Abstract
The Polycomb group (PcG) genes are required to maintain homeotic genes in a silenced state during development in drosophila and mammals and are thought to form several distinct silencing complexes that maintain homeotic gene repression during development. Mutations in the PcG genes result in developmental defects and have been implicated in human cancer. Although some PcG protein domains are conserved between flies and humans, substantial regions of several PcG proteins are divergent and humans contain multiple versions of each PcG gene. To determine the effects of these changes on the composition and function of a PcG complex, we have purified a human Polycomb repressive complex from HeLa cells (hPRC-H) that contains homologues of PcG proteins found in drosophila embryonic PRC1 (dPRC1). hPRC-H was found to have fewer components than dPRC1, retaining the PcG core proteins of dPRC1 but lacking most non-PcG proteins. Preparations of hPRC-H contained either two or three different homologues of most of the core PcG proteins, including a new Ph homologue we have named HPH3. Despite differences in composition, dPRC1 and hPRC-H have similar functions: hPRC-H is able to efficiently block remodeling of nucleosomal arrays through a mechanism that does not block the ability of nucleases to access and cleave the arrays.
The Polycomb group (PcG) genes are essential for maintenance of appropriate expression patterns of developmental master regulators, such as Hox genes, and thus are essential for proper development. Changes in expression of PcG proteins have been associated with cancer, while targeted deletions of members of this family generally have lethal phenotypes (reviewed in reference 20). Two-hybrid and immunoprecipitation studies have found that the PcG proteins form at least two large nonoverlapping protein complexes. The first type of complex, known as class I, contains homologues of the drosophila proteins esc and E(z) (33, 50, 64) and has been shown to associate with histone deacetylase activity (57, 62). The second type of complex (class II) includes homologues of Pc, Psc, Ph, Ring, and Scm (3, 16, 45, 51). No enzymatic activity has been ascribed to this second group of proteins.
The PcG genes were originally identified in drosophila, where homozygous mutant embryos exhibited severe homeotic transformations (reviewed in references 24 and 54). Genetic analysis of these mutants showed extensive derepression of the homeotic genes and suggested that they may play a role in maintenance of silencing. Similarly, targeted deletion of PcG genes in mice results in homeotic transformation of segment identity due to delayed ectopic expression of some Hox genes and is generally lethal (1, 11, 12, 35, 56, 61). The phenotypes of mice lacking single PcG genes are generally milder then those observed in drosophila, at least in part because of the presence of multiple homologues of each drosophila PcG gene in mammals (Table 1). Disruption of both homologues of Psc (Bmi1 and Mel-18), for example, is lethal much earlier in development and causes more extreme Hox gene deregulation than does the disruption of either gene individually (2).
TABLE 1.
Class II PcG genes
| Drosophila | Human | Domain | Reference(s) |
|---|---|---|---|
| Pc | HPC1 (CBX2/M33) | Chromodomain | 37 |
| HPC2 (CBX4) | C terminal | 43 | |
| HPC3 (CBX8/ rectachrome) | 4 | ||
| Ph-p | HPH1 (EDR1/Rae28) | SEP | 18 |
| Ph-d | HPH2 (EDR2) | Zn finger | |
| HPH3 | Homology region I | This study | |
| Scm | SCMH1 | SEP | 5, 58 |
| SCMH2 | Zn finger | ||
| Psc | Bmil | RING finger | 6, 41 |
| Mel-18 | HTH | ||
| dRING | RING1 | RING finger | 29, 43, 48 |
| RNF2 |
PcG complexes are thought to mediate silencing by creating a chromatin configuration that is refractory to transcriptional activation, although the precise mechanisms involved are not understood (reviewed in references 14, 39, and 55). Genetic studies identified the Trithorax group (trxG) genes as suppressors of PcG mutations (reviewed in reference 24). A major function of trxG genes appears to be remodeling of chromatin structure, since several of these genes encode subunits of the SWI/SNF chromatin remodeling complex (13, 22, 36) and others associate with the histone acetyltransferase CBP (38). The presence of specific domains, such as the chromodomain of Pc, and immunolocalization studies originally suggested that the PcG proteins act through effects on chromatin structure. Recent studies that have begun a functional characterization of these complexes lend support to this view, as PcG complexes have been reported to deacetylate histone tails and to block ATP-dependent chromatin remodeling (51, 62). In vivo, these complexes are targeted to Polycomb and Trithorax response elements (PRE/TRE). This targeting is separable from the function of the complex, as artificially targeted complexes are able to repress transcription in vivo (10, 31). Once the complexes are established on the DNA, they are able to maintain silenced transcription long after the targeting factors are removed (reviewed in reference 24).
Gene targeting studies indicate that at least some biological functions of PcG genes, such as regulation of Hox gene expression, have been conserved evolutionarily consistent with conservation of several protein domains between drosophila and mammals. However, many of the PcG genes have also diverged substantially. Furthermore, most of the PcG genes have been duplicated in mammals and different expression patterns and biological functions have been described for different homologues. For example, Bmi1 and Mel-18, the homologues of Psc (9, 63), are less than one-third of the size of Psc (46 and 50 versus 188 kDa) and appear to have opposite effects on cell growth (19, 23, 65). These data suggest that mammalian PcG proteins may interact with different proteins than do drosophila PcG proteins and may have different activities. Comparing composition and functions of complexes purified from human cells to those purified from flies is an important step in investigating these possibilities.
We have recently described the purification and initial functional characterization of the major class II complex from drosophila embryos, drosophila embryonic PRC1 (dPRC1) (45). In parallel with these studies, we have purified and characterized the human counterpart to this complex. A comparison of the components and functions of the class II complexes can reveal core subunits and fundamental activities. We find that only a subset of proteins are conserved between hPRC-H (Polycomb repressive complex from HeLa cells) and dPRC1; most, but not all, of these conserved subunits are PcG genes that emerged from the original developmental screens. Despite the different natures of the complexes, dPRC1 and hPRC-H performed with similar function and efficiency in a variety of protocols that used nucleosomal arrays as template.
MATERIALS AND METHODS
Generation of FLAG-tagged cell lines and tissue culture.
FLAG sequences were added to the C termini of M33 and Bmi1 by PCR and cloned into pBABE. Cell lines were constructed as previously described (53) and grown in Dulbecco modified Eagle medium plus10% fetal calf serum and 1 mg of puromycin per ml. Cells were expanded to large volumes at Cellex Biosciences Inc.
Purification of hPRC-H.
