Abstract
Gene activation mediated by nuclear receptors is regulated in a tissue-specific manner and requires interactions between nuclear receptors and their cofactors. Here, we identified and characterized a tissue-specific coactivator, GT198, that interacts with the DNA-binding domains of nuclear receptors. GT198 was originally described as a genomic transcript that mapped to the human breast cancer susceptibility locus 17q12-q21 with unknown function. We show that GT198 exhibits a tissue-specific expression pattern in which its mRNA is elevated in testis, spleen, thymus, pituitary cells, and several cancer cell lines. GT198 is a 217-amino-acid nuclear protein that contains a leucine zipper required for its dimerization. In vitro binding and yeast two-hybrid assays indicated that GT198 interacted with nuclear receptors through their DNA-binding domains. GT198 potently stimulated transcription mediated by estrogen receptor α and β, thyroid hormone receptor β1, androgen receptor, glucocorticoid receptor, and progesterone receptor. However, the action of GT198 was distinguishable from that of the ligand-binding domain-interacting nuclear receptor coactivators, such as TRBP, CBP, and SRC-1, with respect to basal activation and hormone sensitivity. Furthermore, protein kinase A, protein kinase C, and mitogen-activated protein kinase can phosphorylate GT198 in vitro, and cotransfection of these kinases regulated the transcriptional activity of GT198. These data suggest that GT198 is a tissue-specific, kinase-regulated nuclear receptor coactivator that interacts with the DNA-binding domains of nuclear receptors.
Nuclear receptors are members of a superfamily of DNA-binding transcriptional factors. Hormone-induced gene activation by nuclear receptors involves a variety of biological phenomena such as cell proliferation, differentiation, and development (31, 33). A central question in the field is how a single nuclear receptor molecule elicits complex responses of gene activation or repression in response to a hormonal stimulus in a cell-specific manner. There is evidence that tissue selectivity of gene expression induced by liganded receptors involves the coordination and assembly of an array of coregulatory proteins (9, 15, 26, 33, 53). Together with nuclear receptors, these cofactors are regulated at multiple levels, including tissue-specific distribution (22, 41, 54), variation of the assembly on each subset of hormone response elements (57), and cell-specific interactions between the nuclear receptors and their cofactors (33, 36, 54).
Members of the nuclear receptor family share several structurally related domains (31). The N-terminal transactivation regions of the receptors have variable primary sequences and are largely responsible for the functional differences among the receptor isoforms. The DNA-binding domain (DBD), which targets the receptors to their cognate hormone response elements, is composed of two conserved zinc fingers (31). The C-terminal ligand-binding domain (LBD) is responsible for binding to ligand and enables ligand-dependent disassociation of corepressor and association with coactivators (9). A common LXXLL motif present in most coactivators provides the interface for interaction between conserved LBD and coactivator molecules, including CBP/p300 (24), the SRC-1 family (26, 37), PGC-1 (22, 35, 41), CIA (48), and TRBP/ASC-2/AIB3/RAP250/PRIP/NRC (11, 23, 25, 30, 58). However, whether selective interaction between various nuclear receptors and coactivators occurs to mediate cell-specific action remains to be elucidated (54).
Protein cofactors specialized for interaction with the DBD of nuclear receptors are less characterized or defined. The DBD is the most highly conserved domain among members of the nuclear receptor superfamily (31). It plays an important role in determining the selectivity of DNA binding as well as directing the dimerization of nuclear receptors (6). Extensive studies of the structure and function of the DBD have revealed that receptor dimerization and binding of the DBD to each hormone response element rely on the variation of primary sequences around its zinc fingers and, in turn, contribute to its selectivity (6, 57). However, much less is known about regulation by nuclear cofactors that bind to the DBD. Several cofactors that interact with the DBDs of nuclear receptors have been reported, including SNURF. SNURF is characterized as a small nuclear RING finger protein that activates androgen receptor (AR)-mediated transcription by interaction with the DBD of AR (34, 39). Interestingly, SNURF facilitates nuclear trafficking of AR in a ligand-dependent manner (40). Several other protein factors, including ANPK, ARIP3, and ARIP4, have also been suggested to bind to the AR DBD and to modulate AR-mediated transactivation (21). In addition, DNA-dependent protein kinase was shown to interact with the DBD of progesterone receptor (PR) (45). The DBD of thyroid hormone receptor (TR) was suggested to interact with cofactors with repressive function (28). A recent report also indicates that the DBD of TR and retinoid X receptor (RXR) associates with PSF, a corepressor that recruits Sin3A to the receptor DBD (32). Another group of proteins that influence DNA binding of nuclear receptors is the high-mobility-group (HMG) proteins (7, 8, 38, 43). In addition to facilitating the binding of nuclear receptors to DNA, HMG proteins have diverse regulatory functions in DNA binding and chromatin architecture (16). The DBDs of nuclear receptors have also been shown to interact with other transcriptional factors such as NF-κB and AP-1, with resultant regulation of transcriptional repression (51). Since the DBDs of nuclear receptors are evolutionarily conserved, DBD-binding coactivators might also retain other conserved functions in the action of various DBDs of nuclear receptors.
Nuclear receptor-mediated target gene expression is largely tissue and cell specific, and it is subject to coordinate regulation by cytoplasmic signaling (31). Compared to the extensive studies of the nuclear function of nuclear receptors and cofactors, the regulation of these cofactors by signaling pathways, including phosphorylation, is less clear. However, the cytoplasmic hormonal effects of nuclear receptor ligands, now also referred to as rapid or nongenomic effects, have long been observed in cells. For example, thyroid hormone stimulates the epidermal growth factor pathway (27), estrogen induces mitogen-activated protein kinase (MAPK) and phosphatidylinositol (PI) 3-kinase signaling (29), and tamoxifen affects insulin-like growth factor I receptor action in breast cancer cells (17). In addition to binding of the receptor LBD in the nucleus, ligands may thus stimulate various signaling molecules and ultimately modify protein factors, including the coactivators that are important for nuclear receptor action. In this regard, the regulation of cofactors by hormones via kinase signaling might also contribute to a large extent of the cell-specific effects. Several lines of evidence have supported the notion that nuclear cofactors are kinase regulated. These include phosphorylation of p300 by protein kinase C (PKC) at Ser89 with inhibition of its transactivation (56), phosphorylation and regulation of SRC-1 by MAPK via protein kinase A (PKA) signaling (10), and phosphorylation of TRBP by DNA-dependent protein kinase (23). Thus, posttranslational modification of cofactors may be a common mechanism to mediate, at least in part, the specificity and selectivity of nuclear receptors in transcriptional regulation.
We report here the isolation and characterization of a tissue-specific nuclear receptor coactivator, GT198. GT198 contains a leucine dimerization domain and interacts with nuclear receptors via the DBD. GT198 enhances nuclear receptor-mediated transcription, and coactivation by GT198 may be subject to kinase regulation. The mechanism of its activation is distinguishable from LBD-interacting coactivators such as SRC-1, TRBP, and CBP. GT198 represents a new class of DBD-interacting nuclear receptor coactivators.
MATERIALS AND METHODS
Plasmids.
Mouse CBP, human TRBP, and human SRC-1 plasmids used in this study were previously described (23, 24, 37). Full-length rat GT198 cDNA was inserted into the HindIII/XhoI sites of pcDNA3 (Invitrogen) with an N-terminal Flag tag. GST-GT198 and GST-GR were subcloned into EcoRI/XhoI sites of pGEX-4T-2 (Pharmacia). Yeast expression plasmids were generated by insertion of glucocorticoid receptor (GR)-DBD (amino acids [aa] 411 to 489) or CREB-bZIP (aa 283 to 341) into the pADNS-Sos vector (5). Mouse mammary tumor virus (MMTV) promoter luciferase reporter was generated by inserting the MMTV promoter with HindIII/BglII sites into the PXP2 vector. Full-length human TRβ1, estrogen receptor alpha (ERα), ERβ, AR, GR, and PR were subcloned into pcDNA3. F2/TRE and 2XERE luciferase reporters were previously described (23). A 2XGRE reporter was generated by PCR and inserted with NheI/XhoI sites of pTAL-Luc (Clontech). PKA and MEK kinase plasmids were obtained from Stratagene. The PKCα was inserted into pcDNA3.
Yeast two-hybrid screen and GT198 cDNA cloning.
The Sos-Ras yeast two-hybrid system was employed as previously described (4, 5, 18). Briefly, human GR-DBD (aa 411 to 489) or rat CREB-bZIP (aa 283 to 341) was inserted in frame into the C terminus of human Sos (Ras guanyl nucleotide exchange factor) to produce fusion proteins for interaction. The interacting proteins encoded by a rat pituitary cDNA library were myristoylated and membrane localized. Protein interactions recruit Sos activity to the membrane and, in turn, activate the Ras pathway. This permits the proliferation of a temperature-sensitive yeast strain at a nonpermissive temperature (36°C). Positive clones containing cDNAs for the protein of interest are then detected by the growth of yeast. A partial rat 5′ cDNA of GT198 was isolated, and the full-length cDNA encoding rat GT198 was completed by 3′ rapid amplification of cDNA ends (RACE) with mRNA isolated from rat GC cells. A homology search against GenBank sequences revealed a human GT198 homolog as a transcript mapped to breast cancer susceptibility locus 17q12-q21 (44). A mouse GT198 sequence was previously identified as human immunodeficiency virus (HIV) protein Tat-interacting protein (19, 50). The human and mouse gene structures of GT198 were obtained by comparison of cDNA sequences and genomic DNA sequences derived from a BLAST High Throughput Genome (HTG) search. Introns and exons were identified using GT-AG consensus sequences after comparison of corresponding cDNA and genomic DNAs.
