Abstract
Peroxisomal PTS2-dependent matrix protein import starts with the recognition of the PTS2 targeting signal by the import receptor Pex7p. Subsequently, the formed Pex7p/cargo complex is transported from the cytosol to the peroxisomal docking complex, consisting of Pex13p and Pex14p. In Saccharomyces cerevisiae, the latter event is thought to require the redundant Pex18p and Pex21p. Here we mapped the Pex7p interaction domain of Pex13p to its N-terminal 100 amino acids. Pex18p and Pex21p also interacted with this region, albeit only in the presence of Pex7p. Expression of an N-terminally deleted version of Pex13p in a pex13Δ mutant failed to restore growth on fatty acids due to a specific defect in the import of PTS2-containing proteins. We further show by yeast two-hybrid analysis, coimmunoprecipitation, and in vitro binding assays that Pex7p can bind Pex13p and Pex14p in the absence of Pex18p/Pex21p. The PTS2 protein thiolase was shown to interact with Pex14p but not with Pex13p in a Pex7p- and Pex18p/Pex21p-dependent manner, suggesting that only Pex14p binds cargo-loaded PTS2 receptor. We also found that the cytosolic Pex7p/thiolase-containing complex includes Pex18p. This complex accumulated in docking mutants but was absent in cells lacking Pex18p/Pex21p, indicating that Pex18p/Pex21p are required already before the docking event.
Peroxisomes are ubiquitous organelles of eucaryotic cells and fulfill a number of biochemical functions, including the β-oxidation of fatty acids (51). Peroxisome biogenesis is dependent on a class of genes that is conserved among species, the PEX genes (19, 40). The corresponding gene products, the peroxins, are involved in specific aspects of peroxisome formation, peroxisome proliferation, membrane protein insertion, and matrix protein import (14, 21). Most of the peroxins are required for matrix protein import, which can occur via two different pathways, the peroxisomal targeting signal type 1 (PTS1)-dependent one and the PTS2-dependent one. The majority of matrix proteins possess a PTS1 signal, which is located at the extreme C terminus and is composed of the tripeptide SKL or conservative variants thereof (18, 24). PTS2-containing proteins include 3-ketoacyl-coenzyme A thiolase, which in Saccharomyces cerevisiae is encoded by the FOX3 gene. PTS2 targeting signals are located close to the N terminus and were proposed to follow the consensus sequence H/R-L-X5-H-L (47, 48).
The targeting signals are recognized in the cytosol by the PTS1- and PTS2-specific factors Pex5p and Pex7p, respectively, which in turn deliver the cargo proteins to a docking complex at the peroxisomal membrane. Upon docking, the two pathways seem to merge, as the components of the docking complex, Pex14p and Pex13p, provide binding sites for both Pex5p and Pex7p (1, 16, 32). In the case of Pex5p, interaction with Pex13p and Pex14p has been shown to be direct (2, 41, 50). Since Pex13p and Pex14p are nonredundant peroxins, it follows that they act either together in the same complex or in a consecutive fashion upon protein translocation. Evidence for Pex13p being in a complex that is distinct from the Pex14p-containing complex was recently provided upon purification of both proteins from rat liver peroxisomes (35). In S. cerevisiae, Pex13p is required for the targeting of Pex14p to the peroxisomal membrane (16), and thus, some of the defects in PTS2-dependent import that are observed in pex13Δ mutant cells may in fact be due to the absence of peroxisomal Pex14p from these cells.
PTS2 protein import specifically requires cytosolic factors in addition to Pex7p. These include the redundant Pex18p/Pex21p in S. cerevisiae (34); Pex5pL, the long isoform of Pex5p in higher eukaryotes (5, 31); and Pex20p of Yarrowia lipolytica (49). These PTS2-specific proteins possess only a weak overall sequence similarity, but recent reports suggest that they might perform a similar function. Pex5pL and Pex18p/Pex21p are capable of interacting with Pex7p via a conserved short motif and are thought to deliver cargo-loaded receptor to the peroxisomal membrane, whereas the binding of the PTS2 signal is accomplished by Pex7p (7, 27, 30). Support for a conserved function of these proteins was presented recently by the partial complementation of a pex18Δ pex21Δ mutant with Pex20p (8). However, Pex7p has not yet been identified in Y. lipolytica, and thus, Pex20p may contain the activities of both Pex7p and Pex18p/Pex21p. For Y. lipolytica, it has been shown that Pex20p is involved in the cytosolic oligomerization of thiolase, which seems to be a prerequisite for import. However, Pex20p might also be involved in later events, as it can directly interact with the intraperoxisomal Pex8p (46).
Here we present a functional analysis of the PTS2-specific peroxins Pex7p, Pex18p, and Pex21p and their docking factors Pex13p and Pex14p. The Pex7p-binding domain in Pex13p was mapped to the extreme N terminus of Pex13p, which fails to interact with Pex5p and Pex14p. The physiological relevance of this domain for PTS2-dependent protein import was addressed. We further demonstrate that Pex7p and not Pex18p/Pex21p actually docks to the peroxisomal docking complex and provide evidence that Pex14p represents the initial docking protein. The function of Pex18p and Pex21p will be addressed, and our findings will be discussed in terms of a sequence of events for the early steps in PTS2 protein import.
MATERIALS AND METHODS
Strains, media, and general methods.
Escherichia coli strain DH5α was used for all plasmid amplifications and isolations, and E. coli strain C41 (DE3) (obtained from J. Walker, Medical Research Council, Cambridge, United Kingdom) was used for heterologous expression of recombinant fusion proteins. The yeast strains used in this study are listed in Table 1. PEX18 was deleted from strains HF7cpex21Δ and UTL-7Apex21Δ by using a kanMX4-based disruption cassette that had been amplified from genomic DNA of strain HF7cpex18Δ with primer pair RE221/222. PEX14 was similarly deleted from strain yKat110 by amplifying a LEU2-based cassette with primer pair RE87/88 and genomic DNA of strain UTL-7Apex14Δ as a template. The authenticity of each gene deletion was confirmed by PCR. EcoRV-linearized pHPR131 (expressing PTS2-DsRed) was stably integrated into the trp1 locus of strains UTL-7A (yHPR251), UTL-7Apex7Δ (yHPR252), UTL-7Apex13Δ (yKAT92), and UTL-7Apex18Δpex21Δ (yKat111). Plasmid pHPR132 (expressing DsRed) was similarly integrated into the trp1 locus of UTL-7A (yHPR255). PEX18 was genomically tagged with the TAP tag as described previously (38). In brief, primer pair RE301/302 was used to amplify the TAP tag cassette plus the Kluyveromyces lactis TRP1 marker gene from plasmid pBS1479. The resulting PCR product was then used to transform strains UTL-7Apex7Δ (yAS3) and UTL-7Apex21Δ (yAS4). Correct integration of the cassette was verified by PCR.
TABLE 1.
S. cerevisiae strains used
| Strain | Description | Source or reference |
|---|---|---|
| UTL-7A | MATα leu2-3,112 ura3-52 trp1 | 10 |
| HF7c | MATaura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::GAL1-HIS3 URA3::(GAL4 17-mers)3-CYC1-LacZ | 11 |
| PJ69-4A | MATaura3-52 his3-200 trp1-901 leu2-3,112 gal4Δ gal80Δ LYS2::GAL1-HIS3 met2::GAL7-lacZ GAL2-ADE2 | P. Jamesa |
| UTL-7Apex7Δ | pex7Δ::LEU2 | 26 |
| UTL-7Apex13Δ | pex13Δ::kanMX4 | 23 |
| UTL-7Apex14Δ | pex14Δ::LEU2 | 1 |
| UTL-7Apex18Δ | pex18Δ::kanMX4 | W. Kunaub |
| UTL-7Apex21Δ | pex21Δ::loxP | W. Kunau |
| HF7cpex7Δ | pex7Δ::LEU2 | W. Kunau |
| HF7cpex21Δ | pex21Δ::loxP | W. Kunau |
| yKat36 | UTL-7Apex18Δ::kanMX4 pex21Δ::loxP | This study |
| yKat110 | UTL-7Apex14Δ::LEU2 pex18Δ::kanMX4 pex21Δ::loxP | This study |
| yKat12 | HF7cpex18Δ::kanMX4 pex21Δ::loxP | This study |
| yHPR251 | UTL-7A [pHPR131] | This study |
| yHPR252 | UTL-7Apex7Δ::LEU2 [pHPR131] | This study |
| yHPR255 | UTL-7A [pHPR132] | This study |
| yKat92 | UTL-7Apex13Δ::kanMX4 [pHPR131] | This study |
| yKat111 | UTL-7Apex18Δ::kanMX4 pex21Δ::loxP [pHPR131] | This study |
| yAS3 | UTL-7Apex7Δ PEX18-TAP TRP1 | This study |
| yAS4 | UTL-7Apex21Δ PEX18-TAP TRP1 | This study |
Madison, Wis.
Bochum, Germany.
Standard media for the cultivation of yeast and bacterial strains were prepared as described previously (10, 42). For the induction of peroxisomes, cells were grown in synthetic dextrose medium (SD) containing 0.3% glucose to mid-log phase, shifted into oleic acid medium (YNO), and incubated for 13 h. YNO contained 0.1% oleic acid, 0.05% Tween 40, 0.1% yeast extract, 0.67% yeast nitrogen base, and amino acids as required and was adjusted to pH 6.0 with KOH. For induction of the CUP1 promoter, 25 mg of CuSO4/liter was added to the YNO medium. Oleic acid plates were prepared as described previously and contained 0.1% (wt/vol) oleic acid, 0.5% (wt/vol) Tween 80, 0.1% yeast extract, 0.67% yeast nitrogen base, amino acids, and 0.5% potassium phosphate buffer (pH 6). Standard recombinant DNA techniques, including enzymatic modification of DNA, transformation, and cultivation, were performed essentially as described previously (42). Yeast whole-cell extracts were prepared from 30 mg of cells according to a published protocol (55).
Plasmids and oligonucleotides.
The plasmids and oligonucleotides used are listed in Tables 2 and 3, respectively. Unless otherwise stated, all PCRs were performed with genomic DNA of a wild-type strain as a template and products were subcloned into EcoRV-cut pBluescript SK(+) (Stratagene, La Jolla, Calif.). To construct the PEX13-GFP fusions, the complete PEX13 open reading frame and a truncated form (representing amino acids [aa] 151 to 386) were amplified with primer pairs RE 432/435 and RE 550/435, respectively, cut with BamHI and EcoRI, and cloned into the appropriately digested pUG35 (29), resulting in pMS9 and pKAT136.
