Abstract
Lysophosphatidic acid (LPA), a bioactive lipid produced by several cell types including postmitotic neurons and activated platelets, is thought to be involved in various biological processes, including brain development. Three cognate G protein-coupled receptors encoded by lpa1/lpA1/Edg-2/Gpcr26, lpa2/lpA2/Edg-4, and lpa3/lpA3/Edg-7 mediate the cellular effects of LPA. We have previously shown that deletion of lpa1 in mice results in craniofacial dysmorphism, semilethality due to defective suckling behavior, and generation of a small fraction of pups with frontal hematoma. To further investigate the role of these receptors and LPA signaling in the organism, we deleted lpa2 in mice. Homozygous knockout (lpa2(−/−)) mice were born at the expected frequency and displayed no obvious phenotypic abnormalities. Intercrosses allowed generation of lpa1(−/−) lpa2(−/−) double knockout mice, which displayed no additional phenotypic abnormalities relative to lpa1(−/−) mice except for an increased incidence of perinatal frontal hematoma. Histological analyses of lpa1(−/−) lpa2(−/−) embryonic cerebral cortices did not reveal obvious differences in the proliferating cell population. However, many LPA-induced responses, including phospholipase C activation, Ca2+ mobilization, adenylyl cyclase activation, proliferation, JNK activation, Akt activation, and stress fiber formation, were absent or severely reduced in embryonic fibroblasts derived from lpa1(−/−) lpa2(−/−) mice. Except for adenylyl cyclase activation [which was nearly abolished in lpa1(−/−) fibroblasts], these responses were only partially affected in lpa1(−/−) and lpa2(−/−) fibroblasts. Thus, although LPA2 is not essential for normal mouse development, it does act redundantly with LPA1 to mediate most LPA responses in fibroblasts.
Lysophosphatidic acid (LPA) is a bioactive lipid component of serum produced by many cell types, such as activated platelets (17) and postmitotic neurons in culture (19). LPA induces a variety of cellular responses in most cell types, including intracellular calcium mobilization, stress fiber formation, cell rounding, neurite retraction, proliferation, and survival (30, 34, 37, 38; reviewed in references 9, 20, and 33). Three cognate G protein-coupled receptors encoded by lpa1/Edg-2/Gpcr26, lpa2/Edg-4, and lpa3/Edg-7 have been shown to mediate the cellular effects of LPA in mammals (3, 5, 7, 22, 24, 26, 27; reviewed in references 10, 15, and 20). All three receptors can mediate LPA-induced phospholipase C (PLC) activation and calcium mobilization, whereas only LPA1 and LPA2 can mediate LPA-induced Rho activation required for morphological effects (3, 4, 7, 22, 26, 27).
Based on the presumed locales of LPA production and its effects on neuroblast cells, it has been hypothesized that LPA plays a role in brain development (9). In embryonic cerebral cortex, for example, LPA is thought to mediate numerous aspects of progenitor behavior, including proliferation and cell cycle-associated morphological changes involving process retraction and nuclear movement (19, 21, 23, 24). In general, however, hypotheses regarding the role of LPA in cerebral cortex development have been difficult to assess due to the lack of LPA agonists and antagonists specific to each LPA receptor.
One way of overcoming such difficulties is to delete each LPA receptor gene in mice. Mice lacking the lpa1 gene [lpa1(−/−) knockout mice] were expected to have nervous system defects, because lpa1 is abundantly expressed in progenitor cells of the embryonic cerebral cortex and myelinating glial cells of both the peripheral and central nervous systems (1, 2, 24, 38, 39). Indeed, the most severe phenotype of lpa1(−/−) mice was approximately 50% neonatal lethality due to defective suckling, attributable to defective olfaction (14). Although this was likely related to the loss of LPA responses in neuroblasts of the embryonic cortex or olfactory bulb, no specific abnormalities could be discerned in histological sections of brains (14). Other phenotypes were also observed in the surviving mice, including decreased size, craniofacial dysmorphism, increased apoptosis in peripheral nerve, and a low incidence of frontal hematoma in perinatal pups (approximately 2.5%) (14). These results supported a role for LPA signaling through the LPA1 receptor in nervous system development, as well as in development of other regions of the body.
Further information regarding physiological roles of LPA signaling could be obtained through deletion of additional receptor genes. The lpa2 gene was the second such gene identified, initially through sequence similarity searches using the lpa1 sequence (3, 11). Expression of the mouse lpa2 transcript was found to be most abundant in kidney, testis, and embryonic brain, with low levels found in numerous other organs (13). When the LPA2 receptor was expressed in mammalian cells, it mediated many of the same responses to LPA as LPA1 (4, 27), suggesting a functional redundancy in cells that express both receptors. Such cells might be rather widespread, since the lpa1 and lpa2 transcripts are coexpressed in many mouse organs and cells (e.g., testis, embryonic brain, sciatic nerve, and Schwann cells [13, 14]). Thus, it was expected that lpa1(−/−) lpa2(−/−) double knockout mice would display much more severe phenotypes than lpa1(−/−) single knockout mice. Here we describe the generation and phenotype of lpa2(−/−) single and lpa1(−/−) lpa2(−/−) double knockout mice as well as the LPA responses of cells derived from these mice.
MATERIALS AND METHODS
Materials.
LPA (1-oleoyl-2-hydroxy-sn-glycero-3-phosphate) and sphingosine-1-phoshate (S1P) were purchased from Avanti Polar Lipids (Alabaster, Ala.). R1 embryonic stem (ES) cells were generous gifts from Jamey D. Marth (University of California, San Diego). Trizol and all cell culture reagents were purchased from Life Technologies, Inc. Forskolin, 3-isobutyl-1-methylxanthine (IBMX), and other reagents were purchased from Sigma, unless otherwise noted.
Generation of lpa2 mutant mice.
Isolation and characterization of an lpa2 clone from a 129/SvJ mouse genomic DNA library was described previously (12). The 5.0-kb XhoI/NotI genomic subclone containing exons 1, 2, and part of 3 (XN5.0) was used as starting material. A 599-bp fragment containing the second half of exon 2 (with part of the adjacent intron) was replaced with 1,651 bp of the neomycin resistance gene, creating the lpa2 knockout targeting vector. Twenty micrograms of NotI-linearized lpa2 knockout vector was electroporated into R1 ES cells. Six homologous recombinant clones were isolated from 384 G418-resistant colonies screened, and the correct integration was confirmed by Southern blot analyses. Two of these recombinant clones were injected into C57BL/6 blastocysts, producing chimeric male mice. They were bred with C57BL/6 females. All analyses reported here were from their progenies on a mixed 129/SvJ and C57BL/6 background (with 25 to 50% 129/SvJ). All methods used in this study were approved by the Animal Subjects Program of the University of California, San Diego, and conform to National Institutes of Health guidelines and public law.
Genotyping of lpa2 mutant mice.
Genotyping for lpa2 alleles was done by Southern blot analysis or PCR. For Southern genotyping, BamHI-digested genomic DNA (isolated from embryonic, neonatal, or weanling tails) was probed with two radiolabeled probes, by methods similar to those detailed previously (11). Probe P1/P5 was located outside of the recombination site and was obtained from the XN5.0 clone by PCR amplification with the following two primers: P1 (5′-TCTTGTCTGTTCTTGCACATTTGTC-3′) and P2 (5′-CCACTCGTGCCGCACTACCTT-3′). Probe e2b/e2b′ was located entirely within the deleted region of exon 2 and was obtained from the same clone by PCR amplification with the following two primers: e2b (5′-GGCCGTGTGGTCACACTC-3′) and e2b′ (5′-CCCAGAATGATGACAACCGTCTT-3′). For PCR genotyping, genomic DNA was used as a template in the PCR (35 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 2 min) using the following three primers: lpA2e2mh (5′-CCTACCTCTTCCTCATGTTC-3′), lpA2e2m2 (5′-TGTGCAGGTAGCAACCCCAGA-3′), and Neo2b (5′-CAGCTGGGGCTCGACTAGAGGAT-3′). Expected product sizes for wild-type and targeted alleles were 249 and 207 bp, respectively.
