Abstract
The INK4a/ARF tumor suppressor locus is implicated in the senescence-like growth arrest provoked by oncogenic Ras in primary cells. INK4a and ARF are distinct proteins encoded by transcripts in which a shared exon is decoded in alternative reading frames. Here we analyze dermal fibroblasts (designated Q34) from an individual carrying independent missense mutations in each copy of the common exon. Both mutations alter the amino acid sequence of INK4a and functionally impair the protein, although they do so to different degrees. Only one of the mutations affects the sequence of ARF, causing an apparently innocuous change near its carboxy terminus. Unlike normal human fibroblasts, Q34 cells are not permanently arrested by Ras or its downstream effectors Ets1 and Ets2. Moreover, ectopic Ras enables the cells to grow as anchorage-independent colonies, and in relatively young Q34 cells anchorage independence can be achieved without addition of telomerase or perturbation of the p53 pathway. Whereas ARF plays the principal role in Ras-induced arrest of mouse fibroblasts, our data imply that INK4a assumes this role in human fibroblasts.
Germ line mutations in the CDKN2A (INK4a/ARF) tumor suppressor locus on human chromosome 9p21 are associated with familial predisposition to malignant melanoma and other tumor types (35). The locus has the unusual capacity to encode two structurally and functionally distinct proteins, p16INK4a and p14ARF (p19ARF in the mouse), specified by alternative first exons spliced to a common second exon that is translated in different reading frames (reviewed in references 35, 42, and 43). The p16INK4a product is the prototype of a family of cyclin-dependent kinase (Cdk) inhibitors that interact directly with Cdk4 and Cdk6, the cyclin D-dependent kinases that initiate the phosphorylation of the retinoblastoma protein pRb (38). By preventing the inactivation of pRb, overexpression of p16INK4a causes a G1-phase cell cycle arrest (35). In contrast, p14ARF interacts directly with the MDM2 protein, whose principal function is to associate with and promote the ubiquitin-mediated destruction of the p53 tumor suppressor (1). Thus, overexpression of p14ARF stabilizes p53 and causes a G1/G2-phase cell cycle arrest via the upregulation of the Cdk inhibitor p21CIP1 (42, 43).
In addition to sharing an exon, the INK4a and ARF proteins have been implicated in the same physiological processes, particularly the phenomenon known as replicative senescence (5, 19, 43). The term senescence was originally used to describe the irreversible growth arrest that occurs when somatic cells reach the end of their proliferative life span in tissue culture (12). For human diploid fibroblasts (HDFs) the arrest is primarily attributed to telomere erosion (reviewed in references 19, 37, and 52), but a similar phenotype occurs irrespective of population doublings (PDs) when HDFs are challenged with oncogenic Ras (40). Primary mouse embryo fibroblasts (MEFs) also undergo Ras-induced arrest, but their long telomeres and endogenous telomerase spare them from telomere-triggered arrest (37, 44, 52). MEFs nevertheless have a finite life span, and the present model attributes this to the stresses imposed by tissue culture, which may be in part akin to the stresses imposed by oncogenic Ras (44, 52). Importantly, a series of elegant genetic studies in which individual Ink4a/Arf exons were ablated in transgenic mice (13, 14, 39, 41) have established a preeminent role for Arf rather than Ink4a in both Ras-induced and culture-associated senescence of the corresponding MEFs. Immortalized mouse cells generally show loss of Arf or, more commonly, its downstream target, p53 (13).
This pivotal role for Arf has to be set against the fact that p16INK4a also accumulates in cultured MEFs and that for some mouse cell types immortalization is dependent on loss of Ink4a, irrespective of Arf status (31, 47). Moreover, Ink4a expression is induced by Ras (40). In human fibroblasts, it has been possible to trace the signaling pathway from Ras to the binding of Ets family transcription factors to specific sites in the INK4a promoter (16, 27, 54), whereas human ARF is not significantly induced by oncogenic Ras or by prolonged culture (3, 6, 51). It has been estimated that approximately 75% of established human cell lines lack functional INK4a due to promoter methylation, mutation, or homozygous deletion. In many instances the deletions affect both ARF and INK4a, although a significant minority exclusively affect INK4a (35). Similarly, a substantial proportion of the missense mutations occur in exon 1α and, therefore, have no effect on ARF, whereas deletions or mutations that specifically incapacitate ARF are relatively rare (30, 32, 35).
As part of an ongoing survey of melanoma kindreds in the United Kingdom (10, 11, 20, 26), we have identified a compound heterozygous individual carrying two distinct sequence variants of CDKN2A. Significantly, we find that both forms of p16INK4a encoded by this individual are functionally compromised, albeit to different extents, while neither alteration appears to affect the function of p14ARF. Dermal fibroblasts from this patient are resistant to the senescence-like arrest elicited by oncogenic Ras and by the transcription factors Ets1 and Ets2, suggesting that this arrest is p16INK4a dependent. Conversely, while Ras enables the cells to form anchorage-independent colonies, Ets1 and Ets2 do not. Anchorage independence does not require telomerase expression and can be achieved without discernible perturbation of the p53 pathway. These findings support our contention that in human fibroblasts, p16INK4a rather than p14ARF provides the primary defense against aberrant proliferative signals (3).
MATERIALS AND METHODS
DNA extraction, PCR amplification, and sequencing.
The subject presented with a melanoma in situ on the elbow at the age of 23 and developed a second superficial spreading melanoma 5 years later. Peripheral blood lymphocyte DNA was prepared and analyzed as previously described (10, 11, 26). PCR amplification was performed with oligonucleotide primer pairs specific for CDKN2A exons 1β, 1α, 2, and 3. Sequencing reactions were carried out with the ABI PRISM dye terminator cycle sequencing system on an ABI 373 sequence analyzer (Perkin Elmer). DNA sequencing was performed on both strands.
Construction of vectors encoding mutant forms of p16INK4a.
Selected missense mutations were introduced into the wild-type p16INK4a cDNA sequence by using synthetic oligonucleotides containing the required base substitution and the QuikChange mutagenesis procedure according to the manufacturer's protocols (Stratagene). All the mutations were verified by DNA sequencing. The respective p16INK4a coding sequences, flanked by BamHI and EcoRI restriction sites, were transferred into the pRSET vector (Invitrogen) to facilitate in vitro translation of His6-tagged proteins and into the pBABEpuro retrovirus vector (24) to allow infection of primary fibroblasts. Two copies of the hemagglutinin (HA) tag were incorporated at the amino terminus of the wild-type p16INK4a construct in pBABEpuro by ligating complementary synthetic oligonucleotides as described previously (34).
