Abstract
The RING domain protein Sina, together with Phyllopod and the F-box protein Ebi, forms a Ras-regulated E3 ubiquitin ligase complex that activates photoreceptor cell differentiation in the eye of Drosophila melanogaster. The expression of Phyllopod is induced upon Ras activation, allowing the complex to degrade the transcription repressor Tramtrack and removing its block of neuronal development in photoreceptor precursors. We show that Phyllopod functions as an adaptor in the complex, physically linking Sina with Tramtrack via separate binding domains. One 19-amino-acid domain in Phyllopod interacts with a region of Sina's SBD domain. Another domain in Phyllopod interacts with a C-terminal helix in the POZ domain of Tramtrack. This interaction is specific to the Tramtrack POZ domain and not to other POZ domain proteins present in photoreceptor precursors. Degradation of Tramtrack is dependent upon association of Sina with its cognate binding site in Phyllopod. These results illustrate how Ras signaling can modulate an E3 ligase activity not by the phosphorylation of substrate proteins but by regulating the expression of specific E3 adaptors.
Conditional degradation of signaling proteins by ubiquitin-mediated proteolysis plays a central role in several signal transduction pathways that control cell proliferation and differentiation. Ubiquitination involves three transfer proteins that act in series (20). The E1 ubiquitin-activating protein transfers ubiquitin to an E2 ubiquitin-conjugating enzyme. The E3 ubiquitin ligase facilitates transfer of ubiquitin from E2 to a substrate protein. Once the substrate protein is polyubiquitinated, it interacts with the proteasome and is degraded. E3 ligases are primarily responsible for providing specificity to ubiquitin conjugation and, as such, play a key regulatory role in many cell processes. Most known E3 ligases are multisubunit protein complexes, of which one essential subunit is a protein bearing a RING domain (25). Various RING domains have been shown to associate with E2 conjugating enzymes in vitro, and cocrystal structure determination of the cCbl RING-UbcH7 complex indicates that the interaction is direct (26, 32, 46). It appears that the RING domain facilitates direct transfer of ubiquitin from E2s to substrate, although the precise mechanism is not known.
E3 proteins containing RING domains are structurally diverse outside of their RING domains. This allows them to make specific associations with particular substrate proteins or other components of an E3 complex. For example, Rbx-1 associates in a complex with Skp-1, CUL-1, and various F-box proteins (SCF) to ubiquitinate substrate proteins that are recruited by the F-box protein subunit (13, 42). Rbx-1 also associates with Von Hippel-Lindau protein, CUL-2, and elongins to form a VCB E3 complex that targets different substrate proteins. Other RING domain proteins form complexes with different combinations of proteins to provide diverse substrate docking sites. The RING protein Siah-1 associates with SIP, Skp-1, and the F-box protein Ebi to form a SCF-like complex that ubiquitinates β-catenin (33). β-Catenin is recruited into the complex by association with Ebi and the scaffold protein APC, which also interacts with Siah-1 (31). Thus, specific coupling of a RING domain protein with substrate is often mediated by other components in the E3 complex.
Siah-1 is a member of a family of highly conserved RING domain proteins (22). The majority of the primary structure appears unique to the family, with the RING domain, followed by an SBD domain composed of a cysteine-rich region and a larger C-terminal region. The crystal structure of the Siah-1 SBD domain was determined, and it shows that the cysteine-rich region forms a pair of zinc fingers and that the C-terminal region self-dimerizes (37). Together, this domain forms a tertiary structure not seen in other solved E3 structures. This may account for how Siah-1 forms certain E3 complexes since the SBD domain corresponds to the region in Siah-1 shown to interact with APC and SIP (31, 33).
Many substrates for ubiquitination are components of signal transduction pathways. A widespread mechanism by which such substrates are recognized by E3 ligases is that they are phosphorylated by protein kinases as a result of signaling pathway activities (25). Substrate recognition is often mediated by WD-40 and LRR containing F-box proteins, which bind to phosphoproteins. The Siah-1 family of proteins are involved in two well-characterized signaling pathways. In response to DNA damage and p53 activation, expression of Siah-1 is induced in cell culture (2, 38). This is linked with concomitant degradation of β-catenin and arrest of the cell cycle in these cells, in a SIP- and Ebi-dependent manner (7, 31, 33, 38). Thus, Siah-1 mediates a β-catenin degradation pathway linking p53 activation to cell cycle control. A second signaling pathway involving a Siah family member occurs in the developing eye of Drosophila melanogaster. The Drosophila homolog of Siah-1, called Sina, has also been implicated to act as an E3 ligase (29, 40). Sina binds the Drosophila E2 enzyme UbcD1and a substrate for ubiquitination, the transcription repressor protein Tramtrack (Ttk). Sina is required for downregulation of Ttk protein in a subset of photoreceptor cells in the eye. Two other factors are also required for this reaction: a novel nuclear protein, Phyllopod (Phyl), and the F-box protein Ebi (15, 29, 40). If Sina, Phyl, or Ebi are missing or reduced in photoreceptors, then Ttk is not degraded in these cells, and it blocks their differentiation into neurons. Ttk is downregulated via the Ras signal transduction pathway, which is triggered by receptor tyrosine kinase activation (29). Ras signaling induces the expression of Phyl in the photoreceptors R1, R6, and R7 (10, 14). Thus, targeted proteolysis is a critical event in the Ras signaling pathway and its regulation of cell fate in the fly eye.
Altogether, Sina, Phyl, and Ebi form a complex with Ttk in cells (6, 40). However, the relationship of subunits within the complex is unknown, and the identity of the subunit that recruits Ttk into the complex is unclear. This latter point is exemplified by finding that Ebi, Phyl, and Sina can individually associate with Ttk in two-hybrid or coprecipitation assays (6, 29, 40). Several possible models might explain these results. In one, Sina recruits Ttk into the complex while Phyl and Ebi perform other activities that are necessary for Ttk proteolysis. In another model, Phyl and Ebi function as scaffolds to optimally position Ttk and Sina for ubiquitin transfer. In a third model, Phyl or Ebi act as adaptors to recruit Ttk into a stable complex with Sina. If Ebi functions in this manner, it might specifically recognize Ttk in a phosphorylated state given the WD-40 repeats present within Ebi. Here, we show that Phyl functions as an adaptor protein in the complex, recruiting Ttk to stably associate with Sina. Phyl contains two separate interaction domains that are individually responsible for binding either Ttk or Sina. The Ttk-binding domain resides near the C terminus, and it interacts specifically with a POZ domain in Ttk. The Sina-binding domain is a 19-amino-acid peptide, and it interacts with the SBD domain in Sina. By directed mutagenesis, we identify regions within the POZ and SBD domains that likely direct Phyl association. Thus, Phyl represents a unique class of E3 adaptor protein that couples ubiquitination substrates to RING domain proteins.
MATERIALS AND METHODS
Drosophila genetics.
The following mutant lines were used: UbcD11462, UbcD13527, UbcD16535 (Bloomington Stock Center), UbcD1Δ73, UbcD1Δ112 (9), sina4 (8), and sev-Phyl (14). To induce mutant clones, a FRT82B UbcD1Δ73 chromosome was placed in trans over FRT82B P[w+]90E. Heterozygotes were heat shocked at late L1 stage to induce hs-FLP expression and mitotic recombination.
Histology and immunohistochemistry.
Mosaic clones in adult eyes were identified by the absence of pigment. Adult eyes were fixed and sectioned as described previously (45). Nonmosaic sections were stained with toluidine blue (8). Pupal retinas were dissected, fixed, and stained for antibody localization as described previously (43). Flies were synchronized at the white prepupal stage and were aged a further 40 h at 21°C before dissection. Primary antibody was mouse anti-Cut monoclonal antibody 2B10 (1:100; Developmental Studies Hybridoma Bank University of Iowa). Secondary goat anti-mouse antibodies conjugated to biotin were reacted with avidin-horseradish peroxidase (Vector Labs).
