Abstract
Translation of terminal oligopyrimidine tract (TOP) mRNAs, which encode multiple components of the protein synthesis machinery, is known to be controlled by mitogenic stimuli. We now show that the ability of cells to progress through the cell cycle is not a prerequisite for this mode of regulation. TOP mRNAs can be translationally activated when PC12 or embryonic stem (ES) cells are induced to grow (increase their size) by nerve growth factor and retinoic acid, respectively, while remaining mitotically arrested. However, both growth and mitogenic signals converge via the phosphatidylinositol 3-kinase (PI3-kinase)-mediated pathway and are transduced to efficiently translate TOP mRNAs. Translational activation of TOP mRNAs can be abolished by LY294002, a PI3-kinase inhibitor, or by overexpression of PTEN as well as by dominant-negative mutants of PI3-kinase or its effectors, PDK1 and protein kinase Bα (PKBα). Likewise, overexpression of constitutively active PI3-kinase or PKBα can relieve the translational repression of TOP mRNAs in quiescent cells. Both mitogenic and growth signals lead to phosphorylation of ribosomal protein S6 (rpS6), which precedes the translational activation of TOP mRNAs. Nevertheless, neither rpS6 phosphorylation nor its kinase, S6K1, is essential for the translational response of these mRNAs. Thus, TOP mRNAs can be translationally activated by growth or mitogenic stimuli of ES cells, whose rpS6 is constitutively unphosphorylated due to the disruption of both alleles of S6K1. Similarly, complete inhibition of mammalian target of rapamycin (mTOR) and its effector S6K by rapamycin in various cell lines has only a mild repressive effect on the translation of TOP mRNAs. It therefore appears that translation of TOP mRNAs is primarily regulated by growth and mitogenic cues through the PI3-kinase pathway, with a minor role, if any, for the mTOR pathway.
Cell proliferation involves two processes: cell growth (increase in cell size) and cell division, which are normally intermingled, to the extent that cells must attain a minimal size to progress in the cell cycle. The dependence of DNA replication and cell division on cellular growth appears to enable accumulation of cellular resources to ensure daughter cell survival. Growth is characterized by elevated production of the translational apparatus needed to cope with the increasing demand for protein synthesis (42). Indeed, according to one estimate, most of the energy consumed during cellular growth is utilized for generating the components of the protein synthesis machinery (53).
TOP mRNAs, which encode many components of the translational apparatus [ribosomal proteins, elongation factors eEF1A and eEF2, and poly(A)-binding protein], are translationally regulated by mitogenic signals through their 5′ terminal oligopyrimidine tract (5′TOP) (35). Translational repression of TOP mRNAs is apparent when proliferation of vertebrate cells is blocked by a wide variety of physiological signals (terminal differentiation, contact inhibition, and serum starvation) or by cell cycle inhibitors (aphidicolin and nocodazole). The shift of TOP mRNAs from polysomes into mRNP particles (subpolysomal fraction) under such circumstances clearly indicates that the translational repression results from a block at the translational initiation step (37).
PI3-kinase is a receptor-proximal component of a mammalian growth-regulating pathway. It has a number of downstream effectors and has been implicated in a wide variety of cellular responses (reviewed in reference 10). Stimulation of a variety of growth factor receptors leads to enhanced PI3-kinase activity and elevated levels of its products, phosphatidylinositol-3,4,5-P3 and phosphatidylinositol-3,4-P2. Increased levels of these lipids are also apparent by the loss of function of PTEN (phosphatase and tensin homolog deleted from chromosome 10), which cleaves the D3 phosphate of phosphatidylinositol-3,4,5-P3 and phosphatidylinositol-3,4-P2 (reviewed in reference 34). These second-messenger lipids participate with 3-phophoinositide-dependent kinase 1 (PDK1) in activation of protein kinase B (PKB), which appears to mediate downstream events controlled by PI3-kinase (reviewed in references 8 and 32).
In mammals, the three isoforms PKBα, PKBβ, and PKBγ are encoded by distinct genes. PKBα-deficient mice are viable, but their growth is retarded (14). PKBβ-null mice are viable but impaired in their ability to maintain normal blood glucose homeostasis (13). Likewise, Drosophila eye cells which are PKB (Dakt1) deficient exhibit a reduction in size (65). Overexpression of PKBα in mouse pancreatic β-cells leads to an increase in their size (64). Taken together, it appears that PKBα is a potent determinant of cell size in mammals.
One of the downstream targets of PI3-kinase is the ribosomal protein S6 (rpS6), whose phosphorylation is carried out by two closely related kinases, S6K1 and S6K2, very early following mitogenic stimuli (reviewed in reference 20). The activation of S6K relies, in addition to the PI3-kinase-medaited pathway, on the mammalian target of rapamycin (mTOR; also known as FRAP or RAFT) (reviewed in reference 35). The fact that this event precedes the translational activation of TOP mRNAs has led to a model which attributes the translational efficiency of these mRNAs to S6K activity and rpS6 phosphorylation (26, 27, 62). This model has been based on two lines of evidence. First, inhibition of S6K1 and S6K2, and consequently of rpS6 phosphorylation by the immunosuppressant rapamycin (an mTOR-specific inhibitor) led to suppression of the mitogenic activation of TOP mRNA translation in a selected set of cell lines (reviewed in reference 23). Second, overexpression of a dominant-interfering mutant of S6K1 has been shown to exert a minor inhibitory effect on the translational activation of a chimeric TOP mRNA following mitogenic stimulation (25).
It has recently been shown that in addition to their regulation by mitogenic stimulation, TOP mRNAs are translationally controlled by amino acid sufficiency (58). However, this mode of regulation requires neither S6K1 activity nor rpS6 phosphorylation (58). Moreover, inhibition of mTOR by rapamycin led to fast and complete repression of S6K1, as judged from the lack of rpS6 phosphorylation, but to only partial and delayed repression of translational activation of TOP mRNAs (58). These findings, together with the rapamycin resistance of TOP mRNAs in some cell lines (26, 28), have cast doubts on the role of the mTOR-mediated pathway in the translational control of TOP mRNAs.
Here we show for the first time that TOP mRNAs are translationally activated when cells are induced to grow, even in the absence of cell proliferation. Experiments based on multiple strategies have disclosed that transduction of both growth and mitogenic signals into translational efficiency of TOP mRNAs is absolutely dependent on the PI3-kinase-mediated pathway. In contrast, S6K activity and rpS6 phosphorylation are fully dispensable for this mode of regulation, and the inhibition of mTOR has only a minor, if any, repressive effect on the translational efficiency of TOP mRNAs.
MATERIALS AND METHODS
Cell culture and DNA transfection.
PC12 cells were grown in 100-mm plates in Dulbecco's modified Eagle's medium supplemented with 8% fetal calf serum, 8% donor horse serum, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml. Neuronal differentiation was induced with 25 ng of mouse nerve growth factor per ml (Alomone Labs). Cells were starved for serum by removal of the growth medium, washing once with 5 ml of phosphate-buffered saline and incubation with serum-free medium for the indicated times. Rapamycin (Sigma) and LY294002 (Sigma) were added at 20 nM and 50 μM, respectively.
Akt1/PKBα+/+ and Akt1/PKBα−/− mouse embryo fibroblasts (MEF) were derived as described previously (63) and grown in 100-mm plates in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml. R1 mouse embryonic stem (ES) cells and their S6K1-deficient derivative cells (p70S6K−/−) were kindly provided by Naohiro Terada and grown as described previously (58).
Neuronal differentiation of S6K−/− ES cells was performed as previously described (5, 6). Briefly, cells were trypsinized and transferred into bacterial plates, in which they spontaneously aggregated into embryoid bodies. Four days of incubation in the absence of retinoic acid were followed by 4 days of incubation in its presence (0.5 μM), and then cells were transferred into tissue culture plates. The proliferation of dividing or differentiating S6K−/− ES cells was quantified by the methylene blue staining protocol (41). Human embryonic kidney 293 cells were grown in 100-mm plates and transfected as described previously (22). Mitotic arrest of MEF and S6K−/− ES cells was achieved by incubation in serum-free medium for 44 h. 293 cells were serum starved by first transferring cells into bacterial plates and then incubating in serum-free medium for 26 h.
Mitotic stimulation of these cells was carried out by serum refeeding after they had been transferred back into cell culture plates. WHI1660, WHI1086, WHI1249, and WHI1615 are human lymphoblastoid cell lines (kindly provided by Andrew Zinn) derived by transformation with Epstein-Barr virus and represent a normal female, a normal male, a female with XY chromosomes without Turner syndrome, and a female with XY chromosomes with Turner syndrome, respectively. Cells were grown in suspension (generation time, 26 h) as described previously (4) and harvested for polysomal or Western blot analyses at mid-log phase (5 × 105 to 7 × 105 cells/ml). P1798.C7 mouse lymphosarcoma cells were grown as suspension cultures and mitotically arrested by treatment with 0.1 μM dexamethasone (Sigma) for 24 h as previously described (4).
