Abstract
RecQ DNA helicases, including yeast Sgs1p and the human Werner and Bloom syndrome proteins, participate in telomere biology, but the underlying mechanisms are not fully understood. Here, we explore the protein sequences and genetic interactors of Sgs1p that function to slow the senescence of telomerase (tlc1) mutants. We find that the S-phase checkpoint function of Sgs1p is dispensable for preventing rapid senescence, but that Sgs1p sequences required for homologous recombination, including the helicase domain and topoisomerase III interaction domain, are essential. sgs1 and rad52 mutations are epistatic during senescence, indicating that Sgs1p participates in a RAD52-dependent recombinational pathway of telomere maintenance. Several mutations that are synthetically lethal with sgs1 mutation and which individually lead to genome instability, including mus81, srs2, rrm3, slx1 and top1, do not speed the senescence of tlc1 mutants, indicating that the rapid senescence of sgs1 tlc1 mutants is not caused by generic genome instability. However, mutations in SLX5 or SLX8, which encode proteins that function together in a complex that is required for viability in sgs1 mutants, do speed the senescence of tlc1 mutants. These observations further define roles for RecQ helicases and related proteins in telomere maintenance.
INTRODUCTION
Saccharomyces cerevisiae Sgs1p is a member of the RecQ family of 3′–5′ DNA helicases (1–4). Human RecQ helicases include WRN, BLM and RTS, which are deficient in the Werner, Bloom and Rothmund-Thomson syndromes, respectively (5–7). These are recessive disorders characterized by genome instability, cancer predisposition and—particularly in the case of Werner syndrome—by premature features of aging (8–11). RecQ-family proteins maintain the genome through several mechanisms (3,4,12). For example, Sgs1p stabilizes stalled replication forks and facilitates the proper resumption of DNA synthesis from stalled forks (13–19). Sgs1p inhibits crossover events during homologous recombination (HR) (20), and certain phenotypes in sgs1 mutants can be suppressed by deletion of genes in the RAD52 epistasis group, indicating that Sgs1p normally prevents inappropriate recombination or facilitates resolution of recombination intermediates (21,22). Sgs1p, similar to WRN and BLM, can unwind a variety of DNA structures that may be important during replication or recombination, including replication fork-like substrates, Holliday junctions and G-quadruplexes (23–27). Sgs1p also functions in parallel with Rad24p to ensure a complete S-phase checkpoint response to DNA damage (28). Failure of these activities in sgs1 mutants leads to aberrant recombination in repeated sequences, chromosome loss, gross chromosomal rearrangements, unequal sister chromatid exchange, sensitivity to methyl methanesulfonate (MMS) and hydroxyurea (HU), as well as to defects in ribosomal DNA replication and to the premature cessation of budding by yeast mother cells (1,13,15,29–32).
Sgs1p also participates in telomere function. Yeast telomeres are maintained naturally in part through the actions of telomerase, which is expressed constitutively (33,34). Telomerase can be inactivated genetically, for example by deleting TLC1, the gene encoding the RNA template component of the enzyme (35). Telomeres shorten with cell division in tlc1 mutants and this leads eventually to induction of a DNA damage response and G2/M cell cycle arrest, a process termed senescence (33,36–38). Rare cells escape senescence and these survivors use RAD52-dependent recombination mechanisms to maintain their telomeres (39–41). There are two such survivor mechanisms, type I involving the amplification primarily of subtelomeric Y′ elements along with telomere repeat DNA and type II involving primarily the amplification of the telomere repeat DNA alone. Although deletion of SGS1 does not affect steady state telomere length in cells containing telomerase, mutants lacking both SGS1 and telomerase senesce more rapidly than mutants lacking telomerase alone, and only type I survivors emerge in the absence of SGS1 (29,36,42,43). Thus, Sgs1p slows senescence and is required for initiation of the type II survivor recombination mechanism. Recent findings also support a role for the mammalian WRN and BLM proteins in telomere function. For example, each protein interacts physically and functionally with the telomere chromatin protein TRF2 (44,45), which itself plays a critical role in maintaining telomeres in a protected or ‘capped’ state (46). The capacity of purified TRF2 to catalyze T-loop assembly is thought to be related to this capping function (47), and therefore demonstrations that the WRN helicase and exonuclease activities process telomeric T-loop structures in vitro support the idea that WRN and TRF2 cooperate to maintain telomeres (48,49). Moreover, Werner mutant cells suffer from an elevated level of S-phase-dependent telomere loss (50,51). Furthermore, BLM overexpression lengthens telomeres in cells that use recombination to maintain their telomeres (52), and Wrn and Blm mutations each synergize with telomerase (Terc) mutation to accelerate telomere dysfunction and cause pathology in mice (53,54). Our yeast model provides a relatively simple system in which to probe mechanisms by which RecQ helicases participate in telomere function.
The rapid senescence of tlc1 sgs1 mutants is due, at least in part, to a synergistic interaction between the mutations that renders the cells more prone to arrest at a given average extent of telomere shortening (36). This observation might be explained by a role for Sgs1p in processes that are parallel to or downstream from telomere defects and thus affect the sensitivity of cells to telomere dysfunction. One such possibility is that rapid senescence might result from an elevated level of global genome instability caused by sgs1 mutation, which could sum with telomere dysfunction to yield an overall DNA damage signal that leads to premature cell cycle arrest. A second such possibility is that rapid senescence might instead be due to loss of SGS1-dependent intra-S-phase checkpoint function, which may in turn preclude normal repair of telomere damage and thus cause premature telomere dysfunction. Alternatively, the premature arrest of tlc1 sgs1 mutants at longer mean telomere lengths might reflect a direct role for Sgs1p in maintaining telomere integrity, perhaps via recombinational repair of shortened telomeres. Its requirement for type II survivor formation is consistent with a role for Sgs1p in recombination involving the G-rich telomere sequences (36,42,43). One dysfunctional telomere is sufficient to cause cell cycle arrest (55), and so only a rare telomere might require recombinational repair, thus explaining the premature arrest of tlc1 sgs1 cells at longer mean telomere lengths. Such defects could be repaired by telomerase, when present, and obviate a requirement for Sgs1p, thus explaining the normal telomere lengths and absence of G2/M arrest in TLC1 sgs1 cells (36).
Here, we present evidence that neither loss of the SGS1-dependent intra-S-phase checkpoint nor increased global genome instability explains the rapid senescence of tlc1 sgs1 mutants. Our findings rather support a role for Sgs1p in the recombinational repair of telomeres.
MATERIALS AND METHODS
Yeast strains
All yeasts were grown at 30°C using standard yeast media and techniques (56). All strains are described in Table 1 and were isogenic derivatives of JKM111 (57). The sgs1 deletion in YBJ133 is a null allele (58) but retains the first 481 nt of the open reading frame (ORF) which might generate an N-terminal Sgs1p fragment that, although inert itself, could conceivably function in trans with other Sgs1p derivatives (59). To avoid such potential trans complementation from the various SGS1 derivatives that were examined, the entire SGS1 ORF except for the start and stop codons was deleted by PCR-mediated gene disruption to generate YMA75b. rad52::LEU2 was generated with pSM20 (58). The top1, mus81, slx1, slx5, slx8, est2, rrm3 and srs2 mutations were complete PCR-mediated ORF deletions, using pRS405, pUG6 (kanR) or pAG32 (hphMX4) as templates for marker gene amplification (60). The top3 deletion extended from nucleotides 10 to 1666 of the ORF, to avoid disruption of the overlapping ORFs YLR235C and YLR236C. Deletions were confirmed by PCR amplification of fragments that crossed both mutation breakpoints. Oligonucleotide sequences are available on request.
