Abstract
Yra1p/REF participates in mRNA export by recruiting the export receptor Mex67p to messenger ribonucleoprotein (mRNP) complexes. Yra1p also binds Sub2p, a DEAD box ATPase/RNA helicase implicated in splicing and required for mRNA export. We identified genetic and physical interactions between Yra1p, Sub2p, and Hpr1p, a protein involved in transcription elongation whose deletion leads to poly(A)+ RNA accumulation in the nucleus. By chromatin immunoprecipitation (ChIP) experiments, we show that Hpr1p, Sub2p, and Yra1p become associated with active genes during transcription elongation and that Hpr1p is required for the efficient recruitment of Sub2p and Yra1p. The data indicate that transcription and export are functionally linked and that mRNA export defects may be due in part to inefficient loading of essential mRNA export factors on the growing mRNP. We also identified functional interactions between Yra1p and the exosome components Rrp45p and Rrp6p. We show that yra1, sub2, and Δhpr1 mutants all present defects in mRNA accumulation and that deletion of RRP6 in yra1 mutants restores normal mRNA levels. The data support the hypothesis that an exosome-dependent surveillance mechanism targets improperly assembled mRNPs for degradation.
mRNAs are exported from the nucleus as messenger ribonucleoprotein (mRNP) complexes which begin to be assembled during transcription. mRNA biogenesis requires multiple processing steps, including the addition of a 5′ cap, splicing, and 3′-end formation, which have to be completed before the mRNA can be exported. Fully mature mRNPs are subsequently recognized by the essential mRNA export receptor Mex67p (TAP in metazoans), which mediates the interaction between the mRNP and components of the nuclear pore complex (42, 51). The recruitment of Mex67p/TAP to the mRNP is facilitated by Yra1p (Aly in metazoans), an essential mRNA export factor which binds RNA and directly interacts with Mex67p (TAP) (47, 48, 52). Yra1p is a member of the evolutionary conserved REF family of RNA and export factor binding proteins, and multiple REF members exist in many organisms (48). In Saccharomyces cerevisiae, overexpression of the nonessential REF family member Yra2p can complement the otherwise lethal deletion of YRA1, indicating that these two REF proteins have redundant functions (47, 52).
Evidence has been accumulating that the different steps of gene expression, from transcription in the nucleus to translation and degradation in the cytoplasm, are intimately linked (37). Many mRNA processing factors are recruited to growing mRNPs through an interaction with the transcription machinery. For example, the yeast mRNA capping enzymes required for the addition of the m7G cap structure associate with the C-terminal domain (CTD) of RNA polymerase II at an early stage of transcription (23). Similarly, components of the splicing or polyadenylation machinery associate with the CTD, thereby positioning them to mediate efficient RNA processing. These interactions are required in vivo for maximum levels of splicing and polyadenylation, as well as for efficient transcription termination in vivo (2, 13, 17, 38).
In yeast, defects in 3′-end processing and/or polyadenylation prevent mRNA export and lead to the retention of transcripts at or close to their site of transcription, indicating a coupling between 3′-end formation and mRNA export in a step that involves the release of mRNA from nuclear foci (3, 20). Mutations in the nuclear exosome abolish the nuclear retention of incorrectly processed transcripts, suggesting that this exonuclease complex participates in a quality control mechanism that prevents the nuclear escape of improperly processed mRNAs and directs them to degradation (4, 16, 39).
A functional link between pre-mRNA splicing and mRNA export has also been established (34). In metazoan systems, splicing results in the deposition of the exon junction complex at a fixed position upstream of the splice junction, which marks the mRNA for subsequent steps, such as mRNA export or cytoplasmic turnover (21, 26, 27, 36). The exon junction complex contains the Yra1p homologue Aly/REF, as well as UAP56, a DECD box ATPase/RNA helicase implicated in an early step of spliceosome formation. UAP56 directly interacts with Aly/REF and was proposed to functionally link splicing to export by enhancing the recruitment of Aly/REF to spliced mRNAs, which would in turn facilitate the binding of the export receptor TAP (12, 14, 26, 32, 35, 54). Sub2p, the yeast functional homologue of UAP56, has also been implicated in splicing (22, 31, 53). Sub2p directly interacts with Yra1p, and both sub2 and yra1 mutants induce poly(A)+ RNA export defects (19, 45). Although splicing is an essential step in the formation of virtually all metazoan mRNAs, only 5% of yeast genes have introns, excluding splicing as a major mode of recruitment for Yra1p. Accordingly, Sub2p/UAP56 is required for the export of spliced mRNAs as well as of mRNAs derived from intronless genes. Sub2p/UAP56 may therefore play a general conserved role in recruiting Yra1p/Aly to mRNPs, irrespective of whether they contain an intron. In vitro experiments suggest that in a subsequent step, the binding of Mex67p to Yra1p induces the release of Sub2p from Yra1p prior to mRNA export (14, 19, 32, 35, 42, 45).
These observations raise the question of how Yra1p and Sub2p are directed to mRNAs derived from intronless genes. A recent study indicates that Yra1p/REF is associated with actively transcribed genes, suggesting that this essential mRNA export mediator is recruited to the mRNA during transcription (9, 28). A link between Sub2p and the transcription machinery was similarly suggested by the ability of high-copy-number SUB2 to suppress the deletion of HPR1, a gene implicated in transcription elongation (11). Genetic interactions have also been described between Sub2p and Rad3p, a bona fide RNA polymerase II component (19).
To further characterize the role of Yra1p in mRNA biogenesis and define when and how Yra1p may be recruited to the newly synthesized transcripts, we have searched for mutations synthetically lethal with a yra1 mutant. In this screen, we identified HPR1 and demonstrated that Hpr1p, Sub2p, and Yra1p physically interact. Chromatin immunoprecipitation (ChIP) experiments show that Hpr1p, Sub2p, and Yra1p are recruited cotranscriptionally at a similar time during elongation and that Hpr1p is required for efficient recruitment of Yra1p and Sub2p. Deletion or mutations in these components lead to diminished mRNA levels, suggesting that improper loading of mRNA export factors results in mRNP instability. These observations, together with those of genetic and physical interactions between Yra1p and the nuclear exosome, suggest that mRNPs undergo an exosome-dependent quality control step which competes with mRNP export.
MATERIALS AND METHODS
Plasmid and strain constructions.
Plasmids and strains used in this study are summarized in Tables 1 and 2. The yra1-8 temperature-sensitive allele was constructed by site-directed mutagenesis, which changed 2 amino acids within the conserved N-box (D10D11 to K10K11) of pGexCS-Yra1 (48), creating pGexCS-yra1-8 (pFS1966). The yra1-8 coding sequence was subsequently amplified by PCR and cloned as a SalI fragment into YCplac33-Gal-GFP (pFS1846). The resulting GFP-yra1-8 fusion was transferred as a XhoI PCR fragment into the YCpLac22HA-YRA1 +/−500 bp cassette, resulting in YCpLac22-HA-GFP-yra1-8 (pFS2557). To obtain YCpLac111-HA-GFP-yra1-8 (pFS2554), the BamHI GFP-yra1-8 +/−500 bp fragment from YCpLac22-HA-GFP-yra1-8 was transferred into YCpLac111. YCpLac111-HA-yra1-8 (pFS2328) was obtained by inserting a SalI PCR fragment of yra1-8 into Ycplac22HA-YRA1 +/−500 and subsequent subcloning of a BamHI HA-yra1-8 +/−500 bp fragment in YCpLac111.
TABLE 1.
Yeast vectors and plasmids used in this study
| Plasmid code | Plasmid name | Description | Reference and/or source |
|---|---|---|---|
| pFS2511 | pCH1122-YRA1 | YRA1 gene +/−500 bp cloned as a BamHI fragment into vector pCH1122 (URA3 ADE3 CEN) | This study |
| pFS2576 | Lac111-HPR1 | HPR1 gene +/−500 bp cloned as a BamHI fragment into vector YCpLac111 (LEU2 CEN) | This study |
| pFS2575 | Lac111-RRP45 | RRP45 gene +/−500 bp cloned as a BamHI fragment into YCpLac111 (LEU2 CEN) | This study |
| pFS2525 | Lac111-YRA1 Gen | YRA1 gene +/−500 bp cloned as a BamHI fragment into YCpLac111 (LEU2 CEN) | This study |
| pFS2327 | Lac111-HA-yra1 ΔRBD | HA-tagged YRA1 cDNA +/−500 bp, deleted for central RBD, subcloned as a BamHI fragment from pFS2268 into YCpLac111 (LEU2 CEN) | 52 |
| pFS2325 | Lac111-HA-yra1-77-227 | HA-yra1-77-227 +/−500 bpBamHI fragment from pFS2156 subcloned into YCpLac111 (LEU2 CEN) | This study |
| pFS2328 | Lac111-HA-yra1-8 | yra1-8 (LEU2 CEN); for details, see plasmid and strain constructions | This study |
| pFS2554 | Lac111-GFP-yra1-8 | GFP-yra1-8 (LEU2 CEN); for details, see plasmid and strain constructions | This study |
| pFS2574 | pFL36-SUB2 | SUB2 gene expressed from its own promoter (LEU2 CEN) | 19 |
| pFS2655 | Lac111-sub2-201 | sub2-201 (LEU2 CEN); for details, see plasmid and strain constructions | This study |
| pFS2656 | Lac111-sub2-206 | sub2-206 (LEU2 CEN); for details, see plasmid and strain constructions | This study |
| pFS2625 | pNOPProtA-SUB2 | SUB2 ORF cloned as a NcoI-BamHI fragment into pNOPPATA1L (LEU2 CEN) | This study, 15 |
| pFS2653 | pNOPProtA-sub2-201 | sub2-201 ORF cloned as a NcoI-BamHI fragment into pNOPPATA1L (LEU2 CEN) | This study |
| pFS2654 | pNOPProtA-sub2-206 | sub2-206 ORF cloned as a NcoI-BamHI fragment into pNOPPATA1L (LEU2 CEN) | This study |
| pFS2015 | pLGSD5 | Galactose-inducible β-galactosidase reporter construct (URA3, 2μm) | 25 |
| pFS2672 | GAL-YAT1 | Galactose-inducible YAT1 gene (URA3, 2μm) | 8 |
| pFS2642 | pGex-HPR1 | HPR1 ORF PCR fragment cloned as a BamHI fragment into pGex 4T-1 (Amersham Pharmacia) | This study |
| pFS1853 | pGex-YRA1 | YRA1 ORF cloned into pGexCS | 48 |
| pFS2113 | pGex-YRA2 | YRA2 ORF cloned into pGex4T-1 | 52 |
| pFS2633 | pET9d+-SUB2 | SUB2 ORF cloned as NcoI-BamHI PCR fragment into pET9d+ | This study |
| pFS2676 | pProEX HT-SUB2 | SUB2 ORF cloned as BamHI PCR fragment into pProEX HTb (Life Technologies) | This study |
TABLE 2.
