Abstract
Small nucleolar RNAs (snoRNAs) are associated in ribonucleoprotein particles localized to the nucleolus (snoRNPs). Most of the members of the box C/D family function in directing site-specific 2′-O-methylation of substrate RNAs. Although the selection of the target nucleotide requires the antisense element and the conserved box D or D′ of the snoRNA, the methyltransferase activity is supposed to reside in one of the protein components. Through protein tagging of a snoRNP-specific factor, we purified to homogeneity box C/D snoRNPs from the yeast Saccharomyces cerevisiae. Mass spectrometric analysis demonstrated the presence of Nop1p, Nop58p, Nop56p, and Snu13p as integral components of the particle. We show that purified snoRNPs are able to reproduce the site-specific methylation pattern on target RNA and that the predicted S-adenosyl-l-methionine-binding region of Nop1p is responsible for the catalytic activity.
One of the major posttranscriptional modifications that occurs on rRNA (18), tRNAs (7, 17), snRNAs (11, 30), and possibly mRNAs (5, 6), is site-specific 2′-O-methylation of the ribose moiety. In eukaryotes, with the exception of tRNAs, this modification is carried out by ribonucleoprotein particles (RNPs) containing small nucleolar RNAs (snoRNAs) belonging to the box C/D family (9). These RNAs contain two conserved sequence elements, boxes C (UGAUGA) and D (CUGA), that are positioned at the 5′ and 3′ ends of the molecules, respectively (2), and that are often flanked by short complementary regions that are thought to bring the two boxes close to each other (31, 33). Box C/D snoRNAs may contain imperfect copies of the boxes C and D, referred to as box C′ and D′, that are located internally (12). All of these conserved sequence elements function as protein-binding sites and are essential for snoRNA stability, biogenesis, subcellular localization, and function (27, 29, 34)
The methylation guide snoRNAs direct site-specific modification by base pairing with the substrate RNA through a region, 10 to 20 nucleotides long, immediately upstream to the box D or D′ (2). It was found, initially by sequence analysis and subsequently by genetic studies, that a specific measuring mechanism is utilized for specifying the appropriate site of modification. The residue in the target RNA, which is located in the hybrid region opposite the fifth nucleotide upstream from the box D or D′ of the snoRNA, is selected for ribose methylation (12).
Although the substrate selectivity requires RNA-RNA interaction, indirect observations suggest that the methyltransferase activity is carried out by one of the snoRNP proteins.
Four proteins that are common components of box C/D snoRNPs have been characterized: Nop1p, Nop58p, Nop56p, and Snu13p (10, 25, 33, 35); all of these proteins are evolutionarily conserved (19, 21) and are essential for the function of the particle (15, 16, 33). Nop1p, Snu13p, and Nop58p are required for snoRNA stability and accumulation, suggesting that they form the basic core complex with the snoRNA, whereas Nop56p has no effect on snoRNAs stability or accumulation and depends on Nop1p for its association with the snoRNP (15, 16).
Several evidences indicated that, of the four components, the most likely candidate to be the methyltransferase is the Nop1p component: (i) the search for conserved methyltransferase motifs in a Saccharomyces cerevisiae database allowed the identification of an S-adenosyl-l-methionine (AdoMet)-binding domain in the Nop1p factor (20); (ii) resolution of the crystal structure of the Nop1p homologue from the archaebacterium Methanococcus jannaschii revealed the presence of a methyltransferase-like domain in the C-terminal region (32); and (iii) mapping of the temperature-sensitive mutations found in Nop1p revealed that two mutations clustered in the core of the methyltransferase-like domain and specifically inhibited rRNA methylation (28).
We show here that purified box C/D snoRNPs are able to reproduce methyltransferase activity and the specificity of substrate recognition and modification. We also show that Nop1p binds AdoMet (S-adenosyl-l-methionine) and that this binding is necessary for the methylation reaction.
MATERIALS AND METHODS
Plasmids and strains.
Strains YAF2 and YAF2-U24 were previously described (23). Strains YSG1-3 (MATa nop1.3::URA3 NOP56::TAP::HIS3 leu2) and YSG1-5 (MATa nop1.5::URA3 NOP56::TAP::HIS3 leu2) were derived from strains D335 and D337 (kindly provided by D. Tollervey), respectively. The genotype of strain CH1462 is MATα ade2 ade3 leu2 ura3 his3 can1-100.
