Abstract
The Saccharomyces cerevisiae Ccr4-Not complex is a global regulator of transcription that is thought to regulate TATA binding protein (TBP) function at certain promoters specifically. In this paper, we show interactions between the essential domain of Not1p, which interacts with Not4p and Not5p, and the N-terminal domain of yTAF1. We isolated a temperature-sensitive nonsense allele of TAF1, taf1-4, which is synthetically lethal at the permissive temperature when combined with not4 and not5 mutants and which produces high levels of a C-terminally truncated yTAF1 derivative. Overexpression of C-terminally truncated yTAF1 is toxic in not4 or not5 mutants, whereas overexpression of full-length yTAF1 suppresses not4. Furthermore, mutations in the autoinhibitory N-terminal TAND domain of yTAF1 suppress not5, and the overexpression of similar mutants does not suppress not4. We find that, like Not5p, yTAF1 acts as a repressor of stress response element-dependent transcription. Finally, we have evidence for stress-regulated occupancy of promoter DNA by Not5p and for Not5p-dependent regulation of yTAF1 association with promoter DNA. Taken together with our finding that Not1p copurifies with glutathione S-transferase-yTaf1 in large complexes, these results provide the first molecular evidence that the Ccr4-Not complex might interact with yTAF1 to regulate its association at promoters, a function that might in turn regulate the autoinhibitory N-terminal domain of yTAF1.
Transcription initiation of protein-coding genes by RNA polymerase II involves a large number of general transcription factors that position the polymerase specifically on the core promoter (for reviews, see references 32 and 47). The recruitment of the TATA binding protein (TBP) is the first step in the assembly of a functional preinitiation complex. TBP is a component of the TFIID multiprotein complex (for a review, see reference 6) that in the yeast Saccharomyces cerevisiae consists of 14 additional TBP-associated factors (TAFIIs) (39, 52, 53). It specifically recognizes TATA elements present in the promoters of many genes transcribed by RNA polymerase II. For promoters that lack a canonical TATA sequence (referred to as TATA-less), TAFIIs are likely to participate in the recognition of the promoter. First, certain TAFIIs contact promoter elements such as the initiator (7) and the downstream promoter element (5), and it is possible that other contacts between the promoter and TAFIIs are still to be discovered. Second, experiments with both mammalian systems (45, 58, 59) and yeast (54) have suggested the importance of TFIID rather than TBP in promoter recognition.
TAFIIs were first thought to be exclusively within the TFIID complex, but recent work has demonstrated the existence of multiple complexes that carry TAFIIs (21, 38, 46, 60). In the yeast Saccharomyces cerevisiae, the only two complexes identified so far are TFIID (49) and SAGA (21; for reviews, see references 22 and 55). In turn, the only well-characterized form of TBP recruited to promoters is TFIID. However, while TBP is generally required for transcription by RNA polymerase II and while the level of TBP occupancy at promoters is strongly correlated with transcriptional activity, recent work has suggested that TBP can be brought to promoters in forms other than TFIID (29). Indeed, in contrast to TBP, TAFIIs are underrepresented at many promoters. It is unclear in which other form(s) TBP may be recruited to promoters. TBP is not a component of SAGA, although it is likely that SAGA can interact with TBP through its Spt3p component (18), and the importance of Spt3p for recruitment of TBP at certain promoters has been documented (17). Furthermore, it is known that SAGA is recruited to promoters by activator proteins (16). Other proteins that can interact with TBP include NC2 (20, 24, 40) and Mot1p (1, 8), initially characterized as repressors of transcription, which could nevertheless potentially play a positive role in recruiting TBP at some promoters. Moreover, the occupancy of promoters by the alpha subunit of NC2 has recently been shown to correlate well with transcriptional activity (19).
Why different forms of TBP are brought to promoters, how this may be regulated, and how this affects transcription initiation at given core promoters are open questions. The role of TFIID has been addressed by several approaches. First, a TBP mutant apparently defective in TFIID formation has only specific effects on transcription in vivo (50). Second, yTAF1 (previously called yTafII130p/yTafII145p), a TFIID-specific TAFII thought to be a core subunit of TFIID (for a review, see reference 6), apparently only functions at a subset of all genes. This was determined by microarray analyses at high temperature using a strain carrying a mutation in the TAF1 gene conferring temperature sensitivity (23). However, the analysis of a variety of temperature-sensitive TAF1 mutants has suggested that different target genes can be revealed by analyzing different mutants (56). Other experiments have shown that transcriptional activation by many activators does not seem to require yTAF1 (42) and that the specificity associated with yTAF1 occurs at the core promoter (54). This may be related to the ability of the very N-terminal domain of yTAF1 (TAND domain) to interact with TBP and thereby inhibit its association with DNA (3, 26, 27). Third, individual depletion of yTAF1 and three other TFIID-specific TAFIIs, namely, yTAF13 (yTafII19p), yTAF11 (yTafII40p), and yTAF7 (yTafII67), has demonstrated a common decrease in transcripts originating from promoters that lack canonical TATA elements (HIS3 and TRP3) (42, 43). Taken together, these results might suggest a particular role for TFIID in recruiting TBP to core promoters that lack a canonical TATA sequence.
The NOT genes were isolated by mutations that increase transcription of the TATA-less promoter of the S. cerevisiae HIS3 gene (9, 10, 44). The five Not proteins are associated with the Ccr4 and Caf1 proteins (35) in 1.2- and 2-MDa complexes (37) referred to as Ccr4-Not complexes. Ccr4p and Caf1p are required for nonfermentative gene expression but also participate in other cellular processes, including mRNA deadenylation (11, 13-15, 57). They interact with the N-terminal region of Not1p (amino acids 1 to 1318), while the Not2 to -5 proteins interact with the C-terminal essential region of Not1p (amino acids 1319 to 2108) (4, 37). Dhh1p, a putative RNA helicase of the decapping complex, is also associated with the Ccr4-Not complex via the N-terminal domain of Not1p (36).
Taken together, all the present results show that, whereas the Ccr4-Not complex might regulate mRNA degradation, it is a global regulator of transcription that affects genes positively and negatively in vivo. For instance, recent work demonstrated that transcription of stress response element (STRE)-dependent genes was increased in not mutants, but the stability of STRE-dependent transcripts was not affected (34). The Ccr4-Not complex has the characteristic of preferentially repressing core promoters that lack a canonical TATA sequence. This characteristic has led to the proposition that the Ccr4-Not complex regulates TFIID function. In agreement with such a model, Not1p, the only component of the Ccr4-Not complex essential for yeast viability, is apparently associated with TBP in vivo (31). Furthermore, Not5p interacts physically and functionally with yTAF13 (33), and recombinant Not5p can associate with TBP and yTAFIIs as long as TFIID is integral (2).
In this work, we have tried to determine how the Ccr4-Not complex might regulate TFIID. We focused on the TFIID-specific yTAF1, since like the Ccr4-Not complex it is thought to have both positive and negative effects on transcription in vivo by interfering with or promoting the association of TBP with DNA. We found that yTAF1 is associated with Not1p in vivo, and we could isolate large complexes that contain yTAF1 and Not1p. By two-hybrid analysis we were able to define a minimal region of Not1p that can interact with the N-terminal half of yTAF1. This same region of Not1p can also interact with Not4p and Not5p. The functional importance of this interaction could be demonstrated by different genetic approaches. First, we isolated an allele of TAF1, taf1-4, that causes the accumulation of a C-terminally truncated form of yTAF1 and that, when combined with not4, not2, caf1, and not5 mutations, was synthetically lethal. Second, overexpression of an N-terminal fragment of yTAF1 was toxic in not4 and not5 mutant backgrounds, while overexpression of the full-length protein suppressed not4. Overexpression of yTAF1 derivatives that were mutated in both TAND domains did not suppress not4, while expression of these same derivatives as sole yTAF1 derivatives had a suppressive effect in not5 mutants. Finally, we could demonstrate stress-regulated association of Not5p with promoter DNA and Not5p-regulated yTAF1 occupancy of promoter DNA. These results are the first suggestion that the Ccr4-Not complex, maybe via Not5p and/or Not4p, interacts with yTAF1 to regulate its association at promoters and thereby to play a role in the regulation of the N-terminal autoinhibitory region of yTAF1 in vivo.
MATERIALS AND METHODS
Strains and media.
Media were all standard. Plates containing 5-fluoroorotic acid (FOA) were minimal drop-in plates with yeast nitrogen base. Strains were created by standard genetic techniques. MLY130 (Table 1) was created by disruption of TAF1 in the MY542 diploid and tetrad analysis. Other taf1 disruption strains were then obtained by crosses and tetrad analysis of the sporulated diploids. To isolate TAF1 mutant alleles, we mutagenized a TAF1-containing HIS3 plasmid (pG000, see below) with hydroxylamine and transformed MLY130 with our library of mutagenized plasmids. Transformants were cured of the wild-type allele by growth on FOA, and the FOA-resistant colonies were tested for growth at 30 and 37°C. We isolated temperature-sensitive taf1-1 and taf1-4 mutants, which grew well at the permissive temperature. To confirm that the phenotype was due to the plasmids (pG001 and pG004), they were recovered and retransformed into MLY130. After growth on FOA, the same temperature sensitivity phenotype was observed, confirming that it was plasmid linked. MY2298 and MY2300 were created by transforming a diploid heterozygous for TAF1 disruption with pMAC264 and dissecting tetrads. taf1-5 mutant cells were obtained by transformation of MY2298 with a TRP1 plasmid carrying taf1-5 (pMAC374, see below) and growth of the transformants on FOA (MY3315). To test for growth of not4Δ transformants on galactose minimal medium, transformants were first grown for 24 h in glucose minimal medium and kept in exponential phase.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| MY3 | MATaura3-52 trp1Δ1 leu2::PET56 gcn4Δ gal2 his3::TRP1 | 11 |
| MY542 | Diploid isogenic to MY3 except his3::TRP1/HIS3 | This work |
| MY600 | Isogenic to EGY48, except not3::URA3 | This work |
| MY2010 | Isogenic to MY3, except taf1::LEU2 pTAF1-URA3 not5-1 | This work |
| MY2017 | Isogenic to MY3, except taf1::LEU2 pTAF1-URA3 HIS3 not5::LEU2 | This work |
| MY2038 | Isogenic to MY3, except taf1::LEU2 pTAF1-URA3 not5-2 | This work |
| MY2085 | Isogenic to MY3, except not1::LEU2 pLex202-NOT11151-2108MATα | This work |
| MY2164 | Isogenic to MY3, except taf1::LEU2 pTAF1-URA3 caf1::LEU2 | This work |
| MY2169 | Isogenic to MY3, except taf1::LEU2 pTAF1-URA3 not2-1 MATα | This work |
| MY2173 | Isogenic to MY3, except not1::LEU2 pLex202-NOT1 MATα | This work |
| MY2195 | Isogenic to MY3, except not1::LEU2 pRS316-NOT1 | This work |
| MY2245 | Isogenic to W303, except not4::LEU2 | This work |
| MY2452 | Isogenic to MY3, except MATα not5::LEU2 pLex202-NOT5-HIS3 | This work |
| MY2298 | Isogenic to MY3, except MATα HIS3 taf1::LEU2 pHA3-taf1-4-URA3-2μ | This work |
| MY2300 | Isogenic to MY3, except taf1::LEU2 pHA3-taf1-4-URA3-2μ | This work |
| MY2335 | Isogenic to W303, except not5::LEU2 | This work |
| MY2350 | Isogenic to MY2300, except MATα HIS3 pGST-taf1-4-TRP1 | This work |
| MY2357 | Isogenic to MY2300, except MATα HIS3 pGST-TAF1-TRP1 | This work |
| MY3315 | Isogenic to MY3, except MATα HIS3 taf1::LEU2 ptaf1-5-TRP1 | This work |
| MY3401 | Isogenic to MY3, except MATα HIS3 taf1::LEU2 pGST-taf1-5-TRP1 | This work |
| MY3388 | Isogenic to GY5, except ura3::STRE7-lacZ-URA3 | This work |
| MY2667 | Isogenic to GY6, except ura3::STRE7-lacZ-URA3 | This work |
| MY2668 | Isogenic to GY7, except ura3::STRE7-lacZ-URA3 | This work |
| GY5 | Isogenic to MY3, except taf1::LEU2 pHA3-TAF1-HIS3 | This work |
| GY6 | Isogenic to MY3, except taf1::LEU2 pHA3-taf1-1-HIS3 | This work |
| GY7 | Isogenic to MY3, except taf1::LEU2 pHA3-taf1-4-HIS3 | This work |
| GY13 | Isogenic to MY3, except taf1::LEU2 pTAF1-HIS3 not5-1 | This work |
| MLY129 | Isogenic to MY3, except HIS3 taf1::LEU2 pTAF1-URA3 MATα | This work |
| MLY130 | Isogenic to MY3, except taf1::LEU2 pTAF1-URA3 | This work |
| MLY134 | Isogenic to MY3, except taf1::LEU2 pTAF1-URA3 not1-2 MATα | This work |
| MLY148 | Isogenic to MY3, except taf1::LEU2 pTAF1-URA3 not4-1 MATα | This work |
| EGY48 | ura3 trp1 his3 LEU2::LexAOp6-LEU2 | 59 |
| W303-1A | MATahis3-11,15 trp1 leu2-3,112 ura3-1 can1-100 ade2-1 suc2 | 53 |
DNAs.