TABLE 2.
Plasmids used
| Plasmid | Description | Primer pair | Source or reference |
|---|---|---|---|
| pJR233 | MLS1pr-GFP-SKL | 24 | |
| YEpmycPex7 | CUP1pr-myc-PEX7 | 36 | |
| pPC86-PEX5 | 1 | ||
| pPC86-PEX14 | 1 | ||
| pPC97-PEX7 | 1 | ||
| pPC97-FOX3 | 36 | ||
| pBS1479 | K. lactis TRP1-marked TAP tag cassette | 38 | |
| pHPR126 | DsRed in SK(+) | RE197/198 | This study |
| pHPR131 | ADH2pr-PTS2-DsRed | This study | |
| pHPR132 | ADH2pr-DsRed | This study | |
| pQE31-PEX7 | His6-PEX7 | RE79/80 | This study |
| pQE31-PEX14 | His6-PEX14 | RE87/88 | This study |
| pKat136 | Pex13p151-386-GFP | RE550/435 | This study |
| pKat46 | PEX13pr in SK(+) | RE98/99 | This study |
| pKat111 | PEX13pr-PEX131-386 in SK(+) | RE421/423 | This study |
| pKat112 | PEX13pr-PEX1356-386 in SK(+) | RE422/423 | This study |
| pKat113 | PEX13pr-PEX131-386 in pRSTERM | This study | |
| pKat122 | PEX13pr-PEX1356-386 in pRSTERM | This study | |
| pKat128 | MBP-PEX7 | RE79/80 | This study |
| pKat118 | GST-Pex13p1-100 | RE298/379 | This study |
| pKat16 | pPC86-PEX13 (1-151) | RE25/32 | This study |
| pKat17 | pPC86-PEX13 (1-166) | RE25/33 | This study |
| pKat18 | pPC86-PEX13 (1-264) | RE25/34 | This study |
| pKat19 | pPC86-PEX13 (1-280) | RE25/35 | This study |
| pKat20 | pPC86-PEX13 (1-310) | RE25/36 | This study |
| pKat21 | pPC86-PEX13 (1-340) | RE25/37 | This study |
| pKat22 | pPC86-PEX13 (1-370) | RE25/52 | This study |
| pKat23 | pPC86-PEX13 (1-386) | RE25/38 | This study |
| pKat24 | pPC86-PEX13 (151-386) | RE26/38 | This study |
| pKat25 | pPC86-PEX13 (166-386) | RE27/38 | This study |
| pKat26 | pPC86-PEX13 (264-386) | RE28/38 | This study |
| pKat27 | pPC86-PEX13 (280-386) | RE29/38 | This study |
| pKat28 | pPC86-PEX13 (310-386) | RE30/38 | This study |
| pKat29 | pPC86-PEX13 (340-386) | RE31/38 | This study |
| pKat30 | pPC86-PEX13 (370-386) | RE53/38 | This study |
| pKat31 | pPC97-PEX13 (1-151) | RE25/32 | This study |
| pKat86 | pPC86-PEX13 (1-55) | RE25/378 | This study |
| pKat87 | pPC86-PEX13 (1-100) | RE25/379 | This study |
| pKat88 | pPC97-PEX13 (1-55) | RE25/378 | This study |
| pKat89 | pPC97-PEX13 (1-100) | RE25/379 | This study |
| pKat90 | pPC86-PEX13 (55-100) | RE380/379 | This study |
| pKat91 | pPC97-PEX13 (55-100) | RE380/379 | This study |
| pKat59 | pPC86-PEX21 | RE173/174 | This study |
| pKat66 | pPC86-PEX18 | RE169/170 | This study |
| pKat60 | pPC97-PEX21 | RE173/174 | This study |
| pKat68 | pPC97-PEX18 | RE169/170 | This study |
| pKat146 | pPC86-FOX3 | This study | |
| pMS9 | Pex13p1-386-GFP | RE432/435 | This study |
| pRB107 | GST-PTS2-GFP-prA | RE188/415 | This study |
TABLE 3.
Oligonucleotides used
| Designation | Sequence (5′ to 3′) |
|---|---|
| RE 25 | GTGAATTCGGATCCATATGTCATCCACAGCAGTACCA |
| RE 26 | GTGAATTCGGATCCATATGTTAATAGAAAGTTTGATAGGC |
| RE 27 | GTGAATTCGGATCCATATGCTGGAATCTACTTATATGGCC |
| RE 28 | GTGAATTCGGATCCATATGCCGCTACTATTTTTTTTGATG |
| RE 29 | GTGAATTCGGATCCATATGAACAAATTTATTACTAAACTACAG |
| RE 30 | GTGAATTCGGATCCATATGTTTGCAAGAGCGTTATATGAT |
| RE 31 | GTGAATTCGGATCCATATGAAAGACCCTCTTGGGAGGGAT |
| RE 32 | GCTCTAGAACTAGTTAACTGAAAGGTGGCCTTCGT |
| RE 33 | GCTCTAGAACTAGTCAGCATTTGTGCAAATCCAGT |
| RE 34 | GCTCTAGAACTAGCGGTTTCCATGAAATTTTCCT |
| RE 35 | GCTCTAGAACTAGTGTTTAGTAGATATGGAAAACC |
| RE 36 | GCTCTAGAACTAGTAAATTCTAACTTCGAAGGATC |
| RE 37 | GCTCTAGAACTAGTTTTCTTACTCAAAATTGCCAT |
| RE 38 | GCTCTAGAACTAGTCTAGTGTGTACGCGTTTCATC |
| RE 52 | GCTCTAGAACTAGTTATGATCTCAATATAGTTATA |
| RE 53 | GTGAATTCGGATCCATATGATAAAAAGACGGAAGAAAATTGAG |
| RE 79 | GCTCTAGAGAGCTCACATATGCTCAGATATCACATGCAAGG |
| RE 80 | GGGGTACCCTCGAGCTGCAGTCAACCTAAGCCGTTCCATAC |
| RE 87 | GCTCTAGAGCTAGCAAGGATCCAATGAGTGACGTGGTCAG |
| RE 88 | GGGGTACCGAATCCCTGCAGCTATGGGATGGAGTCTTC |
| RE 98 | CGGGATCCCTGCAGCTATTTATTCATAGTGTC |
| RE 99 | CGGAATTCGCATGCTGACATCGCAGGTATTGTTATAGT |
| RE 169 | GCGTCGACTATGAATAGTAACCGATGC |
| RE 170 | GCGAGCTCTTAAGCAATTCTGTCTTCAAC |
| RE 173 | GCGTCGACTATGCCCAGTGTCTGCCAT |
| RE 174 | GCGAGCTCTCAATCAAGTATGTCTTTGTG |
| RE 188 | CTGCAGTTAATTCGCGTCTACTTTCGG |
| RE 189 | CATATGTCTCAAAGACTACAAAG |
| RE 197 | GCGGCCGCACCATGGTGAGGTCTTCCAAGAATGTT |
| RE 198 | GGATCCGTCGCGGCCGCTAAAGGAAC |
| RE 221 | GTATAATCAGGTATGTAAGGG |
| RE 222 | CGACAACTAAGTTCCAGAAAG |
| RE 298 | CGGAATTCCATATGTCATCCACAGCAGTACCA |
| RE 301 | AACTGGGCTGGTCTTGAGTTCCATGATGTTGAAGACAGAATTGCTTCCATGGAAAAGAGAAG |
| RE 302 | TCTCTTGAAAGTGGTTAAAAAGTGTTATATATCTGAAATTCATGGTACGACTCACTATAGGG |
| RE 378 | GCTCTAGAACTAGTCTAAGCAGACTCACTTGCACT |
| RE 379 | GCTCTAGAACTAGTCTATATAGAGTTCATACTATACGG |
| RE 380 | GTGAATTCGGATCCATATGCCCGAAGTTTTGCCGCGG |
| RE 415 | GGATCCTCTCAAAGACTACAAAGTATC |
| RE 421 | GCATGCGGCGGCCGCTCATCCACAGCAGTACCA |
| RE 422 | GCATGCGGCGGCCGCCCCGAAGTTTTGCCGCGG |
| RE 423 | AAGCTTCTAGTGTGTACGCGTTTCAT |
| RE 432 | GAGGATCCTGCGATGTCATCCACA |
| RE 435 | TCTGAATTCGTGTGTACGCGTTTCATC |
| RE 550 | GAGGATCCTGCGATGTTAATAGAAAGTTTGATAGGC |
Expression of full-length and truncated Pex13p from the PEX13 promoter was achieved as follows. The PEX13 promoter (base pairs −363 to +6) was amplified (RE 98/99) and cloned as a BamHI/EcoRI fragment into pSK(+) (pKat46). The complete PEX13 open reading frame, amplified with primer pair RE 421/423, and the truncated version (representing aa 56 to 386), amplified with primer pair RE 422/423, were digested with SphI/HindIII and cloned into the SphI/HindIII-digested pKat46, resulting in pKat111 and pKat112. The assembled constructs were excised from pKat111 and pKat112 with BamHI/HindIII and ligated into the appropriately cut yeast expression vector pRS-FOXP-TERM (C. Clayton, Heidelberg, Germany), resulting in pKat113 and pKat122, respectively.
The PTS2-DsRed expression construct (pHPR131) was cloned by assembling the following fragments into XbaI/PstI-cut YIplac204 (15): the ADH2 promoter, excised from plasmid pADH2PIP2, as an XbaI/NdeI fragment (3); the amino-terminal 16 aa of Fox3p, obtained from pPTS2-GFP-prA (unpublished) by using primer pair RE 188/189, as an NdeI/NcoI fragment; and DsRed, amplified from pDsRed (Clontech, Palo Alto, Calif.) with primer pair RE 197/198 and subcloned into pBluescript SK(+) (pHPR126), as an NcoI/PstI fragment. The DsRed expression vector lacking a PTS2 (pHPR132) was similarly assembled in EcoRI/PstI-cut YIplac204 by using the DsRed-containing NcoI/PstI fragment of pHPR126 and the ADH2 promoter from pADH2-OAF1 as an EcoRI-NcoI fragment (39).
