Abstract
Cyclin-dependent kinases (Cdks) were originally identified as regulators of eukaryotic cell cycle progression, but several Cdks were subsequently shown to perform important roles as transcriptional regulators. While the mechanisms regulating the Cdks involved in cell cycle progression are well documented, much less is known regarding how the Cdks that are involved in transcription are regulated. In Saccharomyces cerevisiae, Bur1 and Bur2 comprise a Cdk complex that is involved in transcriptional regulation, presumably mediated by its phosphorylation of the carboxy-terminal domain (CTD) of the largest subunit of RNA polymerase II. To investigate the regulation of Bur1 in vivo, we searched for high-copy-number suppressors of a bur1 temperature-sensitive mutation, identifying a single gene, CAK1. Cak1 is known to activate two other Cdks in yeast by phosphorylating a threonine within their conserved T-loop domains. Bur1 also has the conserved threonine within its T loop and is therefore a potential direct target of Cak1. Additional tests establish a direct functional interaction between Cak1 and the Bur1-Bur2 Cdk complex: Bur1 is phosphorylated in vivo, both the conserved Bur1 T-loop threonine and Cak1 are required for phosphorylation and Bur1 function in vivo, and recombinant Cak1 stimulates CTD kinase activity of the purified Bur1-Bur2 complex in vitro. Thus, both genetic and biochemical evidence demonstrate that Cak1 is a physiological regulator of the Bur1 kinase.
Cyclin-dependent kinases (Cdks) were originally identified as key players in eukaryotic cell cycle progression (29). In Saccharomyces cerevisiae, the cell division cycle is controlled by a single Cdk, Cdc28, that interacts with nine different cyclin subunits in a precise order (28, 39), while in mammalian cells multiple Cdks control cell cycle progression (29). As additional Cdks were identified, many of them were found to play important roles in other cellular processes, in particular as transcriptional regulators (29). In S. cerevisiae at least four Cdk complexes, composed of Kin28-Ccl1-Tfb3, Srb10-Srb11, Ctk1-Ctk2-Ctk3, and Bur1-Bur2, have direct and relatively general roles in transcription (4, 22, 23, 30, 48, 50, 54), with all of them implicated in phosphorylating the carboxy-terminal domain (CTD) of Rpb1, the largest subunit of RNA polymerase II. Phosphorylation of the Rpb1 CTD by these four kinases is believed to drive gene expression by regulating the sequential association of transcription initiation and elongation factors and the RNA processing machinery with the polymerase II CTD (35).
The mechanisms that regulate the Cdks involved in cell cycle progression are well studied (29, 32). The cell cycle Cdks are primarily regulated by the binding of their cyclin subunits, whose protein levels change periodically during the cell cycle. Their activities can be further stimulated, however, by binding of additional subunits (6) or by phosphorylation of a threonine or serine residue within their conserved T-loop domains (18). The binding of cyclin A induces a remarkable conformational change of Cdk2 that reorientates the ATP phosphate-binding residues within the active site to ensure the phosphate-transfer reaction (17), while phosphorylation of the T-loop threonine by the Cdk-activating kinase (CAK) (11, 18, 34, 46, 47) results in a smaller yet critical structural change in the substrate binding surface (42). In mammals, Cak activity is conferred by a Cdk complex containing the Cdk7/p40MO15 catalytic subunit (12), whereas in S. cerevisiae Cak activity is conferred by the monomeric Cak1/Civ1 kinase (7, 19, 49). Cak1 is essential for viability (7, 49), phosphorylates and activates both Cdc28 and Kin28 (8, 21), and is involved in meiotic development and spore formation by activating one or multiple kinases in the SMK1 pathway (44, 52). It is not known how many additional bona fide Cak1 substrates exist in vivo. The cell cycle Cdks can also be negatively regulated by inhibitory phosphorylation of tyrosine and/or threonine residues near the amino-terminal end of the kinase domain (15) or by the binding of Cdk inhibitors such as p27Kip1 and p16INK4a (33, 45). The inhibitory phosphorylation sites lie within the glycine-rich loop that assists ATP binding (29), while p27Kip1 inhibits Cdk2 by causing profound changes in the catalytic center, mediated through extensive contacts with both the kinase and cyclin subunits (41), and p16INK4a inhibits cyclin and ATP binding by inducing a structural change at the catalytic cleft of Cdk6 (43).
In contrast to the cell cycle class of Cdks, much less is known about how the Cdks involved in transcription are regulated. By definition, the transcription Cdks require cyclins for full activity, but their cyclin levels do not fluctuate relative to the kinase subunit in response to any regulatory signal (23, 51). Since the major mechanism for regulating the cell cycle Cdks apparently does not apply to the transcription Cdks, other regulatory mechanisms are therefore likely to assume more important roles. Indeed, the subset of Cdks that are involved in transcription share some of the other regulatory mechanisms used by the cell cycle Cdks (29). For example, Kin28 is stimulated by Cak1-dependent phosphorylation of a threonine residue within its T-loop (8, 20, 21), and Kin28 and Ctk1 associate with additional subunits (9, 10, 48). By contrast, there is no evidence thus far for inhibitory phosphorylation or direct inhibitory subunits of the transcription Cdks. At least two distinct inhibitory mechanisms have been identified for the transcription Cdks: human Cdk7 is inhibited by phosphorylation of its cyclin subunit by Cdk8 (1), and Cdk9/P-TEFb activity is inhibited by association with the 7SK RNA (31, 53). A major remaining challenge for understanding this class of Cdks is to identify how their activities are regulated during the transcription cycle and to elucidate the signals to which they respond.
Our lab has been studying proteins that have relatively general roles in regulating transcription. By searching for mutations that increase transcription of a mutant promoter, we identified several BUR genes including BUR1 and BUR2, which encode a Cdk and its cyclin subunit (36, 54). The Bur1-Bur2 complex can phosphorylate the Rpb1 CTD, and mutations in both BUR1 and BUR2 show extensive genetic interactions with mutations in genes that encode CTD kinases, a CTD phosphatase, and transcription elongation factors, suggesting that the Bur1-Bur2 Cdk complex regulates elongation or the transition from initiation to elongation (30). A phylogenetic sequence comparison (24) indicated that Bur1 is the yeast ortholog of human Cdk9, the catalytic subunit of P-TEFb, which is believed to regulate transcription elongation by phosphorylating the Rpb1 CTD (26, 27, 37), although Bur1 has not yet been demonstrated to have P-TEFb activity in vitro. Investigating the mechanisms that regulate Bur1 kinase activity in yeast might therefore help to elucidate the role and regulation of P-TEFb in mammalian cells and help us to understand the regulatory roles of the other Cdks involved in transcription.