Northern blot analysis and RT-PCR.
Total RNA was isolated using Trizol following the manufacturer's instructions. Northern analysis was done as described previously (27). Northern probes for lpa1 and lpa2 consisted of the deleted regions within the coding regions (748 and 311 bp [probe e2b/e2b′], respectively). The probe for lpa3 consisted of the full coding regions. Reverse transcriptase PCR (RT-PCR) for lpa2 (and β-actin as a control) was done as described previously (12). All analyses were performed at least in triplicate, with representative results shown below in the figures. For the Northern blotting, a phosphorimager was used to quantitate and compare specific band intensities between samples.
Histological analysis.
For lpa2 single knockout studies, lpa2(+/−) heterozygous males and females were bred to obtain all three genotypes [wild type, lpa2(+/−) heterozygous, and lpa2(−/−) knockout] within the litters, and these wild-type and lpa2(−/−) knockout littermates were comparatively analyzed. For lpa1 lpa2 double knockout studies, lpa1(+/−) lpa2(+/−) double heterozygous and lpa1(−/−) lpa2(−/−) double knockout mice were bred to obtain lpa1(−/−) lpa2(−/−) double knockout mice, and these were compared to littermate lpa1(+/−) lpa2(+/−) double heterozygous mice and nonlittermate wild-type mice. In both studies, mice were analyzed at two developmental stages: embryonic day 14 (E14) and 12 weeks. For fixation, adult mice (pregnant and nonpregnant) were anesthetized with pentobarbital sodium (Nembutal) solution (0.75 mg/g of body weight) (Abbott) and perfused through the heart with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde in PBS. Each tissue was dissected, postfixed overnight in 4% paraformaldehyde in PBS at 4°C, and processed for paraffin embedding. Parasagittal sections (5 μm) were cut, processed, and stained with hematoxylin and eosin according to standard protocols. For embryonic cerebral cortex analyses, pregnant dams were intraperitoneally injected with 3.08 mg of bromodeoxyuridine (BrdU) per ml 1 or 4.5 h before killing by cervical dislocation. Embryos (E14) were fresh-frozen, sectioned, and stained for BrdU, Nissl substance, and/or fragmented DNA (indicative of apoptosis), as previously described (8).
Preparation of MEF cells.
Mouse embryonic fibroblast (MEF) cells were prepared from E14 embryos generated by wild-type or knockout [lpa1(−/−) single, lpa2(−/−) single, and lpa1(−/−) lpa2(−/−) double] intercrosses as described previously (28). MEF cells were maintained as a monolayer culture on tissue culture dishes, and cells from the second to fourth passages were used for the Northern blot analysis and all functional assays.
PLC assay.
For the PLC assay, MEF cells on 12-well dishes were prelabeled with [3H]inositol in inositol-free medium and assayed as described previously (27, 28). Briefly, the cells were preincubated in buffer containing 10 mM LiCl and stimulated with LPA or S1P for 20 min. Radioactivity in the inositol phosphate (IP1, IP2, and IP3) fractions of the samples was examined, and the activity was expressed as the fold induction above a basal level. In some experiments, cells were infected with retroviruses to express each of the LPA receptors as described previously (27, 28).
Ca2+ mobilization assay.
MEF cells loaded with Fura 2 were analyzed for Ca2+ fluorescence as described previously (29). Briefly, MEF cells were loaded with 1 μM Fura 2/AM in HEPES-Tyrode's-bovine serum albumin (BSA) buffer for 1 h. Cells were stimulated successively for 3 min with 10 μM LPA and 10 μM S1P in the same buffer containing 1.8 mM CaCl2. Measurements of intracellular Ca2+ concentration were performed using a Hitachi F-2000 fluorescence spectrophotometer at excitation wavelengths of 340 and 380 nm and an emission wavelength of 510 nm. Conversion of the 340 nm/380 nm ratio value into nanomolar intracellular Ca2+ was estimated by comparing the cellular fluorescence ratio with ratios acquired with buffers containing known Ca2+ concentrations.
Adenylyl cyclase assay.
MEF cells on 24-well dishes were analyzed for intracellular cyclic AMP (cAMP) content by using a cAMP enzyme immunoassay system (Amersham Pharmacia Biotech) (28, 31). Briefly, the cells were preincubated in buffer containing 0.5 mM IBMX and stimulated with LPA in the absence or presence of 1 μM forskolin for 20 min. The activity was expressed as the percentage of basal level or forskolin-induced cAMP accumulation.
Proliferation assay.
For proliferation assays, MEFs at passage 3 or 4 were seeded onto 24-well plates and allowed to grow until approximately 80% confluent. After serum starving the cells for 1 day, fresh serum-free medium was added along with various concentrations of LPA, S1P, or fetal calf serum. LPA and S1P were prepared as 10 mM and 1 mM solutions, respectively, in PBS with 1% BSA. Vehicle solution consisted of PBS with 1% BSA. After 24 h of incubation, cells were labeled with BrdU for 30 min and then fixed and processed for BrdU detection using the BrdU Labeling and Detection Kit II (Roche). Cell nuclei were counterstained by incubating with 0.35 μg of 4,6-diamino-2-phenylindole (DAPI) per ml for 10 min. The percentage of cells labeled with BrdU in each well was determined by dividing the total number of BrdU-stained nuclei by the total number of nuclei (including both BrdU-labeled and those fluorescent with DAPI). At least 800 cells were counted in each well. Although basal proliferation rates varied between experiments (approximately 1 to 6%), the mitogen-induced fold induction was relatively constant. Initial experiments indicated that 100 μM LPA yielded the most robust proliferation response of a 0.1-to-100 μM series, and this was therefore used as the treatment concentration for the knockout cell experiment.
Kinase assays.
Confluent MEFs on 10-cm or 15-cm dishes were treated with control BSA solution, 10 μM LPA, or 1 μM S1P for 7.5 min, and then the reaction was stopped by adding ice-cold PBS. Total protein was isolated by incubating cells for 10 min in ice-cold lysis buffer (600 μl/10-cm dish) consisting of 50 mM Tris (pH 7.6), 1% (wt/vol) Triton X-100, 500 mM NaCl, 10 mM MgCl2, 10-μg/ml concentrations each of leupeptin and aprotinin, and 1 mM phenylmethylsulfonyl fluoride. After centrifugation at 13,000 × g for 10 min, supernatants were removed, Laemmli buffer was added to 1×, and the samples were boiled for 5 min and then frozen. Electrophoresis, Western blotting, and detection using electrochemiluminescence were done as previously described (28). Kinase activation was assessed by examining kinase autophosphorylation using phospho-specific antibodies. Polyclonal rabbit antibodies used for the JNK and Erk1/2 assays were phospho-SAPK/JNK (Thr183/Tyr185), SAPK/JNK, phospho-p44/42 mitogen-activated protein (MAP) kinase (Thr202/Tyr204), and P44/42 MAP kinase (all from Cell Signaling Technologies). For the Akt assay, a polyclonal rabbit p-Akt1 (Ser473)-R and a mouse monoclonal Akt1 (B-1) antibody were used (Santa Cruz Biotechnology).
Stress fiber formation assay.
