Abstract
The present study shows that arsenic induces GADD45α (growth arrest and DNA damage inducible gene 45α) mainly through post-transcriptional mechanism. Treatment of the human bronchial epithelial cell line, BEAS-2B, with arsenic(III) chloride (As3+) resulted in a significant increase in GADD45α protein and mRNA. However, As3+ only exhibited a marginal effect on the transcription of the GADD45α gene. The accumulation of GADD45α mRNA is largely achieved by the stabilization of GADD45α mRNA in the cellular response to As3+. As3+ is able to induce binding of mRNA stabilizing proteins, nucleolin and less potently, HuR, to the GADD45α mRNA. Although As3+ was unable to affect the expression of nucleolin, treatment of the cells with As3+ resulted in re-distribution of nucleolin from nucleoli to nucleoplasm. Silencing of the nucleolin mRNA by RNA interference reversed As3+-induced stabilization of the GADD45α mRNA and accumulation of the GADD45α protein. Stabilization of GADD45α mRNA, thus, represents a novel mechanism contributing to the production of GADD45α and cell cycle arrest in response to As3+.
INTRODUCTION
Growth arrest and DNA damage inducible gene 45α (GADD45α) is a widely expressed, inducible nuclear protein that plays critical role in the checkpoint function of cells in response to a wide spectrum of DNA-damaging or stress signals (1). GADD45α has been shown to inhibit cyclin B/CDC2, a key protein kinase complex governing G2/M transition of the cell cycle (2). In addition, GADD45α is an important protein involved in genomic stability by its contributions to DNA excision repair (3). Furthermore, GADD45α has been implicated in cell apoptosis, cell survival and innate immunity (4,5). The human GADD45α is an acidic protein composed of 165 amino acids, with some similarities to GADD45β, GADD45γ and ribosomal protein S12. In addition to binding to cyclin B/CDC2 as originally demonstrated (2), GADD45α is also capable of interacting with proliferating cell nuclear antigen (6), p21 (7), histone proteins (8), TAFII70 (9), p38 (10) and MTK1/MEKK4 (11), a MAPK kinase kinase that can activate JNK and p38 subgroups of MAP kinase.
The transcriptional regulation of GADD45α has been extensively studied during the past several years. The best-studied transcriptional regulator for the expression of GADD45α is the tumor suppressor protein, p53 (6). In response to ionizing radiation or methyl methansulfonate, GADD45α was rapidly up-regulated through a p53-dependent mechanism. A consensus p53 binding site has been identified in the third intron region of the GADD45α gene. Ionizing radiation or certain other DNA-damaging signals induce binding of p53 to this site, followed by the recruitment of acetyltransferase p300/CBP and protein arginine methyltransferases PRMT1 or CARM1 to this region to stimulate the transcription of GADD45α (12). The promoter region of GADD45α lacks a consensus p53 binding site. However, p53 can also stimulate the transcription of GADD45α by forming a complex with WT1 that binds directly to the proximal promoter of GADD45α (13). Other transcription factors that possibly contribute to a p53-independent regulation of GADD45α include FoxO3a (14), Oct1 (15), C/EBPα (16), Egr-1 (17), POU family members (18), and two transcriptional repressors of GADD45α, c-myc (19) and ZBRK (20).
Arsenic is a naturally occurring metalloid that exhibits potent carcinogenic effects in mammals (21,22). It exists in both inorganic and organic forms with different oxidation states (23). The primary forms of arsenic in environment are the inorganic trivalent (As3+) and pentavalent arsenic (As5+). Humans are exposed to arsenic mainly through oral consumption of contaminated water, food or drugs, and inhalation of arsenic-containing dust or smoke in several occupational settings. Paradoxically, arsenic has also been used as an effective single therapeutic agent for several tumors, especially acute promyelocytic leukemia (24). However, the molecular mechanisms of arsenic-induced carcinogenesis or arsenic-induced remissions of tumors are not fully understood. We and others have previously shown that arsenic is a potent inducer of GADD45α expression in human cells (25,26). We have also shown that activation of c-Jun N-terminal kinase (JNK) might be partially responsible for the induction of GADD45α by arsenic (27). The involvement of JNK in GADD45α expression was further confirmed in the cellular response to UV radiation (28) or a PPARγ agonist, troglitazone (29). In an attempt to gain insight into the detailed mechanism of arsenic-induced expression of GADD45α, we examined the transcriptional and post-transcriptional regulations of GADD45α expression in human bronchial epithelial cells subjected to arsenic exposure. The data presented here reveal that the arsenic-induced expression of GADD45α is mainly regulated by post-transcriptional mechanism in which the mRNA of GADD45α was bound and stabilized by the RNA binding proteins, mainly nucleolin.
MATERIALS AND METHODS
Cell culture, transfections and luciferase assays
The human bronchial epithelial cell line, BEAS-2B, was purchased from American Tissue Culture Collection (Manassas, VA) and maintained in DMEM supplemented with 5% fetal calf serum and grown at 37°C, 5% CO2 in a humidified incubator. Transfections were performed using lipofectamine 2000 as suggested by the manufacturer (Invitrogen, Carlsbad, CA). The human GADD45α promoter and intron 3 luciferase reporter constructs were provided by Dr Albert J. Fornace at National Institutes of Health (NIH, Bethesda, MD). In these vectors, the GADD45α promoter region from −994 to +26 and the entire intron 3 region were inserted into the upstream of the luciferase reporter gene, respectively. Cells were harvested at 36 h and analyzed for luciferase activity using the Promega Dual-Luciferase Assay System (Promega, Madison, WI). The data shown are the mean of at least three independent experiments with error bars displaying standard deviations.
Cell treatment and western blotting
The BEAS-2B cells were seeded in 6-well tissue plates at a density of 2 × 105 cells/well and cultured for 60 h. The cells were treated with the indicated concentrations of arsenic(III) chloride (As3+) (Sigma-Aldrich, St Louis, MO) or H2O2 (Sigma, MO) in the absence or presence of 10 mM N-acetyl-L-cysteine (NAC) (Sigma, MO). Total cell lysate was prepared as described previously (30). Twenty-five micrograms of the protein lysate from the cells cultured in the absence or presence of As3+ were analyzed by SDS–PAGE and immunoblotted with the indicated antibodies. The antibodies against GADD45α, actin, nucleolin, HuR and IKKγ were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The antibodies against phospho-FoxO3a, total FoxO3a, phospho-Akt and total Akt were purchased from Cell Signaling (Beverly, MA).