Plasmid pG000 is a HIS3 centromeric plasmid expressing a three-hemagglutinin (HA3)-tagged yTAF1 that was recovered from yeast strain BY8391 (kind gift from Tony Weil). pG001 and pG004 were recovered from yeast strains GY6 and GY7 and were derived from pG000 by hydroxylamine mutagenesis. The NotI-NcoI fragment of pG004 was ligated into pG000 digested with NotI and NcoI, leading to pG1. pG3 is the opposite construct, namely, the NotI-NcoI fragment of pG000 ligated into pG004 digested with NotI and NcoI. pG5 was created by cloning the SnaBI-NotI fragment of pG000 into pG004 digested with SnaBI and NotI. These plasmids were used to map the taf1-4 mutations 5′ of the SnaBI site. Similar constructs were used to map the taf1-1 mutation. pMAC264 was created by cloning the SalI-NotI fragment of pG004 into pRS426. pMAC271, pMAC278, and pMAC423 expressing glutathione S-transferase (GST)-yTAF1, GST-yTAF1-4, and GST-TAF1-5, respectively, were made in several steps. The 5′ region of TAF1 (BamHI site to BglII site) was cloned into pET15b (between the NdeI and BamHI sites), leading to pMAC198. The NcoI-EcoRV fragment of pMAC198 was cloned into pML138, a TRP1 centromeric plasmid derived from pPC86 that expresses the GST entity from the ADC1 promoter, leading to pMAC267. Finally, the region of TAF1, taf1-4, or taf1-5 from the NdeI site to the XhoI site was introduced into pMAC267. pMAC433 expressing HA3-yTAF1-5 was made by cloning the KpnI-BamHI fragment of pG000 and the BamHI-XhoI fragment of pMAC374 into pRS314. pMAC262, made to express the C-terminal part of yTAF1 in bacteria, was made by cloning the SnaB1-XhoI fragment of TAF1 into pET15b first digested with NdeI and then treated with Klenow fragments and digested with XhoI. The clone expressing LexA-Not1p1151-2108 has been described previously (37), and the clone expressing B42-yTAF113-562, a kind gift from Claudio de Virgilio, was obtained from a library of genomic DNA digested partially by HaeIII and cloned with EcoRI linkers into plasmid pJG4-5 (62). To make truncated and full-length derivatives of TAF1 under the control of a GAL1 promoter, several steps were used. First, a fragment encompassing wild-type TAF1 sequences from the BamHI site to the end of the gene was cloned into a pBSSK vector, leading to pMAC365, from which the EcoRI-SnaBI and SnaBI-XhoI fragments were cloned into pJG4-5 (62) digested with EcoRI and SalI (pMAC375). The BamHI-AvrII fragment of mutant taf1 alleles was inserted into pMAC375 to replace the wild-type sequences, leading to pMAC383 (expressing yTAF1Y19AF57A) and pMAC385 (expressing yTAF1ΔTAND1). pMAC383, pMAC375, and pMAC385 were digested with ClaI and NcoI, treated with Klenow fragments, and religated to make, respectively, plasmids pMAC387, pMAC386, and pMAC388, expressing truncated yTAF16-645 derivatives. pET15b derivatives of the entire coding sequences of SPT15, TAF3 (TAF47), TAF4 (TAF48), and TAF8 (TAF65) were created to express the yTafIIs and TBP in bacteria and make polyclonal antibodies. To make LexA fusions to different portions of Not1p, NOT1 sequences were amplified by PCR and cloned in pLex202 (62) digested with EcoRI and BamHI. The oligonucleotides used for the PCR amplification led to the creation of EcoRI sites 5′ (in the correct reading frame) and BamHI sites 3′ of the amplified sequences.
Extract preparation.
Total-protein extracts used only for Western blot analysis were prepared by the alkaline lysis protocol. One milliliter of cells at an optical density at 600 nm (OD600) of 1 were spun down and frozen in liquid nitrogen. The cell pellets were thawed on ice, and 150 μl of a lysis buffer composed of 1.85 M NaOH and 7.4% β-mercaptoethanol was added. After trichloroacetic acid (TCA) precipitation, the proteins were resuspended in 20 μl of 0.1 M NaOH-20 μl of sample buffer concentrated twofold.
Total-protein extracts for TFIID analysis or immunoprecipitation experiments were prepared by bead beating either in 350 mM NaCl-40 mM HEPES, pH 7.2-0.1% Tween 20-10% glycerol-protease inhibitors, followed by clarification by ultracentrifugation as previously described (48), or in Woontner buffer (0.2 M Tris base-0.39 M ammonium sulfate, 10 mM MgSO4, 20% [vol/vol] glycerol, 1 mM EDTA [pH 7.9], 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF]) as previously described (61).
Biorex chromatography.
Fifteen milliliters of wet cell pellets was derived from an 8-liter culture of cells growing in rich medium collected at an OD600 of 2.0 at most. Twenty to 45 ml of whole-cell extract (WCE) was obtained by bead beating as described above in Wootner buffer and dialyzed overnight against buffer A (BA) (20 mM HEPES-KOH [pH 7.6], 10% glycerol, 1 mM dithiothreitol, 0.1 mM PMSF, 1 mM benzamidine) with 300 mM potassium acetate. After dialysis, the WCE was ultracentrifuged for 30 min at 35,000 rpm, and during centrifugation a 10-ml Biorex 70 (Biorex) column was preequilibrated with BA300. The cleared dialysate (total extract) was then loaded on the column. The flowthrough (FT) was collected, and, after being washed with 5 volumes of BA300, the bound proteins were eluted with BA1000.
Gel filtration experiments.
The eluate from the Biorex column (8 ml) was directly added to a 500-ml Sepharose 4B (Pharmacia) gel filtration column (90 cm high) that was equilibrated with 40 mM HEPES, pH 7.6-350 mM NaCl-5% glycerol-0.1% Tween 20. The column flow rate was 24 ml/h, and fractions of 4 ml (10 min) were collected. Thyroglobulin (670 kDa) was injected for calibration of the column and eluted at 366 ml, ferritin (440 kDa) eluted at 394 ml, and aldolase (158 kDa) eluted at 416 ml, and the void volume was determined by injection of calf thymus DNA, which eluted at 160 ml.
Coimmunoprecipitation.
One milligram of total-protein extracts was incubated with 0.125 μl of goat polyclonal anti-LexA antibodies (Santa Cruz Biotechnology) in a volume of 100 μl for 2 h, followed by addition of protein G-Sepharose for 1 h. The immunoprecipitates were washed three times with 1 ml of extract buffer, and one-third of the immunoprecipitates were loaded on a sodium dodecyl sulfate-7% polyacrylamide gel electrophoresis (SDS-7% PAGE) gel for Western blot analysis.
Glutathione-Sepharose chromatography.
WCE prepared as described above was precipitated with 40% ammonium sulfate, and the pellet was solubilized with BA300 and loaded on the 500-ml Sepharose 4B column equilibrated with BA300. Fractions 63 to 85 were pooled and loaded on a 5-ml Biorex column. The BA1000 eluate was diluted twice in BA and was bound for 1 h in batch with glutathione-Sepharose previously equilibrated in BA300. The resin was washed twice with 10 ml of BA300 and eluted overnight with BA300 and 20 mM glutathione.
Mutagenesis of TAF1.
The pG000 plasmid (20 μg) was incubated with 0.4 M hydroxylamine in a 0.1 M phosphate-EDTA buffer (pH 6.0) at 75°C for 60 min. The DNA was separated from hydroxylamine by electroelution, and the DNA was then precipitated with ethanol before transformation into bacteria. For site-directed mutagenesis, the stop codon at position 309 was obtained with the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. TAF1 TRP1 centromeric plasmid pML36 was used as template with the following primers: 5′ CCA TTG ATG AAC TTT TCC CTA TTA AAG AGT AAC AAA 3′ and 5′ TTT GTT ACT CTT TAA TAG GGA AAA GTT CAT CAA TGG 3′. The mutation in plasmid pMAC374 was confirmed by DNA sequencing.
RNA preparation and S1 analysis.
Total RNA was prepared by the hot-phenol method as previously described (10). Thirty micrograms was hybridized to radiolabeled oligonucleotides prior to digestion by S1 nuclease and separation on an 8% polyacrylamide denaturing gel. Hybridizations were always internally controlled: the same amount of RNA was analyzed for the levels of the HIS4, DED1, NOT5, TUB2, CLN2, ACT1, ADH1, RPS8A, HSP104, HSP12, and wtRNA transcripts. The oligonucleotides that have not been previously described (10) are available upon request.
Western blot analysis.
After transfer of the SDS-PAGE gels onto nitrocellulose, the desired proteins were revealed by probing with specific polyclonal antibodies to Not1p at 1:10,000, Not5p at 1:10,000, Ccr4p at 1:10,000, yTAF10 (yTafII25p) at 1:5,000, yTAF5 (yTafII90p) at 1:10,000, yTAF3 (yTafII47p) at 1:5,000, yTAF4 (yTafII48p) at 1:5,000, yTAF8 (yTafII17p) at 1:5,000, yTAF7 (yTafII67p) at 1:5,000, TBP at 1:20,000, C-terminal yTAF1 at 1:5,000, yTAF1 at 1:3,000, TFIIB at 1:5,000, Srb4 at 1:3,000, and LexA at 1:3,000 and with commercial monoclonal antibodies (Santa Cruz Biotechnology) to HA at 1:3,000 and GST at 1:3,000; secondary antibodies conjugated with horseradish peroxidase or alkaline phosphatase (Bio-Rad) were used at 1:10,000 and 1:3,000, respectively.