Northern analysis.
Multiple human tissue and mouse embryonic tissue RNA blots were from Clontech. GT198 probes were randomly primed with 32P-labeled rat GT198 cDNA. Northern analysis was performed according to the manufacturer’s protocol.
Immunoblotting.
Polyclonal anti-GT198 was prepared in rabbits by immunization with GST-rat GT198 fusion protein (Covance). Mouse and rat tissue lysates were from Santa Cruz Biotechnology. Human tissue lysates were from Clontech. Approximately 50 μg of proteins from tissue lysates or from selected cell nuclear extracts was loaded per lane. Human cancer cell lysates were from Santa Cruz Biotechnology. Immunoblots were probed with anti-GT198 at a dilution of 1:200 and detected with the ECL system (Amersham Pharmacia).
Immunofluorescence and immunohistochemistry.
GT198 antibodies were affinity purified by using Affi-Gel 10 according to the manufacturer’s protocol (Bio-Rad). HeLa cells were methanol fixed and double stained with affinity-purified polyclonal anti-GT198 (1:100) and monoclonal anticytokeratin (1:200; Santa Cruz) as a control. Anti-rabbit Cy3- and anti-mouse fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) were applied at a dilution of 1:200. To produce cells expressing GT198, HeLa cells were transfected with Flag-tagged GT198. Cells were methanol fixed and stained with anti-Flag antibody M2 (Kodak) at a dilution of 1:500 and anti-mouse Cy3-conjugated secondary antibody at a dilution of 1:200. Adult mouse testis sections were stained with affinity-purified anti-GT198 at a dilution of 1:100. Antibody binding was detected by biotinylated anti-rabbit immunoglobulin G (IgG) F(ab)2 antibody followed by the detecting reagents (DAKO). The section was counterstained with hematoxylin.
Recombinant protein binding assays.
The glutathione S-transferase (GST), full-length GST-GT198, and GST-GR-DBD (aa 411 to 489) were produced in Escherichia coli BL21(DE3) and purified using glutathione-Sepharose resin (Amersham Pharmacia Biotech). In vitro binding assays were performed by incubating GST resin (20 μl, 2 μg) with [35S]methionine-labeled, in vitro-translated proteins (5 μl) produced by rabbit reticulocyte lysate (Promega). Proteins were incubated at room temperature for 1 h in binding buffer (20 mM HEPES [pH 7.4], 50 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% Triton X-100, 10% glycerol, 1 mM dithiothreitol). Bound proteins were washed three times with binding buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by autoradiography.
Coimmunoprecipitation.
Both anti-GT198 and anti-GR (H300; Santa Cruz Biotechnology) antibodies were characterized by Western blotting before use. GR was to be detected by Western blotting because GR is sufficiently larger than IgG heavy chain. HeLa cells were induced overnight with 100 nM dexamethasone. Isolated nuclear extracts were filtered with a 0.65-μm spin column (Millipore) to completely remove insoluble cellular debris. Anti-GT198 or preimmune antibody (15 μl) captured by protein A beads (20-μl slurry) was washed three times and incubated with 4 ml of diluted nuclear extracts at a concentration of 300 μg/ml in binding buffer (20 mM HEPES [pH 7.4], 50 mM NaCl, 75 mM KCl, 1 mM EDTA, 0.05% Triton X-100, 10% glycerol, 1 mM dithiothreitol) overnight at 4°C. Protease inhibitors, leupeptin, aprotinin, and trypsin inhibitor (Sigma) were added in binding buffer at a concentration of 10 μg/ml. Bound protein was washed three times and detected by immunoblotting with anti-GR antibody (1:200).
Protein kinase phosphorylation assays.
In vitro phosphorylation of recombinant GST-GT198 by protein kinases was assayed using the SigmaTECT Protein Kinase Assay System from Promega with modifications. Briefly, a purified protein kinase was incubated in the supplied buffer system with [γ-32P]ATP and GST or GST-GT198 fusion protein as substrates. Labeled GST-protein beads were washed and the protein was resolved by SDS-PAGE followed by autoradiography. The amount of protein kinase used in each assay (5 to 10 U) was titrated and determined before use. Phorbol myristate acetate (100 nM) was used as a PKCα activator. Purified PKA was from Promega, purified PKC was from Stratagene, and MAPK (ERK2) was from New England Biolabs. GST alone, which is not phosphorylated by PKA and PKC and is very weakly phosphorylated by MAPK, was used as a negative control. In vivo Pi labeling was carried out in 293T cells transfected with Flag-tagged GT198. Cells were incubated in serum-free Dulbecco’s modified Eagle’s medium with 17 mg of bovine serum albumin/ml for 1 h before labeling. Pi labeling was performed with 1 mCi of H332PO4 in phosphate-free medium for 1 h. Cell lysates were immunoprecipitated with anti-Flag antibody M2 beads (Kodak), and precipitates were subjected to SDS-PAGE followed by autoradiography.
Cell culture and transient transfection.
CV-1 cells and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 0.1 μg of penicillin-streptomycin/μl in 5% CO2 at 37°C. Cells were plated in 24-well plates for 2 days before transfection. Cells were transfected using Lipofectamine reagent according to the manufacturer’s protocol (Life Technologies, Inc.). Cells were incubated in fresh media containing the indicated concentrations of ligands for 16 to 24 h after transfection. Total amounts of DNA for each well were balanced by adding vector pcDNA3 (Invitrogen). Relative luciferase activities were measured and are reported as means of triplicate transfections ± standard errors.
Nucleotide sequence accession numbers.
The rat and human GT198 sequences reported in this paper have been submitted to the GenBank/EBI Data Bank with accession numbers AF352812 and AF440240.
RESULTS
Identification of GT198 with a yeast two-hybrid system.
The Sos-Ras yeast two-hybrid system has an advantage for identifying proteins with transcriptional activity, such as nuclear receptor coactivators. The protein interactions are examined in the yeast cytoplasm rather than the nucleus, bypassing the transcriptional readout and reducing the background in the screen (4, 5, 18). We have cloned several nuclear receptor coactivators from a rat pituitary cDNA library, including TRBP, using this system (20, 23). In this way, we recently discovered that the GR, through its DBD, interacts with a protein factor originally termed GT198 with unknown function. Rat GT198 cDNA was cloned in a screen using the basic-leucine zipper (bZIP) domain of the cyclic AMP (cAMP)-inducible transcriptional factors as bait (unpublished results). bZIP domains are highly conserved within proteins, including ATF1, CREB, CREM, and ICER (47). The binding affinity of GT198 to bZIP was high in yeast, since we obtained four independent but overlapping GT198 sequences. The interaction between GT198 and GR-DBD was also strong compared to the other positive controls. When the GenBank database was searched with the rat sequence derived from the screen, a human homolog termed GT198 was found. Human GT198 is a genomic transcript with an open reading frame that maps to human chromosome 17 at the breast cancer susceptibility locus 17q12-q21 (44). The mouse homolog, a protein factor that interacts with the HIV Tat protein (TBP-IP), has also been reported (50). Although the function of GT198 was unclear, it appears that GT198 is capable of interacting with multiple DNA-binding proteins.
The full-length rat GT198 cDNA was completed by 3′ RACE. It encodes a 217-amino-acid protein that has high similarity among the rat, human, and mouse homologs (Fig. 1A). GT198 contains a heptad leucine repeat that resembles the leucine dimerization domain (1). When displayed as an α-helical wheel, the heptad repeat of leucines is aligned at the d position and hydrophobic residues are aligned at the a positions to create the classic interacting surface found in the leucine zipper protein family (Fig. 1B). Notably, amino acids are conserved among species at the interacting surface but not on the opposite side of the helical wheel, suggesting that the function of the leucine zipper domain in GT198 is conserved. Deletion of the leucine zipper region abolished GT198 dimerization (see below). A number of kinase phosphorylation sites were also predicted from the GT198 primary sequence, including those for PKA, PKC, and MAPK.
FIG. 1.
Coactivator GT198. (A) Alignment of amino acid sequences of rat GT198 with human GT198 (44) and mouse GT198 (TBP-IP) (50). Amino acid variations between species are indicated. Identical amino acids (−) are indicated. The heptad repeats of leucine and methionine residues are shaded. (B) An α-helical wheel diagram for the leucine dimerization domain of rat GT198 (aa 86 to 117). Aligned leucines are at the d position, and hydrophobic residues are at the a position. (C) Gene structure of mouse GT198 on chromosome 11. Introns are shown as lines and exons are depicted as boxes. Coding regions are shown in black and noncoding regions are in white.
Based on the human and mouse cDNA sequences, genomic information for GT198 was obtained by a BLAST search with the HTG database. The human GT198 gene is located on chromosome 17q21 and contains eight exons spanning approximately 6 kb. The mouse GT198 gene is located on chromosome 11 and contains eight exons spanning 3.2 kb (Fig. 1C).
The expression of endogenous GT198 is tissue specific.
