Abstract
Inhibitors of DNA methyltransferase (Dnmt) and histone deacetylases (HDAC) synergistically activate the methylated metallothionein I gene (MT-I) promoter in mouse lymphosarcoma cells. The cooperative effect of these two classes of inhibitors on MT-I promoter activity was robust following demethylation of only a few CpG dinucleotides by brief exposure to 5-azacytidine (5-AzaC) but persisted even after prolonged treatment with the nucleoside analog. HDAC inhibitors (trichostatin A [TSA] and depsipeptide) either alone or in combination with 5-AzaC did not facilitate demethylation of the MT-I promoter. Treatment of cells with HDAC inhibitors increased accumulation of multiply acetylated forms of H3 and H4 histones that remained unaffected after treatment with 5-AzaC. Chromatin immunoprecipitation (ChIP) assay showed increased association of acetylated histone H4 and lysine 9 (K9)-acetyl H3 with the MT-I promoter after treatment with TSA, which was not affected following treatment with 5-AzaC. In contrast, the association of K9-methyl histone H3 with the MT-I promoter decreased significantly after treatment with 5-AzaC and TSA. ChIP assay with antibodies specific for methyl-CpG binding proteins (MBDs) demonstrated that only methyl-CpG binding protein 2 (MeCP2) was associated with the MT-I promoter, which was significantly enhanced after TSA treatment. Association of histone deacetylase 1 (HDAC1) with the promoter decreased after treatment with TSA or 5-AzaC and was abolished after treatment with both inhibitors. Among the DNA methyltransferases, both Dnmt1 and Dnmt3a were associated with the MT-I promoter in the lymphosarcoma cells, and association of Dnmt1 decreased with time after treatment with 5-AzaC. Treatment of these cells with HDAC inhibitors also increased expression of the MTF-1 (metal transcription factor-1) gene as well as its DNA binding activity. In vivo genomic footprinting studies demonstrated increased occupancy of MTF-1 to metal response elements of the MT-I promoter after treatment with both inhibitors. Analysis of the promoter by mapping with restriction enzymes in vivo showed that the MT-I promoter attained a more open chromatin structure after combined treatment with 5-AzaC and TSA as opposed to treatment with either agent alone. These results implicate involvement of multifarious factors including modified histones, MBDs, and Dnmts in silencing the methylated MT-I promoter in lymphosarcoma cells. The synergistic activation of this promoter by these two types of inhibitors is due to demethylation of the promoter and altered association of different factors that leads to reorganization of the chromatin and the resultant increase in accessibility of the promoter to the activated transcription factor MTF-1.
Methylation of DNA at position 5 of cytosine in CpG dinucleotides has evolved as an epigenetic mechanism in higher eukaryotes, which is essential for development, genomic imprinting, and inactivation of the X chromosome (49, 63). The major outcome of promoter methylation appears to be long-term silencing of the associated genes (6, 29). Interest in elucidating the molecular mechanisms of this unique process has gained considerable momentum in recent years for two reasons. First, silencing of many tumor suppressor genes in many different primary malignancies is correlated with methylation of their promoters (4). Second, mutations in two key protein factors involved in methylation-mediated silencing, namely, DNA methyltransferase 3b (Dnmt3b) and methyl-CpG binding protein 2 (MeCP2), are responsible for the human diseases ICF (immunodeficiency, centromeric instability, and facial anomalies) and Rett syndromes, respectively (1). There has been dramatic progress in the identification of tissue-specific or ubiquitous enzymes involved in initiating methylation at position 5 of cytosines of CpG dinucleotides, although the factors controlling their targeting to specific regions of the genome are yet to be explored. Four different DNA methyltransferases (Dnmt) that catalyze methylation of CpG dinucleotides have been identified in mammals (5). Dnmt1 exhibits predominantly hemimethylase activity. Once methylation is initiated, Dnmt1 maintains it on successive rounds of DNA replication using hemimethylated DNA as a template. An oocyte-specific isoform of Dnmt1, Dnmto, transcribed from the same gene but with an additional exon, is involved in genomic imprinting (26). Two enzymes, Dnmt3a and Dnmt3b, encoded by different genes, catalyze de novo methylation (44, 59). A recently discovered isoform, DnmtL lacks intrinsic DNA methyltransferase activity but cooperates with Dnmt3a and Dnmt3b to control maternal specific genomic imprinting and gene expression (8, 20). Both maintenance and de novo DNA methyltransferases are essential for development, as null mice are embryonically lethal.
In vitro, Dnmts can methylate CpG base pairs in double-stranded DNA in a sequence-independent manner. The in vivo selective methylation of certain genes that occurs specifically in healthy tissues or tumors is probably due to targeting by specific docking proteins. Alternatively, the chromatin structure of the target genes may act as a signal for methylation by Dnmt3a and Dnmt3b. Recent studies have shown that both Dnmt3a and Dnmt3b can also act as transcriptional repressors that require the ATRX domain of the enzyme, rather than its catalytic domain, for the recruitment of histone deacetylase 1 (HDAC1) as a corepressor (2, 14). Dnmt3a can also act as a sequence-specific transcriptional repressor by virtue of its interaction with Rp58 that binds to a specific recognition site (14). This repressor activity of Dnmt3a is also mediated through its interaction with HDAC. These results clearly demonstrate that these proteins have functions beyond DNA methylation.
DNA methylation can repress gene transcription either by inhibiting binding of positive factors to the promoter or by recruiting transcriptional corepressors. In general, methylation of CpG islands does not impede binding of transcription factors to their cognate elements. The silencing of methylated promoters usually requires methyl-CpG binding proteins (MBDs) that specifically recognize symmetrically methylated CpG. To date, five such MBDs with homologous DNA binding domains have been identified (22-24). MBD1, MBD2, and MeCP2 recruit histone deacetylases (HDACs) to repress the methylated promoters. MBD3 is a component of the nuclear remodeling and HDAC complex Mi-2/NuRD that is recruited to the methylated promoter by interacting with MBD2 (54, 55, 62). MBD4 codes for a uracil DNA glycosylase that repairs methylated CpG/TpG mismatch pairs (3, 25). A recent report has suggested that proteins without an MBD can also bind methylated DNA (45). Kaiso, a protein that interacts with β catenin, is a second methylated DNA binding component of MeCP2. This protein uses its POZ-zinc finger domain to bind methylated DNA.
Local chromatin environments can also strongly influence gene expression. An important determinant of local chromatin structure is the complement of posttranslational modifications, such as acetylation, phosphorylation, and methylation, present on the NH2-terminal tails of the core histones. These modifications can affect chromatin structure by altering the electrostatic interactions between histones and DNA, and perhaps more importantly, by controlling the binding of nonhistone proteins to chromatin. There are multiple residues in the core histone NH2-terminal tails that are subject to posttranslational modification. The specific pattern of modification of these residues is important, as different sets of modifications can promote either transcriptionally competent or transcriptionally repressive chromatin structures (49, 63).
An interesting facet of chromatin structural modification is that different types of histone posttranslational modifications appear to function interdependently. Evidence has begun to emerge that the presence of modification on a particular histone NH2-terminal tail residue can influence, either positively or negatively, the modification of other sites on that histone. For example, acetylation of histone H3 lysine 14 by the histone acetyltransferase Gcn5p is stimulated by prior phosphorylation of histone H3 by Snf1p kinase (31, 32). On the other hand, acetylation of histone H4 blocks the histone methyltransferase activity of the transcriptional coactivator PRMT1 (56). Similarly, lysine 9 of histone H3 can either be acetylated or methylated, and the two modifications are mutually exclusive. Acetylation at this site is associated with active chromatin, whereas methylation results in the formation of heterochromatin, which leads to repression of the associated promoter. The posttranslational modifications can, therefore, function as a complex, interconnected, “histone code” that plays a major role in determining the accessibility of the key transcription factors to their cognate sequences and eventually in the expression of specific genes (28, 49).
Metallothioneins (MT) are a group of highly conserved heavy metal binding proteins that are induced at a very high level in response to various stress conditions, e.g., exposure to heavy metals and reactive oxygen species or bacterial or viral infections (15, 18). Four isoforms of these proteins are expressed in mammals of which MT-I and MT-II are highly inducible and coordinately regulated in response to various inducers. The two noninducible, tissue-specific isoforms are MT-III specifically expressed in the glutaminergic neurons and MT-IV localized to squamous epithelium of skin. Although MT-IIA, the predominant form in humans, is overexpressed in cancer cells treated with anticancer drugs, this gene is repressed in some metastatic tumors (61). The mechanism of this down-regulation is not known. Similarly, compared to the surrounding healthy tissues, the level of MT proteins in human hepatocellular carcinomas was found to be significantly less (9). Recently, we have shown that methylation is responsible for silencing the MT-I and MT-II genes in mouse lymphosarcoma cells (36) and in a rat solid tumor, Morris hepatoma 3924A (17). These genes could be activated in these tumors by exposure to 5-azacytidine (5-AzaC), an inhibitor of Dnmt. We have also shown by bisulfite genomic sequencing that the CpG islands located on the MT-I promoter are completely methylated in the mouse lymphosarcoma cells and in the rat hepatoma (17, 36). While the significance of the silencing of these genes in tumors is not known, it should be noted that the mice overexpressing MT are resistant to hepatic hyperplasia induced by hepatitis B virus (46). These observations have prompted us to raise the possibility that MT either alone or in concert with other factors may function as a growth suppressor at least in some tumors (16, 36).
Recent studies have shown that treating cancer cells with the combined regimen of Dnmt and HDAC inhibitors causes regression in tumor growth, and these agents are currently being tested in clinical trials against different leukemias (12, 21, 52). These agents are known to affect the expression of many cellular genes (10, 50). The exact molecular mechanism of this process, however, remains to be elucidated. In the course of investigating the epigenetic mechanism of MT promoter silencing, we observed that the combined effect of a demethylating agent (5-AzaC) and inhibitors of HDAC synergistically activates the promoter. The present study deals with a detailed investigation of the effects of these agents on histone modifications, levels of MBDs and Dnmts, activity of the key transcription factor MTF-1 on the MT-I chromatin structure, and promoter activity.
MATERIALS AND METHODS
Antibodies.
Antibodies against modified forms of H3 and H4 histones and HDAC1 were purchased from Upstate Biotechnology. Antibodies against MeCP2, MBD2, MBD1, MBD3, Dnmt3a, and Dnmt3b were raised in our lab. Anti-Dnmt1 antibody was a generous gift from Shoji Tajima.
Cell culture and treatment with 5-AzaC, TSA, depsipeptide, and heavy metals.
Lymphosarcoma cells were grown in RPMI 1640 medium containing 20 mM HEPES, 2% sodium bicarbonate, 1 mM glutamine, 10 nM 2-mercaptoethanol, and 5% fetal bovine serum. For treatment with 5-AzaC, cells at a density of 0.5 × 106/ml were treated with a 2.5 μM concentration of the nucleotide analog every 24 h for different periods of time indicated below. Cells were treated with trichostatin A (TSA) (Sigma) or depsipeptide (Fukasawa) dissolved in methanol for 12 h. To induce the MT-I gene, cells were treated for 3 to 6 h with ZnSO4 (50 μM).
Isolation of RNA, Northern blot analysis, and RT-PCR.
