Abstract
Drosophila sine oculis, eyes absent, and dachshund are essential for compound eye formation and form a gene network with direct protein interaction and genetic regulation. The vertebrate homologues of these genes, Six, Eya, and Dach, also form a similar genetic network during muscle formation. To elucidate the molecular mechanism underlying the network among Six, Eya, and Dach, we examined the molecular interactions among the encoded proteins. Eya interacted directly with Six but never with Dach. Dach transactivated a multimerized GAL4 reporter gene by coproduction of GAL4-Eya fusion proteins. Transactivation by Eya and Dach was repressed by overexpression of VP16 or E1A but not by E1A mutation, which is defective for CREB binding protein (CBP) binding. Recruitment of CBP to the immobilized chromatin DNA template was dependent on FLAG-Dach and GAL4-Eya3. These results indicate that CBP is a mediator of the interaction between Eya and Dach. Contrary to our expectations, Dach binds to chromatin DNA by itself, not being tethered by GAL4-Eya3. Dach also binds to naked DNA with lower affinity. The conserved DD1 domain is responsible for binding to DNA. Transactivation was also observed by coproduction of GAL4-Six, Eya, and Dach, indicating that Eya and Dach synergy is relevant when Eya is tethered to DNA through Six protein. Our results demonstrated that synergy is mediated through direct interaction of Six-Eya and through the interaction of Eya-Dach with CBP and explain the molecular basis for the genetic interactions among Six, Eya, and Dach. This work provides fundamental information on the role and the mechanism of action of this gene cassette in tissue differentiation and organogenesis.
Development of the compound eye in Drosophila is regulated by several genes, such as twin of eyeless, eyeless, sine oculis, eyes absent, and dachshund (32). Mutation in any one of these genes leads to loss of the eye or to eye abnormality. Ectopic expression of eyeless, eyes absent, or dachshund leads to ectopic eye formation, and the efficiency of the ectopic eye formation is markedly enhanced by coexpression of sine oculis and eyes absent or of eyes absent and dachshund (6, 24). Yeast two-hybrid analysis indicates the specific interaction between Sine oculis and Eyes absent and between Eyes absent and Dachshund (6, 24). As a regulatory network of genes, eyeless activates eyes absent and sine oculis, while they regulate eyeless expression and activate dachshund expression (6, 9, 19). Misexpression of dachshund induces ectopic expression of eyeless, eyes absent, and sine oculis (6). Thus, eyeless, sine oculis, eyes absent, and dachshund form a functional gene network that directs the formation of the compound eye. Niimi et al. (19) reported that Eyeless binds directly to the eya gene promoter region. Vertebrate homologues of eyeless (Pax6), sine oculis (Six1-6), eyes absent (Eya1-4), and dachshund (Dach1-2) have been identified (reviewed in reference 15). A similar gene network among such genes has been shown to be functional in muscle differentiation in chicken (10). Overlapping expression of Six1, Six4, Eya1, Eya2, and Dach1 in cranial ganglia, otic vesicles, dorsal root ganglia, and developing limb buds in mice during development suggests that the gene network is also involved in the formation of various organs other than muscles in mammals (5, 7, 21, 35). From analysis of mice defective for the Eya1 gene, it is suggested that the Six-Eya-Dach hierarchy is also involved in ear and kidney formation as well as in eye formation (34).
Six genes are characterized by the conserved Six and Homeo domains and their product function as transcription factors with specific DNA binding activity (14, 15). Eya is a coactivator of Six proteins and contains the conserved Eya domain, which is indispensable for coactivation activity (20). One member of the human EYA gene family, EYA1, is responsible for branchio-oto-renal syndrome, characterized by deafness, branchial, and renal abnormalities (1). Recently, EYA4 has also been shown to be involved in deafness (33). While the molecular function of Dach is largely unknown, two highly conserved domains, dachshund domain 1 (DD1) and dachshund domain 2 (DD2), have been identified by sequence comparison of Dachshund proteins from fly and mouse (7). It is reported that DD1 is involved in transcriptional activation activity, while DD2 interacts with Eyes absent in Drosophila (6). Recently, it was reported that Six and Eya cooperate during their target gene activation and that nuclear translocation of Eya induced by Six proteins elicits the activation (8, 20). However, the molecular basis for the synergic interaction among these three proteins (Six, Eya, and Dach) that direct specific gene activation has not yet been addressed.
The present study was designed to characterize the molecular interaction and mechanism of synergy in developmental regulation of these proteins. For this purpose, we analyzed the direct interaction among these proteins by gel retardation assays and glutathione S-transferase (GST)-pulldown assays and used mammalian two-hybrid analyses to examine their mode of interaction. Our results showed a direct interaction between Six and Eya and indicated that CREB binding protein (CBP) is a linker molecule that mediates the interaction between Eya and Dach.
MATERIALS AND METHODS
Construction of expression plasmids.
pGEX6P-1Six1 and pGEX6P-2Eya3, which express GST-Six1 and GST-Eya3, respectively, were constructed as follows: a PCR fragment amplified with primers covering the region encoding the Six and Homeo domains of mouse Six1 (5′-CGGGATCCACCGAAAACAATAACTCCTC and 5′-CCGCTCGAGTTAGGAACCCAAGTCCACCA) and pfSix1 (23) as a template was digested with BamHI and XhoI and ligated into the BamHI/XhoI site of pGEX6P-1 (Amersham Pharmacia Biotech, Piscataway, N.J.). A HindIII fragment of pHM6Eya3 (20) was blunt ended with Klenow and ligated into the SmaI site of pGEX6P-2 (Amersham Pharmacia Biotech). GST-Eya3 deletion proteins were constructed by subcloning PCR fragments amplified with appropriate sets of primers and templates as follows: for pGEX6P-1Eya3-EF1, primer 5′-GGAATTCGAACGGGTATTTCTCTGG (OH0003), primer 5′-CCGCTCGAGTCAGTCCACACCTCCCTGAAC (OH0004), and pHM6Eya3; for pGEX6P-1Eya3EF1EF2, primer OH0003, primer 5′-CCGCTCGAGTCACTCTCCTAGTCCATACAGGAG (OH0005), and pHM6Eya3; and for pGEX6P-1Eya3/62aa+EF1, primer 5′-GGAATTCAAGCCTAGTGCTATGGTGCC (OH0002), primer OH0004, and pHM6Eya3. The PCR fragments were digested with EcoRI and XhoI and ligated into the EcoRI/XhoI site of pGEX6P-1. For pGEX6P-2Eya3CΔ1 and pGEX6p-2Eya3CΔ2, pGEX6P-2Eya3 was digested with NcoI and XhoI (for a vector) and NcoI-XhoI fragments of pGEX6P-1Eya3EF1EF2 and pGEX6P-1Eya3EF were subcloned, respectively. pGEX6P-1Eya3/559-1270 (not described in the text) was constructed by subcloning a PCR fragment amplified with primer 5′-GGAATTCAGTCTGATACCCACTTCATCTG and primer OH0004 into the EcoRI/XhoI site of pGEX6P-1. This plasmid was cut with NcoI and XhoI (for a vector), and the NcoI-XhoI fragment of pGEX6P-2Eya3 was ligated, resulting in a pGEX6P-1Eya3NΔ1. For pGEX6p-1Eya3NΔ2, NcoI- and XhoI-digested pGEX6P-1Eya3 62aa+EF1 was used as a vector and the NcoI-XhoI fragment of pGEX6P-2Eya3 was inserted.
