Abstract
In mammalian systems, the heterodimeric basic helix-loop-helix (bHLH)-PAS transcription hypoxia-inducible factor (HIF) has emerged as the key regulator of responses to hypoxia. Here we define a homologous system in Drosophila melanogaster, and we characterize its activity in vivo during development. By using transcriptional reporters in developing transgenic flies, we show that hypoxia-inducible activity rises to a peak in late embryogenesis and is most pronounced in tracheal cells. We show that the bHLH-PAS proteins Similar (Sima) and Tango (Tgo) function as HIF-α and HIF-β homologues, respectively, and demonstrate a conserved mode of regulation for Sima by oxygen. Sima protein, but not its mRNA, was upregulated in hypoxia. Time course experiments following pulsed ectopic expression demonstrated that Sima is stabilized in hypoxia and that degradation relies on a central domain encompassing amino acids 692 to 863. Continuous ectopic expression overrode Sima degradation, which remained cytoplasmic in normoxia, and translocated to the nucleus only in hypoxia, revealing a second oxygen-regulated activation step. Abrogation of the Drosophila Egl-9 prolyl hydroxylase homologue, CG1114, caused both stabilization and nuclear localization of Sima, indicating a central involvement in both processes. Tight conservation of the HIF/prolyl hydroxylase system in Drosophila provides a new focus for understanding oxygen homeostasis in intact multicellular organisms.
In multicellular organisms, oxygen homeostasis requires precise developmental coordination between the growth of metabolizing tissues and that of systems that supply oxygen. Recent advances have provided new insights into how this complex task is achieved. For instance, in mammals it has long been recognized that local hypoxia is a major stimulus for angiogenesis (27). More recently, the recognition that specific angiogenic growth factors such as the vascular endothelial growth factor are powerfully induced by hypoxia through the action of a DNA binding complex termed hypoxia-inducible factor (HIF) has provided mechanistic insights into the process (44, 45).
The HIF DNA-binding complex consists of a heterodimer of basic-helix-loop-helix-PAS (bHLH-PAS) proteins that binds a core element A/(G)CGTG within hypoxia response elements (HREs) (49, 50). Regulation by oxygen involves stabilization of the α-subunit in hypoxia, whereas the β-subunit, a common partner for several other bHLH-PAS proteins, is constitutively expressed regardless of oxygen tension (19, 25, 40, 55). Normoxic degradation of HIF-α is mediated via ubiquitination and subsequent proteolysis, which requires oxygen-dependent interaction with the Von Hippel-Lindau (VHL) tumor suppressor protein (36). This interaction is regulated by hydroxylation of specific prolyl residues within the HIF-α polypeptides (21, 23, 34), and recent work has identified a series of α-ketoglutarate-dependent non-heme iron-dependent dioxygenases that catalyze HIF-α prolyl hydroxylation and thus regulate stability of the polypeptide in accordance with oxygen availability (4, 15). Other studies of HIF induction in mammalian tissue culture systems have defined additional regulatory steps that involve oxygen-dependent subcellular localization (2, 26) and coactivator recruitment (6, 13), although these processes are so far less well understood.
In addition to angiogenic growth factors, HIF-1 drives the expression of genes involved in a broad array of systemic and cellular adaptive responses to hypoxia, suggesting a central role for HIF-1 as a regulator of oxygen homeostasis (35). However, although targeted inactivation of different HIF-α and HIF-β subunits in the mouse is associated with several severe or lethal phenotypes involving defective vascular development (5, 22, 29, 32, 41), few mechanistic studies of the HIF-1 system have been conducted in vivo, and the critical interfaces between developmental processes and HIF activation remain largely undefined.
To better understand HIF regulation in vivo and the role of hypoxia in developmental processes, we therefore sought to define and characterize the system in Drosophila melanogaster. In insects, air reaches the tissues by passive diffusion through a specialized tubular network termed the tracheal system (33, 43). Moreover, development of tracheal terminal branches is oxygen dependent and shares many features with mammalian oxygen-dependent angiogenesis (37) (52). For instance, oxygen-regulated expression of Drosophila Branchless/FGF guides tracheal migration during development (47) and also drives extension of plastic terminal branches in a manner similar to the function of vascular endothelial growth factor in mammalian angiogenesis (24). The existence of a Drosophila HIF homologue has been inferred from DNA-binding assays with nuclear extracts from normoxic or hypoxic SL2 cells (38), and transfection studies in mammalian cells have suggested that the Drosophila bHLH-PAS protein Similar (Sima) (39) might function as an HIF-α homologue (1). However, the system has not yet been fully defined or characterized in vivo in the fly.
In the present study, we demonstrate a hypoxia-inducible transcriptional response in Drosophila that is homologous to the mammalian HIF system, and we use an HRE reporter system in transgenic flies to characterize the temporal and spatial patterns of the activity during development. We confirm the function of Sima as an HIF-α homologue and demonstrate a conserved regulatory process that involves dual mechanisms of protein stabilization and nuclear localization, which are critically dependent on the function of a Drosophila homologue of the HIF prolyl hydroxylases.
MATERIALS AND METHODS
DNA constructs.
The pCaSpeR-LDH-LacZ plasmid was generated by excising an Xba-BamHI 233-bp fragment containing the murine LDH-A enhancer/promoter from pLDHGH (L1) (16) and inserting the fragment into the same sites in a CaSpeR-AUG-βgal vector (a gift from Carl Thummel). The LDH-Gal4 plasmid was generated by amplifying a 51-bp fragment from the same enhancer with a sense oligonucleotide (5′-CGGGATCCCAGCCTACACGTGGG-3′) bearing a BamHI restriction site, and an antisense oligonucleotide (5′-GAAGATCTGCGCTGACGTCAGAGTGG-3′) with a BglII site. After digestion with BamHI and BglII the fragment was cloned into the BamHI site of a CaSpeR-Gal4 vector (3), and plasmids bearing a dimerised insert were isolated.
SimaΔ692-863 deletion was generated by PCR with full-length pNB40-sima as a template and the divergent primers 5′-GCTTCGGGTACCCATGAGGCCAACCAACATGTCAC-3′ and 5′-GCTTCGGGTACCCTAACGGAGACGGATCTCATGTGG-3′ that include KpnI restriction sites at the 5′ ends. Amplification resulted in a 7.7-kb fragment that included the whole pNB40 vector, as well as the sequence encoding Sima protein, lacking amino acids 692 to 863. The product was digested with KpnI and circularized, resulting in pNB40-simaΔ692-863. The SimaΔ692-863 insert was excised with EcoRI and NotI and subcloned into the same sites of the pCaSpeR-UAS vector.
Fly stocks.
Transgenic lines were generated from the above-mentioned constructs by standard procedures. Two independent LDH-LacZ lines 33 and 34, with insertions in the third chromosome, were obtained. Tissue-specific expression of the reporter was the same in both lines. Line 34 was used in all studies. Nine independent LDH-Gal4 lines were obtained, and the reporter gene expression pattern was reproducible in all of them. One insertion in the second chromosome that showed strong expression of a UAS-GFP reporter was selected and used for generating LDH-Gal4/UAS-nGFP.LacZ or LDH-Gal4/UAS-TAU.GFP recombinant chromosomes that were used in experiments. The tgo5 and simH9 stocks were kindly provided by Steve Crews; btlMZ13 was a gift from Benny Shilo; UAS-nGFP-LacZ stocks were a gift from Shigeo Hayashi and the UAS-TAU.GFP line was kindly provided by Andrea Brand; lines bearing Df(3L)emc-E12 deficiency and en-Gal4 on the second chromosome were provided by the Bloomington Stock Center. The line bearing UAS-sima on the second chromosome was previously described (56). For heat shock induction the K25 sev HS-Gal4 line on the third chromosome was used.
Synchronized collection of embryos, hypoxic treatment, and β-galactosidase (β-Gal) assays.
To obtain synchronized collections, egg-laying agar plates were replaced every half an hour, and embryos were kept at 25°C until the desired stage.
