was incubated in 0.1% toluidine blue in PBS for 5 min at room temperature, briefly destained in PBS for 1 min, and visualized under a dissecting microscope. This method is an adaptation of that described in reference 20 for detecting the absence of epidermal barrier.
(iv) Western blotting.
Epidermis was isolated from 2- to 3-day-old mice by incubating 1-cm2 pieces of skin in 2.5 mM EDTA at 50°C for 3 min and then overnight at 4°C. Epidermis was then separated from the dermis by peeling with fine forceps. For a comparison of soluble and insoluble fractions, epidermis was extracted for 20 min on ice with cytoskeletal buffer (16). The isolates were sonicated in PBS on ice for 30 s, boiled in sodium dodecyl sulfate (SDS) sample buffer for 5 min, centrifuged for 5 min at 13,000 × g, and stored at −20°C. Protein concentrations were estimated by using the bicinchoninic acid protein assay (Pierce). SDS-polyacrylamide gel electrophoresis was performed on 7.5 or 10% gels with the Bio-Rad Protein III system. Proteins were blotted onto nitrocellulose which was blocked in 5% nonfat milk overnight at 4°C before incubation with the primary antibodies for 1 h at room temperature. The bound primary antibodies were detected with peroxidase-labeled secondary antibodies (Jackson Laboratories) followed by the ECL Plus detection system (Amersham Pharmacia).
(v) Electron microscopy.
Fresh skin was dissected into small pieces (approximately 1 by 1 by 0.5 mm) and fixed by immersion in 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M sodium cacodylate buffer containing 0.15 M sucrose and 2 mM calcium chloride (pH 7.3). Tissues were fixed at room temperature and then washed four times with cacodylate buffer. They were postfixed with 1% osmium tetroxide for 2 h, dehydrated through an ethanol series, and embedded in agar 100 resin. Ultrathin sections were contrasted with uranyl acetate and lead citrate and examined on a Philips model 400 transmission electron microscope.
(vi) Immunogold labeling.
Ultrathin cryosections of mouse skin were prepared and immunogold labeled as described in reference 32 for bovine nasal epidermis. Anti-hDsg3 primary antibody was applied at a 1/50 dilution for 1 to 2 h, followed by an application of goat anti-rabbit immunoglobulin G-10-nm gold conjugate (BioCell Research Laboratories). Grids were examined as described above.
RESULTS
Expression of the K1-hDSG3 transgene in mouse epidermis.
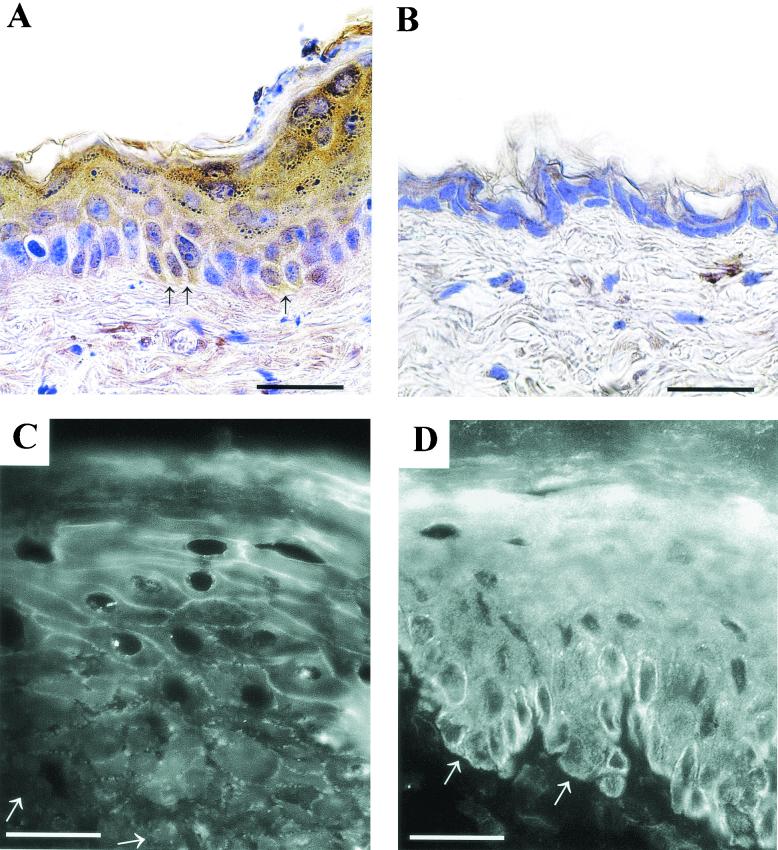
Transgenic mice were generated with the hDSG3 cDNA cloned into the human K1 targeting vector. Potential transgenic mice were screened by PCR using K1- and hDSG3-specific primers (Fig. 1B). Positive animals were crossed with (C57BL/6 × CBA) F1 wild-type mice to establish transgenic lines. Five founder animals were produced, four females and one male. Initial analysis showed that all lines had similar phenotypes, and one line was chosen for the further analysis. The copy number of the transgene in this line, determined by Southern blotting, was 13 to 15 (unpublished data). Expression of the transgene was determined by SDS-polyacrylamide gel electrophoresis and Western blotting of protein extracted from the epidermis of PCR-positive and -negative mice. Western blots with the hDsg3-specific polyclonal antibody demonstrated that PCR-positive mice displayed a band of around 135 kDa corresponding to hDsg3 whereas control, PCR-negative mice did not (Fig. lC). To determine whether the expression of the hDSG3 transgene altered the ratio of the endogenous mDsg3 to mDsg1, Western blotting was performed with mDsg3- and mDsg1-specific antibodies on epidermal extracts from transgenic and wild-type mice. To obtain the most accurate results, the two antibodies were used simultaneously on the same blot and the intensities of the bands were estimated by densitometry. Two bands corresponding in size to Dsg3 (135 kDa) and Dsg1 (160 kDa) were detected on each sample (Fig. 1D). The mean ratio of Dsg3 to Dsg1 was 0.57, and there was no statistically significant difference between transgenic animals and their wild-type littermates. Immunohistochemical staining showed expression of hDsg3 predominantly in the suprabasal layers of the epidermis and its absence from wild-type mice (Fig. 2A and B). Some positively stained cells were found in the basal layer of the transgenic mouse epidermis, consistent with the observation that the K1 targeting vector gives rise to expression in a proportion of basal cells (18). In addition, staining of epidermis with PV patient serum that reacts with Dsg3 but not Dsg1 stained cell peripheries throughout the epidermis in transgenic mice but only the lower layers of the epidermis in wild-type mice (Fig. 2C and D). These data show that the transgene is expressed and that the pattern of Dsg3 expression is altered.