As for plasmids used in mammalian two-hybrid assays, pfDach1 and pMEya1 were described by Ozaki et al. (23). For pMEya2, 5′-GAATTCATGTTAGAAGTGGTGACCT and 5′-TGCTGTACTGTGTCTGG were used as primers for PCR. An EcoRI-NcoI fragment of the PCR product and NcoI-XhoI fragment from pHM6Eya2 (20) were ligated into the EcoRI/SalI site of pM (Clontech Laboratories, Palo Alto, Calif.). For pMEya3, pHM6Eya3 was cut with HindIII and ligated into the HindIII site of pM. For pMEya4, primer 5′-ATAAGCTTGATGGAAGACACCCAGGACCTA (Eya4-FP) and primer 5′-ATAAGCTTACAAATACTCTAATTCCAGTGC (Eya4-RP) were used for PCR to obtain Eya4 cDNA from the mouse skeletal muscle cDNA library (Clontech Laboratories). The HindIII fragment was first subcloned into pKS, verified by DNA sequencing, and subcloned into the HindIII site of pHM6 (pHM6Eya4). This cDNA contains exons 5, 16, and 20 but lacks exon 19. pHM6Eya4 was cut with HindIII and ligated into the HindIII site of pM. For pMDach1, the N-terminal portion of Dach1 was amplified by PCR using primer 5′-TCCCCCGGGCATGGCAGTGCCGGCGG and primer 5′-CCGCTCGAGTCAGGTTGAGTACACGGGTTTCC, digested with SmaI and XhoI, and ligated into the SmaI/XhoI site of pM. The resulting plasmid was digested with SacII and XbaI (for a vector), and a SacII-XbaI fragment from pfDach1 was inserted.
Expression plasmids for deletion mutations of Dach1 were constructed as follows: for pfmDach1A, a 963-bp DNA fragment upstream of amino acid position 607 was amplified by PCR using primers 5′-TCCCCCGGGTCAAAGTGTCACTTCCCCAG and 5′-CCGCTCGAGTCAGCCATCAGGAAACAGAAAGG, digested with SmaI and XhoI, and ligated into the SmaI/XhoI site of pGEX6P-1. The EcoRI-XhoI (blunt-ended) fragment was excised from the resulting plasmid and was ligated into EcoRI/XbaI (blunt-ended)-digested pfDach1. For pfmDach1B and pfmDach1C, a 635-bp DNA fragment upstream of amino acid position 369 was amplified by PCR using primers 5′-TCCCCCGGGCCCCCTCCCTGGGAAAC and 5′-CCGCTCGAGTCAGCCAACACTTGAATTCATGTC, digested with SmaI and XhoI, and ligated into the SmaI/XhoI site of pGEX6P-1. The AccI-XhoI (blunt-ended) fragment and the SmaI-AccI fragment were excised from the resulting plasmid and ligated into AccI/XbaI (blunt-ended)-digested pfDach1 and HindIII (blunt-ended)/AccI-digested pfDach1, respectively. For pfmDach1D, the SmaI-EcoRI fragment from the pGEX6P-1 plasmid for pfmDach1A was ligated into the HindIII (blunt-ended)/EcoRI site of pCMV-Flag-2 and the resultant plasmid was digested with BsmI and XbaI for a vector. The BsmI-XbaI fragment from pfDach1 was ligated into the vector. The expression plasmids for E1A and E1Amut, pBLg-E1A12S and pBLg-E1A12Sdelta CR1(38-65), respectively, were provided by T. Kouzarides (3). The expression plasmid for the N-terminal half of VP16, pVP16-N (16), was donated by D. S. Kessler, and the expression plasmid for CBP, pCMVHACBP (31), was provided by K. K. Yokoyama. Full-length mouse Six5 cDNA was excised from pfSix5 at the NotI and HincII sites (20) and was blunted and cloned into the SmaI site of pSG424 (26). pFA-CHOP (Stratagene) was used as an expression plasmid harboring a transactivation domain with a GAL4 DNA binding domain. All PCR fragments were verified by sequencing.
Gel retardation assay.
Gel retardation assays were performed as described previously (14). C3 oligonucleotide was labeled with [32P]dCTP and was used as a probe (18). The indicated amounts of GST fusion proteins were incubated with the probe. Seventy-five-fold-diluted anti-Eya3 serum (20) was used for a supershift assay.
Mammalian two-hybrid assays.
HEK293 cells (American Type Culutre Collection) were cultured in Dulbecco's modified Eagle's medium containing 4.5 g of glucose (Sigma Chemical Co., St. Louis, Mo.)/liter supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 37°C under 10% CO2. Transfections were performed by the calcium phosphate method according to the protocol recommended by the manufacturer (Cell Phect transfection kit; Amersham Pharmacia). For this purpose, 0.3 μg of the luciferase reporter, pGLMRG5 (13), and 0.01 μg of the internal control plasmid, pEFBOSβ-GAL (11), were transfected into 2 × 105 cells in 3.5-cm-diameter dishes. Luciferase activity in the cell lysate was expressed relative to the β-galactosidase activity of pEFBOSβ-GAL, unless otherwise stated. Three independent transfections of triplet or doublet samples were performed in each experiment, and a typical result showing the average (and standard deviation for triplet) is shown (see below).
Immobilized DNA binding assays and nuclear extracts from 3T3 cells.
