Abstract
Mot1 stably associates with the TATA-binding protein (TBP), and it can dissociate TBP from DNA in an ATP-dependent manner. Mot1 acts as a negative regulator of TBP function in vitro, but genome-wide transcriptional profiling suggests that Mot1 positively affects about 10% of yeast genes and negatively affects about 5%. Unexpectedly, Mot1 associates with active RNA polymerase (Pol) II and III promoters, and it is rapidly recruited in response to activator proteins. At Pol II promoters, Mot1 association requires TBP and is strongly correlated with the level of TBP occupancy. However, the Mot1/TBP occupancy ratio at both Mot1-stimulated and Mot1-inhibited promoters is high relative to that at typical promoters, strongly suggesting that Mot1 directly affects transcriptional activity in a positive or negative manner, depending on the gene. The effect of Mot1 at the HIS3 promoter region depends on the functional quality and DNA sequence of the TATA element. Unlike TBP, Mot1 association is largely independent of the Srb4 component of Pol II holoenzyme, and it also can occur downstream of the promoter region. Mot1 removes TBP, but not TBP complexes or preinitiation complexes, from inappropriate genomic locations. Mot1 inhibits the association of NC2 with promoters, suggesting that the TBP-Mot1 and TBP-NC2 complexes compete for promoter occupancy in vivo. We speculate that Mot1 does not form transcriptionally active TBP complexes but rather regulates transcription in vivo by modulating the activity of free TBP and/or by affecting promoter DNA structure.
The TATA-binding protein (TBP) is the central initiation factor for transcription by all three nuclear RNA polymerases (Pol). TBP is a component of the SL-1, TFIID, and TFIIIB complexes that mediate transcription by Pol I, Pol II, and Pol III, respectively (20, 46). With respect to Pol II transcription in yeast cells, TBP is essential for transcription of all genes (9), whereas the associated factors (TAFs) within the TFIID complex are selectively required (21, 37, 38, 45, 52, 53). Relative to TBP occupancy, TAF association is high at TAF-dependent promoters and is low at TAF-independent promoters, indicating that Saccharomyces cerevisiae has at least two forms of transcriptionally active TBP in vivo (27, 32). The TFIID form is recruited to ribosomal protein promoters by a Rap1-containing activator (35), and it is important at several promoters with weak TATA elements (35, 37, 38, 49). The TAF-independent form(s) of TBP predominates at many strong promoters, and it is preferentially recruited to promoters by most yeast activators (27, 32, 35). It is unclear whether the TAF-independent form(s) is TBP itself, which is present in yeast cell extracts (18), or is a distinct TBP complex(es).
Aside from its presence in TFIID, TBP also forms stable complexes with Mot1 and with NC2, a heterodimer of histone-fold proteins (Bur6 and Ydr1 in yeast). In vitro, NC2 forms a transcriptionally inactive complex with TBP and promoter DNA that physically blocks the incorporation of TFIIA and TFIIB into the preinitiation complex (6, 17, 24, 25, 36). Thus, NC2 biochemically behaves as a general negative regulator, although it can stimulate transcription in vitro of Drosophila melanogaster promoters containing downstream promoter elements by an unknown mechanism (55). In yeast cells, the Bur6 subunit of NC2 positively or negatively affects approximately 17% of the genes in a pattern that resembles the response to environmental stress (16). Interestingly, Bur6 associates with active Pol II promoters in vivo, with Bur6-stimulated promoters showing particularly high levels of Bur6 association (16). Thus, NC2 can play a direct and positive role in Pol II transcription in vivo, although it remains to be shown whether the TBP-NC2 complex is a TAF-independent form of transcriptionally active TBP.
Mot1, a 210-kDa ATPase that is essential for yeast cell viability (14), forms a stable complex with TBP in solution and on promoter DNA (4, 41). In the presence of ATP, Mot1 can dissociate TBP-DNA complexes and hence repress basal and activated transcription in vitro (4). Mot1 repression can be partially overcome by TFIIA and TFIIB, most likely via competitive binding to TBP and/or the increased stability of the resulting TFIIA-TBP-DNA or TFIIB-TBP-DNA complexes. Mot1 can also function as a Leu3-dependent corepressor (51). In accord with these biochemical experiments, Mot1 was identified genetically as a repressor of weak promoters (14, 42) and as a general inhibitor of the Pol II holoenzyme (15). Interestingly, NC2 was also identified in two of these genetic screens (15, 42), indicating that Mot1 and NC2 share phenotypic similarities as general negative regulators of transcription in vivo.
Although originally characterized as a repressor, several lines of evidence indicate that Mot1 can also positively regulate transcription. First, small amounts of Mot1 can stimulate transcription in yeast cell extracts, presumably by regulating the distribution of TBP between promoter and nonpromoter sites (39). In accord with this observation, mot1 mutant strains show higher levels of TBP at nonpromoter regions in yeast cells (33), although the form of TBP at such nonpromoter regions is unknown. Second, mot1 mutations are associated with decreased transcription of certain genes in vivo (7, 34). Interestingly, Mot1, NC2, and TFIID-specific TAFs have a common function in being important for HIS3 transcription from the +1, but not +13, initiation site (7, 31, 37, 38). It is unclear whether the positive functions of Mot1 in vivo arise from TBP redistribution away from nonpromoter sequences, direct action at promoters, or some other indirect effect.
In this paper, we investigate the physiological role of Mot1 by directly measuring the association of Mot1 with yeast promoters in vivo. Our results indicate that Mot1 associates with active promoters and is rapidly recruited by transcriptional activators. Further, Mot1 occupancy is particularly high at promoters that are positively or negatively regulated by Mot1, indicating that Mot1 can directly affect promoter activity in either direction. Finally, Mot1 inhibits the association of NC2 with active promoters, indicating that the TBP-Mot1 and TBP-NC2 complexes compete for promoter occupancy in vivo. From these and other results, we speculate that Mot1 does not form transcriptionally active TBP complexes but rather regulates transcription in vivo by modulating the activity of free TBP and/or by affecting promoter DNA structure. After this work was completed, it was reported that Mot1 activates and represses transcription in vivo by direct ATPase-dependent mechanisms (13), a conclusion that is consistent with that reached here. Similarities and differences between our work and this recent report are discussed.
MATERIALS AND METHODS
Yeast strains.
The experiments for Fig. 1 to 5 were performed in S. cerevisiae strain LK25, which expresses a derivative of TBP containing three copies of the HA1 epitope fused to the N terminus (28) and a derivative of Mot1 containing nine copies of the Myc epitope (10) at the C terminus from their natural promoters at their normal chromosomal locations. For the experiment for Fig. 6, a plasmid expressing Mot1 from the GAL1 promoter was integrated by a one-step disruption (37) of the MOT1 locus of previously described strains that contain HIS3 alleles with various combinations of the TC and TR TATA elements (22). For the experiment for Fig. 7, the Mot1-(Myc)9 derivative was introduced into BYΔ2 containing either a centromeric plasmid expressing the wild type or temperature-sensitive (ts1) TBP as the sole copy of TBP (9). For the experiment for Fig. 8, the Mot1-(Myc)9 derivative was introduced into isogenic wild-type and srb4-138 strains (15). The experiments for Fig. 9 and 10 involved isogenic wild-type and mot1-1 strains (14) that either did or did not express an epitope-tagged version of Bur6 containing three copies of the HA1 epitope at its N terminus (16). Unless stated otherwise, cells were grown in Casamino Acids medium with 2% dextrose to an optical density at 600 nm of 0.6.
