Abstract
Owing to a single missense mutation in the cell proliferation factor HCF-1, the temperature-sensitive tsBN67 hamster cell line arrests proliferation at nonpermissive temperatures, primarily in a G0/G1 state, and displays temperature-sensitive cytokinesis defects. The HCF-1 mutation in tsBN67 cells also causes a temperature-sensitive dissociation of HCF-1 from chromatin prior to cell proliferation arrest, suggesting that HCF-1-chromatin association is important for mammalian-cell proliferation. Here, we report that the simian virus 40 (SV40) early region, in particular, large T antigen (Tag), and the adenovirus oncoprotein E1A can rescue the tsBN67 cell proliferation defect at nonpermissive temperatures. The SV40 early region rescues the tsBN67 cell proliferation defect without restoring the HCF-1-chromatin association, indicating that these oncoproteins bypass a requirement for HCF-1 function. The SV40 early region also rescues the tsBN67 cytokinesis defect, suggesting that the roles of HCF-1 in cell proliferation and proper cytokinesis are intimately linked. The ability of SV40 Tag and adenovirus E1A to inactivate members of the pRb protein family—pRb, p107, and p130—is important for the bypass of HCF-1 function. These results suggest that HCF-1 regulates mammalian-cell proliferation and cytokinesis, at least in part, by either directly or indirectly opposing pRb family member function.
Viruses maintain intimate interactions with the infected host cell to ensure productive virus infection. In some instances, these interactions result in the remodeling of the host cell to create a better environment for viral replication. For example, the DNA tumor viruses, such as simian virus 40 (SV40) and adenovirus, commonly induce cell cycle progression, specifically S phase, to support viral-DNA synthesis (20, 36). To accomplish such goals, these viruses inactivate or modify specific cell cycle regulators. Thus, the early gene products of both SV40 and adenovirus are oncoproteins that inactivate the cell cycle regulators pRb and p53 through direct protein-protein association.
In contrast to the smaller DNA tumor viruses, other viruses such as herpes simplex virus (HSV) do not need to remodel the infected cell in this way to support viral-DNA synthesis. HSV, instead, maintains intimate interactions with the infected host cell to select between two modes of infection: lytic and latent. A key regulator of the lytic pathway is viral immediate-early (IE) gene expression (13, 38). IE gene expression is controlled in part by the VP16-induced complex, a multiprotein-DNA complex that forms on HSV IE gene promoters and consists of the viral protein VP16, the cellular transcription activator Oct-1, and the cell proliferation factor HCF-1 (for host cell factor; reviewed in references 9 and 23).
HCF-1 (also known as HCF, C1, VCAF, and CFF) is a broadly expressed, abundant, chromatin-bound protein (15, 39, 40, 43). In proliferating cells in culture, the majority of HCF-1 exists as a heterodimeric complex of amino-terminal (HCF-1N) and carboxy-terminal subunits derived from the proteolytic processing of a precursor HCF-1 polypeptide (15, 39, 41). HCF-1 interaction with VP16 occurs through a β-propeller domain that resides within the first 380 amino acids of the HCF-1N subunit (10, 16, 42). This VP16 interaction domain is also necessary and sufficient for HCF-1 association with chromatin (43), indicating that VP16 targets an important domain of HCF-1 to stimulate HSV lytic infection.
HCF-1 was first shown to be a cell proliferation factor through the characterization of the tsBN67 cell line (8), a temperature-sensitive derivative of BHK-21 hamster cells (22). This cell line undergoes a reversible and stable cell proliferation arrest after 36 to 48 h at the nonpermissive temperature of 40°C. The gene expression profile of arrested tsBN67 cells suggests a G0/G1 arrest (8, 26). Consistent with a G0/G1 arrest, the pRb protein is in its active G0/G1 hypophosphorylated state in arrested cells (26). Unlike G0/G1 cells, however, a subset of the arrested tsBN67 cells contains two or more nuclei, suggesting a defect in cytokinesis upon exit from mitosis (26).
The temperature-induced proliferation and cytokinesis defects of tsBN67 cells are due to a single missense mutation (P134S) in the HCF-1 VP16 interaction domain, which also disrupts VP16 binding to HCF-1, consistent with VP16 targeting a cell proliferation activity (8, 43). With an asynchronous tsBN67 cell population, HCF-1 dissociates from chromatin after the cells are allowed to proliferate for 12 to 18 h at a nonpermissive temperature, well before the arrest occurs, suggesting that the loss of HCF-1-chromatin association is a primary cause of the cell proliferation defect in tsBN67 cells (43). The requirement for HCF-1 association with chromatin can be bypassed, however, because spontaneous proliferation revertants of tsBN67 cells (called tsBN67rev cells) can proliferate in the absence of HCF-1-chromatin association (26). These tsBN67 cells also display fewer cytokinesis defects, suggesting that the roles of HCF-1 in proliferation and cytokinesis are related (26).
The dispensability of an HCF-1 activity in the tsBN67rev cells suggests that HCF-1 plays a regulatory role in cell cycle progression that can be compensated for by other genes. To identify specific genes that are able to rescue the HCF-1 defect in tsBN67 cells, we have asked whether oncogenes that promote cell proliferation, such as the DNA tumor virus early genes, can overcome the temperature-sensitive HCF-1 defect in tsBN67 cell proliferation and cytokinesis. We have found that, indeed, SV40 and adenovirus early-gene products can rescue long-term tsBN67 cell proliferation and cytokinesis defects at nonpermissive temperatures in the absence of HCF-1-chromatin association. The ability of the SV40 large T antigen (Tag) and adenovirus E1A oncoproteins to inactivate members of the pRb protein family—pRb, p107, and p130—is critical for their rescue of tsBN67 cells.
MATERIALS AND METHODS
Cell culture.
All cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum under 7.5% CO2. Unless otherwise noted, transfected tsBN67 cells and derivatives were selected with 4 or 2 μg of puromycin (Sigma) per ml at 33.5 or 40°C, respectively. The tsBN67 cells used in these studies were of the tsBN67HR1 subclone (26).
Plasmids and coding sequences.
The expression vector used in these analyses, pBABEpuroXBC, is described elsewhere (26). Sequences of potential rescue genes, from the second codon to the stop codon, were cloned into pBABEpuroXBC between the SpeI and BamHI sites. All sequences were amplified by PCR to engineer appropriate restriction sites. The genomic sequence containing the SV40 early-region genes was cloned into pBABEpuroXBC as a 2.4-kb PCR fragment to give pBABEpuroXBC-SV40e, which extends from the second codon to the stop codon of Tag. The same primers were used to amplify the wild-type and mutant Tag cDNAs. The Tag cDNA mutants used were 5110 (34), 3213 (31), and 5031 (24), which are mutated in the J domain, pRb-binding motif, and p53-binding region, respectively. The adenovirus E1A 12S mutants used were the Δ2-36 and the Δ120-140 mutants (11). The yellow fluorescent protein (YFP) expression vector, pEYFP, is described elsewhere (2).
Colony formation assay.
To assay colony formation, 105 cells were seeded onto 10-cm-diameter plates at 33.5°C. The next day, cells were transfected by CaPO4 coprecipitation. Briefly, plasmids were mixed to a final concentration of 80 ng/μl, with pUC119 as the carrier, in a 260-μl solution of 10 mM Tris·HCl-0.1 mM EDTA. This DNA solution was then mixed with 260 μl of 500 mM CaCl2-0.1 mM EDTA-1 mM Tris·HCl, pH 7.2, and combined dropwise into 520 μl of 280 mM NaCl-1.5 mM NaP04-50 mM HEPES·NaOH, pH 7.1. The samples were left to precipitate for 20 min at room temperature, after which 1 ml was added per plate of cells. After 20 h, the medium was removed and the precipitate was rinsed off with 2 mM EGTA in phosphate-buffered saline (PBS). Twenty-four hours after removal of the precipitate, cells from each plate were trypsinized and resuspended in 10 ml of medium. Equal fractions representing 10 to 25% of each suspension were then used to seed a series of 6-cm-diameter plates and maintained under selection conditions for 10 days at 33.5°C or for 14 days at 40°C. Plates were fixed with 4% formaldehyde in PBS, and colonies were visualized by staining with 0.01% crystal violet.