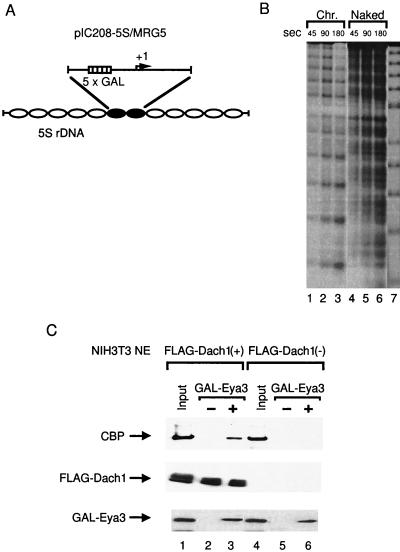
pIC208-5S/MRG5 was constructed as follows: a PCR fragment spanning −183 and +134 (transcription start site at +1) of pGLMRG5 (13) with XhoI sites at both ends was inserted into the XhoI site of pIC208-5S (29). The resultant plasmid, named pIC208-5S/MRG5, contains five GAL4 sites upstream of the synthetic core promoter and 5S histone transfer signals at both ends. pIC208-5S/MRG5 was cut with AflIII and filled in with Klenow in the presence of 40 μM biotin-14-dCTP, followed by digestion with Asp718I. This DNA fragment was isolated from a polyacrylamide gel and was then used for the reconstitution of the nucleosome array (22). The resultant reconstituted chromatin DNA consists of a central dinucleosome-length sequence containing five GAL4 binding sites upstream of the synthetic promoter, flanked on either side by five repeats of a nucleosome-positioning sequence from sea urchin 5S ribosomal DNA (12) (see Fig. 4A). Micrococcal nuclease (MNase) digestion was performed for checking nucleosomal reconstitution (12) (see Fig. 4B). The biotinylated reconstituted chromatin DNA or naked DNA was bound to avidin-conjugated Dynabead kilobaseBINDER (Dynal, Oslo, Norway) using the protocol recommended by the manufacturer. The expressed bacterial GAL4-Eya3 was prepared by purification with a nickel column (Qiagen, Hilden, Germany) followed by a heparin agarose column (Amersham Pharmacia). The resultant purified protein was incubated with DNA-bound beads at room temperature for 30 min in the binding buffer (12), followed by washing with BC-60 (20 mM Tris-HCl, pH 7.3, 60 mM KCl, 0.2 mM EDTA, and 20% [vol/vol] glycerol) containing proteinase inhibitor cocktail (complete mini; Roche Molecular Systems, Inc., Pleasanton, N.J.). Nuclear extracts from transfected NIH 3T3 cells were incubated for 1 h at room temperature and were then washed with BC-60.
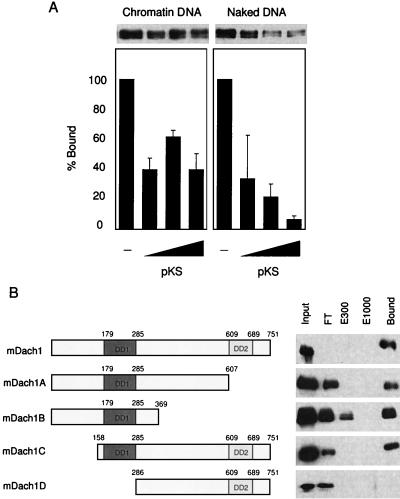
FIG. 4.
Recruitment of CBP to the immobilized chromatin DNA template by GAL4-Eya3 and FLAG-Dach1. (A) Schematic representation of reconstituted chromatin with pIC208-5S/MRG5. rDNA, ribosomal DNA. (B) Reconstituted chromatin (Chr.) or naked DNA (Naked) was end labeled and analyzed by micrococcal nuclease (MNase) digestion. MNase (4 mU for chromatin and 0.8 mU for naked DNA; TaKaRa, Ohtsu, Japan) was added and incubated for the indicated time. The 5S ribosomal DNA repeats were shown by partial EcoRI digestion (lane 7) (12). (C) One-half microgram of chromatin DNA-conjugated Dynabeads was incubated with 20 μg of protein of nuclear extract of NIH 3T3 cells transfected with pfDach1 (lanes 2 and 3) or pCMV-Flag-2 (lanes 5 and 6) in the absence (lanes 2 and 5) or presence (lanes 3 and 6) of 1 μg of GAL4-Eya3. Anti-CBP serum, anti-FLAG antibody, and anti-Eya3 serum (20) were used for detection of recruited CBP, FLAG-Dach1, and GAL4-Eya3, respectively.
For assessing the affinity of Dach1 to chromatin and naked DNA, the washed beads were incubated with pBluescript KS (TOYOBO, Osaka, Japan) for 15 min at room temperature. After washing with BC-60, Dach1 protein on the beads was analyzed by Western blotting using anti-FLAG antibody (Sigma). DNA cellulose (Amersham Pharmacia) was used for mapping of the Dach1 DNA binding domain.
RESULTS
Six and Eya bind directly.
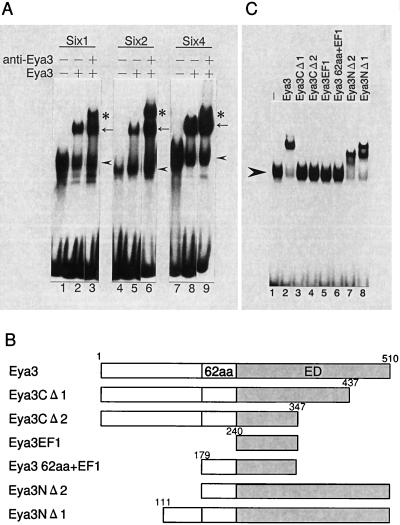
Six proteins can synergize with Eya in activation of their target genes through nuclear translocation of Eya. The presence of the Six-Eya protein complex was evidenced by coimmunoprecipitation in transiently transfected COS7 cells with plasmids expressing Six protein and Eya protein. In this regard, we have previously reported the presence of endogenous complex in nuclear extracts from the rat liver (20). However, it is not clear whether the Six-Eya complex formation is mediated through direct interaction of these proteins or by other proteins. Therefore, we performed a gel retardation assay with recombinant proteins of GST-Six and GST-Eya using C3 oligonucleotide (containing one Six protein-binding site) as a probe. GST-Six1, GST-Six2, and GST-Six4 proteins contain the specific DNA binding domain (Six and Homeo domains) of each Six protein. GST-Eya3 protein contains the whole region of Eya3 protein. As shown in Fig. 1A, supershifted complexes were observed by the addition of GST-Eya3 (lanes 2, 5, and 8). Further supershifted complexes were observed by the addition of anti-Eya3 sera (lanes 3, 6, and 9), indicating that the complexes include Eya3. These results suggest that Eya3 and these Six proteins interact directly and form a complex on the DNA. To identify the region within Eya3 responsible for this interaction, we tested several deletion mutations of Eya3 protein (Fig. 1B). As shown in Fig. 1C, deletion of the Eya domain (lanes 3 and 4) abolished the formation of the Six4-Eya3 supershifted complex, indicating that the Eya domain is essential for the interaction with Six4. The Eya domain alone did not show a supershifted complex (K. Kawakami, unpublished observation). Addition of the adjacent 62-amino-acid portion to the Eya domain (corresponding to the 62-amino-acid region of Eya2, which is necessary for interaction with Six4 in yeast two-hybrid analysis [20]) restored the formation of the Six4-Eya3 complex (lanes 7 and 8). Similar results were obtained with Eya deletion mutations in combination with GST-Six1, GST-Six2, or GST-Six5 (data not shown). These results are consistent with the previous observation that the corresponding region of Eya2 is sufficient for the interaction between Six4 and Eya2 in a yeast two-hybrid assay (20). We concluded that Eya proteins directly bind to Six proteins on the DNA.
