Abstract
A major control point for skeletal myogenesis revolves around the muscle basic helix-loop-helix gene family that includes MyoD, Myf-5, myogenin, and MRF4. Myogenin and MRF4 are thought to be essential to terminal differentiation events, whereas MyoD and Myf-5 are critical to establishing the myogenic cell lineage and producing committed, undifferentiated myogenic stem cells (myoblasts). Although mouse genetic studies have revealed the importance of MyoD and Myf-5 for myoblast development, the genetic targets of MyoD and Myf-5 activity in undifferentiated myoblasts remain unknown. In this study, we investigated the function of MyoD as a transcriptional activator in undifferentiated myoblasts. By using conditional expression of MyoD, in conjunction with suppression subtractive hybridizations, we show that the Id3 and NP1 (neuronal pentraxin 1) genes become transcriptionally active following MyoD induction in undifferentiated myoblasts. Activation of Id3 and NP1 represents a stable, heritable event that does not rely on continued MyoD activity and is not subject to negative regulation by an activated H-Ras G12V protein. These results are the first to demonstrate that MyoD functions as a transcriptional activator in myogenic stem cells and that this key myogenic regulatory factor exhibits different gene target specificities, depending upon the cellular environment.
The molecular and cellular processes associated with myogenesis have served as a paradigm of various schemes that unfold during development. Myogenesis begins at an early stage of embryonic development with the patterning of mesoderm into somites (reviewed in references 4 and 41). The two main substructures that develop from the somite are the dermomyotome and the sclerotome. The sclerotome gives rise to the ribs and accompanying cartilage, while the dermomyotome contributes to both the epaxial musculature (back and intercostal muscles) and the hypaxial musculature (body wall and limb muscles) (9, 13). The emergence of these distinct lineages of muscle cells is associated with the expression of the MyoD, Myf-5, myogenin, and MRF4 transcription factors. These basic helix-loop-helix (bHLH) proteins are referred to as the muscle regulatory factors (MRFs) and are acknowledged to be the driving force behind the specification and differentiation of all myogenic compartments (reviewed in references 32 and 41). Although all four MRFs have similar functional properties in vitro, they exhibit distinct activities during embryogenesis. MyoD or Myf-5 is essential to specification of the muscle lineage (5, 21, 44, 45), whereas myogenin and MRF4 have been implicated in the initial and late stages of differentiation, respectively (17, 38, 40, 52, 54).
The MRFs typically form heterodimers with widely expressed bHLH factors such as E12, E47, HEB, and ITF-1 (25, 30, 37, 49). The MRF-E-protein heterodimer binds to the DNA consensus sequence CANNTG, which is found in the enhancer and promoter regions of most muscle-specific genes. Binding of the MRF-E-protein heterodimer to target genes leads to their transcriptional activation and the subsequent differentiation of myoblasts into functional myotubes. Specificity in DNA binding is mediated by the unique combinatorial interactions of the bHLH regions of individual MRF and E-protein dimers (11, 26, 33, 53), enabling subtle alterations in the targets, as well as the levels of gene transactivation. In addition, cooperative interactions with other DNA binding transcription factors, such as the MEF2 family of transcription factors, is involved in enhancement of the transcription of muscle-specific genes by the MRFs (3, 36, 39). To date, most of the DNA binding and transactivation studies reported for MRFs have focused on terminal differentiation events. Very few studies have examined the transcriptional activity of the MRFs in undifferentiated myoblasts. Most models suggest that the MRFs exhibit no specific function in myoblasts but exist to prime cells for differentiation when the environmental conditions become permissive (i.e., cessation of cellular proliferation signals). The activity of MRFs toward muscle-specific genes is negatively regulated when conditions remain restrictive for differentiation (reviewed in references 32 and 41). Mechanisms mediating this negative regulation include posttranslational modifications leading to reduced heterodimer formation and DNA binding capabilities (16, 29), increased protein turnover (6, 18, 48), and induction of dominant-negative HLH proteins of the Id protein family (25, 33).
One of the remaining challenges in understanding the molecular basis of muscle development is the identification of genes involved in the specification of myogenic stem cells (24). Fundamental differences must exist between the predetermined cells of the epithelial somite and the specified myogenic precursors of the dermomyotome. MyoD and Myf-5 are expressed in myogenic stem cells and thus have long been suspected of specifying these changes. However, no direct target genes have been identified for these MRFs in committed myogenic stem cells and the most attention has been focused on the genes activated by MyoD during the initial stages of differentiation (1, 2, 15, 19, 28, 53).
To identify MyoD transcriptional targets in undifferentiated myoblasts, we utilized C3H10T1/2 (10T1/2) mouse fibroblasts expressing a MyoD protein fused to the estrogen receptor (ER) (19). 10T1/2-MyoD-ER cells are maintained in an undifferentiated state by providing cells with normal growth medium (GM) containing serum. In the presence of β-estradiol, the MyoD-ER protein undergoes a conformational change and becomes available for dimerization with E-proteins. In GM plus β-estradiol, the cells remain undifferentiated myoblasts. When cells are switched to differentiation medium (DM) in the absence of β-estradiol, they remain undifferentiated. However, when β-estradiol is added to DM, the cells rapidly differentiate and activate the expression of an entire set of muscle-specific gene products (19). By establishing tight control over the timing of β-estradiol addition to the 10T1/2 MyoD-ER cell line, it is possible to examine the initial events associated with MyoD activity in proliferating cells.
We utilized suppression subtractive hybridization to compare changes in gene expression associated with the induction of MyoD in undifferentiated cells. Our studies have identified two genes, Id3 and NP1 (neuronal pentraxin 1), that are induced in 10T1/2-MyoD-ER cells when the cells are provided β-estradiol but are not expressed in β-estradiol-treated 10T1/2 fibroblasts or in 10T1/2-MyoD-ER cells grown in the absence of β-estradiol. Id3 and NP1 gene expression is detected in undifferentiated myoblasts from the C2C12, 23A2, and L6A1 myogenic cell lines and can be induced in control 10T1/2 cells following transfection with expression plasmids encoding MyoD or myogenin. Interestingly, MyoD-induced activation of Id3 and NP1 gene expression is not subject to the negative regulation imposed on terminal differentiation events by an activated H-Ras G12V protein. Our results reveal the novel finding that MRFs are transcriptionally active in myogenic stem cells, prior to overt terminal differentiation, and have identified two genetic targets for MRF activity that may be essential to the establishment and maintenance of the myogenic stem cell phenotype.
MATERIALS AND METHODS
Mammalian cell culture.
The C3H10T1/2 (10T1/2), C2C12, and C2 cell lines were obtained from the American Type Culture Collection. The 23A2 myogenic cell line has been described previously (24, 51). 10T1/2 and 23A2 cells were maintained in basal medium Eagle (Gibco BRL) supplemented with 10 or 15% fetal bovine serum (FBS; HyClone Laboratories) and 1% penicillin and streptomycin (Gibco BRL), respectively. C2C12 and C2 cells were maintained in high-glucose Dulbecco's modified Eagle medium (DMEM; Gibco BRL) supplemented with 15% FBS and 1% penicillin and streptomycin. Cells were induced to differentiate in low-glucose DMEM supplemented with 1% ITS (Sigma) containing 1 μg of insulin per ml, 1 μg of transferrin per ml, and 1 ng of selenium per ml plus 1% penicillin and streptomycin. Transient DNA transfections of 10T1/2 cells were performed as previously described (23, 27, 35).