Yeast strains used in this study
| Strain code | Strain name | Genotype | Reference(s) or source |
|---|---|---|---|
| CH1462 background | |||
| CH1462 | wild type | MATα ade2 ade3 his3 leu2 ura3 | 49, 50 |
| FSY1371 | SL screening strain | MATα ade2 ade3 leu2 ura3 his3 yra2::KANryra1ΔRBD 〈pCH1122-URA3-ADE3-YRA1, pFS2511〉 | This study |
| FSY1681 | sl57 | MATα ade2 ade3 leu2 ura3 his3 hpr1-57 yra2::KANryra1ΔRBD 〈pCH1122-URA3-ADE3-YRA1 Gen, pFS2511〉 | This study |
| FSY1682 | sl5 | MATα ade2 ade3 leu2 ura3 his3 rrp45-1 yra2::KANryra1ΔRBD 〈pCH1122-URA3-ADE3-YRA1 Gen, pFS2511〉 | This study |
| W303 background | |||
| W303 | wild type | mata ade2 his3 leu2 trp1 ura3 | |
| FSY1026 | YRA1 shuffle | mata ade2 his3 leu2 trp1 ura3 yra1::HIS3 〈pURA3-YRA1 Gen, pFS1876〉 | 48 |
| FSY1568 | GFP-yra1-8 | mata ade2 his3 leu2 trp1 ura3 yra1::HIS3 〈pLEU2-GFP-yra1-8, pFS2554〉 | This study |
| FSY1655 | YRA1 shuffle Δhpr1 | mata ade2 his3 leu2 trp1 ura3 hpr1::TRP1 yra1::HIS3 〈pURA3-YRA1 Gen, pFS1876〉 | This study |
| FSY1621 | YRA1 shuffle Δrrp6 | mata ade2 his3 leu2 trp1 ura3 rrp6::KANr yra1::HIS3 〈pURA3-YRA1 Gen, pFS1876〉 | This study |
| FSY1624 | Δhpr1 | mata ade2 his3 leu2 trp1 ura3 hpr1::TRP1 | This study |
| DLY23 | SUB2 shuffle | mata ade2 his3 leu2 trp1 ura3 sub2::HIS3 〈pURA3-SUB2, pFS2574〉 | 19 |
| FSY1613 | sub2-201 | mata ade2 his3 leu2 trp1 ura3 sub2::HIS3 〈pLEU2-sub2-201, pFS2655〉 | This study |
| FSY1664 | SUB2 shuffle Δhpr1 | mata ade2 his3 leu2 trp1 ura3 hpr1::TRP1 sub2::HIS3 〈pURA3-SUB2, pFS2574〉 | This study |
| FSY1473 | ProtA-Sub2 | mata ade2 his3 leu2 trp1 ura3 sub2::HIS3 〈pLEU2-ProtA-SUB2, pFS2625〉 | This study |
| FSY1615 | ProtA-sub2-201 | mata ade2 his3 leu2 trp1 ura3 sub2::HIS3 〈pLEU2-ProtA-sub2-201, pFS2653〉 | This study |
| FSY1616 | ProtA-sub2-206 | mata ade2 his3 leu2 trp1 ura3 sub2::HIS3 〈pLEU2-ProtA-sub2-206, pFS2654〉 | This study |
| FSY1656 | ProtA-Sub2 Δhpr1 | mata ade2 his3 leu2 trp1 ura3 hpr1::TRP1 sub2::HIS3 〈pLEU2-ProtA-SUB2; pFS2625〉 | This study |
| FSY1525 | Hpr1-ProtA | mata ade2 his3 leu2 trp1 ura3 sub2::HIS3 〈pURA3-SUB2〉 HPR1-ProtA-KANr | This study, 19 |
| FSY1476 | Rrp45-ProtA | mata ade2 his3 leu2 trp1 ura3 sub2::HIS3 〈pURA3-SUB2〉 RRP45-ProtA-KANr | This study, 19 |
| FSY1657 | Rrp6-ProtA | mata ade2 his3 leu2 trp1 ura3 sub2::HIS3 〈pURA3-SUB2〉 RRP6-TAP-tag-KANr | This study, 19 |
YCpLac111-sub2-201 and -206 (pFS2655 and 2656) were obtained by replacing the wild-type SUB2 coding region by the sub2-201 and -206 mutant sequences into YCplac111-SUB2 +/−500 (OFS2624).
Genomic tagging of HPR1, RRP45, and RRP6 was done as described previously (52). Deletions of HPR1 and RRP6 were obtained as described previously (33). ProtA-Sub2 wild-type and mutant strains were obtained by transforming plasmids pFS2625 (wild type), pFS2653 (sub2-201), and pFS2654 (sub2-206) into the SUB2 shuffle stain (DLY23) or SUB2 shuffle strain Δhpr1 (FSY1664), followed by plasmid shuffling on 5-fluoroorotic acid (5-FOA). ProtA-sub2-201 (FSY1615) and ProtA-sub2-206 (FSY1616) showed a temperature-sensitive growth phenotype similar to that of the nontagged strains (data not shown).
Synthetic lethal screen.
The Yra1 synthetic lethal screen was performed with the screening strain FSY1371 (yra1ΔRBD yra2::KANr) containing plasmid pCH1122-YRA1 (URA3 ADE3; pFS2511), encoding wild-type Yra1p. In brief, cells were mutagenized by UV irradiation to 10% survival, and surviving clones were analyzed for the absence of the red-white sectoring and the inability to grow on 5-FOA-containing medium, as described previously (49, 50). The screen was performed at 30, 34, or 37°C to increase the probability of selecting different synthetic lethal mutants. Mutants were transformed with a genomic library, and transformants were replicated on 5-FOA to select for complementing clones. Strains sl5 and sl57 were identified at 37 and 30°C, respectively.
Yeast extracts and affinity purifications of ProtA fusions.
ProtA-tagged strains were grown in yeast extract-peptone-dextrose (YEPD) medium at 25°C to an optical density at 600 nm (OD600) of 1.2. Cells were collected by centrifugation, washed once with 1× phosphate-buffered saline (PBS), resuspended in lysis buffer (20 mM HEPES-KOH [pH 7.0], 150 mM potassium acetate, 2 mM magnesium acetate, 10% glycerol, 0.1% Tween 20 5 mM β-mercaptoethanol, complete protease inhibitor mix [Roche]) at 80 OD600 of cells/ml, and lysed with glass beads. The soluble fraction was recovered after centrifugation and used for binding on immunoglobulin G (IgG)-Sepharose for 90 min at 4°C. After extensive washing, associated proteins were eluted from the beads by addition of 2 M NaCl-20 mM HEPES-KOH (pH 7.5) for 20 min at 4°C and precipitated with 20% trichloroacetic acid. Typically, 1 of 100 input and 1 of 6 eluted samples were examined by Western blotting.
Antibodies and Western blotting.
Western blotting was performed according to standard procedures. Anti-Yra1p and -Yra2p antibodies were obtained by injecting rabbits with purified glutathione S-transferase (GST) Yra1p and GST-Yra2p proteins. The obtained serum was used at dilutions of 1:3,000 for anti-Yra1p and 1:1,000 for anti-Yra2p in Western blotting. Anti-Hpr1p antibody (a gift from M. F. Christman) was used at 1:3,000. The protein signals were revealed with Super Signal West Pico substrate (Pierce).
Expression, purification of recombinant proteins, and in vitro bindings.