Meninges were dissected from brains of wild-type or knockout [lpa1(−/−) single, lpa2(−/−) single, and lpa1(−/−) lpa2(−/−) double] embryos at E13 and triturated with a fire-polished Pasteur pipette in culture medium. Mouse embryonic meningeal fibroblast (MEMF) cells were seeded onto Cell-Tak-coated glass coverslips (200 to 500 cells per 12-mm coverslip) and cultured at 37°C under 5% CO2 for 24 h. Cells were treated with LPA or S1P and stained for f-actin with tetramethyl rhodamine isocyanate (TRITC)-phalloidin (0.1 μg/ml) as previously described (22).
RESULTS
Targeted deletion of lpa2.
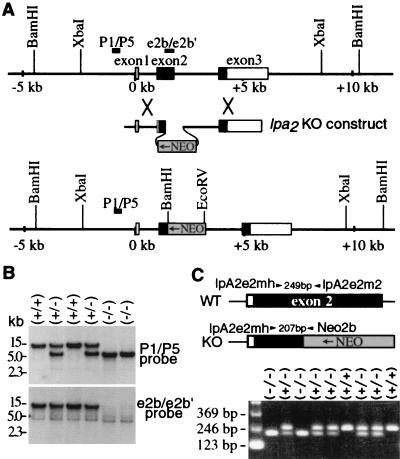
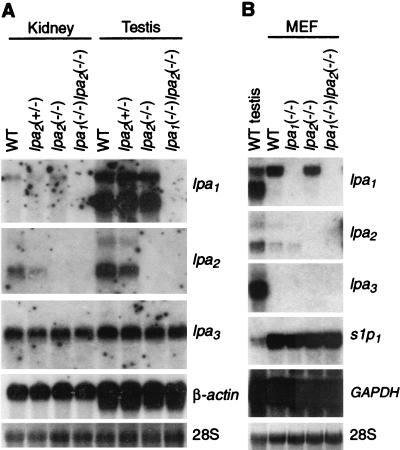
The mouse lpa2 gene consists of three exons, with the start codon and 68% of the coding region (including putative transmembrane domains I to VI) located in exon 2 (12). Therefore, deletions in exon 2 were created in a plasmid construct in which the second half of the exon was replaced with the neomycin resistance gene in reverse orientation (Fig. 1A). Homologous integration of this construct into the mouse genome results in an lpa2 gene with deletions of coding regions for transmembrane domains IV to VI and the intron donor site just after exon 2. Correctly targeted ES cell clones were injected into C57BL/6 blastocysts, producing chimeric mice. Chimeric male mice were mated to C57BL/6 females and the progeny were intercrossed. Correct targeting and deletion in the lpa2 gene was confirmed in lpa2(+/−) heterozygous and lpa2(−/−) homozygous mice through Southern blotting and PCR analyses of genomic DNA (Fig. 1B and C). The absence of lpa2 transcripts from the deleted region in knockout mice was confirmed both by Northern blot analysis (Fig. 2A) and RT-PCR (data not shown) on adult tissues (kidney and testis) in which lpa2 is normally abundantly expressed (12). In each mouse tissue, similar levels of lpa1 or lpa3 expression were observed in all lpa2 genotypes (Fig. 2A).
FIG. 1.
Targeted disruption of the lpa2 gene. (A) Genomic map of the wild-type and homologously recombined lpa2 locus, including exons 1 to 3 (boxes), the targeting construct, and location of the Southern probes (P1/P5 and e2b/e2b′). (B) Southern blot analysis of BamHI-digested genomic tail DNA (10 μg/lane) from mice generated by crossing heterozygotes. The lpa2 genotypes are shown above. The blot was hybridized with both probes P1/P5 and e2b/e2b′. (C) PCR assay with three primers (lpA2e2mh, lpA2e2m2, and Neo2b) detecting the inheritance of the wild-type lpa2 allele (249 bp) and mutant lpa2 allele (207 bp). The relative locations of the three primers are shown above.
FIG. 2.
LPA receptor gene expression in adult kidney and testis, and MEF cells prepared from E14 embryos. Northern blots containing 20 μg of total RNA isolated from adult kidney and testis (A) and MEF cells prepared from embryos (B) were probed with cDNA fragments from the indicated mouse genes. Blots probed with cDNAs of β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ethidium bromide-stained 28S rRNA are shown as loading controls. In panel B, testis blots are shown as a positive control for expression of each gene.
No obvious phenotypic abnormalities were found in lpa2(−/−) mice.
Embryonic and adult lpa2(−/−) mice were grossly indistinguishable from their nonhomozygous siblings in appearance, size, and general behavior. Genotype analysis indicated that lpa2(−/−) mice were born at the expected Mendelian frequency without sexual bias (Table 1). Reproductively, lpa2(−/−) mice were normal in terms of behavior, frequency of mating, and litter sizes. All major internal adult organs were grossly normal. We further analyzed internal adult organs using perfused fixed tissue sections that were stained with hematoxylin and eosin or with cresyl violet. There were no obvious differences between the wild-type and lpa2(−/−) mice in brain, skin, muscle, spleen, heart, lung, kidney, intestine, or stomach. Similarly, no differences were observed in overall appearance, gross anatomy, or histology of embryos at E14. In addition, several of our lpa2(−/−) mice have survived nearly 2 years, indicating that longevity is normal.
TABLE 1.
Inheritance of the lpa2 deletion allele
| Parental lpa2 genotypes | No. (%) of offspring with lpa2 genotypea
|
No. of −/− expected | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Wild-type background | ||||
| +/+ × +/− | 125 (52) | 115 (48) | 0 (0) | 0 |
| +/− × +/− | 52 (24) | 110 (51) | 53 (25) | 54 |
| +/− × −/− | 0 (0) | 133 (53) | 118 (47) | 133 |
| Ratio of sexes (female:male) | 90:85 | 188:170 | 96:93 | |
| lpa1(−/−) background | ||||
| +/− × +/− | 14 (27) | 29 (56) | 9 (17) | 14 |
| +/− × −/− | 0 (0) | 28 (57) | 21 (43) | 28 |
Numbers of individual progeny genotyped from the indicated crosses are shown. In parentheses is the percentage of each genotype from a particular cross. Tailing was done at weaning age (approximately 3 weeks).
Indistinguishable phenotype of lpa1(−/−) lpa2(−/−) mice from lpa1(−/−) mice except for increased incidence of perinatal frontal hematoma.
The complete lack of a gross or histological phenotype in lpa2(−/−) mice led us to examine the effect of a combined loss of both lpa1 and lpa2. Because mouse LPA1 and LPA2 have redundant signaling properties (27), the loss of both receptors should result in a more complete loss of LPA responses in cells that express these two receptors simultaneously. Therefore, the loss of both might result in additional phenotypic defects relative to those observed in lpa1(−/−) mice (semilethality, craniofacial dysmorphism, smaller size, and frontal hematoma). Double knockout [lpa1(−/−) lpa2(−/−)] mice were obtained, and the complete absence of lpa1 or lpa2 transcripts from each of deleted regions was confirmed by Northern blot analyses on RNA isolated from kidney and testis (Fig. 2A). Unexpectedly, lpa1(−/−) lpa2(−/−) mice developed without any apparent additional phenotypes compared with lpa1(−/−) mice. Histological analyses of major adult organs revealed no obvious differences between lpa1(−/−) lpa2(−/−) mice and lpa1(−/−) mice. General behavior, reflexes, and overall motor control appeared normal, indicating no severe defects in the central or peripheral nervous systems. Several parameters in lpa1(−/−) lpa2(−/−) mice were measured that were previously analyzed in the lpa1(−/−) mice, including milk quantities in neonatal stomachs, total body mass, craniofacial dysmorphism, and incidence of perinatal frontal hematoma (14). There were no significant differences in any of these parameters except frontal hematoma, for which a much higher frequency was observed in neonatal lpa1(−/−) lpa2(−/−) pups (26%, or 9 of 34) than in lpa1(−/−) mice (2.5%, or 4 of 160). Also, a slightly increased lethality (30% increase) was associated with the loss of both lpa1 and lpa2 relative to loss of lpa1 alone (Table 1), possibly associated with the hematomas.