For the bacterial expression of a synthetic PTS2 protein (pRB107), PTS2-GFP-prA was amplified from pPTS2-GFP-prA with primer pair RE 188/415 and cloned as a BamHI/EcoRI fragment into pGEX4T-2 (Amersham Pharmacia, Freiburg, Germany). To express Pex7p in E. coli, PEX7 was amplified with primer pair RE 79/80 and cloned as a SacI/PstI fragment into the appropriately cut pQE31 (Qiagen, Hilden, Germany) or pMAL-c2X (New England Biolabs, Beverly, Mass.), yielding QE31-PEX7 and pKat128, respectively. To generate a His6-Pex14p fusion construct (QE31-PEX14), PEX14 was amplified with primer pair RE 87/88 and cloned into pQE31 as a BamHI/PstI fragment. The GST-Pex13p1-100 construct (pKat118) was cloned by amplifying the Pex13p-encoding fragment with primer pair RE 298/379 and ligating the resulting EcoRI/XbaI fragment into pGEX4T-1.
Yeast two-hybrid assays.
The two-hybrid assay was based on the method of Fields and Song (12). Selected PEX genes or truncations thereof were fused to the DNA-binding domain or transcription activation domain of Gal4p in the vectors pPC86 and pPC97 (6). To construct the Gal4p-AD-Pex13p fusions, EcoRI/SpeI fragments of the various PEX13 fragments that had been amplified from genomic DNA were subcloned into the appropriately cut pPC86 (Table 2). To construct the Gal4p-BD-Pex13p fusions, the PEX13 fragments were excised from the pPC86-PEX13 constructs with SmaI/SpeI and subcloned into SmaI/SpeI-digested pPC97 (Table 2). PEX18 and PEX21 were amplified from genomic DNA and subcloned into SalI/SacI-digested pPC86 and pPC97, respectively (Table 2). A SalI/SacI fragment of pPC97-FOX3 containing the FOX3 open reading frame was subcloned into an appropriately cut pPC86, resulting in pPC86-FOX3 (pKat146). Cotransformation of two-hybrid plasmids into HF7c was performed according to the method of Schiestl and Gietz (44). Double transformants were selected on SD synthetic medium without tryptophan and leucine. Histidine auxotrophy of transformed HF7c was determined by growth on selective plates lacking leucine, tryptophan, and histidine but containing 1 or 5 mM 3-aminotriazole.
Antibodies and Western blotting.
To generate polyclonal antibodies against Pex7p, a His6-tagged Pex7p was expressed in E. coli strain C41 (DE3) and purified under denaturing conditions according to the manufacturer's protocol (Qiagen). This protein was subsequently used to immunize rabbits at Eurogentec (Seraing, Belgium). The raised antiserum was specific, as it was able to detect Pex7p in whole-cell extracts from a wild type but not from the respective pex7Δ strain (Fig. 9C, lower panel). The other antibodies used have been described previously, namely, anti-Fox3p (9), anti-Cta1p (20), anti-Pex13p (16), anti-Pex14p (1), anti-maltose-binding protein (MBP; New England Biolabs), anti-protein A (Sigma, St. Louis, Mo.), and anti-myc from 9E10 cell lines (16). Immunoblotting was performed according to standard protocols. Horseradish peroxidase-coupled anti-rabbit or anti-mouse immunoglobulin G (IgG), in combination with the ECL system (Amersham Pharmacia), was used to detect immunoreactive complexes.
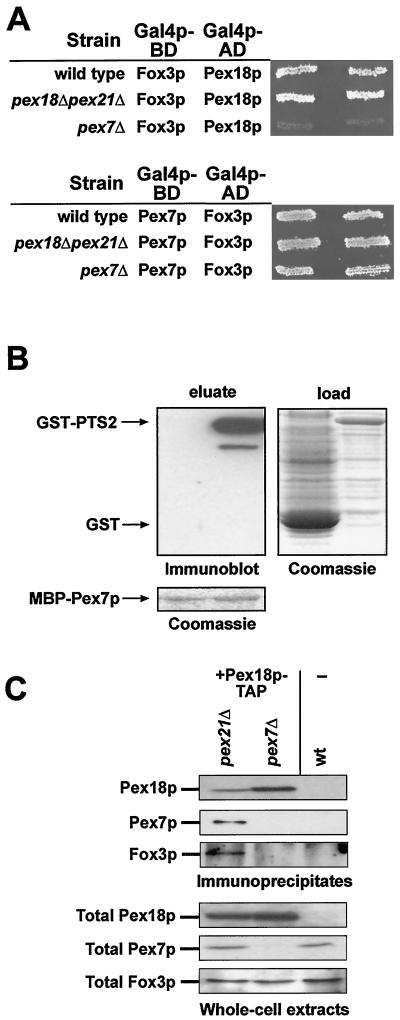
FIG. 9.
Pex18p binds Fox3p only via Pex7p. (A) Fox3p was tested in a two-hybrid assay for interaction with Pex18p (upper panel) and Pex7p (lower panel) in the HF7c wild type as well as the pex18Δ pex21Δ and pex7Δ strains. (B) Binding of Pex7p to a synthetic PTS2 protein in vitro. Equal amounts of MBP-Pex7p were loaded onto affinity columns and incubated with soluble extracts of bacterially expressed GST-PTS2-GFP-prA (GST-PTS2; right lane in each panel) or GST (left lane). The right panel shows a Coomassie blue stain of the loaded fractions. The retention of GST-containing proteins was analyzed by immunoblotting (upper-left panel). The lower panel shows the presence of MBP-Pex7p in both eluates. (C) Coimmunoprecipitation of Fox3p with Pex18p. Strains pex7D PEX18-TAP (yAS3), pex21D PEX18-TAP (yAS4), and the untransformed UTL7-A wild type were induced in oleic acid-containing medium and analyzed for the presence of Pex7p and Fox3p in the precipitate of Pex18p-TAP by immunoblotting (upper panel). The upper and lower panels show 33% of the immunoprecipitated samples and 0.2% of the total cell lysates, respectively.
Coimmunoprecipitation.
Coimmunoprecipitation of myc-Pex7p was performed essentially as described previously (36), with the exception that breakage of the copper-induced cells was achieved by vortexing a suspension of 1 g of cells, 3 volumes of solution A (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 0.2% Triton X-100) that contained the Complete protease inhibitor cocktail (Roche, Mannheim, Germany), and 3 volumes of glass beads (diameter, 0.5 mm; Sigma) for 30 min at 4°C on an IKA-Vibrax VXR (Vibrax, Staufen, Germany). To precipitate Pex18p-TAP, soluble extracts obtained from 3 g of cells (suspended in 2.5 volumes of solution A) were incubated with IgG Sepharose (Amersham Pharmacia) followed by three washing steps with solution A. At that point, aliquots of each sample were removed for analysis of bound Pex18p-TAP. The remainder of the samples were adjusted to 10 mM dithiothreitol and 1 mM EDTA and incubated with 3 U of recombinant tobacco etch virus (TEV) protease (Life Technologies, Karlsruhe, Germany) for 2 h at room temperature with gentle agitation to release Pex18p from the matrix. The resulting eluates were analyzed for the presence of Pex7p and Fox3p.
In vitro binding assays.
To analyze the Pex7p-Pex14p interaction, the soluble fraction of bacterially expressed His6-Pex14p (QE31-PEX14) was incubated with nickel-nitrilotriacetic acid (Ni-NTA) matrix (Invitrogen, De Schelp, The Netherlands) for 2 h at 4°C. After being washed with column buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole [pH 8]) in a column, equal amounts of the soluble fractions of bacterially expressed MBP-Pex7p (pKat128) or MBP alone (pMAL-c2X) were added to the column and incubated with the matrix for 1 h. After being washed with column buffer, the proteins were eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 500 mM imidazole [pH 8]). The eluted samples were analyzed by Coomassie brilliant blue staining or immunoblot analysis.
The in vitro binding of Pex7p to Pex13p was analyzed as follows. The soluble fractions of glutathione S-transferase (GST) and GST-Pex13p1-100 that had been expressed in C41 (DE3) from plasmids pGEX4T-1 and pKat118, respectively, were bound for 2 h at 4°C on a glutathione Sepharose 4B matrix (Amersham Pharmacia). After washing the matrix with 1× phosphate-buffered saline (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4), the soluble fraction of bacterially expressed His6-Pex7p (QE31-PEX7) was added and incubated with the matrix for 1 h at 4°C with gentle rotation. Alternatively, equal amounts of yeast lysates from myc-Pex7p-expressing UTL-7A or UTL-7Apex18Δpex21Δ cells were added. After washing the matrix with 1× phosphate-buffered saline, the proteins were eluted with 10 mM glutathione in 50 mM Tris-HCl (pH 8). The eluted samples were analyzed by Coomassie brilliant blue staining or immunoblot analysis.
The binding of PTS2 protein to Pex7p was tested by loading soluble extracts containing either GST-PTS2 protein (pRB107) or GST (pGEX4T-2) onto MBP-Pex7p that had been purified by using the pMAL purification system from New England Biolabs. After being washed with 10 bed volumes of column buffer (20 mM Tris-HCl [pH 7.4], 200 mM NaCl, 1 mM EDTA), MBP-Pex7p was eluted with column buffer containing 10 mM maltose. The eluates were analyzed for the presence of GST-containing proteins by immunoblotting with an antibody directed against GST-Pex14p (1).
Miscellaneous.
The analysis of live cells for DsRed and green fluorescent protein (GFP) fluorescence was performed according to the method of Westermann and Neupert (53). β-Galactosidase activities were assayed according to manufacturer's instructions and expressed as micromoles of CPRG (chlorophenol red-β-d-galactopyranoside) hydrolyzed per minute per cell (Clontech).
RESULTS
Pex7p interacts with the cytoplasmic N-terminal domain of Pex13p.