Cell culture, retroviral infection, and proliferation assays.
Human diploid fibroblasts expressing the cell surface receptor for mouse ecotropic retroviruses have been described previously, and a similar procedure was used to introduce the receptor into Q34, 904, and Hs68 cells (21). Retroviral stocks were prepared by transient transfection of BOSC-23 cells (28) as described previously (21, 34). For retroviral infections, recipient cells were plated at 2 × 105 cells per 75-cm2 flask and were incubated overnight. The culture medium was replaced with 5 ml of virus supernatant from the BOSC-23 cells, together with 3 ml of fresh medium and 4 μg of polybrene/ml. After 24 h, the medium was replaced and selection in medium containing 1.25 μg of puromycin/ml or 50 μg of bleomycin/ml was initiated on day 2 postinfection.
For proliferation assays, 5 × 103 drug-resistant cells at 5 days postinfection were plated in 24-well plates, and at various times thereafter individual wells were washed twice with phosphate-buffered saline (PBS) and fixed in 10% formaldehyde. Cells were stained with 0.1% crystal violet for 30 min and were extracted with 10% acetic acid, and the amount of intracellular stain was estimated from the optical density at 590 nm (34). Values were normalized to that obtained at day 0, and each time point was assayed in triplicate.
Agar colony assays.
Approximately 2 × 104 cells were suspended in 1 ml of 2× Dulbecco modified Eagle's medium (DMEM) supplemented with 20% fetal calf serum (FCS) and mixed with an equal volume of 0.4% molten agarose, held at 60°C. The mixture was then layered on top of 2 ml of solidified 1% agarose in DMEM plus 10% FCS in a 35-mm-diameter dish. The cells were incubated at 37°C and fed with fresh 0.2% agarose-DMEM-FCS every 7 days. Colonies were counted after 21 days. Multiple fields of >100 cells were observed microscopically, and the number of multicellular colonies was estimated as a percentage of the total number of cells visualized.
Protein analyses.
Following retroviral infection and selection, lysates were prepared from subconfluent cultures as described previously (3). Samples containing 500 μg or 1 mg of total protein were immunoprecipitated with 5 μl of antiserum overnight at 4°C. Alternatively, samples (20 to 30 μg) of total protein were analyzed directly by electrophoresis in sodium dodecyl sulfate polyacrylamide gels (SDS-PAGE).
For immunoblotting, the proteins were transferred to Immobilon-P (Millipore) and the membranes were placed in blocking solution (PBS containing 5% milk powder and 0.2% Tween 20) for 30 min. Membranes were incubated for 1 h at room temperature with monoclonal or polyclonal antibodies diluted in blocking solution and then were thoroughly washed in 0.2% Tween 20 in PBS. Sheep anti-mouse horseradish peroxidase (1:1,000 dilution) and donkey anti-rabbit horseradish peroxidase (1:2,000 dilution) were used as secondary antibodies (Amersham), and antibody binding was visualized by using Amersham ECL reagents.
The monoclonal antibody DO-1 against p53 (sc-126), the rabbit polyclonal antibodies against Cdk4 (sc-601) and Cdk6 (sc-177), and the SMP14 monoclonal against MDM2 (sc-965) were obtained from Santa Cruz. The IF2 (OP46) and 2A10 (OP115) antibodies against MDM2 were from Oncogene Research Products. Polyclonal antisera against p14ARF (JR14) or against a peptide corresponding to residues 54 to 75 (generously provided by David Parry, DNAX Research Institute, Palo Alto, Calif.), the monoclonal antibody 4C6/4 against p14ARF, and the monoclonal antibody JC8 against p16INK4a (from J. Koh and E. Harlow) have been described in earlier reports (3, 17).
UV treatment of cells.
Cells at 60 to 80% confluence in 90-mm-diameter dishes were washed twice in PBS and then overlaid with 2 ml of PBS at 37°C. The lids were removed and the cells were exposed to 254-nm UV light (15 J/m2). The medium was then replaced, and the cells were incubated for a further 17 h before being harvested in SDS sample buffer as described above.
RESULTS
Biallelic CDKN2A mutations in the germ line of a melanoma patient.
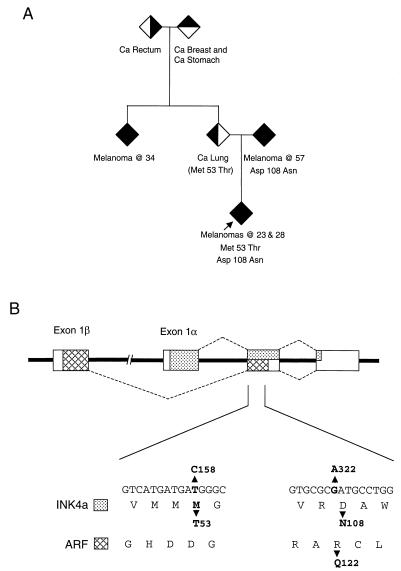
The subject had European parents who were unrelated but who had a strong family history of cancer. In addition to melanoma in both parental lines, family members had carcinomas at other sites, including breast, lung, stomach, colon, and rectum (see Fig. 1A). Two single base changes were identified in the second exon of CDKN2A: T to C at nucleotide 158 of p16INK4a (numbered as in reference 35), which is predicted to cause a Met to Thr change at amino acid 53, and G to A at nucleotide 322, which changes Asp to Asn at residue 108 of p16INK4a (Fig. 1B). Interestingly, the patient carries at least one normal p14ARF allele, since the T158C change would not alter the coding in the −1 reading frame, while the G322A mutation would substitute Gln for Arg at residue 122 of p14ARF. The latter mutation was detected as the only change in one parent, suggesting but not proving that the T158C change was independently inherited from the other parental line. We were unable to address this directly, but haplotype analysis of the proband and siblings, using polymorphic markers on chromosome 9p21, would concur with this conclusion (data not shown).
FIG. 1.
Family pedigree and nature of CDKN2A mutations. (A) The proband is indicated with an arrow, and the shaded symbols refer to individuals diagnosed with cancer (Ca) as follows: melanoma, filled symbol; lung cancer, left half shaded; rectal cancer, right half shaded; breast cancer, top half shaded. To preserve the confidentiality of the patient and family, genders and unaffected siblings are not specified in the pedigree. (B) Schematic representation of the CDKN2A locus with the exons shown as boxes. The sequences encoding p16INK4a are identified by stippling, and those encoding p14ARF are identified by cross-hatches. The nucleotide sequence of the relevant sections of exon 2 and the consequences for translation in the INK4a and ARF reading frames are shown at the bottom.
