Abstract
While regulated transcription requires acetylation of histone N-terminal tails to promote an open chromatin conformation, a similar role for histone acetylation in DNA replication and/or repair remains to be established. Cells lacking the NuA4 subunit Yng2 are viable but critically deficient for genome-wide nucleosomal histone H4 acetylation. We found that yng2 mutants are specifically sensitized to DNA damage in S phase induced by cdc8 or cdc9 mutations, hydroxyurea, camptothecin, or methylmethane sulfonate (MMS). In yng2, MMS treatment causes a persistent Mec1-dependent intra-S-phase checkpoint delay characterized by slow DNA repair. Restoring H4 acetylation with the histone deacetylase inhibitor trichostatin A promotes checkpoint recovery. In turn, mutants lacking the histone H3-specific acetyltransferase GCN5 are similarly sensitive to intra-S-phase DNA damage. The inviability of gcn5 yng2 double mutants suggests overlapping roles for H3 and H4 acetylation in DNA replication and repair. Paradoxically, haploid yng2 mutants do not tolerate mutations in genes important for nonhomologous end joining repair yet remain proficient for homologous recombination. Our results implicate nucleosomal histone acetylation in maintaining genomic integrity during chromosomal replication.
In eukaryotes, DNA is assembled into nucleosomes, the basic structural unit of chromatin. Factors that target nucleosomes to modulate chromatin structure and provide access to nucleosomal DNA are required to facilitate processes such as transcription, replication, and DNA repair. Covalent modification of core histone N-terminal tails by acetylation of conserved lysine residues is thought to relax chromatin structure and allow access of nonhistone factors to nucleosomal DNA (8, 43). Seminal studies by Smerdon and colleagues showed that histone acetylation might be upregulated to facilitate DNA damage repair in mammalian cells after UV irradiation (40, 41, 46). Recent biochemical results support this model. The mammalian TATA-binding protein-free Taf II complex is recruited along with nucleotide excision repair proteins to UV-damaged DNA (5). Importantly, via its Gcn5 histone acetyltransferase (HAT) subunit, the TATA-binding protein-free Taf II complex preferentially acetylates histone H3 in nucleosomes containing UV-damaged DNA. In turn, acetylation of histone H4 also appears to function in DNA damage repair, as expression of a dominant-negative form of the Tip60 H4 HAT in mammalian cells blocks repair of double strand breaks (23).
Studies of histone N-terminal tail acetylation in the budding yeast Saccharomyces cerevisiae also support a role for histone acetylation in DNA repair and genomic integrity. Cells expressing H4 mutants lacking N-terminal acetylation sites activate DNA damage repair signaling and perform a G2/M delay even during unperturbed vegetative growth (33). In turn, conditional mutants in the essential yeast Tip60 homolog Esa1, the catalytic subunit of NuA4, lose nucleosomal histone H4 acetylation and accumulate in G2/M at the nonpermissive temperature. The terminal arrest is partly relieved by abrogating DNA damage checkpoint arrest (13). Analogously, mutants deficient in the Gcn5 or Sas3 histone H3 HAT display a G2/M delay which may also reflect DNA damage checkpoint activation (22, 51).
The molecular pathway leading to DNA damage checkpoint activation in histone acetylation mutants remains poorly understood. Perhaps transcription of genes necessary for DNA metabolism is critically impaired, reflecting the known requirement for targeted histone acetylation in transcriptional activation (6, 28). Nonetheless, transcription defects are unlikely to be the only underlying mechanism. Studies of genome-wide expression in yeast histone H4 acetylation mutants revealed surprisingly few significant changes (10, 42). In addition to gene-specific regulation, Esa1 has been implicated in modification of histone H4 across large segments of chromosomes (50). One interpretation is that the DNA-damage-dependent cell cycle arrest with nonacetylatable H4 and esa1 mutants may arise from the effects of decreased genome-wide acetylation per se rather than decreased expression of specific transcripts.
Like Esa1, virtually every other NuA4 subunit is essential for cell viability (2, 20). However, mutants lacking the NuA4 subunit Yng2 remain viable and yet are specifically compromised in global nucleosomal histone H4 acetylation (10, 30, 35). Mutants lacking Yng2 display a G2/M delay (10) and are sensitized to methylmethane sulfonate (35). Thus, we have used the yng2 mutant as a tool to study the role of NuA4 activity in DNA damage responses. As for esa1, the yng2 G2/M delay is DNA damage checkpoint dependent. Further, we demonstrate that Yng2 is required specifically to respond to DNA damage during S phase, suggesting a critical role for NuA4 in maintaining genomic integrity during DNA replication.
MATERIALS AND METHODS
Strains and media.
All yeast strains used were derived from W303 except for sister-chromatid exchange (SCE) assays. Yeast culture and genetic techniques were essentially carried out as described previously (21). Media were obtained from USBiological, molecular biology reagents were obtained from New England BioLabs, and chemical reagents were obtained from Sigma unless otherwise noted. Yeast were cultured in YPD (1% yeast extract, 2% peptone, 0.3 mM adenine, 2% glucose) or SC (synthetic complete media with 2% glucose) lacking the appropriate amino acids. 5-Fluoroorotic acid (5-FOA) was added to SC medium at a concentration of 1 mg/ml. G418 was added to YPD agar at a concentration of 0.2 mg/ml.
Genetic manipulations.
Complete knockouts were generated in diploid cells by gene replacement using the method of Longtine et al. (31). To determine synthetic lethality with yng2 deficiency, relevant mutant haploids were mated to a yng2 mutant or were deleted in a YNG2/yng2 heterozygous diploid. In each case, diploids were sporulated, dissected, and subjected to meiotic analysis. Genetic interaction was determined by examining predicted double mutant segregants. When double mutants germinated but were unable to form colonies, the genes were considered synthetic lethal or enhanced.
Molecular techniques and flow cytometry.
Cells of the relevant genotypes were grown in YPD overnight at 22°C and then diluted to an optical density at 600 nm of ∼0.05 to 0.1 in fresh YPD and allowed to grow for 3 to 4 h in the presence of 5 μM alpha mating peptide (αf) (Research Genetics) at 22°C. Alternatively, cells were incubated at 22°C in the presence of αf for 2 h and then shifted to 37°C for 1 to 1.5 h. Cells were released from αf arrest by centrifugation, washed with YPD, and then resuspended in liquid YPD at 22°C or YPD prewarmed to 37°C. Cells were collected at 60-min intervals (or as described in Results) for flow-cytometry, Northern, and Western analyses. For flow cytometry, cells were fixed in 70% ethanol, resuspended in 50 mM Tris-HCl (pH 7.5), sonicated, treated with 1 mg of RNase A/ml at 50°C for 1 h, and stained with 0.05 mg of propidium iodide/ml. Approximately 2 × 104 to 3 × 104 cells were analyzed using Cell Quest software and a FACSCalibur flow cytometer (Becton-Dickinson) for each time point. For Northern analysis, 25 ml of culture was collected, and RNA was extracted as described previously (15), separated on a 1% formaldehyde agarose gel, and transferred onto a nylon membrane (Osmonics). Probes labeled with [32P]dATP or [32P]dCTP (ICN) were generated by PCR and used to detect RNR3 mRNA. A PhosphorImager detection system and ImageQuant software (Molecular Dynamics) were used for analysis of blots.
