Abstract
The eukaryotic initiation factor 4E (eIF4E), when dysregulated, transforms cells. A substantial fraction of eIF4E forms nuclear bodies that colocalize with those associated with the promyelocytic leukemia protein PML. Overexpression studies indicate that nuclear eIF4E promotes the transport of cyclin D1 mRNA from the nucleus to the cytoplasm and that PML is a key negative regulator of this function. Since previous studies used overexpression methods, the physiological relevance of eIF4E mRNA transport function or its interaction with PML remained unknown. Therefore, we monitored whether eIF4E-dependent transport could be modulated in response to environmental conditions. Here we report that cadmium treatment, which disperses PML nuclear bodies, leaves eIF4E bodies intact, leading to increased transport of cyclin D1 mRNA and increased cyclin D1 protein levels. Removal of cadmium allows PML to reassociate with eIF4E nuclear bodies, leading to decreased cyclin D1 transport and reduced cyclin D1 protein levels. In contrast, we show that treating cells with interferon increased the levels of PML protein at the PML-eIF4E nuclear body, leading to nuclear retention of cyclin D1 transcripts and reduced cyclin D1 protein levels. Neither interferon nor cadmium treatment altered cyclin D1 levels in PML−/− cells. Consistently, overexpression of a series of PML and eIF4E mutant proteins established that PML eIF4E interaction is required for the observed effects of cadmium and interferon treatment. The present study provides the first evidence that physiological factors modulate the mRNA transport functions of eIF4E and that this regulation is PML dependent.
The eukaryotic translation initiation factor eIF4E is a critical regulator of cellular growth (28). The eIF4E gene is essential since its genomic disruption is lethal in Saccharomyces cerevisiae and overexpression of antisense oligonucleotides to eIF4E is lethal in HeLa cells (1, 9). eIF4E promotes growth, and its overexpression leads to oncogenic transformation of immortalized cell lines (15-17). Increased levels of eIF4E are correlated with a poor clinical outcome in a variety of human cancers including breast cancer and several non-Hodgkin's B-cell lymphomas (22, 34). eIF4E functions in the rate-limiting step of translation initiation, where it binds the 7-methylguanosine (m7G) cap of mRNA and recruits the given transcript to the translation machinery (28). Its functions in translation are conserved from yeast to humans (28). The oncogenic potential of eIF4E lies, at least in part, in its ability to inappropriately translate growth-promoting transcripts (28).
Intriguingly, up to 68% of eIF4E is found in the nucleus, where it forms discrete multiprotein structures that we refer to as eIF4E nuclear bodies (10, 16). We and others have shown that eIF4E nuclear bodies are found in a variety of organisms including yeast, Drosophila, Xenopus, mice, and humans (8, 12, 14, 18, 29). Nuclear eIF4E promotes transport from the nucleus to the cytoplasm of a subset of transcripts including cyclin D1 mRNA but not of products of housekeeping genes such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and actin (8, 14, 26). The specificity of transport is thought to lie in the structure of the untranslated regions (UTRs) of these transcripts (26). Cyclin D1 has a complex, highly structured UTR, whereas GAPDH and actin have relatively short, unstructured UTRs. eIF4E overexpression stimulates the transport of cyclin D1 transcripts from the nucleus to the cytoplasm (26). In these studies, alterations in cyclin D1 mRNA levels were not observed and cyclin D1 transcripts were not loaded onto ribosomes more efficiently than other transcripts (26). Further, eIF4E requires its cap binding activity for its transport function (8). Importantly, this transport activity is correlated with its ability to oncogenically transform cells (8).
The majority of eIF4E nuclear bodies colocalize with those associated with the promyelocytic leukemia protein PML (8, 14). PML and eIF4E colocalize and coimmunoprecipitate in a variety of cell lines (8, 14). PML nuclear bodies, also known as ND10s or PML oncogenic domains, are multiprotein complexes found in all mammalian cell lines reported (19, 21, 23). Over 50 proteins have been reported to associate with these structures (19). PML plays well-established roles in growth suppression, where it induces G1 arrest, apoptosis, and suppression of oncogenic transformation (21). Thus, its activities contrast with the growth-promoting characteristics of eIF4E. PML nuclear bodies are disrupted as a result of a chromosomal translocation in acute promyelocytic leukemia (21). Here, treatment with all-trans-retinoic acid results in reassociation of PML with nuclear bodies, and this correlates with remission in patients. In general, disruption of PML nuclear bodies is correlated with a loss of the growth-suppressive properties of PML (21).
The integrity of PML nuclear bodies is sensitive to a variety of pathogenic conditions. PML nuclear bodies are disrupted by a wide range of viruses including herpesvirus, human foamy virus, and rabies virus (24). PML bodies are relocated to the cytoplasm by lymphocytic choriomeningitis virus (LCMV) and human immunodeficiency virus infection (4, 32). Studies with LCMV and human foamy virus indicate that the PML protein is required, at least in part, for the interferon response of cells to these viruses (10, 25). In uninfected cells, alpha, beta, and gamma interferons (IFN-α, IFN-β, and IFN-γ) stimulate PML production, resulting in PML nuclear bodies approximately twice the size of those found in untreated cells (20, 24). Conversely, treatment with the heavy metal cadmium leads to dispersion of PML nuclear bodies (20). These findings led to the suggestion that PML plays key roles in response to environmental stress and to viral infection (19).
PML negatively regulates the mRNA transport and oncogenic transformation activities of eIF4E (8, 13, 14). The PML protein, through its RING domain, directly interacts with eIF4E, and this association induces a conformational change in the eIF4E protein that reduces the affinity of eIF4E for the m7G cap of mRNA (8, 13). This results in suppression of eIF4E-dependent cyclin D1 mRNA transport (8, 13). Thus, PML overexpression leads to retention of cyclin D1 mRNA in the nucleus and subsequently lower cyclin D1 protein levels, with no alterations in cyclin D1 transcript levels (8, 14). PML-induced suppression of mRNA transport is correlated with its ability to suppress eIF4E-mediated oncogenic transformation (8).
Unlike eIF4E, PML expression is limited to higher eukaryotes and no PML gene is found in S. cerevisiae (http://genome-www.stanford.edu), Drosophila (http://www.fruitfly.org), or Arabidopsis (http://www.arabidopsis.org) genomes (2, 8, 29). Further searches in the protein architecture database SMART (http://smart.embl-heidelberg.de), using adjacent signature motifs of PML and the RING and B boxes as a criteria, indicated that no protein with both of these motifs was encoded in either the S. cerevisiae or the Arabidopsis genome. This lack of evolutionary conservation led us to suggest that PML was a mammalian modulator of an evolutionarily older organelle composed of, in part, eIF4E (2, 8). However, these previous studies relied on overexpression of PML and eIF4E proteins.
In the present study, we monitored the ability of PML to regulate eIF4E function under physiological conditions. In addition, it was not known whether eIF4E-dependent transport could be modulated in response to environmental conditions. To investigate this, we monitored the ability of eIF4E to act in mRNA transport under these conditions and assessed the importance of PML for these stress responses. Our findings strongly suggest that PML is a key stress-responsive regulator of eIF4E mRNA-dependent transport and that the ability of PML to inhibit eIF4E is dependent on its association with the eIF4E nuclear body.
MATERIALS AND METHODS
Cell culture and treatments.
Cells were maintained in 5% CO2 at 37°C in Dulbecco's modified Eagle's medium (DMEM) (Gibco BRL, Life Technologies, Grand Island, N.Y.) supplemented with 10% fetal bovine serum (FBS) for NIH 3T3 cells or 20% FBS for PML−/− cells (28) (Gibco BRL, Life Technologies), 100 U of penicillin per ml, and 100 U of streptomycin per ml. When indicated, the cells were treated with 50 μM CdCl2 for 6 h or with 1000 U of mouse IFN-γ (PBL Biomedical Laboratories) per ml for 18 h (20). For recovery experiments, CdCl2-treated cells were washed four times with 1× phosphate-buffered saline (PBS) (pH 7.4) and incubated for 24 h in CdCl2-free medium.
Antibodies.
Well-characterized polyclonal antibodies to PML were a kind gifts of Gerd Maul and of Paul Freemont (3), the monoclonal antibody to PML, MAb 5E10 was a kind gift from L. de Jong (30), and the eIF4E MAb (Transduction Laboratories) was previously characterized (6, 8, 14). Identical results were obtained with either PML polyclonal antibody.
Constructs.
Full-length PML (69-kDa isoform) and eIF4E constructs were as described (reference 8 and references therein). Vector controls included the empty vector for eIF4E (pMV) and for PML (pEFPLINK) as described previously (8, 15). No differences were seen between these empty vectors on transfection or when both vectors were transfected in these assays.
