Abstract
Rad21 is one of the major cohesin subunits that holds sister chromatids together until anaphase, when proteolytic cleavage by separase, a caspase-like enzyme, allows chromosomal separation. We show that cleavage of human Rad21 (hRad21) also occurs during apoptosis induced by diverse stimuli. Induction of apoptosis in multiple human cell lines results in the early (4 h after insult) generation of 64- and 60-kDa carboxy-terminal hRad21 cleavage products. We biochemically mapped an apoptotic cleavage site at residue Asp-279 (D279) of hRad21. This apoptotic cleavage site is distinct from previously described mitotic cleavage sites. hRad21 is a nuclear protein; however, the cleaved 64-kDa carboxy-terminal product is translocated to the cytoplasm early in apoptosis before chromatin condensation and nuclear fragmentation. Overexpression of the 64-kDa cleavage product results in apoptosis in Molt4, MCF-7, and 293T cells, as determined by TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) and Annexin V staining, assaying of caspase-3 activity, and examination of nuclear morphology. Given the role of hRad21 in chromosome cohesion, the cleaved C-terminal product and its translocation to the cytoplasm may act as a nuclear signal for apoptosis. In summary, we show that cleavage of a cohesion protein and translocation of the C-terminal cleavage product to the cytoplasm are early events in the apoptotic pathway and cause amplification of the cell death signal in a positive-feedback manner.
Normal development and homeostasis require the orderly regulation of both cell proliferation and cell survival. Cell cycle progression and control of apoptosis are thought to be intimately linked processes. Activation of the cell cycle plays a significant role in the regulation of apoptosis (16); in some cell types and under certain conditions, apoptosis has been shown to occur only at specific stages of the cell cycle (24). Mitosis and apoptosis are also closely interrelated (25), and the mitotic index is the most important determinant of the apoptotic index (25). Although proteins that regulate apoptosis have been implicated in the restraint of cell cycle entry (14) and the control of ploidy (29), the effector molecules at the interface between cell proliferation and cell survival have remained elusive.
Studies with yeast and higher eukaryotes, including humans, have indicated that an evolutionarily conserved protein complex, called cohesin, and its subunit, Mcd1/Scc1/hRad21, are required for appropriate arrangement of chromosomes during normal cell division (11, 28; for a review, see references 20, 30, 31, and 36). Analyses of Rad21 function in fission yeast, Schizosaccharomyces pombe, and of Scc1/Mcd1 function in budding yeast, Saccharomyces cerevisiae, have demonstrated that the nuclear phosphoprotein is required for appropriate chromosomal cohesion during the mitotic cell cycle and double-strand-break repair after DNA damage (1, 30). Biochemical analysis of cohesin indicates that it acts as a molecular glue, and human cohesin can promote intermolecular DNA catenation, a mechanism that links two sister chromatids together (26). In budding yeast, loss of cohesion at the metaphase-anaphase transition is accompanied by proteolytic cleavage of the Scc1/Mcd1 protein (11, 28, 30, 37) followed by its dissociation from the chromatids (28, 30). Cleavage depends on a CD clan endopeptidase, Esp1 (also known as separin/separase) (37, 38), which is complexed with its inhibitor, Pds1 (securin), before anaphase (23, 39). In metaphase, ubiquitin-mediated degradation of the securin protein by APC/C-Cdc20 ubiquitin-ligase releases separin protein, which proteolytically cleaves cohesin Rad21, thereby releasing the sister chromatids (6, 7, 10, 18, 42). In budding yeast, fission yeast, and human cells, Rad21 has two mitotic cleavage sites for separase (12, 37, 38), and cleavage by separase appears to be essential for sister chromatid separation and for the completion of cytokinesis (12). In contrast to the simultaneous release of cohesin from the chromosome arms and centromere region in budding yeast by separase cleavage, most cohesin in metazoans is removed in early prophase from chromosome arms by a cleavage-independent mechanism (12, 39, 40). Only residual amounts of cohesin are cleaved at the onset of anaphase, coinciding with its disappearance from centromeres. Thus, Scc1/Mcd1/Rad21 plays a critical role in the eukaryotic cell division cycle by regulating sister chromatid cohesion and separation at the metaphase-to-anaphase transition.
Our results indicate that in addition to establishing and maintaining sister chromatid cohesion during mitosis, hRad21 plays a role in apoptosis, and its cleavage during apoptosis may act as a nuclear signal to initiate cytoplasmic events involved in the apoptotic pathway.
MATERIALS AND METHODS
Plasmids.
Full-length hRAD21 cDNA plasmid (KIAA 0078) in pBluescript SK(+) vector was obtained from Kazusa DNA Research Institute, Chiba, Japan. Full-length hRAD21 cDNA was subcloned into several mammalian expression plasmids, including pFLAGCMV2, pCS2MT, and pCDNA6Myc-HisC, to produce epitope-tagged proteins where applicable. hRad21 cDNA was also subcloned in frame upstream of the myc epitope in pCMV/myc/Nuc and pCMV/myc/Cyto (Invitrogen, Carlsbad, Calif.) to direct the expression of hRad21 protein to the nucleus and cytoplasm, respectively. The following plasmids were used for transfection: pCS2MT-hRAD21 was constructed by in-frame ligation of the 2,331-bp NcoI/DraI fragment bearing the hRAD21 cDNA to the end of the sixth myc epitope in pCS2MT (B. Kelley, Fred Hutchinson Cancer Center, Seattle, Wash.); pFLAGCMV2-hRAD21 was generated by cloning the full-length hRAD21 gene contained on a 2,578-bp MscI/StuI fragment from pSKKIAA0078 into pFLAGCMV2 (Kodak) that was digested with SmaI.
Site-directed mutagenesis of hRad21.
pCS2MT-hRAD21 apoptotic cleavage site (ACS) mutants I (PDSPD279S to PDSPA279S) and II (PD276S277PD279S280 to PA276A277PA279A280) were generated using a PCR-based site-directed mutagenesis protocol as previously described (33). The PCR resulted in a 550-bp internal hRAD21 fragment containing the mutations. A 221-bp piece of wild-type (WT) hRAD21 (from the BsgI to PFLFI sites) was replaced with the comparable mutated fragment. The resulting plasmids, pCS2MT-hRAD21-ACS-mut-I and pCS2MT-hRAD21-ACS-mut-II, were verified by DNA sequencing. The amino-terminal (N-hRad21, encoding amino acids [aa] 1 to 279) and carboxy-terminal (C-hRad21, encoding aa 280 to 631) cleavage products were cloned into myc epitope-tagged pCS2MT vectors by using PCR amplification of the fragments from the hRAD21 cDNA. These constructs were also verified by DNA sequencing.
Generation of hRad21 pAb and mAb.
Rabbit polyclonal antibody (pAb) was raised commercially (Covance, Denver, Pa.) against synthetic peptides corresponding to the sequence of the 14 carboxy terminal aa of hRad21 (SDIIATPGPRFHII). Immunization and affinity purification of antibodies were performed according to the manufacturer's protocol. Monoclonal antibody (mAb) against a partial recombinant hRad21 protein (aa 240 to 631) was also raised commercially (Imgenex, San Diego, Calif.). Both antibodies had very high titers, as determined by enzyme-linked immunosorbent assay. Both antibodies recognized the WT hRad21 protein as a specific 122-kDa band in Western blot analysis and effectively immunoprecipitated endogenous hRad21 from various human and rodent cell lines and tissue lysates. Immunodetection of the 122-kDa band was blocked competitively by pretreatment of the lysates with recombinant hRad21 protein or synthetic C-terminal peptides. Both antibodies were also effective in immunohistochemistry and immunofluorescence staining of both paraffin-embedded and tissue culture slides.
Antisera.
The monoclonal antisera were obtained as follows: human poly(ADP-ribose) polymerase (PARP) from PharMingen, San Diego, Calif.; Flag epitope and mouse β-actin from Sigma, St. Louis, Mo.; c-myc epitope (9E10), bacterial trpE, caspase-3, caspase-7, tubulin, and lamin from Oncogene Research Products, Cambridge, Mass. hRad21 N-terminal antibody was a gift from J.-M. Peters (Research Institute of Molecular Pathology, Vienna, Austria).
Cell cultures and transfection.
MCF-7 breast carcinoma cells, human choriocarcinoma JEG3 cells, and IMR90 primary lung fibroblast cells were obtained from the American Type Culture Collection (ATCC) and were maintained per ATCC protocol. Human Molt4 and Jurkat T-cell leukemia cells (both obtained from ATCC) were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum and maintained at 37°C, 95% humidity, and an atmosphere of 5% CO2. EL-12 mouse mammary epithelial cells were obtained from the Medina Laboratory (Baylor College of Medicine) and maintained as previously described (27). Cells were transfected with appropriate plasmids in 100-mm-diameter dishes using Superfect or Effectene reagents from Qiagen (Valencia, Calif.) according to the manufacturer's protocol. A fixed amount of plasmid DNA was used in any given experiment. The total amount of expression vector DNA was equalized by the addition of blank vectors to control for promoter competition effects. When necessary, transfection efficiency was monitored by use of 1 μg of pDsRedi-Mito plasmid (Clontech, Palo Alto, Calif.) per transfection. Transfection efficiency was determined by counting the percentage of red fluorescent cells in five random fields under a microscope with appropriate fluorescent channels.