S-phase assay.
tsBN67 cells were seeded onto glass coverslips on 3-cm-diameter plates at 104 cells per plate. The next day, cells were transfected by CaPO4 coprecipitation (see above) with 40 ng of pEYFP and 10 μg of the appropriate pBABEpuroXBC expression vector. Immediately after removal of the precipitate, plates were transferred to 40°C and maintained for 72 h, at which time the medium was changed to medium containing 100 μg of bromodeoxyuridine (BrdU) (Sigma) per ml and returned to 40°C for 20 min. Detection of BrdU incorporation was by a modification of protocols described in the work of Spector et al. (30). Briefly, cells were fixed for 15 min at 20°C with a 2% solution of paraformaldehyde in PBS, permeabilized on ice for 5 min with 0.1% Triton X-100 in PBS, and blocked with 2% bovine serum albumin (BSA; Sigma) in PBS-0.1% Tween 20 (PBS-T). BrdU was then immunologically labeled with concurrent nuclease digestion and antibody incubation in a solution of PBS-T, 0.5% BSA, 2.5 mM MgCl2, 100 U of DNase I per ml, and a 1:50 dilution of anti-BrdU antibodies (Pharmingen) for 1 h at 37°C. After washes with PBS-T, the anti-BrdU antibody was detected by probing with a 1:100 dilution of Texas Red-conjugated anti-mouse immunoglobulin G (Amersham) in PBS-T-0.5% BSA for 1 h at 20°C. Slides were subsequently maintained at −70°C until fluorescence visualization. A minimum of 200 YFP-positive cells was counted for each sample.
Growth curves.
Cell lines were seeded onto 6-cm-diameter plates at 2 × 104 cells per plate and maintained at 33.5°C for 48 h. Growth curve analysis was subsequently initiated with the addition of fresh medium and transfer of half the plates to 40°C. At the given times, adherent cells were trypsinized and counted. For comparison between antibiotic-resistant and antibiotic-sensitive cell lines, medium was not supplemented with puromycin.
Immunoblotting.
Lysates from transfected tsBN67 cells were prepared after 2 days growth at 40°C. Lysates of untransfected cells were prepared after the given time at 40°C. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes by semidry transfer. Membranes were probed with monoclonal antibody PAB419 (4) for Tag expression with anti-HCF-1 N18 peptide antiserum (8) or with monoclonal antibodies against cyclin A (Sigma) or PCNA (Transduction Labs). Protein levels were then detected after probing with secondary horseradish peroxidase-conjugated-anti-immunoglobulin G antiserum (New England Biolabs).
Quantification of cell nucleation.
Trypsinized cell suspensions were seeded onto 10-cm-diameter plates at a 1:1,000 dilution from confluent plates. Plates were maintained at 33.5°C for 1 day and then transferred to 40°C for 48 h. Plates were subsequently fixed with 4% formaldehyde in PBS and examined with an inverted phase-contrast microscope at a ×200 magnification. In each sample, cells whose nucleation status was not clearly discernible were eliminated from analysis. More than 200 cells were counted for each sample.
Chromatin enrichment fractionation.
Small-scale biochemical fractionation was performed as described previously (19, 43).
RESULTS
The SV40 early region stimulates tsBN67 cell proliferation at nonpermissive temperatures.
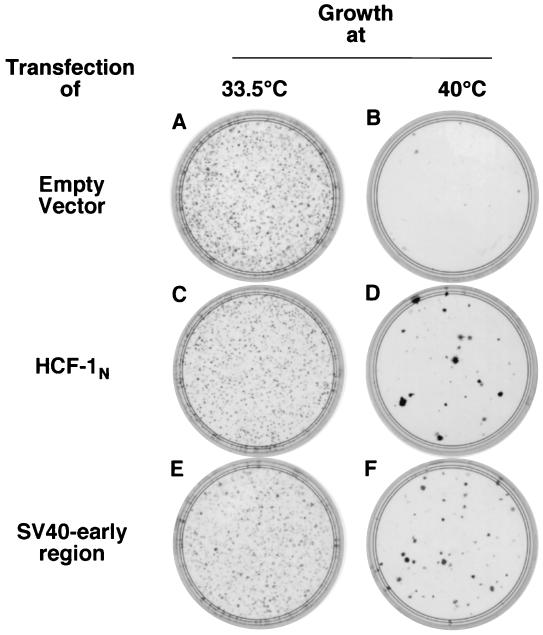
To identify heterologous genes that might overcome the temperature-induced tsBN67 cell proliferation defect, we selected genes known to stimulate proliferation in other mammalian cells. We first analyzed whether the SV40 early region, which encodes both the SV40 large (Tag) and small (tag) tumor antigens, can rescue tsBN67 growth in a 14-day colony formation assay at a nonpermissive temperature. Figure 1 shows the results of such an assay, where colony formations at the permissive temperature (plates A, C, and E) and the nonpermissive temperature (plates B, D, and F) were assayed for cells from the same transfection. After transfection with an empty vector (plates A and B) or a vector directing synthesis of the HCF-1N subunit (plates C and D) or the SV40 early-region gene products (plates E and F), transfected cells were selected by growth in the presence of puromycin (see Materials and Methods). For comparison of the relative abilities of genes to rescue the tsBN67 defect, transfections that gave equal transformation efficiencies as gauged by colony formation at the permissive temperature of 33.5°C (plates A, C, and E) were examined (Fig. 1, legend).
FIG. 1.
Long-term suppression of tsBN67 cell proliferation arrest by SV40 early genes. Shown are the results of a colony formation assay of tsBN67 cells after transfection of 50 fmol (160 ng) of empty pBABEpuroXBC (plates A and B), 50 fmol (235 ng) of pBABEpuroXBCHCF-1N (plates C and D), or 200 fmol (950 ng) of pBABEpuroSV40e (plates E and F) and growth at either 33.5°C (plates A, C, and E) or 40°C (plates B, D, and F). Plates were fixed and stained with crystal violet.
Consistent with previous results (42), the HCF-1N subunit was sufficient to induce colony formation at 40°C (Fig. 1, compare plates B and D). The SV40 early-region gene products also induced colonies at the nonpermissive temperature (plate F). The relative efficiency at which the HCF-1N subunit and the SV40 early-region gene products induce colonies can vary considerably between experiments; in this experiment the relative level of rescue by the HCF-1N subunit was low. In contrast to the HCF-1N subunit-rescued colonies, SV40 early-region-rescued colonies are generally small, suggesting that the SV40 early region either cannot fully complement the growth arrest phenotype of tsBN67 cells or has deleterious effects on cell survival such as through induction of apoptosis, in addition to having a positive effect on cell proliferation. Nevertheless, the SV40 early-region suppression of the tsBN67 cell proliferation arrest demonstrates that the HCF-1 defect can be overcome in a long-term growth assay by a specific set of heterologous genes.
SV40 large Tag is sufficient to rescue tsBN67 cell proliferation at the nonpermissive temperature.