Polysomal fractionation and RNA analysis.
Polysomal fractionation and RNA analysis were performed as previously described (58).
Molecular probes.
The isolated fragment probes used in the Northern blot analysis were a 0.97-kb fragment bearing the rpL32 processed gene, 4A (16); a 1.15-kb PstI fragment containing mouse α-actin cDNA (39); a 1.8-kb BglI fragment containing mouse EF1α cDNA (kindly provided by L. I. Slobin); a 0.8-kb HindIII fragment containing human growth hormone cDNA (kindly provided by T. Fogel, Bio-Technology General); a 0.74-kb NcoI-HindIII fragment containing green fluorescent protein (GFP) cDNA from pEGFP-C1 (Clontech); and a 0.6-kb EcoRI fragment containing the PKBα-specific probe (14).
Western blot analysis.
Immunoblotting was performed as described previously (43) with anti-rpS6, anti-phospho-rpS6 (Ser235/236), or anti-phospho-rpS6 (Ser240/244), anti-PKB, anti-S6K1, anti-phospho-S6K1 (Thr389), and mouse-specific anti-phospho-4E-BP (Thr70) antibodies (Cell Signaling Technology, Beverly, Mass.). The preparation and specificities of the antibodies against rpS6 and its phosphorylated derivatives have been described (58). The use of longer gels in some experiments enabled the detection by anti-rpS6 of two bands, representing unphosphorylated and phosphorylated rpS6 (at least at positions Ser235/236), which were designated α and β, respectively.
RESULTS
TOP mRNAs are translationally activated by growth stimulation even without cell division.
Cells double their ribosome content to ensure attainment of an appropriate size prior to division as well as survival of the daughter cells (1). Indeed, it has previously been shown that TOP mRNAs are translationally activated upon mitogenic stimulation (reference 37 and references therein). However, we were intrigued by the possibility that growth (increase in cell mass) induction might be sufficient for translational activation of TOP mRNAs even if not accompanied by cell division.
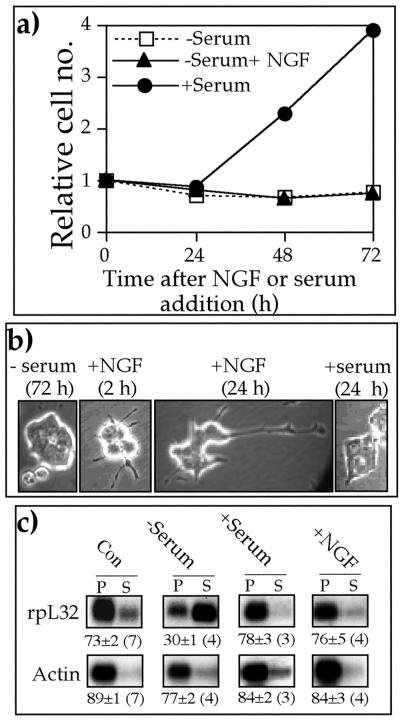
To directly address this issue, rat pheochromocytoma PC12 cells were serum starved for 72 h and then kept without serum, refed with serum, or treated with nerve growth factor (NGF) for an additional 72 h. Figure 1a shows, as reported previously (51), that cells resumed their proliferation only following serum refeeding. However, neurite outgrowth was readily detectable already 2 h after NGF introduction, with multiple long neurites apparent after 24 h (Fig. 1b). Polysomal analysis of a typical TOP mRNA encoding rpL32 showed that its translation was selectively repressed in serum-starved cells, as judged from its exclusion from polysomes. This mRNA, however, was rapidly (within 0.5 h) recruited into polysomes upon mitogenic or growth stimulation by serum refeeding or NGF treatment, respectively (Fig. 1c). It therefore appears that NGF-stimulated growth even without cell division suffices for translational activation of TOP mRNAs.
FIG. 1.
NGF induces translational activation of TOP mRNAs in nondividing PC12 cells. (a) PC12 cells were serum starved for 72 h (zero time) and then either serum refed (+Serum) or further serum starved without (−Serum) or with 25 nM NGF (−Serum + NGF). At the indicated times, cells were trypsinized and counted. Numbers of cells (average of at least two repetitions for each time point) were normalized to the number at time zero. (b) PC12 cells were serum starved for 72 h (−serum) and then either serum refed (+serum) or treated with 25 nM NGF (+NGF). At the indicated times, cells were microscopically visualized at the same magnification. (c) Untreated (Con), 72-h serum-starved (−Serum), 72-h serum-starved and then 0.5-h serum-refed (+Serum), and NGF (25 nM)-treated (+NGF) PC12 cells were harvested, and cytoplasmic extracts were prepared. These extracts were centrifuged through sucrose gradients and separated into polysomal (P) and subpolysomal (S) fractions. RNA from equivalent aliquots of these fractions was analyzed by Northern blot hybridization with cDNAs for actin and rpL32. The radioactive signals were quantified by phosphorimager, and the relative translational efficiency of each mRNA is presented numerically beneath the autoradiograms as a percentage of the mRNA engaged in polysomes. These values are expressed as the average ± standard error of the mean, with the number of determinations shown in parentheses, or as the average, with the individual values in parentheses, if only two determinations were available.
PI3-kinase-mediated pathway is indispensable for growth and mitotic activation of TOP mRNA translation.
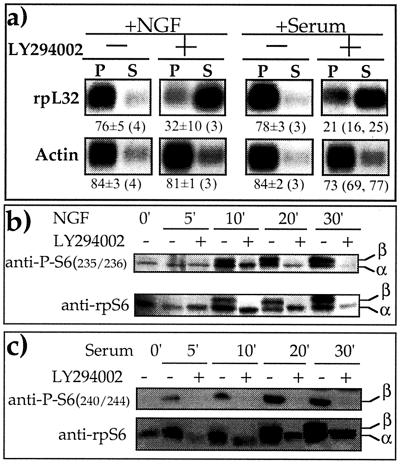
It has previously been shown that epidermal growth factor and NGF induce PI3-kinase in PC12 cells (11). Furthermore, we have demonstrated that translational control of TOP mRNAs by amino acid sufficiency requires the active PI3-kinase-dependent pathway (58). Hence, we set out to examine the role of PI3-kinase in translational activation of TOP mRNAs upon growth or mitogenic stimulation. Figure 2a shows that LY294002, a specific inhibitor of PI3-kinase (66), completely blocked the translational activation of rpL32 mRNA in NGF-treated and serum-refed PC12 cells (Fig. 2a). Furthermore, this agent acted in parallel to completely inhibit the phosphorylation of rpS6 (Fig. 2b and 2c).
FIG. 2.
LY294002 inhibits the phosphorylation of rpS6 as well as the translational activation of TOP mRNAs upon growth or mitogenic stimulation. (a) PC12 cells were serum starved for 72 h and then either treated with 50 ng of NGF per ml (+NGF) or serum refed (+Serum) for 30 min without (−) or with (+) 50 μM LY294002. Subsequently, cells were harvested and subjected to polysomal analysis, as described in the legend to Fig. 1. (b and c) PC12 cells were serum starved for 72 h (0 min) and then either treated with 50 ng of NGF per ml (b) or serum refed (c) without (−) or with (+) 50 μM LY294002 for the indicated times. The cytoplasmic proteins were subjected to Western blot analysis with the indicated antibodies. α and β represent hypophosphorylated and hyperphosphorylated forms of rpS6, respectively.
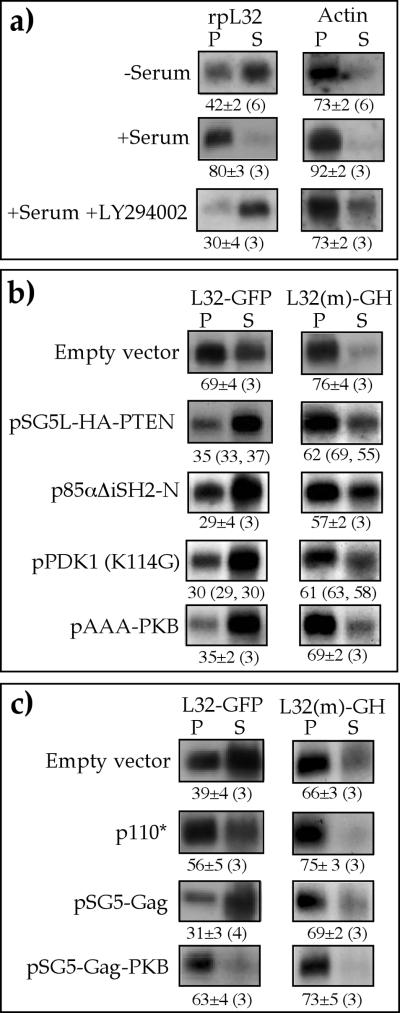
To further establish the role of a PI3-kinase pathway in translational activation of TOP mRNA, we selected 293 cells because of their high transfection efficiency and the fact that the translation of their TOP mRNAs is as sensitive to LY294002 as demonstrated for PC12 cells (Fig. 3a). These cells were cotransfected with two plasmids coding for L32-GFP mRNA and L32(−1C->A)-GH mRNA. The former starts with the 5′TOP motif of mouse rpL32 mRNA, whereas an extra A residue precedes this motif in the latter, and therefore these mRNAs behave essentially as TOP and non-TOP mRNAs, respectively (4, 58).
