Abstract
Most human cancer cells are thought to acquire the ability to divide beyond the capacity of normal somatic cells through illegitimately activating the gene hTERT, which encodes the catalytic subunit of telomerase. While telomerase reverse transcriptase (TERT) is conserved in most eukaryotes, mounting evidence suggests that the C terminus of the human protein may have functions unique to higher eukaryotes. To search for domains responsible for such functions, we assayed a panel of tandem substitution mutations encompassing this region of human TERT for in vitro and in vivo functionality. We found four clusters of mutations that inactivated the biochemical and biological functions of telomerase, separated by mutations that had little or no effect on enzyme activity. We also identified a region where mutations generate catalytically active but biologically inert proteins. This C-terminal region that dissociates activities of telomerase (C-DAT) does not appear to be involved in nuclear localization or protein multimerization. Instead, it appears that the C-DAT region is involved in a step of in vivo telomere synthesis after the assembly of a catalytically active enzyme. Intriguingly, all of the described regions reside in a portion of TERT that is dispensable for cellular viability in yeast, arguing for a divergent role of the C terminus in higher eukaryotes.
Telomerase is a ribonucleoprotein complex that adds telomeric repeats to the ends of telomeres de novo and is essential for complete replication of chromosomes in most eukaryotes. In a variety of organisms, the loss of telomerase activity results in telomere shortening and ultimately a period of cell death or growth arrest (53). In humans, the enzyme plays an important role in cancer. Most somatic cells lack telomerase activity and correspondingly lose telomeric DNA, thus limiting their proliferative capacity. Cancer cells can overcome this proliferative block through the illegitimate transcriptional up-regulation of the gene encoding the catalytic subunit of the enzyme (41).
The core of telomerase is composed of a catalytic protein subunit, termed telomerase reverse transcriptase (TERT), that reverse transcribes a template region of an accompanying RNA subunit (TR) onto the ends of telomeres as DNA. The TERT protein is conceptualized as consisting of three regions. The amino terminus (N terminus) is rather basic and contains at least four domains that appear to be evolutionarily conserved. In a number of eukaryotes, the N terminus is essential for telomerase activity, binding to the RNA subunit or multimerization (3, 5-7, 9, 16, 26, 49). The N terminus of human TERT (hTERT) also contains a domain, termed the region that dissociates activities of telomerase (DAT), which is essential for telomere elongation but dispensable for catalytic activity (3). A single mutation in the same region of the yeast catalytic subunit gives rise to a similar phenotype (16), suggesting an evolutionarily conserved function of telomerase aside from catalysis. The central region of TERT from a variety of organisms contains seven evolutionarily conserved motifs that define the catalytic region of reverse transcriptases and in some instances have been shown to be essential for telomerase enzyme activity (33).
The carboxyl terminus (C terminus) is the least-characterized region of TERT. For humans, this region contains a CRM1 and a 14-3-3 binding site, which appear to regulate nuclear export of the protein (42). A region of the C terminus is also required for protein multimerization (2, 6, 7). Alignments of the C-terminal hTERT amino acid sequence with the TERTs of other, more distant organisms show only weak conservation (38). These sequence divergences could underlie differences in the biological function of telomerase in higher versus lower eukaryotes. Indeed, similar although not identical changes to the sequence of the human and yeast catalytic subunits can give rise to different phenotypes. For example, the addition of a hemagglutinin (HA) epitope tag to the C terminus of hTERT has little effect on telomerase in vitro enzyme activity but impedes the ability of the enzyme to elongate telomeres in a number of human cells (13, 21, 36, 54). On the other hand, addition of the same tag to the C terminus of the yeast protein (also containing a deletion of a terminal repeat not found in other TERT molecules) has no detectable effect on its function (15). Different phenotypes also arise from experimental truncations made to the yeast and human TERT proteins. A Δ153 mutation that truncates the entire C terminus of yeast TERT (termed Est2) five amino acids downstream from the motif Ei (or RT7) (38) and adds four amino acids before reaching a stop codon does not compromise cell viability (16). In contrast, a deletion of 186 amino acids from the C terminus of hTERT that truncates the protein 10 amino acids downstream of motif E (or RT7) (38) was unable to reconstitute enzyme activity in rabbit reticulocyte lysate (5). Taken together, these results suggest that the C terminus of hTERT may contain domains required for a fully functional enzyme that are not conserved in lower eukaryotes. Identifying such domains represents a key step in elucidating novel functions of the human enzyme.
To search for novel domains of human telomerase involved in the biological function of the enzyme, we took advantage of the fact that ectopic expression of hTERT can immortalize human cells, a recognizable biological phenotype. Indeed, relying on cell death as a measure of telomerase function has been a successful means of characterizing mutants of the catalytic subunit of the enzyme in yeast (15, 16, 34, 49) and humans (3). Capitalizing on this approach, we screened a panel of 41 tandem substitution mutants in the C terminus of hTERT for biological and biochemical activity. Specifically, each mutant of hTERT was stably introduced into transformed human cells prior to crisis. Cell extracts were prepared from the resultant cell lines and assayed for biochemical telomerase catalytic activity in vitro, while the corresponding cultures were monitored for signs of crisis. From this analysis we identified mutations in the C terminus that completely inactivated the enzyme, had no effect on telomerase function, or, most interestingly, resulted in the catalytically active, biologically dysfunctional DAT phenotype. Compiling these mutants, we identified discrete regions required for the biochemical and biological functions of the human telomerase enzyme.
MATERIALS AND METHODS
Plasmid constructions.
NAAIRS substitution was performed on the BamHI-SalI fragment of hTERT cloned into the same sites of pBluescript SK(−) (Stratagene, La Jolla, Calif.). In brief, second-strand synthesis was done on the minus strand by annealing oligonucleotides that correspond to the desired mutations, following the manufacturer's protocol (Muta-Gene; Bio-Rad, Hercules, Calif.). Each oligonucleotide was designed to have 15 nucleotides identical to the desired plus-strand sequence on either side of the sequence AATGCTGCTATACGATCG, which corresponds to the amino acid sequence NAAIRS. Using this strategy, 41 oligonucleotides were used to systematically substitute every six amino acids, beginning at amino acid +890 and ending at the stop codon. The six-amino-acid region mutated was confirmed to be correct in all cases by direct sequencing. The mutated BamHI-SalI fragments were then cloned into the same sites of pUC19-FLAG-hTERT (3), after which the new FLAG-hTERT cDNA was removed by digestion with EcoRI and SalI and ligated to the same sites in pBabehygro (for retroviral infections) (31) or pCMV5 (for overexpression in 293 cells) (1). Mutations in the C-DAT region were made by site-directed mutagenesis as described above with primers encoding the following mutations: F1127KT to NAA, I1130LD to IRS, F1127 to A, K1128 to A, and T1129 to A. A FLAG-tagged C-terminal BamHI SalI fragment of hTERT cloned into pBluescript SK(-) was used as a template to create NAAIRS mutants by site-directed mutagenesis as previously described (3). These fragments were then used to replace the BamHI SalI fragment of pCIneoFLAG-hTERT, creating the plasmids pCIneo-FLAG-hTERT-FLAG-NAAIRS +998, +1034, +1088, and +1118. The yellow fluorescent protein (YFP)-hTERT expression plasmids were created by cloning the EcoRI/SalI FLAG-hTERT or FLAG-hTERT-NAAIRS+1127 open reading frame into the same sites in the expression vector pEYFP-C1 (Clontech, Palo Alto, Calif.).
Cell culture.
Using standard protocols as previously described (3, 13), HA5 cells and HA1 cells (46) were infected at population doubling (pd) ∼45 to 50 with amphotrophic retroviruses derived from the above-described pBabehygro constructs. Stable polyclonal populations were selected in medium supplemented with 100 μg of hygromycin/ml. The first plate to reach confluence under selection was arbitrarily defined as pd 0. All cell lines were cultured until crisis or until the cells had divided at least 20 pds beyond telomerase-negative cells. Crisis was defined as the period when plates failed to become confluent after 20 to 30 days and exhibited massive cell death.
RT-PCR of hTERT mRNA, telomerase activity, and telomere length assays.
For reverse transcriptase (RT) PCR analysis of total or endogenous hTERT mRNA or the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, total cellular RNA was prepared with the reagent RNazol (Teltest B, Friendswood, Tex.) according to the manufacturer's instructions, and RT-PCR was carried out with primers specific for the aforementioned transcripts as previously described (20). Reaction products were resolved on 10% polyacrylamide gels, dried, and exposed to a phosphorimager screen.
