Abstract
Methylation of histone H3 has been linked to the assembly of higher-order chromatin structures. Very recently, several examples, including the Schizosaccharomyces pombe mating-type region, chicken β-globin locus, and inactive X-chromosome, revealed that H3-Lys9-methyl (Me) is associated with silent chromatin while H3-Lys4-Me is prominent in active chromatin. Surprisingly, it was shown that homologs of Drosophila Su(var)3-9 specifically methylate the Lys9 residue of histone H3. Here, to identify putative enzymes responsible for destabilization of heterochromatin, we screened genes whose overexpressions disrupt silencing at the silent mat3 locus in fission yeast. Interestingly, we identified two genes, rhp6+ and ubcX+ (ubiquitin-conjugating enzyme participating in silencing), both of which encode ubiquitin-conjugating enzymes. Their overexpression disrupted silencing at centromeres and telomeres as well as at mat3. Additionally, the overexpression interfered with centromeric function, as confirmed by elevated minichromosome loss and antimicrotubule drug sensitivity. On the contrary, deletion of rhp6+ or ubcX+ enhanced silencing at all heterochromatic regions tested, indicating that they are negative regulators of silencing. More importantly, chromatin immunoprecipitation showed that their overexpression alleviated the level of H3-Lys9-Me while enhancing the level of H3-Lys4-Me at the silent regions. On the contrary, their deletions enhanced the level of H3-Lys9-Me while alleviating that of H3-Lys4-Me. Taken together, the data suggest that two ubiquitin-conjugating enzymes, Rhp6 and UbcX, affect methylation of histone H3 at silent chromatin, which then reconfigures silencing.
In eukaryotic cells, chromosomes can be partitioned into two structurally and functionally distinct domains, called euchromatic and heterochromatic regions (5, 33). Unlike the case for euchromatin, heterochromatic regions are condensed even during interphase, and nearby or embedded genes are transcriptionally repressed (called position effect variegation, or silencing). Assembly of these chromatin structures has been linked to posttranslational modification of histone N-terminal tails, including acetylation and phosphorylation (19). In general, heterochromatin contains hypoacetylated histone H3 and H4 compared to the case for euchromatin (7).
Owing to the findings that mammalian and Schizosaccharomyces pombe homologs of Drosophila Su(var)3-9 encode enzymes that specifically methylate histone H3 on lysine 9, histone methylation has emerged as another important modification that distinguishes heterochromatin from euchromatin (30). Methylation of H3 at Lys4 or Lys9 was shown to be reciprocally associated with euchromatic regions and heterochromatic regions, respectively (8, 12, 21, 22, 27, 29). Recently the mechanism by which the H3 methylations are translated into transcriptional states is delineated by the observation that HP1 proteins can bind to Lys9-methylated H3 via their chromo domains (4, 21).
In fission yeast, at least four loci (centromeres, telomeres, silent mating-type loci, and ribosomal DNA) are silenced by heterochromatin-like structures (3, 36). Of the common silencing factors, Clr4, a homolog of Su(var)3-9, has intrinsic H3 Lys9-specific methyltransferase (HMTase) activity both in vitro and in vivo (25). Furthermore, Clr4 recruits Swi6, a fission yeast homolog of HP1, to heterochromatins, suggesting that heterochromatin formation of fission yeast resembles that of higher eukaryotes (9). In addition, Clr3, an H3-specific deacetylase, and Rik1 are required for H3-Lys9 methylation (25). H3-specific deacetylases, such as Clr3 and Clr6, create circumstances favoring methylation at H3-Lys9 by the Clr4/Rik1 complex. Then, methylation induces binding of Swi6, leading to the establishment of a silent chromatin. Once bound to methylated H3, Swi6 serves as an epigenetic imprint for the inheritance of silent chromatin, possibly by recruiting HMTase or other enzymes required for heterochromatin formation after the completion of DNA replication (26). Supporting this model, it was recently shown that Swi6 remains associated with the heterochromatic mat2/3 region throughout the cell cycle, and the mouse homolog of Swi6, M31, physically interacts with Su(var)3-9 (1, 26).
At present, although knowledge of a robust linkage between histone methylation patterns and heterochromatin formation is massively accumulating, it still remains to be understood how the methylation process itself is regulated. Namely, while self-reinforcing mechanisms might be advantageous for the maintenance of silent chromatin, indeed cells may require reconfiguration of silenced chromatin, such as removal of the methyl marker from histone for proper cellular functions, including DNA replication, and mating-type switching, etc. Since histone demethylases are not found yet, the methyl markers of H3 might be removed through the proteolytic pathway (17, 37).
In this report, we demonstrate that two ubiquitin-conjugating enzymes (Ubc or E2), Rhp6 and UbcX, are required for reconfiguration of silenced chromatin in fission yeast. Expressions of RNA Pol II-transcribed genes at heterochromatin are dependent on the dosage of Rhp6 and UbcX. Interestingly, reconfigured silencing induced by altered dosage of the Ubc correlates with the H3 methylation patterns, suggesting a mechanistic link between ubiquitin conjugation and histone H3 methylation.