All recombinant proteins were expressed in Escherichia coli BL21, except for His6-tagged Sub2, which was expressed in E. coli DH5α. Cells were grown in Luria broth plus ampicillin (or ampicillin plus kanamycin for coexpressions) to an OD600 of 0.6 to 0.8 and induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C, except for GST-Hpr1, which was induced for 5.5 h at 28°C. Cells were collected by centrifugation and washed once with PBS, and the pellets were resuspended in universal buffer (20 mM HEPES-KOAc [pH 7.0], 100 mM KOAc, 2 mM MgOAc, 10% glycerol, complete protease inhibitor mix [Roche]) and lysed with a one-shot cell disrupter (Constant System Ltd.) set to 1.75 kPa. Triton X-100 was added to 0.2%, and lysates were incubated on a rotating wheel for 15 min at 4°C. Cell debris was removed by centrifugation, and total extracts were used for purification on either glutathione-Sepharose (Amersham Pharmacia) for GST fusions or Ni-nitrilotriacetic acid agarose (Qiagen) for His6-Sub2. Bindings were performed for 90 min at 4°C on a rotating wheel. Bound proteins were eluted by addition of sodium dodecyl sulfate (SDS) sample buffer, boiled, and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Coomassie staining.
For binding of His6-Sub2 to GST-Yra1 and -Yra2, an E. coli extract expressing His6-Sub2 was incubated with purified GST-Yra1 and -Yra2 immobilized on glutathione beads and washed extensively, and associated proteins were analyzed as described above.
RNA extractions, primer extensions, and Northern blotting.
RNA extractions and primer extensions were done as described previously (10). The following 5′ 32P-labeled primers were used: OFS506 (5′ CACCAGTGAGACGGGC 3′) for β-galactosidase, OFS728B (5′ GCGCTCGTCCTGCAGGGGTT 3′) for YAT1, OFS751(5′ GCTCGGGCACTTTTCGGCCAA 3′) for GAL1, and OFS645 (5′ GGACTTCTTGATCTCCTCTG 3′) for U1 snRNA. Extension products were fractionated on a 6% polyacrylamide-urea gel, autoradiographed, quantified with an Instant Imager apparatus (Packard), and normalized to U1 snRNA levels to correct for loading. Northern blotting was performed according to standard procedures. PMA1 mRNA levels were quantified with an Instant Imager apparatus and normalized to the 18S rRNA internal control.
In situ hybridizations.
In situ hybridizations for poly(A)+ RNA were done as described previously (44), except that a 3′ Cy3-labeled oligonucleotide dT55 probe was used. HSP104-specific in situ hybridization was performed as described previously (20).
ChIPs.
ChIPs were basically done as described previously (24). Cultures (250 ml) were grown at 25°C in YEPD medium to an OD600 of 1.2, formaldehyde was added to a final concentration of 1%, and incubation continued for 25 min. When heat shock was performed, cells were incubated at 37°C for 20 min before addition of formaldehyde and were left at 37°C during the cross-linking reaction. Cross-linking was subsequently quenched by the addition of glycine to a concentration of 240 mM. Cells were collected, washed once with 1× PBS, and resuspended in 3 ml of FA lysis buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, complete protease inhibitor mix [Roche]). Cells were lysed with glass beads, and SDS was added to 0.5%. Extracts were then sonicated to obtain chromatin fragments with an average size between 250 and 450 bp. Insoluble material was removed by centrifugation for 20 min at 12,000 × g in a tabletop centrifuge, and the soluble material was used as input material. For immunoprecipitation, 200 μl of input material was mixed with 800 μl of FA lysis buffer, 30 μl of protein A Sepharose (Amersham Pharmacia), and the appropriate antibodies and incubated overnight at 4°C on a rotating wheel. Six microliters of anti-Yra1p and anti-Yra2p, 5 μl of anti-Cbp80p (gift from Dirk Göhrlich), 5 μl of anti-TATA-binding protein (anti-TBP) (gift from M. Collart), and 9 μl of anti-CTD (Babco 8WG16) were used. For strains containing ProtA fusions, 30 μl of packed IgG-Sepharose (Amersham Pharmacia) was used. Beads were then washed (1 ml each time) once with FA lysis buffer, once with buffer FA500 (50 mM HEPES-KOH [pH 7.5], 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate), once with buffer III (10 mM Tris HCl [pH 8.0], 1 mM EDTA, 250 mM LiCl, 1% NP-40, 1% sodium deoxycholate), and once with 1× Tris-EDTA. Immunoprecipitated material was eluted from the beads by heating for 15 min at 65°C in 200 μl of elution buffer (50 mM Tris HCl [pH 7.5], 1% SDS, 10 mM EDTA). Cross-links were reversed by incubating the eluted fragments for 5 h at 65°C in the presence of 0.75 mg of proteinase K/ml, and DNA was subsequently purified on a QIAquick PCR purification column (Qiagen).
Real-time PCR.
Quantification of the immunoprecipitated DNA was done by quantitative real-time PCR using a Eurogentec qPCR core kit for SYBR Green I and an Applied Biosystems 7000 or 7700 sequence detection system. Immunoprecipitated DNA and different dilutions of sonicated and de-cross-linked input DNA were used for quantification and correction for different PCR efficiencies. PCRs were performed in a volume of 25 μl. Standard PCR conditions were used, i.e., 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. The following primer pairs were used: for the PMA1 promoter, OFS739 (5′ TCCTATCATTATCGTCTAACATCTAAT 3′) and OFS740 (5′ AAATTTATCAACGAGGTTGATAGAAAA 3′); for PMA1 5′, OFS705 (5′ TCAGCTCATCAGCCAACTCAAG 3′) and OFS706 (5′ CGTCGACACCGTGATTAGATTG 3′); for PMA1 middle, OFS741 (5′ TTGCCAGCTGTCGTTACCAC 3′) and OFS742 (5′ TCGACACCAGCCAAGGATTC 3′); for PMA1 3′, OFS708 (5′ TACTGTCGTCCGTGTCTGGATCT 3′) and OFS709 (5′ CCTTCATTGGCTTACCGTTCA 3′); for the PMA1 3′ untranslated region (3′ UTR), OFS759 (5′ TCTCTGGATGGTACTTTTTCTTTCTTG 3′) and OFS760 (5′ TGCGTGTTGTGAATTGTGCTC 3′); and for the intergenic region, OFS710 (5′ CGCATTACCAGACGGAGATGT 3′) and OFS711 (5′ CAAGCAAGCCTTGTGCATAAGA 3′). The intergenic region lies on the right arm of chromosome IV (nucleotides 1516119 to 1516229). Primers were used at a final concentration of 100 nM, except OFS739 and OFS740, which were used at 300 nM. Each PCR was run in duplicate to control for PCR validation, and all immunoprecipitations were repeated at least twice with different chromatin extracts. To express the severalfold increase in signal of the different PMA1 regions compared to that of the nontranscribed intergenic region, the absolute quantities obtained by the quantitative PCR were divided by the quantity of intergenic region immunoprecipitating with the different proteins.
RESULTS
Mutations in Yra1p are synthetically lethal with a component of the transcription machinery.
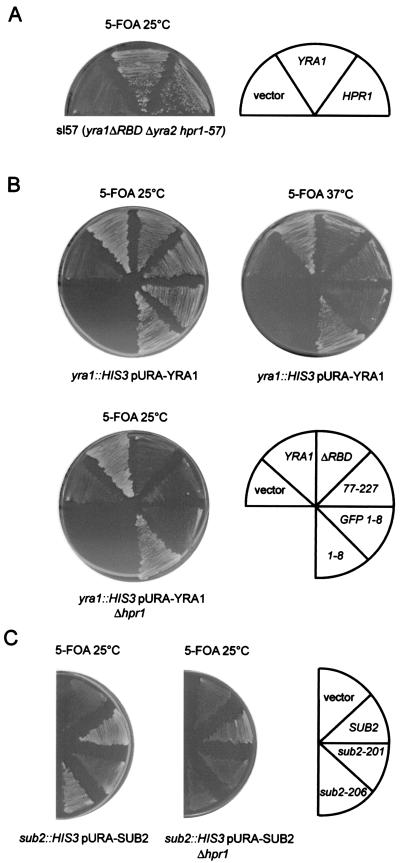
To further characterize Yra1p function, we performed a synthetic lethal screen with strain FSY1371 (yra1ΔRBD yra2::KANr, pCH1122-YRA1), in which the endogenous YRA1 gene was replaced by the mutant yra1ΔRBD gene encoding a protein lacking the central RNA binding domain (RBD). The deletion of the central RBD is viable and induces only a mild growth phenotype at 37°C. To increase the stringency of the screen, we also deleted the redundant, nonessential YRA2 gene (52).
