Abstract
Dickkopfs (Dkks) are secreted developmental regulators composed of two cysteine-rich domains. We report that the effects of Dkks depend on molecular context. Although Wnt8 signaling is inhibited by both Dkk1 and Dkk2 in Xenopus embryos, the same pathway is activated upon interaction of Dkk2 with the Wnt coreceptor LRP6. Analysis of individual Dkk domains and chimeric Dkks shows that the carboxy-terminal domains of both Dkks associate with LRP6 and are necessary and sufficient for Wnt8 inhibition, whereas the amino-terminal domain of Dkk1 plays an inhibitory role in Dkk-LRP interactions. Our study illustrates how an inhibitor of a pathway may be converted into an activator and is the first study to suggest a molecular mechanism for how a ligand other than Wnt can positively regulate β-catenin signaling.
Determination of cell fates in the developing embryo depends on inductive interactions between neighboring tissues. In many cases, these interactions involve secreted proteins of the Wnt family (6, 35). Wnt proteins signal through multiple molecular pathways to control cell fate, polarity, and proliferation in all metazoan embryos, and the regulation of these pathways has been a subject of many studies. Downstream components of the canonical Wnt signaling pathway are involved in specification of the dorsoventral body axis in vertebrates (17). Wnt ligands are thought to bind to and signal through members of the Frizzled family of seven transmembrane domain receptors and the low-density lipoprotein receptor-related proteins LRP5 and LRP6, which function as Wnt coreceptors (3, 28, 37, 46, 47). This signaling is modulated by heparan sulfate proteoglycans (36). Additionally, Wnt signaling is regulated by several extracellular antagonists, including Frizzled-related proteins (38, 49, 50), Cerberus (4), and WIF-1 (19). These inhibitors bind Wnt ligands and may restrict their signaling within a tissue. Another class of Wnt antagonists with a novel mode of action, the Dickkopfs (Dkks), have been described (12, 14, 24). Recent work suggests that Dkks interact with the Wnt coreceptors LRP5 and LRP6 (2, 27, 41) and that Dkk1 inhibits Wnt signaling by disrupting the binding of LRP6 to the Wnt/Frizzled ligand-receptor complex (41).
Dkks are composed of two cysteine-rich domains separated by a variable-length spacer region. Both domains are well conserved among all four members of the Dickkopf family (14, 24). In particular, Dkk1 and Dkk2 share 50% identity in their N-terminal cysteine-rich region and 70% identity in their C-terminal regions. The structure of the C-terminal domain has a weak similarity to colipases (1); however, the functional significance of this observation is unclear. Dkk family members are expressed throughout development in a tissue- and stage-restricted manner. Their transcripts are found in the brain, heart, lungs, limbs, and other tissues in which epithelial-mesenchymal interactions occur (16, 24, 31), suggesting that these proteins modulate a number of important developmental processes.
Dkk1, the most extensively studied Dickkopf family member, is a potent Wnt antagonist (14, 24). In vertebrate embryos, Dkk1 is involved in head development (14, 18, 23, 32, 42), a process that has been postulated to involve inhibition of Wnt signaling (13, 21). Xenopus embryos overexpressing Dkk1 develop enlarged heads and shortened tails, whereas injections of anti-Dkk1 antibodies lead to microcephaly (14). In mice lacking the Dkk1 gene, anterior neural structures are missing, including the telencephalon, diencephalon, and part of the midbrain (32). Dkk1−/− mice also lack most head structures anterior of the otic vesicle, including eyes, olfactory placodes, frontonasal mass, and the mandibular processes (32). Dkk1 has also been shown to induce heart tissue in Xenopus embryos (29, 40) and has recently been implicated in regulation of cell proliferation and programmed cell death in the interdigital spaces (32).
Other Dkks have been studied less extensively. Dkk4 is also a Wnt antagonist, whereas Dkk3 does not appear to modulate Wnt signaling (24). Dkk2 has been reported to cooperate with Frizzled receptors in promoting body axis development (48). The molecular mechanisms underlying the opposite properties of Dkk1 and Dkk2 are not known. To investigate the molecular basis for functional differences between Dkk1 and Dkk2, we examined how the individual domains of these Dkks contribute to their functions. In our study, we assessed the effects of each cysteine-rich region of Dkk1 and Dkk2 on Wnt signaling in Xenopus embryos, both morphologically and with a reporter assay. Our results demonstrate that the individual domains of Dkk1 and Dkk2 possess distinct functional activities. We show that the C-terminal domains of Dkks are both necessary and sufficient for Wnt inhibition. Moreover, both C-terminal domains associate with LRP6 and stimulate LRP6-dependent embryonic axis induction. In contrast, the N-terminal domains appear to play a regulatory role in these interactions.
MATERIALS AND METHODS
DNA constructs.
pCS2-Dkk1-Flag, pCS2-Dkk2-Flag, and pCS2-Dkk3-Flag have been previously described (24). Individual Dkk domain constructs, except for N2 and N2C1, were generated by fusing the signal peptide of Dkk1 to the N-terminal (N1 and N1C2) or C-terminal (C1, C2, and C3) cysteine-rich domains of Dkk1, Dkk2, and Dkk3, respectively. N2 and N2C1 contain the Dkk2 signal peptide fused to the N-terminal cysteine-rich region of Dkk2. The N1 construct was amplified from pCS2-Dkk1 by PCR with the SP6 primer (Promega) and 5′-CCGCTCGAGCTAAGCGTAATCTGGAACATCGTATGGATACCCATCCAAGGTGCT-3′, encoding a hemagglutinin (HA) tag. The PCR product was subcloned into pCS2 by using EcoRI and XhoI sites. This construct was used in all studies, except for the analysis of protein expression levels, for which a Flag-tagged N1 construct was utilized. pCS2-N1-Flag was synthesized with Pfu 1 polymerase (Stratagene), with pCS2-Dkk1 as a template and the primer 5′-CCATCACTGAAAGCTTTGAATTCGACTACAAGGACGACGA-3′, as described previously (26).
The C1 construct was generated by ligating together EcoRI-Asp718-digested pCS2, the HindIII-Asp718-digested C-terminal half of Dkk1, and an EcoRI-HindIII-digested DNA fragment derived from PCR of pCS2-Dkk1 with the SP6 primer and the oligonucleotide 5′-TCCAAGCTTACTGCAGAGCCTGG-3′. The N2 construct was made by PCR with pCS2-Dkk2 as a template, the SP6 primer, and the oligonucleotide 5′-GATGGTACTCGGCACAGAAGCTTGCG-3′. The PCR product was digested with HindIII and subcloned into pCS2-N1 digested with HindIII to remove the N1 fragment. C2 was constructed by PCR amplification of the C-terminal half of Dkk2 from pCS2-Dkk2 with the T3 primer (Stratagene) and the oligonucleotide 5′-CGCAAGCTTAAACCACGGTCATTAC-3′. The PCR fragment digested with HindIII and Asp718 was subcloned into pCS2-C1 digested with HindIII and Asp718 to remove C1. C3 was constructed by PCR of the C-terminal half of Dkk3 by using the T3 primer and the oligonucleotide 5′-CGCAAGCTTGGCCACCAGGGGCAGCA-3′. This fragment was digested with HindIII and XbaI and cloned into pCS2-C1 digested with HindIII and XbaI to remove the C1 fragment. pCS2-N1C2 was constructed by PCR of the C-terminal half of Dkk2 with the T3 primer and the primer used for construction of the C2 construct (described above). This fragment was digested with HindIII and Asp718 and ligated to pCS2-Dkk1 cut with HindIII and EcoRI and Asp718-EcoRI-digested pCS2. pCS2-N2C1 was constructed by PCR of the N-terminal half of Dkk2 with the SP6 primer and the primer used for construction of the N2 construct (described above). This fragment was digested with HindIII and EcoRI and ligated to pCS2-Dkk1 cut with Asp718 and HindIII and Asp718-EcoRI-digested pCS2.