MATERIALS AND METHODS
Media.
Media were used as described previously (24). For low-adenine medium, YE (2% glucose, 0.5% yeast extract) plates not supplemented with adenine and minimal plates containing only 10% of the required amount of adenine were used. FOA medium contained 0.8 g of 5-fluoroorotic acid (FOA) (an antiuracil selection agent) and 50 mg of uracil in 1 liter of minimal medium.
Plasmids and strains.
All strains used in the experiments are shown in Table 1. To assay silencing, we used reporter strains which contain ade6+ gene inserted at the outer repeat of centromere 1, adjacent to mat3 and nearby telomere of minichromosomes (Hu50, Hu51, and Hu60, respectively). Strain PG9 (h90 mat3-Mint::ura4+ leu1-32 ura4-d18 ade6-M216) was used for screen of silencing regulators. To disrupt ubcX+, a 2.3-kb fragment containing the full-length ubcX+ gene was generated by PCR using primers 1 (5′-GGTTTCTTTGGCGCCTTTCTCTTTG-3′) and 2 (5′-CATGTTTACTGC AGAACGCTGTGC-3′) and subcloned into the SmaI site of pBluescript IIKS (Stratagene, La Jolla, Calif.) to produce pBS-ubcX. A 1.8-kb end-filled HindIII fragment of ura4+ was inserted into pBS-ubcX that was digested with StyI and BclI. A 3.6-kb fragment carrying the ubcX::ura4+ construct was used for transformation of each reporter strain. Mutants carrying null alleles of rhp6+ were constructed by transforming each reporter strain with a rhp6::ura4+ construct. The plasmids pREP3X-rhp6+ and pREP3X-ubcX+ were originally isolated from screening of a S. pombe expression library for silencing regulators, as described in the Results section. pREP2X-rhp6+ was constructed by ligation of a PstI-SmaI fragment of pREP3X-rhp6+ with pREP2 digested with the same enzymes. Similarly, pREP2X-ubcX+ was constructed by subcloning the PstI-SmaI fragment of pREP3X-ubcX+ into pREP2.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| PG9 | h90 ade6-M210 leu1-32 ura4-D18 mat3 (EcoRV)::ura4+ | G. Thon |
| Hu50 | h+ade6-DN/N leu1-32 ura4-D18 otr1R(SphI)::ade6+ | K. Ekwall |
| Hu51 | h90 ade6-DN/N ura4-D18 mat3 (EcoRV)::ade6+ | K. Ekwall |
| Hu60 | h+ade6-DN/ N ura4-D18 Ch16-LEU2-ade6+-tel | K. Ekwall |
| ED666 | h+ade6-M210 leu1-32 ura4-D18 | P. Fantes |
| HM248 | h−ade6-M210 his2 Ch16-LEU2-ade6-M216 | M. Yanagida |
| AL91 | h90 ade6-216 ura4-D18 swi6 ::ura4+ | H. Schmidt |
| JS21 | h+ade6-DN/N leu1-32 ura4-D18 otr 1 R(SphI)::ade6+rhp6::ura4+ | This study |
| JS22 | h90 ade6-DN/N ura4-D18 mat3(EcoRV)::ade6+rhp6::ura4+ | This study |
| JS23 | h+ade6-DN/N ura4-D18 Ch16-LEU2-ade6+-tel rhp6::ura4+ | This study |
| JS161 | h+ade6-M210 leu1-32 ura4-D18 Ch16-ade6-M216 | This study |
| JS1 | h90 leu1-32 ura4-D18 rhp6::ura4+ | This study |
| JS2 | h−ade6-M210 leu1-32 ura4-D18 rhp6::ura4 | This study |
| HJ21 | h+ade6-DN/N leu1-32 ura4-D18 otr 1 R(SphI)::ade6+ubcX::ura4+ | This study |
| HJ22 | h90 ade6-DN/N ura4-D18 mat3(EcoRV)::ade6+ubcX::ura4+ | This study |
| HJ23 | h+ade6-DN/N ura4-D18 Ch16-LEU2-ade6+-tel ubcX::ura4+ | This study |
| HJ199 | h−ade6-M210 leu1-32 ura4-D18 his3-D1 ubcX::ura4+ | This study |
| HJ1 | h90 leu1-32 ura4-D18 ubcX::ura4+ | This study |
| JS31 | h+ade6-DN/N ura4-D18 otr 1 R(SphI)::ade6+rhp6::ura4+ubcX::Kanr | This study |
| JS32 | h90 ade6-DN/N ura4-D18 mat3(EcoRV)::ade6+rhp6::ura4+ubcX::Kanr | This study |
| JS33 | h+ade6-M210 leu1-32 ura4-D18 otr 1 R(SphI)::ade6+rhp6::ura4+ubcX::Kanr | This study |
RNA isolation and RT-PCR.
RNA was isolated as described previously (15). The cDNA sequence was synthesized using reverse transcriptase (RT) (Promega). The cDNAs derived from wild-type ade6+ and the truncated ade6-DN/N were PCR amplified using ade6+ primers (upper, 5′-TGAAAAAGCAGGCC AAGAG-3′; lower, 5′-ACCGGGAATGGACAGAGAAC-3′). The PCR products were analyzed using a 1.4% agarose gel.
