Abstract
In CBFβ-SMMHC, core binding factor beta (CBFβ) is fused to the α-helical rod domain of smooth muscle myosin heavy chain (SMMHC). We generated Ba/F3 hematopoietic cells expressing a CBFβ-SMMHC variant lacking 28 amino acids homologous to the assembly competence domain (ACD) required for multimerization of skeletal muscle myosin. CBFβ-SMMHC(ΔACD) multimerized less effectively than either wild-type protein or a variant lacking a different 28-residue segment. In contrast to the control proteins, the ΔACD mutant did not inhibit CBF DNA binding, AML1-mediated reporter activation, or G1 to S cell cycle progression, the last being dependent upon activation of CBF-regulated genes. We also linked the CBFβ domain to 149 or 83 C-terminal CBFβ-SMMHC residues, retaining 86 or 20 amino acids N-terminal to the ACD. CBFβ-SMMHC(149C) multimerized and slowed Ba/F3 proliferation, whereas CBFβ-SMMHC(83C) did not. The majority of CBFβ-SMMHC and CBFβ-SMMHC(149C) was detected in the nucleus, whereas the ΔACD and 83C variants were predominantly cytoplasmic, indicating that multimerization facilitates nuclear retention of CBFβ-SMMHC. When linked to the simian virus 40 nuclear localization signal (NLS), a significant fraction of CBFβ-SMMHC(ΔACD) entered the nucleus but only mildly inhibited CBF activities. As NLS-CBFβ-SMMHC(83C) remained cytoplasmic, we directed the ACD to CBF target genes by linking it to the AML1 DNA binding domain or to full-length AML1. These AML1-ACD fusion proteins did not affect Ba/F3 proliferation, in contrast to AML1-ETO, which markedly slowed G1 to S progression dependent upon the integrity of its DNA-binding domain. Thus, the ACD facilitates inhibition of CBF by mediating multimerization of CBFβ-SMMHC in the nucleus. Therapeutics targeting the ACD may be effective in acute myeloid leukemia cases associated with CBFβ-SMMHC expression.
The core binding factor (CBF) transcription factors contain one of three CBFα subunits, CBFα1/AML3/RUNX2, CBFα2/AML1/RUNX1, or CBFα3/AML2/RUNX3, and a common CBFβ subunit (3, 14, 21, 35, 50). The CBFα subunits contact the consensus DNA binding site, 5′-(Pu)ACCPuCA-3′, via their 127-amino-acid Runt homology domains (3, 30). CBFβ does not bind DNA but increases the DNA affinity of the CBFα subunits via allosteric interaction with the Runt domain (36, 47, 50). Amino acids 1 to 137 of the 182-residue CBFβ are sufficient for increasing the DNA affinity of CBFα subunits (16, 19).
CBFβ is widely expressed, whereas AML1 is largely restricted to hematopoietic cells (43, 50). Mice lacking AML1 or CBFβ do not develop definitive hematopoiesis (33, 37, 42, 51). A role for AML1 during maturation of pluripotent stem cells along the lymphoid and myeloid lineages has been inferred from its ability to transactivate promoters of lineage-restricted genes (34, 41, 46, 58). AML1 possesses only weak intrinsic transactivating potential but cooperates with additional transcription factors to activate genes in hematopoietic cells (7, 40, 46). AML1 is found in the cell nucleus, whereas the large majority of CBFβ is cytoplasmic, reflecting its expression in excess of CBFα and its affinity for the actin cytoskeleton (26, 48).
Chromosomal and mutational abnormalities affecting the genes encoding AML1 and CBFβ are common in acute leukemia cases (14). CBFβ-SMMHC, encoded by inv(16) (p13;q22) or t(16;16) (p13;q22) in 8% of acute myeloid leukemias, inhibits CBF activities via its interactions with AML1 (9, 10, 22, 23). In contrast, AML1-ETO, another CBF oncoprotein, directly represses AML1 target genes (31).
The phenotypes of AML1-ETO and CBFβ-SMMHC knock-in mice are similar to those lacking AML1 or CBFβ, indicating that these oncoproteins interfere with CBF activities in vivo (11, 38, 57). A direct role for CBFβ-SMMHC in the formation of human acute myeloid leukemias can be inferred from the formation of acute myeloid leukemias in chimeric mice exposed to ethylnitrosourea (12).
In addition to stimulating differentiation, CBF is require for the G1 to S transition in 32D cl3 myeloid and Ba/F3 lymphoid cells (9, 25). Deletion of 11 N-terminal CBFβ residues required for interaction with AML1 prevents CBFβ-SMMHC from slowing Ba/F3 proliferation, and exogenous AML1 accelerates G1 progression via transactivation (4, 10, 25, 45). Exogenous AML1, cdk4, cyclin D2, and c-Myc prevent CBFβ-SMMHC from slowing Ba/F3 proliferation (5, 25). During leukemogenesis, genetic alterations which accelerate G1 may potentiate expression of CBFβ-SMMHC, enabling increased blockade of differentiation. Consistent with this model, deletion of the overlapping p16INK4A and p19ARF genes or expression of E7 cooperates with CBFβ-SMMHC or TEL-AML1 to induce acute leukemia in mice (6, 56).
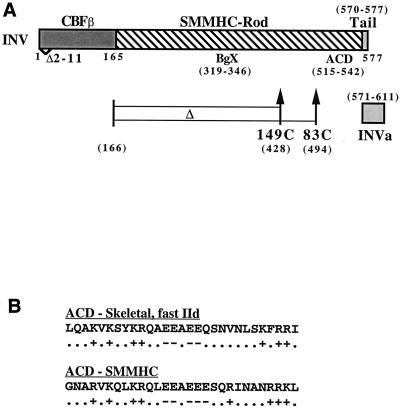
In CBFβ-SMMHC, the majority of CBFβ is linked to the rod domain of smooth muscle myosin heavy chain (SMMHC) (Fig. 1A). The SMMHC rod domain is α-helical and consists of 28 related amino acid regions. Each of these regions contains four heptads in which the a and d residues are often hydrophobic. As a result, the rod domain has a hydrophobic face which mediates dimerization. The other face of the α-helix has alternating positively and negatively charged zones and mediates staggered multimerization (55). Alternative RNA splicing leads to the expression of two CBFβ-SMMHC isoforms, which have either an 8- or a 42-amino-acid nonhelical, C-terminal tailpiece (32).
FIG. 1.
Diagrams of CBFβ-SMMHC (INV), the INV variants studied herein, and the ACD. (A) INV contains CBFβ residues 1 to 165, an α-helical SMMHC rod domain, and an 8-residue nonhelical tailpiece. An alternatively spliced form of INV, INVa, contains a 42-residue tailpiece. Deletion of residues 2 to 11 within the CBFβ segment (Δ2-11) prevents interaction with CBFα subunits. BgX denotes the randomly selected 28-residue segment deleted in INV(ΔBgX), and ACD denotes the amino acid segment homologous to the assembly competence domain of skeletal myosin that is deleted in INV(ΔACD) and INVa(ΔACD). INV(149C) lacks residues 166 to 428, and INV(83C) lacks residues 166 to 494. (B) The primary amino acid sequences of the human fast IId skeletal myosin ACD and of the related segment in human SMMHC are shown. Locations of negatively charged amino acids (D and E, −) and positively charged amino acids (K and R, +) are also shown.
Deletion analysis of the rod domain of skeletal muscle myosin identified a 29-amino-acid segment, designated the assembly competence domain (ACD), that was required for multimerization and capable of conferring the ability to multimerize on a 391-residue rod domain segment which was otherwise incapable of multimerization (44). These authors noted that multiple rat and human sarcomeric myosins contain a segment near their C termini that is virtually identical to the human fast IId sarcomeric myosin heavy chain ACD and that nonmuscle and smooth muscle myosins from several species, including Drosophila, Xenopus, chicken, rabbit, rat, and human, contain segments in a similar location which are highly related to the sarcomeric myosin ACD and which are virtually identical to each other. The amino acid sequences of the human sarcomeric fast IId myosin heavy chain and SMMHC ACDs are compared in Fig. 1B. Deletion analysis of the sarcomeric myosin rod domain also demonstrated that a 194-residue α-helical segment containing the ACD multimerized in vitro, whereas a 145-amino-acid segment did not (44). Thus, even with the ACD intact, a minimal length of α-helical coiled coil is required for multimerization of the myosin rod.
CBFβ-SMMHC multimers compete with CBFβ for interaction with AML1, potentially sequestering AML1 away from chromatin (9, 23). As AML1:CBFβ-SMMHC monomeric and multimeric complexes retain DNA binding activity, CBFβ-SMMHC may also be capable of directly repressing CBF-regulated genes. To ascertain whether multimerization is essential for inhibition of CBF activities, we expressed several CBFβ-SMMHC variants from the zinc-responsive metallothionein (MT) promoter in Ba/F3 cells. Herein we refer to the shorter isoform of CBFβ-SMMHC as INV and to the splice variant with a longer C-terminal, nonhelical tail as INVa. Deletion of the ACD homology domain from INV or INVa inhibited multimerization. In contrast to INV, INV(ΔACD) and INVa(ΔACD) were predominantly cytoplasmic and were defective for inhibition of CBF-mediated transactivation of a reporter gene and of endogenous genes required for G1 to S cell cycle progression.
We also expressed internal deletion variants linking the CBFβ domain to either 149 or 83 C-terminal SMMHC residues. The former multimerized, entered the nucleus, and slowed proliferation, whereas the latter did not. When INV was directed to the nucleus with a simian virus 40 nuclear localization signal (NLS), deletion of the ACD still largely prevented inhibition of CBF activities. As the NLS did not localize CBFβ-ACD fusion proteins to the nucleus, we linked the ACD to AML1 or to the AML1 DNA binding domain. In contrast to AML1-ETO, AML1-ACD proteins did not inhibit CBF-regulated genes. These findings indicate that the ACD facilitates inhibition of CBF by enabling CBFβ-SMMHC multimerization in the nucleus.
MATERIALS AND METHODS
Cell culture, transfection, and proliferation assays.
Ba/F3 cells (39) were maintained in RPMI 1640 medium with 10% heat-inactivated fetal bovine serum, 1 ng of IL-3 per ml (R & D Systems), and penicillin-streptomycin. pMT-INV-3 and pMT-INVa-2 cells were described and are designated pMT-INV and pMT-INVa herein (9, 10). pMT-INV(βΔ2-11) and pMT-CBFβ(165) cells were rederived from DNA constructs described previously (10). A total of 15 μg of each DNA construct was linearized by ScaI digestion and electroporated into 5 × 106 Ba/F3 cells in ice-cold phosphate-buffered saline at settings of 250 mV and 950 μF. Stable transfectants were selected by limiting dilution in 96-well dishes in the presence of 1.2 mg total of G418 per ml. To induce the MT promoter, zinc chloride was added at 100 μM.