Cell transfections.
NIH 3T3 and PML−/− cells were plated at 1.5 × 105 cells/35-mm tissue culture dish prior to transfection. Transfections were performed in serum-containing medium (DMEM supplemented with 10% FBS) using GeneJammer transfection reagent (Stratagene) as specified by the manufacturer. In NIH 3T3 cell transfections, 1 μg of pMV (which contains G418 resistance), pMV-eIF4EWT, and pMVeIF4EW73A mutant were used. PML−/− cells were transfected as above and with 1 μg of pEFPLINK vector, PML-pEFPLINK, and PMLRING mutant-pEFPLINK. PML−/− cells were additionally transfected with 0.2 μg of a vector containing a puromycin selectable marker. NIH 3T3 cells were selected in G418-containing medium (DMEM supplemented with 10% FBS, 100 U of penicillin per ml, and 100 U of streptomycin per ml). For PML−/− cells, selection was done in medium (DMEM supplemented with 20% FBS, 100 U of penicillin per ml, and 100 U of streptomycin per ml) containing 1 mg of G418 per ml and 2 μg of puromycin per ml. In both cases, selection agents were added 24 h after transfection. Using GeneJammer, we routinely obtain transfection efficiencies above 85% as assayed by confocal microscopy.
Cellular fractionation.
Preparation of cytoplasm-free nuclei was previously described (14, 26). Cells were trypsinized, rinsed twice in ice-cold 1× PBS (pH 7.4), and resuspended with slow pipetting in lysis buffer B (10 mM Tris [pH 8.4], 140 mM NaCl, 1.5 mM MgCl2, 0.5 % Nonidet P-40, 1 mM dithiothreitol, RNasin [100 U/ml] [Promega, Madison, Wis.]) in the presence of protease inhibitors (Roche Diagnostics GmbH, Indianapolis, Ind.). Nuclear suspensions were centrifuged at 1,000 × g for 3 minutes at 4°C, and the supernatant was saved as the cytoplasmic fraction. Nuclear pellets were resuspended in lysis buffer B. One-tenth volume of the detergent (3.3% [wt/vol] sodium deoxycholate, 6.6% [vol/vol] Tween 40) was added under slow vorterxing, and the nuclear suspension was incubated on ice for 5 min. Nuclei were pelleted by centrifugation at 1,000 × g for 3 min at 4°C, and the supernatant (postnuclear fraction) was saved and added to the cytoplasmic fraction. Together, these are considered the cytoplasmic fraction. The nuclei were rinsed once in lysis buffer B. This protocol yielded intact nuclei as determined by light microscopy, with no significant cytoplasmic contamination as determined from the tRNALys content (see Fig. 5 and 6) (26). Further, no significant contamination of the cytoplasm by nuclei was observed, as determined by the small amount of U6 small nuclear RNA (snRNA) present (see Fig. 5 and 6) (26).
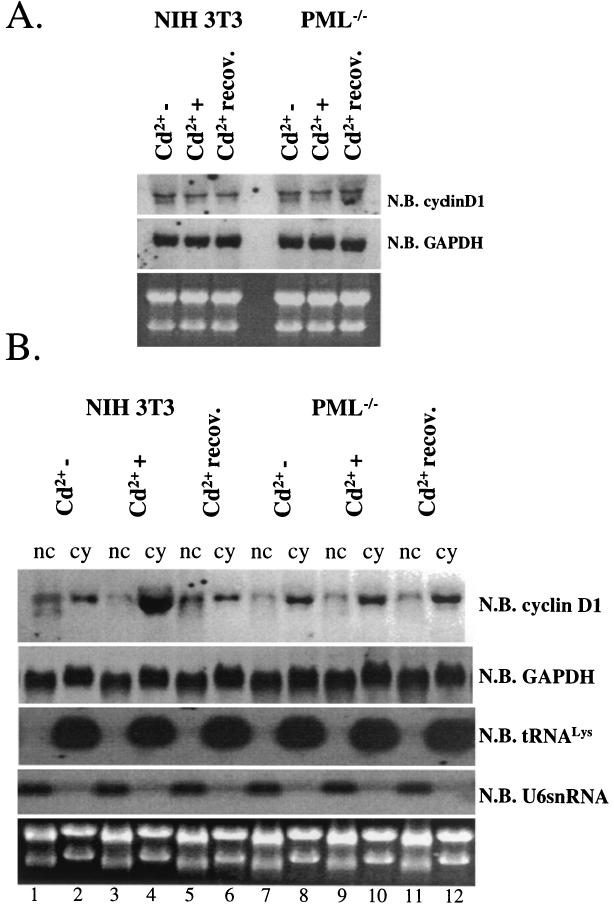
FIG. 5.
Cadmium chloride treatment results in the transport of cyclin D1 transcripts from the nucleus to the cytoplasm in a PML-dependent manner. (A) Total cyclin D1 or GAPDH mRNA was monitored by Northern blot (N.B.) analysis in NIH 3T3 and PML−/− cells. 28S and 18S rRNA show the quality of the RNA. (B) RNA, treated as described in the text, was fractionated into nuclear (nc) and cytoplasmic (cy) fractions and analyzed by Northern blot methods. All RNAs shown were isolated from the same set of experiments. tRNALys, a mainly cytoplasmic RNA, and U6 snRNA, a nuclear RNA, are shown to demonstrate the quality of the fractionation (14, 26).
FIG. 6.
IFN-γ treatment results in the retention of cyclin D1 transcripts in the nucleus in a PML-dependent manner. (A) Total cyclin D1 or GAPDH mRNA was monitored by Northern blot (N.B.) analysis in NIH 3T3 and PML−/− cells. 28S and 18S rRNAs are shown as loading controls. Cells were treated with IFN-γ (IFNγ+) or untreated (IFNγ−) as indicated. (B) RNA, treated as described in the text, was fractionated into nuclear (nc) and cytoplasmic (cy) fractions and monitored by Northern blot analysis. All RNAs shown were isolated from the same set of experiments. tRNAlys, a mainly cytoplasmic RNA, and U6 snRNA, a nuclear RNA, are shown to demonstrate the quality of the fractionation (14, 26). 18S and 28S rRNA demonstrate the quality of the RNA.
Northern blot analysis.
Total RNA and RNA from the cytoplasmic and nuclear fractions were isolated by a Trizol procedure as specified by the manufacturer (Gibco BRL, Life Technologies), RNA from the nuclear fraction was treated with RNase-free DNase I (Promega). Purified RNA was quantified by spectrophotometry. A 5-μg portion of total and fractionated RNA was loaded onto a 1% formaldehyde/agarose gel and subsequently transferred to a positively charged nylon membrane (Roche Diagnostics GmbH). Membranes were prehybridized in ULTRAhyb buffer (Ambion, Austin, Tex.) for 1 h at 45°C and probed with cyclin D1 cDNA probe (20 pM) (14), GAPDH cDNA probe (5 pM) (Ambion) (14), tRNALys antisense oligoprobe (5′-7CTCATGCTCTACCGACTGAGCTAGCCGGGC-3′ [30 pM]) (26), and U6 antisense oligoprobe (5′-7GAATTTGCGTGTCATCCTTGCGCAGGGGCCATGCTAA-3′ [30 pM]) (26) in the same buffer for 16 h at 45°C. cDNA probes were biotinylated using the BrightStar Psoralen-Biotin kit, and signals were detected using CDP Star chemiluminescence (Ambion) as specified by the manufacturer.
Western blot analysis.
Cells were scraped in 1× PBS (pH 7.4), centrifuged and lysed in RIPA buffer supplemented with complete protease inhibitors. Whole-cell protein extracts (20 μg) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7.5 and 15% polyacrylamide) using mouse monoclonal anti-eIF4E Ab (BD Transduction Laboratories, Franklin Lakes, N.J.), mouse monoclonal anti-cyclin D1 Ab (BD PharMingen, Franklin Lakes, N.J.), rabbit polyclonal anti-PML Ab, mouse monoclonal anti-PML (5E10) (30), mouse monoclonal anti-actin Ab (AC 40 [Sigma, St. Louis, Mo.]), and rabbit polyclonal anti-IRF-1 Ab (M-20 [Santa Cruz Biotechnology]). All primary antibodies were used at 1:4,000 except MAb 5E10, which was used at 1:100. Secondary antibodies were used at 1:20,000, and signals were revealed by chemiluminescence (SuperSignal West Pico; Pierce).
Immunofluorescence and laser-scanning confocal microscopy.
