Abstract
Protein kinase Cδ (PKCδ) is a member of the PKC family of phospholipid-dependent serine/threonine kinases and is involved in cell proliferation, apoptosis, and differentiation. Previous studies have suggested that different PKC isoforms might be translationally regulated. We report here that the 395-nt-long 5′ untranslated region (5′ UTR) of PKCδ is predicted to form very stable secondary structures with free energies (ΔG values) of around −170 kcal/mol. The 5′ UTR of PKCδ can significantly repress luciferase translation in rabbit reticulocyte lysate but does not repress luciferase translation in a number of transiently transfected cell lines. By using a bicistronic luciferase reporter, we show that the 5′ UTR of PKCδ contains a functional internal ribosome entry segment (IRES). The activity of the PKCδ IRES is greatest in densely growing cells and during apoptosis, when total protein synthesis and levels of full-length eukaryotic initiation factor 4G are reduced. However, the IRES activity of the 5′ UTR of PKCδ is not enhanced during serum starvation, another condition shown to inhibit cap-dependent translation, suggesting that its potency is dependent on specific cellular conditions. Accumulating data suggest that PKCδ has a function as proliferating cells reach high density and in early and later events of apoptosis. Our studies suggest a mechanism whereby PKCδ synthesis can be maintained under these conditions when cap-dependent translation is inhibited.
Protein kinase C (PKC) is a key mediator of signal transduction pathways that regulate a vast array of physiological functions, including cell proliferation, differentiation, and apoptosis (40). Members of the mammalian PKC family of phospholipid-dependent serine/threonine kinases fall into three subfamilies based on their amino acid sequences and enzymatic properties. The conventional PKCs (PKCα, PKCβI, PKCβII, and PKCγ) are activated by Ca2+ and diacylglycerol (DAG), the novel PKCs (PKCɛ, PKCδ, PKCη, and PKCθ) are DAG dependent and Ca2+ independent, and the atypical PKCs (PKCμ, PKCζ, and PKCι/λ) are Ca2+ and DAG independent (7, 33). Although the heterogeneity of PKC function is postulated to be a consequence of the multiple isoforms of PKC that exist within cells, the specific roles of the various isoforms have not been clearly established.
When overexpressed, the novel PKC isoforms PKCɛ and PKCδ have opposing effects on cell growth and proliferation. Overexpression of PKCɛ stimulates growth and promotes transformation of rat-6 fibroblasts, NIH 3T3 fibroblasts, and rat colonic epithelial cells (2, 36, 42). However, overexpression of PKCδ inhibits growth of NIH 3T3 fibroblasts, results in cell cycle arrest of CHO cells, and is lethal to normal and neoplastic keratinocytes (24, 36, 53). PKCɛ behaves as an oncogene because of its ability to induce tumors in nude mice (2). However, PKCδ is considered a tumor suppressor, as its overexpression reverses the transformed phenotype of D/src rat colonic epithelial cells and depletion of PKCδ promotes c-src-induced transformation of rat 3Y1 fibroblasts (28, 43). Moreover, transgenic mice overexpressing PKCδ are resistant to tumor promotion by phorbol esters (45). PKCδ is also implicated in the induction and execution of programmed cell death, or apoptosis, in response to a number of signals, including ionizing radiation, tumor necrosis factor alpha, agonistic Fas antibody (10), H2O2 (51), etoposide (29, 31), and phorbol esters (13, 24).
Most cellular mRNAs possess 5′ untranslated regions (UTRs) of less than 100 nucleotides (nt) that are likely to allow efficient initiation of translation by cap-dependent ribosomal scanning (21). However, the 5′ UTRs of many regulators of cell proliferation and apoptosis, including PKCɛ, are long and structured and contain upstream AUG codons so that scanning ribosomes are unlikely to efficiently initiate translation (22, 39). Efficient initiation of the translation of a number of these mRNAs is achieved by a cap-independent mechanism mediated by internal ribosome entry segments (IRESs) (reviewed in reference 52). Translation mediated by IRESs is usually favored when cap-dependent initiation is inhibited. The IRESs of XIAP, p97/DAP5/NAT1, and c-myc are active during apoptosis (16, 17, 49). The IRES of vascular endothelial growth factor is active during hypoxia (47), and the IRES of ornithine decarboxylase is active during the G2/M phase of the cell cycle (44).
Nucleotide sequencing of PKCδ isolated from rat and mouse cDNA libraries has revealed that it has a long 5′ UTR (35, 41). We show here that the 395-nt-long 5′ UTR of rat PKCδ can potentially form secondary structures with free energies expected to significantly impede cap-dependent ribosome scanning and may thus be subject to translational regulation. We show that the 5′ UTR of PKCδ can repress luciferase translation in rabbit reticulocyte lysate but not in transiently transfected cells. With a bicistronic reporter assay, we show that the 5′ UTR of PKCδ contains an IRES and that the IRES activity of the 5′ UTR of PKCδ is strongest in cells grown to a high density and during apoptosis, conditions under which cap-dependent translation is reduced. However, the IRES activity of the 5′ UTR of PKCδ is not enhanced during serum starvation, another condition known to inhibit cap-dependent translation.
MATERIALS AND METHODS
DNA constructs.
The PKCδ cDNA clone (41) was derived from rat brain and was a gift from Y. Ono (Kobe University, Kobe, Japan). The luciferase reporter plasmids pGL3-promoter (called pGL here) and pGL3-control (called pGLenh here), the pSV-β-galactosidase plasmid, and cloning plasmid pGEM7Zf(−) were purchased from Promega. Restriction and modifying enzymes were purchased from New England Biolabs or Promega and used in accordance with the manufacturer's instructions. Plasmid DNA was prepared for transfection with a plasmid midi prep kit (QIAGEN).
Plasmid constructs for transient transfection were generated as follows. A 3-kb EcoRI fragment comprising the cDNA of PKCδ was subcloned into pGEM7. The 5′ UTR was cloned by PCR with an SP6 primer (5′ end of the 5′ UTR) and a PKCδ-specific primer from the start codon, introducing an NcoI site (CCATGG) as the start codon. The PCR product was cleaved with HindIII/NcoI to release a 390-bp fragment that was subcloned into the pGL3-promoter plasmid and the pGL3-control plasmid to make pGLδ and pGLenh-δ, respectively. The following plasmids were generously provided by A. Willis (Leicester University, Leicester, United Kingdom) and M. Stoneley (Leeds University, Leeds, United Kingdom): pGL3R (called pR-F here), pGL3RutrH (48), pGL3RHRV (called pR-HRV-F here) (49), and pHpL (called pH here) (50). The 5′ UTR of PKCδ was introduced between the Renilla and firefly luciferase coding regions with the EcoRI and NcoI restriction sites to make pR-δ-F. Bicistronic plasmid pGL3RutrH, containing a stable hairpin 5′ of Renilla luciferase, was used to make pHR-δ-F by exchanging the 5′ UTR of c-myc with the 5′ UTR of PKCδ with the SpeI and NcoI restriction sites. The pHpL monocistronic luciferase plasmid, containing a stable hairpin 5′ of the luciferase start, was used to make pH-δ with the EcoRI and NcoI restriction sites.
For the plasmids used for in vitro transcription/translation, HindIII-to-XbaI fragments from the pGL3-promoter plasmid (containing the entire luciferase coding region) and the same restriction fragment from pGLδ (containing the 5′ UTR of PKCδ linked to the luciferase open reading frame) were treated with the Klenow fragment of DNA polymerase to generate blunt ends and subcloned into the SmaI site of the T7 promoter pA90 vector pBKSpA (a gift from James Malter) to make pT7pA and pT7δpA, respectively. The integrity of all clones was confirmed by DNA sequencing.
In vitro transcription and translation.