Transient transfection was carried out with 293T cells, which were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum; 8 × 104 cells were seeded in 30-mm dishes. The next day, 800 to 900 ng of plasmid DNA and 3 μl of Lipofectamine 2000 (Gibco-BRL) per sample were employed per the manufacturer's instructions. Luciferase assays were carried out 2 days later, as described previously (10). Viable cell counts were determined by enumerating cells which excluded trypan blue dye with a hemacytometer. For cell cycle analysis, cells in log phase were pulsed for 30 min with 30 μM bromodeoxyuridine and then analyzed as described previously (9).
Plasmid construction.
pMT-INV(ΔACD) was constructed by ligating an oligonucleotide lacking the ACD coding sequences (amino acids 515 to 542) between a PstI site 57 bp upstream of the ACD coding segment and a PstI site located 3 bp downstream. In this construct, only the N-terminal glycine of the 29-amino-acid ACD was retained (Fig. 1). pMT-INVa(ΔACD) was prepared by transferring the ACD deletion with unique BglII and NcoI sites surrounding the ACD within both the INV and INVa cDNAs. pMT-INV(ΔBgX) was constructed by inserting an oligonucleotide between a BglII site located at bp 928 of the INV cDNA and an XhoI site located at bp 1028, leading to the deletion of a segment encoding amino acids 319 to 346.
pMT-INV(βΔ2-11ΔACD) and pMT-INV(βΔ2-11ΔBgX) were constructed by transferring the ACD or BgX deletion, respectively, into pMT-INV(βΔ2-11). INV(149C) was prepared by ligating an oligonucleotide between a StuI site located at bp 444 of the CBFβ domain and an XhoI site located at bp 1287 of the INV cDNA. This oligonucleotide encodes all of the CBFβ amino acids downstream of the StuI site and deletes a DNA segment encoding residues 166 to 428 of the INV cDNA. pMT-INV(83C) was constructed by ligating an oligonucleotide between the StuI site and a PstI site located at bp 1485. This oligonucleotide encodes all of the CBFβ amino acids present in INV and downstream of the StuI site and deletes a DNA segment encoding residues 166 to 494 of the INV cDNA.
An oligonucleotide encoding the simian virus 40 nuclear localization signal (NLS) was linked upstream of the cDNA encoding INV and its variants as described previously (10). DNA segments encoding either 83 C-terminal INV residues (83C) or 121 C-terminal INV(ΔACD) residues (149CΔACD) were linked to the human AML1 DNA binding domain (DBD) with a linker oligonucleotide joining either a SmaI site (for AML1-DBDa) or a BamHI site (for AML1-DBDb) to a PstI site in the INV cDNA or to an XhoI site in the INV(ΔACD) cDNA. The 83C and 149CΔACD coding segments were also used to replace the estrogen receptor (ER) segment in AML1-ER, taking advantage of an MluI site that we had positioned just downstream of the AML1B cDNA segment (25), to generate AML1-83C and AML1-149ΔACD.
pMT-AML1-ETO was generated by transferring the AML1-ETO cDNA from pCMV-AML1-ETO (kindly provided by S. Hiebert) into pMTCB6. pMT-AML1ETOmDBD was generated by ligating the oligonucleotide obtained by annealing 5′-AGCTCCGGTACCCCAGAT-3′ and 5′-AGCTATCTGGGGTACCGG-3′ into the HindIII site present in the AML1 DNA binding domain of AML1-ETO. Each construct prepared with a synthetic oligonucleotide was sequenced to confirm that the inserted base pairs were those expected. p(CBF)4TKLUC, containing four CBF-binding sites from the myeloperoxidase gene, pCMV-AML1B, and pCMV-INV have been described (10). Additional cytomegalovirus (CMV) expression vectors were constructed by replacing the INV cDNA with the respective INV, NLS-INV, or AML1-INV variant cDNA.
Western blotting, indirect immunofluorescence, and gel shift assay.
Total cellular extracts corresponding to 106 cells were subjected to Western blotting with rabbit anti-CBFβ antiserum and mouse antiactin antibody, as described previously (25). Antisera specific for AML1B residues 59 to 77 and the INV C-terminal tailpiece were kindly provided by H. Drabkin and R. Adelstein, respectively. Filters were stripped between antibodies as described previously (9).
For analysis of nuclear versus cytoplasmic protein expression, cells exposed to zinc overnight were washed twice with ice-cold phosphate-buffered saline and resuspended on ice in a buffer containing 50 mM KCl, 10 mM HEPES (pH 6.5), 2 mM MgCl2, 0.5 mM dithiothreitol, 2 mM benzamidine, 0.5 mM spermidine, and 0.4 mM phenylmethylsulfonyl fluoride plus 10 μg of leupeptin, 1 μg of pepstatin A, 1 μg of antipain, 1 μg of chymostatin, 2 μg of soybean trypsin inhibitor, and 10 μg of aprotinin per ml. After 5 min, NP-40 was added to 0.1% (vol/vol). After an additional 5 min, the released nuclei were pelleted at 850 × g for 5 min at 4°C. The supernatants containing the cytoplasmic fractions were precipitated with 4 volumes of acetone at −20°C for at least 30 min, centrifuged at 2,000 × g for 15 min, and resuspended in Laemmli sodium dodecyl sulfate sample buffer. The nuclear pellet was washed and then also resuspended in Laemmli sample buffer.
For the gel shift assay, nuclei were prepared with DR-A buffer (10 mM KCl, 10 mM HEPES [pH 7.9], 1 mM dithiothreitol, 1 mM MgCl2, and the above-listed protease inhibitors), and the nuclei were extracted with DR-C buffer (420 mM NaCl, 20 mM HEPES [pH 7.9], 25% glycerol, 1.5 mM MgCl2, and 0.2 mM EDTA) for 30 min at 4°C. The extract was then collected at 16,000 × g for 15 min, aliquoted, snap frozen in liquid nitrogen, and stored at −80°C. Gel shift assay with a 32-bp probe derived from the myeloperoxidase gene and containing a CBF-binding site and oligonucleotide competition with a 50-fold excess of a wild-type probe or of a probe carrying clustered point mutations in the CBF-binding site were carried out as described previously (9). Indirect immunofluorescence on cytospun Ba/F3 cell lines, with a CBFβ antiserum, was also carried out as described previously, with the addition of 0.2 μg of 4′,6′-diamidino-2-phenylindole (DAPI) per ml into the glycerol overlay (9). Cells were visualized by fluorescent microscopy at ×60.
Multimerization assay.
We employed a procedure similar to that used to assess sarcomeric myosin multimerization (44). From 40 × 106 to 50 × 106 Ba/F3 cells expressing INV or its variants were exposed to zinc for 2 days and then lysed by exposure on ice to 2 ml of 300 mM KCl-10 mM HEPES (pH 6.5)-2 mM MgCl2-0.5 mM dithiothreitol-0.1% NP-40, along with benzamidine, spermidine, phenylmethylsulfonyl fluoride, leupeptin, pepstatin, antipain, chymostatin, soybean trypsin inhibitor, and aprotinin as protease inhibitors. This salt concentration extracted the majority of INV from the cells.
To partially purify CBFβ-SMMHC and its variants, the cell lysates were heated to 100°C for 15 min and then clarified by microcentrifuge centrifugation at 16,000 × g for 5 min. Then 0.5-ml aliquots of the lysate were dialyzed overnight at 4°C, with 12,000- to 14,000-Da exclusion 0.25-in. (ca. 1 cm) tubing (Gibco-BRL), against 50, 100, 200, or 300 mM KCl, HEPES (pH 6.5), 2 mM MgCl2, phenylmethylsulfonyl fluoride, benzamidine, and spermidine. The clear dialysates were collected by vigorous washing of the bags and ultracentrifuged at 100,000 × g for 1 h in polyallomer tubes (11 by 34 mm) filled with the corresponding dialysis buffer (Beckman). The supernatants were collected, leaving a small amount above the pellets, concentrated by acetone precipitation, and resuspended in sodium dodecyl sulfate sample buffer. The residual supernatants were removed, and the pellets were solubilized by exposure to 600 mM KCl-10 mM HEPES (pH 7.5)-0.5 mM dithiothreitol overnight at room temperature. The dissolved pellets were then diluted 1:1 with 10 mM HEPES (pH 7.5)-0.5 mM dithiothreitol and resuspended in 2× sodium dodecyl sulfate sample buffer. Samples corresponding to equivalent cell numbers were then subjected to Western blotting with the CBFβ antiserum. Band intensities were quantified with NIH Image 1.62 software.
RESULTS
SMMHC has an assembly competence domain.
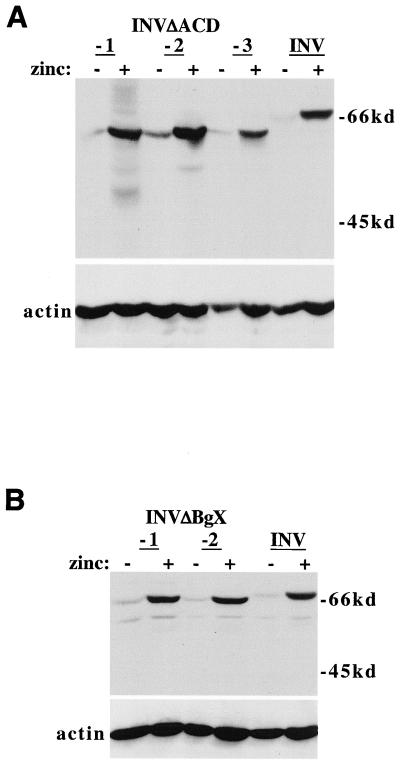
The ACD homology domain was deleted from the INV cDNA to generate INV(ΔACD); 28 residues of the ACD homology region were deleted, rather than all 29, to maintain the repeat structure of the rod domain. As a control, 28 residues further upstream were deleted from INV, between nearby BglII and XhoI sites, to generate INV(ΔBgX). The locations of the ACD and BgX segments within INV are shown in Fig. 1A. These cDNAs were introduced into Ba/F3 cells downstream of the zinc-responsive MT promoter, and Western blotting was employed to identify subclones which produced the corresponding proteins (Fig. 2).
FIG. 2.
