Abstract
Splicing factors of the SR protein family share a modular structure consisting of one or two RNA recognition motifs (RRMs) and a C-terminal RS domain rich in arginine and serine residues. The RS domain, which is extensively phosphorylated, promotes protein-protein interactions and directs subcellular localization and—in certain situations—nucleocytoplasmic shuttling of individual SR proteins. We analyzed mutant versions of human SF2/ASF in which the natural RS repeats were replaced by RD or RE repeats and compared the splicing and subcellular localization properties of these proteins to those of SF2/ASF lacking the entire RS domain or possessing a minimal RS domain consisting of 10 consecutive RS dipeptides (RS10). In vitro splicing of a pre-mRNA that requires an RS domain could take place when the mutant RD, RE, or RS10 domain replaced the natural domain. The RS10 version of SF2/ASF shuttled between the nucleus and the cytoplasm in the same manner as the wild-type protein, suggesting that a tract of consecutive RS dipeptides, in conjunction with the RRMs of SF2/ASF, is necessary and sufficient to direct nucleocytoplasmic shuttling. However, the SR protein SC35 has two long stretches of RS repeats, yet it is not a shuttling protein. We demonstrate the presence of a dominant nuclear retention signal in the RS domain of SC35.
Pre-mRNA splicing takes place within the spliceosome, a macromolecular complex composed of four small nuclear ribonucleoprotein particles (snRNPs) (U1, U2, U4/U6, and U5) and approximately 50 to 100 polypeptides (32). The SR proteins are an extensively characterized family of structurally and functionally related non-snRNP splicing factors that are essential for constitutive splicing and that also influence alternative splicing regulation in higher eukaryotes (5, 19, 58).
The SR proteins are involved in multiple steps of the constitutive splicing reaction. They promote the recruitment of the U1 snRNP to the 5′ splice site (14, 26, 29) and of the U2AF splicing factor and U2 snRNP to the branch site or 3′ splice site (57, 62) and also facilitate the recruitment of the U4/U6-U5 tri-snRNP at later stages of the splicing reaction (51). The SR proteins also bridge pairs of 5′ and 3′ splice sites via RS domain-mediated protein-protein interactions, which can occur across an exon or an intron (exon definition or intron definition) (50, 62). The SR proteins are also important regulators of alternative splicing, and their activity is subject to antagonism by members of the hnRNP A/B family of proteins. Increased levels of SR proteins usually promote the selection of proximal alternative 5′ splice sites both in vitro and in vivo, whereas an excess of hnRNPs A/B proteins usually results in the selection of distal 5′ splice sites (8, 38, 63). Tissue-specific variations in the total and relative amounts of SR proteins and in the molar ratio of SF2/ASF to its antagonist, hnRNP A1, support the notion that the relative levels and activities of these two families of antagonistic factors may be crucial in regulating the patterns of alternative splicing in a tissue-specific or developmentally regulated manner (21, 66).
A class of related RS domain-containing proteins, termed SR protein-related polypeptides or SR-like proteins, some of which also contain RNA recognition motifs (RRMs), are also involved in splicing regulation (16, 19). SR family and SR-like proteins additionally function in the recognition of exonic splicing enhancers, leading to the activation of otherwise inefficient upstream 3′ splice sites (reviewed in references 3, 19, and 22).
The SR proteins have a modular structure consisting of one or two RRMs followed by a C-terminal domain rich in alternating serine and arginine residues (the RS domain). The RS domain consists of simple repeats of arginine and serine, occasionally interrupted by other amino acids, and its length and sequence are highly conserved for individual SR proteins in different species (2, 65). The functional roles of the different domains of SR proteins in constitutive and alternative splicing have been extensively studied. RRM modules determine substrate specificity, whereas RS domains are functionally interchangeable and do not contribute to substrate specificity (9, 41, 59). RS domains direct protein-protein interactions (62) and can function in splicing activation even when fused to a heterologous RNA-binding domain, facts which led to the proposal that RS domains function as activators of splicing (20).
It was originally thought that both the RRMs and the RS domain of SR proteins are essential for constitutive splicing, whereas the RS domain is dispensable for concentration-dependent alternative splice-site switching (4, 69). However, more recent evidence has demonstrated that the RS domain of SF2/ASF is dispensable for the splicing of some substrates, including several constitutive and enhancer-dependent pre-mRNAs; the requirement for this domain is substrate specific and is more pronounced when the 3′ splice site is weak (68). These findings suggested the existence of an RS domain-independent function of SR proteins in constitutive and enhancer-dependent splicing and also suggested that—at least for the RS domain-independent substrates—the role of SR proteins in enhancer-dependent splicing involves functions other than U2AF recruitment.
The RS domains of two Drosophila splicing regulators, SWAP (suppressor of white apricot) and Tra (transformer), direct these splicing factors to the nuclear speckles (23, 36). It was previously determined that the SR protein RS domain acts as a nuclear localization signal (NLS) and that, in the single-RRM-containing SRp20, the RS domain is both necessary and sufficient as a signal for targeting to the nuclear speckles (6). It was also shown that whereas some human SR proteins are confined to the nucleus, three of them—SF2/ASF, SRp20, and 9G8—shuttle continuously between the nucleus and the cytoplasm and that this property is conferred by the RS domain and is affected by RS domain phosphorylation (7). Thus, the RS domain, which is extensively phosphorylated, not only promotes interactions with other splicing factors but also directs the subcellular localization and nucleocytoplasmic shuttling of individual SR proteins (6, 62).
In this report, we analyze the activities of a series of mutant versions of the RS domain of SF2/ASF in constitutive splicing, subcellular localization, and nucleocytoplasmic shuttling. We report that alternating arginine and acidic residues appear to mimic the structural and functional properties of a phosphorylated RS domain that are required for pre-mRNA splicing and for subcellular localization. We also demonstrate the presence of a dominant nuclear retention signal (NRS) in the RS domain of a nonshuttling SR protein, SC35, thereby providing an explanation for why some SR proteins fail to shuttle.
MATERIALS AND METHODS
Expression plasmids.
The SF2/ASF bacterial expression vector, pET9c-SF2 (R/S), and the mammalian expression vector, pCGT7-SF2/ASF, were previously described (6, 31). In pCGT7-SF2/ASF, transcription is driven by the cytomegalovirus enhancer-promoter, and the coding sequence begins with an N-terminal epitope tag, MASMTGGQQMG; this sequence corresponds to the first 11 residues of the bacteriophage T7 gene 10 capsid protein and is recognized by the T7.tag monoclonal antibody (Novagen). The SF2/ASF RS domain mutants RT, RG, GS, KS, ΔRS, and RS10 were previously described (4, 68) and were subcloned into the epitope-tagged expression plasmid vector (6). The RE and RD mutants were constructed by partial gene replacement essentially as described for the other variants (4, 68) and were subcloned into the bacterial or mammalian expression plasmid. Sequence analysis showed that there is a spurious aspartate residue instead of the ninth glutamate in the RE mutant. Recombinant SF2/ASF, RD, and RS10 were also subcloned into a bacterial expression plasmid in which NdeI at the translation initiation site was converted to NcoI; although the sequences of the proteins were not affected, higher expression levels were obtained.