Total RNA was isolated from the cells by the guanidinium acid-phenol method (19). Thirty micrograms of total RNA was separated on a formaldehyde agarose gel, transferred to a nylon membrane, and subjected to Northern blot analysis with 32P-labeled MT-I minigene or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA following published protocols (37). The reverse transcriptase (RT) reaction was performed following the manufacturer's protocol with some modifications (RNA PCR Kit; Perkin-Elmer). Briefly, 1 μg of total RNA from each sample was used in the RT reaction mixture, which contains 5 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl [pH 8.3], 1 mM (each) of the four deoxynucleoside triphosphates, 40 U of RNase inhibitor, 2.5 μM random hexamers, and 50 U of murine leukemia virus RT. The reaction was performed at 42°C for 1 h. For PCR amplification, 1 μl of the RT reaction mixture was used in a 25-μl reaction mixture. The hot-start PCR was performed as follows: (i) 4 min at 94°C and (ii) 25 cycles, with 1 cycle consisting of 30 s at 94°C, 30 s at 64°C, and 1 min at 72°C. The PCR product generated was 234 bp (nucleotides 83 to 316 of cDNA). For mouse MTF-1 PCR, the annealing temperature was 55°C, and the product size was 440 bp (nucleotides 1053 to 1503). As an internal control, the β-actin was amplified using the same conditions except for the annealing temperature of 62°C. PCR products are 540 bp (nucleotides 25 to 564). The PCR primers used were as follows: (i) mMT-I-F (F for forward) (5′-CAACTGCTCCTGCTCCACCG) and mMT-I-R (R for reverse) (5′-AAGACGCTGGGTTGGTCCG), (ii) mMTF1-F (5′-CCACAGCACAATGGATCAGAGG) and mMTF1-R (5′-CACTTCTGGAGGTTGTAAGAGAGCAG), and (iii) mβ-actin-F (5′-GTGGGCCGCTCTAGGCACCAA) and mβ-actin-R (5′-CTCTTTGATGTCACGCACGATTTC).
Isolation of histones.
Cells (5 × 107) were harvested by centrifugation at 2,000 × g for 5 min at 4°C, washed with phosphate-buffered saline (PBS), and resuspended in 5 ml of nuclear isolation buffer (0.25 M sucrose, 10 mM Tris-HCl [pH 7.5], 2 mM ZnSO4, 1.5 mM MgCl2, 1 mM CaCl2, 0.5% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride, 3 μg of aprotinin per ml, 1 mM sodium butyrate, phosphatase inhibitor cocktail [Sigma]) and homogenized in a Dounce homogenizer on ice with a tight pestle until 90 to 95% of the cells were lysed. Nuclei were isolated by centrifugation at 3,000 × g for 5 min at 4°C and washed twice with nuclear isolation buffer without Triton X-100. Histones were extracted from the nuclei with 0.4 N ice-cold H2SO4 for 30 min on ice, and insoluble particles were removed by centrifugation at 15,000 × g for 10 min. Histones were precipitated from the supernatant with 20% trichloroacetic acid (TCA) (final concentration) and collected by centrifugation at 22,000 × g for 30 min. The pellet was washed twice with ice-cold acetone, dissolved in water, and stored at −80°C in small aliquots.
Acid-urea gel electrophoresis of histones.
Acid-urea resolving gels (6 M urea-15% acrylamide-5% acetic acid) were prerun in 5% acetic acid at 200 V overnight at 4°C. A scavenger solution (8 M urea, 5% acetic acid, 0.6 M 2-mercaptoethanol) was layered onto the resolving gel, and then electrophoresis was performed for 1 h at 200 V. The surface of the resolving gel was then rinsed with water before pouring of the stacking gel (6 M urea-7.5% acrylamide-0.375 M potassium acetate). Histone samples were then brought to 4 M urea-4% 2-mercaptoethanol-5% acetic acid-0.001% methylene blue and electrophoresed at 4°C for 17 h at 200 V. The gel was then washed in buffer containing 62.5 M Tris-HCl (pH 6.8), 2.3% sodium dodecyl sulfate (SDS), and 5% 2-mercaptoethanol at room temperature for 30 min. Histones were then transferred to nitrocellulose membrane in the buffer containing 25 mM 3-(1,1-dimethyl-2-hydroxyethyl)amino-2-hydroxypropanesulfonic acid (AMPSO; pH 9.5) and 20% methanol at 0.8 mA/cm2 of gel for 2 h and subjected to Western blot analysis with anti-acetylated H4 antibodies. The antigen-antibody complex was detected using an ECL Kit (Amersham) following the manufacturer's protocol.
Generation of antibodies and immunoblot analysis.
Polyclonal antibodies were raised against recombinant rat MeCP2, human MBD1, mouse MBD2, and MBD3 lacking the N-terminal MBD that is homologous among different isoforms of MeCPs (34). Polyclonal antisera were also raised against the recombinant N-terminal fragment of mouse Dnmt3a and Dnmt3b that lack the homologous catalytic domain (34). The specificity and titer of the antibodies were determined by Western blot analysis. Proteins of interest were separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to nitrocellulose membranes, and subjected to immunoblot analysis with specific antisera or preimmune sera, and the antigen-antibody complex was detected using an ECL Kit (Amersham) following the manufacturer's protocol.
Bisulfite genomic sequencing.
Genomic DNA isolated from lymphosarcoma cells was treated with sodium metabisulfite reagent by using the protocol optimized in our lab (16). The MT-I promoter region was then amplified with gene-specific primers (36) and digested with ApoI and Tsp509I to check complete conversion of unmethylated cytosines to uracils. The amplified product was sequenced with 33P-labeled dideoxynucleoside triphosphate using the Thermosequenase radiolabeled terminator cycle sequencing kit (U.S. Biochemical Corp.).
Immunoprecipitation.
P1798 cells were labeled overnight with [35S]methionine, and the whole-cell extract was prepared by resuspending the cells in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 7.5], 150 mM Nacl, 1% Nonidet P-40 [NP-40], 0.5% deoxycholate, 0.1% SDS) serum and then sonicating them. The extract was precleared with protein A-Sepharose beads. The supernatant was incubated either with preimmune serum or with immune serum overnight at 4°C followed by incubation with protein A-Sepharose beads for an additional 2 h. The beads were then washed sequentially with 1 ml each of low-salt buffer (20 mM Tris [pH 8.1], 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA), high-salt buffer (20 mM Tris [pH 8.1], 150 mM NaCl, 1% SDS, 1% Triton X-100, 2 mM EDTA), LiCl wash buffer (20 mM Tris [pH 8.1], 0.25 mM LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA), and TE (10 mM Tris [pH 8.1], 1 mM EDTA [pH 8.0]), and the immunoprecipitated polypeptides were separated by SDS-PAGE and subjected to fluorography.
Formaldehyde cross-linking and ChIP.
Lymphosarcoma cells at a density of 2 × 106/ml were treated with 1% HCHO at 37°C for 15 min to cross-link proteins to DNA, and the reaction was stopped by adding 0.125 M glycine. The soluble chromatin with an average DNA size of 750 to 1,000 bp was prepared by the protocol of Weinmann et al. (57) and was snap-frozen in small aliquots and stored at −80°C (if not used immediately). Chromatin immunoprecipitation (ChIP) was performed as described previously (34). Antibodies against acetylated histone H3 were raised against a synthetic N-terminal peptide (amino acids 1 to 12) of histone H3 acetylated at lysine 9 (K9). Antibodies against methylated histone H3 were raised against amino acids 6 to 13 of histone H3 where lysine 9 (K9) was dimethylated. Similarly, antibodies against acetylated histone H4 were raised against the N-terminal peptide (amino acids 2 to 19) in which all four lysines were acetylated. One microgram of purifed immunoglobulin G (IgG) or 5 μl of antiserum was used for each ChIP reaction. For ChIP assay with MeCP2, MBD1, Dnmt3a, and Dnmt3b, antisera raised in our laboratory were used with preimmune serum as the control (Dnmt1 antisera was a generous gift from Shoji Tajima). All these antibodies on immunoblot analysis detect only the specific polypeptide in P1798 whole-cell extract. The immunoprecipitated DNA was analyzed by semiquantitative PCR with the following promoter-specific primers: (i) for MT-I, 5′-GATAGGCCGTAATATCGGGGAAAGCAC and 5′-GAAGTACTCAGGACGTTGAAGTCGTGG; (ii) for histone H4-D.1, 5′-CATGGTTGATGGGAGGGATTTG and 5′-GAATGGAAAATGGAGTCAGAGCTG; and (iii) for histone H3-D.1, 5′-CAATGCCACTAAGTTCAAGCTGC and 5′-GCACTTGGAGATGGTGAGTTCAG.
For semiquantitative PCR, 2 pmol of each primer was labeled with [γ-32P]ATP. The annealing temperatures for MT-I, histone H4-D.1, and histone H3-D.1 primers were 60, 52.5, and 52.5°C, respectively, and the sizes of the respective amplified products are 306, 202, and 238 bp, respectively.
In vivo genomic footprinting.
Lymphosarcoma cells were treated with dimethyl sulfate (DMS), and DNA was purified and treated with piperidine as described earlier (35). DNA was dissolved in Tris-EDTA (TE) buffer and treated with piperidine (1 M) for 30 min at 95°C. DNA fragments were collected by ethanol precipitation and analyzed on agarose gels to ensure fragment sizes of <500 nucleotides. Next, an equal amount of DNA (2 μg) from each sample was subjected to ligation-mediated (LM)-PCR with gene-specific primers. To detect LM-PCR products, primer extension reaction was performed with 32P-labeled gene-specific primer. Reaction products were analyzed by urea-acrylamide gel electrophoresis. Several primers were used for LM-PCR of the mouse MT-I promoter. To detect footprinting within first 200 bp of the promoter (that encompasses MREs, MLTF/ARE, and Sp1), we used the primers described earlier (36).
Electrophoretic mobility shift assay.
The DNA binding activity of MTF-1 was measured in the S-100 extract from lymphosarcoma cells following a published protocol (37).
In vivo restriction enzyme mapping.
In vivo restriction enzyme mapping was performed using a modified version of a published protocol (58). Lymphosarcoma cells were harvested at 1,500 × g at 4°C, washed with PBS, and resuspended in lysis buffer (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.25% NP-40, 0.15 mM spermine, 0.5 mM spermidine) at a density of 107/ml. Nuclei were harvested at 800 × g and washed with 1 ml of restriction enzyme digestion buffer (10 mM Tris [pH 7.4], 50 mM NaCl, 10 mM MgCl2, 0.2 mM EDTA, 0.2 mM EGTA, 0.15 mM spermine, 0.5 mM spermidine, 1 mM β-mercaptoethanol). Nuclei (4 × 106/ml) were resuspended in 50 μl of the appropriate restriction enzyme digestion buffer for the specific restriction enzyme and digested with 50 U of the different restriction enzymes at 37°C for 10 min. The nuclei incubated without the restriction enzyme served as the control. Reactions were stopped by adding 50 μl of 2× proteinase K digestion buffer (100 mM Tris [pH 7.5], 200 mM NaCl, 2 mM EDTA, 1% SDS) and incubated at 55°C to inactivate the restriction enzyme. Then, 50 μl each of restriction enzyme digestion buffer and 2× proteinase K (Gibco) buffer and 75 μg of proteinase K was added to the mixture, and the mixture was incubated overnight at 37°C. DNA was purified by successive phenol-chloroform-isoamyl alcohol and chloroform-isoamyl alcohol extractions and ethanol precipitation, and then this DNA was dissolved in TE buffer. One microgram of in vivo restriction enzyme-digested DNA was then cut with a second enzyme, and 250 ng of this DNA was subjected to LM-PCR with strand-specific primers. Naked DNA cut with the same set of enzymes used for digestion of nuclei in vivo was also used as a control. The following sets of primers were to map all the AluI sites: F1 (5′-GAGTTCTCGTAAACTCCAGAGCAGC), F2 (5′-CAGAGCAGCGATAGGCCGTAATATC) F3 (5′-GATAGGCCGTAATATCGGGGAAAGCAC), R1 (5′-GGATAGGACGACCCATGTG), R2 (5′-ACGACCCATGTGACGTGTGG), and R3 (5′-TAGGACGACCCATGTGACG).The annealing temperature for forward (F) primers were 60, 63, and 66°C, respectively, whereas those for the reverse (R) primers were 58, 60, and 64°C, respectively.
RESULTS
Inhibitors of Dnmt (5-AzaC) and HDAC (TSA or depsipeptide) synergistically activate the MT-I promoter in response to heavy metals in lymphosarcoma cells.
