Abstract
Overexpression studies have suggested that Siah1 proteins may act as effectors of p53-mediated cellular responses and as regulators of mitotic progression. We have tested these hypotheses using Siah gene knockout mice. Siah1a and Siah1b were not induced by activation of endogenous p53 in tissues, primary murine embryonic fibroblasts (MEFs) or thymocytes. Furthermore, primary MEFs lacking Siah1a, Siah1b, Siah2, or both Siah2 and Siah1a displayed normal cell cycle progression, proliferation, p53-mediated senescence, and G1 phase cell cycle arrest. Primary thymocytes deficient for Siah1a, Siah2, or both Siah2 and Siah1a, E1A-transformed MEFs lacking Siah1a, Siah1b, or Siah2, and Siah1b-null ES cells all underwent normal p53-mediated apoptosis. Finally, inhibition of Siah1b expression in Siah2 Siah1a double-mutant cells failed to inhibit cell division, p53-mediated induction of p21 expression, or cell cycle arrest. Our loss-of-function experiments do not support a general role for Siah genes in p53-mediated responses or mitosis.
p53 acts as a tumor suppressor by inducing cell cycle arrest, apoptosis, and senescence in response to cellular stresses such as DNA damage or oncogene activation. These effects are mediated largely through the function of p53 as a transcriptional activator or repressor (5, 25). A diverse range of p53 target genes have been identified, for example, p21, an important mediator of G1 phase cell cycle arrest and cellular senescence (10), and several proapoptotic genes including Noxa and Bax (53). However, understanding of the full range of p53 effectors is incomplete and it is likely that multiple signaling pathways are involved in determining cellular responses to p53 activation.
Recent studies have proposed that Siah1, a member of the Siah family of E3 ubiquitin ligases, may act as a downstream effector of p53. Overexpression of p53 induces the transcriptional activation of Siah1 family genes in a variety of mammalian cell lines (1, 20, 32, 35, 43, 44, 47), and overexpression of Siah1 can mimic the effects of p53 activation and induce cell cycle arrest or apoptosis (37, 43, 44).
A molecular mechanism by which Siah1 proteins may mediate p53 function has been proposed. Human SIAH1 functions as a component of an E3 ubiquitin ligase complex including Siah-interacting protein, Skp1, and the F-box protein Ebi, which is proposed to target β-catenin for ubiquitin-mediated degradation in response to activation of p53 (32, 36, 42). Importantly, overexpression of either p53 or SIAH1 can induce β-catenin degradation, and this is blocked by coexpression of an N-terminally truncated SIAH1 protein lacking the RING domain. By this model, p53-mediated induction of SIAH1 following DNA damage induces degradation of β-catenin independently of the GSK3β-mediated degradation pathway. Since overexpression of β-catenin promotes cell cycle progression and inhibits cell cycle arrest induced by gamma irradiation (41), SIAH1-mediated β-catenin degradation may contribute to p53-dependent cell cycle arrest. Interestingly, accumulation of a β-catenin mutant protein that is resistant to degradation by the GSK3β pathway induces activation of p53 (7). Since overexpression of SIAH1 can induce degradation of wild-type or mutant forms of β-catenin, SIAH1 may function downstream of p53 in a pathway that senses and degrades oncogenic β-catenin and thereby contributes to tumor suppression.
Mice have three unlinked Siah genes: Siah1a, Siah1b (collectively Siah1), and Siah2, while humans have single SIAH1 and SIAH2 genes (9, 20). The mammalian Siah proteins are highly homologous to one another. Siah1a and Siah1b proteins are 98% identical, while Siah1 and Siah2 proteins diverge significantly only at their N termini. Siah proteins interact via their RING domains with the E2 ubiquitin conjugating enzymes UbcH5 (36) and UbcH8 (54) and with UbcH9, which catalyzes conjugation of the ubiquitin-like protein SUMO-1 (22). Overexpression of Siah proteins induces the ubiquitination and proteasome-dependent destabilization of diverse substrate proteins. These include DCC, Kid, β-catenin, c-myb, Obf-1, Numb, and TIEG-1 for Siah1 (3, 15, 21, 22, 26, 32, 36, 47, 49, 51), and DCC, Bag-1, N-CoR and synaptophysin for Siah2 (22, 46, 54, 55). Siah1 and Siah2 proteins function equivalently to induce the degradation of some substrates (e.g., DCC), while other substrates (e.g., Bag-1) appear to be specific targets of either Siah1 or Siah2.
In addition to their suggested role as mediators of p53 function, Siah1 proteins have also been implicated in mitosis and meiosis. Siah1a is required for progression past metaphase during meiosis I of spermatogenesis (11), and overexpression of SIAH1 in MCF7 cells induces multinucleation and mitotic abnormalities (4, 15). These findings are consistent with studies showing that overexpression of SIAH1 induces the degradation of the kinesin Kid (15), a DNA-binding microtubule motor protein that forces chromosome arms away from mitotic spindle poles to facilitate chromosome alignment at the metaphase plate (30). Proteasome-mediated degradation of X-Kid is required for chromosome segregation in Xenopus oocytes (2, 13). It has therefore been hypothesized that degradation of Kid by Siah proteins may be necessary to allow chromosome segregation during mitosis and/or meiosis.
To date, the functions of the Siah gene family have been investigated largely through overexpression studies, often in transformed or immortalized cell lines. To begin to characterize the physiological functions of the Siah genes, we have generated and analyzed mice deficient for Siah1a (11), Siah2, or both Siah2 and Siah1a (R. Dickins, I. Frew, V. Hammond, and D. Bowtell, unpublished results). Whereas Siah1a knockout mice are subviable and growth retarded and Siah2 knockout mice are completely viable, Siah2 Siah1a double-mutant mice die at birth. This result supports previous biochemical findings that Siah1 and Siah2 proteins have both distinct and overlapping functions. In this report we describe the generation of embryonic stem (ES) cells and MEFs lacking Siah1b and have used these, and primary cells derived from other Siah knockout mice, to comprehensively investigate whether Siah genes are necessary for p53-mediated responses or for mitosis. We find that Siah genes are not induced by activation of endogenous p53 in mouse tissues or in primary MEFs or thymocytes and are not necessary for p53-mediated senescence, cell cycle arrest, or apoptosis. While Siah1a is clearly required for meiotic progression (11), cellular proliferation and cell cycle progression of MEFs are unaffected by the loss of Siah genes, suggesting that Siah proteins are not necessary for chromosome segregation in somatic cells.
MATERIALS AND METHODS
Targeted mutation of Siah1b.
The Siah1b targeting construct was generated by using the vector pPNTloxP (gift of Paul Orban). DNA for the targeting arms was derived from a 129Sv genomic library λ phage clone encompassing the Siah1b locus (9). PCR was used to generate the 2.6-kb 5′ and 7.6-kb 3′ targeting arms (see Fig. 2A). Transfection of ES cells and selection of drug-resistant colonies were as previously described (11). Genomic DNA from resistant clones was digested with BamHI and Southern blotted. Blots were probed with a 400-bp KpnI-HindIII fragment (probe 4) located 5′ of the region of DNA used as the 5′ targeting arm. Homologous recombination introduces a new BamHI site, leading to a size shift of the hybridizing band from 3.7 to 3.3 kb. A single targeted clone was isolated from 452 clones screened. Probing with a 560-bp SspI fragment (probe 8) of the Siah1b gene confirmed specific deletion of the Siah1b coding region. Chimeras were derived by injection of targeted ES cells into C57Bl/6J blastocysts. Live chimeras were born at low frequency (four were obtained from 320 blastocyst injections, compared with approximately 50% frequency for other ES cell clones injected in parallel experiments) and failed to transmit the agouti coat color to progeny when mated to C57Bl/6J mice.
FIG. 2.