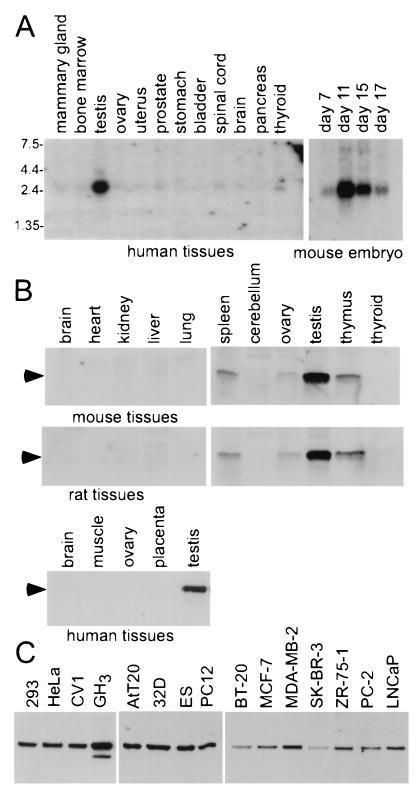
To examine the endogenous expression pattern of GT198, Northern analyses with multiple human tissue and mouse embryo blots were performed. Human GT198 mRNA was highly expressed in testis, but lower levels of signal were also detected in other adult tissues. In the mouse embryo, GT198 was up-regulated at day 11 (Fig. 2A). In order to detect the expression patterns of endogenous GT198 protein, a polyclonal anti-GT198 antibody was generated. The antibody was highly specific for GT198 when characterized with cell-expressed or in vitro-translated GT198. Both transfected and endogenous GT198 have an apparent molecular mass of 34 kDa. In both mouse and rat tissues, GT198 was highly expressed in testis and was also present in the spleen, ovary, and thymus (Fig. 2B). The signals were absent in adult mouse and rat brain, heart, kidney, liver, lung, cerebellum, and thyroid. It was also undetectable in human skeletal muscle, ovary, and placenta (Fig. 2B). These expression patterns were consistent with the report that mouse GT198 (termed TBP-IP) is highly expressed in mouse testis and lymphocytes (50). Among the cell lines examined, the pituitary cell line GH3 had higher levels of GT198 protein expression, whereas other cell lines, including mouse embryonic stem cells and human cancer cell lines, exhibited variable expression of GT198 (Fig. 2C). Several commonly used cultured cell lines, such as 293, HeLa, and CV-1 cells, also had significant amounts of protein expression, suggesting that GT198 may exist in many, or perhaps all, immortalized cell lines. Further, immunohistochemical staining with anti-GT198 antibody showed that GT198 is expressed in mouse adult testis (Fig. 3C) as well as in multiple mouse embryonic tissues (results not shown). Taken together, these data indicate that endogenous GT198 is a tissue-specific protein factor.
FIG. 2.
Endogenous expression pattern of GT198. (A) Northern analysis of mRNA isolated from human tissues and mouse embryo at different stages. Blots were probed with rat GT198 cDNA. Human tissues examined are as shown on top of the panel. Mouse embryos were from day 7 to day 17, as indicated. (B) Western analysis using polyclonal anti-GT198 antibody. Results shown are for whole-cell lysates (50 μg/lane) from mouse, rat, and human tissues. (C) Western analysis with selected cell lines. Nuclear extract (50 μg) isolated from various cell lines was used (left and middle panels). Results for samples from whole-cell lysates of human cancer cells are shown in the panel on the right. ES, embryonic stem cells.
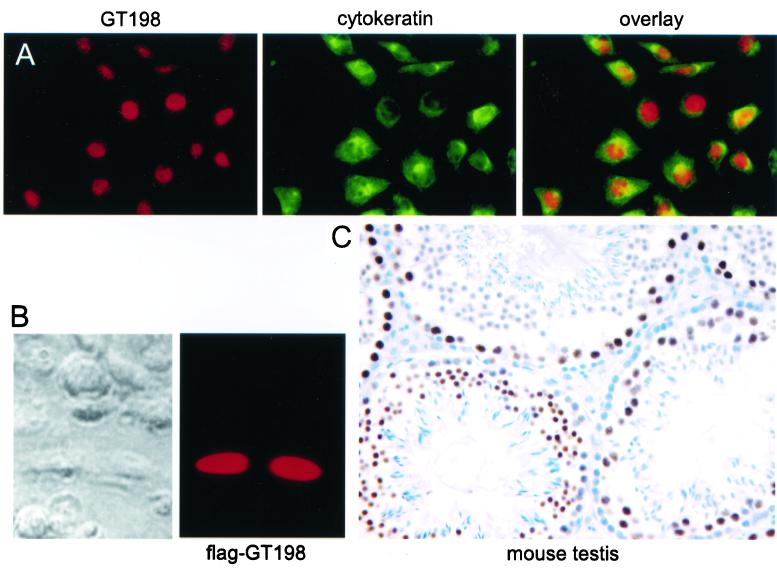
FIG. 3.
GT198 is a nuclear protein. (A) HeLa cells were methanol fixed and double stained with affinity-purified polyclonal anti-GT198 (left panel, red) and monoclonal anticytokeratin (middle panel, green), which was used as a cytoplasmic control. Secondary antibodies were anti-rabbit Cy3- and anti-mouse FITC-conjugated antibodies. An overlay of the two panels is shown on the right. (B) HeLa cells transfected with Flag-tagged GT198. Transfected cells were methanol fixed. Nuclear staining of GT198 was visualized by fluorescence with anti-Flag antibody (M2) and Cy3-conjugated secondary antibody. A phase-contrast view of the same field is shown at the left. (C) Immunohistochemical staining of the seminiferous tubules from mouse testis. The section was stained with affinity-purified anti-GT198 (1:100) and counterstained with hematoxylin.
GT198 is a nuclear protein.
Endogenous GT198 was exclusively detected in nuclear but not cytoplasmic fractions (data not shown). Immunofluorescence staining also demonstrated that GT198 was nuclear in localization. When cytokeratin was used as a control for cytoplasmic staining, endogenous GT198 was clearly observed in the nucleus of HeLa cells (Fig. 3A). The expression pattern of GT198 appeared to be cell cycle regulated. In dividing cells, GT198 was more evenly distributed within the nucleus, whereas a more punctated pattern could be detected at the other stages of the cell cycle (data not shown). The Flag-tagged GT198 was also seen exclusively in the nucleus when expressed in HeLa cells, suggesting that GT198 is a nuclear protein (Fig. 3B). Since the highest level of GT198 expression was detected in adult testes by immunoblotting (Fig. 2B), immunohistochemical staining of mouse testis was performed to determine the detailed expression pattern. Interestingly, endogenous GT198 was highly expressed only in the nucleus of spermatocytes and not in spermatogonia or spermatids (Fig. 3C). GT198 expression was also absent in Leydig cells. Although the physiological relevance of this specific expression in germ cells remains to be further elucidated, the data nevertheless strongly confirm that GT198 is a cell-specific nuclear protein.
GT198 specifically interacts with nuclear receptors via the DBD.
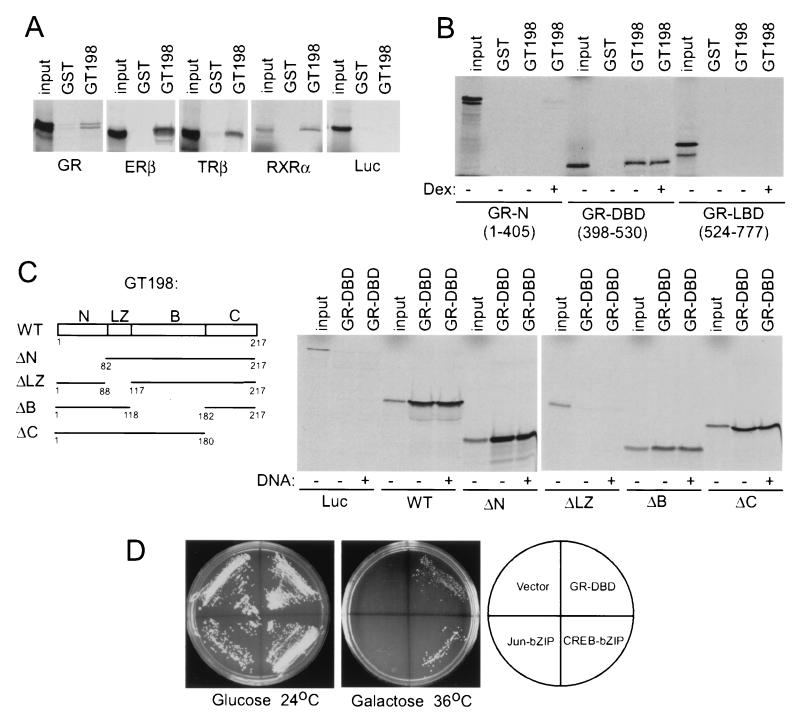
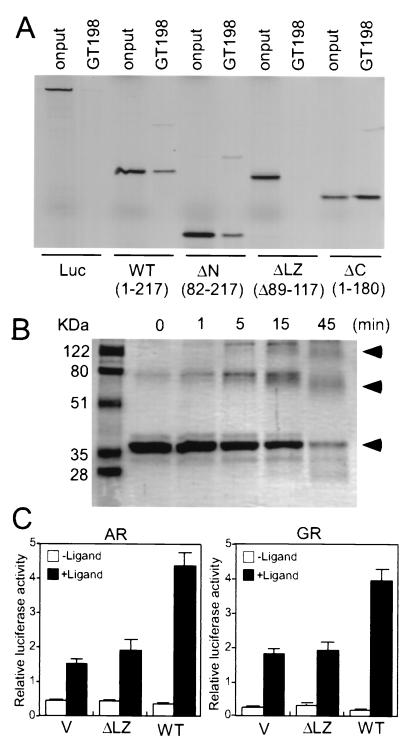
In vitro pull-down analysis was performed with recombinant GST fusion proteins and [35S]methionine-labeled in vitro-translated full-length nuclear receptors. GST-GT198, but not GST alone, interacted with nuclear receptors GR, ERβ, TRβ1, and RXRα (Fig 4A). To map the region within nuclear receptors that might be responsible for this interaction, each region in GR was individually tested. The interaction was specifically localized to the DBD but not to the LBD or N terminus of the nuclear receptor when examined with various GR fragments (Fig. 4B). This in vitro interaction was not affected by the GR ligand dexamethasone. To examine which region located in GT198 might contribute to this interaction, deletion mutants of GT198 were tested with GST-GR-DBD. Deletion of the basic region of GT198 reduced, and deletion of the leucine zipper almost abolished, the interaction between GT198 and GR-DBD (Fig. 4C). However, the GRE-containing double-stranded DNA was not required for this interaction in vitro (Fig. 4C). The above data suggest that the basic region as well as the leucine zipper region of GT198 might be responsible for its interaction with receptor DBD. Since the leucine zipper is required for GT198 dimerization (see below), the interface for binding might be located between nuclear receptor DBD and GT198 dimer rather than the leucine zipper itself. Consistent with the in vitro binding results, GT198 also interacted with the GR-DBD in yeast two-hybrid assays. The interaction of GT198 was specific for GR-DBD and CREB-bZIP but not c-Jun-bZIP, which was used as a negative control (Fig. 4D). Taken together, these data suggest that GT198 may specifically interact with nuclear receptors at the DBD. Thus, GT198 could interact with a subset of DNA-binding proteins, such as nuclear receptors and cAMP-responsive transcription factors.
