Abstract
The 5′ stem-loop of the U4 snRNA and the box C/D motif of the box C/D snoRNAs can both be folded into a similar stem-internal loop-stem structure that binds the 15.5K protein. The homologous proteins NOP56 and NOP58 and 61K (hPrp31) associate with the box C/D snoRNPs and the U4/U6 snRNP, respectively. This raises the intriguing question of how the two homologous RNP complexes specifically assemble onto similar RNAs. Here we investigate the requirements for the specific binding of the individual snoRNP proteins to the U14 box C/D snoRNPs in vitro. This revealed that the binding of 15.5K to the box C/D motif is essential for the association of the remaining snoRNP-associated proteins, namely, NOP56, NOP58, fibrillarin, and the nucleoplasmic proteins TIP48 and TIP49. Stem II of the box C/D motif, in contrast to the U4 5′ stem-loop, is highly conserved, and we show that this sequence is responsible for the binding of NOP56, NOP58, fibrillarin, TIP48, and TIP49, but not of 15.5K, to the snoRNA. Indeed, the sequence of stem II was essential for nucleolar localization of U14 snoRNA microinjected into HeLa cells. Thus, the conserved sequence of stem II determines the specific assembly of the box C/D snoRNP.
The synthesis and processing of eukaryotic rRNAs take place in the nucleolus and involve a complex series of processing steps that includes the covalent, posttranscriptional modification of the rRNA (20). The nucleolus contains an extensive number of small nucleolar RNAs (snoRNAs) that are involved in the processing of the pre-rRNA. Most of the snoRNAs function as sequence-specific guides in the modification of rRNA; however, some are also essential for the rRNA folding and cleavage events (1, 9, 16, 20, 40, 52). There are two major families of snoRNAs, namely, the H/ACA and box C/D snoRNAs, that are categorized based on conserved sequence motifs and their association with common core proteins. The majority of H/ACA snoRNAs function in the site-specific isomerization of uridine to pseudouridine, while the box C/D family of snoRNAs direct the 2′ O methylation of ribose moieties within rRNA and certain spliceosomal snRNAs (1, 9, 16, 20, 40, 52). The box C/D snoRNAs contain two evolutionarily conserved sequence elements, termed box C (box C′ in U3) and box D, that are often flanked by a short 4-bp stem. This flanking stem (stem I in Fig. 1A) and the box C/D motif have recently been shown to be able to fold into a conserved stem-internal loop-stem structure (Fig. 1A) (51). A second conserved element, termed the box C′/D′ motif, is found in the majority of but not in all members of this snoRNA family and is homologous but not identical to the box C/D motif. The site-specific modification of RNA is directed by extensive base pairing between the snoRNA and the target RNA. The base pairing is arranged such that the target nucleotide is positioned 5 bp downstream of either the box D or D′ element (Fig. 1A) (17, 18). A subset of the box C/D snoRNAs, which includes U3, U8, and U14, uses extensive complementary regions to the rRNA and/or pre-rRNA not to direct site-specific modification but to orchestrate the folding and cleavage of the precursor molecule (1, 9, 16, 20, 40, 52). These RNAs are proposed to function as molecular chaperones in the folding and cleavage of the pre-rRNA.
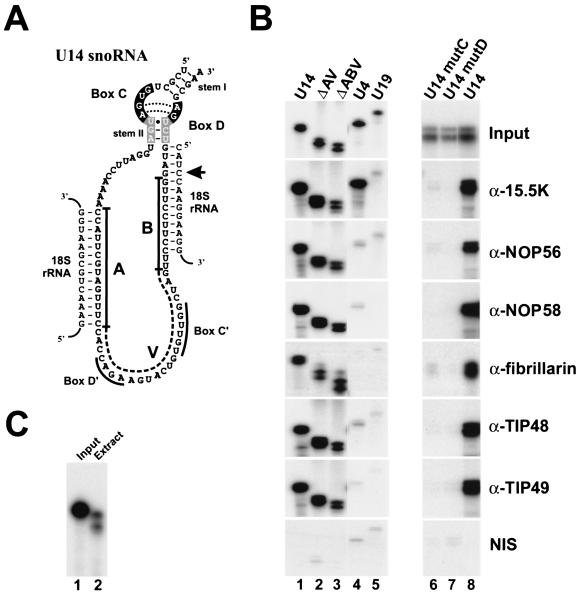
FIG. 1.
The binding of box C/D snoRNP-specific proteins is solely dependent on the box C/D motif. (A) Schematic representation of the U14 box C/D snoRNA. The conserved nucleotides of the box C/D motif are shown in white on a black background for the internal loop and white on a grey background for conserved stem II. The tentative box C′/D′ motif is indicated. Base-pairing interactions necessary for 18S rRNA modification and processing are shown. An arrow indicates the 2′ O methylation site directed by the U14 snoRNA. The three regions of the U14 snoRNA, namely, A, B, and V, are labeled (49). The mutations ΔAV and ΔABV involve the deletion of regions A and V and of A, B, and V, respectively. Mutations U14 mutC and U14 mutD involve the replacement of the conserved GA dinucleotides in boxes C and D, respectively, with CC (50). (B) Coimmunoprecipitation of in vitro-assembled snoRNPs with antibodies specific to the box C/D snoRNP proteins. SnoRNP complexes were assembled onto 32P-radiolabeled RNAs in HeLa nuclear extract (see Materials and Methods). RNP complexes formed during this reaction were then immunoprecipitated, and the copurifying RNAs were analyzed on an 8% polyacrylamide-7 M urea gel. The RNA used is indicated above each lane. Antibodies used for immunoprecipitation are indicated on the right. Input, 10% of the RNA after incubation in nuclear extract. NIS, nonimmune serum. (C) U14 snoRNA transcripts undergo limited trimming upon incubation in nuclear extract. U14 RNA before (lane 1, Input) or after (lane 2, Extract) incubation in nuclear extract. RNA was recovered from the nuclear extract and separated on an 8% polyacrylamide-7 M urea gel.
Most mammalian snoRNAs are encoded within the introns of pre-mRNA genes. The majority of snoRNAs are released from the pre-mRNA via a splicing-dependent pathway, while some are processed via endonucleolytic cleavage of the pre-mRNA. The remaining mammalian snoRNAs, such as U3, U8 and U13, are expressed from independent genes and contain an m3G cap structure (9, 30, 43, 52). The biogenesis of box C/D snoRNAs takes place in the nucleoplasm, where the nascent transcribed RNAs are processed, assembled into RNPs, and transported to the nucleolus. The box C/D motif has been shown to be essential for each of these steps in snoRNP biogenesis. This RNA element is a protein binding site that has been proposed to participate in both the biogenesis and function of snoRNAs via the selective recruitment of specific box C/D binding factors (9, 41, 52).
Four common core proteins are associated with the mature snoRNP, namely, fibrillarin (Nop1p in yeast), NOP56, NOP58, and the 15.5K protein (Snu13p in yeast) (21, 22, 28, 32, 38, 42, 51, 54). The 15.5K protein belongs to a family of RNA binding proteins that includes the ribosomal proteins L7a, S12, and L30 and the H/ACA snoRNP protein Nhp2p (19, 33). Fibrillarin shares sequence and structural similarity with known methyltransferases and is therefore likely to be the enzymatic subunit responsible for RNA modification (47). The proteins NOP56 and NOP58 are homologous to each other and belong to a family of conserved proteins that also includes the U4/U6 protein 61K (PRPF31 or Prp31p in yeast) (10). The members of this family of proteins all share a conserved central NOP domain that is proposed to function in the binding of these proteins to their respective RNP complexes. The human 61K protein has been linked to autosomal dominant retinitis pigmentosa and is essential for the formation of the spliceosomal U4/U6.U5 tri-snRNP (29, 46). Two additional proteins, namely, TIP48 (also known as TIP49b, RUVBL2, p50, TAP54β, and Reptin42) and TIP49 (also known as TIP49a, RUVBL1, p55, TAP54α, and Pontin52), have been shown to be associated with box C/D snoRNPs assembled in mouse nuclear extracts (32). These nucleoplasmic proteins are involved in a number of cellular processes, including DNA repair and transcription, and are not associated with the mature box C/D snoRNPs (32, 34, 36).