CHIP.
For chromatin immunoprecipitation (CHIP) we used a protocol previously described (12, 30) with modifications described by Patrick Schaeffer and Michel Strubin (see Strubin website: www.genmi.unige.ch/STRUBIN_LABb.htm). The yeast strains were grown in 150 ml of yeast extract-peptone-dextrose at 30°C to an OD600 of 1. For the heat shock experiments, the cells were transferred at 39°C for 10 min or kept at 30°C. The cells were then fixed in 1% formaldehyde for 20 min at room temperature (RT). One percent (wt/vol) glycine was added for 5 min at RT. The cells were washed two times with cold Tris-buffered saline (20 mM Tris-HCl, 200 mM NaCl) and resuspended in 600 μl of FA lysis buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 1 mM PMSF). Three hundred microliters of glass beads was added to the cells, and the suspension was vortexed for 40 min at 4°C. The cell lysate was transferred to a new Eppendorf tube, and 15 μl of 20% SDS was added. The cell lysate was sonicated 6 times for 10 s each and centrifuged for 15 min at 16,000 × g at 4°C. The supernatant was sonicated again three times for 10 s each. Thirty microliters of this solution, referred as the input DNA, was taken and stored at −20°C until further use. One milliliter of FA lysis buffer was added to 180 μl of the clear cell lysate, and mixture was incubated with or without the antibodies (0.5 μl of purified immunoglobulin G from a polyclonal anti-yTAF1 or anti-TBP antibody or 2 μl of a polyclonal Not5p antibody) and 40 μl of protein A- and protein G (50/50)-Sepharose beads overnight at 4°C with mild shaking. The Sepharose beads were pelleted by centrifugation at 322 × g and washed once with FA lysis buffer, once with FA lysis buffer-350 mM NaCl, once with buffer III (10 mM Tris-HCl [pH 8], 1 mM EDTA, 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate), and twice with Tris-EDTA (10 mM Tris-HCl [pH 8], 1 mM EDTA). The protein G-Sepharose bead-bound complexes were eluted with 200 μl of buffer IV (50 mM Tris-HCl [pH 7.5], 10 mM EDTA, 1% SDS). From this step on, the input was treated the same way as the eluted complexes. Two hundred microliters of Tris-EDTA (or 400 μl for inputs) and 3 μl of proteinase K at 10 mg/ml were added to the eluted complexes, followed by incubation for 5 h at 65°C. The DNA was extracted two times with phenol-chloroform and once with chloroform and precipitated by ethanol with 2 μg of glycogen as the carrier. After centrifugation, the input DNA was dissolved in 100 μl of water and the immunoprecipitated DNA was dissolved in 25 μl of water.
Quantitative PCR.
For quantitative real-time PCR we used the SYBR green PCR master mixture (PE Applied Biosystems, Branchburg, N.J.) according to a protocol developed for CHIP experiments by Patrick Schaeffer (see Strubin website). The DNA recovered in the precipitate could be expressed relative to the amount of DNA. These values are arbitrary and were always at least an order of magnitude higher when the experiment was performed with an antibody than the background values in the absence of an antibody.
RESULTS
yTAF1 is associated with Not1p in vivo.
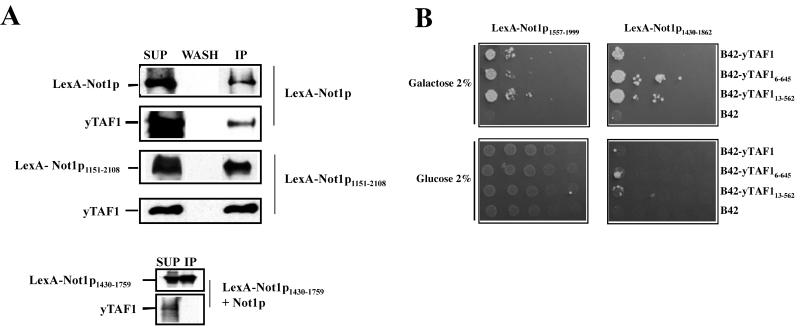
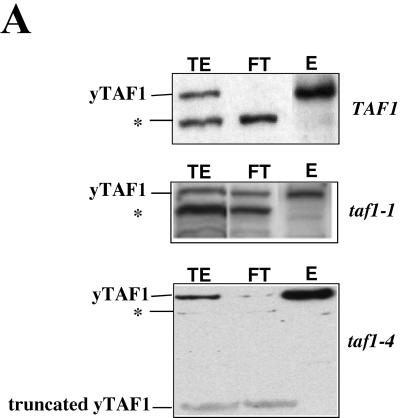
To determine whether yTAF1 might be associated with Not1p in vivo, we transformed a plasmid expressing LexA-Not1p into a not1 null strain carrying a URA3 plasmid with the NOT1 gene (MY2195; Table 1). After passage over FOA, a drug that kills cells expressing a functional URA3 gene, we obtained a strain carrying LexA-Not1p as the sole Not1p derivative (MY2173). By immunoprecipitation experiments we found that yTAF1 was associated with Not1p (Fig. 1A). yTAF1 was also able to coimmunoprecipitate with a viable truncated derivative of Not1p, Not1p1151-2108 (MY2085; Table 1) (Fig. 1A). The immunoprecipitation of yTAF1 was specific, since yTAF1 was not immunoprecipitated from cell extracts by antibodies against LexA in the absence of a LexA-Not1p fusion protein or if the fragment of Not1p fused to LexA was too small (Fig. 1A, bottom). These results correlate well with results of other experiments in which we have found that yTAF1 copurifies with Not1p over a Not1p affinity column (data not shown).
FIG. 1.
yTAF1 is associated with Not1p in vivo. (A) Extracts (1 mg) from cells expressing LexA-Not1p (MY2173), LexA-Not1p1151-2108 (MY2085), Not1p, and LexA-Not1p1430-1759 as indicated were incubated with anti-LexA antibodies followed by protein G-Sepharose. One-fourth of the immunoprecipitate (IP) and last wash (WASH) and 1/10 of the supernatant (SUP) was loaded on SDS-7% PAGE gel, and the presence of the Not1p derivatives or yTAF1, as indicated on the left, was revealed by Western blotting. Exposures of all blots were not identical. (B) The indicated LexA-Not1p derivatives were coexpressed with the indicated B42 fusions to yTAF1 derivatives. Cells were serially diluted and plated on minimal medium lacking leucine, with glucose or galactose as a carbon source as indicated. Growth on galactose but not glucose medium indicates a two-hybrid interaction.
To investigate the association of yTAF1 with Not1p further, we performed two-hybrid experiments. A full-length derivative (amino acids 6 to 1024) and two C-terminally truncated derivatives (amino acids 6 to 648 or 13 to 562) of yTAF1 fused to the B42 activation domain were coexpressed with a number of Not1p fragments fused to LexA. We found that the full-length and C-terminally truncated derivatives of yTAF1 could interact with LexA-Not1p1151-2108 and also with LexA-Not1p1557-1999 and LexA-Not1p1430-1862 (Fig. 1B), suggesting that yTAF1 can interact with a domain of Not1p comprising amino acids 1557 to 1862 (results summarized in Table 2). Interestingly, we found that this same region of Not1p is apparently sufficient for interaction of Not1p with Not5p and Not4p (Table 2). In contrast, the interaction of Not1p with Not2p requires a different domain of Not1p, between amino acids 1854 and 2108 (Table 2). We have also found that LexA-Not1p1151-2108 participates in a two-hybrid interaction with B42 fusions to TBP and other yTAFIIs tested, namely, yTAF3, yTAF4, yTAF6, yTAF10, and yTAF11, suggesting that it might interact with TFIID (data not shown).
TABLE 2.
Two-hybrid interactions between fusions of LexA to Not1p derivatives and fusions of B42 to yTAF113-562 and other Not proteinsa
| LexA-Not1p fusion | Interaction with:
|
||||
|---|---|---|---|---|---|
| B42 | B42- Not2p | B42- Not4p | B42- Not5p | B42- yTAF113-562 | |
| LexA-Not1p1-1150 | − | − | − | − | − |
| LexA-Not1p1151-2108 | − | + | + | + | + |
| LexA-Not1p1430-1862 | − | − | + | + | + |
| LexA-Not1p1430-1999 | − | − | + | + | + |
| LexA-Not1p1557-1999 | − | − | + | + | + |
| LexA-Not1p1854-2108 | − | + | − | − | − |
The indicated derivatives were cotransformed into strain EGY48, and a two-hybrid interaction was determined to be positive (+) or negative (−) depending on whether or not transformants could form colonies on galactose plates lacking leucine.
Thus, yTAF1 and Not1p are associated in vivo, and the N-terminal part of yTAF1 (amino acids 13 to 562) interacts with the same portion of Not1p (amino acids 1557 to 1862) as Not5p and Not4p.
Characterization of genetic interactions between the taf1-4 allele and ccr4-not mutants.
We used a genetic approach to determine whether the interaction between yTAF1 and Not1p was functionally relevant. We isolated two temperature-sensitive alleles of TAF1 (see Materials and Methods) (Fig. 2). We transformed these two mutant alleles, as well as the wild-type TAF1, into a number of strains that had a disrupted TAF1, carried a wild-type TAF1 allele on a URA3 plasmid, and had a mutant CCR4-NOT gene. The objective was to determine whether genetic interactions could be measured. After passage on FOA, the growth phenotypes were investigated. All mutant strains with the taf1-1 allele grew as well as mutants with wild-type TAF1. In contrast, the taf1-4 allele was synthetically lethal in combination with the mutants tested, except the not1-2 mutant, since no colonies were able to form on the FOA plates (Table 3).
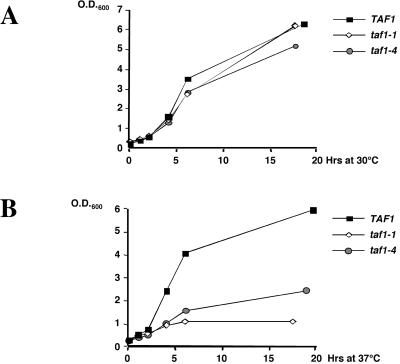
FIG. 2.
Comparative growth of wild-type, taf1-1, and taf1-4 cells. The strains carrying the indicated TAF1 alleles were diluted from an overnight culture in glucose-rich medium at an OD600 of 0.2 and grown at 30°C (A) or were grown to an OD600 of 0.4 at 30°C and shifted to 37°C (B). The OD600 was measured at the indicated time points thereafter.
TABLE 3.
Many taf1 ccr4-not double mutants are inviablea
| TAF1 allele | Viability of transformant carrying:
|
|||||
|---|---|---|---|---|---|---|
| wt | not1-2 | not2-1 | not4-1 | not5-1 | caf1Δ | |
| TAF1 | + | + | + | + | + | + |
| taf1-1 | + | + | + | + | + | + |
| taf1-4 | + | + | − | − | − | − |
Strains carrying a disruption of the chromosomal copy of TAF1 and an episomal copy of TAF1 on a URA3 plasmid and additionally bearing the indicated ccr4-not mutation were transformed with a HIS3 plasmid carrying the indicated TAF1 allele. +, growth of transformant on FOA; −, no growth. Absence of growth on FOA indicates inviability of the double mutants. wt, wild-type allele.