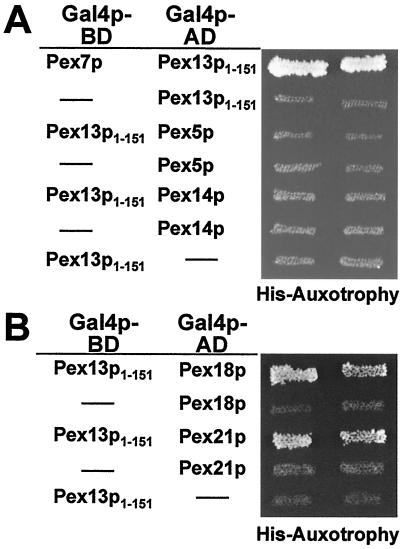
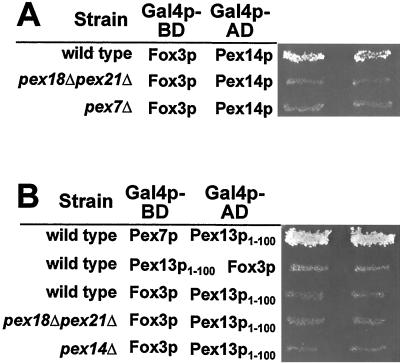
We have shown previously that Pex13p functionally interacts with the PTS2 cargo receptor Pex7p (16). To further characterize the binding of Pex7p to Pex13p, a series of N- and C-terminal deletion constructs of Pex13p were tested in a yeast two-hybrid assay for interaction with Pex7p. The full-length Pex13p (aa 1 to 386) as well as the two C-terminal truncations that lacked part of the SH3 domain (aa 1 to 340) or the complete domain (aa 1 to 310) were able to interact with Pex7p (Fig. 1), thereby demonstrating that the SH3 domain of Pex13p is dispensable for the interaction of Pex13p with Pex7p. Interaction was also observed for even shorter Pex13p variants that lacked the cytosolically oriented C-terminal domain in total, with or without transmembrane domain 2 (TMD 2; aa 1 to 280 and 1 to 264, respectively). By further removing the internal loop (aa 1 to 166) and TMD 1 (aa 1 to 151) of Pex13p, binding to Pex7p remained proficient. On the other hand, removing these 151 N-terminal amino acids from Pex13p (aa 151 to 386) abolished its interaction with Pex7p. In accordance with this observation, all Pex13p fragments that harbored even larger N-terminal truncations also tested negative for Pex7p binding (Fig. 1). Thus, the N-terminal domain of Pex13p, which is exposed to the cytosol (16), is the only binding site for Pex7p. This novel protein interaction module was also tested for interaction with Pex5p and Pex14p, the other known binding partners of Pex13p. However, these proteins failed to bind aa 1 to 151 of Pex13p in a two-hybrid assay (Fig. 2A).
FIG. 1.
Identification of the Pex7p-binding region in Pex13p. (A) Known features of PEX13. The cytosolically exposed SH3 domain (checked box) of the S. cerevisiae peroxisomal membrane protein Pex13p (386 aa) is bound by Pex5p and Pex14p at distinct sites as indicated by the arrows. The two predicted TMDs are marked in dark gray. (B) Analysis of PEX13 fragments for interaction with PEX7 in the yeast two-hybrid assay. The indicated regions of PEX13 were fused to the GAL4 activation domain of plasmid pPC86 and cotransformed with a pPC97-derived plasmid expressing a GAL4 DNA-binding domain-PEX7 fusion into the wild-type strain HF7c. The resulting prototrophs on histidine dropout plates containing 5 mM aminotriazole were considered positive for interaction (+). Numbers flanking the bars indicate the Pex13p amino acid residues that delimit the fragments.
FIG. 2.
Only the PTS2-specific peroxins interact with the N terminus of Pex13p. (A) Analysis of Pex5p and Pex14p for their interaction with Pex13p1-151. The interaction of Pex7p with Pex13p1-151 in a two-hybrid assay is indicated by the histidine prototrophy of two independent clones of the respective transformant. The corresponding PEX5 and PEX14 transformants failed to grow on the same plate. (B) Pex18p and Pex21p interaction with Pex13p1-151. Coexpression of PEX18 and PEX21 fusions to the GAL4 activation domain (AD) with the Pex13p1-151 GAL4 DNA-binding domain (BD) fusion resulted in the formation of histidine prototrophs.
Pex18p, Pex21p, and Pex7p interact with the same region in Pex13p.
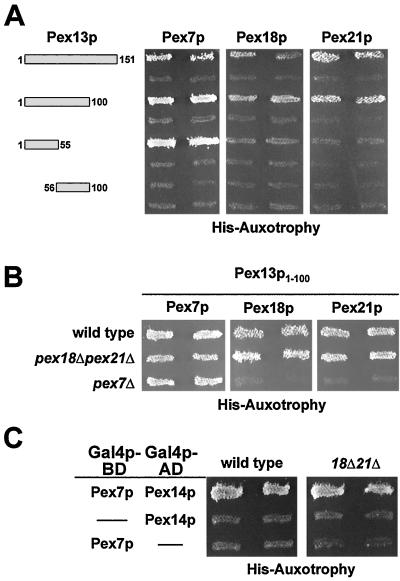
Translocation of proteins with a PTS2 targeting signal specifically requires, in addition to Pex7p, the redundant, largely cytosolic Pex18p and Pex21p (34). In the absence of both proteins, PTS2-dependent import is abolished. Both proteins can interact with Pex7p and are thought to target cargo-loaded Pex7p to the peroxisomal membrane (34). We therefore tested whether Pex18p and Pex21p are able to contact the Pex7p interaction domain of Pex13p. When fused to the Gal4p DNA-binding domain, both proteins exhibited autoactivity in the two-hybrid assay (data not shown). However, the respective activation domain fusions of Pex18p and Pex21p caused growth of the tester strain only when coexpressed with the amino terminus of Pex13p (Fig. 2B). This result indicated that Pex18p and Pex21p can also interact with the N terminus of Pex13p. To narrow down the binding regions in the Pex13p N terminus for the PTS2-specific proteins, smaller fragments of Pex13p were tested in the two-hybrid assay (Fig. 3A). Amino acids 1 to 100 proved even more efficient than aa 1 to 151 in binding the three proteins, while the amino-terminal 55 aa of Pex13p were efficiently recognized only by Pex7p. The construct comprising aa 55 to 100 of Pex13p failed to give rise to a positive signal with any of the proteins analyzed (Fig. 3A).
FIG. 3.
Pex18p and Pex21p contact Pex13p and Pex14p only in the presence of Pex7p. (A) Confinement of the Pex13p-binding region for Pex7p, Pex18p, and Pex21p. The indicated fragments of Pex13p were tested in a two-hybrid assay for interaction with Pex7p, Pex18p, and Pex21p in the HF7c wild-type strain. (B) Dependence of the Pex13p1-100 interactions with Pex7p, Pex18p, and Pex21p on endogenous PEX7 and PEX18/PEX21. The assays with Pex13p1-100 were repeated in the otherwise isogenic HF7c strains pex7Δ and pex18Δ pex21Δ. Histidine prototrophs were not observed for Pex18p/Pex21p in the absence of Pex7p. (C) Dependence of the Pex7p-Pex14p two-hybrid interaction on endogenous PEX18/PEX21. The respective Pex7p and Pex14p fusions were transformed into HF7c and the corresponding pex18Δ pex21Δ strain. In both strains, coexpression of Pex7p and Pex14p caused growth on histidine dropout plates.
Pex13p1-100 and Pex13p1-55 interacted more strongly with Pex7p than with Pex18p or Pex21p, as the Pex7p-containing strains grew faster on histidine dropout plates than those containing Pex18p and Pex21p (data not shown). This result was also verified by quantification of the two-hybrid data by means of a β-galactosidase assay in the strain background of PJ69-4A, which harbors a sensitive GAL7 promoter-lacZ reporter gene. Activities were about threefold higher in the strain expressing Pex7p-Pex13p1-100 (2,509 ± 123 U) than in those expressing Pex7p-Pex13p1-55 (811 ± 137 U), whereas in the strain harboring Pex7p and Pex13p55-100, activities reached only background levels (<50 U). None of the Pex18p/Pex21p-containing strains turned out to yield activities above the detection limit of the assay (<50 U). These data were interpreted to mean that despite the N-terminal 55 aa of Pex13p being essential for the Pex7p interaction, a somewhat larger region of Pex13p, possibly up to its first 100 aa, constitutes the full interaction site for Pex7p. The association of Pex18p/Pex21p with this optimal site could be explained by the assumption of a Pex7p-mediated interaction of Pex18p/Pex21p with Pex13p.
Pex18p and Pex21p bind Pex13p via Pex7p.
The resemblance in binding sites opened the possibility that Pex18p and Pex21p bound to Pex13p in a complex with Pex7p. This was addressed by testing the interaction of Pex7p with Pex13p1-100 in a strain devoid of both Pex18p and Pex21p (Fig. 3B). The interaction was unaffected, suggesting that Pex7p forms contacts with Pex13p that are independent of Pex18p/Pex21p. In contrast, the presence of Pex7p was important for the Pex18p/Pex21p interaction with Pex13p1-100 (Fig. 3B), thereby supporting the notion that Pex7p serves a bridging protein for the interaction of Pex18p/Pex21p with Pex13p.
We then analyzed the possibility that Pex18p and Pex21p are required for the binding of Pex7p to the other docking protein, Pex14p, but this binding event occurred in the pex18Δ pex21Δ mutant background (Fig. 3C). Pex18p/Pex21p interacted only very weakly with Pex14p, and this interaction depended on Pex7p (data not shown), suggesting that these redundant factors do not contact Pex14p directly, either.
Pex7p docking in the absence of Pex18p and Pex21p.