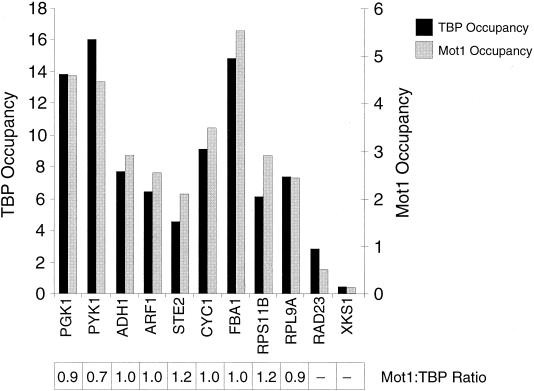
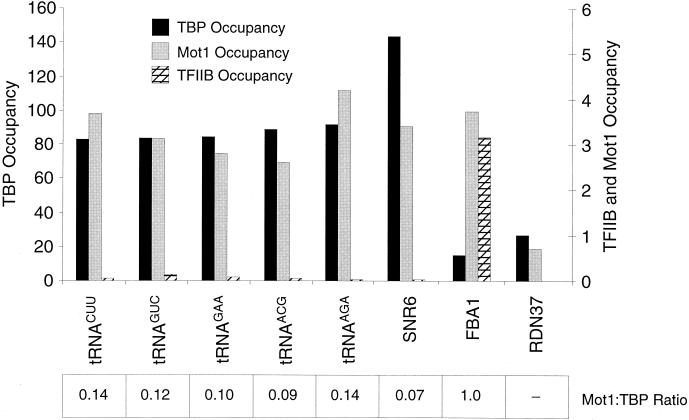
FIG. 1.
Association of Mot1 with Pol II promoters strongly correlates with association of TBP. Cross-linked chromatin was immunoprecipitated with antibodies against the HA (for TBP) or Myc (for Mot1) epitopes, and immunoprecipitated and input material was analyzed by quantitative PCR with primers corresponding to the indicated promoters. TBP and Mot1 occupancy units were calculated as described in Materials and Methods, and the Mot1/TBP occupancy ratios are indicated, with the average value being set arbitrarily to 1. −, Mot1/TBP ratio could not be determined accurately due to low occupancy by one or both factors.
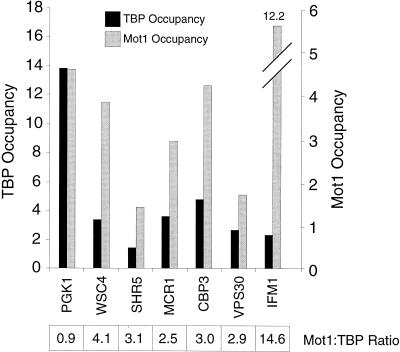
FIG. 5.
Increased Mot1 occupancy relative to TBP at promoters positively or negatively regulated by Mot1. An analysis of genes that are positively (VPS30 and IFM1), negatively (WSC4, SHR5, MCR1, and CBP3), or not (PGK1) regulated by Mot1 as defined by microarray analysis is shown. Occupancy units and Mot1/TBP ratios are indicated as described in the legend for Fig. 1.
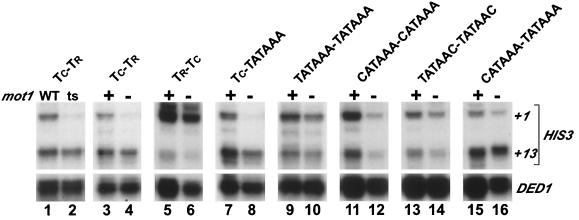
FIG. 6.
Mot1 dependence of HIS3 transcription is affected by the quality and sequence of the TATA element. HIS3 and DED1 RNA levels in wild-type (lane 1), mot1-1 (lane 2), and GAL1-MOT1 (lanes 3 to 16, with + indicating growth in 2% galactose and − indicating growth in a mixture of 1% glucose and 1% galactose) strains containing the indicated HIS3 alleles (see Materials and Methods) are shown. TC-TR, natural HIS3 locus; TR-TC, inversion of the natural elements. Functionally defined TATA elements are indicated by their DNA sequences (22).
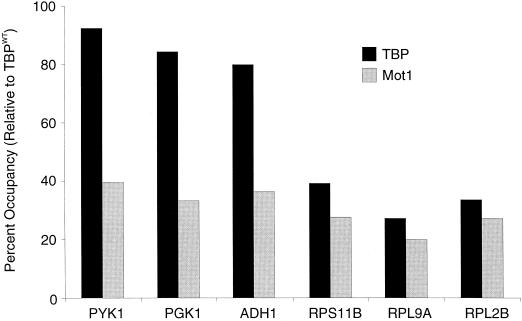
FIG. 7.
TBP is important for Mot1 association with promoters. The percentage of Mot1 and TBP occupancy in cells expressing the ts-1 derivative of TBP with respect to isogenic cells expressing wild-type TBP is shown. Wild-type and mutant cells were analyzed after a 45-min shift from 24 to 37°C.
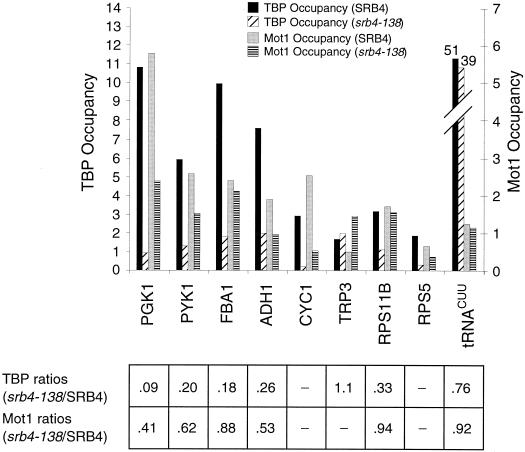
FIG. 8.
Mot1 association with promoters is largely independent of the Srb4 component of Pol II holoenzyme. Mot1 and TBP occupancy at the indicated promoters in isogenic wild-type or srb4-138 cells following a 60-min shift from 24 to 37°C is shown. Occupancy units and Mot1/TBP ratios are indicated as described in the legend for Fig. 1.
FIG. 9.
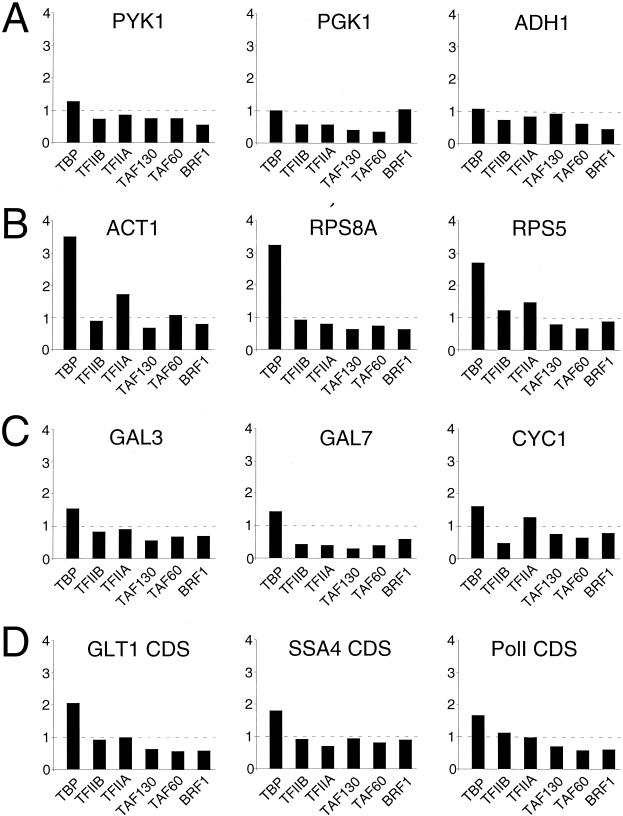
Mot1 removes TBP, but not general or TBP-associated factors, from promoters and protein-coding regions. The fold increase of occupancies of TBP, TFIIB, TFIIA, TAF130, TAF60, and Brf1 in the mot1-1 strain with respect to the isogenic wild-type strain at the indicated TAF-independent promoters (A), TAF-dependent promoters (B), weak promoters (C), and protein-coding regions (D) is shown.