Many synthetic mutants were identified, most of which were found only once, indicating that the screen was not saturated. The synthetic lethal phenotype of mutant sl57 was rescued by plasmids containing the HPR1 gene (Fig. 1A). HPR1 encodes a protein with a proposed role in transcription elongation (1). The HPR1 mutation in strain sl57 has not been determined, but the protein could not be detected by Western blotting with a polyclonal antibody against Hpr1p, indicating that at least a part of the protein is deleted (data not shown). Since HPR1 is not essential, we tested whether the complete disruption of HPR1 would be lethal in combination with different yra1 mutations. The YRA1 shuffle or YRA1 shuffle Δhpr1 strain was transformed with wild-type or mutant YRA1 constructs (LEU2 CEN) and tested for growth on 5-FOA (Fig. 1B). Three yra1 mutants were tested in addition to yra1ΔRBD. One was the deletion mutant yra1-77-227, which lacks the N-terminal domain of Yra1p but has only a mild growth phenotype (52). The second, yra1-8, contains two amino acid substitutions within the conserved N-terminal box (D10D11 to K10K11) and has no obvious growth phenotype on its own. However, this mutant becomes temperature sensitive at 37°C when GFP is fused to its N terminus (GFP-yra1-8). The growth phenotypes of all these yra1 mutants were substantially enhanced in the absence of Hpr1p, which is consistent with a synthetic interaction between yra1 mutants and Δhpr1.
FIG. 1.
(A) yra1ΔRBD is synthetically lethal with a mutation in HPR1. Strain sl57 (yra1ΔRBD hpr1-57 〈pCH1122 YRA1 URA ADE3〉) was transformed with empty vector or YRA1 and HPR1 gene constructs (pFS2525 and pFS2576). Transformants were streaked on 5-FOA plates to select against wild-type pCH1122-YRA1 and examine rescue of the synthetic phenotype. (B) Various yra1 mutants are synthetically lethal with Δhpr1. The YRA1 shuffle (FSY1026) or YRA1 shuffle Δhpr1 (FSY1655) strains were transformed with vector or plasmids encoding wild-type or mutant Yra1p (LEU2 CEN). Transformants were streaked on 5-FOA plates and incubated at 25 or 37°C for 4 days. (C) sub2 mutants are synthetically lethal with Δhpr1. The SUB2 shuffle (DLY23) or the SUB2 shuffle Δhpr1 (FSY1664) strains were transformed with empty vector or plasmids encoding wild-type or mutant Sub2p (LEU2 CEN). Transformants were analyzed as described for panel B.
Since Sub2p directly interacts with Yra1p and acts as a high-copy-number suppressor of Δhpr1, synthetic lethality between sub2 mutants and Δhpr1 was also investigated. The growth of two described temperature-sensitive mutants, sub2-201 and sub2-206 (31), was examined in the presence or absence of Hpr1p (Fig. 1C). The phenotype of both sub2 mutants was enhanced at 25°C in the absence of Hpr1p, indicating a synthetic lethal relationship between sub2 mutants and Δhpr1.
Physical interactions between Hpr1p, Sub2p, and Yra1p and Yra2p.
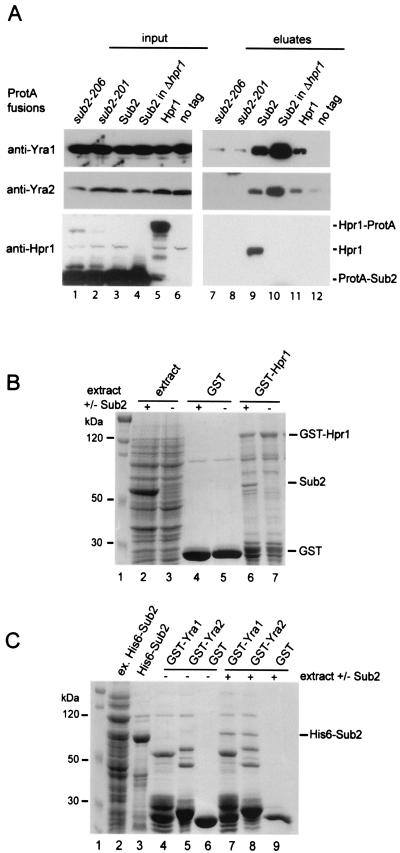
To further characterize the functional relationship between Yra1p, Sub2p, and Hpr1p, we looked for physical interactions among these proteins (Fig. 2A). Strains were constructed in which the endogenous SUB2 gene was deleted and replaced by plasmids expressing N-terminally ProtA-tagged wild-type (ProtA-Sub2p) or mutant (ProtA-sub2-201 and ProtA-sub2-206) Sub2p proteins. A strain expressing ProtA-Sub2 in a Δhpr1 background was also engineered (ProtA-Sub2p, Δhpr1). Finally, a strain expressing Hpr1-ProtA was obtained by genomically tagging the 3′ end of HPR1 with ProtA (Hpr1-ProtA). Total extracts were prepared from the five ProtA-tagged strains and affinity purified on IgG beads. Copurifying proteins eluted with high levels of salt were analyzed by Western blotting for the presence of Yra1p, Yra2p, and Hpr1p. Hpr1p very efficiently copurified with ProtA-Sub2, and this interaction was insensitive to an RNase A treatment (Fig. 2A, lane 9, and data not shown). ProtA-Sub2 also selected Yra1p, which is consistent with the direct interaction described between these two proteins (45). Similarly, Yra2p purified with ProtA-Sub2, confirming the Yra1p and Yra2p functional redundancy. Yra1p and Yra2p were also selected by Hpr1-ProtA (Fig. 2A, lane 11), suggesting that Hpr1p, Sub2p, and Yra1p/Yra2p may be part of a common complex. Mutations in Sub2p (sub2-201 and sub2-206) strongly decreased its ability to interact with Hpr1p and Yra1p and Yra2p (Fig. 2A, lanes 7 and 8). The association of Sub2p with Yra1p and Yra2p was not affected by the deletion of HPR1, and even larger amounts of Yra1p and Yra2p were associated with ProtA-Sub2 in the absence of Hpr1p (Fig. 2A, compare lanes 9 and 10). This observation may indicate a regulatory role for Hpr1p in the interaction of Yra1p with Sub2p.
FIG. 2.
(A) Hpr1p, Sub2p, and Yra1p physically interact in vivo. Total extracts were prepared from the indicated ProtA-tagged strains and purified on IgG beads. Copurifying proteins were eluted with high salt levels. Total extracts (input; lanes 1 to 6) or eluted proteins (eluates; lanes 7 to 12) were examined by Western blotting with antibodies against Yra1p, Yra2p, or Hpr1p. A nontagged (no tag) strain was used as a control for nonspecific binding to IgG beads. The antibodies against Hpr1p also recognize the ProtA-Sub2 and Hpr1-ProtA fusions present in input samples. (B) Sub2p interacts with GST-Hpr1p. GST alone or GST-Hpr1p (pFS2642) was coexpressed in E. coli with plasmid pFS2633 encoding nontagged Sub2p (lanes 4 and 6) or an empty vector (lanes 5 and 7). Total extracts were purified on glutathione beads, and the selected material was fractionated by SDS-PAGE (lanes 4 to 7) and Coomassie stained. A size marker (lane 1) and aliquots from total lysates expressing GST-Hpr1p plus Sub2p (lane 2) or GST-Hpr1p alone (lane 3) were run in parallel. (C) Sub2p interacts with Yra1p and Yra2p in E. coli. GST-Yra1p (pFS1853), GST-Yra2p (pFS2113), or GST alone was immobilized on beads and incubated with extracts from E. coli transformed with vector (lanes 4 to 6) or plasmid pFS2676 expressing a His6-Sub2p-tagged protein (lanes 7 to 9). Material associated with the beads (lanes 4 to 9) and aliquots of His6-Sub2p extract (lane 2) or His6-Sub2p purified on Ni-nitrilotriacetic acid beads (lane 3) were fractionated by SDS-PAGE followed by Coomassie staining.
In vitro interactions between Hpr1p, Sub2p, and Yra1p.
To further characterize the physical interactions identified in vivo, Hpr1p, Sub2p, and Yra1p were expressed as recombinant proteins in E. coli and examined for direct interaction in vitro (Fig. 2B). First, plasmids encoding GST-Hpr1 or GST alone were coexpressed in E. coli with a construct encoding nontagged Sub2p. Total extracts from E. coli strains expressing the GST fusions in the presence or absence of Sub2p were purified on glutathione beads. Affinity-purified proteins were fractionated on SDS-PAGE gels and Coomassie stained (Fig. 2B). Sub2p efficiently copurified with GST-Hpr1p but not with GST alone (Fig. 2B, compare lanes 4 and 6), indicating a direct interaction between Hpr1p and Sub2p. Next we examined the ability of GST-Yra1p and GST-Yra2p immobilized on beads to select a His6-Sub2-tagged protein from an E. coli extract (Fig. 2C). His6-Sub2 specifically copurified with Yra1- and Yra2-GST fusions but not GST alone (Fig. 2C, lanes 7 to 9). These observations show that Yra2p, like Yra1p (45), directly interacts with Sub2p.
A direct interaction between Hpr1p and Yra1p recombinant proteins could not be examined because of tag incompatibilities. In vitro-translated Yra1p or Yra2p showed no specific binding to GST-Hpr1p immobilized on beads (data not shown), suggesting that Yra1p and Yra2p do not directly interact with Hpr1p. For these reasons, it is likely that the interaction between Hpr1p and Yra1p detected in vivo (Fig. 2A) is bridged by Sub2p.
Hpr1p, Sub2p, and Yra1p are recruited during transcription elongation.