To detect telomerase activity, cell lysates were prepared in 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate buffer (25). Samples were normalized according to protein concentration and were diluted in the same buffer to a concentration of 0.1 μg/ul prior to assaying for activity. One half of the sample was heated at 85°C for 2 min to inactivate telomerase, whereas the other half was directly assayed or serially diluted in 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate buffer for dilution experiments. Two microliters of extracts was assayed using a PCR-based telomerase assay, after which the reaction products were resolved on 10% polyacrylamide gels, dried, and exposed to a phosphorimager screen as previously described (25). Activity was calculated as the radioactive counts derived from the experimental extract, minus the heat-treated control, divided by the internal standard as previously described (25). Percent activity of NAAIRS mutants was determined by dividing extract activity by the activity detected in extracts prepared from HA5 cells stably expressing transgenic FLAG-hTERT and multiplying by 100.
Telomeres were visualized by Southern hybridization of HinfI and RsaI restriction enzyme-digested genomic DNA with the 32P-labeled telomeric (C3TA2)3 oligonucleotide exactly as previously described (12), with the exception that 3 μg of digested DNA was resolved and the washes were performed with 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Telomere length was determined by analyzing the modal signal via phosphorimager analysis and correlating the peak position with a molecular weight ladder.
Immunoblotting.
To assay for FLAG-hTERT protein expression, 293 cells were transiently transfected with the pCMV5 vector or derivatives encoding hTERT, FLAG-hTERT, or FLAG-hTERT NAAIRS mutants using the reagent Lipofectamine Plus in accordance with the manufacturer's protocol (GibcoBRL, Gaithersburg, Md.). Cells were harvested 48 h after transfection and lysed in the presence of 1% Triton X-100, 10 mM Na2HPO4-Na2HPO4 (pH 7.0), 150 mM NaCl, and 5 mM EDTA (pH 8.0), boiled, and then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, Mass.) and immunoblotted with the anti-FLAG M2 (Sigma, St. Louis, Mo.) or the anti-actin I-19 (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.) antibody. Blots were washed three times for 10 min in 1× Tris-borate EDTA supplemented with 0.02% Tween 20 at room temperature and then treated with ECL reagent following the manufacturer's protocol (Amersham Pharmacia Biotech, Piscataway, N.J.).
hTR-hTERT and hTERT-hTERT coimmunoprecipitations.
32P-labeled human TR (hTR) and U6 RNAs were produced with the T7 coupled maxiscript system (Ambion, Austin, Tex.) using 1 μg of linearized pBSK-hTR or pBSK-U6. Unincorporated 32P nucleotides were removed by G-25 gel filtration minispin columns (Amersham Pharmacia Biotech). 35S-labeled protein was synthesized in vitro using the T7 quick-coupled TNT system by incubating 1 μg of plasmid pCIneo-FLAG-hTERT-FLAG, pCIneo-FLAG-hTERT-FLAG-NAAIRS +512, +998, +1034, +1088, +1118, or, as a control, pCMV-HDAC in rabbit reticulocyte lysate in the presence or absence of 3 μl of labeled RNA following the manufacturer's instructions (Promega). For experiments showing binding specificity between hTR and U6, 1.5 μl of labeled RNA was used.
Immunoprecipitations were done as described previously (3) using either 9 μg of the anti-FLAG M2 monoclonal antibody (Sigma) prebound to 25 μl of GammaBind G-Sepharose (Amersham Pharmacia Biotech) or 10 μl of anti-FLAG M2 affinity gel. In some cases, 0.5 M urea was added to all final washes. The immunoprecipitate was then resolved by SDS-PAGE. Gels were dried and then exposed to film. When required, a blocking screen was placed between the gel and film to differentiate 32P from 35S signals. Immunoprecipitation of multimeric hTERT complexes was performed as previously described (3). FLAG-tagged proteins and glutathione S-transferase (GST)-tagged proteins were transcribed and translated separately in the presence of either 4 μl of [35S]methionine or 0.5 μl of [35S]methionine and 1 μl of cold methionine, respectively, in the TNT rabbit reticulocyte lysate system (Promega). The plasmids used for the TNT reaction included those mentioned above, as well as pCIneo-FLAG-hTERT-FLAG-NAA +1127 and pCIneo-GST-hTERT-NAAIRS +1127. Reaction mixtures were incubated for 40 min at 30°C, mixed with the appropriate reactions, and then incubated for an additional 60 min at 30°C. Complexes were then immunoprecipitated as described above.
Cytogenetics.
Cells were arrested in metaphase using 10 ng of Colcemid (Gibco Life Technologies, Long Island, N.Y.)/ml for 90 min. Cells were then incubated in hypotonic solution (0.75 M KCl) and fixed in methanol-acetic acid (3:1). Slides were prepared and stained using G-banding. Metaphase spreads were scored for telomere fusions irrespective of ploidy.
YFP localization.
To assay for YFP-hTERT protein localization, the human osteosarcoma cell line U2OS was grown on poly-d-lysine (molecular mass, >300,000 kDa; Sigma)-coated coverslips. Cells were then transiently transfected with the pEYFP-C1 vector or derivatives encoding YFP-FLAG-hTERT, YFP-FLAG-hTERT NAAIRS1034, YFP-FLAG-hTERT NAAIRS1127 using the reagent Fugene-6 (Roche, Indianapolis, Ind.) in accordance with the manufacturer's protocol. Forty-eight hours later, cells were observed under phosphate-buffered saline at ×400 magnification.
RESULTS
NAAIRS substitution mutagenesis.
To screen for regions of hTERT essential for enzyme function, we initially created a series of C-terminal truncations. In agreement with similar deletion analyses done by others with hTERT synthesized in rabbit reticulocyte lysate (5, 7), we found that deletions of 200, 150, and 62 amino acids did not generate catalytically active proteins in telomerase-negative human cells (not shown), implying the presence of C-terminal regions essential for a functional human enzyme. To delineate such regions, we created a panel of FLAG epitope-tagged hTERT mutants in which every six amino acids across the entire C terminus were replaced with the amino acid sequence NAAIRS. The NAAIRS sequence is found in both α-helices and β-sheets (48) and is thus useful for mapping domains since it disrupts primary amino acid sequence while being less likely to distort the overall structure of the protein (3, 43). In this manner, a large region of hTERT could be mutated with a manageable number of genetic manipulations. In total, 41 tandem substitution mutations in the C terminus of hTERT were created by site-directed mutagenesis, starting at amino acid +890 and ending at the stop codon (Fig. 1A).
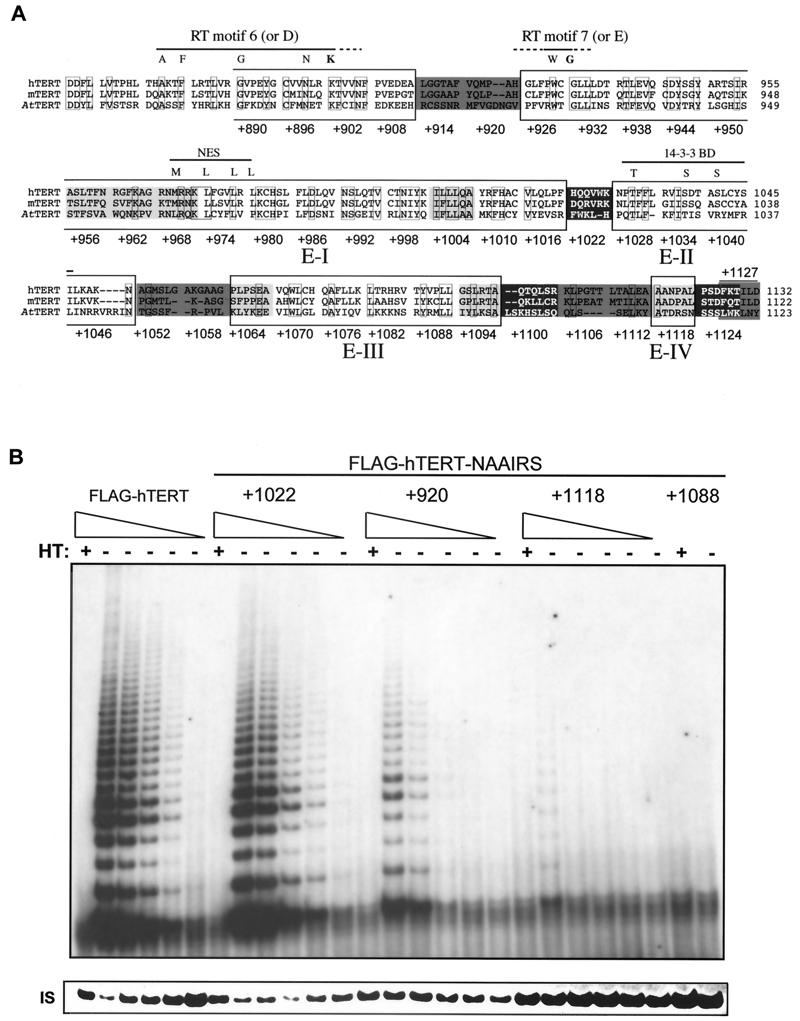
FIG. 1.