The pCS2-Dkk1-, Dkk2-, C1-, and C2-GFP (green fluorescent protein) constructs were generated by PCR of each Dkk with pCS2-Dkk-flag constructs as template, with the primer 5′-GGATCCTTGTCGTCGTGGCC-3′, which contains a BamHI site, and the SP6 primer. These products were subcloned into pEGFP-1 (Clontech) by using BamHI and EcoRI sites. The constructs were then digested with NotI (blunted) and EcoRI and then subcloned into the pCS2 vector. pCS2-N2-GFP was constructed with pCS2-Dkk2-GFP as a template and the primer 5′-GGATGGTACTCGGCAC CTCGAGGACTACAAGGACGACG-3′ as described previously (26). All constructs were verified by DNA sequencing. The pSia-Luc, pCS2-LRP6, pcDNA3.1-LRP6N-IgG (immunoglobulin G), pRK5-IgG, pSP64T-XWnt8, and pSP64T-tBMPR (tBR) plasmids have been described previously (8, 10, 15, 20, 41, 46).
mRNA microinjections and embryo analysis.
For RNA synthesis, DNA templates were linearized with the following restriction enzymes: pSP64T-XWnt8 with EcoRI; pCS2-LRP6 with Asp718; pSP64T-tBMPR with EcoRI; pCS2-Dkk1 with NotI; pCS2-NI and C1 with Asp718; and pCS2-N2, C2, Dkk2, N1C2, and N2C1 with KpnI. Capped synthetic mRNAs were generated by in vitro transcription with SP6 polymerase, by using the mMessage mMachine kit (Ambion) as described by the manufacturer. RNA was quantified by measuring the optical density at 260 nm and by comparing it to that in a gel with a standard of known concentration.
Embryos were obtained from Xenopus females and cultured in 0.1× Marc's modified Ringer's medium (MMR) as described previously (33). Embryonic stages were determined according to the work of Nieuwkoop and Faber (34). For microinjections, four-cell embryos were transferred to 3% Ficoll 400 (Pharmacia) in 0.5× MMR and injected with 10 nl of the specified amount of RNA. Embryos were injected into a single ventro-vegetal blastomere unless otherwise specified. Embryos were cultured until stages 36 to 38 and then fixed with MEMFA (0.1 M morpholinepropanesulfonic acid [MOPS], 2 mM EGTA [pH 8.0], 1 mM MgSO4, 3.7% formaldehyde) for scoring morphological changes and imaging. All experiments were reproduced at least three times. Representative embryos are shown.
Luciferase activity measurements.
Embryos were injected twice ventro-animally with 20 pg of pSia-Luc (10) DNA, along with the indicated amounts of mRNA. At stage 10, sets of five embryos were lysed in 100 μl of 50 mM Tris (pH 7.4) and spun at 12,000 × g for 5 min. Supernatants were assayed for luciferase activity as previously described (11).
Cell culture, transfections, fluorescent microscopy, and IgG pull-down assays.
HEK293T cells were cultured in 1× Dulbecco's modified Eagle's medium (Gibco/Invitrogen) supplemented with 10% fetal calf serum (Gibco/Invitrogen) and 5 μg of gentamicin per ml (Sigma). Cells were transiently transfected by the calcium phosphate method, as described previously (7). For experiments with GFP-tagged Dkks, cell culture medium from cells transiently transfected with pCS2-Dkk-GFP constructs was collected 48 h after transfection and was added to coverslips seeded with HEK293T cells transiently transfected with pCS2 or pCS2-LRP6 for 1 h at 37°C. For competition experiments, media from HEK293T cells transfected with pCS2-C1, pCS2-C2, or pCS2-N1 were added to LRP6-expressing HEK293T cells for 30 min at 37°C. Next, half the original volume of medium was added for another 30 min from cells transfected with pCS2-Dkk-GFP constructs. Coverslips were then washed twice with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde, washed twice with PBS, and visualized by fluorescence microscopy.
For IgG pull-down assays, HEK293T cells were transfected with pCS2-Dkk constructs, pcDNA3.1-LRP6N-IgG, or pRK5-IgG. Forty-eight hours after transfection, cell culture media were collected, and LRP6N-IgG or IgG supernatants were mixed with Dkk-containing media in equal volumes. Protein A-Sepharose CL-4B beads (Pharmacia) were added for 2 h at room temperature. The beads were washed four times with PBS and incubated at 85°C with sample buffer (500 mM Tris-HCl [pH 6.8], 10% sodium dodecyl sulfate [SDS], 20% glycerol, 0.05% bromophenol blue, 1% β-mercaptoethanol) for 8 min. Proteins were subjected to Western analysis as described below.
Western analysis.
Six stage 9 embryos, which had been injected in all four blastomeres at the four-cell stage with 5 ng of Dkk mRNAs, were lysed in 300 μl of lysis buffer (1% Triton X-100, 50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 0.1 mM phenylmethylsulfonylfluoride, 10 mM NaF, 1 mM Na3VO4). After spinning at 16,000 × g for 5 min, samples were heated at 85°C in sample buffer for 8 min and electrophoresed on SDS-12% polyacrylamide gels. The equivalent of 0.25 embryo was loaded per lane. Proteins were transferred to Immobilon-P membranes (Millipore) and probed with M2 anti-Flag antibodies (Sigma), 12CA5 anti-HA tag antibodies (Babco), or anti-human IgG (Fc) antibodies (Jackson Immunoresearch) as previously described (22).
RESULTS
Different biological properties of Dkk1 and Dkk2.
To study the molecular mechanisms underlying the action of Dkks, we assessed the development of Xenopus embryos microinjected with human Dkk1 and Dkk2 RNAs. As previously reported, Dkk1 enhanced anterior development, as evidenced by enlarged cement glands and shortened tails, whereas Dkk2 induced rudimentary secondary axes with ectopic cement glands (Fig. 1A) (14, 48; data not shown). We did not observe complete secondary axes in embryos injected with Dkk2 RNA, as reported by Wu et al. (48), although the reason for this difference is unclear. Thus, despite a high degree of structural similarity, Dkk1 and Dkk2 exhibit distinct biological properties.
FIG. 1.