Ba/F3 lines expressing INV(ΔACD) and INV(ΔBgX). (A) Total cellular proteins were prepared from three Ba/F3 lines expressing INV(ΔACD) or from Ba/F3-INV cells, each cultured with and without 100 μM zinc chloride for 24 h. A total of 106 cell equivalents of each extract were analyzed by Western blotting with a CBFβ antiserum (top) and by an actin antibody (bottom). The locations of the INV and INV(ΔACD) proteins are shown. (B) Total cellular proteins from two Ba/F3-INV(ΔBgX) subclones and from Ba/F3-INV cells were analyzed similarly.
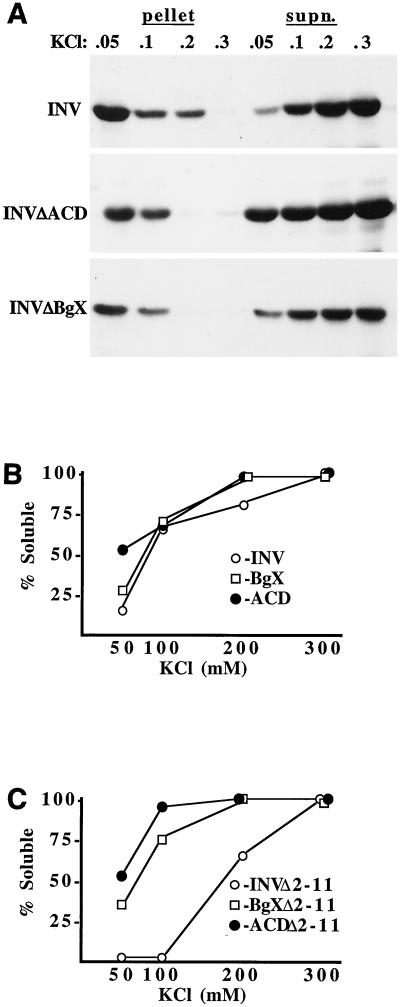
INV, INV(ΔACD), and INV(ΔBgX) cell extracts prepared with a buffer containing 300 mM KCl were heated to 100°C for 15 min, clarified, and dialyzed overnight against a buffer containing either 50, 100, 200, or 300 mM KCl. The dialysates were then collected, with vigorous rinsing of the dialysis tubing, and subjected to ultracentrifugation at 100,000 × g for 1 h. The pellets were solubilized with a buffer containing 600 mM KCl, and the pellets and supernatants were analyzed by Western blotting with a CBFβ antiserum (Fig. 3A). Densitometric analysis of these data are presented in Fig. 3B. At 50 mM KCl, INV(ΔACD) was threefold more soluble than INV and twofold more soluble than INV(ΔBgX). If the extracts were not boiled, a procedure commonly employed to purify myosins and other proteins with coiled-coil domains (29), a significant nonspecific precipitate formed during dialysis, preventing distinction between multimerization and adherence to the precipitate. Nevertheless, INV(ΔACD) was severalfold more soluble than the other two constructs under these conditions (not shown).
FIG. 3.
Deletion of the ACD inhibits INV multimerization. (A) Total cellular extracts were prepared from Ba/F3 lines expressing INV, INV(ΔACD), or INV(ΔBgX) with a buffer containing 0.3 M KCl. After exposure to 100°C for 15 min and microcentrifuge clarification, the resulting supernatants were dialyzed against 0.05, 0.1, 0.2, or 0.3 M KCl. Multimerized proteins were collected by ultracentrifugation at 100,000 × g for 1 h, followed by solubilization of the pellet with 600 mM KCl. The remain proteins were recovered from the supernatants by acetone precipitation. Pellet and supernatant fractions were subjected to Western blotting with a CBFβ antiserum. (B) Densitometric analysis of the data in A. The ratio of protein in the supernatant to the sum of the proteins in the supernatant and pellet is shown as % Soluble for each data point. (C) Extracts prepared from Ba/F3 lines expressing INV(βΔ2-11), INV(βΔ2-11ΔACD), and INV(βΔ2-11ΔBgX) were assayed similarly.
To determine whether a bound CBFα subunit affects ACD-mediated INV multimerization, the ACD and BgX segments were deleted from INV(βΔ2-11), which does not bind CBFα subunits due to lack of a 10-amino-acid CBFβ segment whose location is indicated in Fig. 1A (10). These constructs were introduced into Ba/F3 cells, and the encoded proteins were analyzed for multimerization (Fig. 3C). Deletion of the ACD in this context again greatly reduced multimerization, and deletion of the BgX segment had an intermediate effect.
Thus, deletion of the ACD potently, and to some extent specifically, deters multimerization via the SMMHC domain. This conclusion was further supported by our findings, presented below, that deleting the ACD from INVa also prevented multimerization, that multimerization of both INV and INV(ΔBgX) was detected in a gel shift assay, and that deleting the ACD prevented nuclear localization, which may depend upon multimerization. Deleting the BgX segment altered INV multimerization to some extent in the ultracentrifugation assay; nevertheless, these results justify using INVΔACD to determine whether INV must multimerize to perturb CBF activities.
Deletion of ACD prevents INV from inhibiting CBF activities.
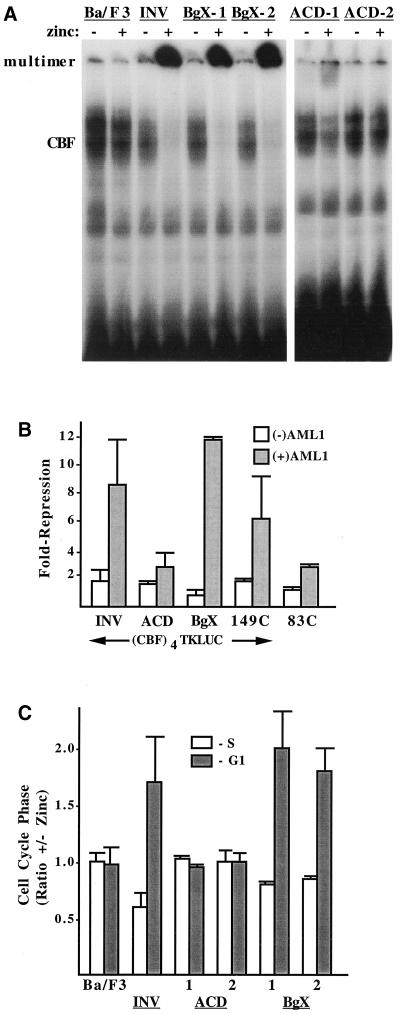
Nuclear extracts from parental Ba/F3 cells or lines expressing INV, INV(ΔBgX), or INV(ΔACD) were subjected to the gel shift assay with a radiolabeled CBF binding site derived from the myeloperoxidase gene (Fig. 4A ). As shown previously, treatment with zinc did not affect CBF DNA binding in parental Ba/F3 cells but greatly reduced CBF DNA binding in INV cells while increasing the expression of a slowly migrating DNA binding species. The latter species likely represents multimeric INV bound to CBFα subunits and to DNA. Similarly, exposure of both INV(ΔBgX) lines to zinc markedly reduced CBF DNA binding and greatly increased binding of multimeric INV(ΔBgX) to the probe.
FIG. 4.
Deletion of the ACD prevents INV from being detected as a nuclear monomer or multimer in a gel shift assay, from inhibiting CBF DNA binding and transactivation, and from slowing G1 to S cell cycle progression. (A) Nuclear extracts prepared from parental Ba/F3 cells and from Ba/F3-INV, -BgX-1, -BgX-2, -ACD-1, and -ACD-2cells, cultured with and without zinc for 48 h, were subjected to the gel shift assay with a radiolabeled probe from the myeloperoxidase gene containing a consensus CBF DNA-binding site. A total of 12 μg of nuclear extract was employed for each binding assay. The locations of gel shift species containing endogenous CBF or multimeric INV or INV variants are shown. (B) 293T cells were transiently transfected with 750 ng of p(CBF)4TKLUC, 50 ng of pCMV (open bars), or pCMV-AML1B (shaded bars), and 50 ng of pCMV, pCMV-INV, pCMV-INV(ΔACD), pCMV-INV(ΔBgX), pCMV-INV(149C), or pCMV-INV(83C). Luciferase activities were determined 2 days later. pCMV-AML1B activated the reporter 3.1- to 6.6-fold (not shown). Repression by the indicated proteins represents activity relative to the reporter in the presence of either 100 ng of pCMV (open bars) or 50 ng of pCMV and 50 ng of pCMV-AML1B (shaded bars). Results represent mean and standard error from four determinations. (C) The indicated cell lines were subjected to cell cycle analysis after culture with and without zinc for 48 h. The proportion of cells in S or in G1 in the presence of zinc divided by the proportion in each cell cycle phase in the absence of zinc was determined by bromodeoxyuridine-propidium iodide analysis and is shown for each cell line (mean and standard error of two determinations). A higher ratio indicates a greater proportion in the presence of zinc.
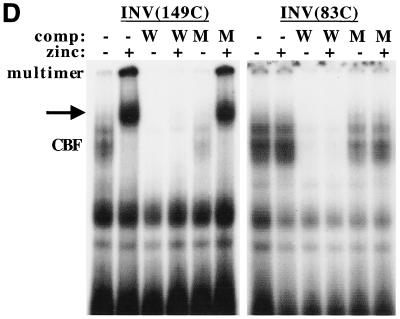
We previously demonstrated that interaction of CBF and multimeric INV with the myeloperoxidase oligonucleotide is prevented by wild-type probe but not if it carries mutations in the CBF consensus site (9) (see Fig. 6D). In contrast to these findings, exposure of Ba/F3-INV(ΔACD) cells to zinc did not generate a multimeric DNA binding species and either mildly reduced or did not reduce endogenous CBF DNA binding. As we will show below, this may reflect defective nuclear localization of INV(ΔACD). Although the extract conditions might allow a portion of cytoplasmic INV(ΔACD) to pellet with the nuclei during isolation, such polypeptides are apparently not detected by this gel shift assay, perhaps because they were not bound to CBFα subunits at the time of cell lysis.
FIG. 6.