Hypoxia was applied by regulating the proportions of oxygen and nitrogen in a Forma Scientific 3131 incubator at 25°C. β-Gal activity determinations were performed as described previously (57). Briefly, embryos were homogenized in 200 to 500 μl of lysis buffer (50 mM Tris HCl [pH 7.8], 2 mM EDTA, 10% glycerol, 2 mM dithiothreitol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride) in a Teflon-glass Potter and centrifuged at 2,500 × g for 3 min at 4°C. Supernatant was recovered, and the protein concentration was determined. Typically, 50 μg of protein extract was used in each assay. Enzymatic reactions were performed by incubating 20 μl of extract with 180 μl of reaction buffer containing 80 mM Na3PO4 (pH 7.3), 102 mM β-mercaptoethanol, 9 mM MgCl2, and 4 mM Chlorophenol Red β-d-galactopyranoside (Roche Diagnostics, Mannheim, Germany) at 37°C, and the optical density at 574 nm was read at 10, 30, 60, and 120 min. Rate of color development was linear throughout this time period. Endogenous background was substracted by using a sample inactivated at 100°C for 10 min. After checking on the linear progression of the reaction, values at 2 h were used.
Antibody staining, in situ hybridization, and RNAi.
Rabbit anti-β-Gal antibodies (Cappel), rat anti-Sima antibody (1), rat anti-Trh antibody (51), and mouse monoclonal 2A12 antitracheal lumen antibody (Developmental Studies Hybridoma Bank, Iowa University) were used. All secondary antibodies (conjugated to Cy2, Cy3, or horseradish peroxidase) were from Jackson Laboratories. Standard procedures for X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and antibody staining were used. Embryos, larvae, and adults were visualized by fluorescent or Nomarski optics in a BX-60 Olympus microscope or by confocal microscopy in a Zeiss LSM510 microscope. Homozygous mutant embryos for tgo, trh, sim, and btl genes were identified by absence of balancer chromosome β-Gal staining. Sense digoxigenin-labeled RNA probe for CG1114 mRNA in situ hybridization was synthesized with T7 RNA polymerase (Roche) with pOT2-CG1114 digested with XhoI as a template. For the antisense probe a PCR product bearing a T7 promoter on the 3′ end was used as a template. CG1114 interference RNA (RNAi) was injected into embryos ca. 30 min after egg laying. Embryos were then maintained at 18°C for 2 h, transferred to 25°C until stage 16, and fixed for staining as described above. RNAi was synthesized by using a PCR product with polymerase T7 promoters at the 5′ ends of both DNA strands. Full-length CG1114 cloned in pOT2 was used as a template for PCR amplification by using two primers (5-GAATTAATACGACTCACTATAGGGAGAAGATCTATGATAACCTCCACGACCACGGACTAC-3′ and 5′-GAATTAATACGACTCACTATAGGGAGAAGATCTTTACGTGCATATTTCGCTGCTGGTG-3′) bearing T7 RNA polymerase promoter sequences at the 5′ ends. The PCR product was used as a template for generating RNAi with T7 polymerase. Synthesis was performed at 37°C for 3 h. After quantification, RNAi was divided into aliquots and stored at −70°C.
Western blot and RNA slot blot.
For Western blots, ca. 400 embryos were rapidly homogenized in 50 μl of loading buffer in a Kontes 1.5-ml polypropylene pestle/tube and immediately incubated at 95°C for 3 min. After centrifugation samples were loaded on a gel and subjected to polyacrylamide gel electrophoresis. Blotting and antibody-ECL detection (New England Biolabs) were performed by standard procedures. The anti-β1-tubulin antibody was a gift from Ricardo Ramos. For RNA slot blotting, RNA was prepared from ca. 400 embryos by the Trizol (Gibco) method. RNA was quantified by absorbance at 260 nm and blotted onto a Zeta-Probe (Bio-Rad) membrane by using a Hybri-Slot Manifold (BRL). [32P]dCTP DNA probes were prepared by random priming (Prime-a-Gene; Promega) by using an 842-bp BamHI fragment from the sima gene, a 1,523-bp EcoRV fragment from β-Gal gene or the complete rRNA 18S sequence. Hybridization, washes, and autoradiography were performed by using standard procedures.
RESULTS
A hypoxia-inducible transcriptional response in Drosophila that is homologous to the mammalian HIF system.
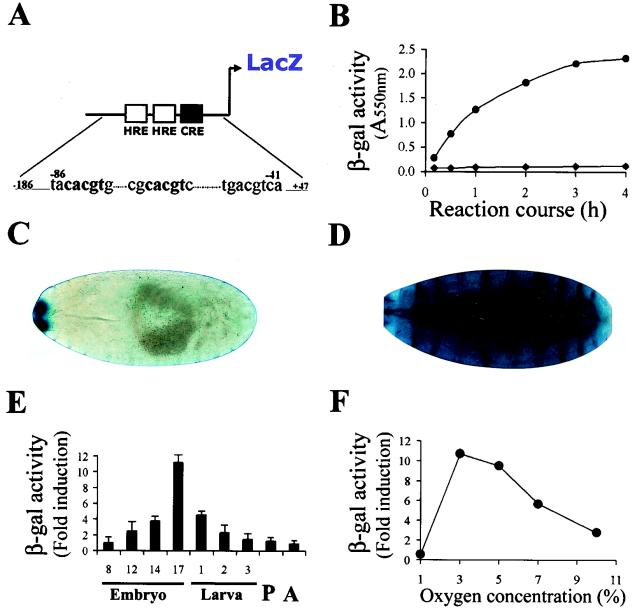
Based on the observation that an HIF homologue present in extracts of Drosophila SL2 cells can bind a mammalian HRE, we sought to characterize the transcriptional system predicted by these experiments by the generation of transgenic flies bearing mammalian HREs linked to a LacZ reporter gene. We first tested a transgenic line bearing a pentamer of an 18-bp sequence from the murine erythropoietin HRE (Epo-LacZ), that is known to be induced by both Trachealess and Single minded (56). However, no significant induction of β-Gal expression was observed when these embryos were exposed to hypoxia. Mutational analysis of the erythropoietin HRE in mammalian cells has indicated that, while this 18-bp sequence is sufficient for binding to HIF-1, an adjacent site that binds an as-yet-unknown factor is also critical for the transcription activity (18). The erythropoietin gene is not conserved in flies, and we reasoned that the additional factor(s) binding to this adjacent site might be missing in flies. Therefore, for the design of a new HRE-LacZ reporter, we selected the mammalian lactate dehydrogenase A (LDH-A) gene, which is hypoxia responsive and well conserved in flies. The mammalian LDH-A hypoxic enhancer includes two HREs separated by an 8-bp spacer and a cyclic AMP responsive element (CRE) located 16 bp further downstream. Both HREs and the CRE consensus have been shown to be necessary for strong hypoxic induction in mammalian cells (16). We generated transgenic lines with an LDH-LacZ reporter based on a 233-bp fragment from the murine LDH-A enhancer bearing the three relevant boxes described above (Fig. 1 -A). In two independent lines carrying this element we observed approximately 10-fold β-galactosidase induction in hypoxic embryos (Fig. 1B to D). These findings demonstrate the operation in Drosophila embryos of a conserved HRE-dependent transcriptional response to hypoxia and provided an opportunity to characterize the transcriptional response to hypoxia in developing flies.
FIG. 1.
A hypoxia-inducible transcriptional response in Drosophila. Physiological characterization of the hypoxic response in vivo. (A) Schematic representation of the LDH-LacZ transcriptional reporter: A 233-bp fragment of the murine LDH-A enhancer from bp −186 to +47 (relative to the transcription initiation site) controls β-Gal expression. The sequences correspond to HRE and CRE elements present in the enhancer. Bases in bold lettering mark the HRE consensus, and the numbers above indicate the position relative to the transcription initiation site. (B) The LDH-LacZ reporter is induced in embryos maintained at 5% O2 for 8 h (•) compared to normoxia (⧫). The x axis represents time course of the β-Gal reaction. (C and D) Stage 17 transgenic embryos carrying the LDH-LacZ reporter maintained in normoxia (C) or hypoxia (8 h at 5% O2) (D) were stained with X-Gal in a reaction developed overnight. Whereas in embryos exposed to hypoxia the X-Gal staining was ubiquitous, in normoxic individuals the expression of the reporter was restricted to a small anterior domain. (E) Modulation of the hypoxic response during development. Individuals bearing the LDH-LacZ reporter were synchronized at different stages of development and were subjected to hypoxia (5% O2) for 4 h. The hypoxia/normoxia β-Gal activity ratio is shown. Hypoxic induction of the reporter is maximal at late embryogenesis, although it is also high at larval stages 1 and 2. P, pupa; A, adult. (F) Effect of oxygen concentration. Stage 16 to 17 embryos bearing the LDH-LacZ reporter were exposed to different oxygen concentrations for 4 h, the β-Gal activity was compared with that of normoxic controls, and the ratios of the activities were calculated. Triplicate determinations of the activity were performed. Induction of the reporter was maximal between 3 and 5% oxygen concentration.