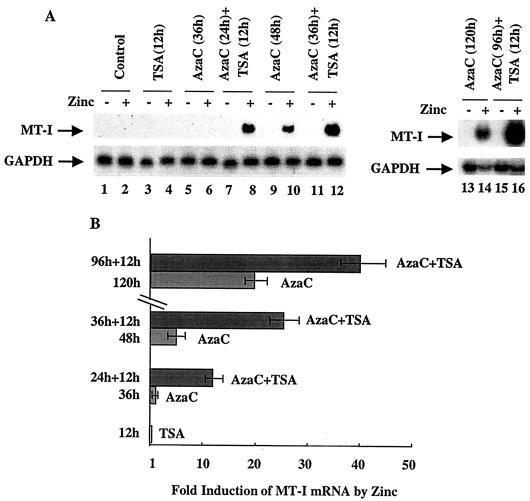
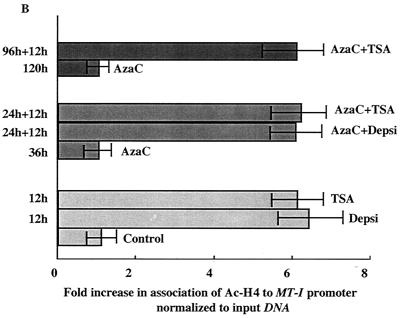
The MT-I promoter is silenced in lymphosarcoma cells (36). Prolonged exposure of these cells to the DNA methyltransferase inhibitor 5-AzaC allows reactivation of MT-I. We explored the possibility that the chromatin structure contributes to MT-I silencing. Acetylation of core histones is known to modulate gene expression by altering chromatin structure. Promoters with methylated CpG islands are hypoacetylated and repressed. To test whether histone hyperacetylation could reverse MT-I promoter silencing in lymphosarcoma (P1798) cells, these cells were treated with the HDAC inhibitors TSA and depsipeptide for 12 h. TSA is a hydroxamic acid derivative that chelates zinc ions present in the active sites of the enzymes. Depsipeptide is a bicyclic tetrapeptide that inhibits HDAC by an undefined mechanism (52). Treatment of cells with inhibitors of HDAC increases acetylation of histones, which, in turn, could alter chromatin structure and induce gene expression. Northern blot analysis showed that TSA treatment alone could not render MT-I inducible by zinc (Fig. 1A, lane 4). Similarly, after treatment with 5-AzaC alone for 36 h, MT-I expression was very low or undetectable in response to zinc (lane 6). Interestingly, when the cells were exposed to 5-AzaC for 24 h and then to TSA for 12 h, MT-I expression increased 12-fold after Zn2+ treatment compared to the cells treated with zinc alone (compare Fig. 1A, lane 8, and Fig. 1B). Exposure of cells to 5-AzaC alone for 48 h resulted in fivefold increase in expression of MT-I in response to zinc (compare Fig. 1A, lane 10, and Fig. 1B), but the induction rose to an even higher level (28-fold) after treatment of cells with 5-AzaC for 36 h followed by an additional 12 h of treatment with TSA (compare Fig. 1A, lane 12, and Fig. 1B). When the cells were treated with 5-AzaC for 120 h, the metal-induced expression increased to 22-fold, which increased further to 40-fold if cells were exposed to TSA following treatment with 5-AzaC for 96 h (compare Fig. 1A, lanes 14 and 16, and Fig. 1B). These results demonstrated the persistence of the synergistic effect of these agents on MT-I expression, although the extent of stimulation diminished with 5-AzaC exposure time. This experiment was repeated at least three times, and reproducible results were obtained. We also performed this experiment with depsipeptide (Fig. 1C), and the quantitative data on fold induction after treatment with these inhibitors are presented in Fig. 1D. Like TSA, treatment with depsipeptide alone for 12 h did not render the MT-I gene inducible by heavy metals (Fig. 1C, lanes 7 and 8), whereas 5-AzaC treatment for 36 h in this experiment produced only very low but detectable levels of expression after heavy metal treatment (lane 6). The exposure of the 5-AzaC-treated cells to depsipeptide, however, increased MT-I mRNA level significantly in a dose-dependent manner (lanes 9 and 10) in response to heavy metals. Depsipeptide was effective at a much lower concentration (as low as 0.1 nM) than TSA was, and MT-I expression at 1 nM was comparable to that at 300 nM TSA (compare Fig. 1B and D). These results demonstrate that HDAC inhibitors alone cannot render the methylated MT-I promoter responsive to heavy metals but that the MT-I gene is synergistically induced in response to heavy metals when the cells are treated first with an inhibitor of DNA methyltransferase and then with HDAC inhibitors.
FIG. 1.
Northern blot analysis of MT-I expression in lymphosarcoma cells treated with 5-AzaC and inhibitors of HDAC. (A) Cells (0.5 × 106/ml) were either untreated (−) or treated with ZnSO4 (50 μM) (+) for 3 h after exposure to TSA (300 nM), 5-AzaC (2.5 μM) for different times, or 5-AzaC followed by TSA. Induction of the MT-I gene was measured by Northern blotting with 32P-labeled mouse MT-I cDNA as the probe. The blot was reprobed with GAPDH cDNA to show equal loading of RNA. (C) Cells were treated with depsipeptide (Depsi) (12 h), 5-AzaC (2.5 μM), or 5-AzaC followed by treatment with depsipeptide for 12 h. MT-I was then induced with zinc as described above for panel A. (B and D) Results of quantitative analysis of the increase in MT-I mRNA level from three different experiments normalized to GAPDH. Mean values ± standard errors (error bars) are shown.
Acetylated histones H4 and H3 accumulate after treatment with inhibitors of HDAC.
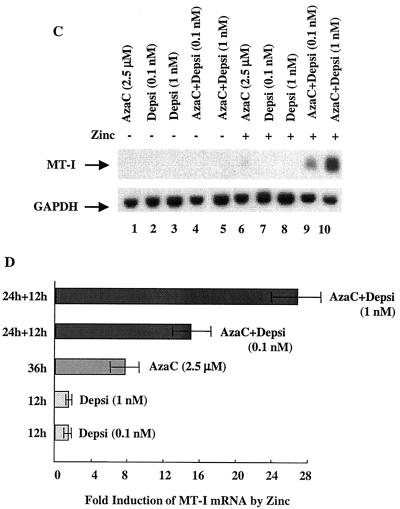
Next we sought to understand the mechanisms by which the inhibitors of Dnmts and HDACs act synergistically in transcriptional activation of the MT-I promoter. We reasoned that these agents induce promoter activation by altering chromatin structure. To address this issue, we determined the acetylation status of the core histones H3 and H4 in lymphosarcoma cells following treatment with these two classes of inhibitors. The rationale for this experiment was that simultaneous treatment with these two inhibitors might accentuate the acetylation of these histones, leading to opening of the chromatin structure, increased binding of transcription factors to their respective cis elements, and promoter activation. Histones were purified from the nuclei of control and treated cells and analyzed by acid-urea gel electrophoresis, which can separate the acetylated isoforms of the histones (Fig. 2). In the untreated cells, only nonacetylated and monoacetylated H4 were detectable (Fig. 2A, lane 1), whereas in cells treated with depsipeptide or TSA, mono-, di-, tri- and tetra-acetylated histone H4 were generated (lanes 2 and 3, respectively). Treatment of cells with 5-AzaC for 36 h did not change the acetylation status of histones (compare lane 4 with lane 1), whereas exposure to both 5-AzaC and depsipeptide or TSA generated the same acetylation profile of histone H4 as treatment with depsipeptide or TSA alone (compare lanes 5 and 6 with lanes 2 and 3, respectively). We confirmed these results by immunoblot analysis of the same proteins with an antibody against acetylated histone H4 (Fig. 2B). Di-, tri-, and tetra-acetylated histone H4 accumulated in cells after treatment with depsipeptide or TSA (lanes 2 and 3, respectively) which did not change after combined exposure of the cells to TSA-depsipeptide and 5AzaC (lanes 5 and 6). Unlike H4, most of the H3 histones in untreated lymphosarcoma cells existed as un- and monoacetylated forms (∼40% of each), and a small fraction (∼20%) was in the diacetylated form (Fig. 2A, lane 1), which was not altered after treatment with 5-AzaC (lane 4). After treatment with depsipeptide or TSA, the majority of H3 was converted to di- and triacetylated forms (lanes 2 and 3), which remained unaffected if the cells were initially exposed to 5-AzaC (lanes 5 and 6). These results clearly demonstrate that HDAC inhibitors caused the accumulation of hyperacetylated histones that is not enhanced by treatment with a DNA-demethylating agent.
FIG. 2.
Analysis of the acetylation status of histones H3 and H4 after treatment of P1798 cells with inhibitors of HDAC (depsipeptide [Depsi] and TSA) alone or in combination with 5-AzaC. Cells were treated with the inhibitors under the same conditions as described in the legend to Fig. 1. Nuclei were isolated, and histones were extracted with 0.4 N H2SO4, precipitated with TCA, and dissolved in water. (A) Equal amounts of protein (100 μg) were run on acid-urea gels to separate the different acetylated forms of histones. The gel was stained with Coomassie blue to visualize the proteins. (B) The protein from an identical gel was transferred to a nitrocellulose membrane and subjected to immunoblot analysis with antibodies against acetylated H4 histone (Ac-H4). For controls, some cells were not treated with an HDAC inhibitor (−). The numbers 0 to 4 on the ordinates indicate un-, mono-, di-, tri-, and tetra-acetylated histones.
Treatment of lymphosarcoma cells with 5-AzaC results in demethylation of the MT-I promoter.
Next, we explored the methylation status of the MT-I promoter following treatment with 5-AzaC and TSA. To determine whether the synergistic effects on MT-I promoter activity observed after treatment with 5-AzaC for a shorter period (36 h) along with TSA for 12 h is due to enhanced demethylation of the promoter, we performed bisulfite genomic sequencing (Fig. 3A). For this purpose, chromosomal DNA from lymphosarcoma cells was denatured and treated with sodium metabisulfite. This treatment converts cytosines to uracils in single-stranded DNA but does not react with methyl cytosines. During subsequent PCR with strand-specific primers, the uracils are amplified as thymines, whereas methyl cytosines are amplified as cytosines. Following sequencing of the PCR product by the dideoxy chain termination method, quantitation of the 33P signal in the C lanes and T lanes at a particular position allowed us to determine the ratio of methylated to unmethylated CpG at each site in the genomic DNA. A segment of the autoradiographs of the sequencing gels that span the MT-I promoter of control and treated lymphosarcoma cells are depicted in Fig. 3A. The results (lanes 3 and 4) demonstrated that almost all the CpG dinucleotides were methylated in the untreated cells (indicated by the closed circles), and only a few CpGs were partially methylated (indicated by the hatched circles). In TSA-treated cells, the methylation status of these CpG was very similar to that of the control cells (lanes 7 and 8, closed circles). As expected, treatment of cells with 5-AzaC for 36 h resulted in partial demethylation of the promoter, as evident by the occurrence of C and T in the positions that were followed by a G (lanes 11 and 12, hatched circles). When the cells were treated with 5AzaC for 24 h followed by 12 h of exposure to TSA, the extent of demethylation at most CpGs was very similar to that observed after treatment with 5-AzaC alone for 36 h (compare lanes 15 and 16 with lanes 11 and 12, respectively). Bisulfite sequencing of the MT-I promoter in cells treated for 120 h with 5-AzaC revealed demethylation (denoted by open circles) of almost all the CpG dinucleotides in the promoter (lanes 19 and 20). The methylation status of 18 CpG dinucleotides located in the proximal promoter region of this gene in cells treated with these inhibitors is presented in Fig. 3B. These results show that 5-AzaC alone induces demethylation of the MT-I promoter, whereas TSA alone or in combination with 5-AzaC does not affect this process.
FIG. 3.
(A) Bisulfite genomic sequencing of the MT-I promoter in lymphosarcoma cells treated with different inhibitors. Genomic DNA isolated from cells was treated with sodium metabisulfite reagent to convert unmethylated cytosines to uracils. The MT-I promoter was then amplified by nested PCR with gene-specific primers and sequenced directly with an internal primer specific for the upper strand. The closed, hatched, and open circles indicate completely methylated, partially methylated, and unmethylated cytosines, respectively. (B) Schematic representation of the methylation status of 18 CpG dinucleotides located within −190 bp of the MT-I transcription start site in cells treated with the inhibitors.