In vitro-transcribed, capped RNA was made with T7 RNA polymerase (Gibco BRL) in a 20-μl reaction mixture comprising 1 μg of HindIII-linearized pT7pA or pT7δpA plasmid DNA; 0.5 mM (each) CTP, UTP, and ATP; and 0.05 mM GTP with 0.5 mM 7mG cap analogue (Promega) in 1× T7 transcription buffer (40 mM Tris/HCl [pH 8.0], 8 mM MgCl2, 2 mM spermidine, 25 mM NaCl), 5 mM dithiothreitol, and 30 U of RNasin (Promega) at 37°C for 2 h. The reaction mixture was treated with DNase I and purified through G-50 Sephadex columns (Pharmacia) to remove unincorporated nucleotides and free 7mG cap. The RNA was quantified spectrophotometrically, and its quality was verified on nondenaturing agarose gels. Equal amounts of RNA (typical final concentration, 20 ng/μl) were used in a 25-μl in vitro translation reaction mixture comprising 70% rabbit reticulocyte lysate (Promega), 0.02 mM each amino acid, and 30 U of RNasin. The in vitro translation was allowed to proceed at 28°C for 90 min, and the reaction mixture was finally treated with 1 μl of RNase A at 10 mg/ml for 10 min at 28°C. One microliter of each reaction mixture was used in a luciferase assay (with 50 μl of luciferase assay reagent [described below]). A 1-μl sample was taken at the start and end of the translation reaction, and the RNA was extracted for Northern blot analysis. The translation reactions were done at least five times with different batches of in vitro-transcribed RNA and rabbit reticulocyte lysate.
Cell culture and transient transfection.
The 3T3 Swiss albino (mouse fibroblast), 3T6 Swiss albino (mouse fibroblast), BALB/3T12-3 (mouse tumorigenic fibroblast), C6 (rat glioma), HeLa (human cervical carcinoma), and MCF7 (human breast carcinoma) cell lines were purchased from ECACC (Salisbury, United Kingdom) and maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle medium (DMEM; Gibco BRL, Life Technologies) containing penicillin/streptomycin and supplemented with 10% fetal bovine serum (FBS; Gibco BRL, Life Technologies). For a typical transient transfection, cells were grown to 50 to 70% density in six-well plates and then cotransfected with 1 μg of pSV-β-galactosidase plasmid and 1 μg of the luciferase reporter plasmid with 10 μl (30 μg) of Superfect reagent (QIAGEN) in accordance with the manufacturer's instructions. Low, medium, and high cell densities were achieved by seeding cells at 4 × 103/cm2 (approximately 10% density), 2 × 104/cm2 (approximately 50% density), or 4 × 104/cm2 (approximately 100% density), respectively. Cells were transfected at these densities and then harvested after 24 h. For serum starvation experiments, cells growing at 50 to 70% density in 10% FBS-containing DMEM were transfected, and then 6 h later, the cells were washed twice with serum-free DMEM and grown for a further 24 h in serum-free DMEM before being harvested. To induce apoptosis, 0.5 μg of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) was added per ml of medium 24 h after transfection and the cells were harvested 6 h later. For the RNA analysis, a 75-cm2 flask of cells was transfected for each construct with 10 μg of plasmid DNA and 15 μl of FuGene 6 (Roche) in accordance with the manufacturer's instructions.
Enzyme assays.
For the monocistronic luciferase assays, cells were harvested by being scraped into 120 μl of reporter lysis buffer and assayed with the Luciferase Assay System (Promega) in accordance with the manufacturer's instructions. For the bicistronic luciferase assays, cell were harvested with 120 μl of passive lysis buffer and assayed with the Dual Luciferase Reporter Assay System (Promega) in accordance with the manufacturer's instructions. The peak light emission for 10 s at 25°C due to luciferase activity was measured as luciferase relative light units (LRLUs) with a Berthoid luminometer. β-Galactosidase activity was measured with the substrate o-nitrophenyl-β-D-galactopyranoside (ONPG). β-Galactosidase activity absorbances were measured at 420 nm. For each transfection, the luciferase activity is expressed relative to the β-galactosidase activity to control for transfection efficiency. The mean values of relative luciferase activity shown in Results were derived from the analysis of at least five independent cotransfections of each luciferase plasmid with pSV-β-galactosidase with at least two different preparations of plasmid DNA.
RNA analysis.
RNA secondary structures were predicted with mFOLD (32), which is available at http://bioinfo.math.rpi.edu/∼mfold/rna/form1.cgi. Total RNA was extracted with the Trizol reagent (Gibco BRL, Life Technologies) and treated with RQ1 DNase (Promega). Polyadenylated RNA was purified with oligo(dT) cellulose with the PolyA-spin mRNA isolation kit (New England Biolabs). RNA from transiently transfected cells [total: 20 μg of luciferase RNA, 10 μg of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA, or 1 μg of poly(A) RNA] was denatured with formamide/formaldehyde and subjected to electrophoresis on a 1% agarose-6% formaldehyde-morpholinepropanesulfonic acid (MOPS) gel. The RNA was transferred onto Hybond N+ membranes (Amersham). The membranes were hybridized with [α-32P]UTP (Amersham Radiochemicals)-labeled single-stranded RNA antisense probes derived from the entire coding regions of firefly luciferase or GAPDH (12) in ULTRAhybe hybridization buffer (Ambion). The luciferase and GAPDH RNA signals were quantified by phosphorimaging with a Molecular Dynamics phosphorimager with ImageQuant (version 5.1) software.
PKCδ and GAPDH RNA levels were assessed in cells growing at different densities with semiquantitative reverse transcription (RT)-PCR. One microgram of total RNA was reverse transcribed with 0.5 μg of 5′-pd(T)12-18-3′ (Pharmacia) with SuperScript II reverse transcriptase (Gibco BRL, Life Technologies) in a 25-μl reaction volume in accordance with the manufacturer's instructions. The PCR was performed in a 50-μl volume with 2 μl of the RT reaction mixture with Taq polymerase (Promega) in accordance with the manufacturer's instructions. The number of amplification cycles chosen for analysis of PKCδ and GAPDH RNA levels by semiquantitative PCR was determined for each primer pair to be within the exponential range for cells growing at different densities. PKCδ was amplified with primers specific to the C-terminal variable region (sequences: 5′ GAC ATC ATG GAG AAG CTA TTC and 5′ TCG ATG AGG TTC TTG TCA CTG) with 20 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, and GAPDH was amplified with specific primers (sequences: 5′ AGA CAG CCG CAT CTT CTT GTG C and 5′ CTC CTG GAA GAT GGT GAT GG) with 15 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C, for 1 min. Ten microliters of the PCR mixture was electrophoresed on 2% agarose gels and stained with ethidium bromide. The intensity of ethidium bromide staining was quantified with the AlphaImager 1200 documentation and analysis system (Alpha Innotech Corporation). The specificity of the PCR products was confirmed by Southern blotting with oligonucleotide probes designed to hybridize to internal sequences of the predicted PCR products (data not shown).
Metabolic labeling of cells.
Cells were grown in serum-free methionine/cysteine-free DMEM for 30 min prior to the addition of 60 μCi of [35S]Met/Cys (Promix; Amersham) per ml and incubation for 1 h. Cells were then washed twice with phosphate-buffered saline and harvested in lysis buffer A (150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl, 1 mM EDTA, pH 8.0) containing 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, and 2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF). Total protein concentrations were estimated with the bicinchoninic acid assay (Pierce), and equal amounts of protein were added to Laemmli loading buffer and heated to 100°C for 5 min before being subjected to denaturing polyacrylamide gel electrophoresis with 10% polyacrylamide gels. Proteins were transferred to Hybond-C (Amersham) membranes. The membranes were stained with Ponceau (Sigma) to visualize the protein, individual lanes were excised, and the amount of incorporated 35S in each lane was quantified by scintillation counting.
Immunoblotting.