FIG. 4.
GT198 interacts with DBD of nuclear receptors. (A) GST-GT198 fusion proteins were incubated with in vitro-translated, 35S-labeled full-length nuclear receptors (GR, ERβ, TRβ1, and RXRα). Bound proteins were resolved by SDS-PAGE and detected by autoradiography. GST alone was used as a control. Each nuclear receptor and luciferase (Luc) used as a negative control are indicated at the bottom. (B) The N-terminal region, DBD, and LBD of GR were in vitro translated and tested for binding GT198 in the presence (+) or absence (−) of 1 μM dexamethasone (Dex), as indicated. (C) Schematic representation of the deletion constructs of GT198 (left panel). Numbers indicate amino acids. The wild-type (WT) and deletion mutants of GT198 were in vitro translated and tested for the binding of GST-GR-DBD in the presence or absence of 30 ng of GRE-containing double-stranded DNA/ml as indicated. Input was 10%. (D) GR-DBD interacted with GT198 in a yeast two-hybrid assay. Full-length rat GT198 in pYes2 vector was cotransfected with bait plasmids, including GR-DBD, CREB-bZIP, c-Jun-bZIP, and pADNS empty vector, as indicated. Positive clones were detected at 36°C on a galactose plate. The same clones grown at 24°C on glucose plates were used as controls.
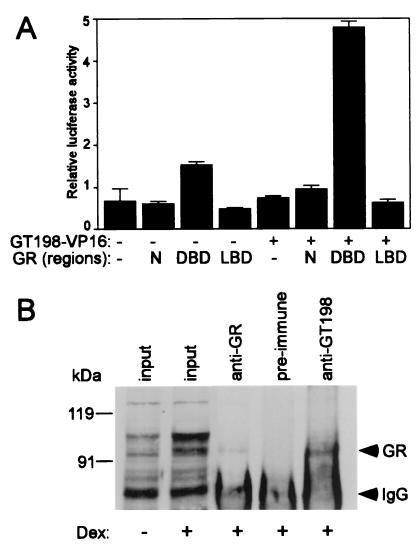
GT198 interacts with GR in vivo.
The interaction between GT198 and GR was further analyzed in CV-1 cells by using a mammalian one-hybrid assay. GT198-VP16 fusion protein stimulated transcription on a GRE reporter in the presence of GR-DBD but not the N terminus or LBD of GR, suggesting GT198 interacted with GR-DBD in cells (Fig. 5A). The N-terminal region and LBD of GR did not stimulate the reporter and served as negative controls. The DBD had a lower level of stimulation in the absence of GT198, presumably because the DBD was able to interact with an endogenous coactivator complex. However, the presence of GT198-VP16 on DBD strongly increased transactivation, whereas GT198-VP16 alone did not, suggesting GT198 interacts with the DBD and recruits VP16. To detect the interaction in vivo, a coimmunoprecipitation assay with anti-GT198 and anti-GR antibodies was also performed. The interaction between endogenous GT198 and GR could be detected in HeLa cells (Fig. 5B). When HeLa cells were treated with dexamethasone, the level of endogenous GR was up-regulated. A specific band of GR protein around 97 kDa could be detected by the anti-GR antibody. Although a higher-mobility band was also induced, it failed to be precipitated by the anti-GR antibody, indicating that it is not likely the GR protein. The GR protein was precipitated by the anti-GR antibody but not the preimmune serum. Importantly, the GR protein was also coimmunoprecipitated by the anti-GT198 antibody. These data indicate that the interactions between GT198 and GR exist in vivo, at least under the induction of dexamethasone. As hormone-stimulated signaling may regulate GT198, evaluation of its regulated interaction will be interesting for future studies. Collectively, these results show that GT198 interacts with GR through its DBD.
FIG. 5.
GT198 interacts with GR in vivo. (A) GT198 interacts with GR-DBD in a mammalian one-hybrid assay. CV-1 cells were transfected in 24-well plates with GRE-luciferase reporter (100 ng) and the following GR fragments: N (aa 1 to 405), DBD (aa 398 to 530), LBD (aa 524 to 777), or a vector control (100 ng). GT198-VP16 or control vector (100 ng) was also cotransfected. Luciferase activity was measured and is shown as the mean of triplicate transfections ± standard error. (B) Coimmunoprecipitation of endogenous GT198 and GR from HeLa cells. HeLa cells were treated with 100 nM dexamethasone (Dex) for 16 h. Nuclear extracts were isolated and incubated with anti-GT198 antibody, which was captured by protein A beads. Anti-GR antibody was used as a positive control and preimmune serum was used as a negative control. Bound GR was detected by immunoblotting with anti-GR antibody. Input of nuclear extracts (30 μg) from induced or uninduced cells is shown on the left. Arrows indicate GR and IgG.
GT198 coactivates nuclear receptor-mediated transcription.
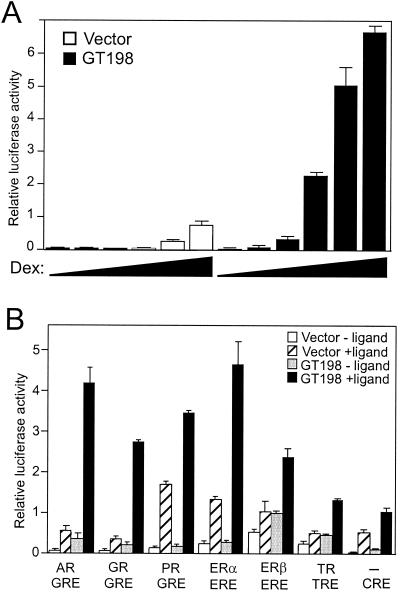
Since GT198 interacts with the DBDs of nuclear receptors, we performed transient-transfection studies in CV-1 cells to investigate the potential coactivation function of GT198 with nuclear receptors. An MMTV luciferase reporter was cotransfected along with GR and GT198. GT198 potently coactivated GR-dependent transcription of the MMTV promoter induced by dexamethasone (Fig. 6A). The level of induction was significant compared to that of the vector control. The activity of GT198 was ligand dependent, suggesting that GT198 activated the reporter through GR. In order to test individual nuclear receptors, luciferase reporters containing minimal hormone response elements were cotransfected along with each nuclear receptor. GT198 was able to activate transcription mediated by all of the tested nuclear receptors, including AR, GR, PR, ERα, ERβ, and TRβ1, in a ligand-dependent manner (Fig. 6B). GT198 also activated CRE-mediated transcription when PKA was used as an inducer (Fig. 6B). The results of the transactivation experiments were consistent with those of the binding assays (Fig. 4) and indicated that GT198 acts on nuclear receptors. Together, these data suggest that GT198 serves as a coactivator in nuclear receptor-mediated transcription.
FIG. 6.
GT198 is a nuclear receptor coactivator. (A) CV-1 cells were cotransfected in 24-well plates with MMTV luciferase reporter (100 ng), GR (10 ng), and GT198 (200 ng), or empty vector as control. Cells were grown in the presence of increasing amounts of dexamethasone (Dex) for GR (0, 10−11, 10−10, 10−9, 10−8, and 10−7 M). (B) CV-1 cells were transfected with the indicated luciferase reporters containing different minimal enhancer elements (2XGRE, 2XERE, F2 TRE, and 5XCRE) (100 ng). GT198 (200 ng), AR, GR, PR, ERα, ERβ, or TRβ1 (10 ng) and inducer plasmid containing the catalytic subunit of PKA (20 ng) for CRE were also cotransfected. For nuclear receptors, each cognate ligand (100 nM) was added to the medium 24 h after the transfection (mibolerone for AR, dexamethasone for GR, progesterone for PR, 17β-estradiol for ERα or ERβ, and T3 for TRβ1). Ligand-dependent activities are shown as the means of triplicate transfections ± standard errors.
GT198 differs in function from the coactivators TRBP, CBP, and SRC-1.