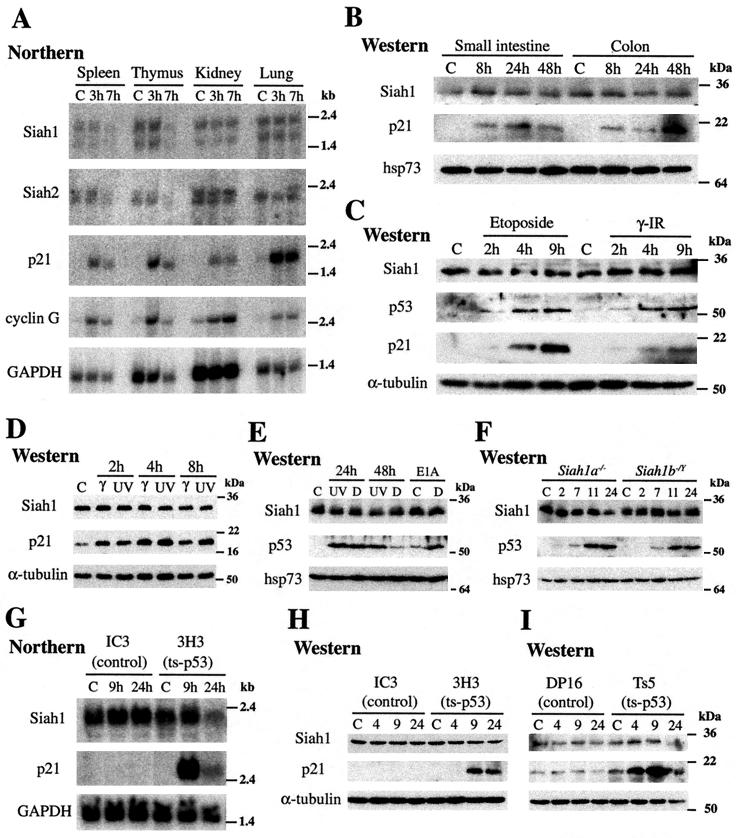
Activation of endogenous p53 does not induce Siah1 expression. In all panels, C refers to untreated control mice or cells. (A) Northern blot analysis of mRNA (10 μg) isolated from spleen, thymus, kidney, or lung from mice 3 or 7 h after treatment with gamma irradiation (10 Gy). Blots were probed with DNA fragments from the 3′ UTR of mouse Siah1a or Siah2 genes or coding region fragments of mouse cyclin G, human p21, or GAPDH genes. The Siah1 probe detects Siah1a and Siah1b. The upper band includes both Siah1a and Siah1b transcripts. The lower band corresponds to a shorter Siah1a transcript. (B to F, H, and I) Western blot analyses of total protein lysates prepared from the indicated tissues or cell types. Siah1, p53, and p21 protein levels were determined by use of specific antibodies. The anti-Siah1 monoclonal antibody recognizes both Siah1a and Siah1b. Probing with antibodies against α-tubulin or hsp73 verified equal loading and transfer. (B) Small intestine and colon from mice 8, 24, or 48 h after gamma irradiation (8 Gy). (C) Mouse thymocytes 2, 4, and 9 h after etoposide (2 μg/ml) addition or gamma irradiation (5 Gy). (D) Primary MEFs 2, 4, and 8 h after gamma (10 Gy) or UV-C (50 J/m2) irradiation. (E) Primary MEFs 24 and 48 h after UV-C irradiation (50 J/m2) or doxorubicin (D) treatment (1 μg/ml). The final two lanes represent lysates from MEFs transformed with E1A 12S, either untreated or treated with doxorubicin (D) (0.1 μg/ml) for 24 h. (F) Siah1a- and Siah1b-null MEFs 2, 7, 11, or 24 h after treatment with UV irradiation (50 J/m2). (G) CEM cells lacking functional p53 (IC3) or expressing a ts-p53 mutant (3H3) were cultured either at 37°C (control) or at 32°C for 9 or 24 h to induce p53 activity. Total RNA (10 μg) was probed with the coding regions of mouse Siah1b or human p21 or GAPDH. (H) Western analysis of total protein lysates from CEM cells (as in panel G) at times after shifting to 32°C. (I) Control erythroleukemic cells lacking functional p53 (DP-16) or expressing a ts-p53 mutant (Ts5) were cultured and analyzed as described for panel H.
Generation of MEFs lacking Siah genes.
MEFs were derived from embryonic day 13.5 (E13.5) embryos according to standard protocols. Siah1a−/−, Siah2−/−, or Siah2−/− Siah1a−/− MEFs were derived from intercrosses of 129Sv.C3-+c+p background Siah1a+/−, Siah2+/−, or Siah2−/− Siah1a+/− mice, respectively. Appropriate control cultures were derived from littermate embryos. Siah1b−/Y MEFs were obtained from chimeric embryos produced via injection of Siah1b-targeted ES cells into blastocysts and reimplantation into foster mothers. Mutant MEFs were selected by culturing in the presence of G418 (200 μg/ml; Gibco-BRL) for 2 passages (5 to 7 days), and the elimination of wild-type cells was confirmed by Southern blotting using probe 4. Cells were frozen at passage 2 and were assayed at passage 4 or 5.
Cell culture and MEF proliferation assays.
MEFs were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin (500 IU/ml), streptomycin (500 μg/ml), and 200 μM β-mercaptoethanol. For proliferation assays, passage 4 MEFs were seeded at 2 × 105 cells per plate in replicate 60-mm-diameter dishes and cell numbers were determined daily for 7 days. Media were replaced every second day. For 3T3 assays, passage 4 MEFs were seeded at 3 × 105 cells per 60-mm-diameter dish, cell numbers were determined after 3 days, and cells were reseeded for the next passage at the starting density. Cell cycle synchronization experiments using HeLa cells were undertaken as described previously (29). CEM (IC3, 3H3) and erythroleukemic (DP-16, Ts5) cell lines were maintained at 37°C in DMEM supplemented with 10% FBS, 250 μM l-asparagine, 13 μM folic acid, penicillin (500 IU/ml), streptomycin (500 μg/ml), and 50 μM β-mercaptoethanol. Temperature shift was achieved by placing cells in media prewarmed to 32°C and by subsequent incubation at 32°C.
p53 activation.
p53 activation was induced by gamma irradiation of mice or cultured cells (137Cs source, 0.75 Gy/min) or by treatment of cells with UV-C irradiation (Stratalinker 1800; Stratagene), disodium N-(phosphonacetyl)-l-aspartic acid (PALA; Developmental Therapeutics Program, National Cancer Institute), doxorubicin (Pharmacia), or etoposide (Sigma). Retrovirus (pME2 SVpuro) (23) expressing E1A 12S was a gift of Dobrila Nesic. Phoenix cells were transfected (Lipofectamine Plus) with the retroviral construct. After 2 days, the retrovirus-containing media were harvested and polybrene (4 μg/ml) was added. MEFs were incubated overnight with this mixture, washed to remove polybrene, and cultured for 24 h before addition of puromycin (3 μg/ml). Transformed MEFs were cultured with the drug for 4 days before the assay.
RNA and protein analysis.
Northern blots were probed with coding region DNA fragments from mouse cyclin G, human p21 or GAPDH genes, an 850-bp EcoRV fragment from the 3′ untranslated region (UTR) of mouse Siah2, or a 680-bp EcoRI-HindIII fragment from the 3′ UTR of mouse Siah1a. Protein extracts from mouse tissues were prepared as previously described (11), and whole-cell extracts were generated from cultured cells by lysis in 1% sodium dodecyl sulfate-10 mM Tris (pH 7.4) followed by boiling for 5 min. Protein lysates were quantitated by the Bio-Rad Dc protein assay, and 20 to 40 μg of protein was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using antibodies against Siah1 (11), p21 (F-5; Santa Cruz,), p53 (FL-393; Santa Cruz), α-tubulin (B-5-1-2; Sigma), hsp73 (spa-815; StressGen), E1A 12S (M58; BD PharMingen), and cyclin B1 (H-433; Santa Cruz).
Cell cycle analysis and cell death assays.
S-phase cells were labeled by incubation with BrdU (30 μM; Sigma) for 30 min (or 4 h; see Fig. 5). Cells were harvested by trypsinization, fixed in 70% ethanol, and treated with 2N HCl-0.5% Triton X-100 for 30 min followed by 0.1 M sodium tetraborate (pH 8.5). Incorporated BrdU was detected by using anti-BrdU-fluorescein isothiocyanate (BD PharMingen) and DNA stained with propidium iodide (PI) in the presence of RNase A (40 μg/ml). Flow cytometry (FACSCalibur) and WinMDI analysis software were used to determine cell cycle distribution after gating to exclude cellular debris and fixation artifacts (e.g., doublets).
FIG. 5.
