Abstract
Skp1 is involved in a variety of crucial cellular functions, among which the best understood is the formation together with Cul1 of Skp1-cullin-F-box protein ubiquitin ligases. To investigate the role of Skp1, we generated transgenic (Tg) mice expressing a Cul1 deletion mutant (Cul1-N252) able to sequestrate and inactivate Skp1. In vivo interference with Skp1 function through expression of the Cul1-N252 mutant into the T-cell lineage results in lymphoid organ hypoplasia and reduced proliferation. Nonetheless, after a period of latency, Cul1-N252 Tg mice succumb to T-cell lymphomas with high penetrance (>80%). Both T-cell depletion and the neoplastic phenotype of Cul1-N252 Tg mice are largely rescued in Cul1-N252, Skp1 double-Tg mice, indicating that the effects of Cul1-N252 are due to a sequestration of the endogenous Skp1. Analysis of Cul1-N252 lymphomas demonstrates striking karyotype heterogeneity associated with c-myc amplification and c-Myc overexpression. We show that the in vitro expression of the Cul1-N252 mutant causes a pleiotrophic phenotype, which includes the formation of multinucleated cells, centrosome and mitotic spindle abnormalities, and impaired chromosome segregation. Our findings support a crucial role for Skp1 in proper chromosomal segregation, which is required for the maintenance of euploidy and suppression of transformation.
The selective degradation of many short-lived proteins in eukaryotic cells is carried out by the ubiquitin-proteasome pathway (reviewed in reference 23). Two distinct ubiquitin conjugation pathways mediate cell division by affecting the transition from G1 to S phase, the separation of sister chromatids during anaphase, and the exit from mitosis. The first event in G1/S requires the ubiquitin-conjugating enzyme Cdc34 (or Ubc3) and a ubiquitin-protein ligase complex termed SCF complex (Skp1-Cullin-F-box protein) in order to activate DNA replication (reviewed in reference 11). The two mitotic events involve a large ubiquitin-ligase complex called the anaphase-promoting complex-cyclosome (APC/C) in combination with one of two distinct ubiquitin-conjugating enzymes (Ubc10 or Ubc4). APC/C regulates mitosis by affecting chromosome and spindle dynamics and by regulating the activity of mitotic cyclin-dependent kinase (Cdks) (reviewed in reference 53).
SCF ligases consist of three invariable subunits (Skp1, Cul1, and Rbx1/Roc1) and a variable component known as the F-box protein. SCF complex targets include cell cycle regulators such as G1-phase cyclins, Cdk inhibitors, and DNA replication and transcription factors, as well as non-cell cycle-specific substrates (27). F-box proteins bind to Skp1 through their F-box motif and serve as the substrate recognition subunit (2, 15, 44). More than 50 mammalian F-box proteins, which are involved in the recruitment of specific substrates, have been identified to date (6, 49). So far, only four SCFs have been well characterized in mammals: SCFβTrCP1, SCFβTrCP2, SCFSkp2, and SCFhCDC4/Fbw7 (reviewed in references 12, 42, and 47).
Skp1 is an adapter subunit that links the F-box protein to Cul1 (33, 35, 44, 51). In yeast, different Skp1 mutants arrest cells either in G1 or in G2, suggesting an involvement of Skp1 in different stages of the cycle (2, 8). Interestingly, in mammalian cells, both Skp1 and Cul1, besides being localized within the cytoplasm and the nucleus, can associate with centrosomes (16, 22). Indeed, it has been suggested that Skp1 forms an extended pericentriolar structure that may serve to organize the centrosomes (16) and could therefore be involved in chromosome segregation. Several studies in yeast indicate that, rather than exclusively linking F-box proteins and Cul1, Skp1 might have additional roles outside the SCF complexes. In particular, Skp1p activates Ctf13p by promoting its phosphorylation, thereby allowing Ctf13p to activate the CBF3 kinetochore complex (24). Moreover, Skp1p has been found to bind Rcy1p to facilitate SNARE recycling (18) and Rav1p has been found to regulate V-ATPase assembly (43). However, these functions have not been confirmed in other organisms.
In humans, six CUL genes have been identified (26). While all of these gene products are capable of binding to Rbx1/Roc1, only Cul1 interacts with Skp1 to form SCF complexes (33, 35). Cul1 has three domains that mediate its association with other components of the SCF complex. The N-terminal region, which in Cul1 mediates binding to Skp1, is the least conserved domain among cullin members (35). The ability to ubiquitinylate substrates depends on two elements at the COOH terminus that are independently required for Cul1 to interact with the E2 enzyme Cdc34 and the RING finger protein Rbx1/Roc1, respectively. The third and most highly conserved domain present in the extreme C terminus of all cullins mediates the attachment of a small ubiquitin-like protein, Nedd8 (31). The conjugation of Nedd8 to the arginine residue at position 720 of Cul1 appears to enhance the ubiquitin-ligating activity of SCF ligases (50) by increasing their affinity for some E2 enzymes (25).
In Caenorhabditis elegans, null mutations of Cul1 generate sterile animals with hyperplastic larval tissues (26). In addition, somatic and germ line cells are smaller, suggesting that Cul1 is involved in cell cycle withdrawal and size regulation of nematode cells. The requirement for the Cul1 gene in development is even more striking in mammalians cells. Mice carrying a Cul1 deletion die in utero around embryonic day 6.5 (10, 48). Unlike C. elegans, Cul1-null blastocysts have limited proliferative capacity in spite of elevated cyclin E levels and contain abnormally large trophoblast cells.
Recently, it has been determined that multiple Skp1-related proteins are expressed in C. elegans, of which only a fraction is able to interact with CUL1 (37, 52). Experiments of Skp1-related RNA interference showed embryonic and larval hyperplasia as observed in Cul1-null animals. However, additional phenotypes were also described, suggesting the existence of Cul1-independent functions (37, 52). Thus, although these studies have shown that Cul1 and Skp1 are indispensable for the early development of nematodes and mammals, their function in postnatal and adult organs has not been demonstrated.
To investigate the role of Skp1 complexes in vivo, we generated transgenic (Tg) mice expressing in the T-cell lineage a Cul1 deletion mutant (Cul1-N252) capable to sequestrate Skp1. Cul1-N252 mice showed hypoplastic lymphoid organs whose T cells were unresponsive to in vitro mitogenic stimulation. Notably, forced expression of Cul1-N252 caused the formation of multinucleated cells, defects in centrosomes and mitotic spindles, impaired chromosome segregation, and chromosomal instability and also resulted in neoplastic transformation with high penetrance. These findings support a crucial role for Skp1 in the preservation of genetic stability.
MATERIALS AND METHODS
Constructs and plasmids.
Cul1 mutant constructs were generated by recombinant PCR. A Flag tag was added to the 3′ end of wild-type (wt) Cul1 and the Cul1 mutants encoding 252 (N252) and 385 (N385) amino-terminal residues. Cul1 K720R mutation was generated by oligonucleotide-directed mutagenesis by PCR with the QuikChange site-directed mutagenesis kit (Stratagene). pRC/cyclin E was kindly provided by J. Lukas. All constructs were subcloned (BamHI-NotI) into an episomal green fluorescent protein retroviral expression vector (Pallino) (21) and subsequently sequenced. His-tagged Skp1 was subcloned in pcDNA3 (4). The Cul1 mutant (residues 324 to 776) was provided by Z. Q. Pan (50). Cul1-N252 was cloned (BamHI-NotI) into the Tet-inducible pcDNA5TO vector (Invitrogen).
Cell cultures.
Transfections of 293T, HeLa, U2-OS, and NIH 3T3 cells were performed with Effectene reagent according to the manufacturer's instructions (Qiagen). Cells were cultured in Dulbecco's modified Eagle medium plus 10% calf serum or fetal calf serum for 24 h, selected with puromycin (1 μg/ml) for 5 days, and lysed. Cytosolic extracts were used for immunoprecipitation or Western blotting. Induction of Cul1-N252 in transfected 293Trex cells (Invitrogen) was achieved with doxycycline (1 μg/ml). Cells were harvested after 72 h of culture.
Extract preparation, immunoprecipitation, Western blotting, and antibodies.
