Abstract
The runt family transcription factor core-binding factor α1 (Cbfa1) is essential for bone formation during development. Surprisingly, transgenic mice overexpressing Cbfa1 under the control of the 2.3-kb collagen type I promoter developed severe osteopenia that increased progressively with age and presented multiple fractures. Analysis of skeletally mature transgenic mice showed that osteoblast maturation was affected and that specifically in cortical bone, bone resorption as well as bone formation was increased, inducing high bone turnover rates and a decreased degree of mineralization. To understand the origin of the increased bone resorption, we developed bone marrow stromal cell cultures and reciprocal coculture of primary osteoblasts and spleen cells from wild-type or transgenic mice. We showed that transgenic cells of the osteoblastic lineage induced an increased number of tartrate-resistant acid phosphatase-positive multinucleated cells, suggesting that primary osteoblasts as well as bone marrow stromal cells from transgenic mice have stronger osteoclastogenic properties than cells derived from wild-type animals. We investigated the candidate genes whose altered expression could trigger this increase in bone resorption, and we found that the expression of receptor activator of NF-κB ligand (RANKL) and collagenase 3, two factors involved in bone formation-resorption coupling, was markedly increased in transgenic cells. Our data thus suggest that overexpression of Cbfa1 in cells of the osteoblastic lineage does not necessarily induce a substantial increase in bone formation in the adult skeleton but has a positive effect on osteoclast differentiation in vitro and can also dramatically enhance bone resorption in vivo, possibly through increased RANKL expression.
In the adult skeleton, bone integrity is maintained through bone remodeling, which consists of bone resorption, carried out by osteoclasts, followed by bone formation, performed by osteoblasts. These two processes are in balance to maintain a constant bone volume. Both osteoblasts and osteoclasts are derived from progenitor cells originating from the bone marrow. Osteoclast differentiation is in part triggered by factors expressed by the osteoblasts such as osteoprotegerin (OPG) and RANKL, also called osteoclast differentiation factor or TRANCE. OPG, a secreted protein, belongs to the tumor necrosis factor receptor family, and RANKL, an osteoclast-differentiating factor, is part of the tumor necrosis factor ligand family. Both are produced by cells of the osteoblastic lineage and have been identified as playing an important role in osteoclastogenesis. RANKL, together with macrophage colony-stimulating factor (M-CSF), can induce osteoclast-like cell differentiation in vitro. Under physiological conditions, RANKL binds to its receptor, RANK, which is expressed on the cell surface of osteoclast precursor cells from the hematopoietic lineage. The RANK/RANKL interaction induces osteoclast differentiation, and OPG, through its binding to RANKL, inhibits this process (25, 45).
Core-binding factor α1 (Cbfa1), also known as AML-3, Pebp2αA, or more recently Runx2, is a transcription factor of the Runt domain gene family that is essential for bone formation during embryogenesis. This factor was identified as a transcriptional activator of osteoblast differentiation and has been proposed to be a master gene for bone (11, 20). Cbfa1 is expressed in mesenchymal condensations during early development at day 11.5 postconception (dpc) and acts as an osteoblastic differentiation factor (12). Recent studies showed that Cbfa1 is already expressed in mesenchymal stem cells prior to cell differentiation (41). Cbfa1 is crucial for osteoblast development and is also expressed in terminally differentiated osteoblasts, where it regulates expression of bone matrix proteins (4, 12, 40). In addition, Cbfa1 has also been shown to be expressed in prehypertrophic chondrocytes (14). In humans, Cbfa1 shows haploinsufficiency, as mutation of one allele has been found in patients affected with the skeletal disorder cleidocranial dysplasia (26, 31). A similar phenotype is observed in mice having a mutation in one allele of the gene (30, 34). Mice deficient for Cbfa1/Runx2 develop a cartilaginous skeleton (24, 34). Endochondral and intramembranous bone formation is nearly completely suppressed in these animals, and chondrocyte maturation is arrested before the stage of hypertrophy (18, 21). In these Cbfa1 knockout mice, forced expression of Cbfa1 driven by the collagen type II (Col1a2) promoter, which targets expression to chondrocytes, rescued differentiation into terminal hypertrophic chondrocytes (46), thus demonstrating that Cbfa1 is a positive regulator of terminal chondrocyte differentiation. Mice were also generated that express a dominant-negative form of Cbfa1: these animals were found to be transiently osteopenic, confirming the importance of Cbfa1 for postnatal bone formation as well (13).
Although it is well established that Cbfa1 is absolutely required for bone formation during development, the role of this factor in the adult skeleton and in the regulation of bone turnover is still not fully understood. To address this question, we have generated transgenic mice which overexpress Cbfa1 under the control of the rat collagen type 1 (Colla 1) promoter. Colla1 is an early marker of osteoblast commitment and the major protein in the bone extracellular matrix. The 2.3-kb fragment of the Colla1 promoter used here has been shown previously to direct transgene expression specifically to cells of the osteoblastic lineage, with a maximum level of expression in osteoblasts and odontoblasts (3, 5, 39).
In this report we describe the bone phenotype of these transgenic mice, which unexpectedly presented severe osteopenia. We show that in these mice, bone formation and resorption are elevated at some skeletal sites during adulthood and aging, with resorption exceeding formation. We also provide evidence that the maturation of osteoblasts is affected in these transgenic mice and that their osteoblasts have increased osteoclastogenic properties, suggesting that elevated Cbfa1 expression in osteoblastic cells can enhance bone resorption in vivo.
MATERIALS AND METHODS
Generation of Cbfa1/Runx2 transgenic mice.
A HindIII-XbaI 2.3-kb fragment of the rat collagen alpha 1 type I promoter was cloned in a pBluescript plasmid containing the Cbfa1/Runx2 cDNA. The Cbfa1 cDNA we used contains the two major translation start sites and thus encodes the type II (MASNSL) and type III (MLHSP) isoforms. At the 3′ end of the Cbfa1/Runx2 cDNA, a XbaI-SfiI-blunted fragment was introduced which contained the 3′ untranslated region of the rabbit β-globin gene, including intronic sequences and polyadenylation signal. An XbaI-XhoI 3-kb fragment of the matrix attachment region from the chicken lysozyme locus which was blunted at the ends was cloned upstream of the promoter in the SalI site of the plasmid. The whole construct was removed using the two BssHII sites of the vector. Transgenic mice were generated by injection of the DNA construct into pronuclei from C57BL/6 × BALB/c mice embryos. PCR amplification using primers 5′TCCTCCCAGCCCTCCTCCATCAG3′ and 5′CGCTGAAGAGGCTGTTTGACGGC3′ and Southern blot hybridization with a Cbfa1/Runx2 cDNA probe were used to identify mice positive for the presence of the transgene. Eight mice were found to be positive for the presence of the transgene, and founder males were mated to C57BL/6 × BALB/c females to establish the transgenic lines. The genotype of the F1 mice was determined by PCR amplification of genomic tail DNA as described above. F1 mice from six lines were investigated for their bone phenotypes. Heterozygote mice overexpressing Cbfa1/Runx2 under the control of the Colla1 promoter were compared to their wild-type littermates.
The animals were sacrificed under CO2 narcosis by decapitation. Prior to sacrifice, they received two fluorochrome labels by subcutaneous injection for evaluation of bone dynamics (on day 6 prior to necropsy, 20 mg of alizarin complexone [Merck, Zurich, Switzerland]/kg of body weight; on day 2 prior to necropsy, 30 mg of calcein/kg [Fluka, Buchs, Switzerland]).
Northern blot and RT (reverse transcription)-PCR Southern analysis.
Total RNAs were isolated using Trizol reagents (Life Technologies, Basel, Switzerland) according to the manufacturer's instructions. For RNA preparation of long bones, bones were cleaned of surrounding soft tissues. Epiphyses were cut off, and the bone marrow was flushed out with phosphate-buffered saline. cDNA preparation from 1 μg of total RNA [using oligo(dT) as primer] and PCR amplification were performed using standard protocols. For the PCR amplification, the following set of primers were used: for transgene, the upper and reverse primers 5′GCACTACCCAGCCACCTTTAC3′ and 5′GCCTGCACCTGAGGAGTGAATTC3′, respectively; for endogenous Cbfa1/Runx2, 5′AGCCTCTTCAGCCGAGTGACACC3′ and 5′CTGGGCCATGGTTGACGAATTTC3′; for Colla1, 5′ACCTTCCTGCGCCTAATGTC3′ and 5′TTGGGTTGTTCGTCTGTTTC3′; for osteopontin (OPN), 5′GAGCGGTGAGTCTAAGGAGT3′ and 5′CTAAATGCAAAGTAAGGAAC3′; for osteocalcin (OC), 5′AAGCAGGAGGGCAATAAGGT3′ and 5′AGCTGCTGTGACATCCCATAC3′; for bone sialoprotein (BSP), 5′AACGGCACCAGCACCAACTC3′ and 5′ACCCTCGTAGCCTTCATAGC3′; for matrix metalloproteinase 13 (MMP-13), 5′GCCACCTTCTTCTTGTTGAGCTG3′ and 5′ATCAAGGGATAGGGCTGGGTCAC3′; for alkaline phosphatase (ALP), 5′AGGCAGGATTGACCACGG3′ and 5′TGTAGTTCTGCTCATGGA3′; for tartrate-resistant acid phosphatase (TRAP), 5′TGACAAGAGGTTCCAGGA3′ and 5′AGCCAGGACAGCTGAGTG3′; for OPG, 5′CGAGGACCACAATGAACAAG3′ and 5′TCTCGGCATTCACTTTGGTC3′; for RANKL, 5′CAGAAGACAGCACTCACTGC3′ and 5′ATGGGAACCCGATGGGATGC3′; for RANK, 5′GAGGCATTATGAGCATCTCGG3′ and 5′TTTCTTTTGTCAGGTGCTTTTCAG3′; for M-CSF, 5′GCTTTGCTGAATGCTCCAGC3′ and 5′CAGAGGGACATTGGACAAACG3′; and for hypoxanthine-guanine phosphoribosyltransferase (HPRT), 5′AGCGATGATGAACCAGGTTA3′ and 5′GTTGAGAGATCATCTCCACC3′. The conditions of amplification were optimized for each pair of primers as follows: 30 s at 94°C and 30 s at 52°C (HPRT, TRAP, and ALP), 55°C (RANKL and OPG), 58°C (Cbfa1), 59°C (transgene, Col1a1, and MMP-13), or 60°C (RANK, M-CSF, OPN, OC, and BSP) and 30 s or 45 s at 72°C. For the genes, 20 cycles of amplification were used (with the exception of Cbfa1 [30 cycles]) for generation of the probes.