C/D snoRNP purification.
Tandem affinity purification (TAP) tagging and purification was carried out as described by Rigaut et al. (24). Copurified RNAs were recovered from the eluate of the immunoglobulin G column by perchloric acid extraction and ethanol precipitation. snoRNA molecules were identified by Northern hybridization. The untagged isogenic strain (CH1462) was utilized as control. We used 10 liters of yeast culture for snoRNP purification and mass spectrometry analysis. Proteins were recovered from the final eluate of the calmodulin column by trichloroacetic acid precipitation and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 14.5% polyacrylamide gels. Proteins were visualized by Coomassie blue staining. Bands of interest were excised from the gel, reduced, alkylated, and in-gel digested with trypsin (26). Of the total digest solution, 0.3 μl of the supernatant of each band was crystallized with the dried droplet technique with α-cyano-4-hydroxycinnamic acid as the matrix and then analyzed by peptide mass mapping on a Voyager-STR (PE-Biosystems, Framingham, Mass.) matrix-assisted laser desorption ionization-time of flight mass spectrometer. Proteins were unambiguously identified by searching a comprehensive no redundant protein database by using the program ProFound (36).
In vitro methylation assay.
For the determination of in vitro activity, 2 liters culture of the different strains were subjected to TAP fractionation and snoRNP purification. The oligonucleotides oligo1436, 1436-met, and 1437-met were purchased from Dharmacon Research, Inc. (Boulder, Colo.); their sequences correspond to the yeast 25S rRNA region between nucleotides 1425 and 1445.
A substrate 1,436 nucleotides long was transcribed in vitro by using T7 polymerase with cold nucleoside triphosphates from a DNA template obtained by PCR amplification of the corresponding region of yeast 25S ribosomal DNA included between nucleotides 1363 and 1446 and (oligonucleotides 25Sa [5′-GGATCCAGCCUAGACCGUAAGGUC-3′] and 25Sb [5′-GGATCCCTACCTCCTTGATCTGCACT-3′]) and cloned into the BamHI site of the pBluescript vector. A substrate 1,436 nucleotides long was obtained by inverse PCR with the oligonucleotides Fw (5′-UUGGUGGUAGGGATCCACTA-3′) and Rev (5′-TGCACTAGAGGCCGTTCGA-3′). Reactions were carried out in 20 μl in the presence of 1 pmol of substrate RNA, 1 mM dithiothreitol, 5 mM MgCl2, 70 mM NaCl, 10 mM Tris-HCl (pH 7.5), 20 U of RNase inhibitor, 0.7 μCi of 3H-labeled AdoMet, and up to 0.2 pmol of purified snoRNP particles. Incubations were allowed to proceed for 1 h at 30°C. RNA was extracted and loaded on urea-15% polyacrylamide gel or urea-6% polyacrylamide gels and exposed at −70°C to Biomax MS films with a Biomax intensifying screen (Transcreen LE; Kodak), which provides a maximum speed for 3H.
In vitro [3H]AdoMet-binding assay and Western blotting.
This procedure was adapted from a previously reported method (23). Box C/D particles were purified as described above and dialyzed against AdoMet-binding buffer (50 mM Tris-Cl [pH 8], 100 mM NaCl, 2 mM dithiothreitol). Then, 5.5 μCi of [3H]AdoMet (75 Ci/mmol; NEN) was added to 10 μl of purified snoRNPs. The tubes were placed in ice, with the caps open, under a transilluminator table (six 15-W tubes) that was held upside down at 4 cm from the top of the tubes. The samples were UV irradiated for 30 min at 245 nm, analyzed as described above on an SDS-PAGE gel, transferred to a nitrocellulose membrane, and exposed at −70°C to Biomax MS films with an intensifying screen (Transcreen LE; Kodak). The same membrane was analyzed by Western blotting. Western blotting was performed as described elsewhere (23). Membrane was saturated in Tris-buffered saline, 0.1% Tween 20, and 5% nonfat milk. Antibodies were incubated in the same medium for 1 h at room temperature. Primary antibody (monoclonal antibody anti-Nop1p) was added at a 1/100 dilution. Anti-mouse secondary antibody coupled to horseradish peroxidase was obtained from Sigma.
RESULTS
Purification of box C/D snoRNPs by the TAP method.