Total protein was extracted from cells by scraping them into lysis buffer A containing 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, and 2 mM AEBSF for eukaryotic initiation factor 4G (eIF4G) or by scraping them into lysis buffer B (0.5% NP-40, 50 mM Tris, 0.5 mM EDTA, 2 mM EGTA, pH 7.5) containing 10 μg of aprotinin per ml, 10 μg of leupeptin per ml, and 2 mM AEBSF for PKCδ. Samples containing 30 μg of protein were subjected to denaturing polyacrylamide gel electrophoresis and transferred to Hybond-C as described above. The membranes were incubated with polyclonal rabbit antiserum against PKCδ (used at 1:5,000; a gift from N. Groom, Oxford Brookes University), with polyclonal rabbit antiserum against the C terminus (amino acids 920 to 1396) of eIF4GI (used at 1:2,000; a gift from S. Morley, Sussex University), or with polyclonal rabbit antiserum against poly(ADP-ribose) polymerase (PARP), which detects both the 116-kDa full-length PARP molecule and the 89-kDa caspase cleavage product (used at 1:1,000; purchased from Cell Signaling Technology). The membranes were then incubated with an anti-rabbit peroxidase-conjugated secondary antibody (used at 1:5,000; Sigma). Immunoreactive bands were detected by chemiluminescence assay.
RESULTS
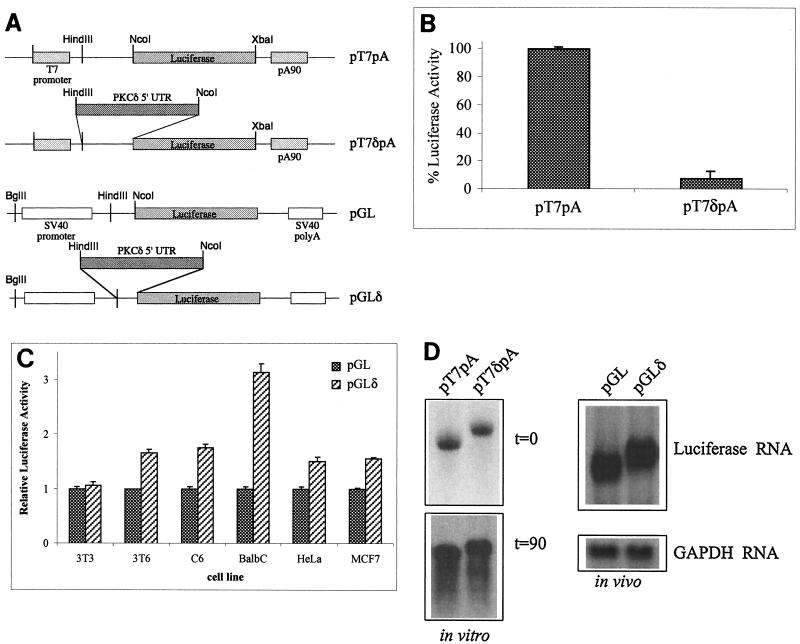
The 5′ UTR of PKCδ is predicted to form highly stable secondary structures and is resistant to cap-dependent translation in vitro but not in vivo.
The 5′ UTR of PKCδ isolated from a rat brain cDNA library is 395 nt long and has a G+C content of 68%. With the computer algorithm of Zuker (32), which estimates the minimum free energy of a folded RNA molecule, the 5′ UTR of PKCδ was predicted to form highly stable secondary structures with free energies (ΔG values) exceeding −170 kcal/mol (data not shown). As an RNA stem-loop structure with a free energy of −50 kcal/mol is inhibitory to cap-dependent ribosomal scanning (20), it was expected that the 5′ UTR of PKCδ would inhibit cap-dependent translation initiation. To test this, the 5′ UTR of PKCδ was introduced immediately upstream of the luciferase start in either T7 RNA polymerase promoter-based plasmid pT7pA or simian virus 40 promoter-based plasmid pGL to create pT7δpA and pGLδ, respectively, as shown in Fig. 1A. Capped and polyadenylated luciferase RNA generated in vitro from pT7δpA with T7 RNA polymerase was poorly translated in rabbit reticulocyte lysate compared to luciferase RNA generated from the control, pT7pA (Fig. 1B). In transiently transfected Swiss 3T3 cells, the levels of luciferase conferred by the pGL and pGLδ constructs were similar (Fig. 1C). However, in transiently transfected Swiss 3T6, C6, BALB/c, HeLa, and MCF7 cells, the pGLδ construct conferred significantly more luciferase activity than did pGL (t test, P < 0.01) (Fig. 1C). Northern blot analysis revealed that comparable amounts of luciferase-specific RNA of the expected size could be recovered after 90 min from the in vitro translation reactions programmed with in vitro-transcribed pT7pA and pT7δpA (Fig. 1D). In transiently transfected cells, the pGL and pGLδ constructs produced similar levels of luciferase mRNA of the expected size. These results suggest that, in contrast to the pT7pA and pGL constructs, the pT7δpA and pGLδ constructs do not affect steady-state levels of luciferase mRNA, nor are they likely to contain splicing sequences. Taken together, these data suggest that the 5′ UTR of PKCδ can repress luciferase translation in rabbit reticulocyte lysate but enhance translation of luciferase in several cell lines.
FIG. 1.
The 5′ UTR of PKCδ represses translation in vitro but not in vivo. (A) Schematic diagram of the T7 RNA polymerase-and pGL3-promoter-based luciferase constructs used for in vitro and in vivo analyses, respectively. The 5′ UTR of PKCδ was introduced immediately upstream of the luciferase start codon in the pGL3-promoter plasmid with the HindIII and NcoI restriction sites to make pGLδ. The luciferase-coding region from pGL and the 5′ UTR of PKCδ linked to luciferase from pGLδ were introduced into the pBKSpA plasmid to make pT7pA and pT7δpA, respectively, as described in Materials and Methods. (B) Capped and polyadenylated in vitro-transcribed RNAs were generated with T7 RNA polymerase from the linearized pT7pA and pT7δpA plasmids. Equal amounts of purified RNA were used to program rabbit reticulocyte in vitro translation reactions, which were subsequently assayed for luciferase activity. The luciferase activity conferred by pT7pA was normalized to 100%. Results shown are the mean and standard error of the mean from at least five independent translation reactions with different in vitro-transcribed RNAs. (C) Rodent and human cell lines were cotransfected with either the pGL or the pGLδ luciferase construct and pSV-β-gal to control for transfection efficiency. For each cell line, the luciferase activity shown is relative to the β-galactosidase activity conferred by pSV-β-gal. The relative luciferase/β-galactosidase activity conferred by pGL is normalized to a value of 1 for each cell line. Results are shown as the mean and standard error of the mean of at least five independent transfections. (D) Northern blot of pT7pA and pT7δpA luciferase RNAs during in vitro translation (top left, 0-min time point; bottom left, 90-min time point). Northern blot of RNA derived from cells transfected (in vivo) with the pGL and pGLδ constructs and hybridized with a 32P-labeled firefly luciferase (top right) or GAPDH (bottom right) riboprobe. SV40, simian virus 40.
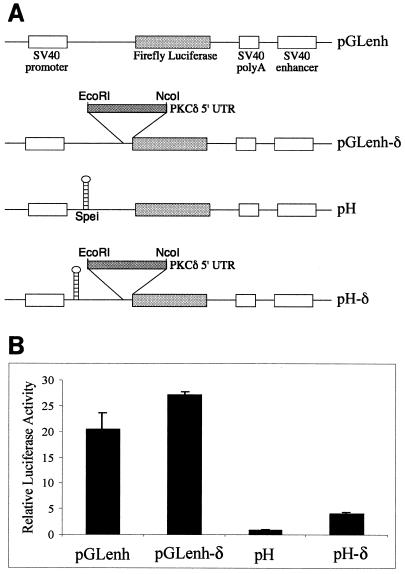
The 5′ UTR of PKCδ enhances translation when cap-dependent ribosomal scanning is inhibited.
HeLa cells were transiently transfected with a construct carrying a stable hairpin structure designed to block cap-dependent translation (Fig. 2A). Luciferase activity conferred by the pH construct containing the stable hairpin structure was 5% of that conferred by the control, pGLenh (Fig. 2B). The pH-δ construct carrying the 5′ UTR of PKCδ between the stable hairpin and the translation start site conferred fourfold more luciferase activity than did pH, suggesting that the 5′ UTR of PKCδ can promote translation initiation when cap-dependent scanning is inhibited (Fig. 2B). However, luciferase activity conferred by the pH-δ construct was around fivefold less than that conferred by the control, pGLenh, and more than sixfold less than that conferred by pGLenh-δ. This suggests that a component of translation initiation by the 5′ UTR of PKCδ must be cap dependent and has been inhibited by the introduced cap-proximal hairpin. Alternatively, the cap-proximal hairpin may have interfered with the function of the 5′ UTR of PKCδ in promoting cap-independent initiation.