For semiquantitative analysis of mRNAs, the PCR products were blotted onto nylon membranes and hybridized, using as probes the corresponding amplified fragments that had been sequenced. Northern blot analysis was performed using standard protocols. The Northern blots were hybridized with PCR-amplified fragments generated for semiquantitative PCR. Quantification was performed using a Storm PhosphorImager.
Radiographs, dual-energy X-ray absorptiometry (DEXA), and peripheral quantitative computed tomography (pQCT).
Whole body radiographs in lateral and frontal positions (Siemens Mammomat [28 kV, 125 mA]; Siemens, Kloten, Switzerland) were taken for all animals after sacrifice. Tibial and femoral bone mineral content (in milligrams) and bone mineral density (in milligrams per square centimeter) were measured using a regular Hologic QDR-1000 instrument (Hologic, Waltham, Mass.). A collimator with 0.9-cm-diameter aperture and an ultrahigh resolution mode (line spacing, 0.0254 cm; resolution, 0.0127 cm) were used.
Following fixation in 70% alcohol, the excised long bones were placed on a resin platform provided by the company for soft tissue calibration. The left tibia and femur were measured in total and in the proximal quarter. Daily scanning of a phantom image controlled the stability of the measurements.
Cortical and cancellous bone mass and geometry were monitored in the proximal tibia metaphysis, 2.5 mm distal from the intercondylar tubercle and 3 mm proximal to the distal femur, using an adapted Stratec-Norland XCT-2000 (Stratec-Norland, Pforzheim, Germany) fitted with an Oxford 50 micro-X-ray tube (Oxford GTA6505 M/LA; Witney, Oxfordshire, United Kingdom) and a collimator with a 0.5-mm-diameter aperture. The bones were scanned in 70% ethanol. Daily scanning of a phantom image controlled the stability of the measurements. Instrument precision and reproducibility had been previously evaluated by calculating the coefficient of variation of repeated DEXA and pQCT measurements. Coefficients of variation were 0.5 to 2% for all evaluated parameters.
Histology and histomorphometry.
After dissection, the calvaria, left femur, tibia, and lumbar and tail vertebrae were placed for 24 h into Karnovsky's fixative, dehydrated in ethanol at 4°C, and embedded in Histodur (Leica, Nussloch, Germany).
The tail vertebra, tibia, and calvaria of a 16-week-old female were processed for further histomorphometric analysis. All parameters were measured and calculated in compliance with the recommendation of Parfitt et al. (35).
Using a Microtome 2050 Supercut (Reichert Jung, Arnsberg, Germany), a set of 4-, 8-, and 10-μm-thick nonconsecutive microtome sections were cut in the frontal midbody plane, and for evaluation of cellular, fluorochrome-label-based dynamic, and structural static parameters, the sections were examined using a Leica DM microscope (Leica, Heerbrugg, Switzerland) fitted with a camera (SONY DXC-950P, Tokyo, Japan) and adapted Quantimet 600 software (Leica, Cambridge, United Kingdom). Three sections per animal were sampled for all sets of parameters. The 4-μm-thick sections were stained with Movat's pentachrome stain, TRAP stain, or modified Giemsa stain for evaluation of bone turnover. Microscopic images of the specimens were digitalized and evaluated semiautomatically on screen (×400 magnification). Osteoclast number values (osteoclast number/bone perimeter [millimeters−1]) and perimeter values (osteoclast perimeter/bone perimeter [percent]) were determined on the TRAP-stained slides, while osteoblast number values (osteoblast number/bone perimeter [millimeters−1]) and perimeter values (osteoblast perimeter/bone perimeter [percent]) were measured on the Giemsa-stained sections. Osteocyte number values were also determined on these sections (osteocyte number/bone area [millimeters−2]).
Fluorochrome labels were evaluated semiautomatically, using the 8-μm-thick sections for determination of bone formation dynamics. Bone perimeter, single- and double-labeled bone perimeter, and interlabel width were measured (×400 magnification). Mineralized perimeter values (percent), mineral apposition rates (micrometers/day) (corrected for section obliquity in the cancellous bone compartment), and daily bone formation rate values (daily bone formation rate/bone perimeter [micrometers/day]) were calculated. All parameters were evaluated in the spongiosa (tail vertebrae, primary spongiosa; tibia, secondary spongiosa) and at the endocortex and the subperiosteal envelope. In addition, osteoclast, osteoblast, and osteocyte parameters were determined in the calvaria.
The mineralized bone at the 10-μm-thick-section surface was silver stained with modified von Kossa stain to facilitate the determination of static structural bone parameters. Images of the stained mineralized surface layer were grabbed and converted into binary images. Bone area per tissue area values (percent), trabecular width (micrometers), and trabecular number (millimeters−1) were determined at ×50 magnification in the primary and secondary spongiosa of the tail vertebrae and the proximal tibial metaphysis according to the plate model (34). Bone area fraction was also evaluated for the cortex of the tail vertebrae.
The tail vertebrae of an additional set of transgenic and wild-type animals (three per group) were decalcified (0.2 M EDTA in 1% paraformaldehyde-phosphate-buffered saline), dehydrated, and embedded in paraffin. A set of 4-μm-thick sections were cut for terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining (APOPDETECT Plus Peroxidase; Qbiogene, Carlsbad, Calif.) using methyl green counterstain.
Backscattered electron imaging.
Plane parallel block samples of embedded tail vertebrae were polished and carbon coated. Standardization was performed by employing halogenated dimethacrylate standards as described previously (6). Bone mineral density was evaluated by the calibrated standardized quantification of backscattered electron gray-level images from a scanning electron microscope (Zeiss DSM 962; Zeiss, Cambridge, United Kingdom) fitted with a four-segment annular solid state backscattered electron detector and interfaced with image analysis software. Whole tail vertebrae were scanned (magnification, ×50; voltage, 20 kV; working distance, 17 mm) to determine average mineralization levels (mean gray value [gray values scaled from 0 to 256]) and mineralization profiles (gray values divided into 16 regions = 16 levels of gray shading per section) (percent).
Osteoblastic cell cultures and cocultures.
Primary osteoblasts were isolated from calvaria of 2- to 4-day-old wild-type and transgenic mice. Calvaria were sequentially digested for 10 minutes in modified Eagle's medium type alpha (MEMα) containing 0.1% collagenase type V and 0.2% dispase. Cells were isolated from fractions 2 to 5 and combined as osteoblastic cell populations. Cells were expanded for 3 to 4 days in MEMα containing 10% fetal calf serum and replated at a density of 0.25 × 105 cells/cm2. After reaching confluency (i.e., after 6 to 8 days), the medium was supplemented with 50 μM ascorbic acid and 100 μM β-glycerophosphate and replaced every 2 to 3 days for 3 or 10 days. Bone marrow cells were collected aseptically from femur and tibia and cultured (2 × 106 cells/well) in 0.5 ml of MEMα containing 10% fetal calf serum, 50 μM ascorbic acid, and 10−8M dihydroxyvitamin D3 (1,25(OH) 2D3) in 24-well plates. Part of the medium (0.4 ml) was replaced every 2 to 3 days for 21 days. For coculture experiments, primary osteoblasts (2 × 104 cells/well) were plated with spleen cells (1 × 106cells/well) in 24-well plates as previously described (33, 44). Medium was changed every 3 days, and osteoclast differentiation was evaluated by TRAP staining (Sigma, Buchs, Switzerland).
Statistical analysis.
The results are expressed as means ± standard errors of the means (SEM). Statistical analysis was carried out using the t test (MicroSoft Excel 97). All statistical tests were two tailed and unpaired. The transgenic mice were tested for difference from the age-matched wild-type group at each evaluated skeletal site.
RESULTS
Transgenic mice overexpressing Cbfa1/Runx2 are osteopenic.
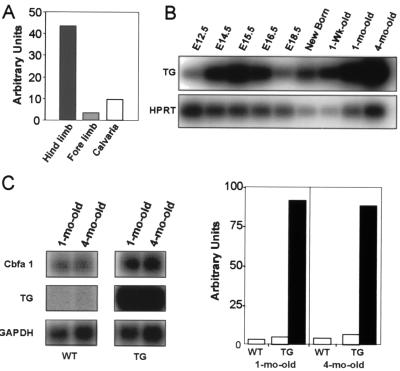
To achieve specific expression of the transgene in osteoblasts, we prepared a construct containing the murine full-length Cbfa1 cDNA fused to the polyadenylation signal of the rabbit β-globin gene and under control of the rat 2.3-kb collagen alpha 1 type I gene promoter. In addition, a 3-kb fragment containing the matrix attachment region of the chicken lysozyme locus was introduced upstream of the promoter. This sequence has previously been shown to ensure expression of a transgene independently from its insertion site (29). In adult mice, the transgene was expressed at high level in mineralized tissues (calvaria, long bones, and teeth). However, by using semiquantitative RT-PCR, we found that the transgene was not expressed at the same level throughout the skeleton. Indeed, in adult mice, expression of the transgene is higher in the hindlimbs than in the forelimbs and the calvaria (Fig. 1A). Hindlimb expression of the transgene starts at 12.5 dpc and shows maximal expression during embryogenesis at 15.5 dpc and in 4-month-old mice, when the peak of bone mass is normally reached in wild-type mice (Fig. 1B). In adult hindlimb bones, the level of expression of the transgene was found to be up to ca. 20 times higher than that of the endogenous Cbfa1 gene. In addition, in adult transgenic mice, expression of the endogenous Cbfa1 gene is ca. two times higher than that in wild-type littermates (Fig. 1C). This finding suggests that Cbfa1 regulates its own expression in a positive manner. This result is compatible with the presence of multiple Cbfa1 binding sites in the proximal promoter and the 5′ untranslated region of the endogenous gene (10, 13, 50), although recent reports examining Cbfa1 gene autoregulation gave discordant results (10, 13).