FIG. 3.
PI3-kinase-dependent pathway mediates mitogenic signals into translational efficiency of TOP mRNAs. (a) 293 cells were serum starved for 26 h (−Serum) or serum refed for 3 h (+Serum) without or with 50 μM LY294002, after which the cells were harvested. The polysomal distribution of mRNAs encoding rpL32 and actin was analyzed and presented as described in the legend to Fig. 1. (b and c) 293 cells were triply cotransfected with 2 μg of vectors encoding L32-GFP and L32(−1C->A)-GH and 16 μg of an empty vector (pSG5) or an expression vector, as indicated at the left. Then, 24 h later, cells were serum starved for 26 h and refed for 3 h (b) or serum starved for 26 h without refeeding (c). The polysomal distribution of L32-GFP and L32(21C−>A)-GH mRNAs was analyzed and presented as described in the legend to Fig. 1.
First, we examined the effect of interference in the delivery of signals along the pathway by overexpression of each of the following proteins: (i) p85αΔiSH2-N, a dominant-negative mutant of the PI3-kinase regulatory subunit p85 which lacks the binding site for the catalytic subunit, p110 (50); (ii) pSG5L-HA-PTEN, which encodes hemagglutinin (HA)-tagged PTEN (48); (iii) PDK1K114G, a dominant-negative mutant of PDK1 (18); and (iv) pAAA-PKB, a kinase-inactive, phosphorylation-deficient PKBα construct, with the mutations K179A, T308A, and S473A (68). Evidently, each of these overproduced proteins was sufficient to block the translational activation of L32-GFP mRNA in serum-refed cells (Fig. 3b). The apparent suppression of L32-GFP mRNA appeared to be selective for the 5′TOP-containing mRNA, as L32(−1C->A)-GH mRNA remained mostly associated with polysomes, although slightly repressed, in the presence of any of the expression vectors examined (Fig. 3b). It is therefore likely that PI3-kinase, PDK1, and PKB are necessary for the serum-induced recruitment of TOP mRNAs into polysomes.
In complementary experiments, we addressed the question of whether overexpression of constitutively active PI3-kinase or its effectors can rescue the translation of TOP mRNAs in serum-starved cells. Indeed, overexpression of p110*, a constitutively active mutant of PI3-kinase (24), or Gag-PKBα, a constitutively active PKB (9), alleviated the translational repression of L32-GFP mRNA exerted by serum starvation (compare p110* and pSG5-Gag-PKBα to empty vector and pSG5-Gag in Fig. 3c).
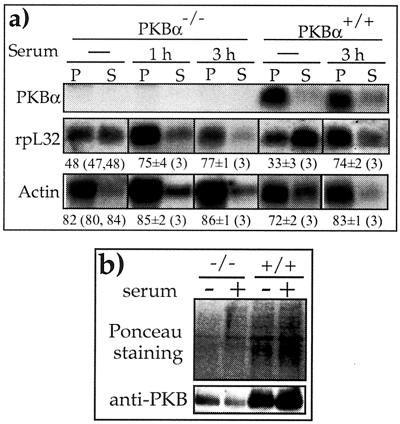
Mice deficient in PKBα are defective in both fetal and postnatal growth (12, 14). Likewise, we observed a slightly slower growth rate of PKBα−/− MEF relative to that of PKBα+/+ MEF (generation times of ≈42 h and 37 h, respectively). To examine whether this disparity in growth rate reflects differences in translation efficiency of TOP mRNAs, we set out to monitor the polysomal association of rpL32 mRNA. Figure 4a shows that despite the complete absence of PKBα mRNA, rpL32 mRNA was as efficiently recruited into polysomes upon serum stimulation of PKBα−/− MEF as of PKBα+/+ MEF. Indeed, PKBα−/− MEF still expressed, albeit to a lesser extent, PKB (β and/or γ isoform), as demonstrated by Western blot analysis with anti-PKB antibody, which can detect all three isoforms (compare PKB to the Ponceau S-stained signals in Fig. 4b). Hence, the redundancy of the PKB activity or the action of other kinases with an overlapping specificity seems to suffice for the requirement for normal translational control of TOP mRNAs, even in PKBα−/− MEF, and the mechanism underlying the slower growth of these cells has yet to be established.
FIG. 4.
Translation of TOP mRNAs is normally regulated in PKBα−/− MEF cells. (a) PKBα+/+ and PKBα−/− MEF were harvested after being serum starved for 44 h without or with 1 or 3 h of serum refeeding. The polysomal distribution of the mRNAs encoding PKBα, rpL32, and actin was analyzed and presented as described in the legend to Fig. 1. (b) Cytoplasmic proteins from PKBα+/+ (+/+) and PKBα−/− (−/−) MEF were harvested after being serum starved for 44 h without (−) or with 1 h of serum refeeding (+) and subjected to Western blot analysis. Bottom panel, immunoblotting with anti-PKB. Top panel, Ponceau S-stained membrane, showing the relative protein loading among samples. Note that the three PKB isoforms contain similar numbers of amino acids (from 466 to 481 residues).
Phosphorylation of rpS6 and S6K1 activity per se cannot support efficient translation of TOP mRNAs.
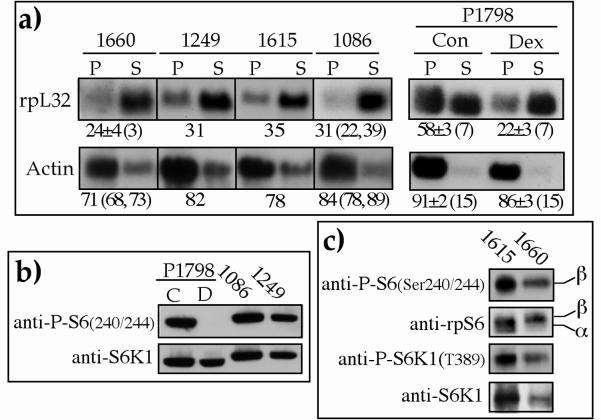
The apparent inhibition of both the phosphorylation of rpS6 and the translational activation of TOP mRNAs by LY294002 (Fig. 2) is consistent with the common dogma, as yet unproved, that the two display a cause-and-effect relationships. To examine this notion, we monitored these two variables in lymphoblastoid cell lines. We have previously shown that the translation of a wide variety of TOP mRNAs is constitutively repressed in the proliferating WHI1249 line of human lymphoblastoid cells (3, 4). Here we show that this exceptional behavior appeared in all four lymphoblastoid cell lines (WHI1660, -1249, -1615, and -1086) examined, as judged by the predominant association of their rpL32 mRNAs with mRNP (mostly in the subpolysomal fraction in Fig. 5a). This commonality is underscored by the fact that these human cell lines were derived from four individuals having different genetic backgrounds.
FIG. 5.
Phosphorylation of rpS6 is not sufficient to enable efficient translation of TOP mRNAs. (a) Cytoplasmic extracts were prepared from four proliferating lymphoblastoid cell lines (WHI1660, WHI1249, WHI1615, and WHI1860) or from P1798 cells which were either dividing (Con or C) or nondividing due to 24 h of dexamethasone treatment (Dex or D). The polysomal distribution of the mRNAs encoding rpL32 and actin was analyzed and presented as described in the legend to Fig. 1. (b and c) Cytoplasmic proteins from the cells mentioned in panel a were subjected to Western blot analysis with the indicated antibodies.
The unique behavior of TOP mRNAs in lymphoblastoid cells is further exemplified by the fact that TOP mRNAs in mouse P1798 lymphosarcoma cells were efficiently translated in untreated cells and repressed only when mitotically arrested (Fig. 5a). Interestingly, translational repression in the lymphoblastoid cell lines occurred despite the apparent phosphorylation of rpS6, which was demonstrated by the phosphorylation-specific Ser240/244 antibody (Fig. 5b and 5c). It is noteworthy that rpS6 was also phosphorylated at Ser235/236 in the lymphoblastoid cells (see band β detected by anti-rpS6 in Fig. 5c). Furthermore, with a phosphorylation-specific antibody, we showed that S6K1 was phosphorylated at Thr389, a critical site for its activity (44), which suggests that the phosphorylation of rpS6 reflects S6K1 activity. These results therefore appear to support the notion that neither of these two suffices to enable efficient translation of TOP mRNAs.
Neither S6K1 activity nor rpS6 phosphorylation is required for translational activation of TOP mRNAs.