To generate the fusion constructs for nuclear retention studies, hnRNP A1 and SF2/ASF were independently amplified with specific primers that introduce an SpeI site at the N terminus and an XbaI site followed by two stop codons and a BamHI site at the C terminus. The resulting PCR products were purified, digested with SpeI and BamHI, and subcloned into the corresponding sites of the pCGT7 vector to generate pCGT7-A1-NRS and pCGT7-SF2-NRS. Subsequently, XbaI-BamHI fragments of pCGT7-SC35, pCGT7-NPc-RSSC35, or NPc-RSSF2 (6) were subcloned into the corresponding sites of the pCGT7-A1-NRS or pCGT7-SF2/ASF-NRS vector to generate pCGT7-A1/SC35, pCGT7-A1/rsSC35, pCGT7-A1/rsSF2, pCGT7-SF2/SC35, and pCGT7-SF2/rsSC35. C-terminal deletions in the RS domain of SC35 were introduced by PCR amplification of an SC35 cDNA with specific primers, and the resulting products were subcloned into the corresponding sites of the pCGT7 expression vector. Putative NRSs identified within the SC35 RS domain were amplified with specific primers and subcloned into the BamHI site of pCGT7-SF2/ASF, resulting in the fusion of the SC35 putative NRSs downstream of the SF2/ASF coding sequence. The correct orientation of the insert was verified by restriction mapping and DNA sequencing.
Oligonucleotides.
The sequences of the specific primers used for PCR are available upon request.
Expression and purification of recombinant proteins.
Untagged wild-type and variant recombinant SF2/ASF proteins were expressed and purified essentially as described previously (68), except that the binding and elution properties were different for the proteins. The peak fractions after chromatography under denaturing conditions were renatured by dialysis as described previously (68).
Cell cultures and transfections.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Transfection was done with 1 μg of plasmid DNA per 60-mm dish of 60 to 75% confluent cells in the presence of 20 μg of Lipofectin (Gibco-BRL) (8). It was previously estimated that the transiently expressed proteins accumulated in the transfected cells at approximately fivefold higher levels than endogenous SF2/ASF (6, 8).
Indirect immunofluorescence.
Cells were fixed and permeabilized for immunofluorescence assays at 24 h after transfection. Fixation was done with 4% p-formaldehyde in phosphate-buffered saline for 15 to 30 min at room temperature, followed by incubation for 10 min in 0.2% Triton X-100. The fixed cells were incubated for 1 h at room temperature with an anti-T7 monoclonal antibody (1:1,000; Novagen), washed with phosphate-buffered saline, and incubated for 1 h at room temperature with fluorescein-conjugated goat anti-mouse immunoglobulin G (IgG) (1:200; Cappel Laboratories). The samples were observed with a Zeiss Axioskop microscope, and the images were acquired with a Photometrics CH250 cooled, charge-coupled device camera by using Digital Scientific Smartcapture extensions in software from IP Lab Spectrum. The immunofluorescence data reported are representative, and each experiment was reproduced in multiple independent transfections.
Heterokaryon assays.
For transient transfections involving interspecies heterokaryons, due to the need for a higher transfection efficiency, transfection was carried out by electroporation with 10 μg of plasmid DNA per 60-mm dish of 70 to 80% confluent cells in the presence of 20 μg of carrier DNA. Transfected HeLa cells were seeded on coverslips, followed by coincubation with an excess of untransfected mouse NIH 3T3 cells for 3 h in the presence of 50 μg of cycloheximide/ml. The concentration of cycloheximide was then increased to 100 μg/ml, and the cells were incubated for an additional 30 min prior to fusion. Cell fusions were done as described previously (49), and the heterokaryons were further incubated for 2 h in medium containing 100 μg of cycloheximide/ml prior to fixation. Immunofluorescence assays with the anti-T7 monoclonal antibody were performed as described above, except that 4′,6′-diamidino-2-phenylindole (DAPI) was included at 5 μg/ml.
In vivo phosphorylation assays.
HeLa cells were harvested 48 h after transfection with Fugene-6. Cells (106) were scraped into 150 μl of NP-40 lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 100 μg of phenylmethylsulfonyl fluoride [PMSF]/ml, 1 μg of aprotinin/ml). The lysates were incubated with or without 500 U of alkaline phosphatase (New England Biolabs, Beverly, Mass.)/ml for 30 min at 37°C. Wild-type and mutant SF2/ASF proteins were detected by Western blotting with the anti-T7 antibody (1:10,000 dilution).
Transcripts.
7CH3-GpppG-capped, 32P-labeled β-globin and IgM M1-M2 pre-mRNA substrates were made by runoff transcription from plasmids pSP64-HβΔ6 and pμM1-M2 linearized with BamHI and XbaI, respectively (39).
In vitro splicing assays.
HeLa cell nuclear and S100 extracts were prepared as described previously (40). Standard conditions were used for the splicing reactions (39). Briefly, 10 fmol of capped transcripts was incubated in 10-μl splicing reaction mixtures. Each reaction mixture contained either 30% nuclear extract or 35% S100 extract complemented with 4 or 8 pmol of wild-type or mutant recombinant SF2/ASF. After incubation at 30°C for 4 h, the RNA was extracted and analyzed on 5.5% polyacrylamide denaturing gels, followed by autoradiography.
RESULTS
To dissect the compositional and primary sequence features of an RS domain that are important for the functional, cellular localization, and shuttling properties of an SR protein, we constructed variants of human SF2/ASF with artificial domains replacing the natural C-terminal RS domain (Fig. 1A). We analyzed a series of mutant proteins in which either Arg or Ser residues within RS or SR dipeptide repeats of the RS domain of SF2/ASF (residues 198 to 248) were substituted to generate RT, RG, GS, and KS domains. Another mutant protein had all of these Arg and Ser residues deleted while retaining other potentially important amino acids present within this domain. These mutant proteins, designated RT, RG, GS, KS, and ΔRS, have been described elsewhere (4). We also analyzed a mutant version of SF2/ASF in which the entire RS domain was replaced by just 10 RS dipeptides (RS10) (68) and two new mutants in which the RS dipeptides present in the RS domain were replaced by RD or RE dipeptides (Fig. 1A). This combination of mutant RS domains allowed us to systematically analyze the specific requirements for Arg and Ser residues and the importance of the interspersed residues, of potential sites of phosphorylation by Ser/Thr protein kinases, and of alternating arginine and acidic residues.
FIG. 1.