FIG. 2.
Expression of hDsg3 in the epidermis. Immunohistochemistry by using hDsg3-specific antibody on the back skin of transgenic (A) and wild-type (B) mice. Arrows in panel A show some positive cells in the basal layer. Scale bars, 30 μm. Immunofluorescence with Dsg3-specific PV serum on frozen sections of transgenic (C) and wild-type (D) mouse skin is shown. Arrows indicate the position of the basement membrane. Scale bars, 20 μm.
K1-hDsg3 mice have epidermal defects.
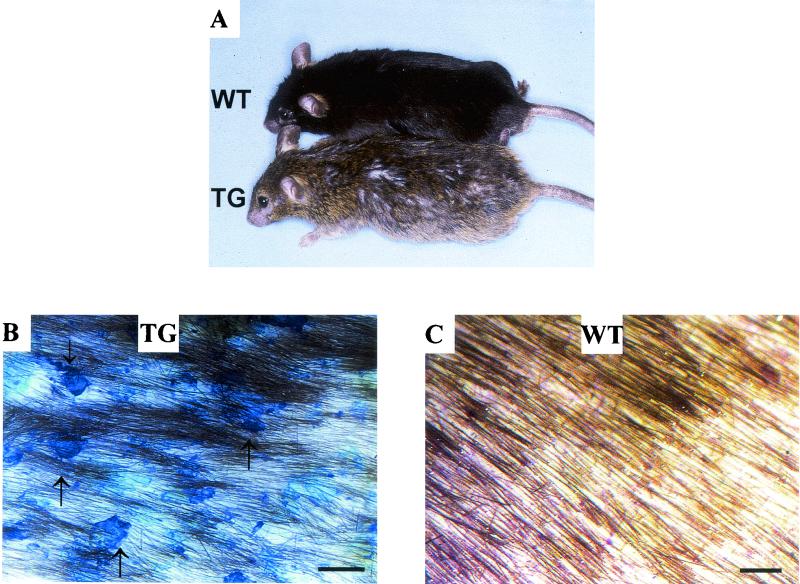
At birth, the skin of transgenic mice appeared normal by both external and histological examination. However, from around 12 weeks, epidermal flaking was detected on the back and belly skin and sometimes on the ears and footpads (Fig. 3). Hair was often sparse in these regions, making the flaking more prominent and revealing inflammation or reddening of the skin and scabbing on the epidermis.
FIG. 3.
K1-Dsg3 mice have epidermal defects. (A) Gross phenotype of transgenic (TG) and wild-type (WT) mice. The transgenic mouse had flaking epidermis on the back and has an untidy coat. (B and C) Surface of transgenic and wild-type mouse back skin after shaving and toluidene blue staining. Wild-type mouse skin remained clear, whereas the lesions of the transgenic mouse skin are visible in blue (arrows). Scale bar, 1 mm.
There was substantial variability in the severity of the phenotype. Some mice looked normal until their hair was shaven, while others were more mildly infected and displayed abnormalities only upon histological analysis. This variation was not linked to the transgene copy number since the variation existed within transgenic lines. There was some regression or healing of the phenotype in many of the mice. These factors made it difficult to judge the actual penetration of the phenotype, but an estimate derived from a histological analysis of 12-week-old mice was that 33% of animals had the severe phenotype, 40% had the milder phenotype, and 27% were normal. Approximately half of the mildly affected 12-week-old mice progressed to the severe phenotype later in life, and about a third of the severely affected mice showed signs of regression. Animals that were housed separately from weaning and so did not fight showed the same phenotype. No epidermal abnormalities occurred in nontransgenic control mice, whether housed individually or communally. We conclude that the phenotype was caused by expression of the transgene.
K1-hDsg3 mice show epidermal hyperproliferation.
Epidermal abnormalities were apparent from examination of hematoxylin- and eosin-stained sections of skin from 12-week-old transgenic mice. For mice with the most severe phenotype, the abnormalities were extensive, always involving the back and belly and sometimes the ears, tail, and footpads; in others with a milder phenotype, lesions were localized to areas of the back and belly skin.
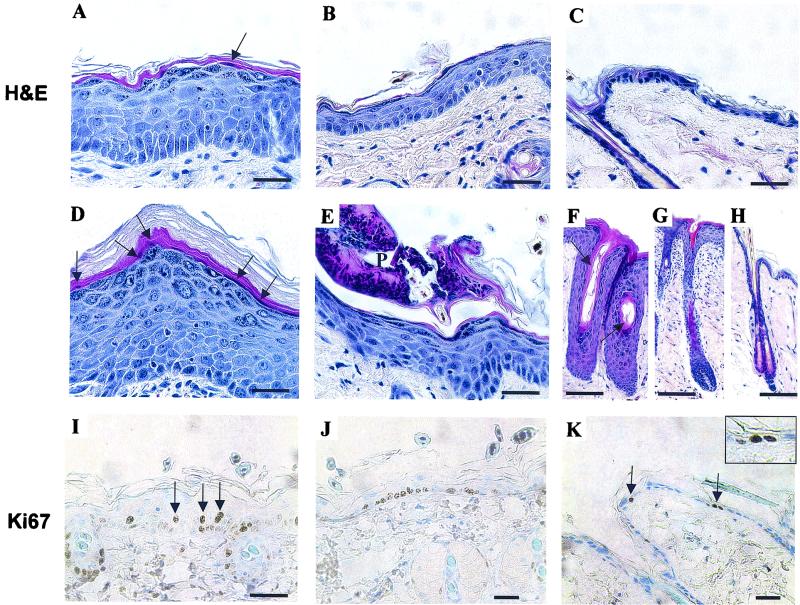
The most obvious change was in the thickness of the epidermis. In transgenic mice, there were as many as 10 cell layers of epidermis compared with 2 or 3 layers in the wild-type animals (Fig. 4A through D). The thickening of the epidermis represented an increase in the number of viable cell layers (acanthosis). The cornified layer showed localized thickening (hyperkeratosis) (Fig. 4D) and occasional retention of nuclei (parakeratosis) (arrowheads in Fig. 4A and D and arrows in Fig. 7G). Hyperproliferation was confirmed by immunohistochemistry for the Ki67 antigen (Fig. 4I through K). In severely affected regions of the transgenic mouse epidermis, a marked increase was seen in the number of positive nuclei in the basal layer (39% positive) (Fig. 4I and J) compared with that of wild-type mice (18% positive) (Fig. 4K). In control mouse skin, all positive nuclei were located in the basal layer of the epidermis or in the outer root sheath or matrix of hair follicles (unpublished data). In transgenic mouse epidermis, however, some Ki67-positive cells were observed in the first suprabasal layer (Fig. 4I), a feature typical of hyperproliferative skin disorders (35).