Nuclear extracts were prepared from 100 liters of tagged HeLa cells (cell lines M33F-1 and BMI1F-17) as described previously (53) with 1.5 M KCl for the high-salt extraction buffer. A 50- to 60-mg sample of undialyzed nuclear extract (5 ml) was diluted in BCN buffer (20 mM HEPES, K+ [pH 7.9], 0.2 mM EDTA, 20% glycerol, 0.1% NP-40, 0.5 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride) containing 300 mM KCl (BCN300) and bound to a 5-ml Hi-Trap heparin column (Amersham-Pharmacia) on an AKTA fast protein liquid chromatography apparatus (Amersham-Pharmacia). Samples were washed with 4 column volumes (CV) of BCN300 and eluted with an 8-CV linear gradient of 300 to 1,000 mM KCl. Fractions from three repeats of this gradient containing the tagged protein (∼350 to 550 mM KCl) were pooled and bound to 1 ml of M2 agarose beads (Sigma) overnight. The beads were washed in a column with 20 CV of BCN300 and BCN450, equilibrated with BCN300, and eluted with 1 mg of FLAG peptide (DYKDDDDK) per ml. For analysis on cation-exchange columns, M2 columns were equilibrated with BCN200 and eluted with FLAG/BCN200 directly onto a 1-ml Hi-Trap S column. The S column was transferred to an AKTA fast protein liquid chromatography apparatus exclusively with PEEK tubing (Upchurch). The column was washed with 5 CV of BCN200, and bound proteins were eluted with a 9-CV linear gradient of 200 to 500 mM KCl. Fractions were either immediately resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for immunoblot analysis or precipitated with trichloroacetic acid (TCA) for immunoblot analysis and silver staining. Yields from this column were approximately 50%.
Estimation of hPRC-H concentration.
Concentrations of the M2-purified fractions were determined by the Bradford assay (Bio-Rad) and estimated by silver staining compared to bovine serum albumin standards. The molecular mass of the complex was determined by adding up the major bands of the complex as if they were stoichiometric. The molecular mass of 500 kDa agreed with gel filtration chromatography estimates generated with a Superose 6 column (Amersham-Pharmacia) in BCN300. Yields from this column were typically less than 1% (data not shown). The protein molecular size markers used were blue dextran (void), thyroglobulin (670 kDa), catalase (230 kDa), and bovine serum albumin (66 kDa).
MS analysis of hPRC-H.
Isolation and mass spectrometry (MS)/MS analysis of the proteins are described in reference 45. The N terminus of the MS-sequenced polypeptide identified as RING1A matched the sequence found in the National Center for Biotechnology Information database under accession number CAC38442. Most of the RING1A sequences in the National Center for Biotechnology Information database (including accession number Q06587) lack 29 amino acids. The additional sequence of RING1A in hPRC-H extends the homology among the known RING1 homologues. To identify HPH3, two peptides, (K/R)MQQPQISVYSGSDR and (K/R)SSLLIEQPVK, were BLAST tested against the full human genome and dbEST. With expressed sequence tags (ESTs) in the database, we generated a contig that contained both peptides in a 2-kb sequence that mapped to chromosome 3. This contig contained the 3.5-kb cDNA CS0DK007YJ17 (Research Genetics). This isolate was sequenced redundantly. Four additional cDNAs tested (American Type Culture Collection) were significantly shorter, and none contained the entirety of the gene. In their cDNA-containing sections, they were identical to the Research Genetics clone.
Immunoblot assays.
The antibodies used in the immunoblot assays were M5 anti-FLAG (Sigma), F6 anti-Bmi1 (3), anti-RING1A (46), anti-HPH1 (18) anti-HPC2 (44), anti-SNF2H (7, 26), anti-BRG1 (47), anti-hsp70 (Santa Cruz), anti-RYBP, anti-E2F6 (59), anti-YY1 (Santa Cruz), anti-ENX, anti-EED (50), anti-p325 (26), anti-WCRF (7), anti-CtBP (49), anti-TAFII250 (Santa Cruz), and anti-TBP (Santa Cruz). Proteins were analyzed by standard SDS-7.5 or 8% PAGE, transferred to Immobilon P membranes, and detected with ECL and ECL+ reagents (Amersham-Pharmacia) in accordance with the manufacturer's recommendations.
Activity assays.
The dPRC1, human Swi/Snf, and histones used in activity assays were prepared as previously described (51). Topological assays were carried out as previously described (25), with chromatin assembled with heat-treated Xenopus egg extracts, except that the reaction volume was reduced to 20 μl. The reaction mixtures contained 100 ng of Swi/Snf, ∼3 ng of chromatinized template, 4 μM ATP, and 2 μM MgCl2 and were electrophoresed on a 1% agarose gel. No change in activity was seen when larger amounts of Swi/Snf were used (data not shown). Assembly and analysis of the 5S array by salt dialysis were done as previously described, and restriction enzyme assays were preformed essentially as previously described (15), with 1 ng of labeled array and 9 ng of HeLa polynucleosomes, as indicated, in 20-μl reaction mixtures. Swi/Snf activity was determined on the basis of the amounts of cutting in reaction mixtures with or without Swi/Snf present (see Fig. 4B, lanes 1 and 2). Micrococcal analysis was performed essentially the same way as the restriction enzyme assay, with 1 ng of end-labeled array. Mononucleosomes were assembled on a TPT-containing DNA sequence (47) by salt dialysis, and reactions were performed under conditions identical to those used for the restriction enzyme assays with 200 ng of Swi/Snf (32).
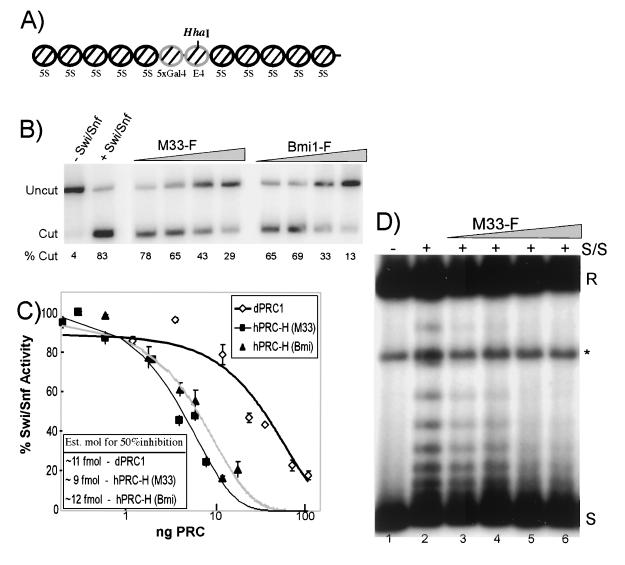
FIG. 4.