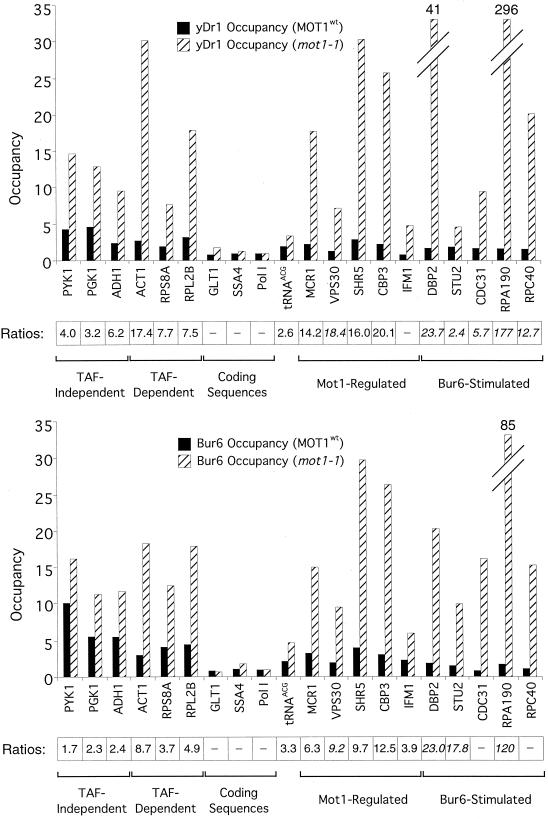
FIG. 10.
Mot1 inhibits association of NC2 with promoters. Association of Ydr1 (top) and Bur6 (bottom) subunits of NC2 at the indicated TAF-dependent, TAF-independent, Mot1-regulated, and Bur6-stimulated promoters and protein-coding sequences in wild-type and mot1-1 mutant strains is shown. The Mot1/TBP ratios are indicated as described in the legend for Fig. 1. Western blotting indicates that the wild-type and mutant strains have comparable intracellular levels of NC2.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed as described previously (16, 27, 35). Immunoprecipitations were performed with monoclonal antibodies to the HA1 epitope (F7; Santa Cruz Biotechnology) and the Myc epitope (9E10; Upstate Biotechnology) as well as polyclonal antisera to the Ydr1 subunit of NC2 (affinity purified and kindly provided by Danny Reinberg); TBP, TFIIB, and TFIIA (16, 27); and TAF130, TAF60, and TAF68 (kindly provided by Michael Green). Quantitative PCR was performed in real time using an Applied Biosystems 7700 sequence detector (16, 35). Mot1/TBP occupancy ratios were calculated by dividing background-subtracted Mot1 binding by background-subtracted TBP binding. Background was defined using the signals from the POL1 and other protein-coding regions. The average of the occupancy ratios for promoters analyzed for Fig. 1 was arbitrarily defined as 1.0; hence, promoters showing ratios of 2.0 and greater (see Fig. 5) contain relatively high Mot1 levels that are clearly beyond those explained by experimental error. Ydr1/TBP and Bur6/TBP occupancy ratios were calculated in a similar manner. Individual values represent the average of at least three independent experiments and have an error of approximately ±25%, except in the cases of very low occupancy, where the error may be greater.
Transcriptional analysis.
Detailed information on experimental procedures for the genome-wide transcriptional profiling of isogenic wild-type and mot1-1 strains can be found at http://www.wi.mit.edu/young/expression/mot1. To analyze the effect of Mot1 on transcription from the various HIS3 derivatives, strains expressing Mot1 from the GAL1 promoter were grown in 2% galactose or a mixture of 1% glucose and 1% galactose. HIS3 RNA levels were determined with respect to DED1 control RNA levels by S1 nuclease mapping (23). The growth conditions were chosen after performing an experiment in which the amounts of glucose and galactose were varied, while maintaining the concentration of the total carbon source at 2%. The pattern of HIS3 initiation sites in cells grown in 2% galactose is indistinguishable from that observed in cells containing a wild-type MOT1 allele. The pattern of HIS3 initiation sites in cells grown in 1% glucose and 1% galactose is indistinguishable from that of mot1-1 cells. We presume that Mot1 levels produced from the glucose-repressed GAL1 promoter under these conditions are roughly comparable to that produced by translational readthrough of the mot1-1 allele. In accord with this presumption, cell growth is inhibited in media containing higher levels of glucose and lower levels of galactose.
RESULTS
Mot1 associates with Pol II promoters in accord with transcriptional activity and TBP occupancy.
In yeast cells, the level of transcriptional activity is strongly correlated with the level of promoter occupancy by TBP, TFIIB, and TFIIA (27, 28, 33). In contrast, association of the TAFs in the TFIID complex is not strictly correlated with TBP occupancy, with the TAF/TBP occupancy ratio varying over a 5- to 10-fold range depending on the promoter (27, 32, 35). The NC2/TBP occupancy ratio is constant at most promoters, although NC2 preferentially associates with the subset of promoters that are positively regulated by NC2 (16).
We first analyzed Mot1 occupancy in wild-type cells at 11 Pol II promoters spanning a range of transcriptional activities and representing mechanistically distinct classes of promoters (Fig. 1). Mot1 associates with all transcriptionally active promoters tested, and the apparent cross-linking efficiency of Mot1 is comparable to that observed with TAFs (27). The Mot1/TBP occupancy ratios at these promoters are indistinguishable, indicating that Mot1 behaves similarly to general initiation factors and NC2 but differently from TAFs. We arbitrarily define the Mot1/TBP occupancy ratio at typical promoters as 1.0, although we stress that these experiments do not address the stoichiometry of Mot1 and TBP at promoters. A normal Mot1/TBP occupancy ratio is also observed at CYC1, an unusual promoter in which significant TBP occupancy occurs under conditions of very low transcription (27, 28, 33). Mot1/TBP ratios are comparable at high-level TAF (e.g., RPS11B and RPL9A) and low-level TAF (e.g., PGK1 and PYK1) promoters. Thus, Mot1 associates with promoters in a manner that is strongly correlated with TBP occupancy and hence with Pol II transcriptional activity.
Rapid association of Mot1 with promoters upon transcriptional induction.