These results show that the synthetic lethality between taf1-4 and ccr4-not mutants is specific to the taf1-4 allele. To determine whether the synthetic lethality might also be specific to the ccr4-not alleles, we took advantage of previous findings with taf13 mutant alleles that produced a synthetic slow-growth phenotype when combined with the not5-1 allele but not when combined with the not5-2 allele (32). These two alleles carry nonsense mutations, but the truncated protein encoded by not5-2 is longer than the one encoded by not5-1. We thus introduced taf1-4 into the not5-2 strain (MY2038) and after plasmid shuffle found that the double mutant was viable, albeit extremely sick (it was only obtained on FOA plates to which all amino acids had been added). Thus, the synthetic lethality between taf1-4 and not5-1 is allele specific, both with regard to the taf1 allele and with regard to the not5 allele.
A C-terminally truncated form of yTAF1 accumulates in taf1-4 mutant cells.
The taf1-4 and not5-1 allele-specific synthetic lethality observed and the physical association of Not1p and yTAF1 suggest that the Ccr4-Not complex and yTAF1 interact functionally in vivo. To further understand this interaction, we characterized the taf1 alleles isolated. We analyzed the levels of a number of components of the TFIID and Ccr4-Not complexes in wild-type and taf1-1 and taf1-4 mutant cells at the permissive temperature and at various times after a shift to the restrictive temperature (Fig. 3).
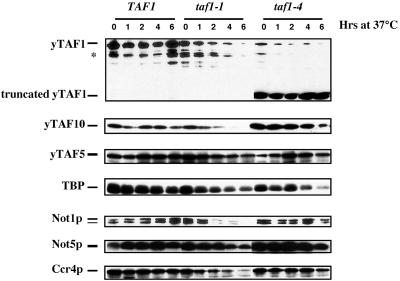
FIG. 3.
Analysis of protein levels in wild-type, taf1-1, and taf1-4 cells. The indicated cells were grown at 30°C to an OD600 of 0.4 (lane 0) and then shifted to 37°C for the times indicated, and equivalent amounts of total-protein extracts from the cells were loaded on SDS-PAGE gels (7% polyacrylamide for yTAF5, Not1p, Not5p, and Ccr4p, 10% for yTAF1, and 15% for TBP and yTAF10). The levels of the different proteins were revealed by Western blot analysis with specific antibodies, except for that of yTAF1, which was revealed by anti-HA antibodies. In wild-type cell extracts yTAF1 is detectable as two forms (yTAF1 and ∗). The amounts of total protein analyzed in all strains at all time points were equivalent, as amounts could be controlled by Coomassie staining of the duplicate gels (data not shown).
yTAF1 is detectable as two forms in wild-type cells that did not change over the time course of the experiment. In taf1-1 and taf1-4 mutant cells, the levels of the two forms of yTAF1 decreased rapidly at the restrictive temperature. Surprisingly, in the taf1-4 mutant, there was additionally a high level of a 50- to 60-kDa C-terminally truncated form of yTAF1, stable at the restrictive temperature at least up to 6 h after the shift (Fig. 3). All forms of yTAF1 were detectable both with monoclonal antibodies against the HA epitope and with polyclonal antibodies against yTAF1. However, as expected, antibodies raised against the C-terminal region of yTAF1 did not recognize the 50- to 60-kDa truncated form in taf1-4 mutant cells (data not shown).
All proteins investigated in wild-type cells remained at a stable level over the time course of the experiment. In taf1-1 mutant cells, yTAF10 decreased to undetectable levels at the restrictive temperature while yTAF5 did not change much, similar to what has been described for other temperature-sensitive taf mutants (33). The levels of Not1p and Ccr4p decreased at the restrictive temperature, as did those of Not5p, although to a lesser extent (Fig. 3). In contrast, in taf1-4 mutants, there was no significant change in the levels of any of the proteins investigated over the time course of the experiment, except for a slight decrease in TBP levels and maybe Not5p and yTAF10 levels after 6 h at the restrictive temperature (Fig. 3).
Importantly, except for the level of yTAF1 itself, no appreciable difference in the levels of any of the proteins analyzed at the permissive temperature was found when the taf1-4 mutant cells were compared to wild-type or taf1-1 mutant cells.
Transcriptional analysis in taf1-1 and taf1-4 mutant cells.
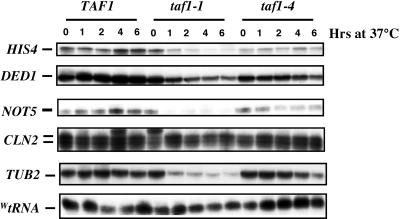
To pursue our characterization of the taf1 mutants, we analyzed a number of transcript levels in wild-type, taf1-1, and taf1-4 cells from the same cultures analyzed above for protein levels (Fig. 4). CLN2 and TUB2 transcripts were included in this analysis, as different taf1 mutants have been shown to differentially affect them (56). In wild-type cells, all transcript levels remained constant during the time course of the experiment. In taf1-1 mutant cells, transcript levels were mostly wild type at the permissive temperature but some transcripts decreased very rapidly at the restrictive temperature (e.g., NOT5 and TUB2). Finally, in taf1-4 mutant cells, of the transcripts measured, only HIS4 was lower than in the wild type at the permissive temperature and at the restrictive temperature none of the transcripts measured varied much over the time course of the experiment. wtRNA levels resulting from RNA polymerase III activity were similar at all times in all strains (Fig. 4).
FIG. 4.
Analysis of transcript levels in wild-type, taf1-1, and taf1-4 cells. The cells collected for protein preparation (Fig. 3) were also used for total-RNA preparation. The levels of the indicated specific transcripts were measured by S1 analysis. All hybridizations were internally controlled, since the levels of DED1 or WtRNA were always measured in the same reaction as that for the other transcripts.
These results suggest an important difference between the two temperature-sensitive taf1 mutants analyzed at the restrictive temperature, since transcription of specific genes rapidly drops to low levels in taf1-1 mutant cells but not in taf1-4 mutant cells. However, at the permissive temperature, the only effect observed in either mutant within this experiment was somewhat decreased HIS4 levels in the taf1-4 mutant.
Characterization of the taf1-4 mutant phenotypes.
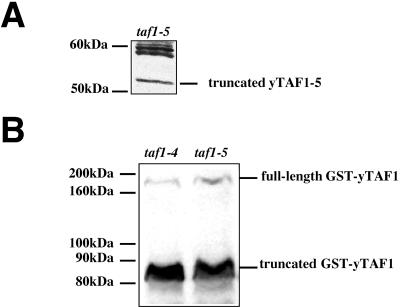
Because of the surprising phenotype in taf1-4 mutant cells, the mutation was localized by creating chimeric clones between the wild-type and mutant alleles (see Materials and Methods). Sequencing revealed a missense mutation (D372N) and a nonsense mutation at codon 309. The presence of a nonsense mutation is in good agreement with the accumulation of a truncated protein in taf1-4 mutant cell extracts, and the observed full-length protein probably results from read-through of the stop codon. However, the position of the stop codon (after 308 amino acids, leading to a truncated protein with a theoretical size of 34 kDa) was unexpected because of the large size of the detected truncated protein (between 50 and 60 kDa).
To determine the role of the nonsense mutation in the taf1-4 mutant phenotypes, we introduced it into a wild-type TAF1 gene by site-directed mutagenesis. The resulting allele, taf1-5, was then introduced into yeast by using a plasmid shuffle assay, which led both to a temperature sensitivity phenotype (data not shown) and to the accumulation of a truncated protein of a surprising large size, namely, larger than 50 kDa (Fig. 5A). By constructing clones that express fusions of GST to yTAF1-4 or yTAF1-5 under the control of the ADH1 promoter, we could determine that the accumulated truncated fusion products had the same apparent size and were both larger than expected (80 to 90 kDa for a fusion protein of 560 amino acids) (Fig. 5B).
FIG. 5.
Introduction of a stop codon at position 309 in TAF1 leads to an allele that produces a truncated protein of more than 50 kDa. (A) Extracts from taf1-5 mutant cells at an OD600 of 1 were made by alkaline lysis and analyzed by Western blotting with polyclonal antibodies against yTAF1. The truncated yTAF1-5 protein migrates with an apparent size of more than 50 kDa. (B) Extracts of cells expressing GST-yTAF1-4 (MY2350) (lane taf1-4) and GST-yTAF1-5p (MY3401) (lane taf1-5) were similarly analyzed with antibodies against GST. The fusion truncated proteins migrate with an apparent size between 80 and 90 kDa, while the fusion full-length proteins migrate with a size bigger than 160 kDa.
taf1-5 was next introduced into not5-1 cells and was found to be synthetically lethal by a plasmid shuffle assay. Thus, the nonsense mutation in taf1-4 is responsible for temperature sensitivity, for the accumulation of a truncated protein of an unexpectedly large size, and for synthetic lethality with not5-1.
The taf1-1 mutation was similarly identified and consists of two missense mutations (P554L and P607T).
Overexpression of the N-terminal part of yTAF1 is toxic in not mutants.
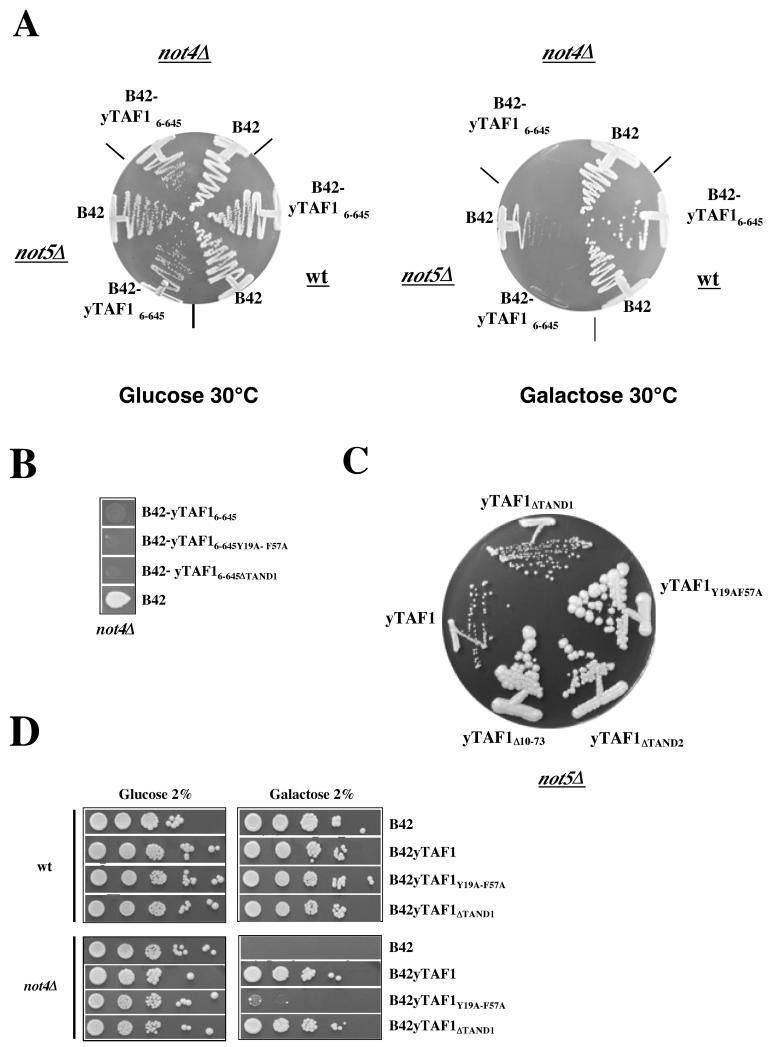
The toxicity of taf1-4 in several not mutant backgrounds might be due to the accumulation of a C-terminally truncated derivative of yTAF1. To determine this, we created not mutant strains in a Gal+ background (MY2245 and MY2335; Table 1) in which we could overexpress derivatives of yTAF1 from the GAL1 promoter, since our usual strain background is gal2. Overexpression of B42-yTAF16-645, but not of B42 alone, impaired the capacity of not4Δ and not5Δ mutant cells to form colonies on minimal galactose medium (Fig. 6A). This phenotype is one that the not mutant cells, particularly not4Δ cells, develop after growing in glucose medium beyond the diauxic shift (Fig. 6D). The N-terminal region of yTAF1 overexpressed in these experiments contains the autoinhibitory TAND domain at its very N terminus; the TAND domain is subdivided into two domains, called TAND1 and TAND2, which can bind TBP in vitro. Introduction of mutations into the TAND domains did not alter the toxicity observed by overproducing yTaf16-645 in not mutants, as shown for not4Δ in Fig. 6B. This experiment, however, has the caveat that in these constructs the B42 activation domain can probably interact with TBP.