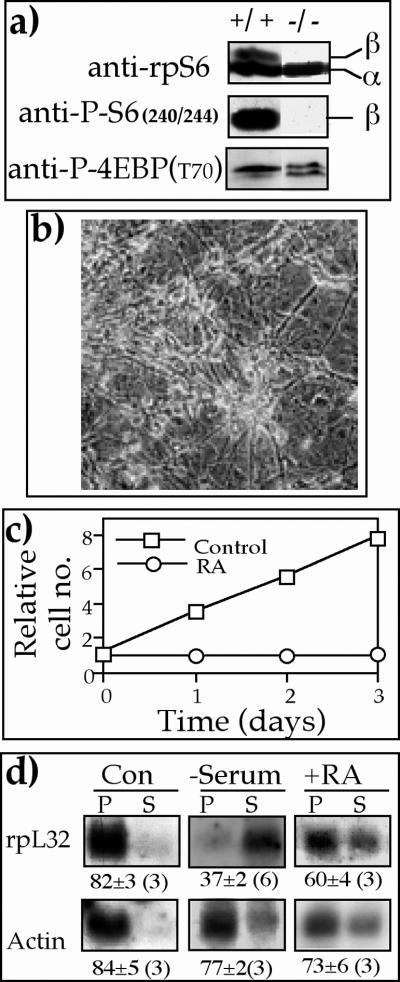
The conflicting reports regarding causal relationships between S6K1 activity and the translational efficiency of TOP mRNAs in serum- or amino acid-stimulated cells (25, 58) prompted us to examine this issue with a mouse diploid ES cell line, p70S6K−/− cells, in which both alleles of S6K1 were disrupted by homologous recombination (28). Figure 6a shows that rpS6 in p70S6K−/− cells, unlike that of the parental cells (R1), was constitutively and selectively unphosphorylated (compare anti-phospho-rpS6 [Ser240/244] with anti-phospho-4E-BP [Thr70]). These results corroborate those originally obtained for these cells (28) but are inconsistent with a later report (33). Apparently, the unphosphorylated status of rpS6 does not diminish the efficient translation of TOP mRNAs when p70S6K−/− ES cells are mitotically active (see rpL32 in dividing cells, Fig. 6d). This observation therefore indicates that neither S6K1 activity nor rpS6 phosphorylation is necessary for efficient translation of TOP mRNAs.
FIG. 6.
Retinoic acid induces translational activation of TOP mRNAs in growing but not dividing p70S6K−/− ES cells. (a) Cytoplasmic proteins from wild-type ES cells (+/+) and p70S6K−/− cells (−/−) were subjected to Western blot analysis with the indicated antibodies. (b) p70S6K−/− ES cells were induced to neuronal differentiation (with retinoic acid), as described in Materials and Methods and visualized microscopically. (c) p70S6K−/− ES cells were either untreated (Con) or induced to neuronal differentiation with retinoic acid (RA), as described in Materials and Methods. Proliferation was monitored by measuring the A650 of the methylene blue dye extracted from stained cells. The absorbance measured 24 h after plating was arbitrarily set at 1, and absorbance measured at later time points (average of at least two repetitions for each time point) was normalized to that value. (d) p70S6K−/− cells were untreated (Con), serum starved for 40 h (−Serum), or induced to neuronal differentiation with retinoic acid (+RA). The polysomal distribution of the mRNAs encoding rpL32 and actin was analyzed and presented as described in the legend to Fig. 1.
It has previously been shown that mouse ES cells treated with retinoic acid are induced to differentiate into neuron-like cells (5). When p70S6K−/− ES cells were similarly treated, they exhibited a typical morphology of differentiating neurons, with extensive neurite outgrowth (Fig. 6b). This differentiation was accompanied by a complete cessation of cell division (Fig. 6c), yet most of rpL32 was associated with polysomes, unlike the situation observed upon serum starvation (Fig. 6d). These results imply that translational control of TOP mRNAs by growth stimulation does not involve S6K1 activity.
Rapamycin inhibits growth- and mitogen-induced rpS6 phosphorylation, with only a minor repressive effect on translational activation of TOP mRNAs.
A putative PKB phosphorylation site (S2448) was observed to be phosphorylated on mTOR in vivo, yet its role in the regulation of mTOR activity is still unclear (reference 49 and references therein). This observation, together with the conflicting reports concerning the ability of rapamycin to inhibit translational activation of TOP mRNAs (reference 23 and references therein) led us to examine whether mTOR is involved in signaling to TOP mRNAs upon growth or mitogenic stimulation.
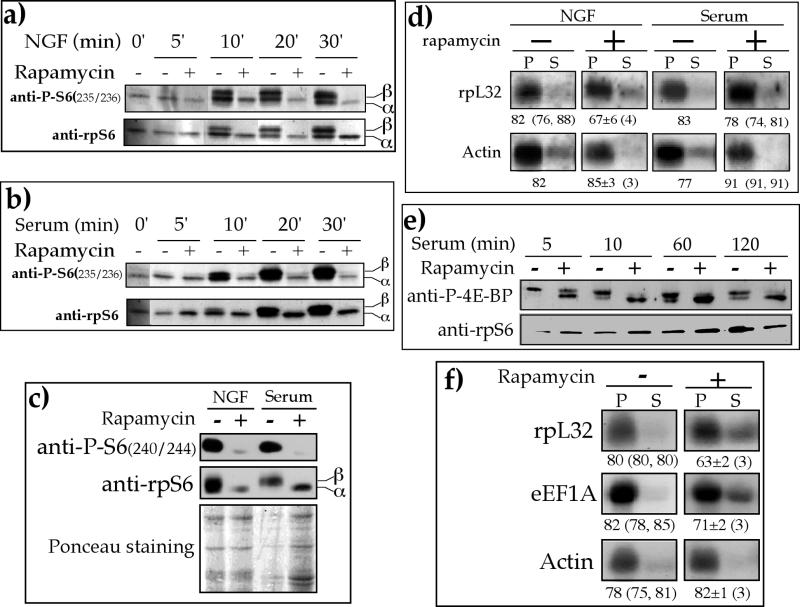
To this end, we monitored the effect of rapamycin on the phosphorylation status of rpS6 and S6K1 as well as the polysome association of rpL32 mRNA in NGF-treated and serum-refed PC12 cells. Figures 7a and 7b show that phosphorylation of rpS6 was induced within 10 min after NGF introduction or serum refeeding. It has previously been shown that rapamycin completely abolished S6K1 activation in NGF-treated PC12 cell (29) and that its inhibitory effect on S6K1 activity was very rapid, with a half-time of about 2 min (15). Indeed, inhibition of the phosphorylation of Ser235/236 (Fig. 7a and 7b) and Ser 240/244 (Fig. 7c) by rapamycin was apparent at the earliest time points measured following NGF treatment and serum refeeding (10 min and 30 min, respectively). This inhibition reflects a block by rapamycin of S6K1 activation, as is evident from the phosphorylation status of its Thr389 (data not shown).
FIG. 7.
Rapamycin has a minor and delayed inhibitory effect on the translation of TOP mRNAs. (a, b, and c) PC12 cells were serum starved for 72 h (0 min) and then either treated with 50 ng of NGF per ml (a and c) or serum refed (b and c) without (−) or with (+) 20 nM rapamycin for the indicated times (a and b) or for 30 min (c), after which the cells were harvested. The cytoplasmic proteins were subjected to Western blot analysis with the indicated antibodies. The bottom panel in c is the Ponceau S-stained membrane, showing the relative protein loading among samples. (d) PC12 cells were serum starved for 72 h and then either treated with 50 ng of NGF per ml or serum refed without (−) or with (+) 20 nM rapamycin for 60 min, after which they were harvested. The polysomal distribution of the mRNAs encoding rpL32 and actin was analyzed and presented as described in the legend to Fig. 1. (e) p70S6K−/− ES cells were serum starved for 40 h and then refed in the absence (−) or presence (+) of 20 nM rapamycin for the indicated times. Cytoplasmic proteins were subjected to Western blot analysis with the indicated antibodies. (f) p70S6K−/− ES cells were serum starved for 40 h and then refed for 3 h in the absence (−) or presence (+) of 20 nM rapamycin. The polysomal distribution of the mRNAs encoding rpL32, eEF1A, and actin was analyzed as described in the legend to Fig. 1. The results in panels d and f are expressed as the average ± standard error of the mean, with the number of determinations in parentheses, or as the average, with the individual values in parentheses, if only two determinations were available.
Nonetheless, despite this efficient inhibition, rapamycin had only a minor repressive effect on the translational activation of rpL32 mRNA. Thus, the proportion of rpL32 mRNA associated with polysomes increased from 30% in serum-starved cells (Fig. 1b) to 82% and 67% following 1 h of NGF treatment without and with rapamycin, respectively (Fig. 7d). Similarly, 83% and 78% of rpL32 mRNA were associated with polysomes 1 h after serum refeeding without and with rapamycin, respectively (Fig. 7d). Our results therefore show that mTOR inhibition cannot prevent translational activation of TOP mRNAs, as monitored within 60 min after NGF or serum addition.