Cells were rinsed twice in 1× PBS (pH 7.4), fixed in 100% methanol at −20°C, blocked, and permeabilized with blocking buffer (10% FBS and 0.1% Tween 20 in 1× PBS [pH 7.4]) as described previously (reference 8 and references therein). Fixed and permeabilized cells were incubated with mouse anti-eIF4E MAb (BD Transduction Laboratories) and rabbit polyclonal anti-PML Ab in blocking buffer for 2 h at room temperature. The antibodies were used at 2μg/ml and 1:100, respectively. After incubation with primary antibodies, the cells were washed three times in 1× PBS (pH 7.4) and incubated with fluorescein isothiocyonate (FITC)-conjugated donkey anti-mouse Ab and Texas Red-conjugated donkey anti-rabbit Ab (Jackson ImmunoResearch Laboratories) for 45 min at room temperature. Transfected cells were incubated in a 1:10 dilution of mouse monoclonal anti-PML MAb 5E10 for the same time and then incubated with Texas Red-conjugated donkey anti-mouse Ab for 45 min. When the FITC-conjugated eIF4E Ab was used, transfected cells were additionally fixed with 3.7% paraformaldehyde for 10 min at room temperature, washed, and then incubated with a 1:20 dilution of FITC-conjugated mouse monoclonal anti-eIF4E Ab (BD Transduction Laboratories) at 4°C overnight. The use of the eIF4E FITC-conjugated Ab gives identical results to experiments which use the unconjugated MAb eIF4E (8). In both cases, secondary antibodies were used at 1:150 in blocking buffer. After secondary-antibody incubation, the coverslips were washed three times in 1× PBS (pH 7.4), mounted in Vectashield with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories Inc.), and sealed. Fluorescence was observed using a 100× objective, further magnified by a zoom of 2, on a Leica inverted scanning confocal microscope (with a UV laser) exciting at 488 nm (green), 568 nm (red), or 351/364 nm (blue). No cross talk was observed between channels. Micrographs represent single sections with a thickness of 300 nm. Experiments were repeated three times with more than 500 cells in each sample.
RESULTS
Environmental factors modulate the PML-eIF4E interaction.
We investigated whether treatments known to alter the subcellular distribution of PML nuclear bodies altered its ability to associate with eIF4E nuclear bodies and therefore to regulate eIF4E-dependent mRNA transport. Previous reports indicated that treatment of cells with IFN increased the production of PML and led to approximately twice the number of PML nuclear bodies, which were approximately twice as large as normal (7, 20). In contrast, treatment with cadmium disperses PML nuclear bodies (20).
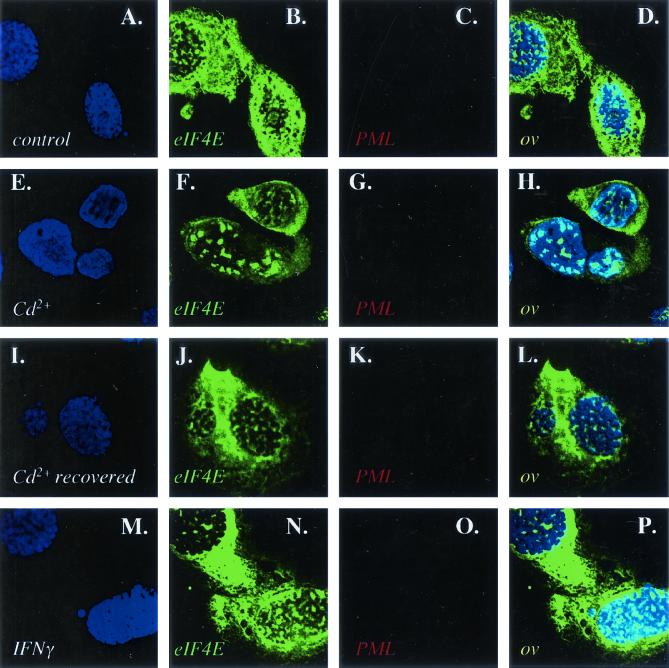
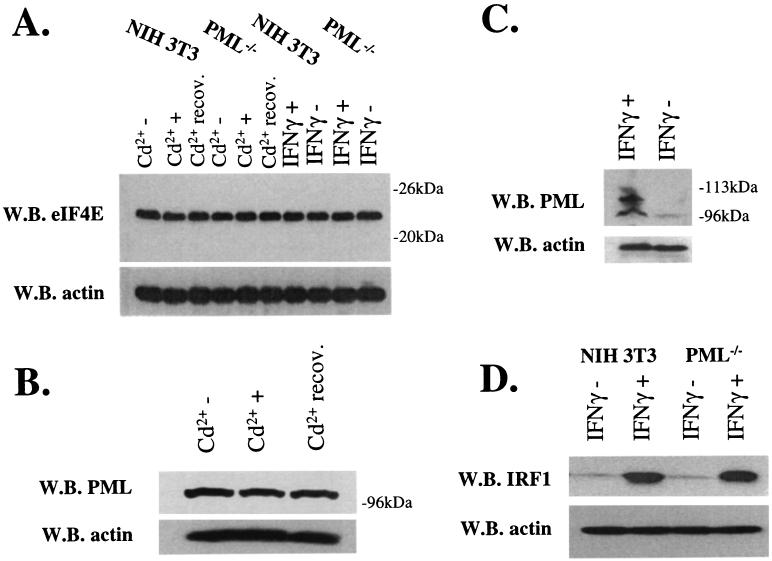
We used immunofluorescence in conjunction with confocal laser microscopy to determine the effects of these treatments on the colocalization of PML and eIF4E in NIH 3T3 cells (Fig. 1). We used well-characterized antibodies to PML and eIF4E (3, 6, 8). Untreated control cells are shown for comparison (Fig. 1A to D). Here substantial colocalization is observed between PML and eIF4E nuclear bodies, consistent with previous reports for NIH 3T3 as well as other cell lines (8, 14). Cadmium treatment for 6 h disrupted PML nuclear bodies, as reported previously (20), moving much of the PML protein to the cytoplasm (Fig. 1E to H). In contrast, eIF4E retained its nuclear localization, with some minor alterations in body morphology, where bodies appeared slightly larger. If the cadmium was removed and the cells were allowed to recover, PML nuclear bodies reappeared and colocalized with eIF4E (Fig. 1I to L). These recovered cells closely resembled untreated controls in terms of the subcellular distribution of both PML and eIF4E proteins. Notably, cadmium treatment only slightly altered the morphology of eIF4E nuclear bodies found in a cell line derived from PML−/− mouse embryo fibroblasts (35), where again eIF4E nuclear bodies appeared slightly larger (compare Fig. 2B and F). In addition, cadmium treatment did not alter PML or eIF4E protein levels in NIH 3T3 or PML−/− cells (Fig. 3A and B). Further, no alterations were observed in eIF4E levels in PML−/− cells (Fig. 3A).
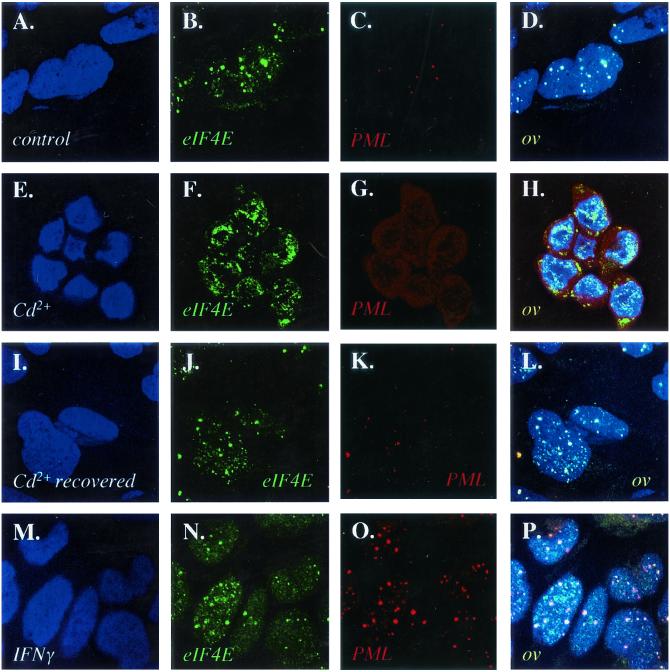
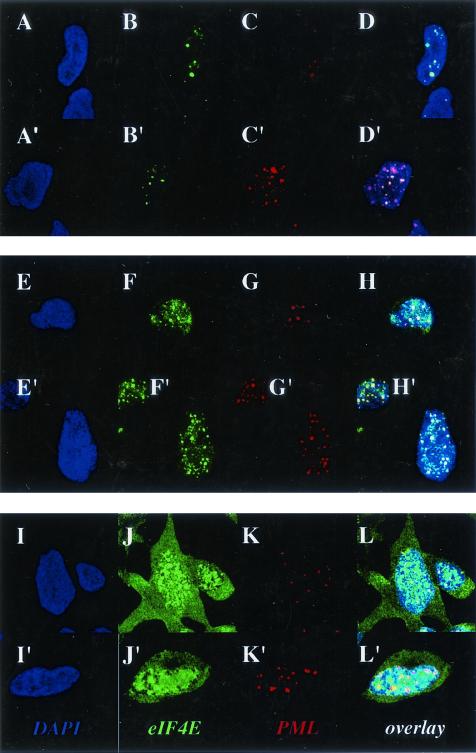
FIG. 1.