Minichromosome loss assays.
Assays were adopted essentially from the work of Allshire et al. (2). Cells from Ade+ colonies were picked and cultured overnight in adenine-free selective media and plated on low-adenine minimal plates. After incubation for 4 to 5 days at 30°C, only red-white half-sectored colonies were counted to determine the loss rate of the ade6+ marker per division. The minichromosome loss rate was calculated by dividing the number of these half-sectored colonies by the total number of white colonies plus half-sectored colonies.
Chromatin immunoprecipitation (ChIP) assay.
Cells (250 ml) were grown to a density of 107 cells/ml and cross-linked with 1% formaldehyde for 20 min at room temperature. Cross-linking was stopped by adding glycine to a concentration of 360 mM for 5 min. Cells were harvested and washed twice with Tris-buffered saline and lysed with glass beads in FA lysis buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na deoxycholate, 0.5% sodium dodecyl sulfate, 1 mM phenylmethylsulfonyl fluoride). The chromatin was sheared by sonication for 20 s eight times. H3 methylated at Lys9 or Lys4 was immunoprecipitated overnight at 4°C with anti-dimethyl Lys9 or Lys4 H3 antibody and protein A-Sepharose beads. From the immunoprecipitates, DNA was released and purified as previously described (20). Purified DNA was PCR amplified with specific primers. PCR was carried with [α-32P]dCTP, and the products were resolved by 6% polyacrylamide gel electrophoresis and exposed to X-ray film.
RESULTS
Identification of cDNAs that disrupt silencing at the mat3 locus when overexpressed.
In fission yeast, prototrophic marker genes placed near the silent mat regions, such as mat2-P and mat3-M, are subject to transcriptional silencing. Since strain PG9 was previously constructed to contain the ura4+ gene adjacent to the mat3 locus (35), the silencing was monitored by using a phenotypic assay for ura4+ expression. Normally, very few cells can grow on uracil-free medium due to the silencing of ura4+ in the reporter cells. Interference with mat3 silencing increases ura4+ expression and then gives rise to mixed clones exhibiting a unique phenotype of Ura+ and FOA sensitivity.
To identify genes whose overexpression reconfigures the mat3 silencing, genetic screening was performed using PG9 and an S. pombe cDNA library. Among 5 × 106 transformants screened, 20 displayed plasmid-linked Ura+ and FOA sensitivity phenotypes. Sequence analysis revealed that 16 of them contained the same sequence encoding Rhp6, which is regarded as an S. pombe homolog of the Saccharomyces cerevisiae E2 enzyme, Ubc2/Rad6 (31). The other four clones contained the same gene encoding a novel putative E2 enzyme that we named ubcX+ (ubiquitin-conjugating enzyme X). The sequence of ubcX+ matches perfectly databases released by the S. pombe Genome Project at Sanger Centre. The ubcX+ sequence has four exons and five introns and encodes a protein of 167 amino acid residues (Fig. 1). The deduced amino acid sequence of UbcX shows 68, 51, and 52% identities with those of human UbcG, S. cerevisiae UBC7, and S. pombe Ubc7, respectively, and also contains a catalytic cysteine residue required for E2-ubiquitin thioester formation (Fig. 1B). The effect of overexpression of each clone on mat3 silencing in PG9 cells was reexamined by measuring cell viability and FOA sensitivity (Fig. 2A). The results showed that overexpression of each gene greatly increased (>100-fold) the ura4+ expression at mat3, possibly due to disruption of heterochromatin structure.
FIG. 1.
Sequence alignment of UbcX. (A) ubcX+ open reading frame. Four introns (open boxes) interrupting the open reading frame and filled boxes indicating exons are shown. The ura4+ gene was used to disrupt the coding region of ubcX+, as indicated. (B) Comparison of the deduced amino acid sequences of UbcX, human UbcG, S. cerevisiae UBC7, and S. pombe Ubc7. The asterisk indicates the putative cysteine residue required for E2-ubiquitin thioester formation. Sequences were aligned using Clustal W, and the output was generated using Genedoc. Identical residues are shown in white text with a black background. Conserved residues are shown with gray shading.
FIG. 2.
Overexpression of each E2 enzyme alleviates silencing at the centromeres and telomeres as well as at the mat3 locus. (A) Overexpression of each gene enhanced the expression of the ura4+ gene in PG9 at least more than 100-fold. The PG9 cells transformed with each cDNA clone or empty vector (pREP3) were spotted onto Ura−, FOA, and nonselective (N/S) plates. Relative amounts of ura4+ expression were measured by the cell viability on uracil-free medium. (B) The effects of overexpression of each ubc gene on mat3 silencing are not gene specific. The overexpression also derepresses the ade6+ expression at mat3 (Hu51, top), at the right-hand side (otr1R) of centromere 1 (Hu50, middle), and at telomeres (Hu60, bottom). Each reporter was transformed with pREP2, pREP2-rhp6+, or pREP2-ubcX+. ade6+ expression was assayed by colony formation ability on adenine-free (−Ade) plates and nonselective plates. (C) Total RNA was prepared from the same strains shown in panel B and subjected to RT-PCR. Each ratio of the level of heterochromatic ade6+ to that of euchromatic ade6-DN/N is also shown.