FIG. 1.
Generation of transgenic mice overexpressing Cbfa1/Runx2. (A) Quantification of the expression of the transgene in the hindlimbs, forelimbs, and calvaria. RNAs were extracted from tissues isolated from 4-month-old mice. The level of expression of the transgene was determined by RT-PCR, using HPRT as internal control. (B) RT-PCR analysis of the temporal expression of the transgene in transgenic mice. RNAs were extracted from long bones of mice at the indicated ages. In all cases, HPRT was used as internal control. E, embryonic day. (C) Northern blot analysis (left pair of panels) showing the levels of expression of the endogenous Cbfa1/Runx2 gene and the transgene in long bones from 4-month-old wild-type (WT) and transgenic (TG) mice. The quantification of the Cbfa1/Runx2 and the transgene expression is shown in the right pair of panels. The results were normalized to the expression level of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) used as internal control. Open bars represent the expression of endogenous Cbfa1/Runx2 in wild-type (WT) and transgenic (TG) mice; black bars represent the expression of the transgene.
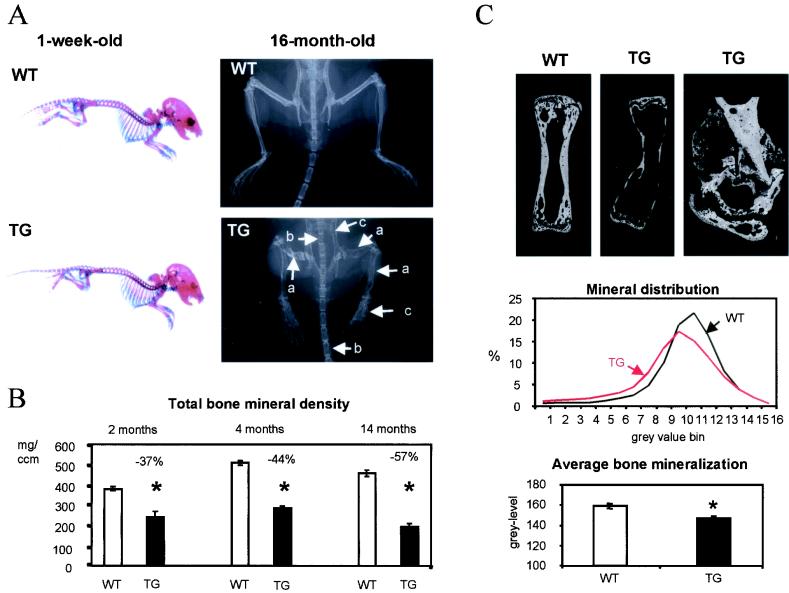
Four independent transgenic mouse lines were analyzed. By Northern blot analysis, three (lines 41, 43, and 44) were found to express the transgene at high levels and one (line 27) was found to express the transgene at a lower level. For each mouse line, the phenotype was independent of the site of observation (tibia or femur) and of the sex of the animal. The animals from line 27 exhibited normal sizes and weights and had a slightly reduced overall bone mass in their hindlimbs. Their total and cortical bone densities were slightly reduced (Table 1). Mice from the lines expressing the transgene at high level (lines 41, 43, and 44) showed a reduced size shortly after birth, which was reflected in most length measurements and in body weight. Bone mineral density in tibia and femur was substantially decreased in these three lines, as indicated by DEXA and pQCT measurements (Table 1). Interestingly, pQCT analysis indicated that differences in the bone phenotype were more dramatic in the cortical than in the cancellous bone compartment. Indeed, the total and cortical cross-sectional volumetric bone mineral densities as well as the bone area were markedly reduced (Fig. 2B) (Table 1), indicating not only a reduction in the amount of cortical bone but also a possible alteration of bone mineralization due either to a modification of material properties or to a high endocortical bone turnover. The cancellous bone compartment was, however, less affected, with only a mild decrease of cancellous bone mineral density and area in transgenic animals (Table 1). As a consequence of the drastic changes in bone density in the transgenic animals, fractures occurred at high frequency in the tail vertebrae, often in the hindlimbs, and occasionally in the ribs, as shown in the X-ray images (Fig. 2A). However, as indicated by Inouye's skeletal staining performed on specimens shortly after birth, the distribution of cartilage and bone appeared to be normal in the skeletons of transgenic animals (Fig. 2A).
TABLE 1.
Bone density and area in tibia from transgenic and wild-type female mice
| Bone parametera | Mouse line 27
|
Mouse line 44
|
||
|---|---|---|---|---|
| WT | TG | WT | TG | |
| BMD (mg/cm2) by DEXA | ||||
| Total | 74.8 ± 1.7 | 65.6 ± 3.5 | 80.4 ± 5.4 | 40.5 ± 5.8b |
| BMD (mg/cm3) by pQCT | ||||
| Cortical | 758.4 ± 13.1 | 707.7 ± 18.9 | 748.6 ± 6.7 | 532.9 ± 38.8b |
| Trabecular | 121.5 ± 5.9 | 95.0 ± 34.2 | 105.9 ± 5.0 | 101.8 ± 4.6 |
| Total | 513.0 ± 14.5 | 458.0 ± 45.0 | 574.5 ± 9.7 | 396.7 ± 25.5b |
| Area (mm2) | ||||
| Cortical | 1.67 ± 0.04 | 1.41 ± 0.11 | 2.03 ± 0.05 | 1.41 ± 0.21 |
| Trabecular | 0.98 ± 0.06 | 0.91 ± 0.06 | 0.76 ± 0.04 | 0.56 ± 0.15 |
| Total | 2.83 ± 0.08 | 2.51 ± 0.04 | 2.78 ± 0.08 | 2.06 ± 0.32 |
BMD, bone mineral density.
P < 0.05.
FIG. 2.
Radiological analysis and mineralization parameters of Cbfa1-overexpressing mice. (A) Skeletal preparation of 1-week-old wild-type (WT) or Cbfa1-overexpressing (TG) mice, followed by Inouye's staining (left panels) and X-ray analysis of 16-month-old wild-type and Cbfa1-overexpressing mice (right panels). At 1 week of age, Cbfa1-overexpressing mice exhibited normal distribution of bone and cartilage tissues, but at 16 months of age, Cbfa1-overexpressing mice exhibited smaller stature and dramatically lower cortical thickness and mineral density (white arrows), which induced multiple fractures in the appendicular (a) and central (b) skeleton and resulted in widened and/or misshapen bone structures (c). Arrows indicate different sites of fracture or bone structure alteration. (B) Total bone mineral density measured in proximal tibia from Cbfa1-overexpressing mice (TG) and their wild-type littermates (WT) at 2, 4, and 14 months of age. The phenotype did not improve with increasing age. (C) BSE images of tail vertebrae from 4-month-old Cbfa1-overexpressing mice (TG) (top middle and top right panels) and from a wild-type littermate (WT) (top left panel). The bone mineralization characteristics, as measured by quantitative BSE imaging, show that the bone mineralization profile (center panel) is shifted towards lower densities in Cbfa1-overexpressing mice (red line) compared to that of wild-type mice (black line) and that the average level of bone mineralization (bottom panel) is reduced in 4-month-old Cbfa1-overexpressing mice (black bar) compared to that of their wild-type littermates (open bar). Magnification of a vertebral fracture site in transgenic mice is presented in the top three panels. The fracture-healing process was normal in transgenic mice, as calcified cartilage was produced normally. Bars represent means ± SEM; statistically significant differences between wild-type and transgenic mice are represented by stars (P < 0.05 by Student 's t test [n = 5 for TG; n = 10 for WT]).
In addition, osteopenia, as evaluated by DEXA and pQCT, increased progressively in the hindlimbs of animals from 2 months of age (growth phase) to 4 months (peak bone mass) and 14 months (aging) (Fig. 2B). As judged from radiographs, decreased bone density was predominantly associated with the hindlimbs and the lumbar and tail vertebrae in 4-month-old animals. Osteopenia was observed at all sites in the skeleton by 16 months (Fig. 2A). These data indicate that no recovery of bone mass occurred over time and suggest that postnatally, bone abnormalities increased in relation to the high levels of transgene expression maintained during adult life.
Transgenic mice present high bone turnover rates, with bone resorption exceeding bone formation.
Histologic and histomorphometric analysis was performed on the tails and tibia of 4-month-old female mice from mouse line 44. The tail vertebrae were chosen as the site displaying the most dramatic bone phenotype at this age, and the proximal tibial metaphysis was chosen as a standard site for bone histomorphometry. In line with the noninvasive X-ray-based measurements, longitudinal sections of tibia and tail vertebrae from 4-month-old transgenic mice revealed a dramatic reduction of cortical thickness and, in the case of the vertebrae, a loss of the sparse secondary trabeculation (Fig. 2C and 3 and unpublished data for static histomorphometry). In spite of this, the fracture-healing capacity appeared to be normal (as shown in Fig. 2C), with normal production of calcified cartilage at the site of fractures.