Effects of IFN and cadmium chloride on the subcellular localization of PML and eIF4E proteins in NIH 3T3 cells. Cells were stained with a PML polyclonal antibody which recognizes mouse PML in red and an eIF4E monoclonal antibody showing eIF4E in green. DAPI staining is shown to identify the nucleus (blue). The PML-eIF4E overlay is shown in light yellow. The objective is 100× with further twofold magnification. Confocal micrographs represent single optical sections through the plane of the cell. Cells were treated as indicated with untreated controls (A to D), cadmium treated (E to H), recovered from cadmium treatment (I to L) and IFN-γ treated (M to P).
FIG. 2.
Effects of IFN and cadmium chloride treatment on eIF4E in PML−/− cells. Cells were treated as described in the legend to Fig. 1. As expected, there is no PML staining in these PML null cells. The objective is 100× with further twofold magnification. Confocal micrographs represent single optical sections through the plane of the cell.
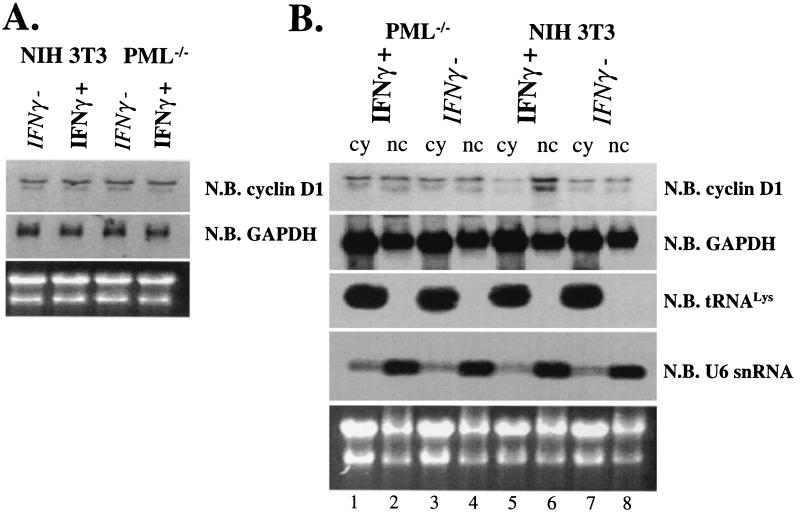
FIG. 3.
IFN and cadmium chloride effects on eIF4E and PML protein levels. Standard Western blot (W.B.) methods were used to monitor the production of proteins as indicated. Actin is shown as a loading control. (A and B) Cd2+−, untreated; Cd2++, treated for 6 h; Cd2++ recov., cells treated with cadmium, washed, and allowed to recover for 24 h. (C and D) IFNγ+ and IFNγ− indicate cells treated with IFN-γ and untreated controls, respectively. Using this PML polyclonal antibody, we observed mainly the isoform which migrates near the 96-kDa marker, probably the 97-kDa isoform, induction of higher-molecular-weight species after IFN treatment (C), similar to those reported previously (7), may represent SUMO modified forms of PML. IRF-1 migrates just above the 52-kDa molecular mass marker, consistent with its known size (D).
In contrast to cadmium treatment, treatment of cells with IFN-γ led to an increase in the number and in size of PML nuclear bodies (Fig. 1M to P), consistent with previous reports (7, 20). After IFN-γ treatment, PML and eIF4E nuclear bodies still colocalized; however, there was more PML per PML-eIF4E nuclear body than in untreated cells (compare Fig. 1D and P). Western analysis demonstrates that PML expression was induced after IFN-γ treatment and that other higher-molecular-weight and/or modified isoforms of PML became expressed, as reported previously (7, 20), whereas eIF4E protein levels were unchanged (Fig. 3A and C). IFN-γ treatment did not alter the levels or morphology of eIF4E nuclear bodies found in PML−/− cells (Fig. 2M to P and 3A). IFN-γ treatment of either NIH 3T3 or PML−/− cells led to an increase in interferon regulatory factor 1 (IRF-1), indicating that both cell types respond to IFN-γ (Fig. 3D).
Localization of PML to eIF4E nuclear bodies reduces cyclin D1 protein levels.
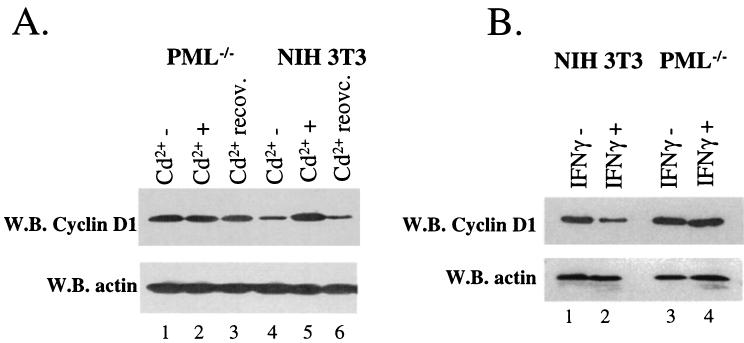
The production of cyclin D1 protein was monitored by Western analysis with cells treated as described above. Cadmium treatment for 6 h resulted in an increase in cyclin D1 protein levels whereas actin levels were unaltered (Fig. 4A, lanes 4 and 5). If the cadmium was removed, and the cells were allowed to recover (see Materials and Methods), cyclin D1 protein levels were reduced to levels similar to those observed prior to treatment (lanes 4 and 6). Interestingly, no effects of cadmium treatment on cyclin D1 levels were observed in PML−/− cells (lanes 1 to 3). These data strongly suggest that this response to cadmium is mediated through the PML protein, probably through the PML nuclear body.
FIG. 4.
Cadmium chloride and IFN treatments alter cyclin D1 protein levels in a PML-dependent manner. Western blot (W.B.) analysis was used to monitor cyclin D1 and actin protein levels as indicated. Cadmium (A) and IFN-γ (B) treatments were as described in the legend to Fig. 3.
Similar studies were conducted to observe the effects of IFN-γ on cyclin D1 protein production. IFN-γ treatment of NIH 3T3 cells led to decreased cyclin D1 protein levels (Fig. 4B, lanes 1 and 2), consistent with other reports (reference 37 and references therein). In contrast, IFN-γ treatment of PML−/− cells did not alter cyclin D1 levels (lanes 3 and 4). Thus, there is a correlation between that higher concentrations of PML at the PML-eIF4E nuclear bodies and reduced production of cyclin D1 protein levels.
Dispersal of PML nuclear bodies by cadmium treatment deregulates transport of cyclin D1 transcripts.
We extended these studies to determine whether the effects of cadmium treatment resulted in alterations in cyclin D1 transcript levels (Fig. 5A). Northern analyses of NIH 3T3 cells treated with cadmium, recovered cells, and untreated controls indicate that cyclin D1 mRNA levels are not altered by cadmium treatment. Similar results were observed in PML−/− cells. GADPH is shown as a loading control, and 28S rRNA and 18S rRNA indicate the quality of the RNA. Thus, the effects of cadmium on cyclin D1 protein production are not transcriptional and, further, the stability of the cyclin D1 mRNA is not altered by this treatment.
As demonstrated above, cadmium treatment disrupts the subcellular distribution of PML, potentially leaving eIF4E nuclear bodies unregulated. Taken together, these findings suggested that disruption of the PML eIF4E interaction resulted in dysregulated transport of cyclin D1 mRNA. To investigate this possibility, NIH 3T3 or PML−/− cells were fractionated into nuclear and cytoplasmic components and the amount of cyclin D1 mRNA in each fraction was monitored by Northern analysis (Fig. 5B). The quality of the fractionation was assessed by monitoring U6 snRNA, a nuclear RNA, and tRNALys, a cytoplasmic RNA, as described previously (14, 26). Note that Northern analyses for all of these RNAs (cyclin D1, GAPDH, U6 snRNA, and tRNALys) were done using RNAs isolated from the same experiments.