For protein extracts, 25 to 50 ml of culture at an optical density at 600 nm of ∼0.1 was pelleted, washed, and resuspended in buffer containing protease inhibitors (50 mM Tris-HCl [pH 7.5], 10% glycerol, 0.5% Nonidet P-40, 2 mM EDTA, 150 mM NaCl, 500 μM benzamidine HCl, 10 μg of aprotinin/ml, 1 μg of leupeptin/ml, 1 μg of pepstatin A/ml, 1 mM phenylmethanesulfonyl fluoride). An equal volume of 0.5-mm-diameter glass beads was added, and the mixture was vortexed at high speed for 30 min at 4°C and centrifuged at 20,000 × g at 4°C to clear the lysate. For Western analysis, 50 μg of protein, measured by the Bradford assay (Bio-Rad), was loaded per lane on a sodium dodecyl sulfate-10% polyacrylamide gel and then transferred to nitrocellulose (ISC Intermountain). Blots were processed as described by the manufacturer for enhanced chemiluminescence (Amersham) and exposed to Hyperfilm ECL (Amersham). Detection of Rad53p-Myc was performed with a 1/1,000 dilution of A14 anti-Myc polyclonal antibody (Santa Cruz).
DNA damage assays.
To test cells for UV and gamma irradiation, hydroxyurea, camptothecin, and MMS sensitivity, freshly grown cells at room temperature were serially diluted 5- or 10-fold, and 3.5 μl was spotted onto a YPD plate and exposed to 100 J of UV irradiation/m2 or 200 to 400 Gy of irradiation or onto YPD plates containing 0.2 M hydroxyurea, 0.006% methyl methanesulfonate, or 10-μg/ml camptothecin and incubated at 25°C for up to 5 days. Plates were photographed with a Pixera camera. For the intra-S-phase checkpoint assay, cells were arrested in αf for 2 to 3 h at 22°C, treated with 0.03% methyl methanesulfonate (MMS) for 45 min, washed, and then resuspended in YPD with 0.03% MMS but in the absence of αf. In experiments with trichostatin A (TSA), cells were grown overnight with 30 μg of TSA (from 5-mg/ml stock solution in methanol)/ml and then treated as described for the intra-S-phase checkpoint except that cells were kept in TSA for the course of the experiments. Cells were collected at hourly intervals after release and processed for flow cytometry as described above.
Synthetic lethal screen.
A haploid yng2::HIS3 knockout strain carrying the centromeric URA3-marked plasmid pCT3-YNG2 was subjected to insertional mutagenesis by transformation with NotI-digested Tn3::lacZ::LEU2 transposon-mutagenized genomic library DNA (7) and selection on SC-Leu. Gene disruptions that conferred a requirement for expression of YNG2 would retain the pCT3-YNG2 plasmid and thus be unable to form colonies on 5-FOA, which counterselects against URA3. From ∼40,000 Leu+ transformants, 65 5-FOA-sensitive candidates were identified by replica plating and characterized further. Each isolate was backcrossed to the parental strain, and segregation of leucine prototrophy and 5-FOA sensitivity were determined. Eight LEU2-marked insertions were found to confer synthetic lethality with yng2. The insertion site of the Tn3::lacZ::LEU2 insertion was amplified by vectorette PCR (http://genome-www.stanford.edu/group/botlab/protocols/vectorette.html). Briefly, genomic DNA digested with RsaI or AluI and ligated to annealed anchor bubble primers was amplified using primers 1 (CGAATCGTAACCGTTCGTACGAGAATCGCT) and I2 (CGAATCGTAACCGTTCGTACGAGAATCGCT). Gel-purified PCR products were sequenced using primer 1 and an ABI cycle sequencing kit (Perkin-Elmer). Sequences were then submitted to the Saccharomyces Genome Database BLAST server (http://genome-www2.stanford.edu/cgi-bin/SGD/nph-blast2sgd) to identify disrupted genes.
Homologous recombination assays.
The assay for HO-endonuclease-induced single-strand annealing (SSA) was performed essentially as described previously (47). Complete knockouts of YNG2 were generated in haploid cells (W1479-11C) (47) by gene replacement using the method of Longtine et al. (31). Cells were grown at 22°C in raffinose overnight, and then galactose (2%) was added to induce HO expression. Cells collected at the indicated times were spread onto SC-Trp to select for cells still bearing the TRP-marked plasmid. Colonies were then replica plated onto SC-Ura to determine the number of cells that had lost the URA3 gene, indicative of SSA-mediated repair of HO-endonuclease-induced breaks. MMS-stimulated SCE events were monitored essentially as described previously (18). Complete knockouts of YNG2 were generated in haploid cells (YB163) (18) by gene replacement using the method of Longtine et al. (31). Asynchronously growing cells were left untreated or treated with 0.03% MMS for 1 h at 22°C and then plated onto YPD or SC-His to select for recombinants.
Chromosomal DNA gel analysis.
Cells were collected prior to and after MMS treatment for processing at the indicated time points from a single culture for each trial. Chromosomal DNA was prepared essentially as described previously (12) and separated in 1% agarose using a contour-clamped homogenous electric field (CHEF) gel electrophoresis apparatus (Bio-Rad). Gels were run for 23 h in 0.5× Tris-borate EDTA buffer kept at 14°C with a 90-s pulse rate at 220 V (6 V/cm).
RESULTS
Cells that are compromised in Yng2-dependent NuA4 HAT activity are supersensitive to DNA damage.
We and others have reported that yng2 mutant cells have reduced NuA4 HAT activity in vitro and in vivo, leading to a global decrease in histone H4 acetylation and phenotypes consistent with defects in DNA metabolism (10, 30, 35). To explore a potential link between NuA4 and DNA damage response, we challenged yng2 mutants with a variety of genotoxic agents. The DNA-damaging agents used, ionizing radiation (IR), UV irradiation, MMS, and hydroxyurea (HU), lead to distinct lesions that directly or via repair can induce DNA double-strand breaks (DSBs) and potentially lethal damage. Via a free-radical mechanism, IR directly generates single-strand breaks and DSBs. UV light induces cyclobutane pyrimidine dimers and 6-4 photoproducts, while MMS alkylates primarily guanine bases. Excision repair of UV or MMS damage may induce single-strand breaks and/or DSBs (9, 19). HU depletes nucleotide pools and inhibits DNA synthesis but at high concentrations leads to DSBs (34).
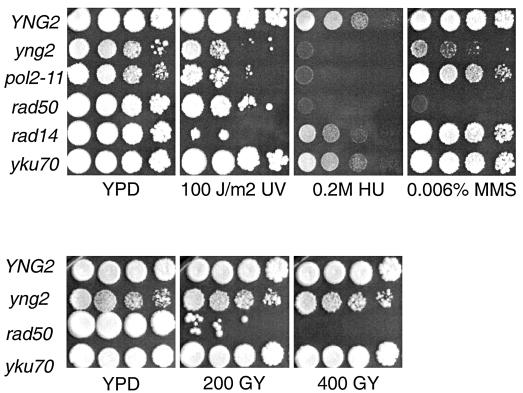
Cells grown in liquid culture were serially diluted, spotted onto YPD rich agar plates, and exposed to IR or UV or spotted onto YPD plates containing MMS or HU (Fig. 1). The viability of haploid wild-type or yng2 mutant cells is not compromised by 200 or 400 Gy of IR (∼15 to 30 DSBs), while a rad50 mutant lacking DSB repair displays marked sensitivity. This suggests that yng2 mutants remain DSB repair competent. By contrast, yng2 mutant cells are more sensitive than wild-type cells to 100 J of UV/m2 but not as sensitive as a rad14 excision repair mutant. Strikingly, the toxicity of 0.2 M HU to yng2 mutants was comparable to the Polɛ pol2-11 mutant. Consistent with a previous report (35), we also observed significant sensitivity for yng2 mutants on 0.006% MMS.