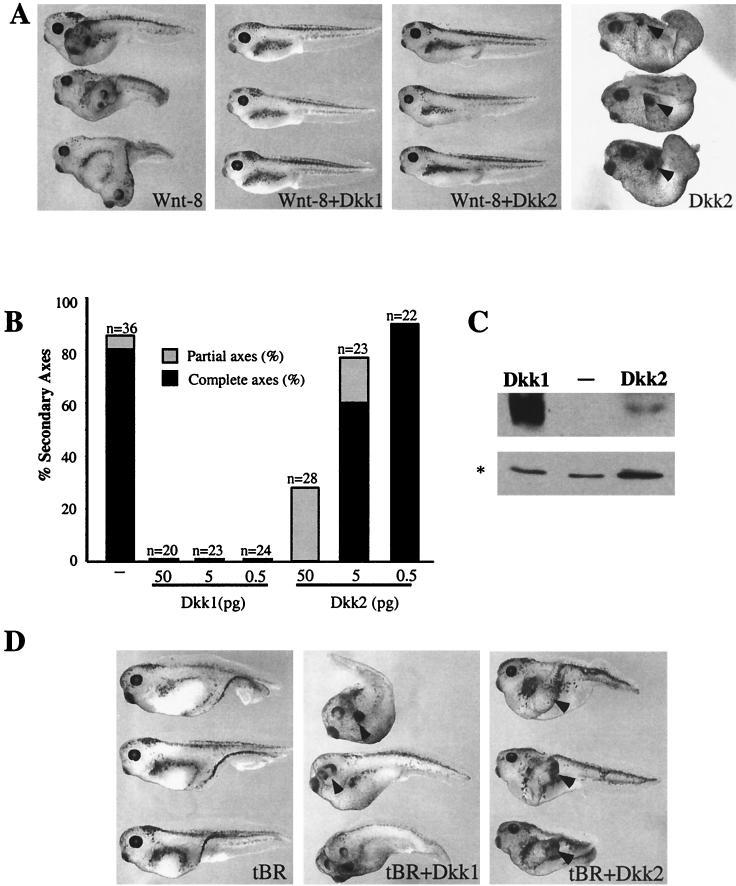
Dkk1 and Dkk2 inhibit Wnt8-dependent secondary axes and synergize with tBR in head induction. (A) Both Dkk1 and Dkk2 inhibit Wnt8-induced secondary axes. Embryos at the four- to eight-cell stage were injected into one ventro-vegetal blastomere with Wnt8 mRNA (1 pg) and Dkk1 (2.5 pg) or Dkk2 (50 pg) mRNAs. The dose of Dkk2 mRNA is higher than that of Dkk1 RNA to compensate for lower protein expression. (B) Dose-dependent inhibition of Wnt8 by Dkk1 and Dkk2. Embryos were injected as described above with Wnt8 mRNA (1 pg) and Dkk1 or Dkk2 mRNAs at the indicated doses. Results are representative of three separate experiments. Complete axes are defined by the presence of a second head with eyes; partial axes are composed of a secondary trunk without anterior head structures. (C) Expression levels of Flag-tagged Dkk1 and Dkk2 in Xenopus embryos. Each blastomere of four-cell embryos was injected with 5 ng of the indicated Dkk RNAs. Embryo lysates prepared at stage 9 were separated in SDS-12% polyacrylamide gels, transferred to Immobilon P membrane, and probed with anti-Flag M2 antibodies. A nonspecific protein band marked by an asterisk reflects loading. (D) Both Dkk1 and Dkk2 synergize with tBR to induce head structures. Embryos were injected with tBR mRNA (20 pg) and Dkk1 (2.5 pg) or Dkk2 (20 pg) mRNA, as indicated. Arrowheads indicate secondary head structures.
To determine whether these distinct phenotypes induced by Dkk1 and Dkk2 reflect their different abilities to regulate Wnt signaling, we compared Dkk1 and Dkk2 in several functional assays. One morphological assay is based on the finding that expression of some Wnt RNAs in ventral-vegetal blastomeres results in induction of a secondary body axis composed of a second trunk and head (43). This activity is likely to mimic the endogenous pathway leading to primary axis specification (30, 43, 44). While 85% of embryos injected with Wnt8 RNA contained duplicated axes, all embryos injected with Wnt8 and Dkk1 RNAs developed a single axis at a range of Dkk1 RNA concentrations (Fig. 1A and B). Dkk2 RNA (50 pg) suppressed secondary axis induction, but at lower doses (0.5 to 5 pg) had little or no inhibitory activity (Fig. 1A and B). Western analysis showed that Dkk2 is expressed in injected embryos at much lower levels than Dkk1 (Fig. 1C and 3C). It is likely that the Wnt-inhibitory activity of Dkk2 has been previously unnoticed due to low levels of protein expression (24, 48). Both Dkks also prevented Wnt-induced activation of Siamois, a direct transcriptional target of Wnts (Fig. 2D) (5, 10; data not shown).
FIG. 3.
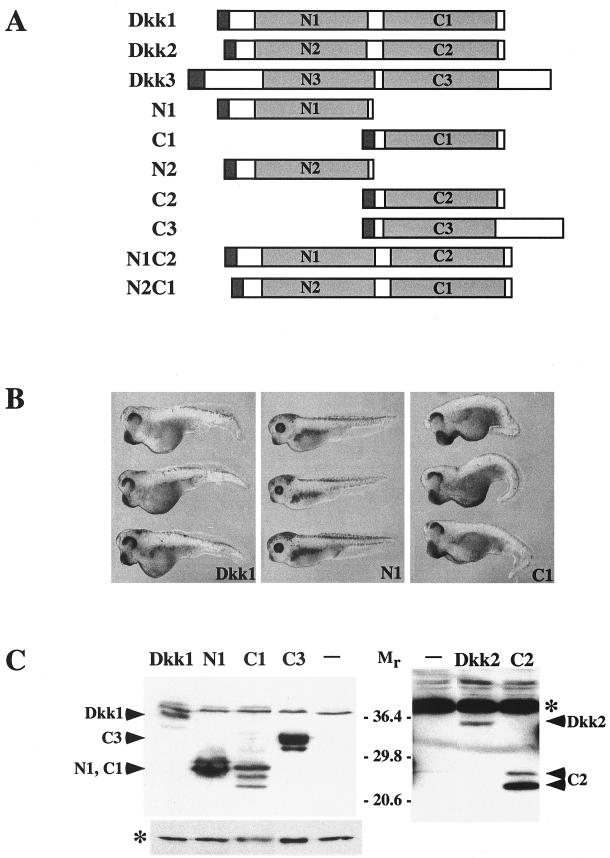
Structure, protein levels, and overexpression phenotypes of Dickkopf constructs. (A) Dkk constructs used in this study. (B) Overexpression phenotypes of Dkk1 constructs. Two dorsal-marginal blastomeres were injected with 50 pg of mRNA encoding Dkk1, N1, or C1, as indicated. Embryos injected with C1 developed enlarged heads and cement glands, impaired eyes, and shortened trunks. Embryos injected with N1, N2, and C2 mRNAs are not significantly different from uninjected controls. (C) Expression levels of Flag-tagged Dkk constructs in Xenopus embryos. Each blastomere of four-cell embryos was injected with 5 ng of the indicated Dkk RNAs. Embryo lysates prepared at stage 9 were separated in SDS-12% polyacrylamide gels, transferred to Immobilon P membrane, and probed with anti-Flag antibodies. The membrane on the right was exposed for 10 times longer than the one on the left to detect lower levels of Dkk2 and C2. A nonspecific protein band marked by an asterisk reflects equal loading.
FIG. 2.
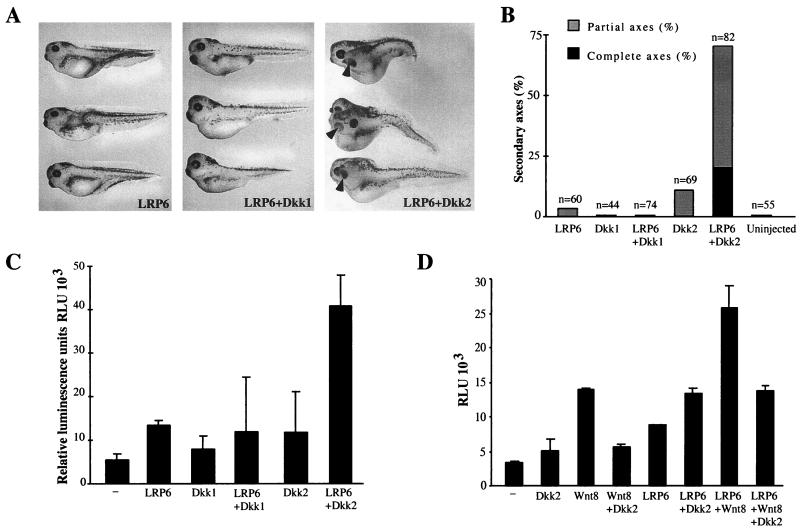
Dkk2, but not Dkk1, synergizes with LRP6 to activate β-catenin signaling pathways. (A and B) Embryos were injected ventro-vegetally with LRP6 mRNA (3 ng) and Dkk1 (5 pg) or Dkk2 (50 pg) mRNAs, as shown. (A) Morphology of injected embryos. (B) Combined results of three separate experiments. (C) Activation of the Siamois promoter by coexpression of LRP6 and Dkk1 or Dkk2. Embryos were injected into two ventral-animal blastomeres with pSia-Luc plasmid (20 pg) and with the following RNAs: LRP6 (5 ng), Dkk1 (20 pg), or Dkk2 (50 pg), as indicated. −, no RNA was coinjected with the pSia-Luc plasmid. (D) Dkk2 inhibits Wnt8, but synergizes with LRP6. Embryos were injected with pSia-Luc plasmid (20 pg), Wnt8 (1 pg), LRP6 (2 ng), and Dkk2 (50 pg), as indicated. (C and D) Luciferase activity was measured in embryonic lysates at stages 10 to 10+. Bars depict the means ± standard errors of triplicate samples, containing five embryos each. Each experiment was performed at least three times; a representative experiment is shown.