FIG. 1.
Direct interaction of Six and Eya on the DNA. (A) Gel retardation assays of Eya3 and Six proteins. Six nanograms of GST-Six1 (lanes 1 to 3), 300 ng of GST-Six2 (lanes 4 to 6), and 6 ng of GST-Six4 (lanes 7 to 9) were incubated with 5 fmol of the C3 probe. One-and-a-half micrograms of GST-Eya3 (lanes 2, 3, 5, 6, 8, and 9) and 0.2 μl of anti-Eya3 sera (lanes 3, 6, and 9) were added. Arrowheads show positions of Six-DNA probe complex. The Eya-Six-DNA complex is shown by arrows, and the supershifted complex is shown by anti-Eya3 by asterisks. (B) Scheme of deletion mutation of Eya3 protein used in Fig. 1C. Positions of amino acids in Eya3 protein deletions are shown. (C) Fifteen-and-a-half picomoles each of full-length GST-Eya3 (lane 2), GST-Eya3CΔ2 (lane 4), GST-Eya3EF1 (lane 5), GST-Eya3/62aa+EF1 (lane 6), GST-Eya3NΔ2 (lane 7), and GST-Eya3NΔ1 (lane 8), and 2.2 pmol of GST-Eya3CΔ1 (lane 3) were added in the presence of 4 ng of GST-Six4 with the C3 probe. Arrowhead shows the position of the GST-Six4-DNA probe complex.
Interaction between Dach and Eya proteins.
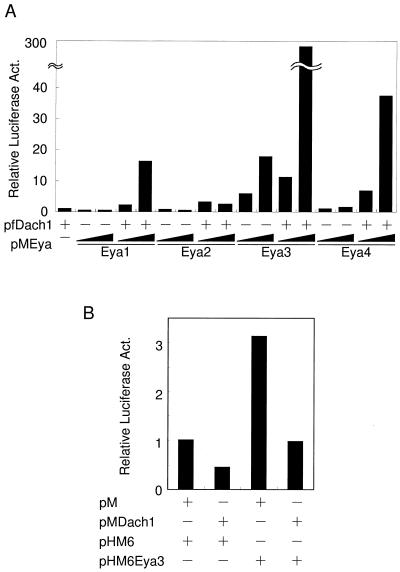
Next, we examined the interaction between Eya and Dach. Interaction has been reported between Drosophila Eyes absent and Drosophila Dachshund by yeast two-hybrid analyses (6). Based on the observation that DD2 is highly conserved between fly Dachshund and mouse Dach and that DD2 has been reported to be responsible for the interaction of Eyes absent (6, 7), it was expected that mouse Dach also interacts with Eya through DD2. We performed mammalian two-hybrid analyses using GAL4-Eya3 as a bait and VP16-Dach1, which contains the entire region of Dach1 as prey in HEK293 cells. Contrary to our expectation, we did not detect transactivation of the GAL4 reporter (pGLMRG5) by VP16-Dach1, although both fusion proteins were produced in the cells (data not shown). We hypothesized that the VP16 moiety could induce yet unknown structural constraints on Dach1 that may perturb activation. We then checked the effects of FLAG-tagged Dach1 protein (pfDach1) with GAL4-Eya1 (pMEya1), GAL4-Eya2 (pMEya2), GAL4-Eya3 (pMEya3), and GAL4-Eya4 (pMEya4) on transactivation of the reporter (Fig. 2A). Cotransfection of pfDach1 and pMEya1 showed 16-fold activation on the reporter, while pfDach1 and pMEya2 showed only a weak activation. pMEya3 alone showed 6- to 17-fold activation, which may be explained by the presence of the activation domain in Eya3. Cotransfection of pfDach1 further boosted the activation up to 300-fold. pMEya4 and Dach1 also showed strong activation of around 40-fold. The simplest explanation for these results is that Dach1 has its own activation domain and that the domain is recruited to the promoter region through interaction with Eya proteins. To check the presence of the Dach1 activation domain in itself, we constructed pMDach1 expressing GAL4-Dach1 fusion protein and tested it for transactivation ability on the GAL4 reporter gene (Fig. 2B). However, it showed no activation but rather repressed transcription of the reporter gene to two- to threefold, regardless of the presence of Eya3 protein. We also used Dach1-GAL4 protein in which the GAL4 DNA binding domain was fused to the carboxyl terminus of Dach1 protein. Furthermore, we dissected the Dach protein into four parts and fused these parts to GAL4. In all cases, we observed negligible activation on the reporter by Dach fusion proteins (data not shown), indicating that forced recruitment of Dach1 protein to DNA by the heterologous DNA binding domain is not sufficient to exhibit the coactivation function of Dach1. Rather, GAL4-Eya3 and FLAG-Dach1 together may form a composite activation domain surface. Consistent with this hypothesis, Eya or Dach itself did not contain its own strong activation domain, as revealed by GAL4-Eya1, Eya2, Eya4, and GAL4-Dach1 in HEK293 cells (Fig. 2A and B). Similar results were obtained in NIH 3T3 cells (data not shown), indicating that the lack of transactivation activity of Dach1 is not cell type specific.
FIG. 2.
Mammalian two-hybrid interaction of Dach1 and Eya proteins. (A) One-half microgram of the reporter, pGLMRG5, was cotransfected with or without pfDach1, and pMEya plasmids. pCMV-Flag-2 and pM were used as a control. Luciferase activity in the cell lysate was normalized with β-galactosidase activity of pEFBOSβ-GAL as an internal control. Increasing amounts (0.1 and 0.3 μg) of pMEya1, pMEya2, pMEya3, and pMEya4 were added. The addition of 0.3 μg of pfDach1 is indicated by +. Data reflect luciferase activity relative to that in the presence of pfDach1. (B) GAL4-Dach1 (pMDach1) showed no activation of the reporter. Combinations of 0.5 μg of pMDach1 and 0.5 μg of pHM6Eya3 are shown. Data represent luciferase activity relative to that in the presence of pM and pHM6. Act., activity.