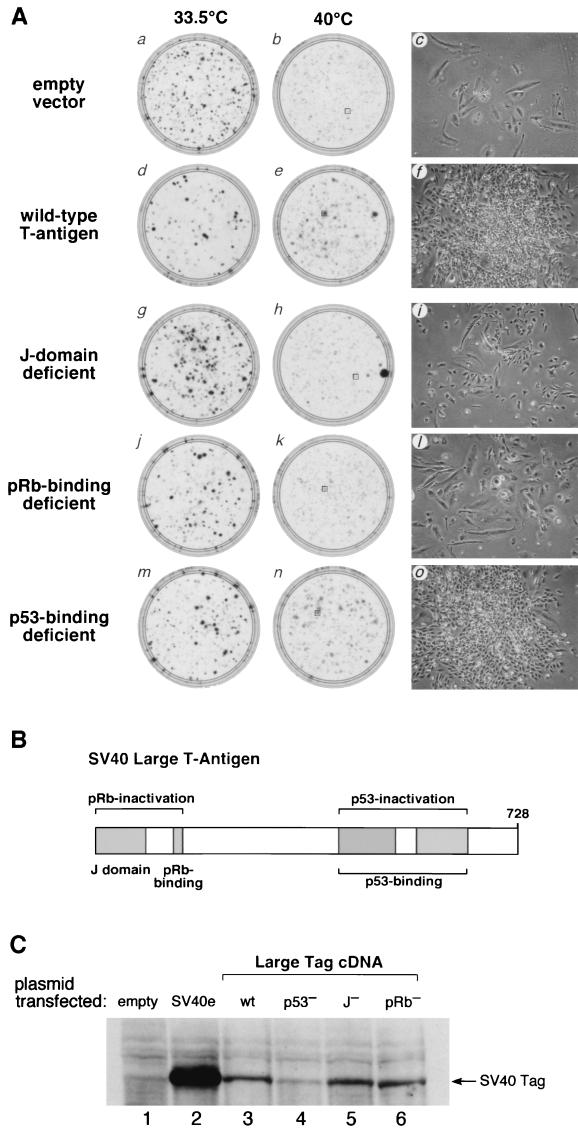
The rescue of tsBN67 cell proliferation at the nonpermissive temperature by the SV40 early region may be due to the activity of SV40 Tag, tag, or both. Because SV40 Tag possesses many growth-promoting functions and is sufficient to transform the parental BHK-21 cells (3), we asked whether it alone is sufficient to rescue the tsBN67 cell proliferation defect in the colony formation assay, as shown in Fig. 2A (panels a to f). Stained plates of cells grown at either 33.5°C (panels a and d) or 40°C (panels b and e) are shown along with an image of a representative unstained colony from the boxed areas in the 40°C plates (panels c and f).
FIG. 2.
The pRb-binding motif and J domain are important for suppression of tsBN67 cell proliferation arrest by SV40 Tag. (A) Panels a, b, d, e, g, h, j, k, m, and n are stained plates after growth at 33.5°C (panels a, d, g, j, and m) or 40°C (panels b, e, h, k, and n) after transfection 18 μg of the empty pBABEpuroXBC vector (panels a and b) or pBABEpuroXBC expressing wild-type Tag (panels d and e), J domain-deficient Tag (panels g and h), pRb-binding-deficient Tag (panels j and k), or p53-binding-deficient Tag (panels m and n). Panels c, f, i, l, and o show images of representative colonies arising from the transfected cells grown at 40°C and shown in panels b, e, h, k, and n, respectively (boxed). (B) Schematic representation of SV40 Tag displaying the approximate sizes and positions of the J domain, pRb-binding motif, and p53-binding region. The diagram was adapted from the work of Stubdal et al. (31). (C) Immunoblot analysis of mutant forms of Tag after transient expression in tsBN67 cells at the nonpermissive temperature. Lysates were prepared from tsBN67 cells after transfection with 20 μg of the empty pBABEpuroXBC vector (lane 1), pBABEpuroXBCSV40e (lane 2), pBABEpuroXBC-Tag (wild type [wt]) (lane 3), p53-binding-deficient pBABEpuroXBC-Tag5031 (lane 4), J domain-defective pBABEpuroXBC-Tag5110 (lane 5), and pRb-binding-deficient pBABEpuroXBC-Tag3213 (lane 6), after maintenance at 40°C for 48 h. Extracts, normalized by determinations of the optical density at 280 nm, were probed with anti-Tag monoclonal antibody 419. The results shown are representative of three independent transient-expression assays.
In this experiment, the empty vector control gave a greater degree of background cell growth than is shown in Fig. 1 (compare Fig. 1B and 2A, panel b). As revealed by the micrograph in panel c, however, the dispersed, large, and flattened morphology of the cells is typical of arrested tsBN67 cells at 40°C (26), indicating that the cells are indeed arrested but at a higher concentration than occurred in the experiment whose results are shown in Fig. 1.
Compared to the control transfection, Tag induced tsBN67 cell proliferation at the nonpermissive temperature (Fig. 2A; compare panels b and e). At 33.5°C, the empty vector control gave a higher number of colonies than transfection of the Tag expression vector (compare plates a and d). Therefore, any stimulation of tsBN67 colony formation by Tag was not due to a higher number of viable cells initially plated. Examination of the representative colony from the Tag expression vector-transfected cells shows that these cells are smaller and much more crowded than the arrested tsBN67 cells (compare panels c and f), consistent with induction of long-term tsBN67 cell proliferation at 40°C. Thus, a specific gene product, SV40 Tag, is sufficient to rescue the tsBN67 cell proliferation defect.
The pRb-binding motif and J domain are important for Tag rescue of tsBN67 cell proliferation at the nonpermissive temperature.
SV40 Tag promotes cell proliferation by inactivating two important cell cycle regulators, the tumor suppressors pRb and p53. Figure 2B shows the regions of Tag involved in pRb and p53 inactivation. A major function of the pRb protein family members—pRb, p107, and p130—is to bind to the transcription factor E2F and repress transcription of S-phase-induced genes. Inactivation of the pRb family members by Tag involves the cooperation of two structurally defined regions, the LXCXE motif and the J domain, which inactivate pRb by a two-step mechanism. In the first step, direct binding of Tag to pRb through the LXCXE motif results in partial inactivation of pRb. In the second step, pRb inactivation is completed by the J domain, which activates Hsc70 to dissociate pRb from E2F (12, 33, 34).
To dissect the requirements of Tag for long-term tsBN67 cell proliferation rescue, we analyzed the activities of three mutant forms of Tag. To examine the role of pRb inactivation, we tested the single-amino-acid substitution D44N in the J domain, which abrogates the ability of Tag to bind to Hsc70 and to dissociate pRb from E2F but does not alter the pRb- or p53-binding activity of Tag (34), and the double-amino-acid LXCXE replacement E107K and E108K, which disrupts Tag binding to pRb but not p53 (31). To examine the contribution of p53 inactivation, we tested a clustered 3-amino-acid substitution, D402N, V404M, and V413M, which abrogates the ability of Tag to bind p53 (24). Figure 2C shows that, after transfection into tsBN67 cells and maintenance at 40°C for 2 days, the wild-type and mutant Tag proteins were synthesized faithfully, albeit at lower levels than with the SV40 early-region expression vector.
In the tsBN67 colony formation assay, the rescue of tsBN67 cell proliferation by the p53-binding-deficient mutant was similar to that of the wild-type Tag protein, both in the number and size of colonies induced at 40°C (Fig. 2A, compare panels e and n) as well as the nature of the colonies and their constituent cells (compare panels f and o). These results suggest that p53 binding and inactivation by Tag is dispensable for rescue of tsBN67 cell proliferation at nonpermissive temperatures. In contrast, the pRb-binding-deficient Tag mutant failed to induce any more robust colony formation than did the empty vector control (Fig. 2A, compare panels b and k) and these colonies contained cells with a dispersed, large, and flattened morphology similar to that of the temperature-arrested tsBN67 cells (compare panels c and l). The J domain mutant displayed an intermediate phenotype: compared to wild-type Tag, it was partially deficient in the rescue of tsBN67 colony formation (compare panels e and h), but unlike with the pRb-binding-deficient Tag mutant, the cells expressing the J domain mutant had the small size more characteristic of wild-type Tag-rescued tsBN67 cells (compare panels c, f, and i). This intermediate phenotype may reflect the possibility that the J domain Tag mutant binds to pRb but that it does not dissociate pRb from E2F (12). Together, these results indicate that inactivation of pRb family members is important for rescue of long-term tsBN67 cell proliferation at nonpermissive temperatures as measured by colony formation.