The role that several of the box C/D snoRNA-associated proteins play in snoRNA accumulation and nucleolar localization has been studied in yeast. Nop58p and Snu13p are necessary for the accumulation of all box C/D snoRNAs (21, 51). In contrast, Nop1p (fibrillarin) is essential for the production of a subset of box C/D snoRNAs, while Nop56p is not essential for snoRNA accumulation (22). Recent work has shown that the yeast orthologue of TIP48 is essential for the production of both the box C/D and the H/ACA snoRNAs (15). In addition, genetic depletion of Snu13p, Nop56p, Nop58p, or Nop1p abolishes nucleolar localization of the box C/D snoRNPs (44).
The 15.5K protein was first characterized as a component of the U4/U6.U5 tri-snRNP that directly binds to the 5′ stem-loop of the U4 snRNA (33, 45). This protein was also shown to directly bind the box C/D motif in vitro. The box C/D motif shows striking similarity in primary and secondary structure to the 5′ stem-loop of the U4 snRNA, and it is assumed that the protein recognizes the snoRNA motif in a similar manner (45, 51). Little is known about how the other box C/D-associated proteins are associated with the snoRNA. Yeast genetic studies have demonstrated that Nop58p and Nop1p (fibrillarin) can bind independently of each other, while the association of Nop56p with the box C/D snoRNAs requires the presence of Nop1p (22). However, it is not yet clear how these proteins associate with the box C/D motif. The fact that 15.5K is associated with both the U4 snRNA and the box C/D snoRNAs makes this point especially interesting. The homology shared between these two RNP complexes extends beyond the similar RNA motifs and 15.5K. The homologous proteins NOP56 and NOP58 and the protein 61K (hPrp31) are associated with the box C/D snoRNPs and the U4/U6 snRNP, respectively. A specific element must be present in the RNAs that enables the box C/D proteins and the U4/U6 factors to distinguish between these two distinct complexes. The crystal structure of 15.5K bound to the U4 5′ stem-loop was recently solved. In this structure the protein is predominantly bound to the internal loop nucleotides, resulting in a severe kink in the phosphodiester backbone of one strand of the RNA (45). Thus, the flanking stem sequences remain free to bind additional associated factors. The similarities seen between the box C/D motif and the U4 5′ stem-loop prompted us to propose that these two RNA motifs bind 15.5K in a similar manner (45, 51). This implies that the two flanking stems of the box C/D motif will also be free to bind additional protein factors. In comparisons of the box C/D motif with the U4 5′ stem-loop, it was previously noted that the sequence of stem II of the snoRNA structure is highly conserved (Fig. 1A) while the corresponding helix in the U4 5′ stem-loop is not (33, 51). Therefore, this short sequence element is the likeliest candidate for the sequence-specific element necessary for the specific binding of the box C/D-associated proteins. We therefore set out to characterize the role that the box C/D motif plays in the binding of the individual snoRNA binding proteins. In this present study we show that 15.5K binding to the box C/D motif is an essential first step in the hierarchical assembly of the box C/D snoRNP. Furthermore, we show that the sequence of stem II in the box C/D motif of the U14 snoRNA is essential for the association of NOP56, NOP58, fibrillarin, TIP48, and TIP49 but not of 15.5K to the box C/D snoRNA. Finally, we show that the sequence of stem II of the box C/D motif is critical for the nucleolar localization of box C/D snoRNAs injected into HeLa cell nuclei.
MATERIALS AND METHODS
DNA oligonucleotides.
Oligonucleotides used were as follows: 1, GAAGATTCGCTGTGAGGATGGATTCCAAAACCATTCG; 2, GCGGATCCCGGGTGTTTCATTCGCTCGGACATCCAAGGAAGGAACTAGC; 3, GAAGATTCGCTGTGATGATGGATTCCAAAACCATTCG; 4, GCGGATCCCGGGTGTTTCATTCGCTCTGACATCCAAGGAAGGAACTAGC; 5, GGAAGATTCGCTGTGAAGATGGATTCCAAAACCATTCG; 6, GCGGATCCCGGGTGTTTCATTCGCTCAGACATCCAAGGAAGGAACTAGC; 7, GAAGATTCGCTGTGATGCTGGATTCCAAAACCATTCG; 8, GCGGATCCCGGGTGTTTCATTCGCTCAGGCATCCAAGGAAGGAACTAGC; 9, GAAGATTCGCTGTGATGGTGGATTCCAAAACCATTCG; 10, GCGGATCCCGGGTGTTTCATTCGCTCAGGCATCCAAGGAAGGAACTAGC; 11, GAAGATTCGCTGTGATCATGGATTCCAAAACCTTCG; 12, GCGGATCCCGGGTGTTTCATTCGCTCACACATCCAAGGAAGGAACTAGC; 13, GAAGATTCGCTGTGATGATGGATTCCAAAACCTTCG; 14, GCGGATCCCGGGTGTTTCATTCGCTCAGACATCCAAGGAAGGAACTAGC; 15, GAAGATTGCGTGTGATGATGGATTCCAAAACC; 16, GCGGATCCCGGGTGTTTCATTGCGTCAGACATCCAAGGAAGGAACTAGCC; 17, GAAGATTCGCTGTGATCGTGGATTCCAAAACCATTCG; 18, GCGGATCCCGGGTGTTTCATTCGCTCACGCATCCAAGGAAGGAACTAGC; 19, CACTCAGACATCCAAGGAAGGTTTACCCAACACTAAGGAAAACCTTTCTGGT-GGAAACTGCGAATG (the 5′ end contains a Cy3 label).
Plasmid construction.
The snoRNA constructs were made by cloning amplified mutant U14 snoRNA sequences into the SmaI-BamHI sites of pSP64T7 (50). Primer sets for each mutant were as follows: U14 stII mut1, oligonucleotides 1 and 2; U14 stII mut2, oligonucleotides 3 and 4; U14 stII mut3, oligonucleotides 5 and 6; U14 stII mut4, oligonucleotides 7 and 8; U14 stII mut5, oligonucleotides 9 and 10; U14 stII mut6, oligonucleotides 11 and 12; U14 stI-1, oligonucleotides 16 and 13; U14 stI-2, oligonucleotides 14 and 15; U14 stI-3, oligonucleotides 15 and 16; U14 stII-1, oligonucleotides 14 and 17; U14 stII-2, oligonucleotides 13 and 18; and U14 stII-3, oligonucleotides 17 and 18. The DNA sequence of each construct was verified by sequencing.
Antibody production.
Anti-NOP58 antibodies used for this paper were described previously (51). For the production of the remaining antibodies, protein-specific peptides were coupled to ovalbumin via a sulfo-succinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate cross-linker (Pierce). The conjugate was used for immunization of rabbits as described in reference 26. The following peptides were used: 15.5K, CTEADVNPKAYPLAD; NOP56, CTVNDPEEAGHRSRSK; fibrillarin, CRGNQSGKNV-MVEPH; TIP48, CTTKVPEIRDVTRIER; and TIP49, DAKSSAKILADQQDKYC. Western blot analysis of total nuclear extract demonstrated that each immunoserum recognized only a single band of the expected size. In addition, the antibodies raised against 15.5K, NOP56, NOP58, and fibrillarin all specifically coprecipitate the box C/D snoRNAs (reference 51 and data not shown).
In vitro assembly of snoRNPs and RNA binding analysis.
Radiolabeled in vitro transcribed U14 snoRNA, mutant U14 snoRNAs, U4 snRNA, and U19 snoRNA were prepared according to references 50 and 51). Native gel analysis of 15.5K-RNA interactions was performed as described in reference 33. In vitro assembly of snoRNPs was performed basically as described previously using HeLa nuclear extract (50). Briefly, 10 μl of HeLa nuclear extract, 10 μg of tRNA, and 50 fmol of 32P-labeled RNA were incubated in a buffer containing 20 mM HEPES (pH 7.9), 150 mM NaCl, 3 mM MgCl2, 0.1% (vol/vol) Triton X-100, 0.5 mM dithiothreitol, and 10% (vol/vol) glycerol for 30 min at 30°C. Heparin was added to a final concentration of 5 mg/ml, and the assembled snoRNPs were immunoprecipitated by incubation with protein A-Sepharose-bound antibodies for 2 h at 4°C. The bound material was washed four times with 20 mM HEPES (pH 7.9), 150 mM NaCl, 3 mM MgCl2, 0.1% (vol/vol) Triton X-100, 0.5 mM dithiothreitol, and 10% (vol/vol) glycerol (48). The immunoprecipitated RNA was recovered from the Sepharose beads by sodium dodecyl sulfate treatment and phenol-chloroform extraction. The RNA was ethanol precipitated and then analyzed on an 8% polyacrylamide-7 M urea gel. Competitor RNA oligonucleotides were incubated with the nuclear extract prior to the assembly reaction as described in reference 32. For addback experiments, recombinant 15.5K protein was bound to the U14 snoRNA transcript as described in reference 33 prior to the assembly reaction.