FIG. 6.
Synthetic phenotypes and suppression effects between taf1 and not mutants, implicating the autoinhibitory N-terminal domain of yTAF1. (A) Wild-type (W303-1A; Table 1) and not4Δ and not5Δ mutant cells (MY2245 and MY2335; Table 1) were transformed with a plasmidoverexpressing activation domain B42 alone or B42-yTAF16-645, as indicated, from the GAL1 promoter. Transformants grown to early log phase were streaked on minimal glucose or galactose plates at 30°C as indicated and left to grow for 4 to 6 days. (B) not4Δ mutant cells (MY2245, see Table 1) were transformed with a plasmid expressing B42 alone or B42 fused to yTAF16-645, either wild type as for panel A or carrying a deletion of TAND1 or point mutations in TAND1 and TAND2 (Y19A and F57A, respectively) (see below), as indicated, from the GAL1 promoter. Transformants were grown to early log phase (OD600 of less than 0.2) in glucose minimal medium selective for plasmid maintenance, and 5 μl of a 10-fold dilution was plated on galactose minimal medium selective for plasmid maintenance. Plates were allowed to grow for 5 days. (C) not5Δ cells (MY2017) were transformed with taf1 alleles expressing the indicated yTAF1 derivatives: yTAF1ΔTAND1 (removes subdomain I, amino acids 10 to 37), yTAF1ΔTAND2 (removes subdomain II, amino acids 46 to 71), yTAF1Δ10-73 (removes both subdomains), and yTAF1Y19A-F57A (abolishes TBP binding to the domain comprising amino acids 1 to 73). After the shuffling of the wild-type TAF1 URA3 plasmid on FOA, transformants were allowed to grow for 10 days at 25°C. (D) Wild-type and not4Δ mutant cells (MY2245, see Table 1) were transformed with a plasmid expressing B42 alone or B42 fused to nearly full-length derivatives of yTAF1 (starting at amino acid 6) with wild-type or mutant sequences, as indicated, from the GAL1 promoter. Transformants were grown to late log phase (OD600 of more than 1.0) in glucose minimal medium selective for plasmid maintenance, and 5-μl portions of 10-fold serial dilutions were plated on glucose or galactose minimal medium selective for plasmid maintenance and left to grow for 4 to 6 days. When not4Δ cells are plated in early log phase, they can grow on galactose minimal medium (see B42 transformant in panel B), whereas, in late log phase, they no longer do (see the B42 transformant in this panel).
Thus, to further investigate the role of the TBP-binding domain at the N terminus of yTAF1 in the functional interactions between yTAF1 and the Ccr4-Not complex, we replaced wild-type TAF1 by various taf1 alleles in several not mutant backgrounds by plasmid shuffle. The introduced alleles carried mutations in the autoinhibitory region of TAF1 (25). Interestingly, we found that replacement of yTAF1 with derivatives lacking TAND2 or carrying point mutations that abolish TBP binding in vitro to the autoinhibitory domain allowed not5Δ cells to form colonies of a wild-type size rather than the usual very small colonies (Fig. 6C). These results suggest that the capacity of the N-terminal domain of yTAF1 to bind TBP might play a role in the mutant growth phenotypes of not5Δ cells, particularly growth when nutrients become limiting.
To characterize these observations further, we transformed not4Δ mutant cells with a plasmid overexpressing a fusion of B42 to most of yTAF1, carrying or not carrying mutations in the TAND domains, from the GAL1 promoter (see Materials and Methods). Interestingly, the overexpression of full-length forms of yTAF1 improved the capacity of not4Δ cells to form colonies on minimal galactose medium after the diauxic shift, unless the autoinhibitory domain was completely impaired in its TBP-binding capacity (Fig. 6D).
Characterization of TFIID in wild-type and taf1 mutant cells.
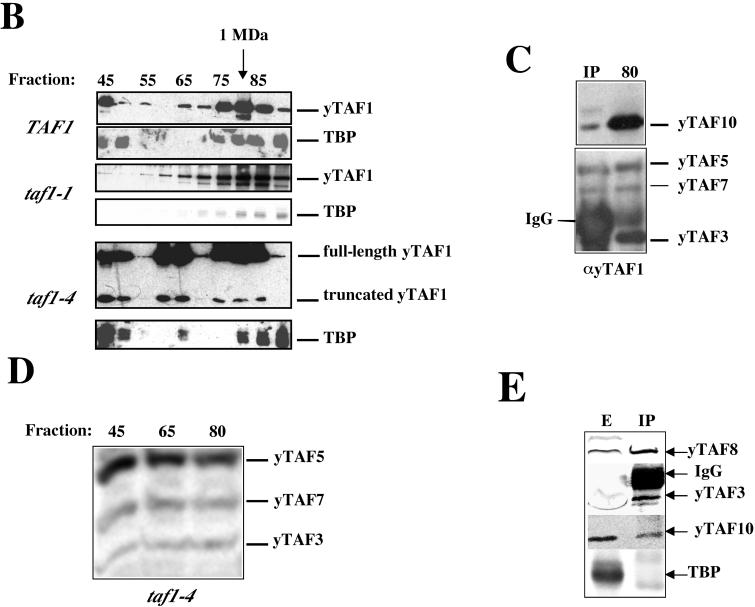
We next characterized yTAF1-containing complexes in wild-type and mutant cells at the permissive temperature, with the hope that this analysis might reveal significant differences between the taf1-4 mutant cells and either wild-type or taf1-1 mutant cells. Indeed, the only significant observation at this point that might be related to the synthetic lethality of ccr4-not mutants with taf1-4 specifically seemed to be the accumulation of a truncated yTAF1 derivative in taf1-4 mutant cells. Since TFIID has been isolated by its capacity to bind a Biorex column, with binding followed by affinity purification (53), we first ran wild-type and mutant extracts over a Biorex column. We analyzed the total extract before binding, the Biorex FT, and the Biorex eluate by Western blotting with anti-HA antibodies for the presence of yTAF1 (Fig. 7A). As expected from previous studies, the slowest-migrating form of yTAF1 bound efficiently the Biorex column (53) and was enriched in the Biorex eluate, except for the taf1-1 mutant, for which much of the yTAF1 was recovered in the FT. In contrast, the faster-migrating form of yTAF1 (either the form in wild-type cells indicated in Fig. 7A or the C-terminally truncated form in taf1-4 mutant cells) was mostly recovered in the FT (Fig. 7A). Longer exposures revealed that some truncated form of yTAF1-4 bound the Biorex column and was present in the eluate (see below).
FIG. 7.
Analysis of yTAF1 in the wild type and taf1-1 and taf1-4 mutants after Biorex and Sepharose 4B gel filtration chromatography. (A) Total protein extract (TE) from wild-type cells and taf1-1 and taf1-4 mutant cells as indicated was loaded on Biorex. Five microliter portions of TE, the FT, and the Biorex eluate (E) were analyzed by Western blotting with antibodies against the HA epitope. The positions of yTAF1 and the truncated protein in the taf1-4 mutant are indicated on the left. Forty-five milliliters of wild-type cells (GY5) led to 70 ml of TE (22 mg/ml), 70 ml of FT (16.5 mg/ml), and 11.5 ml of E (17.2 mg/ml); 20 ml of taf1-1 mutant cells (GY6) led to 50 ml of TE (26 mg/ml), 50 ml of FT (19 mg/ml), and 9 ml of E (16 mg/ml), and 40 ml of taf1-4 mutant cells (GY7) led to 77.5 ml of TE (23 mg/ml), 77.5 ml of FT (17.4 mg/ml), and 10 ml of E (15.6 mg/ml). ∗, form of yTAF1 in wild-type cells. (B) The Biorex eluate from wild-type, taf1-1, and taf1-4 cells was analyzed by Sepharose 4B gel filtration. Every five fractions starting from fraction 45 were precipitated by TCA and analyzed by SDS-PAGE, followed by Western blot analysis for yTAF1 with antibodies against the HA epitope or for TBP with polyclonal antibodies against TBP, as indicated. The void volume corresponds to fraction 40, and thyroglobulin (670 kDa) elutes at 366 ml, ferritin (440 kDa) elutes at 394 ml, and finally aldolase (158 kDa) elutes at 416 ml. Thus fraction 80 (320 ml) corresponds to a size of about 1 MDa as indicated at the top. The absolute amounts in the wild-type and mutant cannot be directly compared, as exposures of the blots were chosen to best show what can be seen for each strain and quantities loaded (8 ml) were not identical for all strains. (C) One milliliter of fraction 80 from wild-type cells was incubated with antibodies (1 μl) against yTAF1. Four hundred microliters of fraction 80 was precipitated by TCA (80) and analyzed together with one-third of the immunoprecipitate (IP) by SDS-PAGE followed by Western blotting for the presence of the indicated yTAFIIs. Additional yTAFIIs (yTAF4 and yTAF8) were also found (data not shown). IgG, immunoglobulin G. (D) Four hundred-microliter portions of fractions 45, 65, and 80 from taf1-4 cells were precipitated by TCA, loaded on SDS-PAGE gel, and analyzed by Western blotting for the presence of the indicated yTAFIIs. yTAF10, yTAF4, and yTAF8 were also present (data not shown). (E) Thirty microliters of the Biorex eluate from wild-type cells (MY1) was immunoprecipitated with antibodies against the C-terminal region of yTAF1. Five-microliter portions of the Biorex eluate (E) and the total immunoprecipitate (IP) were analyzed by Western blotting for the presence of the indicated yTAFIIs and TBP.