RT–PCR
The levels of GADD45α and GAPDH mRNA in cell lysate or immune complex were determined by RT–PCR using the AccessQuick RT–PCR system (Promega, Madison, WI). The cells cultured in 6-well tissue culture plates were washed with phosphate-buffered saline (PBS) and lysed using cell lysis buffer from Cells-to-cDNA II kit (Ambion, Austin, TX) as suggested by the manufacturer. RT–PCR was performed using 3 µl of cell lysate and primer sets as follows: GADD45α sense: 5′-GGAGAGCAGAAGACCGAAA-3′ and GADD45α antisense: 5′-TCACTGGAACCCATTGATC-3′; GAPDH sense: 5′-CTGAACGGGAAGCTCACTGGCATGGCCTTC-3′ and antisense: 5′-CATGAGGTCCACCACCCTGTTGCTGTAGCC-3′.
Real-time RT–PCR
To verify the results of RT–PCR, a quantitative real-time RT–PCR was performed. The GADD45α mRNA levels were measured using TaqMan® primers designed using Universal Probe Library Assay Design Center (http://www.roche-applied-science.com/sis/rtpcr/upl/adc.jsp) with the ABI 7500 Sequence Detector (PE Applied Biosystems, Foster City, CA). The primers for GADD45 (Accession no. L24498) were forward, 5′-TCAGCCCAGCTACTCCCTAC; reverse, 5′-AATCTGCCCTGCTAAAGGAAT, used with Universal Probe #16. The primers for the house-keeping gene GAPDH (NM_002046) were forward, 5′-AGCCACATCGCTCAGACAC; reverse, GCCCAATACGACCAAATCC, used with Universal Probe #60. Total RNA was isolated using RNAqueous™ -4PCR kits (Ambion, Austin, TX) from BEAS-2B cells (∼2 million cells) cultured in the absence or presence of 20 µM As3+ for 1–8 h. One to two micrograms of the DNAse I-treated RNA was reverse transcribed, using Superscript II (Life Technologies, Gaithersburg, MD). The cDNA generated was diluted 1:100 and 15 µl was used to conduct the PCR according to the TaqMan® Master mix PCR kit instructions. The comparative CT (threshold cycle) method was used to calculate the relative concentrations (User Bulletin #2, ABI PRISM® 7700 Sequence Detector, PE Applied Biosystems, Foster City, CA). Briefly, the method involves obtaining the CT values for the GADD45α mRNA, normalizing to a house-keeping gene, GAPDH, and deriving the fold increase compared with control, unstimulated cells.
RNA immunoprecipitation assay
BEAS-2B cells were cultured in the absence or presence of 20 µM As3+ for 4 h and subjected to RNA immunoprecipitation assay as described previously (31,32) with minor modifications. Briefly, cells were lysed in 500 µl of cell lysis buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4 and 1 µg/ml of leupeptin for 30 min at 4°C. Cell debris in the lysates was removed by centrifugation at 14 000 g for 15 min at 4°C. The supernatants were incubated overnight with the indicated antibodies at 4°C under rotation. The protein–mRNA binding complex was immunoprecipitated by incubation of the lysates with Protein A-Agarose for 4 h at 4°C. The immune complex was washed three times in lysis buffer. The mRNA of GADD45α and GAPDH in both the immune complex and supernatant were determined by RT–PCR.
Immunofluorescence staining
BEAS-2B cells were seeded into 24-well tissue culture plate without glass slides at a concentration of 5000 to 10 000 cells/well and cultured for 24 h. The cells were then either untreated or treated with As3+ for an additional 4 h. The cells were fixed directly in the culture plate by 10% formalin and permeabilized with 0.1% Triton X-100 for 10 min at room temperature, respectively. Cells were incubated for 6 h at 4°C with primary antibody diluted (1:100) in PBS containing 5% BSA. After extensive washing with PBS, cells were incubated with Fluorescein (FITC)-conjugated anti-rabbit IgG (Santa Cruz, CA) in 1:100 dilution in PBS containing 5% BSA and 1 µg/ml of propidium iodide (PI) for 1 h at room temperature. Fluorescein images were captured by using a Zeiss Axiovert100 microscope connected with a Pixera Pro150ES digital camera.
RNA interference
The target sequencing of small interference RNA (siRNA) against human nucleolin was selected based on the criteria described by Reynolds et al. (33) using a siRNA design program, Gene-specific siRNA selector, developed by Wistar Bioinformatics (http://biowww.net/detail-574.html). The siRNA targeting region is 983-aaagaaggaaatggccaaaca-1001 (NM_005381). The control siRNA and siRNA transfection was described previously (34).
RESULTS
As3+ induces accumulation of GADD45α protein
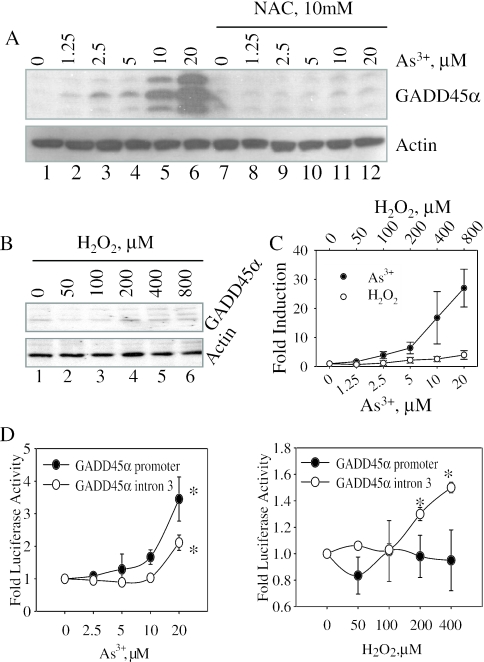
We have previously shown that As3+ induced cell cycle arrest at the G2/M phase, which correlated with the induction of GADD45α protein (25). To obtain insight into the possible mechanism of As3+-induced GADD45α, the cells were pre-treated with 10 mM N-acetyl-cysteine (NAC), a widely used antioxidant that provides cells with exogenous glutathione (GSH) precursor, for 12 h and then treated with 0–20 µM As3+ for an additional 12 h. The expression of GADD45α was barely detectable in the cells without As3+ treatment (Figure 1A). The induction of GADD45α by As3+ was dose-dependent. A plateau of GADD45α induction was reached when the cells were treated with 20 µM As3+. Further elevation of As3+ concentrations (more than 50 µM) did not increase the expression of GADD45α due to cytotoxicity (data not shown). Pre-treatment of the cells with 10 mM NAC completely blocked the induction of GADD45α by As3+ (Figure 1A, lanes 7–12), suggesting that As3+-induced GADD45α expression is possibly through either an oxidative stress response or a direct depletion of GSH. In an additional experimental setting, we pre-treated cells with increasing concentrations of aspirin, another antioxidant which acts as a free radical scavenger, and found that the induction of GADD45α by As3+ was partially inhibited by 10–20 mM aspirin (data not shown).