The 10T1/2-MyoD-ER inducible myogenic cell line (9-5) was a generous gift of Stephen J. Tapscott (Fred Hutchinson Cancer Research Center, Seattle, Wash.) and has been described previously (19). 10T1/2-MyoD-ER cells were maintained in high-glucose DMEM supplemented with 10% FBS, 1% penicillin, and streptomycin plus 250 μg of G418 (Mediatech) per ml. Myogenic differentiation was induced by incubation in low-glucose DMEM supplemented with 1% ITS, 1% penicillin and streptomycin, and 36.7 nM 17-β-estradiol (Sigma) diluted in ethanol.
Immunohistochemistry.
Immunohistochemistry was performed on the 10T1/2-MyoD-ER cell line as described previously (35). Briefly, cells were fixed in 4% paraformaldehyde for 10 min and then permeabilized in phosphate-buffered saline containing 0.1% Triton X-100 (PBS-T; Sigma) for 30 min. Cells were treated with 10% calf serum in PBS-T for 30 min and washed three times for 5 min each time with PBS-T, and then primary antibodies supplemented with 0.1% bovine serum albumin were incubated with the cells for 1 h at room temperature. Cells were washed three times for 5 min each time with PBS-T, fluorescently labeled secondary antibodies were added, and the mixture was incubated for 1 h. Primary antibodies used in these studies were obtained from the University of Iowa Developmental Hybridoma Bank and included the antibodies against embryonic myosin heavy chain (MyHC) (MF-20), troponin T (TnT) (CT3), and tropomyosin (Tm) (CH1).
Western blot analysis.
Whole-cell protein extracts were harvested from cells by aspirating the medium and adding 25 μl of 4× sodium dodecyl sulfate-Laemmli loading buffer per 60-mm-diameter dish. The crude lysate was sonicated for 15 s and then incubated at 95°C for 5 min. Approximately 20 μg of whole-cell lysate was electrophoresed through a sodium dodecyl sulfate-polyacrylamide gel, and the separated proteins were transferred onto Immobilon-P polyvinylidene difluoride membranes (Bio-Rad) with a semidry transfer apparatus. Membranes were blocked for 30 min with 5% BLOTTO (nonfat dry milk in TBS-T), incubated in primary antibodies for 1 h, rinsed, and incubated for 1 h with secondary antibodies. The blot was then developed with an ECL kit (Pierce) and subsequently exposed to film.
PCR-Select suppression subtractive hybridization.
Total RNA was harvested from cell cultures with Trizol (Gibco BRL). Poly(A) mRNA was purified from total RNA with the PolyATract mRNA isolation system (Promega). The poly(A) RNA was used as the starting material for the PCR-Select suppression subtractive hybridization kit (Clontech, Inc.). All reactions were performed exactly as described in the manufacturer's instructions. Briefly, 2 μg of poly(A) RNA from each experimental group (10T1/2-MyoD-ER cells maintained in GM with and without β-estradiol) was used to generate first- and second-strand cDNA synthesis. The generated cDNA was digested with RsaI, and two different adapters were ligated to separate tester cDNA pools (plus β-estradiol; groups A and B). The tester and driver (minus β-estradiol; 100-fold excess) cDNAs were hybridized overnight, and the remaining single-stranded tester A and B cDNAs were hybridized to each other. Double-stranded complexes were amplified by PCR with primers that anneal to the appropriate adapters. Amplified selected cDNAs were cloned into the pT-Adv plasmid (Clontech, Inc.) for further characterization. Inserts were amplified by PCR directly from individual colonies, separated on agarose gels, transferred to nylon membranes, and hybridized with 32P-labeled tester and driver cDNA reaction mixtures that were used in the original subtraction procedure. Potential clones that hybridized specifically to tester cDNAs were characterized further as described in Results.
Expression analysis.
RNA blot hybridizations were performed by separating 20 μg of total RNA through a 1% agarose-6.5% formaldehyde gel. The samples were then transferred to Hybond-N+ (Amersham) membranes and hybridized with [α-32P]dCTP-labeled cDNA probes at 42°C in 50% formamide by following standard procedures. RNA expression also was detected with reverse transcriptase (RT)-coupled PCR. Briefly, 2 μg of total RNA was used for each RT reaction and an aliquot of this material was amplified by PCR with gene-specific primers. All primers crossed intron junctions, and all products were verified by DNA sequencing, nucleic acid hybridization, and restriction enzyme digestion. The DNA primers used in the PCR amplifications are described in Table 1.
TABLE 1.
Primers used for PCR amplification
| Primer | Sequence (5′ → 3′) | Expected cDNA size (bp) |
|---|---|---|
| 5′ MyHC | TGGTGGTTAAACCAGAGGACG | 290 |
| 3′ MyHC | GGTAGGCGTTGTCAGAGATGG | |
| 5′ β-Actin | GCATCCTGACCCTGAAGTACC | 450 |
| 3′ β-Actin | GCTCATAGCTCTTCTCCAGGG | |
| 5′ GAPDHa | ATGGTGAAGGTCGGTGTGAACG | 1,049 |
| 3′ GAPDH | CTCTCTCTTGCTCTCAGTATCC | |
| 5′ MyoD | GAGCAAAGTGAATGAGGCCTT | 330 |
| 3′ MyoD | CACTGTAGTAGGCGGTGTCGT | |
| 5′ Myogenin | AGTGAATGCAACTCCCACAGC | 450 |
| 3′ Myogenin | TCAGAAGAGGATGCTCTCTGC | |
| 5′ Id1 | ATGAAGGTCGCCAGTGGCAGTG | 521 |
| 3′ Id1 | TCAGCGACACAAGATGCGATCG | |
| 5′ Id2 | ATGAAAGCCTTCAGTCCGGTG | 404 |
| 3′ Id2 | TTAGCCACAGAGTACTTTGCT | |
| 5′ Id3 | ATGAAGGCGCTGAGCCCG | 400 |
| 3′ Id3 | GTGGCAAAAGCTCCTCTTGTC | |
| 5′ Id4 | ATGAAGGCGGTGAGCCCGGTGC | 277 |
| 3′ Id4 | ACCCTGCTTGTTCACGGCGCCG | |
| 5′ NP1 | TTCTCCTCTCCCCTGCGG | 1,000 |
| 3′ NP1 | GCACGTAAGTCCACTGCG |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
RESULTS
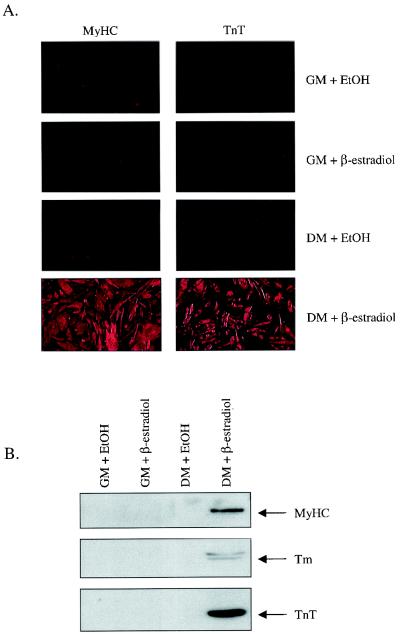
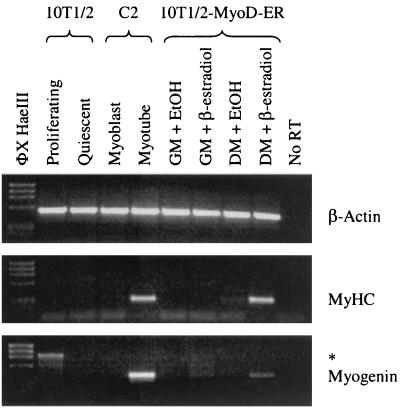
To identify potential MyoD target genes in myogenic stem cells, we utilized a 10T/2 fibroblast cell line expressing a MyoD-ER fusion protein (19). As previously reported, 10T/2-MyoD-ER cells maintained in GM in the presence or absence of β-estradiol remain as undifferentiated fibroblasts and do not express muscle-specific markers such as MyHC or TnT (Fig. 1). Cells maintained in DM lacking β-estradiol also fail to differentiate. However, when cells are provided DM containing β-estradiol, they rapidly fuse into multinucleate myotubes and coordinately express muscle-specific gene products such as MyHC, TnT, and Tm (Fig. 1). This expression pattern is identical to that observed with established myogenic cell lines such as C2. MyHC and myogenin gene transcripts are detected in differentiated C2 myotubes, but no expression is observed in control 10T1/2 cells or in proliferating C2 myoblasts (Fig. 2). Similarly, MyHC and myogenin transcripts are detected in 10T1/2-MyoD-ER cells maintained in DM containing β-estradiol but not in cells maintained in GM with or without β-estradiol (Fig. 2). These results confirm that 10T1/2-MyoD-ER cells fail to express differentiation-specific genes when maintained in GM, regardless of whether the MyoD-ER inducer (β-estradiol) is included in the culture medium.
