Abstract
The Mdm2 protein mediates ubiquitylation and degradation of p53 and is a key regulator of this tumor suppressor. More recently, it has been shown that Mdm2 is highly phosphorylated within its central acidic domain. In order to address the issue of how these modifications might regulate Mdm2 function, putative phosphorylation sites within this domain were substituted, individually or in pairs, with alanine residues. Mutants with serine-to-alanine substitutions between residues 244 and 260 abolished or at least reduced the capacity of Mdm2 to promote p53 degradation. In each case, loss of degradation function was independent of the ability to bind to p53 or p14ARF. Moreover, each of the Mdm2 mutants completely retained the capacity to act as a ubiquitin ligase in vivo. Thus, ubiquitylation and degradation can be uncoupled. Two-dimensional phosphopeptide mapping coupled with the use of phospho-specific antibodies revealed that Mdm2 is phosphorylated physiologically at several sites within this region, consistent with the idea that phosphorylation is important for Mdm2 activity. Strikingly, treatment of cells with ionizing radiation resulted in a significant decrease in the phosphorylation of residues that are important for p53 turnover. This hypophosphorylation preceded p53 accumulation. These findings indicate that Mdm2 contributes an additional function toward the degradation of p53 that is distinct from its ubiquitin ligase activity and is regulated by phosphorylation. Our model suggests that hypophosphorylation of Mdm2 in response to ionizing irradiation inactivates this novel function, thereby contributing to p53 stabilization.
The tumor suppressor protein p53 prevents genomic instability by arresting the cell cycle or initiating programmed cell death upon genotoxic insult. Both options remedy the outgrowth of malignant cells. Loss of p53 therefore enhances the risk of developing malignancies (for a review, see references 2 and 11).
The ability of p53 to induce cell cycle arrest or apoptosis is understood in considerable detail (for a review, see reference 24), but the mechanisms which regulate its abundance are less clear. The antiproliferative activity of p53 necessitates tight control to prevent the onset of cell cycle arrest and apoptosis in cycling cells. This control is achieved largely through the degradation of p53 soon after its synthesis. DNA damage and other forms of cellular stress block p53 degradation, leading to its accumulation and, consequently, to the transcription of p53 target genes. However, some contribution to transcriptional activation through the relief of carboxyl-terminal repression or modification of the amino-terminal transactivation domain cannot be excluded (see references 32 and 35 and references therein). Based on its ability to induce cell death, p53 is considered to be a potential target for therapeutic intervention in the treatment of cancer. Accordingly, an understanding of the mechanisms leading to its accumulation will be of immense value in pursuing this goal.
The stability of p53 is regulated predominantly by the oncoprotein Mdm2, which mediates p53 ubiquitylation and rapid degradation by the 26S proteasome. Mdm2 was first described as one of the genes amplified on the double-minute chromosome of a line of spontaneously transformed BALB/c/3T3 cells (6). The protein gained considerable attention following its identification as the 90-kDa protein that coprecipitates with p53 (30). Mdm2 restrains p53 function by concealing the transcriptional activation domain of p53 and by targeting p53 for rapid degradation (13, 21, 30, 31). The mdm2 gene is amplified in a significant proportion of human soft tissue tumors and osteosarcomas as well as a variety of other tumor types, thereby contributing to tumor development by effectively reducing the availability of functional p53 (31).
The Mdm2 protein can be divided into four major conserved regions: I, an amino-terminal domain (amino acids [aa] 23 through 108); II, a highly acidic region (aa 237 through 260); III, a potential zinc finger (aa 289 through 333); and IV, a ring finger (aa 460 through 489) (8, 18, 22, 29, 31). Conserved region I accommodates the p53-binding pocket, and region IV is required for ubiquitin ligase activity. The function of regions II and III is less clear, although recent investigations showed that p53 can be rescued from degradation by the binding of proteins such as p300, pRb, and p14ARF adjacent to region II or by deletion of the whole domain (1, 12, 19, 33).
In primary cells, p53 is turned over with a typical half-life of less than 20 min yet the half-life is extended to several hours in response to stress signals. Accordingly, cells must exert tight regulation over the interaction of p53 with cellular factors that influence its turnover or stability. Furthermore, these regulatory events must be modulated when specific conditions are met, thereby allowing p53 levels to respond sensitively to changes in the cellular environment. Protein-protein interactions are frequently regulated by phosphorylation, which can alter the affinity of participating proteins for each other. The central domain of Mdm2 is particularly rich in phosphorylation sites, and the degree of conservation in this region between human and murine Mdm2 proteins is very high.
In order to determine whether phosphorylation of conserved region II of Mdm2 regulates p53 degradation, we substituted potential phosphorylation sites in the central domain of Mdm2 with alanine residues and tested the ability of these mutant proteins to target p53 for degradation in cotransfection experiments. Here, we show that the mutation of several serines in the central acidic domain of Mdm2 reduced or abolished the ability to degrade p53. However, neither the ability of these mutants to ubiquitylate p53 nor their ability to interact with p14ARF was affected. These results imply that conserved region II in the central domain of Mdm2 has an important function in p53 turnover that is independent of and downstream of ubiquitylation and which is regulated specifically by phosphorylation. Our data also indicate that this area is constitutively phosphorylated under normal growth conditions but becomes hypophosphorylated in response to ionizing radiation correlated with impairment of p53 degradation. Moreover, the Mdm2 mutants used in our study provide an important tool to identify as-yet-unknown steps in the p53 degradation pathway as they uncouple the molecular events involved in p53 ubiquitylation and p53 destruction.
MATERIALS AND METHODS
Plasmids and mutagenesis.
pcocmdm2X2, which expresses full-length mouse Mdm2, and pcocmdm2ΔXM, in which the p53-binding pocket is deleted (3), were a gift of M. Oren. pcDNA3-mdm2 was obtained by digesting pcocmdm2X2 with EcoRI and ligating the resulting fragment into the EcoRI site of the pcDNA3 vector. pMdm2NeoBam expresses full-length human Mdm2 and was given to us by B. Vogelstein. pDWM659 (14) expresses full-length murine wild-type Mdm2 which is tagged at the amino terminus with the Myc-derived 9E10 epitope. pcDNA3-ARF-FLAG was given to us by Karen Vousden. The plasmid expressing His-tagged ubiquitin (MT107 [38]) was a gift of Sybille Mittnacht. pcDNA3-p53 was obtained by PCR amplification with reversibly transcribed RNA from human cells as a template and the primers 5′-AGGAATTCATGGAGGAGCCGCAGTCAGAT-3′ and 5′-GCCGCTCGAGTTACTATCAGTCTGAGTCAGGCCCTTCTG-3′. The PCR product was ligated into a pGEM-T Easy Vector, digested with EcoRI and XhoI, and cloned into the pcDNA3 vector. The proper wild-type structure was confirmed by sequencing. Site-directed mutations were introduced by PCR with wild-type mdm2 cDNA as a template and primers covering the desired mutations. Mutagenesis was carried out using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The cDNAs were sequenced in order to verify the mutations and were found to have no additional alterations.
Cell lines and their treatments.
H1299 lung carcinoma cells (37) were purchased from the American Type Culture Collection, and GM1604 primary human fibroblasts were purchased from NIGMS, Camden, N.J. H1299 cells were grown in RPMI medium (Sigma) supplemented with 10% fetal calf serum (FCS) and 10 mg of gentamicin/ml. Prior to transfection, RPMI medium was replaced by Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% FCS and 10 mg of gentamicin/ml. GM1604 cells and COS-7 cells were maintained in DMEM supplemented with 10% fetal bovine serum, 100 μg of penicillin-streptomycin/ml, and 2 mM glutamine. All cells were grown at 37°C in a humidified 5% CO2 atmosphere.
For transfection, 5 × 105 cells were seeded into 10-cm-diameter plates and incubated in normal growth medium (DMEM) at 37°C for 24 h. Transfection of COS-7 cells was carried out using Lipofectamin reagent (Gibco BRL) according to the manufacturer's instructions. The cells were then incubated at 37°C for 5 h before adding the same amount of complete growth medium. The following day, cells were washed three times with normal growth medium and incubated in normal growth medium for a further 24 h before they were prepared for [32P]orthophosphate labeling. H1299 cells were transfected by calcium phosphate-DNA coprecipitation (7).
For radiolabeling, cells were preincubated with 8 ml of phosphate-free DMEM (Sigma) containing 10% dialyzed FCS at 37°C for 1 h. The medium was replaced with the same volume containing 4 mCi of [32P]orthophosphate per plate and incubated at 37°C for an additional 3 h.
Ionizing irradiation was carried out in a cesium-137 γ-source with a dose rate of 2.8 Gy/min. Cells were irradiated with 10 Gy in culture medium.