Overexpression of rhp6+ or ubcX+ disrupts silencing at other heterochromatic regions.
To determine whether the derepression is gene specific, mat3 silencing was assayed using ade6+ as a reporter gene. A reporter strain with ade6+ at mat3 (Hu51) was used as a host, and then ade6+ expression upon overexpression of rhp6+ or ubcX+ was measured. As seen in Fig. 2B (top), the overexpression also derepressed ade6+ expression, confirming that the effect is not gene specific.
Next, we questioned whether the action of each ubc gene was limited to mat3 silencing. When rhp6+ or ubcX+ was overexpressed, the repression of ade6+ was significantly reduced in telomere reporter cells, as in Hu51, but the effect was slight in centromere reporter cells (Fig. 2B). In addition, this derepression was confirmed by competitive RT-PCR between wild-type ade6+ mRNA from each heterochromatin and a truncated ade6-DN/N mRNA from the endogenous ade6 locus. As shown in Fig. 2C, ade6+ transcript from the mat3 locus or telomeres was barely detectable in the wild-type cells containing vector only, but overexpression of each ubc gene increased the level significantly, up to 30% of the ade6-DN/N level. However, there was little effect on derepression of ade6+ at the centromere, indicating that the derepression was not enough to be detected in our hands.
In summary, these observations demonstrate that overexpression of rhp6+ and ubcX+ affects silencing at all heterochromatic regions, suggesting that they act as general silencing regulators, although the effect is somewhat moderate at centromere.
Overexpression of rhp6+ or ubcX+ impairs centromeric functions in a dosage-dependent manner.
Ade+ cells form white colonies on low-adenine medium, while Ade− cells form red colonies, thus allowing the use of colony color as a test of silencing. Thus, a wild-type reporter strain with ade6+ placed near heterochromatin produces predominantly red colonies. Overexpression of rhp6+ or ubcX+ was sufficient to turn the red colonies of mat3 or telomere reporters white (data not shown). In a centromere reporter strain, the overexpressions resulted in very little change in colony color (Fig. 3A) owing to strong silencing at centromeres. However, when the expression of rhp6+ or ubcX+ was enhanced by the introduction of another overexpression vector, derepression of ade6+ was dramatically increased in a dosage-dependent manner (Fig. 3A). Additionally, growth defects were observed in proportion to the level of derepression of the ade6+ gene; the viability of the overexpression cells was decreased to about 10% of the wild-type level (data not shown).
FIG. 3.
Overexpression of rhp6+ and ubcX+ not only affects centromeric silencing but also confers sensitivity to TBZ. (A) A centromere reporter, Hu50, was transformed with pREP2/pREP3, pREP2/pREP3-rhp6+, pREP2/pREP3-ubcX+, pREP2-rhp6+/pREP3-rhp6+, and pREP2-ubcX+/pREP3-ubcX+. Each transformant was grown on low-adenine medium, and the colony colors were compared. (B) TBZ sensitivity test. Wild-type CF199 transformed with the same vectors used in panel A was spotted onto plates containing TBZ or dimethyl sulfoxide for a mock control.
It was previously reported that mutations involved in silencing defects at centromeres are accompanied by defective centromeric function (2, 6, 28). To investigate effects of the overexpression of rhp6+ or ubcX+ on centromeric function, the rate of loss of minichromosome Ch16 was measured. The overexpression enhanced the loss rates up to 0.12 and 0.17% per division, which are 6- and 8.5-fold higher than with the wild type (Table 2). When ubc expression was enhanced by introduction of another overexpression vector, the loss rate was further increased to 135- and 165-fold, respectively.
TABLE 2.
The Effect of Rhp6 and UbcX overexpression on loss of Ch16 minichromosome
| Background | No. of half-sectored colonies/no. of total colonies | Loss rate (%) | Relative loss ratea (fold) |
|---|---|---|---|
| WTb | 0/1418, 1/2137, 0/1369 | 0.02 | 1 |
| pREP3-rhp6+ | 0/1769, 2/510, 2/1013 | 0.12 | 6 |
| pREP2-ubcX+ | 0/1208, 2/900, 3/826 | 0.17 | 8.5 |
| pREP3-rhp6+, pREP3-rhp6+ | 6/158, 5/197, 8/341 | 2.7 | 135 |
| pREP2-ubcX+, pREP2-ubcX+ | 5/178, 4/102, 5/143 | 3.3 | 165 |
Increase in rate compared with that of the wild-type control.
WT, wild type.
Mutations interfering with centromeric function affect the interaction of microtubules with kinetochore and show sensitivity to the microtubule-destabilizing drug thiabendazole (TBZ) (9, 10). To determine whether ubc overexpression has a similar effect, we examined TBZ sensitivity. As expected, the overexpression conferred TBZ sensitivity to the host cells in a dosage-dependent manner (Fig. 3B). The results suggest that overexpression of rhp6+ and ubcX+ impairs centromeric function by interfering with heterochromatin formation at centromeres (6, 28). Together, the data argued that cellular levels of Rhp6 and UbcX are critical for both centromeric silencing and function.