FIG. 4.
Expression of hDsg3 results in epidermal hyperproliferation. Histological analysis by hematoxylin and eosin staining revealed increased skin thickening in severely affected transgenic mice (A) and, to a lesser extent, in the mildly affected regions (B) compared with that in wild-type controls (C). Other defects included regional hyperkeratosis (D), parakeratosis (arrowheads in panels A and D), and pustules (P in panel E). Hair follicles were often enlarged in transgenic mice (F and G) compared with those in wild-type (H) mice and in the most severe regions showed abnormal keratinization (arrows in panel F). Immunohistochemistry for the Ki67 antigen showed an increase in the numbers of positive cells in the basal layer of transgenic mice (I and J) and also several positive suprabasal cells (arrows in panel I) compared with wild-type controls (K) (arrows in panel K indicate positive cells in the basal layer). Inset in panel K is a magnification of the arrow on the right. Scale bars in panels: A through E and I through K, 40 μm; F through H, 20 μm.
FIG. 7.
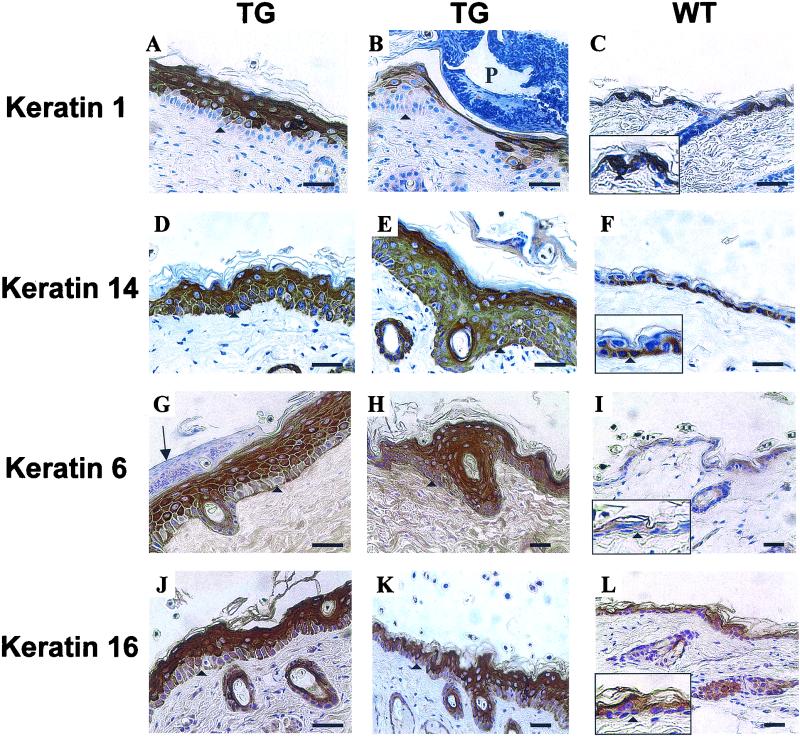
Keratin expression. Back skin of control (WT) and transgenic (TG) mice. For details, see the text. Arrowheads mark the positions of the basement membrane. The arrow in panel G indicates localized parakeratosis. P, pustule. Scale bars, 40 μm. Width of boxes in panels: C and F, 60 μm; I and L, 80 μm.
The granular layer was also affected. This normally consists of a single cell layer but in the transgenic mouse skin was increased to three to five layers (Fig. 4A and D and 8A), with cells showing an increase in both the number and the size of the granules (Fig. 4A and D and 5A and C). In the basal and lower spinous layers, there was some evidence of widening of intercellular spaces (spongiosis) (Fig. 4, compare panels A, B, and D with panel C). Hair follicles within the affected regions were often enlarged and sometimes highly keratinized, with an abnormal morphology (Fig. 4F through H). In the most severely affected regions of the transgenic mouse epidermis, there were pustules within or above the cornified layers containing a large number of inflammatory cells including polymorphs and neutrophils (Fig. 4E and 7B).
FIG. 8.
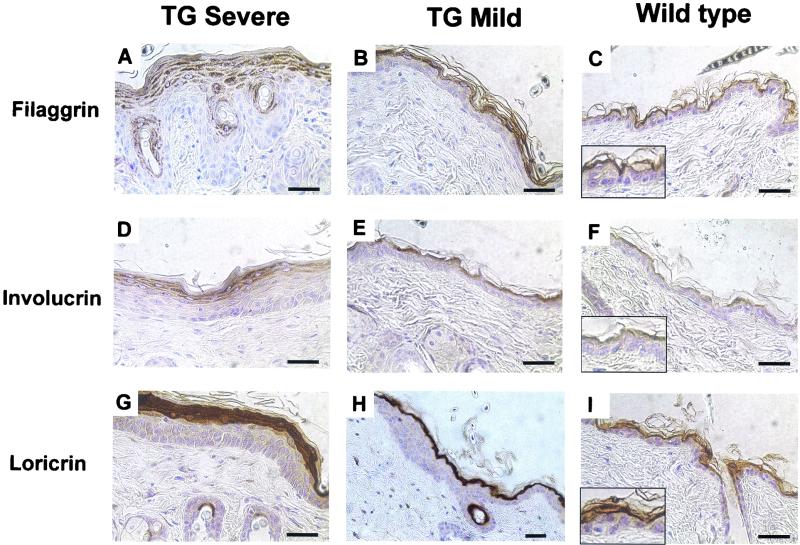
Filaggrin, involucrin, and loricrin expression. Back skin of transgenic (TG) and control mice. Sections of transgenic mouse skin were chosen to illustrate severe and mild phenotypes. Type of staining in is indicated to the left of the panels. Scale bars, 40 μm. Width of boxes in panels C, F, and I is 52 μm.
FIG. 5.