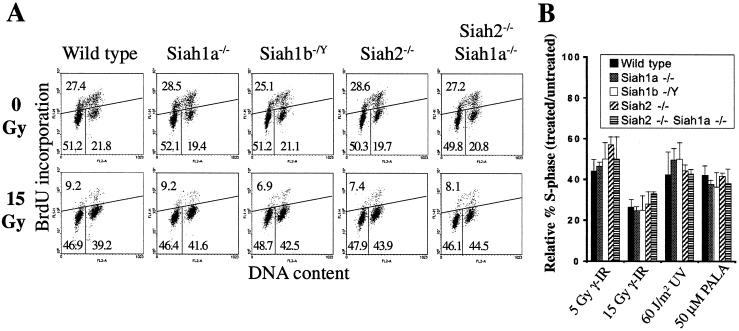
p53-dependent cell cycle arrest does not require Siah genes. (A) Asynchronously growing wild-type, Siah1a−/−, Siah1b−/Y, Siah2−/−, or Siah2−/− Siah1a−/− MEFs were subjected to gamma irradiation (0 or 15 Gy). Cell cycle distributions were assessed 24 h after treatment by BrdU labeling and flow cytometry. The percentage of cells in each phase of the cell cycle (lower left, G1; upper, S; lower right, G2/M) is shown. Distributions shown are representative of duplicate determinations of each of two or more independent MEF preparations. (B) MEFs were harvested 24 h after gamma irradiation (5 or 15 Gy) or UV-C irradiation (60 J/m2) or after 48 h of culture in the presence of PALA (50 μM) and analyzed as described for panel A. The extent of G1 cell cycle arrest is expressed as relative percent S phase, calculated by the following formula: (% BrdU positive in untreated cultures/% BrdU positive in treated cultures) × 100. The means ± standard deviations are derived from duplicate determinations of each of three independent MEF preparations of each genotype. Similar results were obtained in an independent experiment.
ES cells were grown on fibroblast feeder layers, and adherent and nonadherent cells were harvested by trypsinization 24 h after UV irradiation and fixed in 70% ethanol; apoptosis was quantitated by flow cytometry and PI staining to measure the percentage of cells with sub-2N DNA content.
Adherent and nonadherent E1A-transformed or -nontransformed MEFs were harvested by trypsinization 24 h after treatment with doxorubicin, and the percentage of viability was quantitated by flow cytometry and exclusion of PI by live cells.
Thymocytes were isolated from 6- to 10-week-old mice by crushing thymi through a wire mesh. Cells were washed twice in culture medium (DMEM supplemented with 10% FBS, 250 μM l-asparagine, 13 μM folic acid, penicillin [500 IU/ml], streptomycin [500 μg/ml], and 50 μM β-mercaptoethanol) prior to culture at 2 × 106 cells/ml. Percent viability was determined by trypan blue exclusion 24 h after gamma irradiation. Siah2−/− Siah1a−/− thymocytes were obtained 13 weeks after adoptive transfer of E14.5 Siah2−/− Siah1a−/− fetal liver cells (19). Briefly, embryos from timed matings of Siah2−/− Siah1a+/− mice were genotyped by PCR, as described previously (11), and 106 fetal liver cells were injected into the tail veins of B6.SJL-Ptprca (CD45.1) mice (Animal Resources Centre, Perth, Australia) that had been gamma irradiated (5 Gy followed 3 h later by 4.5 Gy). Flow cytometry using anti-CD45.2-FITC (BD PharMingen) showed that >98% of thymocytes were derived from donor fetal liver cells.
Antisense treatment.
Passage 4 MEFs were seeded in 12-well plates at 4 × 104 cells per well the day before transfection with phosphothioate- and 2′-O-methyl-modified oligonucleotides (120 nM) by using cationic peptoid L1 (750 nM) and C (750 nM) transfection reagents (Chiron Corp.). Transfection media were removed after 16 h. Oligonucleotide sequences were as follows: Siah1b AS, 5′-GCTGTGCAATGCTGGTGTCAAACAC; Siah1b RC, 5′-CACAAACTGTGGTCGTAACGTGTCG; Siah1a AS, 5′-ACCGAGGAGTCGCTTCCCAAGTCA; Siah1a RC, 5′-ACTGAACCCTTCGCTGAGGAGCCA.
RESULTS
Generation of Siah1b-null ES cells and of MEFs lacking Siah genes.
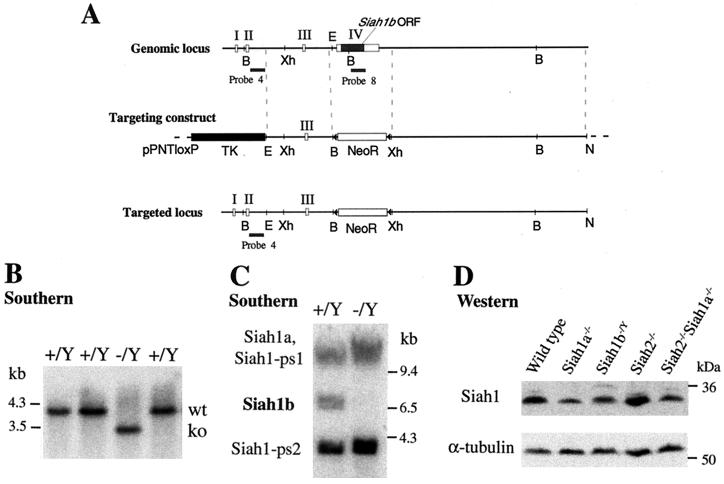
To complete the disruption of the Siah gene family, we attempted to generate Siah1b knockout mice. We designed a targeting vector to completely remove the single coding exon of the Siah1b gene (Fig. 1A) and screened ES cells by using a positive (neomycin resistance) and negative (thymidine kinase) selection strategy to enrich for homologous recombinant ES cell clones. Since the W9.5 ES cell clone used in these experiments is genotypically male (48) and the Siah1b gene resides on the X chromosome (18), homologous recombinant clones are hemizygous (null) for Siah1b. Southern blotting identified a single targeted clone (designated −/Y) from 452 drug-resistant colonies (Fig. 1B). The low targeting frequency likely reflects difficulty in targeting the locus rather than selection against ES cells that had lost Siah1b expression through gene targeting, since we failed to generate targeted clones with a loxP targeting vector designed to retain the Siah1b gene after homologous recombination (0 of 254 clones screened; data not shown). Specific deletion of the Siah1b coding region was confirmed by Southern blotting using coding region probe 8 that also hybridizes to bands corresponding to Siah1a and the two Siah1 pseudogenes (Fig. 1C) (9). The targeted clone was karyotypically normal (data not shown) but failed to generate chimeras that could transmit the mutation to progeny. Live chimeras were born at low frequency (four were obtained from 320 blastocyst injections), and many embryos exhibited developmental abnormalities or were resorbed during pregnancy (data not shown). These data suggest that Siah1b may be necessary for the viability of certain tissues during embryonic development.
FIG. 1.
Disruption of Siah1b in ES cells by gene targeting. (A) Exons I, II, III, and IV of Siah1b are depicted as open rectangles; the coding region of Siah1b is a filled rectangle. The targeting construct in the vector pPNTloxP is depicted. DNA fragments used as the left and right arms of homology are represented by dotted lines. Homologous recombination yields the targeted locus in which the entire coding region is replaced by a loxP-flanked neomycin resistance gene cassette. B, BamHI; Xh, XhoI; E, EcoRI; triangle, loxP element. (B) Southern analysis of BamHI-digested genomic DNA from wild-type (+/Y) or targeted (−/Y) ES cells, probed with probe 4 (located 5′ of the 5′ targeting arm) to identify the targeted locus. (C) Southern analysis (as for panel B) with coding region probe 8 that hybridizes to the four murine Siah1 genes (including the two pseudogenes Siah1-ps1 and Siah1-ps2), to confirm the loss of the Siah1b gene. (D) Western blot analysis of total protein lysates derived from wild-type, Siah1a−/−, Siah1b−/Y, Siah2−/−, or Siah2−/− Siah1a−/− MEFs, using a monoclonal anti-Siah1 antibody that recognizes both Siah1a and Siah1b.