Loss of function of Rhp6 or UbcX enhances silencing.
Since overexpression of rhp6+ or ubcX+ influenced silencing at all heterochromatic regions tested, the effect of deficiency in each gene on silencing was investigated. Deletion of rhp6+ caused reduced cell growth on adenine-free medium, indicating markedly enhanced repression of the ade6+ gene at mat3, centromeres, and telomeres (Fig. 4A). The ΔubcX cells showed a very slight increase in silencing at the heterochromatic regions tested compared with wild-type cells (Fig. 4B).
FIG. 4.
Silencing is enhanced by Δrhp6 or ΔubcX mutation at the mat3 locus, telomeres, and centromeres. (A) A wild-type or null allele of rhp6+ was introduced into each silencing reporter strain (Hu51, Hu50, and Hu60). The resultant cells were spotted onto Ade-free medium, and then the photograph was taken. (B) The effects of ubcX+ deletion on silencing were monitored as described for panel A.
Next, we performed a transition assay to measure the effect of Δrhp6 or ΔubcX on inheritance of the repressed state. Interestingly, the ubcX deletion as well as the rhp6 deletion caused a significant decrease in Ade− (red)-to-Ade+ (white) conversion (Table 3), indicating increased silencing. These results suggest that rhp6+ and ubcX+ both act as a negative regulator of silencing, although the effect of UbcX is very slight.
TABLE 3.
Effect of Δrhp6 or ΔubcX mutation on inheritance of repressed state
| Background | Genotype | Rate of half-sectoring (per cell division)a | Relative transition rateb |
|---|---|---|---|
| mat3::ade6+ | WTc | 1.0 × 10−2 (85/8,333) | 1 |
| mat3::ade6+ | rhp6::ura4+ | 2.3 × 10−4 (1/4,293) | 1/44 |
| mat3::ade6+ | ubcX::ura4+ | 4.7 × 10−4 (4/8,460) | 1/21 |
| otr1R::ade6+ | WT | 2.3 × 10−3 (23/9,975) | 1 |
| otr1R::ade6+ | rhp6::ura4+ | 1.4 × 10−4 (4/7,040) | 1/16 |
| otr1R::ade6+ | ubcX::ura4+ | 2.3 × 10−4 (2/8,548) | 1/10 |
| ade6+-tel | WT | 1.1 × 10−2 (85/7,821) | 1 |
| ade6+-tel | rhp6::ura4+ | <1.4 × 10−4 (0/7,270) | <1/79 |
| ade6+-tel | ubcX::ura4+ | 1.1 × 10−4 (1/9,333) | 1/100 |
Number of half-sectored colonies divided by the total number of colonies.
Increase in rate compared with that of each wild-type control.
WT, wild type.
Rhp6 and UbcX affect H3 methylation patterns at heterochromatic regions.
Since histone demethylase has not been reported yet, proteolytic degradation of the entire H3 histone was proposed to be a potential mechanism for the removal of the methyl marker required for reconfiguration of silencing (17, 37). Interestingly, our finding that two E2 enzymes negatively regulate silencing fits well with this idea. To test this hypothesis, we investigated whether Rhp6 or UbcX affects methylation of H3 by using a ChIP assay. To observe changes in histone methylation in cells overexpressing or lacking rhp6+ or ubcX+, we performed the ChIP assay using antibodies against H3 methylated at Lys9 or Lys4. Because the effects of rhp6+ or ubcX+ on silencing were shown at centromeres, the mat3 locus, and telomeres, DNA amplification was carried out with primers specific for ade6+, which is inserted at these regions. Consistent with the silencing phenotype mentioned above, the results showed a dramatic decrease in the level of H3-Lys9-Me at ade6+ with rhp6+ overexpression at the mat3 locus (Fig. 5A, lane 5, top) and moderate decreases at centromeres and telomeres (Fig. 5A, lanes 2 and 8, top). A much weaker decrease in the level of H3-Lys9-Me at ade6+ was also observed in cells overexpressing ubcX+ (Fig. 5A, lanes 3, 6, and 9, top). On the contrary, the level of H3-Lys4-Me was significantly increased at the mat3 locus and telomere in cells overexpressing rhp6+ or ubcX+ (Fig. 5A, lanes 5, 6, 8, and 9, middle). However, the level of H3-Lys4-Me at the centromere was not significantly changed by overexpression of rhp6+ or ubcX+. Unlike the overexpression results, an opposite result was displayed by deletion of the ubc genes. Deletion of rhp6+ greatly increased the level of H3-Lys9-Me at centromeres, the mat3 locus, and telomeres (Fig. 5B, lanes 2, 5, and 8, top). Although deletion of rhp6+ does not cause a detectable change in the level of H3-Lys4-Me at the mat3 locus (Fig. 5B, lane 5, middle), a slight decrease was induced at centromeres and telomeres in Δrhp6 cells (Fig. 5B, lanes 2 and 8, middle). As expected from its weak silencing phenotype, ΔubcX did not affect methylation patterns as greatly as Δrhp6. However, we could observe a significant increase in the level of H3-Lys9-Me at centromeres and telomeres (Fig. 5B lanes 3 and 9, top) and decrease in the level of H3-Lys4-Me at centromeres, the mat3 locus, and telomeres (Fig. 5B, lanes 3, 6, and 9, middle), a result consistent with the increased silencing found in ΔubcX cells. To test if the effects of the ubc genes on H3 methylation are due to the altered ade6+ gene expression, we performed a ChIP assay with primers specific to the nontranscribed mat2-adjacent region, indicated as mat2-r in Fig. 5C (top). Deletion of rhp6+ greatly enhanced the level of H3-Lys9-Me and reduced the level of H3-Lys4-Me, while overexpression reduced H3-Lys9-Me and enhanced H3-Lys4-Me. Deletion of ubcX+ enhanced H3-Lys9-Me and reduced H3-Lys4-Me, but the effect was not as dramatic as that of rhp6+ and we couldn't detect significant changes in H3 methylation in ubcX+-overexpressing cells at this region (Fig. 5C). This result suggests that the effects of the ubc genes on H3 methylation at heterochromatic regions are not due to the altered expression of ade6+ inserted at these regions.