Association of acetylated histone H4 with the MT-I promoter increases after treatment with HDAC inhibitors.
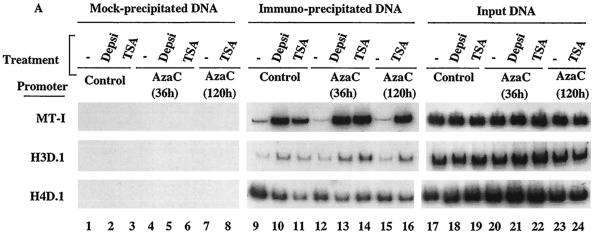
In general, methylated promoters are associated with hypoacetylated histones and are transcriptionally inactive. While inhibitors of Dnmt and HDAC did not exert a synergistic effect on the overall acetylation status of histones H3 and H4, we reasoned that the enhanced expression of MT-I in lymphosarcoma cells after treatment with 5AzaC and TSA may be due to localized hyperacetylation of histones at this promoter. To test this possibility, chromatin from the control and treated cells was immunoprecipitated (ChIP) with anti-acetylated histone H4 antibodies (acetylated at K5, K8, K12, and K16). DNA precipitated by the antibodies was then subjected to semiquantitative PCR with gene-specific primers under conditions when the amplification was linear (see Materials and Methods). The results demonstrate a very low level association of acetylated histone H4 with the MT-I promoter in lymphosarcoma cells that was significantly elevated after treatment of cells with TSA or depsipeptide (Fig. 4A, compare lanes 10 and 11 with lane 9). PCR with mock-precipitated DNA did not yield any amplification (lanes 1 to 8), indicating specific pull-down of MT-I promoter by anti-acetylated H4 antibodies. Association of acetylated H4 histones was not significantly altered after treatment with 5-AzaC for 36 h (lane 12) or 120 h (lane 15). Similarly, amplification of the promoter was the same, irrespective of the treatment of the cells with HDAC inhibitors alone or sequentially with 5-AzaC (compare lanes 13, 14, and 16 with lanes 10 and 11, respectively). For a control for this experiment, we also amplified two other gene promoters, e.g., histone H3D.1 and H4D.1 (Fig. 4A, middle and bottom blots). There was a small but reproducible increase in association of acetylated H4 with the H3D.1 promoter after treatment with depsipeptide or TSA that did not alter after exposure of the cells to 5-AzaC. Association of acetylated H4 with the H4D.1 promoter actually decreased after treatment with HDAC inhibitors. This experiment was repeated three times, and highly reproducible results were obtained. The results of quantitative analysis of the acetylated H4 histone associated with the MT-I promoter are shown in Fig. 4B. These data demonstrate that treatment of cells with HDAC inhibitors result in accumulation of hyperacetylated H4 histones as well as their increased association with the MT-I promoter.
FIG. 4.
(A) PCR amplification of different promoters from the input (not precipitated) DNA and DNA immunoprecipitated with anti-acetylated H4 histone antibody from the formaldehyde cross-linked chromatin prepared from P1798 cells treated with different inhibitors (depsipeptide [Depsi] and TSA). Immunoprecipitation was performed with formaldehyde cross-linked chromatin containing the same amount of DNA isolated from cells treated with different inhibitors. After immunoprecipitation, DNA was de-cross-linked and purified by proteinase K digestion, phenol extraction, and ethanol precipitation. DNA was dissolved in 10 μl of TE buffer, and 1 μl was subjected to semiquantitative PCR with primers specific for different promoters as described in Materials and Methods. The input DNA used for PCR was 1/50 of the amount used for immunoprecipitation. Chromatin subjected to the same treatment without any antibody was used as a negative control (lanes 1 to 8). (B) Results of quantitative analysis of the association of H4 histone to the MT-I promoter normalized to the amplified product from the input DNA. The values are the means of three experiments ± standard errors (error bars).
The association of K9-methyl histone H3 and HDAC1 with the MT-I promoter decreases after treatment with 5-AzaC and TSA, whereas that of K9-acetyl H3 increased after TSA treatment.
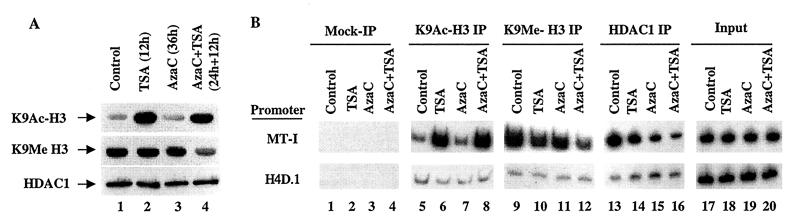
Recent studies have shown that K9-methyl histone H3 is a hallmark of repressed chromatin (27, 39). We therefore determined whether K9-methyl histone H3 was associated with the MT-I promoter in lymphosarcoma cells and whether this was altered after treatment with 5-AzaC and/or TSA. For this purpose, we first performed immunoblot analysis with antibodies specific for K9-methyl or K9-acetyl histone H3 in the whole-cell extract prepared from P1798 cells treated with the inhibitors. As expected, the level of K9-acetyl histone in untreated cells was low and increased at least fivefold after treatment with TSA, while 5-AzaC had no effect (Fig. 5A, top blot, lanes 1 to 4). In contrast, the level of K9-methyl histone H3 was relatively high in untreated cells and was not affected by treatment with TSA or 5-AzaC (middle blot, lanes 1 to 3). After treatment with both inhibitors, there was a significant decrease in the level of K9-methyl histone H3 (lane 4).
FIG. 5.
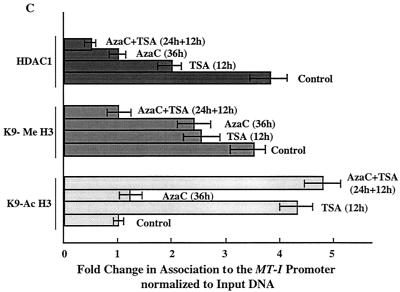
(A) Immunoblot analysis with antibodies specific for K9-acetyl, K9-methyl histone H3, and HDAC1 in the chromatin isolated from lymphosarcoma cells treated with different inhibitors. Equal amounts of the protein (100 μg) from each group were separated by SDS-PAGE on a 15% acrylamide gel, transferred to a nitrocellulose membrane, and subjected to Western blot analysis with antibodies specific for lysine-9 acetylated histone H3 (K9Ac-H3), lysine-9 methylated histone H3 (K9Me H3), and HDAC1. (B) The chromatin isolated from 107 cells was precleared with protein A-Sepharose beads for 1 h at 4°C. The supernatant was incubated with 5 μg of specific antibodies overnight at 4°C. The immune complexes were pulled down with protein A-beads and washed extensively as described in Materials and Methods, and purified DNA was subjected to semiquantitative PCR with gene-specific primers. Equal amounts of the chromatin precipitated with protein A alone (Mock-IP) was used as a control. (C) Results of quantitative analysis of association of K9-acetyl H3, K9-methyl H3, and HDAC1 to the MT-I promoter normalized to that of input DNA. The values are the means of three different ChIP assays ± standard errors (error bars).
Next, we performed ChIP assay of formaldehyde cross-linked chromatin isolated from the control and inhibitor-treated P1798 cells with antibodies against K9-acetyl or K9-methyl histone H3 and amplified the MT-I promoter from the precipitated DNA. As observed with acetylated H4 histones, the association of K9-acetyl H3 histone with the promoter dramatically increased after treatment with TSA, whereas 5-AzaC did not have any effect (Fig. 5B, lanes 5 to 8). The association of K9-acetyl histone H3 with the MT-I promoter correlated with their overall cellular levels (Fig. 5A). ChIP assays with anti-K9 methyl H3 antibody demonstrated a robust association of K9-methyl H3 histone with the MT-I promoter in untreated cells, which decreased to some extent after treatment of cells with TSA or 5-AzaC. After exposure to 5-AzaC in combination with TSA, this association decreased significantly (70%) (Fig. 5B, lanes 9 to 12), which correlated with the decrease in K9-methyl H3 level (Fig. 5A, lane 4). The results of quantitative analysis of the association of the modified H3 histone are shown in Fig. 5C. These results indicate that the association of K9-methyl and K9-acetyl H3 to the MT-I promoter is mutually exclusive and that hyperacetylated K9 histone H3 accumulated after TSA treatment probably displaced methylated K9 histone H3 from the promoter. The concomitant loss of K9-methyl histone H3 and demethylation of the promoter by HDAC inhibitors and 5-AzaC treatment may potentiate the gene activation.
We also explored the effects of HDAC inhibitors on the stability of HDAC1 protein. For this purpose, we performed Western blot analysis with anti-HDAC1 antibodies. The results showed that the level of HDAC1 in these cells was not affected significantly by treatment with TSA or 5-AzaC (Fig. 5A, bottom blot, lanes 1 to 4). HDAC1 is a corepressor associated with MBD as well as Dnmt complexes (2, 7, 14, 40, 54). We therefore investigated whether HDAC1 was associated with the methylated MT-I promoter in P1798 cells and examined the effect of 5-AzaC or TSA on this association. The results of ChIP assay using the anti-HDAC1 antibody showed that HDAC1 was indeed associated with the promoter. After treatment with TSA and 5-AzaC, this association was reduced by 50 and 75%, respectively (Fig. 5B, compare lanes 14 and 15 with lane 13). When cells treated with 5-AzaC for 36 h were exposed to TSA for an additional 12 h, the association decreased to 90% (Fig. 5B, lane 16, and Fig. 5C). These results demonstrate the recruitment of HDAC1 to the MT-I promoter, perhaps by interaction with one or more repressor complexes, and the correlation between promoter activation and dissociation of HDAC1 after 5-AzaC and TSA treatment.
Association of methyl-CpG binding protein MeCP2 with the MT-I promoter increases after treatment with inhibitors of HDAC.
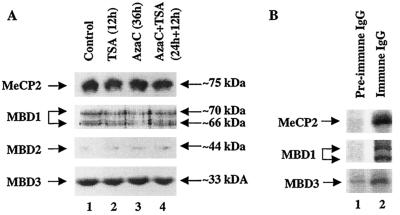
Before we embarked on identifying the MBD associated with the MT-I promoter in P1798 cells, we explored the expression of these proteins in lymphosarcoma cells by immunoblot analysis with antibodies specific for MeCP2, MBD1, MBD2, and MBD3. As these antibodies were raised against the recombinant polypeptides lacking a methyl CpG binding domain, they did not cross-react with one another and specifically detected only the corresponding MBD in immunoblot analysis (Fig. 6A). The results demonstrated that MeCP2 and MBD3 were highly abundant compared to MBD1 and MBD2 was almost undetectable. Treatment with 5-AzaC or TSA alone or in combination did not significantly change the levels of MBD1 and MBD3 (Fig. 6A, lanes 1 to 4), but there was a small but reproducible decrease in the level of MeCP2 after TSA treatment (compare lanes 2 and 4 with lanes 1 and 3, respectively).
FIG. 6.