Immunoprecipitation. (i) Two-dimensional peptide mapping.
After labeling, the cells were washed twice with 5 ml of ice-cold phosphate-buffered saline (PBS) and lysed in 1 ml of ice-cold NP-40 phosphate buffer (10 mM NaH2PO4 [pH 7], 2 mM EDTA, 0.1 M NaCl, 1% NP-40, 1 mM benzamidine). The samples were centrifuged at 13,000 × g for 10 min, and the supernatants were collected. Immunoprecipitation of Mdm2 was performed at 4°C overnight on a rotating wheel with 75 μl of protein A-Sepharose and 10 μl of 9E10 antibody (1.7 μg/μl). The protein A-Sepharose beads were subsequently washed three times each with 1 ml of ice-cold radioimmunoprecipitation assay buffer (10 mM NaH2PO4 [pH 7], 2 mM EDTA, 0.1 M NaCl, 1% NP-40, 1 mM benzamidine, 1% sodium desoxycholate, 0.1% sodium dodecyl sulfate [SDS]) and once with 1 ml of ice-cold 50 mM Tris (pH 7.5). Prior to heat denaturing, 25 μl of 2× SDS sample buffer (4% SDS, 0.16 M Tris [pH 6.8], 20% glycerol, 10% β-mercaptoethanol, 0.002% bromophenol blue) was added and the proteins were separated on an SDS-10% polyacrylamide gel.
(ii) Coimmunoprecipitation.
The cells were washed with ice-cold PBS, scraped into PBS, and centrifuged at 1,000 × g for 5 min. Cells were lysed by incubation in NP-40 lysis buffer (150 mM NaCl, 50 mM Tris [pH 8], 5 mM EDTA, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride) on ice for 30 min. The protein extract was cleared by centrifugation at 13,000 × g at 4°C for 20 min, and the protein concentration of the supernatant (protein extract) was determined by the method of Bradford (Bio-Rad). A 1:1 mixture of precoupled protein A plus protein G (protein A+G) beads (20 μl) was added to 350 μg of protein, and immunoprecipitation was carried out on a rotating wheel at 4°C for 1 h. The Sepharose beads were subsequently washed three times, each with 1 ml of NP-40 lysis buffer. Then, 25 μl of 2× SDS sample buffer was added and the samples were heat denatured before being separated on an SDS-10% polyacrylamide gel.
Two-dimensional phosphopeptide mapping.
Phosphopeptide analysis of Mdm2 was carried out as described previously for p53 (26) with three exceptions. First, the gel was dried and a fragment of the gel containing the radiolabeled Mdm2 was excised. The fragment was rehydrated with 500 μl of an ammonium bicarbonate solution (4 mg/ml) and fractionated with a douncer pestle. β-Mercaptoethanol (10 μl) and 10% SDS (10 μl) were added, and the samples were boiled at 95°C for 5 min before they were incubated on a rotating wheel at room temperature for several hours. Carrier protein was then added, and the proteins were precipitated by the addition of 125 μl of 100% trichloroacetic acid. After washing, the proteins were oxidized with performic acid as described previously. Second, the protein was digested using chymotrypsin in place of trypsin. Third, the solvent used for the chromatographic separation consisted of isobutyric acid (62.5%, vol/vol), n-butanol (1.9%, vol/vol), pyridine (4.8%, vol/vol), and glacial acetic acid (2.9%, vol/vol) in deionized water.
SDS-PAGE and Western blotting.
Cells were washed twice in ice-cold PBS, scraped into PBS, and centrifuged at 1,000 × g for 5 min. Cells were lysed by incubation in NP-40 lysis buffer (150 mM NaCl, 50 mM Tris [pH 8], 5 mM EDTA, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride) on ice for 30 m. The protein extract was cleared by centrifugation at 13,000 × g at 4°C for 20 min, and the protein concentration of the supernatant (protein extract) was determined by the method of Bradford (Bio-Rad). SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting were performed as described previously (4) with the anti-p53 monoclonal antibody DO-1 (40) or Pab1801 (Oncogene Science), the anti-Mdm2 monoclonal antibody 4B2 (8), the monoclonal anti-Flag antibody M1 (Sigma), the phosphorylation-specific Mdm2 antibodies, and the anti-proliferating nuclear antibody PC-10 (41).
Phosphatase treatment.
Proteins were blotted onto a nitrocellulose membrane. After transfer, the membrane was washed in PBS and phosphatase buffer (50 mM Tris [pH 8], 10 mM EDTA) and incubated with 100 U of calf intestinal phosphatase in phosphatase buffer at 37°C for 2 h. The membrane was then washed with PBS plus 0.2% Tween 20 and blocked with 5% dry milk in PBS plus 0.2% Tween 20.
In vivo ubiquitylation assay.
H1299 cells were transfected with plasmids encoding p53, His-tagged ubiquitin, and Mdm2 or with vector DNA and harvested 36 h after transfection of cells. The cells were washed twice in ice-cold PBS and 7.5 × 106 cells were lysed in 6 ml of guanidinium lysis buffer (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4 [pH 8], 0.01 M Tris [pH 8], 5 mM imidazole, 10 mM β-mercaptoethanol). Ni2+-nitrilotriacetic acid (NTA)-agarose beads (75 μl) were added to the lysate, and the mixture was incubated by end-over-end rotation at room temperature for 4 h. The beads were successively washed with the following buffers: guanidinium buffer (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4 [pH 8], 0.01 M Tris [pH 8], 10 mM β-mercaptoethanol), urea buffer (pH 8; 8 M urea, 0.1 M Na2HPO4/NaH2PO4 [pH 8], 0.01 M Tris [pH 8], 10 mM β-mercaptoethanol), buffer A (8 M urea, 0.1 M Na2HPO4/NaH2PO4 [pH 6.3], 0.01 M Tris [pH 6.3], 10 mM β-mercaptoethanol), buffer A plus 0.2% Triton X-100, and buffer A plus 0.1% Triton X-100. Elution was carried out with 200 mM imidazole in 5% SDS-0.15 M Tris [pH 6.7]-30% glycerol-0.72 M β-mercaptoethanol. The eluate was diluted 1:1 with 2× SDS-PAGE sample buffer and subjected to SDS-PAGE. The proteins were transferred to a nitrocellulose membrane and probed with the anti-p53 antibody DO-1 or the Mdm2 antibody 4B2.
Immunofluorescence staining.
H1299 cells (2.5 × 105 cells/6-cm-diameter dish) grown on coverslips were transfected with 5 μg of pcDNA3-mdm2 DNA with or without 2 μg of pcDNA3-p53 DNA and fixed 24 h posttransfection. Cells transfected with p53 and Mdm2 were incubated with the rabbit polyclonal anti-p53 antibody CM-1 (28) and the mouse monoclonal anti-Mdm2 antibody 4B2 diluted 1:250 in DMEM plus 10% FCS as primary antibodies. Cells transfected with only Mdm2 were incubated solely with the 4B2 antibody. The coverslips were washed five times in PBS plus 0.2% Tween 20 and incubated in fluorescein isothiocyanate-conjugated anti-mouse immunoglobulin G (IgG) diluted 1:80 in DMEM plus 10% FCS and in tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit IgG diluted 1:500 in DMEM plus 10% FCS. The cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI; 0.5 mg/ml) for 2 min and washed five times in PBS plus 0.2% Tween 20. The coverslips were mounted on microscope slides with Hydromount (National Diagnostics) supplemented with 2.5% 1,4-diazabicyclo[2,2,2]octane (Sigma) and analyzed by confocal microscopy.
RESULTS
Substitution of putative phosphorylation sites in conserved region II by alanine prevents p53 degradation.
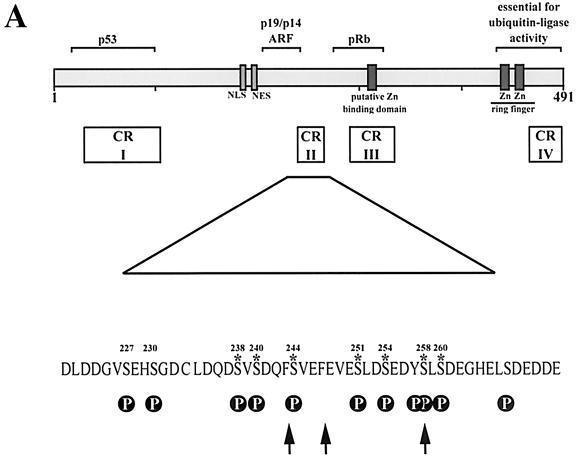
In order to investigate whether conserved domain II (Fig. 1A) plays a role in regulating the ability of Mdm2 to mediate the degradation of p53, we generated a series of Mdm2 mutants in which potential phosphorylation sites were substituted with alanine residues. These changes were focused on the area of Mdm2 bound by p14ARF (aa 219 through 240) and on the contiguous conserved domain II (aa 235 through 260). Here, we replaced all serines between aa 219 and 265 of mouse Mdm2 with alanine. To reduce the number of constructs, we introduced preferably double-point mutations. The region on the N-terminal flank of the p14ARF-binding site was not studied, as mouse and human Mdm2 proteins differ significantly in this region due to the insertion or deletion of 4 to 5 amino acids at two close-by positions.
