Abstract
The kinetics of pre-mRNA processing in living cells is poorly known, preventing a detailed analysis of the regulation of these reactions. Using tetracycline-regulated promoters we performed, during a transcriptional induction, a complete analysis of the maturation of two cellular mRNAs, those for LT-α and β-globin. In both cases, splicing was appropriately described by first-order reactions with corresponding half-lives ranging between 0.4 and 7.5 min, depending on the intron. Transport also behaved as a first-order reaction during the early phase of β-globin expression, with a nuclear dwelling time of 4 min. At a later time, analysis was prevented by the progressive accumulation within the nucleus of mature mRNA not directly involved in export. Our results further establish for these genes that (i) splicing components are never limiting, even when expression is induced in naive cells, (ii) there is no significant RNA degradation during splicing and transport, and (iii) precursor-to-product ratios at steady state can be used for the determination of splicing rates. Finally, the comparison between the kinetics of splicing during transcriptional induction and during transcriptional shutoff reveals a novel coupling between transcription and splicing.
In eukaryotic cells pre-mRNA maturation is a complex process that can entail the removal of multiple introns before a mature transcript can be exported to the cytoplasm. Moreover, the fidelity of processing and the control of alternative pathways are crucial for the correct execution of a genetic program. While our knowledge of the biochemical processes behind splicing and transport has greatly increased over the last years, we still know very little about their kinetics in vivo. This lack is all the more serious since the current view of nuclear RNA processing is that of a large number of regulators competing for the same substrates. This stresses the importance of the local concentration of regulators but also of the kinetics of the corresponding reactions. In addition, it is usually assumed that the RNA species generated during pre-mRNA processing are very unstable (7, 21). If this is true, the level of expression should be critically dependent on the kinetics of processing.
Our lack of knowledge of the kinetics of splicing and transport reflects the paucity of adequate experimental approaches. In vivo labeling studies have been used to investigate mRNA metabolism in mammalian cells. While this approach should provide access to the processing rates of pre-mRNA, two factors seriously limit its usefulness. (i) Studies on the fate of a specific RNA are hampered by the lack of sensitivity. (ii) The equilibration of the labeled compound with the intracellular nucleotide pool requires several minutes, which is the time scale anticipated for splicing and transport reactions. Thus the labeling is too slow for a stop flow type of kinetic analysis and too rapid for an approach to equilibrium analysis. Consequently, only some general information on pre-mRNA processing can be extracted from these studies. First, transcripts can be detected in the cytoplasm within 7 min of labeling, which has been taken as an indication that pre-mRNA processing is rapid (2, 24). Note, however, that if splicing and transport obey first- or second-order kinetics, there should not be any fixed delay in the appearance of transcripts in the cytoplasm. Second, mRNA metabolism in proliferating cells appears to be significantly different from that in quiescent cells (21). Other approaches have indicated that the functional diameter of the nuclear pore, as assessed by its capacity to import nucleoplasmin-coated gold particles, is significantly smaller in quiescent cells (12). mRNA processing may also be affected, as some hnRNP and SR proteins vary in abundance (5, 32, 42). Moreover, the overexpression of a kinase involved in signal transduction, src, affects both splicing and transport (14, 28).
Although the biochemical analysis of splicing has greatly benefited from the existence of an in vitro splicing system, it is obvious that the limited efficiency of the system and its very slow kinetics cannot reflect the characteristics of splicing in vivo. In intact cells, splicing rates for various introns have been derived from pre-mRNA half-lives following arrest of transcription (8, 11, 44). This approach does not allow determination of whether the measured half-life is due to splicing or degradation or a combination of both. Moreover, the transcription block is often achieved with a general inhibitor of transcription, which may affect the cellular physiology. Direct visualization of introns can be achieved in some cases by electronic microscopy and, by comparison with the elongation of transcription, it can be used to estimate a minimal time required for cotranscriptional splicing. In Drosophila melanogaster, this approach yielded an estimate of about 3 min (4).
Transport has been mostly studied by microinjection of in vitro-transcribed RNA into Xenopus laevis oocytes. While this approach has been useful for a biochemical characterization of the transport process, nuclear residence half-lives of the order of 1 h have been reported in most studies (20). Again, such a slow kinetics of export cannot represent the in vivo situation. Since the protein cover of mRNA provides most of the export signals, the injection of naked RNA introduces an additional step corresponding to the assembly of an RNP on an exogenous transcript. Moreover, splicing has been shown to contribute significantly to the recruitment of export factors. Indeed, injection of RNP assembled in an in vitro splicing reaction increased the total number of exported molecules (23). However, it should be noted that it did not improve the rate of export in Xenopus oocytes. This could reflect both the excessive number of molecules injected and the importance of a proper localization of transcripts for their interaction with the transport machinery. Other experimental approaches include the use of nuclear envelope vesicles (36) and isolated nuclei (39). In the first case, it should be possible to investigate specifically the translocation of mRNA through the nuclear pore complex (NPC), provided a suitable environment is re-created. In spite of the harsh preparative steps involved, use of isolated nuclei has the advantage over the other approaches to deal with endogenously synthesized transcripts. In these experiments, however, the export is usually inefficient, potentially reflecting an inadequate recycling of the transport machinery.
In spite of the lack of data, mRNA export to the cytoplasm is usually assumed to be more rapid than splicing, leading to the prediction that in the nuclear compartment the molecules in the process of being exported should constitute a minor population compared with the primary and partially spliced transcripts. This is in conflict with the results of biochemical fractionation. Indeed, not only does mature mRNA accumulate to similar concentrations in the nuclear and cytoplasmic compartments for many genes, but also in several instances this accumulation greatly exceeds that of the corresponding precursors (13). In an attempt to resolve this issue we have previously used a tetracycline-regulated promoter (tet-off) to investigate the fate of the nuclear mRNA population upon a specific inhibition of transcription (45). In these studies, no export of mRNA could be visualized, indicating that the nuclear pool of mRNA is not simply composed of the molecules in the process of being exported. This observation left open the kinetics of mRNA export while indicating that a detailed analysis was likely to be hampered by the existence of the nuclear pool of mRNA.
To investigate the kinetics of pre-mRNA processing in living cells, we have analyzed the accumulation of nuclear and cytoplasmic RNA during transcriptional induction. Two genes were used, the LT-α gene, for which we have extensively characterized the maturation pathway (29, 46) and the β-globin gene, which has been frequently used as an example of efficient splicing. For LT-α, the reinduction of a tet-off promoter was sufficiently efficient for a detailed kinetic analysis while for β-globin a tetracycline-inducible promoter (tet-on) was required. A detailed analysis of splicing for the three introns of LT-α and the two introns of β-globin could be performed throughout the induction, while the analysis of transport was feasible only during the early phase of β-globin induction.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 cell lines expressing the murine LT-α and rabbit β-globin genes under the control of the Tet promoter in the presence of the tetracycline/VP16 activator have been previously designated tTA-LT-α and tTA-β-glo, respectively (45). Cells were routinely maintained in Dulbecco's modified minimal essential medium (DMEM) supplemented with 10% fetal calf serum and 0.8 μg of puromycin, 125 μg of hygromycin, and 800 μg of Geneticin sulfate/ml. For expression studies, puromycin, hygromycin, and Geneticin were omitted and 80 ng of tetracycline/ml was added to the culture medium for 24 and 72 h for LT-α and β-globin, respectively. For the removal of tetracycline, cells were washed three times with phosphate-buffered saline and once with DMEM supplemented with 10% fetal calf serum before incubation in fresh culture medium devoid of antibiotics.
HeLa rtTA HR5 cells (Clontech) were routinely maintained in DMEM supplemented with 10% fetal calf serum and 100 μg of Geneticin sulfate/ml. Transfection was performed with 9 μg of tTA-β-glo plasmid and 1 μg of pY3 plasmid DNA per 85-mm-diameter dish by a standard calcium phosphate procedure (37). The tTA-β-glo plasmid contains the rabbit β-globin gene under the control of a Tet promoter and has been described previously (45). pY3 contains the hygromycin B phosphotransferase gene (6). After 48 h the culture medium was replaced with fresh medium supplemented with 100 μg of Geneticin sulfate and 200 μg of hygromycin/ml. After 15 days hygromycin-resistant clones were isolated and plated in 24-well plates before being routinely maintained in 85-mm-diameter dishes in the presence of Geneticin and hygromycin. For expression studies, Geneticin and hygromycin were omitted and 0 to 20 μg of doxycycline/ml was added to the culture medium for the periods of time indicated in the legends for Fig. 3 to 5 and 7 to 9. Cytochalasin B (Sigma) was used at 2 μg/ml.