FIG. 2.
The 5′ UTR of PKCδ enhances translation when cap-dependent scanning is inhibited in a monocistronic construct. (A) Schematic diagram of the constructs used. The 5′ UTR of PKCδ was introduced immediately upstream of the luciferase open reading frame in the pGL3-control plasmid (called pGLenh) to create pGLenh-δ. The stable hairpin structure with a free energy (ΔG) of −55 kcal/mol was introduced 5′ of the firefly luciferase start in the pGLenh plasmid to create pH as previously described (50). The 5′ UTR of PKCδ was introduced between the hairpin and the luciferase start to create pH-δ as shown. (B) Luciferase activities conferred by the monocistronic hairpin constructs in transiently transfected HeLa cells relative to the β-galactosidase activity conferred by pSV-β-gal to control for transfection efficiency. The relative luciferase or β-galactosidase activity conferred by the pH construct is normalized to a value of 1. The results shown are the mean and standard error of the mean of at least five independent transfections. SV40, simian virus 40.
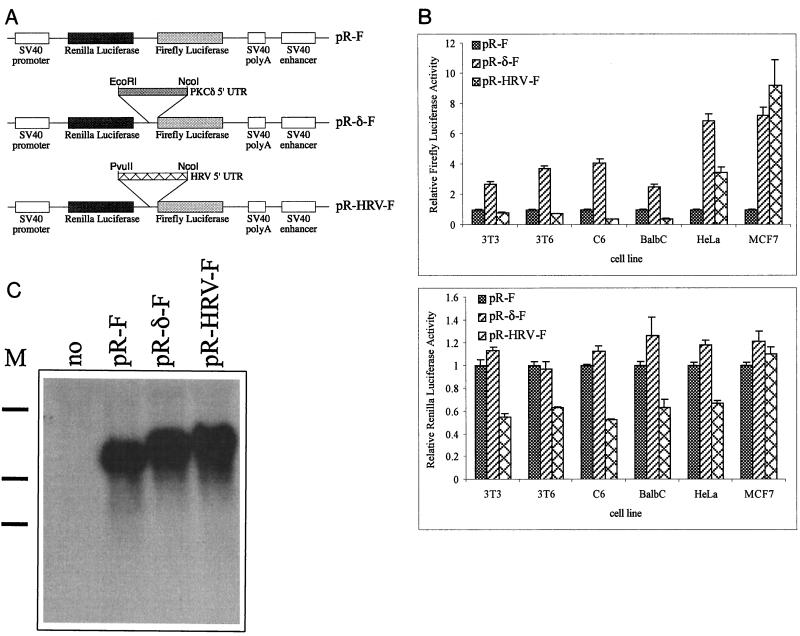
The 5′ UTR of PKCδ functions as an IRES in a bicistronic construct.
The 5′ UTR of PKCδ was introduced between the Renilla and firefly luciferase coding regions in a bicistronic reporter plasmid to test if it could function as an IRES in vivo (Fig. 3A). The human rhinovirus (HRV) IRES was included in the transfections as a positive control (1, 49). In all of the cell lines tested, the presence of the 5′ UTR of PKCδ in the intergenic region resulted in downstream (firefly) luciferase activity two- to sevenfold greater than that of the control, pR-F (Fig. 3B). There were no significant differences in the activity of the upstream (Renilla) luciferase conferred by the pR-F and pR-δ-F constructs in any of the transiently transfected cells (Fig. 3B). The 5′ UTR of PKCδ was at least as efficient as the HRV IRES in initiating translation of the downstream luciferase in cells of human origin and was at least twofold more efficient in cells of rodent origin. The lack of activity of the HRV IRES in nonhuman cells is consistent with previous studies (1). Northern blot analysis of mRNA isolated from HeLa cells transiently transfected with the control, PKCδ, and HRV bicistronic constructs revealed an intact single species of luciferase-specific mRNA migrating at the expected size (Fig. 3C). This supports the notion that the 5′ UTRs of PKCδ and HRV neither alter the steady-state levels of the bicistronic RNA nor result in cleavage of the bicistronic RNA into a monocistronic firefly luciferase.
FIG. 3.
The 5′ UTR of PKCδ acts as an IRES in a bicistronic construct. (A) Schematic diagram of bicistronic Renilla and firefly luciferase constructs used to test for IRES activity in transiently transfected cells. The 5′ UTR of PKCδ was inserted between the coding regions of the Renilla and firefly luciferases in the pR-F vector to make pR-δ-F as detailed in Materials and Methods. The pR-HRV-F construct containing the HRV IRES was used as a positive control (49). (B) Firefly and Renilla luciferase activities conferred by the bicistronic constructs in transiently transfected rodent and human cell lines relative to the β-galactosidase activity conferred by pSV-β-gal. For each cell line, the relative firefly and Renilla luciferase activities conferred by pR-F are normalized to a value of 1. The results shown are the mean and standard error of the mean from at least five independent transfections. (C) Northern blot analysis of 1 μg of poly(A) RNA isolated from HeLa cells transiently transfected with no DNA or the pR-F, pR-δ-F, or pR-HRV-F construct and hybridized with a firefly luciferase-specific 32P-labeled riboprobe. The approximate positions (from top of gel to bottom) of 5-, 3-, and 2-kb RNA markers (M) are indicated on the left.
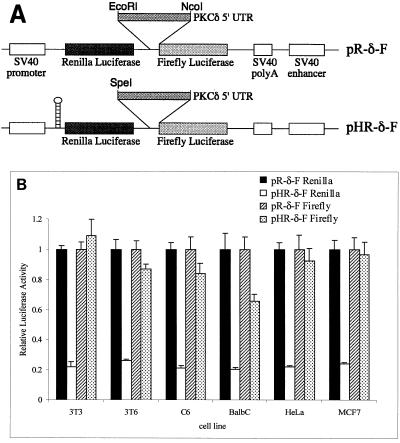
The 5′ UTR of PKCδ does not promote ribosomal readthrough in the bicistronic construct.
The 5′ UTR of PKCδ may function in the bicistronic construct by stimulating readthrough of ribosomes through the intergenic region. If this were the case, it would be expected that, in the presence of a stable hairpin that inhibits translation from the upstream cistron, the level of luciferase activity from the downstream cistron would be proportionally decreased in constructs carrying the 5′ UTR of PKCδ as the intergenic spacer. We found that in the control bicistronic construct, which lacks an intergenic spacer, a stable hairpin structure inserted 5′ of the Renilla coding region reduced translation of both luciferase cistrons to around 20% of the levels conferred by the control lacking the hairpin (reference 48 and data not shown). We compared the Renilla and firefly luciferase activities in the bicistronic construct carrying the 5′ UTR of PKCδ as the intergenic spacer in the absence and presence of a stable hairpin 5′ of the upstream cistron (Fig. 4A). Although Renilla luciferase activity was significantly reduced in all of the cell lines tested, there was no significant reduction in firefly luciferase activity (Fig. 4B). This confirms that the 5′ UTR of PKCδ does not function by enhancing readthrough of the bicistronic construct and supports the notion that the 5′ UTR of PKCδ contains a bona fide IRES.
FIG. 4.
The 5′ UTR of PKCδ does not promote ribosomal readthrough in the bicistronic construct. (A) Schematic diagram of the bicistronic constructs used for transient transfections. The stable hairpin structure with a free-energy of −55 kcal/mol introduced 5′ of the Renilla luciferase start (48) was used to create pHR-δ-F. (B) Renilla and firefly luciferase activities conferred by the bicistronic pR-δ-F and pHR-δ-F constructs in transiently transfected rodent and human cell lines relative to the β-galactosidase activity conferred by pSV-β-gal. For each cell line, the relative Renilla and firefly luciferase activities conferred by the pR-δ-F construct were normalized to a value of 1. The results shown are the mean and standard error of the mean from at least five independent transfections.