To investigate whether Dach1 interacts directly with Eya, we performed a GST pulldown assay using GST-Dach1 protein and in vitro translated Eya proteins. The results showed no specific interactions between the two proteins (data not shown). These results suggest that the interaction between DNA-bound Eya and Dach1 is mediated through other proteins. Accordingly, we performed further analyses using GAL4-Eya1 or GAL4-Eya3 and FLAG-Dach1, because the combination of the two showed strong transactivation.
Involvement of CBP in synergy between Eya and Dach.
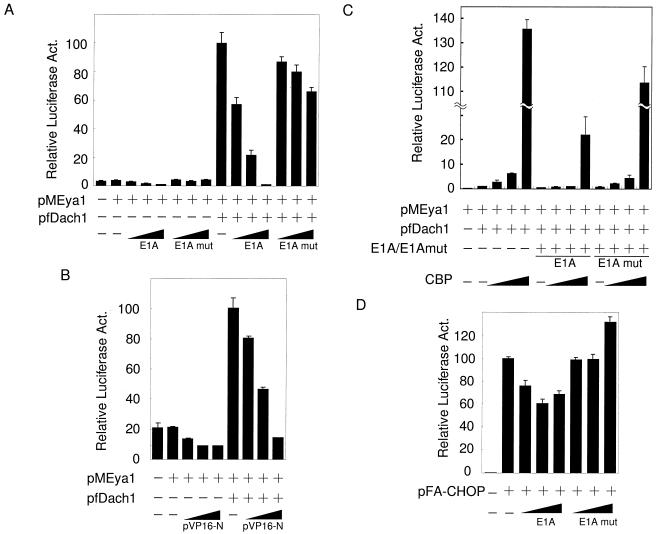
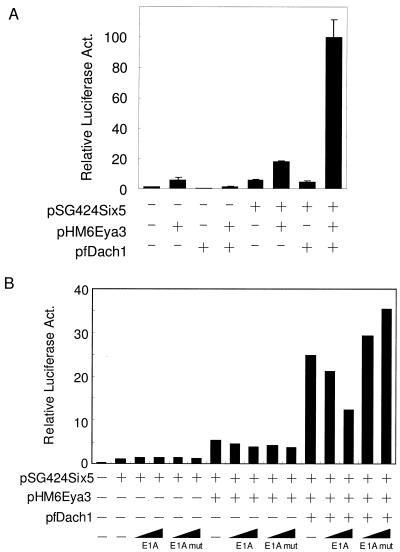
CBP is a well-known coactivator that functions as a key integrator as a result of its large size in various transcriptosomes (2, 28). To determine the possible involvement of CBP in the Dach-Eya transcription complex, we examined the effects of E1A and VP16 (3, 13), both of which tightly bind CBP directly and therefore squelch CBP from other transcriptional complexes. As shown in Fig. 3A, E1A abrogated the transactivation of the reporter by pMEya1 and pfDach1 in a dose-dependent manner (columns 10 to 12), while it showed marginal effects on the reporter when pMEya1 alone was cotransfected (columns 3 to 5). The E1A deletion mutation, which abolishes the specific interaction with CBP, showed little effects on the activation (Fig. 3A, columns 6 to 8 and 13 to 15). E1A also reduced the transactivation of the reporter by pMEya3 and pfDach1 but only moderately, probably due to the presence of the intrinsic transactivation domain in Eya3 (data not shown). VP16 also abrogated the activation by pMEya1 and pfDach1 (Fig. 3B, columns 7 to 9), though in this case, pVP16 also slightly affected transcription by pMEya1 alone (Fig. 3B, columns 3 to 5). In either case, the results suggest that activation through Eya1 or Eya3 and Dach1 is mediated by CBP. In fact, overexpression of CBP markedly enhanced the activation induced by pMEya1 and pfDach1 by 137-fold (Fig. 3C, columns 3 to 5) and that by pMEya3 and pfDach1 (data not shown). Consistent with the above finding, this enhanced activation by CBP was also specifically diminished by E1A (columns 6 to 9) but not by the E1A mutation (columns 10 to 13).
FIG. 3.
Effects of E1A and VP16 on Eya1-Dach1 synergistic activation. (A) One-half microgram of reporter, pGLMRG5, was cotransfected with or without 0.3 μg of pfDach1 and with 0.3 μg of pMEya1 plasmids. Increasing amounts of E1A and its deletion mutation E1Adelta CR1, denoted as E1Amut (5, 50, and 500 ng), were cotransfected. Relative luciferase activity (Act.) was normalized as the protein concentration of each nuclear extract, because E1A influenced the β-galactosidase activity of the internal control pEFBOSβ-GAL. The activity in the presence of pMEya1 and pfDach1 was set at 100. (B) One-half microgram of reporter pGLMRG5 was cotransfected with or without 0.3 μg of pfDach1 and with 0.3 μg of pMEya1 plasmids. Increasing amounts of pVP16-N (4, 40, and 400 ng) were cotransfected. The activity in the presence of pMEya1 and pfDach1 was set at 100. (C) The reporter pGLMRG5 (0.3 μg) was cotransfected with increasing amounts of pCMVHACBP (0.083, 0.25, and 0.75 μg) in the presence of 0.15 μg of pMEya1 and 0.15 μg of pfDach1 (columns 2 to 13) in the presence of 0.01 μg of E1A or E1Amut (columns 6 to 9 or 10 to 13). The luciferase activity in the presence of pMEya1 and pfDach1 (column 2) was set at 1. (D) pGLMRG5 and pFA-CHOP were cotransfected with increasing amounts of E1A and its deletion mutation E1Amut (5, 50, and 500 ng). The luciferase activity in the presence of pFA-CHOP was set at 100.
The involvement of CBP in Eya-Dach activation is factor specific, because transactivation of the GAL4 reporter by another activation domain, such as CHOP (30), was only slightly reduced with the cotransfection of E1A (Fig. 3D, columns 3 to 5). These results suggest that the large mediator CBP is involved in synergistic activation by Eya and Dach1.
Recruitment of CBP to chromatin DNA in the presence of GAL4-Eya3 and FLAG-Dach1.
