Abstract
DNA N-glycosylase/AP (apurinic/apyrimidinic) lyase enzymes of the endonuclease III family (nth in Escherichia coli and Nth1 in mammalian organisms) initiate DNA base excision repair of oxidized ring saturated pyrimidine residues. We generated a null mouse (mNth1−/−) by gene targeting. After almost 2 years, such mice exhibited no overt abnormalities. Tissues of mNth1−/− mice contained an enzymatic activity which cleaved DNA at sites of oxidized thymine residues (thymine glycol [Tg]). The activity was greater when Tg was paired with G than with A. This is in contrast to Nth1, which is more active against Tg:A pairs than Tg:G pairs. We suggest that there is a back-up mammalian repair activity which attacks Tg:G pairs with much greater efficiency than Tg:A pairs. The significance of this activity may relate to repair of oxidized 5-methyl cytosine residues (5meCyt). It was shown previously (S. Zuo, R. J. Boorstein, and G. W. Teebor, Nucleic Acids Res. 23:3239-3243, 1995) that both ionizing radiation and chemical oxidation yielded Tg from 5meCyt residues in DNA. Thus, this previously undescribed, and hence novel, back-up enzyme activity may function to repair oxidized 5meCyt residues in DNA while also being sufficient to compensate for the loss of Nth1 in the mutant mice, thereby explaining the noninformative phenotype.
Escherichia coli endonuclease III and its eukaryotic homologs initiate the process of DNA base excision repair (BER) of C-5, C-6 ring saturated pyrimidines (31). The endonuclease III-like enzymes are bifunctional enzymes which first effect release of the damaged base from the DNA backbone and then effect strand cleavage at the resulting abasic (apurinic/apyrimidinic [AP]) site via β-elimination (19, 20, 32). Among the modified bases recognized by endonuclease III-like enzymes are 5,6-dihydroxy-5,6-dihydrothymine (thymine glycol [Tg]), cytosine glycol, and cytosine hydrate. Tg is an oxidative product formed in DNA in vitro by reaction with chemical oxidizing agents such as KMnO4 and OsO4 as well as via the indirect action of ionizing radiation under aerobic conditions. In cellular DNA, Tg is formed as a product of exposure to aerobic ionizing radiation and other oxidative stresses such as H2O2 in the presence of added Fe2+. Ionizing irradiation under anaerobic conditions yields the reduced derivative 5,6-dihydrothymine (4, 9, 28). Exposure of cytosine to oxidative stress yields cytosine glycol, which is in equilibrium with its dehydrated form, 5-hydroxycytosine (5ohCyt). 5ohCyt is prone to deamination yielding 5ohUra. In addition to inducing formation of dimeric photoproducts in DNA, UV irradiation also induces hydration of the 5,6 double bond of pyrimidines, primarily forming 5,6 dihydro-6-hydroxy cytosine (cytosine hydrate) (2, 3). Like 5ohCyt, cytosine hydrate is prone to deamination, which yields uracil hydrate (26).
Our laboratory first documented the existence of a family of endonuclease III homologs in several species (11) and isolated the cDNA of the human homolog of endonuclease III (hNth1) (12). In this study, we produced targeted deletion of the murine Nth1 to determine its role in preventing sequelae of unrepaired oxidative or reductive stress as well as UV-induced damage to the pyrimidine bases of genomic DNA. mNth1−/− mice were observed for fertility, viability, development, longevity, and spontaneous tumor formation. After 15 months, we saw no phenotypic differences. Therefore, we began to search for back-up enzyme activity(s) which might mitigate the sequelae of the loss of mNth1. We demonstrate such an enzyme activity and offer a hypothesis as to its nature and function.
MATERIALS AND METHODS
Construction of the mNth1 targeting vector, pGTN28/DRE.
Mouse Nth1 (mNth1) cDNA was cloned and completely sequenced by using reverse transcriptase PCR (RT-PCR), rapid amplification of 3′ and 5′ cDNA ends, and two primers from GenBank sequence (accession no. H33255). Murine Nth1 genomic clones were isolated from a mouse 129/SvJ genomic bacterial artificial chromosome library (Incyte Genomics [formerly Genome Systems], St. Louis, Mo.) by hybridization with mNth1-specific cDNA probes. Restriction enzyme maps of the mNth1 locus were determined using bacterial artificial chromosome clones and genomic DNA from mouse tail clippings. One subclone (E2) contained a 7.4-kb EcoRI restriction fragment of genomic mNth1 sequence, including exons 1 through 4, ∼2.2 kb of sequence upstream of exon 1, and ∼2.5 kb of intronic sequence downstream of exon 4. The other subclone (K1) contained a 4.2-kb KpnI restriction fragment of genomic mNth1 sequence, including exon 5.
The HindIII subclone E2 and the Acc65i subclone K1 genomic fragments were cloned into vector pZERO (Invitrogen). The genomic organization of the mNth1 gene was disrupted by inserting the vector-neo cassette (KpnI-HindIII fragment), derived from New England BioLabs pGT-N28 Vector [containing a phosphoglycerate kinase promoter and poly(A)] into the HindIII site between exons 2 and 3 of mNth1 and the KpnI site in front of exon 5. The cassette contains the neomycin phosphotransferase gene as a selectable marker flanked 5′ by the thymidine kinase promoter and 3′ by the thymidine kinase polyadenylation sequence. The targeting vector contained ∼3.8 kb and ∼4.5 kb of 5′ and 3′ homology regions, respectively.
Generation of mice harboring the mNth1 mutation.
129/SvJ embryonic stem (ES) cells were electroporated with NotI-linearized targeting vector and selected with Geneticin (G418; Gibco). A total of 196 ES clones resistant to G418 were analyzed for homologous recombination by Southern blotting using two genomic DNA fragments as probes located in the 5′ and 3′ ends of intronic sequence. Recombinant ES cell clones were injected into C57BL/6 blastocytes and transferred to pseudopregnant foster mothers. Chimeric offspring were bred with C57BL/6 females, and progeny were screened by PCR. mNth1+/− progeny were crossed to generate mNth1−/− mice. In the interest of time and to generate as many mNth1−/− mice as possible, animals were bred as hybrid C57BL/6-129/SvJ. They were maintained in a barrier facility which was in compliance with federal regulations for animal care.
Southern blot analysis of ES cells.
Genomic DNA was extracted from ES cells, digested with EcoRI and EcoRV, electrophoresed on agarose gels, transferred, and hybridized with a radiolabeled 5′-flanking probe. The wild-type allele yielded a 7.4-kb band, while the recombinant allele produced a 4.5-kb band, due to the insertion of a new EcoRV site in the targeting vector. In addition, genomic DNA was also digested with SpeI, electrophoresed on agarose gels, transferred, and hybridized with a radiolabeled 3′ flanking probe. The wild-type allele yielded an 18.0-kb band, while the recombinant allele produced a 14.4-kb band, due to the insertion of a new SpeI site in the targeting vector.
Genotyping by PCR.
PCR-based analysis was used to genotype mice. Each reaction mixture consisted of deoxynucleoside triphosphates, MgCl2, a forward primer specific for mNth1 exon 2 (5′-CCGCATCATGAGAAGCAAGA-3′), a reverse complementary primer specific for mNth1 exon 3 (5′-CAGGATGCTCTCCACAGTCA-3′), a Neo cassette primer (5′-GCGATACCGTAAAGCACGAG-3′), Taq polymerase, and genomic DNA. These primers were demonstrated to anneal and to generate the appropriate fragment length with the Amplify 1.2 program using two different mNth1 DNA sequences from GenBank (accession numbers AJ00167 and AB009371). Conditions for PCR were as follows: (i) 5 min at 94°C; (ii) 35 cycles, with 1 cycle consisting of 30 s at 94°C, 30 s at 60°C, and 60 s at 72°C; and (iii) an elongation period of 7 min at 72°C. Amplified products were analyzed by electrophoresis in 1% agarose gels with a combination of HindIII-digested λ DNA fragments (λDNA/HindIII Fragments; Gibco) and HaeIII-digested replicative-form DNA fragments of pφX174 (pφX174 RF DNA/HaeIII Fragments; Gibco) as molecular size markers.