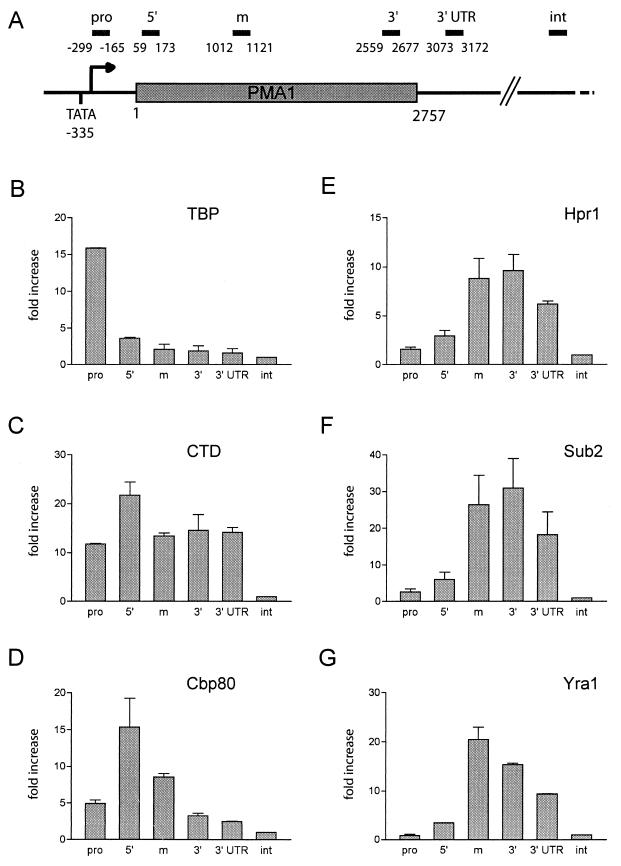
Hpr1p has a proposed role in transcription elongation and is therefore expected to associate with transcriptionally active genes. Earlier ChIP experiments have shown that the recruitment of Yra1p to the mRNP occurs during transcription (28). These observations, together with those of the genetic and physical interactions between Hpr1p, Sub2p, and Yra1p, strongly suggested that Hpr1p could aid the cotranscriptional recruitment of Sub2p and Yra1p to growing mRNP complexes. We have used a similar ChIP approach to analyze the association of Hpr1p, Sub2p, and Yra1p with an actively transcribed gene and to define whether the recruitment of Sub2p and Yra1p depends on Hpr1p. In these experiments, we examined the association of these proteins with different regions of the constitutively expressed 3-kb-long PMA1 gene. PMA1 was chosen because analysis of this gene would allow for comparison with earlier published studies (23, 28).
To validate our experimental conditions, we first analyzed three proteins whose distribution on the PMA1 gene is known or can be anticipated. These included the TBP, RNA polymerase II, and the cap binding protein Cbp80p. Cross-linked and sonicated extracts prepared from wild-type cells grown at 25°C were immunoprecipitated with antibodies specific for TBP, the CTD of RNA polymerase II, or Cbp80p. Coprecipitating DNA segments were amplified with a quantitative real-time PCR assay using primer pairs specific for the promoter (pro) and the 5′, middle, and 3′ coding regions, as well as the 3′ UTR, of the PMA1 gene (Fig. 3A). The precipitated DNA samples were also amplified with primers specific for an intergenic, nontranscribed DNA segment, which is devoid of an open reading frame (ORF) and which served as an internal control for the background. The relative amounts of each fragment in the coprecipitated DNA were defined as described above (see Materials and Methods) and were expressed as a severalfold increase with respect to the intergenic control region, whose value was arbitrarily set to 1.
FIG. 3.
Hpr1p, Sub2p, and Yra1p associate with the PMA1 gene during transcription elongation. (A) Diagram of the constitutively expressed PMA1 gene. Primer sets were designed to amplify 100- to 130-bp-long fragments corresponding to the PMA1 promoter (pro), to the 5′, middle (m), and 3′ coding regions, or to the 3′ UTR. An additional primer pair specific for a nontranscribed intergenic region (int) devoid of an ORF served as the internal background control. As shown in panels B, C, D, and G, a cross-linked and sonicated extract from wild-type cells grown at 25°C was immunoprecipitated with antibodies specific for TBP, the CTD of RNA polymerase II, the 80-kDa cap binding protein (Cbp80), and Yra1p, respectively. Coprecipitating DNA was amplified by quantitative real-time PCR with the primers indicated in panel A, as described in Materials and Methods. The relative abundance of the different PMA1 gene segments in each immunoprecipitate was expressed as a severalfold increase with respect to the nontranscribed intergenic region value, arbitrarily set to 1. The indicated values correspond to the means of at least two independent experiments. (E and F) Cross-linked and sonicated extracts were prepared from strains expressing Hpr1-ProtA (FSY1525) or ProtA-Sub2p (FSY1473) and affinity purified on IgG beads. Copurifying DNA fragments were quantified as described above.
As shown earlier, TBP was uniquely detected at the PMA1 promoter (23, 28). The absence of TBP within the coding region indicates that the conditions used allow good resolution between the PMA1 gene segments (Fig. 3B). Except for a slight increase at the 5′ end of the gene, RNA polymerase II (CTD) was evenly distributed over the PMA1 promoter, coding regions, and 3′ UTR (Fig. 3C). The slight increase at the 5′ end may correspond to stalling of the polymerase at the transition from transcription initiation to elongation (23, 41). Cbp80p, in complex with Cbp20p, binds the m7GpppN cap structure added to mRNA 5′ ends at an early time point during transcription (23, 29). Consistently, we found Cbp80p mainly associated with the 5′ coding region of the PMA1 gene (Fig. 3D), which indicates cotranscriptional recruitment of Cbp80p to the mRNA at a very early stage of transcription, perhaps through an initial interaction with the transcription complex. The weaker cross-linking of Cbp80p to more downstream PMA1 regions may reflect the increasing distance between the mRNA 5′ end and the DNA template.
Next, we examined the association of Yra1p, Sub2p, and Hpr1p with the PMA1 gene. The distribution of Yra1p was analyzed in wild-type extracts with a polyclonal antibody specific for Yra1p, whereas the association of Hpr1p or Sub2p was determined in strains expressing these two proteins as Hpr1-ProtA or ProtA-Sub2 fusions (Fig. 3E, F, and G). Hpr1p, Sub2p, and Yra1p showed very similar distribution patterns over the PMA1 gene. None of these proteins was detectable at the promoter. The association of the three proteins with the 5′ coding region was very weak but clearly strengthened in the middle and the 3′ end of the gene. The association pattern of Yra2p, as defined by immunoprecipitation with a polyclonal antibody against Yra2p, was comparable to that of Yra1p (data not shown). These profiles are consistent with a parallel recruitment of Hpr1p, Sub2p, and Yra1p and Yra2p to the PMA1 gene during transcription elongation. Cross-linking of both Hpr1p and Sub2p started to decrease in the PMA1 3′ UTR, whereas the interaction of Yra1p and Yra2p with the PMA1 gene weakened within the 3′ coding region earlier than that of Sub2p and Hpr1p. The reduced binding at the 3′ end of the gene may reflect mRNP remodeling or changes in the interactions of these proteins with the transcription machinery.
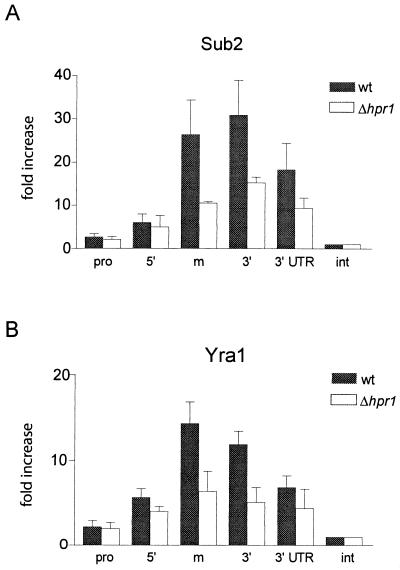
Hpr1 is required for efficient association of Sub2p and Yra1p to the transcribing PMA1 gene.
To define whether Hpr1p influences the cotranscriptional recruitment of Sub2p and Yra1p to the PMA1 gene, we compared the association of these two proteins with PMA1 in the presence or absence of Hpr1p (Fig. 4). The binding of ProtA-Sub2p to PMA1 was substantially decreased in a Δhpr1 strain background, indicating that Hpr1p is important for efficient recruitment of Sub2p to the growing mRNP (Fig. 4A). The association of Yra1p with PMA1 in the absence of Hpr1p was lowered to a comparable extent (Fig. 4B). Deletion of HPR1 did not affect the levels of ProtA-Sub2p or Yra1p in the extracts, nor did it modify the level of association of TBP with the PMA1 gene (data not shown). Furthermore, the decreased association of Sub2p with the PMA1 gene in Δhpr1 cells grown at 25°C (Fig. 4A) cannot be due to a lower transcription rate, since the PMA1 transcript levels are comparable to those of the wild type in these cells (see below, Fig. 7A). All these observations support the hypothesis that the decreased cross-linking of Yra1p and Sub2p to PMA1 represents a direct effect of HPR1 deletion.
FIG. 4.
The efficient recruitment of Sub2p and Yra1p to the transcribing PMA1 gene requires Hpr1p. (A) Strains expressing ProtA-Sub2p in the presence (FSY1473) or absence (FSY1656) of Hpr1p were grown at 25°C and analyzed as described for Fig. 3F. (B) W303 wild-type and Δhpr1 (FSY1624) strains were shifted to 37°C for 20 min prior to cross-linking. Sonicated extracts were immunoprecipitated with antibodies against Yra1p, and coprecipitating DNA fragments were analyzed as described for Fig. 3G.