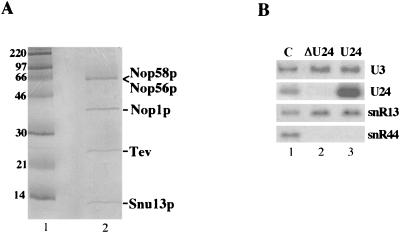
To test whether site-specific methyltransferase activity could be reproduced with purified particles, we undertook the purification of box C/D snoRNPs through the TAP procedure conceived by Seraphin and coworkers (24). The Nop56p protein, which is an integral component of box C/D particles (10), was epitope tagged by the insertion of the TAP tag at the 3′ end of the NOP56 gene (strain YAF2 [8]). This construction was under the control of the endogenous NOP56 promoter that ensures physiological levels of expression. The TAP tag modification was carried out in a genetic background in which the endogenous U24 snoRNA coding sequence was deleted (13). In this strain the U24-dependent rRNA methylation is abolished (13). Figure 1A shows the protein pattern obtained from massive preparation of TAP-tagged snoRNPs from the YAF2 strain. The purified proteins were separated on a SDS-PAGE gel, the bands were visualized by Coomassie blue staining, excised, and subjected to in-gel trypsin digestion; the eluted fragments were subjected to mass spectrometry peptide sequencing (see Materials and Methods). The identified products are reported on the side of the gel, besides the three already well-characterized snoRNP components (Nop1p, Nop58p, and Nop56p), a fourth component, initially identified as an U3 snoRNP-associated factor and subsequently shown to be a more general box C/D-specific factor (33), was also detected. This protein, Snu13p, was demonstrated by biochemical and genetic analyses to be a more general box C/D-specific factor (33). Our purification procedure indicates that Snu13p can be efficiently copurified with the other snoRNP factors on non-U3 snoRNP. The proteins present in substoichiometric amounts were identified as derivative products of the Nop1, Nop56, and Nop58 proteins. The same protein pattern was obtained with strain YAF2 transformed with plasmid GAL/HG-U24 (8) carrying the entire transcriptional unit of the U24 snoRNA host gene (1) under the control of the GAL1 promoter (not shown). In this strain (YAF2-U24), the U24-dependent modification of rRNA is restored (8). Northern analysis of the RNA content of the particles purified from the two strains revealed that they contain box C/D snoRNAs, whereas H/ACA box snoRNAs are not present; in addition, the data confirm that U24 is absent in the YAF2 strain (Fig. 1B).
FIG. 1.
Characterization of box C/D snoRNP components. (A) Coomassie blue staining of a SDS-polyacrylamide gel: lane 1, marker; lane 2, TAP-purified box C/D snoRNPs. The proteins are indicated on the side. The comigration between Nop58p and Nop56p is due to the increase in size of the tagged Nop56p protein that still carries the calmodulin-binding peptide of the TAP tag (24). The Tev band identifies contaminating Tev protein utilized in the purification procedure. (B) RNA was extracted from the control CH1462 yeast strain (C, lane 1) or from the pellets of immunoprecipitations of Nop56-TAP-associated RNAs from the YAF2 (ΔU24, lane 2) and the YAF2-U24 (U24, lane 3) strains. Northern analysis was performed with probes specific for U3, U24, and snR13 box C/D snoRNAs and for the snR44 box H/ACA snoRNA.
Purified box C/D snoRNPs direct site-specific methylation in vitro.