RT-PCR.
Total RNA was extracted from fresh murine tissue using TRI Reagent (Molecular Research Center) according to the manufacturer's instructions. First-strand cDNA was synthesized using SuperScript II RNase H Reverse Transcriptase and oligo(dT) according to the manufacturer's directions (Gibco BRL, Life Technologies). PCR was then performed using the cycling conditions described above, with the exception that only 30 cycles were performed. PR1, the primer pair specific for the mNth1 open reading frame, included the forward primer (5′-TACAGCCCAAGCATGAACTC-3′) for nucleotides 28 to 47 and the reverse primer (5′CCCCCACAAACGTTATTTGG-3′) for nucleotides 1009 to 1028. PR2, the primer pair specific for exons 2 to 4 of mNth1 included the forward primer (5′-CCGCATCATGAGAAGCAAGA-3′) specific for nucleotides 300 to 319 as well as the reverse primer (5′-TAGCCAAGTGTGCCATCTTG-3′) specific for nucleotides 660 to 679. For the β-actin control reactions, the forward primer was (5′-TGACGGGGTCACCCACACTGTGCCCATCTA-3′) specific for nucleotides 390 to 419, while the reverse primer was (5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG-3′) specific for nucleotides 1021 to 1050. Amplified products were analyzed on 1% agarose gels.
Enzymatic assays.
Enzymatic assays of mNth1 activity were of two types. Direct measurement of DNA N-glycosylase activity was performed using a UV-irradiated alternating poly(dG-[3H]dC) oligonucleotide as a substrate and measuring enzymatically mediated release of photochemically induced cytosine hydrate by liquid scintillation counting (2, 3). A more sensitive assay utilized a 32P-labeled 2′-deoxyribose oligonucleotide substrate containing a single Tg residue at a unique site. DNA N-glycosylase activity was assayed by measuring putrescine-induced strand cleavage, and AP lyase activity was assayed by measuring strand cleavage without pretreatment with putrescine. Quantitative analysis of polyacrylamide gel electrophoresis was done with a phosphorimager using a Molecular Imager FX System with Quantity One Software (Bio-Rad) (22, 33).
Preparation of substrates. (i) DNA N-glycosylase assay.
Alternating poly(dG-dC) (Amersham Pharmacia) was nick translated with [3H]dCTP (Moravek) using a nick translation system (Gibco BRL, Life Technologies). The radiolabeled oligonucleotide was separated from unincorporated 2′-deoxyribose mononucleotides by using a G-50 Sephadex column (Amersham Pharmacia). Cytosine hydrate was generated by exposing alternating poly(dG-[3H]dC) to UV radiation from a germicidal bulb. Cytosine hydrate content was determined by using Escherichia coli endonuclease III in a limit digest to measure release of all of the modified base. For each enzyme assay, 100 ng of substrate (150 pmol of 2′-dCMP; 8 × 104 to 10 × 104 cpm/pmol) was used, of which approximately 5% was in the form of cytosine hydrate (2, 3).
(ii) Cleavage assay.
2′-Deoxyribose oligonucleotides were prepared as described previously (22). Substrate containing an AP site was generated from a 30-mer uracil-containing 2′-deoxyribose oligonucleotide d(GATCCTCTAGCGUCGACCTGCAGGCATGCA [the uracil is underlined]). The AP site was made by treatment with uracil DNA N-glycosylase (Life Technologies) (1 U/μl). A 30-mer 2′-deoxyribose oligonucleotide substrate containing a Tg residue (shown underlined) at position 13 d(GATCCTCTAGAGTgCGACCTGCAGGCATGCA) was also used. 2′-Deoxyribose oligonucleotides were 5′ end labeled using [γ-32P]ATP (Perkin-Elmer Life Sciences) and T4 polynucleotide kinase (Life Technologies). To obtain duplex DNA, 2′-deoxyribose oligonucleotides were annealed to the corresponding complementary strand in a 1:1.5 molar ratio. When opposite an AP site, all complementary strands contained an adenine residue. When opposite the Tg residue, complementary strands contained either an adenine or a guanine.
Preparation of cell or tissue sonicates. (i) Sonicates for DNA N-glycosylase assay.
To obtain cellular protein extracts, ES cells were harvested, washed in 1× cold phosphate-buffered saline (PBS), and centrifuged at 1,650 rpm for 10 min in an Allegra 6KR centrifuge (Beckman) with a swinging bucket rotor at 4°C. Cells were suspended in sonication buffer (20 mM HEPES [pH 7.6], 5 mM EDTA, 50 mM NaCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) and lysed with four 20-s bursts of sonication with the microtip of the Sonicator Ultrasonic Processor (Misonix) at 20% output, using conical plastic tubes (Becton Dickinson) for the samples. Sonication tubes were placed in an ice bath to prevent thermal denaturation of proteins. Sonicates were then transferred to Eppendorf tubes and centrifuged at 20,800 × g for 5 min in a 5417C centrifuge (Eppendorf). Cellular protein concentration was estimated by the Bradford assay. To obtain extracts from murine tissues, animals were sacrificed by CO2 asphyxiation in compliance with International Animal Care and Use Committee guidelines. Protein extracts from adult murine tissues and whole embryos were similarly prepared, with the following exceptions. For adult tissue sonicates, organs were removed from sacrificed mice, placed in 1× cold PBS on ice, and then homogenized using a Potter-Elvehjeim homogenizer attached to a hand drill at a final concentration of 20% (wt/vol) before sonication. For preparation of whole-embryo sonicates, embryos were harvested from pregnant females at day 11 or 12 and prepared as 20% (wt/vol) homogenates prior to sonication.
(ii) Sonicates for cleavage assay.
Two-month-old mice were sacrificed, and tissues were quickly obtained and placed in cold 1× PBS. Tissues were weighed and homogenized to a 20% (wt/vol) homogenate in buffer (50 mM HEPES [pH 7.6], 50 mM KCl, 10 mM EDTA, 0.1 mM dithiothreitol, protease inhibitor cocktail). This buffer yielded sharper resolution on gels than the above-described NaCl-containing buffer used for the release assay. Tissue homogenates were subjected to four bursts of sonication as described above for ES cells. The sonicates were transferred to new Eppendorf tubes and spun in an SS-34 rotor at 14,000 rpm at 4°C for 15 min in a Sorvall RC-5B centrifuge. Supernatants were collected, and protein content was estimated.
DNA N-glycosylase assay.
The DNA N-glycosylase base release assay was performed exactly as previously described (2, 3). The specific mNth1 activities of adult murine tissues are approximations due to the long reaction time required to detect base release. Reaction mixtures were incubated for up to 14 h before significant activity was detectable. Therefore, there are too few points to determine the constancy of the rate of the reaction. In contrast, whole-embryo and ES cell extracts released detectable amounts of cytosine hydrates within 2- and 4-h incubations, so the specific activity of these crude extracts could be measured with reasonable precision.
Cleavage assay.
Individual reaction mixtures contained 100 nM [32P]-5′-end-labeled 2′-deoxyribose oligonucleotide duplex and various amounts of cellular protein in a final volume of 20 μl. Assays were performed at 37°C in reaction buffer (50 mM HEPES [pH 7.6], 100 mM KCl, 10 mM EDTA, 0.1 mg of bovine serum albumin per ml, 0.1 mM dithiothreitol). To measure strand cleavage, reaction tubes were removed at the indicated times, and the reaction mixtures were treated with an equal volume of formamide loading dye (95% deionized formamide, 10 mM EDTA, 0.05% bromphenol blue, 0.05% xylene cyanol). To measure DNA N-glycosylase activity, reaction tubes were removed at the indicated times, and reaction mixtures were treated with 5 μl of 0.5 M putrescine (pH 8.0) (33). The treated assay mixtures were then heated at 95°C for 5 min, and 25 μl of loading dye was added to each sample. Samples were then denatured at 95°C for 5 min, and products were separated by 20% polyacrylamide gel electrophoresis in 8 M urea and Tris-borate-EDTA (TBE). Products were quantitatively analyzed via phosphorimaging.
Apoptosis assay.
The apoptotic response of cells of mNth1−/− mice was assessed both by ex vivo and in vitro assays.