No obvious cerebral cortical development abnormalities in lpa1(−/−) lpa2(−/−) mice.
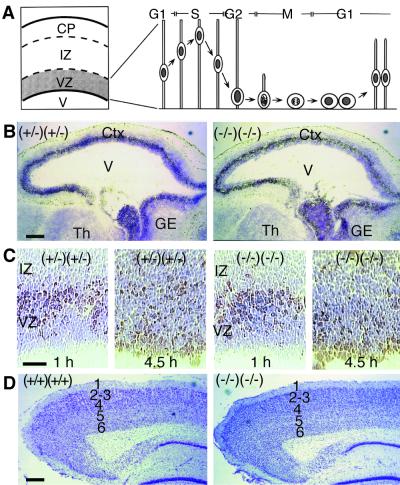
Since LPA signaling through LPA1 and LPA2 receptors is thought to be involved in neuronal development, we analyzed several histological parameters of the cerebral cortex during embryonic and postnatal ages in knockout animals. In addition to neural progenitor proliferation, LPA may be influencing “interkinetic nuclear migration,” a process in which the nucleus of proliferating neuroblasts moves away from and toward the ventricle at various stages of the cell cycle (Fig. 3A) (24). To assess the proliferating population and nuclear movements in embryonic cortex, S-phase nuclei were labeled with BrdU by injection into pregnant dams. After a 1-h pulse of BrdU, histological analysis of lpa1(−/−) lpa2(−/−) embryonic cerebral cortices revealed a normal pattern of BrdU labeling in an outer band of nuclei in the ventricular zone (Fig. 3B). By analyzing the BrdU labeling pattern at later time points, the movement of initially labeled nuclei could be tracked (because within 1 h after injection, most of the BrdU is degraded). In sections prepared 4.5 h after BrdU injection, the majority of BrdU-labeled nuclei in lpa1(−/−) lpa2(−/−) embryos had migrated inward toward the ventricle, similar to control animals (Fig. 3C). In addition, no differences in general histology, thickness, or cell counts (BrdU-labeled or total per section) were found in lpa1(−/−) lpa2(−/−) embryonic or adult cerebral cortices relative to controls (Fig. 3B, D). While other aspects of cortical development may be defective, these results indicated that general proliferation and interkinetic nuclear migration are intact in the lpa1(−/−) lpa2(−/−) embryonic cerebral cortex.
FIG. 3.
Cerebral cortex development in lpa1(−/−) lpa2(−/−) mice. (A) Schematic cross section of developing embryonic cerebral cortex, indicating cortical plate (CP), intermediate zone (IZ), ventricular zone (VZ), and lateral ventricle (V). Diagrammed to the right are the cell morphological changes (interkinetic nuclear migration) that occur during the cell cycle in the VZ. (B) Sagittal sections of E14 frontal brain areas of control lpa1(+/−) lpa2(+/−) double heterozygous [(+/−) (+/−)] and sibling lpa1(−/−) lpa2(−/−) double knockout [(−/−) (−/−)] embryos. Embyros were frozen 1 h after BrdU injection, and then sections were stained for BrdU (brown) and counterstained for Nissl substance (blue). Ctx, cortex; Th, thalamus; GE, ganglionic eminance. (C) Magnified view of sections of the cerebral cortex from E14 embryos frozen 1 or 4.5 h after injection of BrdU, showing S-phase nuclei at the outer part of the VZ after 1 h and the location of such nuclei at the inner part of the VZ after 4.5 h. (D) Cresyl violet-stained sagittal sections of the posterior cerebral cortex from a wild-type and double knockout animal, demonstrating normal cortical lamination (layers 1 to 6 are indicated). In the lower right of each panel is the dentate gyrus of the hippocampus. Bars, 200 μm (B and D) or 50 μm (C).
LPA receptor gene expression in MEF cells.
To be certain that LPA responses were diminished and/or abolished in the lpa2(−/−) and lpa1(−/−) lpa2(−/−) animals, we assessed responses to LPA in cells cultured from these animals. Fibroblast cells have robust LPA responses and have commonly been used to dissect LPA signaling pathways (18, 25, 34-37). We first determined which of the three LPA receptors were expressed in cultured MEFs (derived from E14 embryos). Using Northern blots prepared with wild-type MEF RNA, we detected high levels of lpa1 transcript and low levels of lpa2 transcript, but we did not detect lpa3 transcript (Fig. 2B). The complete absence of transcripts for lpa1, lpa2, or both was confirmed in MEF cells prepared from each type of knockout mouse [lpa1(−/−), lpa2(−/−), and lpa1(−/−) lpa2(−/−), respectively]. In addition, a related S1P receptor gene (s1p1) was also expressed at relatively high levels. The loss of function of a particular gene might result in compensatory increases in the expression of related genes that mediate the same function. However, in each of these knockout MEF cells, no differences in transcript levels were observed other than for each of the targeted genes (Fig. 2B).
PLC activation in MEF cells.
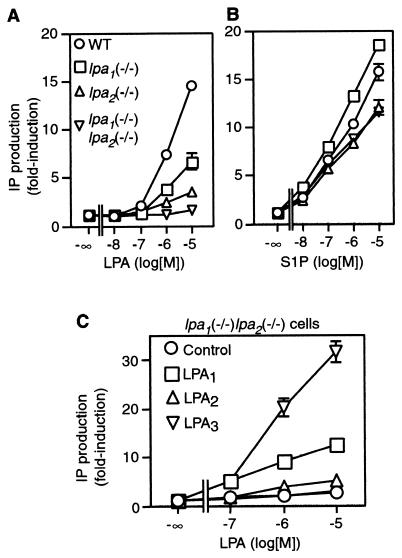
A primary response of most cell types to LPA is PLC activation, which results in IP3 and diacylglycerol production with consequent intracellular calcium mobilization and protein kinase C activation. When expressed heterologously in the cell lines, each of the three LPA receptors (LPA1, LPA2, and LPA3) could mediate this LPA response (27). Therefore, LPA-induced PLC activation was examined in wild-type and knockout MEF cells. LPA activated PLC in the wild-type cells in a concentration-dependent manner, giving a marked response at concentrations between 100 nM and 10 μM (Fig. 4A). In contrast, LPA treatment led to only modest PLC activation in either lpa1(−/−) or lpa2(−/−) MEF cells, inducing 40% or 20%, respectively, of the response achieved in the wild-type cells at concentrations up to 10 μM. LPA-induced PLC activation was abolished in lpa1(−/−) lpa2(−/−) double knockout MEF cells except at the highest concentration of LPA (10 μM), at which a slight but significant response (3% of the response achieved in the wild-type cells with 10 μM) was observed. MEF cells also express three S1P receptor genes, s1p1, s1p2 and s1p3, and are highly responsive to S1P-induced PLC activation (28). S1P-induced PLC activation was roughly comparable in the wild-type and double knockout MEF cells (Fig. 4B), indicating that bioactive lipid receptor coupling to PLC is not generally impaired. Thus, in MEF cells, nearly all PLC activation in response to LPA is dependent on endogenous expression of LPA1 and LPA2, with LPA2 playing a somewhat larger role.
FIG. 4.