FIG.5.
The altered silencing in cells overexpressing or lacking rhp6+ or ubcX+ correlates with the H3-Lys9-Me patterns at silent regions. Schematic representation of silent mating-type region and the positions of primers used in ChIP (top) are shown. (A) Levels of Lys4-Me or Lys9-Me of H3 at centromeres, mat3 locus, and telomeres were determined in cells containing pREP2, pREP2-rhp6+, or pREP2-ubcX+ by ChIP with anti-dimethyl H3 antibody. DNA extracted from ChIP or whole-cell extracts (WCE) was amplified by competitive PCR using primer sets specific to ade6+. Endogenous ade6-DN/N fragment serves as an internal control. The ratios of ade6+ and control ade6-DN/N signals present in WCE were used to calculate relative precipitated fold enrichment show underneath each line. (B) Levels of Lys4-Me or Lys9-Me of H3 in Δrhp6 or ΔubcX cells were monitored as described for panel A. (C) Physical map of the mating-type locus indicating the site of PCR amplification (mat2-r) (top). Levels of Lys4-Me or Lys9-Me of H3 at mat2-r were monitored in cells overexpressing or lacking rhp6+ or ubcX+. Endogenous leu1+ fragment serves as an internal control.
To test the possibility that these ubc genes directly conjugate ubiquitin to H3-Lys9-Me, we investigated whether H3-Lys9-Me is ubiquitinated using a Western blot. However, we failed to detect H3-Lys9-Me, probably due to its extremely low abundance (data not shown). Thus, at this stage we cannot conclude whether the effects of these ubc genes on histone methylation are direct or indirect.
Collectively, these results suggest that rhp6+ and ubcX+ negatively regulate histone H3-Lys9 methylation either directly or indirectly, which can then affect silencing at heterochromatic regions.
DISCUSSION
In this report, we show that two ubiquitin-conjugating enzymes, Rhp6 and UbcX, are negative regulators of heterochromatin structure. Their overexpression disrupted silencing, while a deficiency of each gene enhanced heterochromatic silencing, indicating a dosage-dependent regulation of silencing by Ubc enzymes.
Over the past few decades, several reports strongly argued that components of the ubiquitin pathway are involved in heterochromatin silencing. Deletion of UBP3, which encodes a deubiquitinating enzyme interacting with Sir4, greatly increases silencing at both the telomeres and HML, suggesting that UBP3 antagonizes silencing (23). Similarly, mutation in a gene encoding a putative Drosophila UBP enhances transcriptional repression at centric heterochromatin (13). On the contrary, loss of Dot4, another UBP, resulted in a partial loss of silencing (18). Mutation of S. cerevisiae RAD6 disrupted silencing at HM loci and at telomeres (14). Despite these observations, there had been no clear demonstrations on the molecular role of ubiquitin in regulating silencing. However, recently one of the molecular roles of ubiquitin in silencing was demonstrated by Sun and Allis with S. cerevisiae (34). In this study, they reported that ubiquitination of histone H2B at Lys123 by Rad6 is required for the methylation of histone H3 at Lys4 and for telomeric silencing in S. cerevisiae. If this is also the case for S. pombe, we can interpret our results in view of this mechanism. Since Rhp6 and UbcX negatively regulate silencing and H3-Lys9-Me, we can assume that overexpression of these enzymes would lead to more H2B ubiquitination in heterochromatic regions, blocking Lys9 methylation and promoting Lys4 methylation. Conversely, deletion of rhp6+ or ubcX+ would abolish H2B ubiquitination and Lys4 methylation in heterochromatic regions, increasing Lys9 methylation and silencing. Although there has been no report on Rhp6-mediated H2B ubiquitination in S. pombe, this model fits very well with our results. However, while deletion of RAD6 abolishes whole Lys4 methylation in S. cerevisiae, deletion of rhp6+ affects Lys4 methylation only at heterochromatic regions in S. pombe (Fig. 5 and unpublished results). Therefore, it is assumed that Rhp6 and UbcX are localized exclusively at heterochromatic regions. However, in our ChIP analysis, neither Rhp6 nor UbcX associated with heterochromatic regions (data not shown).