Figure 1.
As3+ induces expression of GADD45α protein. (A) BEAS-2B cells were pre-treated with 10 mM NAC for 12 h and then incubated in the absence or presence of different concentrations of As3+ for an additional 12 h. Total cell lysates were subjected to western blotting for the determination of GADD45α (upper panel) and actin (bottom panel), respectively. (B) Cells treated with the indicated concentrations of H2O2 for 12 h and then subjected to western blotting for GADD45α (upper panel) or actin (lower panel). Data are representative of at least four experiments. (C) Densitometry scanning of the GADD45α bands induced by As3+ and H2O2 in four separate experiments. (D) Cells transfected with GADD45α promoter- or intron3-luciferase reporter construct for 36 h and then treated with the indicated concentrations of As3+ (left panel) or H2O2 (right panel) for an additional 12 h. Total cell lysates were used for luciferase activity analysis. Asterisks indicate statistically difference with a value of P < 0.05. Data are representative of at least three experiments.
The inhibition of As3+-induced GADD45α by NAC and aspirin implies a possible involvement of reactive oxygen species in this process. Indeed, our previous report had demonstrated a substantial accumulation of H2O2 in the cells treated with As3+ (30). To determine whether H2O2 itself is able to induce GADD45α, the cells were treated with 50–800 µM H2O2 for 12 h. Figure 1B indicates that the induction of GADD45α by H2O2 is very marginal in comparing with the cells treated with As3+. An appreciable induction of GADD45α could be observed only in the cells treated with 400–800 µM H2O2 (Figure 1B, lanes 5 and 6, upper panel). At this concentration, however, the cells showed cytotoxic responses as indicated by the notable cell death determined microscopically (data not shown). Densitometry analysis of the GADD45α protein bands in four separate experiments indicated a more than 20-fold induction of the GADD45α by 20 µM As3+ and a 3- to 4-fold induction of the GADD45α by 800 µM H2O2, respectively (Figure 1C).
As3+ has a weak effect on the transcription of GADD45α gene
Earlier studies have indicated that the consensus p53 binding site in the third intron region of the human GADD45α gene is critical for the genotoxic stress-induced expression of GADD45α (12). It is unclear whether As3+ induces GADD45α expression through transcriptional regulation in a manner of either p53-dependent or p53-independent. By the use of GADD45α promoter- and intron3-based luciferase reporter gene vectors, we noted that As3+, at 20 µM, only induced 3- and 2-fold increase of GADD45α promoter-luciferase activity and GADD45α intron3-luciferase activity, respectively (Figure 1D, left panel). Similar to that of immunoblotting (Figure 1B), H2O2 exhibited no significant induction on the GADD45α promoter-luciferase activity at each dose point tested (Figure 1D, right panel). Only about 1.5-fold induction of intron3-luciferase activity was observed in the cells treated with 400–800 µM H2O2 (Figure 1D, right panel).
There is considerable limitation in reporter gene-based transcriptional analysis due to the absence of distant transcription enhancer elements in the reporter constructs. To address whether As3+ truly regulates the transcription of the GADD45α gene, we next performed a RT–PCR-based nuclear run-on assay. Since we had demonstrated that the accumulation of the GADD45α mRNA was peaked by a 4 h As3+ treatment (following), we incubated the cells with 20 µM As3+ for 4 h in this nuclear run-on assay. Exposure of the cells to As3+ did not induce an appreciable transcription in this assay (data not shown). Thus, these data indicate that it is unlikely that transcriptional regulation is the main mechanism of As3+-induced expression of the GADD45α.
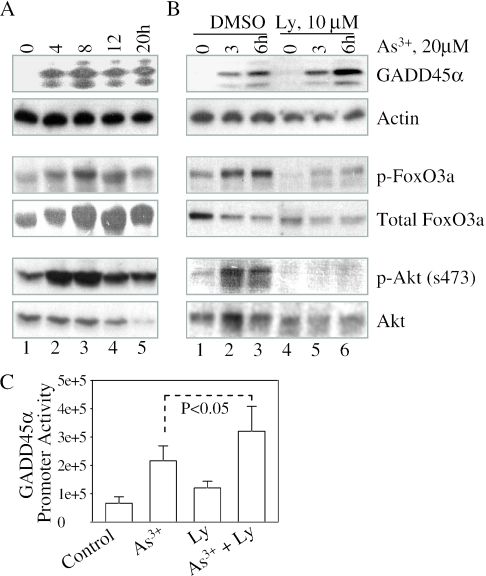
Inhibition of Akt has marginal effect on the expression of GADD45α induced by As3+
Akt signaling pathway is best known for its ability to counteract stress responses that lead to growth arrest or cell apoptosis (35). As a serine-threonine kinase, Akt is able to phosphorylate and inactivate proteins involved in cell cycle arrest or apoptosis. These proteins include FoxO3a, GSK3, Bad, eNOS and procaspase-9 (36). In response to DNA damage signals, FoxO3a appears to be the key transcription factor that up-regulates the transcription of GADD45α (14). Phosphorylation of FoxO3a by Akt suppresses the transcriptional activity of FoxO3a on the expression of GADD45α gene. Thus, inhibition of Akt, a negative regulator of FoxO3a, might indirectly contribute to the induction of GADD45α. To test whether As3+-induced GADD45α is through its effect on Akt-FoxO3a pathway in human epithelial cells, the phosphorylation status of Akt and FoxO3a was investigated in the cells treated with As3+ for different time periods. Induction of GADD45α occurred at 4–20 h of As3+ treatment (Figure 2A). A significant increase, rather than decrease of phosphorylation of FoxO3a and Akt, was observed at these time points. Thus, these results suggest that As3+-induced GADD45α is not through the inhibition of Akt in the human epithelial cells. In contrast, As3+ induces activation of Akt that subsequently phosphorylates and inactivates FoxO3a, which offsets the effect of As3+ on the induction of GADD45α.