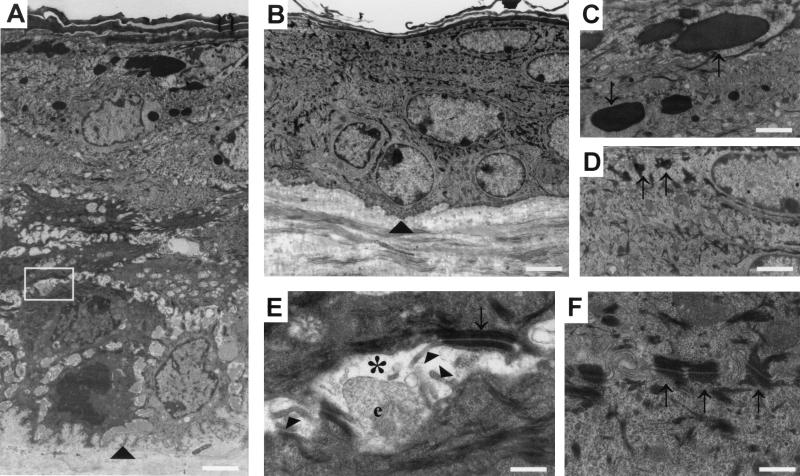
Ultrastructure of K1-hDsg3 epidermis. Back skin samples from a control mouse and a transgenic mouse of severe phenotype is shown. (A) Low magnification of transgenic mouse back skin revealing an increase in the number of cell layers and increased intercellular space in the basal and lower spinous layers. (B) Control mouse skin at the same magnification as in panel A. Arrowheads in A and B mark the position of the basement membrane. (C) Higher magnification of the transgenic mouse granular layer showing an increase in the size and number of granules (arrows) compared with control mouse granules (arrows in panel D). (E) Higher magnification of transgenic mouse skin (box in panel A) showing an intercellular space (asterisk) containing filopodial projections (arrowheads), extracellular material of unknown origin (e), and a desmosome (arrow). (F) Control mouse skin at the same magnification as used in panel E showing normal desmosomes (arrows) and lack of intercellular spaces. Scale bars in panels: A and B, 3.4 μm; C and D, 1.7 μm; E and F, 0.5 μm.
Transgenic mouse epidermis shows ultrastructural abnormalities.
To examine the skin lesions at a higher resolution, electron microscopy was conducted on several transgenic mice and wild-type littermates. Epidermal acanthosis was readily visible, with at least double the thickness and number of layers in the transgenic mouse epidermis (Fig. 5A and B). Abnormalities in the organization of the basal and spinous layers were also more obvious. Transgenic mouse basal and lower spinous cells were separated by wider intercellular spaces than the control mouse cells (Fig. 5A and B). The cells appeared to be held together by desmosomes, but between the desmosomes were large intercellular spaces containing numerous filopodia and often extracellular material of unknown origin (Fig. 5A and E). This material did not stain for mucopolysaccharide by the periodic acid-Schiff staining method (unpublished data). The desmosomes in the transgenic mouse epidermis appeared normal in structure (Fig. 5E and F). However, they were slightly larger (mean, 0.23 μm) and less frequent (0.33 desmosome per μm of membrane) in the transgenic mice than in the wild-type mice (mean, 0.20 μm; 0.37 desmosome per μm of membrane; P = 0.007). In each case, the proportion of membrane occupied by desmosomes was the same (wild type, 7.4%; transgenic, 7.6%). This may indicate some clustering of desmosomal material, possibly due to separation of the cell membranes. There were also differences in the granular layers. The granules were much more abundant and generally much larger in the transgenic mouse epidermis (Fig. 5C and E).
hDsg3 in the suprabasal layers is localized to desmosomes.
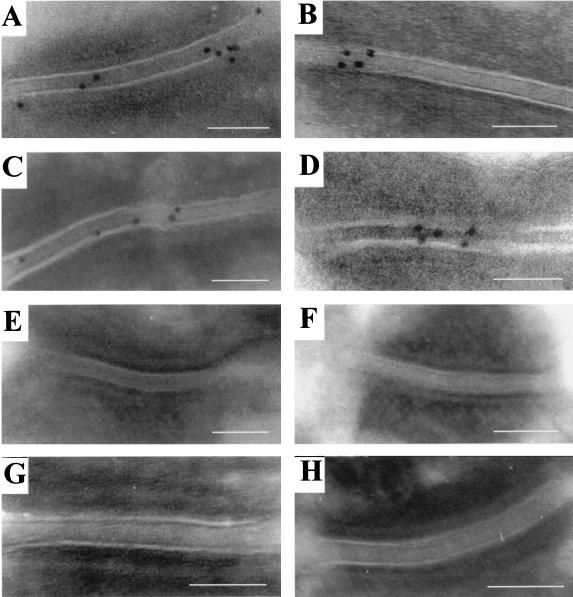
To determine the distribution of Dsg3 in the transgenic mouse epidermis and demonstrate its incorporation into desmosomes, immunogold labeling of sections from normal and transgenic mouse skin was performed with the hDsg3-specific antibody (Fig. 6). In the control mouse epidermis, desmosomes were not labeled with 10-nm gold in either the basal or suprabasal layers, confirming that the antibody does not react with endogenous Dsg3 (Fig. 6G and H). In the transgenic mouse epidermis, the basal layer was mainly negative (only about 5% of desmosomes had labeling) (Fig. 6E and F) but the suprabasal layers, corresponding to where the K1 promoter is active, showed gold labeling of about 80% of the desmosomes (Fig. 6A through D). The majority of gold particles appeared over the desmosomal intercellular space, consistent with the reactivity of the antibody with the Dsg3 extracellular domain. These results demonstrate that desmosomes in the suprabasal layers of transgenic mouse epidermis contain hDsg3.
FIG. 6.
Immuno-electron microscopy for hDsg3. Back skin from transgenic (A through F) and control (G and H) mice is shown. Panels A through D show desmosomes from the suprabasal layers of transgenic mouse epidermis with labeling for hDsg3 predominantly in the desmoglea. Panels E and F depict desmosomes showing the absence of Dsg3 from the basal layer of a transgenic mouse. Desmosomes from the basal layer (panel G) and suprabasal layer (panel H) of a control mouse with no Dsg3 labeling are shown. Scale bars, 100 nm.
Terminal differentiation is altered in the transgenic mouse epidermis.
To address whether the hyperproliferative areas of transgenic mouse epidermis have altered terminal differentiation, staining for the epidermal keratins K1, K14, K6, and K16 was performed (Fig. 7). In the control mouse and mildly affected epidermis samples, K1 was expressed by all keratinocytes except those in the basal layer (Fig. 7C). In the severely affected epidermis, K1 expression was more patchy, with all basal cells and some suprabasal cells appearing negative (Fig. 7A). Notably, regions of epidermis immediately underlying pustules had only occasional K1-positive cells (Fig. 7B). These regions stained for hDsg3, indicating that the K1 targeting vector was active. K14, a basal cell marker, was confined to the basal layers of the control mouse epidermis (Fig. 7F) but in severe regions of the transgenic mouse epidermis extended well into the suprabasal layers (Fig. 7D and E). K6 and K16 are not normally expressed in the interfollicular epidermis but are induced postmitotically in disease states associated with tissue hyperproliferation (41). In addition, staining for K6 and K16 was greatly increased in thickened regions of transgenic mouse epidermis (Fig. 7G through L).