HDFs carrying biallelic CDKN2A mutations.
In view of the rarity and experimental potential of this genetic background, we prepared fibroblast cultures from a region of normal skin in a clinical biopsy from the patient. The fibroblasts (designated Q34 cells) were propagated under standard conditions for growing HDFs to replicative exhaustion, involving regular passaging on a 1:4 split ratio, equivalent to 2 PD per passage. At 4 PD we infected the cells with an amphotropic retrovirus encoding the mouse basic amino acid transporter (21, 40), rendering them sensitive to further infection by ecotropic retroviruses encoding agents known to impinge on the expression or functions of p16INK4a and p14ARF. Dermal HDFs from a normal adult donor (designated 904) and the Hs68 strain of neonatal foreskin fibroblasts were infected and studied in parallel.
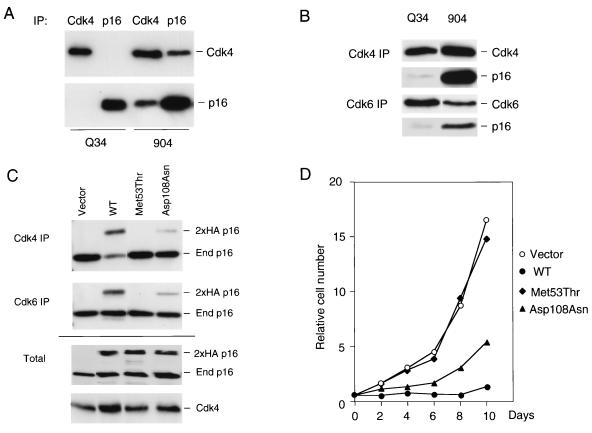
Our initial interest was to determine whether the p16INK4a variants expressed in Q34 cells were functional. To enhance the expression of endogenous p16INK4, we infected the cells with a retrovirus encoding simian virus 40 T antigen to ablate pRb. As previously reported, this causes an approximately fivefold increase in the basal levels of p16INK4a irrespective of the accrued number of PDs (reference 9 and data not shown). Lysates of Q34 and control 904 cells were then immunoprecipitated with an antiserum against p16INK4a and were immunoblotted with monoclonal antibodies against p16INK4a or Cdk4 (Fig. 2A). Although the Q34 and 904 cells expressed similar levels of p16INK4a, there was a substantial difference in the amount of Cdk4 present in the p16INK4a immunoprecipitates, with almost no signal detectable in Q34 cells at the exposure shown. In reciprocal immunoprecipitations with Cdk4 antiserum there was a similar reduction in the proportion of p16INK4a that coprecipitated in Q34 cells. Upon prolonged exposure of the films it was possible to detect a trace amount of p16INK4a associated with both Cdk4 and Cdk6 (Fig. 2B), and on the basis of these types of analyses we estimated that the combined effect of the biallelic mutations in Q34 cells reduces the Cdk binding capacity of p16INK4a to less than 5% of that seen with wild-type p16INK4a in age-matched HDFs.
FIG. 2.
Functional analyses of p16INK4a variants in Q34 cells. (A) Lysates from Q34 cells and the control fibroblasts, 904, previously infected with a retrovirus encoding simian virus 40 T antigen, were immunoprecipitated with polyclonal antibodies against Cdk4 or p16INK4a as indicated. Following SDS-12% PAGE the amounts of Cdk4 and p16INK4a in each precipitate were compared by immunoblotting with the corresponding monoclonal antibodies. IP, immunoprecipitation. (B) Longer exposures are shown of Cdk4 and Cdk6 immunoprecipitates that have been immunoblotted for p16INK4a and the respective Cdks. (C) Normal HDFs were infected with recombinant retroviruses encoding 2× HA-tagged p16INK4a (wild type [WT]), and the indicated variants and pools of infected cells were selected in puromycin. Seven days after infection, cell lysates were prepared and equivalent amounts (500 μg) of protein were immunoprecipitated with antibodies against Cdk4 or Cdk6 (upper two panels). Immunoprecipitated proteins were fractionated by SDS-12% PAGE and were immunoblotted with a monoclonal antibody against p16INK4a. The positions of exogenous (2×HA p16) and endogenous p16INK4a (End p16) are indicated. Samples (20 μg) of total lysate from the same cells were analyzed directly by SDS-PAGE and immunoblotting for p16INK4a and Cdk4 (lower panels). (D) Aliquots of each cell pool, at 7 days postselection, were transferred into 24-well plates (5 × 103 cells/well), and their proliferation was monitored for 10 days. At each time point cells were fixed in 10% formaldehyde and viable cells were stained with crystal violet. Relative numbers of cells were determined by measurements of the optical density at 590 nm. The data present the averages of triplicate measurements.
Functional evaluation of each p16INK4a variant in Q34 cells.
As these in vivo assays did not discriminate between the different mutations, the relevant single-nucleotide changes were introduced into wild-type p16INK4a cDNA by site-directed mutagenesis and recombinant retroviruses were used to express the variant p16INK4a sequences in normal HDFs. In transferring the relevant sequences into the pBABEpuro retroviral vector, two copies of the HA epitope tag were added to the amino terminus of the protein so that exogenous and endogenous p16INK4a could be distinguished (34). After selection of the infected cell pools in puromycin, cell lysates were prepared and either analyzed directly or immunoprecipitated with polyclonal antibodies against Cdk4 and Cdk6. The amount of p16INK4a coprecipitating with each Cdk was assessed by immunoblotting with a monoclonal antibody that is known to be capable of detecting all the p16INK4a variants under study. As illustrated in Fig. 2C, the endogenous p16INK4a present in the host cells was readily detected in every Cdk4 and Cdk6 immunoprecipitate and served as an internal loading control. Of the exogenous HA-tagged versions of p16INK4a, only the wild type and Asp108Asn variants appeared in the Cdk immunoprecipitates (Fig. 2C, lanes 2 and 4). Direct immunoblotting confirmed that the two variants were expressed at levels equivalent to those of wild-type p16INK4a, indicating that they are not inherently unstable. These data implied that the Met53Thr variant did not bind significantly to Cdk4 and Cdk6 and that the affinity of the Asp108Asn variant for Cdks was significantly reduced compared to that of wild-type p16INK4a.