In this work, we identify CAK1 as a high-copy-number suppressor of a bur1 mutation, with additional genetic evidence functionally linking CAK1 and BUR1 in vivo. Furthermore, Bur1 is a phosphoprotein, its phosphorylation is abolished either by bur1 T-loop mutations or by CAK1 deletion, and Bur1 CTD kinase activity is stimulated by recombinant Cak1 in vitro. Thus, both genetic and biochemical evidence indicate that Cak1 is a physiological regulator of the Bur1 kinase in vivo.
MATERIALS AND METHODS
Yeast strains, plasmids, and media.
The S. cerevisiae strains used in this study are listed in Table 1. The cak1-23 mutant strain (pKY271) (7) was provided by M. Solomon, and the Cak1-independent cdc28 strains (1834-1B and 1834-2A) (5) were provided by F. Cross. Plasmids used in this study are as follows: pGP111 (URA3 CEN BUR1), pGP112 (URA3 2μm BUR1), pGP132 (URA3 2μm CAK1), pRU8 (URA3 2μm FLAG-BUR1 6HIS-BUR2), pSY27 (URA3 CEN bur1-240A), pSY28 (URA3 CEN bur1-240E), pSY34 (URA3 2μm FLAG-BUR1-TAP 6HIS-BUR2), pSY40 (LEU2 CEN bur1-240A), pSY41 (LEU2 CEN bur1-240E), pSY51 (URA3 2μm FLAG-bur1-240A-TAP 6HIS-BUR2), pSY52 (URA3 2μm FLAG-bur1-240E-TAP 6HIS-BUR2), and pCDC28-169-43244B (TRP1 CEN cdc28-169-43244B). The bur1-240A (pSY27) and bur1-240E (pSY28) alleles were generated by oligonucleotide-directed mutagenesis. Carboxy-terminal tandem affinity purification (TAP)-tagged BUR1 (pSY34) was constructed on pRU8 (30) by PCR as described previously (38). All media used, including rich (yeast extract-peptone-dextrose [YPD]), sucrose plate (YPSuc) synthetic complete (SC), synthetic dropout (e.g., SC-Ura), minimal (SD), and sporulation media, were made as described elsewhere (40). Formamide (FA) plates contained 2% FA, caffeine plates contained 15 mM caffeine, and 6-azauracil (6-AU) plates contained SC-Ura dropout mix and 50 μM 6-AU.
TABLE 1.
Yeast strains
| Strain | Genotype |
|---|---|
| GY100 | MATahis4-912δ lys2-128δ suc2Δuas(−1900/−390) ura3-52 leu2Δ1 bur1-2 |
| GY167 | MATahis4-912δ lys2-128δ suc2Δuas(−1900/−390) ura3-52 leu2Δ1 bur1-1 |
| GY168 | MATα his4-912δ lys2-128δ suc2Δuas(−1900/−390) ura3-52 trp1Δ63 bur1-1 |
| GY170 | MATα his4-912δ lys2-128δ suc2Δuas(−1900/−390) ura3-52 trp1Δ63 bur1-2 |
| GY458 | MATahis4-912δ lys2-128δ suc2Δuas(−1900/−390) ura3-52 trp1Δ63 |
| GY469 | MATahis4-912δ lys2-128δ suc2Δuas(−1900/−390) leu2Δ1 |
| GY742 | MATα his4-912δ lys2-128δ suc2Δuas(−1900/−390) ura3-52 bur2-1 |
| GY842 | MATα his4-912δ lys2-128δ suc2Δuas(−1900/−390) ade8 bur1-2 |
| OY118 | MATahis4-912δ lys2-128δ suc2Δuas(−1900/−390) leu2Δ1 cak1-23 |
| 1834-1B | MATacdc28Δ::HIS3 trp1 leu2 ura3 ade2 his3 (pCDC28-169-43244B = TRP1 CEN cdc28-169-43244B) |
| 1834-2A | MATα cdc28Δ::HIS3 cak1Δ::LEU2 trp1 leu2 ura3 ade2 his3 (pCDC28-169-43244B = TRP1 CEN cdc28-169-43244B) |
| SY106 | MATα his4-912δ lys2-128δ suc2Δuas(−1900/−390) ura3-52 trp1Δ63 leu2Δ1 bur1Δ1::TRP1 (pSY40 = LEU2 CEN bur1-240A) |
| SY107 | MATα his4-912δ lys2-128δ suc2Δuas(−1900/−390) ura3-52 trp1Δ63 leu2Δ1 bur1Δ1::TRP1 (pSY41 = LEU2 CEN bur1-240E) |
| SY137 | MATα his4-912δ lys2-128δ suc2Δuas(−1900/−390) leu2Δ1 cak1-23 bur1-2 |
| SY148 | MATahis4-912δ lys2-128δ suc2Δuas(−1900/−390) trp1 leu2 ura3 cak1Δ::LEU2 (pCDC28-169-43244B = TRP1 CEN cdc28-169-43244B) |
| SY149 | MATahis4-912δ lys2-128δ suc2Δuas(−1900/−390) trp1 leu2 ura3 (pCDC28-169-43244B = TRP1 CEN cdc28-169-43244B) |
| SY160 | MATα his4-912δ lys2-128δ suc2Δuas(−1900/−390) ura3-52 trp1Δ63 leu2Δ1 kin28Δ1::TRP1 (pSY68 = LEU2 CEN HA-kin28-T162A) |
Protein extract preparation and TAP.