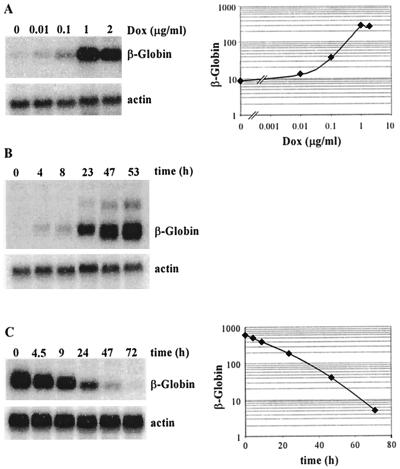
FIG. 3.
β-globin mRNA induction by doxycycline in HeLa rtTA cells. (A) Dose response to doxycycline. HeLa-β-glo cells were grown with the indicated doxycycline concentrations for 48 h. Equal amounts of total RNA were analyzed by Northern blotting with a β-globin probe and reanalyzed with a β-actin probe. The β-globin signals were quantified, corrected for equal loading by using β-actin signals, and plotted as a function of doxycycline concentration. Note that both axes are on a logarithmic scale. (B) Kinetics of β-globin mRNA accumulation following induction. Cells were treated with 2 μg of doxycycline/ml for the indicated periods of time. β-Globin mRNAs were analyzed as described for panel A. (C) Half-life of β-globin mRNA following arrest of transcription. Cells were cultivated for 3 days with 1 μg of doxycycline/ml, extensively washed, transferred to new dishes, and grown in the absence of doxycycline for the indicated periods of time. β-Globin mRNAs were analyzed and quantified as described for panel A, and levels were plotted as a function of time. Note that the y axis is on a logarithmic scale.
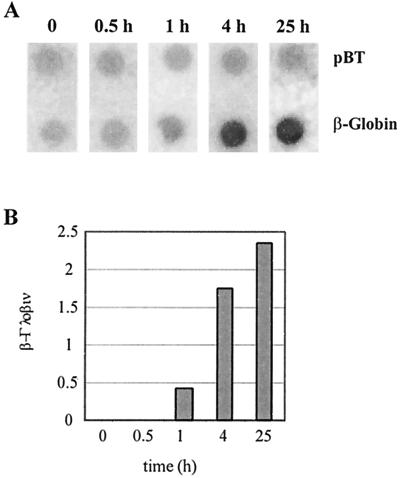
FIG. 5.
Transcriptional induction of β-globin by doxycycline. (A) Nuclear run-on transcription assay. HeLa-β-glo cells were treated with 2 μg of doxycycline/ml for the indicated periods of time. Nuclei were used for in vitro run-on transcription, and the product of the reaction was hybridized to β-globin and the Bluescript plasmid (pBT) DNA. (B) Quantification of the run-on experiment. β-Globin signals were quantified, corrected for background and relative hybridization efficiency using pBT signals, and plotted as a function of time.
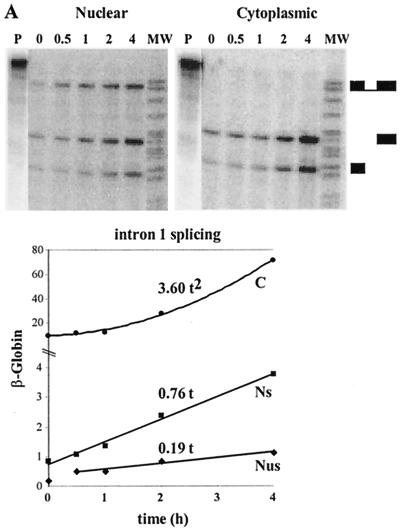
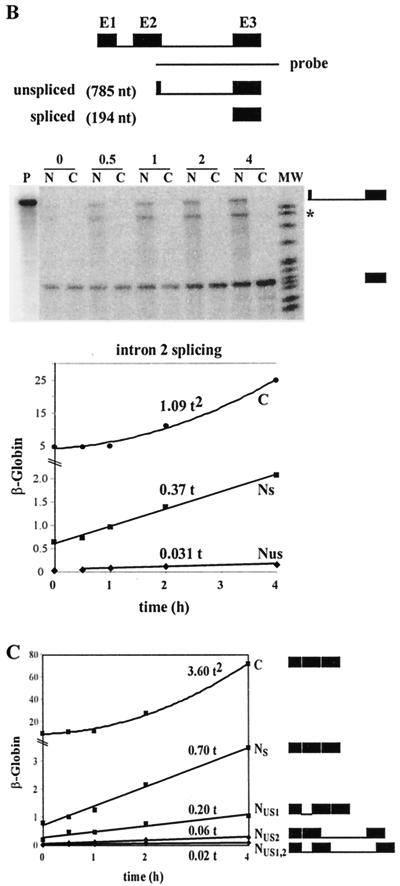
FIG. 7.
Determination of β-globin splicing and transport rates during a short-term induction by using a polynomial description. (A) Intron 1 splicing rate and transport rate. HeLa-β-glo cells were treated with 2 μg of doxycycline/ml for up to 4 h. β-Globin transcripts were analyzed by RNase protection assay with the probe encompassing intron 1, as in Fig. 4. Nuclear spliced (NS) and unspliced (NUS) and cytoplasmic (C) transcripts were plotted as a function of time. Curve-fitting functions, linear for NS and NUS and binomial for C, were applied to the data. The best fits (lines) are displayed, the first term of the equation being indicated above. The correlation coefficients of the binomial fittings are indicated in Materials and Methods. (B) Intron 2 splicing rate and transport rate. The same samples were analyzed by RNase protection assay with a β-globin probe encompassing intron 2. The principle of the protection by spliced and unspliced transcripts is shown schematically at the top. Migration of the protected fragments is indicated at the right of the gel. ∗, migration expected for released linear intron 2. The experimental results and their fit by linear and binomial curves were plotted as for panel A. (C) Combined quantification of all the nuclear and cytoplasmic β-globin transcripts. The ratio between intron 2 unspliced and spliced RNA measured in panel B was used to calculate their participation to intron 1 unspliced and spliced RNA in panel A. These data and their curve fits were plotted as in panel A. The structure of all RNA species is shown on the right.
FIG. 9.
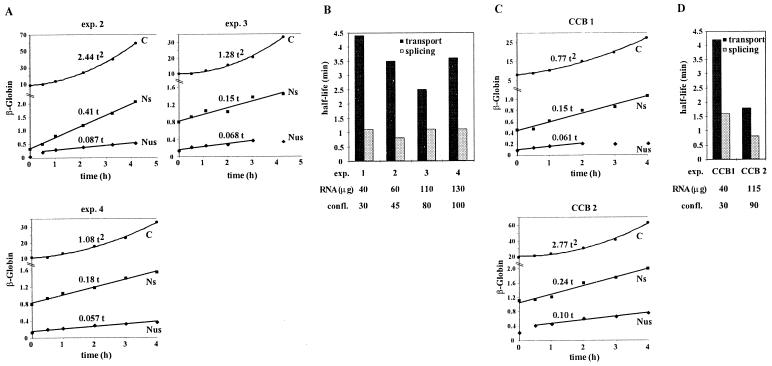
Effects of cell density and cytochalasin B (CCB) on β-globin splicing and transport rates. (A) Kinetics of accumulation of β-globin transcripts at different cell densities. HeLa-β-glo cells plated at various cell densities were treated with 2 μg of doxycycline/ml for up to 4 h. The amounts of RNA extracted per culture dish were 60, 110, and 130 μg for experiments 2, 3, and 4, respectively, corresponding to approximate levels of confluence of 45, 80, and 100%. β-Globin transcripts were analyzed by RNase protection assay with the β-globin probe encompassing intron 1, as in Fig. 7A. (B) Transport and splicing half-lives as a function of cell density. Intron 1 splicing and transport rates were calculated from data presented in Fig. 7A (experiment 1) and Fig. 9A (experiments 2, 3, and 4), as detailed in Results. The corresponding splicing and transport half-lives were plotted. (C) Kinetics of accumulation of β-globin transcripts following treatment with CCB. After a 1-h incubation with 2 μg of CCB/ml, HeLa-β-glo cells were treated with 2 μg of doxycycline/ml in the presence of CCB for up to 4 h. The amounts of RNA extracted per culture dish were 40 and 115 μg for experiments CCB1 and CCB2, respectively. β-Globin transcripts were analyzed as for panel A. (D) Transport and splicing half-lives after treatment with CCB. Splicing and transport half-lives were calculated and plotted as for panel B.
RNA extraction and analysis.
Nuclear and cytoplasmic fractions were obtained by NP-40 lysis in the presence of 10 mM EDTA to dissociate polysomes from the nuclear envelope, as described previously (45). RNA was extracted by denaturation in guanidine thiocyanate prior to ultracentrifugation over a cesium chloride cushion (3). The concentration of purified RNA was determined by spectrophotometry and confirmed by a Northern blot analysis using a β-actin probe.