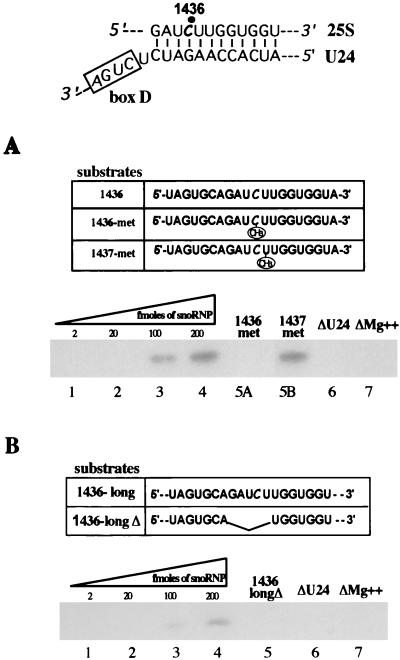
Purified particles were tested in vitro for the ability to direct site-specific methylation utilizing the transfer of tritiated CH3 groups from AdoMet to different substrate RNAs as an assay. Figure 2A (lanes 1 to 4) shows that box C/D snoRNPs purified from strain YAF2-U24 are able to transfer tritiated methyl groups to the oligo1436, which contains one of the sites modified by the U24 snoRNA (site at position +1436 of rRNA [13]). No radioactivity is detected if 1436-met is utilized as substrate (lane 5A); this oligonucleotide contains the C1436, substrate for the modification, which is already methylated by the manufacturer at the 2′-O position. The presence of a methylated group per se does not inhibit the transfer reaction to the oligonucleotide, since a control substrate (1437-met), which contains a 2′-O-methyl group in the nucleotide 3′ to the C1436, undergoes methylation (lane 5B). No activity is found with snoRNPs purified from strain YAF2, depleted of the U24 snoRNA (ΔU24, lane 6) and when Mg2+ was omitted from the reaction (lane7). Altogether, these results indicate that the methyltransferase activity observed depends on the presence of U24 snoRNA and that it is site specific. Figure 2B (lanes 1 to 4) shows that snoRNPs purified from strain YAF2-U24 are also able to transfer tritiated methyl groups onto the C1436 when this site is embedded in an 83-nucleotide RNA context (substrate 1436-long). As control, an RNA substrate carrying five nucleotide substitutions in the U24 pairing region (1436-longΔ) is not methylated (longΔ, lane 5), indicating that specific RNA-RNA recognition is necessary for the transfer reaction. snoRNPs purified from the YAF2 strain, which is devoid of U24 snoRNA, are totally unable to direct methylation of substrate RNAs (ΔU24, lane 6). Given the same number of substrate molecules, lower methylation efficiency of the longer RNA, in comparison to the short oligonucleotide, was observed. This is likely to be due to the higher sequence complexity of longer RNA substrate that can interfere with efficient snoRNA recognition. It is conceivable that in vivo helicases must cooperate with these particles by allowing local unfolding of the RNA substrates.
FIG. 2.
In vitro methylation activity of box C/D snoRNPs. One of the two antisense elements present in U24 snoRNA (13) and the corresponding rRNA target are indicated above. (A and B) In panel A, oligo1436, 1436-met, and 1437-met were utilized as substrates, whereas in panel B methylation was analyzed on 1436-long and 1436-longΔ RNAs. Lanes 1 to 4: reactions with 2, 20, 100, and 200 fmol of box C/D snoRNPs purified from the YAF2-U24 strain. A total of 200 fmol of snoRNPs from this strain was also reacted with oligo1436-met (panel A, lane 5A), 1437-met (panel A, lane 5B), and the 1436-longΔ substrate (panel B, lane 5). Lanes 6, reactions with 200 fmol of snoRNPs purified from the YAF2 strain (ΔU24); lanes 7, same as lanes 4 but without Mg2+. RNA was extracted and run on urea-15% polyacrylamide (A) or urea-6% polyacrylamide (B) gels.
Box C/D snoRNP methyltransferase activity requires a functional Nop1p protein.
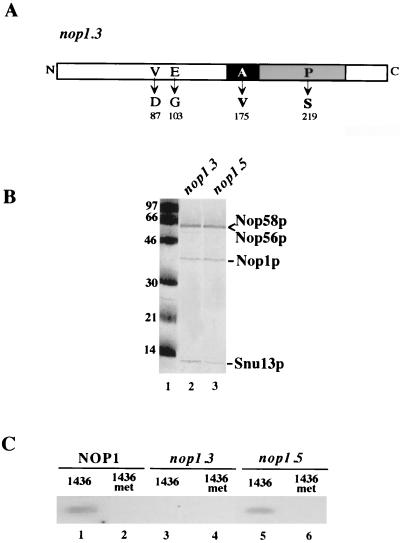
Nop1p was previously indicated as the putative methyltransferase (20, 32), but no direct biochemical evidence of such an activity, with either the purified protein (32) or snoRNPs from cell extracts, was ever reported. A number of temperature-sensitive mutants that affect rRNA maturation have been isolated in Nop1p (28). One in particular, the nop1.3 allele, displays a methyltransferase-deficient phenotype; this mutant contains four amino acid substitutions. Tollervey et al. (28) reported that the mutations causing temperature-sensitive growth could only be attributed to two of the four amino acid substitutions: P219S and A175V (see schematic representation in Fig. 3A). These two mutations are in the putative methyltransferase domain (28); P219 corresponds to a highly conserved proline, while A175 maps in the α1-β1 loop motif that, in all known methyltransferases, is important for AdoMet interaction. According to Wang et al. (32), the latter mutation should interfere with the AdoMet-binding site. We took advantage of our in vitro assay in order to test whether snoRNPs purified from the strain carrying the nop1.3 mutation (28) are able to carry out the methyltransferase reaction; as a control, snoRNPs were purified from the strain carrying the nop1.5 mutation (28). This temperature-sensitive mutant is impaired in rRNA processing but