While we were unable to produce Siah1b knockout mice, we successfully generated MEFs lacking Siah1b from chimeric embryos (see Materials and Methods). To investigate the proposed roles of Siah genes in p53-mediated responses and in chromosome segregation, we also generated a panel of isogenic primary MEFs deficient for Siah1a or Siah2 or both Siah2 and Siah1a. Western blotting, using a monoclonal antibody that recognizes both Siah1a and Siah1b (11), demonstrated that total Siah1 protein levels are decreased by the loss of either Siah1a or Siah1b and that Siah1b protein expression in Siah1a−/− or Siah2−/− Siah1a−/− cells represents approximately one-half the level of total Siah1 expression seen in wild-type cells (Fig. 1D). Antibodies that detect endogenous Siah2 are not available.
Activation of endogenous p53 does not induce Siah expression.
Based on overexpression evidence, current models propose that Siah1 genes are transcriptionally activated by p53. These studies have not investigated Siah gene induction in response to activation of endogenous p53. To address this question, we analyzed the levels of Siah1a, Siah1b, and Siah2 mRNA in tissues harvested from wild-type mice in which p53 activation was induced by whole-body gamma irradiation (Fig. 2A). While Siah gene expression was not increased, established p53 target genes p21 (34) and cyclin G (40) were strongly activated in these tissues. As Siah1 proteins are proposed to mediate p53-induced β-catenin degradation in the gut (32, 36, 42), we analyzed Siah1 protein expression in mouse small intestine and colon for up to 2 days following p53 activation by gamma irradiation. Siah1 protein levels were unaltered by this treatment, yet sustained accumulation of p21 confirmed that p53 was transcriptionally active (Fig. 2B). Thus, expression of Siah genes is not induced in various mouse tissues, including gut, following activation of endogenous p53 by gamma irradiation.
Since gene expression changes mediated by p53 depend on the nature of the inducing stimulus, the level of p53 expression, and the cell type (56), we treated primary thymocytes and MEFs with a range of p53-activating stimuli. Siah1 protein expression in thymocytes was unaltered during p53-mediated apoptosis induced by gamma irradiation or etoposide treatment (Fig. 2C) (6). Activation of p53 in MEFs by treatment with gamma or UV irradiation did not alter Siah1 protein levels at early time points (2, 4, or 8 h) after treatment (Fig. 2D). Similarly, sustained (24 and 48 h) activation of p53 by treatment of MEFs with UV irradiation (Fig. 2E), DNA-damaging drugs including doxorubicin (Fig. 2E) and cisplatin (data not shown), or the disruption of microtubules with nocodazole (data not shown) also failed to increase Siah1 protein levels. Furthermore, Siah1 abundance was not increased in MEFs in which p53 was stabilized as a result of transformation with E1A 12S oncogene (45) or when p53 was further activated in these cells by doxorubicin treatment, leading to induction of p53-mediated apoptosis (Fig. 2E) (12). Since the anti-Siah1 monoclonal antibody detects both Siah1a and Siah1b, we treated Siah1a- or Siah1b-null MEFs with UV irradiation to examine the effect of p53 activation on the expression of each gene individually. Neither Siah1b nor Siah1a expression increased in Siah1a- and Siah1b-null MEFs, respectively, in response to p53 activation (Fig. 2F). Stabilization of p53 and/or induction of p21 confirmed p53 activation in these assays. Thus, we conclude that activation of endogenous p53 in primary MEFs and thymocytes does not induce Siah1 gene expression.
As we observed no induction of Siah1 genes in response to activation of endogenous p53, we sought to determine whether p53 overexpression is sufficient to induce Siah1 expression. Activation of temperature-sensitive p53 (ts-p53) by temperature shift has been reported to induce Siah1 expression in MEF and myeloid leukemia cell lines (1, 20, 43, 44, 47). Here we show that p53-mutant human T-cell lymphoma CEM cells expressing ts-p53 (14) do not induce SIAH1 mRNA or protein expression upon shifting to the permissive temperature (Fig. 2G and H). Similarly, activation of ts-p53 in a p53-null mouse erythroleukemic cell line (27) also failed to increase SIAH1 protein levels (Fig. 2I). Induction of p21 mRNA and/or protein in these cell lines verified that p53 was functionally activated. Finally, in contrast to previous reports (32, 35, 37), we found that transfection of p53 in 293 or 293T cell lines had no effect on SIAH1 expression (data not shown). We conclude from these findings that induction of Siah1 expression is not a general feature of p53 activation and may be restricted to specific cell types or stimuli or dependent on the level of p53 overexpression.
Loss of Siah genes does not alter mitotic progression.
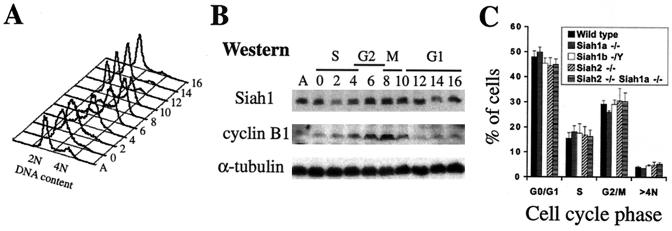
Since Siah1 genes have been linked to negative regulation of the cell cycle and also to mitotic and meiotic progression, we investigated whether SIAH1 protein expression is regulated during the cell cycle. HeLa cells synchronized in early S phase by a double thymidine block were released into the cell cycle, and cultures were harvested at various time points for flow cytometric analysis of DNA content (Fig. 3A) and Western blotting of total protein lysates (Fig. 3B). SIAH1 protein levels initially decreased as cells entered S phase and then increased during the G2 and M phases before again decreasing as the cells exited into G1. Upregulation of SIAH1 expression during the G2 and M phases may reflect an important role in mitosis.
FIG. 3.
Siah genes are not required for cell cycle progression (A) Flow cytometry analysis (DNA content) of HeLa cells after release from double thymidine block. A, asynchronous culture. Cells were released from early S phase at time zero and collected every 2 h for 16 h. Progression through mitosis occurred after 8 to 10 h. (B) Western blot analysis of total protein extracts prepared from the HeLa cells described for panel A. The accumulation and degradation of cyclin B1 (high in G2, degraded at the end of mitosis) provided an independent marker of progression through the cell cycle. (C) Asynchronous cell cycle distribution of passage 4 wild-type, Siah1a−/−, Siah1b−/Y, Siah2−/−, or Siah2−/− Siah1a−/− MEFs. Data represent means ± standard deviations of duplicate assays of MEF preparations from three independent embryos of each genotype.
We have previously shown that Siah1a is necessary for proper progression past metaphase during meiosis I of spermatogenesis (11). While Siah1a−/− MEFs do not display cell cycle or proliferative defects (11), it is possible that Siah1b and/or Siah2 may compensate for loss of Siah1a in somatic cells and allow mitotic progression. It is significant that chromosome segregation defects are seen only during spermatogenesis in Siah1a−/− mice. As Siah2 expression is detected only in postmeiotic spermatocytes (8) and the Siah1b gene resides on the X chromosome and is thus likely to be silenced by X inactivation during spermatogenesis (17), it is possible that this setting reflects a true Siah gene loss-of-function phenotype. To begin to examine this possibility, we have analyzed the cell cycle properties of MEFs lacking Siah1a, Siah1b, or Siah2 or lacking both Siah2 and Siah1a. MEFs of all genotypes displayed unaltered asynchronous cell cycle distribution profiles and did not accumulate cells with greater than 4N DNA content (Fig. 3C), suggesting that mitotic progression was unimpaired.
Normal proliferation, contact inhibition and senescence in MEFs lacking Siah genes.