We then ran the Biorex eluate from the wild-type and the mutants on a Sepharose 4B gel filtration column to see how many different complexes of yTAF1 there were (Fig. 7B). In the wild-type strain, two major complexes of yTAF1 were detectable, eluting in fractions 45 and 80, the latter corresponding to the size expected for TFIID and the former corresponding to much larger complexes. A more precise analysis of fraction 80 revealed that other yTAFIIs were present and furthermore coprecipitated with antibodies against yTAF1 (Fig. 7C). Furthermore, yTAF1 and other yTAFIIs coimmunoprecipitated with TBP in this fraction (data not shown). Taken together, these results suggest that TFIID elutes indeed in fraction 80. All yTAFIIs investigated were also present in fraction 45 (data not shown).
In the taf1-1 mutant, a profile similar to that for the wild-type was observed, although there was less yTAF1 since some yTAF1 did not bind the Biorex (as shown in Fig. 7A). Furthermore, the ratio of the amount of yTAF1 in fraction 45 to that in fraction 80 was less than the ratio for the wild-type. In the taf1-4 mutant, three distinct complexes of yTAF1 were detectable: the same two complexes found in the wild type and a third complex that eluted with an intermediate size. We tested the three peak fractions (45, 65, and 80) for other yTAFIIs and found them in all three fractions (Fig. 7D). Furthermore, yTAF1 coimmunoprecipitated with TBP in all three fractions (data not shown).
It is worth noting that in the taf1-4 mutant the full-length protein cofractionated on the gel filtration column with the small amount of truncated protein present in the Biorex eluate (Fig. 7B). This suggested that either the two forms of yTAF1 were associated or they formed similar complexes. Although we repeatedly tried to demonstrate the association, we failed (data not shown), leading us to believe instead that the two forms of yTAF1 associate in similar-size complexes.
For both mutants and for the wild type, we also analyzed the elution profile of TBP (Fig. 7B). In most fractions containing yTAF1, TBP was present, except in the taf1-4 mutant, where apparently some fractions containing yTAF1 did not carry detectable TBP (fractions 60 and 75). Furthermore, the elution profile of TBP was somewhat shifted in this mutant. Interestingly, by immunoprecipitating yTAF1 from the Biorex eluate of either wild-type or taf1 mutant cell extracts with antibodies raised against the C-terminal region of yTAF1, we could not coimmunoprecipitate any TBP (in contrast to immunoprecipitation with antibodies raised against the entire yTAF1 protein). We could, however, immunoprecipitate other yTAFIIs (Fig. 7E). This finding suggests that there are complexes of yTAF1 with other yTAFIIs that do not carry TBP.
Not1p copurifies with GST-yTAF1.
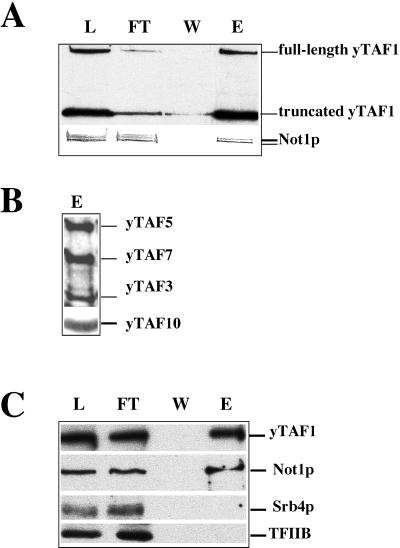
The finding of fractions much larger than TFIID carrying yTAF1, TBP, and other yTAFIIs in wild-type and mutant cells led us to believe that TFIID might be associated with other proteins in larger complexes. The immunoprecipitation and two-hybrid experiments presented above suggested that the Not proteins might be components of these higher-order complexes, especially since both Not1p and Not5p were present in the larger fractions carrying yTAF1 (data not shown). Furthermore, we could efficiently immunoprecipitate yTAF1 from the Biorex eluate with antibodies against either Not3p or Not5p (data not shown).
To investigate this in more detail, we purified GST-yTAF1-4 in several steps from a strain where it replaced chromosomally encoded yTAF1 (see above). We wanted to avoid purifying any possible aggregated forms of GST-yTAF1-4 and thus started with gel filtration to select fractions 63 to 85 and not any fractions close to the void volume (Fig. 7B). For this, we concentrated our extract by ammonium sulfate precipitation prior to the gel filtration. The pooled fractions from the gel filtration were enriched for GST-yTAF1-4 by Biorex chromatography before purification of GST-yTAF1-4 by glutathione-Sepharose chromatography. We found that Not1p bound to, and was eluted from, the glutathione-Sepharose column, together with GST-yTAF1-4 (Fig. 8A) and several other yTAFIIs (Fig. 8B) as well as TBP (data not shown). A similar purification was performed with GST-yTAF1 to investigate whether large complexes of wild-type yTAF1 and Not1p could be demonstrated. Indeed, even though in wild-type cells within fractions 63 to 85 no peak of yTAF1 other than that at fraction 80 was detectable, the elution of yTAF1 was quite broad (Fig. 7B) and yTAF1 was detectable already in fraction 65. We found that Not1p copurified together with GST-yTAF1 (Fig. 8C) as did Not3p and Not5p, yTAFIIs, and TBP (data not shown). In contrast, other proteins, such as Srb4p and TFIIB, did not copurify (Fig. 8C).
FIG. 8.
Not1p copurifies with GST-yTAF1-4 and GST-yTAF1. Total extract from strains expressing the indicated GST fusion proteins was precipitated with 40% ammonium sulfate, and the pellet was solubilized with BA300 and loaded on a Sepharose 4B gel filtration column. Fractions 63 to 85 were pooled and loaded on a Biorex column, and the Biorex eluate was bound to glutathione-Sepharose. The resin was washed twice with 10 ml of BA300 and eluted overnight with 1.5 ml of BA300 and 20 mM glutathione. Equivalent amounts (15 μl) of column load (L), FT, last wash (W), and column eluate (E) were loaded on an SDS-10% PAGE gel, followed by Western blot analysis. The yTAF1 derivatives were revealed with monoclonal antibodies against GST (A) or polyclonal antibodies against the C-terminal domain of yTAF1 (C). All other proteins were revealed by polyclonal antibodies. The analyzed fractions come from the purification of GST-yTAF1-4 (A and B) or of GST-yTAF1 (C).
Transcription mediated by the STRE element is derepressed in taf1 mutants.
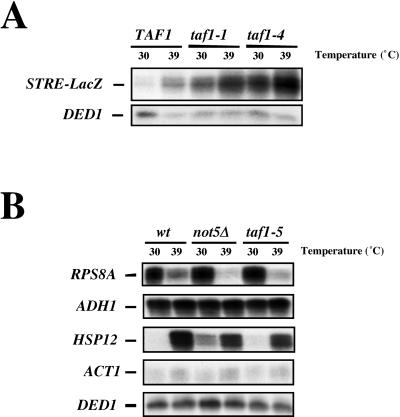
The results presented so far are compatible with an essential function mediated by an appropriate interaction between yTAF1 and the Ccr4-Not complex. Recent results from our laboratory (34) have shown that an important target for transcriptional repression by the Ccr4-Not complex is the family of genes controlled by the Msn2 and Msn4p transcription factors, which bind the STRE promoter element. To determine whether yTAF1 might also be involved in regulation of transcription of STRE-controlled genes, we introduced a reporter construct containing seven STRE elements fused to a lacZ reporter into wild-type cells and the taf1 mutants isolated in this study. Figure 9A shows that STRE-dependent transcription was derepressed in both taf1 mutants, even at the permissive temperature, and that it was superinduced upon heat shock. This finding is compatible with a previous genome-wide study of a conditional taf1 mutant in which at least two STRE-containing genes, CTT1 and HSP26, were upregulated in the mutant (23). Thus, STRE-dependent transcription is apparently also under the direct or indirect repressive control of yTAF1.
FIG. 9.
STRE-dependent transcription is derepressed in taf1 mutants. (A) Total cellular RNA was extracted from the indicated cells grown to early log phase and then shifted or not, as indicated, for 10 min to 39°C. The mRNA levels transcribed from an integrated STRE-lacZ reporter construct were measured by S1 analysis together with the DED1 transcript as an internal control. (B) Wild-type (wt) (MLY130) and taf1-5 (MY3315) and not5Δ (MY2017) mutant cells were grown to early log phase and transferred or not for 10 min to 39°C. Cells were then split in two, one half for S1 analysis of the indicated transcripts, with DED1 always measured as an internal control, and the other half for cross-linking to perform CHIP experiments (Table 4). HSP26 and RPS9 are modulated like HSP12 and RPS8A, respectively, and are thus not included.
Appropriate recruitment to promoters of yTAF1 is Not5p dependent, and Not5p itself can be cross-linked to promoter DNA.
The results presented to this point are highly supportive of a transcriptional control that might be mediated by an interaction between the Ccr4-Not complex and yTAF1. So far, however, no experiments have addressed whether this control may take place on promoters. Preliminary CHIP experiments suggested to us that Not5p, the protein of the Ccr4-Not complex for which there is the most evidence of its possible interaction with TFIID (2, 33) (see above), could be cross-linked to promoters (data not shown). We also found that promoters could be cross-linked to the forms of yTAF1 revealed by using antibodies against the C-terminal region of yTAF1 (see above).
We thus investigated whether Not5p and yTAF1 might interact at promoters by doing CHIP experiments with wild-type cells, cells devoid of any Not5p (not5Δ cells), and cells carrying a mutant allele of the essential TAF1 gene (taf1-5). We looked at occupancy of several promoters by yTAF1 and Not5p before and after a 10-min heat shock. For example, we looked at STRE-containing promoters, such as HSP12 and HSP26, that should be under repressive control by yTAF1 (Fig. 9A) and promoters previously described as yTAF1 dependent, such as RPS8A and RPS9 (23, 54). Samples analyzed by CHIP were also analyzed for transcript levels as shown in Fig. 9B. This showed that in all strains heat shock led to increased HSP12 mRNA levels and decreased RPS8A mRNA levels and had only very slight effects or no effects on ADH1, ACT1, and DED1 mRNA levels. Strikingly, after heat shock, repression of RPS8A was more pronounced, and activation of HSP12 was less pronounced, in both mutants than in the wild type. Furthermore, basal HSP12 mRNA levels were higher in both mutants than in the wild type (more evident for the taf1-5 mutant upon longer gel exposure [data not shown]).
For the CHIP experiments, first, in wild-type cells, immunoprecipitation with antibodies against Not5p demonstrated that Not5p occupancy of all promoters analyzed increased upon heat shock (Table 4). In the control, no increase in any promoter DNA was found in the Not5p immunoprecipitate from not5 null cells after heat shock (data not shown). Furthermore a similar increase could be observed in cells expressing Not5p fused to an epitope by immunoprecipitation with antibodies against the epitope (data not shown). Second, immunoprecipitation of yTAF1 with antibodies raised against its C-terminal region demonstrated that regulation of yTAF1 promoter occupancy correlated quite well with regulation of transcript levels, increasing upon heat shock on stress-inducible promoters (e.g., HSP12) but decreasing on genes repressed by heat shock (e.g., RPS8A).
TABLE 4.