Figure 2.
As3+ induces activation of Akt and phosphorylation of FoxO3a. (A) Cells treated with 20 µM As3+ for the indicated times. Total cell lysates were subjected to western blotting for GADD45α, actin, phospho-FoxO3a, total FoxO3a, phospho-Akt and total Akt. (B) Cells were pre-treated with vehicle solution, DMSO, or 10 µM PI3K inhibitor, Ly294002 (Ly), for 2 h and then treated with 20 µM As3+ for the indicated times. Total cell lysates were used for the detection of expression of GADD45α, phosphorylation of FoxO3a and activation of Akt. (C) Cells transfected with a GADD45α promoter-luciferase reporter for 36 h and then treated with 20 µM As3+ in the absence or presence of 10 µM Ly294002 for an additional 12 h. Luciferase activity was calibrated by protein concentrations and the cell viability. Data show means ± standard deviations of three experiments.
We observed an increase in the phosphorylation of Akt and FoxO3a in the cellular response to As3+. Thus, it is worth testing whether inhibition of Akt amplifies the As3+-induced expression of GADD45α. Ly294002, a relatively specific inhibitor for phosphatidylinositol 3 kinase (PI3K), could completely block the activation of Akt and substantially, decrease the phosphorylation of FoxO3a (Figure 2B). However, only about 1- to 2-fold increase of GADD45α induction by As3+ was observed in the cells pre-treated with 10 µM Ly294002 (Figure 2B). Similarly, in a GADD45α promoter-based luciferase activity analysis, only a marginal amplification of As3+-induced luciferase activity could be seen in the cells pre-treated with Ly294002 (Figure 2C). Thus, these observations suggest that although As3+ is capable of stimulating the activation of Akt, a negative regulator for FoxO3a and the subsequent transcription of GADD45α gene, inhibition of Akt has a very weak effect on the induction of GADD45α by As3+.
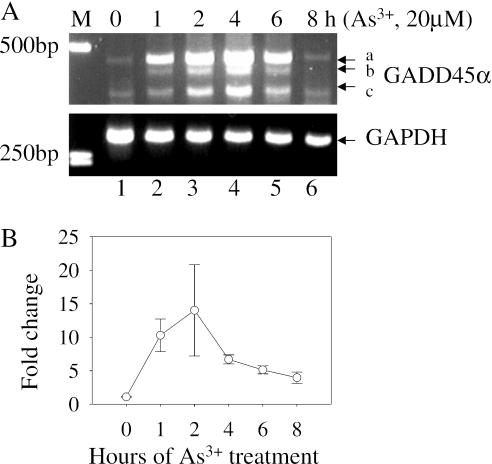
As3+ induces accumulation GADD45α mRNA
To demonstrate the correlation between the levels of protein and gene expression, the effect of As3+ on the induction of GADD45α mRNA was determined by RT–PCR. A substantial induction of GADD45α mRNA was observed in the cells treated with 20 µM As3+ for 1–6 h (Figure 3A). After 8 h, the GADD45α mRNA was declined to the basal level, indicating turnover of mRNA. The PCR primers we used correspond to the exon1 and exon4 region of the GADD45α gene, respectively, which amplify a fragment of GADD45α mRNA with a size of 453 bp (Figure 3A, fragment a). Interestingly, two additional fragments with size of ∼430 bp and ∼350 bp were observed in this RT–PCR analysis (Figure 3A, fragments ‘b’ and ‘c’), possibly resulted from mRNA alternative splicing. DNA sequencing indicated that the fragment ‘a’ is indeed the full-length GADD45α mRNA as expected. The fragment ‘c’ was resulted from the splicing out of the entire exon2 region (GenBank ID DQ008445), whereas the sequencing of fragment ‘b’ was inclusive (data not shown). The accumulation of the GADD45α mRNA induced by As3+ was further verified by a quantitative real-time RT–PCR. In fully agreement with the results of the traditional RT–PCR, a 6- to 10-fold induction of the GADD45α mRNA was observed in the cells treated with As3+ for 1–4 h (Figure 3B).
Figure 3.
As3+ induces accumulation of GADD45α mRNA. (A) The levels of GADD45α mRNA and GAPDH mRNA were determined by RT–PCR using cell lysates from the cells treated with 20 µM As3+ for the indicated times. The sizes of PCR products were estimated by the DNA molecular marker (M) in base pair (bp). The possible alternatively spliced fragments of GADD45α mRNA were indicated as ‘b’ and ‘c’. Data are representative of at least three experiments. (B) Fold changes in GADD45α expression were determined by real-time RT–PCR. BEAS-2B cells were treated with 20 µM As3+ for the indicated times and fold induction of the GADD45α mRNA was measured by real-time RT–PCR as described in ‘Materials and Methods’.
As3+ does not affect the degradation of GADD45α protein
We have observed a more than 10- to 20-fold induction of GADD45α protein and 6- to 10-fold increase of GADD45α mRNA by As3+ in our western blotting and RT–PCR experiments, respectively (Figures 1–3). However, we have failed to observe a significant transcriptional induction of the GADD45α gene by As3+ in both promoter/intron3 luciferase activity assay and nuclear run-on assay (Figure 1D and data not shown). In several other experimental settings, we have also tested the effect of As3+ on some different GADD45α promoter constructs that contain 1–2 kb promoter regions. In these experiments, we have failed to observe a more than a 3-fold induction of these promoter-luciferase activities by As3+ (D. Bhatia, V. Castranova and F. Chen, manuscript in preparation). Thus, we assume that As3+-induced GADD45α might be mainly through post-transcriptional mechanisms including alterations in mRNA or protein stability. We have failed to determine the protein stability of GADD45α by using cycloheximide (data not shown), since the GADD45α protein was barely detectable in the cells without As3+ treatment (Figures 1 and 2).