Drug treatments.
Etoposide (VP-16) (20-mg/ml injections) and camptothecin were purchased from GensiaSicor Pharmaceuticals (Irvine, Calif.) and Sigma, respectively. Camptothecin was dissolved in dimethyl sulfoxide (DMSO) and stored in aliquots at −20°C. Cells were plated at a concentration of 6 × 106 cells/ml and treated with appropriate concentrations of drugs. Molt4 cells were treated with etoposide, while Jurkat cells were treated with camptothecin for 8 h unless otherwise indicated. Controls were treated with equivalent dosages of vehicle. The caspase inhibitor z-VAD-FMK was also dissolved in DMSO and stored at −20°C. Peptide aldehydes MG115 and MG132 were obtained from Peptide Institute, Inc. (Lexington, Ky.) and dissolved at a concentration of 10 mM in DMSO. Cells were treated with a 0.025 mM concentration of proteasome inhibitors for 8 h before harvesting. 15-Deoxy-delta(12,14)-prostaglandin J2 (15dPGJ2) was purchased from Cayman Chemical Co. (Ann Arbor, Mich.). Induction of apoptosis in JEG3 cells by using 15dPGJ2 was carried out as previously described (19).
Protein analysis and IP.
Cells were pelleted by low-speed centrifugations (800 × g for 5 min) and lysed in RIPA buffer (phosphate-buffered saline [PBS], 1% Nonidet P-40, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate) or PBSTDS buffer (PBS, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate) containing protease and phosphatase inhibitors (1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin per ml, 30 μl of aprotinin per ml, 0.5 μg of leupeptin per ml, 100 mM sodium orthovanadate, 100 mM sodium fluoride) (all from Sigma) for 10 to 15 min on ice, followed by passage through a 21-gauge needle. When appropriate, additional phosphatase inhibitor cocktails I and II (Sigma) were added to the lysis buffer at a dilution of 1:100. Lysates were then centrifuged at 1,000 × g for 20 min, and the supernatants were aliquoted and frozen at −80°C until use. Protein samples were also made from the cytoplasmic and nuclear fractions of apoptosis-induced Molt4 cells by using protocols previously described (5). The purities of the cytosolic and nuclear fractions were verified by Western blotting with antibodies to tubulin and nuclear lamin, respectively. After protein quantification (using detergent-compatible protein dye and bovine serum albumin from Bio-Rad as standards) and normalization, 10 to 40 μg of protein extracts was electrophoresed on SDS-polyacrylamide gel electrophoresis (PAGE) gels and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, Mass.). The filters were initially blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 for 1 to 2 h at room temperature and then probed with hRad21 mAb or hRad21 pAb at a 1:1,000 dilution, 1.5 μg of myc epitope/ml, 2.5 μg of Flag epitope/ml, β-actin at a 1:100,000 dilution, or PARP antiserum at a 1:2,000 dilution. The bound antibodies were visualized by the enhanced chemiluminescence detection system (Amersham, Buckinghamshire, England), in combination with the horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies as appropriate, and the intensity of the specific bands in the exposed films was quantified. In some of the later experiments, bound primary antibodies were detected with IRD800 dye-labeled, appropriate species-specific secondary antisera and the signal was visualized on a Li-Cor (Lincoln, Nebr.) Odyssey infrared scanner. Immunoprecipitation (IP) was performed as follows. A 1.0-ml sample of cell lysate was precleared by incubation with 10 μl of normal mouse immunoglobulin G (IgG) and 20 μl of protein G plus agarose (Oncogene Research Products) at 4°C for 1 h on a rotator. The precleared lysate was collected after centrifugation at 800 × g for 15 min. A 0.5- to 1.0-ml sample of precleared lysate normalized for protein concentrations was incubated at 4°C for 1 h with primary antibodies followed by the addition of 20 μl of protein A and protein G plus agarose. The mix was then incubated at 4°C for another 12 to 16 h on a rotator. Precipitates were then washed four times with 1 ml of ice-cold PBS, with a final wash in the lysis buffer before electrophoresis and Western blot analysis.
Mapping of hRad21 apoptotic cleavage sites.
Apoptosis was induced in Molt4 T cells by treatment with 10 μM etoposide for 8 h. Protein lysates were subjected to IP using hRad21 mAb or a control bacterial trpE mAb. The immunoprecipitated samples were run on SDS-6% PAGE gels that included 0.1 mM sodium thioglycolate (Sigma) as a scavenger in the upper running buffer. Electrophoresed samples were then electroblotted onto PVDF membranes at 400 mA for 45 min at room temperature (∼25°C) using CAPS [3-(cyclohexylamino)propanesulfonic acid] buffer (10 mM CAPS, 10% methanol, pH 11). At the end of the transfer, the blotted membranes were rinsed with water for 2 to 5 min, stained with 0.05% Coomassie blue in 1% acetic acid-50% methanol for 5 to 7 min, destained in 50% methanol until the background was pale blue (5 to 15 min), and finally rinsed with water for 5 to 10 min. Appropriate bands were cut out and air dried and sent to the protein chemistry core laboratory at Baylor College of Medicine for N-terminal sequencing.
Immunocytochemistry and detection of apoptosis.
EL-12 cells were grown on Falcon culture slides. Medium was poured off before the cells were treated with UV (0, 50, 100, or 200 J/m2). Fresh medium was added immediately after UV radiation. Cells were fixed with cold methanol after 6 h of UV treatment unless otherwise noted. Double staining of hRad21 was performed by incubating anti-hRad21 mAb and rabbit anti-C-terminal-hRad21 pAb. The signals of mAb and pAb were visualized by the addition of rhodamine-labeled goat anti-mouse IgG (1:100) and fluorescein-labeled goat anti-rabbit IgG (1:800) (Molecular Probes), respectively. Slides were mounted with Vectashield mounting medium with DAPI (4′,6′-diamidino-2-phenylindole; H-1200, Vector) and sealed with nail makeup. Images were obtained with a Zeiss inverted fluorescence microscope coupled to an Axiocam high-resolution digital camera operated with Axiovision 3.0 software (Carl Zeiss Inc., Thornwood, N.Y.).
For detection of apoptosis in transiently transfected 293T and MCF-7 cells, 1 million cells were seeded onto 100-mm-diameter culture dishes 2 days prior to transfection. Cells were transfected with the indicated hRad21 constructs when they reached 60% confluence. For DAPI staining, cells were cotransfected with 5 μg of pDsRed1-Mito (Clontech) and 5 μg of pCS2MT, pCS2MT Rad21 wild type, pCS2MT Rad21 N terminus, or pCS2MT Rad21 C terminus by using the calcium phosphate method. At 16, 24, and 48 h posttransfection, cells were detached with trypsin and collected by centrifugation at 1,000 × g for 5 min. The samples were fixed with 4% paraformaldehyde in PBS (pH 7.2), mounted with Vectashield mounting medium with DAPI (H-1200, Vector), and examined by fluorescence microscopy. The intact and degraded nuclei of cells coexpressing pDsRed1 (with red fluorescence) were counted. About 50 fluorescent nuclei from each treatment group were screened and counted for normal morphology (rounded chromatin) or for apoptotic nuclei (fragmented and condensed chromatin). Data were expressed as the percentage of apoptotic cells among total counted cells. Each treatment was replicated three times. For Annexin V and TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling) staining, 293T cells were transfected with 10 μg of pCS2MT, pCS2MT Rad21 wild type, pCS2MT Rad21 N-terminus, or pCS2MT Rad21 C-terminus by using the calcium phosphate method. After 16, 24, and 48 h, the cells were detached and collected as described above. Annexin V (Annexin V-FITC [fluorescein isothiocyanate] apoptosis direction kit) and TUNEL staining (MEBSTAIN apoptosis kit direct) were performed according to the manufacturer's protocol (MBL, Watertown, Mass.). Staining of the cells with Annexin V-FITC and propidium iodide (PI) was used to distinguish between cells undergoing apoptosis (PI negative) and those that were necrotic or dead (PI positive). Apoptotic cells were identified with TUNEL staining using fluorescein-dUTP as the substrate.
The caspase-3 activities in Molt4 cells were measured using a caspase-3 assay kit from Clontech according to the manufacturer's protocol.
Proteolytic cleavage assay of the in vitro transcribed and translated hRad21.