Immunoprecipitation assays and Western blotting were performed as described previously (28, 38). Monoclonal antibodies (MAbs) to human Cul1, Skp2, Skp1, and cyclin E and MAbs to rabbit cyclin A, Cdk2, and Roc1 were previously described (4, 5, 28). MAbs to p21 (catalog number C24420), p27 (catalog number K25020), and β-catenin (catalog number P46020) were purchased from Transduction Laboratories; MAbs to Flag (catalog number F3165) and α-tubulin (catalog number T5168) were purchased from Sigma; and MAbs to cyclin D3 (catalog number MS-215-P) were purchased from Neomarkers. Rabbit polyclonal antibodies to Flag were purchased from Zymed (catalog number 71-5400), and rabbit polyclonal antibodies to cyclin B (catalog number sc-245), E2F-1 (catalog number sc-193), c-Myc (catalog number sc-746), cyclin E (catalog number sc-481), and His (catalog number sc-803) were purchased from Santa Cruz.
Immunohistochemical staining.
For immunohistochemical staining, anti-p27 MAb (1:1,000, catalog number K25020; Transduction Laboratories) was used. Immunostainings were performed on formalin-fixed, paraffin-embedded tissues by the avidin-biotin-peroxidase complex method and by using a semiautomated immunostainer (Ventana Systems) as described previously (7).
Tg mice.
Human Cul1-N252 mutant or human wt Skp1 proteins were cloned (SacI-SalI) into a vector containing the minimal CD4 enhancer (339 bp), the minimal murine CD4 promoter (487 bp), the transcription initiation site, and 70 bp of the untranslated first exon and part of the first intron of the murine CD4 gene (41). Tg mice were generated as described previously (7). Screening of founder animals was performed by PCR and confirmed by Southern hybridization on genomic DNA from tail biopsy samples. Screening of the offspring was performed by PCR amplification of tail DNA. Double-Tg mice were generated by crossing CD4-Cul1-N252 mice with CD4-Skp1 mice. Mice were housed in the Skirball Institute Animal Facilities of the New York University under National Institutes of Health (NIH) guidelines. Animals were monitored daily. Necropsies were performed on all animals that died spontaneously or were killed during the observation period. A portion of each sample was fixed in formalin, embedded in paraffin, and sectioned for staining with hematoxylin and eosin stain, while another portion was frozen. Histological analysis of the thymus, spleen, and lymph nodes was performed as described previously (7). For immunophenotyping, fluorochrome-conjugated antibodies against CD4 (fluorescein isothiocyanate), CD8 (phycoerythrin), and CD90/Thy-1 (fluorescein isothiocyanate) from Pharmingen were used. Overtime survival was calculated, and the statistical significance was calculated by using the log rank test.
Tumorigenicity in nude mice.
NCr nude mice (nu/nu) were purchased from Taconic. NIH 3T3 cells transfected with wt Cul1, the Cul1-N252 mutant, or empty vector were grown in selective medium (1 μg of puromycin/ml) for 30 days. Subconfluent cells were harvested and resuspended in phosphate-buffered saline (PBS; pH 7.2). One hundred microliters of cells (106) was inoculated subcutaneously in nude mice previously treated with cyclophophamide (150 mg/kg of body weight). Tumor growth was monitored weekly.
T-cell suspension and proliferation assay.
Thymi were dissected, washed in PBS, cut into small pieces, and put in a 60-mm-diameter petri dish containing complete medium (RPMI 1640, 10% fetal bovine serum, 50 μM β-mercaptoethanol, 2 mM l-glutamine, 0.1% penicillin-streptomycin). Single-cell suspensions were mechanically prepared by crushing the pieces of thymus or lymph nodes with the bottom of a 3-ml syringe plunger. Viable cells were then pelleted, resuspended in complete medium, seeded in a 96-well dish (0.25 × 106 cells per well), and then activated with concanavalin A (5 μg/ml) in the presence of interleukin-2 (IL-2; 50 ng/ml) or with phorbol 12-myristate 13-acetate (50 ng/ml) and ionomycin (1 μM) or phytohemagglutinin (PHA; 5 μg/ml) in the presence of IL-1 (2 ng/ml), IL-4 (10 ng/ml), IL-7 (20 ng/ml), and β-mercaptoethanol (50 μM). After a 48-h incubation at 37°C, [3H]thymidine (1 μCi) was added for an additional 24-h period and proliferation was assessed by [3H]thymidine incorporation.
Southern blot analysis.
For Southern blot analysis, 3-μg aliquots of genomic DNA were digested with EcoRI, electrophoresed, denatured, and transferred to a Hybond N+ membrane (Amersham Pharmacia Biotech). Blots were hybridized with 32P-labeled cDNA probes specific for murine c-myc (XhoI fragment) and β-actin.
Karyotype and FISH analysis.
Cell cultures were incubated in medium containing Colcemid (0.1 μg/ml) for 2 to 18 h and harvested by standard cytogenetic procedure. Metaphase spreads were stained with 4′,6′-diamidino-2-phenylindole (DAPI; Sigma). Chromosomal distributions included the analysis of 100 metaphase spreads for each experiment. Fluorescent in situ hybridization (FISH) analysis was performed on 293T cells with centromeric probes specific for chromosomes X, 7, 12, and 18 as described by the manufacturer (Vysis). At least 100 cells were scored for each experiment. For c-myc FISH analysis, metaphases were hybridized with a bacterial artificial chromosome probe kindly provided by M. J. Difilippantonio. The c-myc genomic probe was nick translated with SpectrumGreen dUTP (Vysis) and hybridized by following standard procedures.
Centrosome and mitotic spindle staining.
Transfected NIH 3T3 cells were grown in selective medium (1 μg of puromycin/ml) for 5 days onto glass coverslips, rinsed in PBS, and fixed with ice-cold methanol for 20 min at −20°C, followed by permeabilization for 10 min with 0.25% Triton X-100 in PBS at room temperature. Immunofluorescence stainings were performed with mouse antibodies to α-tubulin or γ-tubulin (Sigma) and detected with anti-mouse biotinylated antibodies, followed by streptavidin-Cy3 (Sigma). Samples were counterstained with DAPI. At least 100 cells were analyzed for each experiment.
RESULTS
Expression of Cul1 mutants increases cellular levels of SCF substrates.
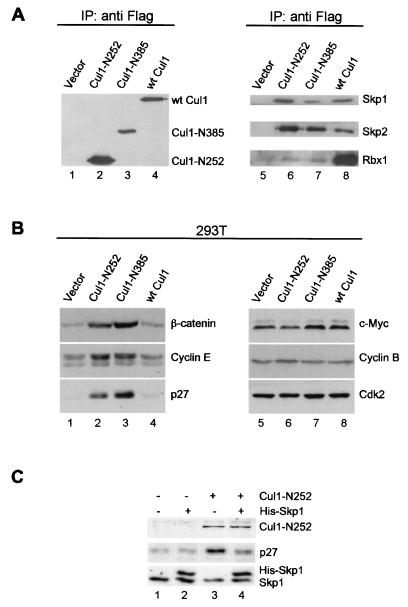
Since Cul1-Skp1 interaction is mediated by the NH2-terminal domains of both proteins and does not require the central and COOH-terminal domains of Cul1, we reasoned that the function of Skp1 complexes could be impaired by expressing the NH2-terminal mutants of Cul1 able to bind Skp1 but lacking the docking sites for Rbx1/Roc1 and Cdc34. wt Cul1 and two Cul1 mutants encoding 252 and 385 NH2-terminal amino acid residues (Cul1-N252 and Cul1-N385, respectively; all 3′ Flag tagged) were transfected into 293T cells. Immunoprecipitation experiments demonstrated that, together with wt Cul1 and the Cul1 mutants (Fig. 1A, lanes 1 through 4), the anti-Flag antibody coprecipitated both Skp1 and Skp2, whereas Rbx1/Roc1 associated with only wt Cul1 (Fig. 1A, lanes 5 through 8). These data show that the Cul1-N252 and Cul1-N385 mutants are able to bind Skp1 and Skp2 and likely other F-box proteins but cannot interact with Rbx1/Roc1.
FIG. 1.