To examine later stages of terminal differentiation, immunohistochemistry was performed with antibodies to filaggrin, loricrin, and involucrin. Filaggrin is an intermediate filament-associated protein expressed specifically in the granular cells of the epidermis. A large increase in the number of filaggrin-positive granular layers was readily apparent in the severe regions of the transgenic mouse epidermis compared with that of the control mouse or unaffected regions (Fig. 8A through C). Antibodies against loricrin and involucrin, major cornified envelope precursors, revealed an increased expression in the transgenic mouse epidermis corresponding to the increase in the number of granular layers (Fig. 8D through H).
Localization of desmosomal proteins in the epidermis of K1-hDsg3 mice.
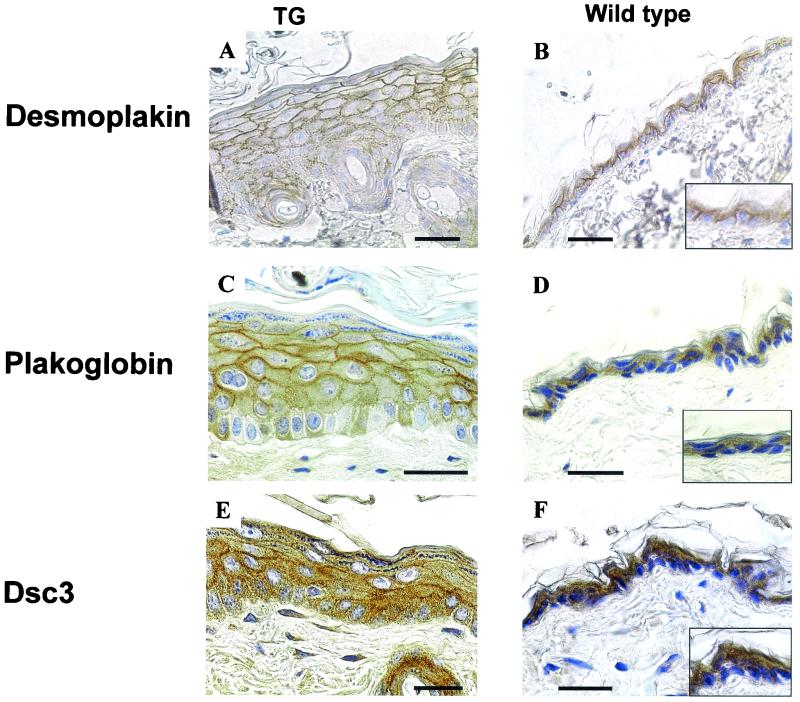
The finding of abnormal proliferation and terminal differentiation, together with widened intercellular spaces between lower epidermal keratinocytes, suggested that the organization of desmosomal components might be perturbed in K1-hDsg3 transgenic mice. To examine this possibility, immunohistochemistry with antibodies against a number of desmosomal proteins was conducted (Fig. 9). The epidermis of transgenic mice was thicker than that in the wild type, and the overall expression of desmosomal components consequently increased in the lower layers of transgenic mouse epidermis. The distributions of desmoplakin, plakoglobin, and Dsc3 appeared more punctuated than those in the suprabasal layers, possibly due to the enlarged intercellular spaces in this area (Fig. 9). Similar changes in the distributions of E-cadherin and β-catenin were also seen (unpublished data), indicating that the location of adherens junctions was also affected. Western blots carried out on the Triton X-100-soluble and -insoluble fractions from the epidermis showed the presence of Dsc3 and plakoglobin in both fractions whereas desmoplakin was limited to the insoluble fraction. There were no differences in these distributions between the wild-type and transgenic samples (unpublished data).
FIG. 9.
Expression of desmosomal proteins. Back skin of transgenic (with severe phenotype) (TG) and control mice is shown. Type of staining is indicated to the left of the panels. Scale bars, 40 μm. Width of boxes in panels: B, 40 μm; D, 55 μm; F, 55 μm.
DISCUSSION
In this study, we have misexpressed the hDSG3 gene in the suprabasal layers of the epidermis of transgenic mice by using the K1 targeting vector. The resulting mice displayed a striking phenotype of hyperproliferation and abnormal epidermal differentiation, suggesting a role for this desmosomal cadherin in the regulation of epidermal morphogenesis.
The epidermal phenotype of these mice is entirely different from that obtained by expressing mDsg3 under the control of the involucrin promoter (15). In involucrin-mDsg3 mice, the stratum corneum of the epidermis resembles the outer layer of oral mucosal epithelium and has an impaired barrier function that is lethal during the first few days of life. This is believed to be caused by an increase in the Dsg3/Dsg1 ratio from epidermal like to oral mucosal like (15, 23). The reason for the difference between the involucrin-mDsg3 and K1-hDsg3 mice is unclear. Table 1 compares the strategies used to produce these two transgenic strains. It seems unlikely that the addition of a flag tag to the C terminus of mDsg3 could result in such a difference in phenotype. Differences may have also arisen from the use of human and murine genes, but the proteins are very similar, showing a 73% amino acid identity. Of more significance in generating different phenotypes was the use of different genetic backgrounds and different promoters. Although genetic backgrounds are well known to affect the phenotypes of mice with similar genetic modifications, the precise nature of and reasons for the differences are generally unclear. K1 was expressed earlier in epidermal differentiation than involucrin (compare Fig. 7A and C with Fig. 8G and H). Thus it may be that our use of the K1 promoter gave earlier overexpression of Dsg3, providing a greater opportunity for this to affect the differentiation process, whereas use of the involucrin promoter may direct effects towards the upper epidermis, producing consequent abnormalities of the stratum corneum and interruption of barrier function.
TABLE 1.