has wild-type rRNA methylation activity (28). In order to do this, the TAP tag was introduced in the NOP56 gene of both mutant strains, giving rise to YSG1-3 and YSG1-5 strains. In both of them the overexpression of U24 snoRNA was obtained by transformation with the GAL1-U24 plasmid (8). The two strains were grown at permissive conditions; TAP purification of snoRNPs was carried out as indicated above, and the purified proteins were separated by SDS-PAGE. As shown in Fig. 3B, the four snoRNP-specific factors are all present in the purification of YSG1-3 and YSG1-5 strains; this indicates that the Nop1p mutations do not affect particle assembly and stability. The levels of snoRNAs tested in both mutant strains were previously shown to be unchanged, as well as the overall stability of the snoRNPs (28). Figure 3C shows that box C/D snoRNPs purified from the strain carrying the nop1.5 allele are able to methylate the substrate oligo1436 (lane 5) similarly to the wild type (lane 1); on the contrary, box C/D particles purified from the strain carrying the nop1.3 mutation are not (lane 3). Since the relevant mutations of the nop1.3 allele fall in the putative methyltransferase domain, these data suggest a direct role of Nop1p protein in the methylation reaction.
FIG. 3.
In vitro methylating activity of snoRNPs from wild-type and NOP1 mutant strains. (A) The amino acid sequence of NOP1 is schematically represented as a box ranging from residue 1 (N terminus) to residue 327 (C terminus). The mutated amino acids and their corresponding position numbers within the Nop1p are shown for the temperature-sensitive nop1.3 allele. Tollervey et al. (28) attributed to A175V and P219S the temperature-sensitive growth defect. Both mutations are localized in the putative methyltransferase domain (gray box). The black box containing the A175V substitution correspond to the motif that interact with the AdoMet. (B) Coomassie blue staining of an SDS-14.5% polyacrylamide gel loaded with marker (lane 1) and with TAP-purified box C/D snoRNPs from strains carrying the nop1.3 (lane 2) and nop1.5 (lane 3) mutant alleles. (C) A total of 200 fmol of snoRNPs purified from the YAF2-U24 strain (lanes 1 and 2) or from the strains carrying the nop1.3 (lanes 3 and 4) and nop1.5 (lanes 5 and 6) alleles was incubated with oligo1436 (lanes 1436) or with the methylated version of it (lanes 1436-met). RNA was extracted and run on a urea-6% polyacrylamide gel.
As control, we tested the ability of the particles purified from the three strains to carry out methylation activity on the 1436-met that contains the nucleotide substrate of the modification, which is already methylated by the manufacturer at the 2′-O position (1436-met). Figure 3 (lanes 2, 4, and 6) shows that the substrate is not methylated in all cases.
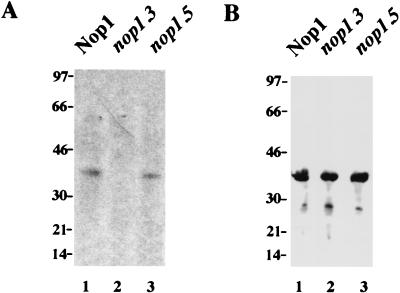
Since sequence and structural data suggested that Nop1p is a methyltransferase (20, 32), we tested whether Nop1p was able to bind in vitro [3H]AdoMet. Box C/D snoRNPs purified from the YAF2, YSG1.3, and YSG1.5 strains were incubated with [3H]AdoMet for 5 min and cross-linked by using UV light irradiation, as described by Pintard et al. (23). Proteins were resolved on a SDS-PAGE gel, transferred to a nitrocellulose membrane and exposed to autoradiography for the presence of tritiated molecules. Figure 4A shows that only one protein, corresponding by size to Nop1p, was revealed after autoradiography when purified particles from wild-type (lane Nop1) and YSG1-5 (lane nop1.5) strains were utilized in this test. On the contrary, no radioactive signal was detected with snoRNPs derived from the YSG1-3 strain (lane nop1.3). Interestingly, the nop1.5 allele was able to bind [3H]AdoMet with the same efficiency as the wild-type protein. Western blotting, performed with Nop1-specific antibodies on the same membrane, allowed us to confirm unambiguously the identity of the protein of interest (Fig. 4B). Western blotting analysis, as well as protein staining, showed that the amount of Nop1p in the wild-type allele and in the nop1.5 and nop1.3 alleles is the same (Fig. 3B and 4B); the lower bands correspond to Nop1p degradation products. When Nop1p was not treated with the UV light, no signal was visible, indicating that protein labeling was not formed in the absence of the UV cross-linking (data not shown). From these data we concluded that Nop1p is able to bind specifically in vitro to [3H]AdoMet and that the binding depends from the integrity of the region considered putative AdoMet interacting domain.