To confirm that the CBP is recruited to the template DNA by these molecules, we used the immobilized, reconstituted chromatin DNA fragments containing the promoter region of pGLMRG5 on magnetic beads (Fig. 4A and B). The immobilized chromatin DNA fragments were incubated with or without bacterially expressed GAL4-Eya3 as the activator, followed by nuclear extracts from NIH 3T3 cells transfected with pfDach1 or pCMV-Flag-2. The beads were recovered and washed, and then bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting. Endogenous CBP was efficiently detected on the beads in the presence of GAL4-Eya3 using Dach1-expressed nuclear extract (Fig. 4C, lanes 2 and 3). A small amount of CBP was detected in the presence or absence of GAL4-Eya3 using nuclear extract without Dach1 (Fig. 4C, lanes 5 and 6). These results indicate the existence of CBP on the promoter in the presence of GAL4-Eya3 and FLAG-Dach1. Interestingly, FLAG-Dach1 was recovered onto immobilized chromatin DNA regardless of the presence of GAL4-Eya3 (Fig. 4C, lanes 2 and 3), indicating that Dach1 is tethered to the chromatin DNA template independent of GAL4-Eya3. On the other hand, we could not detect the formation of the CBP-Dach1 complex in the solution (data not shown). Together with the observations that Dach1 potentiates the transactivation by GAL4-Eya and that GAL4-Dach1 alone could not activate transcription, it is suggested that the transcriptosome with Dach, Eya, and CBP is formed on the target chromatin DNA template.
Dach binds both chromatin and naked DNA.
Dach has never been reported as a DNA binding protein. However, the observations above imply that Dach1 has DNA binding activity. To address whether the FLAG-Dach1 binds naked DNA as well as chromatin DNA, we prepared two immobilized templates on the magnetic beads; one was the chromatin DNA and the other was naked DNA. Nuclear extracts containing Dach1 were incubated with the beads and washed, and then the bound Dach1 protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blotting. As shown in Fig. 5A, FLAG-Dach1 was detected on the beads with naked DNA (column 5) as well as chromatin DNA (column 1). To compare the binding affinity of Dach1 to chromatin and that to naked DNA, we added competitor DNA (pBluescript KS plasmid) to a reaction with chromatin beads (columns 2 to 4) or naked DNA beads (columns 6 to 8). About half of FLAG-Dach1 was dissociated from the chromatin DNA with 1 μg of DNA (Fig. 5A, compare columns 1 and 2); the residual half of FLAG-Dach1 was resistant to competitor DNA and remained on the chromatin in the presence of the maximum 25 μg of DNA (Fig. 5A, columns 2 to 4). In contrast, the FLAG-Dach1 was dissociated from the naked DNA in the presence of competitor DNA in a dose-dependent manner (Fig. 5A, columns 6 to 8). The results confirmed the DNA binding activity of Dach1 to naked DNA as well as to chromatin DNA, the former with lower affinity. We further addressed the binding region of Dach1 to DNA using DNA cellulose (Fig. 5B). DNA cellulose beads were incubated with the nuclear extracts containing FLAG-Dach1 in the presence of 100 mM NaCl. Then the beads were eluted with various salt-containing buffers (from 300 to 1,000 mM NaCl), followed by extensive washing with corresponding elution buffers. Flowthrough, the elution, and washed beads were checked by Western blotting with anti-FLAG antibody. As shown in Fig. 5B (top line), FLAG-Dach1 remained bound on the DNA cellulose even in a buffer containing 1,000 mM NaCl, indicating that FLAG-Dach1 strongly binds to DNA. To localize the portion of Dach1 responsible for the DNA binding, we constructed various deletion mutations of Dach1 that were transfected into NIH 3T3 cells and prepared nuclear extracts. Although a certain amount of each Dach1 deletion mutation protein was observed in the flowthrough fraction, mDach1A, mDach1B, and mDach1C were recovered in the bound fractions after washing with 1,000 mM NaCl. In contrast, the mDach1D that contains the DD2 domain but lacks the DD1 domain showed no binding to DNA (Fig. 5B, bottom line). These results indicate that Dach1 strongly binds to DNA through conserved DD1. Although we cannot exclude the possibility that the observed Dach1 binding is mediated through other protein in the nuclear extracts, we were able to detect DNA binding of bacterially expressed and purified DD1 (data not shown).
FIG. 5.
Binding of Dach1 to naked and chromatin DNA. (A) One hundred nanograms each of chromatin DNA-conjugated Dynabeads (left panel) and naked DNA-conjugated beads (right panel) was incubated with the nuclear extract of NIH 3T3 cells transfected with pfDach1 and washed, and then an increasing amount of pBluescript KS (1, 5, and 25 μg) was added and incubated. After washing, FLAG-Dach1 protein was analyzed by Western blotting (upper panel). The relative amount of FLAG-Dach1 protein was quantitated by densitometry, and those without pBluescript KS (columns 1 and 5) were set at 100. Three exposures were quantitated, and the average and standard deviation are shown. (B) pfDach1 and its deletion constructs (the region in each construct is shown in the left panel) were transfected into NIH 3T3 cells, and nuclear extracts of these cells were incubated with 50 μl of natural DNA cellulose. After centrifugation, supernatant was collected as flowthrough (FT). DNA cellulose beads were eluted in a buffer containing 300 mM NaCl (E300). After washing in the buffer containing 300 mM NaCl, beads were eluted in a buffer containing 1,000 mM NaCl (E1000). Each supernatant and proteins bound to the resin after a washing in 1,000 mM NaCl (Bound) were analyzed by Western blotting using anti-FLAG antibody.
Six-Eya-Dach synergy.
We next examined whether synergistic activation by Eya and Dach is also observed on DNA-bound Six protein. This is critical to dietermine whether Six-Eya synergy and Eya-Dach synergy work independently or are tightly coupled. We used pSG424Six5, which produces GAL4-Six5, and tested the effects in combination with either pHM6Eya3 and/or pfDach1 on pGLMRG5, as shown in Fig. 6A. Coproduction of GAL4-Six5 and HA-Eya3 showed threefold activation of the reporter gene, while that of GAL4-Six5 and FLAG-Dach1 showed no activation. In contrast, coproduction of GAL4-Six5, HA-Eya3, and FLAG-Dach1 greatly activated transcription of the reporter gene (Fig. 6A, lanes 5 to 8). Only a marginal activation was observed with cotransfection of HAEya3 and FLAG-Dach1 in the absence of GAL4-Six5 (Fig. 6A, lanes 1 to 4). These results clearly indicate that activation by Eya and Dach is also relevant when Eya is tethered through Six protein to DNA. Inhibition of activation by E1A and VP16 was also observed in a similar fashion when pSG424Six5, pHM6Eya3, and pfDach1 were cotransfected (Fig. 6B and data not shown), indicating synergy of these proteins through the conserved linker molecule of CBP.