FIG. 1.
Cells deficient in Yng2 are highly sensitive to replication associated DNA damage. The wild type (YNG2), yng2 mutants, and several well-characterized DNA damage mutants were treated with DNA-damaging agents. While MMS and HU predominantly induce S-phase lesions, both UV and gamma irradiation can generate DNA breaks in all phases of the cell cycle. Strikingly, yng2 mutants are most sensitive to S-phase lesions. Mutants defective in several DNA damage repair pathways were used for comparison: rad50 mutants are defective for DSB repair; yku70 mutants are deficient in nonhomologous end-joining; pol2-11 mutants are defective in the intra-S-phase checkpoint response; rad14 mutants have defects in excision repair of UV-induced damage. Cells were serially diluted 10-fold and spotted onto plates. Photographs were taken after 4 to 5 days of growth at 25°C.
The spectrum of sensitivities to DNA-damaging agents displayed by yng2 mutants is distinct from that of previously characterized DNA replication or DNA repair mutants. In the response to UV, HU, and MMS, it is hypothesized that replication forks perform an important role in revealing or exacerbating the damage induced by these agents. If, rather than repair or replication, yng2 mutants are defective in the intra-S-phase checkpoint response, such a spectrum of sensitivities might be observed. These and other considerations turned our attention to S phase as a critical stage for Yng2-dependent DNA damage tolerance.
Cells deficient in Yng2 display a persistent intra-S-phase checkpoint arrest.
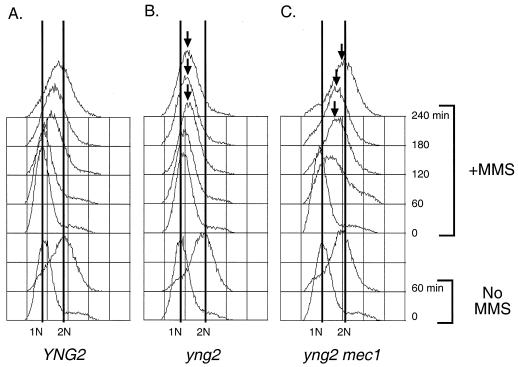
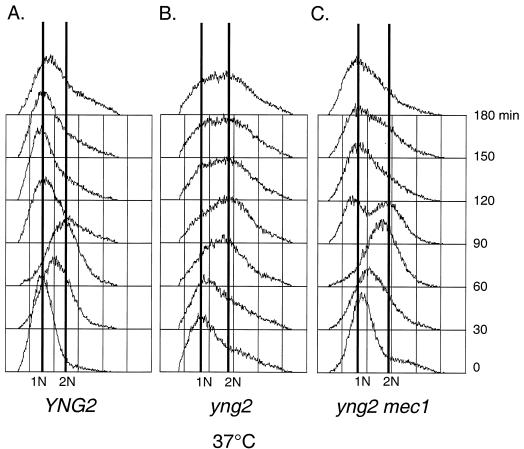
To implicate Yng2 in S phase DNA damage response, we first assessed the integrity of the intra-S-phase checkpoint in yng2 mutant cells. This checkpoint can be readily observed by following DNA replication kinetics in cells treated with MMS (37). Cultures synchronized in G1 with αf were released in the presence of 0.03% MMS, and cell cycle progression was analyzed by flow cytometry. Wild-type cells exposed to 0.03% MMS leave G1 on schedule and slowly proceed through S phase (Fig. 2A). Though yng2 mutant cells also begin replication on schedule, subsequent progress stalls after the initial rightward shift (Fig. 2B). These cells maintain a single, undivided nucleus and remain budded for the duration of the experiment (data not shown). Lower concentrations of MMS that do not slow wild-type S phase (e.g., 0.015%) still have a dramatic effect on yng2 mutants, although these cells can eventually complete replication (data not shown). These results suggest that yng2 mutants are not checkpoint deficient. Rather, the intra-S-phase checkpoint in yng2 mutants appears inappropriately sensitive. Alternatively, yng2 mutants may sustain greater damage than the wild type at the same dose of MMS.
FIG. 2.
Cells deficient in Yng2 display a persistent Mec1-mediated intra-S-phase delay. To examine the intra-S-phase checkpoint, the wild type (YNG2) and yng2 and yng2 mec1 mutants were arrested in G1 with αf, treated with 0.03% MMS for 45 min, and then released into the presence of 0.03% MMS at 22°C. Cells were collected at the indicated times and processed for flow cytometry using propidium iodide. 1N indicates haploid DNA content, and 2N indicates replicated DNA. YNG2 cells (A) display the characteristic slowed S phase in the presence of MMS, while yng2 single mutants (B) arrest in S phase. In contrast, yng2 mec1 double mutants (C) progress through and complete S phase. As a control, cells which were not treated with MMS are shown (No MMS).
Mec1 is required for persistent checkpoint arrest in yng2 mutant cells.
Mec1 is a phosphatidylinositol 3-kinase homolog and the yeast ATM/ATR equivalent that acts as a mediator of the DNA damage checkpoint response by inducing cell cycle arrest, activating transcription of DNA damage repair genes, and preventing late origin firing (16, 32, 48). To determine if the intra-S-phase arrest of yng2 mutants treated with 0.03% MMS represents a bona fide checkpoint response, we tested the dependence of arrest on Mec1. Double mutants between yng2 and mec1 were viable, suggesting that yng2 mutants do not require DNA damage checkpoint activity for life. We performed MMS experiments as described above with mec1 and yng2 mec1 mutants. Unlike yng2 mutants (Fig. 2B), the yng2 mec1 double mutants (Fig. 2C) entered and completed S phase, only slightly delayed by the DNA damage. These results suggest that the S-phase arrest in yng2 mutants reflects a block to fork progression and/or origin firing mediated by DNA damage checkpoint signaling.
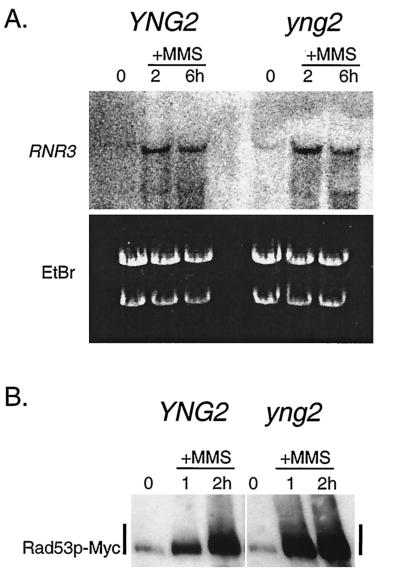
Transcriptional activation of repair genes, such as RNR3, is a hallmark of activation of the Mec1-dependent DNA damage checkpoint (16). Considering that acetylation is a critical determinant of transcriptional activation, one explanation for the persistent S-phase arrest in MMS and sensitivity towards DNA damage might be a loss of inducible transcription of DNA damage repair genes in yng2 mutant cells. We performed Northern analysis to examine expression of RNR3, a transcript known to be specifically induced after DNA damage and important for damage repair (17). We found that wild-type and yng2 mutant cells treated with MMS were able to up-regulate RNR3 to a similar extent (Fig. 3A). Notably, RNR3 is slightly induced in yng2 mutant cells even in the absence of exogenous DNA damage, indicating that these cells may accumulate DNA lesions during unperturbed vegetative growth. Moreover, in previous work using microarray analysis of genome-wide transcription, we found no loss in transcription of known DNA damage repair genes in yng2 mutant cells (10). Taken together, this suggests that defects in transcription are not likely to be the underlying basis for the persistent intra-S-phase arrest.