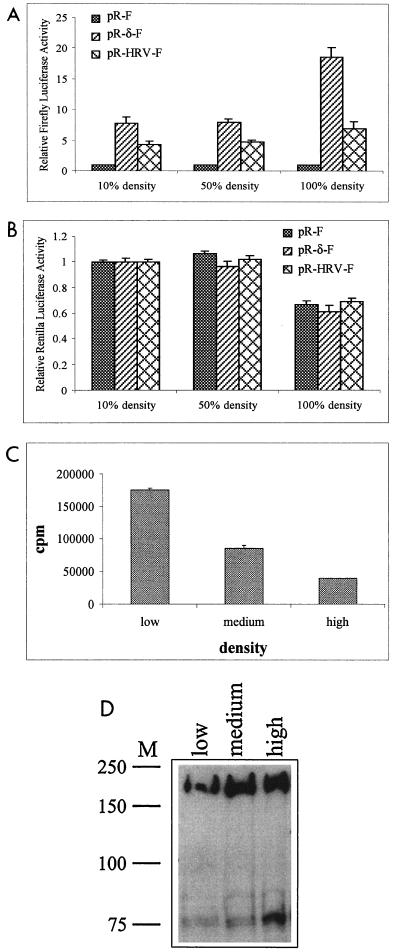
The IRES activity of the 5′ UTR of PKCδ is greatest in cells growing at a high density, when cap-dependent translation is reduced.
Previous studies have indicated that IRES activity of a 5′ UTR can promote translation initiation during times of cellular stress and other occasions when cap-dependent translation is inhibited. Cells grown to a high density, or confluency, enter G0 and growth arrest, which is marked by changes in the expression of many cellular factors (46, 54). PKCδ protein levels are maximal when RPTE cells, C6 rat glioma cells, and 3T6 fibroblasts are grown to a high density rather than a subconfluent cell density (9, 27, 38). We therefore speculated that the IRES activity of the 5′ UTR of PKCδ might be required to translate the mRNA when cells are grown to a high density. To test this, HeLa cells were transiently transfected with the control, PKCδ, and HRV bicistronic constructs at low, medium, and high cell densities. At low and medium cell densities, the level of firefly luciferase activity conferred by pR-δ-F was around eightfold greater than that of the control, pR-F (Fig. 5A). However, when cells were transfected at a high density, the level of firefly luciferase activity conferred by pR-δ-F was 18-fold greater than that conferred by the control. Elevated firefly luciferase activity conferred by pR-δ-F compared to the control was also observed when the cells were transfected at a medium density and then harvested after growth to a high density (data not shown). There was a modest, but not statistically significant, improvement in the IRES activity of the HRV construct in cells growing at a high density compared to that of cells growing at a medium density. These data therefore suggest that the IRES activity of the 5′ UTR of PKCδ is specifically elevated in cells growing at a high density. It is likely that cap-dependent translation is reduced in cells growing at a high density, as the relative amount of the upstream (Renilla) luciferase was significantly reduced under these growth conditions compared to that in low- and medium-density cells (Fig. 5B). However, it is also possible that the half-life of Renilla luciferase is shorter in cells growing at a high density and that cap-dependent translation is not reduced in densely growing cells. To test whether cap-dependent translation is reduced in densely growing cells, HeLa cells growing at different densities were labeled with [35S]methionine for 1 h prior to being harvested (Fig. 5C). Analysis of the cell lysates revealed reduced incorporation of [35S]methionine at a high cell density, supporting the idea that global cell protein synthesis is reduced. A significant reduction in global protein synthesis was also observed in 3T6 cells grown to a high density (data not shown). Proteolytic cleavage of eIF4G has previously been observed under conditions in which global translation is reduced (5, 30). As HeLa cells grow to a high density, immunoblotting for eIF4G revealed a predicted 76-kDa cleavage product (Fig. 5D). Taken together, these results suggest that when cells are growing at a high density, global translation is reduced and the IRES activity of the 5′ UTR of PKCδ is significantly enhanced.
FIG. 5.
The IRES activity of the 5′ UTR of PKCδ is greatest in densely growing cells. (A) Firefly luciferase activities conferred by the bicistronic pR-F, pR-δ-F, and pR-HRV-F constructs in HeLa cells transiently transfected at approximately 10, 50, and 100% cell density relative to the β-galactosidase activity conferred by pSV-β-gal. Relative firefly luciferase activity conferred by pR-F is normalized to a value of 1 for each cell density. (B) Relative Renilla luciferase activity conferred by the bicistronic constructs, normalized to a value of 1 for each construct at 10% cell density. The results shown are the mean andstandard error of the mean from at least five independent transfections. (C) Global protein synthesis was determined by labeling HeLa cells growing at a low (10 to 20%), medium (50 to 70%), or high (90 to 100%) density with [35S]methionine as described in Materials and Methods. (D) Total protein extracted from cultures parallel to those in panel C were electrophoresed through SDS-10% acrylamide gels, and the proteins were transferred to nitrocellulose and probed with antisera raised against eIF4G. The approximate locations of protein molecular size markers (M) in kilodaltons are indicated at the left.
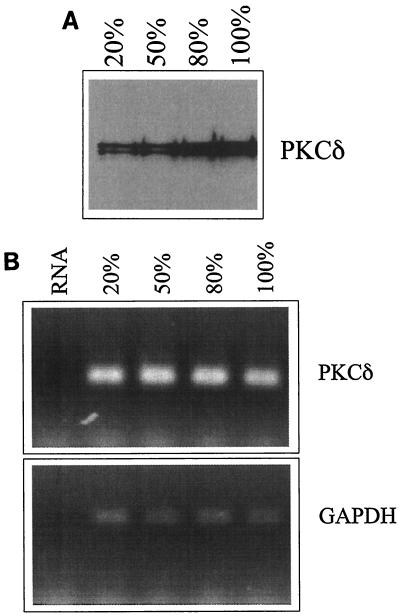
Endogenous PKCδ protein levels are elevated as cells reach a high density, while its mRNA levels do not change.
To determine the status of endogenous PKCδ in HeLa cells growing at different densities, we performed immunoblotting and semiquantitative RT-PCR. Immunoblotting revealed that PKCδ protein levels peak in cells growing at a maximal density (Fig. 6A), while there was no change in the PKCδ mRNA levels (Fig. 6B). This suggests that as HeLa cells grow to a high density, endogenous PKCδ protein levels are modulated posttranscriptionally.
FIG. 6.
Levels of endogenous PKCδ protein, but not its mRNA, increase in densely growing cells. (A) Equal amounts of total protein extracted from HeLa cells growing at approximately 20, 50, 80, and 100% density were electrophoresed through SDS-10% acrylamide gels and then transferred to nitrocellulose and probed with antiserum against PKCδ. (B) PKCδ RNA levels were determined with semiquantitative RT-PCR with total RNA isolated from HeLa cells growing at approximately 20, 50, 80, and 100% density (upper band, PCR product; lower band, primer). The same reverse-transcribed RNA samples were amplified with primers specific for GAPDH. A negative control of amplified RNA is shown in the left lane of each gel.
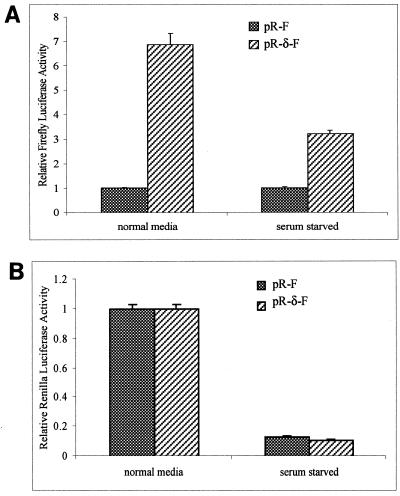
The IRES activity of the 5′ UTR of PKCδ is not enhanced by serum starvation but is enhanced during apoptosis.