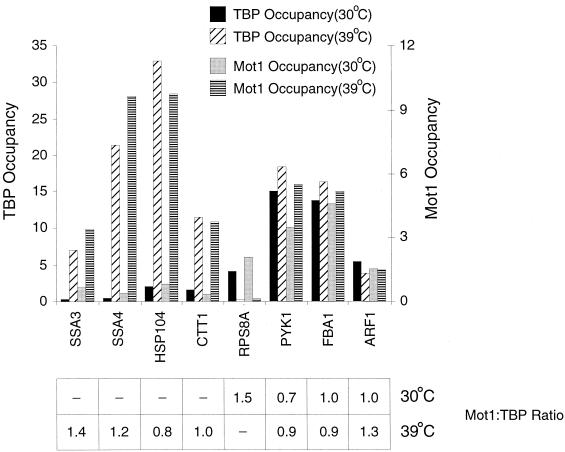
In response to heat shock, transcription of many genes is rapidly induced by the Hsf1, Msn2, and Msn4 activators, whereas transcription of ribosomal protein genes is rapidly inhibited. Promoter occupancies by TBP, general transcription factors, TAFs, and NC2 behave in accord with these transcriptional responses (16, 27, 28, 33, 43). Similarly, Mot1 association parallels that of TBP at all promoters tested (Fig. 2), with Mot1 being rapidly recruited to heat shock promoters (SSA3, SSA4, HSP104, and CTT1) and rapidly dissociating from a ribosomal protein promoter (RPS8A). The Mot1/TBP ratios at heat shock promoters are comparable to those of non-heat-shock promoters (PYK1, FBA1, and ARF1), whose Mot1 and TBP levels are unchanged in response to heat shock. Thus, Mot1 is rapidly recruited to promoters under conditions of transcriptional activation.
FIG. 2.
Mot1 is rapidly recruited to promoters by transcriptional activators. Mot1 and TBP occupancy at heat shock-inducible and uninducible promoters is shown. Cells were grown at 30°C or were heat shocked for 15 min at 39°C after previous growth at 24°C. SSA3 and SSA4 are activated by heat shock factor (Hsf1), CTT1 is activated by the Msn2 and Msn4 activators, and HSP104 is activated by both classes of activator. Heat shock inhibits the RPL8A promoter but does not affect the other promoters tested. Occupancy units and Mot1/TBP ratios are indicated as described in the legend for Fig. 1.
Mot1 associates with Pol III promoters, although the Mot1/TBP occupancy ratio is low.
Although biochemical analysis indicates that repression by Mot1 is specific for Pol II promoters (4), Pol III promoters often have consensus TATA elements that affect transcription via the specific DNA-binding activity of TBP (19, 54). Interestingly, Mot1 associates with the SNR6 and five tRNA promoters at a level that is comparable to that observed at FBA1, a strong Pol II promoter (Fig. 3). However, the Mot1/TBP occupancy ratios at these Pol III promoters are only about 10% the level observed for Pol II promoters, a result that reflects the very high (and nearly complete) TBP occupancy at Pol III promoters (28). Mot1 occupancy at the Pol I promoter (RDN37) is barely above the background level. Thus, highly transcribed Pol III promoters are accessible to Mot1, presumably in the form of a Mot1-TBP complex, but Mot1 is unlikely to significantly regulate TBP activity at Pol III promoters because the overwhelming majority of TBP complexes at those promoters lack Mot1.
FIG. 3.
Association of Mot1 with Pol III, but not Pol I, promoters. Mot1, TBP, and TFIIB occupancy at the indicated Pol III promoters, the FBA1 Pol II promoter, and the RDN37 Pol I promoter is shown. Occupancy units and Mot1/TBP ratios are indicated as described in the legend for Fig. 1. TFIIB occupancy at the RDN37 promoter was not tested.
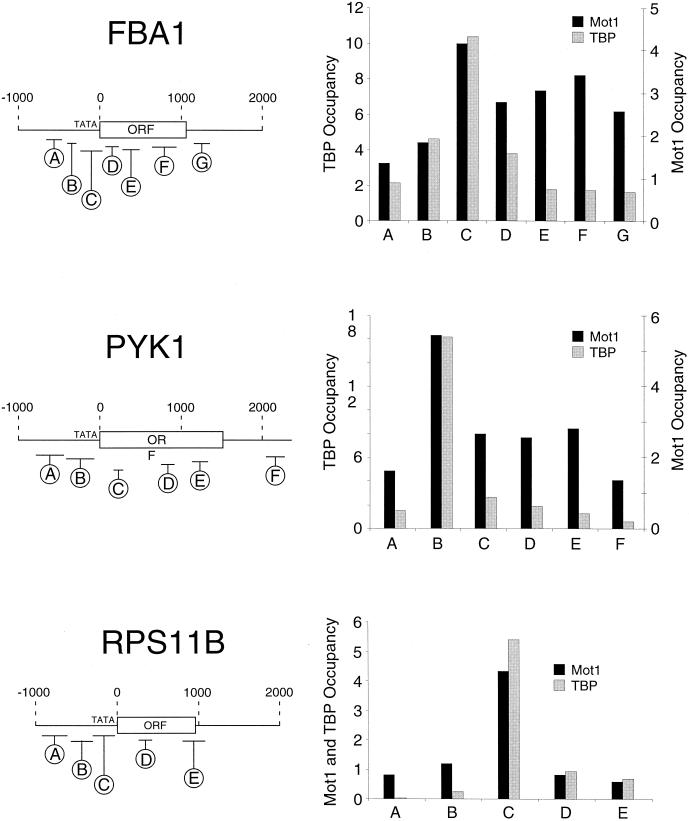
Mot1 associates with the protein-coding regions of some, but not all, transcribed genes.
TBP and other general transcription factors strongly associate with promoters, but not with protein-coding regions. Unexpectedly, Mot1 appears to associate with multiple positions within the FBA1 and PYK1 protein-coding regions (Fig. 4). This observation suggests the possibility that Mot1 might associate with these protein-coding regions in the absence of TBP. Alternatively, the low amount of TBP at these protein-coding regions might be preferentially in the form of a Mot1-TBP complex as opposed to other forms of TBP. In contrast to the situation at the FBA1 and PYK1 protein-coding regions, Mot1 does not associate with the RPS11B protein-coding region. We do not understand why Mot1 associates with some, but not all, protein-coding regions, although it is interesting that transcription of RPS11B, but not FBA1 and PYK1, is TAF dependent.
FIG. 4.
Mot1 associates with some, but not all, protein-coding regions. Mot1 and TBP occupancy at the indicated regions of the FBA1, PYK1, and RPS11B genes.
Mot1 positively and negatively affects the transcription of approximately 15% of yeast genes.
Because Mot1 is an essential gene, genome-wide transcriptional profiling was performed in isogenic wild-type and mot1-1 strains (complete data sets can be found at http://www.wi.mit.edu/young/expression/mot1). The mot1-1 allele is a nonsense mutation that produces approximately 5% of the wild-type protein as a consequence of readthrough translation. The mot1-1 strain shows a modest growth defect at 30°C but is unable to grow at 37°C. Comparison of the wild-type and mot1-1 mutant strains at 37°C indicates that Mot1 has a twofold or greater effect on the transcription of approximately 15% of the yeast genes tested. Funcagenic analysis (11) suggests that Mot1-regulated genes do not fall into clear functional classes.
Surprisingly, genes showing decreased transcription outnumber those displaying higher transcription by a nearly 2-to-1 margin. This result differs from recent transcriptional profiling results for the mot1-14 allele, in which 97% of the 182 Mot1-regulated genes were inhibited and only 3% were stimulated (13). However, when these published data are interpreted with a slight lowering of the confidence level (from 99 to 95%), there is a significant increase in the number of Mot1-regulated genes, especially those stimulated by Mot1 (http://www.people.Virginia.EDU/∼dta4n/auble_lab/Dasgupta_et_al_supplement.html). Direct comparison of the Mot1-repressed genes from the two transcriptional profiling experiments reveals 38 genes in common, a number that is far higher than that expected by chance (P = 3 × 10−6) yet represents only a 20% overlap. We suspect that many of the differences in the two datasets may be attributable to differences in growth conditions and/or strain backgrounds (see Discussion).