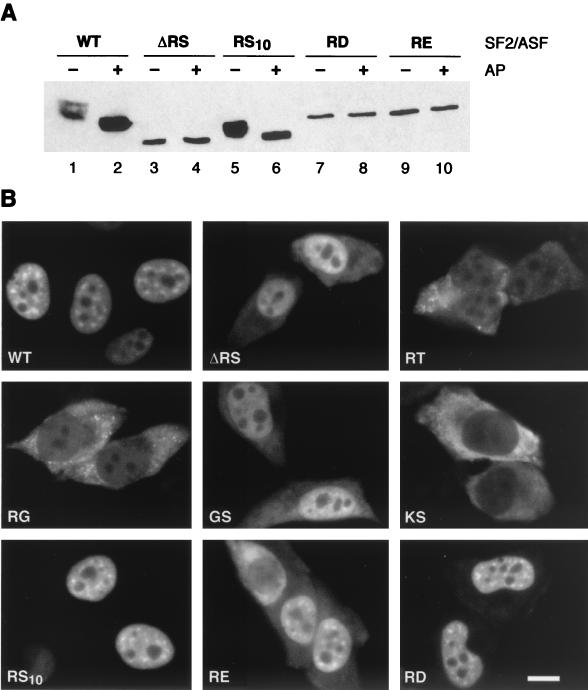
In vitro splicing activities of SF2/ASF proteins with variant RS domains. (A) Sequences of SF2/ASF wild-type (wt) and variant RS domains. All the Arg-Ser or Ser-Arg dipeptides in the RS domain of SF2/ASF were deleted (broken lines) to generate the ΔRS mutant protein. An additional mutant form in which the entire RS domain was deleted (RRM1/RRM2) behaved in a manner similar to that of the ΔRS mutant form. In addition, either Arg or Ser residues within RS or SR dipeptides were substituted with Thr, Gly, Lys, Asp, or Glu to generate RT, RG, GS, KS, RD, or RE mutant proteins. The simplified RS10 domain consists of 10 consecutive RS dipeptides. RS or SR dipeptides as well as the corresponding mutant dipeptides are indicated in bold. The ΔRS, RRM1/RRM2, RT, RG, GS, KS, and RS10 mutant proteins were previously described (4, 68). (B) In vitro splicing activities of the RS10, RE, and RD mutant proteins. Shown is the in vitro splicing of β-globin pre-mRNA (left panel) and IgM M1-M2 pre-mRNA (right panel) in HeLa cell nuclear extract (NE) (lanes 1), S100 extract alone (lanes 2), and S100 extract complemented with 4 and 8 pmol of recombinant SF2/ASF (wild type [WT]) (lanes 3 and 4, respectively), ΔRS (lanes 5 and 6, respectively), RS10 (lanes 7 and 8, respectively), RD (lanes 9 and 10, respectively), or RE (lanes 11 and 12, respectively).
RE and RD domains function in the in vitro splicing of RS domain-dependent splicing substrates.
We first examined whether the new RE and RD domains, which mimic a hyperphosphorylated RS domain, can function as splicing activation domains in vitro (Fig. 1B). To this end, we expressed these proteins in bacteria, purified them, and compared their splicing activities to those of recombinant wild-type SF2/ASF, ΔRS, and RS10. The IgM pre-mRNA substrate was previously shown to require the RS domain for splicing, whereas the β-globin pre-mRNA does not (68). We confirmed that the RS domain is required for IgM but not β-globin pre-mRNA splicing (Fig. 1B, lanes 1 to 6) and that the minimal RS domain—which is rapidly phosphorylated by kinases in the extract (68)—is also functional (Fig. 1B, IgM panel, lanes 7 and 8). We also found that splicing could take place—albeit to reduced extents—when the mutant RD and RE domains replaced the natural domain (Fig. 1B, IgM panel, lanes 9 to 12). Thus, acidic residues (either phosphorylated serines in RS10, aspartates in RD, or glutamates in RE) alternating with basic residues (arginines) can mimic the structural and functional properties of a natural phosphorylated RS domain that are required for splicing in vitro. We do not know if the reduced splicing efficiencies of the variant domains reflect suboptimal mimicry of the natural RS domain or suboptimal folding after renaturation of the proteins expressed in Escherichia coli. However, the latter explanation is consistent with the observation that the proteins with the artificial RD and RE domains also showed reduced splicing efficiency for an RS domain-independent pre-mRNA (Fig. 1B, β-globin panel, lanes 9 to 12).
Structural requirements for the RS domain of SF2/ASF in subcellular localization and targeting to nuclear speckles.
We next used the set of SF2/ASF variants to analyze the roles of compositional features and specific residues within the RS domain that are important for nuclear localization and targeting to the nuclear speckle compartment. We transiently overexpressed epitope-tagged versions of the wild-type or mutant cDNAs in HeLa cells and determined the subcellular distribution and subnuclear localization of the tagged proteins by indirect immunofluorescence microscopy. All constructs encoded proteins with a bacteriophage T7 gene 10 epitope tag at the N terminus, allowing detection of the exogenous proteins with a monoclonal antibody that recognizes this epitope. The expression of the transfected cDNAs was analyzed by Western blotting of whole-cell lysates and showed that all transiently expressed proteins accumulated to similar levels and appeared to be full length (Fig. 2A and data not shown).
FIG. 2.
Role of the RS domain of SF2/ASF in subcellular localization and distribution in nuclear speckles. (A) Immunoblot analysis of the phosphorylation state of SF2/ASF wild-type (WT) and variant proteins. The indicated T7-tagged constructs were transfected into HeLa cells, total cell lysates were incubated in the presence (+) or absence (−) of alkaline phosphatase (AP), and tagged SF2/ASF was detected by Western blotting. Wild-type SF2/ASF and the RS10 mutant protein were phosphorylated in vivo, as indicated by the increase in mobility observed upon treatment with alkaline phosphatase. In contrast, the ΔRS, RD, and RE mutant proteins were not detectably phosphorylated in vivo. (B) HeLa cells were transfected with plasmids encoding the indicated T7-tagged proteins and fixed at 24 h posttransfection. The localization of the expressed proteins was determined by indirect immunofluorescence analysis with anti-T7 monoclonal antibody and fluorescein isothiocyanate-conjugated secondary antibody. Bar, 5 μm.
The phosphorylation status of the transiently expressed epitope-tagged proteins was analyzed by comparing the electrophoretic migration profiles of the respective proteins after incubation in the presence or in the absence of alkaline phosphatase. An increase in the migration of wild-type SF2/ASF as well as of the RS10 mutant protein after treatment with alkaline phosphatase indicated that these proteins were phosphorylated in HeLa cells (Fig. 2A). In contrast, the ΔRS, RE, and RD mutant proteins were not detectably phosphorylated in vivo. It was previously shown that the GS mutant protein becomes phosphorylated but that the RG mutant protein does not; we confirmed these results and also found that the RT and KS mutant proteins did not display any change in mobility, suggesting that they were not phosphorylated in vivo (45) (data not shown).