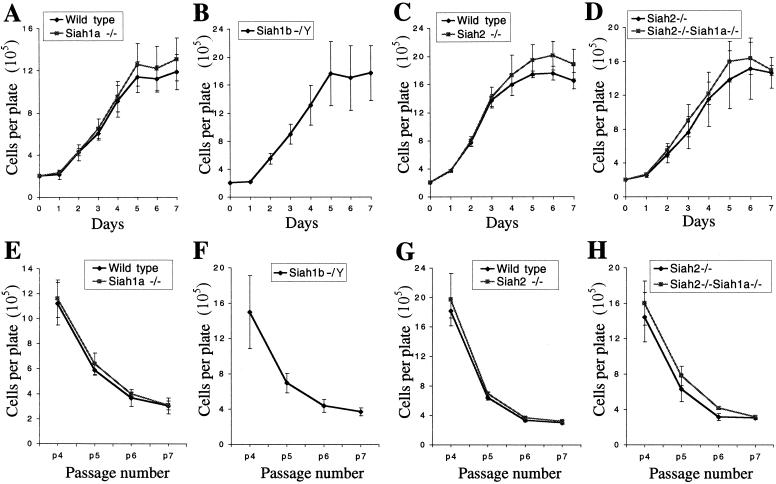
To further assess the integrity of the cell cycle of MEFs lacking Siah genes, we undertook short-term and long-term (3T3) proliferation assays (52). Since Siah1 proteins have been proposed to mediate p53 function and p53 is required for senescence and contact inhibition in MEFs (16), these experiments also tested p53 function in the absence of Siah genes. The rate of proliferation and saturation density of passage 4 MEFs were not altered by the loss of Siah genes (Fig. 4A through D). All genotypes reached cellular densities similar to those of their littermate controls at all passages in 3T3 assays and underwent senescence by passage 7 (Fig. 4E through H). The rates of proliferation, saturation densities, and entry into senescence of Siah1b−/Y MEFs were similar to those of MEFs of the other genotypes. Thus, Siah1a, Siah1b, or Siah2 alone or both Siah2 and Siah1a in combination are not required for p53-mediated contact inhibition or senescence.
FIG. 4.
Normal proliferation, saturation density, and senescence in MEFs lacking Siah genes. (A to D) Proliferation and saturation density of passage 4 MEFs; (E to H) 3T3 analysis of MEF proliferation and senescence. All genotypes entered senescence after passage 7. Due to variations in proliferation rates among MEFs prepared from different pregnancies, comparisons were made only between groups of MEFs derived from littermate embryos, with the exception of Siah1b−/− MEFs, which were derived from chimeric blastocysts by G418 selection and therefore do not have littermate controls. (A and E) Wild type (n = 2) versus Siah1a−/− (n = 3); (B and F) Siah1b−/Y (n = 3); (C and G) wild type (n = 2) versus Siah2−/− (n = 2); (D and H) Siah2−/− (n = 3) versus Siah2−/− Siah1a−/− (n = 3). Data points depict the means ± standard deviations of duplicate assays of MEF preparations from n independent embryos.
Siah genes are not necessary for p53-mediated cell cycle arrest.
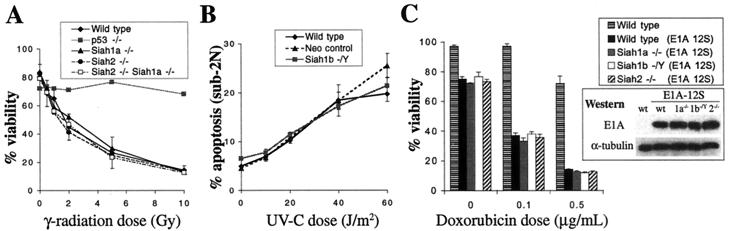
As well as mediating senescence and contact inhibition, p53 is required in MEFs for G1 phase cell cycle arrest in response to DNA damage or cellular stress (10). To investigate whether Siah genes are necessary for p53-mediated G1 arrest, we treated MEFs lacking Siah1a, Siah1b, or Siah2 or both Siah2 and Siah1a with gamma irradiation (0 or 15 Gy) and analyzed cell cycle distribution after 24 h (Fig. 5A). MEF cultures of all genotypes exhibited a decreased percentage of cells in S phase following irradiation, indicating that the p53-dependent G1 checkpoint was unaffected. Like wild-type cells, MEFs lacking Siah genes also accumulated in the G2/M phase after DNA damage, indicating that the p53-independent G2 checkpoint (28) does not require Siah genes. MEFs of all genotypes also displayed a p53-dependent G1 arrest in response to submaximal doses of gamma irradiation (5 Gy), UV irradiation, or ribonucleotide depletion (PALA treatment) (Fig. 5B) (10). In summary, we find that p53-mediated cell cycle arrest induced by a variety of stimuli is unaffected by mutation of Siah1a, Siah1b, or Siah2 alone or by combined mutation of Siah2 and Siah1a.
Siah genes are not necessary for p53-mediated apoptosis.
In some cell types, p53 induces apoptosis in response to DNA damage. Thymocyte apoptosis in response to gamma irradiation is abolished by the loss of p53 (6) (Fig. 6A). Thymocytes isolated from Siah1a or Siah2 knockout mice or from mice in which the hematopoietic system was reconstituted with fetal liver cells from Siah2−/− Siah1a−/− embryos underwent apoptosis in a manner equivalent to that of thymocytes isolated from wild-type mice (Fig. 6A). Similarly, Siah1b hemizygous ES cells underwent normal p53-dependent apoptosis following UV irradiation (Fig. 6B) (5). While primary MEFs are normally resistant to apoptosis induced by DNA damage, MEFs transformed by the E1A 12S oncogene are highly sensitive to apoptosis in response to p53 activation (33). Wild-type, Siah1a−/−, Siah1b−/Y, and Siah2−/− MEFs transformed by retroviral infection with E1A 12S were equally sensitive to p53-mediated apoptosis induced by doxorubicin treatment (Fig. 6C) (12). Western blotting confirmed equal levels of expression of E1A 12S for these genotypes. Thus, Siah1a, Siah1b, or Siah2 alone, or Siah2 and Siah1a together, are dispensable for p53-mediated apoptosis.
FIG. 6.
Siah genes are not required for p53-mediated apoptosis (A) Thymocytes were isolated from wild-type (n = 3), Siah1a−/− (n = 2), Siah2−/− (n = 2), or p53−/− (n = 1) mice or from lethally irradiated mice (n = 2) that had been reconstituted with Siah2−/− Siah1a−/− fetal liver cells. Cell viability was assessed 24 h after gamma irradiation. Duplicate cultures from each of n mice of each genotype were analyzed. Means (± standard deviations [SD] where applicable) are shown. (B) Susceptibility to UV-induced apoptosis of the parental ES cell clone (wild type), a clone containing a random insertion of the Siah1b targeting vector (Neo control), and the Siah1b-targeted clone (Siah1b−/Y). Apoptosis was assessed 24 h after treatment. Means ± SD of triplicate assays are shown. Data are representative of three independent experiments. (C) Wild-type, Siah1a−/−, Siah1b−/Y, or Siah2−/− MEFs were infected with retrovirus expressing E1A 12S. The viability of E1A-expressing MEFs or of noninfected wild-type MEFs was assessed 24 h after treatment with doxorubicin (0.1 or 0.5 μg/ml). Means ± SD of triplicate assays are shown. Western blotting (inset) confirmed equal levels of E1A 12S expression among the genotypes.
Inhibition of Siah1b expression in Siah2−/− Siah1a−/− MEFs does not alter cell cycle progression or p53-mediated G1 arrest.
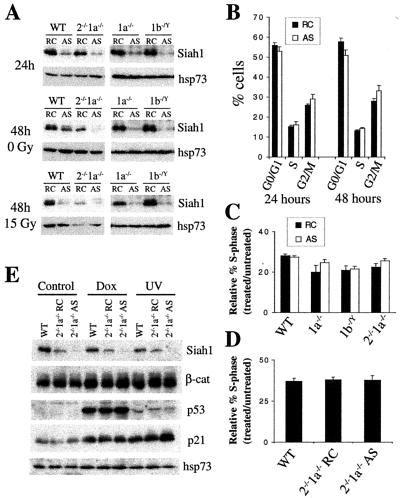
Since the Siah proteins are highly homologous to one another, and Siah1 and Siah2 can function equivalently to induce the degradation of some substrates (22), it is possible that Siah1b provides sufficient Siah activity in Siah1a−/− or Siah2−/− Siah1a−/− cells to allow normal p53 responses and mitosis. To analyze the effects of inhibition of the expression of different combinations of Siah proteins, MEFs were transfected with antisense or reverse control oligonucleotides directed against Siah1a or Siah1b 24 h prior to induction of cell cycle arrest by gamma irradiation or doxorubicin treatment. Siah1 protein levels were efficiently decreased 24 and 48 h after antisense transfection of wild-type, Siah1a−/−, Siah1b−/Y, or Siah2−/− Siah1a−/− MEFs, in both treated and untreated cultures (Fig. 7A and E). Densitometry revealed that, in comparison to wild-type cells, levels of Siah1 protein in antisense-treated Siah2−/− Siah1a−/− MEFs were reduced by 80 to 90%. Stabilization of p53 and induction of p21 protein expression in response to doxorubicin treatment or UV irradiation were unaffected by antisense treatment of Siah2−/− Siah1a−/− MEFs (Fig. 7E). Furthermore, the expression level of β-catenin, a putative target of degradation by Siah1 proteins, was not altered by the loss of Siah protein expression (Fig. 7E). Flow cytometry showed that antisense treatment did not alter cell cycle progression in unirradiated cells (Fig. 7B) or impair p53-mediated G1 cell cycle arrest in response to gamma irradiation (Fig. 7C) or doxorubicin (Fig. 7D). Thus, mitotic progression, p53-mediated induction of p21 expression, and p53-mediated cell cycle arrest occur normally in Siah2−/− Siah1a−/− cells in which total Siah1 protein levels have been markedly reduced.