FIG. 1.
10T1/2-MyoD-ER cells express muscle-specific gene products only when induced to differentiate in the presence of β-estradiol. 10T1/2-MyoD-ER cells were maintained in GM or DM for 18 h in the presence of β-estradiol or ethanol (EtOH) and then subjected to immunofluorescence staining (A) or immunoblot analysis (B) with antibodies specific to embryonic myosin (MyHC), TnT, and Tm. Cells express all three differentiated cell markers only when provided DM containing β-estradiol. No differentiation occurs in cells maintained in GM in the presence of β-estradiol.
FIG. 2.
RT-PCR analysis confirms that 10T1/2-MyoD-ER cells differentiate only when maintained in DM containing β-estradiol. RNA was isolated from control 10T1/2 cells, C2 myoblasts, and myotubes and from 10T1/2-MyoD-ER cells maintained for 18 h in GM or DM supplemented with ethanol (EtOH) or β-estradiol. RT-PCR reveals that the differentiation-specific transcripts from the MyHC and myogenin genes are detected only in C2 myotube cultures and in 10T1/2-MyoD-ER cells maintained in DM containing β-estradiol. β-Actin transcripts served as internal controls. The asterisk denotes a contaminating genomic myogenin fragment found in some samples.
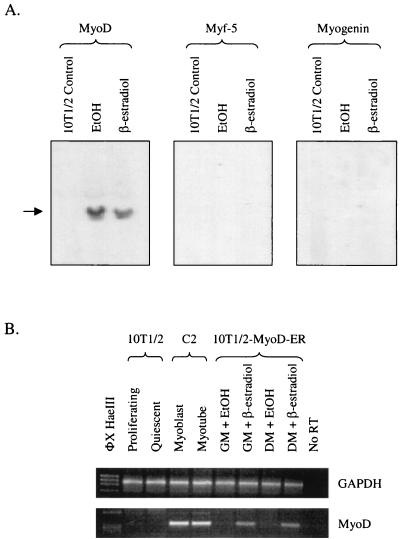
Given that the MyoD-ER fusion protein remains inactive in cells grown in GM, we next examined if other bHLH MRFs were expressed in these cells. Northern blot analysis of total RNA revealed the expected constitutive expression of the MyoD-ER mRNA regardless of the culture medium conditions (Fig. 3A). However, we failed to detect endogenous Myf-5, myogenin, or MRF4 mRNA in these same samples (Fig. 3A and data not shown). These observations confirm that proliferating 10T/2-MyoD-ER cells express MyoD-ER transcripts while other members of the endogenous MRF gene family remain transcriptionally inactive.
FIG. 3.
10T1/2-MyoD-ER cells do not express endogenous MyoD, myogenin, or Myf-5 transcripts in the absence of β-estradiol. (A) Northern hybridization analysis of control 10T1/2 cells and 10T1/2-MyoD-ER cells maintained in GM containing ethanol (EtOH) or β-estradiol. The MyoD-ER transgene transcript is constitutively expressed in 10T1/2-MyoD-ER cells, whereas no Myf-5, myogenin, or MRF4 (data not shown) transcripts are detected. (B) MyoD-ER is capable of activating the endogenous MyoD gene in 10T1/2-MyoD-ER cells maintained in GM containing β-estradiol. RT-PCR analysis confirms that the endogenous MyoD gene is transcribed in 10T1/2-MyoD-ER cells that are provided β-estradiol, suggesting that the MyoD-ER protein autoregulates expression of the endogenous MyoD gene locus.
Previous reports have shown that MyoD regulates its own expression (50). To examine if the 10T1/2-MyoD-ER cell line exhibits this autoregulation phenomenon, the expression of the MyoD-ER transgene and the endogenous MyoD gene was examined under different culture conditions. While the MyoD-ER transcript is present under all culture conditions (data not shown), transcripts from the endogenous MyoD gene are detected in 10T1/2-MyoD-ER cells maintained in DM containing β-estradiol but not in cells lacking β-estradiol (Fig. 3B). Importantly, endogenous MyoD gene transcripts are also found in 10T1/2-MyoD-ER cells maintained in GM in the presence of β-estradiol (Fig. 3B). These data confirm that the MyoD protein regulates its own expression, even in cells maintained in high levels of growth factors, and provide experimental evidence that the MyoD protein remains transcriptionally active in myogenic stem cells.
PCR-Select subtractive hybridization.
Precise control of MyoD-ER activity in the 10T1/2 MyoD-ER cell line provides a system for investigation of the initial stages of myogenesis. Previous groups have used this system to identify immediate targets of MyoD during the first few hours of differentiation (2, 14, 19). However, little information is available regarding the genetic targets of MyoD activity in cells maintained in GM. To identify potential MyoD gene targets in proliferating myoblasts, we compared gene expression profiles of 10T1/2-MyoD-ER cells cultured for 18 h in GM plus ethanol (driver RNA) or cultured in GM containing β-estradiol (tester RNA). PCR-generated products that survived the subtractive hybridization procedure were cloned. The initial procedure yielded 5,352 individual colonies containing inserts, of which 557 were chosen for further analysis. Inserts were amplified by PCR, and the DNA fragments were resolved on four duplicate agarose gels, transferred to Hybond N+ nylon membranes, and hybridized with radiolabeled probes representing each subtraction. Inserts that displayed specific hybridization to the forward subtraction, but not to the reverse subtraction, were chosen as representing transcripts whose expression was up-regulated as a result of MyoD-ER transactivation. Of the 30 clones identified as being up-regulated in the MyoD-ER cells exposed to β-estradiol, several contained overlapping fragments corresponding to the mouse Id3 and NP1 genes. Id3 encodes an HLH nuclear protein that is known to serve as a negative regulator of bHLH factors (7, 31, 34), while NP1 encodes a secreted glycoprotein that previously has been thought to be expressed primarily in neuronal cells (12, 47).
Characterization of Id3 and NP1 gene expression.