Physiological characterization of the transcriptional response to hypoxia.
First, we studied the manner in which the overall response to hypoxia is influenced by development. For this purpose, we used synchronized populations of insects that were subjected to 5% oxygen for 8 h at different stages of development and tested their ability to induce the reporter. As can be seen in Fig. 1E, hypoxia-inducible β-Gal activity rises dramatically at late embryogenesis, although a relatively high induction of the reporter was recorded at all larval stages. Thus, the response is strongly modulated by developmental parameters, reaching peak levels at late embryogenesis.
Second, we wanted to know the range of ambient oxygen concentrations over which the reporter is induced in the whole animal. The response to different oxygen concentrations was studied in stage 16 to 17 embryos. As depicted in Fig. 1F, hypoxic induction was found to be maximal between 3 and 5% oxygen, decreasing gradually as the oxygen concentration rises. This result indicates that activation by low oxygen is strikingly concentration dependent although, presumably because of oxygen gradients within the developing flies, the point of maximal activation is shifted relative to the concentrations of 0.1 to 1.0% oxygen that are maximally active in tissue culture cells. Remarkably, at 1% oxygen, expression of the reporter was below that of the normoxic controls, most probably reflecting general arrest of cell metabolism in very severe hypoxia (10, 11, 17, 54).
Spatially restricted induction of the hypoxic transcriptional response.
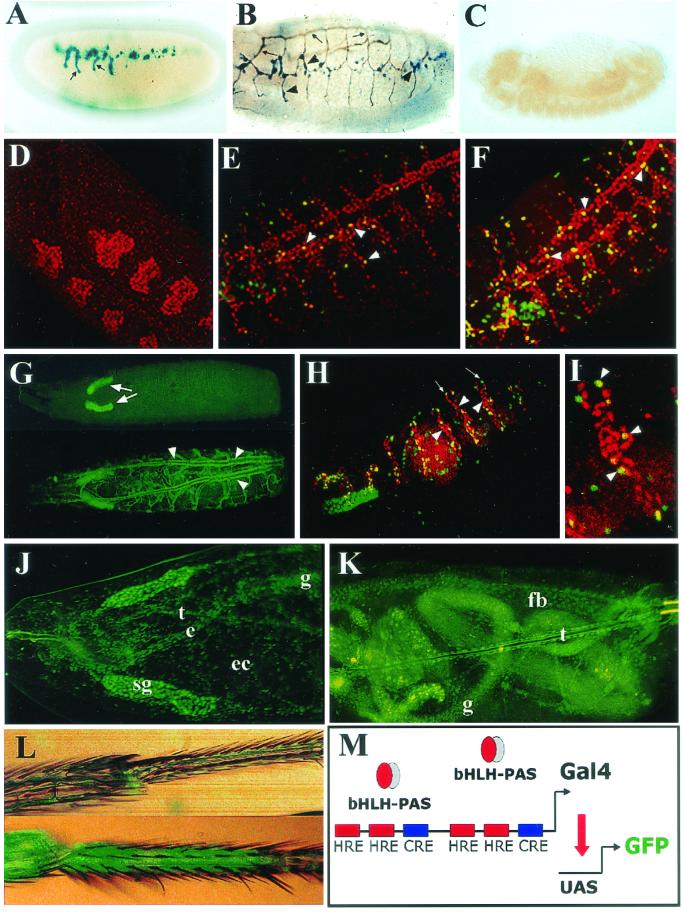
Although the first experiments with X-Gal staining suggested ubiquitous reporter expression (Fig. 1D), this result was obtained by using long incubation times on stage 17 embryos, where the cuticle is beginning to be secreted, and we surmised that very high levels of reporter product may have obscured spatial localization. By using X-Gal staining for 30 min on stage 15 to 16 embryos, a distinct expression pattern that appeared to correspond to tracheal branches was observed (Fig. 2A). Indeed, double staining of hypoxic embryos with an antitracheal lumen antibody revealed hypoxia-inducible β-Gal reporter in the tracheal system, with stronger expression in the lateral trunks and dorsal branches (Fig. 2B and C) (33).
FIG.2.
Spatially restricted induction of the transcriptional response to hypoxia. The expression pattern of LDH-LacZ (A to C) and LDH-Gal4 (D to L) reporters was examined in transgenic embryos or larvae exposed to hypoxia. (A) X-Gal staining of a stage 15 LDH-LacZ embryo subjected to 5% O2 for 4 h. Some branches of the tracheal system (arrows) are stained. (B) Double immunostaining showing the tracheal lumen in brown (arrows) and β-Gal expression in blue (arrowheads). The reporter is expressed at some branches of the tracheal system. (C) In normoxia, no expression of the reporter is seen. (D to F) An unsynchronized population of embryos was exposed to 5% oxygen for 4 h and induction of the reporter was analyzed. Double immunofluorescent confocal image shows that the β-Gal reporter (green) colocalizes with the tracheal marker Trachealess (red). Note that some extratracheal cells express the reporter as well. Expression of the reporter cannot be detected at embryonic stage 11 (see panel D). By stage 13 (panel E), scattered tracheal cells (arrowheads) begin to express the reporter, with further cells responding to hypoxia at stage 14 (arrowheads) (F). (G) By the end of embryogenesis and throughout larval stages, the LDH-Gal4/UAS-TAU.GFP reporter is expressed in the whole tracheal system upon hypoxia (arrowheads, lower panel), but in normoxia no expression is seen in the tracheae (upper panel), although expression of the reporter can be detected in salivary glands (arrows). (H and I) In breathlessMZ13 homozygous embryos tracheal cells (red) fail to migrate (arrowhead), but the hypoxic reporter (UAS-nGFP.LacZ) is induced normally (arrows) in hypoxia. (I) Higher magnification of panel H. (J and K) First-instar larvae subjected to 4% O2 for 16 h express the UAS-nGFP reporter in nontracheal tissues, as well as in the esophagus (e), gut (g), ectoderm (ec), fat body (fb), and tracheae (t). Expression in the salivary glands (sg) is constitutive. (L) Hypoxic induction of the UAS-nGFP reporter is seen in the legs of adult flies maintained at 5% O2 for 24 h (lower panel) compared with normoxia (upper panel). (M) Schematic representation of the LDH-Gal4 transcriptional reporter. A dimerized 51-bp fragment of the murine LDH-A enhancer including the HREs and CRE was constructed as a dimer controlling expression of Gal4.
To study the expression of the reporter in more detail and to assess postembryonic stages, we developed an additional reporter construct based on the binary Gal4/UAS system (3) that would allow more sensitive detection in vivo particularly in the presence of the developing cuticle. We engineered a 51-bp fragment from the core of the LDH-A enhancer (16) as a dimer controlling expression of Gal4 (LDH-Gal4, Fig. 2M) and generated several transgenic fly lines with single insertions in all of the three chromosomes. These lines were crossed with different UAS-green fluorescent protein (GFP) or UAS-GFPn.LacZ lines (42), and expression of the reporter was monitored in normoxia and hypoxia (5% oxygen for 4 h). Unlike the LDH-LacZ lines, these lines showed constitutive reporter expression in salivary glands. However, hypoxia-inducible expression was remarkably similar between the different reporter lines. Double immunofluorescence experiments with antibodies to β-Gal and the tracheal marker Trachealess (53) demonstrated that hypoxia-dependent transcription is indeed elicited mostly in tracheal cells but also in scattered patches on the ectoderm (Fig. 2F). More detailed studies of developmental regulation showed that, whereas stage 11 embryos showed no induction of the reporter (Fig. 2D), scattered cells of the tracheal system began to express the reporter during stages 12 and 13 (Fig. 2E), with the number of cells expressing the reporter increasing gradually from this stage onward (Fig. 2F). By the end of embryogenesis and throughout the larval stages all of the tracheal cells showed a strong response to hypoxia (Fig. 2G, lower panel). To answer the question of whether enhanced hypoxia responsiveness in the tracheal system reflects some microenvironmental signal connected with the position or integrity of the developing tracheal system, we studied hypoxic induction of the reporter in embryos homozygous for a mutation in the gene breathless (btl) that encodes a Drosophila FGF receptor, which is required for tracheal cell migration (28). As is shown in Fig. 2H and I, tracheal cells fail to migrate in btlMZ13 mutants, and the tracheal system is not developed. In these mutants, however, the nonmigrating tracheal cells expressed the hypoxic reporter normally, clearly indicating that tracheal integrity is not required for the hypoxic response.