It can be argued, yet with very low likelihood, that PC12 cells, being an established cell line, may have acquired abnormalities in translational control of TOP mRNAs that have effectively rendered it rapamycin resistant. To examine this possibility, we measured the effect of rapamycin on the translation of TOP mRNAs in serum-refed p70S6K−/− ES cells, which are murine diploid cells. The sensitivity of mTOR to rapamycin in these cells was exemplified by the suppressed phosphorylation of 4E-BP, a direct substrate of mTOR (21), which was nearly complete just 10 min after addition of rapamycin (Fig. 7e), yet translation of two TOP mRNAs, encoding rpL32 and eEF1A, was only mildly repressed after 3 h of rapamycin treatment (Fig. 7f).
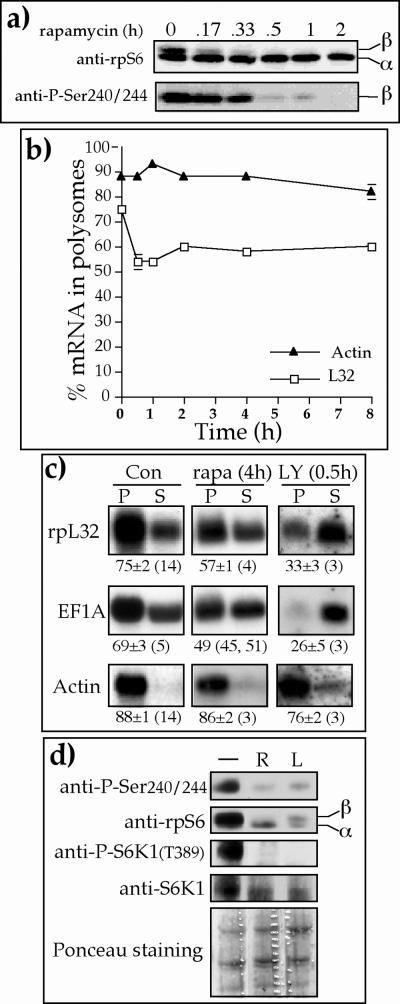
It has previously been shown for some cell lines that translational activation of TOP mRNAs was efficiently blocked if rapamycin was present during mitogenic stimulations which lasted up to 4 h (28, 56, 59, 60). Other cell lines, however, exhibited complete (26, 28) or nearly complete (25) resistance to such treatment. In light of these conflicting results, we set out to monitor the effect of rapamycin on proliferating 293 cells for increasing durations of time. Figure 8a shows that rpS6 in 293 cells was completely dephosphorylated at Ser235/236 (see band β in the upper panel of Fig. 8a) and Ser 240/244 (lower panel in Fig. 8a) within 30 to 60 min following exposure to rapamycin. Nevertheless, most (60 ± 2% [n = 3]) of the pL32 mRNA in these cells remained associated with polysomes even 8 h after the addition of rapamycin (Fig. 8b). Likewise, rapamycin induced a rapid dephosphorylation (within 30 min) of rpS6 in PC12 cells, yet had only a minor effect on the translational efficiency of rpL32 mRNA (decreased from 73% ± 2% [n = 7] to 67% ± 5% [n = 3] in polysomes within 8 h) (data not shown).
FIG. 8.
Prolonged rapamycin treatment only mildly represses the translation of TOP mRNAs. (a and d) Proliferating 293 cells were treated with 20 nM rapamycin for the indicated time (a) or for 1 h (lane R in d) or with 50 mM LY294002 for 30 min (lane L in d), after which the cells were harvested. The cytoplasmic proteins were subjected to Western blot analysis with the indicated antibodies. The bottom panel in d is the Ponceau S-stained membrane, showing the relative protein loading among samples. (b and c) Proliferating 293 cells were treated with 20 nM rapamycin for the indicated times (b) or for 4 h (c) or with 50 mM LY294002 for 30 min. The polysomal distribution of rpL32, eEF1A, and actin mRNAs was analyzed as described in the legend to Fig. 1. The relative amount of the mRNAs in polysomes is presented graphically in b and numerically in c.
The discrepancy between the fast inhibitory effect of rapamycin on the phosphorylation of rpS6 and the relative prolonged resistance of TOP mRNAs was highlighted by the differential response of the latter to rapamycin and LY294002 treatment. Thus, while 0.5 h of treatment of 293 cells with LY294002 led to selective and pronounced repression of mRNAs encoding rpL32 and eEF1A, 4 h of rapamycin treatment had only a mild inhibitory effect on these mRNAs (Fig. 8c). Nevertheless, this relative rapamycin resistance cannot be attributed to residual S6K1 activity, since 1 h of rapamycin treatment completely abolished S6K1 activity, as judged by the dephosphorylation of its Thr389 (Fig. 8d). These results therefore imply that the mTOR pathway does not have a direct role in regulating the translational efficiency of TOP mRNAs.
Clearly, even though rpS6 remained slightly phosphorylated in LY294002-treated cells (see band β in Fig. 8d), it was not sufficient to protect TOP mRNAs from rapid translational repression. Notably, the relative rapamycin resistance of TOP mRNAs, despite the complete inhibition of S6K1, also refutes causal relationships between the translational repression of TOP mRNAs and the inactivation of S6K1 (Fig. 8d), which are exerted by LY294002. In summary, Fig. 7 and 8 indicate that the mTOR pathway plays a minor role, if any, in the translational control of TOP mRNAs in all three cell lines examined (PC12, p70S6K−/− ES, and 293), which represent three mammalian species (rat, mouse, and human, respectively) as well as different tissue origins and ploidy status.
DISCUSSION
Translational regulation of TOP mRNAs by growth and other signals.
Previous studies have shown that mitogenic arrest at the G0/G1, S, or M phase of the cell cycle by a wide variety of means leads to translational repression of TOP mRNAs (37). In an attempt to identify a common denominator for all these stimuli, we reasoned that arresting cells at various stages along the cell cycle should also block their ability toprogress into G1 or G2, where growth occurs. Hence, we have been intrigued by the question of whether it is the ability of cells to grow, rather than their ability to progress through the cell cycle, which matters for the translational control of TOP mRNAs. Indeed, our present results clearly show that PC12 cells can resume the translation of TOP mRNAs when they are induced to grow by NGF even if they remain mitotically arrested in the absence of serum (Fig. 1).
It therefore appears that cells are programmed to restrain the production of the translational apparatus, unless doubling its amount is required during cell divisions or for supporting the extensive protein synthesis needed upon growth stimulation. Likewise, cells limit the synthesis of their translational apparatus by repressing the translation of TOP mRNAs when they sense a shortage in the amino acid supply (58). It is worth noting that inducing cell cycle arrest by inhibiting DNA synthesis with aphidicolin or hydroxyurea also led to increased cell size (17, 19). Nonetheless, such treatments also resulted in repressed translation of TOP mRNAs (3, 4, 55). It is therefore important to determine whether the enlarged cell volume which is induced by inhibitors of the cell cycle involves an elevation in protein content or simply reflects swelling due to increased intracellular osmolarity.
TOP mRNAs are not confined to vertebrates, as many of the ribosomal protein mRNAs in Drosophila melanogaster have been shown to contain the 5′TOP motif and to be translationally controlled (reviewed in reference 36). Interestingly, copulation in the fly leads to an abrupt elevation in the synthesis rate of ribosomal proteins in the paragonial gland, which is not accompanied by increased steady-state levels of the corresponding mRNAs or mitogenic activity. This observation suggests that translation of ribosomal protein mRNAs is activated after copulation to augment the protein synthesis capacity and thus to enable rapid replenishment of the secreted proteins (54). It therefore appears that enhanced translation of TOP mRNAs is attained under any physiological condition in which extensive protein synthesis is necessary (increase in cell mass, secretion, etc.).
It has previously been proposed that efficient translation of TOP mRNAs upon mitogenic stimulation plays a regulatory role in halting cell cycle progression until the growth response is completed (57, 61). However, the continuous proliferation of lymphoblastoid cells despite the constitutively repressed translation of TOP mRNAs (Fig. 5) (3, 4) indicates that efficient translation of these mRNAs is not a determinant in the control of the cell cycle and is not a prerequisite for cell division. Nevertheless, we cannot exclude the possibility that the synthesis of TOP mRNA-encoded proteins is still as efficient in lymphoblastoid cells as in any other dividing cells because of a compensatory increase in the abundance of the respective mRNAs, for example. Finally, translation of TOP mRNA is repressed upon inhibition of DNA synthesis by aphidicolin or prevention of chromosomal segregation by nocodazole even in the presence of a sufficient supply of growth factors and nutrients. This implies that the translation of these mRNAs responds primarily to the ability of cells to progress through the cell cycle rather than to external mitogenic or nutritional cues.