Unusually high level of Mot1 association at Mot1-stimulated and Mot1-inhibited promoters.
To address whether the positive and/or negative effects of Mot1 are direct, we analyzed the Mot1/TBP occupancy ratios at Mot1-stimulated and Mot1-inhibited genes identified by the transcriptional profiling analysis described above. For all six Mot1-regulated genes for which Mot1 and TBP occupancies can be reliably measured, Mot1/TBP occupancy ratios are substantially higher than those observed for Mot1-independent genes (Fig. 5). Interestingly, comparably high Mot1/TBP occupancy ratios are observed for promoters that are positively (VPS30, IFM1, and CBP3) or negatively (WSC4, SHR5, and MCR1) regulated by Mot1. Mot1 occupancy at the IFM1 promoter is extremely high. These observations strongly suggest that Mot1 directly stimulates or inhibits transcription at particular promoters. As such, Mot1 behaves differently from the Bur6 subunit of NC2, which shows increased association at Bur6-stimulated, but not Bur6-inhibited, promoters (16).
Mot1 dependence of HIS3 transcription is affected by the quality and sequence of the TATA element.
The HIS3 core promoter region contains a noncanonical TATA-like element (TC) that mediates initiation from +1 in a TAF-dependent manner and a consensus TATA element (TR) that mediates initiation from +13 in a TAF-independent manner (22, 37, 38). Loss of Mot1 strongly reduces TC-dependent +1, but not TR-dependent +13, HIS3 transcription (7, 22), and Mot1 strongly stimulates TATA-independent, but not TATA-dependent, HIS4 transcription (7).
To address the basis of Mot1-dependent HIS3 transcription in more detail, we analyzed a variety of derivatives in which the TC and TR elements are replaced by TATA sequences of defined functional quality (22, 56). As expected (7, 22), loss of Mot1 results in a strong decrease in +1 transcription from the wild-type HIS3 promoter (Fig. 6, lanes 1 to 4). This effect of Mot1 is due to a functional difference between TC and TR and not to a difference between the HIS3 initiation sites, because Mot1 has little effect on +1 transcription when the order of TC and TR is reversed. Furthermore, loss of Mot1 does not affect transcription from a canonical TATA element (TATAAA) located at the position of TC or TR in any circumstance tested (lanes 7 to 10 and 15 to 16). However, loss of Mot1 reduces transcription from a weakened TATA element (CATAAA) that is located in either the upstream or downstream position (lanes 11 to 12 and 15 to 16), although the effect is less pronounced than that observed for the natural TC element. Mot1 has a marginal effect on another TATA sequence (TATAAC; lanes 13 to 14), whose transcriptional activity is only slightly lower than that of the CATAAA derivative (22, 56). The observation that TC, TATAAC, and CATAAA have different sensitivities to Mot1, despite having similar transcriptional activities and other properties with respect to HIS3 transcription (22), suggests that Mot1 dependence of HIS3 transcription is affected both by the functional quality and by the specific sequence of the TATA element region.
TBP is important for promoter occupancy by Mot1.
Mot1 interacts with DNA only in the presence of TBP (1), and its association with promoters in vivo strongly correlates with TBP association (Fig. 1), suggesting that TBP is required for promoter occupancy by Mot1. We tested whether Mot1 can associate with promoters in a strain containing a temperature-sensitive allele (TBP-ts1) that confers severe transcriptional defects at the restrictive temperature (9). In comparison to the wild-type strain, TBP occupancy in the mutant strain decreases at TAF-dependent promoters but not at TAF-independent promoters (Fig. 7), as observed elsewhere (35). In contrast, Mot1 occupancy decreases significantly (2.5- to 4-fold) at all promoters examined. The simplest explanation for these observations is that the ts1 mutation diminishes the ability of TBP to interact with Mot1 (and presumably with TAFs) but still preserves its TATA-binding function at TAF-independent promoters. In any event, these results indicate that TBP is important for Mot1 to associate with promoters in vivo.
Mot1 promoter occupancy is only modestly reduced by inactivation of Pol II holoenzyme.
Srb4 is an essential component of the Pol II holoenzyme, and it is required for essentially all Pol II transcription in yeast cells (48). The Pol II holoenzyme is important for stability of transcriptionally active TBP at promoters, because loss of Srb4 function via a temperature-sensitive srb4 allele significantly reduces TBP occupancy at promoters (28, 33) (Fig. 8). In contrast, Mot1 association with these promoters is only modestly affected (except for more significant effects at CYC1), such that the Mot1/TBP occupancy ratios are two- to fivefold higher than in the wild-type strain. Thus, formation and/or stability of Mot1-TBP complexes is largely independent of Pol II holoenzyme.
Mot1 removes free TBP from promoters and protein-coding regions.
Although loss of Mot1 can increase TBP occupancy at promoter and coding sequences (33), it is unknown what form of TBP is affected. We therefore analyzed occupancy of TBP and various general factors at TAF-dependent and TAF-independent promoters as well as protein-coding regions in wild-type and mot1-1 mutant strains. TBP occupancy at strong, TAF-independent promoters is largely unaffected by Mot1 (Fig. 9A), whereas it is significantly increased at TAF-dependent promoters (Fig. 9B). Poorly expressed genes (Fig. 9C) and coding sequences (Fig. 9D) exhibit a slight increase in TBP occupancy in a mot1 mutant strain. Thus, although Mot1 removes TBP from many genomic locations, it preferentially removes TBP from TAF-containing promoters. In all cases tested, loss of Mot1 does not significantly increase association of TFIIA, TFIIB, TAF130, TAF60, or Brf1 (Fig. 9). Thus, Mot1 does not affect the association of the TFIID or TFIIIB forms of TBP, nor does the Mot1-TBP complex compete with TFIID for promoter occupancy. In addition, Mot1 removes transcriptionally inactive TBP that is not in the context of a preinitiation complex.
Mot1 competes with NC2 for promoter occupancy.
Mot1 and NC2 both form TBP complexes on transcriptionally active promoters (16) (this work), and they have related functions with respect to transcriptional inhibition in vivo (15, 42). Interestingly, loss of Mot1 results in a striking increase in Bur6 and Ydr1 occupancy at essentially all promoters tested but has little effect at protein-coding regions (Fig. 10). On average, TAF-dependent, Mot1-regulated, and Bur6-stimulated promoters appear to have a greater increase in NC2 occupancy than promoters that are Mot1 and TAF independent. These results strongly argue that Mot1-TBP and NC2-TBP complexes compete for association with yeast promoter regions, presumably by competing for association with TBP.
DISCUSSION
Mot1 associates with transcriptionally active Pol II promoters in vivo.
Although Mot1 removes TBP from promoters in an ATP-dependent manner and inhibits transcription in vitro (4), we show that Mot1 associates with transcriptionally active Pol II promoters in yeast cells (Fig. 1 and 2). Mot1 behaves similarly to general transcription factors in that its association with promoters correlates well with TBP occupancy and transcriptional activity (except at Mot1-regulated genes; see below). It also behaves similarly to NC2, another TBP-interacting factor that associates with active promoters in vivo (16), even though the TBP-NC2-DNA complex has been typically considered to be transcriptionally inactive in vitro (6, 17, 24, 25, 36).