For RNase protection assays, 4 μg of RNA was hybridized with 500 pg of 32P-labeled riboprobe, digested with RNases A and T1, and electrophoresed through 5% urea-polyacrylamide gels as described previously (29). LT-α probes a and b contain nucleotides 1631 to 1967 and 1733 to 2226 of the murine LT-α gene (GenBank accession no. Y00467), respectively. Protection of fully and partially spliced transcripts has been described in detail previously (29). The β-globin probes used for intron 1 and intron 2 splicing analysis contain nucleotides 67 to 556 and 554 to 1336 of the rabbit β-globin gene (GenBank accession no. K03256), respectively. The exon 1-intron 1-exon 2 and exon 2 protected fragments contain 107 and 45 uridines, respectively. The exon 2-intron 2-exon 3 and exon 3 protected fragments contain 185 and 40 uridines, respectively. The β-actin probe contains nucleotides 1090 to 1301 of a murine β-actin cDNA (GenBank accession no. X03672). Protection of human β-actin gives rise to two fragments about 120 nucleotides long. Quantification of the radioactive signals was performed with a Molecular Dynamics Storm analyzer.
For Northern blots, 4 μg of RNA was electrophoresed through 1.5% agarose-formaldehyde gels, transferred onto an uncharged nylon membrane, and hybridized with 32P-labeled riboprobes, as described previously (29). β-Globin and β-actin probes have been described previously (45).
Nuclear run-on.
Isolation of nuclei and in vitro elongation of the transcripts were performed as described previously (16). Purification of labeled RNA and DNA blotting (pBT, β-globin) were carried out as described previously (9). Hybridization was for 72 h at 42°C in 50% formamide-4× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])-0.1% sodium dodecyl sulfate-1× Denhardt's reagent (Appligene Oncor)-0.1 mg of salmon sperm DNA/ml. The β-globin plasmid contains the rabbit β-globin genomic sequence cloned in Bluescript (pBT). Quantification of the radioactive signals was performed with a Molecular Dynamics Storm analyzer.
Modeling of splicing and transport reaction.
The maturation pathways presented in Results were described by a set of first-order reactions as discussed below. Equations were derived for each processing step by balancing the corresponding incoming and outgoing fluxes of molecules. Each of these equations contained only one unknown, which could be determined by a numerical fitting of the data. Thus, for LT-α processing the equation for intron 1 splicing was d(N2 + N1 + N0 + C1 + C0)/dt = kS1N3 − kD0C0 − kD1C1, that for intron 2 splicing was d(N1 + N0 + C1 + C0)/dt = kS2N2 − kD0C0 − kD1C1, that for intron 3 splicing was d(N0 + C0)/dt = kS3N1 − kD0C0, and that for transport was d(C0)/dt = kT0N0 − kD0C0, where kS, kT, and kD are splicing, transport, and degradation rates, respectively. The degradation rates for C0 and C1, kD0 and kD1, were measured previously to be 0.47 and 0.35 h−1, respectively (45). Values for Ni(t) and Ci(t) were experimental values described by a linear interpolation between the data points. An iterative numerical calculation was conducted with a time increment of 0.1 min to determine the best fit for the splicing and transport rates. For instance, for intron 1 the equation was P(t + dt) = P(t) + [kS1N3(t) − 0.47C0(t) − 0.35C1(t)]dt, where equation P(t) = N2(t) + N1(t) + N0(t) + C1(t) + C0(t) describes the sum of all the species generated by intron 1 splicing.
Similarly for β-globin, the following equations with a single unknown were derived: for intron 1 splicing, d(NS + C)/dt = kSNUS − kDC; for transport, dC/dt = kTNS − kDC.
The measured degradation rate, kD, was 0.054 h−1, and the numerical calculation was performed as for LT-α. The equations were as follows: for intron 1 splicing, P(t + dt) = P(t) + [kSNUS(t) − 0.054 C0(t)]dt; for transport, P(t + dt) = P(t) + [kTNS(t) − 0.054C0(t)]dt.
For the analysis of short-term inductions the polynomial fit of the data was performed with Excel software (Microsoft). For binomial fittings of C(t), values of the correlation coefficient, R, were 0.9978, 0.9911, and 0.9978 for experiment 1 in Fig. 7A, B, and C, respectively, 0.9995, 0.9962, and 0.9955 for experiments 2, 3, and 4, respectively, in Fig. 9A, and 0.9986 and 0.9938 for experiments CCB1 and CCB2, respectively, in Fig. 9B.
RESULTS
Kinetic analysis of LT-α induction.
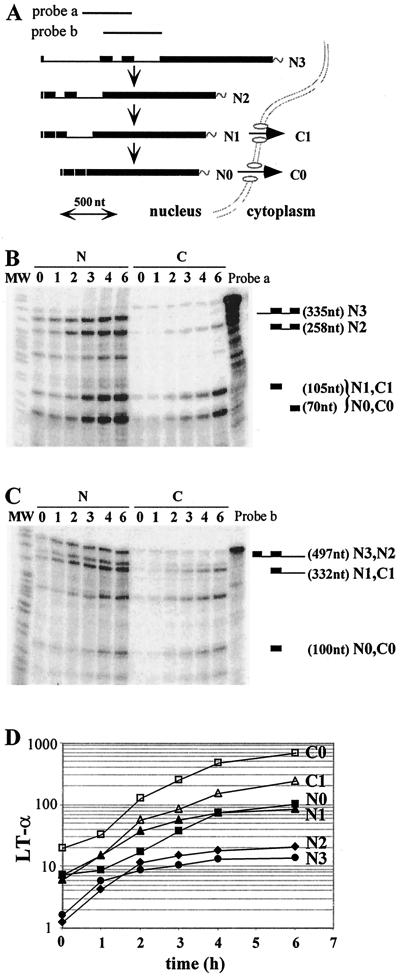
The LT-α gene contains three introns, which are sequentially removed from a polyadenylated primary transcript according to a 5′-to-3′ polarity. Both fully spliced and intron 3-containing RNAs are exported to the cytoplasm, and therefore a total of six RNA species accumulate to significant levels in the nuclear (N3, N2, N1, and N0) and cytoplasmic (C1 and C0) compartments (46) (Fig. 1A ). To analyze the kinetics of splicing and transport of LT-α transcripts, we investigated the accumulation of all the RNA species during a transcriptional induction. We have previously described an NIH 3T3 cell line expressing the LT-α gene under the control of a tetracycline-regulated promoter (45). In this tet-off promoter/regulator system, maximal inhibition of expression could be achieved with 80 ng of tetracycline/ml. As the half-life of all the LT-α transcripts is shorter than 2 h, a 24-h incubation in the presence of tetracycline was sufficient to clear off the preexisting RNA pools (45). Extensive washing of the cells with tetracycline-free medium allowed for a rapid resumption of LT-α transcription. LT-α expression was monitored over 6 h, which corresponds to three mRNA half-lives. Nuclear and cytoplasmic fractions were prepared by biochemical fractionation, and a set of two RNase protection assays, one encompassing intron 1-exon 2-intron 2-exon 3 and the other encompassing exon 2-intron 2-exon 3-intron 3, was used to identify and quantitate the various LT-α RNAs. As shown previously, the reproducibility of this method of quantitation is within 10% (29). Figure 1B and C present the results obtained for one experiment. Signals were quantified and used to calculate the accumulation of the various LT-α transcripts. Results are presented on Fig. 1D, and the calculation is described in the legend. A 9- to 33-fold increase in the accumulation of the six LT-α RNA species took place during the 6-h time course. The pre-mRNA level (N3) reached a plateau at 4 h, suggesting that transcription was fully induced by then. Some indications of a precursor-to-product relationship could be found by visual inspection of the curves. For instance, the delay between N3 and N2 accumulation, as well as that between N1 and N0 accumulation, is in agreement with the sequential removal of introns that we have previously established (46).
FIG. 1.
Kinetics of LT-α RNA accumulation following resumption of transcription. (A) Schematic representation of LT-α mRNA processing. Pre-mRNA (N3), experimentally observed partially spliced RNA (N2, N1, and C1) and fully spliced mRNA (N0 and C0) are represented. Probes a and b, LT-α genomic probes used for RNase protection assays. The scale is indicated at the bottom. (B and C) Analysis of LT-α transcripts by RNase protection assay. After a 24-h culture in the presence of 80 ng of tetracycline/ml, NIH 3T3-LT-α cells were extensively washed and incubated in the absence of tetracycline for 0 to 6 h. Equal amounts of nuclear (N) and cytoplasmic (C) RNA were hybridized with LT-α probes a (B) and b (C) in order to analyze the presence of introns 1 and 2 and introns 2 and 3, respectively. Migration of the riboprobe fragments protected by partially and fully spliced transcripts is indicated on the right. The molecular weight marker (MW) is pBr322 DNA digested with MspI. (D) Quantification of nuclear and cytoplasmic LT-α transcripts. LT-α signals were quantified and corrected for the number of labeled uridines in each fragment. N0, C0, N1, and C1 values were directly obtained from RNase mapping assay with probe b. The ratio between N2 and N3 was obtained from the RNase mapping assay with probe a and was used to calculate the participation of N2 and N3 in the longest fragment protected with probe b. Results were corrected for the relative sizes of the nuclear and cytoplasmic compartments and expressed with arbitrary units. Note that the y axis is on a logarithmic scale and that the basal level of expression has not been subtracted.