LPA-induced inositol phosphate production in MEF cells. (A and B) LPA (A) and S1P (B) induced inositol phosphate production in wild-type (WT), lpa1(−/−) single, lpa2(−/−) single, and lpa1(−/−) lpa2(−/−) double-knockout MEF cells. MEF cells were prelabeled with [3H]inositol and were stimulated with LPA or S1P for 20 min. The radioactivity in the inositol phosphate fraction of the cell extract was determined. (C) LPA-induced inositol phosphate production in lpa1(−/−) lpa2(−/−) MEF cells infected with retroviruses expressing each of the LPA receptors. Activity is expressed as the fold induction above control levels. Data shown are the means ± standard errors of triplicate samples from the representative experiment. Standard error bars are not shown when bars are smaller than the size of the data points.
It is possible that very low LPA3 receptor expression (below our Northern blotting detection limit) might mediate the residual PLC activation in the lpa1(−/−) lpa2(−/−) MEF cells. To determine whether LPA3 is able to mediate this response, as well as to explore the usefulness of a reexpression approach to restore LPA responsiveness, we measured PLC activation in lpa1(−/−) lpa2(−/−) MEF cells infected with retroviruses to express each of the LPA receptors (Fig. 4C) (28). The lpa1(−/−) lpa2(−/−) cells infected to express LPA1, LPA2, or LPA3 responded to 10 μM LPA with a 4.7-fold, 2.0-fold, and 11.8-fold induction of PLC activation relative to the control, respectively (Fig. 4C). The robust coupling of LPA3 to PLC activation in MEF cells suggests that very low LPA3 receptor levels might mediate the residual PLC response in the lpa1(−/−) lpa2(−/−) cells. The lower-than-expected response mediated by the LPA2 retroviral vector (compared to that mediated by LPA1 and LPA3) was likely due to the lower LPA2 receptor expression levels resulting from this virus (previously demonstrated with Western blotting [27]).
Calcium mobilization in MEF cells.
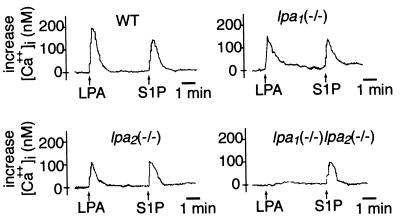
The experiments with PLC indicated that both LPA1 and LPA2 mediated LPA-induced intracellular IP production. Since IP3 is responsible for releasing Ca2+ from intracellular storage sites, we examined LPA-induced Ca2+ responses in MEFs. While moderately reduced responses were observed in lpa1(−/−) or lpa2(−/−) MEFs relative to that of wild-type MEFs, the response in lpa1(−/−) lpa2(−/−) MEFs was completely abolished (Fig. 5). All cells showed normal Ca2+ responses to S1P, indicating intact signal transduction components required for these responses.
FIG. 5.
LPA-induced intracellular calcium mobilization in MEF cells. MEF cells of the indicated genotype were loaded with Fura-2/AM and stimulated successively with 10 μM LPA and 10 μM S1P. The increases in nanomolar intracellular calcium ([Ca++]i) from the basal levels (≈150 nM) are shown. Data are representative of three independent experiments.
Adenylyl cyclase inhibition in MEF cells.
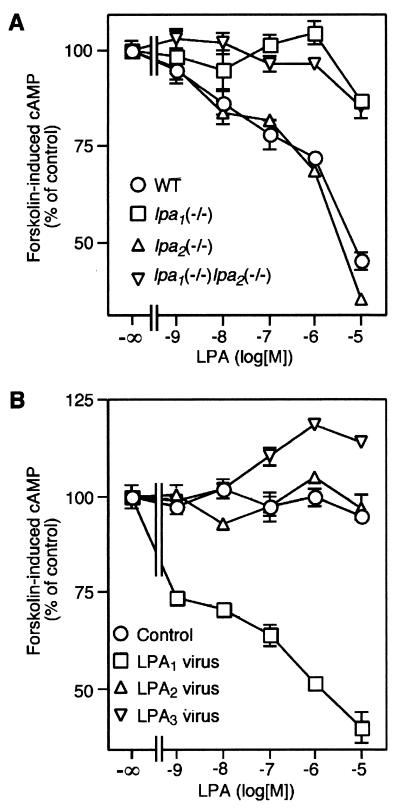
Another major cell response to LPA is adenylyl cyclase inhibition. Previous heterologous expression studies have shown that all three LPA receptors could mediate the inhibition of adenylyl cyclase in mammalian cells, although LPA1 was most effective (4, 27). LPA inhibited forskolin-induced cAMP accumulation in wild-type and lpa2(−/−) single knockout cells in a concentration-dependent manner, giving a marked response at concentrations between 10 nM and 10 μM (Fig. 6A). In contrast, LPA did not inhibit forskolin-induced cAMP accumulation in lpa1(−/−) single knockout and lpa1(−/−) lpa2(−/−) double knockout cells, except for a small residual response at 10 μM LPA. When lpa1(−/−) lpa2(−/−) MEF cells were infected with retroviruses to overexpress each of the LPA receptors (Fig. 6B), LPA-induced inhibition was observed only in LPA1-expressing cells. These results indicate that LPA1, but not LPA2, plays a major role in adenylyl cyclase inhibition in MEF cells.
FIG. 6.
LPA-induced inhibition of forskolin-induced cAMP accumulation in MEF cells. MEF cells were stimulated with increasing concentrations of LPA for 20 min in the presence of 1 μM forskolin and 0.5 mM IBMX. Intracellular cAMP contents were measured. (A) LPA-induced inhibition of forskolin-induced cAMP accumulation in wild-type (WT), lpa1(−/−) single, lpa2(−/−) single, and lpa1(−/−) lpa2(−/−) double knockout MEF cells. (B) LPA-induced inhibition of forskolin-induced cAMP accumulation in lpa1(−/−) lpa2(−/−) MEF cells infected with retroviruses expressing each of the LPA receptors. In all panels, forskolin-induced cAMP accumulation was expressed as 100%. Data shown are the means ± standard errors of triplicate samples from the representative experiment. Standard error bars are not shown when bars are smaller than the size of the data points.
Proliferation, MAP kinase, JNK, and Akt activation in MEF cells.
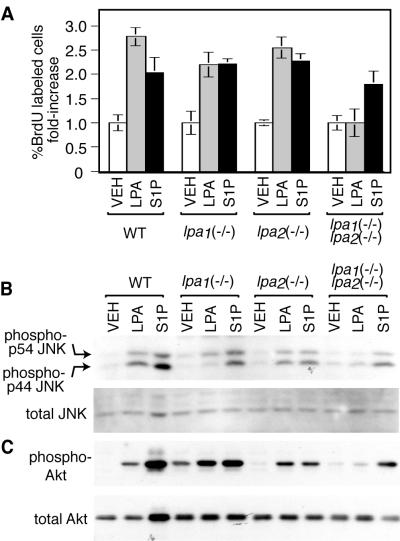
One of the initially discovered responses to LPA was increased proliferation, demonstrated in various fibroblast cell types (37). In order to determine the role of LPA1 and LPA2 receptors in this response, we analyzed proliferation of MEF cells by using a BrdU incorporation assay. While wild-type as well as lpa1(−/−) or lpa2(−/−) cells responded to LPA with 2.2- to 2.8-fold increases in the percentage of cells incorporating BrdU, lpa1(−/−) lpa2(−/−) cells showed no response (Fig. 7A). Various kinases likely to be involved in mediating the proliferative response previously have been shown to be activated by LPA in fibroblasts, including Erk1/2, JNK, and Akt (18, 25, 35, 36). Interestingly, although S1P induced activation of p44/p42 MAP kinase (Erk1/2) in wild-type MEFs, LPA did not (data not shown). However, JNK was activated by LPA in wild-type, lpa1(−/−), and lpa2(−/−) MEFs, a response abolished in lpa1(−/−) lpa2(−/−) MEFs (Fig. 7B). Similar to the activation profile of JNK, we found activation of Akt in wild-type, lpa1(−/−), and lpa2(−/−) MEFs, but no activation in lpa1(−/−) lpa2(−/−) MEFs (Fig. 7C). Thus, JNK activation, Akt activation, and the associated proliferation response to LPA are all redundantly mediated by LPA1 and LPA2 receptors in MEFs, being abolished when these two receptors are deleted.