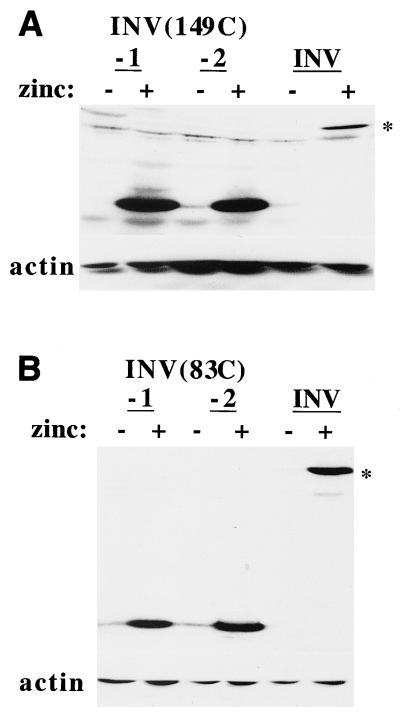
Multimerization via the C terminus of SMMHC inhibits CBF activities. (A) Total cellular extracts corresponding to 106 cells from two Ba/F3-INV(149C) subclones and from Ba/F3-INV cells cultured with and without zinc were subjected to Western blotting with a CBFβ antiserum or an actin antibody. The location of INV is indicated by an asterisk. (B) Extracts from two Ba/F3(83C) subclones were analyzed similarly. (C) Cellular extracts prepared from Ba/F3 cells expressing INV(149C), INV(83C), or CBFβ(165) were heat treated, dialyzed, and subjected to ultracentrifugation and Western blot analysis as described for Fig. 3. (D) Nuclear extracts prepared from INV(149C) or INV(83C) cells were subjected to gel shift assay with a radiolabeled CBF DNA-binding site. A 50-fold excess of unlabeled wild-type (W) or mutant (M) probe was included in some binding assays. The locations of endogenous CBF, slowly migrating multimers, and an additional, potentially monomeric INV(149C) species (arrow) are shown. (E) The relative proportions of each cell line in the S and G1 cell cycle phases, with and without zinc at 48 h, is shown (mean and standard error of two determinations).
The inability of INV(ΔACD) to disrupt AML1:CBFβ DNA binding complexes predicts that INV(ΔACD) will not interfere with CBF-mediated gene activation. We evaluated the affect of INV and its variants on AML1B-mediated gene activation with a transient-transfection assay. p(CBF)4TKLUC contains four CBF-binding sites derived from the myeloperoxidase gene, a TATAA homology and RNA initiation site derived from the herpes simplex virus thymidine kinase (TK) gene, and a luciferase (LUC) cDNA. AML1B is a prominent AML1 isoform and possesses a transactivation domain (3).
In 293T cells, pCMV-AML1B activated p(CBF)4TKLUC 4.5-fold, on average, with a range of 3.1- to 6.6-fold (not shown). INV, INV(ΔACD), and INV(ΔBgX) each reduced the basal activity of p(CBF)4TKLUC about 1.6-fold. On the other hand, AML1B-induced activity of this reporter was reduced 8-fold by INV, 9-fold by INV(ΔBgX), and only 2-fold by INV(ΔACD) (Fig. 4B). Thus, relative to background suppression in the absence of AML1B, INV(ΔACD) or INV(ΔBgX) prevented AML1B transactivation, whereas INV(ΔACD) was ineffective.
Gene activation by CBF is a prerequisite for optimal progression from G1 to S in Ba/F3 cells. Evaluating the affect of INV on endogenous CBF activities offers the advantage of greater in vivo relevance than transient-transfection assays. Therefore, we assessed cell cycle progression in Ba/F3 lines expressing INV or its derivatives as an indirect assay of endogenous CBF activities. Zinc did not affect the proliferation rate of parental Ba/F3 cells and slowed the accumulation of INV cells 3-fold over 72 h, of INV(ΔACD) cells 1.2-fold on average, and of INV(ΔBgX) cells 2-fold (not shown).
The effect of zinc on the proportion of Ba/F3, INV, INV(ΔACD)-1 and -2, and INV(ΔBgX)-1 and -2 cells in the S or G1 cell cycle phase is shown in Fig. 4C. Both INV and INV(ΔBgX) slowed the G1 to S progression similarly, increasing the G1/S ratio about 2.6-fold, whereas induction of INV(ΔACD) for 2 days did not alter the G1/S ratio. These differences occurred even though ACD-1, ACD-2, BgX-1, and BgX-2 cells expressed exogenous protein at a level equal to or greater than that of INV cells (Fig. 2). Thus, INV and INV(ΔBgX) are able to multimerize, to inhibit CBF DNA binding, and to inhibit G1 progression, whereas deletion of the ACD interferes with each of these activities.
ACD is required for INVa to inhibit CBF activities.
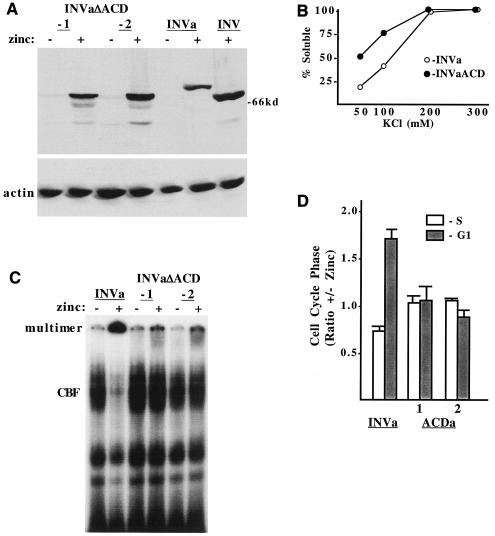
Alternative splicing produces an isoform of INV, designated INVa, with a longer nonhelical tail (Fig. 1A). INVa is more abundant than INV in human acute myeloid leukemia blasts (24). As with INV, expression of INVa in Ba/F3 cells inhibits endogenous CBF DNA binding and slows G1 progression (10). As the ACD is very near the C terminus, its deletion might affect INVa differently than INV. We obtained Ba/F3 lines expressing INVa(ΔACD) at levels similar to or higher than that of INVa (Fig. 5A). For comparison, this Western blot also included an extract from zinc-treated Ba/F3-MTINV cells.
FIG. 5.
ACD homology region is also required for INVa to multimerize, to inhibit CBF DNA binding, and to slow cell cycle progression. (A) Total cellular proteins were prepared from two INVa(ΔACD) subclones or from Ba/F3-INVa cells cultured with and without zinc for 24 h and from INV cells cultured with zinc; 106 cell equivalents were subjected to Western blotting with a CBFβ antiserum and an actin antibody. (B) Cellular extracts were prepared from INVa and INVa(ΔACD) cells with a buffer containing 0.3 M KCl. Each extract was then heat treated, dialyzed against 0.05, 0.1, 0.2, or 0.3 M KCl, and subjected to ultracentrifugation followed by Western blot analysis, as described for Fig. 3. (C) Nuclear extracts prepared from INVa, INVa(ΔACD)-1, and INVa(ΔACD)-2 cells cultured with and without zinc for 48 h were subjected to the gel shift assay with a radiolabeled CBF DNA-binding site. The locations of endogenous CBF and multimeric INVa and INVa(ΔACD) are indicated. (D) The relative proportion of each cell line in the S and G1 cell cycle phases, with and without zinc at 48 h, is shown (mean and standard error of two determinations).
INVa was expressed at a lower level than INV and, as expected, was slightly larger. Multimerization of INVa and INVa(ΔACD) was assessed by ultracentrifugation (Fig. 5B). Deletion of the ACD from INVa interfered with its multimerization in 50 mM or 100 mM KCl. Nuclear extracts prepared from INVa and INVa(ΔACD) cells cultured with and without zinc were subjected to the gel shift assay with a CBF DNA binding region from the myeloperoxidase gene (Fig. 5C). Induction of INVa reduced endogenous CBF DNA binding and increased binding by a slowly migrating species, whereas induction of INVa(ΔACD) did not alter endogenous CBF DNA binding and only minimally increased binding by the slowly migrating, presumably multimeric species. Induction of INVa slowed Ba/F3 proliferation 4-fold over 3 days, whereas INVa(ΔACD) only slowed cell proliferation 1.3-fold, on average (not shown). Consistent with these findings, INVa increased the G1/S ratio almost threefold, whereas INVa(ΔACD) did not affect the relative proportion of cells in the G1 and S cell cycle phases (Fig. 5D).
Multimerization via the C terminus of SMMHC is sufficient to inhibit CBF activities.
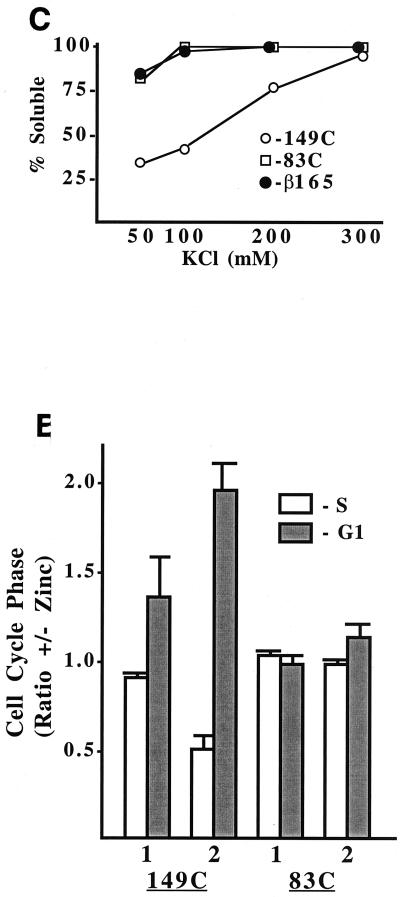
We also generated Ba/F3 cell lines expressing INV variants in which the CBFβ domain was linked to either 149 or 83 C-terminal SMMHC residues (Fig. 6A and 6B). Cells expressing the CBFβ domain alone, CBFβ(165), were rederived as described previously (10). The multimerization of INV(149C), INV(83C), and CBFβ(165) was compared (Fig. 6C). INV(149C) retained the ability to multimerize at 50 or 100 mM KCl, whereas the large majority of INV(83C) and CBFβ(165) remained in the supernatant at these salt concentrations. Nuclear extracts prepared from INV(149C) and INV(83C) cells cultured with and without zinc were subjected to the gel shift assay with a radiolabeled CBF DNA binding site (Fig. 6D).
Induction of INV(149C) strongly inhibited endogenous CBF DNA binding and led to the detection of a slowly migrating multimer and of a novel species (arrow). This novel species likely represents monomeric INV(149C) bound to endogenous CBFα subunits and to DNA; if it was a higher-order multimer, bands of other sizes would have been expected. The appearance of this novel species in this assay but not in the assay of wild-type INV suggests that INV(149C) has weakened multimerization potential, as was expected from its having only 141 α-helical residues. A 50-fold excess of unlabeled wild-type probe (W) inhibited DNA binding by endogenous CBF and by the two species which appeared upon induction of INV(149C), whereas a 50-fold excess of the same oligonucleotide carrying clustered point mutations in the CBF binding site only mildly affected DNA binding by these three species.