Indirect immunofluorescence analysis with the anti-T7 antibody showed that at 24 h posttransfection, as expected, the transiently expressed wild-type SF2/ASF protein was localized exclusively in the nucleus, with a typical speckled pattern; in contrast, the ΔRS protein was localized in the nucleus but also in the cytoplasm (Fig. 2B). In contrast, SF2/ASF variants containing RT, RG, or KS domains, which were not phosphorylated in vivo, were localized aberrantly in the cell. The GS mutant protein was present in nuclear speckles, although its localization was not exclusively nuclear. Thus, a correlation between phosphorylation of the C-terminal domain and proper subcellular localization can be inferred, since the RS10 and GS mutant proteins, which were phosphorylated in vivo, also were localized to the nuclear speckle compartment (Fig. 2). The RS10 domain and, to a lesser extent, the RD and RE domains mediated proper subnuclear localization of the variant SF2/ASF proteins in transfected cells, suggesting that a tract of 10 phosphorylated RS dipeptides is necessary and sufficient to direct proper subcellular and intranuclear localization. In approximately 50% of transfected cells, SF2/ASF variants containing RE and RE domains were localized in the nucleus but also in the cytoplasm (Fig. 2B and data not shown). These results show that alternating arginine and acidic residues apparently mimic, at least to some extent, the structural and/or functional properties of a natural RS domain that are required for proper subcellular localization.
The minimal RS10 domain is sufficient to confer nucleocytoplasmic shuttling in the context of SF2/ASF.
It was previously demonstrated that a bipartite signal consisting of RNA binding mediated by RRMs and a specific RS domain determines the nucleocytoplasmic shuttling properties of a subset of SR proteins (7). We used transient transfection in conjunction with interspecies heterokaryons to assay for nucleocytoplasmic shuttling of the RS10 mutant protein (Fig. 3). HeLa cells were transfected with epitope-tagged cDNAs coding for wild-type SF2/ASF or the RS10 variant and then were fused to mouse NIH 3T3 cells to form heterokaryons. Prior to fusion, the cells were treated with cycloheximide, so that no further protein synthesis could take place in the heterokaryons. At 2 h postfusion, the cells were fixed and stained to examine the distribution of the tagged proteins by immunofluorescence microscopy with a monoclonal antibody directed against the epitope tag. To distinguish the human and mouse nuclei, the cells were stained with DAPI, which gives a characteristic staining of intranuclear bodies in mouse nuclei (49). The RS10 version of SF2/ASF shuttled between the nucleus and the cytoplasm in the same manner as the wild-type protein (Fig. 3), demonstrating that 10 RS dipeptides are necessary and sufficient to confer nucleocytoplasmic shuttling in the context of the RNA-binding domain of SF2/ASF. For the ΔRS, RD, and RE proteins, the incomplete nuclear localization at steady state precluded a similar analysis of their shuttling properties.
FIG. 3.
Analysis of nucleocytoplasmic shuttling of SF2/ASF and RS10 by transient expression in interspecies heterokaryons. Wild-type (WT) SF2/ASF and the RS10 mutant protein were transiently expressed in HeLa cells. At 24 h posttransfection, the cells were treated with cycloheximide and subsequently fused with mouse NIH 3T3 cells in the presence of polyethylene glycol to form heterokaryons. The cells were further incubated for 2 h in the presence of cycloheximide, followed by fixation. (Left panels) The localization of the epitope-tagged proteins in the heterokaryons was determined by indirect immunofluorescence analysis with anti-T7 monoclonal antibody (mAbT7) and fluorescein isothiocyanate-conjugated secondary antibody. (Middle panels) The cells were simultaneously incubated with DAPI for differential staining of human and mouse nuclei within heterokaryons. (Right panels) Phase-contrast images of the same heterokaryons. Arrows indicate the mouse nuclei within human-mouse heterokaryons.
NRSs in the RS domain of a nonshuttling SR protein.
Very little was known about the signals that distinguish shuttling from nonshuttling SR proteins, and it was not clear whether nonshuttling SR proteins are retained in the nucleus because they lack a nuclear export signal (NES) or because of the presence of a nuclear retention signal (NRS). Although the RS domain is part of the bipartite signal required for nucleocytoplasmic shuttling and 10 consecutive RS dipeptides are necessary and sufficient for this process, SC35 has two long stretches of RS repeats, yet it is not a shuttling protein (Fig. 4A). This paradox suggested the existence of an NRS in nonshuttling SR proteins, probably located within their C-terminal RS domain (given previous results obtained in RS domain swapping experiments [7]).
FIG. 4.
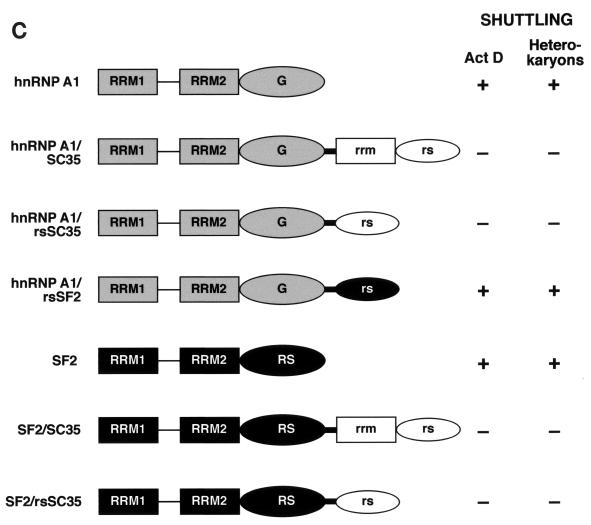
NRS in the RS domain of SC35. (A) Sequences of the SF2/ASF and SC35 RS domains. The long tracts of RS dipeptides are indicated in grey, whereas RS dipeptides outside of these tracts are indicated in bold. (B) Analysis of nucleocytoplasmic shuttling of chimeric forms of hnRNP A1 by transient expression in interspecies heterokaryons. Wild-type hnRNP A1 and chimeric proteins were transiently expressed in HeLa cells, which were then fused with NIH 3T3 cells and analyzed as described in the legend to Fig. 3. Arrows indicate the mouse nuclei within human-mouse heterokaryons. (C) Diagram showing the structures of chimeric proteins in which SC35, the RS domain of SC35, or the RS domain of SF2/ASF was fused to a shuttling protein—either hnRNP A1 or SF2/ASF. The nuclear export properties of these chimeric proteins were analyzed by interspecies heterokaryon assays (panel B) and by transcriptional inhibition assays (data not shown), and the results are summarized. Act D, actinomycin D.
To investigate the presence of a putative NRS in SC35, we constructed chimeric proteins in which SC35 or just its RS domain was fused to a shuttling protein, either hnRNP A1 or SF2/ASF (Fig. 4B and C). The nuclear export properties of these chimeric proteins were analyzed by transcriptional inhibition and interspecies heterokaryon assays (7, 43). The heterokaryon assay showed that chimeric proteins in which SC35 or its RS domain was fused to hnRNP A1 (hnRNP A1/SC35 and hnRNP A1/rsSC35) did not shuttle, as shown by the lack of epitope tag staining in the nuclei of mouse cells (Fig. 4B and C). In contrast, fusion of the RS domain of SF2/ASF to hnRNP A1 (hnRNP A1/rsSF2) did not abolish nucleocytoplasmic shuttling of the resulting chimeric protein. The transcriptional inhibition assay is based on the observation that many shuttling proteins accumulate in the cytoplasm when transcription is inhibited (48). This independent assay also gave very similar results: hnRNP A1, SF2/ASF, and the chimeric fusion hnRNP A1/rsSF2 were localized to the cytoplasm in the presence of actinomycin D, whereas fusion of SC35 or its RS domain to hnRNP A1 or SF2/ASF prevented the cytoplasmic accumulation of the resulting chimeras upon transcriptional inhibition (Fig. 4C and data not shown). These experiments strongly suggest the presence of an NRS sequence in the RS domain of SC35.