Suppression of the tsBN67 S-phase defect by SV40 Tag.
The colony formation assay is a stringent assay for long-term cell proliferation that requires not only cell division but also subsequent cell survival and adhesion to the plate. For example, a heterologous gene could induce tsBN67 cells to proliferate (i.e., enter S phase) but at the same time induce apoptosis such that “proliferating” cells fail to survive. Because there is a paucity of S-phase cells in temperature-arrested tsBN67 cells (8, 26), we used a short-term assay for cells in S phase to directly measure support of cell cycle progression by wild-type and mutant Tag.
In this assay, we transfected the gene of interest along with a small amount of plasmid directing the synthesis of YFP as a transfection marker into tsBN67 cells. We then placed the transfected cells at 40°C for 3 days to induce tsBN67 cell proliferation arrest. At this time, the cells were pulse-labeled for 20 min with BrdU, and following fixation, cells in S phase were visualized by fluorescent BrdU immunostaining. Cells in S phase were quantified in the YFP-positive cell population to eliminate untransfected cells from the analysis.
Table 1 shows the results of three BrdU incorporation assays and the averages thereof, using the SV40 early region and wild-type and mutant forms of Tag. When the YFP-encoding gene was cotransfected with the empty vector, there was an average of 2.2% BrdU-positive S-phase cells. This percentage is similar to the percentage seen in untreated tsBN67 cells or in the untransfected, YFP-negative population in this same sample (data not shown), suggesting that there is no significant effect of successful transfection on the number of tsBN67 cells incorporating BrdU. With transfection of the plasmid encoding the HCF-1N subunit, there was a large increase in BrdU-positive cells (24%). Remarkably, the SV40 early region (22.5%) and Tag alone (23%) rescued S phase to levels similar to those seen with the HCF-1N subunit. These levels of S-phase cells approximate the percentage of S-phase cells in a proliferating tsBN67 cell population at the permissive temperature (26), indicating that, unlike during long-term colony formation, SV40 Tag can suppress the mutant HCF-1-induced tsBN67 S-phase defect at nonpermissive temperatures as well as the HCF-1N subunit. This result also indicates that Tag is at saturating levels in this assay because the plasmid encoding the SV40 early region expresses Tag at much higher levels than the plasmid encoding Tag alone (see Fig. 2C).
TABLE 1.
Transient suppression of the tsBN67 cell S-phase defect
| Introduced gene | % of BrdU+ cellsa
|
|||
|---|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | Avg ± SD | |
| None | 1.5 | 2.0 | 3.0 | 2.2 ± 0.8 |
| HCF-1N1011 | 19.0 | 26.5 | 26.5 | 24.0 ± 4.3 |
| SV40 early region | 13.5 | 27.0 | 27.0 | 22.5 ± 7.8 |
| Large Tag: | ||||
| Wild type | 22.0 | 20.0 | 27.0 | 23.0 ± 3.6 |
| J domain deficient | 14.5 | 19.0 | 19.5 | 17.7 ± 3.9 |
| pRb-binding deficient | 1.5 | 5.0 | 7.0 | 4.5 ± 2.8 |
| p53-binding deficient | 9.5 | 12.5 | 11.5 | 11.2 ± 1.5 |
Percentage of YFP-positive cells that stained for BrdU incorporation as described in Materials and Methods. Two hundred YFP-positive cells were analyzed for each sample for each experiment.
The analysis of the Tag mutants shows that the p53-binding-deficient form of Tag, which induced colony formation to near wild-type levels, stimulated S phase effectively, albeit at an intermediate level compared to that of wild-type Tag (11 versus 23%). This twofold difference may be due to the lower levels of synthesis of this particular Tag mutant in transfected tsBN67 cells (Fig. 2C). Nevertheless, taken together, the short-term S-phase and long-term colony formation assays demonstrate that p53 binding and inactivation by Tag are dispensable for maintenance of tsBN67 cell proliferation at nonpermissive temperatures.
Consistent with the colony formation assay, the pRb-binding-deficient Tag mutant was the most defective in the S-phase induction assay, yielding only an average twofold increase over background (4.5%). Thus, consistent with the inability to rescue tsBN67 colony formation at 40°C, mutation of the LXCXE motif in Tag severely attenuates its ability to rescue short-term tsBN67 cell proliferation at nonpermissive temperatures.
In contrast, the J domain Tag mutant displayed very different activities in the colony formation and S-phase assays. Whereas this mutant gave an intermediate response in the colony formation assay (Fig. 2A), in the S-phase assay, it was very active, yielding an average of 18% S-phase-positive cells. Combined with the results of the pRb-binding-deficient Tag mutant, these results suggest that partial inactivation of pRb family members is sufficient for tsBN67 cells to enter S phase after 3 days at a nonpermissive temperature but that complete inactivation of pRb family members is required for long-term survival of proliferating tsBN67 cells at nonpermissive temperatures.
pRb-binding forms of the adenovirus E1A protein can also suppress the tsBN67 cell proliferation defect at nonpermissive temperatures.
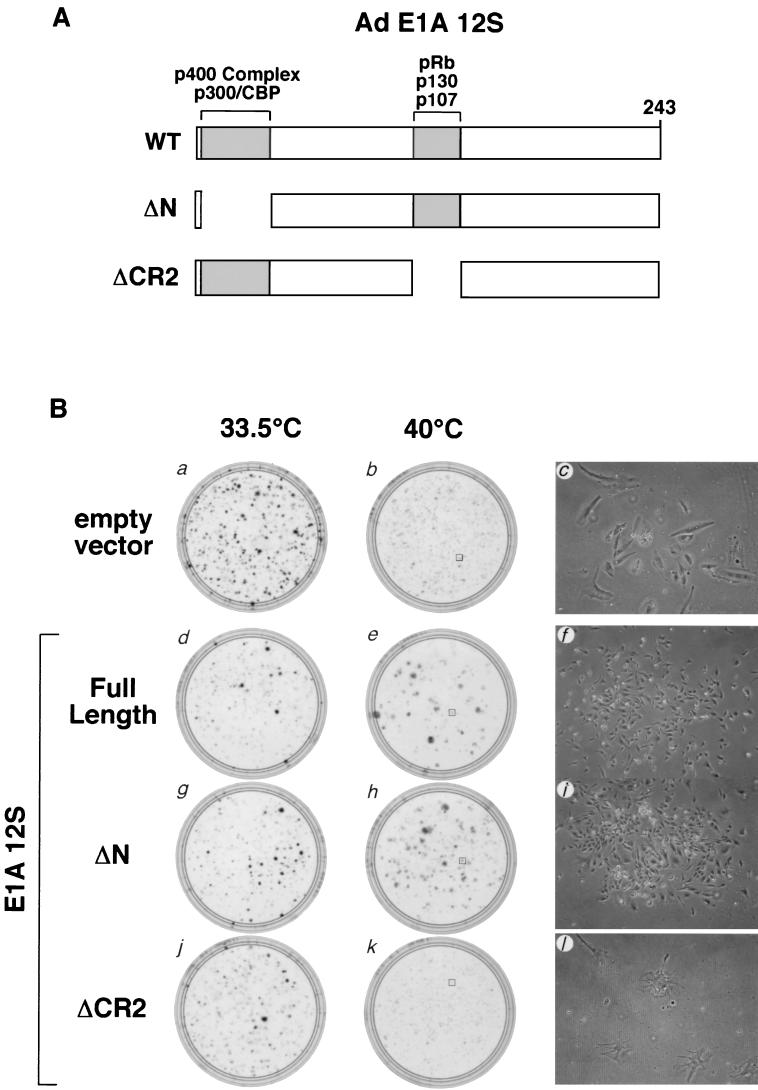
Since pRb inactivation is a common feature of DNA tumor viruses, we tested whether the adenovirus type 5 E1A 12S protein is also able to rescue tsBN67 cell proliferation arrest. To do so, we expressed the full-length wild-type E1A 12S protein as well as two well-characterized deletion mutants. Figure 3A shows a schematic representation of the E1A 12S protein denoting the regions deleted from the two mutants along with the proteins whose association with the E1A 12S protein are disrupted by each deletion. ΔN removes E1A 12S amino acids 2 to 36 and is defective for binding the transcriptional coregulators p300, CBP, and the p400 complex (7, 11, 32), whereas ΔCR2 removes E1A conserved region 2 (CR2), which spans amino acids 120 to 140, and is defective for binding to pRb family members (11).