Enhanced hypoxia-inducible transcription in tracheal cells is of interest since the tracheal system is directly responsible for oxygen delivery in the fly. Nevertheless, current models of oxygen-dependent tracheal plasticity involve a receptor-mediated chemotactic outgrowth of terminal branches that is generated by hypoxia-inducible expression of the Drosophila FGF homologue Branchless in extratracheal metabolizing tissues (24). Therefore, it would be predicted that the hypoxic machinery should operate in nontracheal tissues as well, although perhaps with different sensitivity. To pursue this, we applied more severe hypoxic conditions (4% oxygen for 16 h) to embryos bearing the hypoxic reporter and recorded the GFP expression. Under these conditions, hypoxia-inducible expression was clearly observed outside the tracheae, with approximately one-quarter of late embryos or larvae manifesting widespread reporter expression across the ectoderm, esophagus, gut, fat body, muscles, and gonads (Fig. 2J and K), as well as the trachea. However, tracheal expression remained dominant, and responses in extratracheal cells were more patchy in the majority of embryos. When embryos were placed at an oxygen concentration of 3% or less, arrest of embryonic development occurred, and tissue-specific expression of the reporter could not be recorded. Taken together with the analysis of responses to graded hypoxia in LDH-LacZ flies, this suggests that most if not all tissues in the developing fly can respond to this system, although in many nontracheal tissues this appears to be at a threshold that is close to that which produces metabolic and developmental arrest. In contrast, the tracheal system appears to respond with greater sensitivity. Tracking inducible expression precisely in adult flies was more difficult because of cuticle development. Nevertheless, strong hypoxia-inducible expression was evident in the legs (Fig. 2L).
The bHLH-PAS proteins Similar and Tango mediate the hypoxic response.
The experiments described above established that the HIF response is conserved and strongly active in developing flies, suggesting that genetic studies in Drosophila may be used to explore the upstream and downstream connections of the pathway in an in vivo system.
As a first step in understanding the upstream pathway we wished to define the role of particular Drosophila bHLH-PAS proteins in the pathway. The Drosophila bHLH-PAS protein Tango (Tgo) is a partner for several bHLH-PAS proteins and has been proposed to be homologous to the mammalian protein ARNT (for aryl hydrocarbon receptor nuclear translocator) (46, 56) that functions as the HIF-β subunit. To test this proposed role for Tgo in the hypoxic response, we examined embryos that were homozygous for a strong tgo mutant allele (tgo5). These embryos failed to induce the reporter in hypoxia (Fig. 3A and B), strongly supporting the role of Tgo as the HIF-β subunit and indicating that, as in mammalian cells, this protein is absolutely required for the hypoxia response.
FIG. 3.
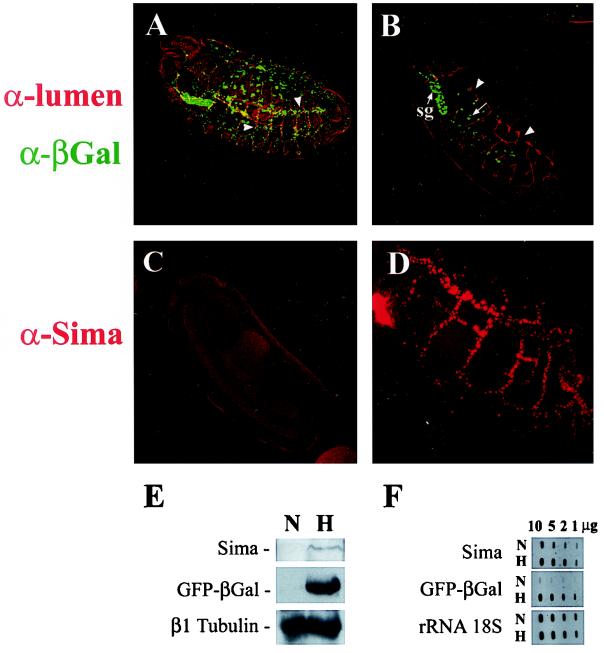
Role of bHLH-PAS proteins in the transcriptional response to hypoxia. (A) Wild-type expression of the β-Gal reporter in the tracheal system of LDH-Gal4/UAS-nGFP.LacZ transgenic embryos subjected to hypoxia (arrowheads). (B) Absence of reporter induction in tango5 homozygous embryos subjected to hypoxia. Only constitutive reporter expression is seen in the salivary glands (sg) and cells of the gut (arrow). Typical tracheal defects are observed in tango mutants (B, arrowheads). (C and D) Sima cannot be detected in normoxic wild-type embryos (C) but is induced upon hypoxia (5% O2 for 14 h) in a pattern that follows the tracheal tree (D). (E) Western blot analysis of lysates of embryos bearing the LDH-Gal4/UAS-GFP.LacZ reporter subjected to hypoxia (5% O2 for 14 h) or maintained in normoxia showing hypoxic induction of both Sima and LDH-Gal4/UAS-GFP.LacZ. (F) Slot blot analysis of RNA extracted from hypoxic or normoxic embryos treated as in panel E. β-Gal mRNA exhibited strong hypoxic induction (9.7- to 10.4-fold), whereas Sima mRNA was induced only slightly (1.3- to 1.5-fold).
We next sought to identify the Drosophila HIF-α homologue(s). Several bHLH-PAS proteins exist in the fruitfly (8, 9), but the best candidate HIF-α homologue is Similar (Sima) since it shows the highest amino acid identity (39), and protein levels are increased at 1% O2 in Drosophila SL2 cells (1). Nevertheless, two other Drosophila bHLH-PAS proteins, Single minded (Sim), a regulator of differentiation of the embryonic central nervous system midline cells (7), and Trachealess (Trh), a key regulator of tracheal development (20, 53), show high identity with HIF-1α in the basic and HLH domains and were also candidates for mediation of the hypoxic transcriptional response. To examine for a role of Sim or Trh in hypoxia-dependent transcription, we crossed the LDH-Gal4/UAS-GFPn.LacZ reporter into flies bearing a sim strong mutant allele (simH9) or a chromosomal deficiency that includes the trh gene and then studied expression of the reporter in homozygous mutant embryos. Homozygous embryos for the Df(3L)emc-E12 chromosomal deficiency that includes the trh gene do not develop a tracheal system and are viable until late embryogenesis (20). After these embryos were challenged with severe hypoxia, strong expression of the reporter was observed, indicating that trh is not required for the hypoxic response (not shown). Similarly, homozygous simH9 embryos subjected to moderate hypoxia (5% oxygen for 4 h) exhibited normal expression of the hypoxic reporter, showing that Single minded is also dispensable for the response (not shown).