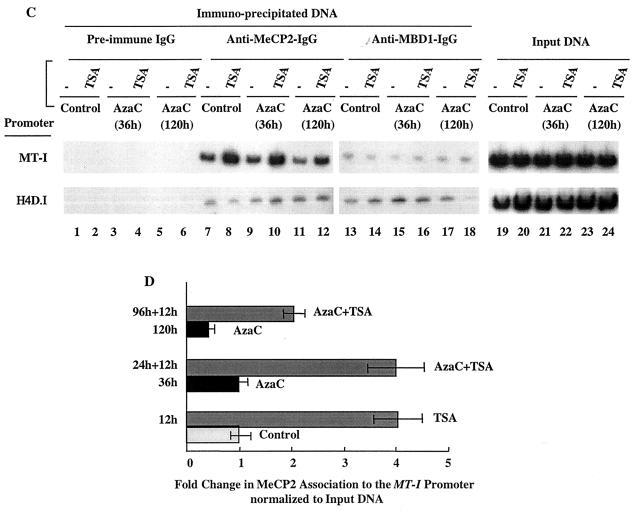
(A) Immunoblot analysis of MBDs in the chromatin isolated from lymphosarcoma cells treated with different inhibitors. Equal amounts of protein (100 μg) from extracts from the control and inhibitor-treated cells were separated by SDS-PAGE and subjected to Western blot analysis with specific antibodies against four different MBD isoforms. (B) [35S]methionine-labeled cell extract was immunoprecipitated with specific antibodies and analyzed by autoradiography after SDS-PAGE. (C) Formaldehyde cross-linked chromatin was immunoprecipitated with antibodies specific for Dnmt3a (lanes 7 to 12), Dnmt1 (lanes 13 to 18), and preimmune IgG (lanes 1 to 6). The purified DNA was amplified with gene-specific primers. The input DNA (lanes 19 to 24) used for PCR was 50-fold less than that of the amount used for immunoprecipitation. (D) Results of quantitative analysis of fold change in association of MeCP2 to the MT-I promoter normalized to input DNA from three different ChIP assays. The means ± standard errors (error bars) are shown.
Next we investigated which of these MBDs were associated with the MT-I promoter in P1798 cells. We first performed immunoprecipitation with specific anti-MBD IgG (that was prepared by purification through recombinant MBD-affinity column) to test whether MeCP2, MBD1, and MBD3 could be precipitated from [35S]methionine-labeled P1798 extract (Fig. 6B). The polypeptides of specific sizes were precipitated by immune IgG but not by preimmune IgG. Next, formaldehyde cross-linked chromatin containing the same amount of DNA from the control cells and the cells treated with inhibitors was precipitated with MBD-specific IgG or preimmune IgG, and the precipitated DNAs were amplified with promoter-specific primers as described above. The results showed that in P1798 cells the MT-I promoter was indeed associated with MeCP2 and this association increased three- to fourfold after treatment of the cells with the TSA (Fig. 6C, lanes 7 and 8, and Fig. 6D). Treatment with 5-AzaC for 36 h did not significantly change this association, whereas prolonged treatment with the nucleotide analog disrupted this association by 50% (Fig. 6C, compare lanes 9 and 11 with lane 7, and Fig. 6D). When these cells were treated with 5-AzaC and then exposed to TSA, the association of MeCP2 with the MT-I promoter increased three- to fourfold, irrespective of the time of exposure to 5-AzaC (Fig. 6C, compare lanes 10 and 12 with lanes 9 and 11, and Fig. 6D). The specificity of the association was demonstrated by the absence of PCR products in DNA pull-down by preimmune IgG (Fig. 6C, lanes 1 to 6). We also used chromatin from cells treated with depsipeptide alone or in combination with 5-AzaC for ChIP assay with anti-MeCP2 antibodies. The association with the MT-I promoter significantly increased compared to the results with untreated or 5-AzaC-treated cells (data not shown). These results indicate that MeCP2, the most abundant MBD, was indeed associated with the MT-I promoter in P1798 cells. This association was not abolished after demethylation of the promoter and increased under the conditions when the promoter was synergistically activated. These results indicate that association of MeCP2 alone cannot repress MT-I promoter. The results were not affected by changing the time and temperature of formaldehyde cross-linking. We detected very low level amplification of the MT-I promoter with the DNA pull down by anti-MBD1 IgG, which was not significantly affected by 5-AzaC and/or TSA treatment (Fig. 6C, lanes 13 to 18). We did not detect amplification with anti-MBD3 IgG, indicating no significant association of this MBD with the MT-I promoter. From these results, we can conclude that in lymphosarcoma cells recruitment of MeCP2 to the methylated MT-I promoter increases after treatment with HDAC inhibitors, whereas exposure to 5-AzaC for a prolonged time disrupts this association due to significant demethylation of the promoter.
Treatment of lymphosarcoma cells with 5-AzaC results in time-dependent decrease in the level of Dnmt1.
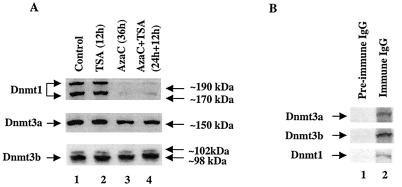
5-AzaC is a potent inhibitor of DNA methyltransferase. When cells are treated with 5-AzaC, it forms an irreversible complex with Dnmts and inactivates them. During replication, newly synthesized DNA strands will not therefore be methylated. After several rounds of replication, this results in demethylation of both strands, leading to transcriptional activation of methylated promoters. We investigated whether treatment of cells with the inhibitors of Dnmts or HDACs affects the level of different isoforms of Dnmts. For this purpose, we treated P1798 cells with 5-AzaC or TSA alone or with both, and the whole-cell extracts were subjected to Western blot analysis with specific antibodies. The results revealed that a single dose of 5-AzaC caused 75 to 80% reduction in the Dnmt1 level within the first 36 h of treatment, whereas the levels of Dnmt3a and Dnmt3b were not significantly altered (Fig. 7A). TSA did alter expression of Dnmts (Fig. 7A). After prolonged treatment with 5-AzaC, the levels of all three Dnmts decreased (data not shown). These results indicate different sensitivities of the three Dnmt isoforms to 5-AzaC in vivo.
FIG. 7.
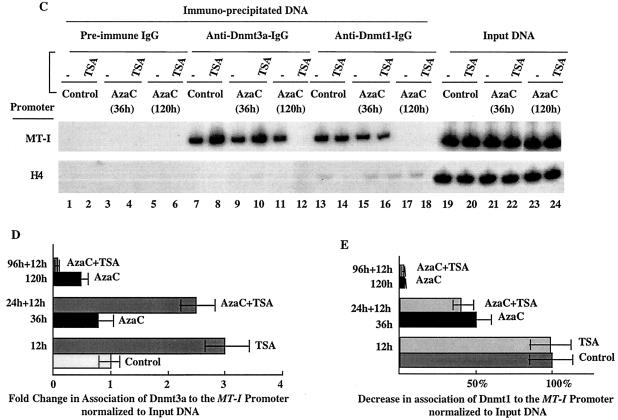
(A) Immunoblot analysis of Dnmts in the chromatin isolated from P1798 cells treated with different inhibitors. Identical amounts (100 μg) of protein of the control or treated cells were separated by SDS-PAGE on a 8% acrylamide gel, transferred to a nitrocellulose membrane, and subjected to Western blot analysis with antibodies specific for Dnmt1, Dnmt3a, and Dnmt3b. (B) P1798 cells were labeled in vivo with [35S]methionine and immunoprecipitated either with the immune IgG or preimmune IgG. Polypeptides pulled down were separated by SDS-PAGE and analyzed by autoradiography. (C) ChIP assay with Dnmt3a and Dnmt1 antibodies. Formaldehyde cross-linked chromatin was immunoprecipitated with antibodies specific for Dnmt3a (lanes 7 to 12), Dnmt1 (lanes 13 to 18), and preimmune IgG (lanes 1 to 6). The purified DNA was amplified. The input DNA (lanes 19 to 24) used for PCR was 50-fold less than that of the amount used for immunoprecipitation. (D and E) Results of quantitative analysis of the fold change in the association of Dnmt3a and Dnmt1 to the MT-I promoter normalized to the input DNA (in three different experiments). The means ± standard errors (error bars) are shown.
5-AzaC treatment affects the association of Dnmt1 and Dnmt3a with MT-I promoter in P1798 cells.
To investigate whether Dnmt1, Dnmt3a, and Dnmt3b are associated with the MT-I promoter, we performed ChIP assay with specific antibodies. First, we determined whether the antibodies can immunoprecipitate the specific proteins from 35S-methionine-labeled whole-cell extracts (Fig. 7B). All three antibodies could pull down the specific isoform that was not precipitated by preimmune IgG. Next we performed ChIP assay with preimmune and immune IgG. The results demonstrated that among the three Dnmt isoforms, Dnmt3a (Fig. 7C, lane 7) and Dnmt1 (lane 13) were associated with the MT-I promoter in P1798 cells. The specificity of the association was demonstrated by the absence of PCR product in DNA pull-down by preimmune IgG (lanes 1 to 6). The association of Dnmt1 was reduced after treatment with 5-AzaC in a time-dependent manner, whereas TSA had no effect (Fig. 7C, lanes 15 to 18, and Fig. 7E). These results correlate with the decrease in the level of Dnmt1 (Fig. 7A). In contrast, there was a three- to fourfold increase in the association of Dnmt3a with the MT-I promoter after treatment with TSA alone or after exposure first to 5-AzaC for 24 h (Fig. 7C, lanes 8 and 10, and Fig. 7D). After prolonged treatment with 5-AzaC, Dnmt3a was also dissociated from the promoter in a TSA-dependent manner (lanes 11 and 12), which correlated with the reduced level of this isoform (data not shown). Anti-Dnmt3b antibody did not pull down MT-I promoter from the control or treated cells (data not shown). The data suggest that activation of MT-I is inversely correlated with recruitment of Dnmt1 but not Dnmt3a.
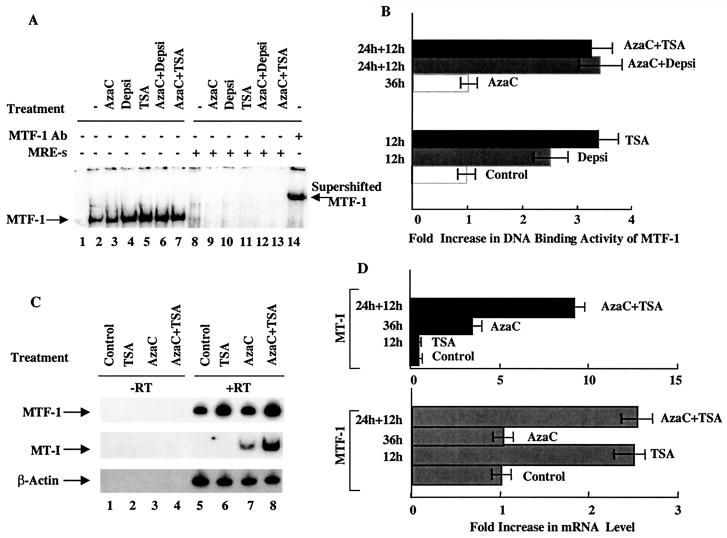
DNA binding activity and expression of the heavy metal-activated transcription factor MTF-1 increase in lymphosarcoma cells treated with inhibitors of HDAC.
MTF-1 is the key transcription factor required for the basal and heavy metal-induced expression of the MT-I gene (47). To test whether HDAC inhibitors modulate the activity of MTF-1 in lymphosarcoma cells, we measured its DNA binding activity with a 32P-labeled MRE-s oligonucleotide to which MTF-1 specifically binds. In extracts from control cells, a specific complex was detected (Fig. 8A, lane 2). The formation of this complex was three- to fourfold higher in extracts from the cells treated with depsipeptide or TSA (Fig. 8A, lanes 4 and 5, and Fig. 8B). Treatment of cells with 5-AzaC alone did not significantly alter MTF-1 activity (Fig. 8A, lane 2), and the activity in extracts treated with both agents was similar to that in the cells treated with HDAC inhibitors (lanes 6 and 7). The specificity of the complex was confirmed by competition of its formation with 50-fold molar excess of unlabeled MRE-s oligonucleotide (lanes 8 to 13) and its supershift with anti-MTF-1 antibodies (lane 14). To test whether HDAC inhibitors up-regulate MTF-1 expression in lymphosarcoma cells, we measured the level of MTF-1 mRNA by RT-PCR with gene-specific primers (Fig. 8C, top panel). The results demonstrated that MTF-1 mRNA was expressed in P1798 cells and its level increased two- to threefold in cells treated with TSA, whereas 5-AzaC did not have any effect (Fig. 8C, lanes 5 to 8, and Fig. 8D). For a control, we measured MT-I mRNA levels that were not affected by TSA treatment but increased two- to threefold after treatment with 5-AzaC and 8- to 10-fold after exposure to both agents (Fig. 5C, middle panel, lanes 5 to 8). These results confirm the Northern blot results presented earlier (Fig. 1).