FIG. 6.
Synergy of Six5, Eya3, and Dach1 and effects of E1A on synergy. (A) Interaction with Six, Eya, and Dach was analyzed. Addition of 0.3 μg of pSG424Six5, 0.3 μg of pHM6Eya3, and 0.3 μg of pfDach1 is indicated by +. Relative luciferase activity (Act.) in the presence of pSG424Six5 was set at 1. (B) Effects of E1A and E1A mutations on the transactivation. Shown is the addition of 0.3 μg of pSG424Six5 (columns 2 to 16), 0.3 μg of pHM6Eya3 (columns 7 to 16), and 0.3 μg of pfDach1 (columns 12 to 16) in the presence or absence of 0.01 μg of E1A (columns 3, 4, 8, 9, 13, and 14) or E1A mutation (columns 5, 6, 10, 11, 15, and 16).
DISCUSSION
Genetic analysis of Drosophila has revealed the synergy among Six, Eya, and Dach, and yeast two-hybrid assays have identified the interactions between Six-Eya and Eya-Dach (4, 6, 24). Our studies indicate that the synergy among mouse homologues is mediated through direct interactions between Six and Eya and that the synergy between Eya and Dach is mediated by CBP, rather than through a direct interaction between the two molecules. Furthermore, Eya-Dach synergy is also observed when Six is bound onto the promoter DNA, indicating a tripartite synergy.
In the present study, gel retardation assays were performed in various combinations of Six and Eya and supershifted complexes were observed at least between Eya3 and Six1, Six2, Six4, and Six5 (Fig. 1). These results indicate that the direct interaction between Six and Eya is conserved between at least the Six1/2 and Six4/5 subfamilies (15).
Mammalian two-hybrid analyses between Eya and Dach yielded unexpected results. The usual two-hybrid assays using GAL4-Eya and VP16-Dach1 showed little activation of pGLMRG5, while coproduction of GAL4-Eya and FLAG-Dach showed strong activation of the reporter. The simple interpretation of these results is that VP16-fused Dach1 protein has an aberrant conformation, which leads to masking of the potential activation domain of Dach1. Alternatively, VP16 moiety may disturb the interaction between Dach1 and CBP, which is necessary for Eya-Dach synergy. This notion is supported by a recent observation that VP16 binds directly to CBP through VP16 domain H2 (13). On the other hand, coproduction of GAL4-Dach1 and HA-Eya3 did not show any activation of the reporter. Considering the observation that FLAG-Dach1 binds to chromatin and naked DNA regardless of the presence of GAL4-Eya (Fig. 4C and 5A), region-specific (the multimerized GAL4 binding sites of the reporter gene) binding through GAL4 moiety may disturb the DNA binding ability of Dach1 moiety itself, which is essential for coactivation activity of Dach1.
CBP is a mediator for various transcription factors with histone acetyltransferase activity. The involvement of CBP is particularly interesting for the synergy in activation of transcription between Eya and Dach. The complex formation between FLAG-Dach and CBP was not detected by coimmunoprecipitation experiments using anti-FLAG antibody (data not shown). This may be explained by one of the following processes: (i) the limited amount of CBP present in a cell is occupied by other endogenous CBP-associated factors, or (ii) a stable complex between CBP and Dach may be formed only on the DNA. In fact, efficient CBP recruitment on the immobilized chromatin DNA template was observed when Dach1 was bound to chromatin DNA (Fig. 4C). This suggests that the latter situation may be the case. The binding of Dach1 to chromatin DNA is not dependent on the presence of activator such as GAL4-Eya3. No sequence-specific DNA binding of Dach has been reported so far, but we observed preferential binding to chromatin. Therefore, Dach may recognize a specific structural form of DNA. We found that well-conserved DD1 is necessary for this interaction of DNA (Fig. 5B). Consistent with this observation, X-ray crystallography of DD1 of human Dach revealed that the domain forms a structure similar to the winged helix, which is known as a DNA binding motif (17). Thus, the previously proposed model of a Six-Eya-Dach synergistic mechanism (6) should be altered so that Dach functions through its DNA binding. Together with the upstream activator of Eya, DNA-bound Dach may assist to form transcriptosome on the specific promoter.
There is sufficient evidence to indicate the involvement of the Six-Eya-Dach gene cassette in organogenesis, suggesting that the cassette directly regulates their target genes during the development of various organs involved (25). Mechanistic analysis of gene regulation by this gene cassette has been hampered by the lack of knowledge of the natural target genes during development. We have identified about 20 putative target genes of Six5 protein using P19 embryonal carcinoma cells (27). These include several genes that encode signaling molecules and transcription factors that are expressed during mesodermal development. Our study is the first to shed some light on the synergistic activation mechanism of the Six-Eya-Dach gene cassette using a model promoter. Future analyses using natural target gene promoters should enhance our understanding of the regulatory mechanism(s) that directs organ differentiation and formation.
Acknowledgments
We thank S. Sato for pHAEya4 construct and useful discussion, H. Ozaki for the constructive discussion, S. Krauss for providing Dach1 cDNA, T. Kouzarides for E1A and E1Amut cDNAs, K. K. Yokoyama for pCMVHACBP, D. S. Kessler for pVP16-N, and H. Mano for HEK293 cells.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan and from the Ministry of Health, Labor and Welfare of Japan.