We have determined that the nature of the RS domain determines nucleocytoplasmic properties (7) and also that 10 consecutive RS dipeptides are necessary and sufficient to direct nucleocytoplasmic shuttling in the context of SF2/ASF (Fig. 3). The Drosophila homolog of human SF2/ASF (dSF2/ASF) lacks a long tract of repeating RS dipeptides at the beginning of its RS domain, which is essential for phosphorylation of the human protein by SR protein kinase 1 (SRPK1) (1). Whereas the absence of these dipeptides has no effect on dSF2/ASF nuclear localization, the protein does not show nucleocytoplasmic shuttling. This finding strongly suggests that the RS dipeptides in the RS domain and/or their phosphorylation are essential for shuttling (1). A tract of eight RS dipeptides is present at the N terminus of the human SF2/ASF RS domain (Fig. 4A), and deletion of these residues results in nuclear retention (see construct hSF2/ASF Δ197-216 in Fig. 4 of Allemand et al. [1]). The RS domain of the nonshuttling protein SC35 has two such tracts (eight and seven consecutive RS dipeptides separated by 33 residues, which also include eight additional RS dipeptides) (Fig. 4A and 5A), yet it is retained in the nucleus. We speculated that a putative NRS may be located within the C terminus of SC35. To delineate more precisely this putative NRS in SC35, we made two C-terminal deletions and tested whether they relieved nuclear retention. The deletion mutant proteins, termed SC35-RS1d and SC35-RS2d and lacking 45 and 30 amino acids, respectively, were able to shuttle between the nucleus and the cytoplasm, as evidenced in heterokaryon assays (Fig. 5B). Thus, deletion of the last 30 amino acids in SC35 (SC35-RS2d) relieved nuclear retention and allowed nucleocytoplasmic shuttling, confirming that this sequence comprises a dominant NRS (Fig. 5C).
FIG. 5.
C-terminal deletions in the RS domain of SC35 relieve nuclear retention. (A) Sequences of the SC35 RS domain and of two C-terminal deletions. The long tracts of RS dipeptides are indicated in grey, whereas RS dipeptides outside of these tracts are indicated in bold. (B) Analysis of nucleocytoplasmic shuttling of SC35 deletion mutant proteins by transient expression in interspecies heterokaryons. Wild-type SC35 and deletion proteins were transiently expressed in HeLa cells, which were then fused with NIH 3T3 cells and analyzed as described in the legend to Fig. 3. Arrows indicate the mouse nuclei within human-mouse heterokaryons. (C) Sequence of the NRS present in the RS domain of SC35.
Finally, we fused the NRS identified in SC35 to SF2/ASF and found that the presence of these 30 amino acids conferred nuclear retention to the otherwise shuttling protein SF2/ASF (Fig. 6). Thus, the NRS identified in the RS domain of SC35 is transferable, dominant, and sufficient to confer nuclear retention on NES-bearing proteins.
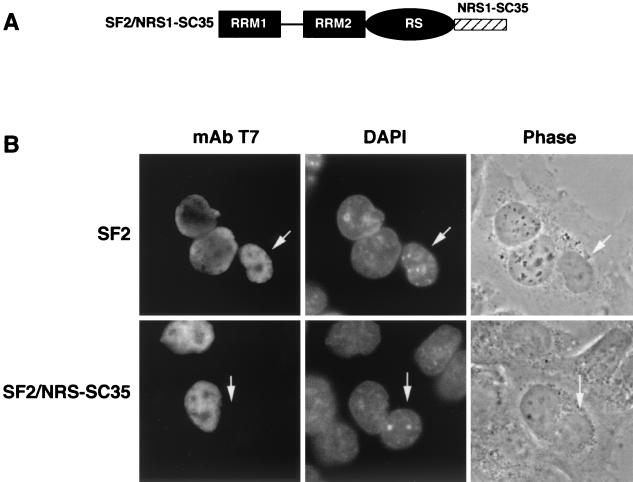
FIG. 6.
The NRS sequence identified in the RS domain of SC35 is transferable. (A) Diagram showing the structure of a chimeric protein in which the NRS identified in the RS domain of SC35 was fused to SF2/ASF. (B) Analysis of nucleocytoplasmic shuttling by transient expression in interspecies heterokaryons. Wild-type SF2/ASF and the chimeric protein SF2/NRS-SC35 were transiently expressed in HeLa cells, which were then fused with NIH 3T3 cells and analyzed as described in the legend to Fig. 3. Arrows indicate the mouse nuclei within human-mouse heterokaryons.
DISCUSSION
The RS domain is a distinctive feature of the SR family of proteins and is also present in a family of related splicing factors known as SR-like proteins. The strong phylogenetic conservation of specific sequences within the RS domains led to the suggestion of specific roles for the RS domains of individual SR proteins. However, the RS domains are functionally interchangeable in splicing and do not contribute to substrate specificity (9, 41, 59). Moreover, chimeric proteins consisting of the RRMs of SF2/ASF fused to RS domains from various SR proteins can rescue cell viability in a chicken DT40 B-lymphocyte cell line in which the endogenous SF2/ASF gene has been inactivated (60). In contrast, analysis of the functional specificity of RS domains in Drosophila demonstrated that not all RS domains are functionally equivalent in their ability to restore the function of the SR-like protein Tra2, which functions in the sex determination alternative splicing cascade (12). Our present results suggest that the high degree of sequence conservation of individual RS domains may not be directly related to their biochemical activities but rather may be related to their ability to direct subcellular localization and influence the ability of SR proteins to shuttle between the nucleus and the cytoplasm.
Role of the RS domain in splicing.
The RS domain of SF2/ASF is not required for the in vitro splicing of all pre-mRNAs (68). It is dispensable for the processing of several constitutively spliced pre-mRNAs with strong splice sites; however, it is required for the splicing of a mutant intron with a weakened polypyrimidine tract and for some, but not all, exonic splicing enhancer-dependent substrates. At least some of these RS domain-dependent substrates also require U2AF35, the small subunit of U2AF that specifically recognizes the AG dinucleotide at the 3′ splice site (22, 42). These experiments suggested that there are two pathways by which SR proteins promote splicing, one of which is independent of the RS domain. Two possible mechanisms for the latter activity of SR proteins can be envisaged. First, other portions of the protein, besides the RS domain, may mediate critical protein-protein interactions. It has been shown that RRM2 of SF2/ASF has a dominant role in both constitutive splicing and alternative splicing, suggesting that this RRM module can mediate protein-protein interactions (9, 11, 41, 59). Second, the binding of SF2/ASF to the pre-mRNA via RRMs may be sufficient to promote splicing by competing with inhibitory factors, such as hnRNPs, that antagonize SR proteins in splice site selection (15, 68).