Activity of the hPRC-H complex. (A) Map of the 5S array used in the restriction enzyme and MNase assays. The HhaI site is indicated. (B) Restriction enzyme assay (REA). One nanogram (8 fmol) of nucleosomes was preincubated with increasing amounts of dPRC1 or hPRC-H from M33- and Bmi1-tagged lines. Lanes: 1, no-Swi/Snf control; 2, Swi/Snf (100 ng) only; 3 to 6, ∼0.4, 1.2, 4, and 12 fmol of hPRC-H (M33); 7 to 10, ∼1.2, 3.6, 12, and 36 fmol of hPRC-H (Bmi1). The percentage of template cut by HhaI is indicated under each lane. (C) Quantification of inhibition of remodeling. Same as panel B but with 80 fmol of nucleosomes. The amount (nanograms) of PRC added was calculated by Bradford analysis and comparative silver staining. Molar amounts were determined with an estimate of 500 kDa as the mass of hPRC-H (see Materials and Methods). Half-maximal repression occurs at ratios of hPRC-H to nucleosomes of approximately 1:8. Est. mol, estimated number of moles. (D) Topological assay. Nucleosomal plasmids were preincubated with hPRC-H for 15 min before being challenged with 100 ng of Swi/Snf and 4 U of topoisomerase I. Remodeled templates are visualized as slower-migrating topoisomers. Increasing amounts of hPRC-H increase the inhibition of Swi/Snf remodeling. Lanes: 1, no-Swi/Snf control; 2, Swi/Snf only; 3 to 6, titration of M33F hPRC-H as in panel B. R, relaxed; ∗, linear; S, fully negatively supercoiled.
RESULTS
Purification of hPRC-H.
To purify a mammalian class II PcG complex, we tagged the murine PcG genes for M33 and Bmi1 at their C termini with the FLAG epitope. Proteins were expressed with a retroviral vector in HeLa cells that have been used extensively for biochemical characterization of complexes. To avoid potential artifactual associations that might result from overexpression of tagged proteins, complexes were purified from cell lines in which the tagged M33 or Bmi1 protein was expressed at less then 25% of the untagged endogenous protein, as indicated by immunoblotting (data not shown). Nuclear extracts from these cell lines were subjected to a two-step purification to isolate proteins complexed with the FLAG-tagged protein (Fig. 1A). More than 80% of the tagged proteins eluted from a heparin column at 0.4 M salt. Western analysis showed that HPH1, Bmi1, and RING1A cofractionated with tagged M33. Fractions containing the FLAG-tagged protein were bound to an anti-FLAG affinity column, washed extensively, and eluted with FLAG peptide. The majority of the endogenous class II PcG proteins flowed through the affinity column, consistent with the low levels of expression of the tagged protein. High-stringency washes (0.6 or 1 M KCl) lowered the overall yield but did not alter the relative stoichiometry of the subunits, as shown by Western blotting and silver staining (data not shown). This procedure resulted in approximately 30 to 50% yields of the tagged proteins and purification of the complex 5,000- to 10,000-fold, as judged by quantification of immunoblots.
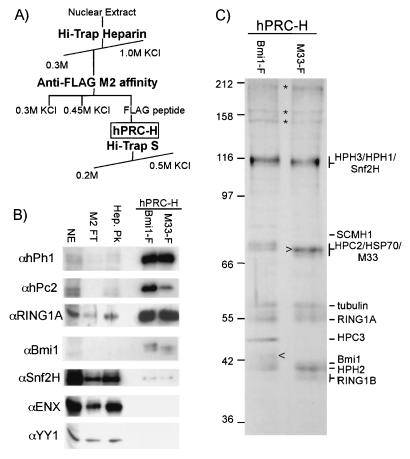
FIG. 1.
Purification of the hPRC-H complex. (A) Schematic for fractionation of hPRC-H. (B) class II PcG proteins cofractionate with FLAG-Bmi1 and FLAG-M33. Nuclear extract (NE; 10 μg), heparin peak fraction (Hep. Pk; 4 μg), M2 flowthrough (∼3 μg), and M2 elutions from Bmi1F and M33F lines (50 ng) were visualized with the indicated antibodies. Most of hPRC-H proteins flow through the M2 column because the tagged proteins are expressed at low levels. (C) Composition of the hPRC-H complex. M33F and Bmi1F M2 eluates (25 μg) were separated by SDS-8% PAGE and stained with silver. Proteins identified by MS sequencing that are present in all of the extracts tested are identified. Changes in the intensity of the bands corresponding to HPC2, HPC3, and Bmi1 between the two complexes likely represent a replacement of Pc homologues by epitope-tagged M33 (>) and a change in mobility of epitope-tagged Bmi1 (<). Asterisks mark proteins not consistently observed from preparation to preparation. The values on the left are molecular sizes in kilodaltons.
Immunoblot assays were performed on fractions containing the tagged proteins. Bmi1, HPC2, HPH1, and RING1A were all detected in significant amounts in both the M33- and Bmi1-based complexes (Fig. 1B). We name this complex hPRC-H. Importantly, class I PcG proteins EED and ENX were not detected in significant quantities. This agrees with previous data (33, 51, 64) suggesting that the two classes form distinct and biochemically separable complexes. Comparison of silver-stained gels of hPRC-H purified from Bmi1- and M33-tagged lines showed very similar banding patterns, with the expected exception of the tagged component and their endogenous paralogs (Fig. 1C, arrowheads indicating tagged protein).
PcG proteins constitute most of hPRC-H.
To identify the components of hPRC-H, we performed MS analyses of the polypeptides isolated from the complex. Purified fractions from M33-tagged cells were separated by SDS-PAGE, and 15 distinct bands were isolated. Peptides from these bands were identified with multidimensional MS, and searches of databases were performed to identify the proteins. Nine of 12 identified bands (excluding the tagged proteins) were identified as PcG genes, including BMI1, RING1A, RING1B, HPH1, HPH2, HPC2, HPC3, and SCMH1 (Fig. 1B). Multiple homologues of the Polycomb protein copurified with the M33-tagged line, suggesting that different Pc homologues may coexist in individual complexes.