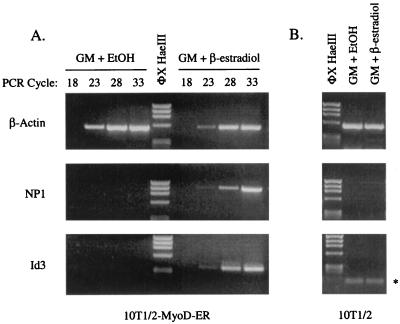
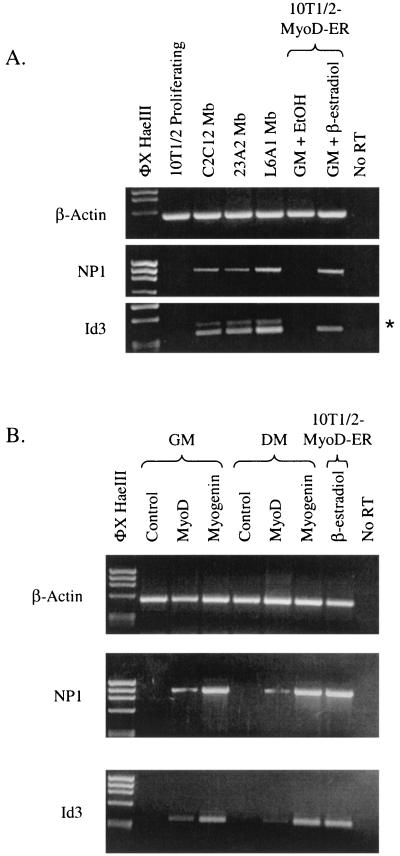
In order to examine and confirm the regulated expression of Id3 and NP1 in 10T1/2-MyoD-ER cells, total RNA isolated from cells maintained in GM for 18 h in the presence or absence of β-estradiol was analyzed by RT-PCR. The results demonstrate that Id3 and NP1 transcripts are not detected in 10T1/2-MyoD-ER cells maintained in GM, even after 33 amplification cycles, while transcripts from Id3 and NP1 are detected in cells treated with GM plus β-estradiol (Fig. 4A). Semiquantitative PCR shows that the expression of these genes is fairly equivalent since the transcripts are not detected after 18 PCR cycles but appear by 23 PCR cycles. A similar profile of expression is observed for the control β-actin-encoding gene. Id3 and NP1 gene expression is specific for cells containing MyoD and is not an artifact of β-estradiol treatment since control 10T1/2 cells do not produce Id3 and NP1 transcripts (Fig. 4B).
FIG. 4.
The Id3 and NP1 genes are expressed in 10T1/2-MyoD-ER cells maintained in GM containing β-estradiol. 10T1/2-MyoD-ER (A) and control 10T1/2 (B) cells were grown for 18 h in GM containing ethanol (EtOH) or β-estradiol, and total RNA was subjected to RT-PCR analysis for 18 to 33 cycles with sequence-specific primers for the mouse Id3 and NP1 genes. Id3 and NP1 transcripts are not detected in control 10T1/2 cells or in 10T1/2-MyoD-ER cells provided ethanol. However, both genes are expressed in 10T1/2-MyoD-ER cells that are provided GM containing β-estradiol, suggesting that the Id3 and NP1 genes are activated by MyoD in undifferentiated myogenic stem cells. The asterisk denotes PCR primers remaining on the gel.
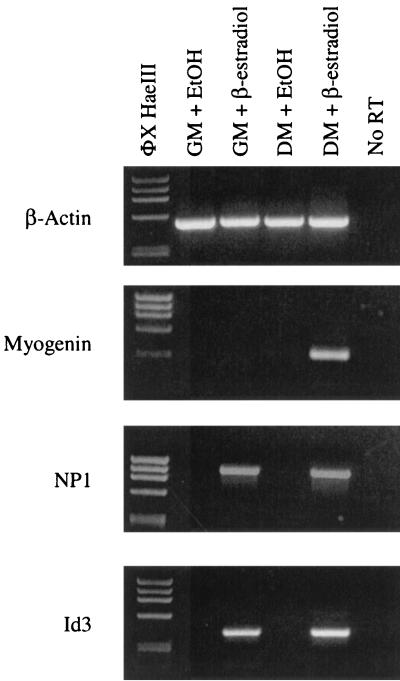
The ability of MyoD to activate transcription of the Id3 and NP1 genes in cells maintained in GM was evaluated further in cells provided GM or DM and then treated with β-estradiol for 18 h. As shown in Fig. 5, NP1 and Id3 transcripts are present in cells treated with GM containing β-estradiol but not in control cells treated with ethanol. Similarly, both genes are expressed in cells maintained in DM plus β-estradiol, which is a treatment that leads to differentiated myotubes (Fig. 1). As expected, transcripts for the differentiation-specific factor myogenin are detected only in fully differentiated cells (Fig. 5). Although MyoD is known to activate the myogenin promoter (14), this activation occurs only when myoblasts are presented with DM. Cells maintained in GM do not express the myogenin gene.
FIG. 5.
MyoD activates expression of the Id3 and NP1 genes in GM or DM. 10T1/2-MyoD-ER cells were provided GM or DM with or without β-estradiol supplementation for 18 h. RT-PCR analysis confirms that the NP1 and Id3 genes are expressed only in cells supplemented with β-estradiol, regardless of whether the cells are maintained in GM or DM. As expected, the differentiation-specific myogenin gene is expressed only in differentiated cells (DM plus β-estradiol). β-Actin transcripts served as positive controls for the RT reactions. EtOH, ethanol.
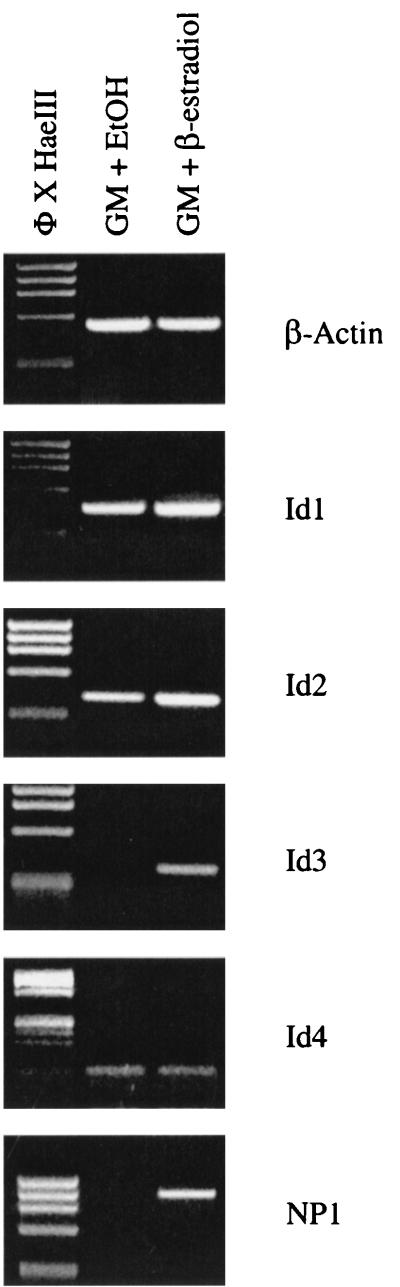
In order to examine if activation of the Id3 gene is unique in the entire Id gene family, we next examined the expression patterns of Id1, Id2, and Id4 in 10T1/2-MyoD-ER cells maintained in GM in the presence or absence of β-estradiol. Whereas Id3 gene transcription occurs only in cells exposed to β-estradiol, expression of the Id1, Id2, and Id4 genes is constitutive in growing cells, regardless of MyoD activity (Fig. 6). These results demonstrate that Id3 is specifically regulated by MyoD in myoblasts and provides further support for the idea that the individual Id family members likely perform distinct biological functions in myogenesis.
FIG. 6.
MyoD transcriptional activity induces Id3 gene expression but does not influence Id1, Id2, and Id4 gene expression. 10T1/2-MyoD-ER cells were maintained in GM containing ethanol (EtOH) or β-estradiol for 24 h, and total RNA was subjected to RT-PCR with Id gene-specific primer sets. Whereas Id3 gene expression is induced in 10T1/2-MyoD-ER cells treated with β-estradiol, the Id1, Id2, and Id4 genes are constitutively active regardless of MyoD activity.