FIG. 4.
UV cross-linking with [3H]AdoMet. Box C/D particles were purified from strains YAF2 (lanes 1), YSG1.3 (lanes 2), and YSG1.5 (lanes 3). The samples were incubated with [3H]AdoMet before being UV cross-linked for 30 min; they were subsequently subjected to SDS-PAGE and then transferred to a nitrocellulose membrane. (A) The membrane was exposed for autoradiography with an intensifying screen to reveal the tritiated molecules. (B) The same membrane was analyzed by Western blotting with a Nop1-specific antibody.
DISCUSSION
The results presented in this study show that purified box C/D snoRNPs are able to direct specific 2′-O-methylation of substrate RNA: the in vitro activity reproduced the in vivo reaction being dependent on correct RNA-RNA pairing and being able to select the specific target nucleotide.
The 2′-O-methylation of the ribose moiety is one important cellular process involved in the maturation of almost all classes of RNAs. Although protein-only enzymes have been shown to carry out this modification in tRNAs (7, 17), for rRNAs, snRNAs, and possibly mRNAs, site-specific methylation requires the intervention of RNP complexes containing small RNAs, i.e., the box C/D snoRNPs (12). Genetic data indicated that in these RNPs the RNA component is necessary to confer the site specificity of substrate modification, whereas the catalytic activity is supposed to reside in one of the protein components. More than 100 different snoRNP complexes have been identified so far in eukaryotic cells (27). Although they share a common set of proteins, they differ in the RNA component: each individual particle containing its specific snoRNA. This feature allows the recognition and modification of a large repertoire of substrates without changing the catalytic component. Thus, box C/D snoRNPs represent quite an interesting example of how a specific enzymatic function has evolved in response to the increasing number of substrates to be modified.
The old origin and the relevance of the 2′-O-methylation process are testified by the evolutionary conservation of box C/D snoRNP components and in particular Nop1p from archaebacteria to mammals (19, 21). Wang and collaborators have reported the crystal structure of the Nop1p homologue from the hyperthermophile M. jannaschii. The protein contains a folding domain that is structurally homologous to the catalytic domain common to many AdoMet-dependent methyltransferases (32). This domain includes a short consensus amino acid sequence, “S-adenosyl-l-methionine-binding motif,” that in all reported methyltransferase enzymes is essential for AdoMet interaction (32). The region containing the AdoMet-binding motif of archaeal Nop1p is highly conserved at the primary amino acid sequence level in all organisms, showing 60% identity (80% similarity [14, 32]). Such high sequence identity among Nop1p homologues suggests that they all contain a methyltransferase domain.
Even if the presence of methyltransferase-like domain in Nop1p suggests that it could be an enzyme, attempts in reproducing in vitro methylation with the protein alone were unsuccessful (32; S. Galardi, unpublished data). Interestingly, the protein-only methyltransferases of rRNAs from Escherichia coli and yeast mitochondria, which recognize their targets without a guide RNA, are active in vitro only when their substrate RNA is assembled with proteins into specific RNPs (3, 4, 23). Consequently, it is very likely that Nop1p is able to carry out specific RNA methylation only when it is assembled into a functional snoRNP. The results presented in here suggest that this might be the case; the TAP-purified native particle is able to reproduce both the methyltransferase activity and the specificity of substrate recognition. The purified particles reveal only the presence of the well-known box C/D protein components, i.e., Nop1p, Nop58p, Nop56p, and Snu13p, indicating that the reaction does not require any other factor. In our in vitro assay, we utilized snoRNPs purified from a strain overexpressing the U24 snoRNA; in these cells the U24 snoRNA is ca. 15-fold more abundant than the endogenous transcript. Our data indicate that this overexpression is essential in order to detect activity with our procedure. In fact, when the methylation assay was carried out with comparable amounts of snoRNPs purified from a strain carrying only the chromosomal copy of the U24 snoRNA, no activity was observed (not shown).