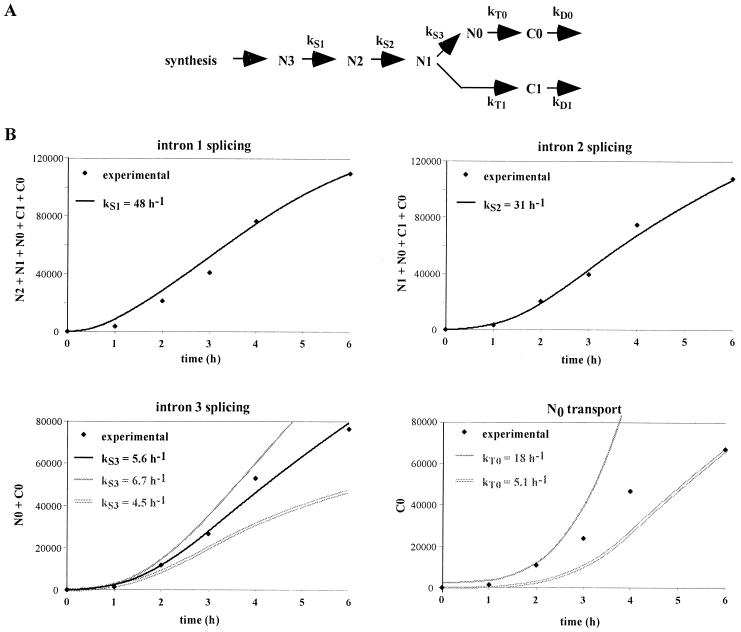
To extract reaction rates from these data, we performed a computer simulation of LT-α RNA maturation. We made two assumptions on the splicing and transport processes: (i) splicing and transport reactions were described by first-order reactions and (ii) degradation of nuclear RNA was slower than splicing and transport. The validity of these hypotheses is discussed below. Note that we make no assumption on the degradation of the primary transcript since we use its accumulation (i.e., the result of both its synthesis and degradation) as the starting point of our modeling. Under these hypotheses the incoming flux of molecules can be described as kS1N3 (i.e., the molecules generated by splicing the primary transcript) and the final output can be described as kD0C0 + kD1C1 (i.e., the cytoplasmic end products), where kS1 designates the splicing rate of intron 1 and kDi designates the degradation rate of Ci (Fig. 2A). Hence the global equation is d(N2 + N1 + N0 + C1 + C0)/dt = kS1N3 − (kD0C0 + kD1C1).
FIG. 2.
Determination of LT-α splicing and transport rates by computer simulation. (A) Representation of splicing and transport reactions. Reactions were assumed to follow first-order kinetics, with kS and kT being the rates for splicing and transport reactions, respectively. (B) Computer simulation of the production of LT-α transcripts. For intron 1 splicing, the kinetics of accumulation of all the transcripts generated from the N3 precursor was calculated for different values of kS1, taking into account the experimental pool of N3, as described in Materials and Methods. Results were plotted along with the experimentally observed accumulation of these transcripts (N2 + N1 + N0 + C1 + C0). Only the best fit, obtained with a splicing rate of 48 h−1, is presented. Similarly, for the following splicing and transport reactions, the accumulation of products was calculated taking into account the experimental pool of precursor and various splicing and transport rates. Black lines, best fits; grey lines (intron 3 splicing), simulations obtained by increasing or decreasing by 20% the splicing rate. For N0 transport, no satisfying fit could be found and two examples fitting either the early or the late time points are presented.
The cytoplasmic degradation rates have been determined by a tetracycline chase in previous experiments (45). The half-lives of 2.0 and 1.5 h for C1 and C0, respectively, correspond to kD1 and kD0 values of 0.35 and 0.47 h−1. Computer simulations were performed to determine a kS1 value which could account for the observed accumulation of transcripts resulting from intron 1 splicing (N2 + N1 + N0 + C1 + C0; see Materials and Methods for more details on the modeling). The best fit is presented in Fig. 2B, with kS1 = 48 h−1. The same approach can be used to search for kS2 and kS3 by modeling the corresponding products: (N1 + N0 + C1 + C0) and (N0 + C0). The best values were 31 and 5.6 h−1 for kS2 and kS3, respectively (Fig. 2B). These results can be expressed as half-lives for the corresponding introns: t1/2 = 0.9, 1.4, and 7.5 min for introns 1, 2, and 3, respectively (these half-lives refer to the time required for half of the molecules to eliminate a given intron). Importantly, in each analysis a single value could account for the six experimental data points, indicating that a first-order kinetics provides an appropriate description of the reaction. To evaluate the precision of the kS values determined with this approach, the impact of a 20% variation of kS3 on the accumulation of spliced products is presented in Fig. 2B. It is apparent that the constraint on the splicing rates increases with the duration of the experiment, so that a high precision is achieved over a 6-h time course. We have previously used similar hypotheses on pre-mRNA processing to derive kS rates from the relative accumulation at equilibrium of the precursor and product RNA (29). For this NIH 3T3 transfectant the relative accumulations are as follows: N3, 1; N2, 1.6; N1, 7; N0, 16.6; C1, 22.2; C0, 77 (45); these yield a kS1 value of 28.6 h−1 (t1/2 = 1.5 min), a kS2 value of 17.6 h−1 (t1/2 = 2.4 min), and a kS3 value of 5.2 h−1 (t1/2 = 8.1 min). Thus, there is an excellent agreement between the splicing rates derived from the kinetic analysis during a transcriptional induction and those which can be derived from the relative accumulations at equilibrium.
The same kinetic modeling can be used to analyze the transport process by deriving the accumulation of cytoplasmic molecules (C0) from the nuclear pool of mature RNA (N0). However (Fig. 2B), no single transport rate could account for both the early part of the kinetics (kT = 18 h−1) and the 6-h time point (kT = 5.1 h−1). Thus, in contrast to what is found for splicing, a first-order reaction is inappropriate for the description of transport. We have previously described, for fully induced cells, the presence within the nucleus of a pool of mature mRNAs which are not in the process of being exported (45). A progressive accumulation of these nuclear mRNAs could account for the observed decrease in apparent transport between the beginning and the end of the transcriptional induction.
Doxycycline-inducible expression of β-globin.
To extend this analysis to another gene, we turned to the rabbit β-globin gene, which has been widely used as an example of an efficiently processed transcript giving rise to a stable mRNA. In this case no order for intron removal has been determined. As for LT-α, we had previously obtained an NIH 3T3 cell line expressing the β-globin gene under tet-off promoter/regulator control (45). Reinduction of β-globin expression was too sluggish during the first hours to allow a detailed kinetic analysis in these cells. This probably reflects the longer treatment with tetracycline which is required to clear off the β-globin mRNA. We therefore generated cell lines in which β-globin expression is inducible in the presence of tetracycline by using the reverse tet operator (tet-on system) (15). The rabbit β-globin plasmid containing the gene including its two introns under the control of the cytomegalovirus/tet promoter, which has been previously described (45), was stably transfected into the HeLa rtTA HR5 cell line (Clontech). These cells constitutively express the chimeric VP16-rtTA transactivator, allowing for positive regulation by tetracycline or doxycycline. Independent clones were isolated and screened for the inducibility of β-globin expression by doxycycline with a Northern blot analysis performed after 72 h of culture in the absence or presence of 1 μg of doxycycline/ml. One clone with a 150-fold induction was chosen for further study unless otherwise indicated.