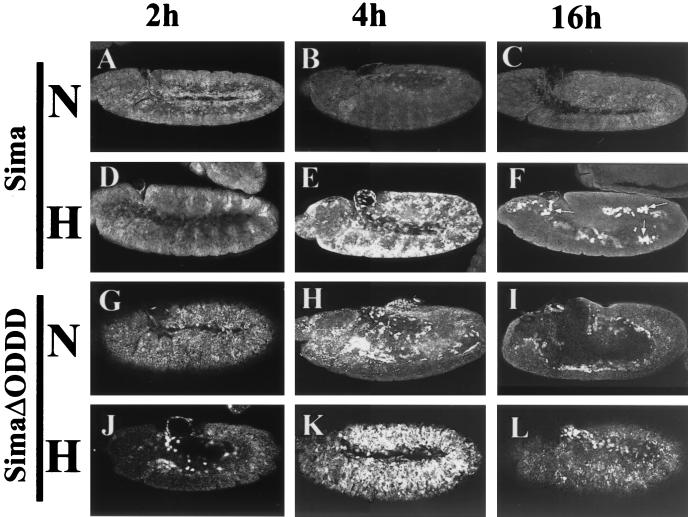
To investigate the role of Sima in the transcriptional response to hypoxia, we performed immunofluorescent detection of Sima in embryos subjected to hypoxia (5% oxygen for 14 h). By performing the fixation immediately (0.5 to 1 min) after opening the hypoxic chamber, strong Sima staining could be detected in stage 15 to 16 embryos subjected to hypoxia, but not in normoxic controls, in a pattern of expression corresponding to the tracheae that strikingly mimics expression of the reporter in hypoxia (Fig. 3C and D). To pursue this further, synchronized stage 8 wild-type embryos bearing the LDH-Gal4/UAS-GFP.LacZ reporter were kept in hypoxia as described above, and protein extracts were analyzed by Western blotting with anti-Sima and anti-β-Gal antisera (1). As shown in Fig. 3E, Sima protein levels are increased in hypoxia, paralleling induction of the β-Gal reporter. However, Sima mRNA increased only slightly in hypoxia (1.3- to 1.5-fold), whereas β-Gal mRNA was upregulated by 9.7- to 10.4-fold (Fig. 3F). These results indicate that Sima is controlled posttranscriptionally, most probably at the level of protein degradation. To test this, we overexpressed the protein in hs-Gal4/UAS-Sima embryos by giving a 37°C heat shock for 20 min and compared the decay of Sima protein levels in hypoxia and normoxia. As depicted in Fig. 4B and E, Sima levels 4 h after heat shock are much higher in hypoxia than in normoxia. At 16 h after heat shock, expression of Sima can still be visualized in patches of cells in hypoxic embryos (Fig. 4F) but is undetectable in embryos kept in normoxia (Fig. 4C), thus confirming that Sima is stabilized in hypoxia.
FIG. 4.
Sima protein is stabilized in hypoxia: identification of the ODDD. Ectopic Sima was expressed by using a heat shock-Gal4 driver (A to F) and detected with an anti-Sima antibody after a 20-min period of heat shock in embryos kept in normoxia (N) or hypoxia (5% O2) (H). At 2 h after heat shock, moderate levels of Sima are detected, but there are no differences between normoxia (A) and hypoxia (D). At 4 h after heat shock, Sima can no longer be detected in normoxia (B), but levels in hypoxia are high (E). At 16 h after heat shock the protein can still be detected in patches of cells in the hypoxic embryos (F, arrows) but not in the normoxic controls (C). (G to L) The Δ692-863 deletion of Sima generated a stable protein. At 2 (G and J), 4 (H and K), or 16 h (I and L) after heat shock, SimaΔ692-863 was detected both in normoxic (G to I) and hypoxic (J to L) embryos, showing that the instability of Sima in normoxia depends on the deleted sequences.
Proteolytic control of mammalian HIF-α is dependent on a central oxygen-dependent degradation domain (ODDD) (19, 40) that contains two sites of oxygen-dependent prolyl hydroxylation (34). Although overall conservation between HIF-1α and Sima outside the basic and HLH domains is low (1, 39), sequence comparison indicates that these prolyl residues are both conserved (Sima Pro747 and Pro850), suggesting that they might define a Sima ODDD. To test this, we generated a deletion of Sima between amino acids 692 and 863 (SimaΔ692-863) and expressed the deleted Sima in UAS transgenic lines through a heat shock-Gal4 driver as described above for full-length Sima. As shown in Fig. 4G to I, this deletion stabilized Sima in normoxia with no further changes in stability when heat-shocked embryos were incubated in hypoxia (Fig. 4J to L). These findings demonstrated that Sima is regulated by oxygen-dependent proteolysis in a manner similar to mammalian HIF-α.
Oxygen-dependent subcellular localization of Sima.
To override the rate of Sima degradation and shed light on an additional mode of regulation, we sought to overexpress the protein by using an engrailed-Gal4 (en-Gal4) driver. The en-Gal4 element was recombined in a chromosome bearing a UAS-nGFP-LacZ element (with a nuclear localization signal) (42) that was used to mark the nuclei. Sima protein, expressed in the characteristic striped engrailed pattern, was easily detected by immunofluorescence in stages 11 to 15 normoxic embryos. Surprisingly, however, we found that the protein was almost exclusively localized in the cytoplasm (Fig. 5A to C). In another experiment, Sima was expressed in embryos bearing the direct LDH-LacZ hypoxic reporter (lacking the UAS-nGFP-LacZ element). In normoxia, the reporter was induced only at very low levels (Fig. 5D). In contrast, when the same experiments were performed with embryos exposed to 5% oxygen, Sima was detected predominantly in the nucleus (Fig. 5E to G), and embryos exhibited strong expression of the reporter in the expected engrailed pattern (Fig. 5H). In contrast, expression of Trachealess or a chimeric protein exhibiting specificity for Single minded target genes (56) under en-Gal4 did not activate the reporter in either normoxia or hypoxia (data not shown).
FIG. 5.
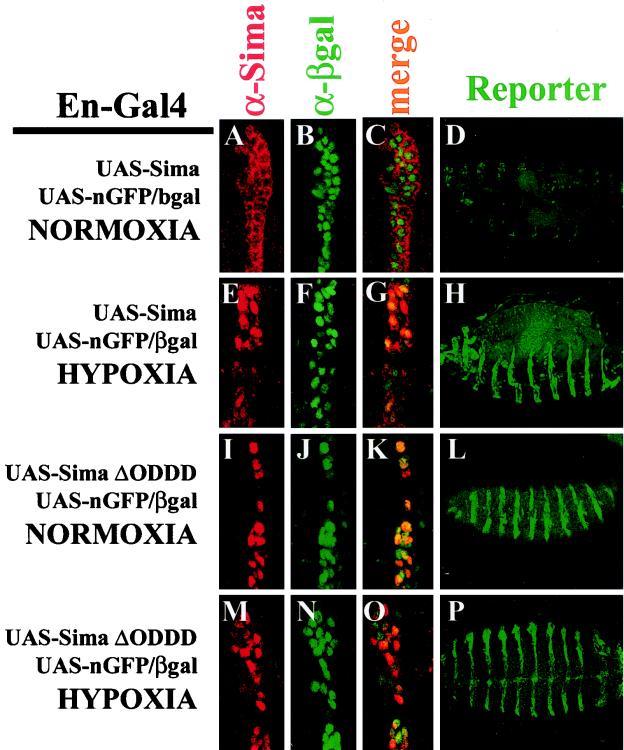
Regulation of Sima subcellular localization by hypoxia. Sima (A and E) or SimaΔ692-863 (I and M) were expressed ectopically through an en-Gal4 driver, and subcellular localization was detected with an anti-Sima antibody. (B, F, J, and N) The embryos also contained a UAS-nGFP.LacZ reporter (bearing a nuclear localization signal) detected with an anti-β-Gal antibody that was used to mark the nuclei. In normoxia, Sima was localized exclusively in the cytoplasm (A to C) and became nuclear in hypoxia (5% O2 for 8 h) (E to G). SimaΔ692-863 was constitutively detected in the nuclei irrespective of oxygen levels (I to K and M to O). (D, H, L, and P) Sima or SimaΔ692-863 ectopic expression driven by en-Gal4 was also performed in embryos bearing the direct LDH-LacZ hypoxic reporter and lacking the UAS-nGFP.LacZ element. Upon ectopic expression of Sima, the LDH-LacZ reporter was strongly induced in hypoxia (H) but only weakly in normoxia (D), a finding consistent with the subcellular localization of Sima. Expression of SimaΔ692-863 produced strong expression of the reporter irrespective of oxygen levels (L and P).
This confirmed that Sima mediates activity of the HRE reporter but also indicated the existence of an additional oxygen-regulated nuclear localization mechanism, as has been proposed in mammalian cells (26, 31). Studies of mammalian HIF-α have demonstrated that stabilization by deletion of the ODDD is associated with constitutive upregulation of transcriptional activity (12, 19). Therefore, we reasoned that the deletion might also affect subcellular localization. The Drosophila system provided an opportunity to compare the localization of SimaΔ692-863 with full-length Sima under similar expression conditions. Accordingly, SimaΔ692-863 was expressed similarly in embryos through the en-Gal4 driver. As depicted in Fig. 5I to K and M to O, deletion of the ODDD caused constitutive localization of Sima in the nucleus irrespective of oxygen levels. Consistent with this, the LDH-LacZ transcriptional reporter showed strong constitutive expression in the expected engrailed pattern after crossing into embryos expressing SimaΔ692-863 (Fig. 5L and P). These results indicate that the Sima ODDD also mediates signals critically required for cytoplasmic localization of Sima in normoxia (see below).