The importance of the Nop1p component in the methylation reaction was indicated by the fact that activity was abolished when snoRNPs were purified from the nop1.3 mutant strain, which has a methylation-deficient phenotype, whereas it was unaffected with particles obtained from a strain carrying the nop1.5 mutation that affects rRNA processing (28). Interestingly, the nop1.3 allele contains a mutation that maps in the putative AdoMet-binding motif (Fig. 3A). Indeed, we showed that Nop1p protein from purified snoRNPs was able to cross-link [3H]AdoMet and that this interaction is abolished in the nop1.3 allele. All of these data suggest a direct role of the Nop1p in the methylation reaction and show that the predicted AdoMet-binding region, conserved among methyltransferases, is involved in the catalytic activity of Nop1p.
In conclusion, we show for the first time the ability to reproduce in vitro the site-specific methylation activity of purified box C/D snoRNPs in eukaryotes. Recent work by Omer et al. (22) showed that, in Archaea, site-specific methylation activity could be reproduced by in vitro reconstitution of snoRNPs. These particles contained a specific snoRNA and the C/D box proteins aFIB, aNOP56, and L7a, shown to be the archaeal counterparts of the eukaryotic proteins Nop1p, Nop56p/Nop58p, and Snu13p, respectively. Altogether, these data corroborate previous observations on the evolutionary conservation of the mechanism and basic components required for site-specific 2′-O-methylation in organisms distantly related such as Archaea and eukaryotes.
Acknowledgments
We are particularly grateful to Laura Nicolini of Istituto Superiore di Sanita for large-scale growth of yeast strains. We thank T. Kiss and D. Tollervey for kindly providing strains, L. Pintard for helpful suggestions in UV cross-linking experiments, and M. Arceci and G. Ricci for skillful technical help.
This work was partially supported by grants from MURST (Biotechnology Program L.95/95, Biotecnologie, PRIN 40%, and Centro di Eccellenza BEMM), from the Ministero della Sanità (Progetto AIDS), and from CNR (Target Project on Biotechnology and Tecnologie di base della post-genomica).
REFERENCES
- 1.Bachellerie, J. P., M. Nicoloso, L. H. Qu, B. Michot, M. Caizergues-Ferrer, J. Cavaille, and M. H. Renalier. 1995. Novel intron-encoded small nucleolar RNAs with long sequence complementarities to mature rRNAs involved in ribosome biogenesis. Biochem. Cell Biol. 73:835-843. [DOI] [PubMed] [Google Scholar]
- 2.Bachellerie, J. P., and J. Cavaille. 1997. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 22:257-261. [DOI] [PubMed] [Google Scholar]
- 3.Bugl, H., E. B. Faumann, B. L. Staker, F. Zheng, S. R. Kushner, M. A. Saper, J. C. Bardwell, and U. Jakob. 2000. RNA methylation under heat shock control. Mol. Cell 6:349-360. [DOI] [PubMed] [Google Scholar]
- 4.Caldas, T., E. Binet, P. Bouloc, A. Costa, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23S ribosomal RNA methyltransferase. J. Biol. Chem. 275:16414-16419. [DOI] [PubMed] [Google Scholar]
- 5.Cavaillé, J., K. Buiting, M. Kiefmann, M. Lalande, C. I. Brannan, B. Horsthemke, J. P. Bachellerie, J. Brosius, and A. Hüttenhofer. 2000. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl. Acad. Sci. USA 97:14311-14316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De los Santos, T., J. Schweizer, C. A. Rees, and U. Francke. 2000. Small evolutionarily conserved RNA, resembling C/D box small nucleolar RNA, is transcribed from PWCR1, a novel imprinted gene in the Prader-Willi deletion region, which is highly expressed in brain. Am. J. Hum. Genet. 67:1067-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Droogmans, L., E. Haumont, S. de Henau, and H. Grosjean. 1986. Enzymatic 2′-O-methylation of the wobble nucleoside of eukaryotic tRNAPhe: specificity depends on structural elements outside the anticodon loop. EMBO J. 5:105-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatica, A., M. Morlando, and I. Bozzoni. 2000. Yeast snoRNA accumulation relies on a cleavage dependent/polyadenylation independent 3′ processing apparatus. EMBO J. 19:6218-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filipowicz, W., P. Pelczar, V. Pogacic, and F. Dragon. 1999. Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim. Pol. 46:377-389. [PubMed] [Google Scholar]