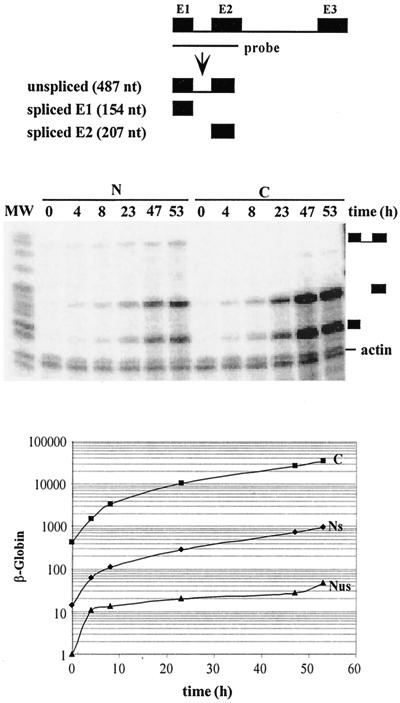
To determine the optimal doxycycline concentration, cells were cultivated in the presence of different doses of doxycycline for 48 h and RNAs were extracted and analyzed by Northern blotting (Fig. 3A). The maximal level of expression was reached at 1 μg of doxycycline/ml. The kinetics of β-globin induction was then investigated. Cells were grown with 2 μg of doxycycline/ml for 0 to 53 h, and cytoplasmic mRNAs were analyzed by Northern blotting (Fig. 3B). Mature β-globin mRNA accumulated progressively and did not approach a steady state after 47 h. In longer experiments, the steady state was still not reached after 60 h (data not shown). Because, once transcription is maximal, a plateau should be reached within a few transcript half-lives (e.g., 75% of maximal expression should be achieved in two half-lives), the absence of an approach to equilibrium could indicate either a very long half-life for β-globin mRNA or a continuous increase in transcription. To address this issue, we measured the β-globin mRNA half-life in these HeLa tet-on cells by washing off doxycycline. A Northern blot analysis and the quantification of the corresponding signals using β-actin as an internal standard are presented in Fig. 3C and indicate a β-globin mRNA half-life of 13 h. Thus, the failure to reach equilibrium after 53 h of induction (i.e., four mRNA half-lives) suggested that transcription kept increasing for at least 2 days after induction by doxycycline.
To further investigate the induction of β-globin expression, an RNase protection assay was performed on nuclear and cytoplasmic RNA with a probe encompassing intron 1 (Fig. 4). In the nucleus, neither the spliced nor the unspliced transcript had reached a steady state at 47 h. This coordinated increase in precursor and product supported an ever-increasing rate of transcription during the time course. To directly assess this hypothesis, we determined the β-globin transcription by a nuclear run-on assay (Fig. 5). Significant transcription was detectable by 1 h and kept increasing between 4 and 24 h. This pattern paralleled the pre-mRNA increase measured with the RNase protection assay (Fig. 4), indicating that most if not all of the pre-mRNA increase could be attributed to a change in transcription rate. This slow induction could have reflected the pharmacokinetics of doxycycline accumulation in these cells. However, increasing the dose of doxycyline up to 20 μg/ml did not accelerate β-globin expression (data not shown). In addition, similar kinetics of induction were observed in two other clones with 3- and 8-fold-lower levels of β-globin expression, with a rapid increase over the first few hours followed by a slower increase over several days.
FIG. 4.
Kinetics of accumulation of the β-globin mRNA and its precursors following induction with doxycycline. HeLa-β-glo cells were treated with 2 μg of doxycycline/ml for the indicated periods of time. Equal amounts of nuclear (N) and cytoplasmic (C) RNA were hybridized with a β-globin probe encompassing intron 1 and analyzed by RNase protection assay. The fragments protected by spliced and unspliced transcripts are shown schematically at the top. Their expected migration is indicated at the right of the gel. A β-actin riboprobe was included in the hybridization to control for equal loading. β-Globin signals were quantified and corrected for the number of labeled uridines, for the relative sizes of the nuclear and cytoplasmic compartments, and for equal loading by using β-actin signals. Nuclear spliced (NS) and unspliced (NUS) and cytoplasmic (C) transcripts were plotted as a function of time. Note that the y axis is on a logarithmic scale.
Kinetic analysis of β-globin expression.
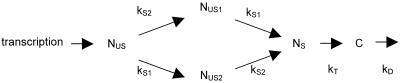
The β-globin processing pathway can be described by the sequence of events shown in Fig. 6. As no sequential removal of introns has been established for β-globin, the existence of two parallel maturation pathways was taken into account.
FIG. 6.
β-Globin processing pathway. NUS, primary transcript; NUSi, partially spliced transcript retaining intron i; NS and C, nuclear and cytoplasmic mature mRNAs, respectively; kSi, splicing rate of intron i; kT and kD, transport and degradation rates, respectively.
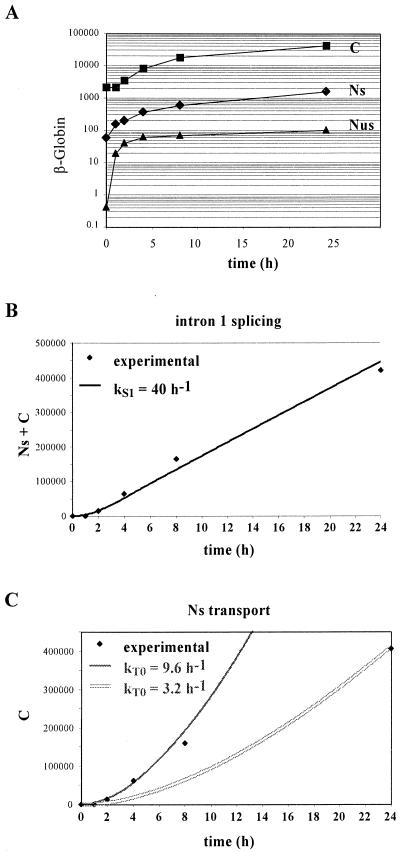
An analysis of nuclear and cytoplasmic RNA accumulation was first performed during a 4-h induction by doxycycline. A set of two RNase protection assays, one encompassing intron 1 (Fig. 7A) and the other encompassing intron 2 (Fig. 7B), with respective flanking exons, was used to identify and quantify the four nuclear RNA species (Fig. 7C). The latter probe detected the unspliced and spliced transcripts, as well as an additional band, which was restricted to the nucleus and whose size was the same as that of intron 2, in accordance with previous studies reporting the accumulation of β-globin-free linear intron 2 in rabbit fetal liver (48). Remarkably, the accumulation of all the nuclear species increased linearly over time, while that of cytoplasmic mRNA followed a quadratic law.
These simple patterns of accumulation made it possible to analyze these results without recourse to computer modeling. As for LT-α, we began by assuming that splicing and transport could be described by first-order reactions. In addition, the cytoplasmic degradation of β-globin mRNA was ignored during the 4-h time course because of its long half-life. Thus, if we first consider transport, the accumulation of cytoplasmic mRNA (C) results only from the summation of the transported molecules, which, for a first-order reaction, is expressed by dC/dt = kTNS. If the accumulation of nuclear mRNA increases linearly over time (NS ≈ aSt), it follows that the cytoplasmic accumulation is given by C ≈ 0.5aSkTt2. As mentioned above, the data of Fig. 7C indicate that indeed C is a second-order polynomial function of time, establishing that cytoplasmic mRNA is generated from the nuclear mRNA by a first-order process. Moreover, since aS is 0.7 h−1 and C is ∼3.6 t2, kT is 10.3 h−1. Such a transport rate corresponds to a nuclear dwell time of mature transcripts of 4 min.
Similarly, splicing rates can be derived from the linear pattern of accumulation of the nuclear RNA. Considering intron 1, the molecules generated by splicing either contribute to the increase in spliced molecules within the nucleus or are exported to the cytoplasm, hence kS1(NUS + NUS1) = d(NUS2 + NS)/dt + kTNS. As the concentration of these molecules increases linearly over time, one can make the following approximations: NUS ≈ aUSt, NUS1 ≈ aUS1t, NUS2 ≈ aUS2t, and NS ≈ aSt. Thus, the previous equation yields kS1t(aUS + aUS1) = aUS2 + aS + kTaSt, and the derivation of this equation with respect to time yields kS1(aUS + aUS1) = kTaS. With the aS, aUS, and aUS1 values of Fig. 7C and the kT value deduced above, this equation yields kS1 = 33 h−1. The same approach for the splicing of intron 2 leads to kS2 = 90 h−1. The corresponding half-lives for splicing introns 1 and 2 are 1.26 and 0.42 min, respectively. Thus the splicing of intron 1 is slower than that of intron 2, leading to the accumulation of NUS1 as the predominant intermediate. Nonetheless, splicing is more rapid than the transport process, and mature mRNA constitutes the most abundant nuclear species (Fig. 7C).
To extend the analysis of β-globin processing to a later phase of transcriptional induction, we analyzed β-globin expression during a 24-h time course (Fig. 8). As the splicing of intron 1 is the limiting step in splicing, we performed the analysis using only an RNase protection assay for intron 1 (Fig. 8A). We then used the modeling approach previously described for LT-α, taking into account the 13-h half-life of β-globin mRNA (Fig. 3). As illustrated in Fig. 8B, a kS1 value of 40 h−1 accounted for the accumulation of mature RNA. The corresponding half-life of intron 1 is 1.1 min, fully in agreement with the value derived in the short-term analysis. Moreover, as in the study of LT-α RNA processing, this first-order process also accounts for the splicing when β-globin is fully induced. Indeed, by using the relative accumulations at 53 h (NUS, 1; NS, 21; C, 752), a kS1 of 40 h−1 is obtained, confirming the validity of a first-order description of splicing throughout the transcriptional induction.