One liter of log-phase yeast (2 × 107 cells per ml) was grown, harvested by centrifugation at 4,000 rpm in a Jovan C412 centrifuge for 10 min, and resuspended in breaking buffer containing 6 mM Na2HPO4, 4 mM NaH2PO4 · H2O, 1% NP-40, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 4 mg of leupeptin/ml, 0.1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1.3 mM benzamidine, and complete protease inhibitor cocktail mix (Boehringer). Cells were mechanically disrupted with a bead beater (Biospec), and extracts were clarified by centrifugation at 16,000 × g for 1 h. TAP-tagged Bur1-containing complexes were purified essentially as described previously (38). One gram of total protein extract was incubated with 500 μl of rabbit immunoglobulin G beads (Sigma) at 4°C on a rotating platform for 2 h. Beads were packed onto a Poly-Prep chromatography column (Sigma), washed sequentially with 10 ml of breaking buffer and 10 ml of tobacco etch virus cleavage buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1.0 mM dithiothreitol), resuspended in 1 ml of cleavage buffer with 30 μl (300 U) of recombinant tobacco etch virus protease (Clontech), and incubated at 4°C on a rotating platform for 2 h. Eluates were adjusted to 2 mM CaCl2, combined with 3 volumes of calmodulin binding buffer [10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM Mg(OAc)2, 1 mM imidazole, 2 mM CaCl2, and 10 mM β-mercaptoethanol] and 400 μl of calmodulin beads (Stratagene), and incubated at 4°C on a rotating platform. The column was washed with 10 ml of calmodulin binding buffer, and proteins were eluted with 3 ml of calmodulin elution buffer [10 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM Mg(OAc)2, 1 mM imidazole, 20 mM EGTA, and 10 mM β-mercaptoethanol].
Phosphatase treatment.
Ten microliters of the purified Bur1-TAP eluate was treated with lambda phosphatase essentially as described previously (8). Eluate was combined with phosphatase buffer (50 mM Tris-HCl [pH 7.8], 5 mM dithiothreitol, 1 mg of bovine serum albumin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml, 1 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride) and either 2 mM MnCl2, 2 mM MnCl2 with 100 U of lambda phosphatase, phosphatase with phosphatase inhibitors (2 mM ZnCl2, 50 mM NaF, and 1 mM Na3VO4), or phosphatase inhibitors alone to a final volume of 50 μl and then incubated at 37°C for 30 min.
Kinase assays.
Five microliters of the TAP-purified Bur1-Bur2-containing complex was incubated with 1 μl of β-galactosidase (β-Gal) CTD substrate (gift from A. Greenleaf) in 30 μl of kinase buffer (7) (10 mM HEPES-NaOH [pH 7.5], 150 mM NaCl, 10 mM MgCl2, 10 μM ATP, 1 μCi of [γ-32P]ATP). Mixtures for assays for stimulation contained 20 ng of recombinant Cak1 (gift from M. Solomon). Reaction mixtures were incubated at 30°C for 30 min, reactions were stopped by addition of gel loading buffer, the mixtures were boiled for 5 min, and products were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and visualized by autoradiography.
RESULTS
Identification of CAK1 as a high-copy-number suppressor of a bur1 mutation.
To study the function and regulation of the Bur1 kinase in vivo, we searched for high-copy-number suppressors of a bur1 mutation. The bur1-1 mutation used for the selection (36) causes a variety of phenotypes, including slow growth at 30°C; lethality at 37°C; sensitivity to FA, caffeine, and 6-AU; and Bur−, Spt−, and Ino− phenotypes that are frequently indicative of general transcription defects. Bur− and Spt− phenotypes refer to the ability of mutations to alter transcription from defective promoters; Bur− mutations increase transcription from a SUC2 promoter mutant that has had its upstream activating sequence deleted, while Spt− mutations alter transcription from HIS4 and LYS2 mutant promoters that contain insertions of transposable elements. Strains containing the his4-912δ, lys2-128δ, and suc2ΔUAS(−1900/−390) mutations are therefore His− Lys− and Suc−, but in the presence of bur1 mutations become His+ Lys+ and Suc+. All strains used in our study contain the suc2Δuas(−1900/−390) allele and the his4-912δ and lys2-128δ promoter insertion mutations to allow evaluation of the Bur− and Spt− phenotypes. A bur1-1 strain was transformed with a high-copy-number 2μm plasmid-based yeast genomic library (2), screening for plasmids that suppressed the recessive bur1-1 temperature-sensitive (Ts) phenotype. After filtering out BUR1-containing plasmids, five additional plasmids that confer suppression were obtained. Each of these plasmids contained a region from chromosome VI, with the overlapping 3.4-kb interval containing two open reading frames (ORFs). One of these ORFs, YFL030w, is uncharacterized and contains no sequence motifs to suggest its function, while the second ORF encodes CAK1. CAK1 was an excellent candidate for the suppressing ORF, as previous studies demonstrated that Cak1 phosphorylates two other Cdks, Cdc28 and Kin28 (7, 8, 19, 21, 49). A plasmid (pGP132) containing CAK1 as the only intact ORF conferred suppression of bur1-1, whereas a subclone that disrupted the CAK1 ORF, leaving YFL030w intact, abolished suppression, confirming that CAK1 was responsible for suppression (Fig. 1 and data not shown).
FIG. 1.
High-copy-number suppression of bur1 by CAK1. (A) A bur1-1 strain (GY168) was transformed with 2μm plasmids containing BUR1 (pGP112), CAK1 (pGP132), and the pRS426 vector. Patches of transformants were replica plated to selective SC-Ura medium and incubated at 30 or 37°C, with SC-Ura medium containing 2% FA or 50 mM 6-AU, SC-Ura plates lacking inositol (Ino), SC-histidine (SC-His), SC-lysine (SC-Lys), and YP-sucrose (YPSuc). The Ts, FAs, caffeine-sensitive, 6-AUs, and Ino− phenotypes were complemented by both BUR1 and 2μm CAK1, whereas Bur− (Suc+ growth) and Spt− (His+ and Lys+ growth) phenotypes were complemented only by BUR1. (B) GY168 (bur1-1) transformants with BUR1 (pGP112), CAK1 (pGP132), and the pRS426 vector were streaked onto an SC-Ura plate and grown at 30°C for 2 days. BUR1 and CAK1 restore vigorous growth to the bur1-1 strain. (C) A bur1-2 strain (GY100) was transformed with BUR1 (pGP112), CAK1 (pGP132), and the pRS426 vector. The 6-AUs and Ino− phenotypes were complemented by BUR1 but not by CAK1.
Genetic interactions among CAK1, BUR1, and BUR2.