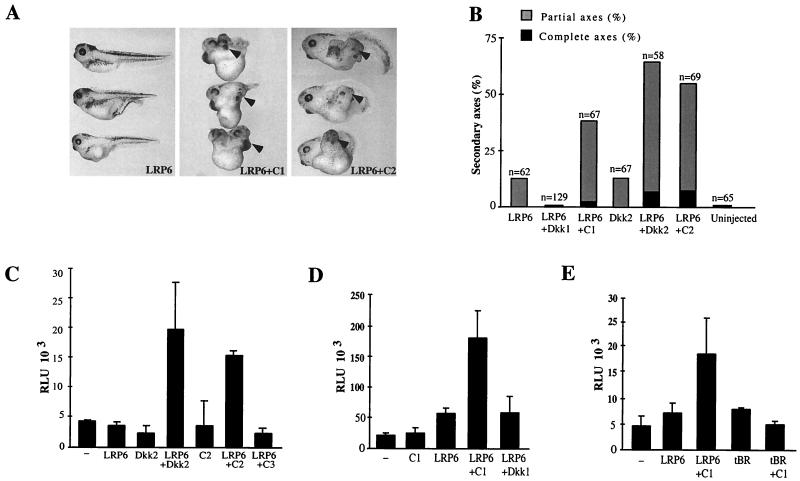
Another assay for Dkk activity is based on the ability of Wnt ligands to suppress head development during gastrulation, after the initial axis specification (9). Consisent with this activity, antagonists of Wnt and BMP (bone morphogenic protein) are known to promote head formation (13). When coexpressed ventrally with the dominant-negative BMP4 receptor (tBR) (15, 45), Dkk1 potentiates head development (14). Whereas ventral injection of tBR RNA resulted in embryos developing partial secondary axes (n = 71, Fig. 1D), both Dkk1 and Dkk2 synergized with tBR to induce complete head-containing axes (72%, n = 59, for Dkk1; and 39%, n = 72, for Dkk2; Fig. 1D). Thus, although Dkk1 and Dkk2 generate different developmental abnormalities, both Dkks are potent Wnt8 inhibitors.
A strikingly different outcome was observed when Dkk1 and Dkk2 were coexpressed with the Wnt coreceptor LRP6, which has been reported to induce partial secondary axes (46). Whereas Dkk1 did not significantly affect LRP6 signaling, Dkk2 strongly cooperated with human LRP6 in induction of secondary axes (Fig. 2A and B) and in activation of the Siamois promoter (Fig. 2C). Furthermore, although Dkk2 prevented activation of the Siamois promoter by Wnt8, in embryos injected with LRP6 and Wnt8, Siamois promoter activity remained elevated, even in the presence of Dkk2 (Fig. 2D). These experiments demonstrate that Dkk2 stimulates LRP-mediated signaling and can act as either an inhibitor or an activator of the Wnt pathway, depending on cellular context.
The C-terminal domains of Dkk1 and Dkk2 are necessary and sufficient for Wnt inhibition.
To identify specific protein regions responsible for the different signaling properties of Dkk1 and Dkk2, we analyzed constructs containing either the amino-terminal or the carboxy-terminal cysteine-rich domains of Dkk1 or Dkk2 (N1 and C1 and N2 and C2, respectively) (Fig. 3A). Embryos expressing the N1 domain had no visible abnormalities and were indistinguishable from uninjected controls (Fig. 3B). In contrast, C1-expressing embryos displayed a phenotype similar to that caused by Dkk1 (14) (Fig. 3B). The difference in embryonic phenotypes was not due to differences in protein levels, since both the C1 and N1 domains were efficiently expressed in injected embryos (Fig. 3C). These observations suggest that the ability of Dkk1 to promote head development and block Wnt signaling is mediated by the C1 domain.
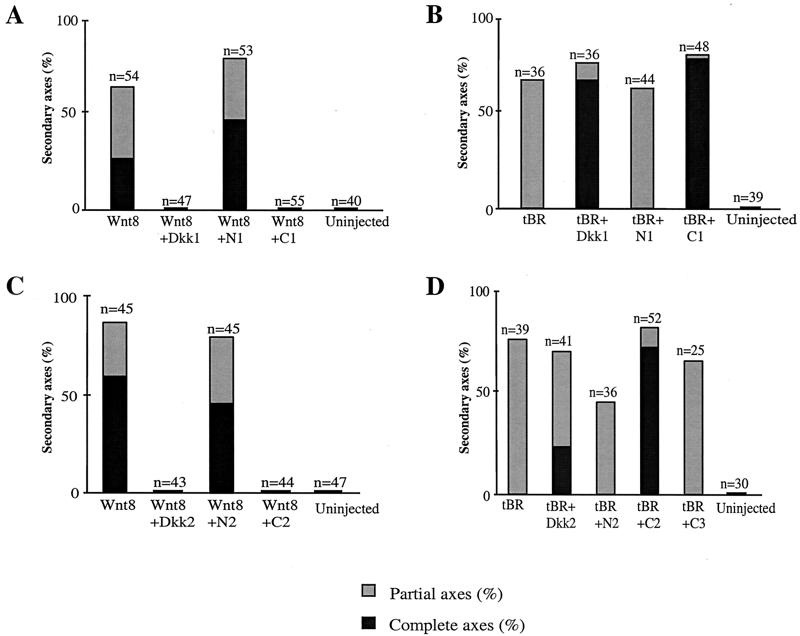
Consistent with the above hypothesis, the C1 construct completely blocked Wnt8-dependent secondary axis induction, while the N1 construct had no significant effect at the same dose of injected RNA (Fig. 4A). Similarly, the C1, but not the N1, domain suppressed Wnt8-mediated activation of the Siamois promoter (data not shown). As a control, we expressed the C-terminal domain of Dkk3 (C3) and found that it did not interfere with Wnt8-mediated secondary axis induction or activation of the Siamois promoter (data not shown). We also evaluated the ability of C1 and N1 to synergize with tBR, since it was possible that distinct domains mediate different Dkk1 activities. C1, but not N1, efficiently cooperated with tBR in head induction, because embryos expressing C1 and tBR developed complete secondary axes with head structures, while only partial axes were induced by N1 and tBR or tBR alone (Fig. 4B). These experiments indicate that the C1 domain of Dkk1 is both necessary and sufficient for repression of Wnt8 signaling by Dkk1 and head induction by a combination of Dkk1 and tBR.
FIG. 4.
The C-terminal domains of Dkks inhibit Wnt8-dependent secondary axes and synergize with tBR in head induction. (A and C) C1 and C2, but not N1 or N2, inhibit secondary axis formation by Wnt8. Embryos were injected into one ventral-vegetal blastomere with Wnt8 mRNA (1 pg) and 2.5 pg of Dkk1, N1, or C1 mRNA (A) or 20 pg of Dkk2, N2, or C2 mRNA (C). Combined results of three separate experiments are shown. (B and D) C1 and C2, but not N1 or N2, induce head structures when expressed with tBR. Embryos were injected with tBR mRNA (20 pg) and 2.5 pg of Dkk1, N1, or C1 mRNA (B) or 20 pg of Dkk2, N2, or C2 mRNA. (D) Combined results of three separate experiments are shown for each graph.