NAAIRS substitution mutagenesis of the C terminus of hTERT identifies five regions essential for enzyme function. (A) Schematic representation of the position and effect on telomerase activity of the NAAIRS substitution mutations of hTERT. The C-terminal amino acid sequences of the human (h), mouse (m), and Arabidopsis (At) TERT protein are aligned. Boxed amino acids denote identical sequence between all three proteins. RT motifs 6 (or D) and 7 (or E), the putative NES and 14-3-3 binding site are denoted with a line. Highly conserved or criticalamino acids of these regions are shown as letters above the alignment. Sequences are divided into six amino acid segments corresponding to the positions of the NAAIRS substitutions in the hTERT protein. Numbers refer to the first amino acid of each substitution, starting with +1 as the initiating Methionine. The six amino acid segments are shaded in dark grey, medium grey, or light grey or are left unshaded to represent high, medium, low and undetectable levels of in vitro telomerase enzyme activity as defined in the text. Large boxes identify novel essential regions E-I, -II, -III, and -IV. (B) NAAIRS mutations with different levels of telomerase activity. Cell lysates (0.2 μg or threefold serial dilutions thereof) prepared from HA5 cells stably infected with retroviruses encoding the wild-type FLAG-hTERT or FLAG-hTERT NAAIRS mutants +1022, +920, +1118, and +1088 were assayed for telomerase activity. HT, heat treated. IS, the internal standard, is shown as a different exposure to more easily visualize the standard.
The panel of FLAG-hTERT mutants and controls, consisting of either no insert or the wild-type version of FLAG-hTERT, were separately cloned into retroviral vectors. Amphotrophic retroviruses were generated from each of these constructs and used to stably infect the cell strain HA5 prior to the onset of crisis. HA5 cells are human embryonic kidney cells transformed with the simian virus 40 T-antigen gene (encoding large and small T-antigen) (46). These cells express hTR but lack detectable levels of hTERT mRNA and telomerase activity (12, 28). Consequently, HA5 cells continue to lose telomeric DNA until a critically short length is reached, at which time genomic instability ensues and the cells enter crisis and die (12). These cells have not been found to spontaneously immortalize (12, 14, 46) but can acquire an unlimited proliferative capacity through the ectopic expression of hTERT (3). Cells infected with the mutant versions of hTERT can therefore serve as a model system for assessing both the biochemical and biological functions of hTERT. Specifically, the resultant cell lines were assayed for telomerase in vitro enzyme activity and for biological function by measuring telomere length and cell life span. From this analysis we identified three mutant phenotypes: (i) catalytically and biologically inactive, (ii) catalytically and biologically active, and most interestingly, (iii) catalytically active but biologically impaired.
Class one: mutations that inactivate biochemical and biological function of hTERT.
To determine the effect of the mutations on biochemical activity, cellular extracts were isolated from all 41 cell lines as well as the two control lines and assayed for in vitro telomerase enzyme activity. Lysates were incubated with a primer that can be elongated by telomerase, and then the reaction products were PCR amplified and resolved to visualize the 6-bp ladder indicative of telomerase activity (25). As a control, duplicate extracts were heat treated to inactivate telomerase prior to assaying for enzyme activity. We found that the mutants displayed different levels of telomerase activity. For example, by dilution of cell lysates, we showed that extracts from HA5 cells infected with FLAG-hTERT or the representative +1022 mutant have similar levels of activity, as characterized by the heat-sensitive 6-bp ladder. Significantly lower levels of telomerase activity were measured in other mutants. For example, telomerase activity was detected in the representative +920 mutant at a protein concentration of 0.03 μg/ul, but barely at a protein concentration of 0.01 μg/ul, corresponding to an approximate fivefold decrease in activity compared to wild-type FLAG-hTERT-infected cells. Enzyme activity was extremely weak in NAAIRS mutants such as the +1118 mutant and undetectable in the representative +1088 mutant, even at the highest protein concentration tested (Fig. 1B).
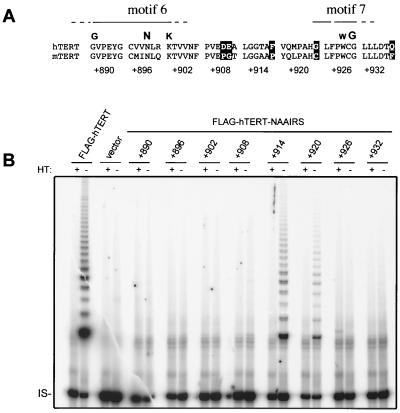
We therefore classified the mutants as reconstituting telomerase in vitro enzyme activity to either ++ levels (high), + levels (medium), +/− levels (low), or −levels (undetectable), based on the signal intensity of telomerase products (Table 1). Combining these data, a pattern emerged for the regions of the C terminus of hTERT essential for telomerase catalytic activity (Fig. 1A). In total, four novel regions, termed essential region I (E-I), E-II, E-III, and E-IV, which encompass most of the C terminus, were found to be required for easily detectable (++ or + levels) in vitro telomerase activity. Consistent with the boundaries of these regions, alignment of the mouse, human, and Arabidopsis TERT protein sequences demonstrate that these areas tend to be more conserved. This type of mutagenesis also successfully pinpointed the RT motifs 6 (or D) and 7 (or E) (22, 24, 28, 34), which are conserved among all reverse transcriptases (39, 50), as being essential for telomerase enzyme activity whereas mutations to the more divergent linker regions were tolerated (Fig. 2). Thus, it is likely that the E regions identified in hTERT represent bona fide regions of the protein required for enzyme function.
TABLE 1.
Characteristics of transformed human cells expressing FLAG-hTERT NAAIRS mutants
Presence (+) or absence (−) of hTERT mRNA as determined by RT-PCR.
Presence (+) or absence (−) of FLAG-hTERT protein in 293 cells as determined by immunoblotting with an α-FLAG antibody.
In vitro telomerase activity for each mutant was determined by normalizing activity to the internal standard and then expressed as a percentage of wild-type FLAG-hTERT activity: ++ (>50%), + (50 to 15%), +/− (<15%), − (undetectable).
Crisis was defined as the point whereby cells failed to reach confluency after a minimum of 20 days. EL denotes an extended lifespan.
FIG. 2.
NAAIRS substitution in RT motifs 6 (or D) and 7 (or E), but not the spacer region, result in a protein that is catalytically inactive in vitro. (A) Alignment of the region encompassing the first eight NAAIRS substitutions between hTERT and mouse TERT (mTERT). Dissimilar amino acids are shaded. The approximate boundaries of RT motif 6 (or D) and motif 7 (or E) are denoted with a line. Highly conserved amino acids are highlighted in bold letters above the alignment. The alignment is shown in blocks of six amino acids, which correspond to the positions of the first eight NAAIRS substitutions. Numbers refer to the first amino acid of each substitution (+1 being the initiating methionine). (B) Cell lysate (0.2 μg) prepared from HA5 cells stably infected with the retroviral vector encoding FLAG-hTERT, no inserted gene (vector), or the FLAG-hTERT NAAIRS substitution mutants shown was assayed for telomerase activity. HT, heat treated; IS, internal standard.