Construction of expression vectors and protein production.
pSina-GST and pANTP-GST have been described previously, and glutathione S-transferase (GST) fusion proteins were produced and purified as described previously (28). Templates for in vitro protein synthesis were either pET-based plasmids or PCR products. Each of the Ttk, Phyl, and Sina deletions were made by PCR, as were the full-length Lola proteins: a T7 promoter sequence (TAATACGACTCACTATAGGGAGACCAC), a translation consensus sequence (GCCACC), and an initiation codon (ATG) were linked to coding sequences in the 5′ oligonucleotide; two anti-stop codons (TTATTA) were linked to the antiparallel sequences as the 3′ oligonucleotide. To fuse Ttk(1-117) with Lola(118-122), the 3′ oligonucleotide linked the corresponding antiparallel Lola sequence with antiparallel Ttk sequence, and the oligonucleotide was used to amplify from a Ttk plasmid template. PCR products were precipitated with ethanol after purification with phenol and chloroform. pET vectors for translation of full-length Phyl, Phyl-hemagglutinin (HA), and Ttk have been described (29). To express full-length Sina in vitro, the Sina coding sequence was inserted into pET3c with a NdeI site at its initiation codon. A triple-HA epitope tag was placed within this NdeI site to generate an amino-terminal tagged Sina. Ttk88 with a carboxy-terminal triple-HA tag had been generated previously for expression in S2 cells as a pMK33 recombinant (29). It was inserted into pET21a for in vitro translation of a full-length Ttk88-HA fusion protein. It was also inserted into pBSKS, and an internal deletion between NdeI and NruI sites within the Ttk coding sequence generated a fusion between the amino-terminal 218 codons of Ttk and the triple-HA tag. Although the plasmid was unable to synthesize a Ttk(1-218)HA fusion protein directly by T7 TNT in vitro transcription, it was used as a PCR template to synthesize a fragment with T7 promoter linked to the coding sequence.
To express β-galactosidase in vitro, the T7 promoter sequence described above was inserted into pWnBE (43) to replace the hsp70 promoter of that vector and create pT7lacZ. A tandem triple repeat of Phyl coding the sequence from residues 109 to 127 with EcoRI ends was generated by annealing six overlapping and complementary oligonucleotides, followed by ligation to create concatemers. These were digested with EcoRI, and a triple-repeat fragment was inserted into the EcoRI site of pT7lacZ, 16 codons upstream of the lacZ termination codon. The final product, pT7lacZ-Phyl, has three tandem copies of Phyl(109-127) fused in frame with the β-galactosidase sequence EPVSIGGIP. Each repeat is separated by a Gly-Gly linker. Alanine substitution mutations were introduced pairwise into the residue 109 to 127 coding sequence of Phyl by site-directed mutagenesis on single-stranded DNA templates as described previously (3). The mutagenic oligonucleotides contained the necessary base substitutions, with all other bases matching the wild-type Phyl sequence. Mutated plasmids were used as PCR templates to synthesize DNA fragments with T7 promoters linked to the altered coding sequence. For S2 cell transfections, mutagenized Phyl coding sequences were inserted into pMK33 (metallothionein promoter).
For in vitro translation, template DNA was mixed with the (T7 RNA polymerase) TNT coupled reticulocyte lysate system (Promega) supplemented with or without [35S]methionine. Proteins were synthesized according to manufacturer's instructions.
Protein interaction assays.
GST pull-down assays were performed as described previously (29) with the following modifications. In 50 μl of Buffer A (50 mM HEPES [pH 7.4], 120 mM NaCl, 0.1%NP-40, 0.5 mg of BSA/ml, 0.2 mM PMSF, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml, 1 mM benzamidine, 50 μg of ethidium bromide/ml), 8 μl of translation mix and 1 μg of purified GST fusion protein were coincubated for 60 min at 25°C. The binding reaction was incubated with glutathione-Sepharose beads for 60 min at 4°C. The beads were then washed three times with 1 ml of cold Buffer A prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
Immunoprecipitation assays were performed as described previously (29) with modifications. Proteins were translated singly or by cotranslation in vitro and labeled by [35S]methionine incorporation as necessary. A total of 8 μl of translation mix with the desired combination of proteins was mixed with 5 μl of Buffer A, followed by incubation at 25°C for 30 min. The binding reaction was precleared by the addition of a 33-μl suspension (10% [vol/vol]) of Pansorbin (Calbiochem) in HIPA Buffer (50 mM HEPES [pH 7.4], 120 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.5 mg of bovine serum albumin/ml, 0.2 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml, 1 mM benzamidine, 50 μg of ethidium bromide/ml) and further incubation for 30 min at 4°C. The precleared supernatant (43 μl) was mixed with 0.2 μg of anti-HA (Roche) and incubated at 4°C for 30 min. Immune complexes were allowed to bind to 3 μl of Pansorbin for 30 min at 4°C and washed with three 1-ml washes (50 mM HEPES [pH 7.4], 120 mM NaCl, 1% NP-40) before denaturation and SDS-PAGE.
SDS-PAGE was done according to standard protocols when we resolved proteins of >25 kDa. To resolve smaller proteins, they were electrophoresed in 15 to 18% polyacrylamide gels containing SDS and 4 M urea.
Glycerol gradient centrifugation.
Equal volumes of 15, 20, 25, and 30% glycerol in Buffer A were overlaid in ultracentrifuge tubes and left overnight to form a density gradient. Proteins were labeled separately with [35S]methionine by in vitro translation and combined together for 60 min at 25°C to allow association. Reactions were diluted with 1.5 volumes of Buffer A and incubated a further 60 min at 25°C, and 100 μl was loaded onto each gradient. The tubes and a separate tube containing protein molecular weight standards were centrifuged in a Beckman SW60 rotor at 50,000 rpm for 3.5 h at 4°C. Fractions of 100 μl were then collected and analyzed by SDS-PAGE.
Pulse-chase measurement of Ttk-HA in S2 cells.
Drosophila S2 cells were grown in M3 medium containing 10% fetal bovine serum. We transfected 20-mm dishes of cells with 1.5 μg of each pMK33 expression vector by using Lipofectin (Invitrogen) as described previously (29). All transfections included vectors that expressed Ttk-HA and Sina (29). To induce expression from the metallothionein promoter of the pMK33 vectors, 0.7 mM CuSO4 was added to the medium for 18 h. Cells were washed in M3M (methionine-free M3 medium supplemented with 10% dialyzed fetal bovine serum and 0.7 mM CuSO4) and incubated for 3.5 h in 0.3 ml of M3M containing 35 μCi of [35S]methionine. The radioactive medium was replaced with M3M supplemented with 10% fetal bovine serum and 3 mg of methionine/ml. Cells were harvested at various times and lysed as described previously (29), and extracts were incubated with 1 μg of anti-HA for 30 min at 4°C. Immune complexes were precipitated with Pansorbin and washed three times with radioimmunoprecipitation assay buffer before denaturation and SDS-PAGE.
RESULTS
Phyl has been proposed to promote the E3 ligase activity of Sina in photoreceptor cells of the developing Drosophila eye. Therefore, we would predict that Phyl interacts with other components of the ubiquitin pathway. Sina physically interacts with the ubiquitin-conjugating E2 enzyme, UbcD1 (40). Sina also interacts genetically with UbcD1. When the activity of the sina gene in R7 precursor cells was reduced with a hypomorphic mutant allele, sina4, ca. 30% of R7 precursors failed to differentiate into R7 photoreceptors (Fig. 1A). When the dosage of the UbcD1 gene was reduced by half, ca. 50% of sina4 R7 precursors failed to develop into R7 photoreceptors (Fig. 1B). A similar enhancement of the sina4 mutant phenotype was observed with three other loss-of-function UbcD1 alleles: UbcD11462, UbcD13527, and UbcD16535. Previously, it was found that reduced UbcD1 gene dosage suppressed a gain-of-function sina mutant phenotype (36). Together, the data suggest that UbcD1 activates Sina in regulating R7 development.