The apparent cross-linking efficiency of Mot1 is roughly comparable to that of four different TAFs and TFIIA (27), which are presumed to be stable components of preinitiation complexes. Since cross-linking efficiencies depend on the properties of individual proteins, they do not provide absolute measurements of promoter occupancy or relative stoichiometry of factors on the promoter. Nevertheless, our results suggest that Mot1 association with promoters in vivo is fairly stable and that ATP-dependent eviction of TBP is relatively inefficient. The typical Mot1/TBP occupancy ratio at Pol II promoters (arbitrarily defined as 1.0) likely represents a situation in which Mot1 is substoichiometric with respect to TBP, because the Mot1/TBP occupancy ratio at certain promoters can be significantly higher. It is unlikely that multiple Mot1 molecules associate with a promoter given the binary interaction of Mot1 and TBP (1) and the importance of TBP for Mot1 association in vivo (Fig. 7).
Our result that Mot1 associates with transcriptionally active promoters appears to conflict with a recent publication that showed minimal, if any, Mot1 association at the highly active ACT1 and RPL5 promoters (13). The basis of this apparent discrepancy is unknown, although we suspect that a higher background signal in the published experiments might have obscured or minimized the ability to observe Mot1 association except in the most favorable cases.
Mot1 can directly stimulate or inhibit the activity of specific promoters.
Transcriptional profiling indicates that Mot1 has twofold or higher transcriptional effects on approximately 15% of yeast genes, with positive effects outnumbering negative effects by a 2-to-1 margin. This observation is surprising in light of the common view that Mot1 is primarily a negative regulator and in comparison to an independent transcriptional profiling experiment in which 97% of Mot1-affected genes were inhibited by Mot1 (13). Despite their differences, we believe that both transcriptional profiling experiments are valid, because all Mot1-regulated genes examined in both studies show unusually high Mot1 association (see below). We suspect that the apparent discrepancy between the transcriptional profiling experiments reflects differences in strain background growth conditions or normalization methods and not in the nature of the mot1 mutations. Both the mot1-1 allele analyzed here and the mot1-14 allele analyzed elsewhere are nonsense mutations that result in severely truncated proteins and low levels of full-length Mot1 due to readthrough translation (13). In any event, expression analysis does not distinguish between direct and indirect effects of Mot1.
Strong evidence that Mot1 directly affects transcription of many genes comes from the observation that Mot1/TBP ratios at Mot1-regulated promoters are substantially higher than the ratios at Mot1-independent promoters (Fig. 5). A similar analysis of NC2 revealed a strict relationship between increased NC2/TBP occupancy ratios and positive NC2 transcriptional effects, indicating that NC2 directly stimulates the activity of certain promoters (16). Unlike NC2, an abnormally high Mot1/TBP occupancy ratio is observed at both Mot1-stimulated and Mot1-inhibited genes, strongly suggesting that direct Mot1 action can either positively or negatively affect transcription, depending on the promoter. As such, our results argue against models in which gene-specific effects of Mot1 are due to redistribution of TBP between promoter and nonpromoter regions (7, 34, 39). Dasgupta et al. (13) have independently obtained similar results and conclusions to those described here and have also shown that the direct positive and negative action of Mot1 requires ATPase activity.
Why do some promoters have abnormally high Mot1/TBP occupancy ratios? One possibility is that Mot1 contacts DNA in the vicinity of the TATA element such that differences in DNA sequence might affect the stability of the Mot1-TBP-TATA complex. Results presented here (Fig. 6) and elsewhere (7) indicate that Mot1 is often important for transcription from promoters lacking canonical TATA elements. In addition to the functional quality of the TATA element, the difference in Mot1 sensitivity between the TC, CATAAA, and TATAAC alleles (Fig. 6) argues that the specific DNA sequence in the vicinity of the TATA element is also important for determining whether the activity of a promoter is influenced by Mot1. In addition to effects mediated by the TATA region, Mot1 stability of the promoter might be affected by interactions (direct or indirect) with activators, repressors, or other proteins that differentially associate with promoters.
Mechanism of Mot1 association with promoters and the function of Mot1-TBP complexes.
There are two basic mechanisms by which Mot1 could associate with Pol II promoters in a manner that strongly correlates with the level of transcription. In one model, the Mot1-TBP complex might be a TAF-independent form of transcriptionally active TBP that forms normal preinitiation complexes and is recruited, directly or indirectly, by activator proteins. In this view, the Mot1-TBP complex is analogous to TFIID, although its specificity for core promoters and/or activators might differ. Alternatively, the Mot1-TBP complex might simply be associated with accessible TATA elements in the absence of a functional preinitiation complex and hence not be transcriptionally active per se. In this model, the Mot1-TBP complex would be in a dynamic equilibrium with transcriptionally active forms of TBP to generate an on-off switch at the promoter.
Although our results do not definitively resolve this question, we favor the view that the Mot1-TBP complex is transcriptionally inactive and hence is in a dynamic equilibrium with active forms of TBP. First, unlike TBP (28, 33), Mot1 association with promoters is not strongly affected by Srb4 (Fig. 8), a component of the mediator subcomplex of Pol II holoenzyme. The simplest interpretation of this result is that association of the Mot1-TBP complex, unlike that of transcriptionally active forms of TBP, occurs in the absence of Pol II holoenzyme and hence a functional preinitiation complex. Second, Mot1 associates with active Pol III promoters (Fig. 3), most of which contain consensus TATA elements that are specifically bound by TBP in the form of the TFIIIB complex (19, 54). As we doubt that Mot1 can functionally substitute for the TBP-associated subunits of TFIIIB that are intimately involved in Pol III transcription, it is very likely that the Mot1-TBP complex simply associates with accessible TATA elements at highly active Pol III promoters. Third, the observation that unusually high Mot1/TBP occupancy ratios can lead to positive or negative effects on transcription (Fig. 5) is not consistent with a simple model in which the Mot1-TBP complex is transcriptionally active. We speculate that Pol II and Pol III forms of transcriptionally active TBP dissociate from promoters at a significant level, thereby leaving an accessible TATA element available for the Mot1-TBP complex.
Mot1 removes free TBP from inappropriate genomic regions.
It has been shown that loss of Mot1 can result in increased TBP occupancy at both promoters and protein-coding regions (33). However, loss of Mot1 does not increase occupancy of TFIIB, TFIIA, TAFs, or Brf1 at these locations (Fig. 9), indicating that increased TBP occupancy is not accompanied by preinitiation complex formation and that Mot1 does not affect TBP occupancy in the form of TFIID or TFIIIB. In addition, NC2 occupancy within protein-coding regions is not increased upon loss of Mot1 (Fig. 10), indicating that increased TBP occupancy is not due to the TBP-NC2 form of TBP (the situation at promoter regions is discussed below). We considered the possibility that the Ccr4-Not complex, which weakly interacts with TBP in vitro (30) and affects TATA-element function in vivo (8), might be involved in increased binding by TBP. We have been unable to cross-link the Ccr4-Not complex to promoters, a result consistent with the identification of this complex as the major cytoplasmic mRNA deadenylase (50). Although we cannot exclude the possibility that Mot1 affects the occupancy of a presently unknown TBP complex, these results strongly suggest that Mot1 removes free TBP from inappropriate genomic regions in vivo. As such, they suggest that free TBP, which can be isolated from yeast cell extracts (18), actually exists in living yeast cells.
Increased TBP occupancy due to loss of Mot1 is selective. Specifically, TBP occupancy is strongly increased at TAF-dependent promoters, modestly increased at weak promoters and protein-coding regions, and not affected at strong TAF-independent promoters (Fig. 9). By analogy with the situation at Pol III promoters, we suggest that free TBP preferentially associates with active promoters over protein-coding regions. At TAF-dependent promoters, free TBP is transcriptionally inactive and hence can be evicted by Mot1. At strong TAF-independent promoters, we suggest that free TBP becomes incorporated into functional preinitiation complexes and hence is immune from Mot1 action. In this regard, TFIIA blocks the TBP-removal function of Mot1 in vitro (3). An implication of this suggestion is that free TBP might be a transcriptionally active form of TBP in vivo.
