Abstract
Diva (also called Boo/Bcl-B) is a member of the Bcl-2 gene family and most likely functions during apoptosis. Diva is highly expressed in the ovary, and both pro- and antiapoptotic functions have been ascribed to this protein. To determine the role of Diva during murine development, we used gene targeting to inactivate Diva. The Diva-null mice are born at the expected ratios, are fertile, and have no obvious histological abnormalities, and long-term survival did not differ from littermate controls. Additionally, Diva was not required for apoptosis occurring from genotoxic insult in the ovaries or other organs. Thus, Diva is not critical for the normal development of the ovaries, or in its absence its function is subserved by another protein.
Cell death that occurs during normal organismal development or results from disease or radiotherapy and chemotherapy usually involves apoptosis, a genetically defined program of cell elimination (1, 8). Apoptosis is critical for homeostasis, and inappropriate control of apoptosis can result in a variety of human pathologies, including cancer and neurodegeneration (18, 22). The molecular basis of apoptosis is evolutionarily conserved, and similar strategies for cell death are used in organisms from nematodes to humans (8). The basic molecular framework involves a stimulus activating one or more of a variety of Bcl-2-related proteins (ced-9 in the nematode) that in turn lead to the activation of caspases (ced-3) that act as proteases to dismantle the dying cell (7, 8, 20).
The Bcl-2 family of proteins is functionally important in apoptosis and often acts in a tissue-specific manner (1, 15, 21). The canonical member of this family, Bcl-2, was first identified as a component of a translocation in B-cell malignancies, and when overexpressed was found to inhibit apoptosis in a variety of biological systems (15). Other Bcl-2 family members act as general effectors of cell death by either promoting or protecting against cell death (1, 3). Pro-apoptotic members include Bax, Bad, Bid, Bak, and Bik, while antiapoptotic members include Bcl-2, Bcl-X, Mcl-1, Bcl-w, and A-1. Bcl-2 family proteins can contain four conserved domains, designated Bcl-2 homology regions (BH1 to BH4) (1, 3, 15). The BH1 and BH2 motifs of the death antagonists (such as Bcl-2 and Bcl-X) and the BH3 domain of the death agonists (such as Bax and Bak) are important for homo- or heterodimerization between family members and facilitate control of apoptosis (1, 3, 15, 30). The BH4 domain, found in several antiapoptotic homologues, is essential for the death-repressing activity (10). Some Bcl-2 family members share sequence homology only with the BH3 domain (11, 26). These BH3-domain-only proteins are thought to activate multidomain Bcl-2 members to initiate apoptosis (4).
As demonstrated in mice with null mutations for Bcl-2 family members, this group of proteins plays important roles during development and homeostasis (21). For example, Bcl-2 inactivation leads to polycystic kidney disease, while inactivation of Bax in the mouse resulted in hyperplasia of thymocytes and male infertility due to spermatocyte hypoplasia (14). Bax−/− animals also show a decrease in normal programmed cell deaths in a number of nervous system tissues, including peripheral ganglia and the trigeminal brainstem nuclear complex, and neuronal cultures derived from Bax−/− animals are resistant to a number of death-inducing agents (5, 6, 28, 29). More dramatic effects are found in Bcl-X-null mice, where embryonic survival requires the presence of this protein (17). Tissue-specific effects are observed in other cases; for example, Bcl-W inactivation results in infertility due to arrested sperm development associated with a gradual loss of germ cells and Sertoli cells from the testis (23, 24). In this report we have investigated the consequences of inactivating the Bcl-2-related protein Diva (12, 25). This gene contains several BH domains (BH1, BH2, and BH4), with some contention existing regarding the presence of a BH3 domain (12, 13, 25), and it can modulate apoptosis in vitro (2, 12, 13, 16, 19, 25). Diva is also relatively restricted in expression, with high levels of expression confined to the ovary (12, 25). Here we report that Diva-null mice are fertile, respond normally to apoptotic stimuli, and do not have any obvious developmental defects.