Table 1.
S.cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| YBJ133 | MATa/α, Δho Δhml::ADE1 Δhmr::ADE1 ade1 ura3-52, leu2-3,112, lys5, TLC1/Δtlc1::kanMX, SGS1/Δsgs1::hisG-URA3 | (36) |
| YJL13b | YBJ133 RAD52/rad52::LEU2 | This study |
| YMA64 | YBJ133 TOP1/Δtop1::LEU2 | This study |
| YBJ161 | YBJ133 SGS1/Δsgs1::hisG′ | This study |
| YMA75b | YBJ161 SGS1/Δsgs1::URA3 | This study |
| YMA76 | YMA75b leu2/LEU2::SGS1* | This study |
| YMA77 | YMA75b leu2/LEU2::SGS1-hd* | This study |
| YMA79 | YMA75b leu2/LEU2::SGS1-ΔN50* | This study |
| YMA90 | YMA75b leu2/LEU2::SGS1-ΔN795* | This study |
| YMA97 | YMA75b leu2/LEU2::SGS1-ΔC200* | This study |
| YMA99 | YMA75b leu2/LEU2::pRS405 | This study |
| YBJ216 | YMA75b leu2/LEU2::TOP3-SGS1-ΔN106* | This study |
| YMA102 | YBJ133 MUS81/Δmus81::HygB | This study |
| YMA104 | YBJ133 SLX8/Δslx8::HygB | This study |
| YMA105 | YBJ133 TOP3/Δtop3::HygB | This study |
| YMA107 | YBJ133 SLX5/Δslx5::LEU2 | This study |
| YMA109 | YBJ133 SLX1/Δslx1::LEU2 | This study |
| YJL22 | MATa/α, Δho Δhml::ADE1 Δhmr::ADE1 ade1 ura3-52, leu2-3,112, lys5, TLC1/Δtlc1::kanMX | This study |
| YJL42 | YJL22 RRM3/Δrrm3::HygB | This study |
| YJL44 | YJL22 SRS2/Δsrs2::HygB | This study |
| YJL68 | MATa/α, Δho Δhml::ADE1 Δhmr::ADE1 ade1 ura3-52, leu2-3,112, lys5, SGS/Δsgs1::hisG-URA3, RAD52/rad52::LEU2 | This study |
| YJL69 | YJL68 EST2/Δest2::kanMX | This study |
Plasmids
The SGS1 deletion and point mutation derivatives (pJM526, pJM511, pJM531, pJM512 and pRL1; kindly provided by S. J. Brill) were cloned downstream a 0.15 kb SGS1 promoter fragment in pRS405, as described previously (59). The TOP3-SGS1-ΔN106 allele was generated by PCR amplification of the TOP3 ORF from pRK500 (kindly provided by J. Wang) with incorporation of NdeI and NcoI sites at the 5′ and 3′ ends, respectively, and insertion of NdeI and NcoI digested product into NdeI and partially-NcoI digested pJM531; this yielded a fusion of the full TOP3 ORF to the SGS1 ORF beginning at amino acid 107, in a fashion similar to that described previously (61). For all SGS1 derivatives, BstEII-digested plasmid DNA was transformed into yeast cells, and proper integration of a single copy of each SGS1* mutant at the LEU2 locus was confirmed by Southern blot analysis of SacI-digested genomic DNA probed with pRS405.
Liquid senescence assays
To ensure that senescence comparisons were made between strains that inherited similar telomeres, senescence assays were performed on the haploid spore products of diploids that were heterozygous for tlc1 and the various test mutations. Liquid senescence assays were performed as described previously (36), starting with spore products from reference plates (estimated population doubling of 30). At the start of each day of growth, 2 × 106 cells were inoculated into 5 ml of YPD followed by growth for 22 h, and the process was repeated until survivors emerged. Cells numbers were determined with a Coulter counter, and population doubling calculated as log2(final cell number/starting cell number). Each genotype was represented by three to six independent spore products, and the mean and standard errors of measurements are shown in figures.
Spot growth assays
Cells were grown overnight on YPD plates and were suspended in sterile water at an OD600 of 3.0. Cells were spotted at this concentration and at four serial 10-fold dilutions on YPAD plates and were grown at 30°C for 60 h.
Telomere Southern analysis
Southern analysis was performed essentially as described previously (36). Briefly, purified genomic DNA was digested with XhoI, separated by electrophoresis in 1% agarose gels, transferred to Hybond-XL membranes, and probed and washed at high stringency with a cloned 256 bp fragment of S.cerevisiae telomere repeat DNA (BamHI–MluI fragment of pJL8-50, Julia Y. Lee and F. Brad Johnson, manuscript in preparation) or with a 784 bp Y′ fragment distal to the XhoI site (36). tlc1 rad51 type II survivor DNA was from (36).
RESULTS
The C-terminus of Sgs1p, required for intra-S-phase checkpoint function, is not required to rescue rapid senescence
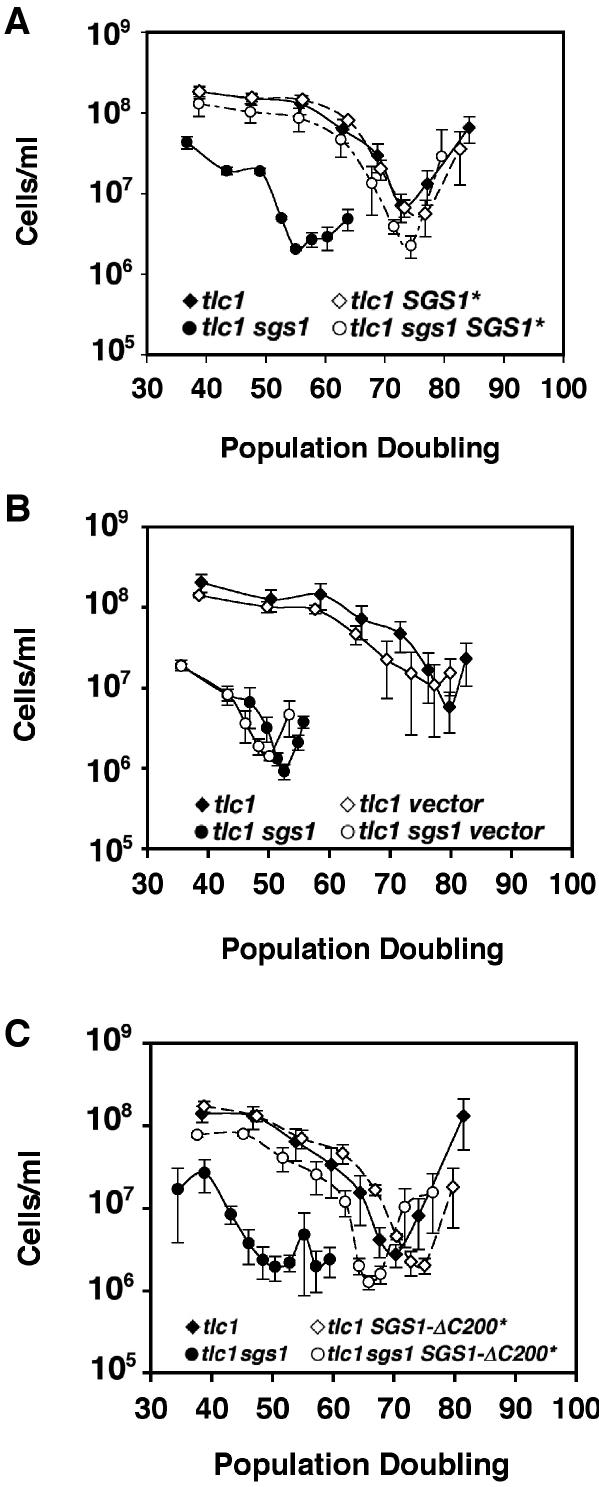
To begin to dissect how Sgs1p slows the rate of senescence of telomerase (tlc1) mutants, we tested different deletion and point mutation alleles of SGS1 (Figure 1) for their ability to rescue the rapid senescence of tlc1 sgs1 mutants. The derivative alleles, designated with an asterisk, were placed downstream of the SGS1 promoter and integrated at the LEU2 locus (59). Diploids heterozygous for tlc1 and sgs1 mutations and an * allele were sporulated, and the rates of senescence of haploid products lacking telomerase were determined using liquid senescence assays. We first confirmed that wild type SGS1 sequences integrated at LEU2 (denoted SGS1*) could rescue an sgs1 mutation during senescence. tlc1 sgs1 mutants carrying SGS1* senesced at a rate equivalent to tlc1 mutants (Figure 2A), while a vector control lacking all SGS1 sequences integrated at LEU2 failed to have any effect (Figure 2B). Furthermore, tlc1 mutants carrying SGS1* senesced at the same rate as tlc1 mutants, indicating that SGS1* did not have effects beyond the rescue of the sgs1 mutation. Thus, integrated SGS1* can function in the senescence assay in the same fashion as the native SGS1 locus.