Translational activation of TOP mRNAs is fully dependent on the PI3-kinase-mediated pathway.
When cells are stimulated by growth factors, they display a wide variety of responses, including increased protein synthesis (2). A well-documented pathway which transduces mitogenic or growth signals involves PI3-kinase and its downstream effector, PKB (32). However, the role of this pathway in activation of protein synthesis has only been established for the activation of initiation factor 2B (eIF2B). Thus, eIF2B is inhibited through its phosphorylation by glycogen synthase 3 and is activated when the latter is phosphorylated, and consequently inactivated, by PKB (69).
In the present report, we demonstrate that both mitogenic and growth stimuli selectively activate the translation of TOP mRNAs in a PI3-kinase-PKB pathway-dependent manner. Thus, blocking the signal through this pathway by LY294002 completely abolished the translational activation of TOP mRNAs by growth or mitogenic signals (Fig. 2a and 3a). Translational activation of a chimeric TOP mRNA was similarly suppressed by overexpression of dominant-negative mutants of PI3-kinase, PDK1, and PKBα as well as overexpression of PTEN (Fig. 3b). It is conceivable, therefore, that the apparent enhanced protein synthesis in growth factor-stimulated cells reflects, at least partially, the activated translation of TOP mRNAs whose products comprise crucial components of the translational apparatus.
It is noteworthy that we have previously shown that a functional PI3-kinase-mediated pathway is essential for translational activation of TOP mRNAs upon amino acids refeeding, even though the activity of neither PI3-kinase nor PKB is compromised by amino acid starvation (58). Finally, the fact that translational repression of TOP mRNAs in serum-starved cells can be only partially relieved by overexpression of constitutively active PI3-kinase or PKBα (Fig. 3c) might reflect insufficient expression of these mutants. Alternatively, the PI3-kinase-mediated pathway is essential for this mode of regulation but is not sufficient.
mTOR-dependent pathway plays a minor role, if any, in translational control of TOP mRNAs.
Activation of S6K and consequently the phosphorylation of rpS6, on the one hand, and translational activation of TOP mRNAs, on the other hand, are apparent under a variety of physiological conditions (35, 58). This tight correlation has laid the foundation for an attractive model according to which rpS6 phosphorylation is a prerequisite for efficient translation of TOP mRNAs. Nevertheless, several lines of evidence, presented in this and other recent reports, indicate that these two variables do not maintain cause-and-effect relationships. Thus, the translation of TOP mRNAs is constitutively repressed in dividing lymphoblastoid cells even though their rpS6 is phosphorylated (Fig. 5). Similarly, overexpression of RSK2 (a nonphysiological S6 kinase) and calyculin A (phosphatase inhibitor) treatment of 293 cells maintained rpS6 in its phosphorylated form, yet they failed to derepress TOP mRNAs upon amino acid starvation. This set of observations implies that rpS6 phosphorylation is not sufficient for recruitment of TOP mRNAs into polysomes.
Finally, TOP mRNAs are efficiently translated in p70S6K−/− ES cells (Fig. 6) and in mouse erythroleukemia cells (7) even though their rpS6 is constitutively unphosphorylated or dephosphorylated, respectively, indicating that rpS6 phosphorylation is not necessary either for efficient translation of TOP mRNAs or for cell division. Moreover, conditional disruption of rpS6 gene in the adult mouse liver did not inhibit hypertrophy of the liver in response to refeeding (67), indicating that not only its phosphorylation status but the entire rpS6 protein is dispensable for cellular growth.
It can be argued that it is the requirement for S6K1 activity, rather than just rpS6 phosphorylation, which is involved in the translational control of TOP mRNAs. However, several lines of evidence are inconsistent with this notion. (i) Elimination of S6K1 by knockout of the corresponding gene had no effect on translational activation of TOP mRNAs upon serum refeeding (Fig. 7f), induction for growth without cell division (Fig. 6), or amino acid replenishment (58). (ii) Mistargeting upstream signals by overexpression of p70S6K,K123 M, a kinase-dead S6K1 mutant, in 293 cells completely abolished any S6K activity, as was evident by the unphosphorylated state of rpS6 (58). Nonetheless, efficient translation of TOP mRNAs was apparent in these cells when they were provided with complete growth medium or refed with amino acids (58). Unlike the lack of inhibitory effect of this mutant, Jefferies and colleagues (25), who used another dominant-interfering mutant, p70S6KA229, reported that its expression led to a modest suppression of the translational activation of a TOP mRNA following mitogenic stimulation. The difference between these results might reflect the number of repetitions used to obtain them (the latter was based on a single-repetition experiment).
(iii) Rapamycin completely inhibits S6K1 and rpS6 phosphorylation (15, 46), yet it has only a minor or no repressive effect on the translational activation of TOP mRNAs following serum or amino acid refeeding (Fig. 7) (25, 27, 28, 58) or in proliferating cells of different genetic backgrounds (Fig. 8 and data not shown). It should be mentioned, however, that suppression of translational activation of TOP mRNAs by rapamycin has been shown upon mitogenic stimulation of MEF (56), lymphoblastoid cells (59), and T cells (60). Nevertheless, in the latter two cases, TOP mRNAs were constitutively repressed even without exposure to rapamycin, and therefore the significance of the effect of this drug is questionable. The minor inhibitory effect, if any, of rapamycin on the translation of TOP mRNAs in most cell lines examined so far is underscored by the rapid and complete repression of TOP mRNAs by amino acid starvation (within 1 h) ([58]) or LY294002 treatment (within 30 min) (Fig. 8c). Taken together, these data clearly attest to the lack of any role of S6K1 and a minor role of mTOR in the translational control of TOP mRNAs in response to mitogenic, growth, or nutritional stimuli.
It has previously been suggested that the typical translational control of TOP mRNAs in p70S6K1−/− cells might by attributed to the compensating activity of S6K2 in these cells (56). However, two lines of evidence are inconsistent with the involvement of S6K2 in this mode of regulation. (i) Reports from several laboratories have shown that rapamycin, at the concentration used here (20 nM), also completely inhibits S6K2 in 293, mouse ES, and COS cells (30, 33, 52, 56), but has only a minor effect on translation of TOP mRNAs (58; this study). (ii) It has been shown that S6K2 is quite refractory to inhibition by amino acid withdrawal (38), yet this resistance was insufficient to prevent the repression of TOP mRNA translation upon amino acid starvation in 293 or p70S6K1−/− cells (58). In any case, unequivocal negation of the role of S6K2 in the translational control of TOP mRNAs should await the establishment of S6K1/S6K2 double knockout mice or study of the translational control of TOP mRNAs in dS6K-null flies.
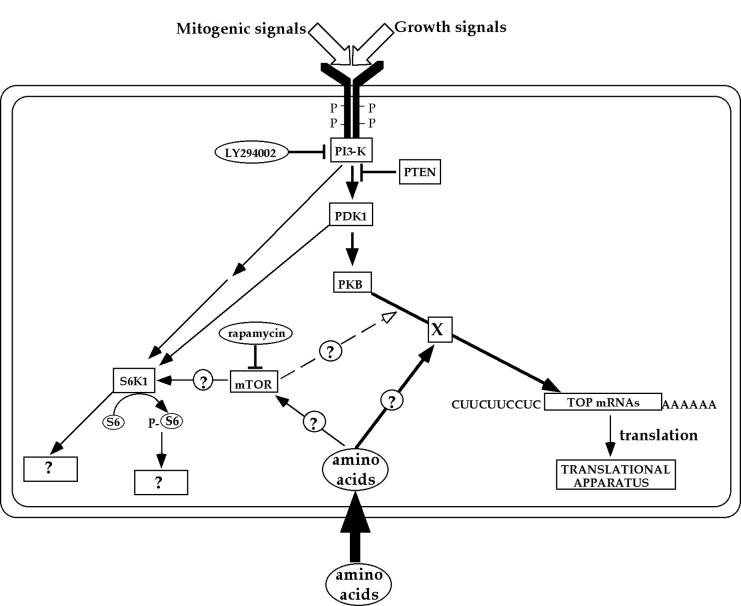
A tentative model depicting the signaling pathways leading to the translational activation of TOP mRNAs by mitogenic, growth, or amino acid sufficiency is presented in Fig. 9. According to this model, mitogenic and growth factors activate the PI3-kinase-dependent pathway, which transduces the respective signals through PDK1 and PKB into translational efficiency of TOP mRNAs. This pathway involves an unknown effector(s) (denoted as X). Signals from amino acids converge with the PI3-kinase-mediated pathway, and their transduction into TOP mRNAs is fully dependent on the integrity of this pathway. However, signaling from all external cues bifurcates prior to the mTOR step, as inferred from the ability of rapamycin to discern between the activity of mTOR and S6K on the one hand and the translational efficiency of TOP mRNAs on the other hand.
FIG. 9.