MATERIALS AND METHODS
Gene targeting.
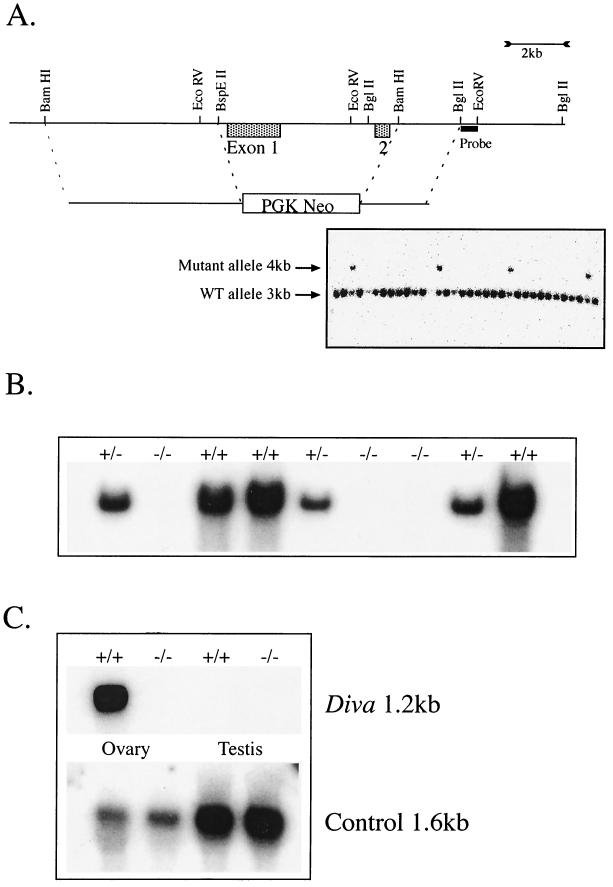
The murine Diva gene contains two exons within a 3.24-kb region of genomic DNA. Diva genomic DNA from strain 129Ola was obtained by isolation of a P1 clone (Genome Systems, St. Louis, Mo.) containing the entire 3.24-kb genomic Diva DNA. A BamHI/BspEII fragment (∼8 kb) immediately 5′ to Diva exon 1 was cloned into the BglII site of pNTK1901 (Stratagene, San Diego, Calif.) to generate pDiva-1, and a 2.5-kb BamHI/BglII fragment immediately 3′ of Diva exon 2 was cloned into the BamHI site of pDiva-1 to generate pDiva-KO. This construct was linearized with SalI and electroporated into W9.5 embryonic stem (ES) cells. Targeted ES cells were identified by Southern blot analysis of EcoRV-digested ES genomic DNA using a genomic BglII/EcoRV fragment 3′ of Diva. The probe, ∼200 bp in length, was generated by PCR using mouse genomic DNA as a template with the following primers: forward, 5′ AGA TCT ACT GAA CTC AGC, and reverse, 5′ ATA TCT GAG AAG CCA AGG. EcoRV digestion of G418-resistant ES cells identified a 4-kb fragment in targeted clones due to loss of an EcoRV site in the mutant allele that was readily distinguishable from the endogenous 2.9-kb Diva genomic EcoRV fragment. Resulting clones were injected into C57BL/6 blastocysts and then implanted into the uteri of pseudopregnant F1 B/CBA foster mothers and allowed to develop to term. Male chimeras were selected for high percentage of agouti coat color and were mated to C57BL/6 females to obtain germ line transmission. The presence of the mutated allele was confirmed by Southern blot analysis or PCR, and heterozygous F1 males and females were interbred to generate F2 animals for subsequent study. Genotyping of Diva mutant mice was done from tail DNA using PCR with the following primers to identify the Diva wild-type (WT) allele: GDP 1, 5′ CAG ACG ATT GCCC CGG C, and GDP 4, 5′ GGT AAC ATC AGC ATC ACA GAA TGC. The Neor marker gene was identified using the following primers: Neo 4, 5′ CGG GAG CGG CGA TAC CGT AAA GC, and Neo 7, 5′ GAA GCG GGA AGG GAC TGG CTG CTA.