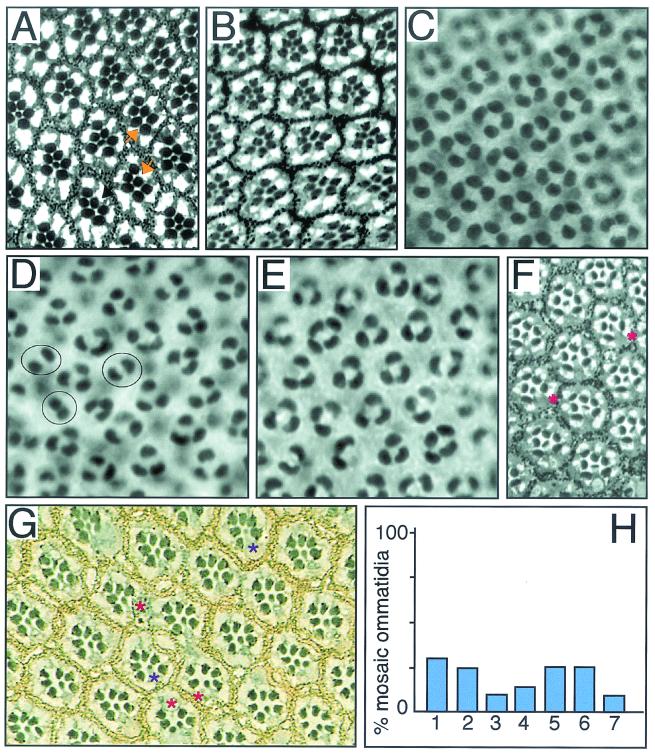
FIG. 1.
Phyl and Sina interact with UbcD1 in Drosophila eye development. All panels have the anterior side to the left. The Drosophila compound eye is comprised of 800 light-sensing cell-clusters called ommatidia. Each ommatidium contains eight photoreceptor cells (R1 to R8) and four nonneuronal cone cells, which assemble by a process of recruitment. First, R8 is determined, and then subsequent photoreceptors are added in pairs: R2 and R5, R3 and R4, and R1 and R6. The final photoreceptor to differentiate is R7 followed by the cone cells. (A) sina4 mutant adult eye. The plane of section only shows the R1 to R7 cells, which are arranged in a trapezoid pattern within each ommatidium. The R7 cell projects to the center of the ommatidium. The sina4 allele causes the transformation of 29% of R7 cells into cone cells (n = 956). Two ommatidia with missing R7 cells are highlighted with arrows. (B) sina4 UbcD1Δ73/+ eye. Reducing the dosage of UbcD1 by half increases the transformation to 47% of R7 cells into cone cells (n = 667). (C) Wild-type 40-h pupal retina stained with anti-Cut. Cut is only present in nuclei of the four cone cells within each ommatidium. A field of 25 ommatidia are seen in this panel. (D) When Phyl is misexpressed in cone cells with a sev-Phyl transgene, many cone cells are transformed into photoreceptors (10, 14), as exhibited by the fewer Cut-positive cells in these pupal retinas. On average, there are 2.9 cone cells per ommatidium (n = 96). Note the ommatidia with two cone cells highlighted. (E) sev-Phyl UbcD1Δ73/+ pupal retina. On average, the number of cone cells per cluster increased to 3.4 (n = 97). No two-cone clusters were observed. (F) UbcD1Δ112 mutant adult eye. This hypomorphic mutation results in occasional missing R1, R6, and R7 cells. Missing R6 cells were most frequently observed (asterisks). (G) Dorsal clones of mutant UbcD1Δ73 cells in adult eye. Mutant cells are marked by the absence of the white gene product that pigments the eye. Pigmentation is visualized as yellow granules in pigment cells and black granules at the base of the rhabdomeres in photoreceptor cells. Mutant clones were very small with only one or two cells in each clone. Mosaic ommatidia had missing photoreceptors (red asterisks) or mutant photoreceptors with abnormal light-sensing rhabdomeres (purple asterisks). There were also mutant cells that appeared normal. (H) Quantitation of UbcD1Δ73 mutant clonal analysis. The frequencies of mosaic ommatidia with missing or abnormal photoreceptors are shown. Numbers on x axis refer to the affected R cell: 1 to 7. Photoreceptor R8 was not examined.
To investigate a possible interaction between Phyl and UbcD1, we redirected cone cells to an R7 cell fate by ectopically expressing Phyl in cone cells, causing their transformation into R7 cells (Fig. 1C and D). If UbcD1 and Phyl function together, then reducing the dosage of the UbcD1 gene might suppress the ability of Phyl to transform cone cells into R7 cells. Indeed, we found this to be the case (Fig. 1E). A second prediction of this model is that UbcD1 is required in the same cells as Phyl (R1, R6, and R7) for their proper development. We examined adult eyes from a weak UbcD1 mutant and observed that R1, R6, and R7 cells were occasionally missing (Fig. 1F). To determine whether UbcD1 was required cell autonomously in R1, R6, and R7, we analyzed UbcD1Δ73 null mutant clones in genetically mosaic flies. Homozygous mutant UbcD1Δ73 tissue contained recognizable ommatidia with several defective features that had also been observed for another UbcD1 allele (36). The number of mutant cells in clones was abnormally small, and some mosaic ommatidia contained one or two fewer photoreceptors than their normal eight-cell contingent (Fig. 1G and H). However, we were able to confidently identify the missing cell in many cases (Fig. 1H). No evidence of non-cell-autonomous effects by UbcD1 on photoreceptor formation was detected. Rather, the results were consistent with a cell-autonomous role for UbcD1. UbcD1 phenotypes were not restricted to R1, R6, and R7 but extended across the entire R1-to-R7 spectrum of cell types. Although this finding suggests that UbcD1 has Phyl-independent activities, it is also consistent with UbcD1 acting with Phyl in determining R1, R6, and R7 cell types.
Earlier interaction studies in our lab and others detected independent physical association between Ttk and either Sina or Phyl (29, 40). The apparent binding affinities of Phyl and Sina for Ttk were quite different (29). Here, we analyze these differences in detail. A GST-Sina fusion protein associated with only 1% of the Ttk protein present in a binding reaction (Fig. 2A). In contrast, it associated with 40% of Phyl protein present in a similar binding reaction, suggesting that it had much higher affinity for Phyl than Ttk. When we examined the affinity of HA-tagged Ttk (Ttk-HA) for Sina versus Phyl, Ttk-HA was associated with <1% of Sina protein and ca. 10% of Phyl protein in a binding reaction (Fig. 2B and D). It was not likely that this difference in binding activities was due to less Sina being able to bind protein. Approximately 10% of the same preparation of Sina interacted with HA-tagged Phyl (Phyl-HA) in a similar binding reaction (Fig. 2B). These data suggest that the different effectiveness in Sina and Phyl binding with Ttk is due to a 10-fold difference in their binding affinities for Ttk. Thus, even though a Sina-Ttk interaction was specific enough that we and others could detect it in vitro (29, 40), it is significantly weaker than the Phyl-Ttk interaction.
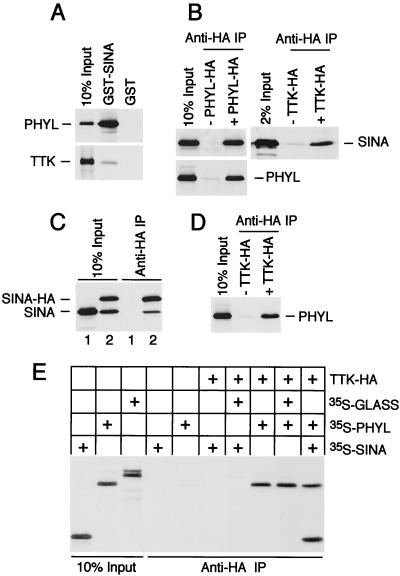
FIG. 2.
Phyl promotes Sina association with Ttk. (A) GST-Sina associates strongly with Phyl and weakly with Ttk. Approximately equimolar quantities of radioactively labeled Phyl and Ttk were used in a GST pull-down binding assay with 1 μg of purified GST-Sina or GST protein. (B) Sina associates strongly with HA-tagged Phyl and weakly with HA-tagged Ttk in vitro. Anti-HA immune complexes were purified from reactions containing unlabeled HA-tagged Phyl or Ttk and equimolar quantities of 35S-labeled Sina or Phyl, as indicated. (C) Sina self-associates in vitro. Sina and tagged Sina-HA proteins were translated in vitro as indicated and immunoprecipitated with anti-HA before separation by SDS-PAGE. (D) Labeled Phyl associates with Ttk-HA immune complexes in vitro. (E) Association of Sina with HA-tagged Ttk is specifically enhanced by Phyl. Binding assays containing unlabeled Ttk-HA were supplemented with the indicated proteins labeled with [35S]methionine from translation reactions. GLASS is a Drosophila zinc finger protein that is unrelated to either Phyl or Sina (35). Bound proteins were purified by anti-HA immunoprecipitation and subjected to SDS-PAGE.