FIG. 1.
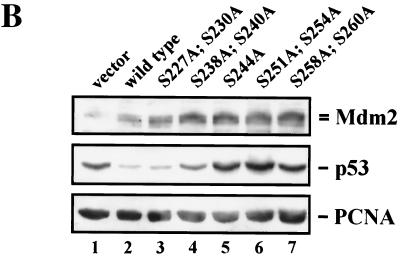
Degradation of p53 by Mdm2 serine→alanine mutants. (A) Diagram of Mdm2 wild-type protein showing the functional domains, conserved regions (CR), and interacting proteins. Arrows point to chymotrypsin cleavage sites. Putative phosphorylation sites (P) are indicated. Asterisks denote serines important for p53 degradation. (B) H1299 cells were transiently transfected with 1 μg of pcDNA3-p53 DNA (lanes 1 through 7) and 5 μg of wild-type pcDNA3-mdm2 DNA (lane 2), the pcDNA3-mdm2 DNA possessing the indicated single- and double-point mutations (lanes 3 through 7) or 5 μg of pcDNA3 vector DNA (lane 1). Thirty-six hours after transfection, cells were harvested, lysed in NP-40 lysis buffer, and analyzed for the presence of p53 and Mdm2. Forty micrograms of crude cellular extract was separated on an SDS-10% PAGE minigel and transferred to a nitrocellulose membrane. The Western blot was serially probed with the anti-p53 antibody DO-1 (ascites, diluted 1:750), the anti-Mdm2 antibody 4B2 (ascites, diluted 1:500), and the anti-PCNA antibody PC-10 (ascites, diluted 1:3,000), developed by enhanced chemiluminescence, and exposed.
Consistent with earlier reports (13, 21), we observed a significant down-regulation of p53 expression upon cotransfection of wild-type Mdm2 compared to expression upon transfection with p53 in the absence of Mdm2 (Fig. 1B). When we cotransfected the Mdm2 mutant S227A;S230A along with p53 into H1299 cells, we found that the activity of this mutant was indistinguishable from that of wild-type Mdm2 (Fig. 1B). However, when we cotransfected the Mdm2 mutants where serines in conserved region II were replaced by alanine, in particular, S238A;S240A, S244A, S251A;S254A, or S258A;S260A, the degradation of p53 was significantly reduced, with the single-point mutation S244A and the double-point mutation S251A;S254A showing the strongest p53 expression. Mdm2 levels remained unchanged under these conditions (Fig. 1B). Expression of p53 in the presence of the Mdm2 double mutant S251A;S254A repeatedly exceeded expression of p53 in the absence of Mdm2. We do not really understand why p53 abundance is higher in the presence of the Mdm2 S251A;S254A double mutant. However, H1299 cells express low levels of endogenous Mdm2 protein (15; C. Blattner, unpublished data), which may degrade p53 at a very low rate. Ectopically expressed Mdm2 mutants may therefore compete with endogenous wild-type Mdm2 for binding to p53, thus interfering with p53 degradation by endogenous wild-type Mdm2. If this were the case, it would suggest that the Mdm2 mutants S238A;S240A, S244A, and S258A;S260A retain some activity to target p53 for degradation.
Defects in p53 degradation were seen over several p53/Mdm2 ratios (compare Fig. 1B and 4). Hybridization with an antibody recognizing proliferating cell nuclear antigen (PCNA) showed that equal amounts of proteins had been loaded onto the gel (Fig. 1B).
FIG. 4.
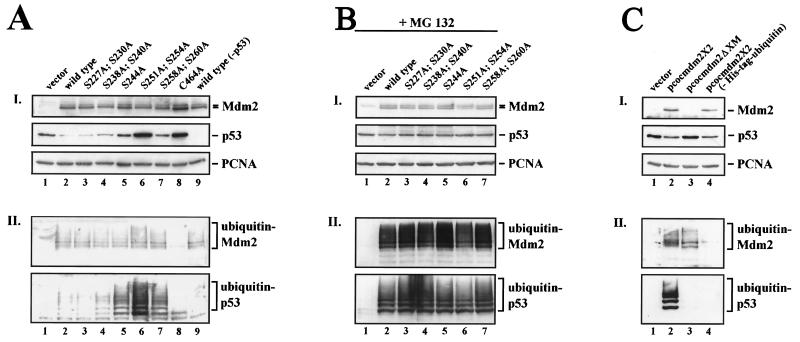
Ubiquitylation is not restrained by Mdm2 serine→alanine mutants. H1299 cells were transiently transfected with 2 μg of His-tagged ubiquitin DNA, 1 μg of pcDNA3-p53 DNA (lanes 1 through 8), and 10 μg of wild-type pcDNA3-mdm2 DNA (lanes 2 and 9), the pcDNA3-mdm2 DNA possessing the indicated single- and double-point mutations (lanes 3 through 8) or 10 μg of pcDNA3 vector DNA (lane 1). Thirty-six hours after transfection, cells were harvested. (A, panel I) Aliquots of the cells were lysed in NP-40 lysis buffer and analyzed for the presence of p53 and Mdm2. Forty micrograms of crude cellular extract were separated on an SDS-10% PAGE minigel and transferred to a nitrocellulose membrane. The Western blot membrane was cut into strips and probed with the anti-p53 antibody DO-1 (ascites, diluted 1:750), the anti-Mdm2 antibody 4B2 (ascites, diluted 1:500), and the anti-PCNA antibody PC-10 (ascites, diluted 1:3,000), developed by enhanced chemiluminescence, and exposed. (A, panel II) The remaining cells were lysed in guanidinium lysis buffer and probed for the presence of ubiquitylated p53 and Mdm2. Ubiquitylated proteins were purified by adsorption to Ni2+-NTA-agarose beads and resolved on an SDS-8% PAGE minigel. After transfer of the proteins to a nitrocellulose membrane, the Western blot was probed with the anti p53-antibody DO-1 (ascites, diluted 1:750) or anti-Mdm2 antibody 4B2 (ascites, diluted 1:500) and developed by enhanced chemiluminescence. (B) H1299 cells were transiently transfected with 2 μg of His-tagged ubiquitin DNA, 1 μg of pcDNA3-p53 DNA (lanes 1 through 8), and 10 μg of wild-type pcDNA3-mdm2 DNA (lane 2), the pcDNA3-mdm2 DNA possessing the indicated single- and double-point mutations (lanes 3 through 7) or 10 μg of pcDNA3 vector DNA (lane 1). Thirty-six hours after transfection, cells were treated with 10 μM MG132 for 6 h, harvested, and processed as described above. (C) H1299 cells were transiently transfected with 2 μg of His-tagged ubiquitin DNA (lanes 1 through 3), 1 μg of pcDNA3-p53 DNA, and 10 μg of wild-type pcocmdm2X2 DNA (lanes 2 and 4), pcocmdm2ΔXM possessing an amino-terminal deletion encompassing the p53-binding site as well as the 4B2 antibody epitope (lane 3) or 10 μg of vector DNA (lane 1). Thirty-six hours after transfection, cells were harvested and treated as described above except that ubiquitylated Mdm2 (panel II) was detected with the anti-Mdm2 antibody 2A10.
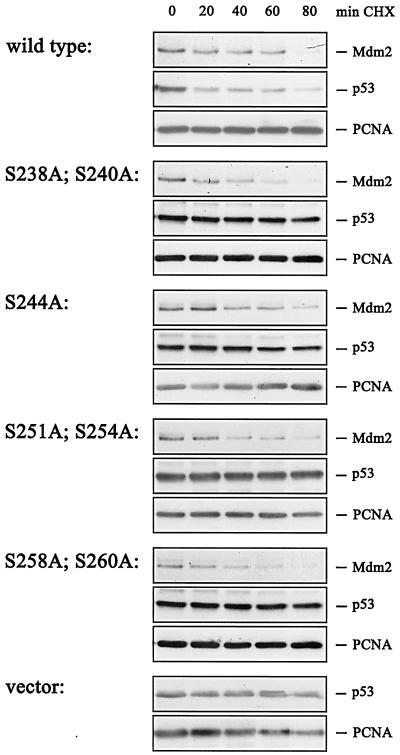
A critical prediction of these results is that the half-life of the p53 protein should be short in the presence of wild-type Mdm2 but should be extended significantly by the serine-to-alanine mutations in the conserved region II that are impaired in their p53 degradation function. To address this question, we transfected cells with plasmids expressing p53 and either wild-type or mutant Mdm2. We then added 20 μg of cycloheximide/ml to block protein synthesis and analyzed the decay of p53 and Mdm2 at defined times after the addition of the protein synthesis inhibitor by immunoblotting. While p53 was degraded by wild-type Mdm2 with a half-life of about 20 to 40 min, the protein was stable when coexpressed with each of the mutant Mdm2 proteins (Fig. 2). In contrast, wild-type and mutant Mdm2 were degraded with equal efficiency, showing a half-life of Mdm2 of about 30 min.