(i) Ex vivo gamma irradiation.
Two-month-old mNth1−/− and mNth1+/+ males were exposed to a dose of 4 Gy of gamma irradiation from a 35Cs source (Gamma-Med Radiator) (34). Animals were sacrificed at various times after exposure. Thymi were obtained from control and irradiated mice, and single-cell suspensions were made in RPMI 1640 medium with 10% fetal bovine serum and penicillin or streptomycin by mechanical disruption of the thymi. Cells were filtered through a 70-μm-pore-size cell strainer and counted with a Coulter Counter. They were maintained at 37°C in RPMI 1640 with 5% CO2 for indicated times. Suspensions were centrifuged at 1,400 rpm for 5 min at 4°C in an Allegra 6KR centrifuge (Beckman) and washed twice with 1× cold PBS. Next, cells were suspended in 1× binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2 [pH 7.4]) and stained with annexin V-FITC (fluorescein isothiocyanate) and propidium iodide using the Annexin V-FITC Apoptosis Detection Kit 1 according to the manufacturer's instructions (PharMingen, Becton Dickinson). Samples were analyzed by flow cytometry with a FACScan (Becton Dickinson).
(ii) In vitro gamma irradiation.
Two-month-old mice were sacrificed, thymi were rapidly obtained, and single-cell suspensions were prepared as described above. Cells were exposed to 4 or 8 Gy of gamma radiation or sham exposed. Cells were then cultured at 37°C with 5% CO2 for 2, 4, or 6 h before harvest, staining, and fluorescence-activated cell sorting analysis, as described above. A zero time point was also obtained.
(iii) In vitro exposure to H2O2.
Single-cell thymocyte suspensions were prepared as described above. H2O2 (Sigma) was added to the medium of test samples at a final concentration of 150, 300, or 600 μM; control cells were cultured in medium only. After 15 min of incubation at 37°C with 5% CO2, cells were centrifuged at 1,400 rpm in an Allegra 6KR centrifuge (Beckman) at 4°C, washed twice with cold 1× PBS, and resuspended in fresh medium (RPMI 1640, 10% fetal bovine serum, and penicillin or streptomycin). Cells were then cultured for another 2 h at 37°C with 5% CO2 before staining and flow cytometry analysis as described above (27).
Histopathological analysis.
Necropsies were performed on mNth1+/+, mNth1+/−, and mNth1−/− adult (2-, 8-, and 10-month-old) male and female mice and embryos. Tissues were fixed in 10% formalin and embedded in paraffin; sections were stained with hematoxylin and eosin.
RESULTS
Targeted disruption of the mNth1 gene and generation of mNth1−/− mice.
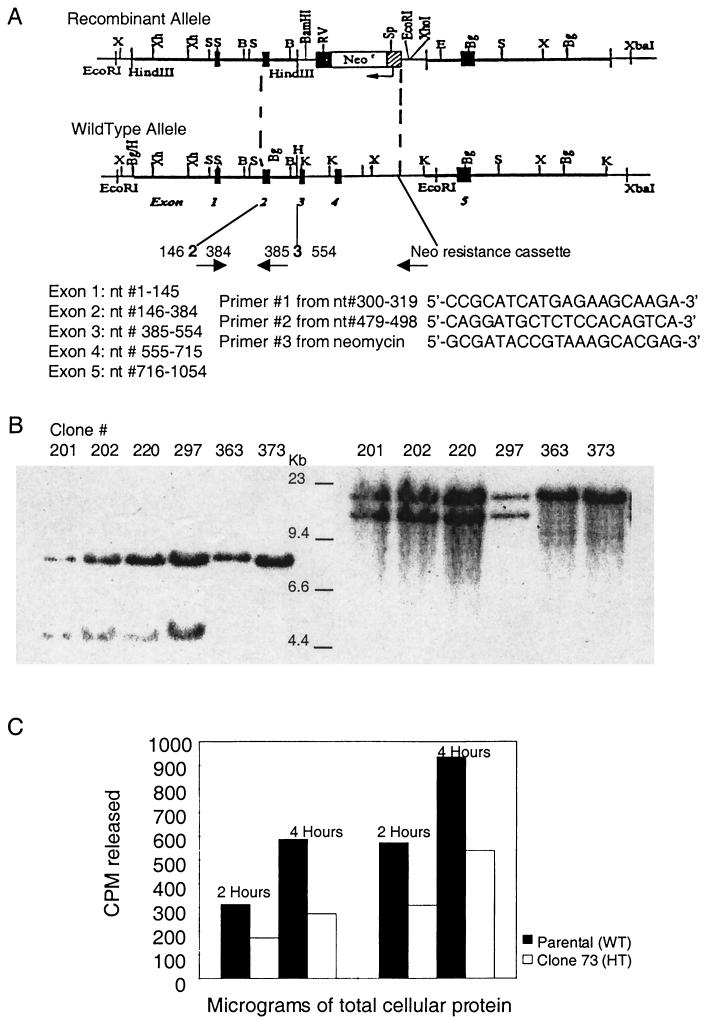
A rodent 3′ expressed sequence tag was used to isolate the full-length cDNA of mNth1. With the mouse cDNA as a probe and twith the assistance of Incyte Genomics Inc., the genomic coding sequence of the enzyme was identified (Fig. 1A ). The targeting vector was generated by replacing exons 3 and 4 of the mNth1 gene with a neomycin resistance cassette. Exon 4 contains a lysine residue at position 208 which is homologous to lysine 212 in hNth1 and lysine 120 in E. coli endonuclease III, both of which are the nucleophiles that initiate formation of the Schiff base ES intermediate (15, 30). Exon 4 also contains the helix-hairpin-helix motif critical for enzyme activity (19, 20, 32).
FIG. 1.
(A) Targeted disruption of mNth1. The mNth1 locus consists of five exons, as represented by the numbered black boxes in the schematic of the wild-type allele. To generate the recombinant allele, exons 2 through 4 were replaced with the neomycin resistance cassette via homologous recombination. The positions of the PCR primers used in routine genotyping are indicated by arrows, and their sequences are numbered based upon their location in the mNth1 cDNA. Abbreviations: B, BamHI; Bg, BglII; E, EcoRI; K, KpnI; RV, EcoRV; S, SacI; Sp, SpeI; X, XbaI;Xh, XhoI. (B) Southern blot analysis of ES cell clones. In the left panel, probing of the 5′ region after EcoRI and EcoRV digestion of genomic DNA yielded a 7.4-kb band for the wild-type allele and a 4.5-kb band for the recombinant allele, due to the insertion of a new EcoRV (RV) site in the targeting vector. In the right panel, probing of the 3′ region after SpeI digestion of genomic DNA produced an 18.0-kb band for the wild-type allele and a 14.4-kb band for the recombinant allele, due to the insertion of a new SpeI (Sp) site in the targeting vector. (C) mNth1 gene dosage effect in ES cells. The enzymatic activity of parental mNth1+/+ (wild type [WT]) and candidate heterozygous (HT) mNth1+/− clones (clone 73 here) were analyzed by DNA N-glycosylase base release assay. ES cells were lysed, and 200 or 400 μg of cellular protein was incubated with 100 ng of UV-irradiated poly(dG-[3H]dC) for 2 or 4 h. Base release was measured by liquid scintillation counting. The results of this experiment are representative of the results obtained with all mNth1+/− clones tested.
ES cells were electroporated with NotI-linearized targeting vector and screened by Southern blot analysis for successful homologous recombination (Fig. 1B). Candidate mNth1+/− clones were assayed for enzyme activity via release of cytosine hydrate from UV-irradiated alternating poly(dG-[3H]dC). Analysis of mNth1+/+ (parental) and putative mNth1+/− clones by use of the DNA N-glycosylase assay revealed that total protein extract from mNth1+/+ ES cells had twice as much specific enzyme activity as the protein extract from mNth1+/− ES cells (Fig. 1C). This gene dosage effect confirmed that the mNth1+/− cells had only one functional copy of the mNth1 gene.