Previous studies have demonstrated that serum starvation and apoptosis reduce global translation (5, 49). We therefore examined whether the IRES activity of the 5′ UTR of PKCδ is improved under these conditions of cellular stress. Cells were transfected with the bicistronic constructs and then subjected to serum starvation or apoptosis induced by TRAIL. During serum starvation, there was a significant reduction in both firefly (cap-independent) and Renilla (cap-dependent) luciferase activities (Fig. 7A and B). This suggests that the IRES activity of the 5′ UTR of PKCδ is diminished during serum starvation.
FIG. 7.
The IRES activity of the 5′ UTR of PKCδ is not enhanced in serum-starved cells. (A) HeLa cells were grown in 10% FBS-containing DMEM to approximately 50% density and transiently transfected with the bicistronic pR-F or pR-δ-F construct and pSV-β-gal to control for transfection efficiency. After 6 h, the cells were washed twice with serum-free DMEM and incubated in serum-free DMEM for a further 24 h before being harvested and assayed for firefly luciferase and β-galactosidase activities. Relative firefly luciferase activity conferred by pR-F is normalized to a value of 1 for each condition. (B) Relative Renilla luciferase activities from the transfected cells described above were determined and normalized to a value of 1 for each construct in cells growing in normal medium. The results shown are the mean and standard error of the mean from at least five independent transfections.
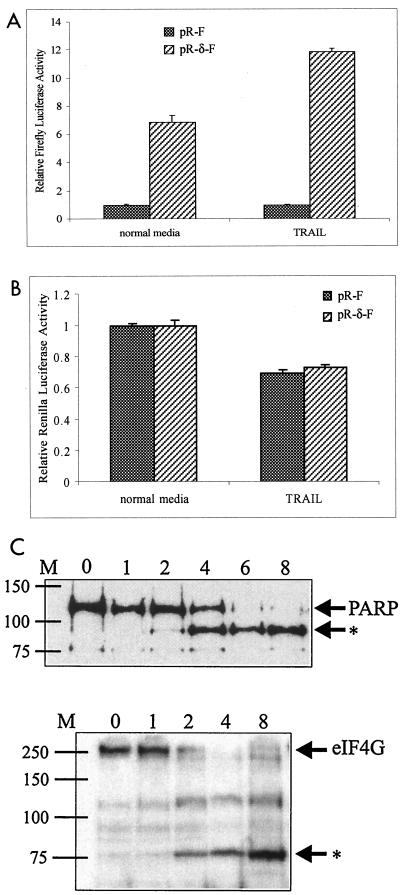
The IRES activity of the 5′ UTR of c-myc is maintained during apoptosis induced by TRAIL (49). In HeLa cells transfected with pR-δ-F, we observed a significant improvement in relative firefly luciferase activity, compared to that of the pR-F control, 6 h after 0.5 μg of TRAIL per ml was added to the culture medium (Fig. 8A). Cap-dependent initiation was modestly but significantly reduced, as shown by analysis of the Renilla luciferase activity (Fig. 8B). Induction of apoptosis by 0.5 μg of TRAIL per ml is clearly evident after 4 h by the appearance of the 89-kDa caspase-specific cleavage product of PARP (Fig. 8C). Further evidence that TRAIL reduces cap-dependent translation is shown by a time-dependent reduction of the full-length eIF4G-specific signal and the appearance of the 76-kDa cleavage product 2 h after the addition of TRAIL to HeLa cells (Fig. 8C). These analyses suggest that the IRES activity of the 5′ UTR of PKCδ is enhanced under specific, rather than general, cellular conditions that reduce cap-dependent translation.
FIG. 8.
The IRES activity of the 5′ UTR of PKCδ is enhanced in cells treated with TRAIL to induce apoptosis. (A) HeLa cells were transiently transfected with the bicistronic pR-F or pR-δ-F construct and pSV-β-gal to control for transfection efficiency. After 24 h, 0.5 μg of TRAIL per ml was added to the medium and the cells were harvested 6 h later. Relative firefly luciferase activities are shown, with the levels conferred by pR-F normalized to a value of 1. (B) Relative Renilla luciferase activities from the transiently transfected cells above were determined and normalized to a value of 1 for each construct in cells in normal medium. (C) Equal amounts of total protein extracted from HeLa cells growing in the presence of 0.5 μg of TRAIL per ml for 0, 1, 2, 4, 6, and 8 h were electrophoresed through SDS-10% acrylamide gels and then transferred to nitrocellulose and probed withantisera against PARP or eIF4G. The positions of full-length PARP and eIF4G are indicated, and the position of specific cleavage products is shown by the asterisk. The approximate locations of protein molecular size markers (M) in kilodaltons are indicated at the left.
DISCUSSION
PKCδ has been implicated in the response of cells to a myriad of signals, including those that promote differentiation, cell transformation, growth arrest, and apoptosis (8, 10, 25, 37, 43). While many studies have addressed the posttranslational activation and action of PKCδ in cells overexpressing native or dominant-negative forms of PKCδ, relatively little is known of the mechanisms that result in de novo PKCδ synthesis.
Most vertebrate transcripts contain 5′ UTRs 20 to 100 nt long that are unlikely to impede translation initiation from the AUG codon (21). We report here that the 395-nt-long 5′ UTR of PKCδ is potentially highly structured and has the ability to significantly repress the translation of a linked luciferase reporter gene with rabbit reticulocyte lysate as a source of translation factors. From this, we speculate that PKCδ might be translationally regulated in cells. In support of this, PKCδ was recently identified as a translationally regulated gene in a differential screen comparing resting and antigen-activated human T lymphocytes (34). An in vivo study suggests that PKCδ might be posttranscriptionally regulated in response to experimentally induced colitis in the rat as PKCδ protein levels increase without an accompanying increase in mRNA abundance (4). We also report here that the levels of PKCδ protein increase as HeLa cells grow to a high density, while its mRNA levels remain unchanged. This is consistent with other studies comparing PKCδ protein and mRNA levels in confluent and subconfluent cultures of RPTE and 3T6 cells (9, 27) and may thus represent a general feature of PKCδ function in response to cell density-dependent growth.
A subset of transcripts that encode growth factors, signaling molecules, and other genes involved in cell proliferation and apoptosis possess long, structured 5′ UTRs that function as IRESs in a bicistronic assay (reviewed in reference 3). We provide evidence here that the 5′ UTR of PKCδ acts as an IRES both in a monocistronic construct, where cap-dependent initiation is blocked by the introduction of a stable hairpin structure, and in a bicistronic construct. The monocistronic and bicistronic luciferase plasmids used in this study have previously been used to study the c-myc and Apaf-1 IRESs in a number of cell lines (6, 48, 49, 50). Compared to the control lacking an intergenic spacer, the IRES activity of the 5′ UTRs of c-myc, Apaf-1, and PKCδ was greatest in HeLa cells and weakest in BALB/c cells, which supports the idea that some cell lines may lack noncanonical factors required for IRES function. However, we found that the IRES activity of the 5′ UTR of PKCδ was comparable in MCF7 and HeLa cells, whereas MCF7 cells do not support the activity of the c-myc or Apaf-1 IRES (6, 50). This suggests that the utilization of cellular factors by different IRESs varies in particular cell types.
Cellular IRESs are considered likely to have an important function in maintaining synthesis of gene products when cap-dependent translation is inhibited, such as during viral infection (18), hypoxia (47), apoptosis (16, 49), gamma irradiation (17), and amino acid starvation (11) and in the G2/M stage of the cell cycle (44). We show here that the IRES activity of the 5′ UTR of PKCδ is greatest during TRAIL-induced apoptosis and when cells grow to a high density. A reduction in global protein synthesis and proteolytic cleavage of eIF4G is well documented during apoptosis (5, 30, 49). With increasing cell density of normal skin fibroblasts, eIF4E protein levels decrease and there is a concomitant increase in the unphosphorylated (eIF4E-sequestering) form of 4E-BP1, suggesting a loss of cap-dependent translation in densely growing cells (14). We also show here that the IRES activity of PKCδ is diminished in response to serum starvation, another condition known to reduce global protein synthesis and promote eIF4G degradation (5). These results suggest that the IRES activity of the 5′ UTR of PKCδ is not enhanced under general conditions of cellular stress but is specifically enhanced in response to particular stimuli.