FIG. 3.
Suppression of tsBN67 cell proliferation arrest with pRb-binding forms of adenovirus type 5 E1A 12S. (A) Schematic representation of adenovirus E1A protein. The N-terminal region and CR2, necessary for binding to p300, CBP, and the p400 complex, and the pRb family members, respectively, are shown. The E1A regions missing in the ΔN (residues 2 to 34) and ΔCR2 (residues 120 to 140) mutant proteins are schematically represented below. (B) Panels a, b, d, e, g, h, j, and k are stained plates after growth at 33.5°C (panels a, d, g, and j) or 40°C (panels b, e, h, and k) after transfection with 18 μg of the empty pBABEpuroXBC vector (panels a and b) or pBABEpuroXBC expressing full-length adenovirus E1A 12S (panels d and e), ΔN E1A 12S (panels g and h), or ΔCR2 E1A 12S (panels j and k) protein. Panels c, f, i, and l show images of representative colonies arising from the transfected cells grown at 40°C and based in panels b, e, h, and k, respectively. Panels a, b, and c are identical to panels a, b, and c in Fig. 2A.
As shown in Fig. 3B, in the same assay used to analyze the Tag mutants (Fig. 2A), at the nonpermissive temperature both wild-type E1A 12S and the ΔN mutant form induced tsBN67 colony formation (compare plate b to plates e and h) as well as the small-cell morphology typical of rescued tsBN67 cells (compare panel c to panels f and i). Thus, these two forms of E1A 12S that possess the ability to associate with and inactivate pRb family members were able to suppress tsBN67 growth arrest in the long-term colony formation assay. Deletion of CR2, however, abrogated the ability of E1A 12S to induce colony formation (panel k) and the small-cell morphology (panel l). Indeed, this mutant displayed a dominant effect on tsBN67 cell growth at the nonpermissive temperature, as illustrated by the decrease in the number of arrested cells and their unique morphology (compare panels c and l). Thus, activities of the ΔCR2 mutant (e.g., p300, CBP, and p400 complex binding) may have deleterious effects on cells that lack HCF-1 activity. In any case, as with SV40 Tag, the ability of E1A 12S to rescue the tsBN67 cell proliferation defect correlates with its ability to inactivate pRb family members.
Persistent and stable growth at the nonpermissive temperature of tsBN67 cells carrying the SV40 early region.
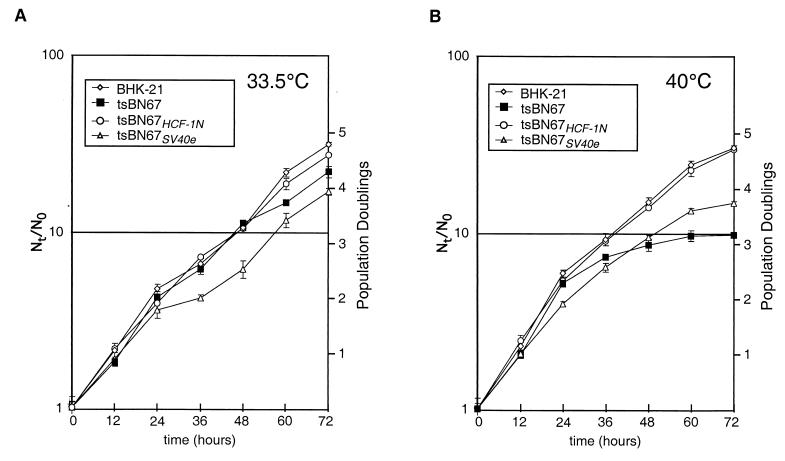
To further characterize the ability of the SV40 early region to rescue tsBN67 cell proliferation at a nonpermissive temperature, we isolated an SV40 early-region-rescued tsBN67 cell line, tsBN67SV40e, which arose in a colony formation assay with the SV40 early-region construct. Additionally, we isolated an HCF-1N subunit-rescued tsBN67 cell line, tsBN67HCF-1N, which similarly arose in a colony formation assay. The growth characteristics of these tsBN67 derivatives, alongside BHK-21 and tsBN67 cells, are shown in Fig. 4. Consistent with previous results, at 33.5°C (Fig. 4A) BHK-21 cells and tsBN67 cells have similar growth rates (8, 22). The tsBN67HCF-1N cells also display wild-type BHK-21 growth characteristics, indicating that synthesis of the HCF-1N subunit alone does not have a deleterious effect on cell growth at permissive temperatures. The tsBN67SV40e cells grew at normal rates during the first 24 h of the analysis but, for unknown reasons, they grew reproducibly more slowly thereafter owing to a decrease in growth rate between 24 and 48 h of this growth curve.
FIG. 4.
Growth characteristics of rescued tsBN67 cell lines. Shown are growth curves of BHK-21, tsBN67, tsBN67HCF-1N, and tsBN67SV40e cells at 33.5°C (A) or 40°C (B) as indicated. Two plates were sampled for each point, and error bars represent 1 standard deviation from the mean.
At 40°C (Fig. 4B), the tsBN67 cells displayed the characteristic tsBN67 cell proliferation arrest (8, 22, 26, 43). For 24 h, they grew identically to BHK-21 cells, after which the growth rate of these cells slowed dramatically until there was a near-complete cell proliferation arrest by 60 h. In contrast, both the BHK-21 and tsBN67HCF-1N cells proliferated similarly throughout the 72-h time course. The tsBN67SV40e cells also proliferated throughout the 72-h time course, although consistently at a rate lower than that of the BHK-21 cells or, indeed, of the tsBN67 cells during the first 24 h of the time course. Importantly, in contrast to the tsBN67rev cells described previously (26), the growth rate of the tsBN67SV40e cells did not change at the time at which the tsBN67 phenotype is initially manifest, i.e., after 24 to 48 h at 40°C, indicating that the SV40 early region can fully complement the short-term tsBN67 HCF-1 defect in tsBN67 cells. There was a decrease in growth rate after 60 h, but this decrease was not unique to the tsBN67SV40e cells and is probably because the cells were not refed during the course of the experiment. Thus, the tsBN67SV40e cell line displays a growth pattern similar to that of the BHK-21 or tsBN67HCF-1N cell line, albeit with slower kinetics. The reason for the slower proliferation (e.g., slower doubling time or increased apoptosis) of tsBN67SV40e cells is not known.
Rescue of E2F-responsive gene expression in the SV40 early-region-rescued tsBN67 cells.