FIG. 7.
LPA-induced proliferation, JNK activation, and Akt activation in MEF cells. (A) LPA and S1P induced increases in the percentage of cells labeled with BrdU. Data shown are the means ± standard errors of at least triplicate samples. Compared to vehicle (VEH) treatment, LPA-induced increases were significant for wild-type (WT), lpa1(−/−), and lpa2(−/−) knockout cells (P < 0.02; paired t tests), but not for lpa1(−/−) lpa2(−/−) cells. Treatment concentrations were 100 μM LPA and 10 μM S1P. The presence of 10% fetal calf serum also led to at least a fourfold increase in the percentage of BrdU-labeled cells in all genotypes (data not shown). (B and C) Western blots showing JNK activation (B) and Akt activation (C) in MEF cells of the indicated genotype, treated for 7.5 min with vehicle, 10 μM LPA, or 1 μM S1P. Blots are representative of at least duplicate experiments.
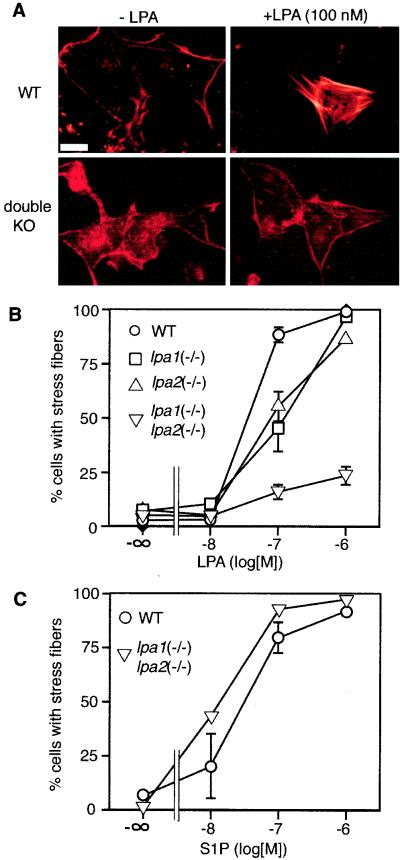
Stress fiber formation in MEMF cells.
Another prominent response of fibroblast cells to LPA is the formation of actin stress fibers (34) through activation of the small GTPase, Rho (22, 34). We previously demonstrated that LPA1 and LPA2, but not LPA3, could induce Rho-mediated cell morphological changes in mammalian cells (22, 27). The quantification of stress fibers is difficult in MEF cells because abundant stress fibers are found in unstimulated cells. We could, however, quantify stress fiber formation in fibroblast cells derived from one particular embryonic source, the meninges. Such MEMFs had a low basal percentage of cells with stress fibers (≈2%) in serum-free media. In addition, they had an expression profile for the lpa1, lpa2, lpa3, and s1p1 transcripts identical to that for MEF cells, with relatively high levels of lpa1 and s1p1, low levels of lpa2, and undetectable expression of lpa3 (data not shown). While stress fiber formation in response to LPA was observed in the wild-type, lpa1(−/−), and lpa2(−/−) MEMFs, it was severely reduced in the lpa1(−/−) lpa2(−/−) MEMFs (Fig. 8A and B). At 100 nM LPA, approximately 95% of wild-type, 45% of lpa1(−/−), 55% of lpa2(−/−), and 13% of lpa1(−/−) lpa2(−/−) cells contained stress fibers (Fig. 8B). At 1 μM LPA, approximately 100% of wild-type, 98% of lpa1(−/−) 92% of lpa2(−/−), and 23% of lpa1(−/−) lpa2(−/−) cells contained stress fibers. To demonstrate that lysophospholipid receptor coupling to stress fiber formation was not generally impaired, the cells were also stimulated with S1P. Both wild-type and lpa1(−/−) lpa2(−/−) cells responded similarly to S1P at all doses, with nearly 100% of cells showing stress fibers with 1 μM S1P (Fig. 8C).
FIG. 8.
LPA-induced stress fiber formation in MEMF cells. (A) Fluorescence microscopy of TRITC-phalloidin-stained wild-type (WT) and lpa1(−/−) lpa2(−/−) double knockout (double KO) meningeal fibroblast cells, either without stimulation (left) or after 15 min of stimulation with 100 nM LPA (right). (B) Percentages of wild-type (WT), lpa1(−/−) single, lpa2(−/−) single, and lpa1(−/−) lpa2(−/−) double knockout meningeal fibroblasts with actin stress fibers after stimulation with increasing concentrations of LPA. (C) Percentages of wild-type (WT) and lpa1(−/−) lpa2(−/−) double knockout meningeal fibroblasts with actin stress fibers after stimulation with increasing concentrations of S1P. Data shown are the means ± standard errors of triplicate samples. Bar, 10 μm.
DISCUSSION
It has long been hypothesized that the cellular and organismal effects of LPA are mediated through specific G protein-coupled receptors. Although several candidate LPA receptors have been proposed, only LPA1, LPA2, and LPA3 have emerged as bona fide LPA receptors through various levels of examination by multiple laboratories. Experiments supporting this have generally consisted of heterologous expression of the receptors in cell lines that have no endogenous LPA responses. The gene deletion experiments reported here provide additional support that these receptors mediate the endogenous LPA responses previously studied. In addition, these experiments have also allowed ascertainment of the organismal role of individual receptors and their associated LPA responses.
Unlike our previous studies on lpa1(−/−) single knockout mice, the present studies on lpa2(−/−) single knockout mice failed to reveal any obvious behavioral, anatomical, or histological defects. Previous overexpression studies demonstrated that LPA2 resembles LPA1 in mediating LPA-induced intracellular calcium mobilization, PLC activation, adenylyl cyclase inhibition, and cell morphological changes (3, 4, 27). Since lpa2 is coexpressed in several organs and cells with lpa1, we expected that deletion of both receptors would result in a more complete loss of LPA signaling and consequently a much more severe phenotype relative to either single knockout alone. Unexpectedly, we could not detect any qualitatively different phenotypes in lpa1(−/−) lpa2(−/−) mice relative to lpa1(−/−) mice. The only differences detected were an increased incidence of frontal hematomas and a slight increase in lethality, which nevertheless confirmed the hypothesis that redundant LPA signaling through LPA1 and LPA2 receptors was occurring.
Other analyses of lpa1(−/−) lpa2(−/−) mice did not reveal abnormalities relative to wild-type controls. Several aspects of cerebral cortex development thought to require LPA signaling were examined and determined to be normal in lpa1(−/−) lpa2(−/−) mice. However, while the initial hypotheses regarding LPA signaling in brain development were tested in a general sense (i.e., proliferation and nuclear movement of cerebral cortical progenitor cells), we did not examine more specific effects or other potential roles for LPA, which are likely operative, based on other studies (21, 23). LPA has also been hypothesized to influence wound healing by stimulating fibroblasts to close cutaneous wounds and possibly by attracting immune system cells to the injury site (6, 16). Although we did not examine immune system function, we did observe normal wound closure in lpa1(−/−) lpa2(−/−) mice relative to control mice (J. J. A. Contos and J. Chun, unpublished data). Thus, the essential in vivo functions of LPA2-mediated LPA signaling still remain to be determined.