Role of the prolyl 4-hydroxylase homologue CG1114 in the hypoxic response in vivo.
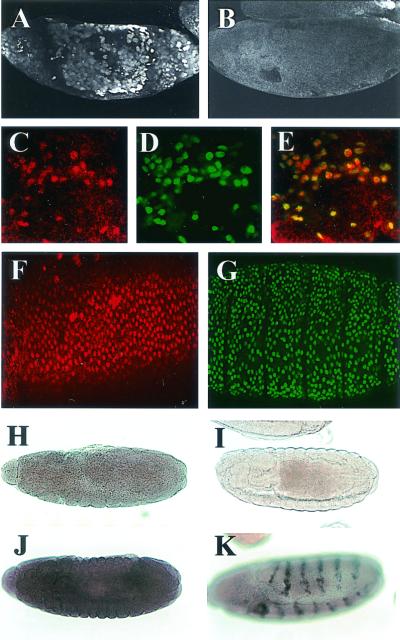
A Drosophila sequence homologous to the HIF prolyl hydroxylases was recently reported (gene CG1114) (4, 48). These enzymes are known to promote proteasomal destruction of HIF-α through the hydroxylation of key prolyl residues in the ODDD. To assess whether CG1114 regulates Sima levels and the transcriptional response to hypoxia in vivo, we abrogated CG1114 expression by injecting double-stranded RNA into early embryos bearing the LDH-Gal4/UAS-nGFP.LacZ reporter gene. As depicted in Fig. 6A, Sima protein was strongly upregulated in normoxic embryos injected with CG1114 RNAi but not in individuals injected with an unrelated double-stranded RNA (Fig. 6B). Sima upregulation (Fig. 6A and C) induced expression of the LDH-Gal4/UAS-nGFP.LacZ nuclear hypoxic reporter (Fig. 6D). Furthermore, complete overlap between Sima protein and the nuclear reporter product demonstrated that Sima was exclusively localized in the nucleus (Fig. 6C to E). To further assess the role of CG1114 in the regulation of Sima, we examined flies that bore mutations predicted to inactivate this gene completely. A lethal P element insertional mutation [l(3)02255], mapping 336 nucleotides upstream of the initiation codon of the CG1114 gene, is available from Drosophila public stock collections. By crossing l(3)02255 heterozygous mutant flies with a strain carrying the Df(3R)3-4 chromosomal deficiency that covers CG1114 gene, we verified that the insertion was indeed causing the lethal phenotype. We analyzed Sima levels and induction of the transcriptional response to hypoxia in l(3)02255 embryos bearing the LDH-Gal4/UAS-nGFP.LacZ reporter. As depicted in Fig. 6F, Sima is ubiquitously upregulated in homozygous mutant embryos in normoxia, and high levels of Sima result in concomitant strong induction of the LDH-Gal4/UAS-nGFP.LacZ reporter in all embryonic tissues (Fig. 6G).
FIG. 6.
Role of the prolyl 4-hydroxylase homologue CG1114 in the hypoxic response in vivo. (A and B) CG1114 expression was abrogated by injecting double-stranded RNA; Sima protein detected with a specific antiserum was strongly upregulated (A) but not when injected with an unrelated double-stranded RNA (B). (C and D) Higher magnification of an embryo injected with CG1114 RNAi revealing Sima stabilization (C) and concomitant expression of the nuclear hypoxic reporter LDH-Gal4/UAS-nGFP.LacZ detected with an anti-βGal antibody (D). (E) The merged confocal image shows that Sima is localized in the nucleus. (F) Sima is upregulated ubiquitously in homozygous mutant l(3)02255 embryos and is localized in the nuclei. (G) These high levels of Sima result in strong, widespread induction of the LDH-Gal4/UAS-nGFP.LacZ reporter. (H) Expression of CG1114 mRNA in embryos in normoxia. (I) Control antisense probe. (J) CG1114 gene expression was strongly upregulated in hypoxia (compare with panel H). (K) Ectopic expression of Sima under control of en-Gal4 induces CG1114 mRNA expression in the typical engrailed pattern.
Ectopic stabilization of Sima could potentially exert a dominant-negative effect on Trachealess or other bHLH-PAS proteins. To investigate a possible effect on Trachealess, the embryonic tracheal system was studied with a specific antitracheal lumen antibody, but no obvious defects were observed (not shown). Taken together, these results imply a general and nonredundant role of CG1114 in the regulation of both subcellular localization and protein stability of Sima. In support of this, we found that CG1114 mRNA is ubiquitously expressed throughout embryogenesis (Fig. 6H and I).
Mammalian HIF prolyl hydroxylase genes are themselves induced by hypoxia (12), suggesting the existence of a feedback response that limits HIF induction in hypoxia. To test whether this aspect of regulation is also conserved in Drosophila and does indeed represent a feedback response dependent on activity in the transcriptional pathway itself, we tested for induction by both hypoxia and Sima overexpression. As shown in Fig. 6J, CG1114 gene expression was strongly upregulated in hypoxia (compare with Fig. 6H) and was strongly induced in the typical engrailed pattern by overexpression of Sima by using an en-Gal4 driver (Fig. 6K). In contrast, the branchless/FGF gene was not induced in embryos exposed to hypoxia or overexpressing Sima (not shown), suggesting that the hypoxic regulation of branchless (24) starts at larval stages.
DISCUSSION
In this work we have used transgenic flies to demonstrate and characterize in vivo the operation of a hypoxia inducible transcription response in Drosophila that is homologous to mammalian HIF. The work confirms the candidacy of Sima and Tgo as the Drosophila homologues of mammalian HIF-α and HIF-β, respectively, defines a conserved multistep mode of regulation for Sima and provides new insights into the mechanisms regulating HIF proteins, as well as into the spatial and temporal operation of the hypoxia-responsive system during Drosophila development.
By tracking reporter gene activation in developing flies, we analyzed the oxygen concentration dependence, developmental regulation, and spatial distribution of the transcriptional response. Serial studies of the hypoxia response during development indicated that induction by hypoxia is modest in early embryogenesis and mid-embryogenesis and then rises sharply to peak levels at the end of embryogenesis, thereafter remaining relatively high throughout the larval stages. This developmentally restricted capacity fits well with the adaptive requirements of Drosophila larvae. After eclosion larvae usually dig into the substrate, while feeding actively, and are probably subjected to major variations in environmental oxygen tension so that enhanced activity of the HIF system is likely to be of critical importance at this stage.
Interestingly and somewhat unexpectedly, analysis of reporter expression patterns in developing flies showed enhanced hypoxia-inducible activity in the cells of the tracheal system. Although experiments using severe hypoxia and genetic inactivation of Sima proteolysis demonstrated a widespread potential for transcriptional activation by this system, exposure to more moderate hypoxia clearly demonstrated enhanced activity in tracheal cells. This was reflected both in higher expression levels of Sima and in higher activity of different HRE-linked reporter genes and, moreover, was shown to be a cell autonomous function that was preserved in cells of tracheal fate even in the face of mutations that disrupt tracheal architecture.
The existence of enhanced responses to hypoxia in cells composing the organ of oxygen delivery is clearly of interest and raises questions as to its function, particularly since current models indicate that the regulation of tracheal development by oxygen is guided by signals arising in the metabolizing tissues outside the tracheae (24). Interestingly, some of the branches of the tracheal system run alongside the Drosophila nervous system (14), and one possibility is that the tracheae function as sensory organs for hypoxia, as the carotid body does in mammals. A hypoxia pathway affecting behavioral responses has been described in flies (54), and it will be interesting to determine whether hypoxia-induced behavioral responses share a regulatory mechanism with the HIF system.
In the current work we also utilized the HRE transgenic reporter system to define upstream control mechanisms operating on the Drosophila HIF system. Our studies identify Sima as the regulatory Drosophila HIF subunit and demonstrate a major mode of regulation through oxygen-dependent proteolysis that involves a central ODDD. Interestingly, both of the sites of prolyl hydroxylation that operate in mammalian HIF-α subunit ODDD (34) appear to be conserved in Sima. Furthermore, genetic ablation of the Drosophila HIF prolyl hydroxylase homologue CG1114 results in striking upregulation of both Sima and reporter gene activity in vivo. This strongly supports a conserved mode of proteolytic regulation of Sima following prolyl hydroxylation at one or both of these sites.