Prior to cadmium treatment, the amount of cyclin D1 mRNA was nearly equal in the nuclear and cytoplasmic fractions (Fig. 5B, compare lanes 1 and 2). However, cadmium treatment caused a substantial accumulation of cyclin D1 mRNA in the cytoplasm, consistent with the finding that cadmium treatment increases cyclin D1 protein levels. After recovery from cadmium treatment, the relative amounts of cyclin D1 mRNA in the nucleus and the cytoplasm were similar to those observed in untreated cells (compare lanes 1 and 2 with lanes 5 and 6). This redistribution of cyclin D1 mRNA is correlated with re-formation of PML nuclear bodies (Fig. 1I to L). In contrast, cadmium treatment of PML−/− cells did not alter the subcellular distribution of cyclin D1 mRNA (Fig. 5B, lanes 7 to 12). Note that none of these treatments altered the distribution of GAPDH mRNA in either cell line. Taken together, these data strongly suggest that disruption of the subcellular distribution of the PML protein, in particular disruption of the PML eIF4E interaction, results in dysregulation of eIF4E dependent transport of cyclin D1 mRNA from the nucleus to the cytoplasm.
IFN treatment inhibits eIF4E-dependent transport of cyclin D1 mRNA.
IFN treatment increases the amount of PML and the PML-eIF4E nuclear body, potentially inhibiting the transport functions of the eIF4E nuclear body. Similar to the above studies with cadmium, we monitored the distribution of cyclin D1 mRNA in IFN-γ-treated and untreated NIH 3T3 and PML−/− cells. Northern analysis of total-cell lysates indicated that IFN-γ treatment did not alter the overall levels of cyclin D1 or GAPDH mRNA in either cell line (Fig. 6A). Fractionation studies indicated that IFN-γ treatment of NIH 3T3 cells led to a nuclear accumulation of cyclin D1 transcripts (Fig. 6B, compare lanes 6 and 8). This is consistent with the downregulation of cyclin D1 protein levels observed in Fig. 4B. IFN-γ treatment did not alter the subcellular distribution of cyclin D1 mRNA in PML−/− cells (lanes 1 to 4). IFN-γ treatment of either NIH 3T3 or PML−/− cells resulted in no change in the GAPDH mRNA distribution. The quality of the fractionation is demonstrated by the distributions of U6 snRNA, which is enriched in the nuclear fraction, and tRNALys, which is enriched in the cytoplasmic fraction, as expected (14, 26). As with the cadmium treatments, Northern analyses for all of these RNAs (cyclin D1, GAPDH, U6 snRNA, and tRNALys) were done using RNAs isolated from the same experiments.
Direct interaction between PML and eIF4E is required for the effects of cadmium on cyclin D1 transport.
To establish a causal link between the observed effects on eIF4E dependent cyclin D1 transport and the localization of PML we conducted a series of overexpression studies with PML and eIF4E mutants. The initial experiments involved the transfection of PML−/− cells with PML, where we observed the effects on cyclin D1 protein levels (Fig. 7A). Similar results were To establish that these effects occur through the PML-eIF4E interaction, we carried out a series of overexpression studies. In these experiments, we monitored the effect of overexpressing wild-type and W73A eIF4E in NIH 3T3 cells on this process, where we could monitor the IFN response of endogenous PML. Similar to studies with endogenous eIF4E, levels of overexpression of either W73A or wild-type eIF4E were not altered by interferon treatment (Fig. 9B). Further, induction of endogenous PML by IFN was not altered by overexpression of these proteins (Fig. 9C). Finally, overexpression of either wild-type or W73A eIF4E did not alter the induction of IRF-1, and similar results were obtained to those of studies not involving transfection (compare Fig. 9C with Fig. 3).
FIG. 7.
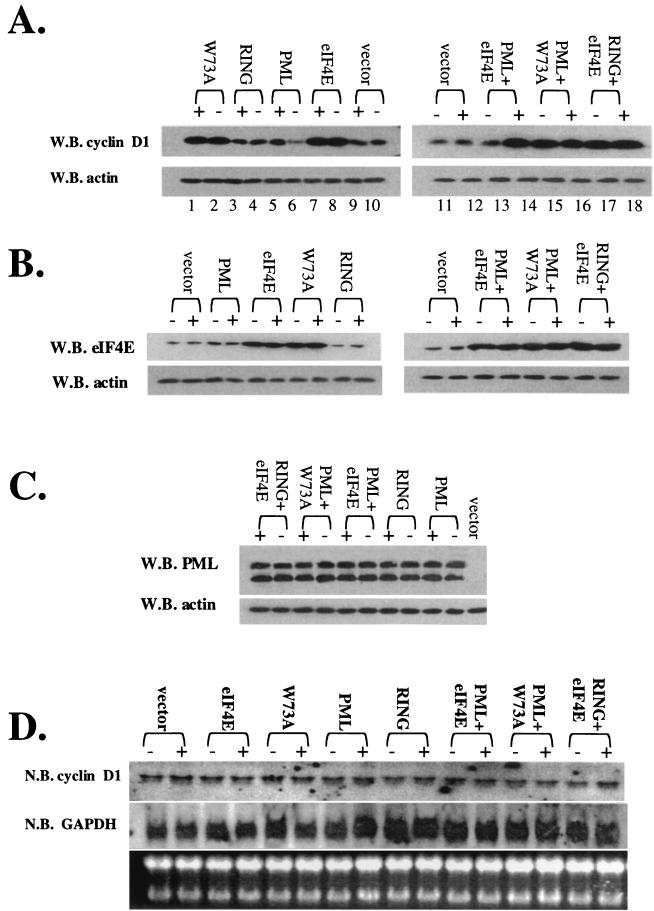
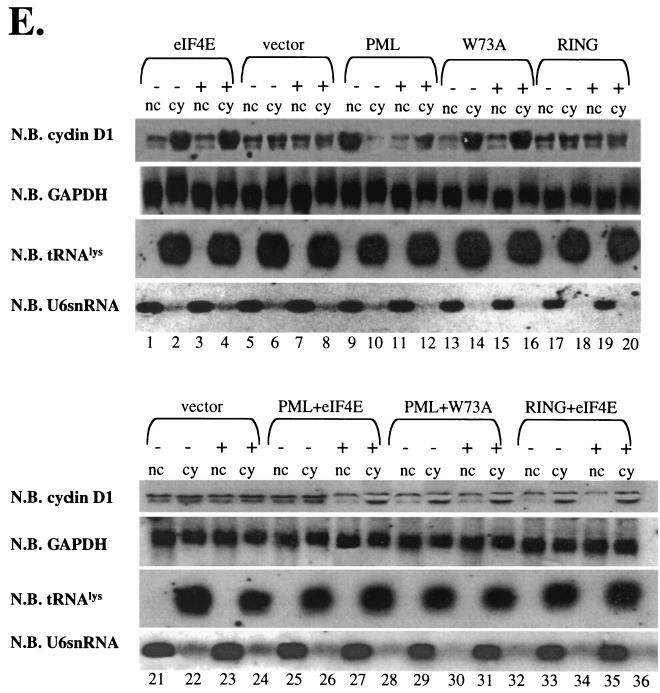
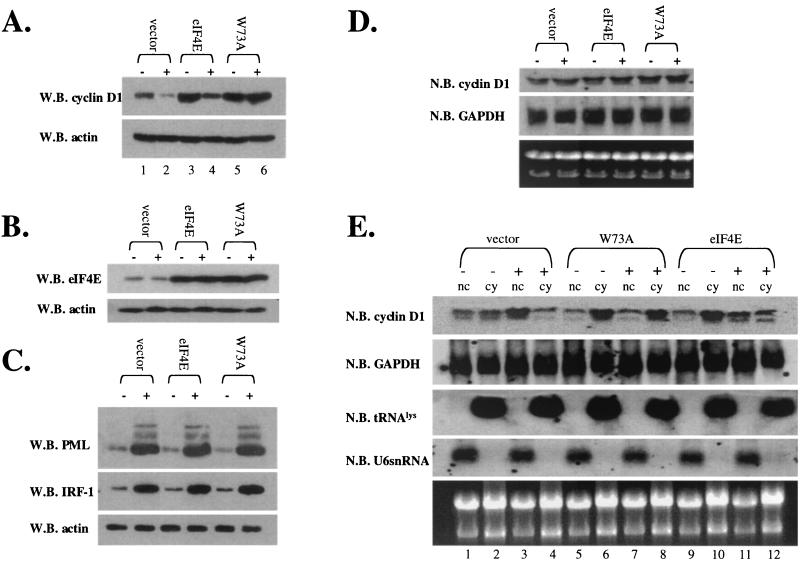
Effects of overexpression of PML and eIF4E wild-type and mutant proteins on the cadmium response in PML−/− cells. Cells were treated for 6 h with cadmium (+) or left untreated (−). (A) Cyclin D1 protein levels were monitored using standard Western blot (W.B.) methods. (B and C) Cadmium treatment does not alter PML or eIF4E overexpression. The MAb 5E10 anti-PML antibody was used to detect expression of the 69-kDa isoform of PML used in these overexpression studies. The upper band represents the full-length isoform, and the lower band represents a degradation product. (D) Total cyclin D1 mRNA levels were monitored by Northern blot (N.B.) analysis for the given transfection experiment. (E) RNA treated as described was fractionated into nuclear (nc) and cytoplasmic (cy) fractions. All RNAs shown were isolated from the same set of experiments. tRNAlys, a mainly cytoplasmic RNA, and U6 snRNA, a nuclear RNA, are shown to demonstrate the quality of the fractionation (14, 26). RING indicates the construct with the point mutation in the RING of full-length PML (PML RING) as described in the text. W73A indicates the eIF4E W73A mutant.