Our experiments in embryonic fibroblast cells demonstrated that LPA1 and LPA2 had redundant functions in mediating multiple endogenous LPA responses, including PLC activation, Ca2+ mobilization, proliferation, JNK activation, Akt activation, and stress fiber formation. These responses are signaled through multiple types of G proteins, including Gi, Gq, and G12/13 (20). In contrast, another response (adenylyl cyclase inhibition), requiring primarily Gi, was mediated by only LPA1. These results are consistent with our previous studies, where expression of either LPA1 or LPA2 conferred PLC activation and morphological responses and where only LPA1 conferred a robust AC inhibition response (27). The observation that LPA1 mediates most of the AC inhibition response in MEFs suggests that the phenotype seen in the lpa1(−/−) mice is more related to deficient LPA-induced AC inhibition (or Gi coupling) than to the other LPA responses. Overall, the current results support the hypothesis that both LPA1 and LPA2 couple well to Gq and G12/13, and that LPA1 couples more strongly to Gi.
Although all LPA responses examined here were drastically reduced in lpa1(−/−) lpa2(−/−) cells, some significant responses still remained at the highest LPA concentrations (Fig. 4A, 6A, and 8B). There are several possible explanations for this. First, although we could not detect lpa3 transcript by Northern blotting or RT-PCR in MEF or MEMF cells, very low levels of lpa3 expression (undetectable by our methods) might be sufficient to result in small significant responses. Second, additional LPA receptors (e.g., LPA4) might exist which could mediate these responses. Although there appears to be no additional lpa gene family members in the human genome, the full mouse genome sequence has yet to be published. Our findings of a duplicated lpa1 exon 4 in Mus spretus (11) and two similar yet distinct lpa1 genes (xlpa1-1 and xlpa1-2) in Xenopus laevis (31) suggest the existence of duplicated lpa genes in various animal species. Third, the S1P receptor S1P1 might mediate some residual responses, because it has been proposed to act as a low-affinity LPA receptor (32) and our data indicate that s1p1 is expressed in fibroblast cells (Fig. 2B). However, when S1P1 was expressed using a retroviral vector in lpa1(−/−) lpa2(−/−) MEFs, S1P responses were potentiated but no additional LPA-mediated responses were observed, arguing against this explanation (I. Ishii and J. Chun, unpublished data). Fourth, non-receptor-mediated effects of LPA (e.g., direct activation of G proteins or membrane-bound effectors) might account for some responses. Sorting out the mechanism of the residual responses must await additional experiments, such as deletion of lpa3 in mice, determination of the complete mouse genome, and experiments examining non-receptor-mediated effects of LPA.
The present study demonstrated that lpa2(−/−) mice have no apparent phenotypes and that lpa1(−/−) lpa2(−/−) mice have no additional phenotypes relative to lpa1(−/−) mice, except for a higher frequency of pups with frontal hematoma. The significant losses of LPA cellular signaling observed in fibroblast cells derived from lpa1(−/−) single, lpa2(−/−) single, and lpa1(−/−) lpa2(−/−) double knockout mice demonstrate redundant and nonredundant roles for the receptors. Such information is essential to fully understanding cellular LPA signaling and the role it plays in the organism.
Acknowledgments
Isao Ishii and James Contos contributed equally to this work.
We thank Forrest Liu at the UCSD Core facility for ES cell work and blastocyst injection, Carol Akita for the maintenance of mice, and Matthew McCreight for help in counting cells in the proliferation assay.
This work was supported by the National Institute of Mental Health (grant no. K02MH01723 to J.C.), a research grant from Allelix Biopharmaceuticals (to J.C.), a grant from the Mitsubishi Pharma Research Foundation (to I.I.), and an unrestricted gift from Merck Research Laboratories (to J.C.).
REFERENCES
- 1.Allard, J., S. Barrón, J. Diaz, C. Lubetzki, B. Zalc, J. C. Schwartz, and P. Sokoloff. 1998. A rat G protein-coupled receptor selectively expressed in myelin-forming cells. Eur. J. Neurosci. 10:1045-1053. [DOI] [PubMed] [Google Scholar]
- 2.Allard, J., S. Barrón, S. Trottier, P. Cervera, C. Daumus-Duport, E. Leguern, A. Brice, J. C. Schwartz, and P. Sokoloff. 1999. Edg-2 in myelin-forming cells: isoforms, genomic mapping, and exclusion in Charcot-Marie-Tooth disease. Glia 26:176-185. [DOI] [PubMed] [Google Scholar]
- 3.An, S., T. Bleu, O. G. Hallmark, and E. J. Goetzl. 1998. Characterization of a novel subtype of human G protein-coupled receptor for lysophosphatidic acid. J. Biol. Chem. 273:7906-7910. [DOI] [PubMed] [Google Scholar]
- 4.An, S., T. Bleu, Y. Zheng, and E. J. Goetzl. 1998. Recombinant human G protein-coupled lysophosphatidic acid receptors mediate intracellular calcium mobilization. Mol. Pharmacol. 54:881-888. [DOI] [PubMed] [Google Scholar]
- 5.An, S., M. A. Dickens, T. Bleu, O. G. Hallmark, and E. J. Goetzl. 1997. Molecular cloning of the human Edg2 protein and its identification as a functional cellular receptor for lysophosphatidic acid. Biochem. Biophys. Res. Commun. 231:619-622. [DOI] [PubMed] [Google Scholar]
- 6.Balazs, L., J. Okolicany, M. Ferrebee, B. Tolley, and G. Tigyi. 2001. Topical application of the phospholipid growth factor lysophosphatidic acid promotes wound healing in vivo. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280:R466-R472. [DOI] [PubMed] [Google Scholar]
- 7.Bandoh, K., J. Aoki, H. Hosono, S. Kobayashi, T. Kobayashi, K. Murakami-Murofushi, M. Tsujimoto, H. Arai, and K. Inoue. 1999. Molecular cloning and characterization of a novel human G-protein-coupled receptor, EDG7, for lysophosphatidic acid. J. Biol. Chem. 274:27776-27785. [DOI] [PubMed] [Google Scholar]
- 8.Blaschke, A. J., K. Staley, and J. Chun. 1996. Widespread programmed cell death in proliferative and postmitotic regions of the fetal cerebral cortex. Development 122:1165-1174. [DOI] [PubMed] [Google Scholar]
- 9.Chun, J. 1999. Lysophospholipid receptors: implications for neural signaling. Crit. Rev. Neurobiol. 13:151-168. [DOI] [PubMed] [Google Scholar]
- 10.Chun, J., E. J. Goetzl, T. Hla, Y. Igarashi, K. R. Lynch, W. Moolenaar, S. Pyne, and G. Tigyi. 2002. Lysophospholipid receptor nomenclature. Pharmacol. Rev. 54:265-269. [DOI] [PubMed] [Google Scholar]
- 11.Contos, J. J., and J. Chun. 1998. Complete cDNA sequence, genomic structure, and chromosomal localization of the LPA receptor gene, lpA1/vzg-1/Gpcr26. Genomics 51:364-378. [DOI] [PubMed] [Google Scholar]
- 12.Contos, J. J., and J. Chun. 2000. Genomic characterization of the lysophosphatidic acid receptor gene, lpA2/Edg4, and identification of a frameshift mutation in a previously characterized cDNA. Genomics 64:155-169. [DOI] [PubMed] [Google Scholar]