Microinjection of fluorescently labeled RNA into HeLa cell nuclei.
Fluorescent RNA transcripts were synthesized in the presence of Chroma Tide Alexa 546-14-UTP as described by the manufacturer (manual; Molecular Probes, Eugene, Oreg.). The RNA was separated from the free nucleotides using a G-50 spin column followed by ethanol precipitation. The samples were resuspended in 1× phosphate-buffered saline (pH 7.4) for microinjection. The RNAs contained on average two fluorescent groups per transcript, and 10,000 molecules were injected per cell. All injections were performed as described previously (7) using an Eppendorf 5242 microinjector and 5171 micromanipulator. The coverslips containing injected cells were prepared and visualized as in reference 12. For the fluorescence in situ hybridization experiment, unlabeled U14 mutC transcript was microinjected into HeLa cells. The injected transcript was then visualized using a Cy3-labeled antisense U14 oligonucleotide (oligonucleotide 19) as described previously (39).
RESULTS
Binding of box C/D snoRNP proteins to the U14 snoRNA is solely dependent on the box C/D motif.
To investigate in detail the role that the box C/D motif plays in the assembly of the multiprotein snoRNP complex, we made use of the nuclear extract assembly system that was recently employed to characterize and identify proteins that bind the box C/D and H/ACA snoRNAs (8, 32, 50). By using this system, the binding of the core snoRNP proteins, namely, 15.5K, NOP56, NOP58, and fibrillarin, as well as that of two additional nucleoplasmic proteins, TIP48 and TIP49, can be analyzed. In order to specifically assess the binding of individual proteins, antibodies were raised against each of the box C/D snoRNP-associated polypeptides (see Materials and Methods). Each of the antibodies is highly specific and recognizes a single band of the correct size in a Western blot of total nuclear proteins. In addition, the antibodies raised against NOP56, NOP58, fibrillarin, and 15.5K specifically coprecipitate the box C/D snoRNAs from HeLa nucleolar extract (reference 51 and data not shown).
We chose the U14 snoRNA as our model RNA with which to study snoRNP assembly. The U14 box C/D motif has been extensively studied and is an exact match to the consensus sequence for this family of RNAs (11, 49, 50, 55). To assay box C/D snoRNP assembly, 32P-labeled U14 box C/D snoRNA transcripts were incubated in nuclear extract and the association of individual proteins was determined by immunoprecipitation using protein-specific antibodies. The immunoprecipitated RNAs were then analyzed by polyacrylamide gel electrophoresis. Using this approach, it was observed that U14 could be immunoprecipitated by antibodies raised against the snoRNP proteins 15.5K, NOP56, NOP58, and fibrillarin as well as the two nucleoplasmic proteins TIP48 and TIP49 (Fig. 1B, lanes 1 and 8). The U14 transcripts used in these experiments are 28 nucleotides longer than the mature RNA (50). As seen in Fig. 1C, the comparison of transcripts before (lane 1) and after (lane 2) the incubation period shows that the RNAs undergo limited trimming in the nuclear extract. Interestingly, both species of trimmed RNA were immunoprecipitated to approximately the same efficiency by each of the antibodies used in these experiments (Fig. 1B). Importantly, only background levels, significantly lower than those observed for the individual antibodies, were coprecipitated by using the preimmune serum, thus demonstrating that the signal observed for each of the antibodies is specific. Similar background coprecipitation levels were observed with each of the antibodies when 32P-labeled H/ACA snoRNA U19 transcripts were used (Fig. 1B, lane 5). In addition, 32P-labeled U4 snRNA transcripts were immunoprecipitated only by the anti-15.5K antibodies and not by the snoRNP-specific antibodies (lane 4). Therefore, by using the nuclear extract system and the antibodies that we have produced, the binding of individual box C/D snoRNP proteins can be analyzed.
The box C/D and the less conserved box C′/D′ motifs are the only highly conserved elements common to this family of snoRNAs. SnoRNP assembly has been proposed to be primarily dependent on the box C/D motif; however, it has also been proposed that other regions within the snoRNA may participate in core snoRNP formation (2, 4, 49). We therefore used mutant U14 snoRNA transcripts to determine which regions of the snoRNA are involved in the recruitment and binding of individual snoRNP proteins (Fig. 1A). As seen in Fig. 1B, deletion of the majority of the U14 snoRNA (mutants ΔAV and ΔABV [lanes 2 and 3]) had little or no effect on the ability of each antibody to precipitate the RNA. Therefore, neither of the two base-pairing regions, involved in either rRNA modification (region B) or rRNA processing (region A), is necessary for the stable association of these proteins. The central region between the two rRNA base-pairing domains (region V) has been proposed to contain a C′/D′ motif (18). However, this region is also not required for the stable binding of any of the common box C/D snoRNA-associated proteins. In contrast, mutation of the highly conserved GA in either box C or box D (U14 mutC and mutD [Fig. 1B, lanes 6 and 7]) resulted in RNAs that were not immunoprecipitated by any of the tested antibodies and thus not bound by any of the snoRNP proteins. Therefore, the binding of all of the snoRNP proteins to the U14 snoRNA appears to be solely dependent on the box C/D motif.
The binding of 15.5K to the box C/D motif is necessary for the assembly of the box C/D snoRNP.
The conserved GA dinucleotides present in the box C/D motif have been shown to be essential for the direct binding of 15.5K (51). The fact that the mutation of these sequences results in the disruption of all protein binding to the snoRNA (Fig. 1B) suggests that 15.5K plays an essential role in the recruitment of the additional associated factors. Since this protein is the likely candidate for the primary RNA binding protein, we decided to determine whether the association of 15.5K is essential for the binding of the other box C/D snoRNP proteins. To achieve this, we made use of the fact that the 15.5K protein binds the 5′ stem-loop of the U4 snRNA (33, 45). In contrast to the U14 snoRNA, the U4 snRNA does not bind any of the additional snoRNP-specific proteins (see above). Therefore, the binding of the 15.5K in the nuclear extract can be blocked using a competitor oligonucleotide corresponding to the 5′ stem-loop of the U4 snRNA (SL1 [Fig. 2A ]). Nuclear extract was incubated with an increasing amount of either the U4 5′ stem-loop oligonucleotide SL1, which binds the 15.5K, or a mutant oligonucleotide, SL17 (Fig. 2A), containing a point mutation abolishing the association of 15.5K (33). Using glycerol gradient centrifugation, followed by Western blot analysis, we have estimated the amount of free 15.5K present in the nuclear extract. The highest amount of competitor oligonucleotide used in these studies corresponds to an approximately 400-fold excess over the unbound protein. The oligonucleotide-saturated extracts were then used to assemble the U14 snoRNP with the binding of specific snoRNP proteins determined by immunoprecipitation (as described earlier).
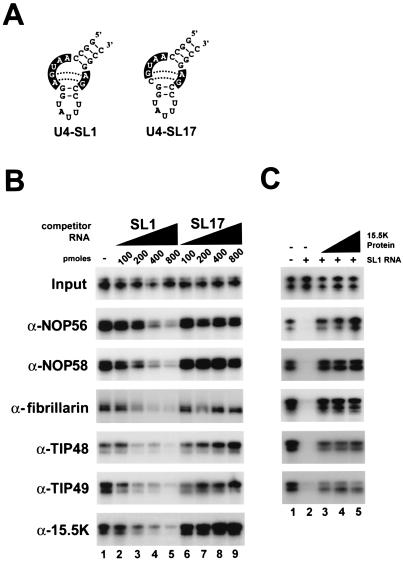
FIG. 2.