Altered promoter occupancy by yTAF1 and Not5p in not5Δ and taf1-5 cellsa
| Immuno- precipitate | Promoter | Wild-type cells
|
taf1-5 cells
|
not5Δ cells
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Promoter DNA level at:
|
Fold change | Promoter DNA level at:
|
Fold change | Promoter DNA level at:
|
Fold change | |||||
| 30°C | 39°C | 30°C | 39°C | 30°C | 39°C | |||||
| yTAF1 | HSP12 | 1 | 10 | 10 | 4.4 | 101.2 | 23 | 10 | 11.9 | 1.19 |
| HSP26 | 1 | 10 | 10 | 3.6 | 214.8 | 59.7 | 4 | 17.2 | 4.3 | |
| RPS8A | 1 | 0.04 | 0.04 | 7.2 | 17.3 | 2.4 | 4 | 1.52 | 0.38 | |
| RPS9 | 1 | 0.06 | 0.06 | 1.9 | 2.01 | 1.1 | 0.86 | 0.16 | 0.19 | |
| ACT1 | 1 | 2.2 | 2.2 | 4.6 | 18.4 | 4 | 3.2 | 3.2 | 1 | |
| ADH1 | 1 | 2.38 | 2.38 | 5.9 | 44.8 | 7.6 | 11 | 8.8 | 0.8 | |
| Not5p | HSP12 | 1 | 3.5 | 3.5 | 2.1 | 2.73 | 1.3 | |||
| HSP26 | 1 | 4.2 | 4.2 | 2 | 2.8 | 1.4 | ||||
| RPS8A | 1 | 1.95 | 1.95 | 3.3 | 0.79 | 0.24 | ||||
| RPS9 | 1 | 2.7 | 2.7 | 3.85 | 1.35 | 0.35 | ||||
| ACT1 | 1 | 2.3 | 2.3 | 3.6 | 3.5 | 0.96 | ||||
| ADH1 | 1 | 1.95 | 1.95 | 3.15 | 2.4 | 0.76 | ||||
Wild-type cells were grown to exponential phase. One half of the cells were collected, while the other half were transferred for 10 min to 39°C. All cells were then cross-linked, and yTAF1 (with antibodies against its C-terminal region) or Not5p, as indicated, was immunoprecipitated from the total-cell extract. The levels of each indicated promoter DNA in the immunoprecipitates relative to its presence in the total input at 39 and 30°C were calculated. For each promoter and immunoprecipitate, the value obtained for the wild-type cell at 30°C was arbitrarily brought to 1 and the other values were expressed relative to this value. The results presented are the averages of three experiments, and each individual experiment showed the same modulation (increase or decrease) of promoter occupancy.
Third, in taf1-5 mutant cells, promoter occupancy by Not5p was generally increased but heat shock mostly did not lead to a further occupancy of promoters by Not5p. On the contrary, heat shock even led to decreased Not5p occupancy of some promoters (Table 4). This situation was quite in contrast to that of yTAF1 itself, which was recruited to promoters by heat shock to a much larger extent in taf1-5 mutant cells than in wild-type cells (yTAF1 increased even on the RPS8A and RPS9 promoters in the mutant; Table 4). Finally, occupancy of promoters by yTAF1 was mostly increased and much less regulated by heat shock in not5Δ mutant cells (Table 4).
Taken together, these experiments show that regulation of promoter occupancy by Not5p is inappropriate in taf1-5 mutant cells. More importantly, they show that appropriate regulation of yTAF1 promoter occupancy requires Not5p.
DISCUSSION
The Ccr4-Not complex functionally interacts with the N-terminal region of yTAF1 to mediate an essential function.
In this work, we show that Not1p is physically associated with yTAF1. First, yTAF1 coimmunoprecipitates with Not1p (Fig. 1) and copurifies with Not1p over a Not1p affinity column (data not shown). Second, Not1p copurifies with GST-yTAF1 over a glutathione-Sepharose column after several chromatographic steps. Third, the essential C-terminal region of Not1p coimmunoprecipitates with yTAF1 and interacts (amino acids 1557 to 1862 are apparently sufficient) with the N-terminal region of yTAF1 (amino acids 13 to 562) in the two-hybrid assay. Interestingly, this same domain of Not1p is sufficient for Not1p to interact with Not4p and Not5p in the two-hybrid assay, indicating that Not4p and/or Not5p may contribute to the interaction of Not1p with yTAF1. Furthermore, yTAF1 coimmunoprecipitates with Not3p and Not5p and both Not3p and Not5p copurify with GST-yTAF1 (data not shown). These results suggest that the Ccr4-Not complex or several of its components might interact with yTAF1. Finally, because the C-terminal region of Not1p can interact with several other yTAFs and TBP in the two-hybrid assay (data not shown), it is likely that Not1p interacts with yTAF1 within the TFIID complex.
Genetic experiments suggest that this interaction is functionally relevant. First, alleles of TAF1 with mutated coding sequences for the autoinhibitory N-terminal domain suppress the early growth arrest of not5Δ cells under limiting nutrient levels. This can be seen by increased colony size on plates left to grow for several days. Second, the overexpression of yTAF1 suppresses the impaired capacity of not4Δ mutant cells to form colonies on galactose minimal medium after they have grown beyond the diauxic shift. This suppression requires the autoinhibitory N-terminal domain of yTAF1. Third, the overexpression of the first half of yTAF1 (amino acids 6 to 645) exacerbates the impairment in the capacity of not4Δ mutant cells to form colonies on galactose minimal medium, since they lose this capacity when growing exponentially under these conditions. Finally, we isolated an allele of TAF1, taf1-4, that is lethal in many ccr4-not mutant backgrounds. This allele overexpresses an N-terminal portion of yTAF1 (amino acids 1 to 308) even in glucose medium.
Taken together these results are compatible with a model in which the Ccr4-Not complex, via the C-terminal region of Not1p and possibly both Not4p and Not5p, interacts with yTAF1/TFIID via the N-terminal half of yTAF1. This interaction mediates an essential function that most likely involves at least in part the N-terminal autoinhibitory domain of yTAF1.
Does the N-terminal region of yTAF1 support transcription?
Transcription does not rapidly decrease in the taf1-4 mutant at high temperature as it does in the taf1-1 mutant, yet the levels of full-length yTAF1 in the two mutants drop apparently equally rapidly (Fig. 3 and 4). One explanation could be that lower levels of full-length yTAF1 can sustain transcription in the taf1-4 mutant. Indeed, we found by CHIP experiments that certain forms of yTAF1 are more efficiently brought to promoters in the taf1-5 mutant (and also in the taf1-4 mutant, data not shown). An alternative explanation could be that the truncated form of yTAF1 supports transcription to some extent, since a small portion of this form that is stable at high temperature for several hours associates into complexes similar to those carrying full-length yTAF1 (Fig. 7). In this regard, it is interesting that a recent paper by Menica and Struhl (41) reports the characterization of C-terminally truncated derivatives of yTAF1 that have dominant-negative growth effects. In their work, they suggest that the C-terminally truncated derivatives can assemble into TFIID-like complexes but that these complexes cannot bind DNA. However, our finding that the proportion of truncated yTAF1 that associates into TFIID-like complexes is very small (Fig. 7) suggests that, even if these complexes can bind DNA, they may not have been detectable in the study by Menica and Struhl. Thus, it remains possible that TFIID-like complexes formed with the truncated derivative support transcription. We are currently trying to isolate and characterize the complexes containing the C-terminally truncated derivative of yTAF1 in order to determine whether they can support transcription.
We made the interesting finding in this work that the accumulated truncated derivative of yTAF1 in taf1-4 mutant cells (308 amino acids) migrates with an aberrantly large size (more than 50 kDa). While we have no explanation for this observation at the present time, one possibility is that this region of yTAF1 is posttranslationally modified. In this regard, it is interesting that, in wild-type cells, yTAF1 migrates as two forms with an apparent size difference of 15 to 20 kDa. The larger form is the one found in TFIID, while the faster-migrating form is not associated with TBP and other yTAFIIs (data not shown). In previous work (42), the faster-migrating form has been considered a stable proteolytic-degradation form of the full-length protein. However, in light of our present observations, it could be that the slower-migrating form is a posttranslationally modified form of the faster-migrating yTAF1. An intriguing extension of this idea is that this is a modification apparently necessary for yTAF1 to form TFIID complexes. More work will be needed to investigate this issue.
Role of the Ccr4-Not complex in regulating TFIID function.
The only known function of yTAF1 is to associate in TFIID complexes. It is thought to play various roles: scaffold for TFIID (for a review, see reference 51), promoter recognition (54, 56), and also inhibition of TBP binding to DNA (3, 25-27). How these roles fit together is not entirely clear. One model suggests that activator proteins act as antirepressors of the autoinhibitory activity of TATA box binding transcription factor TFIID (28). It is still unclear why the cell needs such an autoinhibitory function within TFIID, but this may contribute to the specific recruitment of TFIID to certain core promoters, for instance, those which lack a canonical TATA sequence. In this work, our immunoprecipitation experiments suggest that there are forms of yTAF1 that are associated with other yTAFII proteins but not with TBP. While we have no idea what the nature and function of these forms of yTAF1 exactly are, their occupancy of promoter DNA correlates exceedingly well with transcription levels from the promoters we looked at (Fig. 9B and Table 4). In taf1-5 mutant cells, surprisingly, the occupancy of promoters by these forms of yTAF1 is higher than that in wild-type cells (Table 4) and is more inducible than that in wild-type cells. These findings suggest that the recruitment of yTAF1 is more efficient in the mutant cells. One possible explanation could be that the overexpressed N-terminal domain of yTAF1 might disrupt the regulation of the proportion of full-length yTAF1 that can be recruited to promoters. In this regard, it is interesting that, in wild-type cells, in addition to TFIID, there is a larger complex that contains yTAF1. In cells that express high levels of a truncated yTAF1 (taf1-4 mutant cells), there is an additional third complex of intermediate size (Fig. 7B). One could speculate that the larger complexes in wild-type cells (fraction 45) are regulated pools of yTAF1/TFIID (for instance, Ccr4-Not-TFIID complexes, since the Not proteins [data not shown] and all of the yTAFIIs that we looked for are indeed present in these fractions). Furthermore, we were able to isolate large complexes carrying yTAF1 and Not1p. The appearance of complexes of intermediate size in the mutant (fractions 60 to 65) would reflect their “partial” disruption. And indeed, as already discussed above, it is easy to imagine that the overexpression of a complex containing TBP and a truncated derivative of yTAF1 would disrupt the interaction between TFIID and the Ccr4-Not complex because Not1p can associate with TBP and the N-terminal region of yTAF1. Finally, in strong support of a model in which the Ccr4-Not complex may contribute to the regulation of promoter occupancy by yTAF1 is our finding that Not5p is necessary for the appropriate regulation of promoter occupancy by yTAF1.
How this regulation might occur is still unclear at the present time. In this regard, we have found stress-regulated cross-linking of Not5p to promoters. The interesting observation was that the appropriate regulation of promoter occupancy by Not5p was itself disrupted in a cell overexpressing a truncated form of yTAF1. One can speculate that this was due, or is related, to the deregulation of promoter occupancy by yTAF1 itself in the taf1-5 mutant. Whatever the answer to this question is, these results demonstrate that an appropriate interaction between Not5p and yTAF1 is required for the correct regulation of yTAF1 recruitment in its various forms to promoters. It is likely that the Ccr4-Not complex rather than Not5p alone plays a role in this regulation, considering all of our results, but the role of the individual subunits still remains to be investigated. In turn, this appropriate regulation of yTAF1 at promoters may be important because of the autoinhibitory function of the N-terminal region of yTAF1.