In contrast to these findings and consistent with the ultracentrifuge multimerization assay, induction of INV(83C) did not alter endogenous CBF DNA binding or lead to the appearance of a multimeric gel shift species. INV(149C) inhibited activation of p(CBF)4TKLUC by AML1B 6-fold, on average, in 293T cells, whereas INV(83C) only inhibited activation by 2-fold (Fig. 4B). Induction of INV(149C) slowed cell proliferation 2.5-fold over 72 h and inhibited G1 to S progression, whereas induction of INV(83C) did not affect cell accumulation or the proportion of cells in the G1 and S cell cycle phases (Fig. 6E and not shown). Thus, CBFβ residues 1 to 165 linked to 149 C-terminal SMMHC residues retained the ability to multimerize and inhibit CBF activities, albeit less efficiently than intact CBFβ-SMMHC.
Multimerization is required for INV nuclear localization.
Gel shift assay of Ba/F3-MTINV(149C) nuclear extracts generated a species of intermediate mobility likely representing monomeric INV(149C)-CBFα complexes, but similar assays of extracts containing INV(ΔACD), INVa(ΔACD), or INV(83C) did not. As each of these INV variants has reduced multimerization capacity compared with INV and INV(149C), we considered the possibility that multimerization is required for their nuclear localization and so their detection as complexes with CBFα in nuclear extracts.
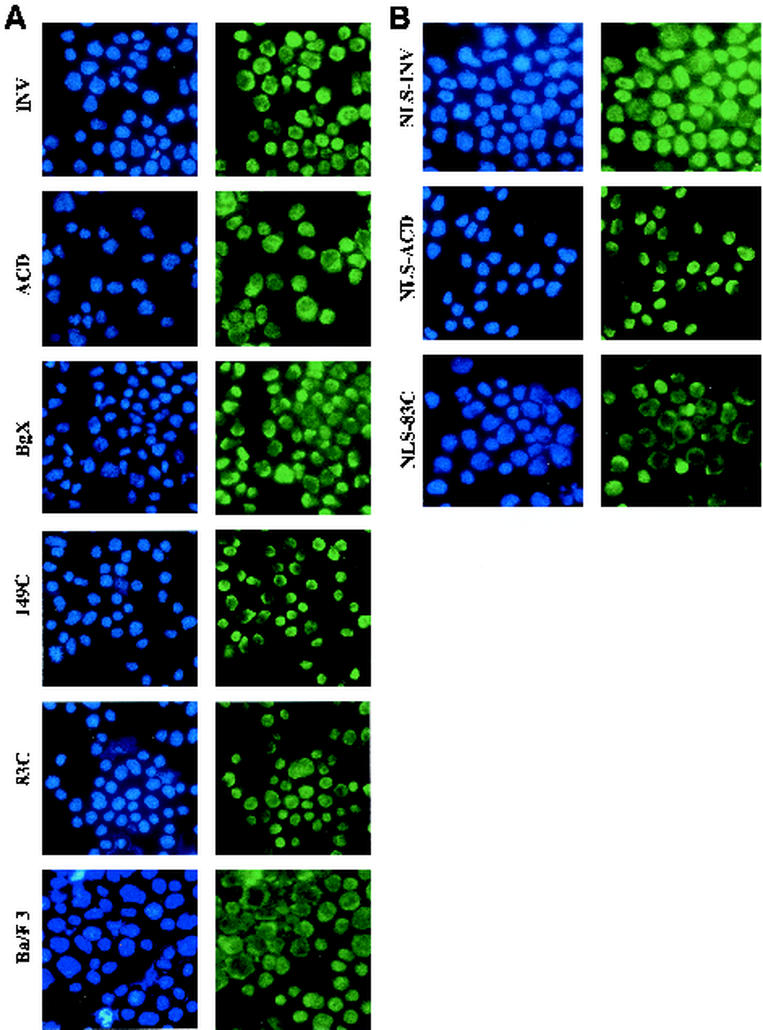
Cytospins of parental Ba/F3 cells or Ba/F3 cells expressing INV, INV(ΔBgX), INV(ΔACD), INV(149C), or INV(83C) were subjected to indirect immunofluorescence with a rabbit anti-CBFβ antiserum (Fig. 7A ). Simultaneous DAPI staining (blue) localized the nucleus. In this assay, there is significant background staining, in part due to endogenous cytoplasmic CBFβ. Nevertheless, INV, INV(ΔBgX), and INV(149C) were detected in the nuclei of the majority of the cells, whereas INV(ΔACD) and INV(83C) were expressed predominantly in the cytoplasm.
FIG. 7.
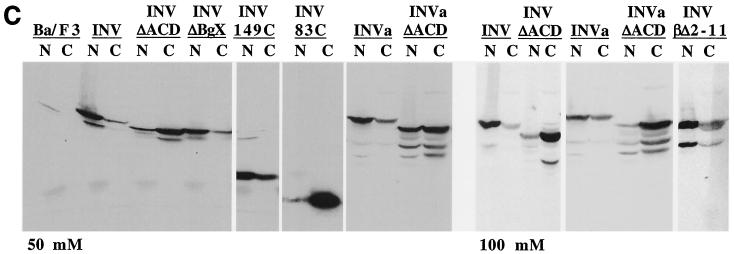
ACD is required for nuclear retention of INV and INVa. (A) Parental Ba/F3 cells and subclones expressing INV, INV(ΔACD), INV(ΔBgX), INV(149C), and INV(83C) were cytospun and subjected to indirect immunofluorescence with a CBFβ antiserum (green). Nuclei were visualized simultaneously with DAPI (blue). Representative fields are shown at ×60. (B) Ba/F3 cells expressing NLS-INV, NLS-INV(ΔACD), and NLS-INV(83C) were analyzed similarly. (C) The plasma membranes of the indicated cell lines, cultured in zinc for 48 h, were lysed with 0.1% NP-40 in a buffer containing either 50 mM or 100 mM KCl. Nuclear (N) and cytoplasmic (C) proteins were then isolated and subjected to Western blot analysis with a CBFβ antiserum to detect INV, INVa, and their variants.
To obtain quantitative data more representative of the entire population of cells expressing INV or its variants, we prepared nuclear and cytoplasmic extracts with a lysis buffer containing 50 mM KCl. This salt concentration reproducibly allowed the majority of INV, initially extracted with 300 mM KCl, to remain in the supernatant after centrifugation at 850 × g for 5 min, the conditions used to fractionate nuclei from the cytoplasm. Analysis of nuclear and cytoplasmic fractions prepared in the presence of 50 mM KCl indicated that the majority of INV, INVa, INV(ΔBgX), and INV(149C) was nuclear, whereas the majority of INV(ΔACD), INVa(ΔACD), and INV(83C) was cytoplasmic (Fig. 7C, top). Also, when cell fractionation was carried out in 100 mM KCl, the majority of INV and INVa still remained in the nuclear fraction (Fig. 7C, bottom). At this salt concentration, cytoplasmic INV and INVa almost certainly did not pellet artifactually with the nuclei due to multimerization. Thus, multimerization is required for the majority of INV to be retained efficiently in the nucleus.
We also evaluated the cellular localization of INV(βΔ2-11) by Western blotting with extracts prepared in 100 mM KCl (Fig. 7C, bottom). The majority of this isoform was found in the nucleus, indicating that interaction with CBFα subunits is not necessary to initiate nuclear localization of INV.
ACD acts in the nucleus to allow CBFβ-SMMHC to inhibit CBF.
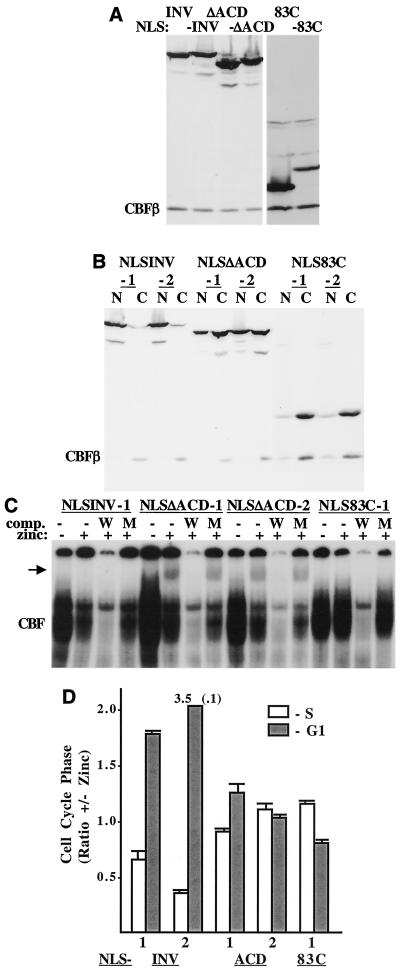
In an effort to express a greater proportion of INV(ΔACD) and INV(83C) in the nucleus, we introduced the simian virus 40 nuclear localization signal (NLS) at their N termini. As a control, the NLS was also linked to INV. These constructs were linked to the MT promoter and introduced into Ba/F3 cells. The presence of the NLS was confirmed by comparing the mobility of the encoded proteins with the corresponding proteins lacking the NLS (Fig. 8A ). Indirect immunofluorescent analysis demonstrated that NLS-INV and NLS-INV(ΔACD) were nuclear in a large majority of the cells, whereas INV(83C) remained cytoplasmic in most cells (Fig. 7B). Western analysis of nuclear and cytoplasmic extracts indicated that 30 to 50% of the INV(ΔACD) protein was now nuclear, whereas INV(83C) was cytoplasmic (Fig. 8B).
FIG. 8.
ACD is required in the nucleus for INV to inhibit CBF activities. (A) Western blot, with CBFβ antiserum, comparing the mobilities of INV, INV(ΔACD), and INV(83C) (lanes 1, 3, and 5, respectively) with those of NLS-INV, NLS-INV(ΔACD), and NLS-INV(83C) (lanes 2, 4, and 6, respectively). The location of endogenous CBFβ is indicated. (B) Two subclones expressing each NLS-linked protein were separated into nuclear (N) and cytoplasmic (C) fractions, and these were subjected to Western blotting with CBFβ antiserum. (C) Nuclear extracts prepared from the indicated subclones were sub-jected to gel shift assay as in Fig. 6D. The arrow indicates the location of a potential monomeric CBFα-INV(ΔACD) complex. (D) The relative proportions of each cell line in the S and G1 cell cycle phases, ± with and without zinc at 48 h, are shown (mean and standard error of two determinations).