We also monitored the proliferation of the infected cell pools over a period of 10 days following selection in puromycin. Cells infected with the empty vector or with the Met53Thr variant grew rapidly, increasing 15- to 20-fold in number over the monitoring period (Fig. 2D), consistent with the complete loss of function of this variant. In contrast, cells infected with wild-type p16INK4a showed markedly reduced proliferation, only increasing about twofold in 10 days. Significantly, the Asp108Asn variant produced an intermediate effect on cell proliferation (Fig. 2D) consistent with the partial reduction in Cdk binding. In comparison to previous mutants surveyed by these methods (34), Met53Thr would score as a totally impaired mutant, and Asp108Asn would score as a partially impaired mutant. The low residual p16INK4a activity detected in Q34 cells is therefore likely to be attributable to the Asp108Asn variant.
Impact of the Arg122Gln mutation on p14ARF function.
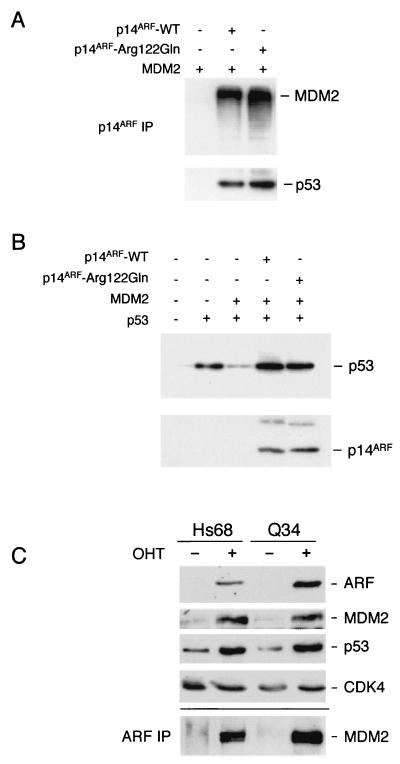
Only one of the CDKN2A mutations affects the sequence of p14ARF, and as the alteration occurs close to the carboxy terminus of p14ARF and is distal to any of the known functional motifs in the protein (Fig. 1B), it seemed conceivable that the mutated allele retains normal ARF functions. To address this possibility we introduced the Arg122Gln alteration into wild-type p14ARF cDNA by site-directed mutagenesis and determined if the resultant protein was capable of interacting with MDM2 and p53. The U20S osteosarcoma cell line, which contains wild-type pRb and p53, was transiently transfected with plasmids encoding MDM2 and either wild-type p14ARF or the Arg122Gln variant. After immunoprecipitation with a polyclonal antibody against p14ARF, the immune complexes were immunoblotted with monoclonal antibodies against MDM2 and p53 (Fig. 3A). Similar amounts of MDM2 and endogenous p53 were present in the p14ARF immunoprecipitates, suggesting that within the limits of this assay there was no significant difference between the Arg122Gln variant and wild-type p14ARF.
FIG. 3.
Functional analysis of p14ARF in Q34 cells. (A and B) U20S cells were transfected with plasmids encoding MDM2, p53 and p14ARF as indicated. (A) Samples (500 μg of protein) of cell lysate were immunoprecipitated with a polyclonal antibody against p14ARF (JR14) and were immunoblotted with antibodies against p14ARF (4C6/4), MDM2 (IF2), and p53 (DO-1). (B) The p53 and MDM2 levels were analyzed by direct immunoblotting of cell lysate. (C) Control (Hs68) and Q34 HDFs expressing E2F1-ER were treated with (+) or without (−) 4-hydroxy tamoxifen (OHT) for 24 h. Samples (20 μg) of cell lysate were fractionated by SDS-PAGE and were immunoblotted with an antiserum raised against amino acids 54 to 75 of human p14ARF and with monoclonal antibodies against MDM2 and p53. Cdk4 served as a loading control. Samples (1 mg) of each lysate were immunoprecipitated with the same p14ARF antiserum and were immunoblotted for MDM2. IP, immunoprecipitation; WT, wild type.
As well as forming ternary complexes with MDM2 and p53, p14ARF has been shown to protect p53 from MDM2-mediated ubiquitination and degradation. This can be readily demonstrated by cotransfection of p53 and MDM2 in U20S cells. In the absence of p14ARF the p53 signal is significantly reduced by the coexpression of MDM2, but this effect is not seen in the presence of p14ARF (Fig. 3B and references 15 and 46). The Arg122Gln variant again proved to be indistinguishable from the wild type in this assay, irrespective of the amounts of plasmid DNA used (Fig. 3B and data not shown). With the caveat that these assays are based on overexpression of p14ARF, the Arg122Gln variant did not appear to be functionally compromised.
To assess endogenous ARF function in Q34 cells, they were infected with a retrovirus encoding E2F-1, a known activator of ARF (2, 33). In the experiment shown, we used a virus in which E2F-1 was fused to the ligand binding domain of the estrogen receptor (25). Upon addition of 4-hydroxy tamoxifen and releasing active E2F-1, the expression of ARF increased substantially, accompanied by upregulation of both MDM2 and p53 (Fig. 3C). The levels of ARF in Q34 cells were comparable and, if anything, were higher than those in the Hs68 control cells, and this was reflected in the amounts of endogenous MDM2 that could be coprecipitated from these lysates by using a polyclonal antibody against human ARF (Fig. 3C). Although these data suggested that Q34 cells have near-normal ARF function, the available methods are not quantitative enough to distinguish between the presence of one versus two functional alleles, and it is also unclear what should be regarded as the normal baseline level of ARF in HDFs from different individuals.
Resistance of Q34 cells to Ras-Mek-Ets-mediated arrest.