FIG. 8.
(A) EMSA of MTF-1 in S-100 extracts of P1798 cells treated with different inhibitors. Identical amounts of extracts (10 μg of protein) prepared from the control cells, cells treated for 12 h with depsipeptide (Depsi) (1 nM) or TSA (300 nM), cells treated for 36 h with 5-AzaC (2.5 μM) or for 24 h with 5-AzaC followed by 12 h of exposure to TSA or depsipeptide were incubated with 32P-labeled MRE-s oligonucleotide under optimum binding conditions. The DNA-protein complex was separated on a 4% acrylamide gel with 0.25× TBE (Tris-borate-EDTA) as the running buffer. The gel was dried and subjected to autoradiography and PhosphorImager analysis. Ab, antibody. (B) Results of quantitative analysis of DNA binding activity of MTF-1. The values are mean of three independent experiments ± standard errors (error bars). (C) RT-PCR analysis of MTF-1, MT-I, and β-actin in P1798 cells. Total RNA (1 μg) of each treatment group was reverse transcribed (+RT), and 100 ng of cDNA was amplified with gene-specific primers. The same amount of RNA incubated without reverse transcriptase (−RT) was used in PCR to rule out the possible DNA contamination of the RNA samples. (D) Results of quantitative analysis of the fold increase in MT-I and MTF-1 expression in three different experiments, normalized to β-actin. The means ± standard errors (error bars) are shown.
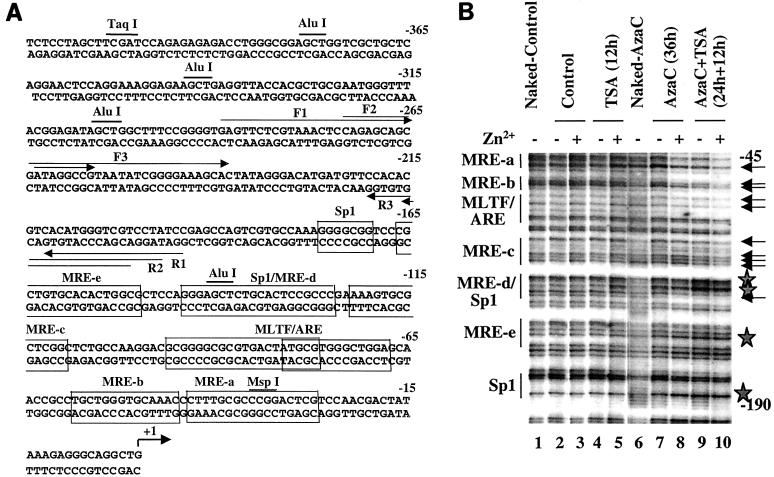
Results of in vivo genomic footprinting studies reveal zinc-induced occupancy of the metal response elements on the MT-I promoter only after treatment of lymphosarcoma cells with 5-AzaC and TSA.
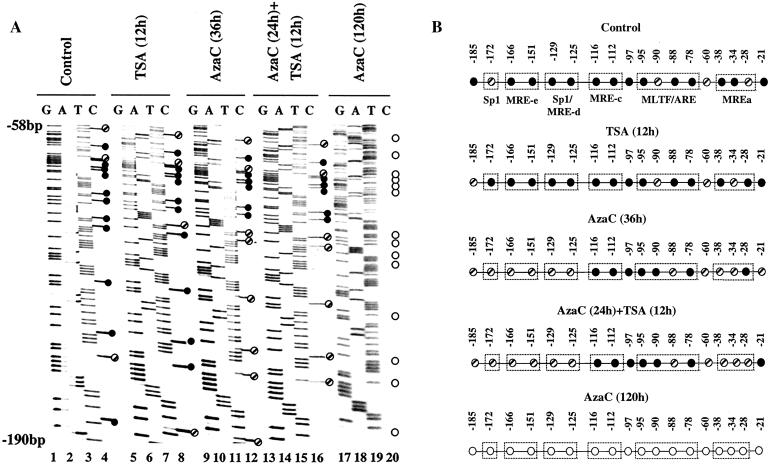
The activation of the MT-I gene in response to heavy metals is mediated through metal-responsive elements (MREs). Several MREs located in the promoter immediately upstream (Fig. 9A) are occupied by MTF-1 (metal regulatory transcription factor-1) in response to activation by heavy metals in vivo (35). Our earlier studies have shown that the methylated MT-I promoter is refractory to the binding of transcription factors (36). To explore whether accessibility of the promoter increased during cooperative activation by 5-AzaC and TSA, we performed in vivo genomic footprinting. Chromosomal DNA, isolated from cells treated with DMS in vivo, was cleaved with piperidine and subjected to LM-PCR with gene-specific primers. The G ladder obtained with naked DNA treated with DMS in vitro served as a control. Figure 9B shows the footprinting profile in the lower strand of the MT-I promoter. In the control and TSA-treated cells, the G ladder (Fig. 9B, compare lanes 2 and 4 with lane 1) is very similar to that of the naked DNA and did not change after treatment with zinc in vivo (lanes 3 and 5, respectively). These results indicate that TSA treatment alone could not open the MT-I promoter to transcription factors, correlating with the expression data (Fig. 1). After treatment with 5-AzaC alone for 36 h followed by zinc treatment, there was no detectable occupancy of any cis element (Fig. 9B, lanes 7 and 8). MRE-d was occupied in cells treated with 5-AzaC and TSA even in the absence of zinc (lane 9). This result was consistent with the higher level of MTF-1 activity in these cells and the RT-PCR data (Fig. 8C) (36). After exposure of these cells to zinc, increased footprinting was evident not only at MRE-d but also at MRE-c, MRE-b, MRE-a, and MLTF/ARE (lane 10). These results suggest that treatment with both inhibitors allows the chromatin structure spanning the MT-I promoter to adopt a conformation that is accessible to the transcription factors. The alteration in chromatin structure facilitates the binding of transcription factors to their cognate cis elements, leading to gene activation in response to heavy metals (Fig. 1). We could not perform footprinting in cells treated for 96 to 120 h with 5-AzaC, as some cells undergo apoptotic cell death after prolonged treatment with the inhibitor, resulting in poor-quality DNA for the analysis.
FIG. 9.
In vivo genomic footprinting of the MT-I promoter in P1798 cells treated with different inhibitors. (A) Location of cis elements, the restriction enzymes (AluI, MspI, and TaqI), and LM-PCR primers (primers F1, F2, F3 and primers P1, P2, P3) on the mouse MT-I promoter. (B) Cells were treated with 5-AzaC (2.5 μM) for 48 h and then exposed to TSA (300 nM) for 10 h. For induction of the MT-I gene, cells were treated with ZnSO4 (50 μM) (+) for 2 h or not treated with ZnSO4 (−). Cells (107 cells in 10 ml) were treated with 0.1% DMS for 2 min at 37°C and washed with PBS containing 1 M β-mercaptoethanol, and DNA was purified after proteinase K digestion. Methylated DNA was then cleaved with piperidine (0.1%) at 95°C for 30 min and lyophilized. Naked DNA was also treated with DMS and piperidine to generate the control G ladder. Identical amounts of DNA (2 μg) from each sample were subjected to LM-PCR with primers F1, F2, and F3 to detect footprinting in the lower strand. Lanes 1 and 6 contain naked DNA from control and 5-azaC-treated cells, respectively.
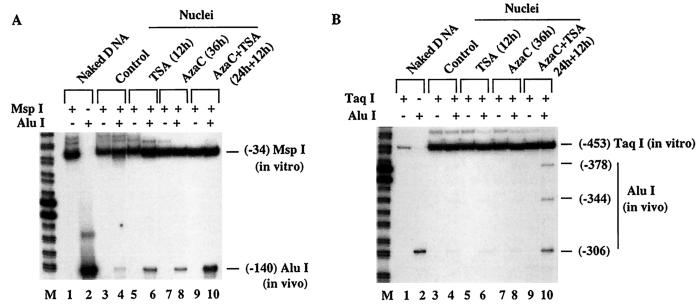
Restriction enzyme accessibility assay results demonstrate that the MT-I promoter forms an open chromatin structure in lymphosarcoma cells after treatment with 5-AzaC and TSA.
Accessibility of the transcription factors to a promoter depends to a significant extent on the chromatin structure of the particular promoter. To characterize in more detail the alteration of chromatin structure of the MT-I promoter in lymphosarcoma cells following treatment with 5-AzaC and/or TSA, we studied the accessibility of restriction enzymes to the MT-I promoter in intact nuclei. Nuclei from the untreated control and treated lymphosarcoma cells were digested with 50 U of AluI to allow limited cleavage of the MT-I promoter without disrupting the integrity of the nuclei. We selected AluI because the MT-I promoter contains multiple sites and the enzyme is not methylation sensitive. DNA digested with AluI was then purified and further digested to completion with a second enzyme, MspI, the site of which is located downstream of the AluI site at −140 bp (Fig. 9A). To analyze the alteration of AluI accessibility at this site, identical amounts of purified and MspI-digested DNA from treated and untreated cells were subjected to LM-PCR with primer sets F1, F2, and F3 specific for lower strand amplification (see Materials and Methods). Intact naked DNA was also digested to completion with either AluI or MspI and subjected to LM-PCR. The amplified DNA generated by single digestion with MspI and AluI is shown in Fig. 10A (lanes 1 and 2). DNA from the control nuclei showed minimal amplification of the AluI fragment (lane 4), which increased to a small but significant extent upon treatment with TSA or 5-AzaC alone (lanes 6 and 8). The AluI fragment, however, was amplified more than twofold when the cells were treated with both drugs (lane 10). To show that the AluI fragment was indeed generated by in vivo digestion, nuclei from each sets of treatment were mock treated with AluI before MspI digestion. Absence of any AluI fragment amplification (lanes 3, 5, 7, and 9) proves the authenticity of in vivo cleavage. Amplification of the MT-I promoter extended to the MspI site (MspI fragment) where in vivo AluI cleavage did not occur. The presence of the MspI fragment in all the samples including the 5-AzaC- and TSA-treated samples was due to digestion with AluI in vivo. We also analyzed the accessibility of AluI at three other sites located between positions −215 and −300 by using TaqI and a different set of primers, R1, R2, and R3 (see Materials and Methods), which specifically amplified the upper strand. A significant cleavage occurred at all three AluI sites only in the nuclei from cells treated with both 5-AzaC and TSA (Fig. 10B), whereas the digestion at these sites in the nuclei from the control or TSA- or 5-AzaC-treated cells was barely detectable (compare lane 10 with lanes 4, 6, and 8). The specificity of AluI digestion was confirmed by lack of amplification at these sites in nuclei incubated without the enzyme (lanes 3, 5, 7, and 9). Similarly, accessibility of the promoter to MspI in nuclei isolated from cells treated with both inhibitors was significantly higher than in the cells treated with either inhibitor alone (data not shown). These results indicate that the MT-I promoter in lymphosarcoma cells treated with 5-AzaC and TSA acquired an open conformation accessible to restriction enzymes.
FIG. 10.
Restriction enzyme accessibility assay in lymphosarcoma cells after treatment with 5-AzaC and TSA. Nuclei isolated from identical numbers of cells (4 × 106) from each group were incubated with 50 U of AluI at 37°C for 10 min (lanes 4, 6, 8, and 10). Nuclei incubated in the same buffer without restriction enzyme were used as controls (lanes 3, 5, 7, and 9). DNA was purified from the nuclei, and 1 μg was digested in vitro with a second enzyme, e.g., MspI in panel A or TaqI in panel B. Identical amounts (250 ng) of DNA from each group and naked DNA digested with respective restriction enzymes were subjected to LM-PCR with strand-specific primers as described in Materials and Methods (the third primer is labeled with 32P). Lanes 1 and 2 contain naked DNA digested with MspI and AluI (A) and TaqI and AluI (B). The reaction product was separated on a sequencing gel and subjected to autoradiography and PhosphorImager analysis. Lanes M contain the 20-bp ladder.