To further investigate the relationship between CAK1 and BUR1, we performed several genetic tests examining the specificity of suppression. Since bur1-1 causes multiple mutant phenotypes, we first investigated whether CAK1 overexpression suppresses those other phenotypes. In addition to suppressing the bur1-1 Ts defect, CAK1 overexpression also suppressed the slow-growth defect (Fig. 1B) and the Ino−, FAs, and 6-AUs phenotypes (Fig. 1A) but was unable to suppress the Bur− or Spt− phenotypes. To determine whether the high-copy-number suppression was allele specific, we transformed a 2μm CAK1 plasmid into a bur1-2 strain, but no suppression of the bur1-2 phenotypes was observed (Fig. 1C). Both alleles contain mutations within the kinase domain, but bur1-1 strains have a more severe growth defect and are Ts−. Finally, 2μm CAK1 was tested for suppression of two different mutations in BUR2, which encodes the cyclin subunit of Bur1 (54), but no suppression of any bur2 phenotypes was observed (data not shown). In summary, CAK1-dependent suppression was allele specific, selectively suppressing a spectrum of phenotypes caused by a bur1 mutation, and did not compensate for defects in the Bur2 cyclin.
Identification of CAK1 as a bur1 high-copy-number suppressor could be explained by two reasonable mechanisms: Cak1 could phosphorylate and directly stimulate Bur1, or Cak1 could stimulate another Cdk, indirectly compensating for the defect caused by the bur1 mutation. We considered the direct mechanism to be more likely, as overexpression of CAK1 cannot bypass a deletion of BUR1, and overexpression of two other CTD kinases, Kin28 and CTDK-1 (Ctk1 + Ctk2 + Ctk3), did not suppress bur1-1. In addition, Bur1 contains a threonine residue (T240) within its conserved T-loop domain at the position that is known to be phosphorylated in other Cak1-responsive kinases. If Cak1 directly stimulates Bur1, we would predict that cak1 mutant phenotypes might overlap with those caused by bur1 mutations. We found that the cak1-23 allele, which was isolated previously in a generic screen for Ts cak1 mutations (8), causes a Bur− phenotype even at the permissive temperature of 30°C and additional caffeine-sensitive and partial Spt− phenotypes that overlap with those caused by bur1-1 at the semipermissive temperature of 33°C (Fig. 2A). The cak1 Bur− and Spt− phenotypes are not mediated through Kin28, since a kin28-T162A allele is Bur+ and Spt+ (Fig. 2B). This suggests that CAK1 function overlaps partially with BUR1, but the full extent of overlap is masked by its essential function. Although CAK1 is essential for viability in an otherwise wild-type background, Cross and Levine isolated cdc28 alleles that bypass the essential function of CAK1 (5), allowing analysis of cak1Δ phenotypes. Using one of these viable cak1Δ cdc28-169-43244B double mutant strains, we now observed more substantial overlap between cak1Δ and bur1 phenotypes, including strong Bur−, Spt−, and caffeine-sensitive phenotypes (Fig. 2B). These phenotypes were caused by cak1Δ and not by the cdc28-169-43244B mutation, since they were not observed in CAK1+ cdc28-169-43244B control strains.
FIG. 2.
Similarity between cak1 and bur1 mutant phenotypes. (A) Patches of yeast strains with the indicated relevant genotypes were replica plated to YPD complete medium, YPSuc sucrose medium, SC-Lys and SC-His selective media, and SC medium containing 15 mM caffeine (Caff) and incubated at 30 or 33°C for 3 days. (B) Patches of yeast strains with the indicated relevant genotypes were replica plated to media indicated on the left. The cak1Δ cdc28-169-43244B double mutant (SY148) shares Ts−, Bur− (Suc+ growth), Spt− (His+ and Lys+ growth), and caffeine-sensitive phenotypes with the bur1-1 strain. These phenotypes are not observed in the cdc28-169-43244B or kin28-T162A strain.
To further investigate the relationship between Cak1 and the Bur1-Bur2 Cdk complex, we examined double mutant phenotypes between the cak1-23 Ts− allele and mutations in either BUR1 or BUR2. Unlike the individual parental strains, bur1-2 cak1-23 double mutants grew extremely poorly on complete medium and were now sensitive to FA (Fig. 3). The bur2-1 cak1-23 phenotype was even more severe, resulting in double mutant lethality (Fig. 3A). In addition to causing Bur− and Spt− phenotypes by themselves, cak1 mutations therefore exacerbated defects caused by mutations in either the Bur1 catalytic subunit or Bur2, its cyclin partner. Combined, these genetic results strongly connect Cak1 and the Bur1-Bur2 Cdk complex in vivo.
FIG. 3.
Synthetic phenotypes of bur1-2 cak1-23 and bur2-1 cak1-23. (A) A cak1-23 strain (OY118) was crossed with bur1-2 (GY842) (left panel) and bur2-1 (GY742) (right panel) strains to access the double mutant phenotypes. A single tetratype tetrad from each cross is shown after growth on YPD medium at 30°C for 4 days. The genotype of each spore is indicated on the left. The bur1-2 cak1-23 double mutants grew significantly slower than single mutants, and the bur2-1 cak1-23 double mutants are lethal. (B) Yeast strains with the indicated genotypes were replica plated to YPD and to YPD containing 2% FA. Unlike wild type and single mutants, the bur1-2 cak1-23 double mutants were unable to grow in the presence of 2% FA.
Cak1 is required for the phosphorylation of Bur1 at threonine 240 in vivo.
The genetic results presented above suggest that Cak1 directly phosphorylates Bur1 within its T-loop domain. To investigate whether Bur1 is phosphorylated on T240 in vivo, we first purified the Bur1-Bur2 complex by the TAP tag purification method (38). Bur1 was tagged with the TAP cassette at its carboxy terminus, BUR1-TAP and BUR2 were overexpressed from a 2μm plasmid in a wild-type yeast strain, and the complex was purified over protein A and calmodulin affinity columns. Control experiments showed that TAP-tagged BUR1 complemented bur1 mutations and did not cause any mutant phenotypes in a BUR1+ background, and the TAP tag therefore did not interfere with BUR1 function. Purified Bur1 migrated as a doublet in SDS-polyacrylamide gels, as determined both by silver staining (Fig. 4A) and by Western blotting (Fig. 4B). The upper band of the doublet shifted to the position of the lower band after incubation with lambda phosphatase but not when lambda phosphatase was added with phosphatase inhibitors (Fig. 4B, lanes 1 to 4), indicating that the upper band was due to phosphorylation of Bur1 in vivo.