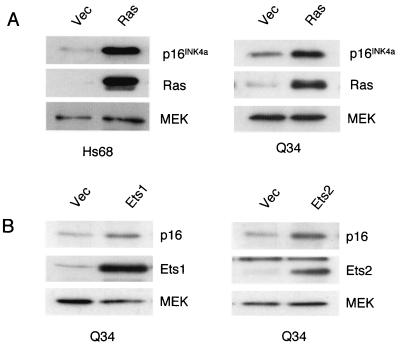
Despite these uncertainties, our functional assessment of the p16INK4a and p14ARF variants in Q34 cells suggested that they are specifically deficient for p16INK4a. They therefore provided an opportunity to investigate Ras signaling to p16INK4a while avoiding the downstream consequences of p16INK4a induction. Young Q34 cells (at ∼30 PD) and Hs68 control HDFs were infected with a retrovirus encoding the oncogenic G12V variant of H-Ras, and after 7 days of selection in bleomycin lysates were prepared and analyzed by immunoblotting. As predicted from previous studies (16, 27, 40, 50, 54), the levels of endogenous p16INK4a protein increased significantly in the cells infected with the Ras virus compared to those in the vector-only control cells (Fig. 4A). Similar increases were noted in Q34 and Hs68 cells, with MEK serving as a loading control. However, whereas the control HDFs underwent a senescence-like growth arrest, manifested as an enlarged flattened appearance (Fig. 5A) and expression of senescence-associated β-galactosidase activity (data not shown), the Q34 cells continued to proliferate. After a transient period of slower growth they eventually resumed the same proliferative behavior as that of the vector-only controls.
FIG. 4.
Induction of p16INK4a by Ras, Ets1, and Ets2. Q34 HDFs and the Hs68 controls infected with retroviruses encoding Ras, Ets1, or Ets2 as indicated or with the empty vector (Vec) were selected in puromycin for 5 days. Samples (20 μg) of cell lysate were analyzed by SDS-12% PAGE and were immunoblotted with antisera against p16INK4a, Ras, Ets1, Ets2, and MEK as indicated.
FIG. 5.
Contrasting effects of Ras, Ets1, and Ets2 in Hs68 and Q34 cells. Pools of Hs68 and Q34 cells infected with retroviruses encoding Ras, Ets1, or Ets2 were photographed at 10 days postinfection. Whereas the Hs68 cells had undergone a senescence-like growth arrest (upper panels), the Q34 cells continued to proliferate, with only rare cells adopting an enlarged phenotype (lower panels).
We previously established that the Ras-Raf-MEK signaling pathway activates p16INK4a expression via the Ets2 transcription factor, whereas Ets1 may activate p16INK4a in senescent HDFs (27). Pools of Q34 HDFs were therefore infected with recombinant retroviruses encoding Ets1 or Ets2. After selection in puromycin for 7 days, the levels of endogenous p16INK4a were again analyzed by immunoblotting, with MEK as a loading control. As illustrated in Fig. 4B, ectopic expression of either Ets1 or Ets2 resulted in a significant increase in p16INK4a expression. However, whereas Ets1 and Ets2 were both able to cause a senescence-like growth arrest in normal HDFs, the Q34 cells continued to proliferate (Fig. 5B). As we have no reason to suspect Q34 cells carry additional genetic defects (see below), these data imply that the ability of Ras, Ets1, and Ets2 to induce growth arrest in HDFs is solely dependent on p16INK4a.
Ras-expressing Q34 cells acquire anchorage independence.
Given the precedent in MEFs, where an inability to undergo Ras-mediated arrest equates with sensitivity to Ras-mediated transformation, we next asked whether the Q34 cells expressing Ras, Ets1, or Ets2 showed any phenotypic manifestations of transformation, such as an ability to form colonies in soft agar. Equal numbers of Q34 and Hs68 cells infected with the relevant retroviruses and empty vector controls were seeded in medium containing 0.2% agarose (see Materials and Methods), and colonies were allowed to develop over a period of 3 to 4 weeks. As illustrated in Table 1, a significant proportion of the Ras-expressing Q34 cells formed multicellular colonies, as determined by direct counting of representative fields. In contrast, few (if any) multicellular colonies were detected with Hs68 cells, with or without Ras (Table 1). Ets1 and Ets2 did not give rise to anchorage-independent colonies in either cell strain (data not shown).
TABLE 1.
Ability of Q34 cells to form anchorage-independent coloniesa
| Cell strain and/or virus | % Cells forming agar colonies |
|---|---|
| Q34 + pBABE | 4 |
| Q34 + RAS | 29 |
| Q34 + hTERT + pBABE | 1 |
| Q34 + hTERT + RAS | 26 |
| Hs68 + pBABE | 1 |
| Hs68 + RAS | 0 |
| Hs68 + hTERT + pBABE | 0 |
| Hs68 + hTERT + RAS | 0 |
Cells were suspended in medium containing 0.2% agarose, and colonies were counted after 21 days. Multiple fields of >100 cells were observed microscopically, and the number of multicellular colonies was estimated as a percentage of the total number of cells visualized.
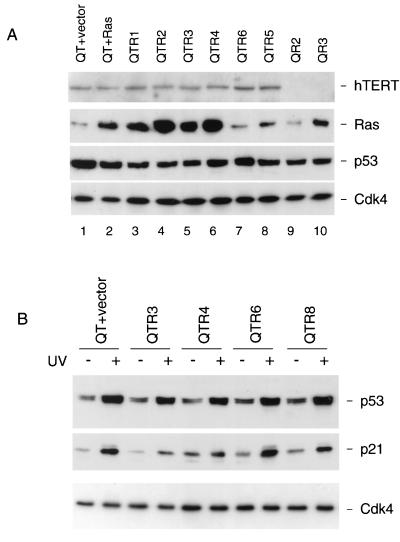
It is important to stress that the colonies were relatively small and could only be observed by microscopy. However, the frequency with which they occurred in repeat experiments suggested that they did not arise by clonal expansion of spontaneously mutated cells. To try to address this possibility, multiple colonies were recovered from the agarose and were propagated as monolayer cultures for further analysis. In general, these colonies had a very limited proliferative potential and after a small but variable number of PDs developed the phenotypic characteristics of M1 senescence. Of 60 colonies picked, 21 were reestablished in culture but only 8 of these grew enough to fill a 30-mm-diameter culture dish. This excludes the possibility that anchorage independence reflected spontaneous immortalization, such as by reactivation of telomerase. In two of the colonies which grew enough to allow biochemical analyses (QR2 and QR3), we found no evidence that they expressed hTERT, the catalytic component of telomerase (Fig. 6A). Conversely, deliberate expression of hTERT did not increase the frequency with which the agar colonies developed in the Ras-expressing Q34 cells and did not enable Hs68 control cells to achieve anchorage independence (Table 1). It did, however, facilitate the continued propagation of the agar colonies and a number of representative examples (QTR1, QTR2, QTR3, etc.) were recovered for further analyses. All the colonies tested grew vigorously in monolayer culture and expressed similar levels of hTERT and p53. Interestingly, the levels of Ras in the anchorage-independent clones were quite variable regardless of whether or not the cells were expressing ectopic hTERT (Fig. 6A). As a first approximation, the larger agar colonies expressed higher levels of Ras. We have not been able to determine whether any of the anchorage-independent colonies can form tumors in nude mice, because the donor from which the Q34 cells were obtained expressly prohibited their use in animal experimentation.