Chimeric C57BL/6 males were born after blastocyst injection of 129/SvJ ES cells. These chimeric males were then bred with C57BL/6 females. DNA from F1 progeny was analyzed by PCR, and both mNth1+/− hybrid males and females were detected, confirming that the chromosome containing the deleted mNth1 allele had been transmitted to the F1 generation.
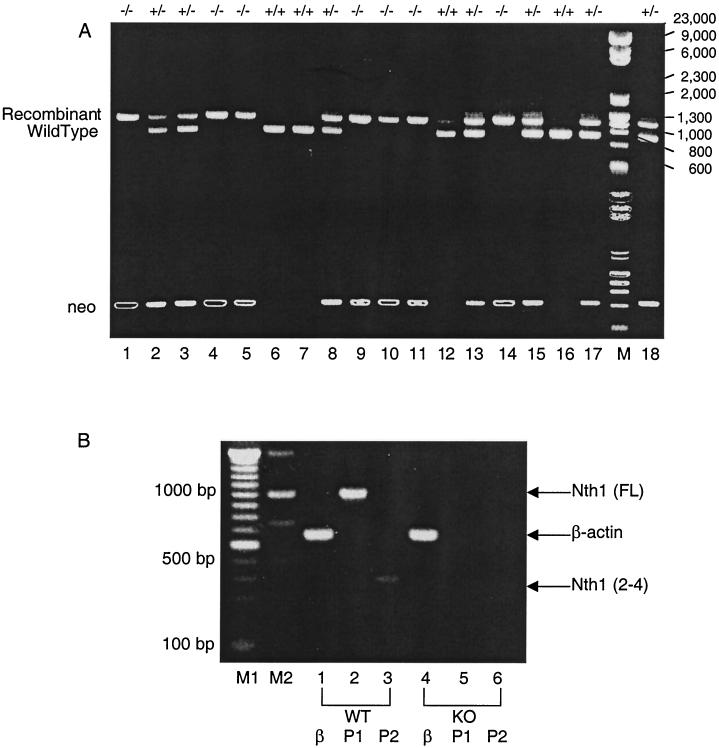
To genotype the mice, PCR-based analysis was employed (Fig. 2A, top part of gel). Primers specific for mNth1 and the neomycin resistance cassette were used to amplify genomic DNA from mice and to visualize wild-type and recombinant alleles. The wild-type genomic DNA fragment is approximately 750 bp, while the recombinant fragment migrates between 1,000 and 1,300 bp, as indicated by the combination of HindIII-digested λ DNA fragments (λDNA/HindIII Fragments; Gibco) and HaeIII-digested replicative-form DNA fragments of pφX174 (pφX174 RF DNA/HaeIII Fragments; Gibco). A second round of PCR analysis was also conducted with forward and reverse primers specific for the neomycin resistance cassette (Fig. 2A). Only mNth1−/− mice showed a saturated band after 30 cycles of PCR (Fig. 2A, bottom part, lanes 1, 4, 5, 9, 10, 11, and 14). This confirmed that two copies of the neomycin resistance cassette were present in the mNth1−/− mice, whereas mNth1+/− mice had only a single copy, and mNth1+/+ mice did not contain the recombinant allele. This second PCR strategy was particularly useful in verifying the genotype when results from the first round of PCR were ambiguous and shadow bands were seen (Fig. 2A, top part, lane 12).
FIG. 2.
(A) PCR-based analysis for genotyping mice. In the top part of the gel, genomic tail DNA samples from mice were amplified using primers 1 to 3 (listed in Fig. 1). The upper band is the recombinant allele which migrates between 1,000 and 1,300 bp, while the lower band is the wild-type allele which is approximately 750 bp. In the lower part of the gel, genomic DNA was amplified with primers specific for the neomycin resistance cassette. Only mNth1−/− mice show saturated neo band patterns (lanes 1, 4, 5, 9, 10, 11, and 14) due to the presence of two copies of the neo resistance cassette. Meanwhile, mNth1+/− mice have less-intense bands resulting from a single copy of the neo gene. mNth1+/+ mice possess two wild-type alleles and show no incorporation of the neo gene. Lane M contains molecular size markers. The numbers to the left of the gel are sizes (in base pairs). (B) Confirmation of mNth1 disruption by RT-PCR analysis of total mRNA. Total RNA was extracted from mNth1+/+ and mNth1−/− murine thymi. RT-PCR was performed with primer pairs specific for mNth1 or β-actin. Abbreviations: WT, wild type; KO, knockout (mNth1−/−); P1, primer pair for mNth1 open reading frame; P2, primer pair for mNth1 exons 2 to 4; β, primer pair for β-actin cDNA; Nth1 (FL), full-length mNth1 cDNA fragment (1,001 bp); β-actin, 661-bp cDNA fragment; Nth1 (2-4), mNth1 cDNA fragment from exon 2 to exon 4 (380 bp).
To confirm that the mNth1 gene had indeed been knocked out, mRNA levels were measured. RT-PCR analysis with mNth1-specific open reading frame primers showed that mNth1+/+ mice had an approximately 1,000-bp mRNA species corresponding to the full-length mNth1 cDNA (Fig. 2B, lane 2). In contrast, the mNth1−/− mice lacked this mNth1 mRNA species (Fig. 2B, lane 5). This was not due to mRNA degradation because β-actin controls confirmed the presence of intact mRNA (Fig. 2B, lanes 1 and 4). RT-PCR analysis was also performed with primers specific for a short fragment of mNth1 (exons 2 to 4), and, once again, a band corresponding to the appropriate 380-bp fragment of mNth1 was detected for mNth1+/+ tissue, but not for the mNth1−/− sample (Fig. 2B, lanes 3 and 6). Therefore, both PCR and RT-PCR analysis confirmed the absence of mNth1 DNA or mRNA in mNth1−/− tissues.
Viability of mNth1−/− mice.
Mice lacking mNth1 activity proved to be viable and fertile. Heterozygote crosses resulted in offspring of the expected Mendelian ratios (+/+, 132 [26.8%]; +/−, 229 [46.4%]; −/−, 132 [26.8%]). No differences in birth weight, litter size, or growth rates were observed between mutant mice and wild-type counterparts. mNth1−/− mice did not exhibit an increased incidence of tumorigenesis, and histopathological analysis of adult tissues (brain, thymus, heart, lungs, liver, kidney, spleen, and intestine) as well as embryos revealed neither gross nor microscopic abnormalities. To date, the oldest mice are 24 months. Thus, deletion of the mNth1 gene did not interfere with development of either gender, and it did not appear to predispose mutants to any obvious pathological sequelae.
Functional analysis of mNth1−/− mice.
(i) Apoptosis and gamma irradiation. Gamma radiation under aerobic conditions is known to form oxidative base damage that is repaired by endonuclease III (4, 9, 28). In the absence of mNth1, it is possible that failure to repair damage may induce apoptosis. To determine the functional consequences of knocking out mNth1, ex vivo and in vitro analyses of gamma radiation-induced apoptosis was performed. For ex vivo assays, 2-month-old adult male mice were exposed to 4 or 8 Gy of gamma radiation or sham exposed, and thymi were obtained after 0, 2, 4, or 6 h. Single-cell thymocyte suspensions were prepared from the control or irradiated thymi, and the extent of apoptosis was quantitated by flow cytometry. mNth1+/+ and mNth1−/− mice showed no significant difference in radiation-induced apoptosis (data not shown).
For in vitro analysis, thymi were obtained from mice, and thymocyte suspensions were prepared. The suspended cells were gamma irradiated, and the extent of apoptosis was quantitated by flow cytometry. Under these conditions, mNth1+/+ and mNth1−/− thymocytes also underwent apoptosis to a similar extent (data not shown).
(ii) Apoptosis and H2O2.
To determine the contribution of mNth1 to the cellular response to chemical oxidative stress, apoptosis was also measured after treatment of thymocyte suspensions with various concentrations of H2O2 (27). A high degree of variability was seen from mouse to mouse. We attribute this difference, at least in part, to the fact that the mice were hybrids, and the differences in genetic background were manifest in the high degree of variability between preparations. However, there were no differences between mNth1+/+ and mNth1−/− animals (data not shown).
(iii) DNA N-glycosylase assay.