In contrast with the mammalian system, where HIF prolyl hydroxylase activity is represented by the three PHD isoforms (4, 15), survey of the Drosophila genome revealed only one homologue (48), raising questions about the potential of this activity to regulate precisely tuned physiological responses. Interestingly, however, we found that the CG1114 gene is itself a Sima target, demonstrating the operation of a conserved feedback control with the potential to contribute to the complex demands of physiological oxygen homeostasis.
Studies of Sima regulation also demonstrated an additional regulatory step. Transgenic overexpression of Sima in normoxic embryos resulted in cytoplasmic accumulation of the protein and little transcriptional activity. In contrast, similar levels of overexpression in hypoxia resulted in nuclear accumulation and a strong transcriptional response, demonstrating the presence of a second oxygen-regulated mechanism controlling Sima subcellular localization. An oxygen-regulated nuclear localization step has previously been demonstrated for mammalian HIF-α (2, 26, 31), although not in every study. Our demonstration of conservation of this mode of regulation in Drosophila Sima, however, provides strong support for the physiological relevance of this process. Our findings suggest that Sima subcellular localization is controlled by an active mechanism that maintains the protein in the cytoplasm in normoxia as opposed to an hypoxia-dependent machinery that mediates nuclear import. Although the strong transcriptional activity of mammalian HIF-α that is observed after deletion of the ODDD (12, 19), mutation of the VHL binding sites (34), or inactivation of VHL (36) is consistent with a role for this domain in cytoplasmic localization in normoxia, this has not been tested in studies of mammalian HIF-α that have examined subcellular localization directly. Moreover, although induction of nuclear localization by iron chelators and cobaltous ions (26) suggests a similar mode of regulation to proteolytic regulation, neither the source of the oxygen-sensitive signal nor the mechanism of transduction have been defined. Our in vivo studies in flies show induction of nuclear Sima after inactivation of CG1114 either by RNAi or by mutation, thus clearly implicating this gene product in the process of cytoplasmic localization in normoxia. Moreover, Sima nuclear localization was also observed in flies bearing the SimaΔ692-863 transgene, indicating that this sequence is absolutely required for cytoplasmic localization.
Very recently nonproteolytic regulation of mammalian HIF-α subunits involving the C-terminal transactivation domains has been shown to be regulated by hydroxylation of a specific asparaginyl residue by an enzymatic activity that, like the prolyl hydroxylases involved in HIF proteolysis, demonstrates the properties of an α-ketoglutarate-dependent dioxygenase (30). Thus, regulatory hydroxylation of HIF-α residues by this class of enzyme appears to extend to both specific asparaginyl and prolyl residues. Currently, the precise substrate requirements of the CG1114 gene product are not defined, and it is not clear whether effects on nuclear localization are mediated through the conserved prolyl residues, possibly reflecting additional functions of the VHL ubiquitylation complex, or whether other sequences within the Sima ODDD mediate this process. Further biochemical and genetic studies should clarify these new insights into the HIF system.
Overall, the high degree of conservation in the Drosophila system indicates that genetic studies in this organism should be highly informative in analyses of both the upstream pathways regulating the HIF system, and the downstream physiological effects in an intact organism.
Acknowledgments
We thank Andrea Brand, Steve Crews, Shigeo Hayashi, Luis Quesada-Allué, Ricardo Ramos, Benny Shilo, Carl Thummel, the Bloomington Stocks Center, and the Developmental Studies Hybridoma Bank for plasmids, antibodies, and fly stocks. We thank Benny Shilo and Osvaldo Podhajcer for critical reading of the manuscript.
This work was supported by the Wellcome Trust grant HHBR6 and Fundación Antorchas grant A-13740/1-119 to P.W. and P.J.R. S.L.-L. and D.M.R. are fellows of the Argentinean National Council of Scientific Research (CONICET), S.N.B. received a postdoctoral fellowship from The Wellcome Trust, L.C. is a fellow of the FONCyT, M.I. and M.M. have FOMEC fellowships, and P.W. is a career investigator of CONICET.
REFERENCES
- 1.Bacon, N. C., P. Wappner, J. F. O'Rourke, S. M. Bartlett, B.-Z. Shilo, C. W. Pugh, and P. J. Ratcliffe. 1998. Regulation of the Drosophila bHLH-PAS protein Sima by hypoxia: functional evidence for homology with mammalian HIF-1α. Biochem. Biophys. Res. Commun. 249:811-816. [DOI] [PubMed] [Google Scholar]
- 2.Berra, E., D. Roux, D. E. Richard, and J. Pouyssegur. 2001. Hypoxia-inducible factor-1α (HIF-1α) escapes O2-driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO Rep. 2:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 4.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet, P., Y. Dor, J. M. Herbert, D. Fukumura, K. Brusselmans, M. Dewerchin, M. Neeman, F. Bono, R. Abramovitch, P. Maxwell, C. J. Koch, P. Ratcliffe, L. Moons, R. K. Jain, D. Collen, and E. Keshet. 1998. Role of HIF-1α in hypoxia-mediated apoptosis, cell proliferation, and tumour angiogenesis. Nature 394:485-490. [DOI] [PubMed] [Google Scholar]
- 6.Carrero, P., K. Okamoto, P. Coumailleau, S. O'Brien, H. Tanaka, and L. Poellinger. 2000. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1α. Mol. Cell. Biol. 20:402-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crews, S. T., R. Franks, S. Hu, B. Matthews, and J. Nambu. 1992. Drosophila single-minded gene and the molecular genetics of CNS midline development. J. Exp. Zool. 261:234-244. [DOI] [PubMed] [Google Scholar]
- 8.Crews, S. T. 1998. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 12:607-620. [DOI] [PubMed] [Google Scholar]
- 9.Crews, S. T., and C. M. Fan. 1999. Remembrance of things PAS: regulation of development by bHLH-PAS proteins. Curr. Opin. Genet. Dev. 9:580-587. [DOI] [PubMed] [Google Scholar]
- 10.DiGregorio, P. J., J. A. Ubersax, and P. H. O'Farrell. 2001. Hypoxia and nitric oxide induce a rapid, reversible cell cycle arrest of the Drosophila syncytial divisions. J. Biol. Chem. 276:1930-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douglas, R. M., T. Xu, and G. G. Haddad. 2001. Cell cycle progression and cell division are sensitive to hypoxia in Drosophila melanogaster embryos. Am. J. Physiol. Regul. Integr. 280:R1555-R1563. [DOI] [PubMed] [Google Scholar]
- 12.Elson, D. A., G. Thurston, L. E. Huang, D. G. Ginzinger, D. M. McDonald, R. S. Johnson, and J. M. Arbeit. 2001. Induction of hypervascularity without leakage or inflammation in transgenic mice overexpressing hypoxia-inducible factor-1α. Genes Dev. 15:2520-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ema, M., K. Hirota, J. Mimura, H. Abe, J. Yodoi, K. Sogawa, L. Poellinger, and Y. Fujii-Kuriyama. 1999. Molecular mechanisms of transcription activation by HLF and HIF1α in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 18:1905-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Englund, C., A. E. Uv, R. Cantera, L. D. Mathies, M. A. Krasnow, and C. Samakovlis. 1999. adrift, a novel bnl-induced Drosophila gene, required for tracheal pathfinding into the Cns. Development 126:1505-1514. [DOI] [PubMed] [Google Scholar]
- 15.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. Caenorhabditis elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 16.Firth, J. D., B. L. Ebert, and P. J. Ratcliffe. 1995. Hypoxic regulation of lactate dehydrogenase A: interaction between hypoxia-inducible factor 1 and cAMP response elements. J. Biol. Chem. 270:21021-21027. [DOI] [PubMed] [Google Scholar]
- 17.Haddad, G. G. 2000. Enhancing our understanding of the molecular responses to hypoxia in mammals using Drosophila melanogaster. J. Appl. Physiol. 88:1481-1487. [DOI] [PubMed] [Google Scholar]