FIG. 4.
Bur1 is phosphorylated in vivo. (A) Plasmid pSY34 expressing 2μm BUR1-TAP and BUR2 was transformed into a wild-type strain (GY458). Bur1 is FLAG tagged at its N terminus and TAP tagged at its C terminus. The Bur1-Bur2-containing complex was purified by the TAP purification method (38). A 100-μl sample was loaded onto an SDS-12.5% polyacrylamide gel and silver stained. Bur1 migrates as a doublet. Numbers at left are molecular masses in kilodaltons. (B) Ten microliters of the TAP-tagged purified sample from panel A was incubated with phosphatase buffer (lane 1), treated with lambda phosphatase (λ PP'ase) in the absence (lane 2) or the presence (lane 3) of phosphatase inhibitors, or treated with phosphatase inhibitors (lane 4). TAP-tagged Bur1 purified from a cak1Δ strain (1834-2A) (lane 5) and Bur1-240A (pSY50) (lane 6) and Bur1-240E (pSY51) (lane 7) purified from a BUR1+ strain (GY458) contain only the lower band of the doublet. Proteins were resolved by SDS-polyacrylamide gel electrophoresis in a 7.5% polyacrylamide gel and subjected to Western blotting with anti-FLAG M2 antibody (Sigma).
To examine whether this phosphorylation occurs on threonine 240 within the T-loop, we generated two mutations that replaced T240 with either alanine or glutamic acid. Both mutations abolish the suspected phosphorylation site, but substitution of glutamic acid might mimic the phosphorylated threonine. Western blots of purified TAP-tagged Bur1-240A and Bur1-240E showed that Bur1-240A and Bur1-240E each migrated as a single band that corresponds to the lower band of the Bur1+ doublet (Fig. 4B, lanes 6 and 7, and Fig. 5 A). Similarly, purification of Bur1-TAP from a cak1Δ strain also resulted in loss of the upper phosphorylated Bur1 band (Fig. 4B, lane 5). These results therefore indicate that both CAK1 and T240 are required for phosphorylation of Bur1.
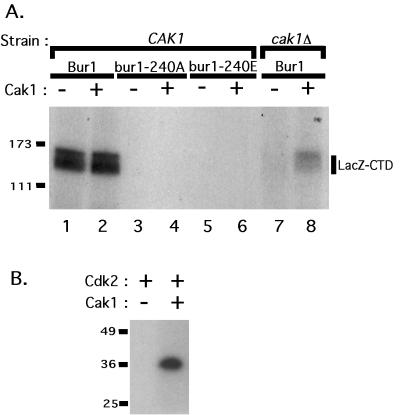
FIG. 5.
Bur1 T-loop mutants are inactive for CTD kinase activity. (A) Bur1, Bur1-240A, and Bur1-240E complexes purified from a wild-type strain and the Bur1 complex purified from a cak1Δ cdc28-169-43244B strain were separated in an SDS-polyacrylamide gel and immunoblotted with anti-FLAG M2 and Bur2 polyclonal antibodies. Each sample contains comparable amounts of Bur1 and Bur2 proteins. (B) The samples shown in panel A were assayed for CTD kinase activity by incubation with kinase buffer alone (lanes 2, 4, 6, and 8) or with recombinant β-Gal-CTD (lanes 1, 3, 5, and 7). Reaction products were loaded onto a 7.5% polyacrylamide gel, and phosphorylated products were visualized by autoradiography. Bur1 purified from the CAK1+ strain phosphorylated the CTD fusion protein in this assay, but no activity was detected for Bur1-240A, Bur1-240E, or Bur1 purified from the cak1Δ cdc28-169-43244B strain.
The effects of T240 mutations on Bur1 function were tested both in vitro and in vivo. When purified Bur1-240A and Bur1-240E were assayed for CTD kinase activity, they were unable to phosphorylate the LacZ-CTD fusion substrate (Fig. 5B). The defective CTD kinase activity was not simply due to the absence of Bur2 in the purified samples, since Bur1 and Bur2 protein levels were comparable to that of the Bur1+ sample (Fig. 5A). Plasmids containing these bur1 T240 mutations were next tested for their ability to complement bur1 mutations in vivo. As expected, bur1-240A completely lacked complementing activity, confirming that the alanine substitution inactivated Bur1, but somewhat surprisingly, the bur1-240E plasmid complemented the Spt−, Bur−, FAs, and 6-AUs phenotypes of bur1-2 nearly as well as the BUR1+ plasmid (Fig. 6A) despite its lack of detectable activity in the in vitro kinase assay. One explanation is that the in vivo assay is more sensitive and that bur1-240E provides a low level of activity that is sufficient to reverse the bur1-2 defects. This was tested by transforming the bur1-240A and bur1-240E mutants into a bur1Δ background, where they provided the only source of Bur1 in the cell. Under these conditions both the bur1-240A and bur1-240E strains were viable but caused Spt− and Bur− phenotypes (Fig. 6B). Bur1-240A and Bur1-240E therefore each provide enough Bur1 activity to allow viability, but as expected, the alanine substitution caused more severe transcription defects than did the glutamic acid substitution.
FIG. 6.
Threonine 240 is important for Bur1 function in vivo. (A) A bur1-2 strain (GY170) was transformed with CEN plasmids containing BUR1 (pGP111), bur1-240A (pSY27), bur1-240E (pSY28), and the pRS416 vector. Patches of transformants were replica plated to the media indicated on the left. BUR1 and bur1-240E complemented all the bur1-2 phenotypes except for caffeine sensitivity, while bur1-240A was completely unable to complement bur1-2. (B) Patches of yeast strains with indicated genotypes were replica plated to SC complete medium, YPSuc sucrose medium, and SC-Lys and SC-His selective media and incubated at 30°C for 3 days. Both bur1-240A (SY106) and bur1-240E (SY107) strains cause Bur− (Suc+ growth) and Spt− phenotypes (His+ and Lys+ growth), as do bur1-2 (GY 170) and bur2-1 (GY742) strains.