FIG. 7.
Inhibition of Siah1a and Siah1b expression does not alter cell cycle progression or p53-mediated G1 arrest. Wild-type (WT), Siah1a−/− (1a−/−), or Siah2−/− Siah1a−/− (2−/−1a−/−) MEFs were transfected with antisense (AS) oligonucleotides directed against Siah1b or with reverse control (RC, reverse sequence of antisense) oligonucleotides. Siah1b−/Y (1b−/Y) MEFs were transfected with AS or RC oligonucleotides directed against Siah1a. Twenty-four hours after transfection, cells were treated with gamma irradiation (15 Gy), doxorubicin (1 μg/ml), or UV-C irradiation (50 J/m2). (A) Siah1 protein levels were assessed by Western blotting of total protein lysates prepared at the time of gamma irradiation (24 h) or 24 h after gamma irradiation (48 h). Probing with antibodies against hsp73 verified equal loading and transfer. (B) AS treatment of Siah2−/− Siah1a−/− MEFs does not alter cell cycle distribution. Cultures were analyzed by BrdU labeling (30 min) 48 h after transfection. Data represent means ± standard deviations (SD) of triplicate transfections. (C and D) G1 phase cell cycle arrest was assayed (as for Fig. 5B) by BrdU labeling (30 min) at the 48 h time point (24 h after gamma irradiation [C] or doxorubicin treatment [D]). Data represent means ± SD of triplicate oligonucleotide transfections. (E) Siah1, β-catenin, p53, and p21 protein levels were assessed by Western blotting of total protein lysates prepared 6 h after treatment with doxorubicin or UV irradiation. Probing with antibodies against hsp73 verified equal loading and transfer.
DISCUSSION
Many studies have proposed that Siah1 genes are induced by p53 activation and may function as negative regulators of cell cycle progression, mediators of apoptosis, or tumor suppressors. Evidence for these hypotheses derives almost exclusively from gain-of-function studies in which p53 or Siah1 genes were overexpressed in transformed cell lines. In this study we have tested these models using a loss-of-function approach. We find no evidence to support a general role for Siah genes in a range of p53-mediated responses.
Previous reports have shown that transient overexpression of p53 in several different cell lines (32, 35) or activation of ts-p53 in immortalized MEF and mouse myeloid leukemic cell lines (1, 20, 43, 44, 47) induces Siah1 gene expression. Additionally, stable transfection of U937 cells with p21 increased SIAH1 mRNA levels (31, 32, 35) and enforced expression of SIAH1 or p21 induced the expression of an overlapping set of genes, supporting a model whereby Siah1 proteins function downstream of p53 and p21 (44). However, it has yet to be shown that Siah1 genes are induced by stimuli that activate endogenous p53. In this study we demonstrate that Siah1a and Siah1b mRNA and protein levels are not increased by activation of endogenous p53 in mouse tissues or in primary MEFs or thymocytes. Furthermore, we show that activation of ts-p53, in two different cell lines that lack functional endogenous p53, does not increase expression of Siah1 genes. The discrepancy between previous reports and our findings may be reconciled by the fact that gene expression changes induced by p53 are dependent on the inducing stimulus, the level of p53 induction, and the cell type (56). It is noteworthy that in some, but not all, experimental settings, p53 overexpression induces cell cycle arrest in G2 (50). As Siah1 protein levels are high in G2 (Fig. 3B), previously observed induction of Siah1 by p53 overexpression may be a secondary consequence of altered cell cycle distribution. We cannot exclude that Siah1 genes may be targets of endogenous p53 in a restricted range of cell types or in response to restricted stimuli. It is significant, however, that Siah1 gene expression was not induced in diverse tissue and cell types (spleen, thymus, lung, kidney, gut, embryonic fibroblasts, thymocytes) under conditions where p53-mediated cell cycle arrest or apoptosis was induced by a wide range of p53-activating stimuli, including DNA damage (gamma irradiation, UV irradiation, etoposide, doxorubicin, cisplatin), oncogenic transformation (E1A 12S), disruption of microtubules (nocodazole), or depletion of ribonucleotide pools (PALA).
A number of observations have been used to support the hypothesis that Siah1 proteins may be effectors of p53-mediated responses. Siah1 proteins have been linked to negative regulation of cell cycle progression. SIAH1 transfection into 293 cells or GM701 immortalized fibroblasts induced growth arrest and blocked S-phase entry without inducing apoptosis (37). Furthermore, stimulation of human fibroblasts with serum induced marked repression of SIAH1 mRNA expression with kinetics similar to those of transcripts encoding proteins that inhibit the cell division cycle (24). Siah1 genes have also been linked to p53-induced apoptosis. Coexpression of Siah1a and Pw-1/Peg-3, a p53-inducible Siah1-binding protein, induced apoptosis in immortalized fibroblasts (43). SIAH1 overexpression also induced apoptosis in U937 cells (4, 44). Significantly, apoptosis induced by activation of ts-p53 in a myeloid leukemia cell line was reduced by expression of antisense Siah1a (47). Thus, overexpression of Siah1 genes can mimic the effects of p53 activation and induce cell cycle arrest and apoptosis.
In this study we have utilized a genetic approach to assess whether Siah genes participate in p53-mediated responses. We show that (i) p53-mediated contact inhibition and senescence in MEFs does not require Siah1a, Siah1b, or Siah2 alone, or Siah2 and Siah1a in combination; (ii) p53-mediated apoptosis of thymocytes occurs normally in the absence of Siah1a, Siah2, or both Siah2 and Siah1a; (iii) p53-mediated apoptosis of ES cells does not require Siah1b; (iv) p53-mediated apoptosis of E1A 12S-transformed MEFs does not require Siah1a, Siah1b or Siah2; and (v) p53-mediated G1 phase cell cycle arrest in MEFs is normal in the absence of Siah1a, Siah1b, or Siah2 or of both Siah2 and Siah1a. These data clearly demonstrate that the loss of Siah genes individually or the loss of two of the three highly homologous Siah family genes (Siah2 and Siah1a) does not impair p53 function in growth arrest and apoptosis. We also used antisense oligonucleotides to markedly decrease Siah1b expression in Siah1a−/− or Siah2−/− Siah1a−/− cells or to decrease Siah1a expression in Siah1b mutant cells. Despite attenuation of Siah1 protein levels by 80 to 90%, cell cycle progression, p53-mediated induction of p21 expression, and p53-mediated cell cycle arrest were unaffected. These findings, based on loss-of-function evidence, are inconsistent with existing hypotheses that Siah genes are necessary for mitotic progression or for p53 function.
SIAH1 has been reported to be the limiting factor of a novel E3 ubiquitin ligase complex that targets β-catenin for degradation (32, 36). It is hypothesized that p53 activation increases SIAH1 protein levels and thus induces the degradation of β-catenin. This process has been proposed to contribute to p53-mediated cell cycle arrest and tumor suppression. Our data does not support this model. Notably, we fail to observe induction of Siah1 protein levels following p53 activation in the gut, a tissue in which β-catenin functions as an oncogene during tumorigenesis (42), nor do we see defects in p53-mediated cell cycle arrest in cells lacking Siah genes. Moreover, total cellular β-catenin levels are not elevated by loss or inhibition of Siah2, Siah1a, and Siah1b protein expression. While other correlative evidence (39, 44) has been used to advance the argument that Siah genes function as tumor suppressors, Siah1a−/−, Siah2−/−, and Siah2−/− Siah1a−/− mice do not develop tumors or display tissue hyperplasia (11; also unpublished results). Previous studies have also failed to detect loss of SIAH1 expression or SIAH1 gene mutations in human cancers and cancer-derived cell lines (20, 38), suggesting that there is no selective pressure to lose SIAH1 gene expression or function during tumorigenesis.