DISCUSSION
The acetylation status of histones associated with a gene promoter is a marker of transcriptional activity of the gene. Hypoacetylated promoters are transcriptionally repressed, and treatment of cells with inhibitors of histone deacetylases results in accumulation of hyperacetylated histones and activation of repressed genes. Genes with methylated CpG islands are, in general, hypoacetylated and silent. It is, therefore, logical to expect activation of the methylated promoters after treatment with HDAC inhibitors such as TSA or depsipeptide. Hypoacetylated, methylated promoters of chromosomal origin are not, however, usually turned on after treatment with these inhibitors alone (6, 13, 53). Interestingly, some methylated promoters, e.g., MLH1, TIMP3, INKN2A, INKN2B, ER, and MDR1, can be robustly activated with TSA after partial demethylation with 5-AzaC (50, 60). With the exception of MDR1, the mechanism(s) of the synergistic effect of these two inhibitors on the expression of the methylated genes has not been explored (13). Activation of methylated MDR1 by 5-AzaC and TSA involves facilitated release of MeCP2 and HDAC1 from the promoter (13). The molecular mechanism of activation of methylated promoters by these two agents may differ depending on the specific gene or cell type. To determine the mechanism by which these two inhibitors cooperatively activate the MT-I promoter in lymphosarcoma cells, we explored in detail a number of aspects of chromatin structure. These aspects include the following: the characterization of the modification state of core histones associated with the MT-I promoter; association of MBDs, Dnmts, and HDAC1 with the promoter; methylation status of MT-I promoter DNA; the chromatin structure of the MT-I gene; and the ability of the metal-responsive transcription factor MTF-1 to bind to the MT-I promoter in the chromatin context. In light of the postulated role of MT as an antioxidant, the regulation of its expression at the chromatin level is of considerable importance.
Tails of core histones undergo different posttranslational modifications, e.g., acetylation at lysine, phosphorylation at serine, and methylation at lysine or arginine. The histones associated with the MT-I promoter in the untreated lymphosarcoma cells are hypoacetylated. Treatment with HDAC inhibitors facilitates hyperacetylation of MT-I-associated histones H3 and H4 but cannot activate the promoter. It should be emphasized that this hyperacetylation of the core histones alone cannot activate the methylated promoters located on chromosomes, e.g., MDR1 and MT-I, while methylated promoters harbored in the plasmids can be activated by TSA treatment (11, 30, 41). These observations indicate that the mechanisms of repression of methylated promoters located within the chromosome are distinct from that of the extrachromosomal DNA.
Recent studies with Neurospora crassa have demonstrated that methylation of histone H3 at K9 is essential for DNA methylation (51). Methylation of H3 at this residue is a primitive mechanism for the repression of promoters in organisms such as Saccharomyces cerevisiae that lack DNA methylation as well as mammalian promoters repressed by mechanisms not involving methylation. In this context, the tumor suppressor protein Rb recruits histone deacetylase as well as histone methyltransferase (SET domain proteins) that methylates lysine-9 of histone H3. This methylation, in turn, is recognized by the heterochromatin protein HP1, resulting in the repression of transcription from Rb-sensitive promoters (43). These observations prompted us to investigate whether K-9 methyl histone H3 plays any role in the repression of MT-I promoter in P1798 cells. Indeed, ChIP assays with specific antibodies demonstrated decreased association of this modified histone H3 with the promoter after treatment with TSA and 5-AzaC. The diminished association of the MT-I promoter with K9-methyl histone H3 and the concomitant increase in association with K9-acetyl histone H3 correlate well with the activation of the MT-I gene. This result suggests that the methylation of histone H3 at K9 may be critical for silencing the methylated MT-I promoter in P1798 cells. After prolonged treatment with 5-AzaC, MT-I can be activated even when the promoter is hypoacetylated. These results suggest that probably dissociation of K9-methyl histone H3 from the promoter rather than association with K9-acetyl H3 is critical for activation of the demethylated MT-I gene.
While it has been reported that TSA treatment can affect DNA methylation states (38, 48), two results suggest that the synergy between HDAC inhibitors and 5-AzaC in activating the MT-I promoter is not the result of a synergistic effect on DNA methylation. First, bisulfite genomic sequencing demonstrated that the extent of MT-I promoter demethylation is comparable whether the cells are treated for the same time period with 5-AzaC or with 5-AzaC and TSA. Second, the synergistic effect of these two inhibitors persists even after complete demethylation of the MT-I promoter.
Another potential mechanism by which TSA plus 5-AzaC function synergistically is through the activation of a key transcription factor that regulates MT-I expression. Indeed, the present study has shown that the transcription factor MTF-1, essential for MT-I expression, was activated following treatment with HDAC inhibitors. We cannot definitively conclude whether this increased activity is due to increased expression of the protein as antibodies against MTF-1 do not detect the denatured protein in Western blots. Alternatively, the increased MTF-1 activity may be due to altered posttranslational modification(s), e.g., phosphorylation or acetylation of MTF-1. However, RT-PCR analysis of MTF-1 mRNA in cells treated with HDAC inhibitors showed increased expression of the MTF-1 gene.
The transcriptional repression of the majority of methylated promoters appears to be mediated through MBDs. Among the MBDs, MeCP2 and MBD3 are expressed at high levels in P1798 cells. ChIP assay results demonstrated that MeCP2 is specifically associated with the MT-I promoter. Recently we observed that MeCP2 is also associated with the methylated, silenced MT-I promoter in a transplanted rat solid hepatoma (34). By contrast, this association was negligible in the liver of the hepatoma-bearing host, suggesting that the affinity of MeCP2 for the MT-I promoter is methylation dependent, as has been observed for methylated MDR1, and proviral DNA (13, 33). To our surprise, the association of MeCP2 with the MT-I promoter increased after treatment of cells with HDAC inhibitors, and this increased interaction persisted even when the cells were pretreated with 5-AzaC. Extended treatment with the Dnmt inhibitor alone (120 h) disrupted this interaction (Fig. 6C and D). The association of MeCP2 to the MT-I promoter was not completely abolished (Fig. 6C and D), even after significant demethylation as evident from bisulfite sequencing (Fig. 3A and B). The increased association of MeCP2 with the MT-I promoter even after treatment with inhibitors of Dnmt and HDAC in combination indicates that the occupancy of methyl CpGs by MeCP2 alone is not sufficient to repress the promoter. A recent study has shown that MeCP2 associates with methyl CpGs, irrespective of its location either in the promoter or exon 2 of overlapping P14/P16 genes (42). Transcription is, however, inhibited only when the promoter is methylated, suggesting that MeCP2 association with the downstream promoter does not impede transcription (42). These results clearly demonstrate that the in vivo functions of MeCP2 may be more complex than has been currently perceived. The nature of the MeCP2 complex formed on methylated promoters may be distinct from that formed on methylated exons. Similarly, the factors associated with MeCP2 in TSA-treated cells may be distinct from those in the control cells. Purification of the MeCP2 complex from the control and TSA-treated cells and of associated proteins may be helpful in elucidating the underlying mechanism. Studies along this line are in progress. Alternatively, TSA treatment may alter posttranslational modification(s) of MeCP2, facilitating increased association with the MT-I promoter. The phosphorylation state of MeCP2 did not change after treatment with inhibitors of HDACs and Dnmts (J. Datta and K. Ghoshal, unpublished data). These observations suggest that the corepressor complex associated with MeCP2 may be the functional entity, e.g., Sin3a and HDAC1, which is disrupted after exposure of cells to these inhibitors. Although the binding of MeCP2 to the promoter increases, the nature of this association is such that it does not block access of the activator to cis elements, as in vivo footprinting analysis showed occupancy of all MRE following treatment of the cells with HDAC and Dnmt inhibitors (Fig. 9B). Perhaps, the inability of MTF-1 to bind to the methylated MT-I promoter in vivo is due to the promoter occlusion by the bulky repressor-corepressor complex, e.g., MeCP2, Sin3a, and HDAC1. The nature of the MeCP2-methylCpG contact in the absence of associated corepressors appears to be such that it does not impede occupancy of MTF-1 to the MT-I promoter.
The role of Dnmts in the suppression of the MT-I promoter deserves comment. Recent studies have shown that Dnmts can also act as transcriptional repressors that do not require their catalytic activities (14). The results of ChIP assays with specific antibodies demonstrated a specific association of both Dnmt3a and Dnmt1 with the MT-I promoter. While there were relatively high levels of Dnmt3b in these cells, Dnmt3b was not found to be associated with the MT-I promoter. It is not known whether Dnmt1 and Dnmt3a cooperate to repress the MT-I promoter in these cells. Dnmt3a was dissociated from the promoter only after prolonged treatment with 5-AzaC followed by TSA. By contrast, the association of Dnmt1 with the MT-I promoter correlated inversely with the activation state of the promoter. After treatment with 5-AzaC, this association was disrupted in a time-dependent manner, which correlated well with the level of expression of this isoform. It is interesting that 5-AzaC not only inhibits Dnmt1 activity but also activates its degradation in lymphosarcoma cells, as has been observed in breast cancer cells (60). The facilitated disruption of Dnmt1 from the promoter may be one of the mechanisms for the synergistic effect of the two inhibitors on the activation of the MT-I promoter.
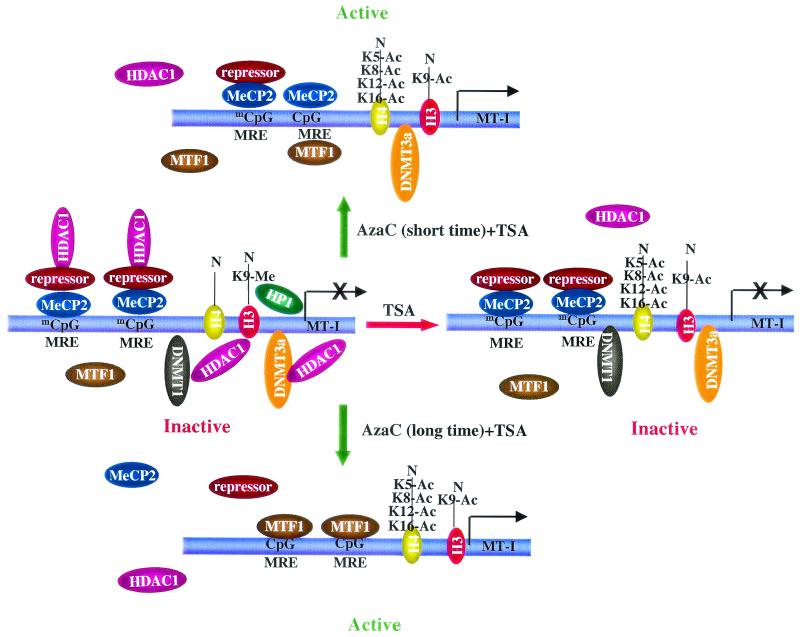
On the basis of the experimental evidence, we propose the following model depicting the roles of histone modifications, HDAC1, MeCP2, and Dnmt1 in controlling the transcriptional state of the MT-I promoter (Fig. 11). In untreated cells the MT-I promoter is silenced by multiple, overlapping mechanisms. These mechanisms are reflected in the MT-I promoter hypermethylation, histone hypoacetylation, histone H3 lysine 9 methylation, and the localization of MeCP2, Dnmt1, Dnmt3a, and HDAC1 proteins. Consistent with this idea, we have observed the positioning of regularly spaced nucleosomes on the MT-I promoter under silencing conditions (data not shown). After TSA treatment, the promoter is hyperacetylated but still repressed due to the association of Dnmt1, Dnmt3a, and MeCP2 with the methylated promoter (Fig. 11). After brief exposure to 5-AzaC, minimal promoter demethylation occurs along with partial dissociation of Dnmt1, K-9-methyl H3, and HDAC1, resulting in low-level expression. Upon subsequent TSA treatment, displacement of K9-methyl histones, hyperacetylation of core histones, and further dissociation of the repressor-corepressor complexes result in synergistic activation (Fig. 11). After prolonged exposure to 5-AzaC, the MT-I promoter is in an active state due to extensive demethylation and dissociation of the repressors. Under these conditions, TSA can further increase MT-I gene expression, by the activation of MTF-1 and probably by hyperacetylation of the promoter (Fig. 11). This model provides a framework for understanding the key molecular mechanisms by which the methylated MT-I promoter is activated following treatment with agents that cause histone hyperacetylation and promoter demethylation.