Alternatively, Rhp6 or UbcX may conjugate ubiquitin directly to H3-Lys9-Me, leading to its proteolysis by the 26S proteasome. Since no histone demethylase has been reported yet, these E2 enzymes may serve as histone demethylation machinery by destabilizing the entire H3 histone. In this model, the overexpression of rhp6+ or ubcX+ is assumed to promote proteolysis of H3-Lys9-Me, leading to disruption of silencing, while their deletions prevent its proteolysis and thus stabilize silencing. This model, which emphasizes a more direct role of ubiquitin, is supported by a report that Rad6 can conjugate ubiquitin to H3 in vitro (11).
Another possible model is that Rhp6 or UbcX may conjugate ubiquitin to a component of the HMTase complex, leading to its proteolysis. This model needs the following speculation: increased activity of HMTase by deletion of the ubc gene would induce increased methylation of H3 at Lys9 and subsequent enhancement in silencing. Supporting this assumption, overexpression of Clr4, an H3-Lys9 HMTase, indeed increases mat3 silencing in fission yeast (26). Histone methylation by Clr4 requires Rik1, a beta propeller domain-containing protein that is assumed to form a complex with Clr4 (25). Thus, given that Rhp6 or UbcX regulates H3-Lys9-Me via the HMTase complex, the most likely candidate for the substrate would be Clr4 or Rik1. However, since the epitope-tagged Clr4 was very stable and Rik1 did not undergo ubiquitination (our unpublished observations), it is unlikely that the ubiquitin pathway degrades Clr4 or Rik1.
Among these models, the first model is the most plausible, but we cannot exclude the other models at this point. To determine which model is correct requires further studies, including a study of whether ubiquitination of H2B by Rhp6 occurs and regulates H3 methylation in S. pombe, whether H3-Lys9-Me is directly ubiquitinated by Rhp6 and UbcX, and whether the 26S proteasome is involved. Indeed, a recent finding reported that mutations in the genes encoding components of the 26S proteasome enhance silencing within centromeres in S. pombe (16).
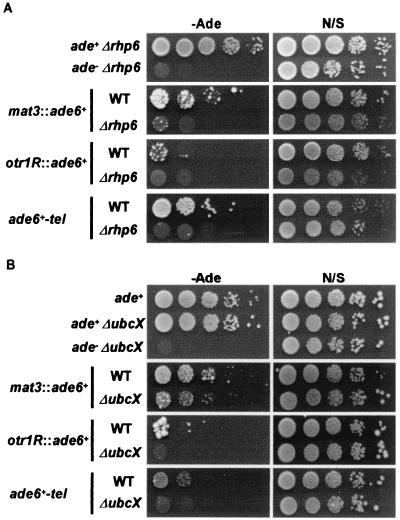
In contrast to a negative role of Rhp6 in silencing, presented here, a recent study suggested its positive role in silencing (32). According to the study, rhp6+ is required to maintain the repression of silent mat genes in switching-competent cells, suggesting its role in the reestablishment of silencing. Consistent with this result, we found that simultaneous deletion of rhp6+ and ubcX+ caused a marked derepression of ade6+ at mat3 in switching-competent cells (Fig. 6). To accommodate this phenotypic discrepancy for the role of Rhp6, we assume that Rhp6 and UbcX may target more than one substrate in regulating silencing.
FIG. 6.
Either rhp6+ or ubcX+ is required to maintain silencing at the mat3 locus. Serially diluted wild-type or Δrhp6ΔubcX cells were spotted onto complete (N/S) and adenine-free (−Ade) plates. The photograph was taken after 4 days of incubation at 30°C.
ACKNOWLEDGEMENTS
We thank K. Ekwall, G. Thon, and M. Yanagida for providing yeast strains and B. Edgar, C. Norbury, L. Prakash, J. Pringle, and R. Rowley for providing S. pombe cDNA library and plasmids. Also, we thank Gwen Sancar and O. Hwang for their critical readings of the manuscript.
This work was supported in part by the Grants for Leading Scientist from the Korea Science and Engineering Foundation (2001) (to S.D.P.), by Research Fellowship BK21 from the Korean Ministry of Education (to E.S.C., H.S.K., S.D.P.), and by Korea Research Foundation (2001; DP0401) (2002; to S.D.P.). This study was also supported in part by a research grant from the National Cancer Center, Ministry of Health & Welfare (to Y.K.J.).