35S-hRad21 or unlabeled (nonisotopic) hRad21 was produced by in vitro transcription-translation using the TNT rabbit reticulocyte lysate system (Promega, Madison, Wis.). Rabbit reticulocyte lysate was combined with 1 μg of plasmid DNA containing either the WT hRAD21 cDNA (pCS2MT-hRAD21) or one of the hRAD21 ACS mutants, ACS-mut-I or ACS-mut-II, and SP6 RNA polymerase. Reaction in the absence of plasmid DNA served as a negative control. Reaction mixtures were incubated at 30°C for 90 min. In vitro cleavage reaction was performed as previously described (9). In brief, 6 μl of in vitro translated 35S-hRad21 (WT), ACS-mut-I, or ACS-mut-II was combined with 30 μl of reaction buffer (20 mM HEPES, pH 7.4, 2 mM dithiothreitol, 10% glycerol) and one of the following enzyme sources: 2 μl (200 U) of recombinant caspase-3, 2 μl (4 U) of caspase-7, or 2 μl (10 μg) of Molt4 cell lysates (treated with DMSO or 10 μM etoposide for 6 h). The cleavage reaction was performed at 37°C for 1 h, after which 8 μl of 6× sample buffer with dithiothreitol was added to stop the reaction. Twenty microliters of this reaction was electrophoresed on SDS-6% PAGE gels, fixed with methanol and acetic acid for 30 min, dried on a gel dryer, and exposed to a Storm imager. Bands were quantified using ImageQuant 5.2 software (Molecular Dynamics, Inc., Sunnyvale, Calif.). Unlabeled (nonisotopic) hRad21 from the TNT reactions was also incubated as described above in the presence or absence of caspase-3 or caspase-7. Samples were then analyzed by SDS-PAGE followed by Western blotting with hRad21 antiserum.
Data analysis.
The differences between the apoptosis levels in cells transfected with various hRad21 constructs were measured using a paired test of proportions based on binomials (8). The results of the caspase-3 activity assay were analyzed using Student's t test.
RESULTS
We report the role of hRad21 in the apoptotic response and cleavage of hRad21 protein in human cells by a caspase-like activity.
Cleavage of hRad21 during apoptosis.
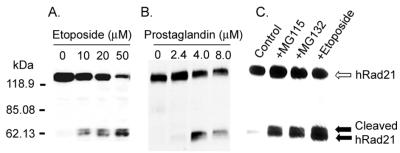
While examining the expression of Rad21 in mammalian cells after DNA damage, we surprisingly identified the cleavage of hRad21 protein after induction of apoptosis. hRad21 was cleaved during etoposide-induced apoptosis in human Molt4 T-cell leukemia cells. Induction of apoptosis resulted in the generation of approximately 64- and 60-kDa cleavage products, as determined with an hRad21 mAb (Fig. 1). The cleavage of hRad21 in Molt4 cells was a function of etoposide dosage (Fig. 1A), as the ratio of cleaved hRad21 to full-length protein appeared to be directly proportional to increasing doses of etoposide over the tested range (10 to 50 μM). hRad21 cleavage products were also detected in a number of other cell lines following induction of apoptosis by DNA-damaging agents (ionizing radiation and topoisomerase inhibitors) (data not shown) and/or non-DNA-damaging agents (prostaglandin [Fig. 1B], proteasome inhibitor [Fig. 1C], cycloheximide treatment, and cytokine withdrawal [data not shown]). In addition, equivalent doses of ionizing radiation in cells that are resistant to apoptosis (Raji lymphoid leukemia and H1299 large-cell lung carcinoma cells) did not generate this band (data not shown); thus, it was not a simple by-product of DNA damage.
FIG. 1.
Apoptosis-induced cleavage of hRad21 in Molt4 T-cell leukemia and JEG3 choriocarcinoma cells treated with etoposide, prostaglandin, and proteasome inhibitors. (A) Dose-related cleavage of hRad21 in Molt4 T-cell leukemia cells treated with increasing concentrations of etoposide (10 to 50 μM) for 6 h. (B) JEG3 cells treated with 15DPGJ2 (2.4 to 8 μM) for 16 h. (C) Molt4 cells were also treated with a 0.025 mM concentration of proteasome inhibitors, MG115, and MG132 for 8 h. Lysates of these samples were resolved on an SDS-PAGE (4 to 20% acrylamide) gel, transferred to a nitrocellulose membrane, and analyzed by Western blot using a monoclonal hRad21 antibody. Induction of apoptosis resulted in the generation of approximately 64- and 60-kDa hRad21 cleavage products (shown by the closed arrows). Full-length hRad21 (122 kDa) is indicated by the open arrow.
Translocation of the carboxy-terminal hRad21 fragment to the cytoplasm after induction of apoptosis.
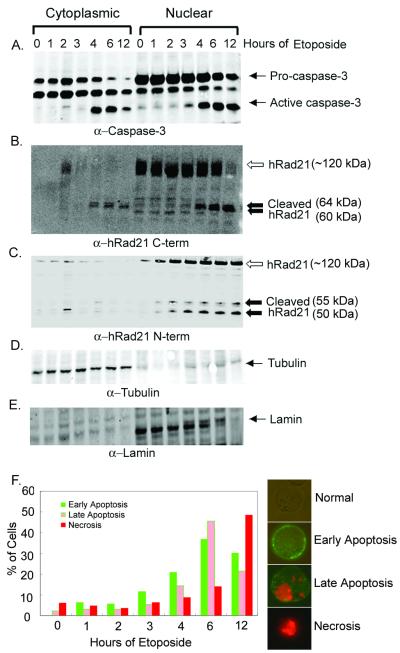
Molt4 cells were treated with 10 μM etoposide for 0, 1, 2, 3, 4, 6, and 12 h. Induction of apoptosis was verified by determination of caspase-3 activity (Fig. 2A), Annexin V staining (Fig. 2F), the cleavage of PARP, and the morphology of DAPI-stained nuclei (data not shown). Western blot analysis of cytoplasmic and nuclear fractions by using a C-terminal hRad21 antibody detected a 122-kDa protein band in the noninduced cells (0 h), and as reported before (12), full-length hRad21 was found exclusively in the nuclear fractions (Fig. 2B). However, induction of apoptosis resulted in the early (4 h after induction) generation of approximately 64- and 60-kDa cleavage products, as determined by a C-terminal hRad21 antibody (Fig. 2B). As indicated by Annexin V staining, hRad21 cleavage shows a clear temporal relationship with the early events of apoptosis when the cell membrane remains intact. Annexin V is a calcium-dependent, phospholipid binding protein with high affinity for phosphatidylserine (PS) and can be used to identify apoptotic cells with exposed PS. As PS exists in the inner face of the cell membrane in normal cells, Annexin V cannot bind to the cell membrane. Early in apoptosis, however, PS is translocated from the inner to the outer surface of the cell membrane. As Annexin V has high affinity for PS, it can then bind to the cell surface via interaction with PS. In the late stages of apoptosis, Annexin V continues to bind PS, and as the membrane permeability is increased, PI can enter the cell and bind DNA. In addition to our results with Annexin V staining, we found that the progressive increase in the cleavage of hRad21 correlates with the level of active caspase-3 (Fig. 2A).
FIG. 2.
Time course of etoposide-induced hRad21 cleavage. Molt4 cells were incubated in the absence (0 h) or presence of 10 μM etoposide for 1, 2, 3, 4, 6, and 12 h. At the end of the incubation period, lysates from cytoplasmic and nuclear fractions were made. (A) Induction of apoptosis was verified by determination of caspase-3 activity in a Western blot analysis using anti-caspase-3 mAb. Pro-caspase-3 and active caspase-3 are indicated. (B and C) The time course of cleavage of hRad21 protein in etoposide-induced cytoplasmic and nuclear fractions was examined using C-terminal (B) and N-terminal (C) Rad21 pAbs. Full-length hRad21 (122 kDa; open arrow) and C-terminal cleaved 60- and 64-kDa fragments and N-terminal 50- and 55-kDa fragments (closed arrows) are indicated. (D and E) The purities of the cytoplasmic (D) and nuclear (E) fractions were verified with antibodies to tubulin and nuclear lamin, respectively. (F) Quantitative measurement of Annexin V staining showing early and late events of apoptosis. Cells stained with Annexin V (green fluorescence in the outer cell membrane) represent early stages of apoptosis, while cells with both PI-stained nuclei (red fluorescence) and Annexin V staining (green fluorescence in the outer cell membrane) represent late stages of apoptosis. Cells with PI-stained red fluorescence represent necrotic cells only. Normal cells lack any staining. Examples from each category are shown at right. Values represent the percentage of green (early stage of apoptosis), green plus red (late stage of apoptosis), or red (necrotic) fluorescent cells, with 120 cells being used at each time point.