In summary, we do not observe induction of Siah1 genes under conditions in which established p53 target genes and p53-mediated cell cycle arrest or apoptosis are clearly induced. Furthermore, the loss of Siah genes, singly or in combination, does not alter p53-mediated responses in primary MEFs, thymocytes, or ES cells. While we cannot exclude that Siah proteins may function to regulate p53 responses in cell types that were not examined in this study, such a requirement is clearly not a general feature of p53 signaling. We suggest that widely held models that place Siah1 proteins downstream of p53 in growth arrest, apoptosis, and tumor suppression should be reassessed. It is possible that they are based on nonspecific consequences of overexpression of Siah or p53 proteins.
Acknowledgments
We thank Fiona Christensen for blastocyst injections, Patrick Humbert for fetal liver reconstitution experiments, Nadia Traficante for assistance with generation of MEFs, Dobrila Nesic and Paul Orban for providing reagents, and Helena Richardson, Patrick Humbert, and members of the Bowtell lab for critical reading of the manuscript and helpful discussions.
This work was supported by grants from the NHMRC. I.J.F. is a Grimwade Scholar (University of Melbourne); A.R.C. and M.O. are a Fellow and a Scholar, respectively, of the Leukemia and Lymphoma Society.
REFERENCES
- 1.Amson, R. B., M. Nemani, J. P. Roperch, D. Israeli, L. Bougueleret, I. Le Gall, M. Medhioub, G. Linares-Cruz, F. Lethrosne, P. Pasturaud, L. Piouffre, S. Prieur, L. Susini, V. Alvaro, P. Millasseau, C. Guidicelli, H. Bui, C. Massart, L. Cazes, F. Dufour, H. Bruzzoni-Giovanelli, H. Owadi, C. Hennion, G. Charpak, A. Telerman, et al. 1996. Isolation of 10 differentially expressed cDNAs in p53-induced apoptosis: activation of the vertebrate homologue of the drosophila seven in absentia gene. Proc. Natl. Acad. Sci. USA 93:3953-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonio, C., I. Ferby, H. Wilhelm, M. Jones, E. Karsenti, A. R. Nebreda, and I. Vernos. 2000. Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102:425-435. [DOI] [PubMed] [Google Scholar]
- 3.Boehm, J., Y. He, A. Greiner, L. Staudt, and T. Wirth. 2001. Regulation of BOB.1/OBF.1 stability by SIAH. EMBO J. 20:4153-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruzzoni-Giovanelli, H., A. Faille, G. Linares-Cruz, M. Nemani, F. Le Deist, A. Germani, D. Chassoux, G. Millot, J. P. Roperch, R. Amson, A. Telerman, and F. Calvo. 1999. SIAH-1 inhibits cell growth by altering the mitotic process. Oncogene 18:7101-7109. [DOI] [PubMed] [Google Scholar]
- 5.Chao, C., S. Saito, J. Kang, C. W. Anderson, E. Appella, and Y. Xu. 2000. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 19:4967-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke, A. R., C. A. Purdie, D. J. Harrison, R. G. Morris, C. C. Bird, M. L. Hooper, and A. H. Wyllie. 1993. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362:849-852. [DOI] [PubMed] [Google Scholar]
- 7.Damalas, A., S. Kahan, M. Shtutman, A. Ben-Ze'ev, and M. Oren. 2001. Deregulated β-catenin induces a p53- and ARF-dependent growth arrest and cooperates with Ras in transformation. EMBO J. 20:4912-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Della, N. G., D. D. Bowtell, and F. Beck. 1995. Expression of Siah-2, a vertebrate homologue of Drosophila sina, in germ cells of the mouse ovary and testis. Cell Tissue Res. 279:411-419. [DOI] [PubMed] [Google Scholar]
- 9.Della, N. G., P. V. Senior, and D. D. Bowtell. 1993. Isolation and characterisation of murine homologues of the Drosophila seven in absentia gene (sina). Development 117:1333-1343. [DOI] [PubMed] [Google Scholar]
- 10.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675-684. [DOI] [PubMed] [Google Scholar]
- 11.Dickins, R. A., I. J. Frew, C. M. House, M. K. O'Bryan, A. J. Holloway, I. Haviv, N. Traficante, D. M. de Kretser, and D. D. Bowtell. 2002. The ubiquitin ligase component Siah1a is required for completion of meiosis I in male mice. Mol. Cell. Biol. 22:2294-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flores, E. R., K. Y. Tsai, D. Crowley, S. Sengupta, A. Yang, F. McKeon, and T. Jacks. 2002. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416:560-564. [DOI] [PubMed] [Google Scholar]
- 13.Funabiki, H., and A. W. Murray. 2000. The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102:411-424. [DOI] [PubMed] [Google Scholar]
- 14.Geley, S., B. L. Hartmann, R. Hattmannstorfer, M. Loffler, M. J. Ausserlechner, D. Bernhard, R. Sgonc, E. M. Strasser-Wozak, M. Ebner, B. Auer, and R. Kofler. 1997. p53-induced apoptosis in the human T-ALL cell line CCRF-CEM. Oncogene 15:2429-2437. [DOI] [PubMed] [Google Scholar]
- 15.Germani, A., H. Bruzzoni-Giovanelli, A. Fellous, S. Gisselbrecht, N. Varin-Blank, and F. Calvo. 2000. SIAH-1 interacts with alpha-tubulin and degrades the kinesin Kid by the proteasome pathway during mitosis. Oncogene 19:5997-6006. [DOI] [PubMed] [Google Scholar]
- 16.Harvey, M., A. T. Sands, R. S. Weiss, M. E. Hegi, R. W. Wiseman, P. Pantazis, B. C. Giovanella, M. A. Tainsky, A. Bradley, and L. A. Donehower. 1993. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene 8:2457-2467. [PubMed] [Google Scholar]
- 17.Heard, E., and P. Avner. 2000. Trans-Siberian X press report. International Symposium on X Chromosome Inactivation in Mammals, Institute of Cytology and Genetics, Novosibirsk, Russia 6-12 September 1999. Trends Genet. 16:64-65. [DOI] [PubMed] [Google Scholar]
- 18.Holloway, A. J., N. G. Della, C. F. Fletcher, D. A. Largespada, N. G. Copeland, N. A. Jenkins, and D. D. Bowtell. 1997. Chromosomal mapping of five highly conserved murine homologues of the Drosophila RING finger gene seven-in-absentia. Genomics 41:160-168. [DOI] [PubMed] [Google Scholar]
- 19.Horwitz, B. H., M. L. Scott, S. R. Cherry, R. T. Bronson, and D. Baltimore. 1997. Failure of lymphopoiesis after adoptive transfer of NF-κB-deficient fetal liver cells. Immunity 6:765-772. [DOI] [PubMed] [Google Scholar]
- 20.Hu, G., Y. L. Chung, T. Glover, V. Valentine, A. T. Look, and E. R. Fearon. 1997. Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics 46:103-111. [DOI] [PubMed] [Google Scholar]
- 21.Hu, G., and E. R. Fearon. 1999. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol. Cell. Biol. 19:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, G., S. Zhang, M. Vidal, J. L. Baer, T. Xu, and E. R. Fearon. 1997. Mammalian homologs of seven in absentia regulate DCC via the ubiquitin-proteasome pathway. Genes Dev. 11:2701-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Innes, K. M., S. J. Szilvassy, H. E. Davidson, L. Gibson, J. M. Adams, and S. Cory. 1999. Retroviral transduction of enriched hematopoietic stem cells allows lifelong Bcl-2 expression in multiple lineages but does not perturb hematopoiesis. Exp. Hematol. 27:75-87. [DOI] [PubMed] [Google Scholar]
- 24.Iyer, V. R., M. B. Eisen, D. T. Ross, G. Schuler, T. Moore, J. C. F. Lee, J. M. Trent, L. M. Staudt, J. Hudson, Jr., M. S. Boguski, D. Lashkari, D. Shalon, D. Botstein, and P. O. Brown. 1999. The transcriptional program in the response of human fibroblasts to serum. Science 283:83-87. [DOI] [PubMed] [Google Scholar]