- 10.Gautier, T., T. Berges, D. Tollervey, and E. C. Hurt. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jady, B. E., and T. Kiss. 2001. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 20:541-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiss, T. 2002. Small nucleolar RNA: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 13.Kiss-László, Z., Y. Henry, J. P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077-1088. [DOI] [PubMed] [Google Scholar]
- 14.Koonin, E. V., R. L. Tatusov, and K. E. Rudd. 1995. Sequence similarity analysis of Escherichia coli proteins: functional and evolutionary implication. Proc. Natl. Acad. Sci. USA 92:11921-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafontaine, D. L. J., and D. Tollervey. 1999. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNP stability. RNA 5:455-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lafontaine, D. L. J., and D. Tollervey. 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 20:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto, T., K. Nishikura, H. Hori, T. Ohta, K. Miura, and K. Watanabe. 1990. Recognition sites of tRNA by a thermostable tRNA(guanosine-2′-)-methyltransferase from Thermus thermophilus HB27. J. Biochem. 107:331-338. [DOI] [PubMed] [Google Scholar]
- 18.Maxwell, E. S., and M. J. Fournier. 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 64:897-934. [DOI] [PubMed] [Google Scholar]
- 19.Newman, D. R., J. F. Kuhn, G. M. Shanab, and E. S. Maxwell. 2000. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 6:861-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niewmierzycha, A., and S. Clarke. 1999. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. J. Biol. Chem. 274:814-824. [DOI] [PubMed] [Google Scholar]
- 21.Omer, A. D., T. M. Lowe, A. G. Russell, H. Ebhardt, S. R. Eddy, and P. P. Dennis. 2000. Homologs of small nucleolar RNAs in Archaea. Science 288:517-522. [DOI] [PubMed] [Google Scholar]
- 22.Omer, A. D., S. Ziesche, H. Ebhardt, and P. P. Dennis. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. USA 99:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pintard, L., D. Kressler, and B. Lapeyre. 2000. Spb1p is a yeast nucleolar protein associated with Nop1p and Nop58p that is able to bind S-adenosyl-l-methionine in vitro. Mol. Cell. Biol. 20:1370-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 10:1030-1032. [DOI] [PubMed] [Google Scholar]
- 25.Schimmang, T., D. Tollervey, H. Kern, R. Frank, and E. C. Hurt. 1989. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 8:4015-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 27.Smith, C. M., and J. A. Steitz. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89:669-672. [DOI] [PubMed] [Google Scholar]
- 28.Tollervey, D., H. Lehtonen, R. Jansen, H. Kern, and E. C. Hurt. 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 72:443-457. [DOI] [PubMed] [Google Scholar]
- 29.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 30.Tycowski, K. T., Z. H. You, P. J. Graham, and J. A. Steitz. 1998. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol. Cell 5:629-638. [DOI] [PubMed] [Google Scholar]
- 31.Vidovic, I., S. Nottrott, K. Hartmuth, R. Luhrmann, and R. Ficner. 2000. Crystal structure of the spliceosomal 15.5 Kd protein bound to a U4 snRNA fragment. Mol. Cell 6:1331-1342. [DOI] [PubMed] [Google Scholar]
- 32.Wang, H., D. Boisvert, K. K. Kim, R. Kim, and S.-H. Kim. 2000. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 Å resolution. EMBO J. 19:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watkins, N. J., V. Segault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Luhrmann. 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 34.Weinstein, L. B., and J. A. Steitz. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11:378-384. [DOI] [PubMed] [Google Scholar]
- 35.Wu, P., S. Brockenbrough, A. Metcalfe, S. Chen, and J. P. Aris. 1998. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J. Biol. Chem. 273:16453-16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, W., and B. T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72:2482-2489. [DOI] [PubMed] [Google Scholar]