FIG. 3.
DNA-damage-dependent activation of RNR3 transcription and Rad53 is normal in yng2 mutants. A hallmark of the DNA damage response is transcriptional activation of DNA damage genes, such as RNR3, and phosphorylation of the checkpoint protein Rad53. Asynchronously growing wild-type (YNG2) and yng2 mutant cells treated with 0.03% MMS were collected at the indicated times. Cells were processed for Northern analysis (A) to detect RNR3 expression and for Western analysis (B) to examine Rad53. Both wild-type and yng2 mutant cells induce RNR3 expression in the presence of MMS. Similarly, YNG2 and yng2 mutant cells expressing Rad53-Myc accumulate shifted forms (consistent with phosphorylation) of Rad53-Myc to similar levels when treated with MMS. Note the appearance of RNR3 transcript in yng2 mutants (A) even in the absence of MMS treatment (0-h lane).
A characteristic increase in expression of the Rad53 kinase and a shift to a higher apparent molecular weight provide a second marker for DNA damage checkpoint activation (44). Similar to the RNR3 response, epitope-tagged Rad53 accumulated and shifted after MMS treatment in both wild-type and yng2 mutant cells (Fig. 3B). These results further demonstrate that yng2 mutant cells are proficient in DNA damage checkpoint activation.
Trichostatin A suppresses the persistent checkpoint arrest in yng2 mutant cells.
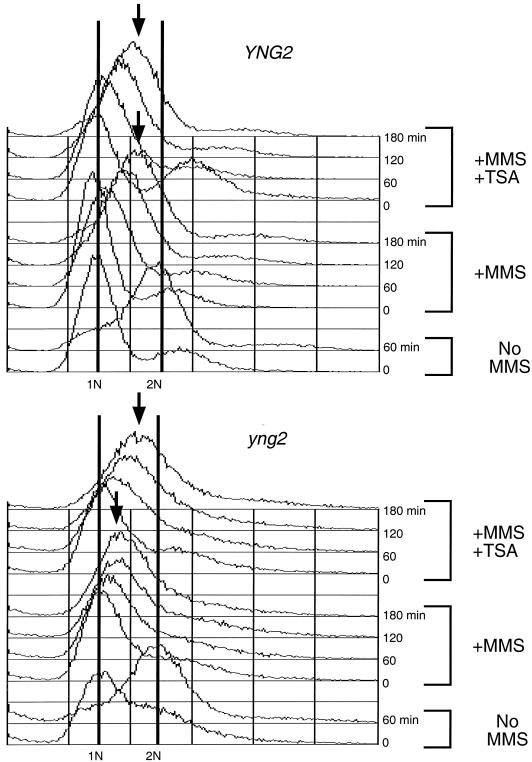
Previously we reported that treating yng2 mutants with the histone deacetylase inhibitor TSA restored genome-wide H4 acetylation (10). By extension, we considered that TSA might similarly suppress the intra-S-phase checkpoint arrest in yng2 mutants. Towards this end, wild-type and yng2 mutant cells were pretreated with TSA, αf synchronized, and released into 0.03% MMS. The TSA did not affect the characteristic checkpoint-induced slow S-phase progression of wild-type cells in MMS (Fig. 4, top). However, TSA treatment of yng2 mutants relieved the S-phase arrest, as evidenced by the continuous rightward progression in DNA content (Fig. 4, bottom). These results lend support to the idea that the persistent intra-S-phase arrest in yng2 mutants might be a result of decreased levels of acetylated chromatin.
FIG. 4.
The deacetylase inhibitor, TSA, alleviates the persistent checkpoint delay in yng2 mutants. Wild-type (YNG2) and yng2 mutant cells were pretreated with 30 μg of TSA/ml overnight (+MMS +TSA) or left untreated (+MMS) prior to addition of αf to induce G1 arrest. MMS was added to a final concentration of 0.03% while cells were in G1 for 45 min, αf was washed away, and cells were allowed to enter the cell cycle in the presence of 0.03% MMS with or without TSA. While YNG2 cells progress through S phase in MMS alone or MMS and TSA, yng2 mutants persist in S phase when treated with MMS but progress through S phase when TSA is present. No MMS, cells not treated with MMS.
Repair of S-phase DNA damage is compromised in yng2 mutants.
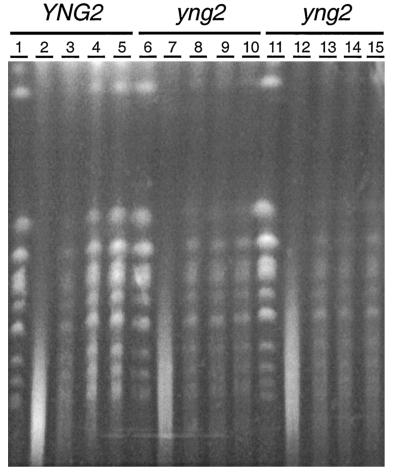
While yng2 mutant cells appear proficient in DNA damage checkpoint functions, such as cell cycle arrest and induction of transcription of repair genes, a defect in DNA repair might underlie the observed DNA damage phenotypes in yng2 mutants. To test this possibility, we used CHEF gels to analyze the integrity of chromosomal DNA from cells treated with MMS. Asynchronously growing wild-type and yng2 mutant cells were treated with 0.03% MMS for 2 h and returned to rich medium without MMS. Prior to the MMS treatment, wild-type and yng2 mutant cells displayed a similar banding pattern characteristic of grossly intact chromosomes (Fig. 5, lanes 1, 6, and 11). After cells were treated with MMS for 2 h, chromosomal DNA was similarly fragmented in both wild-type and yng2 mutant cells, as indicated by the loss of distinct bands and the appearance of a low-molecular-weight smear (Fig. 5, lanes 2, 7, and 12). The similar change in gel mobility suggests that recognition and processing of DNA damage to form DSBs proceeds comparably in wild-type and yng2 mutant cells. By 4 h after washing out MMS, chromosomal DNA bands had reappeared in wild-type cells, consistent with substantial completion of repair (Fig. 5, compare lane 4 to lane 1). In contrast to the wild type, yng2 mutants displayed markedly slower and less complete repair (Fig. 5, compare lanes 9 to 6 and 14 to 11). While the smear at low molecular weight largely disappeared by 2 h (Fig. 5, lanes 8 and 13), consistent with the onset of DSB repair and/or base excision repair, much of the chromosomal DNA did not reappear as bands even 5 h after MMS was removed (Fig. 5, lanes 10 and 15). A drawback of this method is that it does not reliably report the status of chromosomes being actively replicated in S phase or during repair, since their complex structures may prevent entry into the gel. Nonetheless, these results demonstrate an intrinsic defect in the yng2 mutant cell DNA damage response consistent with a deficit in DNA repair.