Expression of Cul1 mutants increases protein levels of SCF complex substrates. (A) Cul1-N252 and Cul1-N385 mutant proteins associate with Skp1 and the F-box protein Skp2 but not with Rbx1/Roc1. 293T cells were transfected with 3′-Flag-tagged Cul1 mutants encoding N-terminal amino acid peptides residues 1 to 252 (Cul1-N252) or residues 1 to 385 (Cul1-N385) or with wt Cul1. Transfected cells were selected for 5 days after the addition of puromycin (1 μg/ml). Lysates were first immunoprecipitated with mouse anti-Flag antibody and then immunoblotted with rabbit anti-Flag antibody (lanes 1 through 4) or the Skp1, Skp2, or Rbx1/Roc1 antibody (lanes 5 through 8). (B) Expression of Cul1-N252 mutants increases protein levels of p27, cyclin E, β-catenin, and p21. 293T cells transfected with empty vector (Pallino) (lanes 1 and 5), Cul1-N252 (lanes 2 and 6), Cul1-N385 (lanes 3 and 7), or wt Cul1 (lanes 4 and 8) were selected with puromycin for 5 days. Lysates from transfected cells were then immunoblotted with the indicated antibodies. (C) Coexpression of Skp1 attenuates the effects of the Cul1-N252 mutant on p27 accumulation. 293T cells were transfected with vector (pcDNA3) alone (2 μg) (lane 1), with a combination of empty vector and His-tagged Skp1 (0.4 and 1.6 μg) (lane 2), with empty vector and the Cul1-N252 mutant (1.6 and 0.4 μg) (lane 3), or with both His-Skp1 and the Cul1-N252 mutant (1.6 and 0.4 μg) (lane 4). Cells were lysed 48 h posttransfection and immunoblotted with the anti-Flag, p27, and Skp1 antibodies.
To test whether the expression of Cul1 mutants in 293T cells interfered with SCF-dependent degradation, we examined the levels of various cell cycle regulators. The abundance of known SCF substrates such as cyclin E, p27, and β-catenin was significantly and specifically increased in cells expressing either the Cul1-N252 or Cul1-N385 mutant, whereas the amounts of other potential substrates, including E2F-1 (data not shown) and c-Myc as well as those of control proteins (cyclin B and Cdk2), did not change (Fig. 1B). Similar effects were observed in other cell lines, including HeLa, U2-OS, and NIH 3T3 (data not shown).
Importantly, coexpression of Skp1 partially blocked the effects of Cul1-N252 on p27 accumulation (Fig. 1C). This strongly suggests that Cul1-N252 effects are specifically mediated by the sequestration of endogenous Skp1-F-box protein complexes. The specificity of the system is further supported by the fact that the expression of an additional Cul1 deletion mutant unable to bind Skp1 (Cul1 from residues 324 to 776) did not result in a detectable increase in p27 levels (data not shown).
Lymphoid organ hypoplasia and reduced T-cell proliferation in CD4-Cul1-N252 mice.
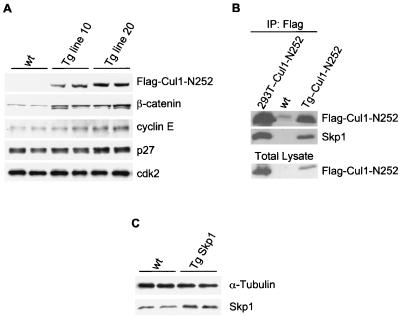
To evaluate the biological effects of the Cul1-N252 mutant in vivo, we generated Tg mice targeting the expression of Cul1-N252 to the T-lymphoid lineage. This model guarantees postnatal development and fertility and represents a good system to study cell proliferation in vivo. Flag-tagged human Cul1-N252 was placed under the murine CD4 minimal promoter in the presence of the CD4 enhancer. This promoter lacks the CD4 silencer region and is transcriptionally active in both single- and double-positive T cells (41). Three independent Cul1-N252 Tg lines of mice (lines 4, 10, and 20) were obtained. All the experiments described herein were performed with the CD4-Cul1-N252 Tg lines 10 and 20 that expressed the highest levels of Flag-tagged Cul1-N252 (Fig. 2A, top panel). Importantly, the human Cul1-N252 mutant was able to bind to murine Skp1 (Fig. 2B), verifying that the exogenous protein could assemble within murine SCF complexes. Moreover, we generated Skp1 Tg mice constitutively expressing Skp1 whose expression was restricted to T lymphocytes (Fig. 2C).
FIG. 2.
Generation of CD4-Cul1-N252 Tg mice. (A) Expression of the Flag-tagged Cul1-N252 mutant and other cell cycle regulatory proteins in CD4-Cul1-N252 Tg mice. Extracts (30 μg of protein) from the thymocytes of two control non-Tg mice (wt), Cul1-N252 Tg line 10 mice, and Tg line 20 mice were immunoblotted with the antibodies to the indicated proteins. (B) Association of the human Flag-tagged Cul1-N252 mutant with endogenous murine Skp1. Thymocytes of a wt (middle lane) and a CD4-Cul1-N252 Tg mouse (right lane) were immunoprecipitated with a rabbit anti-Flag antibody and then immunoblotted with antibodies to Flag or Skp1. 293T cells transfected with Cul1-N252 were used as a positive control (left lane). The bottom panel shows the corresponding total lysates immunoblotted with anti-Flag antibodies. (C) Expression of Skp1 in CD4-Skp1 Tg mice. Thymocytes of wt and Tg mice were immunoblotted with the indicated antibodies.
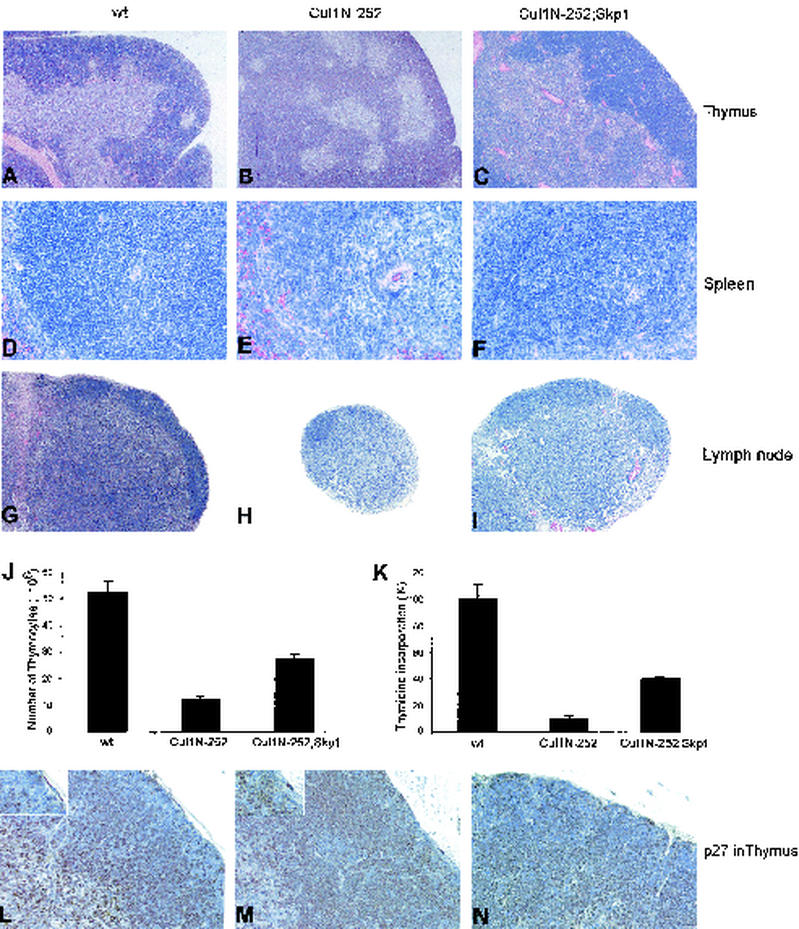
Cul1-N252 Tg mice, born at the expected Mendelian ratio, were viable and fertile. Flow cytometry analysis showed normal proportions of thymic CD4+/CD8+ double positive and CD4+/CD8−, with a minor reduction of CD4−/CD8+-single-positive lymphocytes, demonstrating that T-cell ontogeny and development were largely normal (data not shown). Nevertheless, in Cul1-N252 Tg mice, lymph nodes and the spleen were significantly smaller (∼60% of that of controls) (Fig. 3G and H and data not shown) and the number of thymocytes was substantially decreased (three- to fivefold) (Fig. 3J). Similarly, the number of T cells in the lymph nodes was lower (6- to 10-fold) (data not shown). Histopathological examinations of lymphoid organs of Cul1-N252 Tg mice revealed a prevalence of the cortical layer in the thymus (compare Fig. 3A and B) and lymphoid depletion within the interfollicular areas of peripheral lymphoid organs (Fig. 3E and H). Because T-cell depletion could be due to reduced cell proliferation, we subsequently examined the in vitro growth properties of T thymocytes derived from Cul1-N252 Tg mice in response to different mitogenic stimuli (concanavalin A, CD3, or PHA with IL-1, IL-4, and IL-7) and observed that the rate of proliferation of Cul1-N252 cells was markedly reduced compared to that for the controls (Fig. 3K and data not shown). In contrast, no significant differences between Tg and littermate control mice were observed in studying T-cell susceptibility to spontaneous apoptosis or dexamethasone- or tumor necrosis factor-induced apoptosis (data not shown).