FIG. 8.
Determination of β-globin splicing and transport rates by computer simulation. (A) Experimental data. HeLa-β-glo cells were treated with 2 μg of doxycycline/ml for up to 24 h. β-Globin transcripts were analyzed by RNase protection assay with a probe encompassing intron 1, as in Fig. 4. Nuclear spliced (NS) and unspliced (NUS) and cytoplasmic (C) transcripts were plotted as a function of time. Note that the y axis is on a logarithmic scale. (B) Computer simulation for the determination of intron 1 splicing rate. As in Fig. 2, the accumulation of spliced products was calculated by taking into account the experimental pool of NUS precursors and varying the splicing rate. Only the best fit is presented. Note that the y axis is on a linear scale. (C) Computer simulation for the determination of the transport rate. As in Fig. 2, the accumulation of cytoplasmic mRNA was calculated by taking into account the experimental pool of NS precursors and varying the transport rate. Two examples fitting either the early or the late time point are presented. Note that the y axis is on a linear scale.
As to the transport, a modeling of the generation of cytoplasmic mRNA from the corresponding nuclear pool requires a higher transport rate at the beginning of the induction (kT of 9.6 h−1) than at its end (kT of 3.2 h−1) (Fig. 8C), corresponding to half-lives of 4.4 and 13 min, respectively. The relative accumulation of the nuclear and cytoplasmic transcripts at 53 h indicates that the apparent transport rate decreased further (kT of 1.9 h−1), yielding an apparent half-life of 22 min. Thus, as for LT-α, the progressive accumulation of nuclear mRNA leads to a decrease of the apparent transport rate over time. Consequently, the actual transport rate is only accessible in the early phase of induction, as illustrated in Fig. 7.
Maturation of β-globin transcripts under various conditions of culture.
Having developed a methodology to derive splicing and transport rates from the kinetics of β-globin expression, we sought to determine whether these processes were affected by cell growth. HeLa cells cannot be efficiently led to quiescence by serum deprivation. However, it was possible to compare highly confluent cultures and cells at low density. The kinetics of β-globin expression during the first 4 h of induction in cells grown at various densities was investigated. The appearance of unspliced and spliced transcripts in the nuclear and cytoplasmic compartments was analyzed by an RNase protection assay with the probe encompassing intron 1. The results were quantified (Fig. 9A) and used to calculate kS and kT values. The amount of total RNA extracted per petri dish was used as an indicator of cell density. Including the experiment shown in Fig. 7, it varied from 40 μg when cells did not contact each other to 130 μg when cells were highly confluent. In these four independent experiments, the kT values were 9.5, 12, 17, and 12 h−1, corresponding to half-lives ranging between 2.5 and 4.4 min for mRNA export. The kS values were 38, 56, 38, and 38 h−1, corresponding to half-lives between 0.8 and 1.1 min for intron 1 splicing. Thus, no significant variation in the splicing rates was observed, while some variations in the transport rate were observed. However, no correlation with cell density was found (Fig. 9B).
Recent studies suggested a possible involvement of β-actin in mRNA nuclear processing (34). We investigated whether a drastic change in β-actin metabolism might disturb splicing or transport of β-globin mRNA. In two independent experiments, cells were treated for 1 h with cytochalasin B, an inhibitor of actin polymerization, at 2 μg/ml. β-Globin expression was then induced with doxycycline for up to 4 h in the presence of cytochalasin B. β-Globin transcripts were quantified by an RNase protection assay (Fig. 9C). The calculated kS values were 26 and 55 h−1, indicating half-lives of 1.6 and 0.8 min for intron 1 splicing. The kT values were 10 and 24 h−1, indicating half-lives of 4.2 and 1.8 min for transport. These values, obtained in the presence of cytochalasin B (Fig. 9D), were in the same range as those obtained in untreated cells (Fig. 9C). This is surprising, as this drug is known to induce major changes in the cell architecture. By electron microscopy, it was seen that cytochalasin B dramatically altered the cytoplasm, leading to intense membrane bubbling, but did not affect the morphology of the nucleus (data not shown).
In conclusion, we have tested two factors which might have affected the rates of mRNA maturation: cell density and actin metabolism. Neither of these affected conclusively the splicing or the transport rate of β-globin mRNA. Overall, the measure was very robust, with half-lives ranging between 0.8 and 1.6 min for splicing and between 2.5 and 4.4 min for transport.
DISCUSSION
In this study we present a strategy to analyze the splicing and transport rates of pre-mRNA in living cells that relies on the use of regulated promoters. This approach associates the kinetic dimension of transcription regulation by tetracycline with the sensitivity of the RNase protection assay. Both the reinduction of a tet-off promoter and the induction of the tet-on promoter occurred over hours. While, in the latter case, we don't understand the molecular basis of this slow induction, such a slow kinetics is very useful for the analysis of rapid processes such as splicing and mRNA export. This is because every time point is informative as long as a steady state is not reached. Indeed, in the present work, we were able to accurately derive splicing and transport rates of the order of minutes from kinetics extending over hours. Conversely, a real-time analysis of these processes would require kinetic analysis over a time scale of 1 min, which is unrealistic in these cellular models.
Analysis of mRNA export to the cytoplasm.
While the biochemistry of mRNA export has been the focus of many recent studies, little is known of its kinetics. In this study, we provide the first kinetic analysis of mRNA export in mammalian cells. During the first hours of the transcriptional induction of β-globin, the appearance of mRNA in the cytoplasm can be described by a first-order reaction acting on the nuclear population of mRNA. In five independent experiments the corresponding nuclear residence half-lives ranged between 2.5 and 4.4 min, indicating a rapid but not instantaneous transport of mature mRNA to the cytoplasm. As expected, this is much shorter than what has been observed following microinjection of mRNA into the nuclei of Xenopus oocytes. It falls, however, within a factor of 2 of the results obtained for the export of specific mRNA from isolated nuclei (39).
Since our analysis is based on a modeling of the fate of fully spliced RNA, all the processes intervening downstream of splicing are included in this nuclear residence time: release from the splicing machinery, transit from the site of splicing to the NPC, assembly of export factors, translocation through the NPC, and dissociation from the NPC. mRNA movement through the nucleoplasm has been shown to be as rapid as free diffusion within the interchromatin space, so that an mRNA could reach the nuclear envelope in about 30 s (33, 43). If the transport of proteins can be taken as an indication of the translocation of mRNA through the nuclear pore, this step could take only a few seconds (35). Studies by electron microscopy have failed to reveal an accumulation of mRNA at the NPC, confirming that this step is not rate limiting (30, 43). It is therefore likely that most of the nuclear dwelling time corresponds to dissociation from the splicing machinery and association with the export factors.
When the kinetic analysis was prolonged beyond the first hours of induction, the description of transport as a first-order process could not be maintained. This was partly expected in view of the presence in the nucleus of a pool of mature mRNAs that are not in the process of being exported (45). Thus, at equilibrium there are two distinct populations of mRNAs in the nucleus, those being exported and those that accumulate. We have proposed two mechanisms for the constitution of this pool: either these mRNAs have escaped the transport machinery and are transiently stored in the nucleus or, following their initial export, they have been reimported from the cytoplasm. Our present analysis indicates that the nuclear pool builds up more slowly than the cytoplasmic one. In a nuclear retention mechanism, our results would indicate that the probability that an mRNA escapes the export machinery increases with the expression level, a phenomenon akin to a saturation of the transport machinery. This does not easily account for the previous observation that at equilibrium the nucleocytoplasmic partition is insensitive to the expression level (45). In contrast, such a delayed accumulation could easily be explained by a reimport mechanism.
As the nuclear dwelling time of β-globin mRNA in the process of being exported is of the order of 4 min and the half-life of cytoplasmic mRNA is 780 min (13 h), it follows that, at equilibrium, the nuclear precursors should amount to 4/780 or 0.5% of the cytoplasmic pool of mRNA. Since we observed that 10% of the mRNAs accumulate in the nucleus, the mRNAs in transit represent about 1/20 of the nuclear pool at equilibrium. Such a small proportion of mRNA in transit is in good agreement with our previous lack of detection of a nuclear efflux following transcriptional arrest by tetracycline (45).
Analysis of splicing.