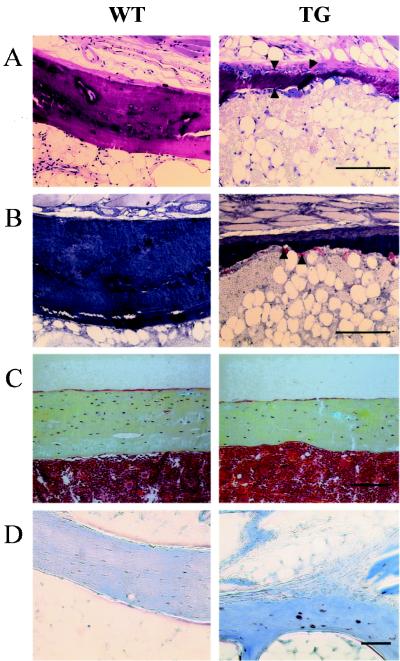
FIG. 3.
Histological analysis of Cbfa1-overexpressing mice. (A) Giemsa staining (bar, 50 μm) of the cortexes of tail vertebrae from a 4-month-old female Cbfa1-overexpressing mouse (TG) and a wild-type littermate (WT). The cortex of the Cbfa1-overexpressing mouse is dramatically thinner than that of the wild-type mouse. Numerous osteoblasts (arrowheads) were present on the cortex of the transgenic bone, especially at the subperiosteal location (top of the cortex). (B) TRAP staining (bar, 50 μm) of the cortexes in tail vertebrae from a 4-month-old female Cbfa1-overexpressing mouse (TG) and a wild-type littermate (WT). The cortex of the transgenic bone is dramatically excavated due to a high level of endocortical bone resorption activity. TRAP-positive osteoclasts are indicated by arrowheads. (C) Movat staining (bar, 50 μm) of longitudinal tibia sections of wild-type (WT) and Cbfa1-overexpressing (TG) mice. The cortex thickness and the number of osteocytes per unit area were markedly reduced in Cbfa1-overexpressing mice compared to their wild-type littermates. (D) Evaluation of osteocyte apoptosis by TUNEL immunostaining (bar, 10 μm) on decalcified vertebrae. More apoptotic cells (stained in brown) were observed in the transgenic vertebrae than in the WT bone.
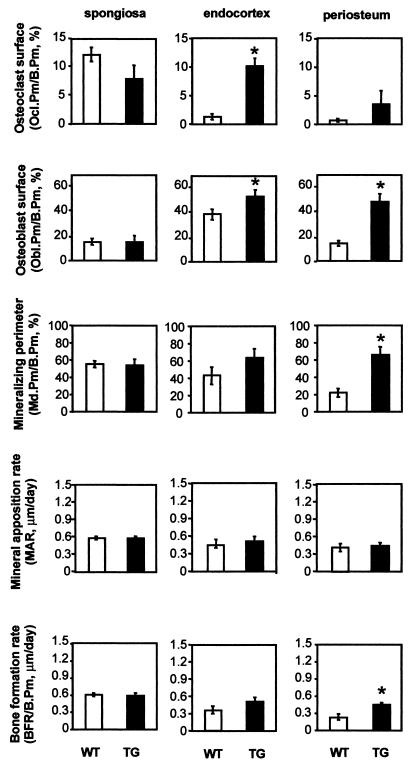
Depending on the bone compartment, bone formation levels as well as bone resorption levels were normal to high in transgenic mice. Normal bone formation and bone resorption occurred in the primary spongiosa of the tail vertebrae (Fig. 4) and in the secondary spongiosa of the tibia (unpublished data). In the cortex, the osteoblast perimeter was increased in the subperiosteal envelope of tail vertebrae of transgenic animals (three- to fourfold) (Fig. 4) as well as in the tibia, although to a smaller degree (about twofold). It was also slightly increased at the endocortex in both skeletal sites. This increase in subperiosteal osteoblast perimeter was also reflected by significant increases in mineralization perimeter and bone formation rate in the tail vertebrae (three- and twofold, respectively) (Fig. 5) and also occurred to some extent in the tibia (unpublished data). Bone resorption was also affected in the cortex. Osteoclast number and perimeter were dramatically increased at the endocortex of tail vertebrae and of the proximal tibial metaphysis of transgenic animals (about 7- to 8-fold and 10-fold, respectively) (Fig. 4). Bone resorption was also elevated in the subperiosteal envelope but to a smaller extent (about five- to sixfold and twofold, respectively) (Fig. 4). Interestingly, the high bone resorption levels observed were achieved independently of sites of high bone turnover due to fracture repair. As a consequence of this increased endocortical bone resorption activity, the cortical bones were in some cases dramatically excavated and the bone marrow cavities were sometimes expanded (Fig. 2A and C).
FIG. 4.
Histomorphometric bone formation and resorption parameters in Cbfa1-overexpressing mice. Parameters of bone formation and bone resorption as well as cell surface areas and numbers were measured in the spongiosa and at the endocortex and the periosteum of tail vertebrae in 4-month-old female Cbfa1-overexpressing mice (black bars, TG) and their wild-type littermates (open bars, WT). Ocl.Pm, osteoclast perimeter; Obl.Pm, osteoblast perimeter; B.Pm, bone perimeter; Md.Pm, mineralized perimeter. Bars represent means ± SEM; statistically significant differences between wild-type and transgenic mice are represented by stars (P < 0.05 by Student 's t test [n = 5 for TG; n = 10 for WT]).
FIG. 5.
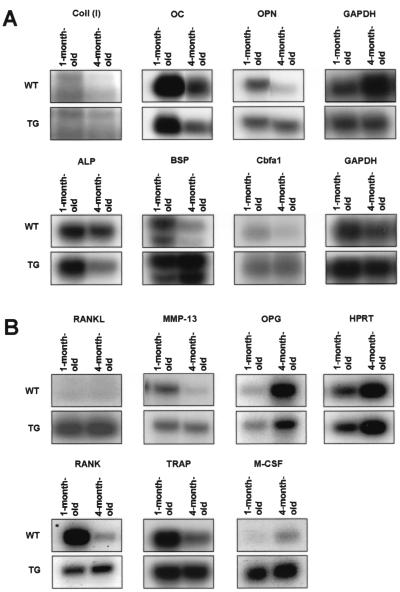
Gene expression in Cbfa1-overexpressing mice. Northern blot (A) or RT-PCR Southern blot (B) analysis of gene expression in long bones of 1- and 4-month-old Cbfa1-overexpressing mice (TG) and their wild-type littermates (WT). Expression of genes involved in bone formation (OPN, BSP, collagen alpha I type I [Coll (I)], Cbfa1/Runx2, OC, and ALP) and in bone resorption (MMP-13, TRAP, M-CSF, OPG, RANK, and RANKL) were analyzed in Cbfa1-overexpressing mice and in their wild-type littermates. RNAs were extracted from hindlimb long bones. GAPDH and HPRT were used as internal controls for Northern blot and RT-PCR Southern blot analysis, respectively.
Histomorphometric analysis of the calvaria of transgenic animals revealed no strong bone phenotype. Subperiosteal osteoblast surfaces were nonsignificantly reduced in size in transgenic animals and endosteal bone resorption was absent both in transgenic and wild-type mice.
Cortical bone modeling was severely disturbed in transgenic animals, while cancellous bone was nearly unaffected. Cortical bone modeling was characterized by expansion of the marrow cavity (due to endocortical bone resorption), which was insufficiently matched by enhanced subperiosteal bone formation, resulting in both an outward drift and substantial thinning of the cortex. Bone formation and bone resorption were unaffected in calvaria, suggesting that the high bone resorption levels observed in the long bones and tail vertebrae are dependent on the level of transgene expression.
Osteocyte number is reduced in transgenic animals irrespective of bone turnover status.
The number of osteocytic lacunae per unit volume of bone matrix was reduced in transgenic bone (Fig. 3C). In tail vertebrae, the osteocyte number per square millimeter was decreased by 50% (146 ± 33 versus 315 ± 27; P < 0.05). In tibia, a similar dramatic decrease in osteocyte number was observed in cortical bone (263 ± 27 versus 433 ± 24; P < 0.05) and to a smaller extent in the secondary spongiosa (322 ± 61 versus 450 ± 16; [P not significant]). These data suggest that the fate of the osteoblastic cells is modified in the transgenic bone. Furthermore, by TUNEL immunostaining, we observed a higher proportion of apoptotic osteocytes in cortical bone of the tail vertebrae of transgenic mice than in those of wild-type animals (Fig. 3D).
Interestingly, the only significant alteration observed in the calvaria of the transgenic mice was again a dramatic decrease in the number of osteocytes (79 ± 21 versus 493 ± 31; P < 0.05). This indicates that the observed decrease in osteocyte number per unit volume of bone matrix is a characteristic of these transgenic animals which is independent of the above-described alterations in bone formation and bone resorption and which occurs even at sites displaying a low level of transgene expression.
Matrix synthesis level per osteoblast is unchanged in transgenic animals, and matrix mineralization is reduced.