The binding of the 15.5K protein is an essential first step in the assembly of the box C/D snoRNP. (A) Sequence and structure of the two U4 5′ stem-loop oligonucleotides used to block 15.5K binding. The conserved internal loop nucleotides are shown in white on a black background. U4-SL1 is the wild-type sequence, while U4-SL17 contains a point mutation in the internal loop that inhibits 15.5K binding (33). (B) An excess of a U4 5′ stem-loop oligonucleotide can specifically block the assembly of the box C/D snoRNP. Radiolabeled U14 snoRNA was incubated in HeLa nuclear extract that had been preincubated with increasing amounts of either U4-SL1 or U4-SL17 RNA oligonucleotides. The binding of individual snoRNP proteins was then assayed by immunoprecipitation (see Materials and Methods). Bound RNAs were recovered and then separated on an 8% polyacrylamide-7 M urea gel. Antibodies used for immunoprecipitation are indicated on the left of the figure. Input, 10% of the RNA after incubation in nuclear extract. The amount as well as the identity of the RNA oligonucleotide used is indicated above each lane. (C) Rescue of 15.5K depletion by the addition of recombinant 15.5K. HeLa nuclear extract was preincubated with either 800 pmol of U4-SL1 RNA oligonucleotide (+) or buffer (−). Radiolabeled U14 snoRNA was subsequently added in the presence (10 pmol in lane 3, 20 pmol in lane 4, and 40 pmol in lane 5) or absence (lanes 1 and 2) of recombinant 15.5K. The binding of individual snoRNP proteins was assayed as for panel B.
The addition of increasing amounts of the wild-type U4 5′ stem-loop oligonucleotide SL1 resulted in an inhibition of the ability of both anti-15.5K antibodies as well as those raised against the snoRNP-specific proteins (NOP56, NOP58, fibrillarin, TIP48, and TIP49) to immunoprecipitate the U14 snoRNA (Fig. 2B, lanes 2 to 5). This inhibition is first seen with the addition of 200 pmol of competitor U4 RNA and is almost complete when 800 pmol of SL1 oligonucleotide is used (lanes 3 and 5, respectively). In contrast, even at the highest concentration, the mutant oligonucleotide SL17 has no noticeable effect on the immunoprecipitation of the U14 snoRNA by any of the antibodies used in this assay (Fig. 2B, lane 9). This implies that inhibiting 15.5K binding to the U14 snoRNA effectively blocks the formation of the complete snoRNP complex.
In order to unequivocally demonstrate that this effect is specifically due to the inhibition of 15.5K binding, it was decided to rescue snoRNP assembly by the addition of recombinant 15.5K protein. For this purpose, 800 pmol of SL1 oligonucleotide was added to the extract to inhibit 15.5K binding. Subsequently, 32P-labeled U14 snoRNA was added to the binding reaction either alone or prebound to recombinant 15.5K. As seen in Fig. 2C, after depletion with the specific oligonucleotide (lane 2), the addition of recombinant 15.5K was sufficient to fully restore the coimmunoprecipitation by each of the antibodies (lanes 3 to 5). This therefore confirms that the SL1 oligonucleotide effectively and specifically blocks the binding of 15.5K to the snoRNA transcript. In addition, this clearly demonstrates that 15.5K binding to the box C/D motif is essential for the assembly of the complete snoRNP complex.
The structure but not the sequence of stems I and II is essential for 15.5K binding.
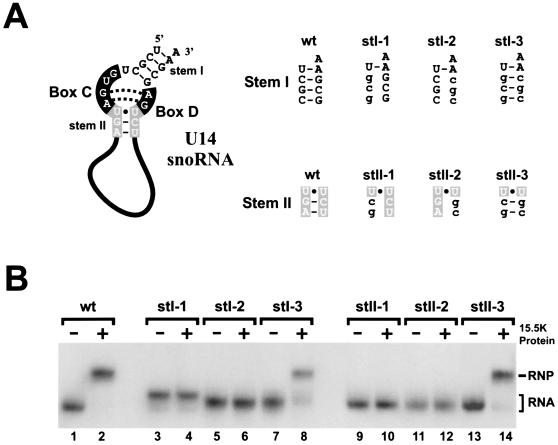
In order to fully understand how the 15.5K-box C/D motif complex specifically recruits the additional snoRNP proteins, it is essential to clarify which of the conserved sequences present in the box C/D motif are responsible for 15.5K binding. The internal loop nucleotides of the box C/D motif have already been demonstrated to be essential for 15.5K binding to the box C/D snoRNA (51). We therefore investigated the role that stems I and II play in the binding of 15.5K to the box C/D motif.
In order to achieve this, a series of mutations were generated in the box C/D motif of the U14 snoRNA that either disrupt base pairing or alter the sequence of stems I and II (Fig. 3A). The mutant transcripts were incubated with recombinant 15.5K, and the resulting complexes were analyzed by native gel electrophoresis. As seen in Fig. 3B, disruption of the Watson-Crick base pairs present in either stem I or II abolished 15.5K binding (Fig. 3B, lanes 4, 6, 10, and 12). However, mutations that altered the sequence but not the base-pairing capability of either stem I or II had little or no effect on protein binding (Fig. 3B, lanes 8 and 14). This therefore clearly demonstrates that the structure but not the sequence of stems I and II is essential for the binding of 15.5K. The data are consistent with the previous mutagenesis analysis of the U4 5′ stem-loop structure and provide further evidence that the box C/D motif folds and binds 15.5K in a manner highly similar to that of the U4 5′ stem-loop (33). This implies that 15.5K primarily contacts the internal loop of the box C/D motif, therefore raising the possibility that stems I and II are available to bind additional snoRNP proteins.
FIG. 3.
Stems I and II of the box C/D motif are essential for 15.5K binding. (A) Sequence and structure of U14 snoRNA stem I and stem II mutations. The conserved nucleotides of the box C/D motif are represented as in Fig. 1A. (B) Gel mobility shift analysis of the interaction of recombinant 15.5K with the stem I and stem II mutants. The wild-type (wt) U14 as well as the mutant transcripts outlined in panel A were incubated with recombinant 15.5K, and the resulting RNA-protein complexes were resolved on a 6% native polyacrylamide gel. The presence (+) or absence (−) of 15.5K is indicated above each lane. The position of the protein-RNA complex (RNP) and the free RNA is indicated on the right. The RNA used is indicated above each lane.
The sequence of stem II of the box C/D motif is essential for the specific assembly of the box C/D snoRNP.
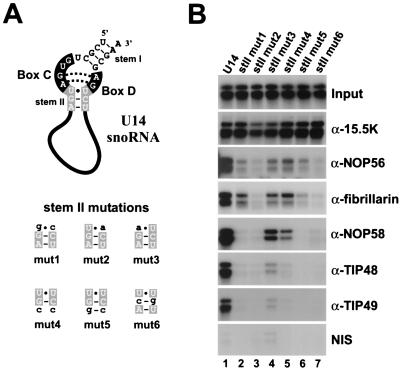
In order to test the role that the conserved sequence of stem II plays in the recruitment of box C/D snoRNP binding proteins, mutations were generated in the U14 snoRNA that altered the sequence of this helix. The mutations were designed in order to systematically analyze each of the conserved base pairs present in stem II (Fig. 4A). Consistent with data presented in Fig. 3, each of these mutant RNAs was shown to effectively bind 15.5K in a gel shift assay (data not shown) and as expected, none of the stem II mutations had any noticeable effect on the ability of each of the transcripts to be coimmunoprecipitated by anti-15.5K antibodies (Fig. 4B). The mutant transcripts were then assayed for their ability to bind the remaining box C/D snoRNA-associated proteins in nuclear extract (as described above).
FIG. 4.
The conserved sequence of stem II is essential for the specific assembly of the box C/D snoRNP. (A) Sequence and structure of the U14 snoRNA stem II mutants. The conserved nucleotides of the box C/D motif are represented as in Fig. 1A. (B) Role of conserved stem II in snoRNP formation in vitro. The binding of individual snoRNP proteins to the mutant transcripts outlined in panel A was assayed by immunoprecipitation (see Materials and Methods) with the bound RNAs being separated on an 8% polyacrylamide-7 M urea gel. The RNA mutant used is indicated above each lane. Antibodies used for immunoprecipitation are indicated on the right. Input, 10% of the RNA after incubation in nuclear extract; NIS, nonimmune serum.