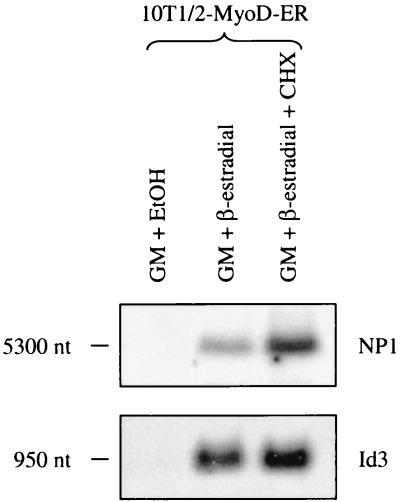
These initial studies suggested that Id3 and NP1 gene expression is a direct result of MyoD activity. To test this hypothesis, cultures were treated with GM containing β-estradiol in the presence or absence of the protein synthesis inhibitor cycloheximide. As shown previously, Id3 and NP1 transcripts are found in 10T1/2-MyoD-ER cells treated with β-estradiol (Fig. 7). However, in cells maintained in GM plus β-estradiol containing cycloheximide, an increase in Id3 and NP1 transcript levels is detected, suggesting that expression of these genes is due to the direct activity of the induced MyoD-ER protein. These results provide the first evidence, aside from MyoD autoactivation, to support the hypothesis that MyoD positively impacts gene expression patterns in undifferentiated myogenic stem cells.
FIG. 7.
Activation of Id3 and NP1 gene expression is directly dependent on the MyoD-ER protein in 10T1/2-MyoD-ER cells. 10T1/2-MyoD-ER cells were maintained in control GM or GM containing β-estradiol with or without cycloheximide (CHX) to inhibit protein synthesis. Northern blot hybridizations reveal that Id3 and NP1 transcripts are only detected in 10T1/2-MyoD-ER cells treated with β-estradiol. Cells provided cycloheximide also accumulate high levels of Id3 and NP1 transcripts, even in the absence of new protein synthesis. These results suggest that MyoD directly activates expression of the Id3 and NP1 genes in this model system. EtOH, ethanol; nt, nucleotides.
Id3 and NP1 gene expression represents a stable, heritable event that is not subject to negative regulation by H-Ras G12V.
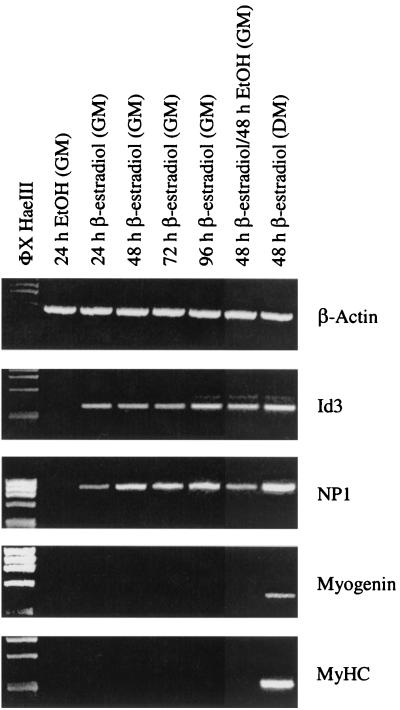
One possibility regarding the MyoD-induced expression of Id3 and NP1 in GM is that the activation of these genes represents an early event in terminal differentiation and therefore is part of a single molecular continuum. In order to examine this in greater detail, we performed a time course experiment in which 10T1/2-MyoD-ER cells were treated with β-estradiol for extended times to see if precocious differentiation would take place. As shown previously, cells maintained in GM and treated with β-estradiol for 24 h express high levels of Id3 and NP1 (Fig. 8). Cells treated with β-estradiol for 48, 72, and 96 h also express Id3 and NP1 transcripts. At each of these time points, there is no evidence of terminal differentiation. Interestingly, cells treated with β-estradiol for 48 h, rinsed extensively, and then maintained in GM without β-estradiol for an additional 48 h continue to express high levels of Id3 and NP1 (Fig. 8). Since the MyoD-ER protein exhibits a very short half-life (∼15 min) (2), these results suggest that MyoD activation of the Id3 and NP1 genes represents a stable, heritable change in gene expression patterns that no longer rely on continued MyoD activity.
FIG. 8.
Activation of the Id3 and NP1 genes by MyoD represents a stable, heritable change in gene expression patterns. 10T1/2-MyoD-ER cells maintained in GM were treated with ethanol (EtOH) for 24 h or with β-estradiol for 24, 48, 72, or 96 h, and total RNA was subjected to RT-PCR analysis. Id3 and NP1 expression is not detected in cells exposed to ethanol. In contrast, cells treated with β-estradiol rapidly activate expression of the Id3 and NP1 genes and expression is maintained throughout the course of the experiment. The expression of Id3 and NP1 represents a stable change in gene expression patterns since cells treated with β-estradiol for 48 h and then maintained in the absence of β-estradiol for an additional 48 h continue to express the Id3 and NP1 genes. Cells treated with β-estradiol in DM were included as a control.
Although it is clear that the Id3 and NP1 genes are expressed following the induction of MyoD in 10T1/2 MyoD-ER cells, it remains possible that this observation is an aberration of this cell line and cannot be duplicated with other myogenic cell model systems. To examine this possibility, we performed RT-PCR analysis on RNA isolated from control 10T1/2-MyoD-ER and parental 10T1/2 cells, as well as from the myogenic cell lines C2C12, 23A2, and L6A1. Once again, Id3 and NP1 transcripts are detected in the 10T1/2-MyoD-ER cells maintained in GM containing β-estradiol but no expression is observed in control 10T1/2 cells (Fig. 9A). Id3 and NP1 transcripts are also readily detected in C2C12, 23A2, and L6A1 myoblasts when the cells are maintained in GM (Fig. 9A). The expression of NP1 and Id3 transcripts in L6A1 cells is of particular interest since these cells express Myf-5 and not MyoD (unpublished results). This observation suggests that other MRFs can substitute for MyoD in activating the NP1 and Id3 genes.
FIG. 9.
The NP1 and Id3 genes are expressed in myogenic stem cell lines and can be induced by transient expression of MRFs in 10T1/2 fibroblasts. (A) RT-PCR analysis of Id3 and NP1 gene expression reveals that these genes are expressed in undifferentiated myoblasts derived from the C2C12, 23A2, and L6A1 cell lines. 10T1/2 and 10T1/2-MyoD-ER cells served as negative and positive controls, respectively. The asterisk denotes a contaminating genomic Id3 fragment found in some samples. (B) 10T1/2 fibroblasts were transfected with a control expression plasmid (pcDNA3) or expression plasmids encoding MyoD and myogenin gene products. RT-PCR analysis reveals that transient expression of MyoD or myogenin in 10T1/2 cells leads to activation of the Id3 and NP1 genes in both undifferentiated and differentiated cells. EtOH, ethanol.
In order to determine if NP1 and Id3 transcripts can be induced by the expression of MRFs in uncommitted cells, control 10T1/2 cells were transfected with expression plasmids encoding MyoD or myogenin and RNA was extracted for analysis by RT-PCR. As expected, 10T1/2 cells transfected with an empty expression plasmid do not express NP1 or Id3 transcripts (Fig. 9B). However, cells expressing MyoD activate expression of the endogenous NP1 and Id3 genes when maintained in GM or when switched to DM. This pattern of gene expression is identical to that observed in the 10T1/2-MyoD-ER cell line when the cells are provided β-estradiol (Fig. 5). Interestingly, expression of myogenin produces the same overall effect in 10T1/2 cells. These results demonstrate that both MyoD and myogenin activate transcription of NP1 and Id3 in 10T1/2 cells under conditions in which the cells are blocked from differentiating, thus identifying NP1 and Id3 as representatives of a potential set of genetic targets transcriptionally regulated by MRFs in committed cells.