FIG. 3.
Characterization of Cul1-N252 Tg mice. (A through I) Cellular depletion in lymphoid organs of Cul1-N252 Tg mice. Histological analysis of thymi and peripheral lymphoid organs from wt, Cul1-N252 Tg (line 20), and Cul1-N252, Skp1 double-Tg mice (6 to 8 weeks old). Sections were stained with hematoxylin and eosin stain. (J) Cul1-N252 Tg mice showing a decreased number of lymphoid cells. Total numbers of thymocytes in wt, Cul1-N252, and Cul1-N252, Skp1 double-Tg mice are shown. Data are the means ± SD representative of six animals for each group. (K) Decreased sensitivity to mitogenic stimulation of Cul1-N252 thymocytes. Thymocytes from wt, Cul1-N252, and Cul1-N252, Skp1 Tg mice were cultured with PHA and IL-1, IL-4, and IL-7 for 72 h. [3H]Thymidine was added to the medium during the last 24 h. The results are shown as the means of [3H]thymidine incorporation representative of three independent experiments with three animals for each group. (L and M) Increased expression of p27 in subcortical thymocytes of Cul1-N252 Tg mice. Anti-p27 was used in immunohistochemistry of wt (L), Cul1-N252 Tg (M), or Cul1-N252, Skp1 Tg mouse thymi. Magnification, ×100 for thymus and lymph node, ×400 for spleen and p27 in thymus.
When protein expression profiles were evaluated by immunoblotting, increased levels of the SCF substrates, cyclin E, and β-catenin were detected in the thymus of Tg line 10 mice and, more considerably, in that of line 20 mice (Fig. 2A). By immunohistochemical staining, we observed a strong increase in p27 protein levels in the subcortical areas of Cul1-N252 thymi, which normally are negative for p27 and have a high proliferation index (Fig. 3M). In contrast, cells in the inner areas of the cortex as well as in the medullar area of the thymus also expressed high levels of p27 in non-Tg mice (Fig. 3L). The lack of a robust accumulation of p27, as detected by immunoblotting (Fig. 2A), is likely due to the limited proportion of cells (subcapsular elements) with higher levels of p27.
To verify the specificity of the Cul1-N252 phenotype in vivo, we generated CD4-Skp1 Tg mice and crossed them with CD4-Cul1-N252 animals (line 20). All CD4-Skp1 mice (Fig. 2C) were viable and fertile and did not display any alterations of the thymus, spleen, or lymph nodes (data not shown). Notably, mice inheriting both the Cul1-N252 and Skp1 transgenes displayed a substantial abrogation of the Cul1-N252 phenotype. Examination of double-Tg mice showed a less severe depletion of T-cell areas in all lymphoid organs (Fig. 3C, F, and I); a normalization of p27 protein expression in double-Tg Clu1-N252, Skp1 mice (Fig. 3N); and an increased sensitivity to mitogenic stimulation compared to that in Cul1-N252 Tg mice (Fig. 3K). Thus, as for the effects of the Cul1-N252 mutant in cell lines (Fig. 1C), the in vivo effects appeared to be mediated by the sequestration of endogenous Skp1 protein.
Cul1-N252 Tg mice develop T-cell lymphomas.
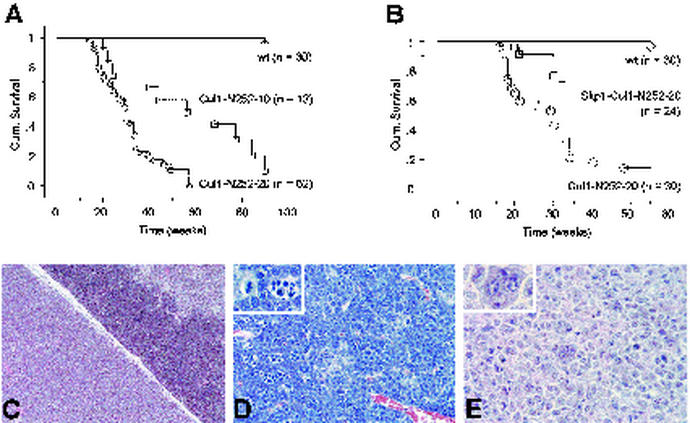
Despite the low index of proliferation and T-cell depletion, more than 80% of Cul1-N252 Tg mice developed T-cell lymphomas and died between 4 and 16 months of age (Fig. 4A). Lymphomas were characterized by atypical cells ranging in size from intermediate to large (Fig. 4C through E), with high mitotic rates and frequent apoptotic bodies (Fig. 4D, inset). Neoplastic cells often infiltrated surrounding perilymphoid tissues and peripheral lymphoid organs. Confirmation of the diagnosis came from flow cytometric analysis of thymic tumors, which revealed that all lymphomas were CD4+/CD8+ double positive and expressed a single T-cell-antigen receptor β-chain indicating their clonal origin (data not shown). Both Cul1-N252 Tg lines developed tumors with a similar penetrance but with a different latency (Fig. 4A). In line 20, 85% of the Cul1-N252 mice developed T-cell lymphomas, with a median survival age of 34 weeks, while in line 10, tumors occurred in 80% of Cul1-N252 Tg mice, with a median survival of 59 weeks. In line 10, tumors originated in peripheral lymphoid organs rather than in the thymus and displayed large pleomorphic, eosinophilic cells with multiple nuclei (Fig. 4E). In agreement with the partial recovery of the Cul1-N252 phenotypes by the coexpression of Skp1 (Fig. 3), double-Tg mice that originated by crossing Cul1-N252 mice (line 20) with Skp1 mice resulted in a decreased incidence of lymphoma and in better survival (Fig. 4B).
FIG. 4.
Lymphomas in Cul1-N252 Tg mice. (A) Percent survival in the progeny of Cul1-N252 Tg mice (lines 10 and 20). Non-Tg control mice are indicated as wt. The percentage of survivors is given against the age in weeks. (B) Increased survival in CD4-Cul1-N252 and CD4-Skp1 double-Tg mice. (C) Hematoxylin-stained section of a thymic lymphoma showing a normal (upper right) and a neoplastic (lower left) thymic lobe. (D) Advanced stage of thymic lymphoma with a high percentage of apoptotic and mitotic cells (inset). (E) Peripheric lymphoma showing pleomorphic and multinucleated neoplastic cells (inset). Magnification, ×100 for panel D, ×400 for panels E and F.
Cul1-N252 lymphomas display c-myc amplification and chromosomal instability.
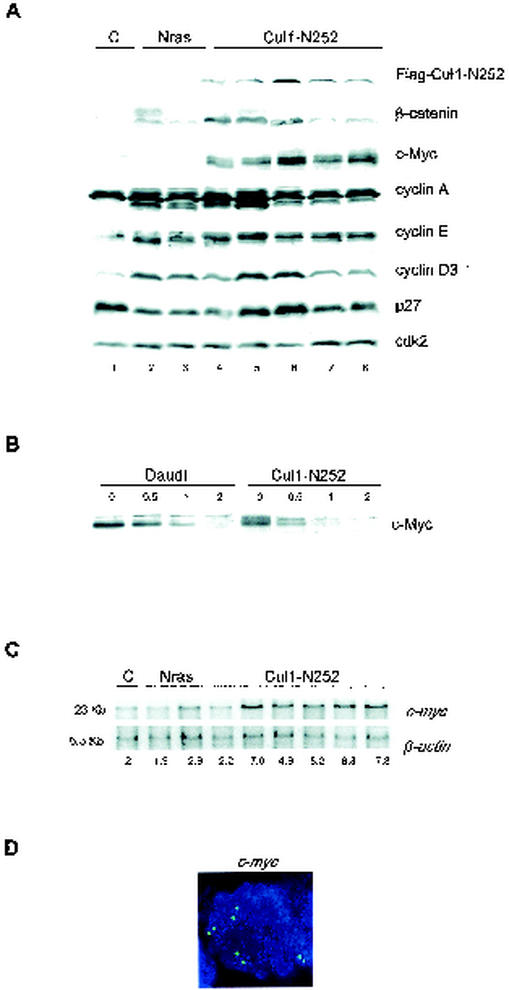
To characterize Cul1-N252 T-cell lymphomas at the molecular level, tumor extracts from CD4-Cul1-N252 (n = 18), CD4-NPM-ALK (n = 5) (54), and TMTV-Nras (n = 5) mice (34) were compared by immunoblotting. Figure 5A shows a representative analysis of these tissue extracts. Overall, compared to that in control thymi and unrelated T-cell lymphomas, most of the Cul1-N252 tumors expressed higher levels of β-catenin. p27 levels were also increased in some Cul1-N252 lymphomas (Fig. 5, lanes 5 and 6). More strikingly, 16 of 18 Cul1-N252 lymphomas displayed very high expression of the c-Myc oncoprotein (Fig. 5A).