The wild-type U14 snoRNA was efficiently immunoprecipiated by the anti-NOP56, -NOP58, -fibrillarin, -TIP48, and -TIP49 antibodies. However, the coimmunoprecipitation of each of the stem II mutant transcripts by these antibodies was either severely reduced or completely abolished (Fig. 4B, lanes 2 to 7). The coimmunoprecipitation level of the mutant transcripts is at most 5% of that seen with the wild-type U14 snoRNA. This therefore clearly demonstrates the role that stem II plays in the recruitment of the box C/D-associated proteins. In addition, this correlates with the highly conserved nature of stem II. Indeed, minor changes in the sequence of this helix render the snoRNA incapable of forming a core RNP particle that is necessary for snoRNP biogenesis. Interestingly, mutations stII mut1 and stII mut5 were weakly coprecipitated by NOP56 and fibrillarin but not by NOP58, TIP48, and TIP49 antibodies, suggesting that the binding of NOP56 and fibrillarin is linked (Fig. 4B, lanes 2 and 6). In addition, the association of TIP48 and TIP49 appears to be dependent on the presence of NOP58 in the snoRNP complex (Fig. 4B, lanes 4 and 5). Mutant stII mut3, a naturally occurring variant of the box C/D motif (55), is weakly precipitated by each of the antibodies (lane 4), suggesting that, in the context of the U14 snoRNA, this nucleotide is essential for efficient snoRNP assembly. The complete disruption of the third base pair (stII mut4) is tolerated more than a mutation that replaces the AU base pair with a GC (stII mut5) in snoRNP assembly. Taken together, the data clearly show that stem II of the box C/D motif provides the sequence specificity necessary to specifically assemble the box C/D-specific proteins. Stem I is present in the majority of box C/D snoRNAs; however, even among the five different U14 snoRNA genes found in humans, the sequence of this helix is not conserved. Consistent with the fact that the conserved sequence of stem II provides the sequence-specific element essential for the binding of the snoRNP-associated proteins NOP56, NOP58, fibrillarin, TIP48, and TIP49, alteration of the sequence but not the structure of stem I had no effect on the binding of the snoRNP-associated proteins (data not shown).
Conserved stem II of the box C/D motif is essential for nucleolar localization.
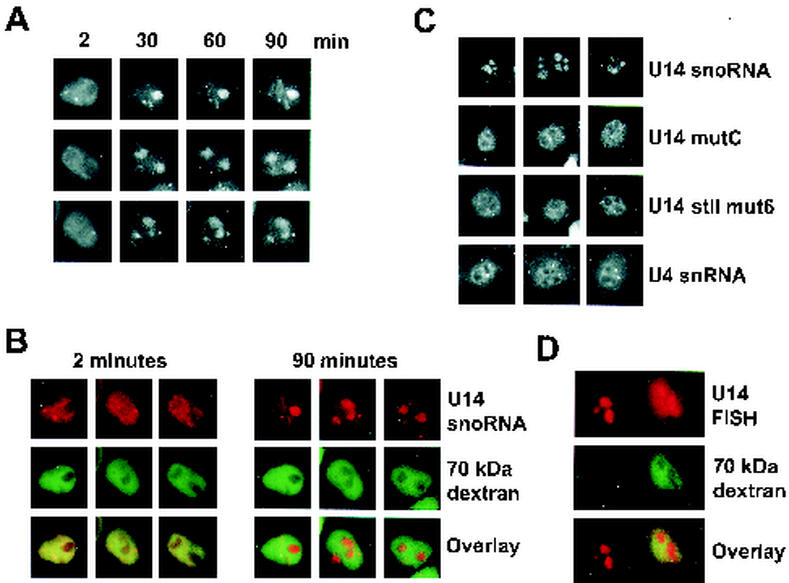
We have demonstrated the role of stem II in the box C/D snoRNA for the specific binding of the proteins NOP56, NOP58, fibrillarin, TIP48, and TIP49 in nuclear extract. However, it is important to confirm the role that this sequence element plays in the snoRNP biogenesis in vivo. Nucleolar localization is one aspect of snoRNP biogenesis that has been studied in detail in vivo. Previous studies have shown that the box C/D motif is essential for the nucleolar localization of box C/D snoRNAs in Xenopus oocytes (23-25, 31, 37). However, these experiments often involved the use of crude mutations that did not take into account the structure and bipartite nature of the box C/D motif as observed in our in vitro assembly experiments. Therefore, in order to be consistent with our assembly studies performed with HeLa nuclear extracts, we analyzed the nucleolar localization of fluorescently labeled U14 snoRNA transcripts injected into the nuclei of cultured HeLa cells. The final localization of these transcripts was monitored directly using fluorescence microscopy. In vitro transcription reactions were performed using Alexa 546-labeled UTP to generate the fluorescently tagged RNA. Approximately 104 molecules, equivalent to the endogenous U14 content, were injected into each cell. As a marker for injection and a control for cell integrity after manipulation, fluorescein isothiocyanate (FITC)-labeled 70-kDa dextran was coinjected with the transcript. Directly after injection, images were taken of manipulated cells. The cells were then incubated at 37°C, and further images of the injected cells were taken at 30, 60, and 90 min after injection.
As can be seen in Fig. 5A, directly after injection the labeled RNA rapidly diffused throughout the nucleoplasm but was not immediately present in the nucleoli. However, after 30 min of incubation the labeled RNA was found predominantly in the nucleoli. Indeed, no change was observed in the distribution of the labeled RNA even 90 min after injection. As a control, we compared the distribution of injected, fluorescently labeled dextran in the cells (Fig. 5B). As expected, and in contrast to the RNA, the dextran remains within the nucleoplasm and appears excluded from the nucleolus even after 90 min of incubation. Importantly, when Alexa 546-labeled UTP alone was injected into the nuclei of HeLa cells, this rapidly diffused throughout the whole cell (data not shown). Therefore, we conclude that the fluorescent signal observed in the nucleoli is indicative of nucleolar localization of the U14 snoRNA and not due to the localization of free nucleotides derived from the degradation of the injected RNA.
FIG. 5.
The conserved sequence of stem II of the box C/D motif is required for nucleolar localization. (A) Time-dependent localization of snoRNPs in HeLa nuclei. Fluorescent U14 snoRNA transcripts were microinjected into the nuclei of cultured HeLa cells. The cells were then incubated at 37°C, and images were taken at 2, 30, 60, and 90 min after injection. Three examples of each time point are shown. The time point is indicated above each set of pictures. (B) Injected fluorescent U14 transcripts and 70-kDa dextran differentially localize upon injection into HeLa nuclei. Fluorescent U14 snoRNA transcripts and FITC-labeled 70-kDa dextran were coinjected into the nuclei of cultured HeLa cells. The cells were then incubated at 37°C, and fluorescent images of the labeled RNA and dextran were taken at 2 and 90 min after injection. Three examples at each time point are shown. The time point is indicated above the panels. The Alexa 546-labeled snoRNA is shown in red, and the FITC-labeled dextran image is shown in green. The overlay of the snoRNA and dextran images is shown at the bottom of the figure. (C) The internal loop and conserved stem II of the box C/D motif are both essential for nucleolar localization. Fluorescent snoRNA and snRNA transcripts were microinjected into the nuclei of cultured HeLa cells. The cells were then incubated at 37°C for 90 min, and fluorescent images were taken. Three examples for each injected RNA are shown. The injected RNA is indicated to the right of the images. (D) Mutant U14 snoRNAs injected into HeLa nuclei are stable. Unlabeled U14 mutC and FITC-labeled 70-kDa dextran were coinjected into the nuclei of cultured HeLa cells. The cells were then incubated at 37°C for 90 min. The U14 snoRNA was subsequently detected by fluorescence in situ hybridization. The Cy3 in situ-hybridized U14 snoRNA (U14 FISH) is shown in red, and the FITC-labeled dextran image is shown in green. The injected cell is shown on the right, and the control, uninjected cell is on the left-hand side of the panel. The overlay of the snoRNA and dextran image is shown at the bottom of the figure.