Competition among distinct TBP complexes at Pol II promoters.
Results presented here and elsewhere (13, 16, 27, 32) indicate that TFIID, the TBP-Mot1 and TBP-NC2 complexes, and perhaps free TBP can all associate with Pol II promoters in vivo. Potential competition between these various TBP forms is unlikely to be due to saturation at the promoter, because TBP occupancy at nearly all Pol II promoters is well below its maximum (28). TAF-dependent and TAF-independent promoters generally show comparable levels of either Mot1 or NC2 occupancy, and loss of Mot1 does not increase TAF occupancy at promoters. These results suggest that TFIID does not compete with the TBP-Mot1 and TBP-NC2 complexes for promoter occupancy. In contrast, NC2 occupancy at promoters increases significantly upon loss of Mot1 (Fig. 10), indicating that the TBP-Mot1 and TBP-NC2 complexes compete for promoter occupancy.
Competition between Mot1 and NC2 likely reduces the association of free TBP with promoters. This view is supported by the observation that increased NC2 occupancy in Mot1-defective strains is most dramatic at TAF-dependent, Mot1-regulated, and Bur6-stimulated promoters (Fig. 10). Because Mot1-regulated genes have high Mot1/TBP occupancy ratios (Fig. 5), loss of Mot1 should result in high levels of free TBP at these promoters, thereby providing a substrate for NC2. Similarly, in mot1 mutant cells, free TBP levels are most significantly increased at TAF-dependent promoters (Fig. 9), again providing a substrate for NC2. Competition could occur between preformed TBP-Mot1 and TBP-NC2 complexes or between Mot1 and NC2 for association with free TBP. We favor the latter possibility because TBP-NC2 complexes are unstable in the absence of DNA and because this explains why such competition does not extend to TFIID, which almost certainly functions as a preformed complex.
The competition between TBP-Mot1 and TBP-NC2 complexes at promoters is in accord with the considerable phenotypic similarities between mutations that inactivate Mot1 and NC2 (15, 30, 31, 42). However, the relative association of Mot1 and NC2 can vary considerably from promoter to promoter, and the transcriptional profiles of mot1 and bur6 mutants are clearly distinct (13, 16; this work). In addition, Mot1 and NC2 strongly affect transcription that is dependent on the his3 TC element (7, 31), a TAF-dependent promoter (37, 38), even though Mot1 and NC2 do not appear to compete with TAFs for promoter occupancy in vivo. Thus, the various TBP-associated factors—TAFs, Mot1, and NC2—have distinct, but in some cases overlapping, transcriptional functions.
How does Mot1 affect transcription?
Results presented here (Fig. 5) and elsewhere (13) indicate that unusually high Mot1 occupancy at specific promoters can directly inhibit or stimulate transcription in an ATPase-dependent manner. Distinct Mot1-dependent effects on transcription could be due to removal of free TBP from the promoter. For promoters for which free TBP is the form primarily responsible for transcriptional activity, Mot1 would act as a negative regulator. On the other hand, free TBP is not active and may even be repressive at certain promoters; in these cases, removal of TBP by Mot1 would increase transcription by removing a repressive form of TBP and allowing the increased association of a transcriptionally active form(s) of TBP.
Alternatively, Mot1 might differentially affect gene expression by causing conformational changes in promoter DNA and/or TBP. Biochemical experiments have suggested that Mot1 affects TBP conformation rather than DNA structure (2, 5, 12), and TBP binding to TATA elements involves a two-step mechanism that includes a conformational change mediated by a solvent-exposed surface of TBP (57). TFIIB (57) and perhaps the nonconserved N-terminal region of TBP (29) can influence this conformational change in the TBP-TATA complex, and TFIIA competes with Mot1 for association with TBP (4). Promoter-specific effects on inherent TFIIB or TFIIA association and/or activator-specific recruitment of TFIIB or TFIIA might underlie the differential effects of Mot1 on transcription. The possibility that Mot1 affects DNA structure has been strengthened by the ability of the RSC nucleosome-remodeling complex, which contains an ATPase domain similar to Mot1, to translocate along the DNA (44). Perhaps the association of Mot1 with certain protein-coding regions of transcribed genes (Fig. 4) reflects movement along the DNA.
The ability of Mot1 to directly inhibit or stimulate transcription is analogous to the situation with ATP-dependent nucleosome-remodeling complexes. Recruitment of the RSC nucleosome-remodeling complex to histone promoters is associated with repression, whereas recruitment to promoters involved in carbohydrate metabolism is mediated by specific activators (40). Similarly, the Swi/Snf complex can positively and negatively affect transcription of particular genes, and neither mode of control is an indirect effect of the other (47). It has been proposed that nucleosome-remodeling complexes all contribute to fluidity of chromatin structure but that the specific transcriptional response depends on the location and conformation of nucleosomes with respect to gene regulatory elements (26). By analogy, we suggest that Mot1 performs the same TBP-dependent ATPase function(s) at all promoters but that this function(s) is differentially interpreted by specific promoters.
Acknowledgments
We thank Frank Holstege and Rick Young for performing the microarray experiment and allowing us to analyze the data, Danny Reinberg and Michael Green for antibodies, and Mario Mencia for useful discussion.
This work was supported by grants to K.S. from the National Institutes of Health (GM30186 and GM53720).