Schematic representation of signal transduction pathways involved in activation of rpS6 phosphorylation and translational control of TOP mRNAs in serum- and amino acid-stimulated cells. Arrows delineate the flow of information. The open arrowhead represents the minor effect of rapamycin on TOP mRNA translation. The site of convergence of this effect with that of other signals is purely speculative. Circled and boxed question marks represent putative links and unknown targets, respectively. See text for details.
D. melanogaster might serve as an appropriate experimental system to resolve the role of the mTOR-dependent pathway in translational control of TOP mRNAs. The major advantage of this organism stems from the conservation of all five phosphorylatable serine residues in its rpS6 and the presence of a single S6K (dS6K) isoform (20). Homozygous flies null for dS6K were developmentally delayed to approximately half the size of wild-type flies. The dramatic reduction in size was due to smaller cells rather than decreased numbers of cells (40). Mutations in the Drosophila homologs of PI3-kinase, PKB, and mTOR have a striking effect on both cell number and cell size (reviewed in reference 31). Unfortunately, the translational efficiency of TOP mRNAs has never been addressed experimentally in any of these mutant flies or cells. Notably, a recent study demonstrated that Drosophila S6K controls cell growth in a Drosophila PKB- and Drosophila PI3-kinase-independent manner, implying that these enzymes reside on distinct pathways (47). If this conclusion is also applicable to mammalian cells, it is not surprising that we observed that translational efficiency of TOP mRNAs is affected by the PI3-kinase pathway in an S6K1-independent fashion.
Our present data and a previous report (58) refuted the only role primarily assigned for S6K and phosphorylated rpS6, translational control of TOP mRNAs. Hence, a new physiological role(s) for this extensively studied kinase should be established. Indeed, recent reports have implicated S6K1 in two other physiological processes, glucose homeostasis, through involvement in insulin secretion (45), and regulation of elongation factor 2 kinase (70) by its phosphorylation. Nevertheless, the function of rpS6 phosphorylation still remains to be elucidated.
Acknowledgments
This work was supported by grants to O.M. from the Israel Science Foundation (grant 460/01-1) and by the United States-Israel Binational Science Foundation (BSF 2000017).
We are grateful to Naohiro Terada for the SK1 knockout ES cells, Anke Klippel for p110*, Julian Downward for the p85 ΔSH2-N, William Sellers for the PTEN construct, Feng Liu for PDK1K114G, James Woodget for pAAA-PKB, and Boudewijn Burgering for pSG5PKBGag. We gratefully acknowledge Maya Schuldiner and Nissim Benvenisty for helping in the neuronal differentiation of ES cells.
REFERENCES
- 1.Aloni, R., D. Peleg, and O. Meyuhas. 1992. Selective translational control and nonspecific posttranscriptional regulation of ribosomal protein gene expression during development and regeneration of rat liver. Mol. Cell. Biol. 12:2203-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antoniades, H., and A. Owen. 1982. Growth factors and regulation of cell growth. Annu. Rev. Med. 33:445-463. [DOI] [PubMed] [Google Scholar]
- 3.Avni, D., Y. Biberman, and O. Meyuhas. 1997. The 5′ terminal oligopyrimidine tract confers translational control on TOP mRNAs in a cell type-and sequence context-dependent manner. Nucleic Acids Res. 25:995-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avni, D., S. Shama, F. Loreni, and O. Meyuhas. 1994. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol. Cell. Biol. 14:3822-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bain, G., D. Kitchens, M. Yao, J. Huettner, and D. Gottlieb. 1995. Embryonic stem cells express neuronal properties in vitro. Dev. Biol. 168:342-357. [DOI] [PubMed] [Google Scholar]
- 6.Bain, G., W. Ray, M. Yao, and D. Gottlieb. 1996. Retinoic acid promotes neural and represses mesodermal gene expression in mouse embryonic stem cells in culture. Biochem. Biophys. Res. Commun. 223:691-694. [DOI] [PubMed] [Google Scholar]
- 7.Barth-Baus, D., C. Stratton, L. Parrott, H. Myerson, O. Meyuhas, D. Templeton, G. Landreth, and J. Hensold. 2002. S6 phosphorylation-independent pathways regulate translation of 5′-terminal oligopyrimidine tract containing mRNAs in differentiating hematopoietic cells. Nucleic Acids Res. 30:1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brazil, D., and B. Hemmings. 2001. Ten years of protein kinase B signalling: a hard Akt to follow. Trends Biochem. Sci. 26:657-664. [DOI] [PubMed] [Google Scholar]
- 9.Burgering, B., and P. Coffer. 1995. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376:599-602. [DOI] [PubMed] [Google Scholar]
- 10.Cantrell, D. 2001. Phosphoinositide 3-kinase signalling pathways. J. Cell Sci. 114:1439-1445. [DOI] [PubMed] [Google Scholar]
- 11.Carter, A., and C. Downes. 1992. Phosphatidylinositol 3-kinase is activated by nerve growth factor and epidermal growth factor in PC12 cells. J. Biol. Chem. 267:14563-14567. [PubMed] [Google Scholar]
- 12.Chen, W., P. Xu, K. Gottlob, M. Chen, K. Sokol, T. Shiyanova, I. Roninson, W. Weng, R. Suzuki, K. Tobe, T. Kadowaki, and N. Hay. 2001. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 15:2203-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho, H., J. Mu, J. Kim, J. Thorvaldsen, Q. Chu, E. r. Crenshaw, K. Kaestner, M. Bartolomei, G. Shulman, and M. Birnbaum. 2001. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ). Science 292:1728-1731. [DOI] [PubMed] [Google Scholar]
- 14.Cho, H., J. Thorvaldsen, Q. Chu, F. Feng, and M. Birnbaum. 2001. Akt1/PKBα is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J. Biol. Chem. 276:38349-38352. [DOI] [PubMed] [Google Scholar]
- 15.Chung, J., C. J. Kuo, G. R. Crabtree, and J. Blenis. 1992. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 kinases. Cell 69:1227-1236. [DOI] [PubMed] [Google Scholar]
- 16.Chung, S., and R. P. Perry. 1989. Importance of introns for expression of mouse ribosomal protein gene rpL32. Mol. Cell. Biol. 9:2075-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conlon, I., G. Dunn, A. Mudge, and M. Raff. 2001. Extracellular control of cell size. Nat. Cell Biol. 3:918-921. [DOI] [PubMed] [Google Scholar]
- 18.Dong, L. Q., R. B. Zhang, P. Langlais, H. He, M. Clark, L. Zhu, and F. Liu. 1999. Primary structure, tissue distribution, and expression of mouse phosphoinositide-dependent protein kinase-1, a protein kinase that phosphorylates and activates protein kinase Cζ. J. Biol. Chem. 274:8117-8122. [DOI] [PubMed] [Google Scholar]
- 19.Fingar, D., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fumagalli, S., and G. Thomas. 2000. S6 phosphorylation and signal transduction, p. 695-717. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Gingras, A., S. Gygi, B. Raught, R. Polakiewicz, R. Abraham, M. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hornstein, E., H. Harel, G. Levy, and O. Meyuhas. 1999. Overexpression of poly(A)-binding protein down-regulates the translation or the abundance of its own mRNA. FEBS Lett. 457:209-213. [DOI] [PubMed] [Google Scholar]
- 23.Hornstein, E., H. Tang, and O. Meyuhas. 2001. Mitogenic and nutritional signals are transduced into translational efficiency of TOP mRNAs. Cold Spring Harb. Symp. Quant. Biol. 66:477-484. [DOI] [PubMed] [Google Scholar]
- 24.Hu, Q., A. Klipple, W. Muslin, L. Fantl, and L. Williams. 1995. Ras-dependent induction of cellular responses by constitutively active phosphatidylinositol-3 kinase. Science 268:100-102. [DOI] [PubMed] [Google Scholar]
- 25.Jefferies, H., S. Fumagalli, P. Dennis, C. Reinhard, R. Pearson, and G. Thomas. 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 16:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jefferies, H. B. J., C. Reinhard, S. C. Kozma, and G. Thomas. 1994. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc. Natl. Acad. Sci. USA 91:4441-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jefferies, H. B. J., G. Thomas, and G. Thomas. 1994. Elongation factor-1α mRNA is selectively translated following mitogenic stimulation. J. Biol. Chem. 269:4367-4372. [PubMed] [Google Scholar]