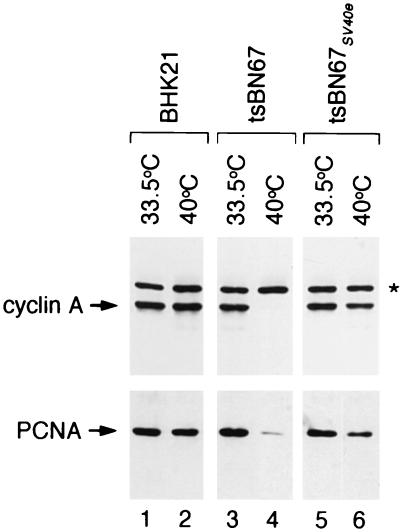
The tsBN67 arrest phenotype is characterized by reduced protein levels of E2F-responsive genes, including cyclin A, PCNA, and CDK2 (5, 14, 26). We, therefore, examined whether the tsBN67SV40e cells demonstrate a rescue of E2F-responsive gene expression consistent with the inactivation of pRb family members. By immunoblot analysis, the results of which are displayed in Fig. 5, we found that the tsBN67SV40e cells had higher levels of cyclin A and PCNA (lane 6) after growth at the nonpermissive temperature than the original tsBN67 cells (lane 4) grown at the nonpermissive temperature. Similar results were obtained for CDK2 (data not shown). These results are consistent with the activation of E2F-responsive promoters in the tsBN67SV40e cells.
FIG. 5.
Rescue of tsBN67 cyclin A and PCNA levels by the SV40 early region. Shown is an immunoblot for cyclin A and PCNA of extracts from BHK-21 (lanes 1 and 2), tsBN67 (lanes 3 and 4), and tsBN67SV40e (lanes 5 and 6) cells maintained at either 33.5°C (lanes 1, 3, and 5) or for 72 h (cyclin A samples) or 190 h (PCNA samples) at 40°C (lanes 2, 4, and 6). The asterisk denotes a cross-reactive band, which acts as a loading control.
The SV40 early region can rescue the temperature-sensitive tsBN67 cytokinesis defect.
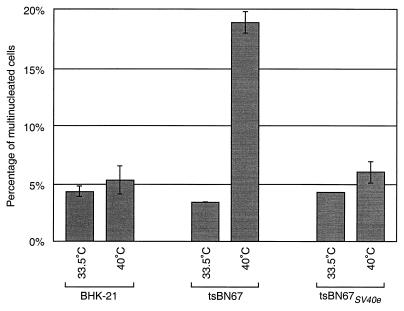
In temperature-arrested tsBN67 cells, loss of HCF-1 function also causes an increase from about 5 to 20% of the cells containing two or more nuclei. To examine whether the SV40 early region can rescue this tsBN67 cytokinesis defect, we examined the number of cells with two or more nuclei arising in populations of BHK-21, tsBN67, and tsBN67SV40e cells maintained at 33.5°C or 40°C for 48 h. The results of such an experiment are shown in Fig. 6. As shown previously (26), BHK-21 cells display similar percentages of multinucleated cells at 33.5°C (4.3%) and 40°C (5.3%) whereas tsBN67 cells display a temperature-sensitive increase in the number of multinucleated—predominantly binucleated—cells, from 3.4% at 33.5°C to 19% at 40°C. In contrast, tsBN67SV40e cells, like BHK-21 cells, had only a slight increase (from 4.3 to 6.0%) in the number of multinucleated cells after incubation at 40°C. These results indicate that expression of the SV40 early region in tsBN67 cells, in addition to rescuing the cell proliferation defect, can also rescue the temperature-induced cytokinesis defect in these cells.
FIG. 6.
The SV40 early region can rescue the temperature-sensitive tsBN67 cytokinesis defect. Cells with two or more nuclei were quantitated as described in Materials and Methods. Plates were maintained at 33.5°C or 40°C for 48 h prior to fixation. Columns represent average percentages of cells with two or more nuclei. Error bars denote 1 standard deviation from the mean.
The SV40 early region bypasses the requirement for HCF-1N subunit chromatin association in tsBN67 cells.
In stimulating tsBN67 cell proliferation at nonpermissive temperatures, the SV40 and adenovirus oncoproteins may be suppressing the HCF-1 defect either by (i) activating an HCF-1 function that is lost in the tsBN67 cells or (ii) bypassing the requirement for this function. Because HCF-1-chromatin association likely represents an essential HCF-1 function in tsBN67 cell proliferation, we asked whether HCF-1 is associated with chromatin in tsBN67SV40e cells proliferating at the nonpermissive temperature. For this analysis, cells were fractionated as described previously (19, 43) into two fractions—a soluble largely cytosolic fraction called S2 and a chromatin-enriched fraction called P3—and analyzed by immunoblot analysis (43).
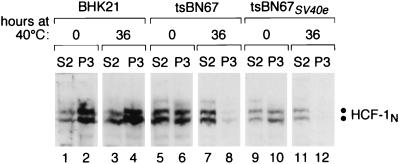
The results of such an experiment are shown in Fig. 7. As described previously, the wild-type HCF-1N subunits in BHK-21 cells are largely found in the chromatin-enriched P3 fraction at both 33.5°C (Fig. 7, lanes 1 and 2) and after 36 h at 40°C (lanes 3 and 4) whereas the mutant HCF-1N subunits in tsBN67 cells are associated with chromatin only at 33.5°C (lanes 5 to 8). Consistent with the hypothesis that HCF-1-chromatin association is important for tsBN67 cell proliferation and with the chromatin association of the HCF-1N subunit in HeLa cells proliferating at 37°C (43), in human HCF-1N-subunit-rescued tsBN67 cells proliferating at 40°C, the HCF-1N subunit was found in the chromatin-enriched P3 fraction (data not shown). In contrast, fractionation of the tsBN67SV40e cells showed that, similar to what occurs in tsBN67 cells, the presence of the endogenous HCF-1N subunits in the chromatin-enriched P3 fraction was temperature sensitive (compare lanes 9 to 12) and the protein was predominantly in the soluble cytosolic fraction when the cells were grown at 40°C for 36 h (compare lanes 11, and 12). The retention of this HCF-1N subunit defect in tsBN67SV40e cells proliferating at the nonpermissive temperature indicates that the SV40 early region can bypass the chromatin association requirement for the HCF-1N subunit.
FIG. 7.
Retention of the temperature-sensitive HCF-1-chromatin association defect in SV40-rescued tsBN67 cells. Shown is an immunoblot for HCF-1N subunit fractionation as described previously (19, 43) from tsBN67 (lanes 1 to 4), tsBN67SV40e (lanes 5 to 8), and BHK-21 (lanes 9 to 12) cells after maintenance at 33.5°C (lanes 1, 2, 5, 6, 9, and 10) or 40°C (lanes 3, 4, 7, 8, 11, and 12) for 36 h. S2 and P3 represent enrichment of cytosol and chromatin fractions, respectively.
DISCUSSION
In the studies described above, we demonstrate that products of DNA tumor virus oncogenes, namely SV40 Tag and adenovirus E1A, are able to rescue tsBN67 cell proliferation and inhibit tsBN67-specific cytokinesis defects at a nonpermissive temperature. Further, we demonstrate that this ability correlates closely with the ability of these proteins to inactivate the pRb family of proteins in both long-term colony formation assays and, for Tag, short-term S-phase assays. Importantly, tsBN67 cells rescued by the SV40 early genes do not reacquire HCF-1-chromatin-binding activity at nonpermissive temperatures, suggesting that Tag bypasses the HCF-1 requirement for tsBN67 cell proliferation and proper cytokinesis.
Heterologous rescue of a thermosensitive mammalian cell line has been previously demonstrated by Sekiguchi et al. (28). They showed that certain genes, including the DNA tumor virus oncogenes E7 and Tag, which function to inactivate pRb family members, are able rescue the TAF1 (previously referred to as TAFII250) defect in tsBN462 cell proliferation. In contrast to the study described here, however, the molecular basis of that rescue, whether through bypass or activation of the mutant TAF1, was not clarified. The combined results of suppression of temperature-sensitive cell proliferation defects in tsBN67 and tsBN462 by pRb-inactivating oncogenes suggest that inactivation of pRb family members may be a common way to rescue temperature-sensitive mammalian-cell proliferation arrest mutations.