Acknowledgments
Nicole James, Laurent Maillet, Miguel Molinete, and Grégory Theiler are equal contributing authors.
We thank members of the laboratory for fruitful discussions. We thank Sandrine Creton for starting the CHIP experiments in the laboratory and George Thireos for receiving her in his laboratory to help her do so. We are also very grateful to Patrick Schaeffer and Michel Strubin for help in using real-time PCR to obtain reproducible and reliable results with CHIP experiments. We thank Eve Lenssen for the not mutant strains created in the W303 background. We thank Yann Nussbaumer for cloning the fusion of B42 to yTAF6. We thank Claudio de Virgilio for the clone expressing B42-yTAF113-562, Sukulyan Chatterjee for the plasmid expressing B42-TBP, Tony Weil for strain BY8391 and for antibodies directed against various yTAFIIs, Steve Hahn for antibodies against Srb4p and TFIIB, Roger Brent for antibodies against LexA, Tetsuro Kokubo for taf1 mutant DNAs, and Ursula Oberholzer for clone pEt15b-TBP. We thank Michel Strubin and Laszlo Tora for a critical reading of the manuscript. Finally, we thank Doris-Beate Kirschner and Laszlo Tora for investigating the presence of several yTAFIIs in our samples.
This work was supported by Swiss National Science Foundation grants (31-39690.93 and 31-49808.96) as well as by the OFES96.0072 TMR grant and a grant from Novartis to M.A.C. for funding of L.M.
REFERENCES
- 1.Auble, D. T., K. E. Hansen, C. G. F. Mueller, W. S. Lane, J. Thorner, and S. Hahn. 1994. MOT1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8:1920-1934. [DOI] [PubMed] [Google Scholar]
- 2.Badarinarayana, V., Y.-C. Chiang, and C. L. Denis. 2000. Functional interaction of CCR4-NOT proteins with TATAAA-binding protein (TBP) and its associated factors in yeast. Genetics 155:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai, Y., G. M. Perez, J. M. Beechem, and A. P. Weil. 1997. Structure-function analysis of TAF130: identification and characterization of a high-affinity TATA-binding protein interaction domain in the N terminus of yeast TAFII130. Mol. Cell. Biol. 17:3081-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai, Y., C. Salvadore, Y.-C. Chiang, M. A. Collart, H.-Y. Liu, and C. L. Denis. 1999. The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol. Cell. Biol. 19:6642-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burke, T., and J. Kadonaga. 1997. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes Dev. 11:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burley, S. K., and R. G. Roeder. 1996. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65:769-799. [DOI] [PubMed] [Google Scholar]
- 7.Chalkley, G. E., and C. P. Verrijzer. 1999. DNA binding site selection by RNA polymerase II TAFs: a TAFII250-TAFII150 complex recognizes the initiator. EMBO J. 18:4835-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chicca, J. J., D. T. Auble, and F. B. Pugh. 1998. Cloning and biochemical characterization of TAF172, a human homolog of yeast MOT1. Mol. Cell. Biol. 18:1701-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collart, M. A., and K. Struhl. 1993. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 12:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collart, M. A., and K. Struhl. 1994. NOT1 (CDC39), NOT2 (CDC36), NOT3, and NOT4 encode a global negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8:525-537. [DOI] [PubMed] [Google Scholar]
- 11.Daugeron, M.-C., F. Mauxion, and B. Séraphin. 2001. The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 29:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dedon, P. C., J. A. Soults, C. D. Allis, and M. A. Gorovsky. 1991. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal. Biochem. 197:83-90. [DOI] [PubMed] [Google Scholar]
- 13.Denis, C. L. 1984. Identification of new genes involved in the regulation of yeast alcohol dehydrogenase II. Genetics 108:833-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denis, C. L., and T. Malvar. 1990. The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics 124:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Draper, M. P., C. Salvadore, and C. L. Denis. 1995. Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol. Cell. Biol. 15:3487-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drysdale, C. M., B. M. Jackson, R. McVeigh, and E. R. Klebanow. 1998. The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID, and the Ada-Gcn5p coactivator complex. Mol. Cell. Biol. 18:1711-1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The SPT components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6:1319-1331. [DOI] [PubMed] [Google Scholar]
- 19.Geisberg, J. V., F. C. Holstege, R. Y. Young, and K. Struhl. 2001. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol. 21:2736-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goppelt, A., G. Stelzer, F. Lottspeich, and M. Meisterernst. 1996. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 15:3105-3116. [PMC free article] [PubMed] [Google Scholar]
- 21.Grant, P. A., D. Schieltz, M. G. Pray-Grant, D. J. Steger, J. C. Reese, J. R. Yates III, and J. L. Workman. 1998. A subset of TAFIIs are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94:45-53. [DOI] [PubMed] [Google Scholar]
- 22.Hahn, S. 1998. The role of TAFs in RNA polymerase II transcription. Cell 95:579-582. [DOI] [PubMed] [Google Scholar]
- 23.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 24.Inostroza, J. A., F. H. Mermelstein, I. Ha, W. S. Lane, and D. Reinberg. 1992. Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell 70:477-489. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi, A., M. Tsuyoshi, Y. Ohyama, M. Kawaichi, and K. Tetsuro. 2001. Mutations in the TATA-binding protein, affecting transcriptional activation, show synthetic lethality with the TAF145 gene lacking the TAF N-terminal domain in Saccharomyces cerevisiae. J. Biol. Chem. 276:395-405. [DOI] [PubMed] [Google Scholar]
- 26.Kokubo, T., M. J. Swanson, J.-I. Nishikawa, A. G. Hinnebusch, and Y. Nakatani. 1998. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell. Biol. 18:1003-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotani, T., K.-I. Banno, M. Ikura, A. G. Hinnebusch, Y. Nakatani, M. Kawaichi, and T. Kokubo. 2000. A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding of transcription factor IID. Proc. Natl. Acad. Sci. USA 97:7178-7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kotani, T., T. Miyake, Y. Tsukihashi, A. G. Hinnebusch, Y. Nakatani, M. Kawaichi, and T. Kokubo. 1998. Identification of highly conserved amino-terminal segments of dTafII230 and yTafII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J. Biol. Chem. 273:32254-32264. [DOI] [PubMed] [Google Scholar]
- 29.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 30.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 31.Lee, T. I., J. J. Wyrick, S. S. Koh, E. G. Jennings, E. L. Gadbois, and R. A. Young. 1998. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 18:4455-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, T. I., and R. A. Young. 2000. Transcription of protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 33.Lemaire, M., and M. A. Collart. 2000. The TATA binding protein-associated factor yTafII19p functionally interacts with components of the global transcriptional regulator Ccr4-Not complex and physically interacts with the Not5 subunit. J. Biol. Chem. 275:26925-26934. [DOI] [PubMed] [Google Scholar]
- 34.Lenssen, E., U. Oberholzer, J. Labarre, C. De Virgilio, and M. Collart. 2001. Saccharomyces cerevisiae Ccr4-Not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol. Microbiol. 43:1023-1037. [DOI] [PubMed] [Google Scholar]
- 35.Liu, H.-Y., V. Badarinarayana, D. C. Audino, J. Rappsilber, M. Mann, and C. L. Denis. 1998. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17:1096-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maillet, L., and M. A. Collart. 2002. Interaction between Not1p, a component of the Ccr4-Not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J. Biol. Chem. 277:2835-2842. [DOI] [PubMed] [Google Scholar]
- 37.Maillet, L., C. Tu, Y. K. Hong, E. O. Shuster, and M. A. Collart. 2000. The essential function of NOT1 lies within the CCR4-NOT complex. J. Mol. Biol. 303:131-143. [DOI] [PubMed] [Google Scholar]
- 38.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 273:23781-23785. [DOI] [PubMed] [Google Scholar]
- 39.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 40.Meisterernst, M., and R. G. Roeder. 1991. Family of proteins that interact with TFIID and regulate promoter activity. Cell 67:557-567. [DOI] [PubMed] [Google Scholar]
- 41.Menica, M., and K. Struhl. 2001. Region of yeast TAF130 required for TFIID to associate with promoters. Mol. Cell. Biol. 21:1145-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moqtaderi, Z., Y. Bai, D. Poon, A. P. Weil, and K. Struhl. 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383:188-191. [DOI] [PubMed] [Google Scholar]
- 43.Moqtaderi, Z., M. Keaveney, and K. Struhl. 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2:675-682. [DOI] [PubMed] [Google Scholar]
- 44.Oberholzer, U., and M. A. Collart. 1998. Characterization of NOT5 that encodes a new component of the NOT protein complex. Gene 207:61-69. [DOI] [PubMed] [Google Scholar]
- 45.Oelgeschlager, T., C. M. Chiang, and R. G. Roeder. 1996. Topology and reorganisation of a human TFIID-promoter complex. Nature 382:735-738. [DOI] [PubMed] [Google Scholar]
- 46.Ogryzko, V. V., T. Kotani, X. Zhang, L. R. Schiltz, T. Howard, X.-J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 47.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657-2683. [DOI] [PubMed] [Google Scholar]
- 48.Peterson, C. L., A. Dingwall, and M. P. Scott. 1994. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. USA 91:2905-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poon, D., and P. A. Weil. 1993. Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J. Biol. Chem. 268:15325-15328. [PubMed] [Google Scholar]
- 50.Ranallo, R. T., K. Struhl, and L. A. Stargell. 1999. A TATA binding protein mutant defective for TFIID complex formation in vivo. Mol. Cell. Biol. 19:3951-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reese, J. C., L. Apone, S. S. Walker, L. A. Griffin, and M. R. Green. 1994. Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature 371:523-527. [DOI] [PubMed] [Google Scholar]
- 52.Reese, J. C., Z. Zhang, and H. Kurpad. 2000. Identification of a yeast transcription factor IID subunit, TSG2/TAF48. J. Biol. Chem. 275:17391-17398. [DOI] [PubMed] [Google Scholar]
- 53.Sanders, S. L., and A. P. Weil. 2000. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 275:13895-13900. [DOI] [PubMed] [Google Scholar]
- 54.Shen, W.-C., and M. R. Green. 1997. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90:615-624. [DOI] [PubMed] [Google Scholar]
- 55.Struhl, K., and Z. Moqtaderi. 1998. The TAFs in the HAT. Cell 94:1-4. [DOI] [PubMed] [Google Scholar]
- 56.Tsukihashi, Y., T. Miyake, M. Kawaichi, and T. Kokubo. 2000. Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol. Cell. Biol. 20:2385-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated proteins, Ccr4p and Caf1p, are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 58.Verrijzer, C. P., K. Yokomori, J.-L. Chen, and R. Tjian. 1994. Drosophila TAFII150: similarity to yeast TSM-1 and specific binding to core promoter DNA. Science 264:933-941. [DOI] [PubMed] [Google Scholar]
- 59.Verrijzer, C. P., J.-L. Chen, K. Yokomori, and R. Tjian. 1995. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell 81:1115-1125. [DOI] [PubMed] [Google Scholar]
- 60.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of a TAFII-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 61.Woontner, M., P. A. Wade, J. Bonner, and J. A. Jaehning. 1991. Transcriptional activation in an improved whole-cell extract from Saccharomyces cerevisiae. Mol. Cell. Biol. 11:4555-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zervos, A. S., J. Gyuris, and R. Brent. 1993. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72:223-232. [DOI] [PubMed] [Google Scholar]