As with the ES cells, various tissues from adult male mice were assayed for enzymatic activity via the DNA N-glycosylase base release assay. Results are summarized in Table 1. Extracts from a wide variety of tissues of mNth1+/+ adult males displayed DNA N-glycosylase activity, with the highest activity being in thymus and brain. However, the specific enzymatic activity of mNth1+/+ adult tissues was 2 orders of magnitude lower than that of mNth1+/+ ES cells. Analysis of extracts from whole mNth1+/+ embryos revealed that the specific activity of mNth1 in embryos was about 1 order of magnitude lower than that of mNth1+/+ ES cells and about 1 order of magnitude higher than that of mNth1+/+ adult tissues.
TABLE 1.
Specific activity of mNth1 in cells and tissues as determined by DNA N-glycosylase base release assaya
| Tissue or cells | mNth1 sp act (fmol/min/mg) from miceb
|
||
|---|---|---|---|
| Wild type | Heterozygote | Knockout | |
| Embryonic | |||
| ES cells | 37.3 ± 2.7 | 24.1 ± 3.0 | NA |
| Embryos (whole; 10-12 days old) | 4.2 ± 1.5 | 0 | |
| Adult tissues | 2.3 ± 0.6 | 0 | |
| C57BL/6-129/SvJ | |||
| Male | |||
| Brain | 0.4 ± 0.2 | 0 | |
| Thymus | 0.6 ± 0.3 | 0 | |
| Liver | 0.7 ± 0.4 | 0 | |
| Female | |||
| Brain | 0.4 ± 0.1 | 0 | |
| Thymus | 0.5 ± 0.4 | 0 | |
| Liver | 0.9 ± 0.4 | 0 | |
| 129/SvJ male | |||
| Brain | 0.5 ± 0.0 | NA | |
| Thymus | 1.2 ± 0.1 | NA | |
| Liver | 0.7 ± 0.1 | NA | |
Cellular or tissue sonicates were incubated with 100 ng of UV-irradiated poly(dG-[3H]dC). Sonicates of ES cells and embryos were assayed for 2 to 4 h. Sonicates of adult tissues were assayed for 14 h.
Each value represents the mean from a minimum of three experiments ± standard deviation. NA, not applicable.
By the DNA N-glycosylase assay, tissues from mNth1−/− adult mice contained no detectable DNA N-glycosylase activity. Likewise, measurement of mNth1 activity from extracts of whole mNth1−/− embryos did not show any detectable enzymatic activity. Gender and strain did not significantly affect specific activity, as determined from assays of activity of tissues from adult female C57BL/6-129/SvJ mice and adult male and female 129/SvJ mice (Table 1).
(iv) Cleavage assay.
Because of the relative insensitivity of the 3H-based DNA N-glycosylase release assay, DNA N-glycosylase activity was measured by the more sensitive method, i.e., cleavage of a Tg residue-containing 2′-deoxyribose oligonucleotide which had been 5′ end labeled with [γ-32P] and paired with a complementary 2′-deoxyribose oligonucleotide containing either an adenine or a guanine opposite Tg. In addition to its increased sensitivity, this assay can distinguish between DNA N-glycosylase activity alone and the sum of DNA N-glycosylase and AP lyase activities (22). DNA N-glycosylase activity cleaves the N-glycosyl bond of the Tg residue, thereby releasing the modified base. However, by itself it does not cleave the newly formed AP site. Thus, the AP site-containing 2′-deoxyribose oligonucleotide remains intact as a 30-mer. Exposure to alkali of such an AP site-containing 2′-deoxyribose oligonucleotide effects strand cleavage 3′ to the AP site via base-catalyzed β-elimination (23). AP lyase enzymatic activity also catalyzes β-elimination of the 3′ phosphate group at an AP site, but this strand cleavage occurs at neutral pH. Thus, differences between the extent of DNA N-glycosylase activity and AP lyase activity can be detected by comparing the extent of strand cleavage under neutral and alkaline conditions. This experimental approach revealed that hNth1 had five- to sevenfold-greater DNA N-glycosylase activity against a Tg residue than AP lyase activity against the resulting AP site generated by the DNA N-glycosylase (22). This difference in the two activities differs markedly from E. coli endonuclease III in which the two enzymatic activities occur virtually simultaneously (30).
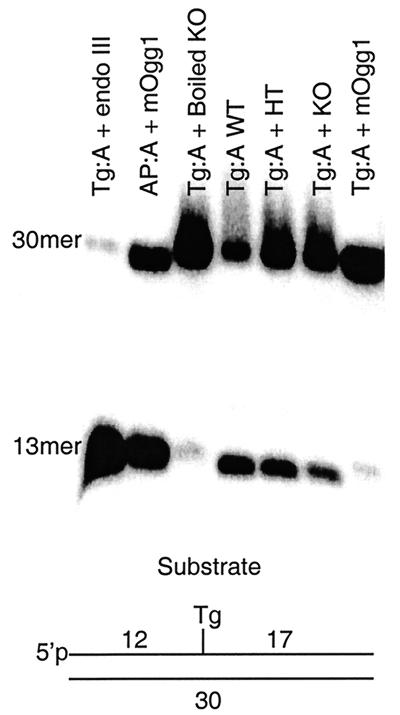
Figure 3 illustrates the results of a cleavage assay with thymic sonicates at neutral pH. Cleavage of the input 30-mer produces a 13-mer product. A gene dosage effect was observed in the analysis of sonicates of mNth1+/+ (Fig. 3, lane Tg:A WT) and mNth1+/− (Fig. 3, lane Tg:A + HT) adult thymic tissue that was consistent with the results obtained with the DNA N-glycosylase base release assay (Fig. 1C and Table 1). Cleavage of Tg:A was detected in mNth1−/− tissue (Fig. 3, lane Tg:A + KO) despite the absence of both alleles of the mNth1 gene.
FIG. 3.
Enzyme activity by DNA cleavage assay. Double-stranded DNA substrate (200 nM) was incubated with pure protein or murine thymic sonicates for 4 h. Samples were run on an 20% polyacrylamide-8 M urea gel. The positions of uncleaved DNA substrate (30mer) and the product of enzymatic strand cleavage (13mer) are shown. Abbreviations: endo III, E. coli endonuclease III; mOgg1, murine homolog of 8-oxoguanine DNA glycosylase; WT, wild-type (mNth1+/+) sonicate; HT, mNth1+/− sonicate; KO, knockout (mNth1−/−) sonicate.
Thus, it seemed that an activity capable of cleaving the Tg-containing 2′-deoxyribose oligonucleotide was present in the tissue of mNth1−/− mice.
Characterization of novel enzyme activity.
To further characterize this novel activity, we conducted additional cleavage assays with 2′-deoxyribose oligonucleotides containing a Tg residue. The back-up activity was present in all tissues examined (brain, thymus, and liver), with the highest specific activity in the thymus (data not shown).
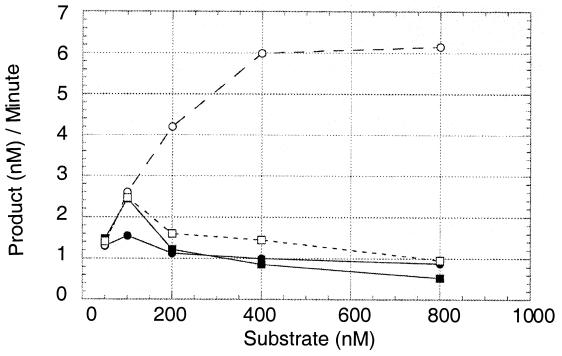
Tg-containing 2′-deoxyribose oligonucleotide was then paired with either adenine or guanine to determine the substrate specificity of the novel activity. The rationale for this approach derived from the data of Fig. 4. The DNA N-glycosylase activity of hNth1 was five- to sevenfold greater than its AP lyase activity when the Tg residue was opposite an adenine residue, but the two activities were the same when the Tg residue was opposite a guanine residue.
FIG. 4.
Differential processing of Tg:A (circles) versus Tg:G (squares) by hNth1 (20 nM). Solid symbols denote AP lyase activity (neutral pH). Open symbols denote combined DNA N-glycosylase/AP lyase activity of hNth1 against the substrate (putrescine, alkaline pH). Reaction time was 30 min.