It was previously observed that Sina self-associates in vitro (24, 29, 40). This result was confirmed by the analysis of Sina association with HA-tagged Sina (Sina-HA) immune complexes (Fig. 2C). The ratio of Sina-HA to Sina that immunoprecipitated was ca. 2:1, suggesting that a small number of subunits was present in these complexes. Indeed, the crystal structure of the Sina homolog Siah-1 indicates that it is a dimeric protein (37). We also found that Phyl associated with anti-Phyl-HA immune complexes (Fig. 2B), indicating that Phyl forms homomeric complexes in vitro.
Evidence of a difference between Sina and Phyl affinity for Ttk binding led us to examine whether Phyl facilitates interaction of Sina with Ttk. Various combinations of HA-tagged Ttk, Phyl, and Sina were coincubated, and anti-HA immune complexes were isolated and analyzed for associated proteins (Fig. 2E). In the presence of all three proteins, anti-Ttk-HA complexes contained equimolar quantities of Sina and Phyl. However, in the absence of Phyl, barely detectable levels of Sina bound Ttk-HA. This result was confirmed through the analysis of Ttk association with GST-Sina complexes (data not shown). Thus, association of Sina with Ttk is enhanced by Phyl. In contrast, Phyl can efficiently associate with Ttk in the absence of Sina (Fig. 2E).
If Ttk, Sina, and Phyl form a stable multiprotein complex, they should have a common size, which is characteristic of the complex. To test this possibility, combinations of Sina, Phyl, and Ttk were sedimented through glycerol gradients, and the resulting fractions were assayed for each protein (Fig. 3). When Sina and Phyl were mixed and sedimented, each protein peaked in gradient fraction 7, corresponding to a Svedberg coefficient of 5.5S. When Sina and Ttk were mixed and sedimented, Sina peaked in gradient fraction 7, whereas Ttk peaked in fraction 21, corresponding to a Svedberg coefficient of 42S. No Sina cosedimenting with Ttk complexes was detected. However, when Phyl was mixed with Sina and Ttk, all three proteins peaked in fractions 21 to 23. Moreover, the sedimentation profile of each protein closely matched the other protein profiles, indicating that the three proteins are present in a common multiprotein complex. This complex was stable at 4°C over the 3.5 h required for centrifugation. Interestingly, in the gradient containing all three proteins, the Sina and Phyl peaks corresponding to the 5.5S complex were greatly reduced or not present (Fig. 3A). Since roughly equivalent quantities of Sina, Phyl, and Ttk were loaded onto the gradient (Fig. 3E), it is possible that the three proteins efficiently associated with one another. If this occurred, it can be inferred that they bind to each other with high affinity.
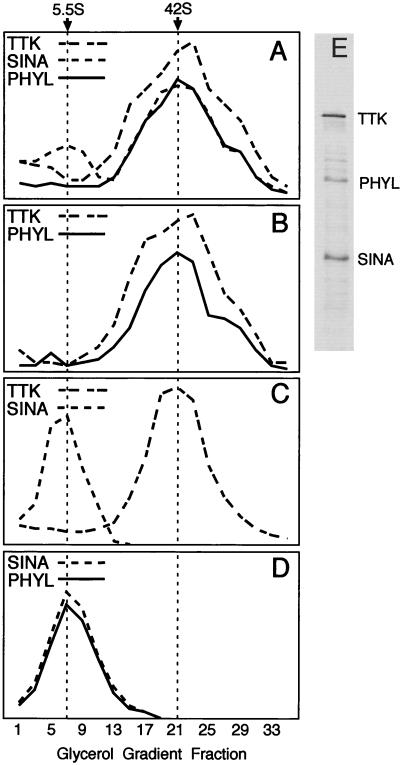
FIG. 3.
Glycerol gradient sedimentation analysis of Sina, Phyl, and Ttk. 35S-labeled proteins were incubated in binding reactions and subsequently sedimented as described in Materials and Methods. Thirty-five fractions were taken from each gradient and subjected to SDS-PAGE, followed by autoradiography. Protein levels in each fraction were quantitated by laser scanning densitometry and are graphed in arbitrary density units. In a parallel gradient, molecular weight standards were sedimented, and portions of each fraction were analyzed by SDS-PAGE. Svedberg coefficients were determined by comparison of the labeled protein peak in a gradient to the sedimentation profile of protein standards. Svedberg coefficients corresponding to gradient peaks are indicated at the top of the figure. (A) Binding reaction containing Sina, Phyl, and Ttk. (B) Binding reaction containing Phyl and Ttk. (C) Binding reaction containing Sina and Ttk. (D) Binding reaction containing Sina and Phyl. (E) SDS-PAGE analysis of labeled Sina, Phyl, and Ttk proteins loaded onto gradients.
The 5.5S value of Phyl-Sina corresponds to a molecular size of ca. 150 to 160 kDa if a globular shape is assumed. The molecular sizes of the Phyl and Sina polypeptides are 45 and 34 kDa, respectively. This would imply that the Sina-Phyl complex is possibly composed of a dimer of Sina and dimer of Phyl, a conclusion consistent with the ability of Sina and Phyl to self-associate. However, since there is no information regarding the shapes of these molecules and since there may be conformational change following their association, the stoichiometry cannot be determined from this experiment. The 42S value of Ttk corresponds to a molecular mass of ca. 1,400 kDa even though the Ttk polypeptide is 88 kDa. This suggests that Ttk multimerizes into large complexes. Consistent with this idea, other proteins with POZ domains adopt homomultimeric complexes (4), and the crystal structure of the POZ domain indicates that this domain mediates oligomerization (1, 30). Since Phyl and Sina associate with the Ttk complex without reducing its apparent size, it indicates that Sina and Phyl bind to Ttk without altering its ability to multimerize. The shift of the Ttk peak in the presence of Phyl and Sina suggests that the multimeric complex is composed of several subunits of Phyl and Sina though the precise stoichiometry cannot be determined.
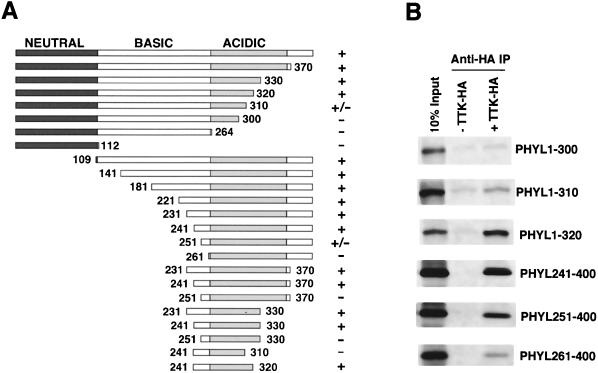
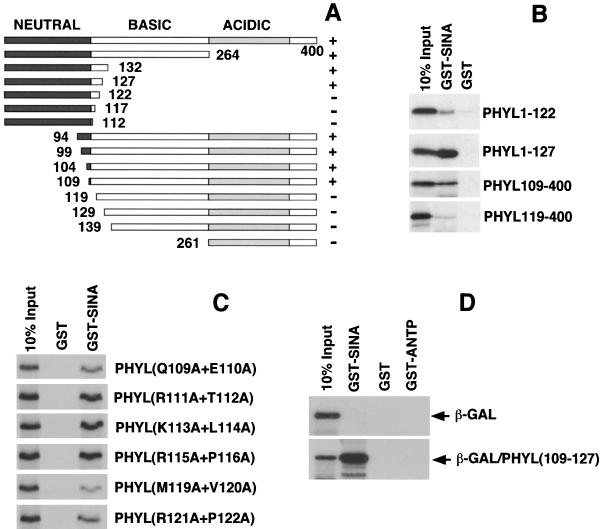
Phyl facilitates association of Sina with Ttk, suggesting that it acts as an adaptor protein. If so, then Phyl would have separate domains for interacting with Sina and Ttk. To physically map the Phyl domain that interacts with Ttk, we generated deletion mutants of Phyl protein and performed a series of binding experiments with HA-tagged Ttk (Fig. 4). Association of Phyl with anti-Ttk-HA immune complexes was abolished by deleting 261 amino acids from the N terminus or by deleting 90 amino acids from the C terminus of Phyl. A fragment of Phyl corresponding to residues 241 to 320 was sufficient to associate with Ttk-HA. No recognizable motifs are evident in this region.
FIG. 4.