FIG. 6.
Characterization of anchorage-independent Q34 cells. (A) Selected anchorage-independent colonies of Q34 cells expressing Ras alone (QR2 and QR3) or hTERT and Ras (QTR1 to QTR6) were analyzed for expression of key proteins. For comparison, similar analyses were conducted on pools of Q34 cells expressing hTERT (QT) and either Ras or empty vector. Samples (20 μg) of lysate were fractionated by SDS-PAGE in 8 and 12% gels and were immunoblotted for hTERT, Ras, p53, and Cdk4. (B) The indicated cell pools and agar colonies were exposed to UV irradiation (15 J/m2), and samples (20 μg) of total protein were subjected to SDS-PAGE and were immunoblotted for p53, p21CIP1, and Cdk4.
p53 and genome status of anchorage-independent colonies.
All of the colonies expressed roughly the same amount of p53 as the original Q34 cell pools. If anything, cells with higher levels of Ras tended to have lower levels of p53 (Fig. 6A). To confirm that the p53 pathway remained operational, we exposed some of the anchorage-independent colonies to UV irradiation. In the various cell pools and colonies tested there was a robust accumulation of p53 following irradiation (Fig. 6B). Similar effects were noted following addition of adriamycin (data not shown). Moreover, UV exposure also resulted in the upregulation of p21CIP1 (Fig. 6B) and cell cycle arrest (data not shown), implying that the induced p53 was functional. We therefore have no reason to suspect any defect in the ARF-p53 pathway in these cells.
We also checked that the Q34 cells remained essentially diploid during these manipulations. Total genomic DNA was prepared from the QTR agar colonies and from pools of Q34 cells before and after the introduction of hTERT. The DNA was then analyzed by comparative genome hybridization (CGH) by using gridded arrays of cloned DNA segments representing the entire human genome (29). There were no significant gains or losses of chromosomal DNA discernible by this approach (data not shown). Thus, at the levels of analyses conducted to date, we would conclude that the Q34 cells achieve anchorage independence as a result of alterations in only two cellular genes, p16INK4a and Ras, but that their continued proliferation requires provision of hTERT.
DISCUSSION
Two previous cases of biallelic mutation of CDKN2A have been reported, occurring as homozygotes in a Dutch familial melanoma kindred with a common founder mutation (7), but to our knowledge the subject described in this study is the first example of a compound heterozygote carrying distinct germ line alterations in each allele of CDKN2A. One allele carries the T158C alteration, resulting in the Met53Thr substitution in p16INK4a but leaving the ARF sequence unchanged (Fig. 1B). This branch of the family has one additional case of melanoma as well as three carcinomas at other sites (Fig. 1A) whose relationship to the mutation remains unclear. By several criteria it is evident that the Met53Thr alteration, which has been recorded once before in a sporadic leukemia (8), causes severe disruption of p16INK4a function (Fig. 2). We previously showed that the Met53Ile variant, which is a more common germ line mutation among United Kingdom melanoma families, also represents a nonfunctional allele (34), consistent with the fact that Met53 is on the surface of p16INK4a and at the interface with Cdk4 and Cdk6 (4, 36).
The situation with the second mutation is less clear-cut because it causes amino acid substitutions in both p16INK4a and p14ARF (Fig. 1B). The Asp108Asn alteration in p16INK4a is relatively conservative and only partially impairs the function of the protein (Fig. 2). Although we have no direct evidence for its inheritance in this branch of the family, it has previously been recorded in a French melanoma kindred (45). Asp108 is involved in stabilizing the hairpin loop between adjacent ankyrin-type repeats, and the mutation is therefore predicted to have an effect on the general structure of the protein (22). In contrast, as most studies conclude that the known functions of p14ARF are attributable to the 64 amino acids encoded by exon 1β, including the ability to localize in the nucleolus, to bind to MDM2 and p53, to stabilize p53, to induce a cell cycle arrest, and to prevent the MDM2-mediated ubiquitination of p53 in vitro (17, 18, 23, 46, 53), it is unlikely that the Arg122Gln mutation would affect any of these functions. The data depicted in Fig. 3 support this conclusion, but there may well be subtle differences or other activities associated with ARF that we are as yet unable to measure. Nevertheless, on the basis of the available evidence, we would conclude that both of the CDKN2A variants in the patient specifically affect p16INK4a function.
Similar analyses of the Dutch founder mutation were recently described (3). In this case, the 19-base-pair deletion in exon 2 results in a frameshift that creates two distinctive fusion proteins. Neither protein has a sufficient number of ankyrin repeats for p16INK4a function, but the product that initiates in exon 1β retains most of the known properties of ARF (3). Importantly, primary dermal fibroblasts (designated Leiden HDFs) from a homozygous carrier of this mutation were also found to be resistant to Ras-induced senescence (3), consistent with the conclusions drawn in the present work. However, the Q34 HDFs that we describe here have the advantage that the missense mutations are more subtle, enabling us to detect the mutant forms of p16INK4a on Western blots, and the cells have a longer proliferative life span than Leiden HDFs (unpublished data), enabling us to conduct experiments without the need for hTERT.
Thus, levels of oncogenic Ras that were sufficient to upregulate p16INK4a caused a senescence-like arrest in normal HDFs but not in Q34 cells (Fig. 4 and 5). Likewise, the Ets1 and Ets2 transcription factors, which are known to activate the p16INK4a promoter, induced senescence in normal HDFs but not in Q34 cells. Importantly, the Q34 cells that resumed proliferation despite expressing oncogenic Ras were able to form anchorage-independent colonies in soft agar. However, the agar colonies were relatively small, and when replated in monolayer culture they showed limited proliferative potential, culminating in M1-like replicative senescence. Provision of hTERT did not increase the frequency of agar colony formation (Table 1) but did allow the propagation and analysis of the anchorage-independent clones. In view of concerns that hTERT might cause transient genetic instability (48, 49), we confirmed that the anchorage-independent colonies were diploid, as judged by CGH on gridded DNA arrays, and that the p53 pathway remained responsive to UV irradiation (Fig. 6B).