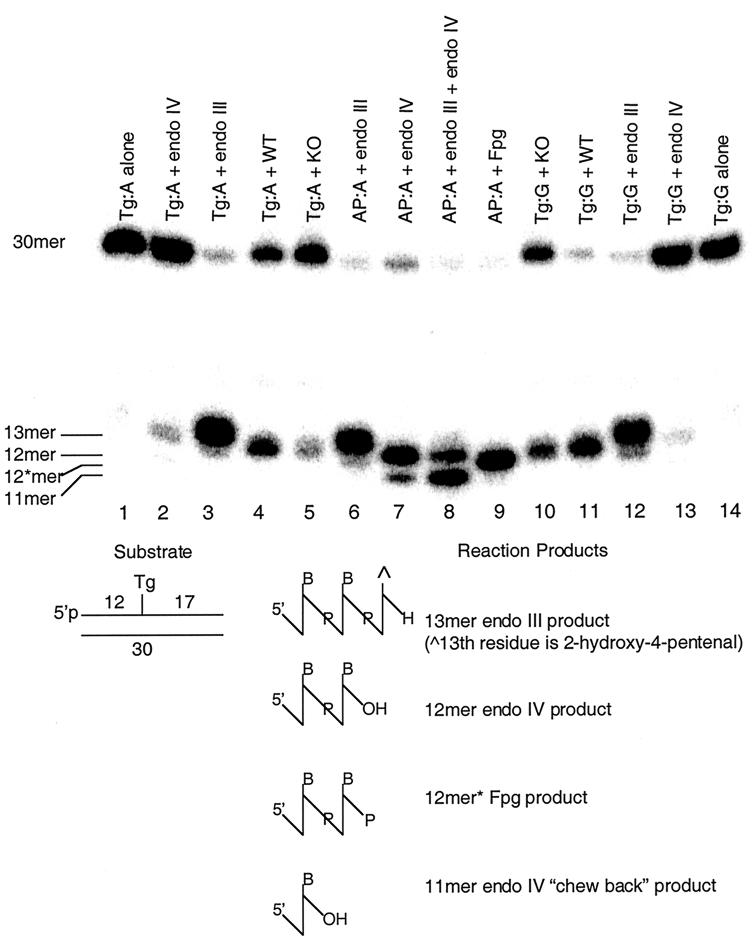
In contrast to results with purified mammalian Nth1 proteins, mNth1+/+ whole-cell sonicates of murine thymi showed a preference for Tg:G rather than Tg:A (Fig. 5, lanes 4 and 11). Under identical conditions in two separate assays, mNth1+/+ sonicates cleaved 77 and 89% of the Tg:G substrate compared to only 40 and 50% of Tg:A based upon quantitation of the cleavage assays with a phosphorimager (22). This difference was also manifest with mNth1−/− thymic sonicates, suggesting that the novel activity recognized Tg:G more efficiently than Tg:A (Fig. 5, lanes 5 and 10). mNth1−/− sonicates, containing the same amount of protein as the paired mNth1+/+ sonicates, cleaved 41 and 73% of Tg:G-containing substrate, whereas cleavage of the Tg:A substrate was only 19 and 21%. Thus, the preference of both mNth1+/+ and mNth−/− sonicates for Tg:G was two- to threefold greater than for Tg:A.
FIG. 5.
Cleavage assay demonstrating differential processing of Tg:G versus Tg:A by mNth1+/+ and mNth1−/− thymic sonicates at neutral pH. The substrate (10 nM) was incubated with purified enzymes or thymic sonicates (175 μg) for 6 h. Abbreviations: endo III, E. coli endonuclease III; endo IV, E. coli endonuclease IV; Fpg, E. coli formamidopyrimidine DNA glycosylase; WT, wild-type (mNth1+/+) sonicate; KO, mNth1−/− sonicate. The positions of uncleaved DNA substrate (30mer), the β-elimination product of endonuclease III (13mer), the product of AP endonuclease activity (12mer), the β/δ cleavage product of Fpg (12∗mer), and the product of exonucleolytic activity by endonuclease IV (11mer) are shown to the left of the gel.
To identify the chemical nature of the cleavage product, we included three markers (Fig. 5, lanes 6 to 9) which illustrated the relative migration of different products of cleavage by different enzymes. The substrate was a 30-mer containing an AP site paired with an adenine residue. The 13-mer of lane 6 represents the β-elimination product characteristic of E. coli endonuclease III. Lane 7 represents the product of reaction with E. coli endonuclease IV, an AP 5′ endonuclease generating a product which is one sugar residue smaller than the β-elimination product generated by the AP lyase activity of endonuclease III in lane 6. The fainter, faster-migrating band is a product of the exonucleolytic activity of endonuclease IV, chewing back 1 more nucleotide to an 11-mer (R. P. Cunningham, unpublished data). Lane 8 demonstrates the reaction of endonuclease III and endonuclease IV together, the product being the 12-mer characteristic of AP endonuclease activity and the 11-mer generated by endonuclease IV exonucleolytic activity. Interestingly, in the presence of endonuclease III, the 11-mer product is increased. The significance of this observation is not understood at this time. Lane 9 is the product of the reaction of the 2′-deoxyribose oligonucleotide with E. coli formamidopyrimidine DNA glycosylase (Fpg) protein which catalyzes both β- and δ-elimination reactions. The product of these two elimination reactions is a 12-mer which differs from that generated by endonuclease IV by the presence of a phosphate residue on its 3′ end as opposed to the OH moiety generated by the phosphoesterase action of endonuclease IV. The retained 3′-phosphate increases the charge-to-mass ratio of the 12-mer, causing it to migrate more rapidly to the cathode than the 12-mer with the 3′OH moiety.
The back-up activity was not attributable to the murine homolog of 8-oxoguanine-DNA glycosylase, mOgg1 (Fig. 3, lane Tg:A + mOgg1). Furthermore, under our conditions E. coli endonuclease IV did not efficiently cleave Tg paired either with adenine or guanine (Fig. 5, lanes 2 and 13), thereby making it unlikely that a mammalian endonuclease IV-like enzyme is responsible for the residual activity (16).
The product of the cleavage by the sonicates of mNth1+/+ and mNth1−/− tissues migrated to the same position as did the product of the reaction of the AP site-containing substrate with endonuclease IV. We suggest that this represents processing of the cleavage product of mNth1 by an endogenous AP endonuclease activity. This activity could not be inhibited by the addition of EDTA in high concentrations. This AP endonuclease processed the product of the endogenous mNth1 reaction (be it the β-elimination product resulting from AP lyase activity or an intact AP site resulting from DNA N-glycosylase activity) to the resulting 12-mer.
DISCUSSION
To determine the role of mNth1 in protecting the genomic integrity of a mammalian organism, we generated a mNth1−/− mouse. Our strategy involved the targeted disruption of those exons critical for the enzymatic activity of the mNth1 protein. The production of mNth1 mRNA, as determined by RT-PCR, was completely absent in the mutant animals.
The phenotype of the mNth1−/− animals was indistinguishable from that of the mNth1+/+ animals by several criteria. Both male and female mNth1−/− mice were viable and fertile, and their litters were of a size and gender ratio equal to those of wild-type litters. Crosses of heterozygotes produced the expected Mendelian ratio of offspring. Moreover, gross and microscopic examination revealed no differences in morphology during development between wild-type and mutant mice. To date, mNth1−/− mice have shown no evidence of spontaneous tumorigenesis, and they have shown no signs of neurological deterioration as judged grossly by observation of their activity, locomotion, and startle response to sudden noise. The oldest mNth1−/− animals are now 24 months old.
In vitro primary mNth1−/− thymocytes did not manifest an increase in apoptotic response after exposure to either aerobic ionizing radiation or H2O2. Thus, our results are similar to those in which DNA N-glycosylases that initiate BER of other modified bases have been deleted. Deletion of the major uracil DNA N-glycosylase (Ung) (8, 25), the Ogg1 protein (which repairs oxidized purines) (24), or the alkylpurine DNA N-glycosylase (Aag) (6-8, 10) did not interfere with the development of the animals, nor was it associated with an increase in spontaneous tumorigenesis. However, mAag cells did show increased susceptibility to the toxic and mutagenic effects of alkylating agents (6-8, 10).