- 28.Kawasome, H., P. Papst, S. Webb, G. M. Keller, G. L. Johnson, E. W. Gelfand, and N. Terada. 1998. Targeted disruption of p70s6k defines its role in protein synthesis and rapamycin sensitivity. Proc. Natl. Acad. Sci. USA 95:5033-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleijn, M., G. I. Welsh, G. Scheper, H. O. Voorma, C. G. Proud, and A. A. Thomas. 1998. Nerve and epidermal growth factor induce protein synthesis and eIF2B activation in PC12 cells. J. Biol. Chem. 273:5536-5541. [DOI] [PubMed] [Google Scholar]
- 30.Koh, H., K. Jee, B. Lee, J. Kim, D. Kim, Y. Yun, J. Kim, H. Choi, and J. Chung. 1999. Cloning and characterization of a nuclear S6 kinase, S6 kinase-related kinase (SRK); a novel nuclear target of Akt. Oncogene 18:5115-5119. [DOI] [PubMed] [Google Scholar]
- 31.Kozma, S., and G. Thomas. 2002. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. BioEssays 24:65-71. [DOI] [PubMed] [Google Scholar]
- 32.Lawlor, M., and D. Alessi. 2001. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J. Cell Sci. 114:2903-2910. [DOI] [PubMed] [Google Scholar]
- 33.Lee-Fruman, K., C. Kuo, J. Lippincott, N. Terada, and J. Blenis. 1999. Characterization of S6K2, a novel kinase homologous to S6K1. Oncogene 18:5108-5114. [DOI] [PubMed] [Google Scholar]
- 34.Leslie, N., and C. Downes. 2002. PTEN: the down side of PI 3-kinase signalling. Cell. Signal. 14:285-295. [DOI] [PubMed] [Google Scholar]
- 35.Meyuhas, O. 2000. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 267:6321-6330. [DOI] [PubMed] [Google Scholar]
- 36.Meyuhas, O., D. Avni, and S. Shama. 1996. Translational control of ribosomal protein mRNAs in eukaryotes, p. 363-384. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Meyuhas, O., and E. Hornstein. 2000. Translational control of TOP mRNAs, p. 671-693. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Minami, T., K. Hara, N. Oshiro, S. Ueoku, K. Yoshino, C. Tokunaga, Y. Shirai, N. Saito, I. Gout, and K. Yonezawa. 2001. Distinct regulatory mechanism for p70 S6 kinase b from that for p70 S6 kinase a. Genes Cells 6:1003-1015. [DOI] [PubMed] [Google Scholar]
- 39.Minty, A. J., M. Caravatti, B. Robert, A. Cohen, P. Daubas, A. Weydert, F. Gross, and M. E. Buckingham. 1981. Mouse actin messenger RNAs: construction and characterization of a recombinant plasmid molecule containing a complementary DNA transcript of mouse α-actin mRNA. J. Biol. Chem. 256:1008-1014. [PubMed] [Google Scholar]
- 40.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase: a regulator of cell size. Science 285:2126-2129. [DOI] [PubMed] [Google Scholar]
- 41.Oliver, M., N. Harrison, J. Bishop, P. Cole, and G. Laurent. 1989. A rapid and convenient assay for counting cells cultured in microwell plates: application for assessment of growth factors. J. Cell Sci. 92:513-518. [DOI] [PubMed] [Google Scholar]
- 42.Pardee, A. 1989. G1 events and regulation of cell proliferation. Science 246:603-608. [DOI] [PubMed] [Google Scholar]
- 43.Parrott, L. A., and D. J. Templeton. 1999. Osmotic stress inhibits p70/85 S6 kinase through activation of a protein phosphatase. J. Biol. Chem. 274:24731-24736. [DOI] [PubMed] [Google Scholar]
- 44.Pearson, R. B., P. B. Dennis, J. W. Han, N. A. Williamson, S. C. Kozma, R. E. Wettenhall, and G. Thomas. 1995. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 14:5279-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pende, M., S. Kozma, M. Jaquet, V. Oorschot, R. Burcelin, Y. Le Marchand-Brustel, J. Klumperman, B. Thorens, and G. Thomas. 2000. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature 408:994-997. [DOI] [PubMed] [Google Scholar]
- 46.Price, D. J., J. R. Grove, V. Calvo, J. Avruch, and B. E. Bierer. 1992. Rapamycin-induced inhibition of the 70-kilodalton protein kinase. Science 257:973-977. [DOI] [PubMed] [Google Scholar]
- 47.Radimerski, T., J. Montagne, F. Rintelen, H. Stocker, J. van Der Kaay, C. Downes, E. Hafen, and G. Thomas. 2002. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell Biol. 4:251-255. [DOI] [PubMed] [Google Scholar]
- 48.Ramaswamy, S., N. Nakamura, F. Vazquez, D. Batt, S. Perera, T. Roberts, and W. Sellers. 1999. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc. Natl. Acad. Sci. USA 96:2110-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raught, B., A. Gingras, and N. Sonenberg. 2001. The target of rapamycin (TOR) proteins. Proc. Natl. Acad. Sci. USA 98:7037-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez-Viciana, P., P. Warne, A. Khwaja, B. Marte, D. Pappin, P. Das, M. Waterfield, A. Ridley, and J. Downward. 1997. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell 89:457-467. [DOI] [PubMed] [Google Scholar]
- 51.Rudkin, B., P. Lazarovici, B. Levi, Y. Abe, K. Fujita, and G. Guroff. 1989. Cell cycle-specific action of nerve growth factor in PC12 cells: differentiation without proliferation. EMBO J. 8:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saitoh, M., P. ten Dijke, K. Miyazono, and H. Ichijo. 1998. Cloning and characterization of p70s6kβ defines a novel family of p70 S6 kinases. Biochem. Biophys. Res. Commun. 253:470-476. [DOI] [PubMed] [Google Scholar]
- 53.Schmidt, E. V. 1999. The role of c-myc in cellular growth control. Oncogene 18:2988-2996. [DOI] [PubMed] [Google Scholar]
- 54.Schmidt, T., P. S. Chen, and M. Pellegrini. 1985. The induction of ribosome biosynthesis in nonmitotic secretory tissue. J. Biol. Chem. 260:7645-7650. [PubMed] [Google Scholar]
- 55.Shama, S., D. Avni, R. M. Frederickson, N. Sonenberg, and O. Meyuhas. 1995. Overexpression of initiation factor eIF-4E does not relieve the translational repression of ribosomal protein mRNAs in quiescent cells. Gene Expr. 4:241-252. [PMC free article] [PubMed] [Google Scholar]
- 56.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. Kozma. 1998. Disruption of the p70s6k/p85s6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stocker, H., and E. Hafen. 2000. Genetic control of cell size. Curr. Opin. Genet. Dev. 10:529-535. [DOI] [PubMed] [Google Scholar]
- 58.Tang, H., E. Hornstein, M. Stolovich, G. Levy, M. Livingstone, D. Templeton, J. Avruch, and O. Meyuhas. 2001. Amino acid-induced translation of TOP mRNAs is fully dependent on PI3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 21:8671-8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Terada, N., H. R. Patel, K. Takase, K. Kohno, A. C. Nairn, and E. W. Gelfand. 1994. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc. Natl. Acad. Sci. USA 91:11477-11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Terada, N., K. Takase, P. Papst, A. C. Nairn, and E. W. Gelfand. 1995. Rapamycin inhibits ribosomal protein synthesis and induces G1 prolongation in mitogen-activated T lymphocytes. J. Immunol. 155:3418-3426. [PubMed] [Google Scholar]
- 61.Thomas, G. 2000. An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2:E71-E72. [DOI] [PubMed] [Google Scholar]
- 62.Thomas, G., and G. Thomas. 1986. Translational control of mRNA expression during the early mitogenic response in Swiss mouse 3T3 cells: identification of specific proteins. J. Cell Biol. 103:2137-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Todaro, G., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tuttle, R., N. Gill, W. Pugh, J. Lee, B. Koeberlein, E. Furth, K. Polonsky, A. Naji, and M. Birnbaum. 2001. Regulation of pancreatic b-cell growth and survival by the serine/threonine protein kinase Akt1/PKBα. Nat. Med. 7:1133-1137. [DOI] [PubMed] [Google Scholar]
- 65.Verdu, J., M. Buratovich, E. Wilder, and M. Birnbaum. 1999. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1:500-506. [DOI] [PubMed] [Google Scholar]
- 66.Vlahos, C., W. Matter, K. Hui, and R. Brown. 1994. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 269:5241-5248. [PubMed] [Google Scholar]
- 67.Volarevic, S., M. Stewart, B. Ledermann, F. Zilberman, L. Terracciano, E. Montini, M. Grompe, S. Kozma, and G. Thomas. 2000. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288:2045-2047. [DOI] [PubMed] [Google Scholar]
- 68.Wang, Q., R. Somwar, P. Bilan, Z. Liu, J. Jin, J. Woodgett, and A. Klip. 1999. Protein kinase B/Akt participates in GLUT4 translocation by insulin in L6 myoblasts. Mol. Cell. Biol. 19:4008-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, X., F. E. Paulin, L. E. Campbell, E. Gomez, K. O'Brien, N. Morrice, and C. G. Proud. 2001. Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon-subunit and their functions in vivo. EMBO J. 20:4349-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, X., W. Li, M. Williams, N. Terada, D. Alessi, and C. Proud. 2001. Regulation of elongation factor 2 kinase by p90RSK1 and p70 S6 kinase. EMBO J. 20:4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]