FIG. 5.
c-myc amplification in Cul1-N252 lymphomas. (A) Representative immunoblotting of tissue extracts from control thymi (C) and tumors developed in TMTV-Nras mice (Nras) or in CD4-Cul1-N252 Tg mice (Cul1-N252). Extracts were immunoblotted with the antibodies to the indicated proteins. (B) Immunoblot analysis of c-Myc in Daudi Burkitt's lymphoma cells and in a representative Cul1-N252 lymphoma cell line after treatment with 100 μg of cycloheximide/ml for the indicated time points (in hours). (C) Southern analysis of lymphoma DNAs. EcoRI-digested DNA from a control Cul1-N252 liver (C) and TMTV-Nras (Nras) and Cul1-N252 T-cell lymphomas was probed with c-myc or β-actin (loading control). Numbers below gels are relative gene dosages based on setting of the liver control (C) to 2. (D) c-myc FISH analysis of a metaphase from a representative Cul1-N252 tumor. Four copies of the c-myc signal (green) are shown on four different chromosomes (stained blue with DAPI).
Since c-Myc is known to be regulated by ubiquitinylation and a link between the failure of c-Myc proteolysis and cancer has been suggested (1, 20, 40), we investigated whether the overexpression of c-Myc in Cul1-N252 lymphoma was the result of protein stabilization. The steady-state level of c-Myc in Cul1-N252 cell lines declined rapidly, suggesting that the proteolytic process was not affected (Fig. 5B). In fact, c-Myc turnover was even shorter than that in the Daudi cell line, which overexpresses a c-Myc protein that is not stabilized (1). This observation is in agreement with the fact that c-Myc is not accumulated in cell lines expressing Cul1-N252 (Fig. 1B).
Gene amplifications and translocations are commonly responsible for the aberrant c-Myc expression observed in many human cancers, including lymphomas (reviewed in reference 9). We performed Southern blot analysis that indicated c-myc gene amplification (five to eight copies) in five of the six tumors examined (Fig. 5C). FISH analysis showed that c-myc amplification was the result of an increased copy number of the c-myc-carrying chromosome (15). We concluded that Cul1-N252 lymphomas overexpress c-Myc protein as a consequence of chromosomal amplification.
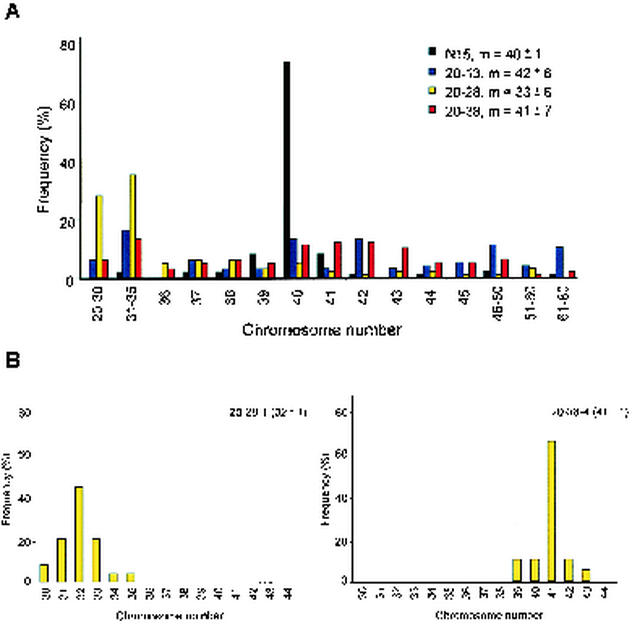
This event was highly suggestive of genetic instability. We therefore analyzed the presence of other karyotype abnormalities. Two Cul1-N252 primary lymphomas and four cell lines derived from Cul1-N252 tumors were subjected to genetic karyotyping. As controls, we used tumors and cell lines from mice expressing oncogenic NPM-ALK under the control of the same CD4 promoter (54). In all Cul1-N252 tumor cells, the chromosome number was highly variable (standard deviation [SD], >6), with only 10 to 15% of cells having a diploid chromosome karyotype (n = 40) (Fig. 6A and data not shown). In contrast, 70% of NPM-ALK cells had normal ploidy (mean ± SD, 40 ± 1.5). Taken together, these data indicate that the expression of Cul1-N252 in T cells promoted c-myc amplification and cellular transformation in vivo, possibly as the result of chromosomal instability.
FIG. 6.
Karyotype heterogeneity in Cul1-N252 lymphomas. (A) Chromosome counts on tumor cells derived from CD4-Cul1-N252 and CD4-NPM-ALK Tg mice after one to five generations are given. Karyotype heterogeneity in Tg-Cul1-N252 lymphomas is shown. Lymphoma cell lines independently derived from Tg-Cul1-N252 tumors (20-13, 20-28, 20-38) constitutively accumulated an increased proportion of aneuploid cells (85 to 90%) compared with that for Tg-NPM-ALK (N15) lymphoma cell lines (30%) and displayed a widespread karyotype heterogeneity within each tumor cell line. (B) Karyotype analysis in established clones (20-28).
Expression of Cul1-N252 induces chromosomal instability and centrosome and mitotic spindle defects and results in cellular transformation.
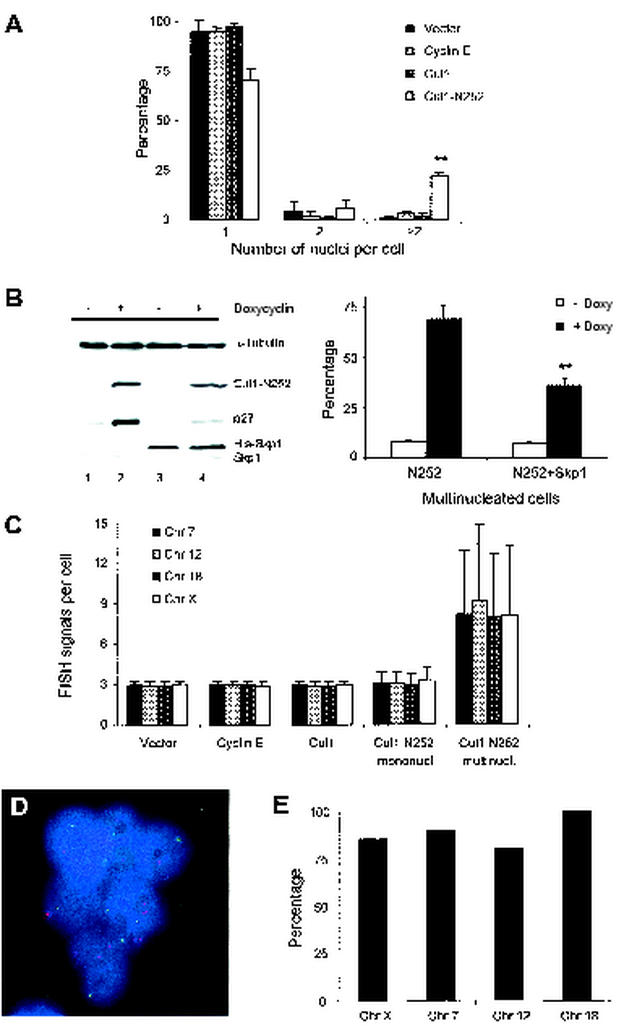
To characterize the chromosomal abnormalities observed in vivo, we expressed the Cul1-N252 mutant in cell lines (NIH 3T3 and 293T) in which morphological and karyotypical alterations could be studied in greater detail than in T cells. Approximately 20 to 25% of cells expressing the Cul1-N252 mutant presented more than two nuclei per cell (Fig. 7A) and had hyperploid DNA content (data not shown). In addition, micronuclei (6%) and enlarged nuclei (11%) were significantly increased in interphase cells expressing Cul1-N252. In contrast, cells enforced to express cyclin E, a known inducer of chromosomal instability (45), showed a profile similar to that of control cells (∼2% of multinucleated cells). To document that these changes were specific and due to the overexpression of Cul1-N252, we generated a Tet Cul1-N252-inducible cassette. Using 293T-transfected cells, we were able to confirm not only that p27 protein levels could be upregulated after doxycycline induction but that the concomitant overexpression of Skp1 could normalize the accumulation of p27 and, more importantly, significantly decrease the number of aberrant, multinucleated cells (Fig. 7B). Furthermore, 293T cells expressing the Cul1-N252 mutant exhibited marked chromosomal instability, as assessed by dual-color interphase FISH with four randomly selected centromeric probes (chromosomes 7, 12, 18, and X) (Fig. 7C). To understand if the changes in DNA content were a mere consequence of the multinucleated phenotype or whether unbalanced chromatid segregation was taking place in Cul1-N252 cells, we scored multinucleated Cul1-N252 cells. Remarkably, almost all Cul1-N252 multinucleated cells exhibited an unequal distribution of one or more chromosomes within different nuclei of the same cell (Fig. 7D and E).