Histology.
Ovaries were obtained from 2-month-old mice 6 h after 18 Gy of whole-body ionizing radiation from a cesium irradiator (delivered at a rate of 1.2 Gy/min) and were placed in 10% formalin. Histology of unirradiated tissues was done using age-matched Diva-null mice and littermate controls. Ovaries were paraffin embedded, sectioned into 8-μm sections with an HM325 microtome (Microm), and hematoxylin and eosin stained according to standard procedures. For studies using nervous system tissues, mice were used 5 days after birth (P5; day of birth is P0) and irradiated with 18 Gy. Nervous system tissues were collected after fixation by transcardial perfusion with 4% paraformaldehyde, cryoprotected in 20% sucrose-phosphate-buffered saline, and cryosectioned (12-μm coronal sections) using an HM500 M cryostat (Microm). Neutral red staining was performed with 1% neutral red (Aldrich Chemical) in 0.1 M acetic acid (pH 4.8) for 1 min followed by dehydration in ethanol and mounting with Permount (Fisher). In all cases, experiments were done in triplicate and comparative studies of Diva-null mice used WT littermates as controls.
RESULTS AND DISCUSSION
Initial reports describing Diva found high expression of this gene in the granulosa cells of the ovary and in the epididymis of the testis, although expression was lower in the testis than in the ovary (12, 25). Additionally, in situ hybridization showed widespread Diva expression in the developing nervous system and the ovary (12). We used Northern blot analysis and PCR to confirm the spatial and temporal distribution of Diva mRNA. Northern blot analysis of a number of adult mouse tissues and various stages throughout mouse development found a detectable signal only in the ovary; no signal was found in any other tissues even after extended exposure (data not shown). However, Diva mRNA was detected using PCR from first-strand cDNA in all tissues examined including developing postnatal day 5 (P5) brain and adult mouse brain, liver, and kidney (data not shown). Therefore, Diva mRNA is abundant in the ovary and at levels only detected by PCR in other tissues.
To determine the biological role of Diva, we used gene targeting to inactivate mouse Diva. This gene (GenBank no. NM013479, NM013479, and AF102501) is located on chromosome 9 and contains two coding exons. Inactivation of Diva was achieved by replacing an ∼3-kb genomic region containing both exons with a Neor selection cassette to delete the entire Diva open reading frame (ORF) (Fig. 1A). Targeting of ES cells occurred at a frequency of approximately 1/25 (Fig. 1A), and two of these targeted ES lines were used to generate chimeras and, subsequently, Diva heterozygous mice. Interbreeding of Diva heterozygotes generated Diva-null mice, which were born at the expected frequency of 1/4. Southern blot analysis using a Diva cDNA probe also showed an absence of Diva coding sequence in Diva−/− mice, while both WT and Diva+/− mice contained Diva ORF sequence (Fig. 1B). We further confirmed that Diva expression was disrupted in the Diva−/− mice using Northern blot analysis; Diva mRNA of 1.2 kb was identified in RNA obtained from WT and heterozygous, but not homozygous Diva−/−, ovaries (Fig. 1C).
FIG. 1.