HCF-1 provides a regulatory activity for mammalian-cell proliferation and cytokinesis.
In this study, we used colony formation and the maintenance of S-phase cells to assay for the ability of genes to suppress the HCF-1 defect in tsBN67 cells. The use of the 14-day colony formation assay has a limitation in that different aspects of cell growth other than cell proliferation, such as cell adhesion or cell motility, may disrupt the physical formation of colonies. Further, the colony formation assay is more sensitive to deleterious effects of introduced genes such as induction of apoptosis, which is a common effect of proliferation-inducing genes (6, 18, 44). The short-term S-phase assay avoided this problem, although it does not measure cell proliferation per se over extended periods. The combined results of these two assays show that the pRb-binding properties of SV40 Tag can promote tsBN67 cell cycle progression and long-term cell proliferation at nonpermissive temperatures.
The role of HCF-1 in cell proliferation has been enigmatic. The viral early proteins used in this study remodel the host cell by altering cell regulation, i.e., by changing gene expression and redirecting cellular resources and not by providing any known metabolic activities. The observation that such regulatory proteins can bypass HCF-1 function supports a model whereby HCF-1 performs a regulatory role in mammalian-cell proliferation.
HCF-1 may function through the pRb family to promote cell proliferation and proper cytokinesis.
The ability of SV40 Tag and adenovirus E1A to suppress the tsBN67 cell proliferation defect correlates with their ability to inactivate the pRb family. In both cases, disruption of the common LXCXE pRb-family-member-binding motif, in Tag (Fig. 2) and E1A (Fig. 3), abrogates the ability of these proteins to rescue tsBN67 cell proliferation. Thus, binding to pRb family members may be essential for these proteins to rescue tsBN67 cell proliferation.
Furthermore, an inactivation of the Hsp70 activation function of the Tag J domain, which is required for the complete inactivation of pRb family members by Tag (12, 33, 34), inhibits rescue of tsBN67 cell proliferation at nonpermissive temperatures. It is not clear why this mutation shows stronger specific defects in colony formation than in maintenance of a proliferative-cell morphology (Fig. 2) or S phase (Table 1) in tsBN67 cells at nonpermissive temperatures. Possibly J domain activation of Hsc70 family chaperone activity is beneficial to cell survival at elevated temperatures. Alternatively, binding of the J domain mutant Tag to pRb family members through the intact LXCXE motif may block complete repression of E2F target genes by histone deacetylases and histone methyl transferases (17, 21) but not completely activate the E2F transcription factors by failing to dissociate the pRb family members from the E2F transcriptional activation domain (12, 33). This circumstance might bring about short-term maintenance of S phase without inducing long-term viability or colony formation. Consistent with this hypothesis, the J domain is dispensable for Tag to induce anchorage-independent growth but is required to induce anchorage-dependent growth at a high density or in a low concentration of serum (35).
Although we have not defined the minimum Tag requirements for tsBN67 rescue, the dramatic loss of activity with the pRb-binding-deficient form of Tag and the efficiency at which the E1A ΔN mutation rescues cell proliferation suggest that pRb family inactivation may be the sole requirement for the bypass of HCF-1 dysfunction in tsBN67 cells. Consistent with the suggestion that tsBN67 cells arrest proliferation because the pRb family of proteins is inappropriately activated, in tsBN67 cells maintained at nonpermissive temperatures, pRb is present at normal levels but there is a reduction in phosphorylation (26), suggesting either reduced phosphorylation of pRb or enhanced dephosphorylation of pRb in these cells. Cellular phosphorylation of the pRb family is controlled by complex mechanisms, which integrate both positive and negative regulatory signals to signal appropriate cell proliferation (25, 29, 37). Thus, it is likely that HCF-1 functions in a regulatory pathway that influences the levels of pRb family protein phosphorylation.
pRb family inactivation leads to coincident suppression of cell proliferation and cytokinesis defects in tsBN67 cells at nonpermissive temperatures.
A coincident partial suppression of cell proliferation and cytokinesis defects has previously been noted in spontaneously revertant tsBN67rev cells, in which multiple uncharacterized genetic or epigenetic changes may bypass HCF-1 function (26). The coincident suppression of both the cell proliferation and cytokinesis defects in the tsBN67SV40e cell line indicates that a single genetic event and likely a single activity, namely, inactivation of the pRb family, can bypass the requirement for HCF-1 in both cell proliferation and execution of faithful mitosis. Thus, the role of HCF-1 in cell proliferation is intimately connected to its role in a cell's exit from mitosis.
Because mitosis is marked by a rapid and regulated dephosphorylation of pRb (27, 35) and because of the important role of the pRb family in G0/G1 control (37), it is likely that timely activation of pRb is critical in the M-to-G0/G1 transition. Precocious pRb family activation, caused by loss of HCF-1 function in tsBN67 cells, may explain the cytokinesis defect as well as the arrest of the majority of tsBN67 cells in the G0/G1 phase of the cell cycle. Consistent with a role for HCF-1 in pRb dephosphorylation, HCF-1 has been shown to associate with the catalytic subunit of protein phosphatase 1 (PP1) (1), which is responsible for pRb dephosphorylation (35). Indeed, Ajuh et al. (1) showed that HCF-1 inhibits the ability of PP1 to dephosphorylate a target protein—phosphorylase a. Although this substrate is unrelated to pRb, these connections suggest at least one means by which HCF-1 may regulate pRb phosphophorylation levels. Through constitutive inactivation of pRb, SV40 Tag and adenovirus E1A may bypass inappropriate activation of pRb during mitosis and thus allow appropriate mitosis and cell proliferation to proceed. Whichever the mechanism, the results described here are consistent with an active role of the pRb family of proteins during mitosis, particularly cytokinesis.
The particular means by which HCF-1 functions to regulate pRb phosphorylation, cell proliferation, and cytokinesis remains to be identified. The requirement for HCF-1 function, however, in mitosis and cell proliferation can be bypassed by viral oncogenes that inactivate the pRb family. This suppression by regulatory proteins supports a model in which HCF-1 is a regulatory molecule in mammalian-cell division and proliferation.
Acknowledgments
We thank Takeharu Nishimoto for providing the tsBN67 cell line; Eric Julien for his help with the tsBN67SV40e nucleation experiment; Paula Sacco-Bubulya, Andrew Samuelson, and James Pipas for providing plasmids; Richard Freiman, J. Peter Gergen, Patrick Hearing, Nouria Hernandez, Scott Lowe, Ute Moll, and William Tansey for helpful discussions; Deborah Aufiero, Martha Daddario, and Regina Whitaker for technical support; Eric Julien and Nouria Hernandez for critical readings of the manuscript; and Bruce Stillman for encouragement and support.
These studies were supported by PHS grants GM54598 and CA13106.