One possibility that As3+ induces accumulation of GADD45α protein is through interfering with either the ubiquitination of or the subsequent proteasome-mediated degradation of GADD45α. We have previously shown that As3+ induced proteasomal degradation of Cdc25C protein (37). Therefore, it is unlikely that As3+ induces GADD45α through inhibiting the proteolytic activity of the proteasome. To test whether As3+ is able to interfere with the process of GADD45α ubiquitination, the cells were pre-treated with a proteasome inhibitor, MG132, for 2 h and then treated with As3+ for 12 h. The ubiquitination of proteins can be visualized as smear high molecular weight bands in immunoblotting using lysates from the cells treated with MG132 or other proteasome inhibitors. The cell lysates were immunoprecipitated using antibody against GADD45α (Supplement Figure 1, lanes 1–4) or ubiquitin (Supplement Figure 1, lanes 5–8) and then the proteins in the immune complexes were immunoblotted with either anti-GADD45α antibody (Supplement Figure 1, upper panel) or re-probed with anti-ubiquitin antibody after stripping (Supplement Figure 1, lower panel). As can be seen in this figure, we did observe induction and ubiquitination of GADD45α in the cells pre-treated with MG132 in the absence of As3+. Treatment of the cells with As3+ did not decrease, but rather increased the ubiquitination of GADD45α protein. Thus, it is unlikely that As3+-induced accumulation of GADD45α is through preventing the ubiquitination of GADD45α protein.
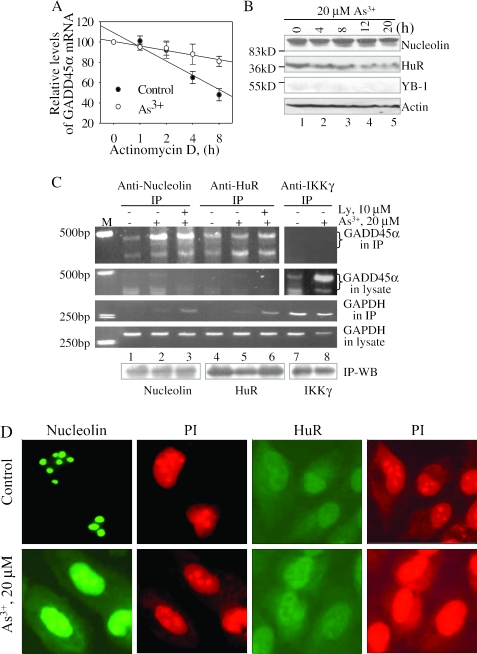
As3+ stabilizes GADD45α mRNA through nucleolin
Next, we tested the possibility that As3+ might be able to regulate the stability of GADD45α mRNA. To this end, cells were incubated with or without 20 µM As3+ for 4 h before that the transcription was blocked by adding 5 µg/ml of actinomycin D. The level of GADD45α mRNA was monitored by quantitative RT–PCR after 0, 1, 2, 4 or 8 h of post-actinomycin D treatment. As indicated in Figure 4A, the GADD45α mRNA from untreated cells displayed a strong reduction by almost 50% in the mRNA level after 8 h of transcription inhibition. In contrast, more than 80% GADD45α mRNA remained at this time point in the cells treated with As3+, indicating that As3+ stabilizes GADD45α mRNA substantially. The stability of the GAPDH mRNA was not affected by As3+. In fact, the GAPDH mRNA appears to be relatively stable (Supplement Figure 2).
Figure 4.
As3+ stabilizes GADD45α mRNA through nucleolin. (A) The stability of GADD45α mRNA was determined by RT–PCR using the cell lysates from the cells pre-treated with 20 µM As3+ for 4 h and then treated with 5 µg/ml of actinomycin D for the indicated times. (B) Cells were treated with 20 µM As3+ for the indicated time and then subjected to western blotting for nucleolin, HuR, YB-1 and actin. The protein standard with the known molecular weights (kD) was used to determine the positions of the indicated proteins on the membrane. (C) Cells were untreated or treated with 20 µM As3+ in the absence or presence of 10 µM Ly294002 (Ly) for 4 h and then disrupted with cell lysis buffer. Immunoprecipitation was performed with the antibodies against nucleolin (lanes 1–3), HuR (lanes 4–6) or IKKγ (lanes 7 and 8) at 4°C for overnight and then treated with protein A-Agarose for an additional 4 h. The mRNAs of GADD45α and GAPDH in the immune complex (IP) and cell lysates were determined by RT–PCR, respectively. The protein levels of nucleolin, HuR and IKKγ in the immune complexes were determined by western blotting (IP-WB). M: DNA marker. Data are representative of at least three experiments. (D) Immunofluorescence staining for the intracellular localization of nucleolin and HuR. The BEAS-2B cells were untreated or treated with 20 µM As3+ for 4 h. The localization of nucleolin and HuR were determined by indirect immunofluorescence using antibody against nucleolin or HuR and FITC-conjugated anti-rabbit IgG. The nuclei were stained by propidium iodide (PI).
The stability for many inducible mRNAs is regulated by a number of RNA-binding proteins that either stabilize or destabilize mRNAs. In mammalian cells, the functional characteristic of several mRNA stabilizing proteins, including nucleolin, HuR and YB-1, has been extensively investigated. To determine the involvement of these RNA-binding proteins in the regulation of GADD45α mRNA, the expression of nucleolin, HuR and YB-1 was investigated. As shown in Figure 4B, the expression of nucleolin and HuR was detectable under the basal condition. Addition of As3+ for 4, 8, 12 or 20 h did not change the level of nucleolin (Figure 4B, top panel), whereas the level of HuR was marginally decreased by As3+ in a roughly time-dependent manner (Figure 4B, the second panel). The expression of YB-1 was undetectable under the conditions tested.