Mapping the Ttk-binding domain within Phyl. (A) Schematic representation of Phyl proteins and summary of their Ttk-binding activities. The 400-amino-acid Phyl protein can be divided into neutral, basic, and acidic regions based on charge distribution. The Phyl amino acids included in each derivative protein are indicated by numbers. Proteins were labeled with [35S]methionine and assayed for Ttk binding by coimmunoprecipitation with unlabeled Ttk-HA. +, Wild-type binding activity; +/−, reduced binding activity; −, no specific binding activity. (B) Analysis of representative binding reactions between Phyl and Ttk-HA. 35S-labeled Phyl proteins were mixed with Ttk-HA and immunoprecipitated with anti-HA before SDS-PAGE.
We had previously mapped the Sina-binding domain in Phyl to a region near the N terminus (29). To further map the Sina-binding domain, we generated deletion mutants of Phyl protein and tested these for their ability to specifically associate with GST-Sina in pull-down assays (Fig. 5A and B). Association of Phyl with GST-Sina was abolished by deleting 119 amino acids from the N terminus but not by deleting 109 amino acids. Deletion of 278 amino acids from the C terminus prevented association with GST-Sina, whereas the deletion of 273 amino acids from the C terminus had little effect. This deletion analysis suggested that Phyl residues 109 to 127 are critical for Sina interaction in vitro. To test this idea, we created alanine substitutions of pairwise amino acids within 109 to 127 and assayed their effect on Phyl protein binding to GST-Sina (Fig. 5C). Phyl association with GST-Sina was reduced twofold when Q109/E110 or R121/P122 were mutated, and association was reduced fourfold when M119/V120 were mutated. In contrast, pairwise mutation of residues 111 to 116 and residues 123 to 126 had no detectable effect on Phyl binding to GST-Sina (Fig. 5C and data not shown). Although these data indicate that a region from residues 109 to 127 is necessary for Sina association, is it sufficient? We swapped the C-terminal 16 amino acids of β-galactosidase with three tandem repeats of the Phyl(109-127) sequence. If the sequence from residues 109 to 127 is sufficient for Sina binding, it should enable the chimeric protein to associate with GST-Sina in a pull-down assay. Indeed, virtually 100% of the chimeric protein specifically associated with GST-Sina, whereas native β-galactosidase displayed no detectable association (Fig. 5D). Thus, a 19-amino-acid sequence from Phyl has high affinity for binding to Sina. Moreover, this motif is physically distinct from the Ttk-binding domain in Phyl. The two interaction domains are some 114 residues apart on the Phyl polypeptide, a finding consistent with the notion that Phyl acts as a high-affinity linker between Sina and Ttk.
FIG. 5.
A 19-amino-acid sequence of Phyl is necessary and sufficient for binding to Sina. (A) Schematic representation of Phyl proteins and summary of their Sina-binding activities. The Phyl amino acids included in each derivative protein are indicated by numbers. Proteins were labeled with [35S]methionine and assayed for Sina binding by copurification with GST-Sina to glutathione beads. +, Wild-type binding activity; +/−, reduced binding activity; −, no specific binding activity. (B) Analysis of representative binding reactions between Phyl and GST-Sina. 35S-labeled Phyl proteins were mixed with GST-Sina or GST and pulled down with glutathione beads before SDS-PAGE. (C) Tandem alanine substitutions in Phyl inhibit its ability to bind Sina. The Phyl amino acid sequence QERTKLRPVAMVRPTVRVQ from positions 109 to 127 was mutated to generate a series of proteins with neighboring pairs of amino acids changed to alanine residues. These mutated proteins (indicated at the right) were 35S labeled and pulled down with GST-Sina or GST coupled to glutathione beads. (D) The Phyl sequence from positions 109 to 127 fused to β-galactosidase produces a fusion protein that can associate with Sina. Three tandem copies of the Phyl sequence QERTKLRPVAMVRPTVRVQ were swapped with the C-terminal 16 amino acids of β-galactosidase, and the resulting fusion protein [β-GAL/Phyl(109-127)] was 35S labeled. Labeled β-galactosidase or fusion protein was incubated with GST-Sina, GST, or GST-ANTP (homeodomain of the Antennapedia protein) and subject to pull-down with glutathione beads.
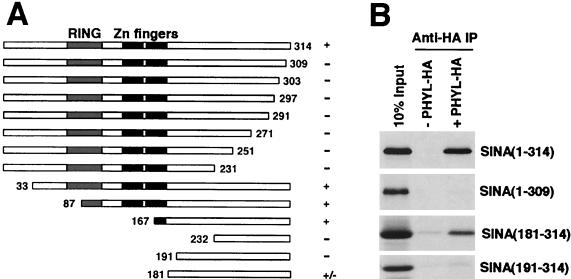
To physically map the Sina domain that interacts with Phyl, we generated deletion mutants of Sina protein and performed a series of binding experiments with HA-tagged Phyl (Fig. 6). Association of Sina with anti-Phyl-HA immune complexes was abolished by deleting 191 amino acids from the N terminus or by the simple removal of 5 amino acids from the C terminus of Sina. A fragment of Sina corresponding to residues 181 to 314 was sufficient to associate with Phyl-HA, although it associated more weakly than a 167- to 314-residue fragment. The region from residues 181 to 314 is part of the highly conserved SBD domain except that it does not contain the two zinc fingers (37).
FIG. 6.
Mapping the Phyl-binding domain within Sina. (A) Schematic representation of Sina proteins and summary of their Phyl-binding activities. The 314-amino-acid Sina protein has a RING domain located near its N terminus, followed by two zinc fingers and a C-terminal region. The Sina amino acids included in each derivative protein are indicated by numbers. Proteins were labeled with [35S]methionine and assayed for Phyl binding by coimmunoprecipitation with unlabeled Phyl-HA. +, Wild-type binding activity; +/−, reduced binding activity; −, no specific binding activity. (B) Analysis of representative binding reactions between Sina and Phyl-HA. 35S-labeled Sina proteins were mixed with Phyl-HA and immunoprecipitated with anti-HA before SDS-PAGE.
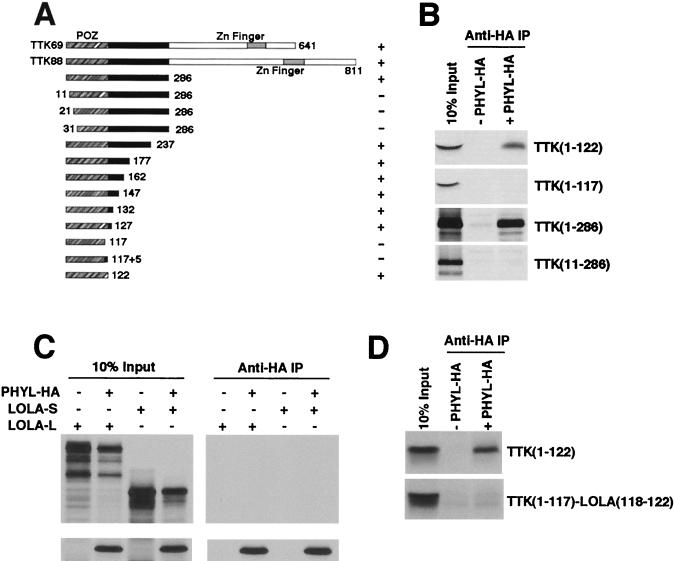
Previous mapping of Ttk had identified the N-terminal 286 amino acids as sufficient to interact with Phyl (29). We further refined the Ttk domain that interacts with Phyl by testing deletion mutants of Ttk protein for association with HA-tagged Phyl (Fig. 7A and B). Deletion of 10 amino acids from the N terminus of Ttk was sufficient to abolish binding to Phyl. Deletion of all but 122 amino acids from the C terminus had no effect on binding to Phyl. Thus, the N-terminal 122 amino acids of Ttk are sufficient for Phyl association. This domain corresponds precisely with the POZ domain of Ttk.
FIG. 7.