We next examined the properties of the corresponding Dkk2 domains by using the same functional assays. Neither N2 nor C2 constructs induced morphological abnormalities in injected embryos (data not shown). Nevertheless, C2 but not N2 efficiently inhibited Wnt8 activity (Fig. 4C) and promoted head development when coinjected with tBR (Fig. 4D). Dkk2 showed a weaker activity in the head induction assay than C2, perhaps due to lower expression levels (Fig. 3C). Together, our results indicate that the C-terminal domains of both Dkks are responsible for Wnt-inhibitory properties of Dkks.
Synergy of C1 and C2 with LRP6.
A key difference in the activities of Dkk1 and Dkk2 is revealed by their interactions with LRP6 (Fig. 2). To determine whether the functional differences observed for the wild-type Dkk proteins could be explained by properties of individual protein domains, we analyzed the ability of the N- and C-terminal regions of Dkk1 and Dkk2 to modulate LRP6 signaling. Neither N1 nor N2 influenced LRP6-dependent axis induction (data not shown). However, similar to Dkk2, C2 cooperated with LRP6 in axis induction (Fig. 5A and B). Surprisingly, C1 also strongly potentiated formation of secondary axes when coexpressed with LRP6 (Fig. 5A and B). These findings show that C1 and C2, like Dkk2, synergize with LRP6.
FIG. 5.
C1 and C2, but not Dkk1, synergize with LRP6 in axis induction. (A and B) C1 and C2 synergize with LRP6 in axis induction. Embryos were injected with LRP6 mRNA (3 ng) and either 10 pg of Dkk1 or C1 or 1 ng of Dkk2 or C2 mRNAs, as shown. (A) Representative embryos. (B) Combined results of three separate experiments. (C to E) Activation of the Siamois promoter by coexpression of LRP6 and the C-terminal Dkk constructs. Embryos were injected into two ventral-animal blastomeres with pSia-Luc DNA (20 pg) and the following RNAs: LRP6 at 2 ng; Dkk2, C2, or C3 at 50 pg; tBR at 20 pg; and Dkk1 or C1 at 10 pg, as shown. Luciferase activity was measured as described in Fig. 2. Each experiment was performed at least three times; representative experiments are shown.
To ensure that this effect was due to direct activation of the early β-catenin signaling pathway rather than an effect on Wnt signaling at later developmental stages, we examined the ability of C1, C2, and C3 to cooperate with LRP6 in activation of the Siamois promoter. In the absence of LRP6, neither C1 nor C2 detectably activated the Siamois promoter (Fig. 5C and D). LRP6 injected with C3 also did not activate the Siamois promoter (Fig. 5C). However, we found that coinjection of LRP6 with either C1 or C2 RNAs efficiently activated the Siamois promoter, while coinjection of C1 and tBR did not (Fig. 5C, D, and E). Therefore, secondary axis potentiation by LRP6 and C1 (or C2) is due to direct activation of β-catenin target genes, which is a mechanism distinct from head induction by tBR and Dkk1 (14).
Because C1 and C2 functioned similarly in both Wnt inhibition and LRP6 potentiation assays (Fig. 4 and 5), the different functional activities of Dkk1 and Dkk2 must be specified by their N-terminal domains. Since intact Dkk1 did not cooperate with LRP6, we conclude that in Dkk1, the N1 domain masks the ability of the C1 region to synergize with LRP6. The N2 domain, in contrast, does not possess this inhibitory activity.
Dkk1/Dkk2 chimeras confirm the importance of the amino-terminal domains in regulating Dkk-LRP6 interactions.
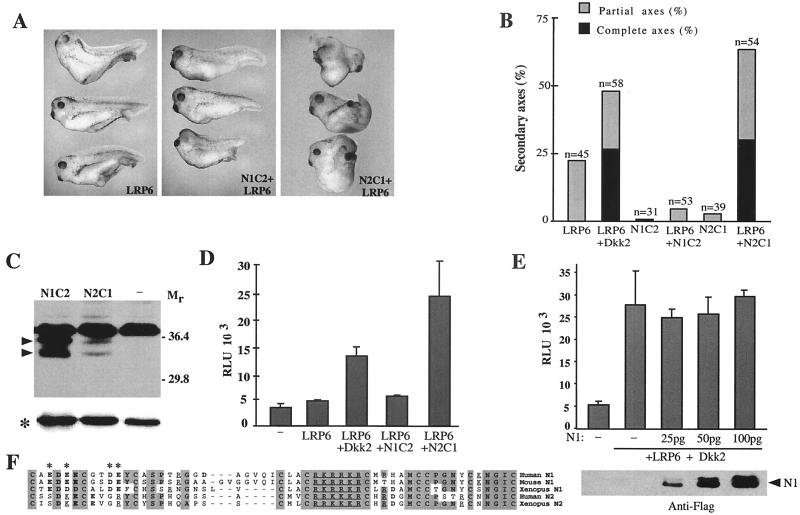
To further investigate the role of the N-terminal domains in specifying the functional properties of Dkk1 and Dkk2, we generated chimeric Dkks (N1C2 and N2C1), in which the N- and C-terminal domains of Dkk1 and Dkk2 were exchanged (Fig. 3A). If the N-terminal domains were essential for regulating the interactions between Dkks and LRP6, a chimeric Dkk that contains N2 but not N1 should synergize with LRP6. Our results supported this prediction. While N2C1 synergized with LRP6 in inducing complete secondary axes and activating the Siamois promoter, N1C2 did not (Fig. 6A, B, and D). This difference in activity was not due to a difference in protein levels, because N1C2 was expressed at higher levels than N2C1 (Fig. 6C). Additionally, although N1C2 did not synergize with LRP6, it retained the ability to inhibit Wnt8-induced secondary axes (data not shown), indicating that the protein is functional. Thus, whereas both C1 and C2 cooperated with LRP6 in axis induction, the N1 domain of Dkk1 negatively regulated the ability of the C1 and C2 domains to synergize with LRP6. These findings support our conclusion that specificity of Dkk effects is due to their N-terminal domains.
FIG. 6.
Role of N-terminal domain in regulation of Dkk signaling. (A, B, and D) Synergistic effects of LRP6 and chimeric Dkk proteins. Fifty picograms of N1C2 and N2C1 RNAs was injected as described in the legend to Fig. 5. (A) Representative embryos. (B) Combined results of three separate experiments. (C) Expression levels of the N1C2 and N2C1 proteins in injected embryos. Arrowheads indicate N1C2 and N2C1 doublets; an asterisk indicates a loading control. (D) Activation of the Siamois promoter by coinjection of LRP6 and N1C2 or N2C1 chimeric RNAs. (E) N1 does not interfere with signaling by LRP6 and Dkk2. RNAs were injected at the following doses: LRP6 at 2 ng, Dkk2 at 50 pg, and N1 as indicated. N1 expression levels in injected embryos are shown below. (F) Comparison of the N-terminal domains of Dkk1 and Dkk2. Shaded areas indicate identical or highly conserved residues. Acidic residues are in boldface; basic residues are underlined. Asterisks mark charged residues that differ between N1 and N2.
N1 may act on the C-terminal domains to inhibit their activation of LRP6. Alternatively, it may interact with a target such as LRP6 to compete for C1 or C2 binding. In the first case, N1 should function only in cis, while in the second case, it could regulate Dkk activity in trans. We examined Dkk2/LRP6 activation in the presence of increasing amounts of the N1 construct, by using the Siamois reporter assay (Fig. 6E). Despite being expressed at high levels, N1 had no inhibitory effect on Dkk2/LRP6 signaling. These results are consistent with the hypothesis that N1 acts on the C-terminal domains of Dkks in cis.