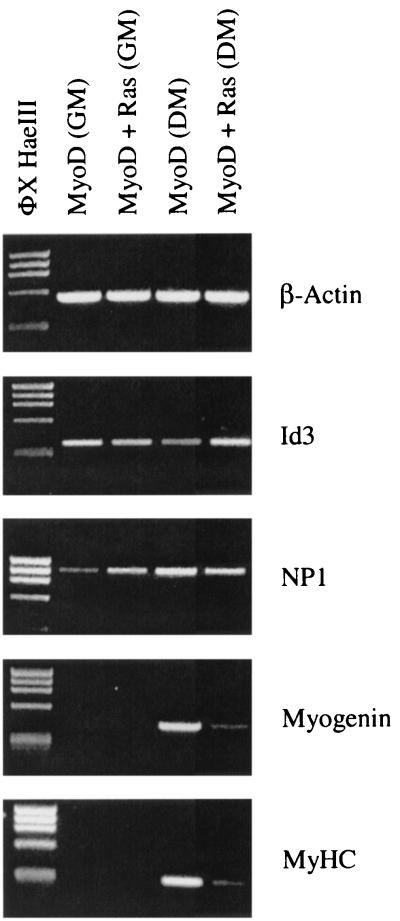
The identity of committed myoblasts versus terminally differentiated muscle cells is an important distinction when defining stem cell populations. Previous studies in our laboratory have shown that expression of a constitutively active H-Ras G-protein can block MyoD-induced terminal differentiation (23, 35), although it was unclear whether H-Ras solely blocks terminal differentiation events or represses the myoblast commitment stage. In order to examine the effect of H-Ras G12V expression on Id3 and NP1 gene activity, 10T1/2 fibroblasts were transfected with expression vectors encoding MyoD, or MyoD plus H-Ras G12V, and then maintained in GM or DM. As shown in Fig. 10, MyoD cells in GM express Id3 and NP1. Similarly, MyoD cells in DM activate expression of the Id3 and NP1 genes, as well as the differentiation-specific genes myogenin and myosin. Interestingly, cells expressing H-Ras G12V continue to express Id3 and NP1 even though H-Ras G12V clearly represses expression of differentiation markers (myogenin and myosin) (Fig. 10), as well as formation of myotubes (data not shown). These results show, for the first time, that H-Ras G12V blocks terminal differentiation events but does not alter the molecular identity of myoblast stem cells.
FIG. 10.
Activation of Id3 and NP1 gene expression by MyoD is not subject to negative regulation by H-Ras G12V. 10T1/2 fibroblasts were transfected with a MyoD or a MyoD plus H-Ras G12V expression plasmid and maintained in GM or DM. RT-PCR analysis confirms that Id3 and NP1 gene expression is induced in MyoD cells in GM or DM. Coexpression of H-Ras G12V has no effect on Id3 and NP1 gene expression. However, H-Ras G12V dramatically inhibits terminal differentiation of MyoD expressing cells maintained in DM, leading to reduced levels of myogenin and myosin gene transcripts, as well as reduced myotube formation (data not shown).
DISCUSSION
The ability of MyoD, Myf-5, myogenin, and MRF4 to convert fibroblasts to skeletal muscle cells stands as one of the most remarkable cellular events described in biology. The concept of a single transcription factor establishing an entirely new developmental program demonstrates the dominant-acting capacity of these regulatory gene products. Genetic, biochemical, and embryological studies have been highly successful in placing the MRFs in a correct developmental scheme, in identifying target DNA sequences, and in providing clues to how the MRF proteins themselves are regulated (reviewed in reference 41).
The main targets for the MRFs are the genes involved in the terminal differentiation process itself, and numerous studies have revealed that activation of these genes occurs only when myoblasts are provided the proper environmental conditions, namely, reduced growth. Myogenic stem cells maintained in a high concentration of serum remain undifferentiated, whereas cells switched to a low concentration of serum generate transcriptionally active MRFs and initiate a complex pattern of gene expression required for sarcomere formation and muscle contraction (1, 15). The tight control of the MRFs is known to occur through a variety of intracellular signal transduction pathways, including regulation of the phosphorylation (16, 29), acetylation (43, 46), and ubiquitination (6, 18, 48) status of the proteins. Posttranslational regulation of the MRFs ensures that myogenic cell differentiation occurs at the correct developmental time and embryological location. Unregulated muscle differentiation can lead to severe abnormalities within the developing embryo.
Given the need to properly control the activity of the MRFs, it remains unclear why MyoD and Myf-5 are expressed in undifferentiated myoblasts. Current models contend that these MRFs are present to allow myoblasts to differentiate rapidly under appropriate environmental conditions. However, it is possible that MyoD (for example) exhibits transcriptional activity in myogenic stem cells and that the correct gene targets for MyoD have not been identified. Indeed, Thayer et al. (50) provided the first evidence that MyoD is transcriptionally active in myoblasts when they reported that exogenously expressed MyoD is capable of activating the endogenous MyoD gene in undifferentiated myoblasts, even though the obvious MyoD target genes (myosin, troponin, and M-creatine kinase) remain transcriptionally silent. The unique activity of MyoD has also been studied by Gerber et al. (14), who showed that MyoD is able to mediate transcriptional activation of muscle-specific genes in repressive chromatin with an efficiency 10-fold greater than that of myogenin. These results demonstrate that the MRFs exhibit distinct molecular activities, as well as unique embryonic expression patterns.
In this study, we have taken advantage of the inducible 10T1/2-MyoD-ER cell line to examine if MyoD is transcriptionally active in myogenic stem cells. Our data demonstrate that the Id3 and NP1 genes are activated by MyoD in undifferentiated myoblasts, prior to overt terminal differentiation. Both genes are transcriptionally silent in control 10T1/2 fibroblasts, in 10T1/2-MyoD-ER cells maintained in the absence of β-estradiol in GM, and in 10T1/2-MyoD-ER cells maintained in the absence of β-estradiol in DM. However, once β-estradiol is provided, effectively activating the constitutively expressed MyoD-ER fusion protein, Id3 and NP1 gene transcription is induced. Expression of NP1 and Id3 is also obtained by transient expression of MyoD or myogenin in 10T1/2 fibroblasts, confirming that the expression of these genes in the 10T1/2-MyoD-ER cell line is not restricted to this experimental system. Indeed, analysis of undifferentiated myoblasts from established myogenic cell lines confirms that the Id3 and NP1 genes are expressed in these myogenic stem cells. Although MyoD is responsible for the initial activation of the Id3 and NP1 genes, continued expression does not require further MyoD activity. These results suggest that activation of the Id3 and NP1 genes represents a stable, heritable event that is associated with myoblast stem cells.
One of the most interesting results of this study is the observation that H-Ras G12V does not alter Id3 and NP1 gene expression although the signaling pathways through which H-Ras functions are potent inhibitors of terminal differentiation events (23, 35). This observation implies that the molecular events by which MyoD activates gene expression programs in myogenic stem cells versus during terminal differentiation are subject to distinct regulatory mechanisms. Indeed, it seems likely that MyoD activity in myoblasts is unaffected by the negative influences of serum growth factors that block terminal differentiation. Understanding these distinct intracellular pathways through which MyoD functions will be an important next step in the characterization of the complete phenotype of myogenic stem cells.