FIG.7.
Chromosomal instability in Cul1-N252 cells. (A) Quantitative analysis of the number of nuclei in 293T cells transfected with empty vector (Pallino), cyclin E, wt Cul1, and the Cul1-N252 mutant is shown. Data are expressed as the percentages of cells that contained the indicated number of nuclei. Data are the means ± SD from three independent experiments. ∗∗, chi-square P < 0.0001. (B) Overexpression of Skp1 overcomes the changes induced by Cul1N252. Cul1-N252 and Cul1-N252-His-Skp1-transfected 293Trex cells were cultured with doxycycline, and protein levels of p27 were analyzed by Western blotting. The expression of Skp1 was measured with anti-His antibody (left panel). Transfected cells were cultured in the presence of doxycycline for 72 h, and the number of multinucleated cells was scored as described above (right panel). (C) FISH analysis of 293T cells with centromeric probes specific for chromosomes X, 7, 12, and 18 is shown. Mean chromosome number and SD are calculated for each transfection after counting at least 100 nuclei. (D) Unequal distribution of chromosomes in a multinucleated Cul1-N252 cell (293T) is indicated. FISH signals were detected with centromeric probes specific for chromosome 7 (red) and chromosome 12 (green). Nuclear DNA was stained with DAPI (blue). (E) Percentage of multinucleated Cul1-N252 cells showing unequal distribution of the indicated chromosomes analyzed by FISH is illustrated. At least 200 cells were counted.
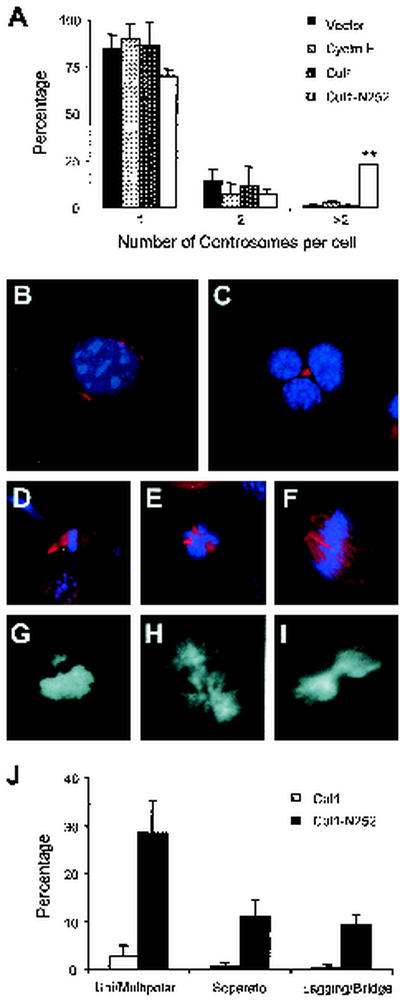
The substantial percentage of abnormally hyperploid cells and the uneven distribution of chromosomes suggested that, in Cul1-N252 cells, the fidelity of chromosome segregation was compromised. Recent studies have proposed that the aberrant replication of centrosomes can result in defective mitotic spindle organization, which leads to aneuploidy (17, 39). To examine whether Cul1-N252 expression affected centrosome duplication, centrosomes of NIH 3T3 cells transfected with the Cul1-N252 mutant were stained with an antibody to γ-tubulin (Fig. 8). One or two centrosomes were detected in cells expressing wt Cul1 or cyclin E. In contrast, about 25% of Cul1-N252 cells had an abnormal number of centrosomes (Fig. 8A). Overduplicated centrosomes as well as centrosomes that duplicated but failed to separate were also frequently seen in only Cul1-N252 cells (Fig. 8B and C).
FIG. 8.
Centrosome amplification and cell division defects in cells expressing Cul1-N252. (A) Quantitative analysis of centrosome number is illustrated. Data are expressed as the percentage of cells that contained the indicated number of centrosomes. Centrosomes were counted for 100 cells per sample. Data are the means ± SD from threeindependent experiments. ∗∗, P < 0.0001. (B and C) NIH 3T3 cells transfected with the indicated constructs were stained for centrosomes with anti-γ-tubulin antibody (red) and counterstained with DAPI (blue) after 5 days of growth in selective medium. (B) Representation of a Cul1-N252 cell with multiple centrosomes is shown. (C) Multinucleated Cul1-N252 cell with nonseparated centrosomes is shown. (D through I) Abnormal mitosis in cells that express Cul1-N252 is shown in the following components and stages: mitotic spindles of unipolar (D), tripolar (E), and tetrapolar (F) mitosis stained with anti-α-tubulin antibody (red) and counterstained with DAPI (blue); metaphase plate with chromosomes not congregated with the bulk of DNA (G); anaphase with lagging chromosomes (H); and defective cytokinesis with bridging chromatin (I). For panels G through I, DNA was stained with DAPI (white). (J) Aberrant mitosis was induced by Cul1-N252 mutant expression and quantified. One hundred cells during mitosis were counted per sample. Data are given as the means ± SD from three independent experiments, ∗∗, P < 0.0001.
If centrosomes fail to duplicate or duplicate more than once in a cell cycle, aberrant spindles are assembled, resulting in unequal chromosome segregation (3). To examine whether Cul1-N252 cells were associated with defects in mitotic spindle organization, we performed immunostaining with α-tubulin (a component of microtubules), which revealed that the Cul1-N252 cells were frequently associated with multipolar as well as unipolar spindles (Fig. 8D through F). Despite the fact that chromatin condensed normally in mitotic cells expressing Cul1-N252, their chromosomes often appeared to congregate improperly with a significant fraction of cells containing one or more chromosomes clearly separated from the bulk of the DNA clustered at the plate (Fig. 8G). Finally, in a large percentage of Cul1-N252 cells, the chromosomes did not segregate cohesively to mitotic poles and lagging chromosomes were often visible during anaphase (Fig. 8H), as were DNA bridges between dividing cells (Fig. 8I). A quantification of aberrant mitosis is shown in Fig. 8J.
The Cul1-N252-dependent mitotic defects were always observed in transfected cells after 5 days of growth in selective medium. These cells also showed a reduction in proliferation (data not shown). Nonetheless, after a variable lag phase, Cul1-N252 NIH 3T3 cells lost contact inhibition and acquired the capability to generate tumors when inoculated into nude mice (7 of 10 injections). In contrast, none of the NIH 3T3 cells transfected with either wt Cul1 or the mock vector (0 of 14 injections) were able to grow in nude mice. These results demonstrated that the abnormal segregation of chromosomes is a common feature of Cul1-N252-expressing cells. Missegregation can cause an unequal nuclear division, leading to the formation of meta-stable genotypes sufficient to result in the neoplastic transformation of Cul1-N252 cells.
DISCUSSION
Here, we show that the in vivo expression of a Cul1 deletion mutant in the T-cell lineage impinges cell proliferation, leading initially to hypoplastic, although fully differentiated, lymphoid organs. Later on, expression of Cul1-N252 induces chromosomal instability and ultimately results in tumorigenesis. These observations provide new in vivo evidence that Skp1 complexes positively regulate proper chromosomal segregation and that the interference with Skp1 is directly responsible for neoplastic transformation.
Inactivation of SCF complexes.
Our data show that expression of the N-terminal deletion mutants of Cul1 increases levels of SCF substrates such as cyclin E, p27, and β-catenin. Moreover, the specificity of the system is confirmed by the fact that coexpression of Skp1 reversed the effects of the Cul1-N252 mutant on p27 accumulation and that mice inheriting both the Cul1-N252 and Skp1 transgenes displayed a substantial inhibition of Cul1-N252 phenotypes. Overall, the effects of the Cul1-N252 deletion mutant are most likely mediated by the sequestration of endogenous Skp1-F-box protein complexes. However, we cannot exclude the possibility that additional non-SCF Skp1 complexes, if existing in mammals, are inactivated by the Cul1 mutant and contribute to this phenotype.