FIG. 9.
Effects of eIF4E overexpression in NIH 3T3 cells on the ability of IFN-induced PML to suppress cyclin D1 protein levels. Cells were transfected with vector, eIF4E, or the W73A eIF4E mutant as indicated. IFN treatment was carried out as described in the text. Cells were treated with IFNγ (+) or left untreated (−) as indicated. (A) Cyclin D1 protein levels were determined by Western blot (W.B.) analysis. (B) Overexpression of eIF4E levels is shown by Western blot analysis. (C) Induction of PML and IRF-1 is shown. PML is detected using a polyclonal antibody that recognizes mouse PML, and the higher-molecular-weight isoforms are observed as described previously (7). Actin is shown as a loading control. IFN treatment does not alter the overexpression of either wild-type or W73A eIF4E as indicated by Western blot analysis. (D) Total cyclin D1 or GAPDH mRNA was monitored by Northern blot (N.B.) analysis. (E) RNA, treated as described in the text, was fractionated into nuclear (nc) and cytoplasmic (cy) fractions and monitored by Northern blot analysis. All RNAs shown were isolated from the same set of experiments. tRNALys, a mainly cytoplasmic RNA, and U6 snRNA, a nuclear RNA, are shown to demonstrate the quality of the fractionation (14, 26). 18S and 28S rRNA demonstrate the quality of the RNA.
Overexpression of either wild-type or W73A eIF4E resulted in increased cyclin D1 protein levels relative to vector controls (Fig. 9A). Cyclin D1 levels were decreased in eIF4E-overexpressing cells in response to IFN treatment (Fig. 9A, compare lanes 3 and 4), consistent with our results with endogenous proteins (Fig. 4B). Further, confocal microscopy data indicate that transfected eIF4E and endogenous PML colocalized before and after IFN treatment (Fig. 10) as observed for endogenous proteins (Figure 1). However, more PML was present at the eIF4E nuclear body after IFN treatment, as observed for endogenous proteins (compare Fig. 1 with Fig. 10E to H and E′ to H′). In contrast to results with wild-type eIF4E, cyclin D1 levels were not lowered in response to IFN in cells overexpressing the W73A mutant (compare lanes 5 and 6). This is consistent with our previous studies showing the PML cannot directly associate with this mutant and therefore cannot repress mRNA transport mediated by it.
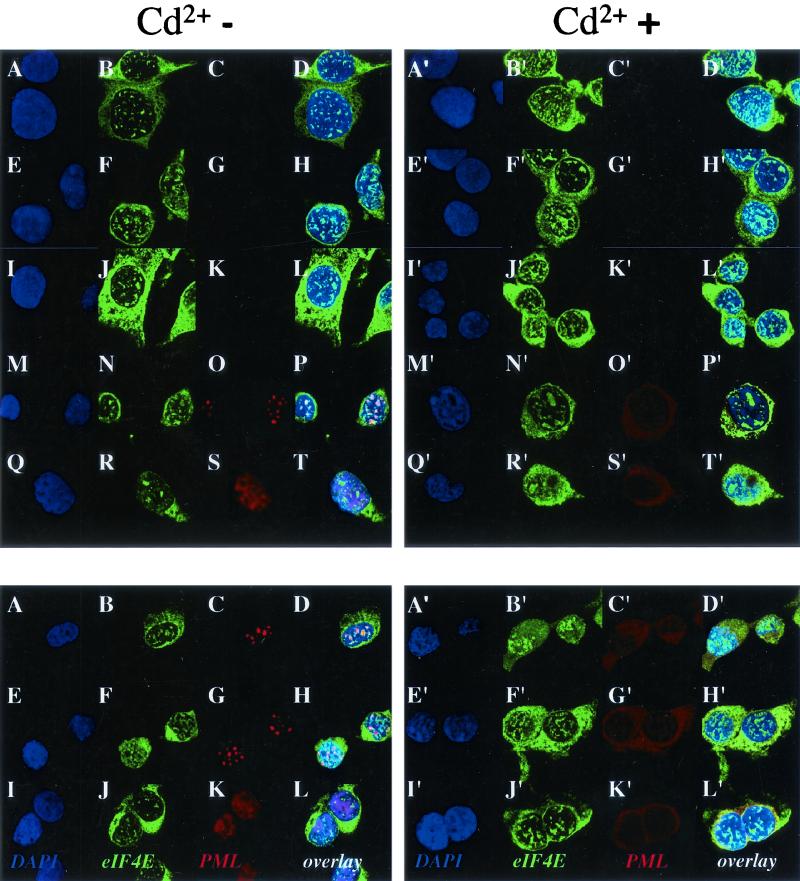
FIG. 10.
Effects of IFN treatment on NIH 3T3 cells overexpressing eIF4E or W73A eIF4E. Cells were stained with a polyclonal PML antibody in red and an eIF4E monoclonal antibody showing eIF4E in green. DAPI staining is shown to identify the nucleus (in blue). The PML-eIF4E overlay is shown in light yellow. The objective is 100× with further 1.6-fold magnification. Confocal micrographs represent single optical sections through the plane of the cell. Cells were transfected as follows: vector (A to D), eIF4E (E to H) and W73A eIF4E (I to L). The prime notation indicates the corresponding cells were treated with IFN, as described in the text.
Consistent with our results with endogenous proteins and previous studies (8, 14, 26), total cyclin D1 mRNA levels were not altered by overexpression of eIF4E or of W73A mutant or by IFN treatment (Fig. 9D). Fractionation studies reveal that for both eIF4E and W73A, there was more cyclin D1 mRNA in the cytoplasmic fraction than for vector controls (Fig. 9E). However, after IFN treatment there was an accumulation of cyclin D1 mRNA in the nucleus of cells overexpressing eIF4E but no change in cyclin D1 mRNA distribution was observed in cells overexpressing the W73A mutant (Fig. 9E, compare lanes 7 and 8 with lanes 11 and 12).
To ensure that the effects observed are indeed due to the presence or absence of PML at eIF4E bodies, we examined the subcellular distribution of overexpressed eIF4E and W73A eIF4E in NIH 3T3 cells before and after treatment with IFN (Fig. 10). In both treated and untreated cells, endogenous PML localized to eIF4E nuclear bodies in vector-transfected controls (Fig. 10A to D and A′ to D′), similar to results observed for endogenous proteins (Fig. 1). Endogenous PML localized to eIF4E nuclear bodies, where IFN treatment led to larger and more abundant PML nuclear bodies, and these bodies still localized to eIF4E (Fig. 10E to H versus E′ to H′). Similar to our results with endogenous eIF4E, the localization of eIF4E in these overexpression studies was not significantly altered by IFN. Finally, the effects of IFN treatment on the W73A eIF4E mutant were examined (Fig. 10I to L and I′ to L′). There was more cytoplasmic W73A than in wild-type overexpression studies, consistent with previous observations (11). IFN treatment did not substantially alter the localization of the W73A eIF4E mutant. PML still localized to eIF4E nuclear bodies, presumably those containing endogenous wild-type eIF4E. However, localization is less easy to determine here because the W73A overexpression resulted in very large bodies that took up the majority of the nucleus. Unlike experiments with wild-type eIF4E, where localization between PML and eIF4E nuclear bodies was nearly complete, a substantial fraction of the W73A eIF4E mutant did not localize to PML. These results are consistent with the above findings, which show that IFN treatment by induction of PML reduced the ability of eIF4E to transport cyclin D1 mRNA. In contrast, induction of PML by IFN treatment did not substantially increase the amount of PML at eIF4E bodies in cells expressing the W73A mutant, consistent with the observations that neither overexpressed PML (Fig. 7A) nor IFN-induced PML (Fig. 9A) can suppress the ability of W73A eIF4E to transport cyclin D1 mRNA.