The isolation of the NP1 gene in this study was surprising since NP1 has been shown to be a neuron- and brain-specific gene (12, 47). At the same time, recent investigation of the Myf-5 gene has demonstrated that, in addition to muscle expression, Myf-5 is also expressed in the brain (10). With this in mind, it remains possible that MyoD is expressed in these regions, stimulating NP1 gene expression in the brain, as well as in myogenic stem cells. Since the exact function of NP1 is not fully understood (12, 22, 47), it seems likely that NP1 also serves an important role in myogenic stem cell development. Future studies examining the activity of NP1 should include analyses of skeletal muscle and neuronal tissues.
The identification of Id3 as a MyoD-regulated gene in undifferentiated myoblasts is an intriguing finding. Id3 is expressed in myoblasts and has been reported to function to block MyoD-induced myogenic differentiation of cultured cells (7, 31). However, there is no direct evidence that Id3 blocks the activities of MyoD in proliferating myoblasts. Indeed, members of the Id family of HLH factors are known to function as repressors of terminal differentiation events by forming DNA binding inactive complexes with the normal MyoD bHLH partner proteins E12, E47, ITF-1, and HEB (8, 20, 33). These Id HLH proteins are also known to enhance cellular proliferation, possibly ensuring that cells remain undifferentiated (31, 42). The ability of MyoD to simultaneously activate expression of the Id3 and NP1 genes suggests that Id3 does not inhibit MyoD transcriptional activity in undifferentiated myoblasts and that the functional complex(es) for MyoD in undifferentiated myoblasts and in differentiated myotubes may be quite different. For instance, the preferred complex within differentiated myotubes is a MyoD-E-protein heterodimer (20, 33), and thus, if Id3 functions to sequester the E-protein partners from MyoD, differentiation events are blocked. In contrast, functional MyoD complexes in undifferentiated myoblasts may be significantly different, possibly existing as MyoD homodimers (20) or as heterodimers with unknown protein partners. These complexes may be resistant to the repressor activity attributed to Id3 in differentiated myotubes. Each of these models suggests a mechanism by which MyoD is unaffected by the signaling events (i.e., H-Ras) that block the ability of MyoD to transcriptionally activate differentiation-specific genes. These models also provide a possible mechanism(s) by which MyoD can specify the myoblast lineage through activation of myoblast-specific gene products and, at the same time, ensure that the proper molecular mechanisms are in place to prevent untimely terminal differentiation events. Further studies are needed to identify the mechanism by which MyoD activates expression of the Id3 gene and to understand the developmental consequences of Id3 gene expression in the context of myogenic stem cells.
The revelation that MyoD initiates the expression of genes in proliferating myoblasts is a fundamental step toward understanding the complete myogenic program. Further analyses must be undertaken to identify additional genes that may help to shape the identity of a proliferating myoblast. More importantly, the true regulation of Id3 and NP1 gene expression needs to be investigated and the molecular interactions, cellular localizations, and functions of these proteins should be defined to clarify their roles in myoblast determination. Characterization of the myoblast phenotype in its earliest stages could yield a wealth of information relevant to other types of cellular determination and differentiation events in which lineage-specific bHLH proteins are involved.
Acknowledgments
We thank Stephen Tapscott and Don Bergstrom for generously supplying the 10T1/2-MyoD-ER cell line.
This work was supported by National Institutes of Health grant AR41115 to S.F.K.
REFERENCES
- 1.Andres, V., and K. Walsh. 1996. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J. Cell Biol. 132:657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergstrom, D. A., B. H. Penn, A. Strand, R. L. S. Perry, M. A. Rudnicki, and S. J. Tapscott. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell 9:587-600. [DOI] [PubMed] [Google Scholar]
- 3.Black, B., and E. N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell. Dev. Biol. 14:167-196. [DOI] [PubMed] [Google Scholar]
- 4.Brand-Saberi, B., and B. Christ. 2000. Evolution and development of distinct cell lineages derived from somites. Curr. Top. Dev. Biol. 48:1-42. [DOI] [PubMed] [Google Scholar]
- 5.Braun, T., M. A. Rudnicki, H. H. Arnold, and R. Jaenisch. 1992. Targeted inactivation of the muscle regulatory factor gene Myf-5 results in abnormal rib development and perinatal death. Cell 71:369-382. [DOI] [PubMed] [Google Scholar]
- 6.Breitschopf, K., E. Bengal, T. Ziv, A. Admon, and A. Ciechanover. 1998. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 17:5964-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, B., B. H. Han, X.-H. Sun, and R. W. Lim. 1997. Inhibition of muscle-specific gene expression by Id3: requirement of the C-terminal region of the protein for stable expression and function. Nucleic Acids Res. 25:423-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, B., and R. W. Lim. 1997. Physical and functional interactions between the transcriptional inhibitors Id3 and ITF-2b. J. Biol. Chem. 272:2459-2463. [PubMed] [Google Scholar]
- 9.Dale, K., and O. Pourquie. 2000. A clock-work somite. Bioessays 22:72-83. [DOI] [PubMed] [Google Scholar]
- 10.Daubas, P., S. Tajbakhsh, J. Hadchouel, M. Primig, and M. Buckingham. 2000. Myf5 is a novel early axonal marker in the mouse brain and is subjected to post-transcriptional regulation in neurons. Development 127:319-331. [DOI] [PubMed] [Google Scholar]
- 11.Davis, R. L., P.-F. Cheng, A. B. Lassar, and H. Weintraub. 1990. The MyoD binding domain contains a recognition code for muscle-specific gene activation. Cell 60:733-746. [DOI] [PubMed] [Google Scholar]
- 12.DeGregorio-Rocasolano, N., T. Gasull, and R. Trullas. 2001. Overexpression of neuronal pentraxin 1 is involved in neuronal death evoked by low K+ in cerebellar granule cells. J. Biol. Chem. 276:796-803. [DOI] [PubMed] [Google Scholar]
- 13.Dockter, J. 2000. Sclerotome induction and differentiation. Curr. Top. Dev. Biol. 48:77-127. [DOI] [PubMed] [Google Scholar]
- 14.Gerber, A. N., T. R. Klesert, D. A. Bergstrom, and S. J. Tapscott. 1997. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 11:436-450. [DOI] [PubMed] [Google Scholar]
- 15.Halevy, O., B. G. Novitch, D. B. Spicer, S. X. Skapek, J. Rhee, G. J. Hannon, D. Beach, and A. B. Lassar. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science 267:1018-1021. [DOI] [PubMed] [Google Scholar]
- 16.Hardy, S., Y. Kong, and S. F. Konieczny. 1993. Fibroblast growth factor inhibits MRF4 activity independently of the phosphorylation status of a conserved threonine residue within the DNA-binding domain. Mol. Cell. Biol. 13:5943-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasty, P., A. Bradley, J. H. Morris, D. G. Edmondson, J. M. Venuti, E. N. Olson, and W. H. Klein. 1993. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature 364:501-532. [DOI] [PubMed] [Google Scholar]