Strategies used to generate transgenic strains involucrin-mDsg3 and K1-hDsg3
| Present study | Reference 15 |
|---|---|
| K1 promoter | Involucrin promoter |
| hDsg3 | mDsg3 |
| C57BL/6 × CBA mice | B6SLF1/J mice |
| No tag | Flag tag |
The major differences between the involucrin-mDsg3 and K1-hDsg3 mice are listed in Table 2. One striking difference is that the transgene copy number is much higher in the involucrin transgenic mice than in K1 transgenic mice. Unfortunately, it is not possible to compare directly the Dsg3/Dsg1 protein ratio in these mice because the exogenous Dsg3 in the K1 transgenic mice was human and recognized by an antibody different from that used to detect endogenous Dsg3. In the involucrin transgenic mice, the exogenous Dsg3 was produced from a murine transgene. However, the Dsg3/Dsg1 ratio in K1 transgenic mice was certainly increased by the addition of exogenous hDsg3 to an unchanged level of endogenous mDsg3. Even though the hDsg3 in our mice was clearly incorporated into desmosomes and slight spongiosis of the epidermis was found, no weakening of the epidermis could be detected by tape stripping (unpublished data), but in the involucrin transgenic mouse, alteration of the structure of the stratum corneum was accompanied by a weakening of epidermal adhesion. The involucrin-mDsg3 mouse exhibits severe loss of epidermal barrier function, demonstrated by an increase in TEWL (15). We compared the levels of TEWL of 2-day-old K1-hDsg3 mice and their wild-type littermates as described by Chidgey et al. (12). TEWL was identical in transgenic and wild-type mice, demonstrating that epidermal barrier function was not impaired in the transgenics (unpublished data). The explanation for this intriguing difference in phenotype merits further investigation.
TABLE 2.
Results for K1-hDsg3 and involucrin-mDsg3 mice
| Parameter | Result for indicated mice
|
|
|---|---|---|
| K1-hDsg3 | involucrin-mDsg3 | |
| Copy no. | 13-15 | 47-51 |
| Dsg3/Dsg1 ratio in epidermis | Increased | Increased from 0.7 to between 1.7 and 2.6 |
| Epidermis scaling by 2-3 days | No | Yes |
| TWEL | Unchanged | Increased 2.4-fold |
| Tape stripping in transgenic animals | Normal | Indicates weakened adhesion |
| Stratum corneum | Normal with localized changes | Compact |
| Epidermal hyperproliferation | Yes | No |
| Epidermal differentiation | Altered | Unchanged |
| Mouse survival | Survival and breeding | Death when a few days old |
It is of great interest that the principal features of the K1-hDsg3 mouse, acanthosis, hyperkaratosis, parakeratosis, hyperproliferation, and abnormal differentiation, are also shown in mice with the epidermally targeted expression of a number of growth factors and cytokines, including bone morphogenetic protein 6 (8), gamma interferon (10), activin A (30), transforming growth factor α (43), keratinocyte growth factor (19), and amphiregulin (13). The most striking resemblance, however, is to the phenotype of the involucrin promoter-β1 integrin transgenic mouse (9). In this mouse, β1 integrin is expressed in the suprabasal layers of the epidermis, resulting in epidermal changes that share many features with human psoriasis, including epidermal thickening and scaling, hyperproliferation, delayed differentiation, superficial pustule formation, inflammatory infiltration, and dilation of dermal blood vessels (9). This phenotype also has a late onset and variable severity and shows regression. The K1-hDsg3 mouse shows all of these abnormalities except epidermal inflammation and dermal blood vessel dilation, two diagnostic features of psoriasis. In addition, the K1-hDsg3 mouse shows hypergranulosis whereas the granular layer is substantially reduced or absent in psoriasis. Thus the K1-hDsg3 mouse does not provide a model for psoriasis but shows a phenotype more closely resembling chronic dermatitis or ichthyosis (14).
The above-mentioned cytokines, growth factors, and integrins are all known to modulate intracellular signaling pathways and thereby gene expression. Since their overexpression gives rise to phenotypic features similar to those found here, it is intriguing to speculate that Dsg3 may also regulate intracellular signaling and gene expression and to consider how this may occur. The only desmosomal component that has been implicated in the regulation of gene expression is plakoglobin, though recent evidence suggests that its role may be indirect via an effect on β-catenin expression (46). Plakoglobin modulates tumorgenicity (40) and causes axis duplication in Xenopus (24). Desmogleins bind plakoglobin (5), so it is possible that overexpression of Dsg3 exerts an effect by modulating the cytoplasmic pool of plakoglobin. It is not clear how large such an effect would need to be in order to produce a change in cell differentation: immunohistochemistry showed no striking change in plakoglobin distribution between wild-type and transgenic mice in our experiments (Fig. 9C and D). Against a role for plakoglobin in epidermal differentiation is the finding that plakoglobin-null mice had no reported differentiative epidermal phenotype (6). On the other hand, overexpression of plakoglobin in the epidermis of mice causes a phenotype affecting mainly the hair (11). It is therefore interesting that our mice showed thinning of hair and abnormal hair follicles. In this context, it has also been shown that desmoglein can bind β-catenin (7), so it is possible that overexpression of Dsg3 may affect β-catenin signaling. A third possibility is that Dsg3 overexpression may stimulate cytokine secretion by keratinocytes and thus influence signaling pathways indirectly.
Overall, data on whether desmosomal cadherins are involved in regulating epidermal differentiation are mixed. Thus, Dsg3-null mice show epidermal acantholysis but no abnormalities of epidermal differentiation (26, 27). By contrast, K14-Dsg3ΔN mutant mice, which express an NH2-terminally truncated Dsg3, exhibit large intercellular spaces, hyperproliferation, and thickening of the epidermis as well as changes in the expression of keratins and desmosomal proteins (1). Mice lacking Dsc1 also show epidermal thickening, overexpression of K6 and K16, and localized hyperkeratosis and parakeratosis (12). However, basal misexpression of Dsc1 is without effect (21). More work on the possible influence of desmosomal cadherins on intracellular signaling as well as a further comparison of the effects of different genetic backgrounds are needed in order to resolve this issue.
We note that two features of K1-hDsg3 mice, hypergranulosis and suprabasal expression of K14, have been found in other types of transgenic and null mice. Hypergranulosis was found in K14-KGF mice (19) and in K10-null and K10T mice (36). The latter phenotype appeared to be caused by defective filaggrin processing (34, 36). Suprabasal K14 expression was also found in K14-KGF mice (19) and in K10-null mice (36). In the latter, this was shown to be due to the delayed degradation of basal keratins during stratification in response to the absence of keratin filaments composed of K1 and K10. Increased expression of K16 in the epidermis also results in aberrant keratinization, acanthosis, and hyperkeratosis (42).
We conclude that overexpression of Dsg3 in the upper epidermis under the control of the K1 promoter results in abnormalities of epidermal differentiation, indicating that Dsg3 may play a role in regulating this process, in addition to its role in desmosomal adhesion.