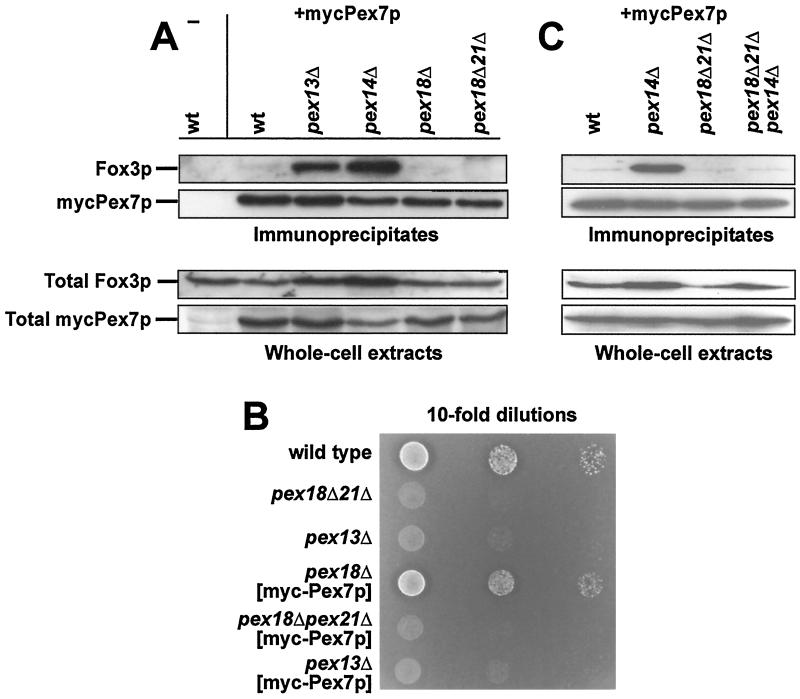
To obtain independent evidence for our results, we set out coimmunoprecipitation experiments with a myc-tagged version of Pex7p. The same protein has previously been used to demonstrate Pex7p binding to Pex13p in the absence of Pex5p and Pex14p (16). The tagged protein was expressed in wild-type and pex18Δ and pex18Δ pex21Δ mutant backgrounds and also in the docking protein mutants pex13Δ and pex14Δ. Expression levels of myc-Pex7p were comparable in all strains (Fig. 4). When myc-Pex7p was immunoprecipitated from whole-cell extracts of the wild-type strain, Pex13p and Pex14p were also found in the precipitate (Fig. 4). As anticipated, in the absence of Pex14p, Pex13p was still found in a complex with Pex7p. Since this was also the case in the pex18Δ pex21Δ double mutant (Fig. 4), this result demonstrated that binding of Pex7p to the docking complex does not require the presence of Pex18p and Pex21p.
FIG. 4.
Docking of Pex7p in the absence of Pex18p and Pex21p. The wild-type strain UTL-7A as well as the indicated, otherwise isogenic set of myc-Pex7p-expressing strains was induced in oleic acid-containing medium and analyzed for the presence of Pex13p and Pex14p in the immunoprecipitate of myc-Pex7p (upper panel). The upper and lower panels show 10% of the immunoprecipitated samples and 0.5% of the cell lysates, respectively, that had been subjected to immunoblotting with antisera against the myc epitope, Pex13p, and Pex14p.
Pex7p binds to Pex14p in vitro.
The ability of Pex7p to bind Pex14p in the absence of Pex18p/Pex21p was also analyzed with bacterially expressed proteins. A His6-tagged version of Pex14p was purified on Ni-NTA affinity columns onto which soluble extracts were loaded that contained either MBP or an MBP-Pex7p fusion protein. After washing, His6-Pex14p was eluted from the columns and the eluates were analyzed for the presence of coeluted MBP-Pex7p. Coomassie brilliant blue staining revealed that Pex7p coeluted with His6-Pex14p (Fig. 5, upper panel, lane 3) whereas MBP did not (lane 4). To ensure that the indicated bands in the Coomassie blue-stained gels indeed represented the MBP fusion protein, the same samples were diluted 20-fold and subjected to immunoblot analysis with an antibody directed against MBP (Fig. 5, lower panel). MBP-Pex7p was clearly detectable (lane 3), whereas little MBP was detected (lane 4). Thus, heterologously expressed Pex7p specifically interacted with Pex14p, demonstrating that these two peroxins can bind each other in a direct manner.
FIG. 5.
Direct binding of Pex7p to Pex14p. Bacterially expressed His6-tagged Pex14p was purified on Ni-NTA affinity columns. Subsequently, soluble extracts containing equal amounts of MBP (lane 1) or MBP-Pex7p (lane 2) were loaded onto these columns. After being washed, the eluates of His6-Pex14p were analyzed for retained MBP-Pex7p (lane 3) or MBP (lane 4) by Coomassie blue staining (upper panel). The same samples were diluted 20-fold and subjected to immunoblot analysis with anti-MBP antibodies (lower panel).
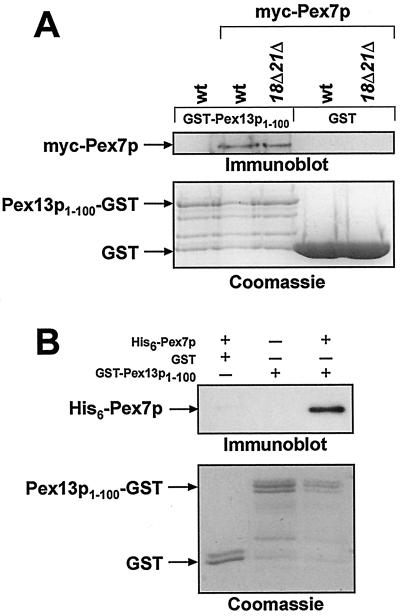
Pex7p binds to Pex13p1-100 in vitro.
To analyze the Pex7p-Pex13p interaction in vitro, the first 100 aa of Pex13p were expressed as a GST fusion protein in E. coli and subsequently purified on GST affinity columns. Subsequently, yeast lysates of wild-type or pex18Δ pex21Δ mutant cells expressing myc-tagged Pex7p were added, and bound GST proteins were visualized by Coomassie blue staining (Fig. 6A, lower panel). myc-Pex7p was detected immunologically by using anti-myc antibodies (Fig. 6A, upper panel). The tagged Pex7p was specifically retained in the presence of Pex13p1-100 even when expressed in cells devoid of Pex18p and Pex21p (lanes 2 and 3). When lysate from an untransformed wild-type strain was applied to the Pex13p1-100-GST-loaded column, no signal was obtained (lane 1). Similarly, GST alone also failed to bind myc-Pex7p (lanes 4 and 5). Thus, it could be inferred that the amino-terminal 100 aa of Pex13p are indeed the binding site for Pex7p and that Pex18p/Pex21p are not required for Pex7p binding to Pex13p1-100.
FIG. 6.
Direct binding of Pex7p to Pex13p1-100. (A) Yeast myc-Pex7p specifically interacts with GST-Pex13p1-100. Equal amounts of GST or GST-Pex13p1-100 were loaded onto affinity columns and incubated with total cell lysates of a wild-type strain or the myc-Pex7p-expressing wild-type or pex18Δ pex21Δ strains. After the columns were washed, bound proteins were eluted and analyzed by immunoblotting for the presence of myc-Pex7p (upper panel). The presence of GST and GST-Pex13p1-100 in these eluates was determined by Coomassie blue staining (lower panel). (B) Binding of Pex7p to Pex13p in vitro. Equal amounts of GST or GST-Pex13p1-100 were loaded onto affinity columns and incubated with soluble extracts of bacterially expressed His6-tagged Pex7p (+) or a mock-transformed E. coli strain (−). The retention of His6-Pex7p was analyzed by immunoblotting (upper panel). The lower panel shows the presence of GST and GST-Pex13p1-100 in these eluates.
To exclude the possibility that another yeast protein contributed to the observed binding of Pex7p to Pex13p, both proteins were bacterially expressed and subjected to in vitro binding studies. When expressed as a His6-tagged version, a large fraction of Pex7p was found in inclusion bodies. Nevertheless, we could obtain a small fraction of His6-Pex7p that was soluble. This fraction was loaded onto columns containing purified GST-Pex13p1-100 or GST (Fig. 6B). Anti-Pex7p antibodies were then used to detect bound Pex7p by immunoblot analysis. His6-Pex7p was only retained in the presence of Pex13p1-100 (Fig. 6B, lane 3), thereby demonstrating that Pex7p can directly bind to the N-terminal region of Pex13p.
The N terminus of Pex13p is required for PTS2-dependent import.
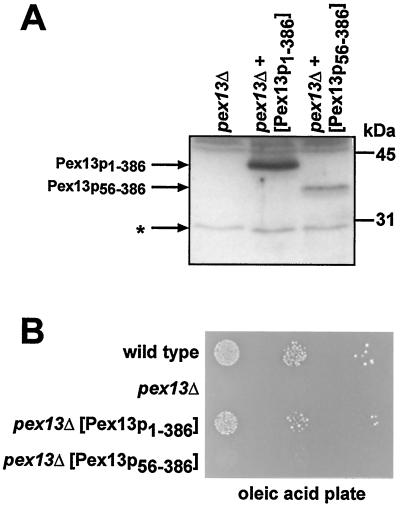
To elucidate the physiological relevance of the Pex7p-binding region in Pex13p, full-length Pex13p as well as a truncated variant of Pex13p lacking the N-terminal 55 aa were expressed from the native PEX13 promoter in a pex13Δ strain and analyzed for their capability to complement the mutant phenotype. Immunoblot analysis demonstrated that truncated Pex13p56-386 was stably expressed, although to a lower degree than full-length Pex13p (Fig. 7A). However, in contrast to full-length Pex13p, Pex13p56-386 was not able to restore the growth defect of a pex13Δ mutant on fatty acids as a sole carbon source (Fig. 7B), indicating that the N-terminal region is important for the function of Pex13p.
FIG. 7.
The Pex7p-binding region of Pex13p is important for its function. (A) Expression of a truncated Pex13p variant in pex13Δ cells. Whole-cell extracts of strain UTL-7Apex13Δ that had expressed plasmid-borne copies of either full-length Pex13p (Pex13p1-386) or an N-terminally truncated version of Pex13p (Pex13p56-386) were subjected to immunoblot analysis with antiserum against Pex13p. The asterisk denotes proteins that had cross-reacted with the antiserum. (B) Complementation analysis of pex13Δ with truncated Pex13p56-386. The indicated strains were spotted as a series of 10-fold dilutions on an oleic acid plate and incubated for 5 days at 30°C.