In summary, the demonstration that Q34 cells fail to arrest in response to oncogenic Ras despite retaining apparently normal ARF and p53 functions reinforces our contention that p16INK4a is the principal effector of premature senescence in HDFs. A very different situation prevails in MEFs, where Ras-induced senescence is indisputably linked to the upregulation of ARF and p53. Thus, MEFs from mice that have a targeted disruption of Ink4a still show limited proliferative potential and undergo a senescence-like arrest when challenged with Ras (14, 41). Mechanistically, there is at least one recognized difference between HDFs and MEFs. Whereas oncogenic Ras will activate the expression of mouse p19ARF, it seems to have little, if any, impact on human p14ARF (3, 6, 51). At present it is not clear whether these differences reflect biases in the regulation of the INK4a/ARF locus in different cell lineages (31, 47) or in different species, and further work will be needed to distinguish between these possibilities. However, the results we have obtained by using cells from melanoma patients reinforce the importance of p16INK4a as a tumor suppressor in human cancers and urge for caution in interpreting the mouse models.
Acknowledgments
We thank our patients for their gift of tissues in the interest of the furtherance of knowledge about melanoma. We also thank Neil McDonald for advice on the structural consequences of the p16INK4a mutations, Richard Seraves and Donna Albertson for CGH analyses, and Caroline Hill, Alan Storey, and Tim Bishop for helpful comments on the manuscript.
REFERENCES
- 1.Ashcroft, M., and K. H. Vousden. 1999. Regulation of p53 stability. Oncogene 18:7637-7643. [DOI] [PubMed] [Google Scholar]
- 2.Bates, S., A. C. Phillips, P. A. Clark, F. Stott, G. Peters, R. L. Ludwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 3.Brookes, S., J. Rowe, M. Ruas, S. Llanos, P. A. Clark, M. Lomax, M. C. James, R. Vatcheva, S. Bates, K. H. Vousden, D. Parry, N. Gruis, N. Smit, W. Bergman, and G. Peters. 2002. INK4a-deficient human diploid fibroblasts are resistant to RAS-induced senescence. EMBO J. 21:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brotherton, D. H., V. Dhanaraj, S. Wick, B. L., P. J. Domaille, E. Volyanik, X. Xu, E. Parisini, B. O. Smith, S. J. Archer, M. Serrano, S. L. Brenner, T. L. Blundell, and E. D. Laue. 1998. Crystal structure of the complex of the cyclin D-dependent kinase Cdk6 bound to the cell-cycle inhibitor p19INK4d. Nature 395:244-250. [DOI] [PubMed] [Google Scholar]
- 5.Drayton, S., and G. Peters. 2002. Immortalisation and transformation revisited. Curr. Opin. Genet. Dev. 12:98-104. [DOI] [PubMed] [Google Scholar]
- 6.Ferbeyre, G., E. de Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 14:2015-2027. [PMC free article] [PubMed] [Google Scholar]
- 7.Gruis, N. A., P. A. van der Velden, L. A. Sandkuiji, D. E. Prins, J. Weaver-Feldhaus, A. Kamb, W. Bergman, and R. R. Frants. 1995. Homozygotes for CDKN2 (p16) germline mutations in Dutch familial melanoma kindreds. Nat. Genet. 10:351-353. [DOI] [PubMed] [Google Scholar]
- 8.Haidar, M. A., X.-B. Cao, T. Manshouri, L. L. Chan, A. Glassman, H. M. Kantarjian, M. J. Keating, M. S. Beran, and M. Albitar. 1995. p16INK4A and p15INK4B gene deletions in primary leukemias. Blood. 86:311-315. [PubMed] [Google Scholar]
- 9.Hara, E., R. Smith, D. Parry, H. Tahara, S. Stone, and G. Peters. 1996. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol. Cell. Biol. 16:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harland, M., E. A. Holland, P. Ghiorzo, M. Mantelli, G. Bianchi-Scarra, A. M. Goldstein, M. A. Tucker, B. A. J. Ponder, G. J. Mann, D. T. Bishop, and J. Newton Bishop. 2000. Mutation screening of the CDKN2A promoter in melanoma families. Genes Chrom. Cancer. 28:45-57. [PubMed] [Google Scholar]
- 11.Harland, M., R. Meloni, N. Gruis, E. Pinney, S. Brookes, N. K. Spurr, A.-M. Frischauf, V. Bataille, G. Peters, J. Cuzick, P. Selby, D. T. Bishop, and J. Newton Bishop. 1997. Germline mutations of the CDKN2 gene in UK melanoma families. Hum. Mol. Genet. 6:2061-2067. [DOI] [PubMed] [Google Scholar]
- 12.Hayflick, L., and P. S. Moorhead. 1961. The serial cultivation of human diploid cell strains. Exp. Cell Res. 25:585-621. [DOI] [PubMed] [Google Scholar]
- 13.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 91:649-659. [DOI] [PubMed] [Google Scholar]
- 14.Krimpenfort, P., K. C. Quon, W. J. Mool, A. Loonstra, and A. Berns. 2001. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 413:83-86. [DOI] [PubMed] [Google Scholar]
- 15.Kubbutat, M. H. G., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 287:299-303. [DOI] [PubMed] [Google Scholar]
- 16.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to consitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llanos, S., P. A. Clark, J. Rowe, and G. Peters. 2001. Stabilisation of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat. Cell Biol. 3:445-452. [DOI] [PubMed] [Google Scholar]
- 18.Lohrum, M. A. E., M. Ashcroft, M. H. G. Kubbutat, and K. H. Vousden. 2000. Contribution of two independent MDM2-binding domains in p14ARF to p53 stabilization. Curr. Biol. 10:539-542. [DOI] [PubMed] [Google Scholar]
- 19.Lundberg, A. S., W. C. Hahn, P. Gupta, and R. A. Weinberg. 2000. Genes involved in senescence and immortalization. Curr. Opin. Cell Biol. 12:705-709. [DOI] [PubMed] [Google Scholar]
- 20.MacGeoch, C., J. A. Newton Bishop, V. Bataille, D. T. Bishop, A.-M. Frischauf, R. Meloni, J. Cuzick, E. Pinney, and N. K. Spurr. 1994. Genetic heterogeneity in familial malignant melanoma. Hum. Mol. Genet. 3:2195-2200. [DOI] [PubMed] [Google Scholar]
- 21.McConnell, B. B., M. Starborg, S. Brookes, and G. Peters. 1998. Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr. Biol. 8:351-354. [DOI] [PubMed] [Google Scholar]
- 22.McDonald, N. Q., and G. Peters. 1998. Ankyrin for clues about the function of p16INK4a. Nat. Struct. Biol. 5:85-88. [DOI] [PubMed] [Google Scholar]