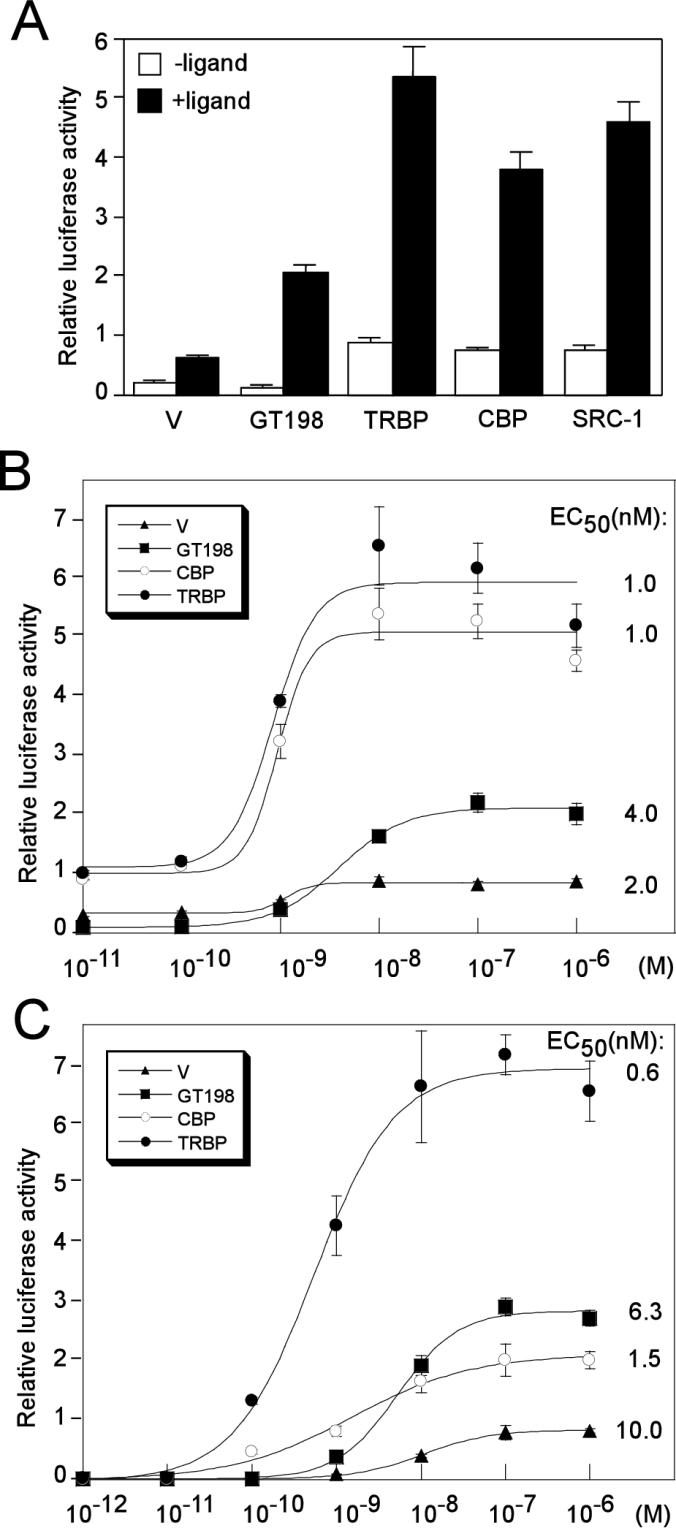
We compared GT198 with LBD-interacting coactivators such as TRBP, CBP, and SRC-1 to determine if a DBD-interacting coactivator might behave differently with respect to its role in transcriptional regulation. We observed several differences. First, coactivators like CBP, TRBP, and SRC-1 exhibited higher basal activation in the absence of ligand. This was clearly observed with the GRE in the presence of AR (Fig. 7A and B). However, GT198 did not significantly increase basal transcription. In fact, a decrease in the basal levels was observed under standard conditions in CV-1 cells. This resulted in a tighter ligand control, which appeared as a greater fold induction by the ligand (Fig. 7A and B). Second, unlike coactivators TRBP and CBP that always showed high activity in transient transfections, the potency of GT198 significantly varied among different cell types (results not shown). One possible explanation for this observation might be the cell-specific regulation of GT198, possibly involving the kinase regulation of GT198 discussed below. Third, when the MMTV promoter was studied, hormone sensitivity was significantly higher with CBP or TRBP than with GT198, especially at lower ligand concentrations (Fig. 7C). TRBP and CBP produced the left shifts of their respective 50% effective concentrations (EC50s) in ligand-dependent activation (Fig. 7B and C). In the presence of CBP or TRBP, hormone sensitivity increased in comparison to that of the vector control. In contrast, GT198 induced a right shift of the EC50 from that of TRBP and CBP. The difference in EC50s modulated by GT198 and CBP or TRBP was as great as 10-fold. Hence, GT198 might facilitate the strict hormonal control of gene activation in a manner distinct from other coactivators. Taken together, these data indicate that the mechanism(s) by which GT198 affects transcription is distinguishable from those of CBP, TRBP, and SRC-1.
FIG. 7.
Coactivator GT198 is functionally distinguishable from TRBP, CBP, and SRC-1. (A) Transcriptional activity of GT198 was compared with that of TRBP, CBP, and SRC-1 together with a vector control (V) (200 ng). CV-1 cells on 24-well plates were cotransfected with a GRE reporter (100 ng) and AR (10 ng) and treated with or without 100 nM mibolerone. (B) Transfections similar to those shown in panel A were performed with a GRE-luciferase reporter and AR, except that increasing amounts of ligand (mibolerone) were used (10−11, 10−10, 10−9, 10−8, 10−7, and 10−6 M). EC50 values are indicated at the right of each curve. Standard errors of EC50s were less than 5% (KaleidaGraph). (C) CV-1 cells were transfected as for panel B, except a MMTV-luc reporter and GR were used in the presence of indicated amounts of the ligand dexamethasone (10−12, 10−11, 10−10, 10−9, 10−8, 10−7, and 10−6 M). EC50 values are indicated on the right. Data shown are means of triplicate transfections ± standard errors. Vector control (filled triangle), GT198 (filled square), CBP (open circle), and TRBP (filled circle) are shown.
GT198 is a functional dimer.
As discussed above, GT198 contains a classic heptad leucine repeat similar to the well-characterized classic leucine dimerization domains (Fig. 1B) (1). In order to determine if the leucine domain is required for GT198 dimerization, mutants of GT198 with deletions of several regions in GT198 were tested. When in vitro-translated GT198 mutants were tested with GST-GT198 in a pull-down assay, the results clearly showed that deletion of the leucine domain, but not other regions of GT198, abolished its ability to homodimerize (Fig. 8A). In addition, glutaraldehyde cross-linking studies of the recombinant His-tagged GT198 indicated that a band with the apparent size of the predicted homodimer was generated after treatment. The dimeric and multimeric forms increased, while the monomeric form decreased with increasing time of treatment with glutaraldehyde (Fig. 8B). These data suggest that GT198 was able to form dimers in solution. Interestingly and importantly, the leucine zipper deletion of GT198 significantly reduced the binding of GT198 to GR-DBD (Fig. 4C), suggesting that the dimerization of GT198 might have functional importance with respect to nuclear receptor interaction as well as transcriptional activation. To confirm this, transient-transfection studies were performed using the leucine zipper deletion of GT198. The data suggested that GR- as well as AR-mediated transcription activation were almost impaired by the deletion of the leucine zipper (Fig. 8C). Together, these results strongly indicated that GT198 is likely a functional dimer in vivo. As many nuclear receptors act as dimers on hormone response elements, the dimerization of GT198 might be important for the action on the DBD of nuclear receptors.
FIG. 8.
GT198 is a functional dimer. (A) Deletion of its leucine zipper abolished GT198 dimerization in vitro. Recombinant GST-GT198 fusion protein was incubated with 35S-labeled GT198 deletion fragments to detect homodimerization. Bound proteins were resolved by SDS-PAGE and detected by autoradiography. Luciferase (Luc) was used as a negative control. GT198 wild-type (WT) and mutants were as follows: WT (aa 1 to 217), N-terminal deletion (ΔN; aa 82 to 217), leucine zipper domain deletion (ΔLZ; Δ89–117) and C-terminal deletion (ΔC; aa 1 to 180). (B) Cross-linking of GT198. His-tagged recombinant GT198 (0.1 μg/μl) was treated with 0.15 mM glutaraldehyde for increasing amounts of time as indicated. Proteins were resolved by SDS-PAGE and detected by Coomassie blue staining. Arrowheads indicate the monomer, dimer, and polymer of GT198. (C) CV-1 cells were cotransfected with a GRE reporter (100 ng) and AR or GR (10 ng), along with wild-type GT198 (WT) or GT198 with the deletion at leucine zipper domain (ΔLZ; Δ89–117) or a pcDNA3 vector (V) as control. Ligand-dependent luciferase activities stimulated by 100 nM mibolerone or dexamethasone are shown as means of triplicate transfections ± standard errors.
GT198 is a kinase-regulated coactivator.