FIG. 2.
Determination of p53 and Mdm2 half-lives in the presence of wild-type or mutant Mdm2. H1299 cells were transfected with 0.5 μg of pcDNA3-p53 DNA and 3.75 μg of wild-type pcDNA3-mdm2 DNA, mutant mdm2 DNA, or vector DNA. Thirty-six hours after transfection, cycloheximide (CHX) was added (20 μg/ml) and cells were harvested at the indicated time points after CHX addition. The cells were lysed in NP-40 lysis buffer. Forty micrograms of cellular extract were separated on an SDS-10% PAGE gel and transferred to a nitrocellulose membrane. Western blot membranes were cut into strips which were probed with the monoclonal antibodies DO-1 (anti-p53), 4B2 (anti-Mdm2), and PC-10 (anti-PCNA) and developed by enhanced chemiluminescence.
Substitution of putative phosphorylation sites in conserved region II with alanine allows interaction with p53 and p14ARF and ubiquitylation of p53.
Ubiquitylation of p53 is a prerequisite for its degradation, as this modification enables the 26S proteasome to recognize the protein as a target for destruction (see reference 10 for an overview). The covalent attachment of ubiquitin to p53 is catalyzed by Mdm2 in vitro, upon overexpression of both proteins in vivo, and presumably also under physiological conditions. The ubiquitylation of p53 requires the interaction of p53 and Mdm2, which occurs via their amino termini and an intact Mdm2 ligase activity. As p53 degradation was prevented by mutant Mdm2, we reasoned that replacing serines in conserved region II of Mdm2 with alanine might either release p53 from the complex, favor the interaction of Mdm2 with p14ARF, alter Mdm2 conformation, or inhibit the ubiquitin ligase activity of Mdm2. All these consequences would prevent the degradation of p53, as was shown earlier by the resistance to degradation of N-terminal mutants of p53 that do not bind Mdm2 (5, 13), by the p14ARF-mediated rescue from degradation (33, 36), and by the stability of p53 in the presence of the C464A mutant of Mdm2 (17) (Fig. 4A, lanes 8).
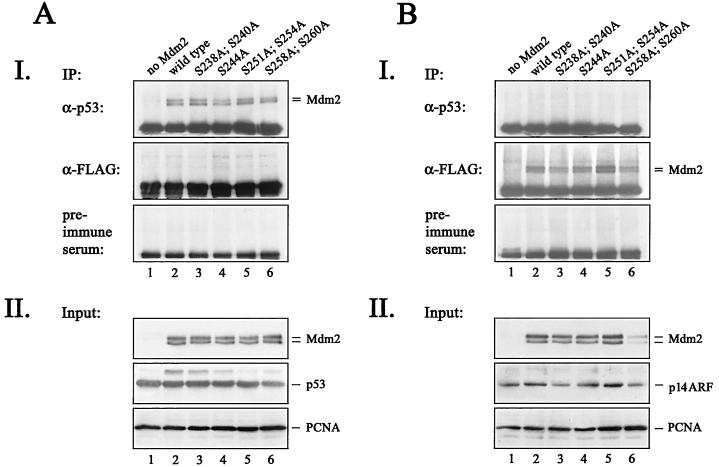
We first tested whether mutant Mdm2 was capable of interacting with p53. We therefore transfected H1299 cells with p53 and wild-type or mutant Mdm2, immunoprecipitated p53 with an anti-p53 antibody, and probed the complexes for the presence of Mdm2 by immunoblotting. To obtain equal levels of p53 and Mdm2, we treated the cells with the proteasome inhibitor MG132 for 6 h prior to cell lysis. When we immunoprecipitated p53 with the p53-specific CM-1 antibody, we detected coprecipitating Mdm2 in each case. As shown in Fig. 3A, wild-type and mutant Mdm2 were bound to p53 equally well. In control analyses, we were unable to detect coimmunoprecipitating Mdm2 when we used an anti-Flag antibody or preimmune serum.
FIG. 3.
Interaction of mutant Mdm2 with p53 and p14ARF. H1299 cells were transfected with 5 μg of pcDNA3-p53 DNA (A) or 5 μg of pcDNA3-ARF-FLAG DNA (B) in the absence (lane 1) or presence (lane 2) of 5 μg of wild-type pcDNA3-mdm2 DNA or the indicated Mdm2 mutants (lanes 3 through 6). Twenty-four hours after transfection, cells were treated with the proteasome inhibitor MG132 for 6 h, prior to lysis. (A, panel I) p53/Mdm2 complexes were immunoprecipitated (IP) out of 350 μg of cellular lysate with either the CM-1 anti-p53 antibody coupled to protein A+G-Sepharose, an anti-Flag antibody, or preimmune serum coupled to protein A+G-Sepharose. (B, panel I) ARF/Mdm2 complexes were precipitated out of 350 μg of cellular lysate with an anti-p53 antibody, an anti-Flag antibody, or preimmune serum coupled to protein A+G-Sepharose The protein/antibody complexes were washed three times in NP-40 lysis buffer and resolved in 1× sample buffer. The complexes were separated on an SDS-10% PAGE gel, transferred to a nitrocellulose membrane, and probed for the presence of Mdm2 with the 4B2 (anti-Mdm2) antibody. Western blots were developed by enhanced chemiluminescence. (A and B, panels II) Forty micrograms of the remaining cell lysate was separated on an SDS-10% (A, panel II) or -13% (B, panel II) PAGE gel, transferred to a nitrocellulose membrane, and probed for the presence of p53 and Mdm2 (A, panel II) by using the DO-1 (anti-p53) and 4B2 (anti-Mdm2) antibodies or for the presence of ARF and Mdm2 (B, panel II) by using 4B2 (anti-Mdm2) and the M1 anti-Flag antibody.
We next used a similar approach to analyze whether the interaction with p14ARF was altered by mutant Mdm2. Thus, we transfected Flag-tagged p14ARF together with wild-type or mutant Mdm2, immunoprecipitated Flag-tagged ARF with an anti-Flag antibody, and probed the complexes with the anti-Mdm2 antibody 4B2. Again, we found no difference between wild-type and mutant Mdm2 in the capacity to bind to p14ARF (Fig. 3B). The absence of Mdm2 from the immunoprecipitates formed by using an antibody against p53 or preimmune serum shows the specificity of the immunoprecipitation. Moreover, the interaction of mutant Mdm2 with p53 and p14ARF indicates that the overall structure is not significantly altered by the alanine substitutions.
To investigate p53 ubiquitylation by mutant Mdm2, we used a well-characterized in vivo ubiquitylation assay in which cells are transfected with His-tagged ubiquitin (34). This form of ubiquitin is fully active but allows the rapid purification of ubiquitylated proteins by adsorption to Ni2+-NTA-agarose beads and elution with the Ni2+ competitor imidazole. His-tagged ubiquitin is covalently attached to target proteins that can be purified, separated on an SDS-PAGE gel, and analyzed by immunoblotting. The ubiquitylated proteins are detected as a ladder of higher-molecular-weight bands due to the increase in their molecular weight by the formation of mono-, di-, and polyubiquitin target protein adducts.