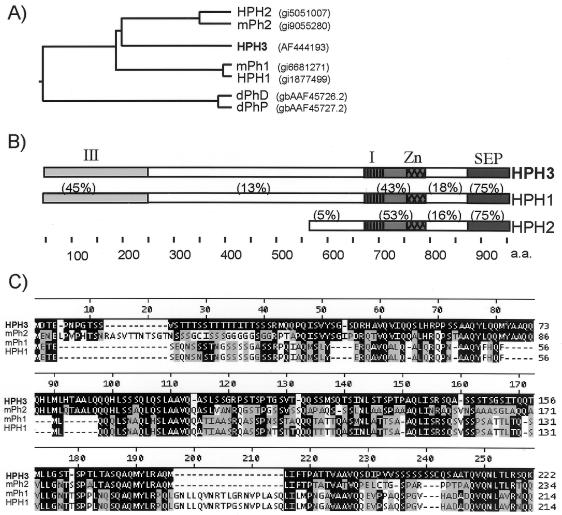
One prominent band (135 kDa) was not found in any nonredundant protein database. Two peptide sequences identified from it by matrix-assisted laser desorption ionization-time of flight MS and electrospray ionization-trandem mass spectrometry were used to search the human EST database. On the basis of several contiguous EST sequences, we obtained an apparent full-length cDNA (Research Genetics) containing both MS-sequenced peptides. This cDNA encodes a putative 931-amino-acid protein that is similar to other mammalian homologues of drosophila Ph. We therefore named this novel gene HPH3. HPH3 contains the conserved SEP domain, Zinc finger, and homology region I previously described (3) and has similar degrees of homology to both HPH1 and HPH2 (Fig. 2A). The N-terminal region of HPH3 has significant degrees of similarity to HPH1 (Fig. 2C). This homology extends to murine Ph2 but is not found in the drosophila polyhomeotic genes.
FIG. 2.
HPH3 is a new PH homologue. (A) Phylogenetic tree of known PH homologues. (B) Domain architecture of HPH proteins. I, homology domain I; Zn, zinc finger; SEP, SEP domain/homology domain II; III, novel homology domain. The values above the sequences refer to percent identity. (C) ClustalW alignment of homology domain III. Sequences identical to HPH3 are highlighted in black. Similar sequences (3 distance units with PAM 62 matrix) are highlighted in gray.
As anticipated from previous studies, one band was identified as HSP70. HSC4 (a drosophila HSP70) was identified as a component of dPRC1 (45). HSC4 shows genetic interactions with PcG genes (30), and HSP70 from SF9 cells appears to copurify in stoichiometric amounts with fly PcG proteins when they are overexpressed with baculovirus vectors (15). These genetic and biochemical interactions are consistent with a biological role for HSP70 in PcG function. However, HSP70 is an abundant protein and further studies are needed to rule out an artifactual association. Similarly, we identified β-tubulin, an abundant protein that is frequently a contaminant in preparations of nuclear proteins. Considerably more characterization is needed to determine the biological relevance of tubulin to hPRC-H function.
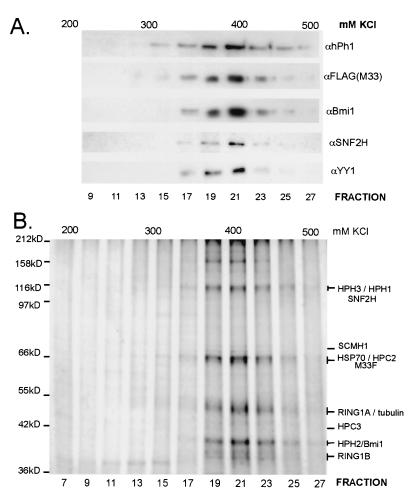
To confirm that these proteins are robustly associated, we subjected the complex to further fractionation. Purified M33-based complex was applied to a Hi-Trap S column and eluted with a shallow 200 to 500 mM KCl gradient. Proteins in the eluted fractions were followed by silver staining when their characteristic migration allowed clear identification (Fig. 3B) or by Western analysis (Fig. 3A). Each of the class II PcG proteins and HSP70 continued to cofractionate on this column, eluting at 400 mM KCl. None of the bands detectable by silver staining (Fig. 3B) fractionated away from the complex. We concluded that the majority of proteins in hPRC-H are known members of the mammalian PcG gene family.
FIG. 3.
Further fractionation of the hPRC-H complex. (A) Immunoblot of 25 μl (∼5%) of each HiTrap-S fraction visualized with the indicated antibody. For SNF2H and YY1, 250 μl was TCA precipitated prior to analysis. (B) Silver staining of HiTrap-S fractions. Fractions (250 μl) were TCA precipitated and separated by SDS-7.5% PAGE.
Other proteins associated with hPRC-H.
It is possible that the hPRC-H fractions we have characterized represent not a single complex but rather a mixture of several highly related complexes. This has previously been seen with the mammalian SWI/SNF family of complexes. The SWI/SNF family of complexes has members that contain a core of BRG1 and certain BAF proteins but differ in whether they contain BAF180, Sin3a, or BAF250 (34, 52, 60, 66). These distinct complexes are difficult to separate, presumably because they have so many shared components and are so large that the small differences in composition do not lead to differences in fractionation. Drosophila PcG proteins have been proposed to reside in distinct complexes that change during development (41), and mammalian PcG proteins change in relative abundance in different tissues (27). It is anticipated, therefore, that the composition of mammalian class II PcG complexes will not be static. Thus, proteins that interact with only a subset of the hPRC-H complexes would be expected to be substoichiometric.
A band in hPRC-H preparations that stained lightly with colloidal Coomassie stain and silver, and thus was inferred to be substoichiometric, was identified as SCMH1. Interestingly, the drosophila homologue Scm is found in substoichiometric amounts in dPRC1. Thus, the association of this protein with class II PcG complexes in small amounts is conserved between drosophila and humans, suggesting that a subset of PRC complexes might contain Scm.
Previously, yeast two-hybrid and immunoprecipitation analyses identified several transcriptional repressors that interact with class II PcG proteins in mammals. These include CtBP, RYBP, and two sequence-specific DNA binding factors, YY1 and E2F6, that appear to associate with the class II proteins via RYBP (17, 59). YY1 is homologous to the drosophila PcG protein Pleiohomeotic (Pho) and has been shown to interact with the class I PcG complex proteins (41, 42). YY1 was not detected in bands at the appropriate molecular mass in hPRC-H preparations that contain HPC2 and HSP70. However, fragments of YY1 were detected by MS analysis at an unexpected molecular mass (∼46 kDa, compared to 68 kDa for intact YY1). Fractions of hPRC-H obtained by Hi-Trap S chromatography were precipitated and tested for the presence of YY1, RYBP, and E2F6. Intact YY1 was detected cofractionating with hPRC-H (Fig. 3A). RYBP and E2F6 were detected with the available antisera in neither the M2 fractions nor the precipitated Hi-Trap fractions. Most of these proteins are not associated with the complex (Fig. 1B and data not shown).