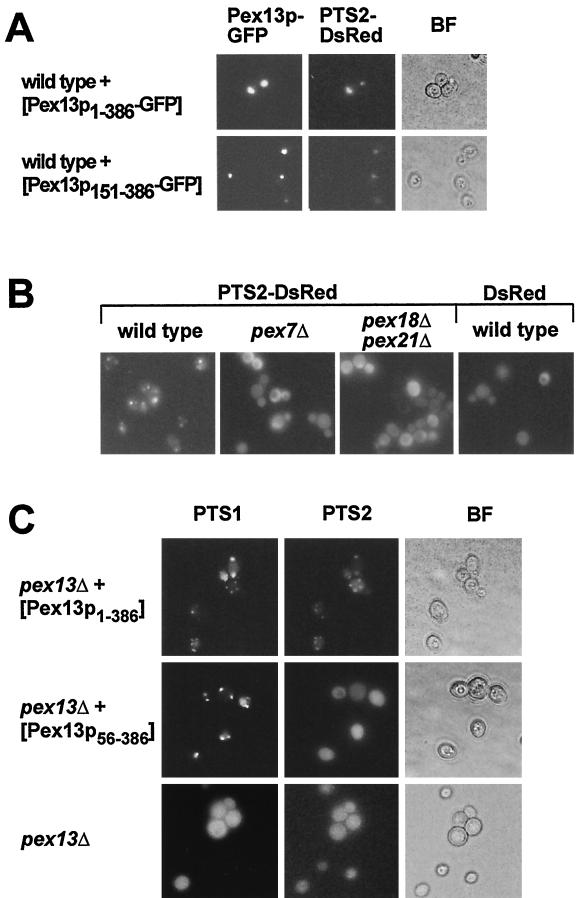
To test whether truncated Pex13p was correctly targeted to peroxisomes, GFP was fused to the C terminus of full-length Pex13p (Pex13p1-386) and to Pex13p that lacked the entire N-terminal cytosolic domain (Pex13p151-386). The GFP fusions of both the truncated and the full-length Pex13p were expressed in a wild-type strain, and their intracellular distribution was analyzed by fluorescence microscopy. Both Pex13p fusion proteins showed a punctate staining pattern (Fig. 8A) that was reminiscent of clustered peroxisomes. This clustering did not affect peroxisomal function, as full-length Pex13p-GFP was able to fully complement a pex13Δ strain (data not shown). Moreover, the staining patterns of both the full-length and the truncated Pex13p coincided with that of the peroxisomal marker protein PTS2-DsRed (Fig. 8A and B). It was therefore concluded that Pex13p1-386 and Pex13p151-386 are correctly targeted to the peroxisomal membrane. We then investigated whether truncated Pex13p56-386 is still functional in peroxisomal matrix protein import. This was achieved by monitoring the subcellular distribution of fluorescent marker proteins for PTS1 and PTS2 import. GFP-SKL was used for analyzing PTS1-dependent import, while PTS2-DsRed, composed of the 16 N-terminal amino acids of Fox3p and the red fluorescent protein DsRed, served as the PTS2 marker protein. This protein was imported into peroxisomes in a wild-type strain but not in a pex7Δ or pex18Δ pex21Δ strain, and DsRed without the PTS2 signal remained cytosolic in a wild-type strain (Fig. 8B). As expected, both GFP-SKL and PTS2-DsRed caused diffuse staining when expressed in the pex13Δ mutant, which is defective in both import pathways (Fig. 8C). Coexpression of full-length Pex13p restored matrix protein import, since both marker proteins led to a punctate fluorescence staining pattern. Most interestingly, upon coexpression of the truncated Pex13p56-386, PTS2-DsRed fluorescence remained diffuse, whereas GFP-SKL gave rise to a punctate staining pattern (Fig. 8C). These results clearly demonstrate that in the absence of the N terminus of Pex13p, PTS1-dependent import can still proceed, while PTS2-dependent import is blocked, probably because the binding of Pex7p to this N-terminal domain is abolished.
FIG. 8.
Only PTS2-dependent import is blocked in the absence of the Pex13p-docking site for Pex7p. (A) Peroxisomal localization of Pex13p151-386. The UTL-7A strains expressing PTS2-DsRed as well as Pex13p1-386-GFP or Pex13p151-386-GFP were examined for GFP (left panels) and DsRed (middle panels) fluorescence. Bright-field microscopy (BF) demonstrates the structural integrity of the cells. (B) Localization of the fluorescing PTS2-DsRed marker protein. The wild-type UTL-7A and the otherwise isogenic pex7Δ and pex18Δ pex21Δ strains expressing PTS2-DsRed were streaked on oleic acid plates. After 2 days, cells were examined for DsRed fluorescence. The expression of a PTS-less DsRed in a wild-type strain was also monitored. (C) Selective complementation of PTS1-dependent import by Pex13p56-386. The pex13Δ strains coexpressing Pex13p1-386 or Pex13p56-386 in combination with GFP-PTS1 (pJR233) and PTS2-DsRed were streaked on oleic acid plates and examined after 2 days for staining patterns of the fluorescing PTS marker proteins.
Pex7p-dependent complex formation of Pex18p/Pex21p and Fox3p.
To determine where in the translocation process Pex18p/Pex21p are required, we reinvestigated the two-hybrid interactions of the PTS2 protein Fox3p with the PTS2-specific peroxins. Interaction of Fox3p with Pex7p was observed in wild-type, pex7Δ, and pex18Δ pex21Δ strains (Fig. 9A, lower panel). On the other hand, the interaction between Fox3p and Pex18p or Pex21p (data not shown) was strictly dependent on Pex7p (Fig. 9A, upper panel), which was in accordance with previous findings (34). This result demonstrated that Pex18p/Pex21p are also not required for the Pex7p-PTS2 interaction. To exclude an involvement of other yeast proteins in the PTS2 recognition event, an in vitro binding assay using bacterially expressed MBP-Pex7p was set out. MBP-Pex7p was bound to an amylose resin onto which soluble extracts from E. coli cells were loaded that had expressed either a synthetic PTS2 protein fused to GST or GST alone. Subsequent analysis of the eluted MBP-Pex7p fractions revealed that the targeting signal-containing protein was specifically retained (Fig. 9B). Thus, Pex7p alone was sufficient to recognize the PTS2 targeting signal.
The apparent requirement for Pex7p in the formation of a complex between Pex18p and Fox3p was analyzed by coimmunoprecipitation. For that matter, Pex18p was chromosomally tagged with the TAP tag at its C terminus in pex21Δ and pex7Δ strains (Fig. 9C, lower panel). Expression of the fusion protein in a pex21Δ strain background revealed that Pex18p-TAP was fully functional, as this strain grew comparably to the wild-type strain on oleic acid plates (data not shown). This fusion protein could be specifically precipitated from whole-cell extracts via its TAP tag by using IgG Sepharose (Fig. 9C, upper panel). The Pex18p-TAP precipitates were released from the column by using TEV protease and analyzed for the presence of Pex7p, which could be detected in the precipitate of the pex21Δ strain but not in those of the pex7Δ or untransformed wild-type strains (Fig. 9C, upper panel). The same precipitates were also analyzed for the presence of Fox3p. While the PTS2 cargo protein was found in the pex21Δ sample, it was absent from the precipitate derived from the pex7Δ strain (Fig. 9C, upper panel). It was therefore concluded that Fox3p can indeed associate with Pex18p in vivo, albeit only in the presence of Pex7p.
Pex7p- and Pex18p/Pex21p-dependent docking of Fox3p to Pex14p.
We then analyzed whether Fox3p would be found in a complex with the docking proteins Pex14p and Pex13p when either Pex7p or Pex18p/Pex21p were absent. This was achieved by testing for an interaction of Fox3p with Pex14p and Pex13p in a two-hybrid assay. Remarkably, a strong interaction between Fox3p and Pex14p was observed in a wild-type strain, which was absent in a pex7Δ or pex18Δ pex21Δ mutant background (Fig. 10A). On the other hand, no interaction between Fox3p and Pex13p1-100, a Pex13p fragment that strongly interacted with Pex7p, was detectable in a wild-type strain or a pex18Δ pex21Δ strain (Fig. 10B). Even in a pex14Δ strain, where Pex7p binding to Pex13p is likely to increase, no interaction between Fox3p and Pex13p was detected (Fig. 10B). These findings indicate that docking of the PTS2 cargo protein is critically dependent on both Pex18p/Pex21p and Pex7p. They further suggest that Pex14p represents the docking protein, whereas Pex13p is involved in the binding of the Pex7p-Pex18p/Pex21p complex without cargo protein.
FIG. 10.
Fox3p interacts with Pex14p in a Pex7p- and Pex18p/Pex21p-dependent manner. The docking proteins Pex14p (A) and Pex13p (B) were tested for interaction with Fox3p in a two-hybrid assay in the indicated strain backgrounds.
Pex18p and Pex21 promote the formation of an import-competent PTS2 substrate complex.
In the absence of a functional docking or translocation machinery, immunoprecipitates of myc-Pex7p contain a drastically increased amount of the PTS2 protein Fox3p (16, 37). This increased binding was interpreted to be due to the cytosolic accumulation of Fox3p in these mutants. Were Pex18p/Pex21p only to mediate the docking of Fox3p to Pex14p, then such an apparent accumulation of cargo-loaded receptor should also occur in a pex18Δ pex21Δ strain. As expected, the myc-Pex7p immunoprecipitates of a wild-type strain contained little Fox3p, whereas those of pex13Δ and pex14Δ strains showed an accumulation of Fox3p (Fig. 11A). However, in the pex18Δ pex21Δ strain, only very little Fox3p could be coprecipitated (Fig. 11A).
FIG. 11.
The Pex7p-Fox3p-containing cytosolic complex does not accumulate in a pex18Δ pex21Δ mutant. (A) Accumulation of a Fox3p-Pex7p complex in peroxin mutants. The indicated myc-Pex7p-expressing strains were analyzed for the presence of Fox3p in the immunoprecipitate of myc-Pex7p (10% of total) by immunoblotting (upper panel). The lower panel shows an immunoblot of the cell lysates (0.5% of total) that were used for precipitation. (B) The effect of overexpressing Pex7p in a pex18Δ pex21Δ strain. Tenfold dilutions of the indicated strains were spotted on oleic acid-containing plates and incubated for 5 days at 30°C. (C) Epistatic analysis of PEX14 and PEX18/PEX21. Precipitation of myc-Pex7p from the indicated oleic acid-induced cells and subsequent analysis of precipitated Fox3p were performed as described for panel A.
Since Fox3p also did not accumulate in a wild-type strain, we analyzed the possibility that in our experiment, ectopic expression of myc-Pex7p suppressed the defect of the pex18Δ pex21Δ strain. In that case, growth of the pex18Δ pex21Δ mutant on oleic acid plates should be restored when it is transformed with the myc-Pex7p expression plasmid. However, as can be seen in Fig. 11B, this strain (pex18Δ pex21Δ [myc-Pex7p]) remained deficient in utilizing oleic acid to a degree similar to that of the untransformed strain (pex18Δ pex21Δ).