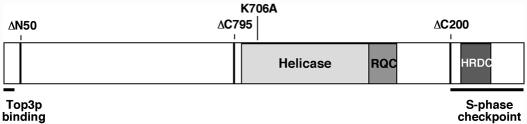
Figure 1.
Map of the 1447 amino acid Sgs1p protein. Indicated by shaded boxes are the helicase domain, the RQC domain (a C-terminal extension from the helicase domain of shared homology among RecQ-family helicases, which is involved in protein–protein interactions) and the HRCD domain (helicase/RNase D C-terminal domain, which appears to be involved in DNA binding). The locations of the N- and C-terminal deletion breakpoints used in this study are indicated by thick vertical lines and labels above the protein, as is the helicase-deficient alanine substitution mutant, K706A. Sequences containing amino acids that are essential for the binding of Sgs1p to Top3p or for the intra-S-phase checkpoint function of Sgs1p are indicated by horizontal bars below the protein.
Figure 2.
Full-length and ΔC200 forms of Sgs1p rescue the fast senescence of tlc1 sgs1 mutants. Full-length SGS1, SGS1-ΔC200 or control vector sequences were integrated at the LEU2 locus of diploids heterozygous for sgs1 and tlc1 null mutations. tlc1 and tlc1 sgs1 spore products without or with the integrated SGS1 alleles (indicated by an asterisk) were compared in liquid senescence assays (see Materials and Methods). The cumulative population doubling since spore germination, and the density of cells after 22 h of growth, from cultures inoculated with 4 × 105 cells/ml are shown. For each time point, the mean and standard errors for at least three independent spore products for each genotype are indicated. For each experiment, filled diamonds indicate tlc1, filled circles indicate tlc1 sgs1, open diamonds indicate tlc1 with an integrated * allele, and open circles indicate tlc1 sgs1 with an integrated * allele, as shown. (A) Rescue by full-length integrated SGS1* (B) Lack of rescue by the pRS405 vector used to integrate the SGS1* alleles. (C) Rescue by SGS1-ΔC200*, which encodes an Sgs1p derivative lacking the C-terminal 200 amino acids.
The C-terminal 200 amino acids of Sgs1p have been shown to be essential for the intra-S-phase checkpoint activity of the protein (28). We therefore tested the ability of SGS1-ΔC200*, which encode an Sgs1p derivative lacking these amino acids, to rescue rapid senescence. Similar to SGS1*, SGS1-ΔC200* slowed the rate of senescence to that observed in tlc1 mutants (Figure 2C). Therefore, the intra-S-phase checkpoint function of SGS1 does not appear to be required to prevent rapid senescence.
Sgs1p slows senescence through an HR-dependent pathway
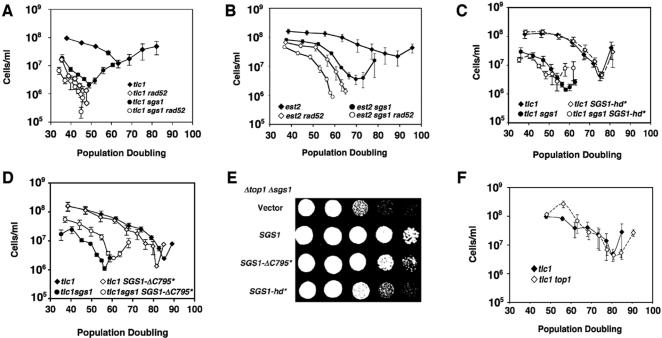
Because the checkpoint function of SGS1 appeared not to be involved in slowing senescence, we considered the possibility that the homologous (or homeologous, in the case of the imperfect yeast telomere repeats) recombination (HR) functions of SGS1 might instead play a role. RAD52 is required for nearly all HR reactions in yeast (62), and so we tested for a possible epistatic relationship between sgs1 and rad52 mutations in the absence of telomerase. As reported previously, tlc1 rad52 mutants senesced rapidly (39,57), consistent with a role for HR in slowing senescence, and at a rate similar to tlc1 sgs1 mutants (Figure 3A); also, as expected, no survivors of senescence were formed in the absence of RAD52 function. Although tlc1 rad52 sgs1 mutants senesced slightly faster than tlc1 rad52 or tlc1 sgs1 mutants, the contribution of the sgs1 or rad52 mutation to the faster senescence of the triple mutant was far less than the contribution of either mutation to the rapid senescence in the double mutants, and thus the rad52 and sgs1 mutations are largely epistatic in our assay. This result was repeated twice using independent sets of spore products and was also observed when telomerase was inactivated via deletion of EST2, rather than TLC1 (Figure 3B). SGS1 therefore appears to function in a RAD52-dependent pathway during senescence. Furthermore, the helicase activity of Sgs1p, which is required for Sgs1p HR activities in other contexts (31,59), was necessary to prevent rapid senescence because the SGS1-hd* allele, which encodes a K706A mutant devoid of helicase activity, failed to rescue the rapid senescence of tlc1 sgs1 mutants (Figure 3C). Similarly, the SGS1-ΔC795* allele, which encodes a protein lacking the helicase domain and C-terminus of Sgs1p, did not rescue rapid senescence (Figure 3D). These observations are consistent with a role for SGS1 in HR during senescence.
Figure 3.