In order to assess the role of the box C/D motif in nucleolar localization in HeLa cells, fluorescently labeled transcripts of U14, U14 mutC (Fig. 1A), U14 stII mut6 (Fig. 4A), and, as a comparison, human U4 snRNA were injected into HeLa nuclei as described above. The two mutants were chosen as U14 stII mut6 binds just 15.5K and as U14 mutC binds none of the proteins tested in the assembly assay. After injection, the cells were incubated for 90 min, and then fluorescent images of the injected cells were taken. As can be seen in Fig. 5B, the U14 snoRNA accumulates in the nucleoli. In contrast, and as expected, the U4 snRNA can be seen to accumulate at various points within the nucleoplasm. Interestingly, even after 90 min of incubation, neither U14 mutC nor U14 stII mut6 localizes to the nucleoli. Interestingly, both of these mutant RNAs localize to the Cajal bodies (data not shown).
Mutation of nucleotides present in either box C or box D has been shown to adversely affect the stability of snoRNA transcripts that are incubated for extended periods in Xenopus oocytes (3, 23, 49, 55). In order to verify whether the injected mutant transcripts were stable in HeLa cells, unlabeled U14 mutC transcript, along with FITC-labeled 70-kDa dextran, was injected into the nuclei of HeLa cells. After 90 min of incubation at 37°C, the injected snoRNA transcript was visualized by in situ hybridization using a fluorescent U14 snoRNA-specific probe. Due to the high sequence conservation of U14, it was not possible to design a probe that differentiated between the injected (mouse) and endogenous snoRNA. As shown in Fig. 5D, only the endogenous, nucleolar U14 snoRNA is detected in the uninjected cell (left side of the panel). However, in addition to the endogenous, nucleolar U14 snoRNA, a nucleoplasmic signal resulting from the U14 mutC RNA was also observed in the injected cell (Fig. 5D, right side of the panel). We can therefore conclude that U14 mutC RNA (and U14 stemII mut6 [data not shown]) is reasonably stable up to 90 min after injection into HeLa nuclei. Therefore, as these mutations do not significantly affect snoRNA stability in this assay, we conclude that both the 15.5K binding site and stem II of the U14 box C/D motif are essential for nucleolar localization. Indeed, these data imply that the binding of the box C/D-associated proteins is essential for the correct localization of these complexes.
DISCUSSION
Hierarchical assembly of the box C/D snoRNP.
The initial analysis of 15.5K led us to propose that this polypeptide, of the four core snoRNP proteins, was the likely primary RNA binding protein (51). In this work we have determined that 15.5K plays an essential role in snoRNP assembly. Competitor oligonucleotides used to block 15.5K binding to the snoRNA specifically inhibit snoRNP assembly. The binding of the snoRNP proteins to the snoRNA could be restored by the addition of recombinant 15.5K, thus clearly demonstrating that this effect was solely due to the block in 15.5K association. Indeed, we show that 15.5K binding is necessary for the binding of all of the core box C/D snoRNP-associated proteins. This therefore implies that snoRNP assembly is a hierarchical event and that the first step in this process is 15.5K binding. Interestingly, 15.5K binding to the U4 snRNA has also recently been shown to be essential for the binding of 61K and the heteromeric 20/60/90 complex to the U4/U6 snRNP (S. Nottrott, H. Urlaub, and R. Lührmann, in press). Therefore, 15.5K binding appears to play a similar role in the assembly of both the U4/U6 snRNP and the box C/D snoRNPs.
How does 15.5K binding to the RNA mediate the assembly of the snoRNP complex? The crystal structure of the 15.5K bound to the U4 5′ stem-loop demonstrated that the binding of this protein has a profound effect on the structure of the RNA. Indeed, the binding of this protein results in a severe kink in one strand of the RNA and a 150° bend in the phosphodiester backbone (45). The box C/D motif can also fold into a stem-internal loop-stem structure homologous to that found in the U4 5 stem-loop. It is therefore reasonable to suggest that the binding of 15.5K to the box C/D motif will also have a dramatic effect on the structure of the RNA. The binding of 15.5K therefore probably results in an essential structural rearrangement in the RNA that generates the binding site for the remaining snoRNP proteins. Thus, the binding of 15.5K can be seen as a nucleation event in the stepwise assembly of the box C/D snoRNP.
The conserved sequence of stem II in the box C/D motif directs the specific assembly of the box C/D snoRNP.
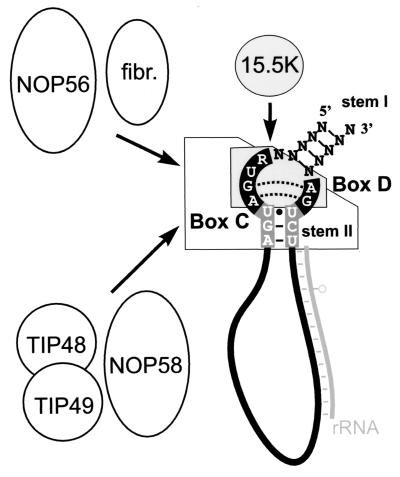
The discovery that 15.5K binding to the box C/D motif is the first step in snoRNP assembly led us to investigate how the remaining proteins bind to the snoRNP. In particular, we were interested in how the snoRNP and U4/U6 snRNP proteins distinguish between the two distinct but homologous complexes. The RNA must play a role in the binding of the additional proteins providing either a structural or sequence-specific binding site. The sequence of stem II is evolutionarily highly conserved in the box C/D motif but not in the U4 snRNA (Fig. 1A) (33). This conserved sequence is not necessary for 15.5K binding and, by comparison to the 15.5K/U4 5′ stem-loop crystal structure, is probably not bound by 15.5K (45). Indeed, mutations that alter the sequence of stem II dramatically affect snoRNP assembly, resulting in a block in the binding of NOP56, NOP58, fibrillarin, TIP48, and TIP49. Therefore, this short conserved helix provides the site for the sequence-specific assembly of the box C/D snoRNP. Based on these results, the box C/D motif can now be visualized as a bipartite protein binding site in which the internal loop nucleotides bind 15.5K and in which conserved stem II specifically recruits the remaining snoRNP factors (Fig. 6). In contrast, the binding of 61K to the U4 snRNA-15.5K complex is dependent on the presence of not only the 5′ stem-loop but also of the 5′ end of the transcript (Nottrott et al., in press). This implies that a specific structural element of the U4 snRNA is required for the association of 61K.
FIG. 6.
Summary of box C/D snoRNP assembly. The box C/D snoRNA, bound to an rRNA target sequence (methylation site is indicated by a circle), is shown here schematically with the conserved nucleotides of the box C/D motif represented as in Fig. 1A. The specific sequence requirements for the hierarchical assembly of the box C/D snoRNP are indicated. The 15.5K protein, the primary snoRNA binding protein, binds to the internal loop, while the remaining box C/D snoRNP proteins bind the 15.5K-snoRNA complex, specifically recognizing the conserved sequence of stem II. Fibrillarin (fibr.) and NOP56 and NOP58, TIP48, and TIP49 are grouped to indicate the connected nature of their association with the snoRNP.
Mutation of the sequence of stem II resulted in a dramatic reduction in protein binding. However, some of the mutations were capable of binding low levels of the stem II-dependent snoRNP proteins. This indicates that the complete stem II sequence is required for efficient assembly of a functional snoRNP. Interestingly, some of the mutations bound just a subset of the stem II-dependent snoRNP proteins. This probably reflects on the differential sequence requirement for the recruitment of individual proteins. The binding of NOP56 and fibrillarin appears to be linked and can occur in the absence of NOP58, TIP48, and TIP49. This is consistent with earlier data in yeast where it was shown that Nop56p binding to the snoRNA occurred only in the presence of Nop1p (fibrillarin) (22). In addition, it was also shown that Nop1p binding to the snoRNA occurs in the absence of Nop58p. Further analysis of the data presented in Fig. 4 suggested that the association of both TIP48 and TIP49 is linked with the binding of NOP58. Thus, these data provide several interesting insights into the binding of the individual proteins.