REFERENCES
- 1.Adamkewicz, J. I., K. E. Hansen, W. A. Prud'homme, J. L. Davis, and J. Thorner. 2001. High affinity interaction of yeast transcriptional regulator, Mot1, with the TATA-box binding protein (TBP). J. Biol. Chem. 276:11883-11894. [DOI] [PubMed] [Google Scholar]
- 2.Adamkewicz, J. I., C. G. Mueller, K. E. Hansen, W. A. Prud'homme, and J. Thorner. 2000. Purification and enzymic properties of Mot1 ATPase, a regulator of basal transcription in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275:21158-21168. [DOI] [PubMed] [Google Scholar]
- 3.Auble, D. T., and S. Hahn. 1993. An ATP-dependent inhibitor of TBP binding to DNA. Genes Dev. 7:844-856. [DOI] [PubMed] [Google Scholar]
- 4.Auble, D. T., K. E. Hansen, C. G. F. Mueller, W. S. Lane, J. Thorner, and S. Hahn. 1994. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 8:1920-1934. [DOI] [PubMed] [Google Scholar]
- 5.Auble, D. T., and S. M. Steggenda. 1999. Testing for DNA tracking by MOT1, a SNF2/SWI2 protein family member. Mol. Cell. Biol. 19:412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cang, V., D. T. Auble, and G. Prelich. 1999. A new regulatory domain on the TATA-binding protein. EMBO J. 18:6662-6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collart, M. A. 1996. The NOT, SPT3, and MOT1 genes functionally interact to regulate transcription at core promoters. Mol. Cell. Biol. 16:6668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collart, M. A., and K. Struhl. 1994. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 8:525-537. [DOI] [PubMed] [Google Scholar]
- 9.Cormack, B. P., and K. Struhl. 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69:685-696. [DOI] [PubMed] [Google Scholar]
- 10.Cosma, M. P., T. Tanaka, and K. Nasmyth. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299-311. [DOI] [PubMed] [Google Scholar]
- 11.Damelin, M., I. Simon, T. I. Moy, B. Wilson, S. Komili, P. Tempst, F. P. Roth, R. A. Young, B. R. Cairns, and P. A. Silver. 2002. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell 9:563-573. [DOI] [PubMed] [Google Scholar]
- 12.Darst, R. P., D. Wang, and D. T. Auble. 2001. MOT1-catalyzed TBP-DNA disruption: uncoupling DNA conformational change and role of upstream DNA. EMBO J. 20:2028-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasgupta, A., R. P. Darst, K. J. Martin, C. A. Afshari, and D. T. Auble. 2002. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl. Acad. Sci. USA 99:2666-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davis, J. L., R. Kunisawa, and J. Thorner. 1992. A presumptive helicase (MOT1 gene product) affects gene expression and is required for viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 12:1879-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gadbois, E. L., D. M. Chao, J. C. Reese, M. R. Green, and R. A. Young. 1997. Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc. Natl. Acad. Sci. USA 94:3145-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geisberg, J. V., F. C. Holstege, R. A. Young, and K. Struhl. 2001. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol. Cell. Biol. 21:2736-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goppelt, A., G. Stelzer, F. Lottspeich, and M. Meisterernst. 1996. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 15:3105-3116. [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn, S., S. Buratowski, P. A. Sharp, and L. Guarente. 1989. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell 58:1173-1181. [DOI] [PubMed] [Google Scholar]
- 19.Heard, D. J., T. Kiss, and W. Filipowicz. 1993. Both Arabidopsis TATA-binding protein (TBP) isoforms are functionally identical in RNA polymerase II and III transcription in plant cells: evidence for gene-specific changes in DNA binding specificity of TBP. EMBO J. 12:3519-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez, N. 1993. TBP, a universal eukaryotic transcription factor? Genes Dev. 7:1291-1308. [DOI] [PubMed] [Google Scholar]
- 21.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 22.Iyer, V., and K. Struhl. 1995. Mechanism of differential utilization of the his3 TR and TC TATA elements. Mol. Cell. Biol. 15:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iyer, V., and K. Struhl. 1996. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5208-5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamada, K., F. Shu, H. Chen, S. Malik, G. Stelzer, R. G. Roeder, M. Meisterernst, and S. K. Burley. 2001. Crystal structure of negative cofactor 2 recognizing the TBP-DNA complex. Cell 106:71-81. [DOI] [PubMed] [Google Scholar]
- 25.Kim, T. K., Y. Zhao, H. Ge, R. Bernstein, and R. G. Roeder. 1995. TATA-binding protein residues implicated in a functional interplay between negative cofactor NC2 (Dr1) and general factors TFIIA and TFIIB. J. Biol. Chem. 270:10976-10981. [DOI] [PubMed] [Google Scholar]
- 26.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 27.Kuras, L., P. Kosa, M. Mencia, and K. Struhl. 2000. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 288:1244-1248. [DOI] [PubMed] [Google Scholar]
- 28.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-612. [DOI] [PubMed] [Google Scholar]
- 29.Lee, M., and K. Struhl. 2001. Multiple functions of the nonconserved N-terminal domain of yeast TATA-binding protein. Genetics 158:87-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, T. I., J. J. Wyrick, S. S. Koh, E. G. Jennings, E. L. Gadbois, and R. A. Young. 1998. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 18:4455-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaire, M., J. Xie, M. Meisterernst, and M. A. Collart. 2000. The NC2 repressor is dispensable in yeast mutated for the Sin4p component of the holoenzyme and plays roles similar to Mot1p in vivo. Mol. Microbiol. 36:163-173. [DOI] [PubMed] [Google Scholar]
- 32.Li, X.-Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 33.Li, X.-Y., A. Virbasius, X. Zhu, and M. R. Green. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 389:605-609. [DOI] [PubMed] [Google Scholar]
- 34.Madison, J. M., and F. Winston. 1997. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:287-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mencia, M., Z. Moqtaderi, J. V. Geisberg, L. Kuras, and K. Struhl. 2002. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast. Mol. Cell 9:823-833. [DOI] [PubMed] [Google Scholar]
- 36.Mermelstein, F., K. Yeung, J. Cao, J. A. Inostroza, H. Erdjument-Bromage, K. Eagelson, D. Landsman, P. Levitt, P. Tempst, and D. Reinberg. 1996. Requirement of a corepressor for Dr1-mediated repression of transcription. Genes Dev. 10:1033-1048. [DOI] [PubMed] [Google Scholar]
- 37.Moqtaderi, Z., Y. Bai, D. Poon, P. A. Weil, and K. Struhl. 1996. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 382:188-191. [DOI] [PubMed] [Google Scholar]
- 38.Moqtaderi, Z., M. Keaveney, and K. Struhl. 1998. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2:675-682. [DOI] [PubMed] [Google Scholar]
- 39.Muldrow, T. A., A. M. Campbell, P. A. Weil, and D. T. Auble. 1999. MOT1 can activate basal transcription in vitro by regulating the distribution of TATA-binding protein between promoter and nonpromoter sites. Mol. Cell. Biol. 19:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poon, D., A. M. Campbell, Y. Bai, and P. A. Weil. 1994. Yeast TAF170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-associated factor complex distinct from transcription factor IID. J. Biol. Chem. 269:23135-23140. [PubMed] [Google Scholar]
- 42.Prelich, G. 1997. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2-alpha homolog that has both positive and negative roles in transcription in vivo. Mol. Cell. Biol. 17:2057-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 44.Saha, R., J. Wittmeyer, and B. R. Cairns. 2002. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16:2120-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen, W. C., and M. R. Green. 1997. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90:615-624. [DOI] [PubMed] [Google Scholar]
- 46.Struhl, K. 1994. Duality of TBP, the universal transcription factor. Science 263:1103-1104. [DOI] [PubMed] [Google Scholar]
- 47.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, C. M., and R. A. Young. 1995. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl. Acad. Sci. USA 92:4587-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsukihashi, Y., T. Miyake, M. Kawaichi, and T. Kokubo. 2000. Impaired core promoter recognition caused by novel yeast TAF145 mutations can be restored by creating a canonical TATA element within the promoter region of the TUB2 gene. Mol. Cell. Biol. 20:2385-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 51.Wade, P. A., and J. A. Jaehning. 1996. Transcriptional corepression in vitro: a Mot1p-associated form of TATA-binding protein is required for repression by Leu3p. Mol. Cell. Biol. 16:1641-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker, S. S., J. C. Reese, L. M. Apone, and M. R. Green. 1996. Transcription activation in cells lacking TAFIIs. Nature 382:185-188. [DOI] [PubMed] [Google Scholar]
- 53.Walker, S. S., W. C. Shen, J. C. Reese, L. M. Apone, and M. R. Green. 1997. Yeast TAFII145 required for transcription of G1/S cyclin genes and regulated by cellular growth state. Cell 90:607-614. [DOI] [PubMed] [Google Scholar]
- 54.Whitehall, S. K., G. A. Kassavetis, and E. P. Geiduschek. 1995. The symmetry of the yeast U6 RNA gene's TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 9:2974-2985. [DOI] [PubMed] [Google Scholar]
- 55.Willy, P. J., R. Kobayashi, and J. T. Kadonaga. 2000. A basal transcription factor that activates or represses transcription. Science 290:982-985. [DOI] [PubMed] [Google Scholar]
- 56.Wobbe, C. R., and K. Struhl. 1990. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol. Cell. Biol. 10:3859-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao, X., and W. Herr. 2002. A regulated two-step mechanism of TBP binding to DNA: a solvent-exposed surface of TBP inhibits TATA box recognition. Cell 108:615-627. [DOI] [PubMed] [Google Scholar]