Evidence that Sgs1p functions via recombination to slow senescence. Senescence rates were measured in the same fashion as Figure 2. (A) rad52 and sgs1 mutations are epistatic during senescence. Haploid spore products were derived from diploids heterozygous for tlc1, sgs1 and rad52 mutations. Curves for tlc1 (filled diamonds), tlc1 rad52 (open diamonds), tlc1 sgs1 (filled circles) and tlc1 sgs1 rad52 (open circles) mutants are shown. This result was repeated two additional times with independently derived spore products (data not shown). (B) est2 and tlc1 mutations are equivalent in the context of sgs1 and rad52 mutations. The experiment was the same as in (A), but with telomerase inactivation via est2 rather than tlc1 deletion (C) Sgs1p helicase activity is required to slow senescence. Diploids heterozygous for tlc1 and sgs1 mutations and with an integrated SGS1-hd* allele, which encodes the helicase-defective K706A point mutant, were sporulated and the senesce rates of tlc1 (filled diamonds), tlc1 sgs1 (filled circles), tlc1 SGS1-hd* (open diamonds) and tlc1 sgs1 SGS1-hd* haploid products were measured. (D) Same as (C), except that the SGS1-ΔC795* allele, which encodes a derivative lacking the C-terminal 795 amino acids of Sgs1p was used instead of SGS1-hd*. (E) SGS1-hd* and SGS1-ΔC795* encode active proteins. The ability of the * alleles to rescue the synthetic slow growth of sgs1 top1 mutants was tested by generating top1 deletions in haploid cells with sgs1 deletion and the indicated * allele. Shown is growth of serial 10-fold dilutions of double mutants containing integrated vector, SGS1*, SGS1-ΔC795* or SGS1-hd* alleles, as indicated. (F) top1 deletion does not speed senescence. Diploids heterozygous for top1 and tlc1 deletions were sporulated and senescence rates of tlc1 (filled diamonds) and tlc1 top1 (open diamonds) haploid spore products were measured.
To confirm that the failure of the SGS1-hd* and SGS1-ΔC795* alleles to rescue rapid senescence was not simply due to lack of their expression, we examined their activities in cells lacking topoisomerase I (top1 mutants). Loss of SGS1 function causes a slight decrease in growth rate, as does loss of TOP1 function, but an sgs1 top1 double mutant has a synthetic growth defect (63). As reported previously (59), the SGS1-hd* and SGS1-ΔC795* alleles partially and fully, respectively, restored the growth rate of sgs1 top1 mutants to that of top1 mutants (Figure 3E), confirming the activities of the alleles. Furthermore, because Sgs1p helicase activity is critical during senescence but less important for growth in top1 mutants, different functions of SGS1 are apparently involved in each case, and this in turn implies that TOP1 function may not play a role during senescence. Indeed, tlc1 top1 null mutants senesce at the same rate as tlc1 mutants (Figure 3F). This is remarkable because, similar to sgs1 deletion, top1 deletion causes genome instability, particularly in the ribosomal DNA (64,65). Therefore, a decrease in genome stability, at least in a generic sense, is not sufficient to speed senescence. This point is explored more fully below.
Sgs1p cooperates with Top3p to slow senescence
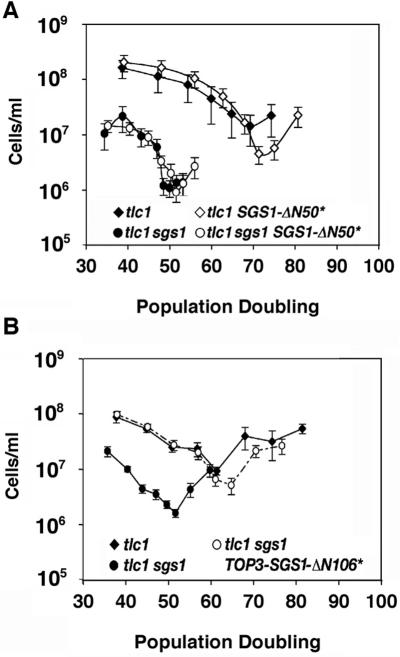
A derivative of Sgs1p lacking the N-terminal 50 amino acids, encoded by SGS1-ΔN50*, was tested and found to be devoid of SGS1 function in the senescence assay (Figure 4A). The deleted N-terminus includes residues that are essential for the functional and physical interaction of the protein with topoisomerase III (Top3p), a type I topoisomerase that cooperates with Sgs1p during homologous recombination (22,61,66–69). Indeed, this may be the only critical function of the N-terminus of Sgs1p because when the TOP3 ORF is fused to a fragment of the SGS1 ORF lacking the first 106 codons, the fusion product (Top3p-ΔN106Sgs1p) is able to rescue the recombination defects and sensitivity of sgs1 mutants to MMS and HU (61). The failure of SGS1-ΔN50* to rescue rapid senescence therefore implies a role for TOP3 during senescence. We investigated this possibility by first examining the rate of senescence of tlc1 top3 null mutants. Haploid spores were derived from diploids with TLC1/tlc1 and TOP3/top3 mutations and tlc1 and tlc1 top3 products compared with each other. top3 and tlc1 top3 mutant spores germinated at very low frequency in our strain background. However, we obtained one tlc1 top3 spore product, and this senesced rapidly, ∼20 generations after germination (data not shown). Additional evidence for the importance of TOP3 during senescence was provided by the ability of the Top3p-ΔN106Sgs1p fusion, encoded by TOP3-SGS1-ΔN106*, to rescue the rapid senescence of tlc1 sgs1 mutants (Figure 4B). Because the fusion lacks all the residues from the N-terminus of Sgs1p that are absent in Sgs1p-ΔN50, this indicates that the inactivity of Sgs1p-ΔN50 was due to its inability to bind productively to Top3p. We conclude that Sgs1p and Top3p cooperate to slow the senescence of tlc1 mutants.
Figure 4.
Sgs1p cooperates with Top3p to slow senescence. Strains were generated and senescence experiments were performed in the same fashion as in Figure 2. (A) SGS1-ΔN50*, which encodes a derivative lacking the N-terminal 50 amino acids of Sgs1p, does not rescue the fast senescence of tlc1 sgs1 mutants. Senescence curves for tlc1 (filled diamonds), tlc1 sgs1 (filled circles), tlc1 SGS1-ΔN50*(open diamonds) and tlc1 sgs1 SGS1-ΔN50* (open circles) haploid spore products are indicated. (B) TOP3-SGS1-ΔN106*, which encodes Top3p fused to an Sgs1p derivative lacking the N-terminal 106 amino acids, rescues rapid senescence. Senescence curves for tlc1 (filled diamonds), tlc1 sgs1 (filled circles) and tlc1 sgs1 TOP3-SGS1-ΔN106* (open circles) haploid spore products are indicated.
Compared with most proteins that function with Sgs1p in other settings, Sgs1p plays a more important role during senescence
A number of mutations, referred to as ‘slx’ mutations, have been found to cause synthetic lethality (or extremely poor growth) in combination with sgs1 null mutations (70–73). Among these are null mutations in SLX1, SLX5, MUS81, SRS2 and RRM3, as well as in TOP1 as described above. Slx1p together with Slx4p forms an endonuclease complex that cleaves 5′ flap structures and is required for normal replication of the ribosomal DNA and for resistance to MMS (13,70,74). Slx5p together with Slx8p forms a complex of undetermined biochemical activity, although the complex is known to promote resistance to HU (70). Mus81p complexes with Mms4p to form an endonuclease that cleaves 3′ flap structures, D-loops and nicked Holliday junctions, and the complex appears to help rescue stalled replication forks (75–77). Srs2p is a 3′–5′ DNA helicase that promotes non-crossover recombination, apparently by inhibiting Rad51p-mediated strand invasion, and that is also required for adaptation following DNA damage (20,78–80). Rrm3p is a 5′–3′ DNA helicase that prevents replication fork stalling in non-nucleosomal chromatin regions, including telomeres and ribosomal DNA sequences (81–83). Similar to SGS1, each of the genes in this group is required for the maintenance of DNA stability throughout the genome.