FIG. 7.
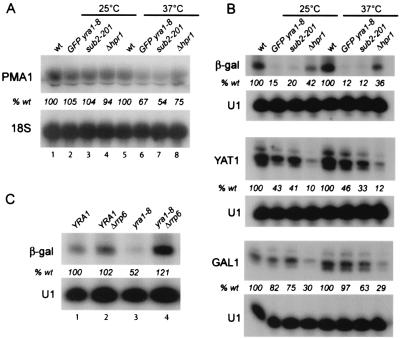
yra1, sub2, and Δhpr1 mutants induce defects in mRNA accumulation. (A) Northern blot analysis of PMA1 mRNAs in wild-type or GFP-yra1-8, sub2-201, and Δhpr1 mutant strains grown at 25°C (lanes 1 to 4) or shifted to 37°C for 45 min (lanes 5 to 8). The PMA1 signals were normalized to 18S rRNA and expressed as a percentage of that of the wild type, as indicated (% wt). (B) Wild-type or GFP-yra1-8 (FSY1568), sub2-201 (FSY1613), and Δhpr1 (FSY1624) mutant strains, transformed with plasmids (2μm URA3) containing the β-galactosidase or YAT1 genes driven by a galactose-inducible promoter, were grown to mid-log phase in selective medium containing 2% lactate-2% glycerol-0.05% glucose and induced with 3% galactose-1% raffinose for 150 min at 25 or 37°C, as shown above the lanes. Total RNA was analyzed by primer extension with 32P-labeled oligonucleotides specific for β-galactosidase, YAT1, or the endogenous GAL1 gene transcripts, as indicated. A primer specific for U1 snRNA was added to the reactions as an internal control for loading. The primer-extended bands were quantified with an Instant Imager apparatus and normalized to U1 snRNA. The transcript levels in mutant strains were expressed as a percentage of that of the wild type, as indicated at the bottom of each gel (% wt). (C) The YRA1 (FSY1026) or YRA1 Δrrp6 (FSY1621) strain was shuffled, as described for Fig. 6B, with plasmid Lac111-YRA1 Gen (pFS2525; lanes 1 and 2) or Lac111-yra1-8 (pFS2328; lanes 3 and 4) and transformed with the galactose-inducible β-galactosidase reporter construct (pLGSD5). Transformants were grown, and RNAs were analyzed as for panel B, except that the galactose induction was performed at 30°C.
The temperature-sensitive GFP-yra1-8 mutant protein is badly recruited to the PMA1 gene and leads to the accumulation of newly synthesized HSP104 transcripts in nuclear foci.
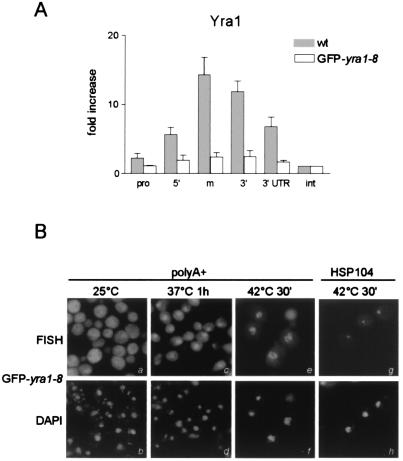
To further investigate the importance of the cotranscriptional recruitment of Yra1p for mRNA export, we next examined the association of the temperature-sensitive GFP-yra1-8 mutant protein with the PMA1 gene after a short heat shock treatment at 37°C (Fig. 5A). Extremely low levels of PMA1 gene fragments were pulled down by anti-Yra1p antibodies, although the mutant protein is recognized by these antibodies in standard immunoprecipitation experiments and is present in amounts similar to those of the wild type (data not shown). These observations show that the cotranscriptional recruitment of this mutant Yra1p protein is severely compromised.
FIG. 5.
GFP-yra1-8 is not recruited to the PMA1 gene, exhibits a poly(A)+ RNA export defect, and sequesters HSP104 transcripts within nuclear foci. (A) W303 wild-type and GFP-yra1-8 strains were shifted to 37°C for 20 min prior to cross-linking. Sonicated extracts were immunoprecipitated with antibodies against Yra1p, and coprecipitating DNA fragments were analyzed as described for Fig. 3G. (B) The GFP-yra1-8 mutant strain was grown at 25°C and shifted to 37°C for 1 h or to 42°C for 30 min. After fixation, the distributions of poly(A)+ RNA or HSP104 mRNAs were examined by in situ hybridization with Cy3-labeled oligo(dT)55 or HSP104-specific oligonucleotide probes, as indicated. DAPI (4′,6′-diamidino-2-phenylindole) staining was used to verify the nuclear localization of the HSP104 and poly(A)+ RNA signals.
Various mRNA 3′ processing and polyadenylation and export mutants induce nuclear retention of bulk poly(A)+ RNA and sequester newly synthesized transcripts in nuclear foci at or close to the site of transcription (20). To determine whether the GFP-yra1-8 mutant strain induces these phenotypes, we examined the distribution of poly(A)+ RNA or HSP104 mRNA in these cells grown at 25°C or shifted to 37°C or 42°C (Fig. 5B). GFP-yra1-8 mutant cells exhibit a poly(A)+ RNA export defect at 37°C, and the nuclear poly(A)+ signal becomes even stronger at 42°C, probably reflecting the strong production of heat shock mRNAs and an even more severe export block. As described for sub2-201 and other mRNA export mutants, GFP-yra1-8 cells retain HSP104 mRNAs induced at 42°C in a nuclear dot. Taken together, these data suggest that the mRNA export defects may be due, at least in part, to poor cotranscriptional recruitment of the mutant Yra1p protein, probably resulting in reduced binding of Mex67p to the mRNP.
Yra1p is synthetically lethal with components of the exosome.
Our synthetic lethal screen identified different genes implicated in mRNA metabolism (data not shown). The synthetic lethal clone of strain sl5 was complemented by RRP45, an essential component of both the cytoplasmic and the nuclear exosome. The exosome is a multiprotein complex of 3′-to-5′ exonucleases involved in the processing and degradation of various cellular transcripts (4). Earlier studies have shown that the phenotype of sub2 mutant strains is enhanced in the absence of Rrp6p, an exonuclease specific for the nuclear exosome (4, 19). To define whether yra1 mutants exhibit a similar synthetic behavior, the YRA1 shuffle or YRA1 shuffle Δrrp6 strains were transformed with wild-type or mutant YRA1 plasmids, as described for Fig. 1B, and tested for growth on 5-FOA (Fig. 6A). The absence of Rrp6p exacerbated the growth phenotypes of all yra1 mutants examined, indicating a strong functional relationship between yra1 mutants and Δrrp6.
FIG. 6.
Yrap1 genetically and physically interacts with components of the exosome. (A) yra1ΔRBD is synthetically lethal with a mutation in RRP45. Strain sl5 (yra1ΔRBD rrp45-1 〈pCH1122-YRA1 URA ADE3〉) was transformed with empty vector or YRA1 and RRP45 gene constructs (pFS2525 and pFS2575). Transformants were streaked on 5-FOA as described for Fig. 1A. (B) Mutations in YRA1 are synthetically lethal with Δrrp6. The YRA1 shuffle (FSY1026) or YRA1 shuffle Δrrp6 (FSY1621) strains were transformed with vector or plasmids encoding wild-type or mutant Yra1p (LEU2 CEN). Transformants were streaked on 5-FOA plates and analyzed as described for panel A. (C) Yra1p physically interacts with components of the exosome. Total extracts were prepared from a nontagged strain (no tag) or strains expressing Rrp45-ProtA (FSY1476) or Rrp6-ProtA (FSY1657) and purified on IgG beads (lanes 1 to 6). Copurifying proteins were eluted with high levels of salt. Total extracts (input; lanes 1 to 3) or eluted proteins (eluates; lanes 4 to 6) were examined by Western blotting with antibodies against Yra1p and Hpr1p.
Yra1p physically interacts with the exosome component Rrp45p.
To further analyze the relationship between Yra1p and the exosome, we examined their physical interaction in vivo (Fig. 6B). Extracts were prepared from strains in which RRP45 or RRP6 had been genomically tagged with protein A and purified as described for Fig. 2A. Copurifying proteins were analyzed by Western blotting for the presence of Yra1p and Hpr1p. Substantial amounts of Yra1p copurified with Rrp45, but we were unable to detect a physical connection between Yra1p and Rrp6p, perhaps because of a lack of sensitivity. Hpr1p was detectable in neither the Rrp45-ProtA nor the Rrp6-ProtA eluate. These observations indicate that at least one component of the exosome is stably associated with Yra1p or the mRNP.
Hpr1, Sub2p, and Yra1p mutants exhibit defects in transcript accumulation.