The box C′/D′ and box C/D motifs share the same consensus sequence, and both have a similar mechanism by which they select the site for 2′ O methylation. In addition, only one protein, namely, fibrillarin, has been proposed to catalyze the snoRNA directed 2′ O methylation (47). This implies that the method of recruiting proteins for the specific modification of RNA is very similar or even identical for both the C/D and C′/D′ motifs. However, the C′/D′ motif is not as well conserved as the box C/D motif. Based on our analysis of the box C/D motif, we would conclude that most of the box C′/D′ motifs contain too many deviations from the consensus sequence to allow stable binding of any of the box C/D-associated proteins. Indeed, several of the box C′ or D′ motifs (e.g., in human U14, U16, U40, U41, and U56) lack either the GA dinucleotide in box D′ or both GA dinucleotides from box C′. These conserved nucleotides are absolutely essential for 15.5K binding. Consistent with this, we observed no stable binding by any of the core box C/D proteins to the box C′/D′ motif of the U14 snoRNA (Fig. 1B) (51). However, it is entirely possible that stable protein binding to the box C′/D′ motif is not essential for the specific modification of RNA. Indeed, it could be that a transient association, possibly assisted by the binding of the substrate RNA, is all that is necessary for this event. In contrast, the box C/D motif requires stable protein association for the biogenesis and stability of the box C/D snoRNPs (3, 49). This difference in binding requirements to the two elements would imply a reduced selection pressure on the box C′/D′ motif, in comparison to the box C/D motif, during evolution, therefore possibly explaining the difference in the sequence conservation of these two RNA motifs.
The structure of the box C/D snoRNP.
The binding of 15.5K to the box C/D motif would, by comparison to the U4 snRNA, introduce a significant bend or kink into the RNA backbone (45). As discussed earlier, the formation of this structure probably creates the binding site(s), which includes conserved stem II, for the stem II-dependent proteins. However, it is unlikely that all of the stem II-dependent proteins directly contact this small helix. Recent in vivo cross-linking experiments, using snoRNPs assembled in Xenopus oocytes, have shown specific interactions between box C/D proteins and the UU pair at the top of stem II (5). Interestingly, it was shown that NOP58 and fibrillarin cross-link to the U in box C and the U in box D, respectively. Therefore, we now begin to see how the box C/D snoRNP assembles and how the box C/D motif functions in directing the site-specific 2′ O methylation of RNA. The highly specific binding of fibrillarin (the putative methyltransferase) to the box C/D motif-15.5K complex positions the active site of this enzyme in order to direct the position-specific 2′ O methylation of the substrate RNA (Fig. 6).
It is likely that the box C/D proteins also contact the nonconserved stem I of the box C/D motif. This can be inferred from the close proximity of this helix to the 15.5K- binding site and stem II. In addition, several pieces of evidence suggest that protein binding to stem I, which comprises the 5′ and 3′ termini of the majority of box C/D snoRNAs, is essential for the processing and stability of the box C/D snoRNAs (49, 55). In yeast it has been shown that Nop58p and Snu13p (15.5K) are essential for the stability of all box C/D snoRNAs (21, 51). In contrast, Nop1p (fibrillarin) is essential only for the stability of a subset of snoRNAs, while Nop56p is not required for snoRNA accumulation (22). As 15.5K is predicted to primarily contact the internal loop nucleotides of the box C/D motif, it is conceivable that NOP58 makes substantial contact with both stems I and II.
Nucleolar localization of box C/D snoRNPs.
We have also investigated the nucleolar localization of the U14 box C/D snoRNAs injected into HeLa cell nuclei. It has been reported that the U3 and U8 snoRNAs accumulate rapidly in the nucleolus, with the majority of the RNA localized to the nucleolus after 20 to 30 s (14). In contrast, in our experiments the majority of the U14 snoRNA was still present in the nucleoplasm 2 min after injection. Indeed, only 30 min after injection was the majority of the U14 snoRNA found in the nucleolus. The U3 snoRNA used in the earlier experiments contained the m7G 5′ cap structure, which was demonstrated to be essential for the rapid localization of this RNA. As U14 does not contain a 5′ cap, no such structure was included in the transcripts used for our localization studies. This may explain the difference in localization kinetics. However, the localization of U3 snoRNA in Xenopus oocytes appears to be independent of cap structure (25, 31). Further investigation is required to determine the mode of localization of the U3 snoRNA in cultured mammalian cells.
The work presented in this paper has used a twofold approach to examine the function of conserved stem II in the box C/D motif in snoRNP biogenesis. We have utilized an in vitro assembly system to analyze the role that this conserved helix plays in the binding of individual snoRNP proteins. In addition, we have analyzed the role of stem II in nucleolar localization by microinjecting fluorescently labeled transcripts into HeLa cell nuclei. As snoRNP formation is proposed to be essential for nucleolar localization, this latter assay provides a means by which we can indirectly study box C/D snoRNP assembly in vivo. Previously, crude mutations that disrupted the complete sequence of either box C or D were used to study the role that the box C/D motif plays in nucleolar localization in Xenopus oocytes (23-25, 31). Here, we have used mutations that take into account the overall structure, as well as the bipartite nature, of the box C/D motif. Indeed, we clearly show that both the 15.5K binding site and the conserved sequence of stem II are required for nucleolar localization of the U14 snoRNA. This is therefore highly suggestive that the complete assembly of the core box C/D snoRNP complex is necessary for the correct subcellular localization of these transcripts.
Recently it has been demonstrated that genetic depletion of either Snu13p (15.5K), Nop56p, Nop58p, or fibrillarin inhibits nucleolar localization of box C/D snoRNPs (44). The nucleolar localization signal could be a composite element derived from multiple core proteins and generated upon completion of assembly. Alternatively, the nucleolar localization signal could reside in one component of the core snoRNP and only be accessible upon completion of the assembly process. Indeed, it is possible that the nucleoplasmic proteins TIP48 and TIP49 could function as chaperones in the assembly process and mask the nucleolar localization signal until the assembly process is complete. TIP48 and TIP49 have been proposed to be involved in multiple cellular processes including histone acetylation and c-Myc activation; however, a defined function in any of these processes has yet to be characterized (6, 13, 27, 35, 53). While it is clear that these proteins are specifically associated with the box C/D snoRNPs, it will be interesting to investigate the role that these proteins play in snoRNP assembly and/or nucleolar localization.
Acknowledgments
We thank Stu Maxwell for the U14 plasmids; Tamas Kiss for the U19 plasmid; Hossein Kohansal and Peter Kempkes for excellent technical assistance; and Claudia Schneider for critically reading the manuscript. We also thank Niamh Cahil and Joan Steitz for communicating results prior to publication.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 523 Teilprojekt A8) and Fonds der Chemischen Industrie (to R. Lührmann) and a postdoctoral fellowship from the Deutsches Krebsforschungszentrum to A. Dickmanns.