When we probed fractions of ubiquitylated proteins for the presence of p53 by Western blotting, we observed a pattern of higher-molecular-weight bands characteristic of ubiquitylated p53 in extracts of cells transfected with p53 and wild-type Mdm2 (Fig. 4AII, lane 2). In the presence of the Mdm2 alanine mutants (tracks 3 through 7,) we also retrieved ubiquitylated p53 from Ni2+-NTA-agarose beads. Moreover, the amount of ubiquitylated p53 corresponded precisely to the amount of total p53 in the crude cell lysate (compare Fig. 4AI with Fig. 4AII). We detected higher p53 expression levels in cells transfected with the Mdm2 alanine mutants S238A;S240A, S244A, S251A;S254A, and S258A;S260A, with the highest p53 abundance observed in the presence of the S251A;S254A double mutant (Fig. 4AI and Fig. 1B). Correspondingly, we isolated relatively more ubiquitylated p53 from cell lysates when the cells had been transfected with p53 and the Mdm2 mutants S238A;S240A, S244A, S251A;S254A, and S258A;S260A, with the highest level of ubiquitylated p53 in the presence of the S251A;S254A double mutant (Fig. 4AII). These results are in clear contrast to those seen with the C464A mutant of Mdm2, where none of the proteins was di- or polyubiquitylated (Fig. 4A, lane 8), demonstrating that all of the serine-to-alanine mutants in conserved region II tested were still able to act as E3 ligases. Immunoblotting of the same purified fraction was used to show that all of the mutant Mdm2 proteins, with the expected exception of the C464A mutant, were themselves polyubiquitylated, supporting the concept that the mutants retained ubiquitin ligase activity (Fig. 4AII). We can therefore exclude the possibility that the alanine mutations interfere with p53 binding or Mdm2 ligase activity since, in the absence of a direct interaction or ligase activity, Mdm2 would be unable to ubiquitylate p53. Our results show, however, that these phosphorylation sites are not obligatory for ubiquitylation but rather are required for subsequent degradation, implying that Mdm2 is involved in an additional step(s) that is necessary for p53 destruction.
To evaluate these observations further, we repeated these experiments in the presence of the proteasome inhibitor MG132. The data (Fig. 4B) show that the level of p53 and of the different serine-to-alanine mutant Mdm2 proteins in the transfected cells was entirely uniform (Fig. 4BI) and, as expected, that ubiquitylated Mdm2 and p53 levels were much higher in the mutants than in the wild type. Again, the capacity to ubiquitylate p53 did not vary between the mutants, establishing that the relatively low levels of p53 and ubiquitylated p53 found in the presence of the wild type and the S227A;S230A mutant (Fig. 1B and 4A) are due to proteasome-dependent degradation of p53. It is striking that in the presence of MG132, the S244A, S251A;S24A, and S258A;S260A Mdm2 mutants mimic the action of wild-type Mdm2 by inducing the accumulation of ubiquitylated p53. This observation is consistent with a model in which the mutants are defective in targeting ubiquitylated p53 to the proteasome.
The surprising finding that Mdm2 mutants could direct ubiquitylation but not degradation of p53 prompted further validation of the in vivo assay. We therefore repeated the assay in the absence of Mdm2 (Fig. 4C, lanes 1), in the absence of His-tagged ubiquitin (Fig. 4C, lanes 4), or with wild-type Mdm2 substituted with the ΔXM N-terminal deletion mutant of Mdm2 (3) that cannot bind p53 (Fig. 4C, lanes 3). Ubiquitylated p53 was detected only in the presence of the full-length Mdm2 protein and His-tagged ubiquitin, fully confirming the assay's specificity. Interestingly, the ΔXM N-terminal deletion mutant, while unable to direct the ubiquitylation of p53, was itself still ubiquitylated. The level of this ubiquitylated ΔXM N-terminal deletion mutant protein was slightly lower than that of the wild-type protein detected with the 2A10 monoclonal antibody. The total level of the ΔXM N-terminal deletion mutant could not, however, be compared with that of wild-type Mdm2, as the ΔXM N-terminal deletion removes the epitope of the 4B2 antibody that is used in the immunoblotting of Mdm2 in whole-cell extracts (the 2A10 monoclonal antibody has too high a level of cross-reactivity for use in whole-cell extracts of H1299 cells).
The Mdm2 mutations are close to the nuclear export (NES) and nuclear localization (NLS) signals, raising the possibility that phosphorylation-dependent modifications of Mdm2 structure could either conceal or expose the NES or NLS sequences and, by this action, regulate Mdm2’s subcellular localization. Alternatively, Mdm2 alanine mutants could favor the interaction with p14ARF. p14ARF binds to Mdm2 between aa 212 and 240 (27), and this interaction rescues p53 from being degraded by Mdm2. Protection of p53 presumably results from several activities of p14ARF which are as follows. (i) p14ARF binds to a domain of Mdm2 that is apparently important for the destruction of p53. (ii) It prevents ubiquitylation of p53, which is a prerequisite for destruction. (iii) It dissociates Mdm2 from p53 by sequestering Mdm2 in the nucleoli (17, 33, 36, 42). Thus, if the Mdm2 alanine mutants favored the interaction with p14ARF, then Mdm2 should be sequestered in the nucleoli.
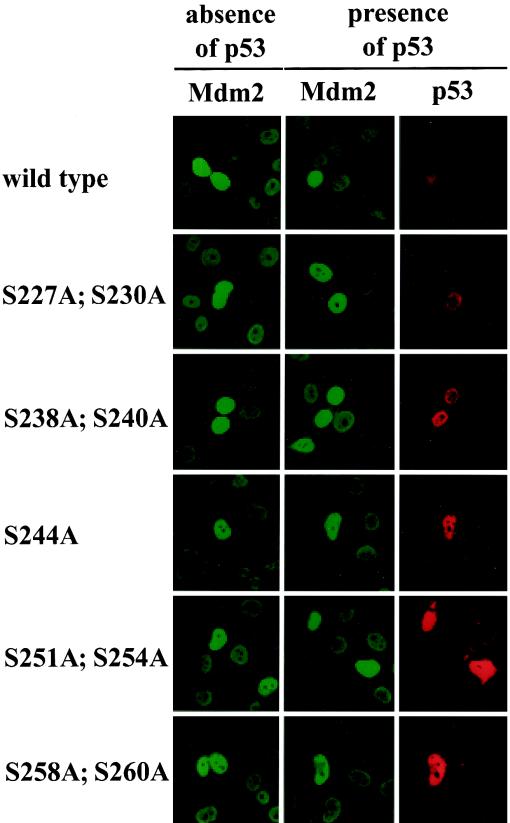
To determine the compartment in which mutant Mdm2 proteins were localized, we transfected plasmids expressing wild-type or mutant Mdm2 into H1299 cells in either the presence or the absence of p53 and examined the cellular localization of the proteins. As shown in Fig. 5, wild-type and mutant Mdm2 were exclusively located in the nucleoplasm with clear nucleolar exclusion. This localization of Mdm2 was independent of the presence of p53. When coexpressed, p53 was also detected exclusively in the nucleoplasm with nucleolar exclusion and indeed colocalized precisely with Mdm2. The expression level of p53 in the presence of the different Mdm2 alanine mutants varied in a manner that reflected the signals obtained by Western blotting (compare Fig. 5 with 1B and 4A) while the expression levels of Mdm2 remained unchanged.
FIG. 5.
Localization of wild-type and mutant Mdm2. H1299 cells were transfected with 5 μg of wild-type or mutant pcDNA3-mdm2 DNA/2.5 × 105 cells in the absence or presence of 1 μg of pcDNA3-p53 DNA. Twenty-four hours after transfection, cells were fixed with acetone-methanol (1:1) and stained for expression of Mdm2 with the mouse monoclonal anti-Mdm2 antibody 4B2 and for p53 with the rabbit polyclonal anti-p53 antibody CM-1. Fluorescein isothiocyanate-coupled anti-mouse IgG and tetramethyl rhodamine isothiocyanate-coupled anti-rabbit IgG were used as secondary antibodies. The two left-hand columns (in green) show expression of Mdm2, and the right-hand column (in red) shows expression of p53.
The nucleolar exclusion of the Mdm2 alanine mutants argues against an enhanced interaction of these mutants with p14ARF and is consistent with the result of the immunoprecipitation (Fig. 3B), where we also did not see a change in affinity for p14ARF by mutant Mdm2.
In summary, our data suggest that the inability of mutant Mdm2 to target p53 for degradation is independent of an increased affinity for p14ARF. This conclusion is based on several observations. (i) Although Mdm2 alanine mutations fail to target p53 for degradation, they are not transported into the nucleolus. (ii) Ubiquitylation of p53, which is blocked upon binding of p14ARF to Mdm2, is permitted by mutant Mdm2. (iii) Wild-type and mutant Mdm2 bind to p14ARF with equal affinities. (iv) p53 is rescued from degradation by p14ARF in the presence of Mdm2 aspartic acid mutants (data not shown).
Mdm2 is hypophosphorylated at several residues in conserved region II in response to ionizing irradiation.
The observed changes in the behavior of the Mdm2 mutants are consistent with a model in which phosphorylation of serine residues within conserved region II is required to mediate degradation of p53. In order to explore this model further, we examined directly the phosphorylation status of this region in a cellular context.
Two-dimensional phosphopeptide mapping allows dissection of the individual and multiple posttranslational modifications of a protein. Coupled with the use of mutant proteins harboring changes at potential or putative phosphorylation sites, this technique provides a means of identifying residues that are phosphorylated in vivo and can reveal whether certain stimuli influence the phosphorylation of any given site(s) (see reference 26 for a detailed description of this approach).