SNF2H, a member of the Swi2/Snf2 family of ATP-dependent chromatin-remodeling factors and a homologue of the drosophila ISWI protein, was also detected in the purified fractions. Comparative Western analysis of SNF2H with RING1A, HPC2, and Bmi1 showed that levels of SNF2H varied significantly from preparation to preparation; even preparations with relatively high levels contained substoichiometric amounts of SNF2H relative to HPH3, as judged by the intensity of colloidal Coomassie staining (data not shown). These levels represent only a small portion of the cellular SNF2H (Fig. 1B). We were unable to detect either p325 or WCRF-180 by immunoblotting, suggesting that hPRC-H does not contain appreciable amounts of the RSF (26) or WCRF (7) complex, both of which contain SNF2H. When TCA-precipitated samples obtained by Hi-Trap S chromatography were analyzed by immunoblotting, the substoichiometric amounts of SNF2H cofractionated with hPRC-H components (Fig. 3A). Together, these data demonstrate that SCM, YY1, and SNF2H can be seen at substoichiometric levels that maintain cofractionation with major hPRC-H components after immunoaffinity and Hi-Trap S fractionation. These proteins are therefore candidates for association with a subset of hPRC-H complexes, and further analysis is required to assess the potential function of these proteins within hPRC-H.
hPRC-H specifically inhibits Swi/Snf remodeling of nucleosomal arrays.
The mechanism of PcG repression of transcription in vivo is unknown. However, genetically, it is clear that PcG proteins act antagonistically to trxG-encoded proteins. We previously took advantage of this to develop in vitro assays for dPRC1 by examining its ability to inhibit the activities of the trxG-related human complex Swi/Snf (51). To determine whether this activity was conserved in the mammalian PcG complex despite the changes in the primary sequence and composition of the complex, we investigated whether hPRC-H is also able to inhibit Swi/Snf-dependent remodeling of nucleosomal templates with a variety of assays.
We first examined the ability of hPRC-H to inhibit Swi/Snf-dependent remodeling on linear nucleosomal arrays (Fig. 4A). In vitro and in vivo, the presence of nucleosomes can reduce the accessibility of restriction endonucleases (40). Addition of Swi/Snf to nucleosomal templates can increase restriction enzyme access in an ATP-dependent manner (28). Embryonic dPRC1 is able to block this Swi/Snf-induced increase in restriction enzyme cutting on a polynucleosomal template (15). We tested whether hPRC-H is also able to analogously block Swi/Snf-stimulated restriction enzyme access with a chromatinized template containing two unpositioned nucleosomes flanked on each side by five nucleosomes positioned by repeats of the Xenopus 5S nucleosomal positioning sequence. hPRC-H blocked Swi/Snf-stimulated cutting by HhaI in a manner similar to that of dPRC1. Furthermore, this inhibition was concentration dependent (Fig. 4B). On the basis of calculations of hPRC-H concentration with an estimated size of 500 kDa (see Materials and Methods), hPRC-H tagged on either the Bmi1 or the M33 subunit and dPRC1 have similar specific activities and both can block Swi/Snf activity at concentrations lower than one PRC per nucleosome (Fig. 4C). The inhibition of Swi/Snf-stimulated cutting is strongly reduced when hPRC-H is not preincubated with the template (data not shown).
In addition to increasing the access of restriction enzymes to nucleosomal templates in vitro, Swi/Snf has also been shown to alter the topology of the DNA on closed circular plasmids in the presence of topoisomerase I (25). Previously, dPRC1 was shown to block these topological changes induced by Swi/Snf. We tested the effect of hPRC-H in this assay by preincubating the template with hPRC-H for 15 min and then allowing it to react with Swi/Snf. Increasing amounts of hPRC-H blocked the ability of Swi/Snf to form remodeled products (Fig. 4D, lanes 3 to 6). Taken together, the restriction enzyme assay and topological data suggest that hPRC-H, like dPRC1, can inhibit chromatin remodeling by Swi/Snf.
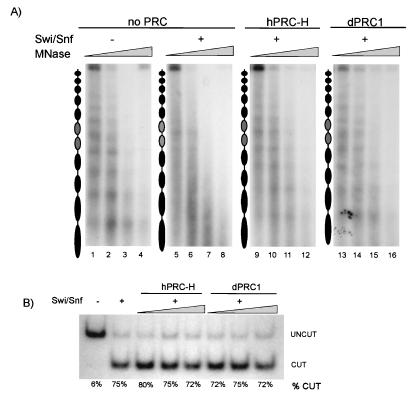
One explanation for the previous data is that hPRC-H nonspecifically restricts the accessibility of the template to both topoisomerases and restriction enzymes. To evaluate this possibility, we tested the effect of hPRC-H on template digestion by micrococcal nuclease (MNase), which cleaves preferentially in the linker region, creating a defined nucleosomal ladder. Incubation of end-labeled 5S array with Swi/Snf has previously been shown to cause randomization of the normally well-spaced MNase digestion pattern of nucleosomes (21) (Fig. 5A, compare lanes 1 to 4 to lanes 5 to 8, respectively). Preincubation of the template with hPRC-H or dPRC1 (15) prevented Swi/Snf from randomizing the array (lanes 9 to 16). In these reaction mixtures, only a slight decrease in MNase sensitivity was observed, suggesting that hPRC-H blocks Swi/Snf-dependent remodeling but not MNase access.
FIG. 5.
hPRC-H activity is specific to polynucleosomal templates. (A) hPRC-H inhibits Swi/Snf without disrupting nucleosome position. End-labeled 5S arrays were treated as described in the legend to Fig. 4B, except that HhaI was not added. Following incubation of the templates with Swi/Snf, increasing amounts of MNase were added to each reaction mixture. Lanes: 1 to 4, control template; 5 to 8, 25 fmol of Swi/Snf; 9 to 12, same as lanes 5 to 8 with ∼12 fmol of hPRC-H; 13 to 16, with ∼12 fmol of dPRC1. All reaction mixtures are from the same experiment; nucleosome positions align when samples are analyzed side by side (data not shown). (B) hPRC-H does not block Swi/Snf remodeling on mononucleosomes. Internally labeled TPT mononucleosomes (1 ng) (47) were incubated as described in the legend to Fig. 4B with 200 ng of Swi/Snf and PstI. Amounts of hPRC-H (M33) and dPRC1 are ∼1.2, 4, and 12 fmol.