FIG. 5.
CHEF gel analysis reveals that yng2 mutants are compromised in DNA damage repair. Chromosomal DNA from wild-type (YNG2) (lanes 1 to 5) and yng2 mutant (lanes 6 to 13) cells was analyzed before, during, and after MMS treatment. Asynchronously growing cells display a characteristic banding pattern of intact chromosomal DNA seen in both YNG2 (lane 1) and yng2 mutant cells (lanes 6 and 11). After cells were treated for 2 h with 0.03% MMS, chromosomal DNA is fragmented, as evidenced by a low-molecular-weight smear in both YNG2 (lane 2) and yng2 (lanes 7 and 12) mutant cells. MMS was washed away, and cells were allowed to grow in rich medium, collected, and analyzed after 2, 4, and 5 h. Chromosomal DNA bands reappear by 2 h in YNG2 and yng2 mutant cells (lanes 3, 8, and 13). By 4 and 5 h, YNG2 cells have substantially repaired their chromosomal DNA, as indicated by the reappearance of even the highest-molecular-weight DNA bands (near tops of lanes 4 and 5). In contrast, the low levels of repair in yng2 mutants seen in lanes 8 and 13 persist for the remainder of the experiment, indicative of a defect in repair.
Mec1-dependent cell cycle delay in yng2 mutants during vegetative growth.
We previously showed that vegetatively growing yng2 mutants had a prolonged mitotic delay after replication that is markedly exacerbated at the nonpermissive temperature of 37°C (10). Based on our observation of elevated RNR3 expression in the absence of exogenous damage and the known checkpoint dependence of esa1 mitotic delay (13), we hypothesized that the yng2 mutant mitotic delay may be DNA damage checkpoint dependent. We constructed yng2 mec1 double mutants and performed αf arrest and release experiments to compare cell cycle kinetics of wild-type cells and yng2 and yng2 mec1 mutant cells at 37°C (Fig. 6). As previously reported, the yng2 mutants completed DNA synthesis with kinetics similar to those for the wild type but then delayed indefinitely after reaching replicated (2 N) DNA content. Like wild-type controls, yng2 mec1 double mutants do not delay in mitosis but return rapidly to G1. An implication of this result is that the mitotic arrest at 37°C in yng2 mutants may derive from DNA lesions and/or defects in chromatin structure that persist after completion of DNA replication.
FIG. 6.
yng2 mutant cells display a Mec1-dependent cell cycle delay during vegetative growth. The wild type (YNG2) and yng2 and yng2 mec1 mutants were arrested in G1 with αf, raised to 37°C for 1 h, and then released into rich medium at 37°C. Cells were collected at the indicated times and analyzed by flow cytometry. In contrast to yng2 single mutants, which remain with a large number of 2 N-containing cells (middle), yng2 mec1 mutants (right) are able to exit mitosis and return to G1 with nearly wild-type kinetics, evidenced by the reappearance of a 1 N DNA peak (compare to YNG2 [left]). αf was added again after cells entered S phase to trap cells in the subsequent G1 phase.
yng2 mutants display genetic interactions with replication mutants.
One interpretation of the pattern of sensitivity towards MMS and HU is that yng2 mutants cannot tolerate DNA breaks that arise during replication. Thus, we examined the genetic interaction between the yng2 mutation and conditional mutations affecting initiation, progression, or completion of replication. CDC7 encodes an essential kinase that activates replication origins (4). CDC8 encodes thymidylate kinase (24). CDC9 encodes the major Okazaki fragment DNA ligase (3). At nonpermissive temperature, the cdc7-1 mutant displays delayed origin firing, the cdc8-1 mutant is blocked during S phase progression, and the cdc9-8 mutant performs a mitotic arrest. We crossed yng2 to cdc7-1, cdc8-1, and cdc9-8 temperature-sensitive mutants and subjected the diploids to meiotic analysis at permissive temperature (Table 1). We found that yng2 cdc7-1 double mutants were fully viable and displayed phenotypes similar to those of yng2 single mutants. However, all predicted yng2 cdc8-1 segregants and the majority of yng2 cdc9-8 segregants were inviable, indicating synthetic enhancement. That we observed a strong genetic interaction with mutants that affected strand synthesis and ligation but not origin firing is consistent with Yng2 functioning during replication fork migration and confirms the supersensitivity towards HU and MMS in yng2 mutants. These data indicate a role for Yng2 in protecting cells from accumulating DNA lesions, specifically during S phase.
TABLE 1.
Genetic interactions between yng2 and mutants in DNA metabolism
| Gene name | Function | Synthetic enhancement with yng2a |
|---|---|---|
| CDC7 | Origin firing | − |
| CDC8 | Maintain nucleotide pools | ++ |
| CDC9 | DNA ligation | + |
| GCN5 | Histone H3 acetyltransferase | ++ |
| YKU70 | Binds DNA ends/NHEJb | ++ |
| RAD50 | Break resection/NHEJ and HRc | ++ |
| MRE11 | Break resection/NHEJ and HR | + |
| RAD52 | Primarily for HR | − |
| RAD57 | Primarily for HR | − |
| TOP1 | Topoisomerase 1 | + |
At least 20 tetrads were analyzed for each test. −, double mutants are phenotypically similar to the single mutant. ++, >90% of predicted double mutants were inviable. +, >60% of predicted double mutants were inviable, while double mutants which formed colonies displayed enhanced growth defects compared to either single mutant.
NHEJ, nonhomologous end joining.
HR, homologous recombination.
Topoisomerase 1 activity is essential in yng2 mutant cells.
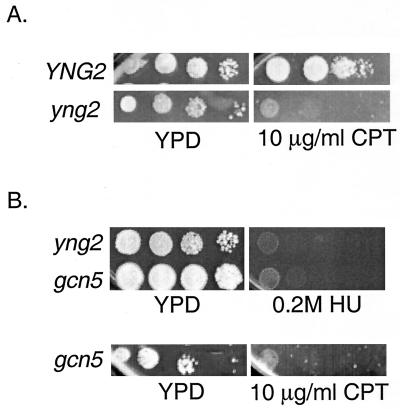
During replication, the type I topoisomerase Top1 acts to relieve superhelical tension which may accumulate ahead of the advancing fork (25). We hypothesized that in yng2 mutant cells, hypoacetylated chromatin might result in abnormally high levels of torsional stress due to increased DNA-histone and/or internucleosomal interactions. To test this possibility, we subjected diploids heterozygous for both yng2 and top1 to meiotic analysis. Consistent with a previous report (25), we find that top1 mutants are viable and have only a subtle growth defect. Strikingly, although virtually all predicted yng2 top1 double mutants germinated, double mutants were unable to form colonies, indicating that in the absence of Yng2, Top1 activity becomes essential (Table 1). Consistent with the synthetic enhancement with top1, yng2 mutant cells treated with the topoisomerase I inhibitor camptothecin (CPT) (11) display marked sensitivity compared to wild-type cells (Fig. 7A). While the supersensitivity towards CPT in yng2 mutants might result from persistent DNA breaks induced by the formation of CPT-Top1-DNA complexes (11), depletion of Top1 activity from the treated cells is also likely to be significant.
FIG. 7.