The lack of an informative phenotype in the mNth1−/− mice may be due to one or more of the following four explanations. (i) The modified pyrimidines repaired by mNth1 may be repaired by previously undescribed back-up activities. (ii) Nucleotide excision repair activity, which is known to remove Tg residues (17, 18, 21), may be sufficient to prevent manifestation of a phenotype. (iii) mNth1 may act to prevent pathological consequences resulting from oxidative stresses that were not modeled in our experiments. (iv) The consequences of nonrepair may accumulate over multiple generations and are not yet apparent in the relatively short time periods of these experiments.
Using a DNA N-glycosylase 3H-labeled base release assay, we could neither conclusively demonstrate nor exclude the existence of a back-up activity in the tissues of mNth1−/− mice. To determine whether such an activity did indeed exist, we assayed extracts of tissues of adult mice for activity using the more sensitive cleavage assay that utilized a Tg-containing 32P-5′-labeled 2′-deoxyribose oligonucleotide as a substrate. This assay revealed what we believe to be a novel enzymatic activity which recognized Tg in DNA, particularly when opposite a guanine residue. The migration of the product 2′-deoxyribose oligonucleotide in an 8 M urea-22% polyacrylamide gel was characteristic of the product of an AP endonuclease cleavage activity. At this time, it remains to be determined whether the novel repair activity is a DNA N-glycosylase, a DNA N-glycosylase/AP lyase, or an endonuclease which nicks 5′ to the Tg-containing site. In our hands, the product of the interaction of crude sonicates of wild-type tissue with a Tg:A-containing substrate similarly yielded a product of AP endonuclease activity. Since such tissue contains abundant mNth1 whose enzymatic products are either an AP site and/or a β-elimination product, it is apparent that either one or both of these products were processed during the reaction by an endogenous AP endonuclease activity. Such an AP endonuclease activity was not suppressed by high concentrations of EDTA, which is consistent with the presence in tissue sonicates of an AP endonuclease activity which was not dependent on chelatable divalent cations for activity. E. coli endonuclease IV contains three nonchelatable Mn2+ residues (13). Therefore, although no endonuclease IV homolog has been identified in searches of the murine or human genome, an analogous activity could conceivably be present in these sonicates.
Recently, a repair endonuclease activity of E. coli endonuclease IV directed against several types of ring-saturated pyrimidine residues was described, and additional evidence for an analogous endonuclease activity in HeLa cells was offered. This report posited the existence of yet another repair pathway which was termed nucleotide incision (16). However, in those experiments, Tg was not used as a substrate. Here we showed that E. coli endonuclease IV has virtually no activity against a Tg-containing 2′-deoxyribose-oligonucleotide, be it Tg:A or Tg:G. Our results in this regard are identical to those of Sarker et al. (30) who similarly detected no activity of endonuclease IV against Tg in DNA. However, the contention that an endonuclease IV-like analogous activity may be present in mammalian cells is supported by our observation that the product of cleavage in both mNth1+/+ and mNth1−/− sonicates is indicative of an AP endonuclease which is not inhibited by EDTA.
It is clear that in vitro, hNth1 does not process Tg:G pairs as efficiently as it processes Tg:A, as shown in Fig. 4. Furthermore, as we have shown previously, the specific activity of the DNA N-glycosylase activity of hNth1 is not linked to its AP lyase activity in a 1:1 ratio (22). Both these observations are in sharp contrast to the in vitro properties of endonuclease III which processes both Tg:G and Tg:A with equal efficiency, yielding the β-elimination cleavage product (30). Asagoshi and colleagues (1) concluded that the activity of mNth1 against a Tg:G pair was identical to its activity against a Tg:A pair, but they measured cleavage only at neutral pH. Here we showed that the DNA N-glycosylase activity of mammalian Nth1 is much greater when the Tg residue is opposite an adenine than when opposite a guanine and that an activity which processes Tg:G pairs more efficiently than Tg:A pairs is present in tissue sonicates. Thus, it appears that during evolution, the substrate specificity of mammalian Nth1 diverged from that of its E. coli homolog.
From a standpoint of completeness, we showed in Fig. 3 (lane Tg:A + mOgg1) that the back-up activity was not mOgg1. Although mOgg1 was not tested against Tg:G, its complete absence of activity against Tg:A is inconsistent with our finding that the novel repair activity also processes Tg:A, albeit not as efficiently as Tg:G.
When does Tg:G occur in DNA? It is generally held that Tg is a product of oxidation of thymine in situ in DNA and that the consequences of its formation are manifest primarily as cytotoxicity due to interference with transcription and replication (5, 14, 29). However, we demonstrated that the exposure of 5-methylcytosine (5meCyt) residues in DNA to either aerobic ionizing radiation or H2O2 yielded Tg almost exclusively as the product of oxidation of 5meCyt. Furthermore, we showed that the G value for formation of Tg from 5meCyt was virtually identical to the formation of Tg from thymine residues in DNA (34). Thus, it is entirely possible that oxidation of 5meCyt to Tg occurs in cellular DNA under the very same conditions which generate Tg from thymine. For reasons that are still not well understood, the oxidation of 5meCyt residues in organisms that use 5meCyt to regulate gene expression may have promoted the development of a repair enzyme whose primary function was to remove such potentially mutagenic lesions and to participate in the process of maintaining correct DNA methylation.
In summary, we have demonstrated that mice can survive despite the complete absence of mNth1 activity. Moreover, in generating these mice, we identified a heretofore undescribed and hence novel repair enzyme activity directed primarily against Tg:G pairs, and we suggest that this repair activity functions in maintaining the integrity of methylated DNA. The fact that this novel activity also repairs Tg:A pairs may explain why the deletion of mNth1 did not result in an informative phenotype. We are now actively characterizing the nature of this novel DNA repair activity.
Acknowledgments
This work was supported in part by National Institutes of Health grants CA 16669 and CA 49869 (to G.W.T.), CA 16087 (to Kaplan Cancer Center), 5T32 CA-09161 (to M.T.A.O.), GM 46312 (to R.P.C.), and NIES-ES 09127 (to A.K.B.).
We thank D. Zharkov for supplying mOgg1, J. Hirst for fluorescence-activated cell sorting analysis, X. Zhang for assistance with RT-PCR, R. Basch and D. Levy for their critical reading of the manuscript, and J. Weider for assistance with graphics.