Although hRad21 is a nuclear protein, the cleaved products are found in both nuclear and cytoplasmic fractions after induction of apoptosis (Fig. 2B). The identities of these two cleavage products were investigated using an N-terminal hRad21 antibody. As expected, the N-terminal antibody could not detect the 64- and 60-kDa cleavage products either in the cytoplasmic or nuclear fraction. In contrast, this antibody detected two other bands (approximately 50 and 55 kDa) only in the nuclear fractions (Fig. 2C). The purities of the cytosolic and nuclear fractions were verified with antibodies to tubulin and lamin, respectively (Fig. 2D and E). These results indicate that hRad21 may potentially be cleaved at two different sites following induction of apoptosis. The C-terminal hRad21 cleavage products but not the N-terminal hRad21 products are found in the cytoplasm after cleavage following induction of apoptosis.
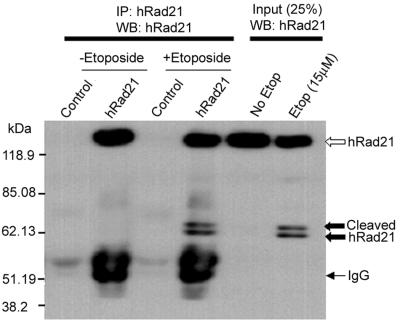
The identities of the cleavage products were confirmed through recognition by mAbs to hRad21 in IP and Western blot analyses (Fig. 3). hRad21 mAb selectively immunoprecipitated both the 60- and 64-kDa hRad21 cleavage products, along with the native 122-kDa full-length hRad21 protein in etoposide-induced Molt4 cells. Analysis of cells treated with vehicle only and control IP with isotype bacterial TrpE antibody did not detect these bands, confirming that the cleaved bands were hRad21 products. Both monoclonal and polyclonal C-terminal antibodies detected the 64- and 60-kDa bands, confirming that these bands were derived from the C-terminal portion of the cleaved protein.
FIG. 3.
IP and Western blot (WB) analyses of the hRad21 cleavage products. Apoptosis was induced in Molt4 T cells by treatment with 10 μM etoposide (Etop) for 8 h. Protein lysates were subjected to IP using hRad21 mAb or a control bacterial trpE mAb. Both monoclonal and polyclonal C-terminal antibodies detected 64- and 60-kDa bands (closed arrows), indicating that these bands are from the C-terminal portion of the cleaved protein.
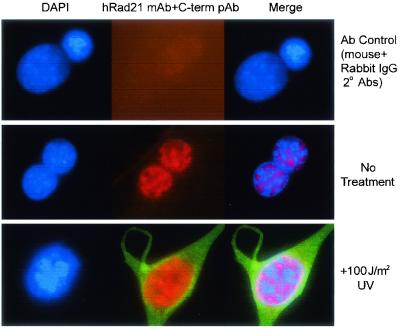
Translocation of hRad21 was further investigated in EL-12 mammary epithelial cells by immunofluorescence staining using the monoclonal and C-terminal hRad21 polyclonal antisera. Unlike Molt4 cells, EL-12 cells have a large cytoplasm to facilitate visualization. In these cells, Rad21 was entirely nuclear (Fig. 4, middle panel). Apoptosis was induced by treatment of EL-12 cells with UV light (100 J/m2) and was verified by the cleavage of Rad21 and PARP (data not shown). Immunofluorescence staining of the UV-treated cells by the C-terminal antibody clearly demonstrated translocation of the cleaved C-terminal Rad21 to the cytoplasm (Fig. 4, bottom panel).
FIG. 4.
C-terminal Rad21 cleavage product translocates to the cytoplasm after induction of apoptosis. Apoptosis was induced by treatment of EL-12 mammary epithelial cells with UV light (100 J/m2). UV-treated (bottom panel) and untreated control (middle panel) cells were subjected to immunofluorescence staining and microscopy using a C-terminal hRad21 pAb (green fluorescence) and hRad21 mAb (red fluorescence), respectively. The signals of mAb and pAb were visualized by the addition of rhodamine-labeled goat anti-mouse IgG and fluorescein-labeled goat anti-rabbit IgG, respectively. The upper panel shows the background staining (negative control) from the fluorescein-labeled secondary antibody in the presence of normal mouse and rabbit IgGs. The nuclear material is visualized by DAPI staining (blue fluorescence). Panels at right show merged images of red, blue, and green fluorescence.
Inhibition of hRad21cleavage by caspase peptide inhibitors.
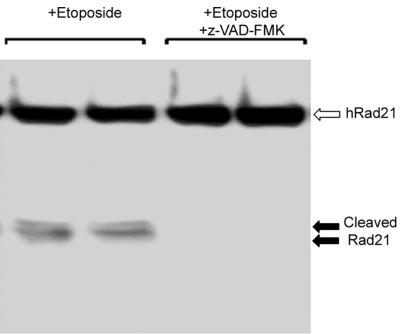
Peptide-based caspase inhibitors inhibited the apoptosis-induced cleavage of hRad21, suggesting the involvement of caspases in hRad21 cleavage (Fig. 5). Molt4 cells were treated with 20 μM z-VAD-FMK, a broad-spectrum caspase inhibitor, 1 h prior to etoposide (10 μM) treatment. As shown in Fig. 5, treatment with z-VAD-FMK completely blocked etoposide-induced hRad21 cleavage. In an in vitro cleavage assay (described below), z-VAD-FMK also inhibited caspase-3-induced cleavage of 35S-hRad21 (data not shown).
FIG. 5.
Caspase peptide inhibitors inhibit etoposide-induced cleavage of hRad21. Molt4 cells were treated with 20 μM z-VAD-FMK, a broad-spectrum caspase inhibitor, 1 h prior to etoposide (10 μM) treatment for 6 h. At the end of the incubation period, protein lysates were analyzed on an SDS-6% PAGE gel followed by Western blot analysis using hRad21 C-terminal pAb.
Identification of the apoptotic cleavage site in hRad21.
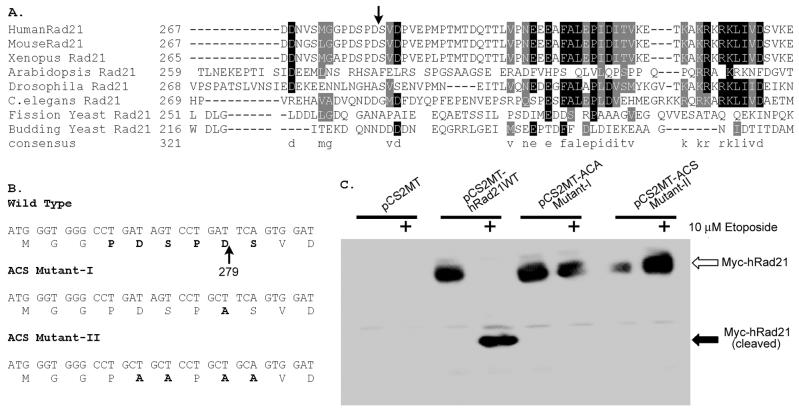
The hRad21 cleavage site was mapped through N-terminal sequencing of the immunoprecipitated hRad21 cleavage products from electroblotted and Coomassie-stained PVDF membranes (Fig. 6). Sequencing of the 64-kDa band revealed that hRad21 was cleaved at Asp-279 (D279). The N-terminal sequence of the 64-kDa band was SVDPVEP. In the full-length protein, the sequence immediately N terminal to the D279 cleavage site was PDSPD279. Thus, there was a repeat of the PDS sequence encompassing the cleavage site, i.e., PDSPD279/SVDPVEP (Fig. 6A). We were not successful in sequencing the 60-kDa band, possibly because of an N-terminal blocking effect. To verify whether specific cleavage occurred at D279 in hRad21 after induction of apoptosis, we introduced a point mutation (mut-I) by substituting an alanine (A) for aspartate (D) at this position of hRad21 (D279A) (Fig. 6B). Furthermore, because of the repetition of the cleavage sequence 275PDSPDS280, we made a second mutant construct (mut-II) by substituting alanine (A) for both aspartate (D) and serine (S) residues, i.e., hRad21 (275PDSPDS280 to 275PAAPAA280). We then transiently transfected Molt4 cells with WT or mutant hRad21 constructs tagged with the myc epitope at the N terminus and treated these cells with etoposide as indicated. As shown in Fig. 6C, antibody against myc tag (9E10) revealed the proteins encoded by the transfected hRad21 WT and hRad21 ACS-mut-I and ACS-mut-II constructs. However, the cleavage fragments were detected only for WT hRad21, not for either of the mutants, indicating that a point mutation at D279 prevented cleavage (Fig. 6C). We reprobed the blot with anti-hRad21 mAb and found that both the 60- and 64-kDa C-terminal fragments were present in all etoposide-treated cells (data not shown), confirming that endogenous hRad21 was cleaved in cells transfected with hRad21 mutants.
FIG. 6.