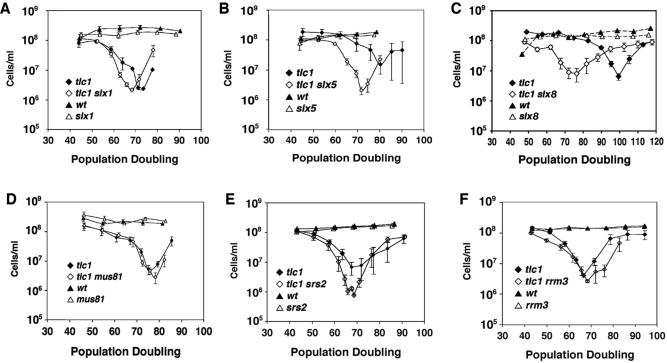
A mutation that is synthetically lethal with sgs1 may identify a gene that accomplishes an essential function through a pathway that is distinct from and parallel to an SGS1-dependent pathway. Thus, either SGS1 or the gene in question can support a critical biochemical pathway at a level that is sufficient for viability, but loss of both genes is lethal. Alternatively, the two genes may each contribute to the efficiency of a single pathway, so that loss of both genes slows the pathway to below a threshold needed for viability. By either model, some of the synthetic lethal genes might possibly function, like SGS1, to slow the senescence of tlc1 mutants. We therefore compared rates of senescence in tlc1 strains with tlc1 strains also containing deletions of SLX1, SLX5, MUS81, SRS2 or RRM3 (Figure 5A, B, D–F). Remarkably, of all the mutations in this group only slx5 had an appreciable effect on the rate of senescence, accelerating senescence by 10 population doublings (PD) (Figure 5A–F). To confirm the effect of the slx5 mutation, we examined the effect of mutating SLX8, which encodes the partner of Slx5p. As expected, tlc1 slx8 mutants senesced more rapidly than tlc1 mutants, by ∼20 PD (Figure 5C). Although srs2 mutation did not speed senescence appreciably, it did delay by 2–3 days the formation of survivors and caused a concomitant depression of the minimum cell density prior to survivor formation (Figure 5E). We conclude that, with the exception of SLX5/8, none of the genes we examined plays a role as important as SGS1 to slow senescence. Furthermore, as detailed in the Discussion, the findings indicate that generalized genome instability is not the cause of rapid senescence in tlc1 sgs1 mutants, but rather that Sgs1p plays a more specific role at telomeres or in response to telomere dysfunction.
Figure 5.
Effects of sgs1 synthetic lethal mutations on senescence. The indicated synthetic lethal mutations were introduced individually into diploids heterozygous for tlc1 deletion. Senescence rates of spore products were measured in the same fashion as Figure 2. In all cases, filled diamonds indicate tlc1 mutants, open diamonds indicate tlc1 plus the synthetic lethal mutation, and filled and open triangles indicate wild-type and the synthetic lethal mutant, respectively. Comparison of tlc1 and tlc1 with (A) slx1 deletion, (B) slx5 deletion, (C) slx8 deletion, (D) mus81 deletion, (E) srs2 deletion and (F) rrm3 deletion. Note the increased number of days (data points) spent in the nadir in tlc1 srs2 mutants.
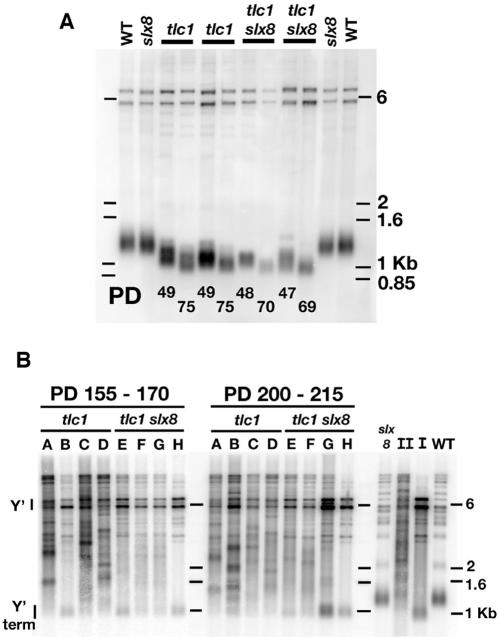
In addition to speeding the senescence of tlc1 cells, sgs1 mutation increases telomere shortening per population doubling and blocks the formation of type II survivors (36,42,43). We therefore asked if slx8 mutation also had these other effects. We examined Y′-containing telomere length by Southern analysis (Figure 6A). slx8 mutation did not affect steady-state telomere length in TLC1+ cells. Comparison of telomere lengths for four tlc1 mutants at days 1 and 4 of growth in liquid (mean PD 49 versus 75) and four tlc1 slx8 mutants at 1 and 5 (mean PD 48 versus 71), yielded telomere shortening rates of 2.3 ± 0.12 bp/PD for tlc1 and 4.0 ± 0.62 bp/PD for tlc1 slx8 cells (P = 0.04), indicating that telomeres shorten faster per population doubling in the absence of SLX8 function (Figure 6A and data not shown; samples were from the cultures shown in Figure 5C). We next examined survivor type by Southern analysis of XhoI-digested genomic DNA. Type I survivors are typified by amplification of subtelomeric Y′ elements of 5.2 and 6.7 kb in size, the presence of ∼1 kb Y′ fragments containing terminal telomere repeat DNA, and by the relative absence of telomere repeat-containing fragments between these two signals. Type II survivors appear as fragments of heterogeneous size that range in size from 1 to >10 kb and they usually have less amplification of Y′ elements. In general, type I survivors predominate early in survivor formation, but because of their faster growth rate type II survivors eventually overtake liquid cultures (40). Three of four tlc1 cultures formed type II survivors by the 17th day after loss of telomerase (PD 155–170) and all four were type II by day 22 (PD 200–215) (Figure 6B). In comparison, two of four tlc1 slx8 cultures had clear type II character at day 17, but there was little further progression to type II by day 22, and one of the cultures (Figure 6B, G) actually developed more type I character. Thus, SLX8 may facilitate the formation or growth of type II survivors, but, in contrast to SGS1, is not absolutely required for type II survivors to form.
Figure 6.
Telomere shortening and survivor formation in tlc1 slx8 cells. (A) Y′-containing telomere lengths from wild type, slx8 and from two independent cultures each of tlc1 and tlc1 slx8 mutants during senescence were measured by digesting genomic DNA with XhoI and probing with Y′ sequences distal to the cut site. The number of population doublings (PD) since the start of spore growth is indicated. (B) Survivor type in four tlc1 (A–D) and four tlc1 slx8 (E–F) cultures was examined by Southern analysis of XhoI-digested genomic DNA visualized with a telomere repeat probe. Samples are from days 17 (PD 155–170) and 22 (PD 200–215) after sporulation, and samples were obtained from the cultures shown in Figure 5C. Y′ and Y′ term indicate the position of tandemly-repeated Y′ elements, and the terminal Y′ fragment, respectively, that are most prominent in type I survivors. On the right, samples from slx8, pure type I (tlc1 sgs1), pure type II (tlc1 rad51) and wild-type cultures are provided for comparison, and the position of DNA markers is indicated with sizes in kilobases (kb).