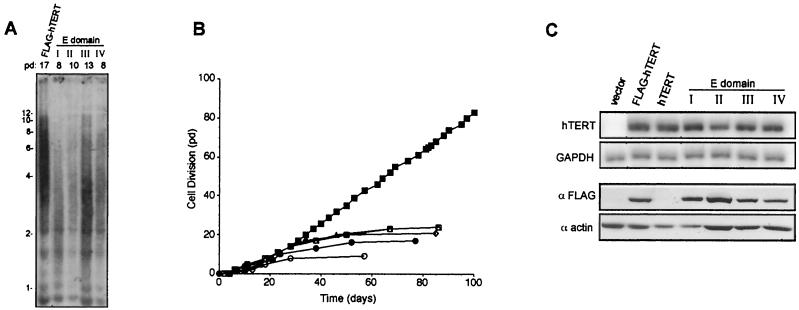
To address whether regions E-I to -IV are essential for the biological function of hTERT, we monitored telomere length and life span of the HA5 cell lines expressing the NAAIRS mutants in these regions. Telomere length was measured near terminal passages in cell lines expressing FLAG-hTERT containing a mutation in one of the four essential regions (+998, +1034, +1088, and +1118) and compared to the telomere length of the FLAG-hTERT-expressing positive-control cells at a similar passage. The positive-control cells had an average telomere length of ∼5 kbp at pd 17 (Fig. 3A), which gradually shortened, and then telomeres maintained a stable length of ∼3 kbp (not shown), as observed in other cell types (37, 51, 54). These cells were monitored in tissue culture for at least 75 pd beyond crisis, corresponding to more than four times the number of cell divisions as with vector control cells (Fig. 3B). Conversely, the telomere length of the four cell lines expressing the mutated versions of FLAG-hTERT had already shortened to a length of ∼3 kbp by pd 8 to 13 (Fig. 3A). Accordingly, these mutants failed to proliferate more than the negative-control cells, entered crisis, and died (Fig. 3B). Indeed, all telomerase-negative cell lines were mortal (Table 1), indicating that the mutations that greatly reduced or abolished telomerase activity failed to immortalize the human cells.
FIG. 3.
Class one mutants are biochemically and biologically inactive. (A) C-terminal regions E-I, -II, -III, and -IV are essential for telomere maintenance. Telomere length of DNA isolated from cells expressing FLAG-hTERT, or NAAIRS mutants +998, +1034, +1088, or +1118 representing each of the four regions E-I, -II, -III, and -IV, was determined by Southern hybridization with a telomeric probe.(B) HA5 cells expressing hTERT containing a NAAIRS mutation in one of the four essential regions are mortal. Cell divisions in pds versus time in days is plotted for HA5 cells infected with FLAG-hTERT (▪), an empty vector (•), or the aforementioned representative FLAG-hTERT-NAAIRS mutants for E regions I (○), II (□), III (▵), and IV (⋄). (C) Mutations in regions E-I, -II, -III, and -IV do not alter RNA or protein levels of hTERT. Total RNA prepared from the same aforementioned cell lines, or the control HA5 cells stably expressing untagged hTERT, was RT-PCR amplified with primers specific for hTERT or GAPDH. Protein lysates prepared from 293 cells transiently transfected with an empty (vector) mammalian expression vector or one encoding FLAG-hTERT, hTERT, or representative FLAG-hTERT NAAIRS mutants for each of the four essential regions were resolved by SDS-PAGE and Western blotted with an anti-FLAG or antiactin antibody.
The mutations described define the first mutant phenotype observed: catalytically and biologically inert. To ascertain if this phenotype was simply due to poor transcription, total RNA was isolated from each of the 41 cell lines expressing various hTERT mutations and RT-PCR amplified with primers specific for hTERT or control GAPDH mRNA. We found that all NAAIRS mutants, including those in regions E-I to -IV, were overexpressed in HA5 cells (Fig. 3C and Table 1). We also confirmed with a subset of these lines that the endogenous hTERT transcript was not detectable (not shown). Thus, poor transcription of the transgenes did not underlie the lack of telomerase activity in cell lines expressing FLAG-hTERT containing mutations in the E regions.
The absence of telomerase enzyme activity in FLAG-hTERT proteins containing NAAIRS mutations in regions E-I through E-IV could potentially have been due to a decrease in protein stability, as observed with some mutants of yeast TERT that display the same phenotype (16). We have been unable to detect ectopic hTERT in cells stably infected with the described hTERT retrovirus by immunoblotting either with an antibody specific for a FLAG tag introduced into the N terminus of hTERT or with an anti-hTERT antibody (not shown). However, FLAG-hTERT can be detected by immunoblot analysis when transiently expressed in 293 cells (3, 7, 30). Expression of FLAG-hTERT in 293 cells should identify any gross defects affecting protein stability. Therefore, epitope-tagged NAAIRS mutants and controls (empty vector or FLAG-tagged or untagged versions of wild-type hTERT) were transiently overexpressed in 293 cells. A protein of ∼130 kDa, corresponding to the size of hTERT (8, 14, 35, 47), was detected by Western blot using an anti-FLAG antibody in cells transfected with the FLAG-hTERT but not the hTERT construct (Fig. 3C). In the case of the representative mutants in regions E-I through E-IV (+998, +1034, +1088, and +1118), the NAAIRS FLAG-hTERT proteins were detected at levels reflecting that observed with the wild-type FLAG-hTERT control (Fig. 3C). Similarly, other mutations in these regions did not abrogate protein expression (Table 1). These data suggest that the total, or near total, loss of telomerase activity upon mutation of region E-I, E-II, E-III, or E-IV was unlikely to be related to a drastic loss of protein expression. Thus, the majority of the C terminus of hTERT contains regions that are essential for enzyme activity, as evidenced by substitutions that inactivate the known functions of telomerase.
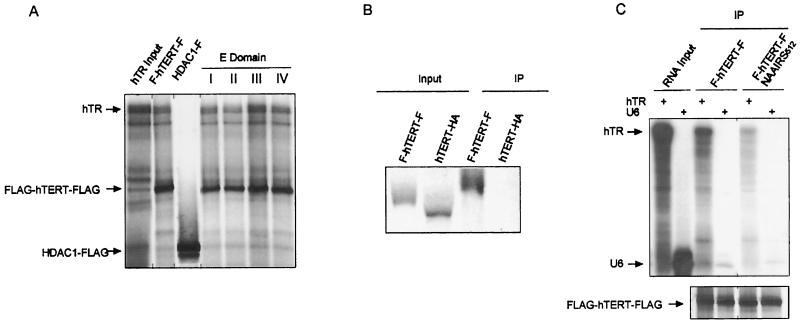
Expression of hTERT and hTR in an environment that facilitates complex formation is sufficient to reconstitute a catalytically active telomerase enzyme in vitro (4, 8, 23, 47). This implies that the loss of in vitro telomerase catalytic activity observed in cells expressing the E-region mutants of hTERT could be attributed to either a defect in the interaction with hTR or some other intrinsic defect in catalysis. To differentiate between these possibilities, we assayed the association of hTR with wild-type and mutant hTERT. 35S-labeled recombinant wild-type FLAG-hTERT-FLAG protein was generated in rabbit reticulocyte lysates in the presence of 32P-labeled in vitro-transcribed hTR RNA. The FLAG-hTERT-FLAG protein was then immunoprecipitated with an anti-FLAG antibody and resolved by SDS-PAGE. In agreement with others (4, 7), we observe that the hTR RNA is coimmunoprecipitated with the recombinant FLAG-hTERT-FLAG protein (Fig. 4A). The specificity of this interaction was demonstrated by four experiments. First, hTR did not associate with an irrelevant FLAG-tagged protein (HDAC), despite the fact that this protein was readily produced and immunoprecipitated with the anti-FLAG antibody (Fig. 4A). Second, we show that the proteins are specifically immunoprecipitated by their FLAG tag, as the HA epitope-tagged version of hTERT was not immunoprecipitated by the anti-FLAG antibody (Fig. 4B). Third, the +512 mutation in one of the known RNA-binding regions of hTERT (3, 26, 32) greatly diminished its in vitro association with hTR (Fig. 4C). Fourth, an irrelevant RNA (U6) showed only negligible binding to both wild-type hTERT and the hTR-binding mutant (Fig. 4C).
FIG. 4.
Mutations to essential regions I to IV do not affect hTR RNA binding. (A) hTR binding to hTERT E-region mutants. The 35S-labeled FLAG-hTERT-FLAG protein and variants containing mutations at positions +998, +1034, +1088, and +1118, as well as the HDAC1-FLAG recombinant protein, were produced in the presence of 32P-labeled hTR and immunoprecipitated with an anti-FLAG antibody. The hTR input lane represents 1/100th of the RNA remaining after immunoprecipitation. (B) FLAG-tagged hTERT is specifically immunoprecipitated. FLAG-hTERT-FLAG and hTERT-HA proteins were immunoprecipitated (IP) with the anti-FLAG antibody. Due to the poor expression of the hTERT-HA construct, the in vitro transcription and translation reaction was scaled up eightfold. Input lanes represent 1/20th of the total protein remaining after immunoprecipitation.(C) hTR-hTERT interactions are specific. Wild-type and hTR binding domain-defective hTERT proteins were synthesized in the presence of hTR or U6 RNA and immunoprecipitated. Input RNA lanes represent 1/100th of the total RNA added to the transcription-and-translation reaction. To more clearly differentiate protein and RNA components, the 35S protein signal has been filtered out in the upper panel by the use of a blocking screen. Immunoprecipitated proteins are shown in the lower panel.