To determine whether serines in conserved domain II of Mdm2 are phosphorylated in vivo, we transfected Myc-tagged wild-type Mdm2 or Mdm2 in which serines of the central domain had been replaced by alanine in COS-7 cells. The cells were subsequently radiolabeled with [32P]orthophosphate, and the Myc-tagged Mdm2 was immunoprecipitated from cell extracts with the tag-specific monoclonal antibody 9E10. The radiolabeled Mdm2 proteins were then digested with chymotrypsin, and the peptides were separated in two dimensions by electrophoresis and chromatography.
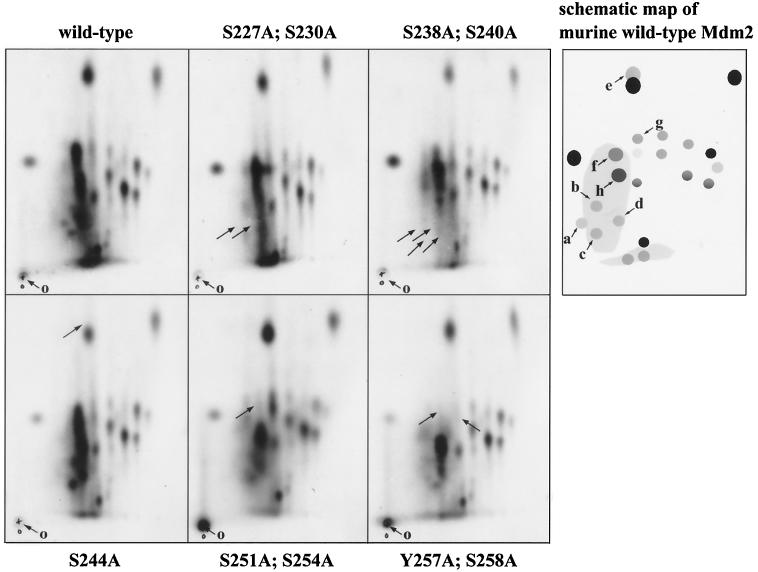
Twenty-three separable phosphopeptides could be discerned in wild-type Mdm2, consistent with the pattern observed previously in COS-7 and MCF-7 cells (14). On the phosphopeptide map of the mutant S227A;S230A, two phosphopeptides were absent from the bottom left-hand side (Fig. 6), which is consistent with the loss of phosphorylation of one or both of these residues. Similarly, an additional two phosphopeptides were absent from the bottom left-hand side in the phosphopeptide map of the mutant S238A;S240A (the finding that the S227A;S230A phosphopeptides were also absent in the S238A;S240A map suggests the possibility that the phosphorylation of S227 and/or S230 may depend on the modification status of S238 and/or S240). The loss of a hydrophobic phosphopeptide is indicated in the S244A map (Fig. 6). Unfortunately, this phosphopeptide overlaps considerably with a more intensely labeled phosphopeptide arising from a different and as-yet-uncharacterized phosphorylation event in Mdm2. The peptide phosphorylated at S244 can be discerned in the wild-type (and other) maps on the upper side of this intense phosphopeptide. The predicted mobility of the chymotryptic phosphopeptide comprising phosphoserine at position 244 (calculated as described in reference 39) is entirely consistent with our identification of the S244A phosphopeptide provided by the mutagenesis approach. It should also be noted that the relative intensity of the comigrating phosphopeptide in the S244A panel was reduced significantly, consistent with the loss of an overlapping phosphopeptide. In the map of the S251A;S254A double mutant, two major phosphopeptides are absent from the middle of the map, again consistent with modification of one or both of these residues. Finally, the occurrence of the phosphorylation of residues 258 and/or 260 has been demonstrated elsewhere (16). Taken together, these data argue very strongly that serines 227 and/or 230, 238 and/or 240, 244, and 251 and/or 254 and tyrosine 257 and/or serine 258 and/or serine 260 of Mdm2 are phosphorylated in cultured cells. These data are also highly consistent with the idea that the attenuated capacity to mediate p53 degradation by Mdm2 proteins with amino acid changes at these residues is a consequence of the loss of phosphorylation at these sites.
FIG. 6.
Mdm2 is phosphorylated in vivo in conserved domain II. Myc-tagged mouse wild-type mdm2 DNA (pDWM659) or mdm2 DNA possessing the indicated mutations was transiently transfected into COS-7 cells. Twelve hours prior to irradiation, cells were incubated with the proteasome inhibitor MG132, and 48 h after transfection, cells were radioactively labeled with [32P]orthophosphate for 3 h. Cells were harvested and lysed in NP-40 lysis buffer, and Myc-tagged Mdm2 was precipitated with the anti-Myc monoclonal antibody 9E10. After washing, the Mdm2-antibody complexes were resolved on an SDS-8% PAGE minigel. The Mdm2 protein was eluted from the gel and digested with chymotrypsin, and the resulting peptides were separated by electrophoresis in buffer (pH 1.9; first dimension) and chromatography buffer (second dimension). The origin (o) where the phosphopeptides were loaded is indicated. Arrows point to changes which resulted from the serine substitutions. The schematic map shows the following phosphopeptides, identified by serine→alanine substitutions: a through d, aa 189 through 243; e, aa 244 through 247; f, aa 248 through 257; g and h, aa 248 through 274.
More recently, it has been shown that ionizing irradiation enhances the migration of Mdm2 through SDS-PAGE (20; Blattner, unpublished). In an attempt to investigate whether Mdm2 is posttranslationally modified upon irradiation, we expressed Myc-tagged wild-type Mdm2 protein in COS-7 cells, pulse-labeled the cells with [32P]orthophosphate, and then irradiated the cells. Following immunoprecipitation of the labeled Mdm2 proteins, we assessed phosphorylation at specific sites by comparing chymotryptic phosphopeptide maps of Mdm2 from nonirradiated and irradiated cells (Fig. 7).
FIG. 7.
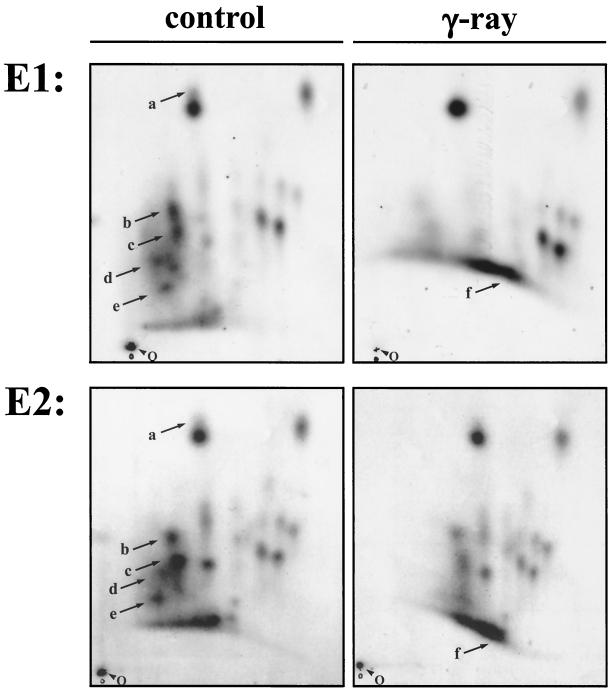
Mdm2 is posttranslationally modified in response to irradiation. Cells were irradiated with 10 Gy 3 h prior to harvest and processed as described in the legend to Fig. 6. Arrows point to irradiation-mediated changes in phosphorylation. E1, experiment 1; E2, experiment 2, O, origin.
We found that Mdm2 is both phosphorylated and dephosphorylated upon irradiation, though at different peptides. One peptide that we did not investigate in greater detail was phosphorylated upon irradiation (Fig. 7, phosphopeptide f). We speculate that, based on its migration properties in two dimensions, this phosphopeptide encompasses serine 393 (corresponding to serine 395 in human Mdm2), a residue that has been shown recently to be phosphorylated by ataxia telangiectasia mutated (ATM) upon irradiation (25). More strikingly, we found that all of the phosphopeptides from the conserved region II became hypophosphorylated upon irradiation (Fig. 7, phosphopeptides a through e). Hypophosphorylation was seen at peptides comprising S244A (Fig. 7, phosphopeptide a) and at the peptides accommodating S227A;S230A (Fig. 7, phosphopeptide d), S238A;S240A (Fig. 7, phosphopeptide e), and S251A;S254A (Fig. 7, phosphopeptide b), as well as at the double peptide which results from impaired chymotryptic cleavage upon phosphorylation of tyrosine 257, harboring serines 251, 254, 258, and 260 and tyrosine 257 (Fig. 7, phosphopeptide c).