Characterization of the apoptotic cleavage site of hRad21. The hRad21 cleavage site was mapped biochemically, as described in Materials and Methods, through N-terminal sequencing of a Coomassie-stained PVDF membrane that was electroblotted with immunoprecipitated hRad21 cleavage products. (A) Comparison of the apoptotic cleavage recognition site and the adjoining sequence of hRad21 with those of other vertebrates (MouseRad21 and Xenopus Rad21) and simpler eukaryotes. C.elegans, Caenorhabditis elegans. The arrow indicates the peptide bond cleaved during apoptosis. (B) Construction of the apoptotic cleavage site mutants, ACS-mut-I and ACS-mut-II, to verify whether specific cleavage occurs at D279 in hRad21 after induction of apoptosis, by introduction of a point mutation to substitute an alanine (A) for aspartate (D) (mut-I) or alanine (A) for aspartate (D) and serine (S) (mut-II). (C) Molt4 cells were transiently transfected with blank vector (pCS2MT), WT hRad21 (pCS2MT-hRAD21), or ACS mutants tagged with myc epitope at their N termini (pCS2MT-ACSmut-I or pCS2MT-ACSmut-II) and treated with etoposide as indicated. Lysates were analyzed with SDS-6% PAGE followed by Western blot analysis using antibody against myc tag (9E10) to distinguish the cleavage products from the native forms of transfected hRad21 WT and hRad21 ACS-mut-I and ACS-mut-II proteins.
Involvement of caspases in hRad21 cleavage.
Closer inspection of the adjoining sequence at the Rad21 apoptotic cleavage site (DSPD279) (Fig. 6A) revealed a putative recognition sequence for a primitive caspase, Ced3. This sequence is conserved Rad21 in vertebrates, including humans, mice, and frogs (Xenopus spp.). The sequence of the putative cleavage site, together with the inhibitory effect of a panel of caspase inhibitors on etoposide-induced apoptosis in Molt4 cells, indicated the possible involvement of a caspase family protease. While the experiments with caspase inhibitors suggested the involvement of a caspase family of protease(s) in the pathway leading to hRad21 cleavage, they did not demonstrate direct internal cleavage of hRad21 by a caspase. We therefore utilized an in vitro cleavage assay as described previously for the retinoblastoma protein (9) to examine the ability of purified caspase to cleave hRad21 (Fig. 7). We used two caspases, caspase-3 and caspase-7, that are major regulators of apoptosis in diverse cell types (35) (Fig. 7A), along with lysates from Molt4 cells treated with etoposide (apoptotic lysate) or vehicle (nonapoptotic lysate), to examine their role in hRad21 cleavage (Fig. 7B). Together, caspase-3 and caspase-7 comprise the caspase-3 subfamily, and both enzymes recognize and cleave after the consensus cleavage site DXXD (E. S. Alnemri, D. J. Livingston, D. W. Nicholson, G. Salvesen, N. A. Thornberry, W. W. Wong, and J. Yuan, Letter, Cell 87:171, 1996). Addition of recombinant caspase-3 or caspase-7 to the in vitro transcribed and translated hRad21 in rabbit reticulocyte lysates clearly resulted in the production of a 64-kDa hRad21 fragment (Fig. 7A and B). The 64-kDa fragment produced by these caspases precisely comigrated with the 64-kDa band produced by apoptotic Molt4 lysates, while the control (nonapoptotic) cell lysate could not cleave hRad21 protein. On the other hand, both of the hRad21 ACS mutants failed to be cleaved by these two caspases or by apoptotic cell lysates in this assay, strongly suggesting that caspase-3- or caspase-7-like enzymes in the apoptotic cells or extracts were responsible for cleavage at the putative caspase recognition site (D279) of hRad21.
FIG. 7.
Cleavage of in vitro translated hRad21 by recombinant caspase-3 and caspase-7. (A) In a cleavage reaction, in vitro translated unlabeled (nonisotopic) WT hRad21 or ACS mutant hRad21 (ACSmut-I or ACSMut-II) in the rabbit reticulocyte lysate were incubated with saline (vehicle) (lanes a), caspase-3 (lanes b), or caspase-7 (lanes c). TNT reactions in the absence of plasmid DNA served as a negative control. Samples were analyzed on an SDS-6% PAGE gel followed by Western blotting with hRad21 C-terminal pAb. Molecular weight markers are shown at left. (B) 35S-hRad21 was incubated in the absence of extract (left lane) or in the presence of Molt4 cell lysates treated with DMSO (middle lane) or 10 μM etoposide (right lane) for 6 h. Samples were resolved on a 4 to 20% gradient SDS-PAGE gel, fixed with methanol and acetic acid for 30 min, dried on a gel dryer, and exposed to a Storm imager.
In addition to the 64-kDa fragment, several other fragments were generated by caspase-3 and -7. It is possible that the 64-kDa fragment was degraded further by these caspases, resulting in the production of smaller fragments. In the presence of caspase-7 but not caspase-3, a 95-kDa fragment of hRad21 was also generated, suggesting another caspase site N-terminal to the D279 cleavage site. In the in vitro cleavage assay, caspase-3 and -7 failed to generate the 60-kDa hRad21 fragment that accompanies the 64-kDa fragment after induction of apoptosis, suggesting that the cleavage site generating the 60-kDa fragment was not recognized by caspase-3 and -7.
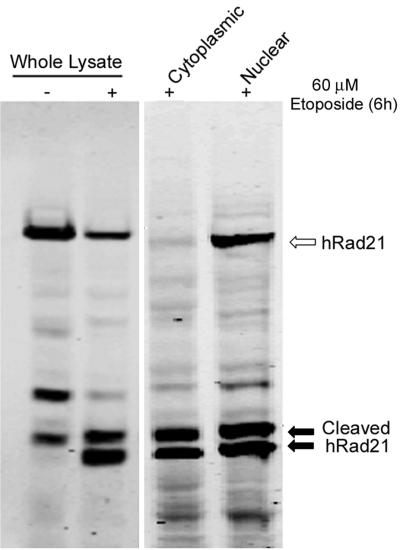
To determine whether caspase-3 is essential for the in vivo cleavage of hRad21, we utilized a caspase-3-deficient MCF-7 breast cancer cell line (21). In experiments using etoposide- or tamoxifen-induced apoptosis in MCF-7 cells, hRad21 cleavage products were detected, indicating that caspase-3 was not essential for hRad21 cleavage (Fig. 8) and that a caspase other than caspase-3 can act upon hRad21 to cause cleavage following induction of apoptosis.
FIG. 8.
Cleavage of hRad21 in caspase-3-deficient MCF-7 breast cancer cells. Apoptosis was induced by treatment of MCF-7 cells with 60 μM etoposide for 6 h. Cells treated with DMSO (vehicle) served as a control. Whole-cell lysates or lysates from the cytoplasmic and nuclear fractions were electrophoresed on an SDS-6% PAGE gel and subjected to Western blot analysis using hRad21 mAb. Arrows indicate the hRad21 products.
hRad21 C-terminal cleavage product promotes apoptosis.