Bone matrix synthesis level per osteoblast was not increased at any bone envelope, as indicated by the normal matrix apposition rate found at all sites in both vertebrae and tibia of transgenic mice (Fig. 4 and unpublished data). An increase in bone mineralization surface size was observed in cortical bone compartments, in keeping with the observation of the increase in osteoblast surface size leading to an enhanced bone formation rate (Fig. 4). Evaluation of bone mineralization by quantitative back-scattered electron (BSE) imaging on block samples of tail vertebrae demonstrated that the average degree of bone mineralization was lowered in transgenic animals compared to that of their wild-type littermates (Fig. 2C). Newly formed bone is less mineralized and less dense than mature bone. In the BSE images, which are used for quantification, less-well-mineralized bone appears in darker shades of gray. Detection of a significant alteration in the average bone mineralization level reflects an alteration in the bone formation characteristics. The density profile represents the distribution of the different mineralization degrees in bone, which normally show a peak in the upper half of the density range (7). Drastic alterations of this profile are associated with pathological alterations of the mineralization pattern (7, 38). In the transgenic animals, no such drastic alteration of the profile had occurred. However, compared to the wild-type animals, a shift of the curve towards the less-mineralized regions was indeed observed, indicating an increased portion of recently formed bone (Fig. 2C) reminiscent of the bone mineralization profile resulting from long-term anabolic parathyroid hormone treatment (23). This is consistent with the reduced cortical volumetric density detected by pQCT (Table 1) and the high cortical bone turnover rates.
In adult transgenic mice, expression of bone formation marker genes is unaffected; however, that of osteoclastic marker genes is increased.
As shown by histomorphometric data, the number of osteoblasts, as well as the number of osteoclasts, is increased in the cortical bone of transgenic mice. Moreover, the amount of matrix secreted and mineralized per osteoblast (i.e., the mineral apposition rate) (Fig. 4) was not significantly modified in transgenic mice, suggesting that overexpression of Cbfa1 has no effect or only a mild effect on osteoblastic function in this mouse model. The organization of the matrix appeared lamellar under polarized light at most skeletal sites; however, at sites of fracture or of newly formed bone, woven bone was observed (unpublished data).
In order to evaluate which molecular disorder(s) was underlying the unexpected phenotype of these mice, we analyzed the level of expression of the major osteoblastic and osteoclastic marker genes in hindlimb bone RNA. In the growing skeleton (1-month-old animals), expression of most of the analyzed genes was close to normal (Fig. 5A and B). Nevertheless, that of OC, a marker of differentiated osteoblasts, was slightly decreased, indicating that osteoblastic maturation is probably affected in young transgenic mice (see also Liu et al. [27]). In addition, expression of RANKL and M-CSF was elevated and RANK expression was reduced. In 4-month-old transgenic mice, the age at which the animals had reached their peak level of bone mass, the expression of the genes encoding the extracellular matrix proteins (Colla1, OC, OPN, BSP, and ALP) was more affected. Except for OC and ALP, the expression of which slightly decreased, that of all the genes analyzed increased two- to threefold (Fig. 5A). In addition, the expression of collagenase 3, a matrix metalloproteinase that has been shown to be positively regulated by Cbfa1 (19, 49), also increased three- to fivefold in adult transgenic mice compared to that in wild-type mice (Fig. 5B). This increase of bone marker gene expression in hindlimb bone probably reflects the increase in osteoblast number observed in the bone cortex and not an increase in osteoblast performance. These data are in agreement with the dynamic parameters presented in Fig. 4. Furthermore, the decrease in OC expression in 4-month-old animals compared to that already observed in 1-month-old mice indicates that osteoblastic cell maturation was not restored in the mature skeleton.
The histomorphometric data showed that bone resorption exceeded bone formation. As expected, expression of osteoclastic genes in long bones also increased: TRAP and RANK mRNA levels increased fivefold (Fig. 5B).
In addition, the expression of OPG and RANKL, two molecules produced by cells of the osteoblastic lineage that play an important role in osteoclastogenesis, was found to be altered in hindlimbs of adult transgenic mice: that of OPG was reduced, whereas that of RANKL and M-CSF was upregulated (Fig. 5B). This expression pattern was also observed in bone marrow stromal cells and primary osteoblasts derived from transgenic newborn mouse calvaria (Fig. 6 and 7), suggesting that these cells have higher intrinsic osteoclastogenic activity than their wild-type counterparts (see below).
FIG. 6.
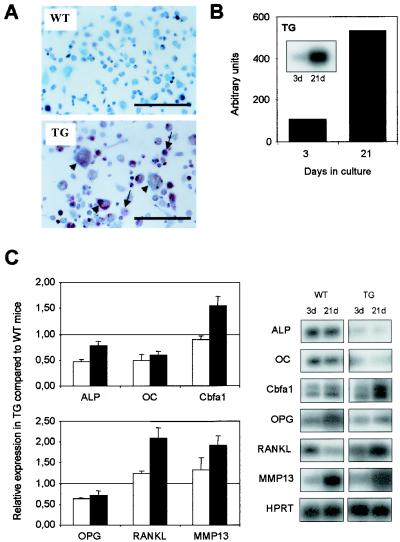
In vitro analysis of bone marrow stromal cells from Cbfa1-overexpressing mice. (A) Bone marrow cells from wild-type (WT) or Cbfa1-overexpressing (TG) mice were cultured in the presence of dihydroxyvitamin D3 (10−8 M) and ascorbic acid (50 μM) to induce osteoclastogenesis. After 21 days in culture, cells were stained for TRAP activity and counterstained with methyl green to visualize the nuclei. Arrows and arrowheads represent mononucleated and multinucleated TRAP-positive cells, respectively. Bar, 50 μm. (B) RT-PCR analysis of the expression of the transgene in bone marrow cells after 3 or 21 days in culture. The results were normalized to the expression level of HPRT used as internal control. (C) Expression of gene markers of osteoblasts (Cbfa1, OC, and ALP) and of genes involved in osteoclastogenesis (MMP-13, OPG, and RANKL) was analyzed by RT-PCR Southern blotting in bone marrow stromal cells from wild-type (WT) and transgenic (TG) mice after 3 (open bars) or 21 days (black bars) in culture (pairs of panels on right). Values were normalized against HPRT expression and are presented as severalfold induction in the transgenic cells relative to that in the wild-type cells. (Left panels; results represent means ± SEM of two independent experiments).
FIG. 7.
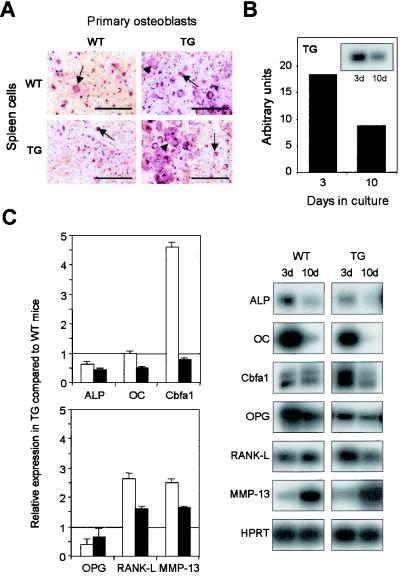
In vitro analysis of primary osteoblasts from Cbfa1-overexpressing mice (TG). (A) Primary osteoblasts prepared from calvaria from 2- to 4-day-old mice and spleen cells from wild-type (WT) or Cbfa1-overexpressing (TG) mice were cocultured in the presence of dihydroxyvitamin D3 (10−8M) and ascorbic acid (50 μM) to induce osteoclastogenesis. After 8 days, cells were stained for TRAP activity. Arrows and arrowheads represent mononucleated and multinucleated TRAP-positive cells, respectively. Bar, 50 μm. (B) RT-PCR analysis of the expression of the transgene in primary osteoblasts isolated from calvaria after 3 or 10 days of culturing in presence of ascorbic acid (50 μM). The results were normalized to the expression level of HPRT used as internal control. (C) Analysis of the expression of osteoblastic marker genes (Cbfa1, OC, and ALP) and of genes involved in osteoclastogenesis (MMP-13, OPG, and RANKL) by RT-PCR Southern blotting with primary osteoblasts from wild-type (WT) and transgenic (TG) 2- to 4 day-old mice after 3 days (open bars) or 10 days (black bars) in culture (pairs of panels on right). Values were normalized against HPRT expression and are presented as fold induction in the transgenic cells relative to that in the wild-type cells (left panels; results represent means ± SEM of two independent experiments).
Osteoblasts from transgenic mice have increased osteoclastogenic properties in vitro.
We examined whether the cells of the osteoblastic lineage isolated from transgenic mice had an increased ability to form TRAP-positive bone-resorbing cells in vitro. For this examination, we developed bone marrow stromal cell culture and reciprocal coculture of primary osteoblasts and spleen cells from wild-type and transgenic mice. We first performed bone marrow stromal cell cultures in the presence of ascorbic acid and vitamin D3. Under these conditions, we observed an increased number of TRAP-positive multinucleated cells with cells of transgenic genotype compared to those observed with wild-type cells, suggesting that bone marrow stromal cells from transgenic mice have stronger osteoclastogenic properties than cells derived from wild-type animals (Fig. 6A). In addition, to determine whether Cbfa1 expression correlates with the increase of TRAP-positive multinucleated cells, the levels of expression of the transgene and the endogenous Cbfa1 were analyzed by semiquantitative RT-PCR. We also examined, in a time course experiment, the expression of osteoblastic markers (OC and ALP) and of factors whose expression was shown to be altered in the bone of transgenic mice (OPG, RANKL, and MMP13).
In wild-type as well as in transgenic cells, expression of most of the osteoblastic marker genes increased over time in culture, reflecting the differentiation of the cells (Fig. 6B and C). However, the expression of RANKL was markedly decreased in wild-type cells but not in transgenic cells. This observation for wild-type cells is in agreement with previous results with human stromal cells, showing that during osteoblastic differentiation, RANKL expression is decreased whereas OPG expression is increased (16). Interestingly, in transgenic cells, the expression levels of OC, a marker of mature osteoblast, and of ALP were lower than in wild-type cells, suggesting an intrinsic maturation defect in the transgenic stromal cells. In addition, after 21 days of culture, the OPG RNA level was lower in transgenic cells than in the wild-type cells, whereas the RANKL and MMP-13 RNA levels were higher, as was initially observed in transgenic bone; this suggests that expression of the transgene influences the regulation of OPG, RANKL, and MMP-13.