Supersensitivity to CPT and HU in yng2 and gcn5 mutants. The wild type (YNG2) and yng2 (A) and gcn5 (B) mutants were serially diluted 10-fold and spotted onto plates containing CPT, a topoisomerase inhibitor, and HU, a replication inhibitor. Strikingly, yng2 and gcn5 mutants display sensitivities towards CPT and HU similar to those displayed by wild-type cells. Photographs were taken after 4 to 5 days of growth at 25°C.
A synthetic lethal screen reveals a requirement for GCN5 in yng2 mutants.
We were prompted by finding specific genetic interactions between yng2 and replication mutants to pursue a genetic screen for other mutations that confer lethality to a yng2 deletion. Synthetic lethality or enhancement between two mutations typically indicates that the two gene products act in a single essential pathway or have compensatory functions. Therefore, this approach should help shed light not only on the mechanism of Yng2 function but also on essential functions of histone H4 acetylation. Thus, we constructed a strain in which a wild-type allele of YNG2 was maintained on an unstable, URA3-marked low-copy-number episome that complemented a deletion of the YNG2 genomic locus. Evicting the YNG2 complementing plasmid by counterselection with 5-FOA revealed the underlying yng2 phenotype. A library of genomic fragments mutagenized by random integration of a LEU2-containing cassette (7) was transformed into this strain to create a population of marked insertional mutants. Eight independent mutations were isolated from more than 40,000 transformants that conferred inviability on 5-FOA medium. Each of these mutations was observed to be synthetic lethal or enhanced, with a yng2 deletion, upon conventional meiotic analysis. Sequence analysis revealed that each insertion had interrupted genes affecting histone physiology and/or DNA damage processing.
We were most struck by two mutations. One insertion interrupted the GCN5 open reading frame. GCN5 encodes an H3-specific HAT, suggesting that histone H3 and histone H4 acetylation may share an essential overlapping function. Indeed, truncation of the N-terminal tails of both histones H3 and H4 is not tolerated (29). Similar to Esa1 function in genome-wide acetylation of histone H4, Gcn5 has been reported to function in global acetylation of histone H3 (27, 50). On the other hand, gcn5 mutants have been found to have a slight decrease in acetylated isoforms of histone H4 (51).
That yng2 gcn5 double mutants were inviable suggested that acetylation of histones H3 and H4 might serve overlapping roles. One possibility is that this shared function is related to DNA damage tolerance. Strikingly, gcn5 mutants were sensitive to hydroxyurea and camptothecin to roughly the same degree as yng2 mutants (Fig. 7B) but less so towards MMS and UV (data not shown). These results suggest overlapping roles for histone H3 and H4 acetylation during replication.
Genetic interactions between yng2 and DSB repair mutations.
A second insertion isolated as a yng2 synthetic lethal or enhancement mutation disrupted the open reading frame encoding the yeast Ku70 homolog YKU70/HDF1. YKU70 is critical for nonhomologous end joining (NHEJ) repair of DSBs and is required for telomere stability but performs a minor role in homologous recombination (HR) repair. One interpretation of the yng2 yku70 synthetic enhancement is that NHEJ may be uniquely able to repair DNA damage arising in yng2 mutants. Alternatively, a lack of normal histone H4 acetylation creates a specific requirement for yKu70 rather than NHEJ per se. To distinguish these possibilities, the yng2 mutant was crossed with other NHEJ mutants (rad50 and mre11) and with HR mutants (rad52 and rad57) to examine genetic interactions (36). Supporting a requirement for NHEJ, yng2 rad50 segregants were all inviable, while most yng2 mre11 double mutants were inviable or displayed enhanced growth defects (Table 1). In contrast, yng2 rad52 and yng2 rad57 double mutants were all viable and were phenotypically similar to yng2 single mutants (Table 1). Together these results strongly suggest that yng2 mutants require NHEJ repair or a specialized function(s) of these genes in the absence of Yng2-dependent NuA4 HAT activity.
Cells deficient in Yng2 are proficient for homologous recombination.
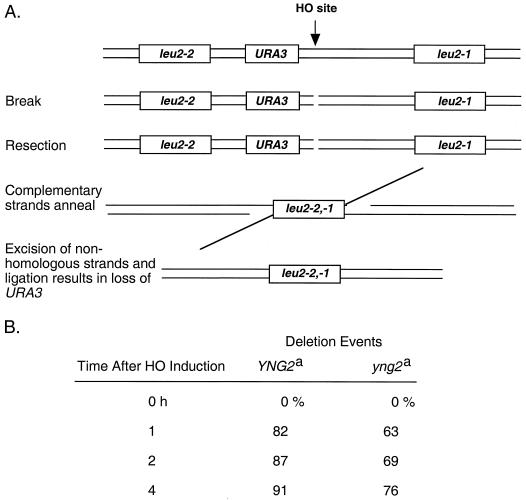
We reasoned that if yng2 mutants are critically defective in homologous recombination, loss of the NHEJ pathway might be lethal while HR mutations would have no consequence. To explore this possibility, we assayed wild-type and yng2 mutant cells for their ability to perform two forms of HR: SSA and SCE. To determine if cells were proficient in SSA, we used strains in which a HO site is placed adjacent to a URA3 gene flanked by two leu2 alleles (Fig. 8A). Galactose-induced expression of HO endonuclease results in a persistent DSB. Resection by 5′-to-3′ exonucleases beginning at the break leads to single-stranded DNA that uncovers complementary leu2 sequences. Annealing between the two complementary strands of DNA at leu2 and subsequent DNA ligation completes repair but excises URA3, allowing growth on 5-FOA (Fig. 8A) (47). Aliquots were collected at 1-h intervals after galactose addition, and cells were spread onto 5-FOA and nonselective plates. The number of 5-FOA-resistant cells relative to the number of total cells plated was used as an estimate of the fraction of DSBs repaired by SSA at that time point (Fig. 8B). We found that wild-type and yng2 mutant cells lost URA3 at a comparable rate, suggesting that the resection, annealing, processing, and ligation events in SSA-mediated repair can each occur in the absence of Yng2.
FIG. 8.
yng2 mutants are able to perform SSA repair. (A) Schematic of the leu2 direct repeat cassette used to measure an HO-induced recombination event measured by loss of the URA3 gene. (B) Wild-type (YNG2) cells and yng2 mutant cells containing this cassette were transformed with a TRP1 plasmid containing a galactose-inducible HO endonuclease gene. Cells were taken at the indicated times post-galactose addition, and the number of cells that had undergone a recombination event and lost the URA3 gene was determined. YNG2 and yng2 mutant cells display similar levels of SSA. Footnote a, approximately 200 to 900 colonies were scored for each time point. The percentage of deletion events is a measure of the total number of colonies that have lost Ura prototrophy relative to the total number of cells plated that were Trp+.