DISCUSSION
We have investigated possible mechanisms by which the Sgs1p helicase slows the senescence of tlc1 mutants. Our findings are most compatible with a role for Sgs1p in HR-mediated repair of telomeres, rather than a role for the Sgs1p-dependent S-phase checkpoint or for a generalized role of Sgs1p in genome stability. In support of an HR role are (i) the epistatic relationship of sgs1 and rad52 mutations in telomerase mutants, (ii) a requirement for Sgs1p helicase activity and (iii) a role for Top3p. Helicase activity and cooperation with Top3p have been shown to be essential for all of the recombination functions of Sgs1p so far investigated (22,59,61,69,84). Furthermore, Rad52p is a component of a protein complex that includes Sgs1p, consistent with a role for Sgs1p in Rad52p-dependent HR (85). The higher rate of recombination among shortened telomeres in Kluyveromyces lactis (86), the direct observation of recombinant sequences at the termini of ∼6% of telomeres in tlc1 mutants (87), and the rapid senescence of tlc1 mutants also lacking RAD52, RAD51, RAD54 or RAD57 (57) support the idea that HR can contribute to telomere maintenance during senescence. We note that a different interpretation of the relationship between RAD52 and SGS1 in cells senescing due to telomerase inactivation was reported previously (43), where an additive effect of rad52 and sgs1 mutations on senescence was observed. However, in the report telomerase was inactivated in haploid strains already carrying the rad52 and/or sgs1 mutations, and it is thus possible that differences in telomere integrity in the different mutant backgrounds prior to telomerase inactivation contributed to senescence rates. Our experimental approach eliminates this confound by comparing haploid spore products derived from diploids that contain one wild-type allele for each mutation being studied and, further, compares cells with telomeres derived from the same diploid parent and is thus less subject to random fluctuations in steady state telomere length. We are therefore confident that SGS1 and RAD52 function in a less-than-additive, i.e. epistatic, fashion during the senescence of telomerase mutants.
A reasonable candidate for an HR-based mechanism of telomere repair is break-induced replication (BIR) (88–90). Such a BIR mechanism could, for example, involve invasion of a critically shortened telomere end into a longer telomere, which would then be used as a template for DNA synthesis to extend the shortened telomere. A role for the Sgs1p–Top3p complex in this context could be to facilitate progression during DNA synthesis, and thus resolution at the telomere terminus, of a Holliday junction formed by such an invasion event. Particularly in a case where the telomere DNA was not free to rotate, as could be the case in telomeric chromatin, the helicase activity of Sgs1p could cooperate with the strand-passage activity of Top3p to enable separation of the newly synthesized strand from the template. This would be mechanistically similar to the branch migration and resolution of double Holliday junctions shown to be catalyzed by the BLM–TOP3α complex (91), and may also be related to the stalling of DNA synthesis during HR that occurs in Drosophila blm mutants (92). A BIR mechanism has been proposed previously to underlie the formation and maintenance of survivors of telomerase deletion (57), and it is possible that BIR operates both during and after senescence. BIR is RAD51-independent when initiated from chromosomal sites cleaved by the HO-endonuclease, but proceeds by a RAD51-dependent mechanism when initiated from a transformed chromosomal fragment (88,90). Because tlc1 rad51 mutants senesce rapidly [(57) and Q. Chen and F.B. Johnson, unpublished], Rad51p is also involved in slowing senescence and so any such telomere BIR mechanism would apparently be of the RAD51-dependent type. Consistent with the chromosome fragment assay reflecting a BIR process similar to that proposed to operate at telomeres during senescence, RAD50 and RAD59 are dispensable for BIR in this assay (despite being required for BIR in the HO-cleavage assay) and tlc1 rad50 or tlc1 rad59 mutants senesce at normal rates (57,90,93). Furthermore, although SGS1 is not required for BIR at an internal chromosomal site cleaved by the HO endonuclease (94), it remains possible that it is required for BIR from non-HO cleaved ends or there could be particular requirements for Sgs1p in recombination involving G-rich telomere repeats, for example the unwinding ofG-quadruplexes.
We were surprised to find that so many different genes involved in maintenance of genome stability had no significant effect on senescence rates. These include five ‘slx’ genes (SLX1, MUS81, SRS2, RRM3 and TOP1) whose loss of function causes synthetic sickness or lethality in combination with sgs1 mutation, and so which might be expected to have some functions similar to Sgs1p. Indeed, one such pair of genes, SLX5 and SLX8, do have effects on the senescence rate (see below). Even though the slx genes have genetic relationships to SGS1, they clearly affect different classes of processes required for genome stability, based on their different genetic interactions with other factors, biochemical activities and mutant phenotypes. Thus, the five mutations sample the importance during senescence of several distinct functions required for genome maintenance. Our findings are consistent with previous demonstrations that mutations in RAD1 or RAD10, which are required for nucleotide excision repair and the single-strand annealing pathway of double-strand break repair, mutations in MSH2, which is required for mismatch repair, the pol3-01 mutation, which leads to an elevated rate of point mutations, and mutations in TEL1 or MEC1, which mediate DNA damage responses, do not speed the senescence of telomerase mutants (38,95). Together with our findings, these results indicate that induction of several different types of genome instability is not sufficient to speed senescence, and they further suggest that Sgs1p plays a relatively specialized role in telomere function.
In contrast to the other sgs1-synthetic lethal mutations we tested, slx5 and slx8 mutations sped the senescence of tlc1 mutants. The molecular function of the Slx5p/8p complex is currently unknown, and so this finding does not yet provide any detailed mechanistic insight. However, it is interesting that the synthetic lethality of sgs1 slx5 mutants is not suppressed by rad52 mutation, unlike the synthetic lethality of sgs1 with several other mutations including mus81, srs2 and rrm3 (17,18,21,22,76). This might reflect an involvement of Slx5p in a pathway for which HR is an essential component; the key role of HR in slowing senescence is consistent with an HR-related role of Slx5p being important during senescence. We also found that SLX8 is not required for the formation of type II survivors of tlc1 deletion, although their appearance appears to have been inhibited. This contrasts with an apparent absolute requirement for SGS1 in type II survivor formation. However, we note that once formed, type II survivors no longer absolutely require the continued presence of SGS1 (36), and so the functional differences between SGS1 and SLX8 in the type II mechanism may be subtle and reflect a particularly important role for SGS1 during the initial stages of the formation of type II survivors.
Although srs2 mutation did not speed the senescence of tlc1 mutants, it did substantially delay the appearance of survivors. We do not yet understand the basis for this delay, but it might be connected with the requirement for srs2 in the process of adaptation. Adaptation enables cells that have arrested their cell cycle in response to DNA damage to resume growth even prior to repair of the offending lesion. srs2 mutants show impaired adaptation after incurring an irreparable double strand chromosomal break (78). Because critically shortened telomeres are interpreted by the cell as a form of DNA damage (37,38), the survivor mechanism might involve an adaptation-like response to enable the resumption of growth. Further experiments will be necessary to determine if other components of the adaptation response are required for the formation of survivors.
Recently, it was reported that telomere sequences that are the products of lagging-strand DNA synthesis are selectively lost in Werner mutant cells (51). This was attributed to a defect in the removal of G-quadruplexes from the replication template caused by the absence of WRN. Our findings are consistent with this possibility. However, our data suggest a role for Sgs1p in a HR-based pathway at telomeres, and because replication forks stall naturally in telomere repeat DNA (81,96), and because HR is critical for the restart of stalled replication forks (97), we also suggest an alternative that RecQ-family helicases might remove G-quadruplexes that would otherwise interfere with HR-based rescue of stalled replication forks at telomeres.