The requirement for preincubation of hPRC-H with the template suggests that hPRC-H acts by binding to the chromatinized template, as has been seen with recombinant subunits (15). To rule out the possibility that hPRC-H inhibits SWI/SNF directly, we examined the ability of the human complex to inhibit remodeling of mononucleosomes instead of nucleosomal arrays. Similar to what is seen on the arrays, remodeling of the mononucleosome by Swi/Snf increases restriction enzyme access (28, 32). hPRC-H is unable to block Swi/Snf activity on the mononucleosome (Fig. 5B) under conditions in which remodeling is inhibited on nucleosomal arrays included as an internal control (data not shown). This implies that hPRC-H does not directly affect SWI/SNF activity and suggests that hPRC-H requires a chromatin template larger then a mononucleosome in order to inhibit Swi/Snf remodeling. This behavior is similar to that previously seen with dPRC1.
DISCUSSION
A comparison of the composition and activity of hPRC-H with dPRC1 leads to he proposal that class II PcG complexes have an evolutionarily conserved core group of subunits with conserved function. The human complex contains homologues of drosophila Pc, Ph, Psc, and dRING and also shares with dPRC1 the apparent association with HSP70 and substoichiometric amounts of Scm. The human complex does not have robust interactions with many other proteins that are tightly associated with dPRC1, such as TAF proteins. The conserved core proteins appear to be sufficient to bestow conserved function: both dPRC1 and hPRC-H inhibit remodeling by Swi/Snf at ratios of less then one PRC per nucleosome (Fig. 4). Both complexes do not significantly alter nucleosomal position or access of nucleases to nucleosomal arrays, suggesting that both complexes preferentially inhibit remodeling without making the template inaccessible. This activity might be a key component of the mechanism of class II PcG complexes.
One major difference between the human and drosophila complexes is the presence of multiple homologues of most of the core proteins in the complex. There are two possible explanations for this result. First, there might be single complexes that contain multiple paralogs of every protein. This possibility is supported by the identification, by both immunoblotting and MS, of HPC2 and HPC3 in fractions purified with tagged M33 (MPC1). Alternatively, there may be multiple highly related complexes that contain single PcG homologues. This possibility is supported by the apparent varied stoichiometry (as judged by Coomassie staining intensity) of the PcG proteins in the complex (Fig. 1) and the significant substoichiometry of SCMH1. The latter hypothesis would help explain a function for some of the specificity seen by the different PcG proteins, such as the binding of CtBP preferentially to HPC2 rather then HPC1 (49). These differences in domains may lead to differential targeting of complexes. Multiple highly related complexes are also seen in other chromatin-modifying factors, such as the Swi/Snf family of complexes (34, 52, 60, 66).
PRC composition is likely to be regulated in both a cell type and developmental manner. Contrary to hPRC-H, dPRC1 associates with stoichiometric amounts of Zeste, most members of the TFIID family, and about a half dozen other non-PcG proteins (45). No members of the TFIID family were found in detectable amounts associated with hPRC-H (data not shown), and mammals have no known Zeste homolog. On the basis of the data in Fig. 4, the role these proteins play in blocking chromatin remodeling appears to be minimal under the conditions that have been tested. This is consistent with the hypothesis that these additional proteins may be involved, instead, in targeting of the complex to specific genes, in stabilizing the association of dPRC1 with template, or in providing additional enzymatic activities. The presence of stoichiometric amounts of non-PcG proteins in the dPRC1 complex may be related to the embryonic state of the extract when there is a transition between establishment of a repressed state on target genes and formation of a structure that can maintain that repressed state. In addition, at early stages of development, repression must be maintained during rapid cycles of replication, which might require a particularly robust mechanism. It remains formally possible that the association of the TAF proteins with dPRC1 occurred during extract preparation and does not reflect an association that is mechanistically significant; however, data from in vivo colocalization studies support the notion that these proteins interact in drosophila embryos (8). While there are already data demonstrating changes in the composition of class II PcG complexes during drosophila and human development, further work on the nature and functional consequences of these changes will be important in explaining the developmental role of these complexes. The identification of core subunits in the PRC family of complexes should allow dissection of the roles of other subunits in changing the capabilities of this family of complexes to maintain a repressed state across the lifetime of an organism.
The importance of the core of the PcG complex in its biochemical activity has recently been demonstrated by studies that have shown that a reconstituted core complex containing four drosophila PcG proteins (Pc, Psc, Ph, and dRING1) has many of the same functions as the complete dPRC1 complex (15). The experiments reported here extend these studies in two important ways. First, we show that a complex exists in HeLa cells that is similar to the reconstituted drosophila core complex in that it is primarily made up of the human homologues of the four core dPRC1 proteins. This suggests that this core complex plays a biologically significant role. Second, the observation (Fig. 4 and 5) that hPRC-H and dPRC1 have similar abilities to inhibit chromatin remodeling shows that, in vitro, this function is conserved despite limited sequence conservation between the human and drosophila PcG proteins. This parallels in vivo experiments in which addition of M33 was able to complement a Pc mutation in the fly (31). The observation that hPRC-H and dPRC1 have similar activities therefore supports the argument that these activities are an important aspect of PcG function.
Given the large differences in composition between the human and drosophila complexes and the additional PcG homologues in humans, it is tempting to speculate on the possible role of the differences. While both organisms use the Polycomb proteins to maintain repression of the Hox genes, mammalian PcG proteins also play key roles in other systems, including the immune system (61). The composition of the complex changes across development as the expression pattern of the paralogs varies. This suggests that hPRC may act more dynamically than its drosophila counterpart. We hypothesize that this variation in composition could lead to changes in targeting of the same biochemical mechanism, leading to distinct biological functions.
Acknowledgments
We thank M. van Lohuizen, A. Otte, J. Lees, R. Shiekhattar, and D. Reinberg for generous gifts of antibodies; J. Muller for the M33 cDNA; Anita Grewal and Arpi Nazarian for help with mass analysis; M. Wildermouth for aid with sequence analysis; N. Francis, I. King, M. Lavigne, G. Narlikar, A. Saurin, J. Dennis, and M. Donohoe for reagents and critical reading of the manuscript; and W. Forrester and members of the Kingston laboratory for discussions and comments.
This work was supported by grants from the National Science Foundation (to S.S.L.), the National Cancer Institute Cancer Center (to P.T.), and the National Institutes of Health (to R.E.K.).