Given that the hTR-hTERT interaction could be recapitulated in vitro, we next assayed whether mutations in any of the four essential regions affected this interaction. Representative FLAG-hTERT-FLAG proteins containing a single NAAIRS mutation in one of the four essential regions (+998, +1034, +1088, and +1118) were expressed in the presence of hTR and immunoprecipitated as described above. We find that hTR was coimmunoprecipitated with all of the E-region mutant hTERT proteins at levels comparable to that for the wild-type protein (Fig. 4A). Thus, mutations in essential regions I to IV render the hTERT protein catalytically inactive but still permit specific interactions with the hTR RNA subunit in vitro.
Class two: mutations that do not affect the biochemical and biological function of hTERT.
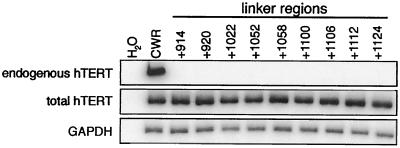
Between regions E-I to E-IV were portions of the protein that were poorly conserved and where NAAIRS substitutions had little or no measurable effect on enzyme activity (Fig. 1A). Correspondingly, mutations to these regions did not diminish the ability of the hTERT protein to immortalize HA5 cells (Table 1). The presence of telomerase activity in these cells was ascribed to the ectopic expression of FLAG-hTERT. First, we sequenced the cDNA derived from the ectopic expressed transcripts and confirmed that only the mutant TERT was present in the cells (not shown). Second, we amplified by RT-PCR RNA isolated at late passage from all nine cell lines that comprised this class of mutants with primers specific for total hTERT mRNA or for the endogenous hTERT transcript. Although a strong signal was detected in all cell lines by using primers for total hTERT mRNA, the endogenous hTERT mRNA could not be detected in these cell lines despite the fact that this transcript was readily amplified in an hTERT-expressing control cancer cell line (Fig. 5 and data not shown). The absence of endogenous hTERT RNA was not due to a limitation of RT-PCR, since we could dilute RNA from telomerase-positive cells 64-fold and still readily detect the transcript (not shown). Thus, the activity detected in cells infected with this class of mutants is derived from the ectopically expressed mutant protein and not activation of the endogenous gene. The regions which tolerate substitutions may therefore act as linkers between the essential regions.
FIG. 5.
Telomerase activity detected in cells expressing class two hTERT mutants is not due to activation of the endogenous hTERT gene. Total RNA was prepared from each of the nine cell lines that had telomerase activity and divided beyond crisis, as well as from vector- and FLAG-hTERT-infected cells and the telomerase-positive cell line CWR22. This RNA was RT-PCR amplified with primers specific for total hTERT (endogenous or ectopic transcripts), endogenous hTERT, or, as a control, GAPDH mRNA. All but samples +914, +920, and +1052 were isolated at late passage.
Class three: a DAT mutation that inactivated the biological function of hTERT without abrogating catalytic activity.
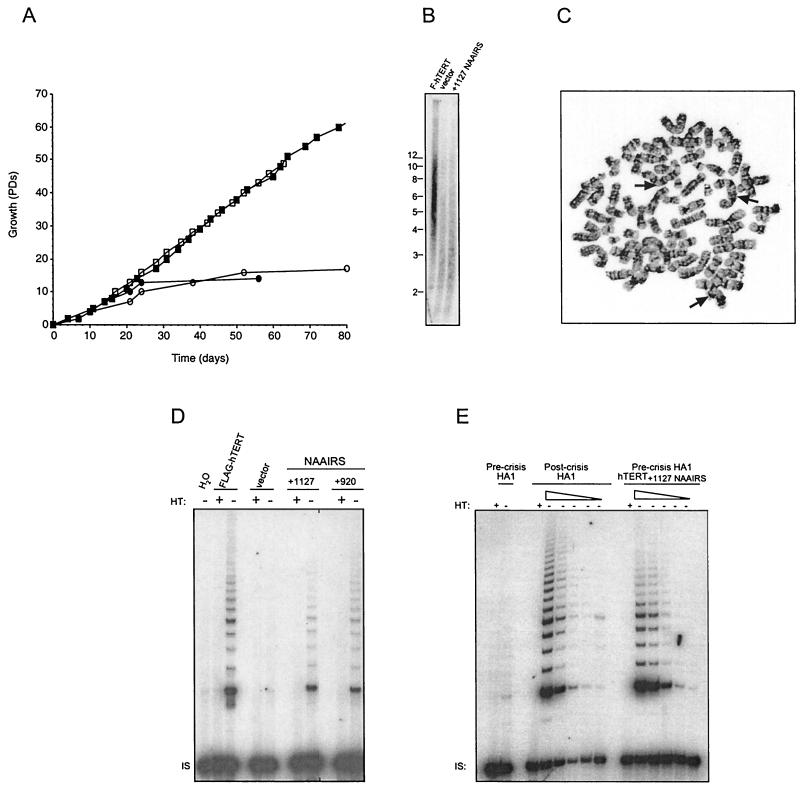
Intriguingly, we also identified a third mutant phenotype. The extreme C-terminal NAAIRS substitution at position +1127 generated a catalytically active but functionally dead protein. Cells expressing this mutant protein failed to immortalize and instead behaved in a fashion similar to that of the vector-infected cells and underwent crisis at pd ∼14 (Fig. 6A). This inability of the +1127 mutant to immortalize cells was attributed to a telomere defect. Similar to vector control-infected cells, cells expressing the +1127 mutant exhibited shortened telomeres of approximately 2.5 kb after nine pds (Fig. 6B). This was in marked contrast to cells expressing wild-type hTERT, which displayed much longer telomeres of ∼6 kb after the same number of pds. Furthermore, in addition to telomere shortening, cells expressing the +1127 mutant demonstrated a high frequency of telomere fusion events. Specifically, the cells expressing the +1127 mutant had an eightfold increase in dicentric chromosomes (Fig. 6C), compared to the same cells expressing wild-type hTERT (83 versus 11 fusion events per 50 metaphase spreads).
FIG. 6.
Mutations to the +1127 C-DAT region render hTERT catalytically active but biologically inert.(A) HA5 cells expressing hTERT +1127 NAAIRS mutant are mortal. Cell division in pds versus time in days is plotted for HA5 cells infected with FLAG-hTERT (▪), an empty vector (•), or the aforementioned FLAG-hTERT-NAAIRS mutants +1127 (○), and +920 (□). (B) Cells expressing the +1127 NAAIRS mutant have shortened telomeres. DNA was isolated from cells expressing FLAG-hTERT, vector control, or the hTERT +1127 NAAIRS mutant, all at pd 9. Digested DNA was resolved and subject to Southern hybridization with a telomeric probe. (C) Cells expressing the hTERT +1127 NAAIRS mutant demonstrate chromosome instability near terminal passage. Representative metaphase spread is depicted with arrows indicating dicentric chromosomes. (D) The NAAIRS substitution at position +1127 in the C-DAT region does not drastically lower telomerase activity. Cell lysates (0.2 μg) prepared from HA5 cells stably infected with retroviruses encoding the wild-type FLAG-hTERT or an empty vector or FLAG-hTERT NAAIRS mutants +1127 or +920 were assayed for telomerase activity. HT, heat treated. IS, internal standard. Water was included as a negative control.(E) Catalytic activity of hTERT +1127 NAAIRS mutant is of a level sufficient for cellular immortalization. Cell lysates (0.2 μg and threefold serial dilutions thereof) were prepared from precrisis and postcrisis HA1 cells and HA1 cells stably infected with FLAG-hTERT+1127 NAAIRS and assayed for telomerase activity. HT, heat treated. IS, internal standard.
While the +1127 mutant left the protein biologically dead, it retained a moderate (+) level catalytic activity, suggesting that this area of the protein may define a DAT region (Fig. 6D). The activity of the +1127 mutant cell line was comparable to that of the +920 mutant, which displayed the lowest amount of telomerase activity consistent with immortalization of the NAAIRS mutant cell lines (Fig. 6D). Further serial dilution of cellular extracts showed the +1127 mutant to actually have slightly less activity than the +920 mutant (data not shown). This raised the possibility that the inability of the +1127 mutant to immortalize human cells could be related to catalytic activity that was below the threshold required for cellular immortalization. To distinguish these possibilities, we stably expressed this mutant in the closely related HA1 cell line which can spontaneously immortalize (12). Dilution analysis of cellular lysates demonstrated the activity of the +1127 mutant to be comparable to that of late-passage spontaneously immortalized HA1 cells (∼140 pds beyond crisis) that activate endogenous hTERT (Fig. 6E). Thus, the catalytic activity of the +1127 mutant was at a level sufficient for cellular immortalization.