REFERENCES
- 1.Bachellerie, J. P., and J. Cavaille. 1997. Guiding ribose methylation of rRNA. Trends Biochem. Sci. 22:257-261. [DOI] [PubMed] [Google Scholar]
- 2.Baserga, S. J., X. D. Yang, and J. A. Steitz. 1991. An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 10:2645-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caffarelli, E., A. Fatica, S. Prislei, E. De Gregorio, P. Fragapane, and I. Bozzoni. 1996. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 15:1121-1131. [PMC free article] [PubMed] [Google Scholar]
- 4.Caffarelli, E., M. Losito, C. Giorgi, A. Fatica, and I. Bozzoni. 1998. In vivo identification of nuclear factors interacting with the conserved elements of box C/D small nucleolar RNAs. Mol. Cell. Biol. 18:1023-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cahill, N. M., K. Friend, W. Speckmann, Z. H. Li, R. M. Terns, M. P. Terns, and J. A. Steitz. 2002. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 21:3816-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho, S.-G., A. Bhoumik, L. Broday, V. Ivanov, B. Rosenstein, and Z. Ronai. 2001. TIP49b, a regulator of activating transcription factor 2 response to stress and DNA damage. Mol. Cell. Biol. 21:8398-8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dickmanns, A., F. R. Bischoff, C. Marshallsay, R. Lührmann, H. Ponstingl, and E. Fanning. 1996. The thermolability of nuclear protein import in tsBN2 cells is suppressed by microinjected Ran-GTP or Ran-GDP, but not by RanQ69L or RanT24N. J. Cell Sci. 109:1449-1457. [DOI] [PubMed] [Google Scholar]
- 8.Dragon, F., V. Pogacic, and W. Filipowicz. 2000. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol. 20:3037-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filipowicz, W., P. Pelczar, V. Pogacic, and F. Dragon. 1999. Structure and biogenesis of small nucleolar RNAs acting as guides for ribosomal RNA modification. Acta Biochim. Pol. 46:377-389. [PubMed] [Google Scholar]
- 10.Gautier, T., T. Berges, D. Tollervey, and E. C. Hurt. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, G. M., A. Jarmolowski, J. C. R. Struck, and M. J. Fournier. 1992. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′, 3′ terminal stem. Mol. Cell. Biol. 12:4456-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber, J., U. Cronshagen, M. Kadokura, C. Marshallsay, T. Wada, M. Sekine, and R. Lührmann. 1998. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J. 17:4114-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson, M. R., and T. Pederson. 1998. A 7-methylguanosine cap commits U3 and U8 small nuclear RNAs to the nucleolar localization pathway. Nucleic Acids Res. 26:756-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King, T. H., W. A. Decatur, E. Bertrand, E. S. Maxwell, and M. J. Fournier. 2001. A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins. Mol. Cell. Biol. 21:7731-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding rNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 17.Kiss-Laszlo, Z., Y. Henry, J. P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077-1088. [DOI] [PubMed] [Google Scholar]
- 18.Kiss-Laszlo, Z., Y. Henry, and T. Kiss. 1998. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 17:797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin, E. V., P. Bork, and C. Sander. 1994. A novel RNA-binding motif in omnipotent suppressors of translation termination, ribosomal proteins and a ribosome modification enzyme? Nucleic Acids Res. 22:2166-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lafontaine, D. L., and D. Tollervey. 2001. The function and synthesis of ribosomes. Nat. Rev. Mol. Cell. Biol. 2:514-520. [DOI] [PubMed] [Google Scholar]
- 21.Lafontaine, D. L., and D. Tollervey. 1999. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 5:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lafontaine, D. L., and D. Tollervey. 2000. Synthesis and assembly of the box C+D small nucleolar RNPs. Mol. Cell. Biol. 20:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange, T. S., A. Borovjagin, E. S. Maxwell, and S. A. Gerbi. 1998. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 17:3176-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lange, T. S., A. V. Borovjagin, and S. A. Gerbi. 1998. Nucleolar localization elements in U8 snoRNA differ from sequences required for rRNA processing. RNA 4:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lange, T. S., M. Ezrokhi, A. V. Borovjagin, R. Rivera-Leon, M. T. North, and S. A. Gerbi. 1998. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol. Biol. Cell 9:2973-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauber, J., G. Plessel, S. Prehn, C. L. Will, P. Fabrizio, K. Groning, W. S. Lane, and R. Lührmann. 1997. The human U4/U6 snRNP contains 60 and 90kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA 3:926-941. [PMC free article] [PubMed] [Google Scholar]
- 27.Lim, C. R., Y. Kimata, H. Ohdate, T. Kokubo, N. Kikuchi, T. Horigome, and K. Kohno. 2000. The Saccharomyces cerevisiae RuvB-like protein, Tih2p, is required for cell cycle progression and RNA polymerase II-directed transcription. J. Biol. Chem. 275:22409-22417. [DOI] [PubMed] [Google Scholar]
- 28.Lyman, S. K., L. Gerace, and S. J. Baserga. 1999. Human Nop5/Nop58 is a component common to the box C/D small nucleolar ribonucleoproteins. RNA 5:1597-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makarova, O. V., E. M. Makarov, S. Liu, H. P. Vornlocher, and R. Lührmann. 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6.U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21:1148-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maxwell, E. S., and M. J. Fournier. 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 64:897-934. [DOI] [PubMed] [Google Scholar]
- 31.Narayanan, A., W. Speckmann, R. Terns, and M. P. Terns. 1999. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol. Biol. Cell 10:2131-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman, D. R., J. F. Kuhn, G. M. Shanab, and E. S. Maxwell. 2000. Box C/D snoRNA-associated proteins: two pairs of evolutionarily ancient proteins and possible links to replication and transcription. RNA 6:861-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nottrott, S., K. Hartmuth, P. Fabrizio, H. Urlaub, I. Vidovic, R. Ficner, and R. Lührmann. 1999. Functional interaction of a novel 15.5kD [U4/U6.U5] tri-snRNP protein with the 5′ stem-loop of U4 snRNA. EMBO J. 18:6119-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogura, T., and A. J. Wilkinson. 2001. AAA+ superfamily ATPases: common structure—diverse function. Genes Cells 6:575-597. [DOI] [PubMed] [Google Scholar]
- 35.Park, J., M. A. Wood, and M. D. Cole. 2002. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol. Cell. Biol. 22:1307-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel, S., and M. Latterich. 1998. The AAA team: related ATPases with diverse functions. Trends Cell Biol. 8:65-71. [PubMed] [Google Scholar]
- 37.Samarsky, D. A., M. J. Fournier, R. H. Singer, and E. Bertrand. 1998. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 17:3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schimmang, T., D. Tollervey, H. Kern, R. Frank, and E. C. Hurt. 1989. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 8:4015-4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taneja, K. L., L. M. Lifshitz, F. S. Fay, and R. H. Singer. 1992. Poly(A) RNA codistribution with microfilaments: evaluation by in situ hybridization and quantitative digital imaging microscopy. J. Cell Biol. 119:1245-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terns, M. P., and R. M. Terns. 2002. Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin. Gene Expr. 10:17-39. [PMC free article] [PubMed] [Google Scholar]
- 41.Tollervey, D., and T. Kiss. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell Biol. 9:337-342. [DOI] [PubMed] [Google Scholar]
- 42.Tyc, K., and J. A. Steitz. 1989. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 8:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tycowski, K. T., and J. A. Steitz. 2001. Non-coding snoRNA host genes in Drosophila: expression strategies for modification guide snoRNAs. Eur. J. Cell Biol. 80:119-124. [DOI] [PubMed] [Google Scholar]
- 44.Verheggen, C., J. Mouaikel, M. Thiry, J. M. Blanchard, D. Tollervey, R. Bordonne, D. L. Lafontaine, and E. Bertrand. 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 20:5480-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vidovic, I., S. Nottrott, K. Hartmuth, R. Lührmann, and R. Ficner. 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell 6:1331-1342. [DOI] [PubMed] [Google Scholar]
- 46.Vithana, E. N., L. Abu-Safieh, M. J. Allen, A. Carey, M. Papaioannou, C. Chakarova, M. Al-Maghtheh, N. D. Ebenezer, C. Willis, A. T. Moore, A. C. Bird, D. M. Hunt, and S. S. Bhattacharya. 2001. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11). Mol. Cell 8:375-381. [DOI] [PubMed] [Google Scholar]
- 47.Wang, H., D. Boisvert, K. K. Kim, R. Kim, and S. H. Kim. 2000. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J. 19:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Lührmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H box/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4:1549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watkins, N. J., R. D. Leverette, L. Xia, M. T. Andrews, and E. S. Maxwell. 1996. Elements essential for processing intronic U14 snoRNA are located at the termini of the mature snoRNA sequence and include conserved nucleotide boxes C and D. RNA 2:118-133. [PMC free article] [PubMed] [Google Scholar]
- 50.Watkins, N. J., D. R. Newman, J. F. Kuhn, and E. S. Maxwell. 1998. In vitro assembly of the mouse U14 snoRNP core complex and identification of a 65-kDa box C/D-binding protein. RNA 4:582-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watkins, N. J., V. Segault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Lührmann. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 52.Weinstein, L. B., and J. A. Steitz. 1999. Guided tours: from precursor snoRNA to functional snoRNP. Curr. Opin. Cell Biol. 11:378-384. [DOI] [PubMed] [Google Scholar]
- 53.Wood, M. A., S. B. McMahon, and M. D. Cole. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5:321-330. [DOI] [PubMed] [Google Scholar]
- 54.Wu, P., J. S. Brockenbrough, A. C. Metcalfe, S. Chen, and J. P. Aris. 1998. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J. Biol. Chem. 273:16453-16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia, L., N. J. Watkins, and E. S. Maxwell. 1997. Identification of specific nucleotide sequences and structural elements required for intronic U14 snoRNA processing. RNA 3:17-26. [PMC free article] [PubMed] [Google Scholar]