To determine the binding of nucleolin and HuR to GADD45α mRNA, we next performed RNA immunoprecipitation, an established method described in the literatures (31,32), by using antibody against either nucleolin or HuR. The mRNAs of GADD45α and GAPDH in the immune complexes and the supernatants post-immunoprecipitation were determined by RT–PCR. In agreement with the western blotting data (Figure 4B), the amount of nucleolin in the immune complexes was unchanged upon treatment of cells with As3+, whereas the level of HuR was marginally reduced after the treatment of As3+ (Figure 4C, bottom panel). Trace amount of GADD45α mRNA in the control cells could be co-precipitated by either anti-nucleolin or anti-HuR antibody, indicating basal association of nucleolin and HuR with GADD45α mRNA. Treatment of the cells with 20 µM As3+ for 4 h increased the association of GADD45α with nucleolin (Figure 4C, the panel of GADD45α in IP, lane 2). As3+ was also capable of inducing the association of GADD45α mRNA with HuR, although in a less potent fashion compared with nucleolin (Figure 4C, the panel of GADD45α in IP, lane 5). Since there are reports indicating interconnection between PI3K-Akt and mRNA stability or nucleolin (38–40), we then tested the possible involvement of Akt signaling in the association of RNA-binding proteins with the GADD45α mRNA. Pre-treatment of the cells with Ly294002 inhibits phosphorylation of Akt (Figure 2). However, Ly294002 showed no effect on the As3+-induced association of GADD45α mRNA with nucleolin or HuR (Figure 4C, lanes 3 and 6). The association of GADD45α mRNA with nucleolin and HuR appeared to be specific, since there was no detectable GAPDH mRNA in the immune complexes (Figure 4C, the panel of GAPDH in IP, lanes 1, 2, 4 and 5). A non-specific association of GAPDH mRNA with either nucleolin or HuR was observed in the cells pre-treated with Ly294002 alone (data not shown) or in the presence of As3+ (Figure 4C, lanes 3 and 6). We also monitored the levels of GADD45α mRNA in the supernatants after immunoprecipitation with anti-nucleolin and anti-HuR antibody, respectively. The GADD45α mRNA was barely detected in these supernatants (Figure 4C, the panel of GADD45α in lysate), indicating that the majority of GADD45α mRNA had been co-precipitated by immunoprecipitation for either nucleolin or HuR. In a control experiment, we used an antibody against IKKγ in immunoprecipitation and found no association of GADD45α mRNA with IKKγ protein in the cells without or with As3+ treatment (Figure 4C, top panel, lanes 7 and 8). The basal and As3+-induced GADD45α mRNAs remained in the cell lysates that had been subjected to IKKγ immunoprecipitation (Figure 4C, the ‘GADD45 in lysate’ panel, comparing lanes 7 and 8 with lanes 1–6). Therefore, these data strongly suggest that the stabilization of GADD45α mRNA by As3+ is through the inducible binding of nucleolin and less potently, HuR to GADD45α mRNA.
As3+ appeared to be very potent in inducing binding of nucleolin to the GADD45α mRNA (Figure 4C). However, As3+ was unable to influence the expression of nucleolin (Figure 4B). Thus, it is worth testing whether the functional aspect of nucleolin was modulated by As3+. For that purpose, we investigated the intracellular location of nucleolin in the cells without or with As3+ treatment by immunofluorescent techniques. In control cells, nucleolin was concentrated in nucleoli (Figure 4D, top panel). Following treatment of the cells with 20 µM As3+ for 4 h, a notable intracellular re-distribution of nucleolin from nucleoli to nucleoplasm was observed (Figure 4D, bottom panel). In addition, some As3+-treated cells showed cytoplasm staining of nucleolin. In both control cells and the cells treated with As3+, the HuR protein was localized throughout nucleoplasm and cytoplasm, but was predominantly stained in nuclei (Figure 4D).
Nucleolin silencing reversed As3+-induced stabilization of the GADD45α mRNA
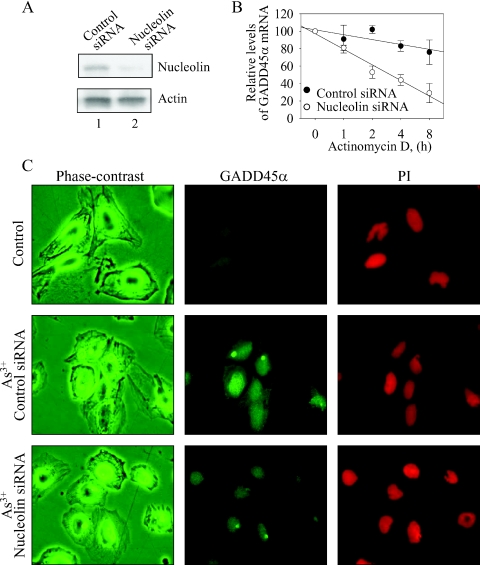
To address the importance of nucleolin in As3+-induced stabilization of the GADD45α mRNA, we next used small interference RNA (siRNA) technique to knockdown nucleolin and determined the mRNA stability of the GADD45α in the cells treated with As3+. As indicated in Figure 5A, nucleolin siRNA effectively reduced the level of nucleolin protein after 36 h of siRNA transfection, whereas the control siRNA against luciferase showed no inhibition on the level of the nucleolin protein. The data of mRNA stability analysis by a quantitative RT–PCR showed a significant decrease in the stability of the GADD45α mRNA induced by As3+ in the cells transfected with nucleolin siRNA (Figure 5B, comparing the control siRNA with the nucleolin siRNA).
Figure 5.
Knockdown of nucleolin reverses As3+-induced stabilization of the GADD45α mRNA. (A) The BEAS-2B cells were transfected with the control siRNA against firefly luciferase and the nucleolin siRNA for 24 h. The protein levels of nucleolin and actin were determined by western blotting. (B) Stability analysis of the GADD45α mRNA in the cells treated with As3+ and transfected with the control or nucleolin siRNA. (C) Immunofluorescence staining for the expression of the GADD45α protein. The BEAS-2B cells were first transfected with either the control siRNA or nucleolin siRNA for 36 h. Then the cells were either untreated or treated with 20 µM As3+ for 4 h. The expression of GADD45α protein was determined by indirect immunofluorescence using antibody against GADD45α and FITC-conjugated anti-rabbit IgG (middle column). The cell morphology was shown as phase-contrast images (left column). The nuclei were stained by propidium iodide (PI) (right column).