Decreased number of T cells, unresponsiveness to mitotic stimuli, and lymphomagenesis in CD4-Cul1-N252 Tg mice.
We found that T cells of CD4-Cul1-N252 mice developed normally and responded to apoptotic stimuli as control cells. However, all lymphoid organs showed a remarkable depletion of T cells, which had impaired proliferation in vitro. The decreased sensitivity to mitogens suggests that a reduced rate of cell growth may be responsible for the hypoplasic lymphoid organs of Cul1-N252 Tg mice. Nevertheless, expression of the Cul1-N252 mutant induced chromosomal instability and ultimately results in tumorigenesis. T-cell lymphomas of CD4-Cul1-N252 mice have a short latency and high penetrance and lead to a significantly decreased survival.
The cellular effects of the Cul1-N252 mutant are reminiscent of those caused by the lack of Skp2. In fact, mice lacking Skp2 showed a reduced growth rate and accumulation of cyclin E and p27, accompanied by multiple centrosomes and endoreplication. However, Skp2−/− mice do not exhibit any predisposition to cancer (36). In addition, these animals do not show uneven chromosomal segregation and mitotic spindle defects (Fig. 7 and 8). Moreover, cells forced to express Cul1-N252 have increased aneuploid DNA content, which is not reminiscent of the polyploid profile of Skp2−/− cells (data not shown). Finally, in Skp2−/− p27−/− double-knockout mice, all the abnormalities of Skp2−/− cells were rescued, showing that p27 accumulation was responsible for endoreplication and centrosome overduplication (K. Nakayama et al., personal communication). In contrast, crossing Cul1-N252 with p27−/− mice did not reverse any of the cellular and histopathologic abnormalities of Cul1-N252 Tg mice and tumors derived from these animals showed the same genetic instability of Cul1-N252 tumors (R. Piva, S. Lin, A. Pellicer, M. Pagano, and G. Inghirami, unpublished results). All these discrepancies clearly indicate that the interference with Skp1 has a broader effect, which cannot simply be recapitulated by the inactivation of Skp2 function and consequent p27 accumulation.
Genetic instability and centrosomal and mitotic spindle defects.
We showed that the inactivation of Skp1 promotes the cellular transformation of T cells and chromosomal instability, resulting in lymphomas with a high rate of aneuploidy and frequent amplification of the c-myc oncogene. Genetic instability is widely recognized to be central in the evolution of cancer (29, 32). Although different forms of genetic instability have been described, the vast majority of solid tumors exhibit chromosomal instability, which consists of chromosome segregation defects leading to an abnormal chromosome number (aneuploidy) (14, 30). Contrary to somatic mutations, which need to be accumulated in a multistep process in order to result in tumor formation, only a single mutational hit involving one of the genes that monitor the fidelity of chromosome segregation is required to produce a chromosomal instability phenotype (29). This event in turn primes a chain reaction that eventually will lead toward cellular transformation. The development of tumors in Cul1-N252 mice appears to fulfill this paradigm, suggesting that Skp1 complexes are master regulators of proper chromosome segregation.
It has been observed that forced expression of cyclin E induces chromosome instability (45). Since expression of the Cul1-N252 mutant resulted in the accumulation of cyclin E, we determined that the overexpression of cyclin E was not sufficient to induce a phenotype similar to that caused by the Cul1-N252 mutant, as increased multinucleated cells or centrosome overduplication were absent in transfected cyclin E cells (45). These observations, therefore, do not support the hypothesis that cyclin E accumulation is primarily responsible for the aberrations produced by the Cul1-N252 mutant.
A critical role for Skp1 in cell division has previously been established in yeast, in which severe defects in progression through mitosis have been described in a number of skp1 mutants by Bai et al. and Connelly and Heiter (2, 8). Furthermore, SKP1 has been found to be associated with kinetochore components (24, 46). In vitro experiments suggest that SCF-dependent proteolysis controls centriole splitting in mammalians and hence might be required for maintaining ploidy and genomic stability (16). Our in vivo results strongly support that theory that Skp1 complexes are required for the proper centrosome cycle. In fact, enforced expression of the Cul1-N252 mutant induced multiple centrosome abnormalities, including overduplication and the failure of duplicated centrosomes to separate. This resulted in aberrant mitotic spindles that might participate in cellular transformation. The discovery of centrosome amplification in most human cancer cells suggests that centrosome abnormalities could indeed initiate the transformation process (3, 19). After the acquisition of centrosome defects, cells initiate the assembly of abnormal spindles that cause mitotic failure or malsegregation of the replicated chromosome complement. It is reasonable to speculate that, out of such chaos, a daughter cell would then acquire, by chance, a gene dosage that confers survival, a mutator phenotype, and tumorigenesis. The surviving daughter cells would subsequently retain mutations that suppress the centrosome overload by assembling a bipolar spindle, a condition that favors mitotic stability and neoplastic growth.
In conclusion, our results show for the first time that deregulation of Cul1-Skp1 stoichiometric balance in mammalian cells can affect the fidelity of chromosome transmission and results in cell transformation in vivo. In this prospect, the Cul1 gene may represent sizable genetic targets for mutational inactivation during tumorigenesis. Indeed, the 7q35 region where CUL1 resides has been found to have been deleted in some malignancies (13). The current challenge is to identify the molecule(s) making up the link. Moreover, Cul1-N252 cells and CD4-Cul1-N252 Tg animals may be a useful model not only to identify still-unknown SCF substrates and the pathological consequences of their deregulated degradation but also to shed light on the mechanisms leading to chromosomal instability and to cellular transformation.
Acknowledgments
We thank J. Lukas, A. Pellicer, M. J. Difilippantonio, and Z. Q. Pan for reagents and R. Dalla Favera, E. Kipreos, D. Levy, and N. Forbes for critical reading of the manuscript.
M.P. is a recipient of the Irma T. Hirschl scholarship. This work was partially supported by grants from the NIH (R01-CA76584 and R01-GM57587) to M.P. and the NIH (RO1-CA64033) to G.I. R.P. was partially supported by a fellowship from the University of Turin (Italy).