Gel shift analysis of nuclear extracts derived from several of these lines is shown in Fig. 8C. As with INV, induction of NLS-INV with zinc increased detection of slowly migrating multimers and reduced detection of CBF complexes. A high background near the top of each lane, potentially representing CBF nonspecifically bound to cellular proteins, precluded clear visualization of multimers in this experiment. Strikingly, and in both NLS-INV(ΔACD) subclones analyzed, exposure to zinc reduced endogenous CBF DNA binding severalfold and allowed detection of a monomeric gel shift species (arrow), while there was no increase in signal above background near the top of each lane. These findings stand in sharp contrast to those for INV(ΔACD), which only minimally affected endogenous CBF and did not enter the nucleus and form a monomeric gel shift species.
As expected from its cellular localization, INV(83C) did not affect CBF DNA binding or produce a monomeric gel shift species. When transiently transfected into 293T cells, NLS-INV inhibited AML1B-mediated reporter activation 4.5-fold, on average, whereas NLS-INV(ΔACD) and INV(83C) were less effective, inhibited activation 3.0- and 2.5-fold, respectively (not shown). The difference in activity between NLS-INV and NLS-INV(ΔACD) was more evident when their effects on cell cycle progression mediated by endogenous CBF were assessed. NLS-INV inhibited proliferation 4- to 9- fold over 72 h, while NLS-INV(ΔACD) and NLS-83C only inhibited cell accumulation 2-fold during this time period (not shown). Similarly, NLS-INV markedly inhibited G1 to S cell cycle progression, whereas NLS-INV(ΔACD) and NLS-INV(83C) were largely ineffective (Fig. 8D). Thus, the ACD is required in the nucleus for INV to inhibit endogenous CBF activities.
ACD does not repress CBF target genes required for proliferation.
The failure of NLS-INV(83C) to enter the nucleus leaves open the possibility that the ACD enables INV to slow cell proliferation by directly attracting corepressors to CBF target genes rather than through its capacity to mediate INV multimerization. To address this issue, we linked the segment 83C, containing the ACD but too short to multimerize, to the AML1 DNA-binding domain, either residues 1 to 216 (AML1DBDa) or residues 1 to 290 (AML1DBDb) of AML1B. Segment 83C was also linked to the C terminus of full-length AML1B. As controls, 149 C-terminal residues from which the ACD had been deleted, 149C(ΔACD), were also linked to AML1DBDb or AML1B. 149C(ΔACD) was not capable of mediating multimerization when linked to CBFβ (not shown). As a positive control, pMT-AML1-ETO was also introduced into Ba/F3 cells.
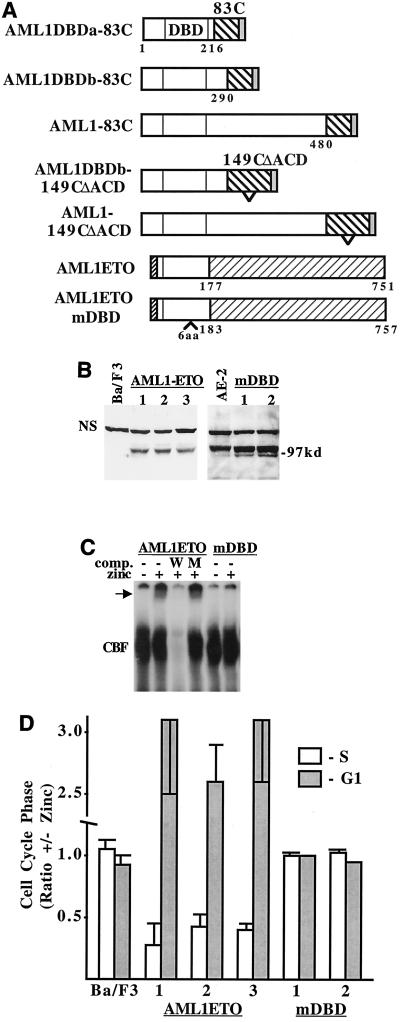
In AML1-ETO, residues 1 to 177 of AML1 are linked to the potent ETO corepressor. AML1-ETO was previously shown to inhibit G1 to S progression in hematopoietic cells (2, 8), but the dependence of this effect on interaction with AML1 binding sites has not been determined. To verify that our positive control acts on AML1 target genes, we introduced six amino acids between residues 145 and 146 of AML1-ETO's DNA-binding domain. These cDNAs are diagrammed in Fig. 9A . Each was introduced into Ba/F3 cells.
FIG. 9.
ETO slows G1 progression when directed to AML1 target genes. (A) Diagram of AML1-INV fusion proteins, AML1-ETO, and AML1ETOmDBD. The 83 C-terminal INV residues, including the ACD, were linked at position 216, 290, or 480 to AML1B, and 121 C-terminal residues of 149C(ΔACD), lacking the ACD, were linked to AML1B at residue 290 or 480. Six amino acids were inserted between amino acids 145 and 146 in the DNA-binding domain of AML1-ETO to generate AML1ETOmDBD. The N terminus of AML1-ETO differs from that of AML1B. (B) Western blot with an AML1 antiserum, detecting AML1-ETO and AML1-ETOmDBD in Ba/F3 subclones.The location of a nonspecific band, which served as a loading control, is shown (NS). AE-2, AML1-ETO clone 2. (C) Gel shift assay with AML1-ETO and AML1ETOmDBD nuclear extracts was carried out as in Fig. 6D. The arrow indicates the AML1-ETO gel shift complex. (D) The relative proportion of each indicated cell line in the S and G1 cell cycle phases, with and without zinc at 48 h, is shown (mean and standard error of two determinations).
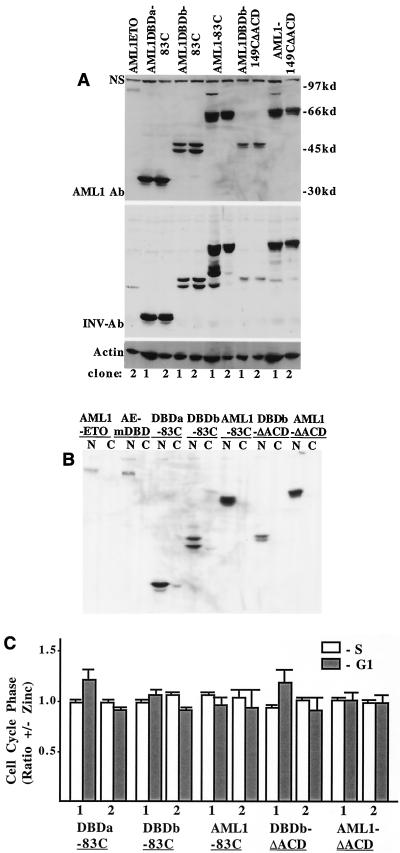
AML1ETOmDBD expression was achieved at higher levels than AML1-ETO (Fig. 9B), and AML1-ETO bound a CBF-binding site in the gel shift assay, whereas AML1ETOmDBD did not (Fig. 9C). AML1-ETO dramatically slowed Ba/F3 G1 to S progression, whereas AML1ETOmDBD was ineffective (Fig. 9D). The expression of each AML1-INV fusion protein was greater than that of AML1-ETO when detected with an AML1 antiserum (Fig. 10A, top panel). When this blot was reprobed with a C-terminal SMMHC antiserum, each of the AML1-INV fusion proteins was detected, verifying the presence of the SMMHC segment (Fig. 10A, bottom panel). AML1-ETO, AML1ETOmDBD, and the AML1-INV fusion proteins were detected only in the nucleus (Fig. 10B). The AML1-INV proteins bound DNA on Electrophoretic mobility shift assay (not shown). Each of the AML1-INV fusions activated the p(CBF)4KTLUC reporter in 293T cells, albeit less efficiently than AML1B (not shown). Most strikingly, in contrast to AML1-ETO, none of the AML1-INV constructs significantly affected G1 to S progression in Ba/F3 cells (Fig. 10C). Thus, the 83 C-terminal residues of SMMHC, containing the ACD, are do not directly repress AML1-regulated genes.
FIG. 10.
ACD does not slow G1 progression when directed to AML1 target genes. (A) Western blot comparing expression of AML1-INV fusions with AML1-ETO. The gel in the top panel was probed with an AML1 antiserum, and that in the bottom panel was probed with an antiserum specific for the nonhelical, C-terminal tail of INV. A prominent nonspecific band (NS) was again seen with the AML1 antiserum. (B) Representative subclones expressing the indicated proteins were fractionated into nuclei (N) and cytoplasm (C) and subjected to Western blotting with the AML1 antiserum. AE-mDBD, AML1ETOmDBD. (C) The relative proportion of each cell line in the S and G1 cell cycle phases, with and without zinc at 48 h, is shown (mean and standard error of two determinations).
DISCUSSION
Our findings indicate that INV must multimerize both to localize to the nucleus and, once there, to inhibit endogenous CBF activities. As repression of CBF is the common feature of several genetic abnormalities associated with acute myeloid leukemia, we expect that multimerization is also required for INV to contribute to leukemic transformation.
The ACD defined for the fast IId isoform of skeletal muscle myosin is highly conserved in other skeletal myosins, and smooth muscle and nonmuscle myosins have a less-related but still highly homologous sequence near their C termini (44). Our findings support the prediction that these segments also function as ACDs. Deletion of the SMMHC ACD from INV, INV(βΔ2-11), INVa, and INV(149C) consistently increased solubility approximately 4-fold at 50 mM KCl and also increased solubility approximately 3-fold at 100 mM and 1.5-fold at 200 mM KCl in several assays. In comparison, deletion of the ACD from a skeletal myosin rod domain increased its solubility 8-fold at 50 or 100 mM KCl and 2-fold at 200 mM KCl (44). Although INVa was more soluble than INV, INV(βΔ2-11) was even more soluble, suggesting that differences observed reflect assay variability.