Comparison of N1 and N2 showed that the amino-terminal halves of N1 and N2 are less conserved than the carboxy-terminal halves (Fig. 6F). In particular, N1 contains a cluster of acidic residues, which have been substituted for basic or neutral residues in N2. This region could be responsible for the difference between Dkk1 and Dkk2.
The C-terminal domains of Dkks physically interact with LRP6.
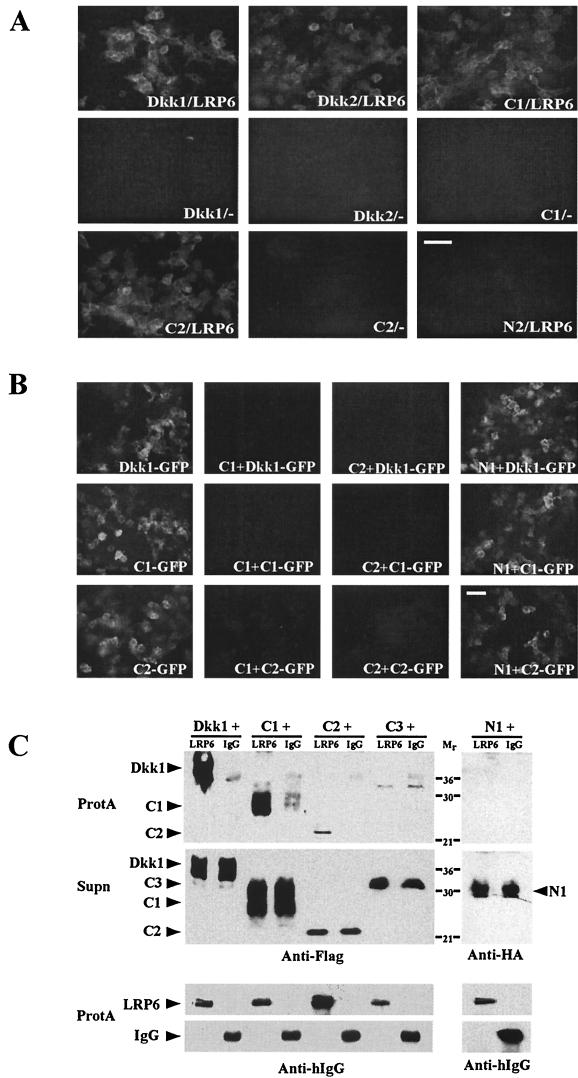
Recent studies have demonstrated that Dkk1 and Dkk2 can associate with LRP6 (2, 27, 41). To evaluate which Dkk domains are involved in these interactions, we fused Dkk domains with GFP to generate proteins that are secreted by transfected HEK293T cells. Cell medium containing Dkk-GFP fusion proteins was incubated with HEK293T cells transiently transfected with LRP6 or a control vector. Efficient binding of Dkk1, Dkk2, C1, and C2 to LRP6-expressing cells was detected by GFP fluorescence (Fig. 7A ). In contrast, N2-GFP did not show any detectable binding. No fluorescence was observed on control vector-transfected HEK293T cells after incubation with Dkk-GFP-containing media (Fig. 7A). Western blot analysis showed that all proteins are efficiently expressed (data not shown). To illustrate specific binding of C1 and C2 to LRP6-expressing cells, we performed competition studies, examining binding of GFP-tagged Dkk1, C1, and C2 in the presence of untagged C1, C2, and N1. Both C1 and C2 efficiently competed with Dkk1-GFP, C1-GFP, and C2-GFP for binding to LRP6-expressing cells, while N1 did not (Fig. 7B). These results suggest that the C-terminal domains of Dkks specifically bind to LRP6 expressed on the cell surface.
FIG. 7.
Association of C1 and C2 with LRP6. (A) Cell culture media with Dkk-GFP proteins were incubated for 1 h with HEK293T cells transfected either with LRP6 (LRP6) or a control vector (−) as indicated. GFP fluorescence reflects Dkk-GFP binding to cells. Bar, 65 μm. (B) LRP6-transfected HEK293T cells were preincubated for 30 min with the culture media containing C1, C2, or N1, followed by additional 30-min incubation with Dkk-GFP-containing media as indicated. Bar, 65 μm. (C) Biochemical association of C1 and C2 with LRP6. Media from HEK293T cells, expressing the extracellular domain of LRP6 fused to the IgG heavy chain (LRP6) or control secreted IgG heavy chain (IgG) were mixed in equal volumes with Dkk-containing media as indicated and incubated with protein A-Sepharose beads (ProtA) to bind IgG or LRP6. The presence of Dkks in ProtA pellets and original supernatants (Supn) was assessed with anti-Flag or anti-HA antibodies, and LRP6 and IgG were detected with anti-human IgG (Fc) (Anti-hIgG) antibody.
To study the association between Dkk domains and LRP6 biochemically, we examined binding of Dkk domains to a soluble form of the LRP6 extracellular domain fused with the IgG heavy chain (41). LRP6- and Dkk-containing cell culture media were mixed, and IgG-tagged LRP6 was isolated by binding to protein A beads. Dkk1, C1, and C2, but not N1 and C3, formed complexes with LRP6-IgG, but not with a control IgG (Fig. 7C). Together, these data provide biochemical evidence for the functional interactions observed between the C-terminal domains of Dkks and LRP6.
DISCUSSION
In our study, we have compared the functional activities of two members of the Dickkopf family, Dkk1 and Dkk2. Both Dkks share the ability to inhibit Wnt8 and cooperate with a dominant-negative BMP4 receptor in inducing head structures. In contrast, only Dkk2 synergizes with the Wnt coreceptor LRP6 to activate β-catenin signaling. We show that these distinct activities are due to differences in the N-terminal domains. Analysis of chimeric Dkks shows that the N1 domain inhibits the ability of C1 and C2 to synergize with LRP6 (Fig. 5 and 6), whereas the N2 domain does not. Thus, the different properties of N1 and N2 underlie the opposing effects of Dkk1 and Dkk2 on LRP6-dependent axis induction.
We find that Dkk2 is an inhibitor of Wnt8, yet it synergizes with LRP6 in the same signal transduction pathway. This paradoxical finding can be explained by postulating that Dkk2 interferes with the binding of Wnt8 to LRP6, but itself functions as a weak activator of LRP6-mediated signaling (Fig. 2D). Consistent with this idea, we show that both C-terminal domains of Dkks form a complex with LRP6 (Fig. 7). Our conclusions are supported by a recent study in which the C-terminal region of Dkk2 inhibited Wnt3a signaling in cultured cells, but activated a Wnt-responsive promoter when coexpressed with LRP6 (25). Therefore, depending on the cellular context (i.e., the presence of Wnt ligands or levels of LRP5 or LRP6), Dkk2 can act as an activator or an inhibitor of the β-catenin pathway.
We also observe that C1 can activate LRP-dependent signaling. Unlike Dkk2, there is no evidence that Dkk1 could function as an activator of Wnt signaling, yet our data suggest that it might be the case, especially if there is a mechanism for the release of the C-terminal domain. Although we do not see major proteolytic fragments of Dkk1 in injected embryos (Fig. 3C), limited evidence for proteolytic processing of Dkks has been reported (24). Moreover, there is a conserved furin site in the N-terminal regions of Dkks (24) (Fig. 6F), suggesting that Dkks may be regulated by proteolysis. A similar situation has been documented for the bipartite zinc finger transcription factor Gli2, which can be activated by the removal of the N-terminal repressor domain (39). More work is necessary to determine whether Dkk1 activity is indeed regulated in the embryo.