Acknowledgments
We thank Susan Andrew, Carolyn Byrne, and Christopher Griffiths for valuable advice and Carl Mousely and Johnathan Humphries and members of the E. M. Department for technical assistance. The K1 promoter cassette was kindly provided by Dennis Roop, and Dsg3 clones and antibodies were provided by John Stanley. Antibody to K16 was kindly supplied by Birgitte Lane.
This work was supported by the Medical Research Council. Mohamed Berika was funded by the Egyptian government.
REFERENCES
- 1.Allen, E., Q.-C. Yu, and E. Fuchs. 1996. Mice expressing a mutant desmosomal cadherin exhibit abnormalities in desmosomes, proliferation, and epidermal differentiation. J. Cell Biol. 133:1367-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amagai, M., V. Klaus-Kovtun, and J. R. Stanley. 1991. Autoantibodies against a novel epithelial cadherin in pemphigus vulgaris, a disease of cell adhesion. Cell 67:869-877. [DOI] [PubMed] [Google Scholar]
- 3.Amagai, M., P. J. Koch, T. Nishikawa, and J. R. Stanley. 1996. Pemphigus vulgaris antigen (desmoglein 3) is localized in the lower epidermis, the site of blister formation in patients. J. Investig. Dermatol. 106:351-355. [DOI] [PubMed] [Google Scholar]
- 4.Arnemann, J., K. H. Sullivan, A. I. Magee, I. A. King, and R. S. Buxton. 1993. Stratification-related expression of isoforms of the desmosomal cadherins in human epidermis. J. Cell Sci. 104:741-750. [DOI] [PubMed] [Google Scholar]
- 5.Bannon, L. J., B. L. Cabrera, M. S. Stack, and K. J. Green. 2001. Isoform-specific differences in the size of desmosomal cadherin/catenin complexes. J. Investig. Dermatol. 117:1302-1306. [DOI] [PubMed] [Google Scholar]
- 6.Bierkamp, C., K. J. McLaughlin, H. Schwarz, O. Huber, and R. Kemler. 1996. Embryonic heart and skin defects in mice lacking plakoglobin. Dev. Biol. 180:780-785. [DOI] [PubMed] [Google Scholar]
- 7.Bierkamp, C., H. Schwarz, O. Huber, and R. Kemler. 1999. Desmosomal localization of beta-catenin in the skin of plakoglobin null-mutant mice. Development 126:371-381. [DOI] [PubMed] [Google Scholar]
- 8.Blessing, M., P. Schirmacher, and S. Kaiser. 1996. Overexpression of bone morphogenetic protein-6 (BMP-6) in the epidermis of transgenic mice: inhibition or stimulation of proliferation depending on the pattern of transgene expression and formation of psoriatic lesions. J. Cell Biol. 135:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll, J. M., M. R. Romero, and F. M. Watt. 1995. Suprabasal integrin expression in the epidermis of transgenic mice results in developmental defects and a phenotype resembling psoriasis. Cell 86:957-968. [DOI] [PubMed] [Google Scholar]
- 10.Carroll, J. M., T. Crompton, J. P. Seery, and F. M. Watt. 1997. Transgenic mice expressing IFN-γ in the epidermis have eczema, hair hypopigmentation, and hair loss. J. Investig. Dermatol. 108:412-422. [DOI] [PubMed] [Google Scholar]
- 11.Charpentier, E., R. M. Lavker, E. Acquista, and P. Cowin. 2000. Plakoglobin suppresses epithelial proliferation and hair growth in vivo. J. Cell Biol. 149:503-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chidgey, M., C. Brakebusch, E. Gustafsson, A. Cruchley, C. Hail, S. Kirk, A. Merritt, A. North, C. Tselepis, J. Hewitt, C. Byrne, R. Fassler, and D. Garrod. 2001. Mice lacking desmocollin 1 show epidermal fragility accompanied by barrier defects and abnormal differentiation. J. Cell Biol. 155:821-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cook, P. W., M. Piepkorn, C. H. Clegg, G. D. Plowman, J. M. DeMay, J. R. Brown, and M. R. Pittelkow. 1997. Transgenic expression of the human amphiregulin gene induces a psoriasis-like phenotype. J. Clin. Investig. 100:2286-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elder, D. 1997. Levers's histopathology of the skin, 8th ed. Lippincott-Raven, Philadelphia, Pa.
- 15.Elias, P. M., N. Matsuyoshi, H. Wu, C. Lin, Z. H. Wang, B. E. Brown, and J. R. Stanley. 2001. Desmoglein isoform distribution affects stratum corneum structure and function. J. Cell Biol. 153:243-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fey, E. G., K. M. Wan, and S. Penman. 1984. Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: three-dimensional organization and protein composition. J. Cell Biol. 98:1973-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdes, J., H. Lemke, H. Baisch, H. Wacker, U. Schwab, and H. Stein. 1984. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 133:1710-1715. [PubMed] [Google Scholar]
- 18.Greenhalgh, D. A., J. A. Rothnagel, X. Wang, M. I. Quintanilla, C. C. Orengo, T. A. Gagne, D. S. Bundman, M. A. Longley, C. Fisher, and D. R. Roop. 1993. Hyperplasia, hyperkeratosis and benign tumor production in transgenic mice by a targeted v-fos oncogene suggest a role for fos in epidermal differentiation and neoplasia. Oncogene 8:2145-2157. [PubMed] [Google Scholar]
- 19.Guo, L., Q. C. Yu, and E. Fuchs. 1993. Targeting expression of keratinocyte growth factor to keratinocytes elicits striking changes in epithelial differentiation in transgenic mice. EMBO J. 12:973-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardman, M. J., P. Sisi, D. N. Banbury, and C. Byrne. 1998. Patterned acquisition of skin barrier function during development. Development 125:1541-1552. [DOI] [PubMed] [Google Scholar]
- 21.Henkler, F., M. Strom, K. Mathers, H. Cordingley, K. Sullivan, and I. King. 2001. Transgenic misexpression of the differentiation-specific desmocollin isoform 1 in basal keratinocytes. J. Investig. Dermatol. 116:144-149. [DOI] [PubMed] [Google Scholar]
- 22.Hogan, B., R. Beddington, F. Constantini, and E. Lacy (ed.). 1994. Manipulating the mouse embryo, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Ishii, K., and K. J. Green. 2001. Cadherin function: breaking the barrier. Curr. Biol. 11:R569-R572. [DOI] [PubMed] [Google Scholar]