Taken together, our mutagenesis and overexpression results are completely consistent with the results of studies using endogenous proteins. These studies strongly suggest that the effects of PML with eIF4E-dependent cyclin D1 mRNA transport rely on the direct association of PML on eIF4E and its ability to localize to eIF4E-associated nuclear bodies. Dispersal of PML by cadmium treatment allowed eIF4E to operate unchecked and the increased association of PML with eIF4E inhibited eIF4E-dependent cyclin D1 transport in IFN-treated cells.
DISCUSSION
PML negatively regulates eIF4E-dependent transport of cyclin D1 transcripts from the nucleus to the cytoplasm when PML is overexpressed in a variety of cell lines (8, 14). Mutational analysis indicates that inhibition of this transport activity is linked to the ability of PML to suppress eIF4E-mediated oncogenic transformation (8). Biochemically, PML reduces the affinity of eIF4E for the m7G cap of mRNA, an activity required by eIF4E for both its transport and transformation functions (8, 13). Since these previous studies depended on in vitro and overexpression methods, the physiological importance of the PML eIF4E interaction was not known. Further, the effect of these treatments on eIF4E nuclear body morphology and mRNA transport functions had not been investigated.
Here, we monitored the effects of two environmental stresses, IFN and cadmium, on the ability of PML to inhibit the nuclear functions of eIF4E. These two treatments were chosen because of their opposing effects on PML nuclear body morphology (20). In the cadmium experiment, we observed that PML nuclear bodies were dispersed, eIF4E nuclear bodies were retained, and transport of cyclin D1 mRNA to the cytoplasm increased, leading to an increase in cyclin D1 protein levels. Further, if cadmium was removed and the cells were allowed to recover, PML nuclear bodies relocalized with eIF4E nuclear bodies and cyclin D1 protein levels decreased. Thus, this response is robust and reversible. In PML−/− cells, cadmium treatment did not alter cyclin D1 production.
In the IFN-γ treatment experiment, we observed more PML protein at the PML-eIF4E nuclear bodies while eIF4E was essentially unaltered, and transport of cyclin D1 transcripts was inhibited, leading to decreased cyclin D1 protein levels. These results are consistent with previous reports showing that IFN-γ is antiproliferative and reduces cyclin D1 protein levels in other cell types (reference 37 and references therein). Again in the absence of PML, IFN-γ had no effect on cyclin D1 production (Fig. 4).
Mutagenesis and overepxression studies demonstrated that the effects of cadmium and IFN on cyclin D1 transport were indeed a result of interactions between PML and eIF4E (Fig. 7 to 10), consistent with our previous studies (8, 14). Further, either overexpressed PML or PML induced by IFN suppresses eIF4E-mediated cyclin D1 mRNA transport. Our data showed that for cells expressing PML or eIF4E mutants that no longer directly interact, these cells no longer responded, at the level of cyclin D1, to these treatments. These findings are similar to those that we observed in PML−/− cells where neither cadmium nor IFN altered cyclin D1 transport. Taken together, these data strongly suggest that PML must directly interact with eIF4E in order to inhibit its cyclin D1 transport function.
These findings have a number of implications. First, eIF4E-dependent mRNA transport is modulated by external factors. Previous studies on eIF4E-dependent transport were based on overexpression methods (8, 14, 26); therefore, this is the first report of these functions being modified as part of a physiological response. Importantly, the specificity of transport reported here is the same as that reported in overexpression studies, where cyclin D1 mRNA, but not actin and GAPDH mRNAs, was affected (8, 14, 26). Second, the PML-eIF4E interaction is physiologically relevant, where PML modifies eIF4E activity in response to external factors. Third, PML is a key mediator of these responses, at least in terms of cyclin D1, since these responses are lost in PML−/− cells. PML−/− cells are resistant to apoptosis induced by a number of factors including IFN (36). This is consistent with our findings that PML−/− cells have lost the ability to modulate cyclin D1 levels in response to IFN.
Previous studies indicated that the presence of PML is important for the IFN response in a variety of viral infections. PML confers resistance to infections by vesicular stomatitis virus, influenza A virus, human foamy virus, and LCMV but not to the IFN-resistant encephelomyocarditis virus (24). In human foamy virus infection, IFN inhibits viral replication in PML+/+ but not PML−/− cells, suggesting that in this infection, PML mediates the antiviral effects of IFN (25). IFN-γ treatment of LCMV-infected cells leads to a reduction in viral replication for both PML+/+ and PML−/− cells. However, IFN-γ decreases the production of viral proteins 30% more in PML+/+ than PML−/− cells (10). During LCMV infection, PML nuclear bodies are relocated to the cytoplasm, whereas the morphology of eIF4E nuclear bodies is not altered (6). Thus, eIF4E is potentially left unregulated under these conditions. Although other factors in addition to PML can mediate the IFN response during LCMV infection, PML appears to amplify these effects (10). Interestingly, in NIH 3T3 cells, LCMV infection prolongs cell survival under serum starvation conditions (5), suggesting that when PML is not associated with the eIF4E nuclear body, the growth promoting properties of eIF4E proceed unchecked. For LCMV, which establishes chronic infections in tolerant hosts (27), clearly this would be advantageous to the viral life cycle. Taken together, these studies suggest that the ability of PML to negatively regulate eIF4E dependent mRNA transport is important to the cellular antiviral response and growth control.
Although these studies focused on cyclin D1 transport, it is likely that other mRNAs, yet to be identified, are also regulated in a similar manner by PML and eIF4E (8, 26). Thus, these mRNAs may play critical roles in the physiological response to modulation of eIF4E-dependent transport. In addition, many studies show that cyclin D1 mRNA levels can be modulated posttranscriptionally. Our studies, consistent with previous reports (14, 26), demonstrate that under these conditions, levels are regulated through transport. Consistently, we observed no alterations in transcript levels (Fig. 5 and 6), indicating that steady-state transcript stability is not significantly altered by these treatments in this system.
eIF4E is overexpressed in a variety of human cancers (28). Previous studies indicate that not only its translation but also its transport functions contribute to its oncogenic potential (8). The present studies suggest that during cadmium treatment when PML is dispersed from the eIF4E nuclear body, the growth-suppressive properties of PML are reduced and the growth-promoting properties of eIF4E are dysregulated. Consistently, cadmium is considered a human carcinogen, and exposure to this metal is linked to lung and prostate cancers (33). Further, PML−/− animals are more likely to get cancers as a result of environmental challenges (35), presumably because they have lost a key stress-responsive regulator of eIF4E nuclear bodies. A critical question is why are the PML−/− mice not prone to spontaneous cancers through dysregulation of eIF4E? There must be other negative regulators of eIF4E, which may be expressed in a cell type-specific manner, such as the proline-rich homeodomain PRH, which binds both PML and eIF4E; however, these regulators do not appear to respond to stress in the same way that PML does (31; Topisirovic et al., submitted).
In summary, we demonstrated that eIF4E-dependent mRNA transport is modulated by environmental challenges in the presence of the PML protein in mammalian cells. Presumably in other organisms where PML is absent, other factors regulate the nuclear functions of eIF4E. The ability of PML to modulate cyclin D1 protein production posttranscriptionally suggests that negative regulation of the eIF4E nuclear body provides a means of responding to environmental challenges or viral infection more quickly than through modulation of transcription. Our findings suggest that PML must be present at the nuclear eIF4E body and that PML nuclear bodies need to be intact to inhibit transport, since removal of PML from these eIF4E structures results in dysregulation of its transport function. Since this association can be modified under these conditions, there are likely to be other situations, such as certain viral infections, or other factors where the association between PML and eIF4E is modulated. These alterations lead to modulation of mRNA transport and eventually alter cellular growth (8). Identifying physiological situations and factors that result in modulation of this interaction is likely to give us new insights into how its dysregulation could contribute to eIF4E-mediated carcinogenesis.
FIG. 8.
Our studies with endogenous proteins strongly suggest that IFN treatment suppresses cyclin D1 mRNA transport and protein production through induction of PML and its subsequent ability to inhibit this activity of eIF4E (Fig. 9).
Acknowledgments
We are grateful for the kind gift of PML polyclonal antibodies from Paul Freemont and Gerd Maul; for the kind gift of MAb5E10 from L. de Jong, I. van der Kraan and R. van Driel; for the wild-type eIF4E pMV construct from Nahum Sonenberg; for technical advice from Gerd Maul and Isabelle Nefkins; and for PML−/− cells from P. P. Pandolfi. We thank Alex Kentsis for critical reading of the manuscript.