A two-domain structure for the family of Dkk proteins was originally proposed based on the conservation of their amino- and carboxy-terminal cysteine-rich regions, which are separated by variable-length spacer sequences (14, 24). The functional significance of this two-domain structure was previously unknown. Our experiments demonstrate that individual Dkk domains operate as discrete functional units. We find that the C-terminal regions C1 and C2 are both necessary and sufficient for the ability of Dkks to inhibit Wnt8 signaling and to synergize with LRP6 in axis induction. In contrast, the N-terminal regions have diverged to confer opposing functions to these two highly similar proteins.
Our data suggest that the N1 domain of Dkk1 acts to suppress the ability of the C1 domain to activate LRP6 signaling. The exact mechanism by which this occurs is unclear. One possibility is that the N1 region interacts with the C1 domain, thereby inhibiting its ability to synergize with LRP6. However, we have not been able to detect an association between the N1 and C1 proteins (data not shown). Another possibility is that the N1 region, tethered to LRP6 by the C-terminal Dkk domain, prevents LRP6 from complexing with Frizzled receptors. This idea is supported by the model of Semenov et al. (41), who have shown that Dkk1 prevents LRP6 from forming complexes with Frizzled and Wnt1. Lack of competition between N1 and Dkk2 (Fig. 6E) also suggests that the N-terminal domain functions only in the context of a complete Dkk molecule or interacts with non-LRP targets. Similar to N1, the N2 domain may interact with other extracellular modulators of Wnt signaling, such as Frizzled receptors or heparan sulfate proteoglycans. This is likely, since the effects of Dkk2 and C2 on embryonic development appear to be different. Further experiments are required to elucidate the role of other regulators of Dkk/LRP signaling.
Dkk1 and Dkk2 have different expression patterns during development. Xenopus Dkk1 is expressed in the organizer (14), a region responsible for specification of head and heart mesoderm. In contrast, the expression of Dkk2 in head mesenchyme, lens, and somites is not detected until organogenesis (48). It has been proposed that the head- and heart-inducing activities of Dkk1 are due to its activity as a Wnt antagonist (14, 23, 29, 40). Because the Wnt-inhibitory activity of our various Dkk constructs correlates with their ability to induce head structures (Fig. 4), our findings lend support to this hypothesis.
In contrast to the effect of Dkk1, embryos injected with Dkk2 RNA do not develop enlarged head structures (14; data not shown). This observation indicates that Dkk2 is unlikely to function solely as a Wnt antagonist in vivo. We have shown that Dkk2 may function as an activator or an inhibitor of Wnt signaling, depending on the cell context. Thus, Dkk2 may have opposite activities in different embryonic tissues. To provide more insight into the molecular mechanisms of Dkk2 action during development, a detailed knowledge of the expression levels of endogenous LRP and other proteins interacting with Dkk2 will be required.
Our experiments have investigated regulation of Wnt signaling by two closely related Dkk family members. We have shown that although both proteins inhibit Wnt signaling, they possess an intrinsic ability to activate the Wnt coreceptor LRP6, which is suppressed in the case of Dkk1 by its N-terminal domain. The C-terminal domains of both Dkks associate with LRP6 and appear to modulate LRP6 signaling. Our studies suggest a mechanism for the regulation of Wnt signaling by Dkk1 and Dkk2 and further contribute to our understanding of this signaling pathway.
Acknowledgments
We thank Xi He and Jonathan Graff for plasmids. We also thank Misha Semenov, Xi He, Mark Mercola, and Ruchika Gupta for extensive discussion and communication of data. We thank Keiji Itoh, Yana Kamberov, and Stephen Baccus for critical reading of the manuscript and Suzanne Ruscitti for secretarial assistance.
This work has been supported by NIH grants to S.Y.S.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 1998. A colipase fold in the carboxy-terminal domain of the Wnt antagonists—the Dickkopfs. Curr. Biol. 8:477-478. [DOI] [PubMed] [Google Scholar]
- 2.Bafico, A., G. Liu, A. Yaniv, A. Gazit, and S. A. Aaronson. 2001. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat. Cell Biol. 3:683-686. [DOI] [PubMed] [Google Scholar]
- 3.Bhanot, P., M. Brink, C. H. Samos, J. C. Hsieh, Y. Wang, J. P. Macke, D. Andrew, J. Nathans, and R. Nusse. 1996. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature 382:225-230. [DOI] [PubMed] [Google Scholar]
- 4.Bouwmeester, T., S. Kim, Y. Sasai, B. Lu, and E. M. De Robertis. 1996. Cerberus is a head-inducing secreted factor expressed in the anterior endoderm of Spemann's organizer. Nature 382:595-601. [DOI] [PubMed] [Google Scholar]
- 5.Brannon, M., M. Gomperts, L. Sumoy, R. T. Moon, and D. Kimelman. 1997. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 11:2359-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cadigan, K. M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 11:3286-3305. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., and H. Okayama. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christian, J. L., J. A. McMahon, A. P. McMahon, and R. T. Moon. 1991. Xwnt-8, a Xenopus Wnt-1/int-1-related gene responsive to mesoderm-inducing growth factors, may play a role in ventral mesodermal patterning during embryogenesis. Development 111:1045-1055. [DOI] [PubMed] [Google Scholar]
- 9.Christian, J. L., and R. T. Moon. 1993. Interactions between Xwnt-8 and Spemann organizer signaling pathways generate dorsoventral pattern in the embryonic mesoderm of Xenopus. Genes Dev. 7:13-28. [DOI] [PubMed] [Google Scholar]
- 10.Fan, M. J., W. Gruning, G. Walz, and S. Y. Sokol. 1998. Wnt signaling and transcriptional control of Siamois in Xenopus embryos. Proc. Natl. Acad. Sci. USA 95:5626-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, M. J., and S. Y. Sokol. 1997. A role for Siamois in Spemann organizer formation. Development 124:2581-2589. [DOI] [PubMed] [Google Scholar]
- 12.Fedi, P., A. Bafico, A. Nieto-Soria, W. H. Burgess, T. Miki, D. P. Bottaro, M. H. Kraus, and S. A. Aaronson. 1999. Isolation and biochemical characterization of the human Dkk-1 homologue, a novel inhibitor of mammalian Wnt signaling. J. Biol. Chem. 274:19465-19472. [DOI] [PubMed] [Google Scholar]
- 13.Glinka, A., W. Wu, D. Onichtchouk, C. Blumenstock, and C. Niehrs. 1997. Head induction by simultaneous repression of Bmp and Wnt signalling in Xenopus. Nature 389:517-519. [DOI] [PubMed] [Google Scholar]
- 14.Glinka, A., W. Wu, H. Delius, A. P. Monaghan, C. Blumenstock, and C. Niehrs. 1998. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391:357-362. [DOI] [PubMed] [Google Scholar]
- 15.Graff, J. M., R. S. Thies, J. J. Song, A. J. Celeste, and D. A. Melton. 1994. Studies with a Xenopus BMP receptor suggest that ventral mesoderm-inducing signals override dorsal signals in vivo. Cell 79:169-179. [DOI] [PubMed] [Google Scholar]
- 16.Grotewold, L., T. Theil, and U. Ruther. 1999. Expression pattern of Dkk-1 during mouse limb development. Mech. Dev. 89:151-153. [DOI] [PubMed] [Google Scholar]