Deletion of a more N-terminal, randomly selected 28-amino-acid segment had less effect on solubility in low KCl concentrations and did not alter detection of multimers in the gel shift assay. Consistent with our findings, an antibody specific for the C terminus of the coiled-coil domain of turkey smooth muscle myosin prevents and even reverses multimerization (18). Questions remain regarding the biophysical interactions and mechanistic steps which mediate myosin multimerization. As SMMHC multimerizes in a staggered, antiparallel manner (54), it will be of interest to identify the more N-terminal protein segment with which it interacts. A 63-residue “extended ACD” has more bulky aliphatic and fewer net acidic residues on its outer surface than the majority of the myosin rod and may contact a similar internal domain to initiate multimerization (13). Of note, deletion of the nonhelical tailpiece slowed but did not prevent multimerization of a nonmuscle myosin (17), the antibody capable of inhibiting turkey smooth muscle myosin multimerization does not interact with the tailpiece (18), and deletion of the tailpiece from INV did not affect its ability to multimerize and inhibit CBF activities in Ba/F3 cells (T. Kummalue, J. Lou, and A. D. Friedman., unpublished data).
Multimerization of INV is required for its optimal localization to the nucleus, reflecting the affinity of intermediate filaments for CBFβ (48). We and others have previously observed INV in the nucleus of primary leukemic samples and Ba/F3 cells (9, 24), whereas CBFβ-SMMHC is predominantly cytoplasmic in adherent cell lines, perhaps reflecting the presence of a greater number of intermediate filaments (1, 20, 49). Nuclear retention may require the formation of larger multimers which cannot easily exit the nucleus. Even though they have reduced capacity for multimerization in vitro, INV(ΔBgX) and INV(149C) apparently retained the ability to multimerize well enough in vivo to localize to the nucleus and inhibit CBF. Addition of an NLS to INV(ΔACD) shifted the balance between these competing effects, perhaps because INV(ΔACD) retains some capacity for multimerization, whereas addition of NLS did not allow entry of CBFβ(83C) into the nucleus.
Monomeric INV(149C)-CBFα nuclear complexes likely were detected by the gel shift assay due to weakened multimerization, a consequence of its having only 141 α-helical SMMHC residues. As INV(βΔ2-11) was predominantly nuclear, affinity for CBFα subunits cannot account for nuclear retention of INV. Consistent with this conclusion, in Ba/F3 lines expressing INV(ΔACD), approximately 30% of nuclear AML1 relocalized to the cytoplasm (not shown).
Deletion of the ACD reduced the ability of INV, INVa, and NLS-INV to inhibit CBF DNA binding and transactivation and obviated their ability to inhibit CBF-mediated G1 to S cell cycle progression. This disruption of INV activities likely resulted from reduced multimerization capacity, as the ACD domain has evolved to mediate this activity. It is also possible that the ACD directly mediates transrepression by CBFα-INV complexes bound to CBF-binding sites in chromatin, as both endogenous mSin3a and exogenous HDAC8 bind INV in Cos7 cells dependent upon the presence of the ACD (K. L. Durst, B. Lutterbach, T. Kummalue, A. D. Friedman, and S. W. Hiebert, submitted for publication).
To address this issue, we expressed AML1DBDa-83C, AML1DBDb-83C, and AML1-83C, which link the ACD and surrounding residues to the AML1 DNA-binding domain or to full-length AML1B. In contrast to AML1-ETO, which dramatically slowed Ba/F3 G1 progression dependent upon the integrity if its DNA-binding domain, these fusion proteins were essentially inactive when expressed at severalfold higher levels than AML1-ETO. We therefore propose that loss of multimerization potential rather than loss of interactions with heterologous proteins accounts for the inability of INV(ΔACD) and INVa(ΔACD) to inhibit CBF activities in Ba/F3 cells. Weakened multimerization capacity also explains the inability of an INVa isoform lacking 95 C-terminal residues, including the ACD, to transrepress the p21 promoter in cooperation with AML1 (28). On the other hand, AML1-ETO binds several corepressors simultaneously (15, 27, 53), and so it remains possible that intact and/or multimeric INV binds mSin3a or another corepressor with greater affinity than an isolated, monomeric ACD domain.
Our investigations do not directly address how nuclear INV multimers inhibit CBF. In the absence of the ACD, INV remains capable of heterodimerizing with CBFα subunits, and this complex binds DNA, as confirmed by our detecting a monomeric CBFα-NLS-INV(ΔACD) gel shift species. Gel shift analysis indicates that the majority of CBFα subunits are bound to these multimers, but whether this results in their sequestration away from CBF-regulated genes or whether CBFα-INV multimeric complexes directly repress gene expression by physically preventing interactions of positive regulators, by interaction with corepressors, or by a combination of these mechanisms remains to be elucidated.
Inhibition of INV multimerization may prove useful in the therapy of acute myeloid leukemia patients with inv(16) or t(16;16) mutations. A 28-amino-acid peptide corresponding to the SMMHC ACD did not perturb INV multimerization in vitro (data not shown). Further mechanistic investigations should assist efforts to identify a more effective reagent.
Acknowledgments
We thank L. A. Leinwand for helpful discussions.
This work was supported by grant R01 HL15388 from the NIH to A.D.F. T.K. was supported by a grant from the Siriraj Hospital, Mahidol University. A.D.F. is a Leukemia and Lymphoma Society Scholar and is generously supported by the Children's Cancer Foundation.
REFERENCES
- 1.Adya, N., T. Stacy, N. A. Speck, and P. P. Liu. 1998. The leukemic protein core binding factor beta (CBFβ)-smooth muscle myosin heavy chain sequesters CBFα2 into cytoskeletal filaments and aggregates. Mol. Cell. Biol. 18:7432-7443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21:6470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, S.-C., Y. Yamaguchi-Iwai, E. Ogawa, M. Maruyama, M. Inuzuka, H. Kagoshima, K. Shigesada, M. Satake, and Y. Ito. 1993. Isolation of PEBP2αB cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene 8:809-814. [PubMed] [Google Scholar]
- 4.Bernardin, F., and A. D. Friedman. 2002. AML1 stimulates G1 to S progression via its transactivation domain. Oncogene 21:3247-3252. [DOI] [PubMed] [Google Scholar]
- 5.Bernardin, F., Y. Yang, C. I. Civin, and A. D. Friedman. c-Myc overcomes cell cycle inhibition by CBFβ-SMMHC, a myeloid leukemia oncoprotein. Cancer Biol. Ther., in press. [DOI] [PubMed]
- 6.Bernardin, F., Y. Yang, R. Cleaves, M. Zahurak, L. Cheng, C. I. Civin, and A. D. Friedman. 2002. TEL-AML1, expressed from t(12;21) in human acute leukemia, induces acute leukemia in mice. Cancer Res. 62:39904-39908. [PubMed] [Google Scholar]
- 7.Britos-Bray, M., and A. D. Friedman. 1997. Core binding factor cannot synergistically activate the myeloperoxidase proximal enhancer in immature myeloid cells without c-Myb. Mol. Cell. Biol. 17:5127-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burel, S. A., N. Harakawa, L. Zhou, T. Pabst, D. G. Tenen, and D. E. Zhang. 2001. Dichotomy of AML1-ETO functions: growth arrest versus block of differentiation. Mol. Cell. Biol. 21:5577-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao, W., M. Britos-Bray, D. F. Claxton, C. A. Kelley, N. A. Speck, P. P. Liu, and A. D. Friedman. 1997. CBFβ-SMMHC, expressed in M4eo acute myeloid leukemia, reduced CBF DNA binding and inhibited the G1 to S cell cycle transition at the restriction point in myeloid and lymphoid cells. Oncogene 15:1315-1327. [DOI] [PubMed] [Google Scholar]
- 10.Cao, W., N. Adya, M. Britos-Bray, P. P. Liu, and A. D. Friedman. 1998. The core binding factor a interaction domain and the smooth muscle myosin heavy chain segment of CBFβ-SMMHC are both required to slow cell proliferation. J. Biol. Chem. 273:31534-31540. [DOI] [PubMed] [Google Scholar]
- 11.Castilla, L. H., C. Wijmenga, T. Stacy, N. A. Speck, M. Eckhaus, M. Marin-Padilla, F. S. Collins, A. Wynshaw-Boris, and P. P. Liu. 1996. Failure of embryonic haematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell 87:687-696. [DOI] [PubMed] [Google Scholar]
- 12.Castilla, L. H., L. Garrett, N. Adya, D. Orlic, A. Dutra, S. Anderson, J. Owens, M. Eckhaus, D. Bodine, and P. P. Liu. 1999. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukemia. Nat. Genet. 23:144-146. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, C., and D. A. D. Parry. 1998. A conserved C-terminal assembly region in paramyosin and myosin rods. J. Struct. Biol. 122:180-187. [DOI] [PubMed] [Google Scholar]
- 14.Friedman, A. D. 1999. Leukemogenesis by CBF oncoproteins. Leukemia 13:1932-1942. [DOI] [PubMed] [Google Scholar]
- 15.Gelmetti, V., J. Zhang, M. Fanelli, S. Minucci, P. G. Pelicci, and M. A. Lazar. 1998. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 18:7185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golling, G., L.-H. Li, M. Pepling, M. Stebbins, and J. Gergen. 1996. Drosophila homologs of the proto-oncogene product PEBP2/CBFβ regulate the DNA-binding properties of runt. Mol. Cell. Biol. 16:932-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodge, T. P., R. Cross, and J. Kendrick-Jones. 1992. Role of the COOH-terminal nonhelical tailpiece in the assembly of a vertebrate nonmuscle myosin rod. J. Cell Biol. 118:1085-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikebe, M., S. Komatsu, J. L. Woodhead, K. Mabuchi, R. Ikebe, J. Saito, R. Craig, and M. Higashihara. 2002. The tip of the coiled-coil rod determines the filament formation of smooth muscle and nonmyscle myosin. J. Biol. Chem. 276:30293-30300. [DOI] [PubMed] [Google Scholar]
- 19.Kagoshima, H., Y. Akamatsu, Y. Ito, and K. Shigesada. 1996. Functional dissection of the α and β subunits of transcription factor PEBP2 and the redox susceptibility of its DNA-binding activity. J. Biol. Chem. 271:33074-33082. [DOI] [PubMed] [Google Scholar]