REFERENCES
- 1.Aagaard, L., G. Laible, P. Selenko, M. Schmid, R. Dorn, G. Schotta, S. Kuhfittig, A. Wolf, A. Lebersorger, P. B. Singh, G. Reuter, and T. Jenuwein. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18:1923-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allshire, R. C., E. R. Nimmo, K. Ekwall, J. P. Javerzat, and G. Cranston. 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9:218-233. [DOI] [PubMed] [Google Scholar]
- 3.Allshire, R. C. 1996. Transcriptional silencing in the fission yeast; a manifestation of higher-order chromosome structure and functions, p. 443-466. In V. E. Russo et al. (ed.), Epigenetic mechanisms in gene regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 4.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A. C., A. G. West, and G. Felsenfeld. 2001. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science 291:447-450. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, P., J. F. Maure, J. F. Patridge, S. Genier, J. P. Javerzat, and R. C. Allshire. 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294:2539-2542. [DOI] [PubMed] [Google Scholar]
- 7.Bjerling, P., R. A. Silverstein, G. Thon, A. Caudy, S. Grewal, and K. Ekwall. 2002. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Mol. Cell. Biol. 22:2170-2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis. 2002. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30:73-76. [DOI] [PubMed] [Google Scholar]
- 9.Ekwall, K., E. R. Nimmo, J. P. Javerzat, B. Borgstrom, R. Egel, G. Cranston, and R. Allshire. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109:2637-2648. [DOI] [PubMed] [Google Scholar]
- 10.Freeman-Cook, L. L., J. M. Sherman, C. B. Brachman, R. C. Allshire, J. D. Boeke, and L. Pillus. 1999. The Schizosaccharomyces pombe hst4+ gene is a SIR2 homologue with silencing and centromeric functions. Mol. Biol. Cell. 10:3171-3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haas, A., P. M. Reback, G. Pratt, and M. Rechsteiner. 1990. Ubiquitin-mediated degradation of histone H3 does not require the substrate-binding ubiquitin protein ligase, E3, or attachment of polyubiquitin chains. J. Biol. Chem. 265:21664-21669. [PubMed] [Google Scholar]
- 12.Heard, E., C. Rougeulle, D. Arnaud, P. Avner, C. D. Allis, and D. L. Spector. 2001. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107:727-738. [DOI] [PubMed] [Google Scholar]
- 13.Henchoz, S., F. De Rubertis, D. Pauli, and P. Spierer. 1996. The dose of a putative ubiquitin-specific protease affects position-effect variegation in Drosophila melanogaster. Mol. Cell. Biol. 16:5717-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, H., A. Kahana, D. E. Gottschling, L. Prakash, and S. W. Liebman. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jang, Y. K., Y. H. Jin, M. K. Kim, R. H. Seong, S. H. Hong, and S. D. Park. 1995. A simple and efficient method for the isolation of total RNA from the fission yeast Schizosaccharomyces pombe. Biochem. Mol. Biol. Int. 37:339-344. [PubMed] [Google Scholar]
- 16.Javerzat, J. P., G. McGurk, G. Cranston, C. Barreau, P. Bernard, C. Gordon, and R. C. Allshire. 1999. Defects in components of the proteasome enhance transcriptional silencing at fission yeast centromeres and impair chromosome segregation. Mol. Cell. Biol. 19:5155-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 193:1074-1080. [DOI] [PubMed] [Google Scholar]
- 18.Kahana, A., and D. E. Gottschling. 1999. DOT4 links silencing and cell growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6608-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 20.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 21.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 22.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 23.Moazed, D., and D. Johnson. 1996. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86:667-677. [DOI] [PubMed] [Google Scholar]
- 24.Moreno, S., A. Klar, and P. Nurse. 1991. Methods. Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama, J., A. J. Klar, and S. I. Grewal. 2000. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101:307-317. [DOI] [PubMed] [Google Scholar]
- 27.Noma, K., C. D. Allis, and S. I. Grewal. 2001. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 293:1150-1155. [DOI] [PubMed] [Google Scholar]
- 28.Nonaka, N., T. Kitajima, S. Yokobayashi, G. Xiao, M. Yamamoto, S. I. Grewal, and Y. Watanabe. 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell. Biol. 4:89-93. [DOI] [PubMed] [Google Scholar]
- 29.Peters, A. H., J. E. Mermoud, D. O'Carroll, M. Pagani, D. Schweizer, N. Brockdorff, and T. Jenuwein. 2002. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30:77-80. [DOI] [PubMed] [Google Scholar]
- 30.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 31.Reynolds, P., M. H. Koken, J. H. J. Hoeijmakers, S. Prakash, and L. Prakash. 1990. The rhp6+ gene of Schizosaccharomyces pombe: a structural and functional homolog of the RAD6 gene from the distantly related yeast Saccharomyces cerevisiae. EMBO J. 9:1423-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh, J., V. Goel, and A. J. Klar. 1998. A novel function of the DNA repair gene rhp6+ in mating-type silencing by chromatin remodeling in fission yeast. Mol. Cell. Biol. 18:5511-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, F. L., and S. C. Elgin. 1999. Putting boundaries on silence. Cell 99:459-462. [DOI] [PubMed] [Google Scholar]
- 34.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 35.Thon, G., A. Cohen, and A. J. Klar. 1994. Three additional linkage groups that repress transcription and meiotic recombination in the mating-type region of Schizosaccharomyces pombe. Genetics 138:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thon, G., and J. Verhein-Hansen. 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155:551-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]