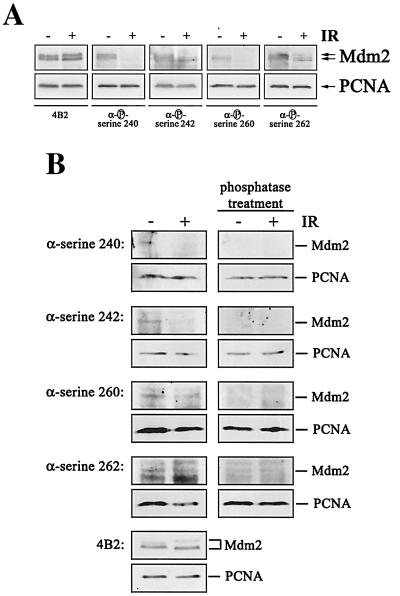
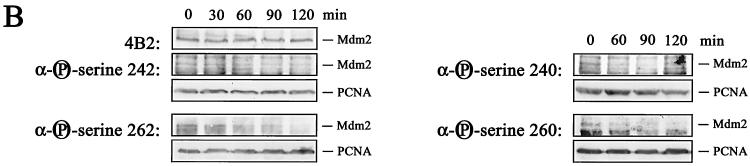
We attempted next to raise phosphorylation-specific antibodies against phosphorylated residues in the central conserved domain of Mdm2. Although we were unable to raise antibodies against all the phosphorylated sites in the central conserved domain of Mdm2, we were able to confirm the results from the two-dimensional peptide analysis. We obtained antibodies recognizing phosphorylated serine 240 of human Mdm2 (corresponding to serine 238 of murine Mdm2), phosphorylated serine 242 (corresponding to serine 240 of murine Mdm2), phosphorylated serine 260 (corresponding to serine 258 of murine Mdm2), and phosphorylated serine 262 (corresponding to serine 260 of murine Mdm2). All of these antibodies recognized Mdm2 from nonirradiated cells. Recognition, however, was abolished or at least reduced when we probed extracts from irradiated cells (Fig. 8A). Hybridization with the antibody 4B2, which recognizes a nonphosphorylated epitope, showed that the levels of Mdm2 protein were not altered in response to irradiation, while probing the membranes with an anti-PCNA antibody showed that equal amounts of proteins were loaded on the gel (Fig. 8A). The phospho-specific antibodies recognize only the phosphorylated form of Mdm2, demonstrated by the absence of a signal when the membranes were treated with calf intestinal phosphatase prior to antibody incubation (Fig. 8B). Hypophosphorylation of Mdm2 was already detectable at 30 to 60 min after irradiation (Fig. 9B), thus clearly preceding p53 accumulation (Fig. 9A). Rehybridization of the membrane with the 4B2 antibody showed that the amount of Mdm2 remained unchanged after irradiation (Fig. 9B).
FIG. 8.
Mdm2 is hypophosphorylated in the central conserved domain upon irradiation (IR). GM1604 cells were treated with MG132 for 10 h. Prior to lysis (1.5 h), cells were irradiated with 10 Gy. (A) Forty micrograms of lysate was separated on an SDS-10% PAGE gel, transferred to a nitrocellulose membrane, and probed with 4B2 or phosphorylation-specific anti-Mdm2 antibodies. Hybridization with an anti-PCNA antibody was used for loading control. Western blots were developed by enhanced chemiluminescence. (B) Forty micrograms of lysate was separated on an SDS-10% PAGE gel and transferred to a nitrocellulose membrane. After transfer, membranes were incubated with calf intestinal phosphatase at 37°C for 2 h prior to antibody incubation. Membranes were then probed with 4B2 or phosphorylation-specific anti-Mdm2 antibodies and with an anti-PCNA antibody for loading control.
FIG. 9.
Hypophosphorylation precedes p53 accumulation. (A) GM1604 cells were irradiated with 10 Gy and harvested at the indicated time points. Forty micrograms of lysate was separated on an SDS-10% PAGE gel, transferred to a nitrocellulose membrane, and probed with Pab1801. Hybridization with an anti-PCNA antibody was used for loading control. Western blots were developed by enhanced chemiluminescence. (B) GM1604 cells were treated with MG132 for 10 h. At the indicated times prior to lysis, cells were irradiated with 10 Gy. Forty micrograms of lysate was separated on an SDS-10% PAGE gel, transferred to a nitrocellulose membrane, and probed with phosphorylation-specific antibodies. Hybridization with the anti-Mdm2 antibody 4B2 shows that Mdm2 levels remained unchanged, and the anti-PCNA antibody was used for loading control. Western blots were developed by enhanced chemiluminescence.
Based on data derived from two-dimensional phosphopeptide mapping and immunoblotting with phosphorylation-specific antibodies, we conclude that the central conserved domain of Mdm2 is hypophosphorylated upon treatment with irradiation. Thus, dephosphorylation (occurring through inactivation of kinases, activation of phosphatases, or both) may provide a physiological mechanism that regulates the activity of Mdm2. Hypophosphorylation of Mdm2 precedes p53 accumulation and may thus contribute to p53 accumulation in response to irradiation.
DISCUSSION
In this study, we attempted to investigate the influence of posttranslational modifications of Mdm2 on its ability to target p53 for destruction. We focused on the domain that interacts with p14ARF and on the contiguous conserved region II, where we replaced putative phosphorylation sites in Mdm2 with alanine residues. Using the approaches of two-dimensional peptide analysis coupled with the use of phosphorylation-specific antibodies, we confirmed that key sites within this region are indeed phosphorylated.
When we replaced particular putative phosphorylation sites in conserved region II of Mdm2 with alanine, the capacity of Mdm2 to target p53 for degradation was significantly reduced. The efficacy of attenuating p53 degradation increased from the amino-terminal mutants S227A;S230A, S238A;S240A, and S244A and then reached a maximum at S251A;S254A and decreased again at S258A;S260A. This variability from the amino-terminal part of the domain to the carboxyl-terminal part would be consistent with the idea that this central domain of Mdm2 associates with another protein (which is as yet not identified) that is crucial for p53 destruction. This interaction could be regulated by phosphorylation. According to this model, alterations of amino acids in the center should have a strong impact while alterations at the boundary such as S227A;S230A, S238A;S240A, and S258A;S260A should be less critical. Interestingly, one of these sites, serine 253 (corresponding to murine serine 251), was mutated in a human breast tumor (D. Crichton and D. P. Lane, unpublished data), substantiating the importance of this particular residue.
Although Mdm2 mutants with alanine substitutions in the central conserved domain failed to target p53 for degradation, they were still able to ubiquitylate p53, clearly indicating that neither the interaction of p53 and mutant Mdm2 nor the ligase activity of Mdm2 was affected by alanine substitutions of the phosphorylation sites. It is normally thought that ubiquitylated proteins are immediately recognized by the 26S proteasome and degraded. We show that p53 ubiquitylation and degradation are independent biochemical processes both requiring Mdm2. Based on the inability of the Mdm2 mutants to target p53 for degradation, we conclude that Mdm2 has a second function in the degradation of p53 that is distinct from directing ubiquitylation. This function depends on phosphorylation of the central acidic domain. The nature of this function is as yet unclear.
One possibility would be that the central domain of Mdm2 is necessary for connecting ubiquitylated p53 with the proteasome. How ubiquitylated proteins reach the proteasome and how they are taken up and processed by the regulatory subunit is as yet ill understood. Our more recent work provides strong evidence that the human homologues of yeast Rad23 are required for p53 degradation (Glockzin et al., unpublished data). Whether the central domain of Mdm2 is essential for this interaction is currently under investigation.
The complexity of ubiquitylation and degradation that we observed for p53 is, however, not unique. NF-κB precursor ubiquitylation, for example, also results in incomplete degradation of the protein (9). Mutant Mdm2 proteins which ubiquitylate p53 but fail to promote its degradation may be of considerable benefit as tools for the identification of as-yet-unknown steps in the degradation process. Biochemical events identified by the use of Mdm2 mutants may highlight factors common to the degradation of proteins by the 26S proteasome and may therefore contribute to our understanding of these processes.
In contrast to p53 degradation, degradation of Mdm2 is apparently independent of phosphorylation within central domain II. Wild-type and mutant Mdm2 possess an identical turnover rate which was also confirmed by invariant Mdm2 expression levels of wild-type and mutant Mdm2. This result suggests that Mdm2 is not degraded simultaneously with p53 and that its degradation is presumably regulated in a different manner.