In order to exclude an increased ability of the osteoblastic progenitors to differentiate, we performed coculture experiments with primary osteoblasts isolated from two- to four-day-old mice calvaria and spleen cells from wild-type or transgenic mice (Fig. 7A). Wild-type primary osteoblasts, when cultured in the presence of spleen cells from wild-type or transgenic mice, supported the differentiation of TRAP-positive cells, with only a few of the cells becoming multinucleated. In contrast, transgenic osteoblasts cultured in the presence of splenic cells (irrespective of their origin) triggered a marked increase in the number of TRAP-positive osteoclasts, including multinucleated cells (Fig. 7A). This indicates that Cbfa1 overexpression in osteoblasts enhances osteoclastogenesis ex vivo.
We also analyzed, in the primary osteoblasts used to induce the osteoclastic differentiation, the expression of the transgene (Fig. 7B) and of the genes involved in the osteoclastogenesis or of markers of osteoblastic maturation (Fig. 7C). As shown for rat and human primary osteoblasts in culture (36, 43), after the phase of growth and proliferation (day 0 of treatment), the osteoblastic markers are expressed at their maximal levels. After 3 days of culture, cells (irrespective of their origin) were still expressing the markers, including OC, ALP, and Cbfa1, at high levels. However, after 10 days in culture, expression of most of the genes decreased, with the exception of that of MMP-13, which increased. Nevertheless, the relative expression levels of RANKL and MMP-13 were higher in transgenic cells than in wild-type cells and that of OPG RNA was lower, as was also observed for the bone marrow cells.
Taken together, these data indicate that the elevated number of osteoclasts found in transgenic bones results from augmented osteoclastogenic activity of the transgenic osteoblasts, possibly reflecting an altered function of stromal cells, and not from an increased ability of transgenic osteoclast precursors to differentiate.
DISCUSSION
Here we have investigated the role of the transcription factor Cbfa1 for the maintenance of the adult bone by generating transgenic mice overexpressing this transcription factor under the control of the Colla1 promoter. Together, our data indicate that Cbfa1 controls not only genes that are important for osteoblast differentiation (12) and function (13) but also genes that are involved in osteoclast differentiation and bone formation-resorption coupling. In the light of these novel findings, Cbfa1 appears to be at the center of the regulatory circuits controlling bone formation and homeostasis.
Cbfa1 overexpression induces progressive osteopenia and high cortical bone turnover during adulthood and aging.
To our surprise, bone mineral density was decreased in the hindlimbs of the transgenic lines and the severity of the phenotype was directly correlated to the level of Cbfa1 expression. Multiple fractures had occurred in the tail vertebrae, hindlimbs, and occasionally in the ribs in these transgenic animals. The consistency of the observed bone phenotype in several independent lines indicates that it was not related to the site of insertion of the transgene. No recovery of bone mass occurred from 2 to 4 months of age, when the animals reach peak levels of bone mass (2). Osteopenia further progressed during aging, as monitored in mice up to the age of 14 months. This indicates that the alterations observed in the adult transgenic mice were most likely a consequence of the high level of transgene expression during adulthood.
Interestingly, pQCT measurements indicated that the bone phenotype was more pronounced in the cortical bone compartment than in cancellous bone. Both the amount of cortical bone and its volumetric density were reduced, while cancellous bone was less affected. Detailed histologic and morphometric analysis showed that bone formation levels as well as bone resorption levels were elevated. Cortical bone modeling was severely disturbed and characterized by expansion of the marrow cavity. This phenotype was most distinct in the tail vertebrae but was also observed in the cortical bone compartment of the tibial metaphysis. Bone resorption exceeded bone formation in the affected envelopes, resulting in the osteopenia described above. Evaluation of bone mineralization demonstrated that the average degree of bone mineralization was lowered in transgenic animals, which was likely due to the elevated bone turnover and modeling rates, with bone being removed before it reached its full maturation. This is reminiscent of the bone mineralization profile resulting from long-term bone anabolic parathyroid hormone treatment, where the large amount of newly formed bone is maintained at high bone turnover rates (23). However, drastic alterations of the mineralization profile, which are usually associated with pathological alterations of the mineralization pattern (7, 38), were not observed here. The osteopenic bone phenotype described here presents some similarities to several bone abnormalities characterized by increased bone turnover, which include estrogen deficiency, hyperparathyroidism, and Paget's disease. In cases involving abnormalities such as these, an increase in osteoclast number and activity relative to osteoblasts has been observed, affecting different sites throughout the skeleton and leading to multiple fractures. In addition, we also note that recent reports indicate that Cbfa1 is a potential regulator in the parathyroid hormone induction of bone resorption (37, 42).
While we were preparing this manuscript, a report of a similar mouse model was published by Liu et al. (27). These authors also generated mice overexpressing Cbfa1 in bone and described a phenotype similar to that presented here, namely, severe osteopenia associated with multiple fractures and decrease of bone mineral density due to reduction of the cortical bone, with trabecular bone nearly unaffected. However, although not entirely distinct, findings regarding the histomorphometric parameters and expression of bone markers were somewhat different in the two models. The mice described by Liu et al. showed decreases in bone formation rate, matrix apposition rate, and mineralized surface area in trabecular bone as well as in cortical bone compared to those of the control mice. In our case, we found that mice with a mature skeleton display bone formation rates comparable to those of their wild-type littermates at most skeletal sites and can even partly compensate for the dramatic endocortical bone excavation due to increased resorption activity by enhanced bone formation. This is, however, not related to an enhanced matrix synthesis per osteoblast but is due to a rise in their number. We believe that the differences between these two mouse models can be largely explained by the fact that we have analyzed fully mature or even aging mice, while Liu et al. mostly examined animals in which the skeleton was still developing. In addition, the regulatory cassettes used in the two studies for expression of the Cbfa1 cDNA were not identical, so the precise levels, timings, and locations of CBFA1 expression may be marginally different. Because of these facts, some aspects of the phenotypes were unique to one or the other model. In particular, the crucial effect of Cbfa1 on osteoclastogenesis was not seen by Liu et al. (27).
Osteoblastic cell maturation is affected in transgenic bone.
Molecular analysis of the adult bone in transgenic animals showed a markedly decreased expression of ALP, an early marker of the cells of the osteoblastic lineage, and OC, a marker of mature osteoblasts. These data suggest that osteoblast maturation is reduced in transgenic mice when the peak bone mass is reached; however, it is not abolished, since an increase in bone formation rate is still observed in these animals at the subperiosteal envelope. The overexpression of Cbfa1 probably enhances osteoblastic differentiation of the mesenchymal progenitors but cannot support complete maturation of the osteoblastic cells into mature osteoblasts. As described above, we also observed a dramatic decrease in osteocyte number at all investigated skeletal sites—namely, those of the vertebrae, long bones, and calvaria—and hence in endochondral, membranous, and intramembranous bone. The decreased osteocyte number per matrix volume observed in these transgenic mice cannot be the trigger for enhanced removal of the bone matrix by increased osteoclastic activity, since in the calvaria, osteocyte number was dramatically reduced while resorption was unchanged. Most likely, the observed decrease in osteocyte number may be the result of the impaired maturation of the osteoblasts. Furthermore, a higher proportion of apoptotic osteocytes was detected by TUNEL immunostaining in the cortical bone of the tail vertebrae of transgenic mice than in that of wild-type animals, suggesting that the few cells that commit to the osteocytic fate have decreased viability.
Cbfa1 is involved in the bone resorption process.
The observed increase in bone resorption was unexpected, particularly because Cbfa1 has been widely assumed to promote bone formation (13). Importantly, no evidence was found in our study that Cbfa1 overexpression leads to an increase in bone matrix synthesis per osteoblast at any bone envelope. Local increase in bone formation was related to a rise in the number of bone-forming cells and not to a change in their performance. This contrasts with an observation made for mice overexpressing a dominant-negative mutant of Cbfa1 (13) under the control of the OC promoter. In this case, Cbfa1 removal (by the action of the dominant-negative protein) was reported to have a negative impact on the level of matrix synthesis per osteoblast, while osteoblast number was unaffected. A likely explanation for the observed differences lies in the choice of the promoter, which determines the time point and cell specificity of transgene expression. While Colla1 is a very early marker of osteoblast commitment expressed in most cells of the osteoblastic lineage, including immature progenitors and bone marrow stromal cells (9), OC is a very late marker of that expressed in fully mature cells (1).
The potential role of Cbfa1 in osteoclastogenesis is so far unclear, but an involvement of this transcription factor in this process has been suggested: in contrast to what is seen in wild-type osteoclasts, in Cbfa1-deficient osteoclasts, expression of RANKL is not increased following treatment with vitamin D3 and dexamethasone, suggesting that Cbfa1 is important for RANKL expression (15). In agreement with this, it has been shown that two putative Cbfa1 binding sites are present in the promoter region of the RANKL gene (reference 22 and C. A. O'Brien, N. Farrar, and S. C. Manolagas, Abstr. ASBMR-IBMS 2nd Joint Meet., abstr. 1003, 1998). However, recent results from transfection experiments have failed to demonstrate a role for these sites in the regulation of the RANKL gene (32). Nevertheless, the elevated RANKL expression observed here (Fig. 5B and 6), as well as the greatly reduced number of osteoclasts present in Cbfa1 null mice (24), indicates that in vivo Cbfa1 is involved, perhaps indirectly, in regulating the RANKL gene. Clearly, additional studies will be required to fully define the relationship between Cbfa1 and RANKL expression.