Here, we show that the in vitro splicing of an RS domain-dependent pre-mRNA can take place when a mutant RD, RE, or RS10 domain replaces the natural domain. Thus, alternating arginine and acidic residues appear to mimic the structural and functional properties of a phosphorylated RS domain that are required for splicing in vitro. Indeed, several splicing factors possess natural RD and RE domains, sometimes in addition to their RS domains (2, 56). Interestingly, some monoclonal antibodies recognize structural features shared by several proteins with domains rich in arginines alternating with glutamates, aspartates, and/or phosphorylated serines (47).
Previous studies suggested that a dynamic phosphorylation state, i.e., both serine hyperphosphorylation and hypophosphorylation of the natural RS domain, is required during different stages of splicing and can also affect the cellular localization and recruitment of SR proteins (for a review, see reference 44). The artificial RD and RE domains analyzed here in the context of SF2/ASF do not undergo detectable phosphorylation within the remaining serines, and the fact that they can still promote splicing argues that the phosphorylation and dephosphorylation of SF2/ASF are not strictly required for splicing. We cannot rule out the possibility that some of the residues that flank the artificial RD or RE repeats may undergo cycles of phosphorylation and dephosphorylation, even though we do not see any changes in electrophoretic mobility. The suboptimal activities of the RD and RE mutant proteins could indicate that a dynamic phosphorylation state is optimal for splicing. In addition, RE and RD repeats may only imperfectly emulate some of the properties of hyperphosphorylated RS repeats, and dephosphorylation of natural RS domains may indeed be essential for splicing.
Nuclear localization of SR proteins.
In this study, we show that an RS domain consisting of 10 consecutive RS dipeptides is sufficient to confer correct nuclear localization, targeting to the nuclear speckles, and nucleocytoplasmic shuttling. We observed a good correlation between the phosphorylation of the RS domain and nuclear localization and also found that alternating positive and negative charges (arginine and aspartic or glutamic acid residues) at least partially mimic the functional properties of a phosphorylated RS domain and direct proper subcellular localization. RS domain-containing proteins do not use the typical importin α/β import receptors but rather interact with a nuclear import receptor that is specific for SR proteins and for which two nearly identical genes, TRN-SR1 and TRN-SR2, have been identified (28, 33). This SR protein-specific import receptor shows sequence similarity to the importin β/transportin family. There is a single homologous gene in the nematode Caenorhabitis elegans, termed CeTRN-SR, which is essential for proper development (37).
The subcellular localization of SR proteins is influenced by their phosphorylation state, which most likely affects both nuclear import and export. For instance, phosphorylation of the RS domain is required for TRN-SR2-mediated nuclear import of RS domain-containing proteins (33, 34). However, it is not clear whether TRN-SR1 discriminates between phosphorylated and unphosphorylated SR proteins as import substrates.
Studies of a yeast SR protein-specific kinase homolog, Sky1p, have suggested that a function of this type of kinase may be to regulate nucleocytoplasmic shuttling of SR proteins. For example, the nuclear import of human SC35 expressed in yeast cells is severely inhibited when Sky1p is deleted, and a GFP-KS mutant form of SF2/ASF that is not phosphorylated in vivo localizes to the cytoplasm (64). Also, phosphorylation by Sky1p of the shuttling protein Npl3p, a yeast hnRNP that contains eight SR or RS dipeptides, promotes its dissociation from exported messenger RNPs and also enhances its binding to the Mtr10p import receptor in vivo (18).
The RS domain NLS may essentially consist of alternating residues of opposite charges after serine phosphorylation; consistent with this idea, we showed here that alternating positive and negative charges—as in the RE and RD domains—are also able to direct nuclear localization, suggesting that these sequences can interact with the transportin-SR import system. Therefore, the direct correlation between phosphorylation of the RS domain and nuclear localization most likely reflects the requirement for alternating charges to allow interactions with the transportin-SR import system (33, 34). Thus, the RS domain NLS may behave as a regulated nuclear import signal, and regulation of the accessibility of the NLS to the transport machinery by phosphorylation is known to be a major mechanism controlling the nucleocytoplasmic transport of proteins (27). Phosphorylation of SR proteins also affects their nuclear export. It has been shown that overexpression of the SR protein kinase Clk/Sty causes a diffuse distribution of SR proteins in the nucleus, probably due to hyperphosphorylation of their RS domains (10, 52), and also results in the cytoplasmic accumulation of SF2/ASF (7). Overexpression of an SRPK2 kinase-inactive mutant also results in the cytoplasmic accumulation of SF2/ASF, apparently due to a stable interaction of SRPK2 with SF2/ASF (30).
NESs and NRSs in the RS domains of SR proteins.
The number of proteins known to shuttle between the nucleus and the cytoplasm is increasingly growing. Although the precise function(s) of nucleocytoplasmic shuttling is not understood, the emerging picture is that shuttling proteins convey information on nuclear and cytoplasmic activities within the cell (17). It is likely that shuttling proteins may have evolved to perform different functions in the nucleus and in the cytoplasm, thus constituting a group of multifunctional regulatory proteins that control gene expression in both cellular compartments (for a review, see references 54 and 61).
The recent findings that Drosophila SF2/ASF—which lacks a continuous tract of RS dipeptides at the N terminus of its RS domain—is a nonshuttling protein and that deletion of this RS tract in human SF2/ASF results in its retention in the nucleus indicated that such a tract of consecutive RS dipeptides is necessary to promote shuttling in the context of human SF2/ASF (1). Our present analysis of the human SF2/ASF RS10 variant confirms and extends these findings. The 20-amino-acid minimal RS domain is both necessary and sufficient for the protein to shuttle between the nucleus and the cytoplasm in the context of the bipartite NES that also includes RNA binding.
We also demonstrated that SC35 is restricted to the nucleus not because it lacks an NES but because it contains a dominant NRS. There is little available information about nuclear retention of proteins. Specific sequences that mediate the retention of proteins in the nucleus have been described, such as the one that directs the localization of the shuttling protein nucleolin to the nucleolus (53) or a region of approximately 200 amino acids that mediates the nuclear retention of the La protein (55). However, in none of these cases has it been shown whether these proteins have transferable NRSs capable of conferring nuclear retention on proteins that contain NESs.
An NRS has been identified in hnRNP C1, a nonshuttling hnRNP (46), supporting the idea that nonshuttling hnRNPs are confined to the nucleus due to signals that actively promote their nuclear retention. The hnRNP C1 NRS was the first sequence shown to retain NES-bearing proteins in the nucleus. It comprises a core NRS sequence of 58 amino acids that requires approximately 10 amino acids at both the amino and the carboxyl termini for full NRS function. This proline-rich NRS sequence contains clusters of basic residues, potential phosphorylation sites for casein kinase II and protein kinase C, and a potential glycosylation site (46).