If CAK1 overexpression suppresses bur1-1 directly by phosphorylating T240 and stimulating Bur1 kinase activity, suppression should require the threonine at position 240 of Bur1. High-copy-number CAK1 was unable to suppress any phenotypes of bur1-240A and bur1-240E strains (data not shown), strongly supporting the model that suppression occurs via direct stimulation of Bur1 by Cak1. Based on the combined results presented in this section, we conclude that T240 is important but not essential for Bur1 function in vivo and serves as a substrate for Cak1-dependent phosphorylation and that CAK1 overexpression suppresses bur1 mutations by phosphorylating T240.
Cak1 stimulates Bur1 kinase activity in vitro.
To analyze whether Cak1 can stimulate Bur1 activity directly, it was necessary to purify the Bur1 complex in the absence of Cak1 activity. The TAP-tagged Bur1-Bur2 complex was therefore purified from a cak1Δ cdc28-169-43244B strain. As shown above (Fig. 4B, lane 5), Bur1 migrated as a single band when purified from a cak1Δ strain, indicating that no other kinases compensated for Cak1 in phosphorylating T240. The Bur1 complex purified from the cak1Δ cdc28-169-43244B strain had reduced activity for phosphorylating the β-Gal-CTD fusion protein relative to the activity of Bur1 purified from either the CAK1+ CDC28+ or CAK1+ cdc28-169-43244B strains. Addition of bacterially expressed recombinant Cak1 stimulated Bur1 purified from the cak1Δ cdc28-169-43244B strain approximately fivefold (Fig. 7) but had no effect on the Bur1 complex purified from the control wild-type and CAK1+ cdc28-169-43244B strains. No stimulation was observed in the equivalently purified Bur1-240A and Bur1-240E samples, indicating that threonine 240 is essential for the stimulation and that the stimulation was mediated through Bur1 and not some copurifying kinase. Upon longer exposure we observed a slight increase in phosphorylation of Bur1 in the presence of recombinant Cak1 (data not shown), further suggesting that Cak1 directly phosphorylates Bur1, but this signal is difficult to detect due to the relatively high background caused by Bur1 autophosphorylation.
FIG. 7.
Cak1 stimulates Bur1 CTD kinase activity in vitro. (A) The TAP-tagged Bur1-Bur2 complex was overexpressed and purified from a CAK1+ cdc28-169-43244B strain (1834-1B) (lanes 1 and 2) and a cak1Δ cdc28-169-43244B strain (1834-2A) (lanes 7 and 8). The TAP-tagged bur1-240A-Bur2 (lanes 3 and 4) and bur1-240E-Bur2 (lanes 5 and 6) were overexpressed and purified from a wild-type strain. The samples contain equal amounts of Bur1 and Bur2 (Fig. 5A). Purified Bur1-containing complexes were incubated with recombinant β-Gal-CTD (lanes 1, 3, 5, and 7) or with β-Gal-CTD and recombinant Cak1 (lanes 2, 4, 6, and 8). Reaction products were loaded onto a 7.5% polyacrylamide gel, and phosphorylated products were visualized by autoradiography. (B) Recombinant Cak1 is active for phosphorylation of human Cdk2. Numbers at left of each panel are molecular masses in kilodaltons.
DISCUSSION
The cyclin-dependent protein kinase family has important roles in regulating both cell cycle progression and gene expression in eukaryotes (29). In S. cerevisiae, four Cdk complexes have been implicated as broadly affecting transcription via phosphorylation of the polymerase II CTD, and yet surprisingly little is known about the proteins and signaling mechanisms that regulate these kinases (35). The goal of this study was to identify factors that regulate the Bur1-Bur2 Cdk complex. Our results show that Bur1 is phosphorylated and stimulated by Cak1 in vivo and in vitro. First, Cak1 is identified as a high-copy-number suppressor of a bur1 mutation. Second, cak1 mutant phenotypes overlap considerably with those caused by bur1 mutations. Third, phosphorylation of Bur1 in vivo requires both Cak1 and threonine 240 within the Bur1 T-loop domain. Fourth, recombinant Cak1 stimulates Bur1 CTD kinase activity in vitro. Taken together, the genetic and biochemical results strongly argue that Cak1 is directly required for maximum Bur1 activity in vivo. Because Kin28 is believed to stimulate promoter escape, while Bur1 is thought to be involved in elongation, these results implicate Cak1 in regulation of two different steps of transcription.
A regulatory role for Cak1 in transcription?
Cak1 phosphorylates and stimulates several Cdks in budding yeast, including Cdc28, Kin28, Bur1, and perhaps additional substrates, but the major question is whether Cak1-dependent phosphorylation is simply a constitutive requirement for activity or whether it has a regulatory role for any of these kinases (18). For Cdc28, Schizosaccharomyces pombe Cdc2, and the metazoan cell cycle kinases Cdk2, Cdk4, and Cdk6, Cak-dependent phosphorylation is required for activity (29), but there is little evidence that it constitutes a regulatory mechanism, as no changes in Cak activity occur during cell cycle progression (7). Cak-dependent phosphorylation of these kinases might still be part of a regulatory system, however, if phosphatase activity toward the T-loop threonine is regulated (3, 16, 25). The significance of the Cak1-dependent phosphorylation in regulating Kin28 also is not completely understood. Cak1 clearly phosphorylates Kin28 in vivo and in vitro and stimulates CTD kinase activity in vitro, and yet mutations that cause substitutions at the Kin28 T-loop threonine 162 cause no detectable phenotypes and no effect on the CTD phosphorylation state in vivo (8, 20, 21). Kin28 kinase-defective mutants, by contrast, are lethal and cause a dramatic decrease in CTD phosphorylation (50). Combined, these results suggested that T-loop phosphorylation has no overt effect on Kin28 activity. More detailed analysis, however, revealed that the kin28 threonine 162 T-loop mutants show defects when combined with secondary mutations in KIN28 or in other genes that encode subunits of the TFIIH complex, suggesting that T-loop phosphorylation is important but has a redundant role with other mechanisms that result in efficient Kin28 activity (20, 21). The requirement for Cak1-dependent phosphorylation in stimulating Kin28 has therefore been firmly established, but the question of whether Kin28 Thr162 phosphorylation changes during the transcription cycle or in response to any physiological stimulus has not been addressed.