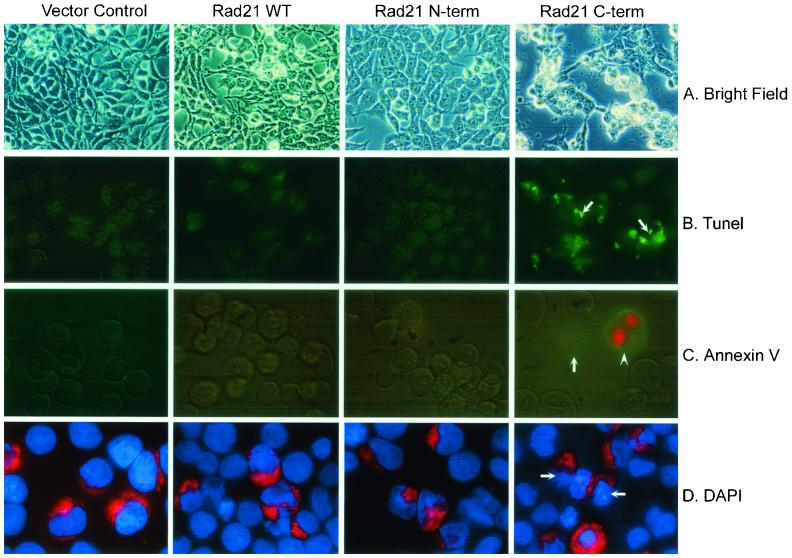
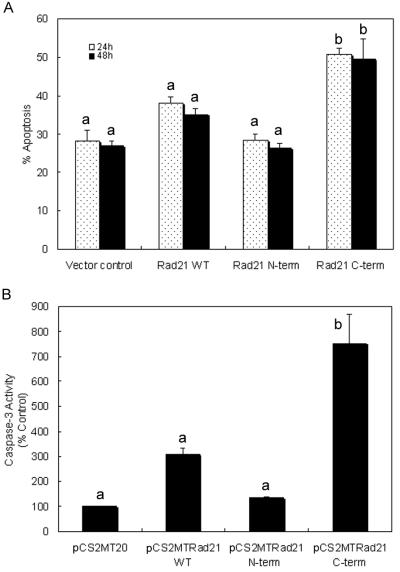
A possible role for hRad21 in the induction of apoptosis was first seen in preliminary experiments in which overexpression of hRad21 in Molt4, 293T, and MCF7 cells resulted in apoptotic phenotypes. However, conclusive evidence for the role of cleaved hRad21 in the promotion of apoptosis was obtained by transient transfection of 293T and Molt4 cells with cytomegalovirus (CMV) promoter-driven myc-tagged mammalian expression plasmids encoding the full-length hRad21 or cDNAs encoding the two cleavage products, hRad21 N-terminal (aa 1 to 279) or hRad21 C-terminal (aa 280 to 631) proteins (Fig. 9 and 10). Analysis of transfected 293T cells by multiple apoptosis assays, including examination of cellular morphology under light microscopy (Fig. 9A) and staining with TUNEL (Fig. 9B), Annexin V (Fig. 9C), and DAPI (Fig. 9D), clearly indicated the ability of the 64-kDa C-terminal hRad21 to induce apoptosis. Quantitative analysis of 293T cells transfected with C-terminal hRad21 plasmids indicated a significant increase (P < 0.05) of nuclear degradation in cells transfected with the C-terminal hRad21 compared with that in cells transfected with vector control (pCS2MT), WT hRad21 (pCS2MT Rad21), or N-terminal hRad21 (pCS2MT hRad21 N-term) (Fig. 10A). In these cells, apoptosis was also assayed by monitoring the phenotype of the DAPI-stained nuclei. As shown in Fig. 9D, cells transfected with hRad21 C-terminal plasmid displayed significantly more (P < 0.05) cellular and nuclear phenotypes typical of apoptosis, such as a round shape with shrunken cell volume, chromatin condensation, and nuclear disintegration, compared with the vector control and cells expressing the full-length and N-terminal hRad21 constructs. Similar results were also obtained with MCF7 and Molt4 cells (data not shown).
FIG. 9.
Analysis of the proapoptotic activity of C-terminal hRad21 in 293T cells by examination of cellular morphology (A) and staining with TUNEL (B), Annexin V (C), and DAPI (D). In panels A to C, 293T cells were transiently transfected with CMV promoter-driven myc-tagged mammalian expression plasmids (pCS2MT) encoding the full-length hRad21 or cDNAs encoding the two cleavage products, hRad21 N-terminal (aa 1 to 279) and hRad21 C-terminal (aa 280 to 631) proteins. In panel D, in addition to the above constructs, cells were also cotransfected with a red fluorescence plasmid (pDsRed1-Mito) to account for the hRad21-transfected cells. Note that C-terminal-hRad21-transfected cells have round morphology, as shown in light micrographs (A) and are positive for TUNEL (B) and Annexin V (C) staining (arrows). PI was used to detect the integrity of cell membranes in Annexin V-FITC staining; hence, the nuclei of some apoptotic cells were red (arrowhead). In panel D, more than 50% of the C-terminal-hRad21-transfected cells with red fluorescence have fragmented nuclei and condensed chromatin, typical of apoptotic cells.
FIG. 10.
Quantitative analysis of the proapoptotic activity of C-terminal hRad21 in 293T and Molt4 T-cell leukemia cells. Cells were transiently transfected with CMV promoter-driven myc-tagged mammalian expression plasmids encoding the full-length hRad21 or cDNAs encoding the two cleavage products, hRad21 N-terminal (aa 1 to 279) and hRad21 C-terminal (aa 280 to 631) proteins. 293T cells were also cotransfected with the red fluorescence plasmid pDSRed1-mito to account for the hRad21-transfected cells. Nuclear degradation of 293T cells (A) and caspase-3 activity in Molt4 cells (B) were measured as described in Materials and Methods. For the nuclear degradation assay, cells were collected and fixed 24 and 48 h after transfection. The samples were stained with DAPI and examined with fluorescence microscopy. Cells with fragmented nuclei or condensed chromatin were counted as apoptotic. For each sample, a total of 50 intact and degraded nuclei of cells coexpressing pDsRed1-mito (with red fluorescence) was counted. Each data point represents the average and standard errors of the mean from three experiments. The data for caspase-3 activity shown in panel B are the averages and standard errors of the mean from two experiments. The results of the apoptosis assay were analyzed using a binomial test of proportions and those for caspase-3 activity were analyzed using Student's t test. Data labeled “a” are significantly different (P < 0.05) from data labeled “b.”
The proapoptotic activity of the C-terminal hRad21 was further strengthened by a significantly increased level (P < 0.05) of caspase-3 activity in Molt4 cells transfected with hRad21 C-terminal plasmids compared with that in cells transfected with the WT hRad21 (pCS2MT hRad21) and N-terminal hRad21 (pCS2MT hRad21 N-term) constructs. As shown in Fig. 10B, C-terminal hRad21 overexpression resulted in a five- to sevenfold increase in caspase-3 activity compared with that of the empty vector control. Although overexpression of WT hRad21 induced moderate but statistically insignificant levels of apoptosis, as determined by caspase-3 activity in Molt4 cells, overexpression of the hRad21 C-terminal cleavage product but not the N-terminal hRad21 cleavage product dramatically increased caspase-3 activity (P < 0.05) in Molt4 cells (Fig. 10B). The transfection efficiency in Molt4 cells was 35%, as determined by cotransfection with a red fluorescence plasmid, pDSRed1-mito. Similar results for caspase-3 activity were also obtained with MCF-7 and 293T cells transfected with hRad21 constructs (data not shown).
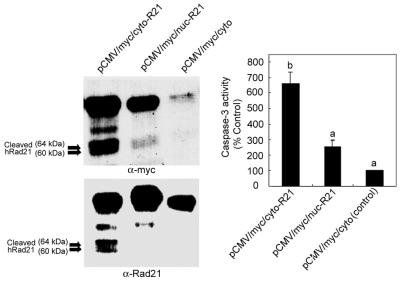
Further experiments using vectors to direct hRad21 expression either to the cytoplasm or nucleus demonstrated that expression of hRad21 in the cytoplasm but not in the nucleus resulted in the cleavage of hRad21 protein and induction of apoptosis, as determined by assaying of caspase-3 activity (Fig. 11). It is interesting that both the myc-tagged (cytoplasmic) hRad21 and the WT (normally nuclear) hRad21 were cleaved in the cells transiently transfected with the pCMV/myc/cyto-hRad21 construct (Fig. 11). In summary, the C-terminal hRad21 cleavage product was proapoptotic, as determined by increased caspase-3 activity and apoptotic morphology, and its translocation to the cytoplasm may play a role in promoting apoptosis. These findings demonstrate the ability of the 64-kDa Rad21 fragment to induce apoptosis.
FIG. 11.
Effect of the cytoplasmic expression of hRad21 on apoptosis and cleavage of hRad21 protein. Molt4 cells were transiently transfected with the constructs pCMV/myc/cyto-hRad21 and pCMV/myc/nuc-hRad21, which direct the expression of the recombinant protein to the cytoplasm and nucleus, respectively. Empty PCMV/myc/cyto vector served as a control. At 48 h posttransfection, protein lysates were made and subjected to Western blot analysis using c-myc epitope (9E10) and hRad21 mAb to detect the fusion and/or native hRad21 proteins, respectively. Lysates were also used for the assaying of caspase-3 activity as described in Materials and Methods. The data for caspase-3 activity shown at right are the averages and standard errors of the mean from two experiments. Individual values were compared using Student's t test, and values labeled “a” are significantly different (P < 0.05) from that labeled “b.”
Apoptotic cleavage of hRad21 is not affected by the status of the p53 tumor suppressor protein in the cell.
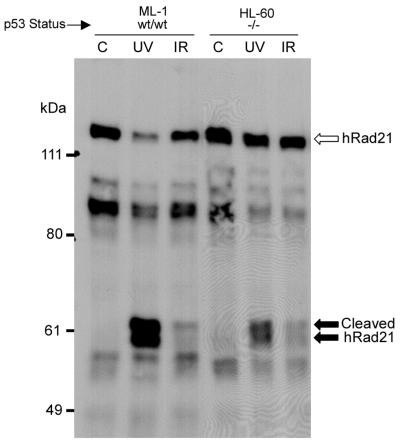
In view of the pivotal role of the p53 gene product in regulation of the cell cycle and apoptosis, we examined the role of p53 in the apoptotic cleavage of hRad21. We used two myeloid leukemia cell lines, ML-1 and HL-60, with WT and null p53 genotypes, respectively (32, 41). Apoptosis was induced using UV (20 J/m2) and ionizing radiation (20 Gy) in these cells. As shown in Fig. 12, the induction of apoptosis resulted in the cleavage of hRad21 protein in both cell lines, indicating the lack of a role for p53 in hRad21 cleavage.