REFERENCES
- 1.Abdelhak, S., V. Kalatzis, R. Heilig, S. Compain, D. Samson, C. Vincent, D. Weil, C. Cruaud, I. Sahly, M. Leibovici, M. Bitner-Glindzicz, M. Francis, D. Lacombe, J. Vigneron, R. Charachon, K. Boven, P. Bedbeder, N. Van Regemorter, J. Weissenbach, and C. Petit. 1997. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat. Genet. 15:157-164. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, A. J., and T. Kouzarides. 1995. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 14:4758-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bui, Q. T., J. E. Zimmerman, H. Liu, and N. M. Bonini. 2000. Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics 155:709-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caubit, X., R. Thangarajah, T. Theil, J. Wirth, H. G. Nothwang, U. Ruther, and S. Krauss. 1999. Mouse Dac, a novel nuclear factor with homology to Drosophila dachshund shows a dynamic expression in the neural crest, the eye, the neocortex, and the limb bud. Dev. Dyn. 214:66-80. [DOI] [PubMed] [Google Scholar]
- 6.Chen, R., M. Amoui, Z. Zhang, and G. Mardon. 1997. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91:893-903. [DOI] [PubMed] [Google Scholar]
- 7.Davis, R. J., W. Shen, T. A. Heanue, and G. Mardon. 1999. Mouse Dach, a homologue of Drosophila dachshund, is expressed in the developing retina, brain and limbs. Dev. Genes Evol. 209:526-536. [DOI] [PubMed] [Google Scholar]
- 8.Fan, X., L. F. Brass, M. Poncz, F. Spitz, P. Maire, and D. R. Manning. 2000. The α subunits of Gz and Gi interact with the eyes absent transcription cofactor Eya2, preventing its interaction with the six class of homeodomain-containing proteins. J. Biol. Chem. 275:32129-32134. [DOI] [PubMed] [Google Scholar]
- 9.Halder, G., P. Callaerts, S. Flister, U. Walldorf, U. Kloter, and W. J. Gehring. 1998. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development 125:2181-2191. [DOI] [PubMed] [Google Scholar]
- 10.Heanue, T. A., R. Reshef, R. J. Davis, G. Mardon, G. Oliver, S. Tomarev, A. B. Lassar, and C. J. Tabin. 1999. Synergistic regulation of vertebrate muscle development by Dach2, Eya2, and Six1, homologs of genes required for Drosophila eye formation. Genes Dev. 13:3231-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda, K., J. P. Halle, G. Stelzer, M. Meisterernst, and K. Kawakami. 1998. Involvement of negative cofactor NC2 in active repression by zinc finger-homeodomain transcription factor AREB6. Mol. Cell. Biol. 18:10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda, K., D. J. Steger, A. Eberharter, and J. L. Workman. 1999. Activation domain-specific and general transcription stimulation by native histone acetyltransferase complexes. Mol. Cell. Biol. 19:855-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda, K., T. Stuehler, and M. Meisterernst. 2002. The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells 7:49-58. [DOI] [PubMed] [Google Scholar]
- 14.Kawakami, K., H. Ohto, K. Ikeda, and R. G. Roeder. 1996. Structure, function and expression of a murine homeobox protein AREC3, a homologue of Drosophila sine oculis gene product, and implication in development. Nucleic Acids Res. 24:303-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawakami, K., S. Sato, H. Ozaki, and K. Ikeda. 2000. Six family genes—structure and function as transcription factors and their roles in development. Bioessays 22:616-626. [DOI] [PubMed] [Google Scholar]
- 16.Kessler, D. S. 1997. Siamois is required for formation of Spemann's organizer. Proc. Natl. Acad. Sci. USA 94:13017-13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, S. S., R. Zhang, S. E. Braunstein, A. Joachimiak, A. Cvekl, and R. S. Hegde. 2002. Structure of the retinal determination protein dachshund reveals a DNA binding motif. Structure 10:787-795. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, M., and K. Kawakami. 1995. ATF-1CREB heterodimer is involved in constitutive expression of the housekeeping Na,K-ATPase α 1 subunit gene. Nucleic Acids Res. 23:2848-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niimi, T., M. Seimiya, U. Kloter, S. Flister, and W. J. Gehring. 1999. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development 126:2253-2260. [DOI] [PubMed] [Google Scholar]
- 20.Ohto, H., S. Kamada, K. Tago, S. I. Tominaga, H. Ozaki, S. Sato, and K. Kawakami. 1999. Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol. Cell. Biol. 19:6815-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliver, G., R. Wehr, N. A. Jenkins, N. G. Copeland, B. N. R. Cheyette, V. Hartenstein, S. L. Zipursky, and P. Gruss. 1995. Homeobox genes and connective tissue patterning. Development 121:693-705. [DOI] [PubMed] [Google Scholar]
- 22.Owen-Hughes, T., R. T. Utley, D. J. Steger, J. M. West, S. John, J. Cote, K. M. Havas, and J. L. Workman. 1999. Analysis of nucleosome disruption by ATP-driven chromatin remodeling complexes. Methods Mol. Biol. 119:319-331. [DOI] [PubMed] [Google Scholar]
- 23.Ozaki, H., Y. Watanabe, K. Ikeda, and K. Kawakami. 2002. Impaired interactions between mouse Eya1 harboring mutations found in patients with branchio-oto-renal syndrome and Six, Dach, and G proteins. J. Hum. Genet. 47:107-116. [DOI] [PubMed] [Google Scholar]
- 24.Pignoni, F., B. Hu, K. H. Zavitz, J. Xiao, P. A. Garrity, and S. L. Zipursky. 1997. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91:881-891. [DOI] [PubMed] [Google Scholar]
- 25.Relaix, F., and M. Buckingham. 1999. From insect eye to vertebrate muscle: redeployment of a regulatory network. Genes Dev. 13:3171-3178. [DOI] [PubMed] [Google Scholar]
- 26.Sadowski, I., and M. Ptashne. 1989. A vector for expressing GAL4(1-147) fusions in mammalian cells. Nucleic Acids Res. 17:7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sato, S., M. Nakamura, D. H. Cho, S. J. Tapscott, H. Ozaki, and K. Kawakami. 2002. Identification of transcriptional targets for Six5: implication for the pathogenesis of myotonic dystrophy type 1. Hum. Mol. Genet. 11:1045-1058. [DOI] [PubMed] [Google Scholar]
- 28.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 29.Steger, D. J., and J. L. Workman. 1997. Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J. 16:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ubeda, M., X.-Z. Wang, H. Zinszner, I. Wu, J. F. Habener, and D. Ron. 1996. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol. Cell. Biol. 16:1479-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ugai, H., K. Uchida, H. Kawasaki, and K. K. Yokoyama. 1999. The coactivators p300 and CBP have different functions during the differentiation of F9 cells. J. Mol. Med. 77:481-494. [DOI] [PubMed] [Google Scholar]
- 32.Wawersik, S., and R. L. Maas. 2000. Vertebrate eye development as modeled in Drosophila. Hum. Mol. Genet. 9:917-925. [DOI] [PubMed] [Google Scholar]
- 33.Wayne, S., N. G. Robertson, F. DeClau, N. Chen, K. Verhoeven, S. Prasad, L. Tranebjarg, C. C. Morton, A. F. Ryan, G. Van Camp, and R. J. H. Smith. 2001. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet. 10:195-200. [DOI] [PubMed] [Google Scholar]
- 34.Xu, P.-X., J. Adams, H. Peters, M. C. Brown, S. Heaney, and R. Maas. 1999. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 23:113-117. [DOI] [PubMed] [Google Scholar]
- 35.Xu, P.-X., I. Woo, H. Her, D. R. Beier, and R. L. Maas. 1997. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development 124:219-231. [DOI] [PubMed] [Google Scholar]