In contrast with the transport process, splicing is readily described by a first-order reaction throughout the transcriptional induction. This is true for both LT-α with its three introns, including the partially spliced intron 3, and β-globin with its two efficiently spliced introns. The corresponding half-lives range between 0.4 and 1.5 min for the efficiently spliced introns and reach 7.5 min for LT-α intron 3. The minimal sequences required for this slow splicing are contained within the intron and the last 3 nucleotides of exon 3 (29), and both the 5′ and 3′ splice sites are implicated (C. Giansante, D. Weil, H. Neel, and F. Dautry, unpublished data). That a single reaction rate can account for the splicing of a given intron at all phases of expression has several important implications for the splicing reaction, the use of equilibrium precursor-to-product ratios to estimate splicing rates, and the degradation of maturation intermediates in the nucleus.
The first inference is that the splicing machinery is never limiting, even when expression is induced in naive cells, as for β-globin. The splicing reaction requires a large number of factors (snRNPs, essential and accessory proteins) which have been shown to have a highly dynamic localization within the nucleus. Indeed, sites of accumulation of splicing components move toward new sites of transcription during the induction of gene expression, suggesting an adaptive response to a high level of expression (26). Our results suggest that the limiting steps in nuclear gene expression are at the transcriptional level rather than the posttranscriptional level. There is increasing evidence that proteins involved in pre-mRNA processing are loaded onto pre-mRNA during transcription (22, 25, 27, 41, 47). This is true not only for capping and polyadenylation but also for splicing and possibly transport. Our results fully support a view in which all of the potentially limiting factors involved in splicing are recruited by the transcription complex.
A second aspect of our results is that the same description of splicing continues to apply once a steady state in expression has been reached. Therefore, it is possible to derive the splicing rates from the precursor-to-product ratios at equilibrium. We have previously used such an approach to demonstrate the regulation of splicing of LT-α and β-globin by cellular signaling pathways (28), but, in the absence of a kinetic analysis, it was not possible to reach a conclusion on the nature of the modification. At this stage, the validity of using precursor-to-product ratios at equilibrium has been established only for LT-α and β-globin, but the robustness of the analysis and the fact that it works equally well for efficiently and partially spliced introns suggest that it could have a wide applicability, providing a simple way to determine the splicing rates.
Nuclear degradation during pre-mRNA processing.
Our modeling of the pre-mRNA processing provides a unique opportunity to investigate the importance of nuclear degradation during pre-mRNA processing. Indeed, we can easily introduce into our analysis degradation rates for all the intermediate species (i.e., partially spliced and mature mRNA), except for the primary transcript since we only use in the modeling its actual accumulation level and not its transcription rate. Computer simulations indicate that, if a significant degradation of nuclear RNA is introduced, it becomes impossible to describe the kinetics of LT-α and β-globin expression. To compensate for degradation, it would be necessary to increase the corresponding flux of molecules. While it is possible to adjust the splicing and degradation rates to account for the steady-state accumulations, increasing both the input and the output of a given intermediate impacts its kinetics of accumulation. In other words, the experimental data are accurately described by one and only one set of first-order reactions, excluding the possibility of a significant nuclear degradation.
With respect to the pool of mRNAs which slowly builds up in the nucleus and which is not taken into account in our modeling, we have previously observed that these mRNAs have the same half-life as the cytoplasmic mRNAs following a transcriptional arrest. Including such a degradation would not significantly alter the splicing and transport rates since it is much slower than these two reactions. In summary, our analysis establishes that for all the LT-α and β-globin processing intermediates the nuclear degradation is in any case much slower than the splicing and transport reactions.
Practicality of the strategy.
The measurement of splicing and transport rates in living cells opens the possibility to investigate the regulation of mRNA maturation. In this paper, we explored two aspects: cell growth and cell architecture. First, we compared dense and sparse cultures. Although overconfluent cells were not quiescent with respect to cell division, their metabolism was slowed down and their morphology changed as they established abundant cell-cell interactions. The β-globin splicing and transport rates were not significantly affected by these changes. Second, we explored a possible role of β-actin in nucleocytoplasmic exchanges. Several studies have shown the involvement of β-actin in the export of dextran and proteins from the nucleus (18, 38). In addition, actin has been detected on nascent transcripts and accompanies the RNA molecules through the nuclear pore to the cytoplasm, suggesting a possible role in mRNA export (31). Actin has also been reported to participate in pre-mRNA binding to the nuclear matrix, suggesting that it may play a role in mRNA splicing (40). We could not specifically target intranuclear actin by injecting antibodies into the nucleus, since our analysis requires a large number of cells. We used cytochalasin B, which inhibits actin polymerization and thus disorganizes fibrillar actin. Although actin does not seem to be polymerized in the nucleus (34), studies which demonstrated a role for actin in nucleocytoplasmic traffic found an effect of cytochalasin (18, 38), which may be attributed to global disturbance of actin metabolism or to the disruption of interactions between the NPC and the cytoskeleton (1, 19). In our study, cytochalasin B had no effect on β-globin splicing and transport rates. Overall these results indicate that mRNA processing is a robust process, insensitive to minor changes in cell growth or modifications of cell adhesion.
Coupling between transcription and splicing.
In this study we have determined half-lives of 0.9, 1.4, and 7.5 min for the splicing of introns 1, 2, and 3 of LT-α, respectively. This hierarchy of splicing rates is in good agreement with our previous analysis at steady state of the maturation pathway of LT-α transcripts in lymphocytes expressing the endogenous LT-α gene (46) and in transfected cells. It cannot account, however, for some of our observations on the fate of LT-α maturation intermediates following a transcriptional shutoff by tetracycline (45). Indeed, in the absence of transcription we observed a disappearance of intron 2-containing transcripts (i.e., N3 and N2) with a half-life of 12 min. We concluded from this that splicing can occur on a previously transcribed molecule. However, this half-life is incompatible with the splicing rates determined in the present study, as in 12 min 99% of intron 2-containing transcripts should have been processed. Thus, although splicing proceeds in the absence of ongoing transcription, it does so at a much-reduced rate. This indicates some novel coupling between transcription and splicing and invalidates the use of transcription inhibition to analyze splicing rates.
As discussed above, several lines of evidence indicate that at least some components of the splicing machinery are brought onto the native transcript by the transcription complex itself. This represents a coupling between transcription and splicing in which splicing factors are loaded onto the molecule being transcribed (i.e., a coupling in cis). It does not, however, explain a requirement for ongoing transcription for molecules, such as LT-α pre-mRNA, which are mostly spliced posttranscriptionally, i.e., an effect in trans of the transcription process. It is currently assumed that splicing takes place in the vicinity of transcription sites and is therefore sensitive to the local concentration of factors. One possible link between transcription and splicing could be the RNA polymerase II (Pol II) itself. Indeed, in in vitro splicing reactions, an hypophosphorylated Pol II inhibits splicing while an hyperphosphorylated Pol II stimulates splicing (17, 47). The carboxy-terminal domain of the polymerase is sufficient for activation and gets transiently associated with pre-mRNA during the early step of exon recognition (49). Thus, following transcriptional arrest, the presence of a local concentration of hypophosphorylated Pol II could interfere with splicing. Alternatively, ongoing transcription could be important for recycling some components of the processing machinery which are necessary at an early phase of the process but which need to be removed from the transcript for the splicing reaction to be completed. It is interesting that during mitosis rRNA maturation is coupled to RNA Pol I transcription. At the beginning of mitosis, RNA Pol I transcription stops and fully transcribed pre-rRNAs are arrested in their maturation pathway, which resumes at the issue of mitosis, along with RNA Pol I transcription (10).
In summary, our data indicate a two-way coupling between transcription and splicing. First, at no point during transcriptional induction is the splicing machinery limiting, supporting the notion that the transcription complex brings in all the essential factors. Second, a specific transcriptional arrest leads to a slowing down of splicing, indicating that ongoing transcription facilitates splicing even when it normally takes place posttranscriptionally. Finally, these results point out that the relative accumulation of processing intermediates at equilibrium, but not the kinetics following an arrest of transcription, can be used to determine the splicing rates.
Acknowledgments
We thank E. Bertrand for critical reading of the manuscript. We are grateful to F. Harper for performing the electronic microscopy analysis and to A. Diaz for technical help.
A. Audibert was supported by the Biotech project 970324. This work was supported by the Centre National de la Recherche Scientifique and by the European Union (Biotech project 970324).