Thus, Pex18p/Pex21p probably act in a step that precedes docking of Pex7p to the membrane. In that case, the observed increase of Fox3p bound to myc-Pex7p in a pex14Δ mutant would be abolished by additionally deleting PEX18 and PEX21. The amount of coimmunoprecipitated Fox3p in a pex14Δ pex18Δ pex21Δ triple-mutant strain was therefore compared to those of the pex14Δ, pex18Δ pex21Δ, and wild-type strains. Strikingly, an accumulation of Fox3p was not observed in the triple-deletion strain (Fig. 11C). This result indicates that the accumulation of the Pex7p-Fox3p complex in a pex14Δ mutant is dependent on Pex18p/Pex21p and, as a consequence, that the function of Pex18p and Pex21p is already required in the import process prior to Pex14p and thus before the Fox3p-Pex7p-Pex18p/Pex21p complex docks at the peroxisomal membrane.
DISCUSSION
Despite the identification of probably all components of the peroxisomal matrix protein import machinery in recent years (40, 47), the mechanism underlying the translocation process is still largely unknown. In this report, we have shown that S. cerevisiae Pex13p plays a direct role in PTS2-dependent peroxisomal protein import and begun to unravel the function of the PTS2-specific peroxins in the early steps of this process.
After their synthesis, PTS2 proteins such as thiolase are specifically recognized by Pex7p (36, 56). The recognition of PTS2 by Pex7p does not require S. cerevisiae Pex18p/Pex21p, whereas the latter proteins interact with Fox3p only in the presence of Pex7p (34). Our demonstration of yeast Pex7p binding a synthetic PTS2 protein in vitro goes beyond that of a previous report, where a Pex7p was used that had been immunoprecipitated from wild-type yeast extracts and thus might have contained other yeast proteins (36). Our first attempts to study in vitro the influence of Pex18p on the Pex7p-PTS2 interaction failed because bacterially expressed Pex18p did not bind Pex7p, even in the concomitant presence of PTS2 protein (data not shown). However, we could show by immunoprecipitation that such a ternary complex of Pex18p, Pex7p, and Fox3p indeed exists, and we substantiated the requirement for Pex7p in the formation of such a complex. Notwithstanding that, we found that the Fox3p-Pex7p-containing complex that accumulates in a pex14Δ strain vanished when PEX18 and PEX21 were additionally deleted. S. cerevisiae thiolase is assembled into its active dimeric form in the cytosol even in the absence of its PTS2 sequence (17), arguing against an essential role for Pex18p/Pex21p in the assembly of enzymatically active Fox3p. Rather, these proteins are required to form an import-competent Fox3p complex.
A similar function in thiolase import was proposed for Y. lipolytica Pex20p. This protein binds thiolase autonomously, in a PTS2 targeting signal-independent fashion. In fact, Pex20p forms hetero-oligomers with Fox3p, an event that is probably mandatory for the generation of an import-competent complex (49). In analogy to Pex20p, Pex18p/Pex21p could physically contact Fox3p in the Pex7p-bound conformation. In that case, the targeting signal would be bound by Pex7p, while Pex18p/Pex21p would contact both Pex7p, via their conserved motif (7), and Fox3p, which would then trigger higher-order oligomerization. Alternatively, Pex18p/Pex21p could assist in forming a Fox3p-Pex7p-containing complex by stabilizing the binding between Pex7p and Fox3p, although the existing two-hybrid data do not support this idea. Still, both cases would explain why Pex20p was able to partially complement a pex18Δ pex21Δ mutant strain (8). Oligomerization as a requirement for import is not just an idiosyncrasy of thiolase. Such a postulate was already raised upon investigating the import of Candida boidinii alcohol oxidase, which seemed to depend on the oligomerization or aggregation at the cytoplasmic side of the peroxisomal membrane (4). Since then, the ability of peroxisomes to import folded or even oligomeric proteins has been amply recorded (28, 52).
Our data also suggest that a complex is formed that contemporaneously contains membrane-associated Pex14p, Pex7p, Pex18p/Pex21p, and Fox3p (Fig. 10A). In the absence of either Pex7p or Pex18p/Pex21p, Fox3p was no longer able to interact with Pex14p. On the other hand, Fox3p failed to interact with the Pex7p-binding domain of Pex13p even in a wild-type strain (Fig. 10B). It is unlikely that the lack of interaction was caused by a masking of the interaction domain in Pex13p with the Gal4p moieties, as the same fusion construct was very efficient in recognizing Pex7p and Pex18p/Pex21p, which in turn were able to interact with the Fox3p fusion proteins. Thus, we believe that our data point to a scenario where Pex14p is the initial docking site in PTS2-dependent protein import. We also considered the possibility of Pex13p constituting the initial docking site for the empty Pex5p and Pex7p, which would only then bind their cargo proteins. However, this is hard to reconcile with the observed formation of a Pex7p-Pex18p/Pex21p-Fox3p complex in the pex13Δ mutant, where this complex must have formed in the cytosol. Future biochemical work will have to demonstrate whether cargo-loaded PTS2 receptor can indeed be found in a complex with Pex14p but not with Pex13p. In mammalian cells, the Pex7p-Pex5pL-PTS2 substrate complex also likely contacts Pex14p prior to Pex13p (30). It is worth noting that a similar conclusion was drawn for the PTS1 receptor Pex5p of Pichia pastoris, since substrate-loaded Pex5p bound Pex14p with higher affinity than Pex5p alone, whereas the opposite was true for Pex13p (50).
In contrast to Fox3p, Pex7p does not require the presence of Pex18p/Pex21p to contact Pex14p (Fig. 3C). Our in vitro binding analysis showed that this binding event can even occur in the absence of any other yeast protein. Pex7p also contacts Pex13p directly (Fig. 6), thereby ruling out the possibility of Pex18p/Pex21p being adapter proteins between Pex7p and the docking complex. Notably, Y. lipolytica Pex20p did bind S. cerevisiae Pex13p and Pex14p in the absence of Pex7p (8). Thus, in Y. lipolytica the function of a putative Pex7p might be restricted to targeting signal recognition, whereas Pex20p might be required to form an import-competent complex as well as to contact the docking machinery. Alternatively, Pex20p might contain the function of both Pex18p/Pex21p and Pex7p. In mammals, the Pex18p-like factor Pex5pL is able to interact with Pex14p via its diaromatic pentapeptide repeats (13, 41, 54). It is therefore imaginable that Pex5pL mediates the docking of Pex7p to Pex14p. However, Shimizu and colleagues demonstrated direct binding of Pex7p to Pex14p (45). In this case, it is also possible that Pex5pL, in analogy to Pex18p, is involved in the formation of a PTS2 cargo import-competent complex.
In light of our results, the role of Pex13p in PTS2 protein import could be substantiated. Deletion of the N-terminal domain of Pex13p, which specifically interacted with Pex7p, affected PTS2-dependent import but left PTS1 protein import intact. The latter observation indicated that Pex14p was correctly targeted to the peroxisomal membrane. As a consequence, the role of Pex13p in PTS2 import is not restricted to the targeting of Pex14p. Rather, Pex13p fulfills an essential step in PTS2 import that is distinct from that governed by Pex14p. This result also meant that although the import of both PTS1 and PTS2 requires Pex13p, the two pathways utilize different regions within Pex13p and therefore do not coincide at the stage of Pex13p in a more narrow sense. Mutations that specifically block one of the two routes have also been found previously for Pex8p and Pex2p, two peroxins that are possibly involved in later steps of the import cascade (22, 25).
The steps following docking are less clear. Peroxisomal import deviates considerably from the well-established import into mitochondria or the endoplasmic reticulum (43). An obvious protein translocation channel is lacking, and the ability to import folded or even oligomeric proteins must also be taken into account. Interestingly, Pex18p and Pex21p are constantly degraded due to ubiquitination, which is apparently restricted to the fraction of peroxisomally localized Pex18p/Pex21p (33). Although it remains to be determined whether the observed degradation of Pex18p/Pex21p is physiologically relevant for PTS2-dependent protein import, it could be that Pex18p/Pex21p are required not only for the formation of an import-competent PTS2 cargo protein complex but also for a step that follows docking.
Acknowledgments
We are grateful to M. Nündel for excellent technical assistance. We thank X. Hong for plasmids His6-PEX14 and His6-PEX7, R. Bahadori for plasmid pRB107, A. Hartig for plasmid pJR233, B. Seraphin for plasmid pBS1479, H. Otto for anti-myc antibodies, and particularly W. H. Kunau for the provision of plasmids, strains, and antibodies.
This work was supported by the Deutsche Forschungsgemeinschaft, grants ER178/2-3 and SFB449, and by the Fonds der Deutschen Chem. Industrie. H.R. was supported by an EMBO long-term fellowship (ALTF255-2000).