The POZ domain of Ttk specifically interacts with Phyl. (A) Schematic representation of Ttk proteins and summary of their Phyl-binding activities. The Ttk69 and Ttk88 isoforms share a common POZ domain (orange) and differ in their C-terminal zinc finger domains. Truncated forms of Ttk are indicated with amino acid positions of endpoints shown. Proteins were labeled with [35S]methionine and assayed for Phyl binding by coimmunoprecipitation with unlabeled Phyl-HA. +, Wild-type binding activity; +/−, reduced binding activity; −, no specific binding activity. (B) Analysis of representative binding reactions between Ttk and Phyl-HA. 35S-labeled Ttk proteins were mixed with Phyl-HA and immunoprecipitated with anti-HA before SDS-PAGE analysis. (C) Phyl does not associate with another Drosophila POZ protein, Lola. Lola has two Drosophila isoforms: Lola-L that contains a C-terminal zinc finger domain, and the smaller Lola-S that is missing the C-terminal region of Lola-L, including the zinc finger domain. Both isoforms contain a POZ domain that bears 62% sequence identity to the Ttk POZ domain. Labeled Lola-L or Lola-S were incubated with labeled Phyl-HA in a binding reaction as indicated, followed by anti-HA immunoprecipitation. Complexes were resolved by SDS-PAGE. (D) The carboxy-most 5-amino-acid residues of the Ttk POZ domain are critical for Phyl association. Deletion analysis from panel A indicated that the Ttk sequence residues 118 to 122 (EVNDD) are necessary for Phyl binding. A chimeric Ttk-Lola protein fragment was made that contained the sequence from positions 1 to 117 of Ttk fused to the sequence from positions 118 to 122 of Lola (DNRTG). 35S-labeled Ttk or Ttk-Lola chimeric protein was incubated with unlabeled Phyl-HA as indicated, and anti-HA complexes were immunoprecipitated and analyzed by SDS-PAGE.
The POZ domain is an evolutionarily conserved protein-protein interaction domain present in 39 distinct genes in the Drosophila sequence database. It is estimated that 5 to 10% of human zinc finger proteins contain these domains (11). The crystal structure of the PLZF POZ domain is dimeric (1, 30). Since the residues that stabilize dimerization and the core of the PLZF POZ monomer are strongly conserved in all POZ proteins, including Ttk, it is likely that the Ttk POZ domain has a homologous tertiary fold. This raised the question of whether Phyl is able to associate with POZ domains of other Drosophila proteins. A sequence comparison between Ttk and other Drosophila POZ proteins indicated that the POZ domain of Lola is most similar to Ttk POZ (62% identical). The lola gene encodes two major protein products (Lola-L and Lola-S) that contain a common N-terminal POZ domain but differ in their C termini (19). Indeed, it has been observed that the longer isoform Lola-L, plays a role in photoreceptor determination (L. Zheng and R. W. Carthew, unpublished data). We tested whether Phyl can associate with Lola by coincubating HA-tagged Phyl with Lola-L or Lola-S. Anti-HA immune complexes were isolated and analyzed for associated proteins (Fig. 7C). No association between Lola and Phyl-HA was detected. We also failed to detect association between GST-Sina and Lola or another Drosophila POZ protein, Fru, in pull-down assays (data not shown). These results are consistent with finding no effect of phyl or sina mutants on Lola protein abundance in photoreceptors (Zheng et al., unpublished). Thus, Phyl and Sina do not associate with all proteins bearing POZ domains, but rather there is specificity to POZ recognition.
Our deletion analysis provided clues as to what features of the Ttk POZ domain are required for Phyl interaction. Deletion of either residues 1 to 10 or residues 118 to 122 abolished Phyl association in vitro (Fig. 7B). Interestingly, these two regions are predicted to pack together in antiparallel fashion along an exposed lip, based on the PLZF crystallographic model (1, 30). Moreover, residues 1 to 10 and residues 118 to 122 correspond to two of the six contiguous sequences that are divergent between Lola and Ttk. If this surface of the Ttk POZ domain is required for Phyl association, then substituting Ttk with Lola sequences in this region would inhibit Phyl association. We substituted Ttk residues 118 to 122 (EVNDD) with the homologous residues from Lola (DNRTG) and tested the substitution mutant for binding to Phyl-HA immune complexes (Fig. 7D). No binding was detected, indicating that residues 118 to 122 are critical for Phyl association.
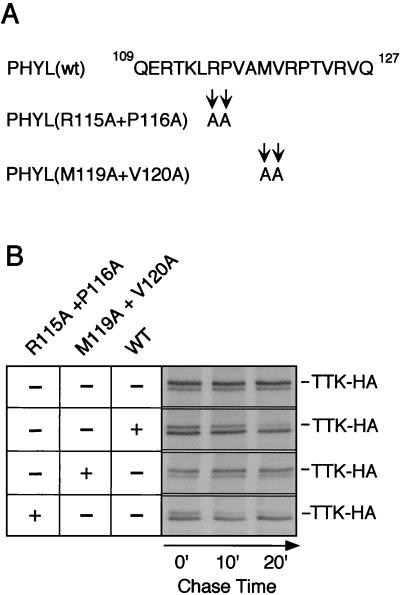
Our analysis of binding interactions in vitro suggests that one critical function of Phyl is to provide a high-affinity physical link between Sina and Ttk. In Phyl's absence, Sina interacts weakly with Ttk. We speculate that the reason for forming a stable complex between Sina and Ttk is to allow the complex to ubiquitinate Ttk and thereby lead to its degradation. If so, then crippling the ability of Phyl to link Sina with Ttk would cripple the ability of Sina to degrade Ttk. To address this, we used a previously characterized S2 cell transfection assay to measure Ttk protein stability dependent upon Sina (29). HA-tagged Ttk was cotransfected with Sina and Phyl into Drosophila S2 cells, and Ttk-HA protein turnover was measured by pulse-chase labeling (Fig. 8). The half-life of Ttk-HA was determined to be 25 min under these conditions. In the absence of Phyl, the half-life of Ttk-HA increased to 8 h (Fig. 8) (29). We had identified alanine substitution mutations in the Sina-binding domain of Phyl that inhibited Sina association with Phyl in vitro (Fig. 5). In particular, the M119/V120 mutant had the most severe effect on Sina binding. We cotransfected this Phyl mutant with Ttk-HA and Sina into S2 cells and observed a slow turnover of Ttk-HA (Fig. 8B). In contrast, transfection of the R115/P116 mutant resulted in rapid turnover of Ttk-HA comparable to that of wild-type Phyl. The R115/P116 mutant did not inhibit Sina association with Phyl in vitro (Fig. 5). One possible explanation for this result is that the different Phyl mutant proteins accumulate to different levels in S2 nuclei. We could not examine this possibility since Phyl antibodies are not available. However, we think this possibility is unlikely since the mutated residues do not reside in any known nuclear localization or destabilization motifs. Instead, this precise correlation between Sina-binding activity in vitro and Ttk-HA degrading activity in vivo suggests that Phyl association with Sina is critical for Ttk degradation.
FIG. 8.
Ttk protein degradation is dependent on Sina associating with Phyl in S2 cells. (A) Sequence of the Sina-binding region of Phyl from residues 109 to 127. Note that the alanine substitutions changing Arg-115 and Pro-116 had little effect on Sina binding to Phyl in vitro, whereas the alanine substitutions changing Met-119 and Val-120 inhibited Sina binding to Phyl (see Fig. 5). (B) Pulse-chase analysis of Ttk-HA protein stability in the presence of wild-type and mutant Phyl. After cotransfection of Ttk-HA, Sina, and the indicated Phyl expression vectors, S2 cells were metabolically labeled with [35S]methionine. This was followed by incubation for various times as indicated, in medium with excess unlabeled methionine. Ttk-HA protein was immunoprecipitated from cell extracts by using anti-HA 12CA5 and visualized by autoradiography after SDS-PAGE. The doublet bands observed correspond to Ttk-HA proteins, with the upper band of the doublet representing full-length Ttk-HA (29). It was previously shown that only this larger Ttk-HA product was susceptible to Phyl-induced ubiquitination and degradation (29).
DISCUSSION
In the present study, we have demonstrated that Phyl acts to stimulate Sina association with Ttk by 10- to 20-fold, thereby allowing Sina to ubiquitinate and promote destruction of Ttk. Moreover, Phyl binds to Ttk and Sina with comparable affinities. The most likely interpretation is that Phyl functions as an adaptor protein to physically link Sina and Ttk together in a high-affinity multiprotein complex. Two alternative hypotheses are that Phyl stabilizes a form of Sina compatible with Ttk binding or that, conversely, Phyl stabilizes a form of Ttk compatible with Sina binding. However, these two hypotheses do not require that Phyl have bivalent binding activities, associating with Sina and Ttk via separate binding domains. Since Phyl contains such domains, the most likely reason is to provide direct physical linkage.