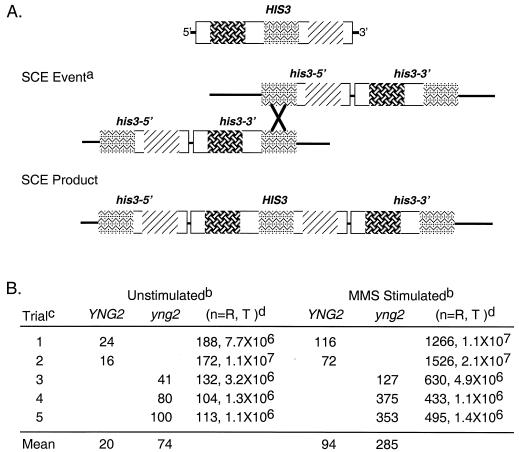
SCE was measured by determining the number of His+ recombinants arising from mitotic recombination between two his3 alleles juxtaposed on a single chromosome (18) (Fig. 9A). The his3-5′ and his3-3′ alleles are truncated at the 5′ and 3′ ends, respectively, so that an unequal crossover event between sister chromatids can generate a partial duplication and a wild-type HIS3 allele (Fig. 9A). Rather than being blocked for SCE, yng2 mutant cells yielded fourfold more spontaneous recombinants than the wild-type population as measured by the number of His+ clones per million cells (Fig. 9B, unstimulated). SCE can be stimulated by treating cells with DNA-damaging agents, leading to an increased accumulation of His+ clones. Treating asynchronously growing wild-type and yng2 mutant cells for 1 h with 0.03% MMS increased the number of His+ recombinants by fivefold in the wild type and fourfold in yng2 mutants (Fig. 9B, MMS stimulated). These observations confirm that yng2 mutant cells remain proficient in homologous recombination.
FIG. 9.
yng2 mutants are proficient in SCE-mediated recombination. (A) Diagram showing the physical arrangement of the his3 alleles (his3-3′ is truncated at the C terminus and his3-5′ is truncated at the N terminus) and the predicted SCE event which gives rise to a wild-type structure of HIS3 conferring histidine prototrophy. (B) Asynchronously growing YNG2 and yng2 mutant cells containing the construct shown in panel A were left untreated (unstimulated) or treated with sublethal levels of MMS (MMS stimulated). The frequency of recombination events was determined based on the number of colonies formed on SC-His relative to the total number of cells plated. YNG2 and yng2 mutants display a similar frequency of induced SCE events (relative to unstimulated, fivefold, and fourfold increase, respectively, based on the mean). Note the ∼fourfold increase in mean unstimulated recombinants for yng2 mutants (unstimulated) relative to results for YNG2 cells. Footnote a, SCE event illustrates a predicted recombination event between two sister chromatids at a region of homology between his3-5 and his3-3: the recombination product (SCE product) on one sister chromatid. Footnote b, unstimulated indicates that cells were asynchronously growing in liquid medium before plating, and MMS Stimulated indicates that asynchronously growing cells were treated for 1 h with MMS and then plated. The number of colonies formed on SC-His plates (indicative of SCE events) per million cells plated is shown for each trial. Footnote c, trials 1 and 2 were performed with the same YNG2 isolate, whereas in each trial for yng2 (trials 3 to 5), independent yng2 mutant isolates were used. Footnote d, (n=R, T) represents the number of recombinants (R) (colonies formed on SC-His plates) and the total (T) number of cells plated.
DISCUSSION
Several studies have shown that cells deficient for the nonessential NuA4 subunit Yng2 have decreased NuA4 activity and an overall reduction in levels of bulk acetylated histone H4 (10, 30, 35). In this report, we exploit yng2 mutants to investigate the potential connection between histone H4 acetylation and DNA damage tolerance. The relative sensitivity to different types of DNA damage and a unique pattern of genetic interactions point to a special deficit in yng2 mutants affecting tolerance of DNA damage during DNA replication. Yng2 had previously been implicated in DNA damage tolerance (35). We find that yng2 mutant cells are no more sensitive to IR than wild-type cells but cannot tolerate even low doses of UV irradiation, hydroxyurea, camptothecin, or MMS, all agents that induce replication-specific blocks or lesions (11, 19, 34). Implicating Yng2 in S-phase progression rather than initiation, we find synthetic enhancement in yng2 mutants combined with conditional mutations in the CDC8 thymidylate kinase (24) and the CDC9 DNA ligase (3) but not the CDC7 kinase that participates in origin activation (4). We infer that the defect is not at the level of sensing DNA damage, since yng2 mutant cells are proficient in activating the Mec1-dependent checkpoint pathway. The yng2 mutant performs a persistent Mec1-dependent intra-S-phase delay in the presence of 0.03% MMS, a dose at which wild-type cells slow S phase but can complete replication. Importantly, CHEF gel analysis reveals that yng2 mutants process MMS-damaged DNA to create DSBs but do not complete DNA repair.
We were surprised to discover that viability of yng2 mutant cells largely depends on TOP1 and several genes known to be critical for NHEJ but not on HR genes. We speculate that defects in transcription, repair, and/or replication are all possible explanations for these requirements. Considering that transcription is intimately coupled to histone acetylation, Yng2 might be important for transcription of genes that function in replication and/or repair. Possibly, in yng2 mutant cells critical genes would be transcriptionally down-regulated, leading to the requirement for TOP1 or NHEJ genes for viability. However, our microarray analysis of genome-wide expression shows no down-regulation of DNA repair genes (10). Moreover, in this report we show that RNR3 transcription remains inducible after MMS treatment. Thus, a transcription defect seems unlikely.
Perhaps, as suggested by the higher basal rate of SCE recombination in yng2 mutants, DNA strand breaks that occur during vegetative growth or that are induced by exogenous agents are repaired slowly in hypoacetylated chromatin, leaving damage that may require Top1 or NHEJ function for proper repair. Alternatively, Yng2 might have other functions beyond its role in NuA4 that affect DNA repair. Like its mammalian homolog, p33ING1b, which has been shown to directly interact with both the PCNA complex and several HAT complexes (45, 49), Yng2 might interact with and thereby modulate activities of yeast factors that play a more direct role in DNA metabolism. On the other hand, acetylation of nonhistone proteins (i.e., p53) by HAT complexes in mammalian cells is well studied and is shown to modulate protein activity. Interestingly, Nourani and colleagues (35) showed that p53 expressed in yeast can interact with NuA4 and that this interaction depends on the presence of Yng2. Thus, Yng2 might help target NuA4 to acetylate and activate nonhistone proteins important for DNA repair.
We cannot rule out a defect in replication itself, considering our finding that yng2 mutants are specifically sensitized to DNA damage in S phase. An intriguing possibility is that chromatin which contains less acetylated histone H4 might be susceptible to lesions in front of the replication fork. These DNA breaks might be exclusively repaired by NHEJ, since a sister chromatid or homolog would not be available for HR-mediated repair (14, 36). Work with other organisms examining requirements for acetylation of histones at the replication fork suggests that a complete block to replication is highly unlikely (38). Nonetheless, a more subtle defect during replication that arises as a result of topological constraints conferred by hypoacetylated chromatin remains a formal possibility (1, 39).
Histone H3 acetylation might also have an important role in DNA damage tolerance. Interestingly, gcn5 mutants are synthetic lethal with yng2 and display similar sensitivities towards S phase DNA-damaging agents. Similar to yng2, esa1, and nonacetylatable histone H4 mutants, cells lacking GCN5 display a mitotic delay during vegetative growth (22, 26, 51). Certainly, acetylation of histones H3 and H4 may have equivalent roles in responding to DNA damage at several levels (i.e., gene expression, recognition of lesions, and repair). Determining the basis for supersensitivity towards HU and CPT in gcn5 mutants will require further study.
This report reveals a novel role for histone acetylation in preserving genomic integrity in the face of DNA damage during S phase. Future studies to understand the connections between histone acetylation, replication, and repair may establish chromatin conformation as a key determinant of DNA metabolism.