REFERENCES
- 1.Akasaka, T., M. Kanno, R. Balling, M. A. Mieza, M. Taniguchi, and H. Koseki. 1996. A role for mel-18, a Polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development 122:1513-1522. [DOI] [PubMed] [Google Scholar]
- 2.Akasaka, T., M. van Lohuizen, N. van der Lugt, Y. Mizutani-Koseki, M. Kanno, M. Taniguchi, M. Vidal, M. Alkema, A. Berns, and H. Koseki. 2001. Mice doubly deficient for the Polycomb group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128:1587-1597. [DOI] [PubMed] [Google Scholar]
- 3.Alkema, M. J., M. Bronk, E. Verhoeven, A. Otte, L. J. van't Veer, A. Berns, and M. van Lohuizen. 1997. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian Polycomb complex. Genes Dev. 11:226-240. [DOI] [PubMed] [Google Scholar]
- 4.Bardos, J. I., A. J. Saurin, C. Tissot, E. Duprez, and P. S. Freemont. 2000. HPC3 is a new human polycomb orthologue that interacts and associates with RING1 and Bmi1 and has transcriptional repression properties. J. Biol. Chem. 275:28785-28792. [DOI] [PubMed] [Google Scholar]
- 5.Berger, J., H. Kurahashi, Y. Takihara, K. Shimada, H. W. Brock, and F. Randazzo. 1999. The human homolog of Sex comb on midleg (SCMH1) maps to chromosome 1p34. Gene 237:185-191. [DOI] [PubMed] [Google Scholar]
- 6.Beuchle, D., G. Struhl, and J. Muller. 2001. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128:993-1004. [DOI] [PubMed] [Google Scholar]
- 7.Bochar, D. A., J. Savard, W. Wang, D. W. Lafleur, P. Moore, J. Cote, and R. Shiekhattar. 2000. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc. Natl. Acad. Sci. USA 97:1038-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breiling, A., B. M. Turner, M. E. Bianchi, and V. Orlando. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412:651-655. [DOI] [PubMed] [Google Scholar]
- 9.Brunk, B. P., E. C. Martin, and P. N. Adler. 1991. Drosophila genes Posterior Sex Combs and Suppressor two of zeste encode proteins with homology to the murine bmi-1 oncogene. Nature 353:351-353. [DOI] [PubMed] [Google Scholar]
- 10.Bunker, C. A., and R. E. Kingston. 1994. Transcriptional repression by Drosophila and mammalian polycomb group proteins in transfected mammalian cells. Mol. Cell. Biol. 14:1721-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Core, N., S. Bel, S. J. Gaunt, M. Aurrand-Lions, J. Pearce, A. Fisher, and M. Djabali. 1997. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development 124:721-729. [DOI] [PubMed] [Google Scholar]
- 12.del Mar Lorente, M., C. Marcos-Gutierrez, C. Perez, J. Schoorlemmer, A. Ramirez, T. Magin, and M. Vidal. 2000. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127:5093-5100. [DOI] [PubMed] [Google Scholar]
- 13.Dingwall, A. K., S. J. Beek, C. M. McCallum, J. W. Tamkun, G. V. Kalpana, S. P. Goff, and M. P. Scott. 1995. The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell 6:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis, N. J., and R. E. Kingston. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell. Biol. 2:409-421. [DOI] [PubMed] [Google Scholar]
- 15.Francis, N. J., A. J. Saurin, Z. Shao, and R. E. Kingston. 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8:545-556. [DOI] [PubMed] [Google Scholar]
- 16.Franke, A., M. DeCamillis, D. Zink, N. Cheng, H. W. Brock, and R. Paro. 1992. Polycomb and polyhomeotic are constituents of a multimeric protein complex in chromatin of Drosophila melanogaster. EMBO J. 11:2941-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia, E., C. Marcos-Gutierrez, M. del Mar Lorente, J. C. Moreno, and M. Vidal. 1999. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 18:3404-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunster, M. J., D. P. E. Satijn, K. M. Hamer, J. L. den Blaauwen, D. de Bruijn, M. J. Alkema, M. van Lohuizen, R. van Driel, and A. P. Otte. 1997. Identification and characterization of interactions between the vertebrate Polycomb-group protein BMI1 and human homologues of Polyhomeotic. Mol. Cell. Biol. 17:2326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haupt, Y., W. S. Alexander, G. Barri, S. P. Klinken, and J. M. Adams. 1991. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in Eμ-myc transgenic mice. Cell 65:753-763. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs, J. J., and M. van Lohuizen. 2002. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim. Biophys. Acta 1602:151-161. [DOI] [PubMed] [Google Scholar]
- 21.Jaskelioff, M., I. M. Gavin, C. L. Peterson, and C. Logie. 2000. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 20:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kal, A. J., T. Mahmoudi, N. B. Zak, and C. P. Verrijzer. 2000. The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev. 14:1058-1071. [PMC free article] [PubMed] [Google Scholar]
- 23.Kanno, M., M. Hasegawa, A. Ishida, K. Isono, and M. Taniguchi. 1995. mel-18, a Polycomb group-related mammalian gene, encodes a transcriptional negative regulator with tumour suppressive activity. EMBO J. 14:5672-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kennison, J. A. 1995. The polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu. Rev. Genet. 29:289-303. [DOI] [PubMed] [Google Scholar]
- 25.Kwon, H., A. Imbalzano, P. A. Khavari, R. E. Kingston, and M. R. Green. 1994. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature 370:477-481. [DOI] [PubMed] [Google Scholar]
- 26.LeRoy, G., A. Loyola, W. S. Lane, and D. Reinberg. 2000. Purification and characterization of a human factor that assembles and remodels chromatin. J. Biol. Chem. 275:14787-14790. [DOI] [PubMed] [Google Scholar]
- 27.Lessard, J., S. Baban, and G. Sauvageau. 1998. Stage-specific expression of polycomb group genes in human bone marrow cells. Blood 91:1216-1224. [PubMed] [Google Scholar]
- 28.Logie, C., and C. L. Peterson. 1997. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 16:6772-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lovering, R., I. M. Hanson, K. L. Borden, S. Martin, N. J. O'Reilly, G. I. Evan, D. Rahman, D. J. Pappin, J. Trowsdale, and P. S. Freemont. 1993. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc. Natl. Acad. Sci. USA 90:2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mollaaghababa, R., L. Sipos, S. Y. Tiong, O. Papoulas, J. A. Armstrong, J. W. Tamkun, and W. Bender. 2001. Mutations in Drosophila heat shock cognate 4 are enhancers of Polycomb. Proc. Natl. Acad. Sci. USA 98:3958-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller, J. 1995. Transcriptional silencing by the Polycomb protein in Drosophila embryos. EMBO J. 14:1209-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narlikar, G. J., M. L. Phelan, and R. E. Kingston. 2001. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell 8:1219-1230. [DOI] [PubMed] [Google Scholar]