Our studies of the relationship between Cak1 and Bur1 presented here reveal results that are strikingly analogous to those just described above for Kin28. Recombinant Cak1 stimulates both Bur1 and Kin28 CTD kinase activity, CAK1 and the T-loop threonines are required for Bur1 and Kin28 phosphorylation in vivo, bur1 and kin28 mutants show synthetic growth defects in combination with cak1 mutations, and mutants with bur1 and kin28 T-loop mutations are defective for CTD kinase activity and yet are viable, despite the fact that null alleles or kinase-inactive mutations are lethal (8, 21). The suppression of bur1 mutations by overexpression of CAK1 and the observation that cak1 mutations and bur1T240 mutations cause similar Bur− and Spt− phenotypes further connect BUR1 and CAK1, but analogous genetic connections between KIN28 and CAK1 have not been described. The identification of transcription-related Bur− and Spt− phenotypes conferred by cak1 and bur1-240A mutations suggests that Cak1 might have a more critical role in stimulating Bur1 than it has in stimulating Kin28. Furthermore, the observation that only about 50% of Bur1, but nearly all of Kin28 (8, 21), in a wild-type extract is shifted by Cak1-dependent phosphorylation suggests that Bur1 is not completely phosphorylated on threonine 240 in vivo and raises the possibility that Bur1 T-loop phosphorylation is regulated. Experiments to address whether Bur1 T240 or Kin28 T162 phosphorylation changes during the transcription cycle will be critical for determining whether Cak1 has a regulatory role during transcription through these kinases.
Dependence of Bur1 and Kin28 for Cak1 in vivo versus that in vitro.
Both Bur1 and Kin28 exhibit different requirements for Cak1 and T-loop phosphorylation in vivo than in vitro. Mutation of threonine 240 or purification of Bur1 from a cak1Δ strain results in similar loss of detectable Bur1 CTD kinase activity in vitro, whereas bur1-240A or bur1-240E mutants are viable and exhibit surprisingly healthy growth on complete medium. Because strains containing either a bur1Δ or a Bur1 catalytic site point mutation (D213A) are inviable, we conclude that T-loop phosphorylation is important in vivo but is not absolutely required for the essential function of Bur1. One trivial explanation is that the in vivo assay is simply more sensitive, but another possibility is that the in vitro kinase assays could be more defective due to loss of additional Bur1 stimulatory factors. Since both Kin28 and Ctk1 purify as trimeric complexes that contain an additional subunit besides the kinase and cyclin, a third subunit that acts redundantly with T-loop phosphorylation might exist for the Bur1-Bur2 complex. Finally, other kinases might partially compensate for the defective Bur1 T-loop mutants in vivo. An excellent candidate would be Ctk1, whose function might partially overlap with Bur1 in vivo (30).
Remaining questions.
CAK1 constitutes one component of the Bur1 signaling pathway, but it is possible that other regulatory components exist. For example, in addition to phosphorylating exogenously added CTD fusion substrate, wild-type Bur1 becomes autophosphorylated in our in vitro kinase assays. The autophosphorylation site and its functional significance are currently unknown. Cdk9, the likely human Bur1 ortholog, is unaffected by Cak but is regulated by autophosphorylation at multiple C-terminal serine and threonine residues, which are required for the assembly of a multicomponent elongation complex during transcription of the human immunodeficiency virus type 1 genome (13, 14). Second, Bur1 contains a threonine residue at position 70, which corresponds to Thr14 of S. pombe cdc2, which is important for inhibiting cdc2 kinase activity (15), suggesting that Bur1 might also be subject to inhibitory phosphorylation. Finally, immunoprecipitation of Bur1 results in coimmunoprecipitation of a subset of Rpb1. The Rpb1 that coimmunoprecipitates with Bur1 does not cross-react with antibodies that recognize the phosphorylated CTD, suggesting that Bur1 associates with a specific subpopulation of Rpb1 in vivo (30). Identifying the factors that regulate the association and dissociation of Bur1 and the other CTD kinases with Rpb1 remains an important goal for future experiments that will be necessary to understand the dynamic changes in CTD phosphorylation that occur during transcription.
Acknowledgments
We thank Mark Solomon, Fred Cross, Arno Greenleaf, Steve Buratowski, and Bertrand Seraphin for strains and reagents.
This work was supported by grant GM52486 from the National Institutes of Health to G.P.
REFERENCES
- 1.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407:102-106. [DOI] [PubMed] [Google Scholar]
- 2.Carlson, M., and D. Botstein. 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28:145-154. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, A., K. E. Ross, P. Kaldis, and M. J. Solomon. 1999. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Genes Dev. 13:2946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cismowski, M. J., G. M. Laff, M. J. Solomon, and S. I. Reed. 1995. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 15:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross, F. R., and K. Levine. 1998. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol. Cell. Biol. 18:2923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devault, A., A. M. Martinez, D. Fesquet, J. C. Labbe, N. Morin, J. P. Tassan, E. A. Nigg, J. C. Cavadore, and M. Doree. 1995. MAT1 (′menage a trois'), a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 14:5027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Espinoza, F. H., A. Farrell, H. Erdjument-Bromage, P. Tempst, and D. O. Morgan. 1996. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science 273:1714-1717. [DOI] [PubMed] [Google Scholar]
- 8.Espinoza, F. H., A. Farrell, J. L. Nourse, H. M. Chamberlin, O. Gileadi, and D. O. Morgan. 1998. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol. 18:6365-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faye, G., M. Simon, J. G. Valay, D. Fesquet, and C. Facca. 1997. Rig2, a RING finger protein that interacts with the Kin28/Ccl1 CTD kinase in yeast. Mol. Gen. Genet. 255:460-466. [DOI] [PubMed] [Google Scholar]
- 10.Feaver, W. J., N. L. Henry, Z. Wang, X. Wu, J. Q. Svejstrup, D. A. Bushnell, E. C. Friedberg, and R. D. Kornberg. 1997. Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH. Homology to human cyclin-dependent kinase activating kinase and IIH subunits. J. Biol. Chem. 272:19319-19327. [DOI] [PubMed] [Google Scholar]