- 18.Hu, B., E. Wright, L. Campbell, and K. L. Blanchard. 1997. In vivo analysis of DNA-protein interactions on the human erythropoietin enhancer. Mol. Cell. Biol. 17:851-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, L. E., J. Gu, M. Schau, and H. F. Bunn. 1998. Regulation of hypoxia-inducible factor 1α is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95:7987-7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isaac, D. D., and D. J. Andrew. 1996. Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes Dev. 10:103-117. [DOI] [PubMed] [Google Scholar]
- 21.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. S. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for oxygen sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 22.Iyer, N. V., L. E. Kotch, F. Agani, S. W. Leung, E. Laughner, R. H. Wenger, M. Gassmann, J. D. Gearhart, A. M. Lawler, A. Y. Yu, and G. L. Semenza. 1998. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. V. Kriegsheim, H. F. Heberstreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the Von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 24.Jarecki, J., E. Johnson, and M. A. Krasnow. 1999. Oxygen regulation of airway branching in Drosophila is mediated by Branchless FGF. Cell 99:211-220. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, B. H., J. Z. Zheng, S. W. Leung, R. Roe, and G. L. Semenza. 1997. Transactivation and inhibitory domains of hypoxia-inducible factor 1α: modulation of transcriptional activity by oxygen tension. J. Biol. Chem. 272:19253-19260. [DOI] [PubMed] [Google Scholar]
- 26.Kallio, P. J., K. Okamoto, S. O'Brien, P. Carrero, Y. Makino, H. Tanaka, and L. Poellinger. 1998. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1α. EMBO J. 17:6573-6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klagsbrun, M., D. Knighton, and J. Folkman. 1976. Tumor angiogenesis activity in cells grown in tissue culture. Cancer Res. 36:110-114. [PubMed] [Google Scholar]
- 28.Klambt, C., L. Glazer, and B.-Z. Shilo. 1992. breathless, a Drosophila Fgf receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes Dev. 6:1668-1678. [DOI] [PubMed] [Google Scholar]
- 29.Kozak, K. R., B. Abbott, and O. Hankinson. 1997. ARNT-deficient mice and placental differentiation. Dev. Biol. 191:297-305. [DOI] [PubMed] [Google Scholar]
- 30.Lando, D., D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelaw. 2002. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295:858-861. [DOI] [PubMed] [Google Scholar]
- 31.Luo, J. C., and M. Shibuya. 2001. A variant of nuclear localization signal of bipartite-type is required for the nuclear translocation of hypoxia-inducible factors (1α, 2α, and 3α). Oncogene 20:1435-1444. [DOI] [PubMed] [Google Scholar]
- 32.Maltepe, E., J. V. Schmidt, D. Baunoch, C. A. Bradfield, and M. C. Simon. 1997. Abnormal angiogenesis and responses to glucose and oxygen deprivation in mice lacking the protein ARNT. Nature 386:403-407. [DOI] [PubMed] [Google Scholar]
- 33.Manning, G., and M. A. Krasnow. 1993. Development of the Drosophila tracheal system, p. 609-685. In A. Martinez-Arias and M. Bate (ed.), The development of Drosophila melanogaster, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Masson, N., C. Willam, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20: 5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maxwell, P. H., C. W. Pugh, and P. J. Ratcliffe. 1993. Inducible operation of the erythropoietin 3′ enhancer in multiple cell lines: evidence for a widespread oxygen-sensing mechanism. Proc. Natl. Acad. Sci. USA 90:2423-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 37.Metzger, R. J., and M. A. Krasnow. 1999. Genetic control of branching morphogenesis. Science 284:1635-1639. [DOI] [PubMed] [Google Scholar]
- 38.Nagao, M., B. L. Ebert, P. J. Ratcliffe, and C. W. Pugh. 1996. Drosophila melanogaster SL2 cells contain a hypoxically inducible DNA binding complex which recognizes mammalian HIF-binding sites. FEBS Lett. 387:161-166. [DOI] [PubMed] [Google Scholar]
- 39.Nambu, J. R., W. Chen, S. Hu, and S. T. Crews. 1996. The Drosophila melanogaster similar bHLH-PAS gene encodes a protein related to human hypoxia-inducible factor 1 alpha and Drosophila single-minded. Gene 172:249-254. [DOI] [PubMed] [Google Scholar]
- 40.Pugh, C. W., J. F. O'Rourke, M. Nagao, J. M. Gleadle, and P. J. Ratcliffe. 1997. Activation of hypoxia-inducible factor-1: definition of regulatory domains within the alpha subunit. J. Biol. Chem. 272:11205-11214. [DOI] [PubMed] [Google Scholar]
- 41.Ryan, H. E., J. Lo, and R. S. Johnson. 1998. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiga, Y., M. Tanaka-Matakatu, and S. Hayashi. 1996. A nuclear GFP/β-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev. Growth Differentiation 38:99-106. [Google Scholar]
- 43.Shilo, B.-Z., L. Gabay, L. Glazer, M. Reichman-Fried, P. Wappner, R. Wilk, and E. Zelzer. 1997. Branching morphogenesis in the Drosophila tracheal system. Cold Spring Harbor Symp. Quant. Biol. 62:241-247. [PubMed] [Google Scholar]
- 44.Shweiki, D., A. Itin, D. Soffer, and E. Keshet. 1992. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature 359:843-845. [DOI] [PubMed] [Google Scholar]
- 45.Shweiki, D., M. Neeman, A. Itin, and E. Keshet. 1995. Induction of vascular endothelial growth factor expression by hypoxia and by glucose deficiency in multicell spheroids: implications for tumor angiogenesis. Proc. Natl. Acad. Sci. USA 92:768-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnenfeld, M., M. Ward, G. Nystrom, J. Mosher, S. Stahl, and S. Crews. 1997. The Drosophila tango gene encodes a bHLH-PAS protein that is orthologous to mammalian Arnt and controls CNS midline and tracheal development. Development 124:4571-4582. [DOI] [PubMed] [Google Scholar]
- 47.Sutherland, D., C. Samakovlis, and M. A. Krasnow. 1996. Branchless encodes a Drosophila Fgf homolog that controls tracheal cell migration and the pattern of branching. Cell 87: 1091-1101. [DOI] [PubMed] [Google Scholar]
- 48.Taylor, M. S. 2001. Characterization and comparative analysis of the EGLN gene family. Gene 275:125-132. [DOI] [PubMed] [Google Scholar]
- 49.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, G. L., and G. L. Semenza. 1995. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270:1230-1237. [DOI] [PubMed] [Google Scholar]
- 51.Wappner, P., L. Gabay, and B.-Z. Shilo. 1997. Interactions between the EGF receptor and DPP pathways establish distinct cell fates in the tracheal placodes. Development 124:4707-4716. [DOI] [PubMed] [Google Scholar]
- 52.Wappner, P., and P. J. Ratcliffe. 2001. Development of branched structures and the cellular response to hypoxia: an evolutionary perspective, p. 91-137. In G. G. Haddad and T. Xu (ed.), Genetic models in cardiorespiratory biology. Lung Biology in Health and Disease series, vol. 156. Marcel Dekker, Inc., New York, N.Y.
- 53.Wilk, R., I. Weizman, and B.-Z. Shilo. 1996. Trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev. 10:93-102. [DOI] [PubMed] [Google Scholar]
- 54.Wingrove, J. A., and P. H. O'Farrell. 1999. Nitric oxide contributes to behavioral, cellular, and developmental responses to low oxygen in Drosophila. Cell 98:105-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wood, S. M., J. M. Gleadle, C. W. Pugh, O. Hankinson, and P. J. Ratcliffe. 1996. The role of the aryl hydrocarbon receptor nuclear translocator (ARNT) in hypoxic induction of gene expression. Studies in ARNT-deficient cells. J. Biol. Chem. 271:15117-15123. [DOI] [PubMed] [Google Scholar]
- 56.Zelzer, E., P. Wappner, and B. Z. Shilo. 1997. The PAS domain confers target gene specificity of Drosophila bHLH/PAS proteins. Genes Dev. 11:2079-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zwiebel, L. J., P. E. Hardin, X. Liu, J. C. Hall, and M. Rosbash. 1991. A posttranscriptional mechanism contributes to circadian cycling of a per-β-galactosidase fusion protein. Proc. Natl. Acad. Sci. USA 88:3882-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]