- 33.Ng, J., C. M. Hart, K. Morgan, and J. A. Simon. 2000. A Drosophila ESC-E(Z) protein complex is distinct from other Polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol. 20:3069-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nie, Z., Y. Xue, D. Yang, S. Zhou, B. J. Deroo, T. K. Archer, and W. Wang. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20:8879-8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Carroll, D., S. Erhardt, M. Pagani, S. C. Barton, M. A. Surani, and T. Jenuwein. 2001. The Polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21:4330-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papoulas, O., S. J. Beek, S. L. Moseley, C. M. McCallum, M. Sarte, A. Shearn, and J. W. Tamkun. 1998. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development 126:3955-3966. [DOI] [PubMed] [Google Scholar]
- 37.Pearce, J. J., P. B. Singh, and S. J. Gaunt. 1992. The mouse has a Polycomb-like chromobox gene. Development 114:921-929. [DOI] [PubMed] [Google Scholar]
- 38.Petruk, S., Y. Sedkov, S. Smith, S. Tillib, V. Kraevski, T. Nakamura, E. Canaani, C. M. Croce, and A. Mazo. 2001. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294:1331-1334. [DOI] [PubMed] [Google Scholar]
- 39.Pirrotta, V. 1998. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell 93:333-336. [DOI] [PubMed] [Google Scholar]
- 40.Polach, K. J., and J. Widom. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130-149. [DOI] [PubMed] [Google Scholar]
- 41.Poux, S., D. McCabe, and V. Pirrotta. 2001. Recruitment of components of Polycomb Group chromatin complexes in Drosophila. Development 128:75-85. [DOI] [PubMed] [Google Scholar]
- 42.Satijn, D. P. E., K. M. Hamer, J. den Blaauwen, and A. P. Otte. 2001. The Polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 21:1360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satijn, D. P. E., D. J. Olson, J. van der Vlag, K. M. Hamer, C. Lambrechts, H. Masselink, M. J. Gunster, R. G. A. B. Sewalt, R. van Driel, and A. P. Otte. 1997. Interference with the expression of a novel human Polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol. Cell. Biol. 17:6076-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Satijn, D. P. E., M. J. Gunster, J. van der Vlag, K. M. Hamer, W. Schul, M. J. Alkema, A. J. Saurin, P. S. Freemont, R. van Driel, and A. P. Otte. 1997. RING1 is associated with the Polycomb group protein complex and acts as a transcriptional repressor. Mol. Cell. Biol. 17:4105-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 46.Saurin, A. J., C. Shiels, J. Williamson, D. P. E. Satijn, A. P. Otte, D. Sheer, and P. S. Freemont. 1998. The human polycomb-group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142:887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnitzler, G., S. Sif, and R. E. Kingston. 1998. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell 94:17-27. [DOI] [PubMed] [Google Scholar]
- 48.Schoorlemmer, J., C. Marcos-Gutierrez, F. Were, R. Martinez, E. Garcia, D. P. E. Satijn, A. P. Otte, and M. Vidal. 1997. Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 16:5930-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sewalt, R. G. A. B., M. J. Gunster, J. van der Vlag, D. P. E. Satijn, and A. P. Otte. 1999. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sewalt, R. G. A. B., J. van der Vlag, M. J. Gunster, K. M. Hamer, J. L. den Blaauwen, D. P. Satijn, T. Hendrix, R. van Driel, and A. P. Otte. 1998. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol. Cell. Biol. 18:3586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C.-T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 52.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sif, S., P. T. Stukenberg, M. W. Kirschner, and R. E. Kingston. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon, J. 1995. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr. Opin. Cell Biol. 7:376-385. [DOI] [PubMed] [Google Scholar]
- 55.Simon, J. A., and J. W. Tamkun. 2002. Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr. Opin. Genet. Dev. 12:210-218. [DOI] [PubMed] [Google Scholar]
- 56.Takihara, Y., D. Tomotsune, M. Shirai, Y. Katoh-Fukui, K. Nishii, M. A. Motaleb, M. Nomura, R. Tsuchiya, Y. Fujita, Y. Shibata, T. Higashinakagawa, and K. Shimada. 1997. Targeted disruption of the mouse homologue of the Drosophila polyhomeotic gene leads to altered anteroposterior patterning and neural crest defects. Development 124:3673-3682. [DOI] [PubMed] [Google Scholar]
- 57.Tie, F., T. Furuyama, J. Prasad-Sinha, E. Jane, and P. J. Harte. 2001. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128:275-286. [DOI] [PubMed] [Google Scholar]
- 58.Tomotsune, D., Y. Takihara, J. Berger, D. Duhl, S. Joo, M. Kyba, M. Shirai, H. Ohta, Y. Matsuda, B. M. Honda, J. Simon, K. Shimada, H. W. Brock, and F. Randazzo. 1999. A novel member of murine Polycomb-group proteins, Sex comb on midleg homolog protein, is highly conserved, and interacts with RAE28/mph1 in vitro. Differentiation 65:229-239. [DOI] [PubMed] [Google Scholar]
- 59.Trimarchi, J. M., B. Fairchild, J. Wen, and J. A. Lees. 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 98:1519-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Underhill, C., M. S. Qutob, S. P. Yee, and J. Torchia. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463-40470. [DOI] [PubMed] [Google Scholar]
- 61.van der Lugt, N. M. T., J. Domen, K. Linders, M. van Roon, E. Robanus-Maandag, H. te Riele, M. van der Valk, J. Deschamps, M. Sofroniew, M. van Lohuizen, and A. Berns. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 8:757-769. [DOI] [PubMed] [Google Scholar]
- 62.van der Vlag, J., and A. P. Otte. 1999. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23:474-478. [DOI] [PubMed] [Google Scholar]
- 63.van Lohuizen, M., M. Frasch, E. Wientjens, and A. Berns. 1991. Sequence similarity between the mammalian bmi-1 proto-oncogene and the Drosophila regulatory genes Psc and Su(z)2. Nature 353:353-355. [DOI] [PubMed] [Google Scholar]
- 64.van Lohuizen, M., M. Tijms, J. W. Voncken, A. Schumacher, T. Magnuson, and E. Wientjens. 1998. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol. Cell. Biol. 18:3572-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Lohuizen, M., S. Verbeek, B. Scheijen, E. Wientjens, H. van der Gulden, and A. Berns. 1991. Identification of cooperating oncogenes in Eμ-myc transgenic mice by provirus tagging. Cell 65:737-752. [DOI] [PubMed] [Google Scholar]
- 66.Xue, Y., J. C. Canman, C. S. Lee, Z. Nie, D. Yang, G. T. Moreno, M. K. Young, E. D. Salmon, and W. Wang. 2000. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 97:13015-13020. [DOI] [PMC free article] [PubMed] [Google Scholar]