Acknowledgments
The authors thank Steven Brill and James Wang for generously sharing plasmids and advice, and Robert Wilson, Erin Pennock, Nina Luning-Prak and the members of the Johnson laboratory for discussions and comments on the manuscript. This work was supported by grants from American Federation for Aging Research and the National Institute on Aging to F.B.J. (5R01AG021521) and a National Research Service Award to J.Y.L. is (F32AG22769). Funding to pay the Open Access publication charges for this article was provided by the Department of Pathology and Laboratory Medicine, University of Pennsylvania School of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gangloff S., McDonald J.P., Bendixen C., Arthur L., Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol. Cell. Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watt P.M., Louis E.J., Borts R.H., Hickson I.D. Sgs1: a eukaryotic homolog of E.coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 3.Khakhar R.R., Cobb J.A., Bjergbaek L., Hickson I.D., Gasser S.M. RecQ helicases: multiple roles in genome maintenance. Trends Cell Biol. 2003;13:493–501. doi: 10.1016/s0962-8924(03)00171-5. [DOI] [PubMed] [Google Scholar]
- 4.Hickson I.D. RecQ helicases: caretakers of the genome. Nature Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 5.Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 6.Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S., et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 7.Kitao S., Shimamoto A., Goto M., Miller R.W., Smithson W.A., Lindor N.M., Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nature Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 8.Epstein C.J., Martin G.M., Schultz A.L., Motulsky A.G. Werner's syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 9.Ellis N.A., German J. Molecular genetics of Bloom's syndrome. Hum. Mol. Genet. 1996;5:1457–1463. doi: 10.1093/hmg/5.supplement_1.1457. [DOI] [PubMed] [Google Scholar]
- 10.Wang L.L., Levy M.L., Lewis R.A., Chintagumpala M.M., Lev D., Rogers M., Plon S.E. Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am. J. Med. Genet. 2001;102:11–17. doi: 10.1002/1096-8628(20010722)102:1<11::aid-ajmg1413>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 11.Monnat R.J., Jr, Saintigny Y. Werner syndrome protein—unwinding function to explain disease. Sci. Aging Knowledge Environ. 2004;2004:re3. doi: 10.1126/sageke.2004.13.re3. [DOI] [PubMed] [Google Scholar]
- 12.Opresko P.L., Cheng W.H., Bohr V.A. Junction of RecQ helicase biochemistry and human disease. J. Biol. Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 13.Kaliraman V., Brill S.J. Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr. Genet. 2002;41:389–400. doi: 10.1007/s00294-002-0319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobb J.A., Bjergbaek L., Shimada K., Frei C., Gasser S.M. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Versini G., Comet I., Wu M., Hoopes L., Schwob E., Pasero P. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 2003;22:1939–1949. doi: 10.1093/emboj/cdg180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weitao T., Budd M., Campbell J.L. Evidence that yeast SGS1, DNA2, SRS2, and FOB1 interact to maintain rDNA stability. Mutat. Res. 2003;532:157–172. doi: 10.1016/j.mrfmmm.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Torres J.Z., Schnakenberg S.L., Zakian V.A. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol. Cell. Biol. 2004;24:3198–3212. doi: 10.1128/MCB.24.8.3198-3212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt K.H., Kolodner R.D. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol. Cell. Biol. 2004;24:3213–3226. doi: 10.1128/MCB.24.8.3213-3226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bjergbaek L., Cobb J.A., Tsai-Pflugfelder M., Gasser S.M. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ira G., Malkova A., Liberi G., Foiani M., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gangloff S., Soustelle C., Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 22.Fabre F., Chan A., Heyer W.D., Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl Acad. Sci. USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bennett R.J., Keck J.L., Wang J.C. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S.cerevisiae. J. Mol. Biol. 1999;289:235–248. doi: 10.1006/jmbi.1999.2739. [DOI] [PubMed] [Google Scholar]
- 24.Sun H., Bennett R.J., Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han H., Bennett R.J., Hurley L.H. Inhibition of unwinding of G-quadruplex structures by Sgs1 helicase in the presence of N,N′-bis[2-(1-piperidino)ethyl]-3,4,9,10- perylenetetracarboxylic diimide, a G-quadruplex-interactive ligand. Biochemistry. 2000;39:9311–9316. doi: 10.1021/bi000482r. [DOI] [PubMed] [Google Scholar]
- 26.Huber M.D., Lee D.C., Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennett R.J., Keck J.L. Structure and function of RecQ DNA helicases. Crit. Rev. Biochem. Mol. Biol. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 28.Frei C., Gasser S.M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 29.Watt P.M., Hickson I.D., Borts R.H., Louis E.J. SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinclair D.A., Mills K., Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 31.Onoda F., Seki M., Miyajima A., Enomoto T. Elevation of sister chromatid exchange in Saccharomyces cerevisiae sgs1 disruptants and the relevance of the disruptants as a system to evaluate mutations in Bloom's syndrome gene. Mutat. Res. 2000;459:203–209. doi: 10.1016/s0921-8777(99)00071-3. [DOI] [PubMed] [Google Scholar]
- 32.Myung K., Datta A., Chen C., Kolodner R.D. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nature Genet. 2001;27:113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 33.Lundblad V., Szostak J.W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 34.Taggart A.K., Zakian V.A. Telomerase: what are the Est proteins doing? Curr. Opin. Cell. Biol. 2003;15:275–280. doi: 10.1016/s0955-0674(03)00040-1. [DOI] [PubMed] [Google Scholar]
- 35.Singer M.S., Gottschling D.E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 36.Johnson F.B., Marciniak R.A., McVey M., Stewart S.A., Hahn W.C., Guarente L. The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase. EMBO J. 2001;20:905–913. doi: 10.1093/emboj/20.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nautiyal S., DeRisi J.L., Blackburn E.H. The genome-wide expression response to telomerase deletion in Saccharomyces cerevisiae. Proc Natl Acad. Sci. USA. 2002;99:9316–9321. doi: 10.1073/pnas.142162499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.IJpma A.S., Greider C.W. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:987–1001. doi: 10.1091/mbc.02-04-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lundblad V., Blackburn E.H. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 40.Teng S.C., Zakian V.A. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:8083–8093. doi: 10.1128/mcb.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lundblad V. Telomere maintenance without telomerase. Oncogene. 2002;21:522–531. doi: 10.1038/sj.onc.1205079. [DOI] [PubMed] [Google Scholar]
- 42.Huang P., Pryde F.E., Lester D., Maddison R.L., Borts R.H., Hickson I.D., Louis E.J. SGS1 is required for telomere elongation in the absence of telomerase. Curr. Biol. 2001;11:125–129. doi: 10.1016/s0960-9822(01)00021-5. [DOI] [PubMed] [Google Scholar]
- 43.Cohen H., Sinclair D.A. Recombination-mediated lengthening of terminal telomeric repeats requires the Sgs1 DNA helicase. Proc. Natl Acad. Sci. USA. 2001;98:3174–3179. doi: 10.1073/pnas.061579598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Opresko P.L., Von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A. Telomere-binding Protein TRF2 Binds to and Stimulates the Werner and Bloom Syndrome Helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 45.Lillard-Wetherell K., Machwe A., Langland G.T., Combs K.A., Behbehani G.K., Schonberg S.A., German J., Turchi J.J., Orren D.K., Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 46.Karlseder J., Smogorzewska A., de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 47.Stansel R.M., de Lange T., Griffith J.D. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Opresko P.L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kolvraa S., May A., Seidman M.M., Bohr V.A. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 49.Machwe A., Xiao L., Orren D.K. TRF2 recruits the Werner syndrome (WRN) exonuclease for processing of telomeric DNA. Oncogene. 2004;23:149–156. doi: 10.1038/sj.onc.1206906. [DOI] [PubMed] [Google Scholar]