Because the deletion of HPR1 affects the expression levels of β-galactosidase and YAT1 or LYS2 transcripts, it was proposed that Hpr1p is preferentially required for transcription through long or G+C-rich DNA sequences (8). Based on the genetic and physical interactions between Hpr1p, Sub2p, and Yra1p, we asked whether mutations in Sub2p and Yra1p would have effects on transcript accumulation similar to those observed in Δhpr1 strains (Fig. 7). First we examined the steady-state levels of the endogenous PMA1 transcripts. The wild-type, GFP-yra1-8, sub2-201, or Δhpr1 strain was grown to mid-log phase in YEPD medium at 25°C and either kept at 25°C or shifted to 37°C for 45 min. Total RNAs were extracted and analyzed by Northern blotting with probes specific for PMA1 or 18S rRNA as an internal control for loading (Fig. 7A). PMA1 transcript levels were comparable in all four strains grown at 25°C but were substantially reduced in the three mutant strains after a shift to 37°C. Next, the levels of β-galactosidase or YAT1 transcripts were examined in a wild-type (W303) strain or in mutant GFP-yra1-8, sub2-201, and Δhpr1 strains transformed with high-copy-number plasmids expressing these genes from a galactose-inducible promoter (8, 25). The strains were grown to mid-log phase at 25°C in selective medium and induced for 150 min with galactose at 25°C or 37°C. Total RNA was extracted and subjected to primer extension analysis with primers specific for β-galactosidase and YAT1 transcripts (Fig. 7B). Endogenous constitutively expressed YAT1 mRNAs were not detectable in this assay (data not shown). As previously reported, the levels of galactose-induced β-galactosidase and YAT1 mRNAs were clearly lower in the absence of Hpr1p compared to those for the wild type. The accumulation of these transcripts was also substantially affected in the GFP-yra1-8 and sub2-201 mutant strains. Because the levels of endogenous induced GAL1 transcripts were moderately affected in the absence of Hpr1p, as described previously (8), and appeared mildly altered in GFP-yra1-8 and sub2-201, we conclude that the decrease of β-galactosidase and YAT1 mRNA levels in these mutant strains is not due to a loss in galactose inducibility. These observations, taken together, indicate that the functionally related Hpr1p, Sub2p, and Yra1p proteins are required for the efficient accumulation of certain transcripts. These experiments did not determine, however, whether mutations in these proteins affect mRNA production, mRNA stability, or both.
The genetic and physical interactions between Yra1p and the nuclear exosome indicate that they participate in a common process. This raises the question of whether the mRNA accumulation defect in the GFP-yra1-8 mutant strain is linked to an exosome-dependent RNA degradation mechanism which recognizes and degrades defective mRNPs resulting from inefficient loading of export factors. We therefore tested whether the low mRNA levels detected in the yra1 mutant strain could be restored by deleting RRP6. The GFP-yra1-8 mutant strain could not be used in this experiment, because it is unviable in the absence of RRP6. We therefore used the milder yra1-8 mutant, which grows when RRP6 is deleted (Fig. 6B). The yra1-8 mutant grows nearly as well as the wild type and shows only weak nuclear accumulation of poly(A)+ RNA at 37°C, and retention of HSP104 transcripts at 42°C in nuclear foci could not be detected (data not shown). However, as for GFP-yra1-8, the β-galactosidase mRNA levels are significantly decreased in yra1-8 and accumulate only to 50% of the amounts present in the wild-type strain. Deletion of RRP6 restored the levels of these transcripts to those present in the wild type (Fig. 7C). Deletion of RRP6 in the wild-type background did not substantially affect the β-galactosidase mRNA levels. These data show that the defect in mRNA accumulation in the yra1-8 mutant strain is not due to reduced mRNA production but rather to the degradation of the transcripts by the nuclear exosome.
DISCUSSION
We have used a synthetic lethal approach to search for additional partners of the essential mRNA export factor Yra1p. We identified HPR1, a gene encoding a protein with a proposed role in transcription elongation (8) and whose deletion induces poly(A)+ RNA export defects (43). Deletion of Hpr1p is synthetically lethal with mutations in Yra1p as well as in Sub2p, and pull-down experiments show that Hpr1p, Sub2p, and Yra1p and Yra2p physically interact in vivo (Fig. 1 and 2A). In vitro binding experiments demonstrate that these interactions are direct (Fig. 2B and C). Finally, ChIP assays show that Hpr1p, Sub2p, and Yra1p are recruited cotranscriptionally and that efficient association of Sub2p and Yra1p with a transcribing gene is dependent on the presence of Hpr1p (Fig. 3 and 4). These results establish a functional link between transcription and mRNA export and indicate that the mRNA export defects in hpr1, sub2, and yra1 mutants are due, at least in part, to poor loading of export factors onto nascent mRNAs. These improperly assembled mRNPs are unstable and are degraded through an exosome-dependent surveillance mechanism (Fig. 5, 6, and 7) (30).
Hpr1p is part of the THO complex, which contains three other proteins, Tho2, Mft1, and Thp2p. These four components behave as a functional entity, as deletion of any of them results in poor growth at 37°C and induces similar gene expression defects (7, 40). Recent studies published while this work was in preparation show that Yra1p, Sub2p, and the THO components can be purified as a complex (18, 46). Synthetic lethality of sub2 and yra1 mutants with all four THO subunits confirms the functional relevance of these interactions. Finally, the poly(A)+ RNA export defect exhibited by yra1, sub2, and THO complex mutants further supports the idea of a role of these proteins in a common function (30, 46).
THO components have been implicated in transcription elongation, but the exact role of the complex in this process has not been defined (6-8, 40). Hpr1p and the other THO component, Tho2p, have been shown to be recruited to active genes (46). Whereas this latter study proposed a transcription-dependent association of Hpr1p and Tho2p over the entire coding region, our ChIP and quantitative PCR analyses allowed us to identify a preferential binding of Hpr1p towards the middle and 3′ end of the PMA1 gene. Genetic and physical interactions functionally link Hpr1p to the Cdc73p/Paf1p/Ccr4p complex, proposed to function in transcription through an association with RNA polymerase II (5). Since Paf1p/Cdc73p complexes are present at the promoter and on coding regions of different classes of genes (41), they could mediate the recruitment of Hpr1p/THO to active genes during transcription elongation.
Our data further show that the association profiles of Sub2p and Yra1p with PMA1 parallel the distribution of Hpr1p, which is consistent with a synchronous recruitment of these factors during elongation. The specific association of Yra1p with PMA1 was proposed earlier to occur preferentially towards the 3′ end of the gene (28). Our observations are consistent with these data. However, the utilization of additional primer sets in the present study shows that the peak of Yra1p association occurs farther upstream, in the middle of the PMA1 coding region, a distribution similar to that observed on the GAL10 transcription unit (28). Importantly, the binding of Sub2p and Yra1p to PMA1 is substantially decreased in the absence of Hpr1p. This reduced association with PMA1 in the absence of Hpr1p is unlikely due to a reduced transcription rate, since the levels of PMA1 transcripts at 25°C are not affected in Δhpr1 (Fig. 4A and 7A). The data therefore support that Hpr1p is required for efficient recruitment of Sub2p and Yra1p to the growing mRNP. It is not possible so far to distinguish whether Sub2p and Yra1p first become associated with the transcription machinery through an interaction with Hpr1p/THO and subsequently bind the mRNP or whether Hpr1p/THO and the nascent transcript stimulate the binding of Yra1p and Sub2p directly to the mRNP. The residual binding of Sub2p and Yra1p in the absence of Hpr1p is consistent with the viable phenotype of Δhpr1 mutants and indicates the existence of alternate binding sites, perhaps provided by the other THO subunits or the mRNA itself. Sub2p was earlier identified as a high-copy-number suppressor of Δhpr1 and becomes associated with an RNA polymerase II subunit in the absence of Hpr1p (11). It will be interesting to determine whether overexpression of Sub2p in Δhpr1 restores optimal recruitment of Yra1p to the nascent mRNP.
The association of Yra1p, Sub2p, and Hpr1p to PMA1 decreases towards the 3′ end of the gene. The reduction in Yra1p and Sub2p cross-linking may reflect remodeling of the mRNP or a decrease in the proximity of the mRNP with respect to the DNA template. Alternatively, it may correspond to the transfer of Yra1p and Sub2p from the transcription complex to the mRNP. Since THO components exhibit RNA binding activity (1), Hpr1p/THO may also become associated with the mRNP, perhaps marking the mRNA for a subsequent step in mRNP biogenesis. Examining the distribution of Yra1p, Sub2p, and Hpr1p/THO on other genes, those of different sizes and with or without introns, will be necessary to better understand the mechanisms underlying the cotranscriptional assembly of export-competent mRNPs.
We found that the phenotypes of yra1 mutants are enhanced or become lethal in combination with rrp45 and rrp6 exosome mutants (Fig. 6A and B). The phenotypes of sub2, Δhpr1, and other THO mutants are similarly exacerbated by the deletion of RRP6, and these synthetic interactions represent an additional common feature of Yra1p, Sub2p, and THO components (19, 30). Recent evidence suggests that the nuclear exosome functions in an mRNA surveillance system that degrades transcripts in response to defects in mRNA processing and export (4). Interestingly, the expression levels of several mRNAs are substantially reduced in yra1, sub2, and Δhpr1 mutant backgrounds (Fig. 7). Importantly, the β-galactosidase mRNA levels, which are very low in the yra1-8 mutant strain, are restored to wild-type levels when Rrp6p is deleted (Fig. 7C). Similarly, the levels of HSP104 transcripts induced at high temperature are severely reduced in sub2 and Δhpr1 and other THO mutants but rescued when Rrp6p is deleted (19, 30). Taken together, these data support the hypothesis that mRNPs assembled in all these mutant backgrounds are unstable and subject to an exosome-dependent quality control function which targets these transcripts to degradation. These observations also suggest that THO components may have a primary role in stable mRNP formation rather than in transcription elongation.