Our present analysis of the shuttling properties of chimeric proteins in which domains of SC35 were fused to either hnRNP A1 or SF2/ASF determined that an NRS was present within the RS domain of SC35. We made C-terminal deletions in the RS domain of SC35 and determined that the NRS mapped to a 30-amino-acid segment at the C terminus of the RS domain, which comprises a proline-rich sequence (Fig. 5C). Interestingly, the Drosophila homolog of human SC35, which unlike its human counterpart has a short RS domain and shuttles between the nucleus and the cytoplasm, lacks this NRS sequence (Jamal Tazi, personal communication). We have also found that the human SC35 NRS sequence is portable and dominant, since it confers nuclear retention to the shuttling protein SF2/ASF (Fig. 6).
Other than the proline-rich composition, there is no discernible primary sequence similarity between the NRS sequences identified in hnRNP C1 and SC35. Both sequences are proline and serine rich and basic in character, at least in the absence of phosphorylation. The only apparent similarity between the NRS regions of SC35 and hnRNP C1 is at the level of their secondary structures: they are both predicted to form exposed loops (data not shown). The basic and serine-rich composition of the SC35 NRS may be mostly a reflection of the similar composition of the RS domain as a whole. Interestingly, SRp40, which is also retained in the nucleus and has multiple consecutive RS repeats (7), does not comprise a sequence with obvious similarity to the SC35 NRS. Most likely, nuclear retention is mediated by interactions of the NRS with nuclear components which are yet to be defined. With regard to the nuclear retention of RNA, it was recently shown that hyperedited, inosine-containing RNAs associate with a complex of three proteins that appear to be structural components of the nuclear matrix (p54nrb, PSF, and matrin 3) and mediate their nuclear retention (67).
The shuttling ability of a subset of SR proteins resembles that of hnRNP A1 and certain other hnRNPs (13, 49). Thus, different SR proteins have unique intracellular transport properties, suggesting that family members that shuttle may have roles in nuclear pre-mRNA splicing but also in mRNA transport and/or in some cytoplasmic events. It has been proposed that shuttling proteins, such as hnRNP A1, are involved in mRNA export (25), and the role of two shuttling SR proteins, SF2/ASF and 9G8, in the export of intronless mRNAs was recently described (24). SF2/ASF has also been shown to modulate mRNA stability (35); however, this effect is highly selective for a PKCI-related mRNA, and it remains to be seen whether SR proteins have a general role in controlling mRNA stability. Conversely, nonshuttling hnRNPs containing NRSs that override NESs may act to prevent the export of mRNA transcripts that have not been fully processed from the nucleus to the cytoplasm, where their translation would be detrimental to the cell. The removal of nucleus-restricted SR and hnRNPs, such as SC35, SRp40, and hnRNP C1, from messenger RNP particles is likely to be critical to allow mRNA to be exported from the nucleus. In contrast, the dissociation of shuttling SR proteins from mature mRNA in vivo occurs in the cytoplasm, where these and other proteins are presumably exchanged for messenger RNPs, some of which are exclusively cytoplasmic.
In summary, we have extensively dissected the compositions and sequences within an RS domain that are important for promoting splicing, nuclear localization, and nucleoctyoplasmic shuttling. We have also shown that the retention of nonshuttling SR proteins in the nucleus is due to the presence of a dominant NRS rather than to the absence of an NES.
Acknowledgments
We are grateful to Tom Misteli and Jeremy Sanford for critical reading of the manuscript. We thank Jamal Tazi for communicating results prior to publication.
We acknowledge support from the Medical Research Council (to D.C., E.H., and J.F.C.) and from the National Cancer Institute and the National Institute of General Medical Sciences (to J.Z., L.M., and A.R.K.).
REFERENCES
- 1.Allemand, E., R. Gattoni, H. M. Bourbon, J. Stévenin, J. F. Cáceres, J. Soret, and J. Tazi. 2001. Distinctive features of Drosophila alternative splicing factor RS domain: implication for specific phosphorylation, shuttling, and splicing activation. Mol. Cell. Biol. 21:1345-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birney, E., S. Kumar, and A. R. Krainer. 1993. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 21:5803-5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106-110. [DOI] [PubMed] [Google Scholar]
- 4.Cáceres, J. F., and A. R. Krainer. 1993. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 12:4715-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cáceres, J. F., and A. R. Krainer. 1997. Mammalian pre-mRNA splicing factors, p. 174-212. In A. R. Krainer (ed.), Eukaryotic mRNA processing. IRL Press, Oxford, United Kingdom.
- 6.Cáceres, J. F., T. Misteli, G. R. Screaton, D. L. Spector, and A. R. Krainer. 1997. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 138:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cáceres, J. F., G. R. Screaton, and A. R. Krainer. 1998. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 12:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cáceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 265:1706-1709. [DOI] [PubMed] [Google Scholar]
- 9.Chandler, S. D., A. Mayeda, J. M. Yeakley, A. R. Krainer, and X. D. Fu. 1997. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc. Natl. Acad. Sci. USA 94:3596-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colwill, K., T. Pawson, B. Andrews, J. Prasad, J. L. Manley, J. C. Bell, and P. I. Duncan. 1996. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO J. 15:265-275. [PMC free article] [PubMed] [Google Scholar]
- 11.Dauksaite, V., and G. Akusjärvi. 2002. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem. 277:12579-12586. [DOI] [PubMed] [Google Scholar]
- 12.Dauwalder, B., and W. Mattox. 1998. Analysis of the functional specificity of RS domains in vivo. EMBO J. 17:6049-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreyfuss, G., M. J. Matunis, S. Piñol-Roma, and C. G. Burd. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289-321. [DOI] [PubMed] [Google Scholar]
- 14.Eperon, I. C., D. C. Ireland, R. A. Smith, A. Mayeda, and A. R. Krainer. 1993. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 12:3607-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eperon, I. C., O. V. Makarova, A. Mayeda, S. H. Munroe, J. F. Cáceres, D. G. Hayward, and A. R. Krainer. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 20:8303-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu, X. D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 17.Gama-Carvalho, M., and M. Carmo-Fonseca. 2001. The rules and roles of nucleocytoplasmic shuttling proteins. FEBS Lett. 498:157-163. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert, W., C. W. Siebel, and C. Guthrie. 2001. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA 7:302-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graveley, B. R., and T. Maniatis. 1998. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol. Cell 1:765-771. [DOI] [PubMed] [Google Scholar]
- 21.Hanamura, A., J. F. Cáceres, A. Mayeda, B. R. Franza, and A. R. Krainer. 1998. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA 4:430-444. [PMC free article] [PubMed] [Google Scholar]
- 22.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13:302-309. [DOI] [PubMed] [Google Scholar]
- 23.Hedley, M. L., H. Amrein, and T. Maniatis. 1995. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc. Natl. Acad. Sci. USA 92:11524-11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Y., and J. A. Steitz. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7:899-905. [DOI] [PubMed] [Google Scholar]