Finally, we examined the effect of nucleolin siRNA on the induction of GADD45α protein induced by As3+. In agreement with the observations in western blotting (Figures 1 and 2), immunofluorescent staining showed that the GADD45α protein was undetectable in the cells without As3+ treatment (Figure 5C, top panels). A substantial elevation of nuclear-stained GADD45α protein was observed in the cells treated with As3+ (Figure 5C, middle panels). Transfection of the cells with nucleolin siRNA partially diminished the increase of GADD45α protein induced by As3+ (Figure 5C, bottom panels).
DISCUSSION
In this report, we have provided evidence that As3+-induced expression of GADD45α is through both transcriptional and more importantly, post-transcriptional mechanisms: stabilization of GADD45α mRNA. We have demonstrated that the accumulation of GADD45α mRNA induced by As3+ is very likely due to the inducible binding of nucleolin, and less potently, HuR, two RNA stabilizing proteins, to the GADD45α mRNA. Silencing of nucleolin by an siRNA specifically targeting nucleolin reversed As3+-induced stabilization of the GADD45α mRNA and elevation of the GADD45α protein.
A number of stress signals can induce accumulation of GADD45α mRNA or protein. Oxidative stress due to the generation of reactive oxygen species appears to be a common feature in cellular responses to a variety of stress signals, such as As3+- or inflammatory cytokine-induced stress responses (30,41). It is plausible, therefore, to assume that the induction of GADD45α by As3+ is mediated by oxidative stress. Indeed, pre-treatment of the cells with antioxidants prevented As3+-induced accumulation of GADD45α protein (Figure 1). However, administration of the cells with the exogenous reactive oxygen species, H2O2, only resulted in a marginal induction of GADD45α (Figure 1B). The reporter gene assay using GADD45α promoter and intron3 constructs indicated that As3+ regulated GADD45α promoter and intron3 activity, whereas H2O2 only exhibited its effect on intron3 (Figure 1D). Thus, these data provide evidence indicating that As3+-induced GADD45α is independent of oxidative stress.
Transcriptional up-regulation appears to be the most important and common mechanism in genes encoding stress response proteins. The majority studies on the expression of GADD45α induced by a variety of stress signals focused on the transcription of the GADD45α gene. The data presented in this study suggest that As3+ has a very limited effect on the transcription of the GADD45α gene, as can be seen in both reporter gene activity analysis and nuclear run-on assay (Figure 1D and data not shown). However, As3+ appeared to be very capable of inducing a nucleolin-dependent stabilization of the GADD45α mRNA. These observations are compensatory to a recent study by Fan et al. (42) who showed an increased ratio of UVC-induced GADD45α mRNA transcript versus UVC-induced transcription in a nuclear run-on assay (Supplementary Table 2, row 938). The involvement of nucleolin in GADD45α mRNA stabilization was further verified by siRNA-mediated gene knockdown of nucleolin, which reversed As3+-induced stabilization of the GADD45α mRNA. Immunoblotting and immunofluorescent staining suggested that nucleolin was constitutively expressed in the cells used in the present studies. Although As3+ exhibited no influence on the expression of nucleolin protein, As3+ was able to induce intracellular re-distribution of nucleolin from nucleoli to nucleoplasm. This could be an indication in the functional up-regulation of nucleolin in response to As3+, which contributes to the stabilization of the GADD45α mRNA.
Several earlier reports suggested that UV, DNA-damaging agents, retinoid CD437 or glutamine deprivation induced GADD45α through stabilization of GADD45α mRNA in Chinese hamster ovary cells or human breast carcinoma cell lines (43–45). It was unclear, however, how the stability of GADD45α mRNA was regulated in these cells under such conditions. The findings that nucleolin and less potently, HuR, bind to GADD45α mRNA in the cellular response to As3+ (Figure 4C) provide a mechanistic explanation for the stress-induced accumulation of GADD45α. Nucleolin is a ubiquitous nucleolar phosphoprotein that consists of four RNA-binding domains that are responsible for the binding of this protein to pre-rRNA or mRNA (46). In addition, nucleolin has also been implicated as the human helicase IV that destabilizes helices of DNA–DNA, DNA–RNA and RNA–RNA (47). Accumulating evidence indicates that nucleolin is a key protein involved in the post-transcriptional regulation of mRNAs. Previous studies by other laboratories suggested that nucleolin was able to stabilize mRNAs of IL-2 (31), β-amyloid precursor protein (APP) (48), bcl2 (49), renin (50) and CD154 (51). In response to T-cell activation, nucleolin stabilizes IL-2 mRNA by interacting with the 5′-untranslated region (UTR) of IL-2 mRNA in a JNK-dependent manner (31). Recently, we have demonstrated an oxidative stress-mediated binding of nucleolin to mouse GADD45α mRNA in mouse fibroblast cells (52). In an in vitro analysis for the selection of mRNA ligands by nucleolin, Yang et al. (53) demonstrated a binding of nucleolin to a number of other mRNAs, such as heat shock protein 90, glutathione peroxidase, peroxiredoxin 1, etc. Several lines of evidence indicate that nucleolin binds to pre-rRNA that contains a consensus sequence, (U/G)CCCG(A/G), in a loop of stem structure with 7–14 bp (54). Although the recognition elements of nucleolin in the 5′- or 3′-UTR of IL-2, APP, bcl2 and CD154 have been identified, no consensus sequence or homology sequence has been found in these mRNAs. Sequence comparison suggested that there is no sequence similarity among the 5′-UTRs of GADD45α mRNA and the mRNAs of IL-2, APP, bcl2 or CD154. However, it is interesting to note that both 5′- and 3′-UTR of human GADD45α mRNA contain a potential stem–loop with sequence, GCCCGG. This sequence matches completely with the nucleolin recognition element, (T/G)CCCG(A/G), in pre-rRNA (54).