REFERENCES
- 1.Albertini, M., P. Rehling, R. Erdmann, W. Girzalsky, J. A. Kiel, M. Veenhuis, and W. H. Kunau. 1997. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89:83-92. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, P., G. Bottger, A. T. Klein, H. F. Tabak, and B. Distel. 2000. The peroxisomal membrane protein Pex13p shows a novel mode of SH3 interaction. EMBO J. 19:6382-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgartner, U., B. Hamilton, M. Piskacek, H. Ruis, and H. Rottensteiner. 1999. Functional analysis of the Zn(2)Cys(6) transcription factors Oaf1p and Pip2p: different roles in fatty acid induction of beta-oxidation in Saccharomyces cerevisiae. J. Biol. Chem. 274:22208-22216. [DOI] [PubMed] [Google Scholar]
- 4.Bellion, E., and J. M. Goodman. 1987. Proton ionophores prevent assembly of a peroxisomal protein. Cell 48:165-173. [DOI] [PubMed] [Google Scholar]
- 5.Braverman, N., G. Dodt, S. J. Gould, and D. Valle. 1998. An isoform of Pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 7:1195-1205. [DOI] [PubMed] [Google Scholar]
- 6.Chevray, P. M., and D. Nathans. 1992. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl. Acad. Sci. USA 89:5789-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodt, G., D. Warren, E. Becker, P. Rehling, and S. J. Gould. 2001. Domain mapping of human PEX5 reveals functional and structural similarities to Saccharomyces cerevisiae Pex18p and Pex21p. J. Biol. Chem. 276:41769-41781. [DOI] [PubMed] [Google Scholar]
- 8.Einwächter, H., S. Sowinski, W. H. Kunau, and W. Schliebs. 2001. Yarrowia lipolytica Pex20p, Saccharomyces cerevisiae Pex18p/Pex21p and mammalian Pex5pL fulfil a common function in the early steps of the peroxisomal PTS2 import pathway. EMBO Rep. 2:1035-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdmann, R., and W. H. Kunau. 1994. Purification and immunolocalization of the peroxisomal 3-oxoacyl-CoA thiolase from Saccharomyces cerevisiae. Yeast 10:1173-1182. [DOI] [PubMed] [Google Scholar]
- 10.Erdmann, R., M. Veenhuis, D. Mertens, and W. H. Kunau. 1989. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86:5419-5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feilotter, H. E., G. J. Hannon, C. J. Ruddell, and D. Beach. 1994. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 22:1502-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 13.Fransen, M., S. R. Terlecky, and S. Subramani. 1998. Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc. Natl. Acad. Sci. USA 95:8087-8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiki, Y. 2000. Peroxisome biogenesis and peroxisome biogenesis disorders. FEBS Lett. 476:42-46. [DOI] [PubMed] [Google Scholar]
- 15.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 16.Girzalsky, W., P. Rehling, K. Stein, J. Kipper, L. Blank, W. H. Kunau, and R. Erdmann. 1999. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J. Cell Biol. 144:1151-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glover, J. R., D. W. Andrews, and R. A. Rachubinski. 1994. Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc. Natl. Acad. Sci. USA 91:10541-10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould, S. J., G. A. Keller, and S. Subramani. 1988. Identification of peroxisomal targeting signals located at the carboxy terminus of four peroxisomal proteins. J. Cell Biol. 107:897-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould, S. J., and D. Valle. 2000. Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet. 16:340-345. [DOI] [PubMed] [Google Scholar]
- 20.Gurvitz, A., H. Rottensteiner, S. H. Kilpelainen, A. Hartig, J. K. Hiltunen, M. Binder, I. W. Dawes, and B. Hamilton. 1997. The Saccharomyces cerevisiae peroxisomal 2,4-dienoyl-CoA reductase is encoded by the oleate-inducible gene SPS19. J. Biol. Chem. 272:22140-22147. [DOI] [PubMed] [Google Scholar]
- 21.Holroyd, C., and R. Erdmann. 2001. Protein translocation machineries of peroxisomes. FEBS Lett. 501:6-10. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Y., R. Ito, S. Miura, T. Hashimoto, and M. Ito. 2000. A missense mutation in the RING finger motif of PEX2 protein disturbs the import of peroxisome targeting signal 1 (PTS1)-containing protein but not the PTS2-containing protein. Biochem. Biophys. Res. Commun. 270:717-721. [DOI] [PubMed] [Google Scholar]
- 23.Huhse, B., P. Rehling, M. Albertini, L. Blank, K. Meller, and W. H. Kunau. 1998. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J. Cell Biol. 140:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lametschwandtner, G., C. Brocard, M. Fransen, P. Van Veldhoven, J. Berger, and A. Hartig. 1998. The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J. Biol. Chem. 273:33635-33643. [DOI] [PubMed] [Google Scholar]
- 25.Liu, H., X. Tan, K. A. Russell, M. Veenhuis, and J. M. Cregg. 1995. PER3, a gene required for peroxisome biogenesis in Pichia pastoris, encodes a peroxisomal membrane protein involved in protein import. J. Biol. Chem. 270:10940-10951. [DOI] [PubMed] [Google Scholar]
- 26.Marzioch, M., R. Erdmann, M. Veenhuis, and W. H. Kunau. 1994. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 13:4908-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumura, T., H. Otera, and Y. Fujiki. 2000. Disruption of the interaction of the longer isoform of Pex5p, Pex5pL, with Pex7p abolishes peroxisome targeting signal type 2 protein import in mammals. Study with a novel Pex5-impaired Chinese hamster ovary cell mutant. J. Biol. Chem. 275:21715-21721. [DOI] [PubMed] [Google Scholar]
- 28.McNew, J. A., and J. M. Goodman. 1994. An oligomeric protein is imported into peroxisomes in vivo. J. Cell Biol. 127:1245-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otera, H., T. Harano, M. Honsho, K. Ghaedi, S. Mukai, A. Tanaka, A. Kawai, N. Shimizu, and Y. Fujiki. 2000. The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J. Biol. Chem. 275:21703-21714. [DOI] [PubMed] [Google Scholar]
- 31.Otera, H., K. Okumoto, K. Tateishi, Y. Ikoma, E. Matsuda, M. Nishimura, T. Tsukamoto, T. Osumi, K. Ohashi, O. Higuchi, and Y. Fujiki. 1998. Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol. Cell. Biol. 18:388-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purdue, P. E., and P. B. Lazarow. 2001. Peroxisome biogenesis. Annu. Rev. Cell. Dev. Biol. 17:701-752. [DOI] [PubMed] [Google Scholar]
- 33.Purdue, P. E., and P. B. Lazarow. 2001. Pex18p is constitutively degraded during peroxisome biogenesis. J. Biol. Chem. 276:47684-47689. [DOI] [PubMed] [Google Scholar]
- 34.Purdue, P. E., X. Yang, and P. B. Lazarow. 1998. Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J. Cell Biol. 143:1859-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reguenga, C., M. E. Oliveira, A. M. Gouveia, C. Sa-Miranda, and J. E. Azevedo. 2001. Characterization of the mammalian peroxisomal import machinery: Pex2p, Pex5p, Pex12p, and Pex14p are subunits of the same protein assembly. J. Biol. Chem. 276:29935-29942. [DOI] [PubMed] [Google Scholar]
- 36.Rehling, P., M. Marzioch, F. Niesen, E. Wittke, M. Veenhuis, and W. H. Kunau. 1996. The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J. 15:2901-2913. [PMC free article] [PubMed] [Google Scholar]
- 37.Rehling, P., A. Skaletz-Rorowski, W. Girzalsky, T. Voorn-Brouwer, M. M. Franse, B. Distel, M. Veenhuis, W. H. Kunau, and R. Erdmann. 2000. Pex8p, an intraperoxisomal peroxin of Saccharomyces cerevisiae required for protein transport into peroxisomes binds the PTS1 receptor Pex5p. J. Biol. Chem. 275:3593-3602. [DOI] [PubMed] [Google Scholar]
- 38.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 39.Rottensteiner, H., A. J. Kal, B. Hamilton, H. Ruis, and H. F. Tabak. 1997. A heterodimer of the Zn2Cys6 transcription factors Pip2p and Oaf1p controls induction of genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Eur. J. Biochem. 247:776-783. [DOI] [PubMed] [Google Scholar]
- 40.Sacksteder, K. A., and S. J. Gould. 2000. The genetics of peroxisome biogenesis. Annu. Rev. Genet. 34:623-652. [DOI] [PubMed] [Google Scholar]
- 41.Saidowsky, J., G. Dodt, K. Kirchberg, A. Wegner, W. Nastainczyk, W. H. Kunau, and W. Schliebs. 2001. The di-aromatic pentapeptide repeats of the human peroxisome import receptor PEX5 are separate high affinity binding sites for the peroxisomal membrane protein PEX14. J. Biol. Chem. 276:34524-34529. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schatz, G., and B. Dobberstein. 1996. Common principles of protein translocation across membranes. Science 271:1519-1526. [DOI] [PubMed] [Google Scholar]
- 44.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu, N., R. Itoh, Y. Hirono, H. Otera, K. Ghaedi, K. Tateishi, S. Tamura, K. Okumoto, T. Harano, S. Mukai, and Y. Fujiki. 1999. The peroxin Pex14p. cDNA cloning by functional complementation on a Chinese hamster ovary cell mutant, characterization, and functional analysis. J. Biol. Chem. 274:12593-12604. [DOI] [PubMed] [Google Scholar]
- 46.Smith, J. J., and R. A. Rachubinski. 2001. A role for the peroxin Pex8p in Pex20p-dependent thiolase import into peroxisomes of the yeast Yarrowia lipolytica. J. Biol. Chem. 276:1618-1625. [DOI] [PubMed] [Google Scholar]
- 47.Subramani, S., A. Koller, and W. B. Snyder. 2000. Import of peroxisomal matrix and membrane proteins. Annu. Rev. Biochem. 69:399-418. [DOI] [PubMed] [Google Scholar]
- 48.Swinkels, B. W., S. J. Gould, A. G. Bodnar, R. A. Rachubinski, and S. Subramani. 1991. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 10:3255-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Titorenko, V. I., J. J. Smith, R. K. Szilard, and R. A. Rachubinski. 1998. Pex20p of the yeast Yarrowia lipolytica is required for the oligomerization of thiolase in the cytosol and for its targeting to the peroxisome. J. Cell Biol. 142:403-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urquhart, A. J., D. Kennedy, S. J. Gould, and D. I. Crane. 2000. Interaction of Pex5p, the type 1 peroxisome targeting signal receptor, with the peroxisomal membrane proteins Pex14p and Pex13p. J. Biol. Chem. 275:4127-4136. [DOI] [PubMed] [Google Scholar]
- 51.van den Bosch, H., R. B. Schutgens, R. J. Wanders, and J. M. Tager. 1992. Biochemistry of peroxisomes. Annu. Rev. Biochem. 61:157-197. [DOI] [PubMed] [Google Scholar]
- 52.Walton, P. A., P. E. Hill, and S. Subramani. 1995. Import of stably folded proteins into peroxisomes. Mol. Biol. Cell 6:675-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westermann, B., and W. Neupert. 2000. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast 16:1421-1427. [DOI] [PubMed] [Google Scholar]
- 54.Will, G. K., M. Soukupova, X. Hong, K. S. Erdmann, J. A. Kiel, G. Dodt, W. H. Kunau, and R. Erdmann. 1999. Identification and characterization of the human orthologue of yeast Pex14p. Mol. Cell. Biol. 19:2265-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yaffe, M. P., and G. Schatz. 1984. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA 81:4819-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, J. W., and P. B. Lazarow. 1996. Peb1p (Pas7p) is an intraperoxisomal receptor for the NH2-terminal, type 2, peroxisomal targeting sequence of thiolase: Peb1p itself is targeted to peroxisomes by an NH2-terminal peptide. J. Cell Biol. 132:325-334. [DOI] [PMC free article] [PubMed] [Google Scholar]