- 25.Jimenez, G. S., M. Nister, J. M. Stommel, M. Beeche, E. A. Barcarse, X. Q. Zhang, S. O'Gorman, and G. M. Wahl. 2000. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat. Genet. 26:37-43. [DOI] [PubMed] [Google Scholar]
- 26.Johnsen, S. A., M. Subramaniam, D. G. Monroe, R. Janknecht, and T. C. Spelsberg. 2002. Modulation of TGFβ/Smad transcriptional responses through targeted degradation of the TGFβ inducible early gene-1 by the human seven in absentia homologue. J. Biol. Chem. 277:30754-30759. [DOI] [PubMed] [Google Scholar]
- 27.Johnson, P., S. Chung, and S. Benchimol. 1993. Growth suppression of Friend virus-transformed erythroleukemia cells by p53 protein is accompanied by hemoglobin production and is sensitive to erythropoietin. Mol. Cell. Biol. 13:1456-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kastan, M. B., O. Onyekwere, D. Sidransky, B. Vogelstein, and R. W. Craig. 1991. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51:6304-6311. [PubMed] [Google Scholar]
- 29.Koniaras, K., A. R. Cuddihy, H. Christopoulos, A. Hogg, and M. J. O'Connell. 2001. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene 20:7453-7463. [DOI] [PubMed] [Google Scholar]
- 30.Levesque, A. A., and D. A. Compton. 2001. The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154:1135-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linares-Cruz, G., H. Bruzzoni-Giovanelli, V. Alvaro, J. P. Roperch, M. Tuynder, D. Schoevaert, M. Nemani, S. Prieur, F. Lethrosne, L. Piouffre, V. Reclar, A. Faille, D. Chassoux, J. Dausset, R. B. Amson, F. Calvo, and A. Telerman. 1998. p21WAF-1 reorganizes the nucleus in tumor suppression. Proc. Natl. Acad. Sci. USA 95:1131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, J., J. Stevens, C. A. Rote, H. J. Yost, Y. Hu, K. L. Neufeld, R. L. White, and N. Matsunami. 2001. Siah-1 mediates a novel β-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol. Cell 7:927-936. [DOI] [PubMed] [Google Scholar]
- 33.Lowe, S. W., H. E. Ruley, T. Jacks, and D. E. Housman. 1993. p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74:957-967. [DOI] [PubMed] [Google Scholar]
- 34.Macleod, K. F., N. Sherry, G. Hannon, D. Beach, T. Tokino, K. Kinzler, B. Vogelstein, and T. Jacks. 1995. p53-dependent and independent expression of p21 during cell growth, differentiation, and DNA damage. Genes Dev. 9:935-944. [DOI] [PubMed] [Google Scholar]
- 35.Maeda, A., T. Yoshida, K. Kusuzaki, and T. Sakai. 2002. The characterization of the human Siah-1 promoter. FEBS Lett. 512:223-226. [DOI] [PubMed] [Google Scholar]
- 36.Matsuzawa, S., and J. C. Reed. 2001. Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 responses. Mol. Cell 7:915-926. [DOI] [PubMed] [Google Scholar]
- 37.Matsuzawa, S., S. Takayama, B. A. Froesch, J. M. Zapata, and J. C. Reed. 1998. p53-inducible human homologue of Drosophila seven in absentia (Siah) inhibits cell growth: suppression by BAG-1. EMBO J. 17:2736-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Medhioub, M., C. Vaury, R. Hamelin, and G. Thomas. 2000. Lack of somatic mutation in the coding sequence of SIAH1 in tumors hemizygous for this candidate tumor suppressor gene. Int. J. Cancer 87:794-797. [PubMed] [Google Scholar]
- 39.Nemani, M., G. Linares-Cruz, H. Bruzzoni-Giovanelli, J. P. Roperch, M. Tuynder, L. Bougueleret, D. Cherif, M. Medhioub, P. Pasturaud, V. Alvaro, H. der Sarkissan, L. Cazes, D. Le Paslier, I. Le Gall, D. Israeli, J. Dausset, F. Sigaux, I. Chumakov, M. Oren, F. Calvo, R. B. Amson, D. Cohen, and A. Telerman. 1996. Activation of the human homologue of the Drosophila sina gene in apoptosis and tumor suppression. Proc. Natl. Acad. Sci. USA 93:9039-9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto, K., and D. Beach. 1994. Cyclin G is a transcriptional target of the p53 tumor suppressor protein. EMBO J. 13:4816-4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orford, K., C. C. Orford, and S. W. Byers. 1999. Exogenous expression of β-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J. Cell Biol. 146:855-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polakis, P. 2001. More than one way to skin a catenin. Cell 105:563-566. [DOI] [PubMed] [Google Scholar]
- 43.Relaix, F., X. Wei, W. Li, J. Pan, Y. Lin, D. D. Bowtell, D. A. Sassoon, and X. Wu. 2000. Pw1/Peg3 is a potential cell death mediator and cooperates with Siah1a in p53-mediated apoptosis. Proc. Natl. Acad. Sci. USA 97:2105-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roperch, J. P., F. Lethrone, S. Prieur, L. Piouffre, D. Israeli, M. Tuynder, M. Nemani, P. Pasturaud, M. C. Gendron, J. Dausset, M. Oren, R. B. Amson, and A. Telerman. 1999. SIAH-1 promotes apoptosis and tumor suppression through a network involving the regulation of protein folding, unfolding, and trafficking: identification of common effectors with p53 and p21(Waf1). Proc. Natl. Acad. Sci. USA 96:8070-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuelson, A. V., and S. W. Lowe. 1997. Selective induction of p53 and chemosensitivity in RB-deficient cells by E1A mutants unable to bind the RB-related proteins. Proc. Natl. Acad. Sci. USA 94:12094-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sourisseau, T., C. Desbois, L. Debure, D. D. Bowtell, A. C. Cato, J. Schneikert, E. Moyse, and D. Michel. 2001. Alteration of the stability of Bag-1 protein in the control of olfactory neuronal apoptosis. J. Cell Sci. 114:1409-1416. [DOI] [PubMed] [Google Scholar]
- 47.Susini, L., B. J. Passer, N. Amzallag-Elbaz, T. Juven-Gershon, S. Prieur, N. Privat, M. Tuynder, M. C. Gendron, A. Israel, R. Amson, M. Oren, and A. Telerman. 2001. Siah-1 binds and regulates the function of Numb. Proc. Natl. Acad. Sci. USA 98:15067-15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szabo, P., and J. R. Mann. 1994. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development 120:1651-1660. [DOI] [PubMed] [Google Scholar]
- 49.Tanikawa, J., E. Ichikawa-Iwata, C. Kanei-Ishii, A. Nakai, S. Matsuzawa, J. C. Reed, and S. Ishii. 2000. p53 suppresses the c-Myb-induced activation of heat shock transcription factor 3. J. Biol. Chem. 275:15578-15585. [DOI] [PubMed] [Google Scholar]
- 50.Taylor, W. R., and G. R. Stark. 2001. Regulation of the G2/M transition by p53. Oncogene 20:1803-1815. [DOI] [PubMed] [Google Scholar]
- 51.Tiedt, R., B. A. Bartholdy, G. Matthias, J. W. Newell, and P. Matthias. 2001. The RING finger protein Siah-1 regulates the level of the transcriptional coactivator OBF-1. EMBO J. 20:4143-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todaro, G., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vousden, K. H. 2000. p53: death star. Cell 103:691-694. [DOI] [PubMed] [Google Scholar]
- 54.Wheeler, T. C., L. S. Chin, Y. Li, F. L. Roudabush, and L. Li. 2002. Regulation of synaptophysin degradation by mammalian homologues of seven in absentia. J. Biol. Chem. 277:10273-10282. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, J., M. G. Guenther, R. W. Carthew, and M. A. Lazar. 1998. Proteasomal regulation of nuclear receptor corepressor-mediated repression. Genes Dev. 12:1775-1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, R., K. Gish, M. Murphy, Y. Yin, D. Notterman, W. H. Hoffman, E. Tom, D. H. Mack, and A. J. Levine. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981-993. [PMC free article] [PubMed] [Google Scholar]