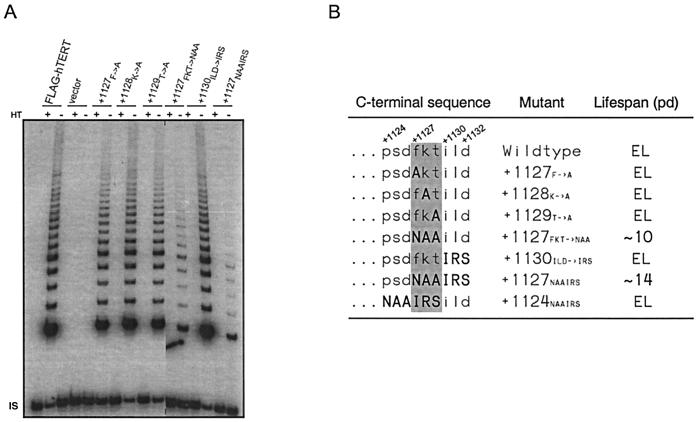
This catalytically active but biologically dead phenotype is analogous to that displayed by mutations to the DAT domain in the N terminus of the protein (3); hence we have named this novel C-terminal region of the protein C-DAT. To more precisely identify the amino acids defining the C-DAT region, we divided the NAAIRS sequence in two, substituting the sequence F1127KT with NAA or replacing the sequence I1130LD with IRS. When these two mutants were introduced into HA5 cells, both restored in vitro telomerase activity (Fig. 7A) but only the F1127KT to NAA substitution recapitulated the phenotype of the +1127 mutant, failing to extend the life span of the cells (Fig. 7B) while only moderately decreasing telomerase activity. We note that not all amino acid substitutions in the F1127KT region led to a DAT phenotype. For example, the +1124 mutant that more conservatively substituted these amino acids did not affect enzyme function (Fig. 7B); similarly single alanine substitutions also did not have any effect on in vitro or in vivo telomerase function (Fig. 7). We therefore define the three-amino-acid-sequence F1127KT as the C-DAT region, being essential for the ability of the enzyme to elongate telomeres in vivo.
FIG. 7.
The sequence F1127KT of hTERT defines the biologically important C-DAT region. (A) Cell lysates (0.2 μg) were prepared from HA5 cells that were stably infected with FLAG-hTERT, vector or hTERT variants with mutations in the +1127 region, and assayed for telomerase activity. HT, heat treated. IS, internal standard. (B) The sequence F1127KT of hTERT is necessary for immortalization of HA5 cells. The last nine amino acids of the various versions of FLAG-hTERT are shown beside the life span of cells expressing these proteins. Amino acids in bold denote changes to primary sequence. The grey box highlights the region necessary for an immortal life span. EL, extended life span.
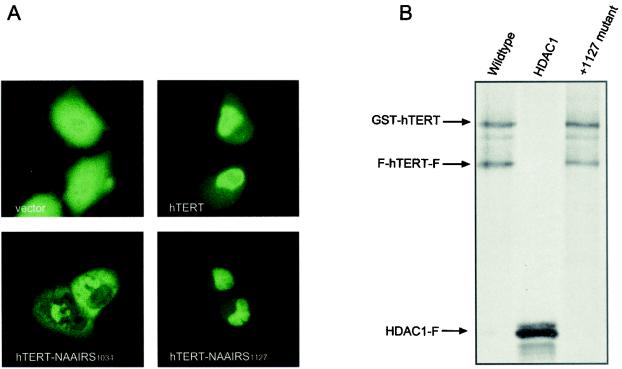
One simple explanation for the defect resulting from mutations to the C-DAT region is that the protein was retained in a functional state in the cytoplasm, where it could not elongate telomeres in vivo. Indeed, the C terminus has been implicated in nuclear localization; a 14-3-3 binding site in this region (Fig. 1A) reportedly masks a putative nuclear export signal (NES) in hTERT, preventing the protein from being exported from the nucleus (42). To address whether a mutation in the C-DAT region disrupts nuclear localization of hTERT, we transiently transfected U2OS cells with plasmids encoding YFP or YFP fused to hTERT in the wild-type or the +1127 NAAIRS configuration and assayed for protein localization. The localization of the wild-type and +1127 NAAIRS versions of YFP-hTERT was similar, being found primarily in the nucleus, although we note that cytoplasmic localization was occasionally observed. On the other hand, the +1034 NAAIRS protein that disrupted the 14-3-3 binding site (42) had a markedly more cytoplasmic distribution, while YFP was found throughout the cell (Fig. 8A). Taken together, these data suggest that mutation of the C-DAT region does not grossly alter the ability of hTERT to enter the nucleus.
FIG. 8.
The +1127 C-DAT region does not appear to be involved in nuclear localization or hTERT multimerization. (A) Mutating the C-DAT region does not perturb the nuclear localization of hTERT. Human U2OS cells were transiently transfected with either YFP, YFP-hTERT, or variants of YFP-hTERT with NAAIRS substitutions in the 14-3-3 binding domain (+1034 NAAIRS) or the C-DAT region (+1127 NAAIRS). Representative cells expressing the aforementioned proteins were viewed as fluorescence images and photographed at a magnification of ×400. (B) Mutating the C-DAT region does not affect hTERT multimerization. FLAG- and GST-tagged proteins were expressed and 35S labeled in vitro, mixed together, further incubated, and immunoprecipitated with an anti-FLAG antibody. hTERT proteins were expressed in wild-type form or with mutations in the C-DAT region. The irrelevant FLAG-tagged protein HDAC1 used as a negative control failed to immunoprecipitate GST-hTERT.
Another plausible function of the C-DAT region is in protein multimerization. Recently, hTERT was shown to be capable of forming multimeric complexes in vitro (2, 3) and in vivo (6, 7). hTERT multimerization is believed to occur via two putative interaction sites in the N and C termini of the protein (2). Although the C-terminal interaction site is thought to be upstream of the C-DAT region (6, 7), we hypothesized that mutations in the +1127 region might allosterically affect complex formation. Potentially, multimerization could be essential for in vivo telomere elongation but dispensable for catalytic activity; thus, loss of this function could give rise to a DAT phenotype. This prompted us to investigate whether the C-DAT region of hTERT might be involved in protein multimerization. To assess the ability of hTERT C-DAT mutants to multimerize, we performed an in vitro coimmunoprecipitation assay. FLAG-tagged and GST-tagged proteins were synthesized, 35S labeled, and allowed to form potential protein complexes in rabbit reticulocyte lysate. Protein complexes were immunoprecipitated with an anti-FLAG antibody and resolved by SDS-PAGE. The multimeric hTERT complex was distinguished by the coimmunoprecipitation of the larger, more slowly migrating GST-hTERT protein, in association with the smaller FLAG-tagged protein. The specificity of the interaction was demonstrated by the inability of GST-hTERT to be coimmunoprecipitated by the irrelevant protein FLAG-HDAC1. In the case of both wild-type and mutant hTERT, the GST-tagged proteins were coimmunoprecipitated by the FLAG-tagged proteins (Fig. 8B), suggesting that the C-DAT mutant proteins retained the capacity to multimerize and that the function of the C-DAT region is distinct from protein multimerization.
DISCUSSION
Circumstantial evidence from functional analysis and sequence alignment has suggested that the function of the C terminus of hTERT may differ from that of its counterpart in yeast. Most notably, while C-terminal deletions and modifications to the yeast protein are rather well tolerated, analogous mutations of the human protein have severe effects on enzyme function. These data also imply that the C terminus of TERT in higher eukaryotes contains important functional domains. To identify such domains in hTERT, we introduced a comprehensive panel of NAAIRS substitution mutants into human cells and assayed for changes in the biochemical and biological function of the enzyme. NAAIRS substitution in the previously identified RT motifs 6 (or D) and 7 (or E) of hTERT inactivated the enzyme, while two out of three similar mutations in the intervening spacer region did not affect enzyme function. These RT motifs are highly conserved among telomerases, containing amino acids that are nearly invariant in all classes of reverse transcriptases (see Fig. 1A). Mutations in these motifs, but not the spacer region, of the RNA-dependent RNA polymerase of the encephalomyocarditis virus ablate catalytic activity (40). NAAIRS substitution therefore appeared sensitive enough to narrowly define regions critical for enzyme activity. Admittedly, since six amino acids are substituted at once, it is possible that the boundaries of these regions may deviate slightly from the map shown in Fig. 1 or that certain amino acids within the regions may not be essential. Nevertheless, this approach has proven highly valuable in generating a high-resolution map defining key regions of the N terminus (3) and now the C terminus of hTERT.