In the absence of Phyl, Sina exhibits weak but specific association with Ttk (29, 40). Our data would suggest that Sina's affinity for Ttk is not sufficient for it to form a stable complex and promote Ttk ubiquitination. The bridging activity of Phyl would augment Sina's association with Ttk, allowing stable interaction to occur. Another possibility is that the weak association between Sina and Ttk reflects participation of a mammalian Phyl homolog present in the reticulocyte extract used to synthesize recombinant Ttk. However, we performed deletion analysis of Sina and Ttk to map the regions required for their interaction (data not shown) and found that Ttk sequences between residues 71 and 197 were sufficient to allow Sina association. This region is overlapping but clearly distinct from the Phyl-binding domain, which corresponds to sequences from residues 1 to 122. Likewise, we found that Sina sequences between residues 66 and 87 (the RING finger) were absolutely required for interaction with Ttk, whereas these sequences are dispensable for Phyl association. Thus, the Sina-Ttk interaction domains are distinct from those for Phyl, arguing that a Phyl-like activity is not responsible for the weak Sina-Ttk association. One unexpected aspect of our mapping Sina and Ttk was that directional deletions sometimes failed to define the interaction domain boundaries (data not shown). For example, progressive deletion from the C terminus of Sina caused progressive reduction in Ttk-binding activity without any precipitous drop-off in activity. This might suggest that some of the binding energy is obtained through nonspecific surface interactions rather than discrete folded surfaces on each protein. The weak Sina-Ttk interaction we observed in vitro is probably not sufficient to allow Sina to target Ttk degradation in vivo. Ttk degradation in S2 culture cells requires the presence of Phyl (Fig. 8), and Ttk downregulation in the Drosophila eye requires the phyl gene (29, 40).
Phyl interacts with two highly conserved domains in Sina and Ttk that have been structurally characterized in related proteins. Phyl specifically associates with the POZ domain in Ttk and not with the POZ domain in Lola. The POZ domain is a multifunctional protein-protein interaction domain. It allows proteins to self-associate into dimers and higher-order oligomeric structures (5, 11). POZ domains recruit transcription corepressor molecules such as N-CoR and Sin3A, which in turn draw histone deacetylases to locally remodel chromatin (12, 21). This feature confers transcription repressor activity to proteins that contain POZ domains. The POZ domain may also affect chromatin structure by oligomerizing and binding multiple DNA sites, leading to DNA bending (16, 27).
Where might Phyl bind to the Ttk POZ domain with such specificity? The crystal structure of the PLZF POZ domain has been determined, and mapping studies suggest that essential structural features are conserved between POZ proteins (1, 30, 34). There is an extensive dimer interface, a charged pocket that mediates transcription repression by recruiting N-CoR/Sin3A, a broad hydrophobic surface opposite the pocket, and a neighboring acidic patch. Most of the hydrophobic surface is predicted to be conserved between Ttk and Lola if their sequences are mapped to the PLZF crystal structure. However, a short α-helix at the C terminus of the POZ domain (residues 118 to 122) that is essential for Phyl association is predicted to act as a lip dividing the hydrophobic and acidic faces. Moreover, the amino acid sequence of this helix is highly variable among POZ proteins, including Ttk and Lola. Substitution of this helix from Ttk with the corresponding Lola helix rendered the domain unable to interact with Phyl (Fig. 7). Thus, the lip of the Ttk POZ domain is a critical specificity feature of Phyl interaction.
Phyl interacts with part of the SBD domain of Sina. This conserved domain interacts with a wide variety of proteins, including ones that are degraded in response to Sina or its mammalian Siah homologs (17, 18, 23, 24, 39, 41, 44). The SBD from Siah-1 also interacts with mammalian cofactors APC and SIP to form an SCF-like complex, which includes Skp-1 and Ebi (31, 33). Recent determination of the Siah-1 crystal structure hints at two possible protein recognition surfaces in the SBD: a deep cleft adjacent to the first zinc finger and a shallow groove centered at the dimer interface (37). Although it is not known whether these structural features are also present in Sina, two lines of evidence argue that this is likely the case. First, the residues that stabilize dimerization and the core of the Siah-1 monomer are completely conserved in Sina, indicating that the Sina SBD domain has a homologous tertiary fold. Second, the primary sequence of the Sina SBD is 95% identical (98.5% similar) to the Siah-1 SBD.
Can we speculate as to where Phyl might bind Sina? Only 19 amino acids of Phyl interact with Sina, making it probable that the Phyl peptide fits within a folded surface. It is unlikely to be in the cleft region of SBD since the first zinc finger within the Sina SBD is not required for Phyl association, and it folds to create one wall of the cleft in Siah-1. In contrast, the shallow groove is contained within the Phyl-binding domain defined by deletion analysis. Moreover, removing five residues from the C terminus completely abolished Phyl association. These residues map to part of a β-strand that constitutes both sides of the groove in the Siah-1 SBD. Point mutagenesis of the SBD combined with cocrystal structure determination should resolve this issue. Comparison of the Sina-binding domain in Phyl to sequences in other Sina or Siah binding proteins did not uncover shared sequence motifs that might mediate their binding activities. This would suggest that the SBD domain, to which they all bind, does not have a single means to interact with its numerous partners.
The Sina/Siah family shares a common function in promoting ubiquitination of certain target proteins, leading to their degradation (24, 29, 40). To perform this function, the RING domain in Sina/Siah recruits E2 conjugating enzymes such as UbcD1 that transfer ubiquitin to substrates (24, 32, 33). The SBD domain in Sina/Siah recruits the ubiquitin substrates. For some substrates, such as DCC and the transcription corepressor N-CoR, Sina or Siah appear to directly bind the substrate proteins that are ubiquitinated (23, 24, 44). For the substrate protein β-catenin, it is recruited to an SCF-like E3 complex composed of Siah, SIP, Skp-1, and Ebi (33). In this complex, SIP and Skp-1 act as a molecular bridge to link Siah with Ebi, which is the subunit to which β-catenin directly binds. We find that Ttk is recruited by the adaptor protein Phyl into a complex containing Sina. It is likely that this complex also contains Ebi based on genetic and biochemical evidence. Mutations in the ebi gene perturb R7 differentiation, and Ebi participates in Phyl-dependent downregulation of Ttk in eye disks and when expressed in S2 cell culture (6, 15). Moreover, Ebi protein associates with Sina and Phyl complexes in cells (6). However, the role of Ebi in this complex is unclear. It is not an obligatory adaptor linking Sina, Phyl, and Ttk, as we have shown currently. However, it might stabilize complexes once formed, or stabilize complex components thereby promoting complex formation. Future experiments should explore how Ebi, and possibly Skp-1 and SIP, are linked to the Sina-Phyl dependent pathway of Ttk degradation.
Acknowledgments
We thank B. Baker for Fru cDNAs, E. Giniger for Lola cDNAs, K. Moses for the Glass cDNA, and M. Kuziora for pGST-ANTP. We also thank the Bloomington Stock Center, M. Gatti, and B. Dickson for fly stocks and the Developmental Studies Hybridoma Bank for antibodies. We are grateful to K. Krock for help in making some of the alanine substitution mutants.
This study was supported by a grant from the NIH to R.W.C. (EY10111) and a Mellon Fellowship to S.L. while at the University of Pittsburgh.