REFERENCES
- 1.Ajuh, P. M., G. J. Browne, N. A. Hawkes, P. T. Cohen, S. G. Roberts, and A. I. Lamond. 2000. Association of a protein phosphatase 1 activity with the human factor C1 (HCF) complex. Nucleic Acids Res. 28:678-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumann, C. T., C. S. Lim, and G. L. Hager. 1998. Simultaneous visualization of the yellow and green forms of the green fluorescent protein in living cells. J. Histochem. Cytochem. 46:1073-1076. [DOI] [PubMed] [Google Scholar]
- 3.Chang, L. S., M. M. Pater, N. I. Hutchinson, and G. di Mayorca. 1984. Transformation by purified early genes of simian virus 40. Virology 133:341-353. [DOI] [PubMed] [Google Scholar]
- 4.Crawford, L., K. Leppard, D. Lane, and E. Harlow. 1982. Cellular proteins reactive with monoclonal antibodies directed against simian virus 40 T-antigen. J. Virol. 42:612-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeGregori, J., T. Kowalik., and J. R. Nevins. 1995. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol. Cell. Biol. 15:4215-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura, V. Ogryzko, W. S. Lane, Y. Nakatani, and D. M. Livingston. 2001. The p400 complex is an essential E1A transformation target. Cell 106:297-307. [DOI] [PubMed] [Google Scholar]
- 8.Goto, H., S. Motomura, A. C. Wilson, R. N. Freiman, Y. Nakabekku, K. Fukushima, M. Fujishima, W. Herr, and T. Nishimoto. 1997. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 11:726-737. [DOI] [PubMed] [Google Scholar]
- 9.Herr, W. 1998. The herpes simplex virus VP16-induced complex: mechanisms of combinatorial transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63:599-607. [DOI] [PubMed] [Google Scholar]
- 10.Hughes, T. A., S. La Boissiere, and P. O'Hare. 1999. Analysis of functional domains of the host cell factor involved in VP16 complex formation. J. Biol. Chem. 274:16437-16443. [DOI] [PubMed] [Google Scholar]
- 11.Kannabiran, C., G. F. Morris, C. Labrie, and M. B. Mathews. 1993. The adenovirus E1A 12S product displays functional redundancy in activating the human proliferating cell nuclear antigen promoter. J. Virol. 67:507-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, H. Y., B. Y. Ahn, and Y. Cho. 2001. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knipe, D. M. 1989. The role of viral and cellular nuclear proteins in herpes simplex virus replication. Adv. Virus Res. 37:85-123. [DOI] [PubMed] [Google Scholar]
- 14.Knudsen, K. E., A. F. Fribourg, M. W. Strobeck, J.-M. Blanchard, and E. S. Knudsen. 1999. Cyclin A is a functional target of retinoblastoma tumor suppressor protein-mediated cell cycle arrest. J. Biol. Chem. 274:27632-27641. [DOI] [PubMed] [Google Scholar]
- 15.Kristie, T. M., J. L. Pomerantz, T. C. Twomey, S. A. Parent, and P. A. Sharp. 1995. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J. Biol. Chem. 270:4387-4394. [DOI] [PubMed] [Google Scholar]
- 16.LaBoissiere, S., S. Walker, and P. O'Hare. 1997. Concerted activity of host cell factor subregions in promoting stable VP16 complex assembly and preventing interference by the acidic activation domain. Mol. Cell. Biol. 17:7108-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai, A., B. K. Kennedy, D. A. Barbie, N. R. Bertos, X. J. Yang, M. C. Theberge, S. C. Tsai, E. Seto, Y. Zhang, A. Kuzmichev, W. S. Lane, D. Reinberg, E. Harlow, and P. E. Branton. 2001. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 21:2918-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leone, G., R. Sears, E. Huang, R. Rempel, F. Nuckolls, C. H. Park, P. Giangrande, L. Wu, H. I. Saavedra, S. J. Field, M. A. Thompson, H. Yang, Y. Fujiwara, M. E. Greenberg, S. Orkin, C. Smith, and J. R. Nevins. 2001. Myc requires distinct E2F activities to induce S phase and apoptosis. Mol. Cell 8:105-113. [DOI] [PubMed] [Google Scholar]
- 19.Mendez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance protein during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moran, E. 1993. DNA tumor virus transforming proteins and the cell cycle. Curr. Opin. Genet. Dev. 3:63-70. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 22.Nishimoto, T., and C. Basilico. 1978. Analysis of a method for selecting temperature-sensitive mutants of BHK cells. Somatic Cell Genet. 4:323-340. [DOI] [PubMed] [Google Scholar]
- 23.O'Hare, P. 1993. The virion transactivator of herpes simplex virus. Semin. Virol. 4:145-155. [Google Scholar]
- 24.Peden, K. W., A. Srinivasan, J. V. Vartikar, and J. M. Pipas. 1998. Effects of mutations within the SV40 large T antigen ATPase/p53 binding domain on viral replication and transformation. Virus Genes 16:153-165. [DOI] [PubMed] [Google Scholar]
- 25.Ravitz, M. J., and C. E. Wenner. 1997. Cyclin-dependent kinase regulation during G1 phase and cell cycle regulation by TGF-beta. Adv. Cancer Res. 71:165-207. [DOI] [PubMed] [Google Scholar]
- 26.Reilly, P. T., and W. Herr. 2002. Spontaneous reversion of tsBN67-cell proliferation and cytokinesis defects in the absence of HCF-1 function. Exp. Cell Res. 277:119-130. [DOI] [PubMed] [Google Scholar]
- 27.Rubin, E., S. Mittnacht, E. Villa-Moruzzi, and J. W. Ludlow. 2001. Site-specific and temporally-regulated retinoblastoma protein dephosphorylation by protein phosphatase type 1. Oncogene 20:3776-3785. [DOI] [PubMed] [Google Scholar]
- 28.Sekiguchi, T., T. Nishimoto, and T. Hunter. 1999. Overexpression of D-type cyclins, E2F-1, SV40 large T antigen and HPV16 E7 rescue cell cycle arrest of tsBN462 cells caused by the CCG1/TAF(II)250 mutation. Oncogene 18:1797-1806. [DOI] [PubMed] [Google Scholar]
- 29.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 30.Spector, D. L., R. D. Goldman, and L. A. Leinwald. 1998. Cells: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 31.Srinivasan, A., A. J. McClellan, J. Vartikar, I. Marks, P. Cantalupo, Y. Li, P. Whyte, K. Rundell, J. L. Brodsky, and J. M. Pipas. 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 17:4761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stein, R. W., M. Corrigan, P. Yaciuk, J. Whelan, and E. Moran. 1990. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J. Virol. 64:4421-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stubdal, H., J. Zalvide, K. S. Campbell, C. Schweitzer, T. M. Roberts, and J. A. DeCaprio. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 17:4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan, C. S., P. Cantalupo, and J. M. Pipas. 2000. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol. Cell. Biol. 20:6233-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamrakar, S., E. Rubin, and J. W. Ludlow. 2000. Role of pRB dephosphorylation in cell cycle regulation. Front. Biosci. 5: D121-D137. [DOI] [PubMed] [Google Scholar]
- 36.Vousden, K. H. 1995. Regulation of the cell cycle by viral oncoproteins. Semin. Cancer Biol. 6:109-116. [DOI] [PubMed] [Google Scholar]
- 37.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 38.Whitley, R. J., and B. Roizman. 2001. Herpes simplex virus infections. Lancet 357: 1513-1518. [DOI] [PubMed] [Google Scholar]
- 39.Wilson, A. C., K. LeMarco, M. G. Peterson, and W. Herr. 1993. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell 74:115-125. [DOI] [PubMed] [Google Scholar]
- 40.Wilson, A. C., J. E. Parrish, H. F. Massa, D. L. Nelson, B. J. Trask, and W. Herr. 1995. The gene encoding the VP16-accessory protein HCF (HCFC1) resides in human Xq28 and is highly expressed in fetal tissues and the adult kidney. Genomics 25:462-468. [DOI] [PubMed] [Google Scholar]
- 41.Wilson, A. C., M. G. Peterson, and W. Herr. 1995. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 9:2445-2458. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, A. C., R. N. Freiman, H. Goto, T. Nishimoto, and W. Herr. 1997. VP16 targets an amino-terminal domain of HCF involved in cell-cycle progression. Mol. Cell. Biol. 17:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wysocka, J., P. T. Reilly, and W. Herr. 2001. Loss of HCF-1 chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell. Biol. 21:3820-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziebold, U., T. Reza, A. Caron, and J. A. Lees. 2001. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 15:386-391. [DOI] [PMC free article] [PubMed] [Google Scholar]