- 18.Hatoum, O. A., S. Gross-Mesilaty, K. Breitschopf, A. Hoffman, H. Gonen, A. Ciechanover, and E. Bengal. 1998. Degradation of myogenic transcription factor MyoD by the ubiquitin pathway in vivo and in vitro: regulation by specific DNA binding. Mol. Cell. Biol. 18:5670-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollenberg, S. M., P. F. Cheng, and H. Weintraub. 1993. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc. Natl. Acad. Sci. USA 90:8028-8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadesch, T. 1993. Consequences of heteromeric interactions among helix-loop-helix proteins. Cell Growth Differ. 4:49-55. [PubMed] [Google Scholar]
- 21.Kaul, A., M. Koster, H. Neulhaus, and T. Braun. 2000. Myf-5 revisited: loss of early myotome formation does not lead to a rib phenotype in homozygous Myf-5 mutant mice. Cell 102:17-19. [DOI] [PubMed] [Google Scholar]
- 22.Kirkpatrick, L. L., M. M. Matsuki, D. C. Dodds, and M. S. Perin. 2000. Biochemical interactions of the neuronal pentraxins. J. Biol. Chem. 275:17786-17792. [DOI] [PubMed] [Google Scholar]
- 23.Kong, Y., S. E. Johnson, E. J. Taparowsky, and S. F. Konieczny. 1995. Ras p21val inhibits myogenesis without altering the DNA binding or transcriptional activities of the muscle regulatory factors. Mol. Cell. Biol. 15:5205-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konieczny, S. F., and C. P. Emerson. 1984. 5-Azacytidine induction of stable mesodermal stem cell lineages from 10T1/2 cells: evidence for regulatory genes controlling determination. Cell 38:791-800. [DOI] [PubMed] [Google Scholar]
- 25.Langlands, K., X. Yin, G. Anandi, and E. V. Prochownik. 1997. Differential interactions of Id proteins with basic helix loop helix transcription factors. J. Biol. Chem. 272:19785-19793. [DOI] [PubMed] [Google Scholar]
- 26.Lassar, A., J. N. Buskin, D. Lockshon, R. L. Davis, S. Apone, S. D. Hauschka, and H. Weintraub. 1989. MyoD is a sequence-specific DNA binding protein requiring a region of myc homology to bind to the muscle creatine kinase enhancer. Cell 58:823-831. [DOI] [PubMed] [Google Scholar]
- 27.Lemercier, C., R. Q. To, R. A. Carrasco, and S. F. Konieczny. 1998. The basic helix-loop-helix transcription factor Mist1 functions as a transcriptional repressor of MyoD. EMBO J. 17:1412-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, H., and Y. Capetanaki. 1992. Regulation of the mouse desmin gene: transactivation by MyoD, myogenin, MRF4, and Myf5. Nucleic Acids Res. 21:335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, L., J. Zhou, G. James, R. Heller-Harrison, M. P. Czech, and E. N. Olson. 1992. FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains. Cell 71:1181-1194. [DOI] [PubMed] [Google Scholar]
- 30.Lin, H., and S. F. Konieczny. 1992. Identification of MRF4, myogenin, and E12 oligomer complexes by chemical crosslinking and two-dimensional gel electrophoresis. J. Biol. Chem. 267:4773-4780. [PubMed] [Google Scholar]
- 31.Loveys, D. A., M. B. Streiff, and G. J. Kato. 1996. E2A basic-helix-loop-helix transcription factors are negatively regulated by serum growth factors and by the Id3 protein. Nucleic Acids Res. 24:2813-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludolph, D. C., and S. F. Konieczny. 1995. Transcription factor families: muscling in on the myogenic program. FASEB J. 9:1595-1604. [DOI] [PubMed] [Google Scholar]
- 33.Massari, M. E., and C. Murre. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meinikova, I. N., and B. A. Christy. 1996. Muscle cell differentiation is inhibited by the helix-loop-helix protein Id3. Cell Growth Differ. 7:1067-1079. [PubMed] [Google Scholar]
- 35.Mitin, N., A. J. Kudla, S. F. Konieczny, and E. J. Taparowsky. 2001. Signaling through NFκB is not required for Ras-induced inhibition of skeletal muscle differentiation. Oncogene 20:1276-1286. [DOI] [PubMed] [Google Scholar]
- 36.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 37.Murre, C., P. S. McCaw, and D. Baltimore. 1989. A new binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777-783. [DOI] [PubMed] [Google Scholar]
- 38.Nabeshima, Y., K. Hanaoka, M. Hayasaka, E. Esumi, S. Li, I. Nonaka, and Y. Nabeshima. 1993. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature 364:532-535. [DOI] [PubMed] [Google Scholar]
- 39.Naya, F., and E. N. Olson. 1999. Mef2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr. Opin. Cell Biol. 11:683-688. [DOI] [PubMed] [Google Scholar]
- 40.Olson, E., H. H. Arnold, P. W. J. Rigby, and B. J. Wold. 1996. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85:1-4. [DOI] [PubMed] [Google Scholar]
- 41.Perry, R. L. S., and M. A. Rudnicki. 2000. Molecular mechanisms regulating myogenic determination and differentiation. Front. Biosci. 5:750-767. [DOI] [PubMed] [Google Scholar]
- 42.Peverali, F. A., T. Ramqvist, R. Saffrich, R. Pepperkok, R. V. Barone, and L. Philipson. 1994. Regulation of G1 progression by E2A and Id helix-loop-helix proteins. EMBO J. 13:4291-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polesskaya, A., I. Naguibneva, L. Fritsch, A. Duquet, S. A.-S. Ali, P. Robin, A. Vervisch, L. L. Pritcahrd, P. Cole, and A. Harel-Bellan. 2001. CBP/p300 and muscle differentiation: no HaT, no muscle. EMBO J. 20:6816-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rudnicki, M., T. Braun, S. Hinuma, and R. Jaenisch. 1992. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71:383-390. [DOI] [PubMed] [Google Scholar]
- 45.Rudnicki, M., P. N. J. Schnegelsberg, R. H. Stead, T. Braun, H. H. Arnold, and R. Jaenisch. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75:1351-1359. [DOI] [PubMed] [Google Scholar]
- 46.Sartorelli, V., P.L Puri, Y. Hamamori, V. Ogryzko, G. Chung, Y. Nakatani, J. Y. Wang, and L. Kedes. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4:725-734. [DOI] [PubMed] [Google Scholar]
- 47.Schlimgen, A. K., J. A. Heims, H. Vogel, and M. S. Perin. 1995. Neuronal pentraxin, a secreted protein with homology to acute phase proteins of the immune system. Neuron 14:519-526. [DOI] [PubMed] [Google Scholar]
- 48.Song, A., Q. Wang, M. G. Goebl, and M. A. Harrington. 1998. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol. Cell. Biol. 18:4994-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, X. H., and D. Baltimore. 1991. An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell 64:459-470. [DOI] [PubMed] [Google Scholar]
- 50.Thayer, M. J., S. J. Tapscott, R. L. Davis, W. E. Wright, A. B. Lassar, and H. Weintraub. 1969. Positive autoregulation of the myogenic determination gene MyoD1. Cell 58:241-248. [DOI] [PubMed] [Google Scholar]
- 51.Vaidya, T. B., S. J. Rhodes, E. J. Taparowsky, and S. F. Konieczny. 1989. Fibroblast growth factor and transforming growth factor β repress expression of the myogenic regulatory gene MyoD1. Mol. Cell. Biol. 9:3576-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venuti, J., J. H. Morris, J. L. Vivian, E. N. Olson, and W. H. Klein. 1995. Myogenin is required for late but not early aspects of myogenesis during development. J. Cell Biol. 128:563-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yutzey, K. E., S. J. Rhodes, and S. F. Konieczny. 1990. Differential trans activation associated with the muscle regulatory factors MyoD1, myogenin, and MRF4. Mol. Cell. Biol. 10:3934-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, W., R. R. Behringer, and E. N. Olson. 1995. Inactivation of the myogenic bHLH gene MRF4 results in up-regulation of myogenin and rib abnormalities. Genes Dev. 9:1388-1399. [DOI] [PubMed] [Google Scholar]