- 23.Midgley, C. A., J. M. P. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARFpeptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 24.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newton Bishop, J. A., M. Harland, D. C. Bennett, V. Bataille, A. M. Goldstein, M. A. Tucker, B. A. J. Ponder, J. Cuzick, P. Selby, and D. T. Bishop. 1999. Mutation testing in melanoma families: INK4A, CDK4 and INK4D. Br. J. Cancer 80:295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohtani, N., Z. Zebedee, T. J. G. Huot, J. A. Stinson, M. Sugimoto, Y. Ohashi, A. D. Sharrocks, G. Peters, and E. Hara. 2001. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409:1067-1070. [DOI] [PubMed] [Google Scholar]
- 28.Pear, W. S., G. P. Nolan, M. L. Scott, and D. Baltimore. 1993. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA 90:8392-8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinkel, D., R. Segraves, D. Sudar, D. Clark, I. Poole, D. Kowbel, C. Collins, W.-L. Kuo, C. Chen, Y. Zhai, S. H. Dairkee, B. Ljung, J. W. Gray, and D. G. Albertson. 1998. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 20:207-211. [DOI] [PubMed] [Google Scholar]
- 30.Randerson-Moor, J. A., M. Harland, S. Williams, D. Cuthbert-Heavens, E. Sheridan, J. Aveyard, K. Sibley, L. Whitaker, M. Knowles, J. Newton Bishop, and D. T. Bishop. 2001. A germline deletion of p14ARF but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum. Mol. Genet. 10:55-62. [DOI] [PubMed] [Google Scholar]
- 31.Randle, D. H., F. Zindy, C. J. Sherr, and M. F. Roussel. 2001. Differential effects of p19Arf and p16Ink4a loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc. Natl. Acad. Sci. USA 17:9654-9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizos, H., S. Puig, C. Badenas, J. Malvehy, A. P. Darmanian, L. Jiménez, M. Milà, and R. F. Kefford. 2001. A melanoma-associated germline mutation in exon 1β inactivates p14ARF. Oncogene. 20:5543-5547. [DOI] [PubMed] [Google Scholar]
- 33.Robertson, K. D., and P. A. Jones. 1998. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down regulated by wild-type p53. Mol. Cell. Biol. 18:6457-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruas, M., S. Brookes, N. Q. McDonald, and G. Peters. 1999. Functional evaluation of tumour-specific variants of p16INK4a/CDKN2A: correlation with protein structure information. Oncogene 18:5423-5434. [DOI] [PubMed] [Google Scholar]
- 35.Ruas, M., and G. Peters. 1998. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim. Biophys. Acta 1378:115-177. [DOI] [PubMed] [Google Scholar]
- 36.Russo, A. A., L. Tong, J.-O. Lee, P. D. Jeffrey, and N. P. Pavletich. 1998. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature 395:237-243. [DOI] [PubMed] [Google Scholar]
- 37.Sedivy, J. M. 1998. Can ends justify the means? Telomeres and the mechanisms of replicative senescence and immortalization in mammalian cells. Proc. Natl. Acad. Sci. USA 95:9078-9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serrano, M., G. J. Hannon, and D. Beach. 1993. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366:704-707. [DOI] [PubMed] [Google Scholar]
- 39.Serrano, M., H.-W. Lee, L. Chin, C. Cordon-Cardo, D. Beach, and R. A. DePinho. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85:27-37. [DOI] [PubMed] [Google Scholar]
- 40.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 41.Sharpless, N. E., N. Bardeesy, K.-H. Lee, D. Carrasco, D. H. Castrillon, A. J. Aguirre, E. A. Wu, J. W. Horner, and R. A. DePinho. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413:86-91. [DOI] [PubMed] [Google Scholar]
- 42.Sharpless, N. E., and R. A. DePinho. 1999. The INK4A/ARF locus and its two gene products. Curr. Opin. Genet. Dev. 9:22-30. [DOI] [PubMed] [Google Scholar]
- 43.Sherr, C. J. 2001. The INK4a/ARF network in tumour suppression. Nat. Rev. 2:731-737. [DOI] [PubMed] [Google Scholar]
- 44.Sherr, C. J., and R. A. DePinho. 2000. Cellular senescence: mitotic clock or culture shock? Cell 102:407-410. [DOI] [PubMed] [Google Scholar]
- 45.Soufir, N., M.-F. Avril, A. Chompret, F. Demenais, J. Bombled, A. Spatz, D. Stoppa-Lyonnet, T. F. F. M. S. Group, J. Bénard, and B. Bressac-de Paillerets. 1998. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. Hum. Mol. Genet. 7:209-216. [DOI] [PubMed] [Google Scholar]
- 46.Stott, F. J., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sviderskaya, E. V., S. P. Hill, T. J. Evans-Whipp, L. Chin, S. J. Orlow, D. J. Easty, C. C. Cheong, D. Beach, R. A. DePinho, and D. C. Bennett. 2002. p16Ink4a in melanocyte senescence and differentiation. J. Natl. Cancer Inst. 94:446-454. [DOI] [PubMed] [Google Scholar]
- 48.Vaziri, H., J. A. Squire, T. K. Pandita, G. Bradley, R. M. Kuba, H. Zhang, S. Gulyas, R. P. Hill, G. P. Nolan, and S. Benchimol. 1999. Analysis of genomic integrity and p53-dependent G1 checkpoint in telomerase-induced extended-life-span human fibroblasts. Mol. Cell. Biol. 19:2373-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, J., G. J. Hannon, and D. H. Beach. 2000. Risky immortalization by telomerase. Nature 405:755-756. [DOI] [PubMed] [Google Scholar]
- 50.Wei, S., W. Wei, and J. M. Sedivy. 1999. Expression of catalytically active telomerase does not prevent premature senescence caused by overexpression of oncogenic Ha-Ras in normal human fibroblasts. Cancer Res. 59:1539-1543. [PubMed] [Google Scholar]
- 51.Wei, W., R. M. Hemmer, and J. M. Sedivy. 2001. The role of p14ARF in replicative and induced senescence of human fibroblasts. Mol. Cell. Biol. 21:6748-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright, W., and J. W. Shay. 2000. Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nat. Med. 6:849-851. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, J., D. Woods, M. McMahon, and J. M. Bishop. 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 12:2997-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]