Confocal laser scanning microscopy was performed at MSSM-LCSM core facility, supported by funding from NIH (1 S10 RR0 9145-01) and NSF (DBI-9724504). K.L.B.B. is a scholar of the Leukemia and Lymphoma Society. Financial support was provided by the NIH (CA 80728 and CA 88991).
REFERENCES
- 1.Altmann, M., P. P. Muller, J. Pelletier, N. Sonenberg, and H. Trachsel. 1989. A mammalian translation initiation factor can substitute for its yeast homologue in vivo. J. Biol. Chem. 264:12145-12147. [PubMed] [Google Scholar]
- 2.Borden, K. L. B. 2002. Pondering the PML puzzle: possible functions for promyelocytic leukemia PML nuclear bodies. Mol. Cell. Biol. 22:•••-•••. [DOI] [PMC free article] [PubMed]
- 3.Borden, K. L. B., M. N. Boddy, J. Lally, N. J. O'Reilly, S. Martin, K. Howe, E. Solomon, and P. S. Freemont. 1995. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 14:1532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden, K. L. B., E. J. Campbell Dwyer, and M. S. Salvato. 1998. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J. Virol. 72:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borden, K. L. B., E. J. Campbell Dwyer, and M. S. Salvato. 1997. The promyelocytic leukemia protein PML has a pro-apoptotic activity mediated through its RING domain. FEBS Lett. 418:30-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell Dwyer, E. J., H. Lai, R. C. MacDonald, M. S. Salvato, and K. L. Borden. 2000. The lymphocytic choriomeningitis virus RING protein Z associates with eukaryotic initiation factor 4E and selectively represses translation in a RING-dependent manner. J. Virol. 74:3293-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chelbi-Alix, M. K., L. Pelicano, F. Quignon, M. H. Koken, L. Venturini, M. Stadler, J. Pavlovic, L. Degos, and H. de The. 1995. Induction of the PML protein by interferons in normal and APL cells. Leukemia 9:2027-2033. [PubMed] [Google Scholar]
- 8.Cohen, N., M. Sharma, A. Kentsis, J. M. Perez, S. Strudwick, and K. L. Borden. 2001. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 20:4547-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Benedetti, A., and R. E. Rhoads. 1990. Overexpression of eukaryotic protein synthesis initiation factor 4E in HeLa cells results in aberrant growth and morphology Proc. Natl. Acad. Sci. USA 87:8212-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djavani, M., J. Rodas, I. S. Lukashevich, D. Horejsh, P. P. Pandolfi, K. L. Borden, and M. S. Salvato. 2001. Role of the promyelocytic leukemia protein PML in the interferon sensitivity of lymphocytic choriomeningitis virus. J. Virol. 75:6204-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dostie, J., M. Ferraiuolo, A. Pause, S. A. Adam, and N. Sonenberg. 2000. A novel shuttling protein, 4E-T, mediates the nuclear import of the mRNA 5′ cap-binding protein, eIF4E. EMBO J. 19:3142-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iborra, F. J., D. A. Jackson, and P. R. Cook. 2001. Coupled transcription and translation within nuclei of mammalian cells. Science 293:1139-1142. [DOI] [PubMed] [Google Scholar]
- 13.Kentsis, A., E. C. Dwyer, J. M. Perez, M. Sharma, A. Chen, Z. Q. Pan, and K. L. Borden. 2001. The RING domains of the promyelocytic leukemia protein PML and the arenaviral protein Z repress translation by directly inhibiting translation initiation factor eIF4E. J. Mol. Biol. 312:609-623. [DOI] [PubMed] [Google Scholar]
- 14.Lai, H. K., and K. L. Borden. 2000. The promyelocytic leukemia (PML) protein suppresses cyclin D1 protein production by altering the nuclear cytoplasmic distribution of cyclin D1 mRNA. Oncogene 19:1623-1634. [DOI] [PubMed] [Google Scholar]
- 15.Lazaris-Karatzas, A., K. S. Montine, and N. Sonenberg. 1990. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345:544-547. [DOI] [PubMed] [Google Scholar]
- 16.Lazaris-Karatzas, A., M. R. Smith, R. M. Frederickson, M. L. Jaramillo, Y. L. Liu, H. F. Kung, and N. Sonenberg. 1992. Ras mediates translation initiation factor 4E-induced malignant transformation. Genes Dev. 6:1631-1642. [DOI] [PubMed] [Google Scholar]
- 17.Lazaris-Karatzas, A., and N. Sonenberg. 1992. The mRNA 5′ cap-binding protein, eIF-4E, cooperates with v-myc or E1A in the transformation of primary rodent fibroblasts Mol. Cell. Biol. 12:1234-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lejbkowicz, F., C. Goyer, A. Darveau, S. Neron, R. Lemieux, and N. Sonenberg. 1992. A fraction of the mRNA 5′ cap-binding protein, eukaryotic initiation factor 4E, localizes to the nucleus. Proc. Natl. Acad. Sci. USA 89:9612-9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129:278-287. [DOI] [PubMed] [Google Scholar]
- 20.Maul, G. G., E. Yu, A. M. Ishov, and A. L. Epstein. 1995. Nuclear domain 10 (ND10) associated proteins are also present in nuclear bodies and redistribute to hundreds of nuclear sites after stress. J. Cell. Biochem. 59:498-513. [DOI] [PubMed] [Google Scholar]
- 21.Melnick, A., and J. D. Licht. 1999. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood 93:3167-3215. [PubMed] [Google Scholar]
- 22.Nathan, C. A., P. Carter, L. Liu, B. D. Li, F. Abreo, A. Tudor, S. G. Zimmer, and A. De Benedetti. 1997. Elevated expression of eIF4E and FGF-2 isoforms during vascularization of breast carcinomas. Oncogene 15:1087-1094. [DOI] [PubMed] [Google Scholar]
- 23.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 24.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 25.Regad, T., A. Saib, V. Lallemand-Breitenbach, P. P. Pandolfi, H. de The, and M. K. Chelbi-Alix. 2001. PML mediates the interferon-induced antiviral state against a complex retrovirus via its association with the viral transactivator. EMBO J. 20:3495-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rousseau, D., R. Kaspar, I. Rosenwald, L. Gehrke, and N. Sonenberg. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 93:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvato, M. S., and K. S. Rai. 1996. Arenaviruses, p. 629-650. In L. H. Collier, and B. W. J. Mahy (ed.), Topley and Wilson's microbiology and microbial infections, 9th ed. Arnold Publishing, London, United Kingdom.
- 28.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10:268-275. [DOI] [PubMed] [Google Scholar]
- 29.Strudwick, S., and K. L. B. Borden. 2002. The emerging roles for nuclear translation factors. Differentiation 70:10-22. [DOI] [PubMed] [Google Scholar]
- 30.Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773-784. [DOI] [PubMed] [Google Scholar]
- 31.Topcu, Z., D. L. Mack, R. A. Hromas, and K. L. Borden. 1999. The promyelocytic leukemia protein PML interacts with the proline-rich homeodomain protein PRH: a RING may link hematopoiesis and growth control. Oncogene 18:7091-7100. [DOI] [PubMed] [Google Scholar]
- 32.Turelli, P., V. Doucas, E. Craig, B. Mangeat, N. Klages, R. Evans, G. Kalpana, and D. Trono. 2001. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication Mol. Cell 7:1245-1254. [DOI] [PubMed] [Google Scholar]
- 33.Waalkes, M. P. 2000. Cadmium carcinogenesis in review. J. Inorg. Biochem. 79:241-244. [DOI] [PubMed] [Google Scholar]
- 34.Wang, S., I. B. Rosenwald, M. J. Hutzler, G. A. Pihan, L. Savas, J. J. Chen, and B. A. Woda. 1999. Expression of the eukaryotic translation initiation factors 4E and 2alpha in non-Hodgkin's lymphomas. Am. J. Pathol. 155:247-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 279:1547-1551. [DOI] [PubMed] [Google Scholar]
- 36.Wang, Z. G., D. Ruggero, S. Ronchetti, S. Zhong, M. Gaboli, R. Rivi, and P. P. Pandolfi. 1998. PML is essential for multiple apoptotic pathways. Nat. Genet. 20:266-272. [DOI] [PubMed] [Google Scholar]
- 37.Xaus, J., M. Cardo, A. F. Valledor, C. Soler, J. Lloberas, and A. Celada. 1999. Interferon gamma induces the expression of p21waf-1 and arrests macrophage cell cycle, preventing induction of apoptosis. Immunity 11:103-113. [DOI] [PubMed] [Google Scholar]