- 13.Contos, J. J., and J. Chun. 2001. The mouse lpA3/Edg7 lysophosphatidic acid receptor gene: genomic structure, chromosomal localization, and expression pattern. Gene 267:243-253. [DOI] [PubMed] [Google Scholar]
- 14.Contos, J. J. A., N. Fukushima, J. A. Weiner, D. Kaushal, and J. Chun. 2000. Requirement for the lpA1 lysophosphatidic acid receptor gene in normal suckling behavior. Proc. Natl. Acad. Sci. USA 97:13384-13389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Contos, J. J. A., I. Ishii, and J. Chun. 2000. Lysophosphatidic acid receptors. Mol. Pharmacol. 58:1188-1196. [DOI] [PubMed] [Google Scholar]
- 16.Demoyer, J. S., T. C. Skalak, and M. E. Durieux. 2000. Lysophosphatidic acid enhances healing of acute cutaneous wounds in the mouse. Wound Repair Regen. 8:530-537. [DOI] [PubMed] [Google Scholar]
- 17.Eichholtz, T., K. Jalink, I. Fahrenfort, and W. H. Moolenaar. 1993. The bioactive phospholipid lysophosphatidic acid is released from activated platelets. Biochem. J. 291:677-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang, X., S. Yu, R. LaPushin, Y. Lu, T. Furui, L. Z. Penn, D. Stokoe, J. R. Erickson, R. C. Bast, Jr., and G. B. Mills. 2000. Lysophosphatidic acid prevents apoptosis in fibroblasts via G(i)-protein-mediated activation of mitogen-activated protein kinase. Biochem. J. 352(Pt. 1):135-143. [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushima, N., J. Weiner, and J. Chun. 2000. Lysophosphatidic acid (LPA) is a novel extracellular regulator of cortical neuroblast morphology. Dev. Biol. 228:6-18. [DOI] [PubMed] [Google Scholar]
- 20.Fukushima, N., I. Ishii, J. J. Contos, J. A. Weiner, and J. Chun. 2001. Lysophospholipid receptors. Annu. Rev. Pharmacol. Toxicol. 41:507-534. [DOI] [PubMed] [Google Scholar]
- 21.Fukushima, N., I. Ishii, Y. Habara, C. B. Allen, and J. Chun. Dual regulation of actin rearrangement through lysophosphatidic acid receptor in neuroblast cell lines. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- 22.Fukushima, N., Y. Kimura, and J. Chun. 1998. A single receptor encoded by vzg-1/lpA1/edg-2 couples to G proteins and mediates multiple cellular responses to lysophosphatidic acid. Proc. Natl. Acad. Sci. USA 95:6151-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukushima, N., J. A. Weiner, D. Kaushal, J. J. Contos, K. Y. Kim, and J. Chun. 2002. Lysophosphatidic acid influences the morphology and motility of young, postmitotic cortical neurons. Mol. Cell. Neurosci. 20:271-282. [DOI] [PubMed] [Google Scholar]
- 24.Hecht, J. H., J. A. Weiner, S. R. Post, and J. Chun. 1996. Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. J. Cell Biol. 135:1071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hordijk, P. L., I. Verlaan, E. J. van Corven, and W. H. Moolenaar. 1994. Protein tyrosine phosphorylation induced by lysophosphatidic acid in Rat-1 fibroblasts. Evidence that phosphorylation of MAP kinase is mediated by the Gi-p21ras pathway. J. Biol. Chem. 269:645-651. [PubMed] [Google Scholar]
- 26.Im, D. S., C. E. Heise, M. A. Harding, S. R. George, B. F. O'Dowd, D. Theodorescu, and K. R. Lynch. 2000. Molecular cloning and characterization of a lysophosphatidic acid receptor, Edg-7, expressed in prostate. Mol. Pharmacol. 57:753-759. [PubMed] [Google Scholar]
- 27.Ishii, I., J. J. A. Contos, N. Fukushima, and J. Chun. 2000. Functional comparisons of the lysophosphatidic acid receptors, LPA1/Vzg-1/Edg2, LPA2/Edg4, and LPA3/Edg7 in neuronal cell lines using a retrovirus expression system. Mol. Pharmacol. 58:895-902. [DOI] [PubMed] [Google Scholar]
- 28.Ishii, I., B. Friedman, X. Ye, S. Kawamura, C. McGiffert, J. J. Contos, M. A. Kingsbury, G. Zhang, J. H. Brown, and J. Chun. 2001. Selective loss of sphingosine 1-phosphate signaling with no obvious phenotypic abnormality in mice lacking its G protein-coupled receptor, LPB3/EDG-3. J. Biol. Chem. 276:33697-33704. [DOI] [PubMed] [Google Scholar]
- 29.Ishii, I., X. Ye, B. Friedman, S. Kawamura, J. J. Contos, M. A. Kingsbury, A. H. Yang, G. Zhang, J. H. Brown, and J. Chun. 2002. Marked perinatal lethality and cellular signaling deficits in mice null for the two sphingosine 1-phosphate receptors: S1P2/LPB2/EDG-5 and S1P3/LPB3/EDG-3. J. Biol. Chem. 277:25152-25159. [DOI] [PubMed] [Google Scholar]
- 30.Jalink, K., T. Eichholtz, F. R. Postma, E. J. van Corven, and W. H. Moolenaar. 1993. Lysophosphatidic acid induces neuronal shape changes via a novel, receptor-mediated signaling pathway: similarity to thrombin action. Cell Growth Differ. 4:247-255. [PubMed] [Google Scholar]
- 31.Kimura, Y., A. Schmitt, N. Fukushima, I. Ishii, H. Kimura, A. R. Nebreda, and J. Chun. 2001. Two novel Xenopus homologs of mammalian LPA1/EDG-2 function as lysophosphatidic acid receptors in Xenopus oocytes and mammalian cells. J. Biol. Chem. 276:15208-15215. [DOI] [PubMed] [Google Scholar]
- 32.Lee, M. J., S. Thangada, C. H. Liu, B. D. Thompson, and T. Hla. 1998. Lysophosphatidic acid stimulates the G-protein-coupled receptor EDG-1 as a low affinity agonist. J. Biol. Chem. 273:22105-22112. [DOI] [PubMed] [Google Scholar]
- 33.Moolenaar, W. H., O. Kranenburg, F. R. Postma, and G. C. Zondag. 1997. Lysophosphatidic acid: G-protein signalling and cellular responses. Curr. Opin. Cell Biol. 9:168-173. [DOI] [PubMed] [Google Scholar]
- 34.Ridley, A. J., and A. Hall. 1992. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70:389-399. [DOI] [PubMed] [Google Scholar]
- 35.Sasaki, T., T. Maehama, T. Yamamoto, S. Takasuga, S. Hoshino, H. Nishina, O. Hazeki, and T. Katada. 1998. Activation of c-Jun N-terminal kinase (JNK) by lysophosphatidic acid in Swiss 3T3 fibroblasts. J. Biochem. (Tokyo) 124:934-939. [DOI] [PubMed] [Google Scholar]
- 36.Seufferlein, T., D. J. Withers, D. Mann, and E. Rozengurt. 1996. Dissociation of mitogen-activated protein kinase activation from p125 focal adhesion kinase tyrosine phosphorylation in Swiss 3T3 cells stimulated by bombesin, lysophosphatidic acid, and platelet-derived growth factor. Mol. Biol. Cell 7:1865-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Corven, E. J., A. Groenink, K. Jalink, T. Eichholtz, and W. H. Moolenaar. 1989. Lysophosphatidate-induced cell proliferation: identification and dissection of signaling pathways mediated by G proteins. Cell 59:45-54. [DOI] [PubMed] [Google Scholar]
- 38.Weiner, J. A., and J. Chun. 1999. Schwann cell survival mediated by the signaling phospholipid lysophosphatidic acid. Proc. Natl. Acad. Sci. USA 96:5233-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiner, J. A., J. H. Hecht, and J. Chun. 1998. Lysophosphatidic acid receptor gene vzg-1/lpA1/edg-2 is expressed by mature oligodendrocytes during myelination in the postnatal murine brain. J. Comp. Neurol. 398:587-598. [PubMed] [Google Scholar]