Different RNA processing and export mutants, including sub2, Δhpr1, and THO mutants, sequester newly made heat shock HSP104 transcripts within nuclear dots at or close to the transcription site. These foci were suggested to be involved in exosome-dependent quality control, since they disappear in the absence of Rrp6p (16, 19, 20, 30). Mutations in Yra1p also induce the accumulation of heat-shock transcripts within nuclear dots (Fig. 5B). However, we were not able to show that these foci disappear upon deletion of RRP6, since the double mutants are unviable (Fig. 6B and data not shown). Using ChIP experiments, we could not detect an association of Rrp45p or Rrp6p with the PMA1 gene (data not shown), suggesting that mRNPs are recognized by the surveillance machinery at a posttranscriptional step, perhaps in the nuclear foci. The observed physical interaction between Yra1p and Rrp45p (Fig. 6C) may therefore correspond to a step subsequent to mRNP release from the template.
Although the possibility cannot be excluded that Yra1p, Sub2p, and Hpr1p contribute to the overall integrity and activity of the transcription-elongating complex, our results and those of Libri et al. (30) support the idea that normal levels of full-length transcripts are produced in yra1, sub2, and hpr1 backgrounds but that these transcripts are subsequently degraded by an Rrp6p-dependent process. mRNPs produced in these mutant strains are likely to be malformed and recognized by a surveillance mechanism which competes with other mRNP biogenesis steps, including mRNA export. It will be interesting to identify which features of these incorrectly assembled mRNP complexes are recognized by this exosome-dependent quality control.
Acknowledgments
Our special thanks go to R. Sahli, R. Imoberdorf, and S. Creton for help and advice in ChIPs and real-time PCR. Our thanks also go to D. Libri and T. H. Jensen for communicating and discussing results before publication. We are grateful to V. Müller for excellent technical assistance. We also thank D. Libri for yeast strains and plasmids, E. Izaurralde and A. Aguilera for plasmids, and M. Collart, D. Görlich, and M. Strubin for anti-TBP, anti-CBP80, and anti-CTD antibodies, respectively. We thank T. H. Jensen, D. Libri, and M. Collart for comments on the manuscript.
This work was supported by a grant from the Swiss National Science Foundation (no. 31-61378-00) to F.S.
REFERENCES
- 1.Aguilera, A. 2002. The connection between transcription and genomic instability. EMBO J. 21:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, D. 1999. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11:347-351. [DOI] [PubMed] [Google Scholar]
- 3.Brodsky, A. S., and P. A. Silver. 2000. Pre-mRNA processing factors are required for nuclear export. RNA 6:1737-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, J. S. 2002. The yin and yang of the exosome. Trends Cell Biol. 12:90-96. [DOI] [PubMed] [Google Scholar]
- 5.Chang, M., D. French-Cornay, H.-Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chavez, S., and A. Aguilera. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 11:3459-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavez, S., T. Beilharz, A. G. Rondon, H. Erdjument-Bromage, P. Tempst, J. Q. Svejstrup, T. Lithgow, and A. Aguilera. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19:5824-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavez, S., M. Garcia-Rubio, F. Prado, and A. Aguilera. 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:7054-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, C. N. 2001. Choreographing mRNA biogenesis. Nat. Genet. 29:6-7. [DOI] [PubMed] [Google Scholar]
- 10.Colot, H. V., F. Stutz, and M. Rosbash. 1996. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 10:1699-1708. [DOI] [PubMed] [Google Scholar]
- 11.Fan, H.-Y., R. J. Merker, and H. L. Klein. 2001. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol. Cell. Biol. 21:5459-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864-1872. [DOI] [PubMed] [Google Scholar]
- 13.Fong, N., and D. L. Bentley. 2001. Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev. 15:1783-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gatfield, D., H. Le Hir, C. Schmitt, I. C. Braun, T. Köcher, M. Wilm, and I. Izaurralde. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11:1716-1721. [DOI] [PubMed] [Google Scholar]
- 15.Hellmuth, K., D. M. Lau, F. R. Bischoff, M. Kunzler, E. Hurt, and G. Simos. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18:6374-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilleren, P., T. McCarthy, M. Rosbash, R. Parker, and T. H. Jensen. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413:538-542. [DOI] [PubMed] [Google Scholar]
- 17.Hirose, Y., and J. L. Manley. 2000. RNA polymerase II and the integration of nuclear events. Genes Dev. 14:1415-1429. [PubMed] [Google Scholar]
- 18.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, T. H., J. Boulay, M. Rosbash, and D. Libri. 2001. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11:1711-1715. [DOI] [PubMed] [Google Scholar]
- 20.Jensen, T. H., K. Patricio, T. McCarthy, and M. Rosbash. 2001. A block to mRNA nuclear export in Saccharomyces cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7:887-898. [DOI] [PubMed] [Google Scholar]
- 21.Kataoka, N., H. Yong, N. V. Kim, F. Velazquez, R. A. Perkinson, F. Wang, and G. Dreyfuss. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6:673-682. [DOI] [PubMed] [Google Scholar]
- 22.Kistler, A. L., and C. Guthrie. 2001. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 15:42-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 25.Legrain, P., and M. Rosbash. 1989. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57:573-583. [DOI] [PubMed] [Google Scholar]
- 26.Le Hir, H., D. Gatfield, E. Izaurralde, and M. J. Moore. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20:4987-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Hir, H., E. Izaurralde, L. E. Maquat, and M. J. Moore. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lei, E. P., H. Krebber, and P. A. Silver. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis, J. D., and E. Izaurralde. 1997. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 247:461-469. [DOI] [PubMed] [Google Scholar]
- 30.Libri, D., K. Dower, J. Boulay, R. Thomsen, M. Rosbash, and T. H. Jensen. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22:8254-8266. [DOI] [PMC free article] [PubMed]
- 31.Libri, D., N. Graziani, C. Saguez, and J. Boulay. 2001. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 15:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linder, P., and F. Stutz. 2001. mRNA export: travelling with DEAD box proteins. Curr. Biol. 11:R961-R963. [DOI] [PubMed] [Google Scholar]
- 33.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 34.Luo, M. J., and R. Reed. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96:14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo, M. L., Z. Zhou, K. Magni, C. Christoforides, J. Rappsilber, M. Mann, and R. Reed. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413:644-647. [DOI] [PubMed] [Google Scholar]
- 36.Lykke-Andersen, J. 2001. mRNA quality control: marking the message for life or death. Curr. Biol. 11:R88-R91. [DOI] [PubMed] [Google Scholar]
- 37.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 38.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell, P., and D. Tollervey. 2000. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 7:843-846. [DOI] [PubMed] [Google Scholar]
- 40.Piruat, J. I., and A. Aguilera. 1998. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 17:4859-4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9:799-809. [DOI] [PubMed] [Google Scholar]
- 42.Reed, R., and E. Hurt. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108:523-531. [DOI] [PubMed] [Google Scholar]
- 43.Schneiter, R., C. E. Guerra, M. Lampl, G. Gogg, S. D. Kohlwein, and H. L. Klein. 1999. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol. Cell. Biol. 19:3415-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strahm, Y., B. Fahrenkrog, D. Zenklusen, E. Rychner, J. Kantor, M. Rosbash, and F. Stutz. 1999. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr255p. EMBO J. 18:5761-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413:648-652. [DOI] [PubMed] [Google Scholar]
- 46.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 47.Strässer, K., and E. Hurt. 2000. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19:410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stutz, F., A. Bachi, T. Doerks, I. C. Braun, B. Séraphin, M. Wilm, P. Bork, and I. Izaurralde. 2000. REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6:638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stutz, F., J. Kantor, D. Zhang, T. McCarthy, M. Neville, and M. Rosbash. 1997. The yeast nucleoporin Rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Genes Dev. 11:2857-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stutz, F., J. Tang, and M. Rosbash. 1998. Synthetic lethal/enhancer screening to identify snRNA-protein and protein-protein interactions in yeast pre-mRNA splicing, p. 161-182. In C. W. J. Smith (ed.), RNA-protein interactions: a practical approach. Oxford University Press, New York, N.Y.
- 51.Zenklusen, D., and F. Stutz. 2001. Nuclear export of mRNA. FEBS Lett. 498:150-156. [DOI] [PubMed] [Google Scholar]
- 52.Zenklusen, D., P. Vinciguerra, Y. Strahm, and F. Stutz. 2001. The yeast hnRNP-like proteins Yra1p and Yra2p participate in mRNA export through an interaction with Mex67p. Mol. Cell. Biol. 21:4219-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, M., and M. R. Green. 2001. Identification and characterization of yUAP/Sub2.p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev. 15:30-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, H., M.-J. Luo, K. Straesser, J. Katahira, E. Hurt, and R. Reed. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407:401-405. [DOI] [PubMed] [Google Scholar]