The four regions denoted E-I, -II, -III, and -IV, identified as being essential for enzyme activity, were separated by spacer regions in which substitutions were less deleterious to telomerase activity. Four lines of reasoning suggest that NAAIRS substitution to these E regions disrupted some precise function of the enzyme, as opposed to globally altering protein folding. First, substitution with the sequence NAAIRS should be rather nonintrusive since the number of amino acids is unchanged and because the sequence NAAIRS can presumably conform to different secondary structures (48). Second, this type of mutagenesis had been used successfully to finely map the binding site of E2F in the pocket domain of Rb (43), RNA binding, and other domains in the N terminus of hTERT (3) and now to identify known RT motifs in hTERT. Third, similar levels of protein were detected in 293 cells expressing the various mutant forms of hTERT, arguing that the mutant proteins were not grossly misfolded and targeted for degradation, although this analysis may not detect more subtle defects when the proteins are expressed in the HA5 cells. Fourth, the association of hTR RNA was not disrupted through NAAIRS substitutions in regions E-I to E-IV.
In an alignment of the human, mouse, and Arabidopsis TERT protein sequences, the four essential regions were found to reside in the most conserved regions of the C terminus (Fig. 1A). Region E-I and RT motif 7 (or E) form one continuous stretch of sequence essential for enzyme activity. In yeast, RT motif 7 (or E) and the downstream C-terminal extension have been implicated in processive nucleotide addition during synthesis of a single telomeric repeat (38). While the present study also suggests that the function of region E-I may be related to RT motif 7 (or E), this particular defect in nucleotide addition was not noted. In addition, a highly conserved threonine residue at position +938 lies in region E-I beyond the defined RT regions, once again suggesting that E-I function may in part be related to the conserved RT motif 7 (or E). The E-I region is also implicated in protein localization and contains a putative NES that has been reported to interact with the protein CRM1 in vitro (42). Region E-II encompasses the reported binding site of 14-3-3 (42), suggesting that the proposed amphipathic helix in this region is essential not only for binding to this class of proteins but also for enzyme activity. The function of the remaining E regions remains to be elucidated. However, mutations to the E-regions did not affect hTR binding, nor did they affect protein stability, suggesting that these regions are important for some step in enzyme catalysis.
Perhaps most interesting is the observation that the +1127 NAAIRS mutation defining the C-DAT region completely abolished the biological function of telomerase while only partially affecting in vitro enzyme activity (Fig. 6). Since the level of enzyme activity was slightly lower than that of mutants that allowed cellular immortality, we investigated whether the inability to maintain telomeres and immortalize cells was related to low catalytic activity or some other yet-unidentified biological activity of hTERT. We compared the activity of the C-DAT mutant in precrisis HA1 cells to endogenous activity of the same cells that had spontaneously immortalized through hTERT expression. Since the telomerase activity of the +1127 C-DAT mutant cell line was comparable to that of the immortal HA1 cells, it was surmised that the C-DAT region was responsible for a crucial telomerase function distinct from catalysis.
Indeed, the phenotype of mutations in this region is identical to that of cells expressing a C-terminal HA-tagged hTERT protein (13, 21, 36, 54). More precise mapping revealed that the mutation of F1127KT to NAA was responsible for the loss of biological function observed in the C-DAT mutant. Since this sequence is just three amino acids away from the position of the HA tag in the hTERT-HA protein, we propose that the sequence F1127KT represents the region that the HA tag exerts its effect upon. Although the exact function of the C-DAT region remains to be resolved, it is apparently unrelated to the ability of the protein to localize to the nucleus or form multimeric complexes. We therefore speculate that the extreme C terminus of hTERT may have evolved to augment some aspect of in vivo telomere elongation, such as binding to telomeres, recognizing the telomere end, or possibly another step in the telomere elongation process itself. We note that the loss of yeast proteins that reside in the telomerase pathway also give rise to telomerase-positive cells which lose telomeric DNA (27). Although speculative, it is possible that the C-terminal region of human TERT defined by mutations to F1127KT may be involved in interactions with mammalian equivalents to these yeast proteins. We also note that the HA tag perturbs binding of hTERT to hnRNP C1 and C2, which are known to colocalize with telomere binding proteins (15a).
While the telomerase-mediated replication of chromosome ends has been a highly conserved theme throughout evolution, the enzyme itself does display differences in its function. For example, the ciliate telomerases are extraordinarily processive (17, 18) and, interestingly, contain specialized motifs not conserved in other organisms (10, 29). We now define several essential regions required for the full biological function of the human enzyme that apparently are not conserved in lower eukaryotes. The human telomerase enzyme does differ, for example, from yeast telomerase in its regulation, fidelity, and association with a much smaller RNA subunit (11, 45). In addition, the telomere repeat sequence, length, and perhaps even configuration differ between these two organisms (19, 52). We speculate that the newly identified domains may encode functions that are unique to higher eukaryotes, which ultimately may be manifested in the replication of telomeres. Since telomerase is activated in the vast majority of human tumors (44), defining such functional domains of the hTERT protein will be important for elucidating how this enzyme functions in malignant cells. Ultimately this may provide insights for the specific inhibition of this pathway for the treatment of human cancers.
Acknowledgments
We thank Nesrin Hamad, Blaine Armbruster, Tso-Pang Yao, Mathew Meyerson, Bill Hahn, John and Ian York, Scott Tenenbaum, James Mitchell, Kathy Collins, and the members of the Xiao-Fan Wang and Ann Marie Pendergast laboratories for helpful discussions and Tony Means and Ann Marie Pendergast for critical reading of the manuscript. We thank Roger Reddel for the plasmid phTRA, Tso-Pang Yao for the plasmid pCMV-HDAC1, Mariano Garcia-Blanco for the plasmid pBSK-U6, and Mike Ward for technical assistance.
S.S.R.B. holds a predoctoral fellowship from the U.S. Department of Defense, and C.M.C. is a Kimmel Scholar. This work was supported by grants from the National Cancer Institute (CA82481) and the V-Foundation.