- 20.Kanno, Y., T. Kanno, C. Sakakura, S.-C. Bae, and Y. Ito. 1998. Cytoplasmic sequestration of the polyomavirus enhancer binding protein (PEBP2)/Core binding factor (CBFα) subunit by the leukemia-related PEBP2/CBFβ-SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol. Cell. Biol. 18:4252-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levanon, D., V. Negreanu, Y. Bernstein, I. Bar-Am, L. Avivi, and Y. Groner. 1994. AML1, 2 and 3. The human members of the runt domain gene family. Genomics 23:425-432. [DOI] [PubMed] [Google Scholar]
- 22.Liu, P., S. A. Tarle, A. Hajre, D. F. Claxton, P. Marlton, M. Freedman, M. J. Siciliano, and F. S. Collins. 1993. Fusion between transcription factor CBFβ/PEBP2β and a myosin heavy chain in acute myeloid leukemia. Science 261:1041-1044. [DOI] [PubMed] [Google Scholar]
- 23.Liu, P. P., N. Seidel, D. Bodine, N. A. Speck, S. Tarle, and F. S. Collins. 1994. Acute myeloid leukemia with inv(16) produces a chimeric transcription factor with a myosin heavy chain tail. Cold Spring Harbor Symp. Quant. Biol. 59:547-553. [DOI] [PubMed] [Google Scholar]
- 24.Liu, P. P., C. Wijmenga, A. Hajra, T. B. Blake, C. A. Kelley, R. S. Adelstein, A. Baggf, J. Rector, J. Cotelingam, C. L. Willman, and F. S. Collins. 1996. Identification of the chimeric protein product of the CBFB-MYH11 fusion gene in inv(16) leukemia cells. Genes Chrom. Cancer 16:77-87. [DOI] [PubMed] [Google Scholar]
- 25.Lou, J., W. Cao, F. Bernardin, K. Ayyanathan, F. J. Rauscher, I. I. I., and A. D. Friedman. 2000. Exogenous cdk4 overcomes reduced cdk4 RNA and inhibition of G1 progression in hematopoietic cells expressing a dominant-negative CBF—a model for overcoming inhibition of proliferation by CBF oncoproteins. Oncogene 19:2695-2703. [DOI] [PubMed] [Google Scholar]
- 26.Lu, J., M. Maruyama, M. Satake, S.-C. Bae, S. Ogawa, H. Kagoshima, K. Shigesada, and Y. Ito. 1995. Subcellular localization of the α and β subunits of the acute myeloid leukemia-linked transcription factor PEBP2/CBF. Mol. Cell. Biol. 15:1651-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutterbach, B., J. J. Westendorf, B. Linggi, A. Patten, M. Moniwa, J. R. Davie, K. D. Huynh, V. J. Bardwell, R. M. Lavinsky, M. G. Rosenfeld, C. Glass, E. Seto, and S. W. Hiebert. 1998. ETO, a target of t(8;21) in acute leukemia, interacts with N-CoR and mSin3 corepressors. Mol. Cell. Biol. 18:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutterbach, B., Y. Hou, K. L. Durst, and S. W. Hiebert. 1999. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc. Natl. Acad. Sci. USA 96:12822-12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNally, E., R. Sohn, S. Frankel, and L. Leinwand. 1991. Expression of myosin and actin in Escherichia coli. Methods Enzymol. 196:368-389. [DOI] [PubMed] [Google Scholar]
- 30.Meyers, S., J. R. Downing, and S. W. Hiebert. 1993. Identification of AML1 and the (8;21) translocation protein AML1-ETO as sequence-specific DNA-binding proteins: the runt homology domain is required for DNA binding and protein-protein interactions. Mol. Cell. Biol. 13:6336-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyers, S., N. Lenn, and S. W. Hiebert. 1995. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol. Cell. Biol. 15:1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai, R., M. Kuro-o, P. Babij, and M. Periasamy. 1998. Identification of two types of smooth muscle myosin heavy chain isoforms by cDNA cloning and immunoblot analysis. J. Biol. Chem. 264:9734-9737. [PubMed] [Google Scholar]
- 33.Niki, M., H. Okada, H. Takano, J. Kujo, K. Tani, H. Hibino, S. Asano, Y. Ito, M. Satake, and T. Noda. 1997. Hematopoiesis in the fetal liver is impaired by the targeted mutagenesis of the gene encoding a non-DNA-binding subunit of the transcription factor, PEBP2/CBF. Proc. Natl. Acad. Sci. USA 94:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nuchprayoon, I., S. Meyers, L. M. Scott, J. Suzow, S. Hiebert, and A. D. Friedman. 1994. PEBP2/CBF, the murine homolog of the human myeloid proto-oncogenes AML1 and PEBP2β/CBFβ, regulates the murine myeloperoxidase and neutrophil elastase genes in immature myeloid cells. Mol. Cell. Biol. 14:5558-5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogawa, E., M. Maruyama, H. Kagoshima, M. Inuzuka, J. Lu, M. Satake, K. Shigesada, and Y. Ito. 1993. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc. Natl. Acad. Sci. USA 90:6859-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogawa, E., M. Inuzuka, M. Maruyamna, M. Satake, M. Naito-Fujimoto, Y. Ito, and K. Shigesada. 1993. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA-binding protein, PEBP2α. Virology 194:314-331. [DOI] [PubMed] [Google Scholar]
- 37.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML-1, the target of multiple chromosomal translocations in human leukemia, is essential for normal murine fetal hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 38.Okuda, T., Z. Cai, S. Yang, N. Lenny, C. Lyu, M. A. van Deursen, H. Harada, and J. R. Downing. 1998. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood 91:3134-3143. [PubMed] [Google Scholar]
- 39.Palacios, R., and M. Steinmetz. 1985. IL3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell 41:727-734. [DOI] [PubMed] [Google Scholar]
- 40.Petrovick, M. S., S. W. Hiebert, A. D. Friedman, C. J. Hetherington, D. G. Tenen, and D.-E. Zhang. 1998. Multiple functional domains of AML1: PU.1 and C/EBPα synergize with different regions of AML1. Mol. Cell. Biol. 18:3915-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Redondo, J. M., J. L. Pfohl, C. Hernandez-Munain, S. Wang, N. A. Speck, and M. S. Krangel. 1992. Indistinguishable nuclear factor binding to functional core sites of the T-cell receptor δ and murine leukemia virus enhancers. Mol. Cell. Biol. 12:4817-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki, K., H. Yagi, R. T. Bronson, K. Tominaga, T. Matsunashi, K. Deguchi, Y. Tani, T. Kishimoto, and T. Komori. 1996. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor β. Proc. Natl. Acad. Sci. USA 93:12359-12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Satake, M., S. Nomura, Y. Yamaguchi-Iwai, Y. Takahama, Y. Hashimoto, M. Niki, Y. Kitamura, and Y. Ito. 1995. Expression of the runt domain-encoded PEBP2α genes in T cells during thymic development. Mol. Cell. Biol. 15:1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sohn, R. L., K. L. Vikstrom, M. Strauss, C. Cohen, A. G. Szent-Gyorgyi, and L. A. Leinwand. 1997. A 29 residue region of the sarcomeric myosin rod is necessary for filament formation. J. Mol. Biol. 266:317-330. [DOI] [PubMed] [Google Scholar]
- 45.Strom, D. K., J. Nip, J. J. Westendorf, B. Linggi, B. Lutterbach, J. R. Downing, N. Lenny, and S. W. Hiebert. 2000. Expression of the AML-1 oncogene shortens the G1 phase of the cell cycle. J. Biol. Chem. 275:3438-3445. [DOI] [PubMed] [Google Scholar]
- 46.Suzow, J. G., and A. D. Friedman. 1993. The murine myeloperoxidase promoter contains multiple functional elements—one element binds a cell type-restricted transcription factor, myeloid nuclear factor 1 (MyNF1). Mol. Cell. Biol. 13:2141-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tahirov, T. H., T. Inoue-Bungo, H. Morii, A. Fujikawa, M. Sasaki, K. Kumra, M. Shiina, K. Sato, T. Kumasaka, M. Yamamoto, S. Ishii, and K. Ogata. 2001. Structural analyses of DNA recognition by the AML1/Runx-1 runt domain and its allosteric control by CBFβ. Cell 104:755-767. [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, Y., T. Watanabe, N. Chiba, M. Niki, Y. Kuroiwa, Y. T. Nishihira, S. Satomi, Y. Ito, and M. Satake. 1997. The protooncogene product, PEBP2/CBFβ, is mainly located in the cytoplasm and has an affinity with cytoskeletal structures. Oncogene 15:677-683. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka, Y., M. Fujii, K. Hayashi, N. Chiba, T. Akaishi, R. Shineha, T. Nishihira, S. Satomi, Y. Ito, T. Watanabe, and M. Satake. 1998. The chimeric protein, PEBP2β/CBFβ-SMMHC, disorganizes cytoplasmic stress fibers and inhibits transcriptional activation. Oncogene 17:699-708. [DOI] [PubMed] [Google Scholar]
- 50.Wang, S., Q. Wang, B. E. Crute, I. N. Melnikova, S. R. Keller, and N. A. Speck. 1993. Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol. 13:3324-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, Q., T. Stacy, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl. Acad. Sci. USA 93:3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Q., T. Stacy, J. D. Miller, A. F. Lewis, T.-L. Gu, X. Huang, J. H. Bushweller, J.-C. Bories, F. W. Alt, G. Ryan, P. P. Liu, A. Wynshaw-Boris, M. Binder, M. Marin-Padilla, A. H. Sharpe, and N. A. Speck. 1996. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell 87:697-708. [DOI] [PubMed] [Google Scholar]
- 53.Wang, J., T. Hoshino, R. L. Redner, S. Kajigaya, and J. M. Liu. 1998. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. USA 95:10860-10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, J.-Q., B. A. Harder, P. Uman, and R. Craig. 1996. Myosin filament structure in vertebrate smooth muscle. J. Cell Biol. 134:53-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yanagisawa, M., Y. Hamada, Y. Katsuragawa, M. Imamura, T. Mikawa, and T. Masaki. 1987. Complete primary structure of vertebrate smooth muscle myosin heavy chain deduced from its complementary DNA sequence. J. Mol. Biol. 198:143-157. [DOI] [PubMed] [Google Scholar]
- 56.Yang, Y., W. Wang, R. Cleaves, M. Zahurak, L. Cheng, C. I. Civin, and A. D. Friedman. 2002. Acceleration of G1 cooperates with CBFβ-SMMHC to induce acute leukemia in mice. Cancer Res. 62:2232-2235. [PubMed] [Google Scholar]
- 57.Yergeau, D. A., C. J. Hetherington, Q. Wang, P. Zhang, A. H. Sharpe, M. Binder, M. Marin-Padilla, D. G. Tenen, N. A. Speck, and D.-E. Zhang. 1997. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat. Genet. 15:303-306. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, D.-E., K.-I. Fujioka, C. J. Hetherington, L. H. Shapiro, H.-M. Chen, A. T. Look, and D. G. Tenen. 1994. Identification of a region which directs the monocytic activity of the colony stimulating factor-1 (macrophage-colony stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol. Cell. Biol. 14:8085-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]