A. Audibert and D. Weil contributed equally to this study.
REFERENCES
- 1.Bassell, G. J., C. M. Powers, K. L. Taneja, and R. H. Singer. 1994. Single mRNAs visualized by ultrastructural in situ hybridization are principally localized at actin filament intersections in fibroblasts. J. Cell Biol. 126:863-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastos, R. N., and H. Aviv. 1977. Globin RNA precursor molecules: biosynthesis and process in erythroid cells. Cell 11:641-650. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L., and A. L. Kimmel. 1987. Guide to molecular cloning techniques. Methods Enzymol. 152:224-226. [PubMed]
- 4.Beyer, A. L., and Y. N. Osheim. 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 2:754-765. [DOI] [PubMed] [Google Scholar]
- 5.Biamonti, G., M. T. Bassi, L. Cartegni, F. Mechta, M. Buvoli, F. Cobianchi, and S. Riva. 1993. Human hnRNP protein A1 gene expression. Structural and functional characterization of the promoter. J. Mol. Biol. 230:77-89. [DOI] [PubMed] [Google Scholar]
- 6.Blochlinger, K., and H. Diggelmann. 1984. Hygromycin B phosphotransferase as a selectable marker for DNA transfer experiments with higher eucaryotic cells. Mol. Cell. Biol. 4:2929-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousquet-Antonelli, C., C. Presutti, and D. Tollervey. 2000. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102:765-775. [DOI] [PubMed] [Google Scholar]
- 8.Clement, J. Q., L. Qian, N. Kaplinsky, and M. F. Wilkinson. 1999. The stability and fate of a spliced intron from vertebrate cells. RNA 5:206-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dautry, F., D. Weil, J. Yu, and A. Dautry-Varsat. 1988. Regulation of pim and myb mRNA accumulation by interleukin 2 and interleukin 3 in murine hematopoietic cell lines. J. Biol. Chem. 263:17615-17620. [PubMed] [Google Scholar]
- 10.Dundr, M., and M. O. Olson. 1998. Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol. Biol. Cell 9:2407-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, D. J., and M. Rosbash. 1996. Yeast pre-mRNA is composed of two populations with distinct kinetic properties. Exp. Cell Res. 229:181-188. [DOI] [PubMed] [Google Scholar]
- 12.Feldherr, C. M., and D. Akin. 1994. Variations in signal-mediated nuclear transport during the cell cycle in BALB/c 3T3 cells. Exp. Cell Res. 215:206-210. [DOI] [PubMed] [Google Scholar]
- 13.Gondran, P., F. Amiot, D. Weil, and F. Dautry. 1999. Accumulation of mature mRNA in the nuclear fraction of mammalian cells. FEBS Lett. 458:324-328. [DOI] [PubMed] [Google Scholar]
- 14.Gondran, P., and F. Dautry. 1999. Regulation of mRNA splicing and transport by the tyrosine kinase activity of src. Oncogene 18:2547-2555. [DOI] [PubMed] [Google Scholar]
- 15.Gossen, M., S. Freundlieb, G. Bender, G. Muller, W. Hillen, and H. Bujard. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268:1766-1769. [DOI] [PubMed] [Google Scholar]
- 16.Greenberg, M. E., and E. B. Ziff. 1984. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature 311:433-438. [DOI] [PubMed] [Google Scholar]
- 17.Hirose, Y., R. Tacke, and J. L. Manley. 1999. Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev. 13:1234-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann, W., B. Reichart, A. Ewald, E. Muller, I. Schmitt, R. H. Stauber, F. Lottspeich, B. M. Jockusch, U. Scheer, J. Hauber, and M. C. Dabauvalle. 2001. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J. Cell Biol. 152:895-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen, R. P. 1999. RNA-cytoskeletal associations. FASEB J. 13:455-466. [PubMed] [Google Scholar]
- 20.Jarmolowski, A., W. C. Boelens, E. Izaurralde, and I. W. Mattaj. 1994. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 124:627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, L. F., R. Levis, H. T. Abelson, H. Green, and S. Penman. 1976. Changes in RNA in relation to growth of the fibroblast. IV. Alterations in the production and processing of mRNA and rRNA in resting and growing cells. J. Cell Biol. 71:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei, E. P., H. Krebber, and P. A. Silver. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo, M. J., and R. Reed. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96:14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mariman, E., A. M. Hagebols, and W. van Venrooij. 1982. On the localization and transport of specific adenoviral mRNA-sequences in the late infected HeLa cell. Nucleic Acids Res. 10:6131-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minvielle-Sebastia, L., and W. Keller. 1999. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 11:352-357. [DOI] [PubMed] [Google Scholar]
- 26.Misteli, T., J. F. Caceres, and D. L. Spector. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387:523-527. [DOI] [PubMed] [Google Scholar]
- 27.Mortillaro, M. J., B. J. Blencowe, X. Wei, H. Nakayasu, L. Du, S. L. Warren, P. A. Sharp, and R. Berezney. 1996. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. USA 93:8253-8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neel, H., P. Gondran, D. Weil, and F. Dautry. 1995. Regulation of pre-mRNA processing by src. Curr. Biol. 5:413-422. [DOI] [PubMed] [Google Scholar]
- 29.Neel, H., D. Weil, C. Giansante, and F. Dautry. 1993. In vivo cooperation between introns during pre-mRNA processing. Genes Dev. 7:2194-2205. [DOI] [PubMed] [Google Scholar]
- 30.Pante, N., A. Jarmolowski, E. Izaurralde, U. Sauder, W. Baschong, and I. W. Mattaj. 1997. Visualizing nuclear export of different classes of RNA by electron microscopy. RNA 3:498-513. [PMC free article] [PubMed] [Google Scholar]
- 31.Percipalle, P., J. Zhao, B. Pope, A. Weeds, U. Lindberg, and B. Daneholt. 2001. Actin bound to the heterogeneous nuclear ribonucleoprotein hrp36 is associated with Balbiani ring mRNA from the gene to polysomes. J. Cell Biol. 153:229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Planck, S. R., M. D. Listerud, and S. D. Buckley. 1988. Modulation of hnRNP A1 protein gene expression by epidermal growth factor in Rat-1 cells. Nucleic Acids Res. 16:11663-11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Politz, J. C., R. A. Tuft, T. Pederson, and R. H. Singer. 1999. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol. 9:285-291. [DOI] [PubMed] [Google Scholar]
- 34.Rando, O. J., K. Zhao, and G. R. Crabtree. 2000. Searching for a function for nuclear actin. Trends Cell Biol. 10:92-97. [DOI] [PubMed] [Google Scholar]
- 35.Ribbeck, K., and D. Gorlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riedel, N., M. Bachmann, D. Prochnow, H. P. Richter, and H. Fasold. 1987. Permeability measurements with closed vesicles from rat liver nuclear envelopes. Proc. Natl. Acad. Sci. USA 84:3540-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schindler, M., and L. W. Jiang. 1986. Nuclear actin and myosin as control elements in nucleocytoplasmic transport. J. Cell Biol. 102:859-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroder, H. C., M. Rottmann, R. Wenger, M. Bachmann, A. Dorn, and W. E. Muller. 1988. Studies on protein kinases involved in regulation of nucleocytoplasmic mRNA transport. Biochem J. 252:777-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroder, H. C., D. Trolltsch, R. Wenger, M. Bachmann, B. Diehl-Seifert, and W. E. Muller. 1987. Cytochalasin B selectively releases ovalbumin mRNA precursors but not the mature ovalbumin mRNA from hen oviduct nuclear matrix. Eur. J. Biochem. 167:239-245. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 14:2435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Screaton, G. R., J. F. Caceres, A. Mayeda, M. V. Bell, M. Plebanski, D. G. Jackson, J. I. Bell, and A. R. Krainer. 1995. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 14:4336-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh, O. P., B. Bjorkroth, S. Masich, L. Wieslander, and B. Daneholt. 1999. The intranuclear movement of Balbiani ring premessenger ribonucleoprotein particles. Exp. Cell Res. 251:135-146. [DOI] [PubMed] [Google Scholar]
- 44.Wang, J., L. Shen, H. Najafi, J. Kolberg, F. M. Matschinsky, M. Urdea, and M. German. 1997. Regulation of insulin preRNA splicing by glucose. Proc. Natl. Acad. Sci. USA 94:4360-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weil, D., S. Boutain, A. Audibert, and F. Dautry. 2000. Mature mRNAs accumulated in the nucleus are neither the molecules in transit to the cytoplasm nor constitute a stockpile for gene expression. RNA 6:962-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weil, D., S. Brosset, and F. Dautry. 1990. RNA processing is a limiting step for murine tumor necrosis factor beta expression in response to interleukin-2. Mol. Cell. Biol. 10:5865-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuryev, A., M. Patturajan, Y. Litingtung, R. V. Joshi, C. Gentile, M. Gebara, and J. L. Corden. 1996. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich pro-teins. Proc. Natl. Acad. Sci. USA 93:6975-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeitlin, S., and A. Efstratiadis. 1984. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell 39:589-602. [DOI] [PubMed] [Google Scholar]
- 49.Zeng, C., and S. M. Berget. 2000. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell. Biol. 20:8290-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]