REFERENCES
- 1.Andersson, S., D. L. Davis, H. Dahlback, H. Jornvall, and D. W. Russell. 1989. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J. Biol. Chem. 264:8222-8229. [PubMed] [Google Scholar]
- 2.Arai, K., K. Masutomi, S. Khurts, S. Kaneko, K. Kobayashi, and S. Murakami. 2002. Two independent regions of human telomerase reverse transcriptase are important for its oligomerization and telomerase activity. J. Biol. Chem. 277:8538-8544. [DOI] [PubMed] [Google Scholar]
- 3.Armbruster, B. N., S. S. Banik, C. Guo, A. C. Smith, and C. M. Counter. 2001. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 21:7775-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachand, F., and C. Autexier. 1999. Functional reconstitution of human telomerase expressed in Saccharomyces cerevisiae. J. Biol. Chem. 274:38027-38031. [DOI] [PubMed] [Google Scholar]
- 5.Bachand, F., and C. Autexier. 2001. Functional regions of human telomerase reverse transcriptase and human telomerase RNA required for telomerase activity and RNA-protein interactions. Mol. Cell. Biol. 21:1888-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 2001. Functional multimerization of the human telomerase reverse transcriptase. Mol. Cell. Biol. 21:6151-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 2000. Polymerization defects within human telomerase are distinct from telomerase RNA and TEP1 binding. Mol. Biol. Cell 11:3329-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beattie, T. L., W. Zhou, M. O. Robinson, and L. Harrington. 1998. Reconstitution of human telomerase activity in vitro. Curr. Biol. 8:177-180. [DOI] [PubMed] [Google Scholar]
- 9.Bryan, T. M., K. J. Goodrich, and T. R. Cech. 2000. Telomerase RNA bound by protein motifs specific to telomerase reverse transcriptase. Mol. Cell 6:493-499. [DOI] [PubMed] [Google Scholar]
- 10.Bryan, T. M., J. M. Sperger, K. B. Chapman, and T. R. Cech. 1998. Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl. Acad. Sci. USA 95:8479-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohn, M., and E. H. Blackburn. 1995. Telomerase in yeast. Science 269:396-400. [DOI] [PubMed] [Google Scholar]
- 12.Counter, C. M., A. A. Avilion, C. E. Le Feuvre, N. G. Stewart, C. W. Greider, C. B. Harley, and S. Bacchetti. 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11:1921-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Counter, C. M., W. C. Hahn, W. Wei, S. D. Caddle, R. L. Beijersbergen, P. M. Lansdorp, J. M. Sedivy, and R. A. Weinberg. 1998. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. USA 95:14723-14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Counter, C. M., M. Meyerson, E. N. Eaton, L. W. Ellisen, S. D. Caddle, D. A. Haber, and R. A. Weinberg. 1998. Telomerase activity is restored in human cells by ectopic expression of hTERT (hEST2), the catalytic subunit of telomerase. Oncogene 16:1217-1222. [DOI] [PubMed] [Google Scholar]
- 15.Counter, C. M., M. Meyerson, E. N. Eaton, and R. A. Weinberg. 1997. The catalytic subunit of yeast telomerase. Proc. Natl. Acad. Sci. USA 94:9202-9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Ford, L. P., J. M. Suh, W. E. Wright, and J. W. Shay. 2000. Heterogeneous nuclear ribonucleoproteins C1 and C2 associate with the RNA component of human telomerase. Mol. Cell. Biol. 20:9084-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedman, K. L., and T. R. Cech. 1999. Essential functions of amino-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 13:2863-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greider, C. W. 1991. Telomerase is processive. Mol. Cell. Biol. 11:4572-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greider, C. W., and E. H. Blackburn. 1985. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43:405-413. [DOI] [PubMed] [Google Scholar]
- 19.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. de Lange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 20.Hahn, W. C., C. M. Counter, A. S. Lundberg, R. L. Beijersbergen, M. W. Brooks, and R. A. Weinberg. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464-468. [DOI] [PubMed] [Google Scholar]
- 21.Halvorsen, T. L., G. Leibowitz, and F. Levine. 1999. Telomerase activity is sufficient to allow transformed cells to escape from crisis. Mol. Cell. Biol. 19:1864-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrington, L., W. Zhou, T. McPhail, R. Oulton, D. S. Yeung, V. Mar, M. B. Bass, and M. O. Robinson. 1997. Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev. 11:3109-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holt, S. E., D. L. Aisner, J. Baur, V. M. Tesmer, M. Dy, M. Ouellette, J. B. Trager, G. B. Morin, D. O. Toft, J. W. Shay, W. E. Wright, and M. A. White. 1999. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev. 13:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilian, A., D. D. L. Bowtell, H. E. Abud, G. R. Hime, D. J. Venter, P. K. Keese, E. L. Duncan, R. R. Reddel, and R. A. Jefferson. 1997. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum. Mol. Genet. 6:2011-2019. [DOI] [PubMed] [Google Scholar]
- 25.Kim, N. W., and F. Wu. 1997. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res. 25:2595-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, C. K., J. R. Mitchell, and K. Collins. 2001. RNA binding domain of telomerase reverse transcriptase. Mol. Cell. Biol 21:990-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lingner, J., T. R. Cech, T. R. Hughes, and V. Lundblad. 1997. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA 94:11190-11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyerson, M., C. M. Counter, E. N. Eaton, L. W. Ellisen, P. Steiner, S. D. Caddle, L. Ziaugra, R. L. Beijersbergen, M. J. Davidoff, Q. Liu, S. Bacchetti, D. A. Haber, and R. A. Weinberg. 1997. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 90:785-795. [DOI] [PubMed] [Google Scholar]
- 29.Miller, M. C., J. K. Liu, and K. Collins. 2000. Template definition by Tetrahymena telomerase reverse transcriptase. EMBO J. 19:4412-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitchell, J. R., E. Wood, and K. Collins. 1999. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 402:551-555. [DOI] [PubMed] [Google Scholar]
- 31.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriarty, T. J., S. Huard, S. Dupuis, and C. Autexier. 2002. Functional multimerization of human telomerase requires an RNA interaction domain in the N terminus of the catalytic subunit. Mol. Cell. Biol. 22:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura, T. M., and T. R. Cech. 1998. Reversing time: origin of telomerase. Cell 92:587-590. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, T. M., G. B. Morin, K. B. Chapman, S. L. Weinrich, W. H. Andrews, J. Lingner, C. B. Harley, and T. R. Cech. 1997. Telomerase catalytic subunit homologs from fission yeast and human. Science 277:955-959. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama, J., H. Tahara, E. Tahara, M. Saito, K. Ito, H. Nakamura, T. Nakanishi, T. Ide, and F. Ishikawa. 1998. Telomerase activation by hTRT in human normal fibroblasts and hepatocellular carcinomas. Nat. Genet. 18:65-68. [DOI] [PubMed] [Google Scholar]
- 36.Ouellette, M. M., D. L. Aisner, I. Savre-Train, W. E. Wright, and J. W. Shay. 1999. Telomerase activity does not always imply telomere maintenance. Biochem. Biophys. Res. Commun. 254:795-803. [DOI] [PubMed] [Google Scholar]
- 37.Ouellette, M. M., M. Liao, B. S. Herbert, M. Johnson, S. E. Holt, H. S. Liss, J. W. Shay, and W. E. Wright. 2000. Subsenescent telomere lengths in fibroblasts immortalized by limiting amounts of telomerase. J. Biol. Chem. 275:10072-10076. [DOI] [PubMed] [Google Scholar]
- 38.Peng, Y., I. S. Mian, and N. F. Lue. 2001. Analysis of telomerase processivity: mechanistic similarity to HIV-1 reverse transcriptase and role in telomere maintenance. Mol. Cell 7:1201-1211. [DOI] [PubMed] [Google Scholar]
- 39.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sankar, S., and A. G. Porter. 1992. Point mutations which drastically affect the polymerization activity of encephalomyocarditis virus RNA-dependent RNA polymerase correspond to the active site of Escherichia coli DNA polymerase I. J. Biol. Chem. 267:10168-10176. [PubMed] [Google Scholar]
- 41.Sedivy, J. M. 1998. Can ends justify the means?: telomeres and the mechanisms of replicative senescence and immortalization in mammalian cells. Proc. Natl. Acad. Sci. USA 95:9078-9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seimiya, H., H. Sawada, Y. Muramatsu, M. Shimizu, K. Ohko, K. Yamane, and K. Tsuruo. 2000. Involvement of 14-3-3 proteins in nuclear localization of telomerase. EMBO J. 19:2652-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sellers, W. R., B. G. Novitch, S. Miyake, A. Heith, G. A. Otterson, F. J. Kaye, A. B. Lassar, and W. G. Kaelin, Jr. 1998. Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 12:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shay, J. W., and S. Bacchetti. 1997. A survey of telomerase activity in human cancer. Eur. J. Cancer 33:787-791. [DOI] [PubMed] [Google Scholar]
- 45.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 46.Stewart, N., and S. Bacchetti. 1991. Expression of SV40 large T antigen, but not small t antigen, is required for the induction of chromosomal aberrations in transformed human cells. Virology 180:49-57. [DOI] [PubMed] [Google Scholar]
- 47.Weinrich, S. L., R. Pruzan, L. Ma, M. Ouellette, V. M. Tesmer, S. E. Holt, A. G. Bodnar, S. Lichtsteiner, N. W. Kim, J. B. Trager, R. D. Taylor, R. Carlos, W. H. Andrews, W. E. Wright, J. W. Shay, C. B. Harley, and G. B. Morin. 1997. Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nat. Genet. 17:498-503. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, I. A., D. H. Haft, E. D. Getzoff, J. A. Tainer, R. A. Lerner, and S. Brenner. 1985. Identical short peptide sequences in unrelated proteins can have different conformations: a testing ground for theories of immune recognition. Proc. Natl. Acad. Sci. USA 82:5255-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia, J., Y. Peng, I. S. Mian, and N. F. Lue. 2000. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 20:5196-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, J., E. Chang, A. M. Cherry, C. D. Bangs, Y. Oei, A. Bodnar, A. Bronstein, C. P. Chiu, and G. S. Herron. 1999. Human endothelial cell life extension by telomerase expression. J. Biol. Chem. 274:26141-26148. [DOI] [PubMed] [Google Scholar]
- 52.Zakian, V. A. 1996. Structure, function, and replication of Saccharomyces cerevisiae telomeres. Annu. Rev. Genet. 30:141-172. [DOI] [PubMed] [Google Scholar]
- 53.Zakian, V. A. 1995. Telomeres: beginning to understand the end. Science 270:1601-1607. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, J., H. Wang, J. M. Bishop, and E. H. Blackburn. 1999. Telomerase extends the lifespan of virus-transformed human cells without net telomere lengthening. Proc. Natl. Acad. Sci. USA 96:3723-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]