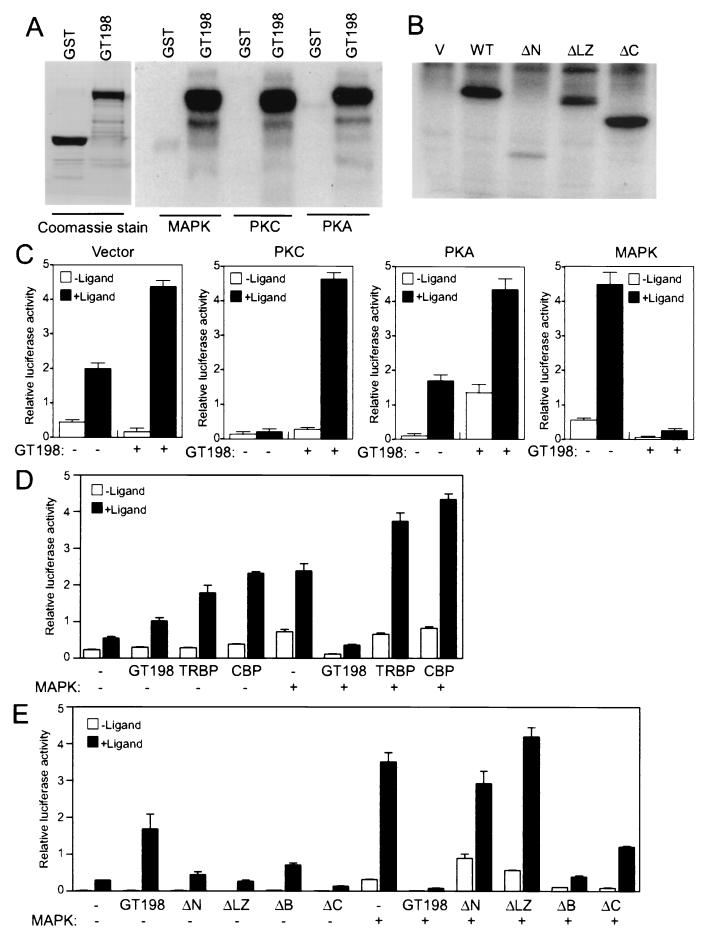
Increasing evidence suggests that nuclear receptor coactivators are regulated in vivo by signaling molecules such as protein kinases (10, 56). The primary sequence of GT198 predicts several phosphorylation sites for several protein kinases, including MAPK, PKA, and PKC. Some of the predicted sites (Thr155, Ser181, and Ser194) are conserved among human, rat, and mouse GT198, implying that phosphorylation regulation of GT198 might be conserved and functionally relevant. In a kinase-phosphorylation study, recombinant GST-GT198 could be markedly phosphorylated in vitro by PKA, PKC, and MAPK (Fig. 9A). The phosphorylation was specific for GT198, since the control protein GST was not phosphorylated. Multiple phosphorylation sites were detected for each kinase when examined using GT198 deletion fragments (data not shown). In vivo Pi labeling also confirmed that GT198 was heavily phosphorylated in cells (Fig. 9B). To examine if phosphorylation by kinases affects the transcriptional capability of GT198, transfection studies were performed by coexpressing each kinase. As shown in Fig. 9C, in the vector control without stimulation of kinase, transcription of the GRE reporter was moderately up-regulated by GT198 in the presence of ligand. Stimulation with PKC significantly promoted ligand-dependent activation by GT198 compared to that of the control without cotransfection of GT198. The basal levels increased rather than decreased with PKC induction and increased remarkably with PKA induction. However, MAPK stimulation reversed the action of GT198, with transcription strongly repressed by GT198 in CV-1 cells (Fig. 9C). The suppressive effects of GT198 were also detected with other nuclear receptors such as ER, GR, PR, and TR when coexpressing MAPK (data not shown). The MAPK effect in CV-1 cells was specific to GT198 but not to the other coactivators such as TRBP or CBP (Fig. 9D). In fact, MAPK stimulation in CV-1 cells was also observed in the vector control. However, GT198 exerted the repressive effect under the same MAPK stimulation, suggesting that MAPK regulation of GT198 was specific. Similar effects for GT198 were observed in HeLa cells, as wild-type GT198 strongly activated transcription in the absence of MAPK induction but repressed transcription in the presence of MAPK induction (Fig. 9E). Interestingly, each deletion mutant of GT198 behaved differently with and without MAPK stimulation (Fig. 9E). In addition, while wild-type GT198 behaved similarly in CV-1 and HeLa cells, the activities of each mutant varied (data not shown), indicating that regulation of GT198 by phosphorylation, which was in part reflected by the activities of mutants, might be cell specific and complex in vivo. Nevertheless, deletion of each region of GT198 including the leucine zipper partially impaired full GT198 function (Fig. 9E). Taken together, the data indicate that protein kinase phosphorylation might trigger multiple signaling pathways that, in turn, directly or indirectly regulate GT198 in vivo. The regulation of GT198 might then be highly cell specific, inasmuch as phosphorylation by each kinase is cell type dependent. These data suggest that kinase stimulation may affect GT198 coactivation in vivo.
FIG. 9.
GT198 is phosphorylated and regulated by PKA, PKC, and MAPK. (A) GST-GT198 was phosphorylated in vitro by PKA, PKC, and MAPK as described in Materials and Methods. After washing, phosphorylated GST-GT198 proteins were resolved by SDS-PAGE and detected by autoradiography. GST alone served as a negative control. Coomassie staining of GT198 and GST proteins, shown in the left panel, was to confirm that equal amounts of protein were used. (B) 293 cells were transfected with Flag-tagged GT198 and its deletion mutants. Abbreviations are as shown in the legend to Fig. 8A. Cells were Pi labeled, and GT198 proteins were immunoprecipitated with anti-Flag M2 antibody beads (Kodak). After washing, the bound protein was separated by SDS-PAGE and visualized by autoradiography. (C) Kinase stimulation regulates GT198 coactivation function. The effects of kinases on GT198 were analyzed in CV-1 cells. Cells were cotransfected in 24-well plates with GRE-luciferase reporter (100 ng) and AR (10 ng). Coactivator GT198 or a vector control (200 ng) was cotransfected with PKA, PKC, or MAPK (20 ng) or a vector control. Cells were treated with or without 100 nM mibolerone. (D) MAPK regulation of GT198 coactivation is specific. The effects of MAPK on GT198 were analyzed in CV-1 cells with GRE-luciferase reporter (100 ng) and AR (10 ng). Coactivator GT198, TRBP, CBP, or vector control (200 ng) was cotransfected with or without MAPK (20 ng) and treated with or without the 100 nM mibolerone. (E) HeLa cells were transfected as CV-1 cells were, except that wild-type and deletion mutants of GT198 (200 ng) were used. Data shown are means of triplicate transfections ± standard errors.
DISCUSSION
In this study, we described the identification and characterization of the coactivator, GT198, that interacts with the DBDs of nuclear receptors. The DBDs of nuclear receptors are highly conserved in structure as well as function (31). Increasing evidence suggests that DNA binding in vivo is a regulated event that involves multiple protein cofactors. An example is the coactivator SNURF, a RING finger protein that stimulates AR-mediated transcription via AR-DBD (34). Although GT198 does not possess a RING finger but instead has a leucine zipper, GT198 displays several characteristics similar to those of the coactivator SNURF. Like SNURF, GT198 stimulates other transcriptional factors in addition to the nuclear receptors. The GT198 protein expression pattern was also correlated with that of SNURF, in that GT198 is highly expressed in rapidly dividing cells, such as germ cells of testis (50), thymus, spleen, embryo, cancer cells, and cultured cell lines (Fig. 2). Similar to SNURF, GT198 by itself did not bind DNA with high affinity (data not shown), but it interacted with the DBDs of nuclear receptors and modulated their transcriptional activities. SNURF has also been suggested to interact with the TATA-binding protein and to stimulate Sp1-dependent transcription (39). It is currently unknown whether GT198 could have a similar function. However, it is noteworthy that GT198 regulates basal activity under the influence of kinase phosphorylation in transient transfections (Fig. 9C).
GT198 is a structured protein with high α-helical content (Jpred/PHD [http://jura.ebi.ac.uk:8888/]). Its structured nature might make it suitable for protein interaction with multiple surfaces. Deletion of limited regions of GT198 only partially impaired or altered its transactivation properties (Fig. 9E), indicating that the entire protein, which is small in size, might be required for its integrity and full function. The leucine zipper is a powerful dimerization domain. Deletion of the leucine dimerization domain of GT198 dramatically reduced its binding to nuclear receptors (Fig. 4B). It is currently unknown if the dimerization is regulated in vivo. However, the dimerization of GT198 is required for the full coactivation function (Fig. 8C and 9E). GT198 also contains a basic region rich in lysine and arginine residues. Deletion of this region reduced the binding between GT198 and GR (Fig. 4B). This basic region might facilitate the binding of negatively charged DNA. Although bacterially expressed GT198 did not bind to the DNA by itself, the binding of DNA in vivo may require cooperation with other DNA-binding factors. Gel-shift analysis with His-tagged GT198 suggested that GT198 promoted CREB binding to CRE but decreased ERβ binding to an ERE (data not shown). Since GT198 is a highly phosphorylated protein in vivo (Fig. 9), recombinant dephosphorylated GT198 protein might not completely reflect its true nature in vivo. Nevertheless, the effects of GT198 on DBD or bZIP binding to DNA were pronounced.