A cap-independent mechanism that would allow continued synthesis of PKCδ when cells grow to a high density and during apoptosis is consistent with PKCδ function under these conditions. Although an increase in PKCδ protein levels appears to be a general phenomenon of cells growing to a high density, its role here is only speculated. PKCδ may be required in densely growing cells to promote growth arrest and/or to prepare the cells for apoptosis. In support of this, many studies have implied a role for PKCδ in the induction of growth arrest (36, 43, 53) and loss of PKCδ is associated with cellular transformation and malignancy of gliomas (28, 29). In most apoptotic cells, PKCδ is activated by caspase 3-mediated proteolysis (10). The active proteolytic product of PKCδ is sufficient to induce apoptosis of HeLa cells (15). However, PKCδ-mediated apoptosis can occur in the absence of proteolytic cleavage (13). Furthermore, in response to mitochondrion-targeting toxins, a dominant-negative form of PKCδ inhibits caspase 3 activity, suggesting that PKCδ is also involved in a pathway leading to caspase 3 activation (31).
In apparent contrast to its role in growth arrest and apoptosis, PKCδ is also implicated in proliferative pathways. PKCδ promotes cellular transformation via the insulin-like growth factor I receptor (25), and elevated PKCδ levels are observed in simian virus 40-transformed rat fibroblasts and in highly metastatic mammary tumors (19, 26). Intriguingly, serum-stimulated PKCδ phosphorylates the repressor of eIF4E, 4E-BP1, in vivo and thus is involved in the activation of cap-dependent translation (23). This perhaps serves to highlight the important roles that PKCδ plays in many aspects of cell life, in promoting cell proliferation in response to some signals and in stimulating growth arrest and apoptosis in response to others. PKCδ may thus represent a switch for cell fate.
Acknowledgments
This work was supported by a grant (049296) from the Wellcome Trust.
We are grateful to Y. Ono (Kobe University, Kobe, Japan) for the generous gift of the PKCδ cDNA, to James Malter (University of Wisconsin, Madison) for the gift of the pBKSpA plasmid, and to Simon Morley (Sussex University) for the gift of eIF4G antiserum. We are especially grateful to Anne Willis (Leicester University) and members of her group for advice and many helpful discussions and for the gifts of TRAIL, the HRV construct, and most of the luciferase reporter plasmids used here.
REFERENCES
- 1.Borman, A. M., P. Le Mercier, M. Girard, and K. M. Kean. 1997. Comparison of picornaviral IRES-driven internal initiation of translation in cultured cells of different origins. Nucleic Acids Res. 25:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cacace, A. M., S. N. Guadagno, R. S. Krauss, D. Fabbro, and I. B. Weinstein. 1993. The epsilon isoform of protein kinase C is an oncogene when overexpressed in rat fibroblasts. Oncogene 8:2095-2104. [PubMed] [Google Scholar]
- 3.Carter, M. S., K. M. Kuhn, and P. Sarnow. 2000. Cellular internal ribosome entry site elements and the use of cDNA microarrays in their investigation, p. 615-635. In N. Sonenberg, J. W. B. Hershey, and M. B. Matthews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Chang, Q., B. D. Soper, B. R. Yacyshyn, and B. L. Tepperman. 2000. Alterations in protein kinase C isoforms in experimentally-induced colitis in the rat. Inflamm. Res. 49:27-35. [DOI] [PubMed] [Google Scholar]
- 5.Clemens, M. J., M. Bushell, and S. J. Morley. 1998. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene 17:2921-2931. [DOI] [PubMed] [Google Scholar]
- 6.Coldwell, M. J., S. A. Mitchell, M. Stoneley, M. MacFarlane, and A. E. Willis. 2000. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 19:899-905. [DOI] [PubMed] [Google Scholar]
- 7.Dekker, L. V., and P. J. Parker. 1994. Protein kinase C—a question of specificity. Trends Biochem. Sci. 19:73-77. [DOI] [PubMed] [Google Scholar]
- 8.Denning, M. F., A. A. Dlugosz, M. K. Howett, and S. H. Yuspa. 1993. Expression of an Oncogenic rasHa gene in murine keratinocytes induces tyrosine phosphorylation and reduced activity of protein kinase C δ. J. Biol. Chem. 268:26079-26081. [PubMed] [Google Scholar]
- 9.Dong, L., J. L. Stevens, D. Fabbro, and S. Jaken. 1994. Protein kinase C isozyme expression and down-modulation in growing, quiescent, and transformed renal proximal tubule epithelial cells. Cell Growth Differ. 5:881-890. [PubMed] [Google Scholar]
- 10.Emoto, Y., Y. Manome, G. Meinhardt, H. Kisaki, S. Kharbanda, M. Robertson, T. Ghayur, W. W. Wong, R. Kamen, R. Weichselbaum, and D. Kufe. 1995. Proteolytic activation of protein kinase Cδ by an ICE-like protease in apoptotic cells. EMBO J. 14:6148-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez, J., I. Yaman, R. Mishra, W. C. Merrick, M. D. Snider, W. H. Lamers, and M. Hatzoglou. 2001. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J. Biol. Chem. 276:12285-12291. [DOI] [PubMed] [Google Scholar]
- 12.Fort, P., L. Marty, M. Piechaczyk, S. el Sabrouty, C. Dani, P. Jeanteur, and J. M. Blanchard. 1985. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 13:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii, T., M. L. Garcia-Bermejo, J. L. Bernabo, J. Caamano, M. Ohba, T. Kuroki, L. Li, S. H. Yuspa, and M. G. Kazanietz. 2000. Involvement of protein kinase Cδ (PKCδ) in phorbol ester-induced apoptosis in LNCaP prostate cancer cells. Lack of proteolytic cleavage of PKCδ. J. Biol. Chem. 275:7574-7582. [DOI] [PubMed] [Google Scholar]
- 14.Galy, B., A. Maret, A. C. Prats, and H. Prats. 1999. Cell transformation results in the loss of the density-dependent translational regulation of the expression of fibroblast growth factor 2 isoforms. Cancer Res. 59:165-171. [PubMed] [Google Scholar]
- 15.Ghayur, T., M. Hugunin, R. V. Talanian, S. Ratnofsky, C. Quinlan, Y. Emoto, P. Pandey, R. Datta, Y. Huang, S. Kharbanda, H. Allen, R. Kamen, W. Wong, and D. Kufe. 1996. Proteolytic activation of protein kinase Cδ by an ICE/CED 3-like protease induces characteristics of apoptosis. J. Exp. Med. 184:2399-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henis-Korenblit, S., N. L. Strumpf, D. Goldstaub, and A. Kimchi. 2000. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol. Cell. Biol. 20:496-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holcik, M., C. Lefebvre, C. Yeh, T. Chow, and R. G. Korneluk. 1999. A new internal-ribosome-entry-site motif potentiates XIAP-mediated cytoprotection. Nat. Cell Biol. 1:190-192. [DOI] [PubMed] [Google Scholar]
- 18.Johannes, G., M. S. Carter, M. B. Eisen, P. O. Brown, and P. Sarnow. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 96:13118-13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiley, S. C., K. J. Clark, M. Goodnough, D. R. Welch, and S. Jaken. 1999. Protein kinase Cδ involvement in mammary tumor cell metastasis. Cancer Res. 59:3230-3238. [PubMed] [Google Scholar]