FIG. 12.
Effect of p53 status on the cleavage of hRad21. Apoptosis was induced in the myeloid leukemia cell lines ML-1 and HL-60, with WT and null p53 genotypes, respectively, by treatment of the cells with UV light (20 J/m2) or ionizing radiation (IR) (20 Gy). Six hours after treatment, protein lysates were made and resolved by SDS-6% PAGE followed by Western blot analysis using hRad21 mAb. Arrows indicate hRad21 cleavage products. C, control without UV or IR treatment.
DISCUSSION
Sister chromatid cohesion during DNA replication plays a pivotal role in accurate chromosomal segregation in the eukaryotic cell cycle. Rad21 is one of the major cohesin subunits that keeps sister chromatids together until anaphase when proteolytic cleavage by separase allows the chromosomes to separate. Mitotic cleavage sites in Rad21 in yeast as well as in humans have been mapped (12, 37, 39). Here we show that hRad21 cleavage occurs during apoptosis and is induced by various agents, including DNA-damaging (ionizing radiation and topoisomerase inhibitors) and non-DNA-damaging agents (cycloheximide treatment, cytokine withdrawal, and treatment with proteasome inhibitors). We have biochemically mapped the apoptotic cleavage site in human Rad21 (PDSPD279/S), which is distinct from the mitotic cleavage sites (DREIMR172/E and IEEPSR450/L) previously described (12). The apoptotic cleavage site is conserved among vertebrate species, and it is likely that cleavage is mediated by a nuclear caspase or caspase-like molecule, as this cleavage site bears the characteristic caspase-3 subfamily recognition motif (DXXD) and hRad21 is cleaved in vitro by the two major apoptosis executioner caspases, caspase-3 and caspase-7. hRad21 cleavage is not restricted to transformed cancer cells, as induction of apoptosis resulted in hRad21 cleavage in the primary cell line IMR90 (data not shown) as well as the nontransformed immortal cell line EL-12.
Cleavage of hRad21 appears to be an early event in the apoptotic pathway. The immunofluorescence experiments and Western blot analysis of nuclear and cytoplasmic fractions of cells undergoing apoptosis demonstrate the translocation of the hRad21 C-terminal cleavage products to the cytoplasm early (3 to 4 h after insult) in apoptosis. Our results clearly show that hRad21 proteolysis by a caspase family protease at D279/S leads to the production of a proapoptotic C-terminal cleavage product. The specific protease that cleaves hRad21 in vivo and promotes hRad21-induced apoptosis is yet to be identified. Nuclear changes determined by Annexin V staining and examination of the morphology of DAPI-stained nuclei indicate a strong temporal relationship between hRad21 cleavage and apoptosis. As determined by Annexin V staining, hRad21 cleavage correlates well with the early events of apoptosis when the cell membrane remains intact. Furthermore, the progressive increase in the cleavage of hRad21 correlates well with the level of caspase activation, as determined by assaying of by caspase-3 activity. Translocation of the 64-kDa hRad21 cleavage product to the cytoplasm early in apoptosis may act as a nuclear signal that promotes and accelerates subsequent events of apoptosis. The specificity of this product was determined further, as the N-terminal hRad21 cleavage product neither translocates nor has the ability to induce apoptosis. We have not explored the role of the 60-kDa hRad21 product generated at a cleavage site other than D279/S in the apoptotic process.
The physiological significance of cohesin hRad21 cleavage in apoptosis is intriguing. The nuclear signal(s) that detects subsequent events of apoptosis in the cytoplasm and mitochondria has remained elusive. It is possible that cleavage of hRad21 at the onset of apoptosis and the translocation of the C-terminal cleavage product to the cytoplasm act as cues to accelerate the apoptotic process. Supporting evidence in favor of this possibility include the following: (i) hRad21 is not normally cytoplasmic; (ii) early in apoptosis, hRad21 is found in the cytoplasm; and (iii) directed expression of either the C-terminal or full-length hRad21 to the cytoplasm induces apoptosis. It is not clear whether localization of C-terminal hRad21 to the cytoplasm is due to an active or a passive transport process following cleavage. The carboxy-terminal fragment contains a putative nucleolar localization signal sequence, which argues against a passive transport process. These findings further strengthen the notion that the translocation of the C-terminal hRad21 protein to the cytoplasm may play a functional role in apoptosis.
We have firmly established the proapoptotic activity of the C-terminal hRad21 cleavage product by several apoptotic assays, including Annexin V staining, TUNEL methods, quantitative measurement of DAPI-stained nuclear morphology, and assaying of caspase-3 activity. However, the exact mechanism by which cleaved hRad21 induces apoptosis requires further investigation. It is interesting that a BLAST search of the apoptosis database (www.apoptosis-db.org) indicated that C-terminal hRad21 possesses a stretch of 80 aa (aa 282 to 362) that has homology to the tumor necrosis factor receptor superfamily and other apoptosis-inducing proteins, including TRAIL-R2 and death receptor 5. However, the functional significance of this domain in apoptosis-inducing proteins is not known.
The caspase-mediated proteolysis of hRad21 and the partial removal of hRad21 from the nucleus may also expose the chromosomal DNA to DNase and other proteins responsible for chromatin condensation and apoptotic DNA fragmentation. hRad21 was originally isolated in fission yeast as an essential protein with a role in the repair of DNA double-strand breaks induced by ionizing radiation (2). It is therefore logical to think that disruption of the DNA repair function of hRad21 may be necessary during the execution of apoptosis. This notion has been strengthened by recent findings that a number of DNA repair enzymes such as Rad51 (15), ATM (13), DNA-PK (4), and PARP (22) and cell cycle regulators such as retinoblastoma protein (9) are cleaved by caspases. Coordinated destruction of the DNA repair machinery and cell cycle regulators by the caspase family of proteases therefore constitutes a physiologically relevant process that promotes and accelerates chromosomal condensation and DNA fragmentation without interference by the cell cycle and DNA repair machinery. Unlike hRad21, however, cleavage products of these other DNA repair proteins have not been reported to play a direct role in promoting apoptosis. In this case, cleavage of hRad21 by caspases may play a unique role in amplifying the apoptotic signal by elevating the level of caspase activity. A similar mechanism for amplifying the apoptotic signal for the caspase substrate vimentin has recently been described (3).
The p53 tumor suppressor protein plays a central role in the regulation of the cell cycle and apoptosis after DNA damage (17, 34). In the event that DNA damage is more severe and not repairable, p53 directs the cells into apoptosis through the Bax/Bcl-2 pathway. p53 status does not appear to have any effect on the apoptotic cleavage of hRad21 after DNA damage (i.e., UV and ionizing radiation), indicating the lack of involvement of the p53 pathway in hRad21 cleavage. It is possible that a parallel p53-independent pathway may regulate the genotoxic-damage-induced cleavage of hRad21.
Finally, it is interesting that cleavage of cohesin hRad21 is carried out by a separase in mitosis and by a caspase in apoptosis at different sites in the protein. Both of these proteases belong to the distantly related CD clan protease family (38), suggesting an evolutionarily conserved mechanism shared by the mitotic and apoptotic machinery. hRad21 may serve as the link between the two key cellular processes of mitosis and apoptosis. In summary, in contrast to the previously described functions of Rad21, i.e., in chromosome segregation and DNA repair, cleavage of the cohesion protein and translocation of the C-terminal cleavage product to the cytoplasm are early events in the apoptotic pathway that amplify the apoptotic signal in a positive-feedback manner, possibly by activating more caspases. These results provide the framework for identification of the physiologic role of hRad21 in the apoptotic response in normal and malignant cells.
ADDENDUM IN PROOF
A similar conclusion regarding the function of RAD21 in apoptosis has been published by F. Chen et al. (J. Biol. Chem 277:16775-16781, 2002).
Acknowledgments
We thank T. Nagase (Kazusa DNA Research Institute, Chiba, Japan) for the KIAA0078 (SK-hRad21) plasmid, J.-M. Peters (Research Institute of Molecular Pathology, Vienna, Austria) for the Rad21 N-terminal pAb, and D. Medina (Baylor College of Medicine) for the EL-12 cell line. We thank Lisa Wang for critically reading the manuscript and Sara Ekhlassi for technical assistance.
This study was supported by grants from the U.S. Army Medical Research and Materiel Command (DAMD-17-00-1-0606, DAMD-01-1-0142, and DAMD 01-1-0143 to D.P. and DAMD-17-97-1-7284 and DAMD-17-98-1-8281 to S.E.P.).