Acknowledgments
We thank A. Amon, M. Fasullo, J. Fitz-Gerald, M. Lisby, R. Rothstein, and M. Snyder for generously providing reagents and for critical discussions. We extend special thanks to C. Sham and D. Bishop for help with CHEF gel analysis and valuable comments. We thank B. Tobe for critical reading of the manuscript and colleagues in the Center for Molecular Oncology for valuable discussions.
These studies have been generously supported by a James S. McDonnell Foundation Scholar Award, a grant from the Ludwig Fund for Cancer Research, and NIH grant RO1 GM60443. S.J.K. is a Leukemia & Lymphoma Society Scholar.
REFERENCES
- 1.Alexiadis, V., L. Halmer, and C. Gruss. 1997. Influence of core histone acetylation on SV40 minichromosome replication in vitro. Chromosoma 105:324-331. [DOI] [PubMed] [Google Scholar]
- 2.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker, D. G., A. L. Johnson, and L. H. Johnston. 1985. An improved assay for DNA ligase reveals temperature-sensitive activity in cdc9 mutants of Saccharomyces cerevisiae. Mol. Gen. Genet. 200:458-462. [DOI] [PubMed] [Google Scholar]
- 4.Bousset, K., and J. F. Diffley. 1998. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 12:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand, M., J. G. Moggs, M. Oulad-Abdelghani, F. Lejeune, F. J. Dilworth, J. Stevenin, G. Almouzni, and L. Tora. 2001. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 7.Burns, N., B. Grimwade, P. B. Ross-Macdonald, E. Y. Choi, K. Finberg, G. S. Roeder, and M. Snyder. 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8:1087-1105. [DOI] [PubMed] [Google Scholar]
- 8.Cheung, P., C. D. Allis, and P. Sassone-Corsi. 2000. Signaling to chromatin through histone modifications. Cell 103:263-271. [DOI] [PubMed] [Google Scholar]
- 9.Chlebowicz, E., and W. J. Jachymczyk. 1979. Repair of MMS-induced DNA double-strand breaks in haploid cells of Saccharomyces cerevisiae, which requires the presence of a duplicate genome. Mol. Gen. Genet. 167:279-286. [DOI] [PubMed] [Google Scholar]
- 10.Choy, J. S., B. T. Tobe, J. H. Huh, and S. J. Kron. 2001. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 276:43653-43662. [DOI] [PubMed] [Google Scholar]
- 11.Christiansen, K., and O. Westergaard. 1996. The effect of camptothecin on topoisomerase I catalysis. Ann. N. Y. Acad. Sci. 803:50-59. [DOI] [PubMed] [Google Scholar]
- 12.Chu, G., D. Vollrath, and R. W. Davis. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582-1585. [DOI] [PubMed] [Google Scholar]
- 13.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19:2515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Critchlow, S. E., and S. P. Jackson. 1998. DNA end-joining: from yeast to man. Trends Biochem. Sci. 23:394-398. [DOI] [PubMed] [Google Scholar]
- 15.Cross, F. R., and A. H. Tinkelenberg. 1991. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell 65:875-883. [DOI] [PubMed] [Google Scholar]
- 16.Elledge, S. J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 17.Elledge, S. J., and R. W. Davis. 1990. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 4:740-751. [DOI] [PubMed] [Google Scholar]
- 18.Fasullo, M., P. Giallanza, Z. Dong, C. Cera, and T. Bennett. 2001. Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics 158:959-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 20.Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Heliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Cote. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5:927-937. [DOI] [PubMed] [Google Scholar]
- 21.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology, vol. 194. Academic Press, Inc., San Diego, Calif.
- 22.Howe, L., D. Auston, P. Grant, S. John, R. G. Cook, J. L. Workman, and L. Pillus. 2001. Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15:3144-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 24.Jong, A. Y., C. L. Kuo, and J. L. Campbell. 1984. The CDC8 gene of yeast encodes thymidylate kinase. J. Biol. Chem. 259:11052-11059. [PubMed] [Google Scholar]
- 25.Kim, R. A., and J. C. Wang. 1989. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J. Mol. Biol. 208:257-267. [DOI] [PubMed] [Google Scholar]
- 26.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 27.Kuo, M. H., E. vom Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 28.Kuo, M. H., J. Zhou, P. Jambeck, M. E. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling, X., T. A. Harkness, M. C. Schultz, G. Fisher-Adams, and M. Grunstein. 1996. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 10:686-699. [DOI] [PubMed] [Google Scholar]
- 30.Loewith, R., M. Meijer, S. P. Lees-Miller, K. Riabowol, and D. Young. 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20:3807-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longtine, M. S., A. R. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 32.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 33.Megee, P. C., B. A. Morgan, and M. M. Smith. 1995. Histone H4 and the maintenance of genome integrity. Genes Dev. 9:1716-1727. [DOI] [PubMed] [Google Scholar]
- 34.Merrill, B. J., and C. Holm. 1999. A requirement for recombinational repair in Saccharomyces cerevisiae is caused by DNA replication defects of mec1 mutants. Genetics 153:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nourani, A., Y. Doyon, R. T. Utley, S. Allard, W. S. Lane, and J. Cote. 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21:7629-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulovich, A. G., and L. H. Hartwell. 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82:841-847. [DOI] [PubMed] [Google Scholar]
- 38.Perry, C. A., C. D. Allis, and A. T. Annunziato. 1993. Parental nucleosomes segregated to newly replicated chromatin are underacetylated relative to those assembled de novo. Biochemistry 32:13615-13623. [DOI] [PubMed] [Google Scholar]
- 39.Quintini, G., K. Treuner, C. Gruss, and R. Knippers. 1996. Role of amino-terminal histone domains in chromatin replication. Mol. Cell. Biol. 16:2888-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramanathan, B., and M. J. Smerdon. 1986. Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis 7:1087-1094. [DOI] [PubMed] [Google Scholar]
- 41.Ramanathan, B., and M. J. Smerdon. 1989. Enhanced DNA repair synthesis in hyperacetylated nucleosomes. J. Biol. Chem. 264:11026-11034. [PubMed] [Google Scholar]
- 42.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 43.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 45.Scott, M., P. Bonnefin, D. Vieyra, F. M. Boisvert, D. Young, D. P. Bazett-Jones, and K. Riabowol. 2001. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J. Cell Sci. 114:3455-3462. [DOI] [PubMed] [Google Scholar]
- 46.Smerdon, M. J., S. Y. Lan, R. E. Calza, and R. Reeves. 1982. Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. J. Biol. Chem. 257:13441-13447. [PubMed] [Google Scholar]
- 47.Smith, J., and R. Rothstein. 1999. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics 151:447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 49.Vieyra, D., R. Loewith, M. Scott, P. Bonnefin, F. M. Boisvert, P. Cheema, S. Pastyryeva, M. Meijer, R. N. Johnston, D. P. Bazett-Jones, S. McMahon, M. D. Cole, D. Young, and K. Riabowol. 2002. Human ING1 proteins differentially regulate histone acetylation. J. Biol. Chem. 277:29832-29839. [DOI] [PubMed] [Google Scholar]
- 50.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408:495-498. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, W., J. R. Bone, D. G. Edmondson, B. M. Turner, and S. Y. Roth. 1998. Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17:3155-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]