Recent in vitro studies provided evidence that Cbfa1 contributes to the expression of OPG, a potent inhibitor of osteoclast differentiation and function (47, 48), indicating a potential role for Cbfa1 in the inhibition of bone resorption. Nevertheless, in our transgenic mice we found no increase in OPG expression but found rather that bone resorption was elevated. Previous reports established a link between the levels of RANKL and OPG expression. In particular, the RANKL/OPG ratio seems to correlate with the differentiation of osteoclastic-like cells in vitro (16, 17). It was shown that a high RANKL/OPG ratio is required for the initiation and support of osteoclastogenesis by undifferentiated bone marrow stromal cells and that mature osteoblasts cannot support osteoclastogenesis in vitro. Here, Cbfa1 overexpression led to downregulation of OPG and to activation of RANKL expression in cells of the osteoblastic lineage. We showed that the alteration of RANKL and OPG expression, leading to an increase in RANKL expression in comparison to that of OPG, correlates with the appearance of TRAP-positive multinucleated cells in bone marrow stromal cell cultures and in coculture of primary osteoblasts with splenic cells. These data are in agreement with the observation of Gori et al. (16), who found that osteoclastogenesis is supported by undifferentiated osteoblast cells and observed that RANKL expression decreases during the course of differentiation, while that of OPG, a biological competitor of RANKL, increases. In our transgenic mice, Cbfa1 expression is driven by a 2.3-kb Colla1 promoter which has recently been shown to become more active concomitant with the expression of BSP and before that of OC. This corresponds to the transition from preosteoblasts to osteoblasts (9). Thus, it is possible that this putative increase in specific Cbfa1 expression in differentiated osteoblasts ensures a sustained expression of RANKL, which would explain, at least in part, the observed increase in bone resorption.
As shown in ex vivo culture experiments, Cbfa1 transgenic cells of the osteoblastic lineage have an increased ability to support osteoclastic differentiation. This is in agreement with the histomorphometric analysis of adult transgenic bones. Hence, our in vivo results extend the reports by Horwood et al. (17) and Gori et al. (16) and suggest that an increase in Cbfa1 expression at an early stage of osteoblast commitment can induce osteoclast recruitment via an increase in RANKL expression and a decrease in OPG expression.
Another observation that could explain the elevated bone resorption observed in these transgenic mice is the increased level of collagenase 3 (MMP-13) expression. Several reports have presented data indicating that expression of osteoblastic matrix metalloproteinases, including MMP-13, is a critical step in the initiation of the resorptive process through its effect on the migration of the osteoclasts through the osteoid (19). Together with the data presented here, this suggests that by facilitating access of osteoclasts to resorbing sites, Cbfa1 is involved in osteoclast differentiation and also (indirectly) in osteoclast function.
As discussed above, the increased number of immature osteoblasts, cells that have been shown earlier to support osteoclastogenesis (28), is probably at the origin of the hyperresorption observed in these mice. This hypothesis is also supported by a previous report showing normal bone resorption after the complete depletion of fully mature osteoblasts (8).
In summary, our study provides evidence that Cbfa1 has a more complex role in bone maintenance and bone turnover than initially expected. In particular, the high bone turnover rates observed in the transgenic animals described here indicate that Cbfa1 is not only involved in bone remodeling and modeling by enhancing bone formation by mature osteoblasts but also in the regulation of bone resorption. In answer to our initial question, namely, that of whether Cbfa1 overexpression is able to trigger additional bone formation in a skeletally mature animal, we have to conclude that high expression of Cbfa1 in cells of the osteoblastic lineage does not necessarily induce substantial increases in bone formation in the adult skeleton but can enhance dramatically bone resorption, probably through elevated levels of RANKL expression. Furthermore, this mouse model of the overexpression of the transcription factor Cbfa1 in bone is the first to demonstrate cross talk between bone formation and bone resorption in animals with a mature skeleton.
Acknowledgments
We thank Alexander Lichtler, University of Connecticut Health Center, Farmington, Conn., for providing the rat Colla1 promoter and Jérome Rossert, hôpital Tenon, Paris, France, for providing the matrix attachment region. H.-J. Keller, Bone Metabolism Unit, Novartis Pharma, Basel, Switzerland, generated the Cbfa1/Runx2 cDNA. We also thank J.-F. Spetz for generating the transgenic mice and A. Studer, R. Cortesi, and M. Bruederlin for their excellent technical assistance. V.G. thanks M.-C. de Vernejoul for critical reading of the manuscript.
This work was supported by a Novartis fellowship to V.G.
REFERENCES
- 1.Aubin, J. E. 1998. Bone stem cells. J. Cell. Biochem. 31:73-82. [DOI] [PubMed] [Google Scholar]
- 2.Beamer, W. G., L. R. Donahue, C. J. Rosen, and D. J. Baylink. 1996. Genetic variability in adult bone density among inbred strains of mice. Bone 18:397-403. [DOI] [PubMed] [Google Scholar]
- 3.Bedalov, A., R. Salvatori, M. Dodig, M. S. Kronenberg, B. Kapural, Z. Bogdanovic, B. E. Kream, C. O. Woody, S. H. Clark, K. Mack, D. W. Rowe, and A. C. Lichtler. 1995. Regulation of COL1A1 expression in type I collagen-producing tissues: identification of a 49 base pair region which is required for transgene expression in bone of transgenic mice. J. Bone Miner. Res. 10:1443-1451. [DOI] [PubMed] [Google Scholar]
- 4.Benson, M. D., J. E. Aubin, G. Xiao, P. E. Thomas, and R. T. Franceschi. 1999. Cloning of a 2.5 kb murine bone sialoprotein promoter fragment and functional analysis of putative Osf2 binding sites. J. Bone Miner. Res. 14:396-405. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanovic, Z., A. Bedalov, P. H. Krebsbach, D. Pavlin, C. O. Woody, S. H. Clark, H. F. Thomas, D. W. Rowe, B. E. Kream, and A. C. Lichtler. 1994. Upstream regulatory elements necessary for expression of the rat COL1A1 promoter in transgenic mice. J. Bone Miner. Res. 9:285-292. [DOI] [PubMed] [Google Scholar]
- 6.Boyde, A., S. J. Jones, J. Aerssens, and J. Dequeker. 1995. Mineral density quantification of the human cortical iliac crest by backscattered electron image analysis: variations with age, sex, and degree of osteoarthritis. Bone 16:619-627. [DOI] [PubMed] [Google Scholar]
- 7.Boyde, A., R. Travers, F. H. Glorieux, and S. J. Jones. 1999. The mineralisation density of iliac crest bone from children with osteogenesis imperfecta. Calcif. Tissue Int. 64:185-190. [DOI] [PubMed] [Google Scholar]
- 8.Corral, D. A., M. Amling, M. Priemel, E. Loyer, S. Fuchs, P. Ducy, R. Baron, and G. Karsenty. 1998. Dissociation between bone resorption and bone formation in osteopenic transgenic mice. Proc. Natl. Acad. Sci. USA 95:13835-13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dacic, S., I. Kalajzic, D. Visnjic, A. C. Lichtler, and D. W. Rowe. 2001. Col1a1-driven transgenic markers of osteoblast lineage progression. J. Bone Miner. Res. 16:1228-1236. [DOI] [PubMed] [Google Scholar]
- 10.Drissi, H., Q. Luc, R. Shakoori, S. Chuva de Sousa Lopez, J.-Y. Choi, A. Terry, M. Hu, S. Jones, J. C. Neil, J. B. Lian, J. L. Stein, A. J. Van Wijnen, and G. S. Stein. 2000. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell. Physiol. 184:341-350. [DOI] [PubMed] [Google Scholar]
- 11.Ducy, P. 2000. Cbfa1: a molecular switch in osteoblast biology. Dev. Dyn. 219:461-471. [DOI] [PubMed] [Google Scholar]
- 12.Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. [DOI] [PubMed] [Google Scholar]
- 13.Ducy, P., M. Starbuck, M. Priemel, J. Shen, G. Pinero, V. Geoffroy, M. Amling, and G. Karsenty. 1999. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 13:1025-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enomoto, H., M. Enomoto-Iwamoto, M. Iwamoto, S. Nomura, M. Himeno, Y. Kitamura, T. Kishimoto, and T. Komori. 2000. Cbfa1 is a positive regulatory factor in chondrocyte maturation. J. Biol. Chem. 275:8695-8702. [DOI] [PubMed] [Google Scholar]
- 15.Gao, Y. H., T. Shinki, T. Yuasa, H. Kataoka-Enomoto, T. Komori, T. Suda, and A. Yamaguchi. 1998. Potential role of Cbfa1, an essential transcriptional factor for osteoblast differentiation, in osteoclastogenesis: regulation of mRNA expression of osteoclast differentiation factor (ODF). Biochem. Biophys. Res. Commun. 252:697-702. [DOI] [PubMed] [Google Scholar]