Inactivation of mouse Diva. (A) Diva was inactivated by replacing both exons 1 and 2 with a neomycin selection cassette driven by the PGK promoter derived from pNTK901. Selected restriction sites relevant to the generation and analysis of Diva inactivation are indicated. Homologous recombination removed an EcoRV site from the Diva locus, resulting in a 4-kb mutant Diva allele fragment after EcoRV digestion of genomic DNA; Southern blot analysis was done using a probe encompassing the genomic region contained in a 3′ BglII/EcoRV fragment (probe). (B) The Southern blot shown was probed with Diva cDNA, is a representative analysis of mice derived from mating Diva heterozygotes, and shows that exons 1 and 2 containing the Diva ORF are absent from Diva−/− mice. (C) Northern blot analysis shows that the Diva message (1.2 kb) is present in the WT but not Diva−/− ovaries, while no Diva signal is detected in the testis. The control probe (reticulon, 1.6 kb; GenBank no. AF133669) was used to ensure RNA integrity in samples used for Northern analysis.
Although gene targeting resulted in the complete removal of the genomic DNA encoding Diva, these mice were fertile and had no obvious behavioral defects or affected organs, and long-term survival was indistinguishable from that of WT littermates. Survival of Diva−/− mice was monitored for up to 2 years and histological analysis of the mice at various ages showed no gross anatomical defects in the Diva−/− ovaries compared to WT ovaries (Fig. 2a and e). As Diva expression was reported in the developing nervous system (12), we performed histological analysis of the nervous system at various ages up to 8 months but found no discernible differences in any brain regions compared to littermate controls. Immunohistochemical studies using a variety of markers failed to reveal any differences between Diva-null and control mice up to 8 months of age in a number of tissues, including the ovaries (data not shown).
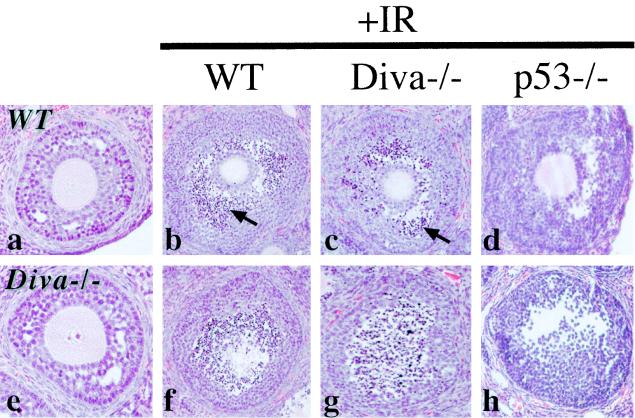
FIG. 2.
Radiation-induced apoptosis in Diva-null ovaries. Ovaries from WT (a) or Diva−/− (e) mice are histologically indistinguishable, as was apoptosis after ionizing radiation treatment (b, c, f, and g). However, p53−/− ovaries (d, h) were completely resistant to radiation-induced apoptosis. Panels b to d and f to h represent two different comparative views through the ovaries. Panels a and e are unirradiated WT and Diva−/− mice, respectively. Arrows identify pyknotic cells indicative of apoptosis. Magnification, ×200.
Because Diva was highly expressed in the ovary and Diva-null animals were fertile and showed no differences from control littermates, we reasoned that Diva−/− mice might be deficient in apoptosis. Diva has been implicated in apoptosis involving Apaf-1 and caspase-9 (12, 25), which are components known to be associated with genotoxic stress-induced apoptosis. Furthermore, apoptosis induced by Diva can be inhibited by a dominant-negative mutant of caspase-9 (12). Consistent with this, Diva can interact with Apaf-1 and displace Bcl-X from the Apaf-1/Bcl-X complex, suggesting that inhibition of Bcl-X function by Diva may occur through competitive binding to Apaf-1 (12, 25).