REFERENCES
- 1.Biggins, S., and A. W. Murray. 1999. Sister chromatid cohesion in mitosis. Curr. Opin. Genet. Dev. 9:230-236. [DOI] [PubMed] [Google Scholar]
- 2.Birkenbihl, R. P., and S. Subramani. 1992. Cloning and characterization of rad21, an essential gene of Schizosaccharomyces pombe involved in DNA double-strand-break repair. Nucleic Acids Res. 20:6605-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byun, Y., F. Chen, R. Chang, M. Trivedi, K. J. Green, and V. L. Cryns. 2001. Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ. 8:443-450. [DOI] [PubMed] [Google Scholar]
- 4.Casciola-Rosen, L. A., G. J. Anhalt, and A. Rosen. 1995. DNA-dependent protein kinase is one of a subset of autoantigens specifically cleaved early during apoptosis. J. Exp. Med. 182:1625-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaturvedi, M. M., R. LaPushin, and B. B. Aggarwal. 1994. Tumor necrosis factor and lymphotoxin. Qualitative and quantitative differences in the mediation of early and late cellular response. J. Biol. Chem. 269:14575-14583. [PubMed] [Google Scholar]
- 6.Ciosk, R., W. Zachariae, C. Michaelis, A. Shevchenko, M. Mann, and K. Nasmyth. 1998. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 93:1067-1076. [DOI] [PubMed] [Google Scholar]
- 7.Cohen-Fix, O., J.-M. Peters, M. W. Kirschner, and D. Koshland. 1996. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 10:3081-3093. [DOI] [PubMed] [Google Scholar]
- 8.Downie, N. M., and R. W. Heath. 1965. Basic statistical methods, 2nd ed. Harper & Row, New York, N.Y.
- 9.Fattman, C. L., S. M. Delach, Q. P. Dou, and D. E. Johnson. 2001. Sequential two-step cleavage of retinoblastoma protein by caspase-3/-7 during etoposide-induced apoptosis. Oncogene 20:2918-2926. [DOI] [PubMed] [Google Scholar]
- 10.Funabiki, H., H. Yamano, K. Kumada, K. Nagao, T. Hunt, and M. Yanagida. 1996. Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature 381:438-441. [DOI] [PubMed] [Google Scholar]
- 11.Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell 91:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauf, S., I. C. Waizenegger, and J.-M. Peters. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science 293:1320-1323. [DOI] [PubMed] [Google Scholar]
- 13.Hotti, A., K. Jarvinen, P. Siivola, and E. Holtta. 2000. Caspases and mitochondria in c-Myc-induced apoptosis: identification of ATM as a new target of caspases. Oncogene 19:2354-2362. [DOI] [PubMed] [Google Scholar]
- 14.Huang, D. C. S., L. A. O'Reilly, A. Strasser, and S. Cory. 1997. The anti-apoptosis function of Bcl-2 can be genetically separated from its inhibitory effect on cell cycle entry. EMBO J. 16:4628-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, Y., S. Nakada, T. Ishiko, T. Utsugisawa, R. Datta, S. Kharbanda, K. Yoshida, R. V. Talanian, R. Weichselbaum, D. Kufe, and Z. M. Yuan. 1999. Role for caspase-mediated cleavage of Rad51 in induction of apoptosis by DNA damage. Mol. Cell. Biol. 19:2986-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter, T. 1997. Oncoprotein network. Cell 88:333-346. [DOI] [PubMed] [Google Scholar]
- 17.Israels, L. G., and E. D. Israels. 1999. Apoptosis. Stem Cells 17:306-313. [DOI] [PubMed] [Google Scholar]
- 18.Jallepalli, P. V., I. C. Waizenegger, F. Bunz, S. Langer, M. R. Speicher, J.-M. Peters, K. W. Kinzler, B. Vogelstein, and C. Lengauer. 2001. Securin is required for chromosomal stability in human cells. Cell 105:445-457. [DOI] [PubMed] [Google Scholar]
- 19.Keelan, J. A., T. A. Sato, K. W. Marvin, J. Lander, R. S. Gilmour, and M. D. Mitchell. 1999. 15-Deoxy-delta(12,14)-prostaglandin J2, a ligand for peroxisome proliferator-activated receptor-gamma, induces apoptosis in JEG3 choriocarcinoma cells. Biochem. Biophys. Res. Commun. 262:579-585. [DOI] [PubMed] [Google Scholar]
- 20.Koshland, D. E., and V. Guacci. 2000. Sister chromatid cohesion: the beginning of a long and beautiful relationship. Curr. Opin. Cell Biol. 12:297-301. [DOI] [PubMed] [Google Scholar]
- 21.Kurokawa, H., K. Nishio, H. Fukumoto, A. Tomonari, T. Suzuki, and N. Saijo. 1999. Alteration of caspase-3 (CPP32/Yama/apopain) in wild-type MCF-7 breast cancer cells. Oncol. Rep. 6:33-37. [DOI] [PubMed] [Google Scholar]
- 22.Lazebnik, Y. A., S. H. Kaufmann, S. Desnoyers, G. G. Poirier, and W. C. Earnshaw. 1994. Cleavage of poly (ADP-ribose) polymerase by a proteinase with properties like ICE. Nature 371:346-347. [DOI] [PubMed] [Google Scholar]
- 23.Leismann, O., A. Herzig, S. Heidmann, and C. F. Lehner. 2000. Degradation of Drosophila PIM regulates sister chromatid separation during mitosis. Genes Dev. 14:2192-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 25.Lipponen, P., S. Aaltomaa, V.-M. Kosma, and K. Syrjanen. 1994. Apoptosis in breast cancer as related to histopathological characteristics and prognosis. Eur. J. Cancer 30A:2068-2073. [DOI] [PubMed] [Google Scholar]
- 26.Losada, A., and T. Hirano. 2001. Shaping the metaphase chromosome: coordination of cohesion and condensation. Bioessays 23:924-935. [DOI] [PubMed] [Google Scholar]
- 27.Medina, D., and F. S. Kittrell. 1993. Immortalization phenotype dissociated from the preneoplastic phenotype in mouse mammary epithelial outgrowths in vivo. Carcinogenesis 14:25-28. [DOI] [PubMed] [Google Scholar]
- 28.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 29.Minn, A. J., L. H. Boise, and C. B. Thompson. 1996. Expression of Bcl-Xl and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 10:2621-2631. [DOI] [PubMed] [Google Scholar]
- 30.Nasmyth, K., J.-M. Peters, and F. Uhlmann. 2000. Splitting the chromosome: cutting the ties that bind sister chromatids. Science 288:1379-1385. [DOI] [PubMed] [Google Scholar]
- 31.Nasmyth, K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673-745. [DOI] [PubMed] [Google Scholar]
- 32.O'Connor, P. M., J. Jackman, I. Bae, T. G. Myers, S. Fan, M. Mutoh, D. A. Scudiero, A. Monks, E. A. Sausville, J. N. Weinstein, S. Friend, A. J. Fornace, Jr., and K. W. Kohn. 1997. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 57:4285-4300. [PubMed] [Google Scholar]
- 33.Shimada, A. 1996. PCR-based site-directed mutagenesis. Methods Mol. Biol. 57:157-165. [DOI] [PubMed] [Google Scholar]
- 34.Strasser, A., L. O'Connor, and V. M. Dixit. 2000. Apoptosis signaling. Annu. Rev. Biochem. 69:217-245. [DOI] [PubMed] [Google Scholar]
- 35.Thornberry, N. A., and Y. Lazebnik. 1998. Caspases: enemies within. Science 281:1312-1316. [DOI] [PubMed] [Google Scholar]
- 36.Uhlmann, F., and K. Nasmyth. 1998. Cohesion between sister chromatids must be established during DNA replication. Curr. Biol. 8:1095-1101. [DOI] [PubMed] [Google Scholar]
- 37.Uhlmann, F., F. Lottspeich, and K. Nasmyth. 1999. Sister chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 400:37-42. [DOI] [PubMed] [Google Scholar]
- 38.Uhlmann, F., D. Wernic, M. A. Poupart, E. V. Koonin, and K. Nasmyth. 2000. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103:375-386. [DOI] [PubMed] [Google Scholar]
- 39.Waizenegger, I. C., S. Hauf, A. Meinke, and J.-M. Peters. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 103:399-410. [DOI] [PubMed] [Google Scholar]
- 40.Warren, W. D., S. Steffensen, E. Lin, P. Coelho, M. Loupart, N. Cobbe, J. Y. Lee, M. J. McKay, T. Orr-Weaver, M. M. Heck, and C. E. Sunkel. 2000. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr. Biol. 10:1463-1466. [DOI] [PubMed] [Google Scholar]
- 41.Zhan, Q., S. Fan, I. Bae, C. Guillouf, D. A. Liebermann, P. M. O'Connor, and A. J. Fornace. 1994. Induction of bax by genotoxic stress in human cells correlates with normal p53 status and apoptosis. Oncogene 9:3743-3751. [PubMed] [Google Scholar]
- 42.Zou, H., T. J. McGarry, T. Bernal, and M. W. Kirschner. 1999. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285:418-422. [DOI] [PubMed] [Google Scholar]