REFERENCES
- 1.Ahmad, K. F., C. K. Engel, and G. G. Prive. 1998. Crystal structure of the BTB domain from PLZF. Proc. Natl. Acad. Sci. USA 95:12123-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amson, R. B., M. Nemani, J. P. Roperch, D. Israeli, L. Bougueleret, I. Le Gall, M. Medhioub, et al. 1996. Isolation of 10 differentially expressed cDNAs in p53-induced apoptosis: activation of the vertebrate homologue of the Drosophila seven in absentia gene. Proc. Natl. Acad. Sci. USA 93:3953-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Ball, H. J., A. Melnick, R. Shaknovich, R. A. Kohanski, and J. D. Licht. 1999. The promyelocytic leukemia zinc finger (PLZF) protein binds DNA in a high molecular weight complex associated with cdc2 kinase. Nucleic Acids Res. 27:4106-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bardwell, V. J., and R. Treisman. 1994. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 8:1664-1677. [DOI] [PubMed] [Google Scholar]
- 6.Boulton, S. J., A. Brook, K. Staehling-Hampton, P. Heitzler, and N. Dyson. 2000. A role for Ebi in neuronal cell cycle control. EMBO J. 19:5376-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruzzoni-Giovanelli, H., A. Faille, G. Linares-Cruz, M. Nemani, F. Le Deist, A. Germani, D. Chassoux, G. Millot, J. P. Roperch, R. Amson, A. Telerman, and F. Calvo. 1999. Siah-1 inhibits cell growth by altering the mitotic process. Oncogene 18:7101-7109. [DOI] [PubMed] [Google Scholar]
- 8.Carthew, R. W., and G. M. Rubin. 1990. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell 63:561-577. [DOI] [PubMed] [Google Scholar]
- 9.Cenci, G., R. B. Rawson, G. Belloni, D. H. Castrillon, M. Tudor, R. Petrucci, M. L. Goldberg, S. A. Wasserman, and M. Gatti. 1997. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev. 11:863-875. [DOI] [PubMed] [Google Scholar]
- 10.Chang, H. C., N. M. Solomon, D. A. Wassarman, F. D. Karim, M. Therrien, G. M. Rubin, and T. Wolff. 1995. phyllopod functions in the fate determination of a subset of photoreceptors in Drosophila. Cell 80:463-472. [DOI] [PubMed] [Google Scholar]
- 11.Collins, T., J. R. Stone, and A. J. Williams. 2001. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 21:3609-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David, G., L. Alland, S. H. Hong, C. W. Wong, R. A. DePinho, and A. Dejean. 1998. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene 16:2549-2556. [DOI] [PubMed] [Google Scholar]
- 13.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 14.Dickson, B. J., M. Dominquez, A. van den Straten, and E. Hafen. 1995. Control of Drosophila photoreceptor cell fates by Phyllopod, a novel nuclear protein acting downstream of the Raf kinase. Cell 80:453-462. [DOI] [PubMed] [Google Scholar]
- 15.Dong, X., L. Tsuda, K. H. Zavitz, M. Lin, S. Li, R. W. Carthew, and S. L. Zipursky. 1999. ebi regulates epidermal growth factor receptor signaling pathways in Drosophila. Genes Dev. 13:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Espinas, M. L., E. Jimenez-Garcia, A. Vaquero, S. Canudas, J. Bernues, and F. Azorin. 1999. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J. Biol. Chem. 274:16461-16469. [DOI] [PubMed] [Google Scholar]
- 17.Germani, A., H. Bruzzoni-Giovanelli, A. Fellous, S. Gisselbrecht, N. Varin-Blank, and F. Calvo. 2000. SIAH-1 interacts with alpha-tubulin and degrades the kinesin Kid by the proteasome pathway during mitosis. Oncogene 19:5997-6006. [DOI] [PubMed] [Google Scholar]
- 18.Germani, A., F. Romero, M. Houlard, J. Camonis, S. Gisselbrecht, S. Fischer, and N. Varin-Blank. 1999. hSiah2 is a new Vav binding protein which inhibits Vav-mediated signaling pathways. Mol. Cell. Biol. 19:3798-3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giniger, E., K. Tietje, L. Y. Jan, and Y. N. Jan. 1994. lola encodes a putative transcription factor required for axon growth and guidance in Drosophila. Development 120:1385-1398. [DOI] [PubMed] [Google Scholar]
- 20.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 21.Hong, S. H., G. David, C. W. Wong, A. Dejean, and M. L. Privalsky. 1997. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARα) and PLZF-RARα oncoproteins associated with acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 94:9028-9033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, G., Y. L. Chung, T. Glover, V. Valentine, A. T. Look, and E. R. Fearon. 1997. Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics 46:103-111. [DOI] [PubMed] [Google Scholar]
- 23.Hu, G., and E. R. Fearon. 1999. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol. 19:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, G., S. Zhang, M. Vidal, J. L. Baer, T. Xu, and E. R. Fearon. 1997. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 11:2701-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 26.Joazeiro, C. A., S. S. Wing, H. Huang, J. D. Leverson, T. Hunter, and Y. C. Liu. 1999. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286:309-312. [DOI] [PubMed] [Google Scholar]
- 27.Katsani, K. R., M. A. Hajibagheri, and C. P. Verrijzer. 1999. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 18:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kauffmann, R. C., S. Li, P. A. Gallagher, J. Zhang, and R. W. Carthew. 1996. Ras1 signaling and transcriptional competence in the R7 cell of Drosophila. Genes Dev. 10:2167-2178. [DOI] [PubMed] [Google Scholar]
- 29.Li, S., Y. Li, R. W. Carthew, and Z. C. Lai. 1997. Photoreceptor cell differentiation requires regulated proteolysis of the transcriptional repressor Tramtrack. Cell 90:469-478. [DOI] [PubMed] [Google Scholar]
- 30.Li, X., H. Peng, D. C. Schultz, J. M. Lopez-Guisa, F. J. Rauscher III, and R. Marmorstein. 1999. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 59:5275-5282. [PubMed] [Google Scholar]
- 31.Liu, J., J. Stevens, C. A. Rote, H. J. Yost, Y. Hu, K. L. Neufeld, R. L. White, and N. Matsunami. 2001. Siah-1 mediates a novel β-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell 7:927-936. [DOI] [PubMed] [Google Scholar]
- 32.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuzawa, S., and J. C. Reed. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 responses. Mol. Cell 7:915-926. [DOI] [PubMed] [Google Scholar]
- 34.Melnick, A., K. F. Ahmad, S. Arai, A. Polinger, H. Ball, K. L. Borden, G. W. Carlile, G. G. Prive, and J. D. Licht. 2000. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 20:6550-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moses, K., M. C. Ellis, and G. M. Rubin. 1989. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature 340:531-536. [DOI] [PubMed] [Google Scholar]
- 36.Neufeld, T. P., A. H. Tang, and G. M. Rubin. 1998. A genetic screen to identify components of the sina signaling pathway in Drosophila eye development. Genetics 148:277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polekhina, G., C. M. House, N. Traficante, J. P. Mackay, F. Relaix, D. A. Sassoon, M. W. Parker, and D. D. Bowtell. 2002. Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-α signaling. Nat. Struct. Biol. 9:68-75. [DOI] [PubMed] [Google Scholar]
- 38.Roperch, J. P., F. Lethrone, S. Prieur, L. Piouffre, D. Israeli, M. Tuynder, M. Nemani, P. Pasturaud, M. C. Gendron, J. Dausset, M. Oren, R. B. Amson, and A. Telerman. 1999. SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21Waf1. Proc. Natl. Acad. Sci. USA 96:8070-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sourisseau, T., C. Desbois, L. Debure, D. D. Bowtell, A. C. Cato, J. Schneikert, E. Moyse, and D. Michel. 2001. Alteration of the stability of Bag-1 protein in the control of olfactory neuronal apoptosis. J. Cell Sci. 114:1409-1416. [DOI] [PubMed] [Google Scholar]
- 40.Tang, A. H., T. P. Neufeld, E. Kwan, and G. M. Rubin. 1997. PHYL acts to downregulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell 90:459-467. [DOI] [PubMed] [Google Scholar]
- 41.Tanikawa, J., E. Ichikawa-Iwata, C. Kanei-Ishii, A. Nakai, S. Matsuzawa, J. C. Reed, and S. Ishii. 2000. p53 suppresses the c-Myb-induced activation of heat shock transcription factor 3. J. Biol. Chem. 275:15578-15585. [DOI] [PubMed] [Google Scholar]
- 42.Tyers, M., and P. Jorgensen. 2000. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev. 10:54-64. [DOI] [PubMed] [Google Scholar]
- 43.Xu, C., R. C. Kauffmann, J. Zhang, S. Kladny, and R. W. Carthew. 2000. Overlapping activators and repressors delimit transcriptional response to receptor tyrosine kinase signals in the Drosophila eye. Cell 103:87-97. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, J., M. G. Guenther, R. W. Carthew, and M. A. Lazar. 1998. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 12:1775-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng, L., J. Zhang, and R. W. Carthew. 1995. frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development 121:3045-3055. [DOI] [PubMed] [Google Scholar]
- 46.Zheng, N., P. Wang, P. D. Jeffrey, and N. P. Pavletich. 2000. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102:533-539. [DOI] [PubMed] [Google Scholar]