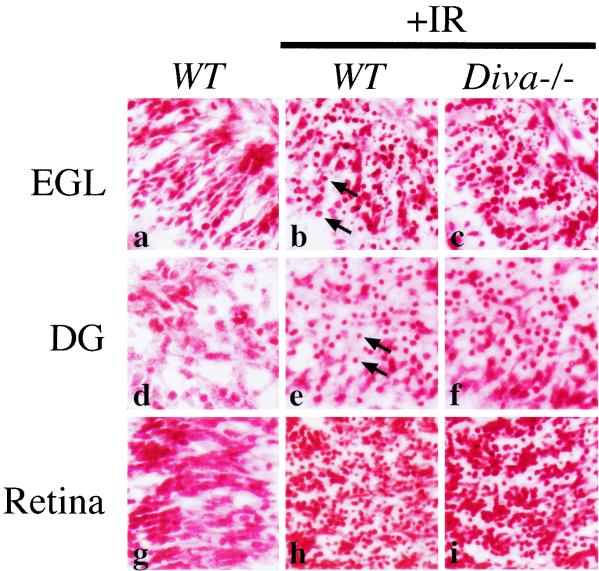
To determine if Diva−/− mice were differentially sensitive to genotoxic stress compared to WT littermates, we examined ionizing radiation (IR)-induced apoptosis in these mice. Pronounced apoptosis as determined histologically was observed at 6 h following IR in granulosa cells in both Diva-null and WT controls (Fig. 2b, c, f, and g). However, while no differences were found between Diva−/− and WT mice, there is a clear genetic basis for ovarian radiation-induced apoptosis as p53−/− null mice were completely resistant to IR-induced apoptosis in the ovary (Fig. 2d and h). Because Diva was detected in the developing brain we also examined IR-induced apoptosis in various developing nervous system tissues of Diva−/− and WT controls. Widespread IR-induced apoptosis was found throughout susceptible regions of the developing nervous system (5), including the cerebellar external granule layer (Fig. 3b and c), the hippocampal dentate gyrus (Fig. 3e and f), and the retina (Fig. 3h and i), and was identical in both Diva−/− and WT controls. Therefore, while other Bcl-2-related members can modulate the response to radiation (5), Diva is not required for IR-induced apoptosis in the developing nervous system.
FIG. 3.
Radiation-induced apoptosis in the Diva-null developing nervous system. The nervous system of Diva−/− mice was histologically indistinguishable from that of WT controls. Apoptosis 6 h after IR in the P5 Diva−/− cerebellar external granule layer (EGL; b and c), dentate gyrus (DG; e and f), and retina (h and i) was similar to that in WT tissue. Apoptosis was assessed by the presence of pyknotic nuclei (arrows) by use of neutral red staining (5). Magnification, ×400.
The Bcl-2 family is important for regulating apoptosis as determined by extensive in vitro analysis and mouse knockout models for many of these molecules (21). Diva (Boo/Bcl-B) has been ascribed both pro- and antiapoptotic roles (2, 12, 13, 16, 19, 25), although because of the relative tissue-restricted expression of this gene, it is likely that cellular context will be important for Diva-regulated apoptosis. Furthermore, as Diva has been shown to interact with a number of different Bcl-2 family members, including Bcl-X and Bax (12, 13), it is likely that these associations also modulate Diva function. In addition to Diva, other Bcl-2 family proteins have been found in the ovary, including Mcl-1, Bok, Bod, and Bad, suggesting the potential for functional modulation by interaction between these various anti- and pro-apoptotic factors (9). Recent data have shown that the interplay between Bcl-2 family members can determine the outcomes of apoptotic signals whereby multidomain Bcl-2-related proteins influence the activity of the pro-apoptotic BH3-domain-only proteins (4). Moreover, activation of either Bax or Bak is a critical determinant for apoptosis in many instances (27). Thus, perhaps the apparent lack of a phenotype in the Diva-null mice and the physiological role of Diva may be understood with further genetic manipulation such as the generation of mice with other apoptotic control genes inactivated in concert with Diva.
Acknowledgments
These studies were supported by the NIH (NS-37956, NS-39867 and CA-21765) and the American Lebanese and Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital.