The in vivo function of GT198 is likely regulated in part through kinase phosphorylation. Furthermore, the regulation might be cell type specific. In transient-transfection studies, the potency of coactivation by GT198 varied among cell types and was also affected by cell density. Immunofluorescence staining in HeLa cells showed that endogenous GT198 was more evenly spread in the nuclei of dividing cells and was distributed in punctated patterns in confluent cells. This might indicate that GT198 is differentially regulated in the cell cycle. Transfections in HeLa cells but not 293 cells generally produced more consistent results. With MAPK stimulation in CV-1 cells, the GT198 effect was completely reversed, in contradistinction to PKC and PKA stimulation (Fig. 9). Thus, the in vivo functions of coactivator GT198, including its binding of nuclear receptors and its transcriptional activities, might be regulated. Similarly, regulated function is evident for other coactivators. For example, the phosphorylation of p300 by PKC at Ser89 inhibits the transactivation potential of p300 (56), and the phosphorylation of SRC-1 by MAPK via PKA signaling modulates SRC-1 function (10). The modification of GT198 by phosphorylation could alter its activities in a tissue-specific and hormone-regulated manner, in spite of its presence in a number of tissues and cell lines.
The Sos-Ras yeast two-hybrid system has proven to be a reliable and powerful method for the detection of protein-protein interactions between transcriptionally active molecules (4, 5, 18). In addition to GT198, we have cloned the coactivators TRBP (23) and CoAA (20) from a rat pituitary cDNA library using this system. Previously, six independent cDNA isolates encoding four overlapping GT198 sequences were identified when CREB-bZIP was used as bait (data not shown). GT198 specifically interacted with CREB-bZIP but not c-Jun-bZIP in yeast (Fig. 4D). Although the functional importance between GT198 and CREB is not clear, the present studies suggest that GT198 may regulate the transcriptional activities mediated by both nuclear receptors and cAMP-responsive transcription factors. The bZIP, a basic region followed by a heptad leucine repeat, is a highly conserved DBD found in a family of cAMP-responsive transcription factors that includes CREB, ATF1, CREM, and ICER (47). They are the key regulators in many important physiological events, including development, endocrine system function, and brain function (46, 47). CREM is highly expressed in spermatocytes and plays a key role in germ cell differentiation (46). The ability of GT198 to interact with bZIP proteins suggests that GT198 might regulate CREM function. Homo- and heterodimerization among CREB and CREMα depend on their bZIP domains for DNA binding (14, 49). Similarly, the DBDs of nuclear receptors are also important for dimerization and the selectivity of these receptors on each hormone response element (57). GT198 might regulate both DBD and bZIP domains in the coordination of their functions on complex promoters in vivo. An example of the coordinate regulation of glucocorticoid stimulation and cAMP signaling is the phosphoenolpyruvate carboxykinase gene promoter, wherein the glucocorticoid response regulated by PKA correlates with the differentiation states of hepatoma FGC4 cells (2). In addition, full activation of the phosphoenolpyruvate carboxykinase gene promoter requires the coordination of multiple response elements present within it (13). Since GT198 can be phosphorylated by multiple kinases, including PKA, and appears to be regulated by hormonal signaling, it is possible that GT198 is an integrating coregulator for both steroid hormones and the cAMP pathways which, in turn, coordinate cell-specific gene activation in vivo.
The human GT198 gene was originally identified through the mapping of the breast cancer susceptibility gene BRCA1 locus at chromosome 17q12-q21 (44). Germ line mutations in the breast cancer susceptibility genes BRCA1 and BRCA2 have been shown to result in human hereditary breast and ovarian cancers (52). In previous mapping of additional genes that might also be involved in breast and ovarian cancers, human GT198 was found to be one of the transcripts within the locus that is located within 320 kb of the BRCA1 gene (44). The GT198 transcript was found to have a tissue-specific expression pattern (44). Interestingly, the expression pattern of GT198 has some similarities to BRCA1 and BRCA2 (42). GT198 mRNA is abundant in adult testis and in the developing mouse embryo (Fig. 2A). GT198 protein is expressed at higher levels in several tissues, including testis, thymus, and spleen (Fig. 2B). In addition, it is also expressed in several human cancer cells (Fig. 2C). The mouse GT198 homolog TBP-IP, which has been identified as an interacting protein for HIV Tat, is present in testis and lymphocytes (50). On the other hand, BRCA1 is a nuclear receptor coactivator that has been shown to affect the ER-responsive promoter (52) and to stimulate AR-mediated transcription in prostate cancer (55). These observations suggest that BRCA1, which is a RING finger protein, functions as a nuclear receptor coactivator and modulates nuclear receptor-mediated transcription. It is currently unclear if GT198 could have functional overlap with BRCA1 or if GT198 could be another candidate breast cancer susceptibility gene. Of note, the amplification or abnormality of genes in cancer is frequently associated with encoding of proteins that have a transactivation function. These include AIB1/SRC-3 (3), AIB3/TRBP (25), MOZ-TIF2 (12), and MOZ-CBP as well as BRCA1 (52). Thus, it will be important for future studies to determine if the coactivator GT198 is involved in human cancers.
In conclusion, we have cloned and characterized a novel tissue-specific coactivator GT198 that stimulates transcription through the interaction of DBDs of nuclear receptors and other transcriptional factors. The action of GT198 in vivo might be dynamic and modulated by protein kinase phosphorylation. A better understanding of GT198 function may provide more insight into the regulation of tissue-specific gene activation mediated by nuclear receptors.
Acknowledgments
We thank Marc Montminy for CREB-related studies; Ami Aronheim for the yeast two-hybrid system; Elizabeth Galbreath and Linda Schirtzinger for immunohistochemistry; Jacques Simard for the human GT198 cDNA; and Mary Makowske for the MMTV and PKC plasmids. We also acknowledge Thomas Burris for helpful discussions and Toshiharu Iwasaki, Christine Reyes, Keith Stayrook, and Xin Bu for their technical assistance.
This work was supported by the Lilly Postdoctoral Research Fellowship Program.
REFERENCES
- 1.Albert, T. 1992. Structure of the leucine zipper. Curr. Opin. Genet. Dev. 2:205–210. [DOI] [PubMed] [Google Scholar]
- 2.Angrand, P.-O., C. Coffinier, and M. C. Weiss. 1994. Response of the phosphoenolpyruvate carboxykinase gene to glucocorticoids depends on the integrity of the cAMP pathway. Cell Growth Differ. 5:957–966. [PubMed] [Google Scholar]
- 3.Anzick, S. L., J. Kononen, R. L. Walker, D. O. Azorsa, M. M. Tanner, X. Y. Guan, G. Sauter, O. P. Kallioniemi, J. M. Trent, and P. S. Meltzer. 1997. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965–968. [DOI] [PubMed] [Google Scholar]
- 4.Aronheim, A. 2000. Protein recruitment systems for the analysis of protein-protein interactions. Biochem. Pharmacol. 60:1009–1013. [DOI] [PubMed] [Google Scholar]
- 5.Aronheim, A., E. Zandi, H. Hennemann, S. J. Elledge, and M. Karin. 1997. Isolation of an AP-1 repressor by a novel method for detecting protein-protein interactions. Mol. Cell. Biol. 17:3094–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beato, M., P. Herrlich, and G. Schutz. 1995. Steroid hormone receptors: many actors in search for a plot. Cell 83:851–857. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi, M. E., and M. Beltrame. 1998. Flexing DNA: HMG-box proteins and their partners. Am. J. Hum. Genet. 63:1573–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boonyaratanakornkit, V., V. Melvin, P. Prendergast, M. Altmann, L. Ronfani, M. E. Bianchi, L. Taraseviciene, S. K. Nordeen, E. A. Allegretto, and D. P. Edwards. 1998. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol. Cell. Biol. 18:4471–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourguet, W., P. Germain, and H. Gronemeyer. 2000. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol. Sci. 21:381–388. [DOI] [PubMed] [Google Scholar]
- 10.Bowan, B. G., N. Garrison, N. L. Weigel, and B. W. O’Malley. 2000. 8-Bromo-cyclic AMP induces phosphorylation of two sites in SRC-1 that facilitate ligand-independent activation of the chicken progesterone receptor and are critical for functional cooperation between SRC-1 and CREB binding protein. Mol. Cell. Biol. 20:8720–8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caira, F., P. Antonson, M. Pelto-Huikko, E. Treuter, and J. Å. Gustafsson. 2000. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. J. Biol. Chem. 275:5308–5317. [DOI] [PubMed] [Google Scholar]
- 12.Carapeti, M., R. C. Aguiar, J. M. Goldman, and N. C. Cross. 1998. A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood 91:3127–3133. [PubMed] [Google Scholar]
- 13.Fass, D. M., J. C. Craig, S. Impey, and R. H. Goodman. 2001. Cooperative mechanism of transcriptional activation by a cyclic AMP-response element modulator α mutant containing a motif for constitutive binding to CREB-binding protein. J. Biol. Chem. 276:2992–2997. [DOI] [PubMed] [Google Scholar]