- 20.Kozak, M. 1986. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA 83:2850-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak, M. 1987. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15:8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak, M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, V., P. Pandey, D. Sabatini, M. Kumar, P. K. Majumder, A. Bharti, G. Carmichael, D. Kufe, and S. Kharbanda. 2000. Functional interaction between RAFT1/FRAP/mTOR and protein kinase Cδ in the regulation of cap-dependent initiation of translation. EMBO J. 19:1087-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, L., P. S. Lorenzo, K. Bogi, P. M. Blumberg, and S. H. Yuspa. 1999. Protein kinase Cδ targets mitochondria, alters mitochondrial membrane potential, and induces apoptosis in normal and neoplastic keratinocytes when overexpressed by an adenoviral vector. Mol. Cell. Biol. 19:8547-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, W., Y.-X. Jiang, J. Zhang, L. Soon, L. Flechner, V. Kapoor, J. H. Pierce, and L.-H. Wang. 1998. Protein kinase C-δ is an important signaling molecule in insulin-like growth factor I receptor-mediated cell transformation. Mol. Cell. Biol. 18:5888-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao, L., K. Ramsay, and S. Jaken. 1994. Protein kinase C isozymes in progressively transformed rat embryo fibroblasts. Cell Growth Differ. 5:1185-1194. [PubMed] [Google Scholar]
- 27.Littlebury, P., J. Watson, T. Williams, G. Beale, and M. Rumsby. 1997. Protein expression of the epsilon subspecies of protein kinase C ceases as Swiss 3T6 fibroblasts increase in cell density even though message for the protein is still present. FEBS Lett. 400:304-308. [DOI] [PubMed] [Google Scholar]
- 28.Lu, Z., A. Hornia, Y.-W. Jiang, Q. Zang, S. Ohno, and D. A. Foster. 1997. Tumor promotion by depleting cells of protein kinase Cδ. Mol. Cell. Biol. 17:3418-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandil, R., E. Ashkenazi, M. Blass, I. Kronfeld, G. Kazimirsky, G. Rosenthal, F. Umansky, P. S. Lorenzo, P. M. Blumberg, and C. Brodie. 2001. Protein kinase Cα and protein kinase Cδ play opposite roles in the proliferation and apoptosis of glioma cells. Cancer Res. 61:4612-4619. [PubMed] [Google Scholar]
- 30.Marissen, W. E., and R. E. Lloyd. 1998. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 18:7565-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matassa, A. A., L. Carpenter, T. J. Biden, M. J. Humphries, and M. E. Reyland. 2001. PKCδ is required for mitochondrial-dependent apoptosis in salivary epithelial cells. J. Biol. Chem. 276:29719-29728. [DOI] [PubMed] [Google Scholar]
- 32.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 33.Mellor, H., and P. J. Parker. 1998. The extended protein kinase C superfamily. Biochem. J. 332:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikultis, W., B. Pradet-Balade, B. Habermann, H. Beug, J. A. Garcia-Sanz, and E. W. Müllner. 2000. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 14:1641-1652. [DOI] [PubMed] [Google Scholar]
- 35.Mischak, H., A. Bodenteich, W. Kolch, J. Goodnight, F. Hofer, and J. F. Mushinski. 1991. Mouse protein kinase C-δ, the major isoform expressed in mouse hemopoietic cells: sequence of the cDNA, expression patterns, and characterization of the protein. Biochemistry 30:7925-7931. [DOI] [PubMed] [Google Scholar]
- 36.Mischak, H., J. Goodnight, W. Kolch, G. Martiny-Baron, C. Schaechtle, M. G. Kazanietz, P. M. Blumberg, J. H. Pierce, and J. F. Mushinsky. 1993. Overexpression of protein kinase C-δ and -ɛ in NIH 3T3 cells induces opposite effects on growth, morphology, anchorage dependence, and tumorigenicity. J. Biol. Chem. 268:6090-6096. [PubMed] [Google Scholar]
- 37.Mischak, H., J. H. Pierce, J. Goodnight, M. G. Kazanietz, P. M. Blumberg, and J. F. Mushinski. 1993. Phorbol ester-induced myeloid differentiation is mediated by protein kinase C-α and -δ and not by protein kinase C-βII, -ɛ, -ζ, and -η. J. Biol. Chem. 268:20110-20115. [PubMed] [Google Scholar]
- 38.Moreton, K., R. Turner, N. Blake, A. Paton, N. Groome, and M. Rumsby. 1995. Protein expression of the α, γ, δ and ɛ subspecies of protein kinase C changes as C6 glioma cells become contact inhibited and quiescent in the presence of serum. FEBS Lett. 372:33-38. [DOI] [PubMed] [Google Scholar]
- 39.Morrish, B. C., and M. G. Rumsby. 2001. The 5′ UTR of protein kinase Cɛ confers translational regulation in vitro and in vivo. Biochem. Biophys. Res. Commun. 283:1091-1098. [DOI] [PubMed] [Google Scholar]
- 40.Nishizuka, Y. 1992. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science 258:607-614. [DOI] [PubMed] [Google Scholar]
- 41.Ono, Y., T. Fujii, K. Ogita, U. Kikkawa, K. Igarashi, and Y. Nishizuka. 1988. The structure, expression, and properties of additional members of the protein kinase C family. J. Biol. Chem. 263:6927-6932. [PubMed] [Google Scholar]
- 42.Perletti, G. P., M. Folini, H. C. Lin, H. Mischak, F. Piccinini, and A. H. J. Tashjian. 1996. Overexpression of protein kinase Cɛ is oncogenic in rat colonic epithelial cells. Oncogene 12:847-854. [PubMed] [Google Scholar]
- 43.Perletti, G. P., E. Marras, P. Concari, F. Piccinini, and A. H. J. Tashjian. 1999. PKCδ acts as a growth and tumor suppressor in rat colonic epithelial cells. Oncogene 18:1251-1256. [DOI] [PubMed] [Google Scholar]
- 44.Pyronnet, S., L. Pradayrol, and N. Sonenberg. 2000. A cell cycle-dependent internal ribosome entry site. Mol. Cell 5:607-616. [DOI] [PubMed] [Google Scholar]
- 45.Reddig, P. J., N. E. Dreckschmidt, H. Ahrens, R. Simsiman, C. P. Tseng, J. Zou, T. D. Oberley, and A. K. Verma. 1999. Transgenic mice overexpressing protein kinase Cδ in the epidermis are resistant to skin tumor promotion by 12-O-tetradecanoylphorbol-13-acetate. Cancer Res. 59:5710-5718. [PubMed] [Google Scholar]
- 46.Singh, R. K., N. Llansa, C. D. Bucana, R. Sanchez, A. Koura, and I. J. Fidler. 1996. Cell density-dependent regulation of basic fibroblast growth factor expression in human renal cell carcinoma cells. Cell Growth Differ. 7:397-404. [PubMed] [Google Scholar]
- 47.Stein, I., A. Itin, P. Einat, R. Skaliter, Z. Grossman, and E. Keshet. 1998. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: implications for translation under hypoxia. Mol. Cell. Biol. 18:3112-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoneley, M., F. E. Paulin, J. P. Le Quesne, S. A. Chappell, and A. E. Willis. 1998. c-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene 16:423-428. [DOI] [PubMed] [Google Scholar]
- 49.Stoneley, M., S. A. Chappell, C. L. Jopling, M. Dickens, M. MacFarlane, and A. E. Willis. 2000. c-Myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 20:1162-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoneley, M., T. Subkhankulova, J. P. C. LeQuesne, M. J. Coldwell, C. L. Jopling, G. J. Belsham, and A. E. Willis. 2000. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, X., F. Wu, R. Datta, S. Kharbanda, and D. Kufe. 2000. Interaction between protein kinase Cδ and the c-Abl tyrosine kinase in the cellular response to oxidative stress. J. Biol. Chem. 275:7470-7473. [DOI] [PubMed] [Google Scholar]
- 52.Vagner, S., B. Galy, and S. Pyronnet. 2001. Irresistible IRES. EMBO Rep. 2:893-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe, T., Y. Ono, Y. Taniyama, K. Hazama, K. Igarashi, K. Ogita, U. Kikkawa, and Y. Nishizuka. 1992. Cell division arrest induced by phorbol ester in CHO cells overexpressing protein kinase C-δ subspecies. Proc. Natl. Acad. Sci. USA 89:10159-10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams, J. G., and S. Penman. 1975. The messenger RNA sequences in growing and resting mouse fibroblasts. Cell 6:197-206. [DOI] [PubMed] [Google Scholar]