REFERENCES
- 1.Asagoshi, K., H. Odawara, H. Nakano, T. Miyano, H. Terato, Y. Ohyama, S. Seki, and H. Ide. 2000. Comparison of substrate specificities of Escherichia coli endonuclease III and its mouse homologue (mNTH1) using defined oligonucleotide substrates. Biochemistry 39:11389-11398. [DOI] [PubMed] [Google Scholar]
- 2.Boorstein, R. J., T. P. Hilbert, R. P. Cunningham, and G. W. Teebor. 1990. Formation and stability of repairable pyrimidine photohydrates in DNA. Biochemistry 29:10455-10460. [DOI] [PubMed] [Google Scholar]
- 3.Boorstein, R. J., T. P. Hilbert, J. Cadet, R. P. Cunningham, and G. W. Teebor. 1989. UV-induced pyrimidine hydrates in DNA are repaired by bacterial and mammalian DNA glycosylase activities. Biochemistry 28:6164-6170. [DOI] [PubMed] [Google Scholar]
- 4.Cadet, J., A.-G. Bourdat, C. D'Ham, V. Duarte, D. Gasparutto, A. Romieu, and J.-L. Ravanat. 2000. Oxidative base damage to DNA: specificity of base excision repair enzymes. Mutat. Res. 462:121-128. [DOI] [PubMed] [Google Scholar]
- 5.Clark, J. M., and G. P. Beardsley. 1986. Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic Acids Res. 14:737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elder, R. H., J. G. Jansen, R. J. Weeks, M. A. Willington, B. Deans, A. J. Watson, K. J. Mynett, J. A. Bailey, D. P. Cooper, J. A. Rafferty, M. C. Heeran, S. W. Wijnhoven, A. A. van Zeeland, and G. P. Margison. 1998. Alkylpurine-DNA-N-glycosylase knockout mice show increased susceptibility to induction of mutations by methyl methanesulfonate. Mol. Cell. Biol. 18:5828-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelward, B. P., G. Weeda, M. D. Wyatt, J. L. Broekhof, J. de Wit, I. Donker, J. M. Allan, B. Gold, J. H. Hoeijmakers, and L. D. Samson. 1997. Base excision repair deficient mice lacking the Aag alkyladenine DNA glycosylase. Proc. Natl. Acad. Sci. USA 94:13087-13092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedberg, E. C., and L. B. Meira. 2000. Database of mouse strains carrying targeted mutations in genes affecting cellular responses to DNA damage. Version 4. Mutat. Res. 459:243-274. [DOI] [PubMed] [Google Scholar]
- 9.Gasparutto, D., A. G. Bourdat, C. D'Ham, V. Duarte, A. Romieu, and J. Cadet. 2000. Repair and replication of oxidized DNA bases using modified oligodeoxyribonucleotides. Biochimie 82:19-24. [DOI] [PubMed] [Google Scholar]
- 10.Hang, B., B. Singer, G. P. Margison, and R. H. Elder. 1997. Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc. Natl. Acad. Sci. USA 94:12869-12874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilbert, T. P., R. J. Boorstein, H. C. Kung, P. H. Bolton, D. Xing, R. P. Cunningham, and G. W. Teebor. 1996. Purification of a mammalian homologue of Escherichia coli endonuclease III: identification of a bovine pyrimidine hydrate-thymine glycol DNAse/AP lyase by irreversible cross linking to a thymine glycol-containing oligoxynucleotide. Biochemistr. 35:2505-2511. [DOI] [PubMed] [Google Scholar]
- 12.Hilbert, T. P., W. Chaung, R. J. Boorstein, R. P. Cunningham, and G. W. Teebor. 1997. Cloning and expression of the cDNA encoding the human homologue of the DNA repair enzyme, Escherichia coli endonuclease III. J. Biol. Chem. 272:6733-6740. [DOI] [PubMed] [Google Scholar]
- 13.Hosfield, D. J., Y. Guan, B. J. Haas, R. P. Cunningham, and J. A. Tainer. 1999. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 98:397-408. [DOI] [PubMed] [Google Scholar]
- 14.Ide, H., Y. W. Kow, and S. S. Wallace. 1985. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 13:8035-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda, S., T. Biswas, R. Roy, T. Izumi, I. Boldogh, A. Kurosky, A. H. Sarker, S. Seki, and S. Mitra. 1998. Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem. 273:21585-21593. [DOI] [PubMed] [Google Scholar]
- 16.Ischenko, A. A., and M. K. Saparbaev. 2002. Alternative nucleotide incision repair pathway for oxidative DNA damage. Nature 415:183-187. [DOI] [PubMed] [Google Scholar]
- 17.Kow, Y. W., S. S. Wallace, and B. Van Houten. 1990. UvrABC nuclease complex repairs thymine glycol, an oxidative DNA base damage. Mutat. Res. 235:147-156. [DOI] [PubMed] [Google Scholar]
- 18.Kuipers, G. K., B. J. Slotman, H. A. Poldervaart, I. M. van Vilsteren, C. A. Reitsma-Wijker, and M. V. Lafleur. 2000. The role of nucleotide excision repair of Escherichia coli in repair of spontaneous and gamma-radiation-induced DNA damage in the lacZalpha gene. Mutat Res. 460:117-125. [DOI] [PubMed] [Google Scholar]
- 19.Kuo, C.-F., D. E. McRee, C. L. Fisher, S. F. O'Handley, R. P. Cunningham, and J. A. Tainer. 1992. Atomic structure of DNA repair [4Fe-4S] enzyme endonuclease III. Science 258:434-440. [DOI] [PubMed] [Google Scholar]
- 20.Kuo, C.-F., D. E. McRee, R. P. Cunningham, and J. A. Tainer. 1992. Crystallization and crystallographic characterization of the iron-sulfur-containing DNA-repair enzyme endonuclease III from Escherichia coli. J. Mol. Biol. 227:347-351. [DOI] [PubMed] [Google Scholar]
- 21.Lin, J. J., and A. Sancar. 1989. A new mechanism for repairing oxidative damage to DNA: (A)BC excinuclease removes AP sites and thymine glycols from DNA. Biochemistry 28:7979-7984. [DOI] [PubMed] [Google Scholar]
- 22.Marenstein, D. R., M. T. A. Ocampo, M. K. Chan, A. Altamirano, A. K. Basu, R. J. Boorstein, R. P. Cunningham, and G. W. Teebor. 2001. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J. Biol. Chem. 276:21242-21249. [DOI] [PubMed] [Google Scholar]
- 23.Mazumder, A., J. A. Gerlt, M. J. Absalon, J. Stubbe, R. P. Cunningham, J. Withka, and P. H. Bolton. 1991. Stereochemical studies of the beta-elimination reactions at aldehydic abasic sites in DNA: endonuclease III from Escherichia coli, sodium hydroxide, and Lys-Trp-Lys. Biochemistry 30:1119-1126. [DOI] [PubMed] [Google Scholar]
- 24.Minowa, O., T. Arai, M. Hirano, Y. Monden, S. Nakai, M. Fukuda, M. Itoh, H. Takano, Y. Hippou, H. Aburatani, K. Masumura, T. Nohmi, S. Nishimura, and T. Noda. 2000. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. USA 97:4156-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsen, H., I. Rosewell, P. Robins, C. M. Skjelbred, S. Andersen, G. Slupphaug, G. Daly, H. E. Krokan, T. Lindahl, and D. E. Barnes. 2000. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell 5:1059-1065. [DOI] [PubMed] [Google Scholar]
- 26.O'Donnell, R. E., R. J. Boorstein, R. P. Cunningham, and G. W. Teebor. 1994. Effect of pH and temperature on the stability of UV-induced repairable pyrimidine hydrates in DNA. Biochemistry 33:9875-9880. [DOI] [PubMed] [Google Scholar]
- 27.Oyama, Y., S. Noguchi, M. Nakata, Y. Okada, Y. Yamazaki, M. Funai, L. Chikahisa, and K. Kanemaru. 1999. Exposure of rat thymocytes to hydrogen peroxide increases annexin V binding to membranes: inhibitory actions of deferoxamine and quercetin. Eur. J. Pharmacol. 384:47-52. [DOI] [PubMed] [Google Scholar]
- 28.Ravanat, J.-L., T. Douki, and J. Cadet. 2001. Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B 63:88-102. [DOI] [PubMed] [Google Scholar]
- 29.Rouet, P., and J. M. Essigmann. 1985. Possible role for thymine glycol in the selective inhibition of DNA synthesis on oxidized DNA templates. Cancer Res. 45:6113-6118. [PubMed] [Google Scholar]
- 30.Sarker, A. H., S. Ikeda, H. Nakano, H. Terato, H. Ide, K. Imai, K. Akiyama, K. Tsutsui, Z. Bo, K. Kubo, K. Yamamoto, A. Yasui, M. C. Yoshida, and S. Seki. 1998. Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J. Mol. Biol. 282:761-774. [DOI] [PubMed] [Google Scholar]
- 31.Scharer, O. D., and J. Jiricny. 2001. Recent progress in the biology, chemistry and structural biology of DNA glycosylases. Bioessays 23:270-281. [DOI] [PubMed] [Google Scholar]
- 32.Thayer, M. M., H. Ahern, D. Xing, R. P. Cunningham, and J. A. Tainer. 1995. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J. 14:4108-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zharkov, D. O., T. A. Rosenquist, S. E. Gerchman, and A. P. Grollman. 2000. Substrate specificity and reaction mechanism of murine 8-oxoguanine-DNA glycosylase. J. Biol. Chem. 275:28607-28617. [DOI] [PubMed] [Google Scholar]
- 34.Zuo, S., R. J. Boorstein, and G. W. Teebor. 1995. Oxidative damage to 5-methylcytosine in DNA. Nucleic Acids Res. 23:3239-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]