- 11.Fesquet, D., J. C. Labbe, J. Derancourt, J. P. Capony, S. Galas, F. Girard, T. Lorca, J. Shuttleworth, M. Doree, and J. C. Cavadore. 1993. The MO15 gene encodes the catalytic subunit of a protein kinase that activates Cdc2 and other cyclin-dependent kinases (CDKs) through phosphorylation of Thr161 and its homologues. EMBO J. 12:3111-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, R. P., P. Jin, H. M. Chamberlin, and D. O. Morgan. 1995. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell 83:47-57. [DOI] [PubMed] [Google Scholar]
- 13.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garber, M. E., T. P. Mayall, E. M. Suess, J. Meisenhelder, N. E. Thompson, and K. A. Jones. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20:6958-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gould, K. L., and P. Nurse. 1989. Tyrosine phosphorylation of the fission yeast cdc2+ protein kinase regulates entry into mitosis. Nature 342:39-45. [DOI] [PubMed] [Google Scholar]
- 16.Gould, K. L., S. Moreno, D. J. Owen, S. Sazer, and P. Nurse. 1991. Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J. 10:3297-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffrey, P. D., A. A. Russo, K. Polyak, E. Gibbs, J. Hurwitz, J. Massague, and N. P. Pavletich. 1995. Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature 376:313-320. [DOI] [PubMed] [Google Scholar]
- 18.Kaldis, P. 1999. The cdk-activating kinase (CAK): from yeast to mammals. Cell. Mol. Life Sci. 55:284-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaldis, P., A. Sutton, and M. J. Solomon. 1996. The Cdk-activating kinase (CAK) from budding yeast. Cell 86:553-564. [DOI] [PubMed] [Google Scholar]
- 20.Keogh, M., E. Cho, V. Podolny, and S. Buratowski. 2002. Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell. Biol. 22:1288-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimmelman, J., P. Kaldis, C. J. Hengartner, G. M. Laff, S. S. Koh, R. A. Young, and M. J. Solomon. 1999. Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol. 19:4774-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, J. M., and A. L. Greenleaf. 1991. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1:149-167. [PMC free article] [PubMed] [Google Scholar]
- 23.Liao, S. M., J. Zhang, D. A. Jeffery, A. J. Koleske, C. M. Thompson, D. M. Chao, M. Viljoen, H. J. van Vuuren, and R. A. Young. 1995. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 374:193-196. [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., and E. T. Kipreos. 2000. Evolution of cyclin-dependent kinases (CDKs) and CDK-activating kinases (CAKs): differential conservation of CAKs in yeast and metazoa. Mol. Biol. Evol. 17:1061-1074. [DOI] [PubMed] [Google Scholar]
- 25.Lorca, T., J. C. Labbe, A. Devault, D. Fesquet, J. P. Capony, J. C. Cavadore, F. Le Bouffant, and M. Doree. 1992. Dephosphorylation of cdc2 on threonine 161 is required for cdc2 kinase inactivation and normal anaphase. EMBO J. 11:2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270:12335-12338. [DOI] [PubMed] [Google Scholar]
- 28.Mendenhall, M. D., and A. E. Hodge. 1998. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1191-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 30.Murray, S., R. Udupa, S. Yao, G. Hartzog, and G. Prelich. 2001. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell. Biol. 21:4089-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 32.Pavletich, N. P. 1999. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J. Mol. Biol. 287:821-828. [DOI] [PubMed] [Google Scholar]
- 33.Polyak, K., M. H. Lee, H. Erdjument-Bromage, A. Koff, J. M. Roberts, P. Tempst, and J. Massague. 1994. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78:59-66. [DOI] [PubMed] [Google Scholar]
- 34.Poon, R. Y., K. Yamashita, J. P. Adamczewski, T. Hunt, and J. Shuttleworth. 1993. The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 12:3123-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prelich, G. 2002. RNA polymerase II carboxy-terminal domain kinases: emerging clues to their function. Eukaryot. Cell 1:153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prelich, G., and F. Winston. 1993. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics 135:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 39.Reed, S. I., J. A. Hadwiger, and A. T. Lorincz. 1985. Protein kinase activity associated with the product of the yeast cell division cycle gene CDC28. Proc. Natl. Acad. Sci. USA 82:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose, M. D., F. Winston, and P. Heiter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Russo, A. A., P. D. Jeffrey, A. K. Patten, J. Massague, and N. P. Pavletich. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382:325-331. [DOI] [PubMed] [Google Scholar]
- 42.Russo, A. A., P. D. Jeffrey, and N. P. Pavletich. 1996. Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat. Struct. Biol. 3:696-700. [DOI] [PubMed] [Google Scholar]
- 43.Russo, A. A., L. Tong, J. O. Lee, P. D. Jeffrey, and N. P. Pavletich. 1998. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 395:237-243. [DOI] [PubMed] [Google Scholar]
- 44.Schaber, M., A. Lindgren, K. Schindler, D. Bungard, P. Kaldis, and E. Winter. 2002. CAK1 promotes meiosis and spore formation in Saccharomyces cerevisiae in a CDC28-independent fashion. Mol. Cell. Biol. 22:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Serrano, M., G. J. Hannon, and D. Beach. 1993. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366:704-707. [DOI] [PubMed] [Google Scholar]
- 46.Solomon, M. J., J. W. Harper, and J. Shuttleworth. 1993. CAK, the p34cdc2 activating kinase, contains a protein identical or closely related to p40MO15. EMBO J. 12:3133-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Solomon, M. J., T. Lee, and M. W. Kirschner. 1992. Role of phosphorylation in p34cdc2 activation: identification of an activating kinase. Mol. Biol. Cell 3:13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sterner, D. E., J. M. Lee, S. E. Hardin, and A. L. Greenleaf. 1995. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol. Cell. Biol. 15:5716-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thuret, J. Y., J. G. Valay, G. Faye, and C. Mann. 1996. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell 86:565-576. [DOI] [PubMed] [Google Scholar]
- 50.Valay, J. G., M. Simon, M. F. Dubois, O. Bensaude, C. Facca, and G. Faye. 1995. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J. Mol. Biol. 249:535-544. [DOI] [PubMed] [Google Scholar]
- 51.Valay, J. G., M. Simon, and G. Faye. 1993. The Kin28 protein kinase is associated with a cyclin in Saccharomyces cerevisiae. J. Mol. Biol. 234:307-310. [DOI] [PubMed] [Google Scholar]
- 52.Wagner, M., M. Pierce, and E. Winter. 1997. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 16:1305-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 54.Yao, S., A. Neiman, and G. Prelich. 2000. BUR1 and BUR2 encode a divergent cyclin-dependent kinase-cyclin complex important for transcription in vivo. Mol. Cell. Biol. 20:7080-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]