- 14.Foulkes, N. S., E. Borrelli, and P. Sassone-Corsi. 1991. CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcript. Cell 64:739–749. [DOI] [PubMed] [Google Scholar]
- 15.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in the transcriptional functions of nuclear receptors. Genes Dev. 14:121–141. [PubMed] [Google Scholar]
- 16.Grosschedl, R., K. Giese, and J. Pagel. 1994. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 10:94–100. [DOI] [PubMed] [Google Scholar]
- 17.Guvakova, M. A., and E. Surmacz. 1997. Tamoxifen interferes with the insulin-like growth factor I receptor (IGF-IR) signaling pathway in breast cancer cells. Cancer Res. 57:2606–2610. [PubMed] [Google Scholar]
- 18.Hubsman, M., G. Yudkovsky, and A. Aronheim. 2001. A novel approach for the identification of protein-protein interaction with integral membrane proteins. Nucleic Acids Res. 29:E18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ijichi, H., T. Tanaka, T. Nakamura, H. Yagi, A. Hakuba, and M. Sato. 2000. Molecular cloning and characterization of a human homologue of TBP-IP, a BRCA1 locus-related gene. Gene 248:99–107. [DOI] [PubMed] [Google Scholar]
- 20.Iwasaki, T., W. W. Chin, and L. Ko. 2001. Identification and characterization of a RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J. Biol. Chem. 276:33375–33383. [DOI] [PubMed] [Google Scholar]
- 21.Janne, O. A., A. Moilanen, H. Poukka, N. Rouleau, U. Karvonen, N. Kotaja, M. Hakli, and J. J. Palvimo. 2000. Androgen-receptor-interacting nuclear proteins. Biochem. Soc. Trans. 28:401–405. [PubMed] [Google Scholar]
- 22.Knutti, D., A. Kaul, and A. Kralli. 2000. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol. Cell. Biol. 20:2411–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ko, L., G. R. Cardona, and W. W. Chin. 2000. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. USA 97:6212–6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger, R. G. Brennan, S. G. Roberts, M. R. Green, and R. H. Goodman. 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223–226. [DOI] [PubMed] [Google Scholar]
- 25.Lee, S. K., S. L. Anzick, J. E. Choi, L. Bubendorf, X. Y. Guan, Y. K. Jung, O. P. Kallioniemi, J. Kononen, J. M. Trent, D. Azorsa, B. H. Jhun, J. H. Cheong, Y. Lee, P. S. Meltzer, and J. W. Lee. 1999. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J. Biol. Chem. 274:34283–34293. [DOI] [PubMed] [Google Scholar]
- 26.Leo, C., and J. D. Chen. 2000. The SRC family of nuclear receptor coactivators. Gene 245:1–11. [DOI] [PubMed] [Google Scholar]
- 27.Lin, H., A. Shih, F. B. Davis, and P. J. Davis. 1999. Thyroid hormone promotes the phosphorylation of STAT3 and potentiates the action of epidermal growth factor in cultured cells. Biochem. J. 338:427–432. [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, Y., A. Takeshita, T. Nagaya, A. Baniahmad, W. W. Chin, and P. M. Yen. 1998. An inhibitory region of the DNA-binding domain of thyroid hormone receptor blocks hormone-dependent transactivation. Mol. Endocrinol. 12:34–44. [DOI] [PubMed] [Google Scholar]
- 29.Lobenhofer, E. K., G. Huper, J. D. Iglehart, and J. R. Marks. 2000. Inhibition of mitogen-activated protein kinase and phosphatidylinositol 3-kinase activity in MCF-7 cells prevents estrogen-induced mitogenesis. Cell Growth Differ. 11:99–110. [PubMed] [Google Scholar]
- 30.Mahajan, M. A., and H. H. Samuels. 2000. A new family of nuclear receptor coregulators that integrates nuclear receptor signaling through CREB-binding protein. Mol. Cell. Biol. 20:5048–5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangelsdrof, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathur, M., P. W. Tucker, and H. H. Samuels. 2001. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 21:2298–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna, N. J., R. B. Lanz, and B. W. O’Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev. 20:321–344. [DOI] [PubMed] [Google Scholar]
- 34.Moilanen, A.-M., H. Poukka, U. Karvonen, M. Hakli, O. A. Janne, and J. J. Palvimo. 1998. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol. Cell. Biol. 18:5128–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monsalve, M., Z. Wu, G. Adelmant, P. Puigserver, M. Fan, and B. M. Spiegelman. 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell 6:307–316. [DOI] [PubMed] [Google Scholar]
- 36.Muller, J. M., U. Isele, E. Metzger, A. Rempel, M. Moser, A. Pscherer, T. Breyer, C. Holubarsch, R. Buettner, and R. Schule. 2000. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 19:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onate, S. A., S. Y. Tsai, M. J. Tsai, and B. W. O’Malley. 1995. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 270:1354–1357. [DOI] [PubMed] [Google Scholar]
- 38.Piekielko, A., A. Drung, P. Rogalla, R. Schwanbeck, T. Heyduk, M. Gerharz, J. Bullerdiek, and J. R. Wisniewski. 2001. Distinct organization of DNA complexes of various HMGI/Y family proteins and their modulation upon mitotic phosphorylation. J. Biol. Chem. 276:1984–1992. [DOI] [PubMed] [Google Scholar]
- 39.Poukka, H., P. Aarnisalo, H. Santti, O. A. Janne, and J. J. Palvimo. 2000. Coregulator small nuclear RING finger protein (SNURF) enhances Sp-1 and steroid receptor-mediated transcription by different mechanisms. J. Biol. Chem. 275:571–579. [DOI] [PubMed] [Google Scholar]
- 40.Poukka, H., U. Karvonen, N. Yoshikawa, H. Tanaka, and J. J. Palvimo. 2000. The RING finger protein SNURF modulates nuclear trafficking of the androgen receptor. J. Cell Sci. 113:2991–3001. [DOI] [PubMed] [Google Scholar]
- 41.Puigserver, P., Z. Wu, C. W. Park, R. Graves, M. Wright, and B. M. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 20:829–839. [DOI] [PubMed] [Google Scholar]
- 42.Rajan, J. V., S. T. Marguis, H. P. Gardner, and L. A. Chodosh. 1997. Developmental expression of Brca2 colocalizes with Brca1 and is associated with proliferation and differentiation in multiple tissues. Dev. Biol. 184:385–401. [DOI] [PubMed] [Google Scholar]
- 43.Romine, L. E., J. R. Wood, L. A. Lamia, P. Prenfergast, D. P. Edwards, and A. M. Nardulli. 1998. The high mobility group protein 1 enhances binding of the estrogen receptor DNA binding domain to the estrogen response element. Mol. Endocrinol. 12:664–674. [DOI] [PubMed] [Google Scholar]
- 44.Rommens, J. M., F. Durocher, J. McArthur, P. Tonin, J.-F. LeBlanc, T. Allen, C. Samson, L. Ferri, S. Narod, K. Morgan, and J. Simard. 1995. Generation of a transcription map at the HSD17B locus centromeric to BRCA1 at 17q21. Genomics 28:530–542. [DOI] [PubMed] [Google Scholar]
- 45.Sartorius, C. A., G. S. Takimoto, J. K. Richer, L. Tung, and K. B. Horwitz. 2000. Association of the Ku autoantigen/DNA-dependent protein kinase homoenzyme and poly(ADP-ribose) polymerase with the DNA binding domain of progesterone receptors. J. Mol. Endocrinol. 24:165–182. [DOI] [PubMed] [Google Scholar]
- 46.Sassone-Corsi, P. 1998. Regulating the balance between differentiation and apoptosis: role of CREM in the male germ cells. J. Mol. Med. 76:811–817. [DOI] [PubMed] [Google Scholar]
- 47.Sassone-Corsi, P. 2000. Transcriptional regulation by cyclic AMP-responsive factors. Prog. Nucleic Acid Res. Mol. Biol. 64:343–369. [DOI] [PubMed] [Google Scholar]
- 48.Sauve, F., L. D. B. McBroom, J. Gallant, A. N. Moraitis, F. Labrie, and V. Giguere. 2001. CIA, a novel estrogen receptor coactivator with a bifunctional nuclear receptor interacting determinant. Mol. Cell. Biol. 21:343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schumacher, M. A., R. H. Goodman, and R. G. Brennan. 2000. The structure of a CREB bZIP-somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J. Biol. Chem. 275:35242–35247. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka, T., T. Nakamura, H. Takagi, and M. Sato. 1997. Molecular cloning and characterization of a novel TBP-1 interacting protein (TBP-IP): enhancement of TBP-1 action on Tat by TBP-IP. Biochem. Biophys. Res. Commun. 239:176–181. [DOI] [PubMed] [Google Scholar]
- 51.Tao, Y., C. Williams-Skipp, and R. I. Scheinman. 2001. Mapping of glucocorticoid receptor DNA binding domain surfaces contributing to transrepression of NF-κB and induction of apoptosis. J. Biol. Chem. 276:2329–2332. [DOI] [PubMed] [Google Scholar]
- 52.Welsch, P. L., K. N. Owens, and M. C. King. 2000. Insights into the functions of BRCA1 and BRCA2. Trends Genet. 16:69–74. [DOI] [PubMed] [Google Scholar]
- 53.Westin, S., M. G. Rosenfeld, and C. K. Glass. 2000. Nuclear receptor coactivators. Adv. Pharmacol. 47:89–112. [DOI] [PubMed] [Google Scholar]
- 54.Xu, J., L. Liao, G. Ning, H. Yoshida-Komiya, C. Deng, and B. W. O’Malley. 2000. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl. Acad. Sci. USA 97:6379–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeh, S., Y. Hu, M. Rahman, H. Lin, C. Hsu, H. Ting, H. Kang, and C. Chang. 2000. Increase of androgen-induced cell death and androgen receptor transactivation by BRCA1 in prostate cancer cells. Proc. Natl. Acad. Sci. USA 97:11256–11261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan, L. W., and J. E. Gambee. 2000. Phosphorylation of p300 at serine 89 by protein kinase C. J. Biol. Chem. 275:40946–40951. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, J., and M. A. Lazar. 2000. The mechanism of action of thyroid hormones. Annu. Rev. Physiol. 62:439–466. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, Y., L. Kan, C. Qi, S. Yashpal, Y. S. Kanwar, V. Anjana, A. V. Yeldandi, M. Sambasiva Rao, and J. K. Reddy. 2000. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J. Biol. Chem. 275:13510–13516. [DOI] [PubMed] [Google Scholar]