Physiological events that control phosphorylation of the central domain of Mdm2 are not entirely clear. Nevertheless, when Mdm2 was derived from cells that had been irradiated with ionizing radiation, we repeatedly found both an increase and a decrease in phosphorylation, though at different parts of the protein. The increase in phosphorylation most likely corresponds to the ATM-dependent phosphorylation of serine 395 (in human Mdm2), which was recently described by Maya and coworkers (25). Hypophosphorylation in response to irradiation, however, corresponds precisely to the phosphorylation sites in the central domain of Mdm2 that are essential for p53 degradation. p53 accumulates in response to ionizing rays, as the protein is protected from degradation. Hypophosphorylation of the central domain of Mdm2 is detectable as early as 30 to 60 min after irradiation, thus clearly preceding p53 accumulation. Interestingly, p53 has been shown previously to remain ubiquitylated even after exposure to irradiation (23). In agreement with these data, our own analyses lead us to conclude that irradiation-mediated hypophosphorylation of Mdm2 contributes to the increase in p53 stability by rendering Mdm2 incapable of targeting p53 for degradation.
Acknowledgments
We thank Peter Herrlich, Jonathan Sleeman, and Deborah Weih for critical reading of the manuscript.
This work was funded by the Cancer Research Campaign, the Medical Research Council (United Kingdom), and the Deutsche Forschungsgemeinschaft BL/526-1-2. D.P.L. is a Gibb fellow of the Cancer Research Campaign. D.W.M. is a senior research fellow of the Medical Research Council (United Kingdom). C.B. is a habilitation fellow of the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Argentini, M., N. Barboule, and B. Wasylyk. 2001. The contribution of the acidic domain of MDM2 to p53 and MDM2 stability. Oncogene 20:1267-1275. [DOI] [PubMed] [Google Scholar]
- 2.Attardi, L. D., and T. Jacks. 1999. The role of p53 in tumor suppression: lessons from mouse models. Cell. Mol. Life Sci. 55:48-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barak, Y., E. Gottlieb, G. T. Juven, and M. Oren. 1994. Regulation of mdm2 expression by p53: alternative promoters produce transcripts with non-identical translation potential. Genes Dev. 8:1739-1749. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, C., A. Sparks, and D. P. Lane. 1999. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol. Cell. Biol. 19:3704-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bottger, A., V. Bottger, A. Sparks, W.-L. Liu, S. F. Howard, and D. P. Lane. 1997. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Biol. 7:860-869. [DOI] [PubMed] [Google Scholar]
- 6.Cahilly-Snyder, L., T. Yang-Feng, U. Francke, and D. L. George. 1987. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat. Cell Mol. Genet. 13:235-244. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., and H. Okayama. 1987. High efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, J., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciechanover, A., H. Gonen, B. Bercovich, S. Cohen, I. Fajerman, A. Israel, F. Mercurio, C. Kahana, A. L. Schwartz, K. Iwai, and A. Orian. 2001. Mechanisms of ubiquitin-mediated, limited processing of the NF-kappaB1 precursor protein p105. Biochimie 83:341-349. [DOI] [PubMed] [Google Scholar]
- 10.Ciechanover, A., A. Orian, and A. L. Schwartz. 2000. The ubiquitin mediated proteolytic pathway: mode of action and clinical implications. J. Cell. Biochem. Suppl. 34:40-51. [DOI] [PubMed] [Google Scholar]
- 11.Evans, S. C., and G. Lozano. 1997. The Li-Fraumeni syndrome: an inherited susceptibility to cancer. Mol. Med. Today 3:390-395. [DOI] [PubMed] [Google Scholar]
- 12.Grossman, S. R., M. Perez, A. L. Kung, M. Joseph, C. Mansur, Z. X. Xiao, S. Kumar, P. M. Howley, and D. M. Livingston. 1998. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell 4:405-415. [DOI] [PubMed] [Google Scholar]
- 13.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 14.Hay, T. J., and D. W. Meek. 2000. Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains. FEBS Lett. 478:183-186. [DOI] [PubMed] [Google Scholar]
- 15.Hengstermann, A., N. J. Whitaker, D. Zimmer, H. Zentgraf, and M. Scheffner. 1998. Characterization of sequence elements involved in p53 stability regulation reveals cell type dependence for p53 degradation. Oncogene 17:2933-2941. [DOI] [PubMed] [Google Scholar]
- 16.Hjerrild, M., D. Milne, N. Dumaz, T. Hay, O. G. Issinger, and D. Meek. 2001. Phosphorylation of murine double minute clone 2 (MDM2) protein at serine-267 by protein kinase CK2 in vitro and in cultured cells. Biochem. J. 355:347-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda, R., and H. Yasuda. 1999. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein Mdm2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh, J.-K., F. Chan, D. J. O'Connor, S. Mittnacht, S. Zhong, and X. Lu. 1999. RB regulates the stability and the apoptotic function of p53 via Mdm2. Mol. Cell 3:1-20. [DOI] [PubMed] [Google Scholar]
- 20.Khosravi, R., R. Maya, T. Gottlieb, M. Oren, Y. Shiloh, and D. Shkedy. 1999. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc. Natl. Acad. Sci. USA 96:14973-14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 22.Leng, P., D. R. Brown, C. V. Shivakumar, S. Deb, and S. P. Deb. 1995. N-terminal 130 amino acids of MDM2 are sufficient to inhibit p53-mediated transcriptional activation. Oncogene 10:1275-1282. [PubMed] [Google Scholar]
- 23.Maki, C. G., and P. M. Howley. 1997. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol. 17:355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.May, P., and E. May. 1999. Twenty years of p53 research: structural and functional aspects of the p53 protein. Oncogene 18:7621-7636. [DOI] [PubMed] [Google Scholar]
- 25.Maya, R., M. Balass, S. T. Kim, D. Shkedy, J. F. Leal, O. Shifman, M. Moas, T. Buschmann, Z. Ronai, Y. Shiloh, M. B. Kastan, E. Katzir, and M. Oren. 2001. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 15:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meek, D. W., and D. M. Milne. 2000. Analysis of multi-site phosphorylation of the p53 tumor-suppressor protein by tryptic phosphopeptide mapping. Methods Mol. Biol. 99:447-463. [DOI] [PubMed] [Google Scholar]
- 27.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 28.Midgley, C. A., C. J. Fisher, J. Bartek, B. Vojtesek, D. P. Lane, and D. M. Barnes. 1992. Analysis of p53 expression in human tumours: an antibody raised against human p53 expressed in Escherichia coli. J. Cell Sci. 101:183-189. [DOI] [PubMed] [Google Scholar]
- 29.Momand, J., H. H. Wu, and G. Dasgupta. 2000. MDM2—master regulator of the p53 tumor suppressor protein. Gene 242:15-29. [DOI] [PubMed] [Google Scholar]
- 30.Momand, J., G. P. Zambetti, D. C. Olson, D. George, and A. J. Levine. 1992. The mdm2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 92:1237-1245. [DOI] [PubMed] [Google Scholar]
- 31.Oliner, J. D., J. A. Pietenpol, S. Thiagalingam, J. Gyuris, K. W. Kinzler, and B. Vogelstein. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362:857-860. [DOI] [PubMed] [Google Scholar]
- 32.Oren, M. 1999. Regulation of the p53 tumor suppressor protein. J. Biol. Chem. 274:36031-36034. [DOI] [PubMed] [Google Scholar]
- 33.Pommerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H.-W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez, M. S., J. M. Desterro, S. Lain, D. P. Lane, and R. T. Hay. 2000. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol. 20:8458-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sionov, R. V., and Y. Haupt. 1999. The cellular response to p53: the decision between life and death. Oncogene 18:6145-6157. [DOI] [PubMed] [Google Scholar]
- 36.Stott, F. J., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi, T., M. M. Nau, I. Chiba, M. J. Birrer, R. K. Rosenberg, M. Vinocour, M. Levitt, H. Pass, A. F. Gazdar, and J. D. Minna. 1989. p53: a frequent target for genetic abnormalities in lung cancer. Science 246:491-494. [DOI] [PubMed] [Google Scholar]
- 38.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the δ-Domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 39.Van der Geer, P., and T. Hunter. 1994. Phosphopeptide mapping and phosphoamino acid analysis by electrophoresis and chromatography on thin-layer cellulose plates. Electrophoresis 15:544-554. [DOI] [PubMed] [Google Scholar]
- 40.Vojtesek, B., H. Dolezalova, L. Lauerova, M. Svitakova, P. Havlis, J. Kovarik, C. A. Midgley, and D. P. Lane. 1995. Conformational changes in p53 analyzed using new antibodies to the core DNA binding domain of the protein. Oncogene 10:389-393. [PubMed] [Google Scholar]
- 41.Waseem, N. H., and D. P. Lane. 1990. Monoclonal antibody analysis of the proliferating cell nuclear antigen (PCNA)—structural conservation and the detection of a nucleolar form. J. Cell Sci. 96:121-129. [DOI] [PubMed] [Google Scholar]
- 42.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell. Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]