- 24.Karnovsky, A., and M. Klymkowsky. 1995. Anterior axis duplication in Xenopus induced by the over-expression of the cadherin-binding protein plakoglobin. Proc. Natl. Acad. Sci. USA 92:4522-4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King, I. A., K. H. Sullivan, R. Bennet, and R. S. Buxton. 1995. The desmocollins of human foreskin epidermis: identification of a third gene and expression pattern of the three isoforms. J. Investig. Dermatol. 105:314-321. [DOI] [PubMed] [Google Scholar]
- 26.Koch, P. J., M. G. Mahoney, G. Cotsarelis, K. Rothenberger, R. M. Lavker, and J. R. Stanley. 1998. Desmoglein 3 anchors telogen hair in the follicle. J. Cell Sci. 111:2529-2537. [DOI] [PubMed] [Google Scholar]
- 27.Koch, P. J., M. G. Mahoney, H. Ishikawa, L. Pulkkinen, J. Uitto, L. Shultz, G. F. Murphy, D. Whitaker-Menezes, and J. R. Stanley. 1997. Targeted disruption of the pemphigus vulgaris antigen (desmoglein 3) gene in mice causes loss of keratinocyte cell adhesion with a phenotype similar to pemphigus vulgaris. J. Cell Biol. 137:1091-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legan, P. K., K. K. M. Yue, M. A. J. Chidgey, J. L. Holton, R. W. Wilkinson, and D. R. Garrod. 1994. The bovine desmocollin family: a new gene and expression patterns reflecting epithelial cell proliferation and differentiation. J. Cell Biol. 126:507-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahoney, M. G., Z. H. Wang, and J. R. Stanley. 1999. Pemphigus vulgaris and pemphigus foliaceus antibodies are pathogenic in plasminogen activator knockout mice. J. Investig. Dermatol. 113:22-25. [DOI] [PubMed] [Google Scholar]
- 30.Muntz, A., H. Simola, F. Engelhardt, K. Bleuel, M. Brauchle, I. Lein, L. W. Evans, D. Huylebroek, R. Balling, and S. Werner. 1999. Overexpression of activin A in the skin of transgenic mice reveals new activities of activin in epidermal morphogenesis, dermal fibrosis and wound repair. EMBO J. 18:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.North, A. J., M. A. J. Chidgey, J. P. Clarke, W. G. Bardsley, and D. R. Garrod. 1996. Distinct desmocollin isoforms occur in the same desmosomes and show reciprocally graded distributions in bovine nasal epidermis. Proc. Natl. Acad. Sci. USA 93:7701-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.North, A. J., W. G. Bardsley, J. Hyam, E. A. Bornslaeger, H. C. Cordingley, B. Trinnaman, M. Hatzfeld, K. J. Green, A. I. Magee, and D. R. Garrod. 1999. Molecular map of the desmosomal plaque. J. Cell Sci. 112:4325-4336. [DOI] [PubMed] [Google Scholar]
- 33.Parrish, E. P., P. V. Steart, D. R. Garrod, and R. O. Weller. 1987. Antidesmosomal monoclonal antibody in the diagnosis of intracranial tumours. J. Pathol. 153:265-273. [DOI] [PubMed] [Google Scholar]
- 34.Porter, R. M., A. M. Hutcheson, E. L. Rugg, R. A. Quinlan, and E. B. Lane. 1998. cDNA cloning, expression, and assembly characteristics of mouse keratin 16. J. Biol. Chem. 273:32265-32272. [DOI] [PubMed] [Google Scholar]
- 35.Potten, C. S. 1981. Cell proliferation in epidermis (keratopoiesis) via discrete units of proliferation. Int. Rev. Cytol. 69:271-318. [DOI] [PubMed] [Google Scholar]
- 36.Reichelt, J., H. Bussow, C. Grund, and T. M. Margin. 2001. Formation of a normal epidermis supported by increased stability of keratins 5 and 14 in keratin 10 null mice. Mol. Biol. Cell 12:1557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothnagel, J. A., M. A. Longely, D. Bundman, D. A. Greenhalgh, A. M. Dominey, and D. R. Roop. 1990. Targeting gene expression to the epidermis of transgenic mice: potential applications to genetic skin disorders. J. Investig. Dermatol. 95:59S-61S. [DOI] [PubMed] [Google Scholar]
- 38.Runswick, S. K., M. J. O'Hare, L. Jones, C. H. Streuli, and D. R. Garrod. 2001. Desmosomal adhesion regulates epithelial morphogenesis and cell positioning. Nat. Cell Biol. 3:823-830. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu, H., T. Masunaga, A. Ishiko, A. Kikuchi, T. Hashimoto, and T. Nishikawa. 1995. Pemphigus vulgaris and pemphigus foliaceus sera show an inversely graded binding pattern to extracellular regions of desmosomes in different layers of human epidermis. J. Investig. Dermatol. 105:153-159. [DOI] [PubMed] [Google Scholar]
- 40.Simcha, I., B. Geiger, S. Yehuda-Levenberg, D. Salomon, and A. Ben-Ze'ev. 1996. Suppression of tumorigenicity by plakoglobin: an augmenting effect of N-cadherin. J. Cell Biol. 133:199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoler, A., R. Kopan, M. Duvic, and E. Fuchs. 1988. Use of monospecific antisera and cRNA probes to localise the major changes in keratin expression during normal and abnormal epidermal differentiation. J. Cell Biol. 107:427-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takahashi, K., J. Folmer, and P. A. Coulombe. 1994. Increased expression of keratin 16 causes abnormalities in cytoarchitecture and keratinization in transgenic mouse skin. J. Cell Biol. 127:505-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vassar, R., and E. Fuchs. 1991. Transgenic mice provide new insights into the role of TGF-α during epidermal development and differentiation. Genes Dev. 5:714-727. [DOI] [PubMed] [Google Scholar]
- 44.Yue, K. K. M., J. L. Holton, J. P. Clarke, J. K. L. Hyam, T. Hashimoto, M. A. J. Chidgey, and D. R. Garrod. 1995. Characterisation of a desmocollin isoform (bovine DSC3) exclusively expressed in lower layers of stratified epithelia. J. Cell Sci. 108:2163-2173. [DOI] [PubMed] [Google Scholar]
- 45.Zhai, W. 1997. Ph.D. thesis. University of Manchester, Manchester, United Kingdom.
- 46.Zhurinsky, J., M. Shtutman, and A. Ben-Ze'ev. 2000. Differential mechanisms of LEF/TCF family-dependent transcriptional activation by beta-catenin and plakoglobin. Mol. Cell. Biol. 20:4238-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]