- 50.Bai Y., Murnane J.P. Telomere instability in a human tumor cell line expressing a dominant-negative WRN protein. Hum. Genet. 2003;113:337–347. doi: 10.1007/s00439-003-0972-y. [DOI] [PubMed] [Google Scholar]
- 51.Crabbe L., Verdun R.E., Haggblom C.I., Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 52.Stavropoulos D.J., Bradshaw P.S., Li X., Pasic I., Truong K., Ikura M., Ungrin M., Meyn M.S. The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 2002;11:3135–3144. doi: 10.1093/hmg/11.25.3135. [DOI] [PubMed] [Google Scholar]
- 53.Chang S., Multani A.S., Cabrera N.G., Naylor M.L., Laud P., Lombard D., Pathak S., Guarente L., DePinho R.A. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nature Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 54.Du X., Shen J., Kugan N., Furth E.E., Lombard D.B., Cheung C., Pak S., Luo G., Pignolo R.J., DePinho R.A., et al. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol. Cell. Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandell L.L., Zakian V.A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 56.Burke D., Dawson D., Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 57.Le S., Moore J.K., Haber J.E., Greider C.W. RAD50 and RAD51 define two pathways that collaborate to maintain telomeres in the absence of telomerase. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park P.U., Defossez P.A., Guarente L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:3848–3856. doi: 10.1128/mcb.19.5.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mullen J.R., Kaliraman V., Brill S.J. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 61.Bennett R.J., Wang J.C. Association of yeast DNA topoisomerase III and Sgs1 DNA helicase: Studies of fusion proteins. Proc. Natl Acad. Sci. USA. 2001;98:11108–11113. doi: 10.1073/pnas.201387098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paques F., Haber J.E. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu J., Mullen J.R., Brill S.J., Kleff S., Romeo A.M., Sternglanz R. Human homologues of yeast helicase. Nature. 1996;383:678–679. doi: 10.1038/383678a0. [DOI] [PubMed] [Google Scholar]
- 64.Christman M.F., Dietrich F.S., Fink G.R. Mitotic recombination in the rDNA of S.cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- 65.Trigueros S., Roca J. Circular minichromosomes become highly recombinogenic in topoisomerase-deficient yeast cells. J. Biol. Chem. 2001;276:2243–2248. doi: 10.1074/jbc.M008930200. [DOI] [PubMed] [Google Scholar]
- 66.Fricke W.M., Kaliraman V., Brill S.J. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J. Biol. Chem. 2001;276:8848–8855. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Onodera R., Seki M., Ui A., Satoh Y., Miyajima A., Onoda F., Enomoto T. Functional and physical interaction between Sgs1 and Top3 and Sgs1-independent function of Top3 in DNA recombination repair. Genes Genet. Syst. 2002;77:11–21. doi: 10.1266/ggs.77.11. [DOI] [PubMed] [Google Scholar]
- 68.Oakley T.J., Goodwin A., Chakraverty R.K., Hickson I.D. Inactivation of homologous recombination suppresses defects in topoisomerase III-deficient mutants. DNA Repair (Amst) 2002;1:463–482. doi: 10.1016/s1568-7864(02)00032-0. [DOI] [PubMed] [Google Scholar]
- 69.Shor E., Gangloff S., Wagner M., Weinstein J., Price G., Rothstein R. Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics. 2002;162:647–662. doi: 10.1093/genetics/162.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mullen J.R., Kaliraman V., Ibrahim S.S., Brill S.J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Page N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 72.Ooi S.L., Shoemaker D.D., Boeke J.D. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nature Genet. 2003;35:277–286. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 73.Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 74.Fricke W.M., Brill S.J. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–1778. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaliraman V., Mullen J.R., Fricke W.M., Bastin-Shanower S.A., Brill S.J. Functional overlap between Sgs1-Top3 and the Mms4–Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bastin-Shanower S.A., Fricke W.M., Mullen J.R., Brill S.J. The mechanism of mus81-mms4 cleavage site selection distinguishes it from the homologous endonuclease rad1-rad10. Mol. Cell. Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Whitby M.C., Osman F., Dixon J. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- 78.Vaze M.B., Pellicioli A., Lee S.E., Ira G., Liberi G., Arbel-Eden A., Foiani M., Haber J.E. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell. 2002;10:373–385. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 79.Krejci L., Van Komen S., Li Y., Villemain J., Reddy M.S., Klein H., Ellenberger T., Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 80.Veaute X., Jeusset J., Soustelle C., Kowalczykowski S.C., Le Cam E., Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 81.Ivessa A.S., Zhou J.Q., Schulz V.P., Monson E.K., Zakian V.A. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002;16:1383–1396. doi: 10.1101/gad.982902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ivessa A.S., Lenzmeier B.A., Bessler J.B., Goudsouzian L.K., Schnakenberg S.L., Zakian V.A. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol. Cell. 2003;12:1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 83.Torres J.Z., Bessler J.B., Zakian V.A. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S.cerevisiae. DNA helicase Rrm3p. Genes Dev. 2004;18:498–503. doi: 10.1101/gad.1154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ui A., Satoh Y., Onoda F., Miyajima A., Seki M., Enomoto T. The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper- recombination in sgs1 disruptants. Mol. Genet. Genomics. 2001;265:837–850. doi: 10.1007/s004380100479. [DOI] [PubMed] [Google Scholar]
- 85.Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 86.McEachern M.J., Iyer S. Short telomeres in yeast are highly recombinogenic. Mol. Cell. 2001;7:695–704. doi: 10.1016/s1097-2765(01)00215-5. [DOI] [PubMed] [Google Scholar]
- 87.Teixeira M.T., Arneric M., Sperisen P., Lingner J. Telomere length homeostasis is achieved via a switch between telomerase- extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 88.Malkova A., Ivanov E.L., Haber J.E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl Acad. Sci. USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kraus E., Leung W.Y., Haber J.E. Break-induced replication: a review and an example in budding yeast. Proc. Natl Acad. Sci. USA. 2001;98:8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davis A.P., Symington L.S. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu L., Hickson I.D. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 92.Adams M.D., McVey M., Sekelsky J.J. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–267. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 93.Chen Q., Ijpma A., Greider C.W. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Signon L., Malkova A., Naylor M.L., Klein H., Haber J.E. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 2001;21:2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rizki A., Lundblad V. Defects in mismatch repair promote telomerase-independent proliferation. Nature. 2001;411:713–716. doi: 10.1038/35079641. [DOI] [PubMed] [Google Scholar]
- 96.Makovets S., Herskowitz I., Blackburn E.H. Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol. Cell. Biol. 2004;24:4019–4031. doi: 10.1128/MCB.24.9.4019-4031.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McGlynn P., Lloyd R.G. Recombinational repair and restart of damaged replication forks. Nature Rev. Mol. Cell. Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]