REFERENCES
- 1.Bahram, F., N. von der Lehr, C. Cetinkaya, and L. G. Larsson. 2000. c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 95:2104-2110. [PubMed] [Google Scholar]
- 2.Bai, C., P. Sen, K. Hofmann, L. Ma, M. Goebl, J. W. Harper, and S. J. Elledge. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86:263-274. [DOI] [PubMed] [Google Scholar]
- 3.Brinkley, B. R. 2001. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell. Biol. 11:18-21. [DOI] [PubMed] [Google Scholar]
- 4.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1:193-199. [DOI] [PubMed] [Google Scholar]
- 5.Carrano, A. C., and M. Pagano. 2001. Role of the F-box protein Skp2 in adhesion-dependent cell cycle progression. J. Cell Biol. 153:1381-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cenciarelli, C., D. S. Chiaur, D. Guardavaccaro, W. Parks, M. Vidal, and M. Pagano. 1999. Identification of a family of human F-box proteins. Curr. Biol. 9:1177-1179. [DOI] [PubMed] [Google Scholar]
- 7.Chiarle, R., A. Podda, G. Prolla, E. R. Podack, G. J. Thorbecke, and G. Inghirami. 1999. CD30 overexpression enhances negative selection in the thymus and mediates programmed cell death via a Bcl-2-sensitive pathway. J. Immunol. 163:194-205. [PubMed] [Google Scholar]
- 8.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang, C. V., L. M. Resar, E. Emison, S. Kim, Q. Li, J. E. Prescott, D. Wonsey, and K. Zeller. 1999. Function of the c-Myc oncogenic transcription factor. Exp. Cell Res. 253:63-77. [DOI] [PubMed] [Google Scholar]
- 10.Dealy, M. J., K. V. Nguyen, J. Lo, M. Gstaiger, W. Krek, D. Elson, J. Arbeit, E. T. Kipreos, and R. S. Johnson. 1999. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat. Genet. 23:245-248. [DOI] [PubMed] [Google Scholar]
- 11.DeSalle, L. M., and M. Pagano. 2001. Regulation of the G1 to S transition by the ubiquitin pathway. FEBS Lett. 490:179-189. [DOI] [PubMed] [Google Scholar]
- 12.Deshaies, R. J. 1999. SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 13.Dohner, K., J. Brown, U. Hehmann, C. Hetzel, J. Stewart, G. Lowther, C. Scholl, S. Frohling, A. Cuneo, L. C. Tsui, P. Lichter, S. W. Scherer, and H. Dohner. 1998. Molecular cytogenetic characterization of a critical region in bands 7q35-q36 commonly deleted in malignant myeloid disorders. Blood 92:4031-4035. [PubMed] [Google Scholar]
- 14.Duesberg, P., D. Rasnick, R. Li, L. Winters, C. Rausch, and R. Hehlmann. 1999. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 19:4887-4906. [PubMed] [Google Scholar]
- 15.Feldman, R. M., C. C. Correll, K. B. Kaplan, and R. J. Deshaies. 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91:221-230. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E., K. R. Lacey, P. Huie, S. A. Lyapina, R. J. Deshaies, T. Stearns, and P. K. Jackson. 1999. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 13:2242-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukasawa, K., T. Choi, R. Kuriyama, S. Rulong, and G. F. Vande Woude. 1996. Abnormal centrosome amplification in the absence of p53. Science 271:1744-1747. [DOI] [PubMed] [Google Scholar]
- 18.Galan, J. M., A. Wiederkehr, J. H. Seol, R. Haguenauer-Tsapis, R. J. Deshaies, H. Riezman, and M. Peter. 2001. Skp1p and the F-box protein Rcy1p form a non-SCF complex involved in recycling of the SNARE Snc1p in yeast. Mol. Cell. Biol. 21:3105-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghadimi, B. M., D. L. Sackett, M. J. Difilippantonio, E. Schrock, T. Neumann, A. Jauho, G. Auer, and T. Ried. 2000. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer 27:183-190. [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory, M. A., and S. R. Hann. 2000. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol. Cell. Biol. 20:2423-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grignani, F., T. Kinsella, A. Mencarelli, M. Valtieri, D. Riganelli, L. Lanfrancone, C. Peschle, G. P. Nolan, and P. G. Pelicci. 1998. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 58:14-19. [PubMed] [Google Scholar]
- 22.Gstaiger, M., A. Marti, and W. Krek. 1999. Association of human SCF(SKP2) subunit p19(SKP1) with interphase centrosomes and mitotic spindle poles. Exp. Cell Res. 247:554-562. [DOI] [PubMed] [Google Scholar]
- 23.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan, K. B., A. A. Hyman, and P. K. Sorger. 1997. Regulating the yeast kinetochore by ubiquitin-dependent degradation and Skp1p-mediated phosphorylation. Cell 91:491-500. [DOI] [PubMed] [Google Scholar]
- 25.Kawakami, T., T. Chiba, T. Suzuki, K. Iwai, K. Yamanaka, N. Minato, H. Suzuki, N. Shimbara, Y. Hidaka, F. Osaka, M. Omata, and K. Tanaka. 2001. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 20:4003-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kipreos, E. T., L. E. Lander, J. P. Wing, W. W. He, and E. M. Hedgecock. 1996. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell 85:829-839. [DOI] [PubMed] [Google Scholar]
- 27.Kipreos, E. T., and M. Pagano. 2000. The F-box protein family. Genome Biol. 1:3002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Latres, E., R. Chiarle, B. Schulman, A. Pellicer, G. Inghirani, and M. Pagano. 2001. Role of the F-box protein Skp2 in lymphomagenesis. Proc. Natl Acad. Sci. USA 98:2515-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 30.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1997. Genetic instability in colorectal cancers. Nature 386:623-627. [DOI] [PubMed] [Google Scholar]
- 31.Liakopoulos, D., T. Busgen, A. Brychzy, S. Jentsch, and A. Pause. 1999. Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc. Natl. Acad. Sci. USA 96:5510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loeb, L. A. 1991. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 51:3075-3079. [PubMed] [Google Scholar]
- 33.Lyapina, S. A., C. C. Correll, E. T. Kipreos, and R. J. Deshaies. 1998. Human CUL1 forms an evolutionarily conserved ubiquitin ligase complex (SCF) with SKP1 and an F-box protein. Proc. Natl. Acad. Sci. USA 95:7451-7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangues, R., W. F. Symmans, S. Lu, S. Schwartz, and A. Pellicer. 1996. Activated N-ras oncogene and N-ras proto-oncogene act through the same pathway for in vivo tumorigenesis. Oncogene 13:1053-1063. [PubMed] [Google Scholar]
- 35.Michel, J. J., and Y. Xiong. 1998. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 9:435-449. [PubMed] [Google Scholar]
- 36.Nakayama, K., H. Nagahama, Y. A. Minamishima, M. Matsumoto, I. Nakamichi, K. Kitagawa, M. Shirane, R. Tsunematsu, T. Tsukiyama, N. Ishida, M. Kitagawa, and S. Hatakeyama. 2000. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 19:2069-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nayak, S., F. E. Santiago, H. Jin, D. Lin, T. Schedl, and E. T. Kipreos. 2002. The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 12:277-287. [DOI] [PubMed] [Google Scholar]
- 38.Pagano, M., S. Tam, A. Theodoras, P. Beer, S. Delsal, I. Chau, R. Yew, G. Draetta, and M. Rolfe. 1995. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269:682-685. [DOI] [PubMed] [Google Scholar]
- 39.Pihan, G. A., A. Purohit, J. Wallace, H. Knecht, B. Woda, P. Quesenberry, and S. J. Doxsey. 1998. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 58:3974-3985. [PubMed] [Google Scholar]
- 40.Salghetti, S. E., S. Y. Kim, and W. P. Tansey. 1999. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 18:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sawada, S., J. D. Scarborough, N. Killeen, and D. R. Littman. 1994. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77:917-929. [DOI] [PubMed] [Google Scholar]
- 42.Schwab, M., and M. Tyers. 2001. Cell cycle. Archipelago of destruction. Nature 413:268-269. [DOI] [PubMed] [Google Scholar]
- 43.Seol, J. H., A. Shevchenko, and R. J. Deshaies. 2001. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3:384-391. [DOI] [PubMed] [Google Scholar]
- 44.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 45.Spruck, C. H., K. A. Won, and S. I. Reed. 1999. Deregulated cyclin E induces chromosome instability. Nature 401:297-300. [DOI] [PubMed] [Google Scholar]
- 46.Stemmann, O., and J. Lechner. 1996. The Saccharomyces cerevisiae kinetochore contains a cyclin-CDK complexing homologue, as identified by in vitro reconstitution. EMBO J. 15:3611-3620. [PMC free article] [PubMed] [Google Scholar]
- 47.Tyers, M., and P. Jorgensen. 2000. Proteolysis and the cell cycle: with this RING I do thee destroy. Curr. Opin. Genet. Dev. 10:54-64. [DOI] [PubMed] [Google Scholar]
- 48.Wang, Y., S. Penfold, X. Tang, N. Hattori, P. Riley, J. W. Harper, J. C. Cross, and M. Tyers. 1999. Deletion of the Cul1 gene in mice causes arrest in early embryogenesis and accumulation of cyclin E. Curr. Biol. 9:1191-1194. [DOI] [PubMed] [Google Scholar]
- 49.Winston, J. T., D. M. Koepp, C. Zhu, S. J. Elledge, and J. W. Harper. 1999. A family of mammalian F-box proteins. Curr. Biol. 9:1180-1182. [DOI] [PubMed] [Google Scholar]
- 50.Wu, K., A. Chen, and Z.-Q. Pan. 2000. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275:32317-32324. [DOI] [PubMed] [Google Scholar]
- 51.Yam, C. H., R. W. Ng, W. Y. Siu, A. W. Lau, and R. Y. Poon. 1999. Regulation of cyclin A-Cdk2 by SCF component Skp1 and F-box protein Skp2. Mol. Cell. Biol. 19:635-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamanaka, A., M. Yada, H. Imaki, M. Koga, Y. Ohshima, and K. Nakayama. 2002. Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with cullins and F-box proteins. Curr. Biol. 12:267-275. [DOI] [PubMed] [Google Scholar]
- 53.Zachariae, W., and K. Nasmyth. 1999. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 13:2039-2058. [DOI] [PubMed] [Google Scholar]
- 54.Zamo, A., R. Chiarle, R. Piva, J. Howes, Y. Fan, M. Chilosi, D. E. Levy, and G. Inghirami. 2002. Anaplastic lymphoma kinase (ALK) activates Stat3 and protects hematopoietic cells from cell death. Oncogene 21:1038-1047. [DOI] [PubMed] [Google Scholar]