REFERENCES
- 1.Adams, J. M., and S. Cory. 1998. The Bcl-2 protein family: arbiters of cell survival. Science 281:1322-1326. [DOI] [PubMed] [Google Scholar]
- 2.Aouacheria, A., E. Arnaud, S. Venet, P. Lalle, M. Gouy, D. Rigal, and G. Gillet. 2001. Nrh, a human homologue of Nr-13, associates with Bcl-Xs and is an inhibitor of apoptosis. Oncogene 20:5846-5855. [DOI] [PubMed] [Google Scholar]
- 3.Chao, D. T., and S. J. Korsmeyer. 1998. BCL-2 family: regulators of cell death. Annu. Rev. Immunol. 16:395-419. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, E. H., M. C. Wei, S. Weiler, R. A. Flavell, T. W. Mak, T. Lindsten, and S. J. Korsmeyer. 2001. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 8:705-711. [DOI] [PubMed] [Google Scholar]
- 5.Chong, M. J., M. R. Murray, E. C. Gosink, H. R. Russell, A. Srinivasan, M. Kapsetaki, S. J. Korsmeyer, and P. J. McKinnon. 2000. Atm and Bax cooperate in ionizing radiation-induced apoptosis in the central nervous system. Proc. Natl. Acad. Sci. USA 97:889-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cregan, S. P., J. G. MacLaurin, C. G. Craig, G. S. Robertson, D. W. Nicholson, D. S. Park, and R. S. Slack. 1999. Bax-dependent caspase-3 activation is a key determinant in p53-induced apoptosis in neurons. J. Neurosci. 19:7860-7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cryns, V., and J. Yuan. 1998. Proteases to die for. Genes Dev. 12:1551-1570. [DOI] [PubMed] [Google Scholar]
- 8.Hengartner, M. O. 2000. The biochemistry of apoptosis. Nature 407:770-776. [DOI] [PubMed] [Google Scholar]
- 9.Hsu, S. Y., and A. J. Hsueh. 2000. Tissue-specific Bcl-2 protein partners in apoptosis: an ovarian paradigm. Physiol. Rev. 80:593-614. [DOI] [PubMed] [Google Scholar]
- 10.Huang, D. C., J. M. Adams, and S. Cory. 1998. The conserved N-terminal BH4 domain of Bcl-2 homologues is essential for inhibition of apoptosis and interaction with CED-4. EMBO J. 17:1029-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, D. C., and A. Strasser. 2000. BH3-only proteins—essential initiators of apoptotic cell death. Cell 103:839-842. [DOI] [PubMed] [Google Scholar]
- 12.Inohara, N., T. S. Gourley, R. Carrio, M. Muniz, J. Merino, I. Garcia, T. Koseki, Y. Hu, S. Chen, and G. Nunez. 1998. Diva, a Bcl-2 homologue that binds directly to Apaf-1 and induces BH3-independent cell death. J. Biol. Chem. 273:32479-32486. [DOI] [PubMed] [Google Scholar]
- 13.Ke, N., A. Godzik, and J. C. Reed. 2001. Bcl-B, a novel Bcl-2 family member that differentially binds and regulates Bax and Bak. J. Biol. Chem. 276:12481-12484. [DOI] [PubMed] [Google Scholar]
- 14.Knudson, C. M., K. S. Tung, W. G. Tourtellotte, G. A. Brown, and S. J. Korsmeyer. 1995. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science 270:96-99. [DOI] [PubMed] [Google Scholar]
- 15.Korsmeyer, S. J. 1999. BCL-2 gene family and the regulation of programmed cell death. Cancer Res. 59:1693S-1700S. [PubMed] [Google Scholar]
- 16.Lee, R., J. Chen, C. P. Matthews, J. K. McDougall, and P. E. Neiman. 2001. Characterization of NR13-related human cell death regulator, Boo/Diva, in normal and cancer tissues. Biochim. Biophys. Acta 1520:187-194. [DOI] [PubMed] [Google Scholar]