FIG. 11.
Model depicting the involvement of different components of the methylation machinery in silencing the MT-I gene in lymphosarcoma cells and their dissociation after treatment with inhibitors of Dnmts and HDACs. Untreated cells, cells treated with TSA with 12 h, cells treated with 5-AzaC for 24 h and then with TSA for 12 h, and cells treated with 5-AzaC for 120 h are shown.
Acknowledgments
We sincerely thank Qin Zhu for expert technical assistance; Aubrey Thompson for providing P1798; Adrian Bird, Walter Schaffner, and Richard Palmiter for providing cDNAs for rat MeCP2, human MTF-1, and mouse MT-I, respectively; Shoji Tajima for providing anti-Dnmt1 antibodies; and Peggy Farnham for sharing the ChIP protocol with us.
This research was supported, in part, by grants ES10874 and CA81024 (S.T.J.).
REFERENCES
- 1.Amir, R. E., I. B. Van den Veyver, M. Wan, C. Q. Tran, U. Francke, and H. Y. Zoghbi. 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23:185-188. [DOI] [PubMed] [Google Scholar]
- 2.Bachman, K. E., M. R. Rountree, and S. B. Baylin. 2001. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276:32282-32287. [DOI] [PubMed] [Google Scholar]
- 3.Bader, S., M. Walker, B. Hendrich, A. Bird, C. Bird, M. Hooper, and A. Wyllie. 1999. Somatic frameshift mutations in the MBD4 gene of sporadic colon cancers with mismatch repair deficiency. Oncogene 18:8044-8047. [DOI] [PubMed] [Google Scholar]
- 4.Baylin, S. B., M. Esteller, M. R. Rountree, K. E. Bachman, K. Schuebel, and J. G. Herman. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10:687-692. [DOI] [PubMed] [Google Scholar]
- 5.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. [DOI] [PubMed] [Google Scholar]
- 6.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 7.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression-belts, braces, and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 8.Bourc'his, D., G. L. Xu, C. S. Lin, B. Bollman, and T. H. Bestor. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536-2539. [DOI] [PubMed] [Google Scholar]
- 9.Cai, L., G. J. Wang, Z. L. Xu, D. X. Deng, S. Chakrabarti, and M. G. Cherian. 1998. Metallothionein and apoptosis in primary human hepatocellular carcinoma (HCC) from northern China. Anticancer Res. 18:4667-4672. [PubMed] [Google Scholar]
- 10.Cameron, E. E., K. E. Bachman, S. Myohanen, J. G. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103-107. [DOI] [PubMed] [Google Scholar]
- 11.Curradi, M., A. Izzo, G. Badaracco, and N. Landsberger. 2002. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol. Cell. Biol. 22:3157-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Croce, L., V. A. Raker, M. Corsaro, F. Fazi, M. Fanelli, M. Faretta, F. Fuks, F. Lo Coco, T. Kouzarides, C. Nervi, S. Minucci, and P. G. Pelicci. 2002. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science 295:1079-1082. [DOI] [PubMed] [Google Scholar]
- 13.El-Osta, A., P. Kantharidis, J. R. Zalcberg, and A. P. Wolffe. 2002. Precipitous release of methyl-CpG binding protein 2 and histone deacetylase 1 from the methylated human multidrug resistance gene (MDR1) on activation. Mol. Cell. Biol. 22:1844-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuks, F., W. A. Burgers, N. Godin, M. Kasai, and T. Kouzarides. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20:2536-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghoshal, K., and S. T. Jacob. 2001. Regulation of metallothionein gene expression. Prog. Nucleic Acid Res. Mol. Biol. 66:357-384. [DOI] [PubMed] [Google Scholar]
- 16.Ghoshal, K., S. Majumder, and S. T. Jacob. 2002. Analysis of promoter methylation and its role in silencing metallothionein-I gene expression in tumor cells. Methods Enzymol. 353:476-486. [DOI] [PubMed] [Google Scholar]
- 17.Ghoshal, K., S. Majumder, Z. Li, X. Dong, and S. T. Jacob. 2000. Suppression of metallothionein gene expression in a rat hepatoma because of promoter-specific DNA methylation. J. Biol. Chem. 275:539-547. [DOI] [PubMed] [Google Scholar]
- 18.Ghoshal, K., S. Majumder, Q. Zhu, J. Hunzeker, J. Datta, M. Shah, J. F. Sheridan, and S. T. Jacob. 2001. Influenza virus infection induces metallothionein gene expression in the mouse liver and lung by overlapping but distinct molecular mechanisms. Mol. Cell. Biol. 21:8301-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghoshal, K., Y. Wang, J. F. Sheridan, and S. T. Jacob. 1998. Metallothionein induction in response to restraint stress. Transcriptional control, adaptation to stress, and role of glucocorticoid. J. Biol. Chem. 273:27904-27910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hata, K., M. Okano, H. Lei, and E. Li. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983-1993. [DOI] [PubMed] [Google Scholar]
- 21.He, L. Z., T. Tolentino, P. Grayson, S. Zhong, R. P. Warrell, Jr., R. A. Rifkind, P. A. Marks, V. M. Richon, and P. P. Pandolfi. 2001. Histone deacetylase inhibitors induce remission in transgenic models of therapy-resistant acute promyelocytic leukemia. J. Clin. Investig. 108:1321-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrich, B., C. Abbott, H. McQueen, D. Chambers, S. Cross, and A. Bird. 1999. Genomic structure and chromosomal mapping of the murine and human Mbd1, Mbd2, Mbd3, and Mbd4 genes. Mamm. Genome 10:906-912. [DOI] [PubMed] [Google Scholar]
- 23.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrich, B., and A. Bird. 2000. Mammalian methyltransferases and methyl-CpG-binding domains: proteins involved in DNA methylation. Curr. Top. Microbiol. Immunol. 249:55-74. [DOI] [PubMed] [Google Scholar]
- 25.Hendrich, B., U. Hardeland, H. H. Ng, J. Jiricny, and A. Bird. 1999. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401:301-304. [DOI] [PubMed] [Google Scholar]
- 26.Howell, C. Y., T. H. Bestor, F. Ding, K. E. Latham, C. Mertineit, J. M. Trasler, and J. R. Chaillet. 2001. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell 104:829-838. [DOI] [PubMed] [Google Scholar]
- 27.Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 29.Jones, P. A., and D. Takai. 2001. The role of DNA methylation in mammalian epigenetics. Science 293:1068-1070. [DOI] [PubMed] [Google Scholar]
- 30.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 31.Lo, W. S., L. Duggan, N. C. Tolga, Emre, R. Belotserkovskya, W. S. Lane, R. Shiekhattar, and S. L. Berger. 2001. Snf1-a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science 293:1142-1146. [DOI] [PubMed] [Google Scholar]
- 32.Lo, W. S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu, C. D. Allis, R. Marmorstein, and S. L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 33.Lorincz, M. C., D. Schubeler, and M. Groudine. 2001. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol. 21:7913-7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majumder, S., K. Ghoshal, J. Datta, S. Bai, X. Dong, N. Quan, C. Plass, and S. T. Jacob. 2002. Role of de novo DNA methyltransferases and methyl C-binding proteins in gene silencing in a rat hepatoma. J. Biol. Chem. 13:16048-16058. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Majumder, S., K. Ghoshal, R. M. Gronostajski, and S. T. Jacob. 2001. Downregulation of constitutive and heavy metal-induced metallothionein-I expression by nuclear factor I. Gene Expr. 9:203-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Majumder, S., K. Ghoshal, Z. Li, Y. Bo, and S. T. Jacob. 1999. Silencing of metallothionein-I gene in mouse lymphosarcoma cells by methylation. Oncogene 18:6287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Majumder, S., K. Ghoshal, Z. Li, and S. T. Jacob. 1999. Hypermethylation of metallothionein-I promoter and suppression of its induction in cell lines overexpressing the large subunit of Ku protein. J. Biol. Chem. 274:28584-28589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Milutinovic, S., Q. Zhuang, and M. Szyf. 2002. Proliferating cell nuclear antigen associates with histone deacetylase activity, integrating DNA replication and chromatin modification. J. Biol. Chem. 277:20974-20978. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 40.Nan, X., S. Cross, and A. Bird. 1998. Gene silencing by methyl-CpG-binding proteins. Novartis Found. Symp. 214:6-21, 46-50. [DOI] [PubMed] [Google Scholar]
- 41.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen, C. T., F. A. Gonzales, and P. A. Jones. 2001. Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res. 29:4598-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 44.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 45.Prokhortchouk, A., B. Hendrich, H. Jorgensen, A. Ruzov, M. Wilm, G. Georgiev, A. Bird, and E. Prokhortchouk. 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quaife, C. J., R. L. Cherne, T. G. Newcomb, R. P. Kapur, and R. D. Palmiter. 1999. Metallothionein overexpression suppresses hepatic hyperplasia induced by hepatitis B surface antigen. Toxicol. Appl. Pharmacol. 155:107-116. [DOI] [PubMed] [Google Scholar]
- 47.Radtke, F., O. Georgiev, H. P. Muller, E. Brugnera, and W. Schaffner. 1995. Functional domains of the heavy metal-responsive transcription regulator MTF-1. Nucleic Acids Res. 23:2277-2286. [DOI] [PMC free article] [PubMed]
- 48.Selker, E. U. 1998. Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc. Natl. Acad. Sci. USA 95:9430-9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki, H., E. Gabrielson, W. Chen, R. Anbazhagan, M. Van Engeland, M. P. Weijenberg, J. G. Herman, and S. B. Baylin. 2002. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 31:141-149. [DOI] [PubMed] [Google Scholar]
- 51.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 52.Ueda, H., H. Nakajima, Y. Hori, T. Goto, and M. Okuhara. 1994. Action of FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum no. 968, on Ha-ras transformed NIH3T3 cells. Biosci. Biotechnol. Biochem. 58:1579-1583. [DOI] [PubMed] [Google Scholar]
- 53.Wade, P. A. 2001. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene 20:3166-3173. [DOI] [PubMed] [Google Scholar]
- 54.Wade, P. A. 2001. Methyl CpG-binding proteins and transcriptional repression. Bioessays 23:1131-1137. [DOI] [PubMed] [Google Scholar]
- 55.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
- 56.Wang, H., Z. Q. Huang, L. Xia, Q. Feng, H. Erdjument-Bromage, B. D. Strahl, S. D. Briggs, C. D. Allis, J. Wong, P. Tempst, and Y. Zhang. 2001. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293:853-857. [DOI] [PubMed] [Google Scholar]
- 57.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weinmann, A. S., S. E. Plevy, and S. T. Smale. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665-675. [DOI] [PubMed] [Google Scholar]
- 59.Xie, S., Z. Wang, M. Okano, M. Nogami, Y. Li, W. W. He, K. Okumura, and E. Li. 1999. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene 236:87-95. [DOI] [PubMed] [Google Scholar]
- 60.Yang, X., D. L. Phillips, A. T. Ferguson, W. G. Nelson, J. G. Herman, and N. E. Davidson. 2001. Synergistic activation of functional estrogen receptor (ER)-alpha by DNA methyltransferase and histone deacetylase inhibition in human ER-alpha-negative breast cancer cells. Cancer Res. 61:7025-7029. [PubMed] [Google Scholar]
- 61.Zhang, L., W. Zhou, V. E. Velculescu, S. E. Kern, R. H. Hruban, S. R. Hamilton, B. Vogelstein, and K. W. Kinzler. 1997. Gene expression profiles in normal and cancer cells. Science 276:1268-1272. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., H. H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]