- 25.Izaurralde, E., A. Jarmolowski, C. Beisel, I. W. Mattaj, G. Dreyfuss, and U. Fischer. 1997. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J. Cell Biol. 137:27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamison, S. F., Z. Pasman, J. Wang, C. Will, R. Lührmann, J. L. Manley, and M. A. Garcia-Blanco. 1995. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res. 23:3260-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jans, D. A., and S. Hubner. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76:651-685. [DOI] [PubMed] [Google Scholar]
- 28.Kataoka, N., J. L. Bachorik, and G. Dreyfuss. 1999. Transportin-SR, a nuclear import receptor for SR proteins. J. Cell Biol. 145:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohtz, J. D., S. F. Jamison, C. L. Will, P. Zuo, R. Lührmann, M. A. Garcia-Blanco, and J. L. Manley. 1994. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368:119-124. [DOI] [PubMed] [Google Scholar]
- 30.Koizumi, J., Y. Okamoto, H. Onogi, A. Mayeda, A. R. Krainer, and M. Hagiwara. 1999. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs). J. Biol. Chem. 274:11125-11131. [DOI] [PubMed] [Google Scholar]
- 31.Krainer, A. R., A. Mayeda, D. Kozak, and G. Binns. 1991. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66:383-394. [DOI] [PubMed] [Google Scholar]
- 32.Krämer, A. 1996. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem. 65:367-409. [DOI] [PubMed] [Google Scholar]
- 33.Lai, M. C., R. I. Lin, S. Y. Huang, C. W. Tsai, and W. Y. Tarn. 2000. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J. Biol. Chem. 275:7950-7957. [DOI] [PubMed] [Google Scholar]
- 34.Lai, M. C., R. I. Lin, and W. Y. Tarn. 2001. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. USA 98:10154-10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemaire, R., J. Prasad, T. Kashima, J. Gustafson, J. L. Manley, and R. Lafyatis. 2002. Stability of a PKCI-1-related mRNA is controlled by the splicing factor ASF/SF2: a novel function for SR proteins. Genes Dev. 16:594-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, H., and P. M. Bingham. 1991. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell 67:335-342. [DOI] [PubMed] [Google Scholar]
- 37.Longman, D., I. L. Johnstone, and J. F. Cáceres. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19:1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayeda, A., and A. R. Krainer. 1992. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell 68:365-375. [DOI] [PubMed] [Google Scholar]
- 39.Mayeda, A., and A. R. Krainer. 1999. Mammalian in vitro splicing assays. Methods Mol. Biol. 118:315-321. [DOI] [PubMed] [Google Scholar]
- 40.Mayeda, A., and A. R. Krainer. 1999. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 118:309-314. [DOI] [PubMed] [Google Scholar]
- 41.Mayeda, A., G. R. Screaton, S. D. Chandler, X. D. Fu, and A. R. Krainer. 1999. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol. 19:1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcárcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-841. [DOI] [PubMed] [Google Scholar]
- 43.Michael, W. M., M. Choi, and G. Dreyfuss. 1995. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell 83:415-422. [DOI] [PubMed] [Google Scholar]
- 44.Misteli, T. 1999. RNA splicing: what has phosphorylation got to do with it? Curr. Biol. 9: R198-R200. [DOI] [PubMed] [Google Scholar]
- 45.Misteli, T., J. F. Cáceres, J. Q. Clement, A. R. Krainer, M. F. Wilkinson, and D. L. Spector. 1998. Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J. Cell Biol. 143:297-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakielny, S., and G. Dreyfuss. 1996. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol. 134:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Neugebauer, K. M., J. A. Stolk, and M. B. Roth. 1995. A conserved epitope on a subset of SR proteins defines a larger family of pre-mRNA splicing factors. J. Cell Biol. 129:899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piñol-Roma, S., and G. Dreyfuss. 1991. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science 253:312-314. [DOI] [PubMed] [Google Scholar]
- 49.Piñol-Roma, S., and G. Dreyfuss. 1992. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature 355:730-732. [DOI] [PubMed] [Google Scholar]
- 50.Robberson, B. L., G. J. Cote, and S. M. Berget. 1990. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol. Cell. Biol. 10:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roscigno, R. F., and M. A. Garcia-Blanco. 1995. SR proteins escort the U4/U6.U5 tri-snRNP to the spliceosome. RNA 1:692-706. [PMC free article] [PubMed] [Google Scholar]
- 52.Sacco-Bubulya, P., and D. L. Spector. 2002. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt-Zachmann, M. S., C. Dargemont, L. C. Kuhn, and E. A. Nigg. 1993. Nuclear export of proteins: the role of nuclear retention. Cell 74:493-504. [DOI] [PubMed] [Google Scholar]
- 54.Shyu, A. B., and M. F. Wilkinson. 2000. The double lives of shuttling mRNA binding proteins. Cell 102:135-138. [DOI] [PubMed] [Google Scholar]
- 55.Simons, F. H., F. J. Broers, W. J. van Venrooij, and G. J. Pruijn. 1996. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp. Cell Res. 224:224-236. [DOI] [PubMed] [Google Scholar]
- 56.Staknis, D., and R. Reed. 1995. Members of a family of proteins (the RD family) detected by a U1 70K monoclonal antibody are present in spliceosomal complexes. Nucleic Acids Res. 23:4081-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarn, W. Y., and J. A. Steitz. 1995. Modulation of 5′ splice site choice in pre-messenger RNA by two distinct steps. Proc. Natl. Acad. Sci. USA 92:2504-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valcárcel, J., and M. R. Green. 1996. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem. Sci. 21:296-301. [PubMed] [Google Scholar]
- 59.van Der Houven Van Oordt, W., K. Newton, G. R. Screaton, and J. F. Cáceres. 2000. Role of SR protein modular domains in alternative splicing specificity in vivo. Nucleic Acids Res. 28:4822-4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, J., S. H. Xiao, and J. L. Manley. 1998. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 12:2222-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkinson, M. F., and A. B. Shyu. 2001. Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. Bioessays 23:775-787. [DOI] [PubMed] [Google Scholar]
- 62.Wu, J. Y., and T. Maniatis. 1993. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell 75:1061-1070. [DOI] [PubMed] [Google Scholar]
- 63.Yang, X., M. R. Bani, S. J. Lu, S. Rowan, Y. Ben David, and B. Chabot. 1994. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl. Acad. Sci. USA 91:6924-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yeakley, J. M., H. Tronchère, J. Olesen, J. A. Dyck, H. Y. Wang, and X. D. Fu. 1999. Phosphorylation regulates in vivo interaction and molecular targeting of serine/arginine-rich pre-mRNA splicing factors. J. Cell Biol. 145:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zahler, A. M., W. S. Lane, J. A. Stolk, and M. B. Roth. 1992. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 6:837-847. [DOI] [PubMed] [Google Scholar]
- 66.Zahler, A. M., K. M. Neugebauer, W. S. Lane, and M. B. Roth. 1993. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science 260:219-222. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, Z., and G. G. Carmichael. 2001. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106:465-475. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, J., and A. R. Krainer. 2000. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 14:3166-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zuo, P., and J. L. Manley. 1993. Functional domains of the human splicing factor ASF/SF2. EMBO J. 12:4727-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]