- 17.Harland, R., and J. Gerhart. 1997. Formation and function of Spemann's organizer. Annu. Rev. Cell. Dev. Biol. 13:611-667. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto, H., M. Itoh, Y. Yamanaka, S. Yamashita, T. Shimizu, L. Solnica-Krezel, M. Hibi, and T. Hirano. 2000. Zebrafish Dkk1 functions in forebrain specification and axial mesendoderm formation. Dev. Biol. 217:138-152. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, J. C., L. Kodjabachian, M. L. Rebbert, A. Rattner, P. M. Smallwood, C. H. Samos, R. Nusse, I. B. Dawid, and J. Nathans. 1999. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature 398:431-436. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh, J. C., A. Rattner, P. M. Smallwood, and J. Nathans. 1999. Biochemical characterization of Wnt-frizzled interactions using a soluble, biologically active vertebrate Wnt protein. Proc. Natl. Acad. Sci. USA 96:3546-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh, K., T. L. Tang, B. G. Neel, and S. Y. Sokol. 1995. Specific modulation of ectodermal cell fates in Xenopus embryos by glycogen synthase kinase. Development 121:3979-3988. [DOI] [PubMed] [Google Scholar]
- 22.Itoh, K., V. E. Krupnik, and S. Y. Sokol. 1998. Axis determination of Xenopus involves biochemical interactions of axin, glycogen synthase kinase-3 and β-catenin. Curr. Biol. 8:591-594. [DOI] [PubMed] [Google Scholar]
- 23.Kazanskaya, O., A. Glinka, and C. Niehrs. 2000. The role of Xenopus dickkopf1 in prechordal plate specification and neural patterning. Development 127:4981-4992. [DOI] [PubMed] [Google Scholar]
- 24.Krupnik, V. E., J. D. Sharp, C. Jiang, K. Robison, T. W. Chickering, L. Amaravadi, D. E. Brown, D. Guyot, G. Mays, K. Leiby, et al. 1999. Functional and structural diversity of the human Dickkopf gene family. Gene 238:301-313. [DOI] [PubMed] [Google Scholar]
- 25.Li, L., J. Mao, L. Sun, W. Liu, and D. Wu. 2002. Second cysteine-rich domain of Dickkopf-2 activates canonical Wnt signaling pathway via LRP-6 independently of dishevelled. J. Biol. Chem. 277:5977-5981. [DOI] [PubMed] [Google Scholar]
- 26.Makarova, O., E. Kamberov, and B. Margolis. 2000. Generation of deletions and point mutations with one primer in a single cloning step. BioTechniques 29:970-972. [DOI] [PubMed] [Google Scholar]
- 27.Mao, B., W. Wu, D. Hoppe, P. Stanne, A. Glinka, and C. Niehrs. 2001. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature 411:321-325. [DOI] [PubMed] [Google Scholar]
- 28.Mao, J., J. Wang, B. Liu, W. Pan, G. H. Farr, C. Flynn, H. Yuan, S. Takada, D. Kimelman, L. Li, and D. Wu. 2001. Low-density lipoprotein receptor-related protein binds to axin and regulates the canonical Wnt signaling pathway. Mol. Cell 7:801-809. [DOI] [PubMed] [Google Scholar]
- 29.Marvin, M. J., G. Di Rocco, A. Gardiner, S. M. Bush, and A. B. Lassar. 2001. Inhibition of Wnt activity induces heart formation from posterior mesoderm. Genes Dev. 15:316-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMahon, A. P., and R. T. Moon. 1989. Ectopic expression of the proto-oncogene int-1 in Xenopus embryos leads to duplication of the embryonic axis. Cell 58:1075-1084. [DOI] [PubMed] [Google Scholar]
- 31.Monaghan, A. P., P. Kioschis, W. Wu, A. Zuniga, D. Bock, A. Poustka, H. Delius, and C. Niehrs. 1999. Dickkopf genes are co-ordinately expressed in mesodermal lineages. Mech. Dev. 87:45-56. [DOI] [PubMed] [Google Scholar]
- 32.Mukhopadhyay, M., S. Shtrom, C. Rodriguez-Esteban, L. Chen, T. Tsukui, L. Gomer, D. W. Dorward, A. Glinka, A. Grinberg, S. P. Huang, C. Niehrs, J. C. Belmonte, and H. Westphal. 2001. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev. Cell 1:423-434. [DOI] [PubMed] [Google Scholar]
- 33.Newport, J., and M. Kirschner. 1982. A major developmental transition in early Xenopus embyros. I. Characterization and timing of cellular changes at the midblastula stage. Cell 30:675-686. [DOI] [PubMed] [Google Scholar]
- 34.Nieuwkoop, P. D., and J. Faber. 1967. Normal table of Xenopus laevis. North-Holland Publishing Co., Amsterdam, The Netherlands.
- 35.Peifer, M., and P. Polakis. 2000. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science 287:1606-1609. [DOI] [PubMed] [Google Scholar]
- 36.Perrimon, N., and M. Bernfield. 2000. Specificities of heparan sulphate proteoglycans in developmental processes. Nature 404:725-728. [DOI] [PubMed] [Google Scholar]
- 37.Pinson, K. I., J. Brennan, S. Monkley, B. J. Avery, and W. C. Skarnes. 2000. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407:535-538. [DOI] [PubMed] [Google Scholar]
- 38.Rattner, A., J. C. Hsieh, P. M. Smallwood, D. J. Gilbert, N. G. Copeland, N. A. Jenkins, and J. Nathans. 1997. A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc. Natl. Acad. Sci. USA 94:2859-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sasaki, H., Y. Nishizaki, C. Hui, M. Nakafuku, and H. Kondoh. 1999. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126:3915-3924. [DOI] [PubMed] [Google Scholar]
- 40.Schneider, V. A., and M. Mercola. 2001. Wnt antagonism initiates cardiogenesis in Xenopus laevis. Genes Dev. 15:304-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Semenov, M., K. Tamai, B. K. Brott, M. Kuhl, S. Sokol, and X. He. 2001. Head inducer Dickkopf-1 is a ligand for Wnt co-receptor LRP6. Curr. Biol. 11:951-961. [DOI] [PubMed] [Google Scholar]
- 42.Shinya, M., C. Eschbach, M. Clark, H. Lehrach, and M. Furutani-Seiki. 2000. Zebrafish Dkk1, induced by the pre-MBT Wnt signaling, is secreted from the prechordal plate and patterns the anterior neural plate. Mech. Dev. 98:3-17. [DOI] [PubMed] [Google Scholar]
- 43.Smith, W. C., and R. Harland. 1991. Injected Xwnt-8 RNA acts early in Xenopus embryos to promote formation of a vegetal dorsalizing center. Cell 67:753-765. [DOI] [PubMed] [Google Scholar]
- 44.Sokol, S., J. L. Christian, R. T. Moon, and D. A. Melton. 1991. Injected Wnt RNA induces a complete body axis in Xenopus embryos. Cell 67:741-752. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, A., R. S. Thies, N. Yamaji, J. J. Song, J. M. Wozney, K. Murakami, and N. Ueno. 1994. A truncated bone morphogenetic protein receptor affects dorsal-ventral patterning in the early Xenopus embryo. Proc. Natl. Acad. Sci. USA 91:10255-10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamai, K., M. Semenov, Y. Kato, R. Spokony, C. Liu, Y. Katsuyama, F. Hess, J. P. Saint-Jeannet, and X. He. 2000. LDL-receptor-related proteins in Wnt signal transduction. Nature 407:530-535. [DOI] [PubMed] [Google Scholar]
- 47.Wehrli, M., S. T. Dougan, K. Caldwell, L. O'Keefe, S. Schwartz, D. Vaizel-Ohayon, E. Schejter, A. Tomlinson, and S. DiNardo. 2000. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature 407:527-530. [DOI] [PubMed] [Google Scholar]
- 48.Wu, W., A. Glinka, H. Delius, and C. Niehrs. 2000. Mutual antagonism between dickkopf1 and dickkopf2 regulates Wnt/beta-catenin signalling. Curr. Biol. 10:1611-1614. [DOI] [PubMed] [Google Scholar]
- 49.Xu, Q., P. A. D'Amore, and S. Y. Sokol. 1998. Functional and biochemical interactions of Wnts with FrzA, a secreted Wnt antagonist. Development 125:4767-4776. [DOI] [PubMed] [Google Scholar]
- 50.Zorn, A. M. 1997. Cell-cell signalling: frog frizbees. Curr. Biol. 7:501-504. [DOI] [PubMed] [Google Scholar]