- 16.Gori, F., L. C. Hofbauer, C. R. Dunstan, T. C. Spelsberg, S. Khosla, and B. L. Riggs. 2000. The expression of osteoprotegerin and RANK Ligand and the support of osteoclast formation by stromal-osteoblast lineage cells is developmentally regulated. Endocrinology 141:4768-4776. [DOI] [PubMed] [Google Scholar]
- 17.Horwood, N. J., J. Elliott, J. Martin, and M. T. Gillespie. 1998. Osteotropic agents that regulate the expression of osteoclast differentiation factor and osteoprotegerin in osteoblastic stromal cells. Endocrinology 139:4743-4746. [DOI] [PubMed] [Google Scholar]
- 18.Inada, M., T. Yasui, S. Nomura, S. Miyake, K. Deguchi, M. Himeno, M. Sato, H. Yamagiwa, T. Kimura, N. Yasui, T. Ochi, N. Endo, Y. Kitamura, T. Kishimoto, and T. Komori. 1999. Maturational disturbance of chondrocytes in Cbfa1-deficient mice. Dev. Dyn. 214:279-290. [DOI] [PubMed] [Google Scholar]
- 19.Jiménez, M. J. G., M. Balbín, J. M. López, J. Alvarez, T. Komori, and C. López-Otín. 1999. Collagenase 3 is a target of Cbfa1, a transcription factor of the runt gene family involved in bone formation. Mol. Cell. Biol. 19:4431-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karsenty, G. 2001. Transcriptional control of osteoblast differentiation. Endocrinology 142:2731-2733. [DOI] [PubMed] [Google Scholar]
- 21.Kim, I. S., F. Otto, B. Zabel, and S. Mundlos. 1999. Regulation of chondrocyte differentiation by Cbfa1. Mech. Dev. 80:159-170. [DOI] [PubMed] [Google Scholar]
- 22.Kitazawa, R., S. Kitazawa, and S. Maeda. 1999. Promoter structure of mouse RANKL/TRANCE/OPGL/ODF gene. Biochim. Biophys. Acta 1445:134-141. [DOI] [PubMed] [Google Scholar]
- 23.Kneissel, M., A. Boyde, and J. A. Gasser. 2001. Bone tissue and its mineralization in aged estrogen-depleted rats after long-term intermittent treatment with parathyroid hormone (PTH) analog SDZ PTS 893 or human PTH(1-34). Bone 28:237-250. [DOI] [PubMed] [Google Scholar]
- 24.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y.-H. Gao, M. Inada, M. Sato, T. Okamoto, Y. Kitamuta, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764. [DOI] [PubMed] [Google Scholar]
- 25.Lacey, D. L., E. Timms, H.-L. Tan, M. J. Kelley, C. R. Dunstan, T. Burgess, R. Elliot, A. Colombero, G. Elliot, S. Scully, H. Hsu, J. Sullivan, N. Hawkins, E. Davy, C. Capparelli, A. Eli, Y.-X. Qian, S. Kaufman, I. Sarosi, V. Shalhoub, G. Senaldi, J. Guo, J. Delaney, and W. J. Boyle. 1998. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93:165-176. [DOI] [PubMed] [Google Scholar]
- 26.Lee, B., K. Thirunavukkarasu, L. Zhou, L. Pastore, A. Baldini, J. Hecht, V. Geoffroy, P. Ducy, and G. Karsenty. 1997. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor osf2/cbfa1 in cleidocranial dysplasia. Nat. Genet. 16:307-310. [DOI] [PubMed] [Google Scholar]
- 27.Liu, W., S. Toyosawa, T. Furuichi, N. Kanatani, C. Yoshida, Y. Liu, M. Himeno, S. Narai, A. Yamagushi, and T. Komori. 2001. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J. Cell Biol. 155:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manolagas, S. C. 2000. Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr. Rev. 21:115-137. [DOI] [PubMed] [Google Scholar]
- 29.McKnight, R. A., A. Shamay, L. Sankaran, R. J. Wall, and L. Henninghausen. 1992. Matrix-attachment regions can impart position-independent regulation of a tissue-specific gene in transgenic mice. Proc. Natl. Acad. Sci. USA 89:6943-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mundlos, S., L. F. Huang, P. Selby, and B. R. Olsen. 1996. Cleidocranial dysplasia in mice. Ann. N. Y. Acad. Sci. 785:301-302. [DOI] [PubMed] [Google Scholar]
- 31.Mundlos, S., F. Otto, C. Mundlos, J. B. Mulliken, A. S. Aylsworth, S. Albright, D. Lindhout, W. G. Cole, W. Henn, J. H. Knoll, M. J. Owen, R. Mertelsmann, B. U. Zabel, and B. R. Olsen. 1997. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89:773-779. [DOI] [PubMed] [Google Scholar]
- 32.O' Brien, C. A., B. Kern, I. Gubrij, G. Karsenty, and S. C. Manolagas. 2002. Cbfa1 doesn't regulate RANKL gene activity in stromal/osteoblastic cells. Bone 30:453-462. [DOI] [PubMed] [Google Scholar]
- 33.Otsuka, E., Y. Kato, S. Hirose, and H. Hagiwara. 2000. Role of ascorbic acid in the osteoclast formation: induction of osteoclast differentiation factor with formation of the extracellular matrix. Endocrinology 141:3006-3011. [DOI] [PubMed] [Google Scholar]
- 34.Otto, F., A. P. Thornell, T. Crompton, A. Denzel, K. C. Gilmour, I. R. Rosewell, G. W. H. Stamp, R. S. P. Beddington, S. Mundlos, B. R. Olsen, P. B. Selby, and M. J. Owen. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia, is essential for osteoblast differentiation and bone development. Cell 89:765-771. [DOI] [PubMed] [Google Scholar]
- 35.Parfitt, A. M., M. K. Drezner, F. H. Glorieux, J. A. Kanis, H. Malluche, P. J. Meunier, S. M. Ott, and R. R. Recker. 1987. Bone histomorphometry: standardization of nomenclature, symbols, and units. J. Bone Miner. Res. 2:595-608. [DOI] [PubMed] [Google Scholar]
- 36.Pockwinse, S. M., L. G. Wilming, D. M. Conlon, G. S. Stein, and J. B. Lian. 1992. Expression of cell growth and bone specific genes at single cell resolution during development of bone tissue-like organization in primary osteoblast cultures. J. Cell. Biochem. 49:310-323. [DOI] [PubMed] [Google Scholar]
- 37.Porte, D., J. Tuckermann, M. Becker, B. Baumann, S. Teurich, T. Higgins, M. J. Owen, M. Schorpp-Kistner, and P. Angel. 1999. Both AP-1 and Cbfa1-like factors are required for the induction of interstitial collagenase by parathyroid hormone. Oncogene 18:667-678. [DOI] [PubMed] [Google Scholar]
- 38.Roschger, P., H. Plenk, Jr., K. Klaushofer, and J. Eschberger. 1995. A new scanning electron microscopy approach to the quantification of bone mineral distribution: backscattered electron image gray, levels correlated to calcium K alpha, line intensities. Scanning Microsc. 9:75-88. [PubMed] [Google Scholar]
- 39.Rossert, J., H. Eberspaecher, and B. de Crombrugghe. 1995. Separate cis-acting DNA elements of the mouse pro-α1(1) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J. Cell Biol. 129:1421-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato, M., E. Mori, T. Komori, H. Kawahatao, M. Sugimoto, K. Terai, H. Shimizu, T. Yasui, H. Ogihara, N. Yasui, T. Ochi, Y. Kitamura, Y. Ito, and S. Nomura. 1998. Transcriptional regulation of osteopontin gene in vivo by PEBP2alphaA/CBFA1 and ETS1 in skeletal tissues. Oncogene 17:1517-1525. [DOI] [PubMed] [Google Scholar]
- 41.Satomura, K., P. Krebsbach, P. Bianco, and P. G. Robey. 2000. Osteogenic imprinting upstream of marrow stromal differentiation. J. Cell. Biochem. 78:391-403. [PubMed] [Google Scholar]
- 42.Selvamurugan, N., M. R. Pulumati, D. R. Tyson, and N. C. Partridge. 2000. Parathyroid hormone regulation of the rat collagenase-3 promoter by protein kinase A-dependent transactivation of core binding factor α1. J. Biol. Chem. 275:5037-5042. [DOI] [PubMed] [Google Scholar]
- 43.Siggelkow, H., K. Rebenstorff, W. Kurre, C. Niedhart, I. Engel, H. Schulz, M. J. Atkinson, and M. Hüfner. 1999. Development of the osteoblast phenotype in primary human osteoblasts in culture: comparison with rat calvarial cells in osteoblast differentiation. J. Cell. Biochem. 75:22-35. [PubMed] [Google Scholar]
- 44.Suda, T., E. Jimi, I. Nakamura, and N. Takahashi. 1997. Role of 1α,25-dihydroxyvitamine D3 in osteoclast differentiation and function. Methods Enzymol. 282:223-235. [DOI] [PubMed] [Google Scholar]
- 45.Suda, T., N. Takahashi, N. Udagawa, E. Jimi, M. T. Gillespie, and T. J. Martin. 1999. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand family. Endocr. Rev. 20:345-357. [DOI] [PubMed] [Google Scholar]
- 46.Takeda, S., J.-P. Bonnamy, M. J. Owen, P. Ducy, and G. Karsenty. 2001. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 15:467-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thirunavukkarasu, K., D. L. Halladay, R. R. Miles, Y. Xuhao, R. J. S. Galvin, S. Chandrasekhar, T. J. Martin, and J. E. Onyia. 2000. The osteoblast-specific transcription factor Cbfa1 contributes to the expression of osteoprotegerin, a potent inhibitor of osteoclast differentiation and function. J. Biol. Chem. 33:25163-25172. [DOI] [PubMed] [Google Scholar]
- 48.Thirunavukkarasu, K., R. R. Miles, D. L. Halladay, X. Yang, R. J. S. Galvin, S. Chandrasekhar, T. J. Martin, and J. E. Onyia. 2001. Stimulation of osteoprotegerin (OPG) gene expression by transforming growth factor-β (TGF-β)—mapping of the OPG promoter region that mediates TGF-beta effects. J. Biol. Chem. 276:36241-36250. [DOI] [PubMed] [Google Scholar]
- 49.Winchester, S. K., R. C. Selvamurugan, R.C. D'Alonzo, and N. C. Partridge. 2000. Developmental regulation of collagenase-3 m-RNA in normal, differentiating osteoblasts through the activator protein-1 and the runt domain binding sites. J. Biol. Chem. 30:23310-23318. [DOI] [PubMed] [Google Scholar]
- 50.Xiao, Z. S., S.-G. Liu, T. K. Hinson, and L. D. Quarles. 2001. Characterization of the upstream mouse Cbfa1/runx2 promoter. J. Cell. Biochem. 82:647-659. [DOI] [PubMed] [Google Scholar]