- 17.Motoyama, N., F. Wang, K. A. Roth, H. Sawa, K. Nakayama, I. Negishi, S. Senju, Q. Zhang, S. Fujii, et al. 1995. Massive cell death of immature hematopoietic cells and neurons in Bcl-x-deficient mice. Science 267:1506-1510. [DOI] [PubMed] [Google Scholar]
- 18.Mullauer, L., P. Gruber, D. Sebinger, J. Buch, S. Wohlfart, and A. Chott. 2001. Mutations in apoptosis genes: a pathogenetic factor for human disease. Mutat. Res. 488:211-231. [DOI] [PubMed] [Google Scholar]
- 19.Naumann, U., S. Weit, J. Wischhusen, and M. Weller. 2001. Diva/Boo is a negative regulator of cell death in human glioma cells. FEBS Lett. 505:23-26. [DOI] [PubMed] [Google Scholar]
- 20.Nunez, G., M. A. Benedict, Y. Hu, and N. Inohara. 1998. Caspases: the proteases of the apoptotic pathway. Oncogene 17:3237-3245. [DOI] [PubMed] [Google Scholar]
- 21.Ranger, A. M., B. A. Malynn, and S. J. Korsmeyer. 2001. Mouse models of cell death. Nat. Genet. 28:113-118. [DOI] [PubMed] [Google Scholar]
- 22.Reed, J. C. 2001. Apoptosis-regulating proteins as targets for drug discovery. Trends Mol. Med. 7:314-319. [DOI] [PubMed] [Google Scholar]
- 23.Ross, A. J., S. P. Amy, P. L. Mahar, T. Lindsten, C. M. Knudson, C. B. Thompson, S. J. Korsmeyer, and G. R. MacGregor. 2001. BCLW mediates survival of postmitotic Sertoli cells by regulating BAX activity. Dev. Biol. 239:295-308. [DOI] [PubMed] [Google Scholar]
- 24.Ross, A. J., K. G. Waymire, J. E. Moss, A. F. Parlow, M. K. Skinner, L. D. Russell, and G. R. MacGregor. 1998. Testicular degeneration in Bclw-deficient mice. Nat. Genet. 18:251-256. [DOI] [PubMed] [Google Scholar]
- 25.Song, Q., Y. Kuang, V. M. Dixit, and C. Vincenz. 1999. Boo, a novel negative regulator of cell death, interacts with Apaf-1. EMBO J. 18:167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strasser, A., H. Puthalakath, P. Bouillet, D. C. Huang, L. O'Connor, L. A. O'Reilly, L. Cullen, S. Cory, and J. M. Adams. 2000. The role of bim, a proapoptotic BH3-only member of the Bcl-2 family in cell-death control. Ann. N. Y. Acad. Sci. 917:541-548. [DOI] [PubMed] [Google Scholar]
- 27.Wei, M. C., W. X. Zong, E. H. Cheng, T. Lindsten, V. Panoutsakopoulou, A. J. Ross, K. A. Roth, G. R. MacGregor, C. B. Thompson, and S. J. Korsmeyer. 2001. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292:727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White, F. A., C. R. Keller-Peck, C. M. Knudson, S. J. Korsmeyer, and W. D. Snider. 1998. Widespread elimination of naturally occurring neuronal death in Bax-deficient mice. J. Neurosci. 18:1428-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiang, H., Y. Kinoshita, C. M. Knudson, S. J. Korsmeyer, P. A. Schwartzkroin, and R. S. Morrison. 1998. Bax involvement in p53-mediated neuronal cell death. J. Neurosci. 18:1363-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zha, H., C. Aime-Sempe, T. Sato, and J. C. Reed. 1996. Proapoptotic protein Bax heterodimerizes with Bcl-2 and homodimerizes with Bax via a novel domain (BH3) distinct from BH1 and BH2. J. Biol. Chem. 271:7440-7444. [DOI] [PubMed] [Google Scholar]