Nucleolin has also been implicated in the cap-independent but internal ribosome entry site (IRES)-dependent translation of hepatitis C virus (55). Analysis of the 5′-UTR region of human GADD45α mRNA revealed a potential IRES domain proximal to the AUG code. We have recently observed that As3+ was also very potent in the induction of GADD45α protein in the growth-arrested cells where the general protein synthesis machinery was inhibited by rapamycin (data not shown). This phenomenon is very likely due to the IRES-dependent translational regulation. Whether nucleolin or other factors participated in this process remains to be investigated.
In summary, our data suggest that elevation in the expression of GADD45α in cellular response to As3+ is mainly through the regulation of mRNA stability of GADD45α. Treatment of the cells with As3+ increased binding of nucleolin and to lesser extent, HuR to the mRNA of GADD45α, which extends the half-life of GADD45α mRNA. It is unknown at present how the association of nucleolin with the GADD45α mRNA is regulated, despite we noted a re-distribution of nucleolin protein from nucleoli to nucleoplasm in the cells treated with As3+. Changes in mRNA stability have been considered important mechanisms in which cells sense stress or damage in concert with transcriptional and/or other mechanisms. The contributions of GADD45α mRNA stabilization to the cell cycle regulation and apoptosis are currently under investigation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Dr Albert J. Fornace (NIH, Bethesda, MD) for providing luciferase reporter vectors containing GADD45α promoter, intron1 or intron3. The authors are grateful to Dr Murali Rao and Mr Terence G. Meighan at National Institute for Occupational Safety and Health for assistance in real-time RT–PCR of GADD45α mRNA. Funding to pay the Open Access publication charges for this article was provided by annual budget of US government agency.
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhan Q., Bae I., Kastan M.B., Fornace A.J., Jr The p53-dependent gamma-ray response of GADD45. Cancer Res. 1994;54:2755–2760. [PubMed] [Google Scholar]
- 2.Wang X.W., Zhan Q., Coursen J.D., Khan M.A., Kontny H.U., Yu L., Hollander M.C., O'Connor P.M., Fornace A.J., Jr, Harris C.C. GADD45 induction of a G2/M cell cycle checkpoint. Proc. Natl Acad. Sci. USA. 1999;96:3706–3711. doi: 10.1073/pnas.96.7.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollander M.C., Sheikh M.S., Bulavin D.V., Lundgren K., Augeri-Henmueller L., Shehee R., Molinaro T.A., Kim K.E., Tolosa E., Ashwell J.D., et al. Genomic instability in Gadd45a-deficient mice. Nature Genet. 1999;23:176–184. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 4.Salvador J.M., Hollander M.C., Nguyen A.T., Kopp J.B., Barisoni L., Moore J.K., Ashwell J.D., Fornace A.J., Jr Mice lacking the p53-effector gene Gadd45a develop a lupus-like syndrome. Immunity. 2002;16:499–508. doi: 10.1016/s1074-7613(02)00302-3. [DOI] [PubMed] [Google Scholar]
- 5.Hildesheim J., Fornace A.J., Jr Gadd45a: an elusive yet attractive candidate gene in pancreatic cancer. Clin. Cancer Res. 2002;8:2475–2479. [PubMed] [Google Scholar]
- 6.Smith M.L., Chen I.T., Zhan Q., Bae I., Chen C.Y., Gilmer T.M., Kastan M.B., O'Connor P.M., Fornace A.J., Jr Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science. 1994;266:1376–1380. doi: 10.1126/science.7973727. [DOI] [PubMed] [Google Scholar]
- 7.Kovalsky O., Lung F.D., Roller P.P., Fornace A.J., Jr Oligomerization of human Gadd45a protein. J. Biol. Chem. 2001;276:39330–39339. doi: 10.1074/jbc.M105115200. [DOI] [PubMed] [Google Scholar]
- 8.Carrier F., Georgel P.T., Pourquier P., Blake M., Kontny H.U., Antinore M.J., Gariboldi M., Myers T.G., Weinstein J.N., Pommier Y., et al. Gadd45, a p53-responsive stress protein, modifies DNA accessibility on damaged chromatin. Mol. Cell. Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W., Nahta R., Huper G., Marks J.R. TAFII70 isoform-specific growth suppression correlates with its ability to complex with the GADD45a protein. Mol. Cancer Res. 2004;2:442–452. [PubMed] [Google Scholar]
- 10.Salvador J.M., Mittelstadt P.R., Bolova G.I., Fornace A.J., Jr, Ashwell J.D. The autoimmune suppressor Gadd45alpha inhibits the T cell alternative p38 activation pathway. Nature Immunol. 2005;6:396–402. doi: 10.1038/ni1176. [DOI] [PubMed] [Google Scholar]
- 11.Mita H., Tsutsui J., Takekawa M., Witten E.A., Saito H. Regulation of MTK1/MEKK4 kinase activity by its N-terminal autoinhibitory domain and GADD45 binding. Mol. Cell. Biol. 2002;22:4544–4555. doi: 10.1128/MCB.22.13.4544-4555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An W., Kim J., Roeder R.G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Zhan Q., Chen I.T., Antinore M.J., Fornace A.J., Jr Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. Mol. Cell. Biol. 1998;18:2768–2778. doi: 10.1128/mcb.18.5.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran H., Brunet A., Grenier J.M., Datta S.R., Fornace A.J., Jr, , DiStefano P.S., Chiang L.W., Greenberg M.E. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi S., Saito S., Ohtani N., Sakai T. Involvement of the Oct-1 regulatory element of the gadd45 promoter in the p53-independent response to ultraviolet irradiation. Cancer Res. 2001;61:1187–1195. [PubMed] [Google Scholar]
- 16.Tao H., Umek R.M. Reciprocal regulation of gadd45 by C/EBP alpha and c-Myc. DNA Cell Biol. 1999;18:75–84. doi: 10.1089/104454999315646. [DOI] [PubMed] [Google Scholar]
- 17.Thyss R., Virolle V., Imbert V., Peyron J.F., Aberdam D., Virolle T. NF-kappaB/Egr-1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death. EMBO J. 2005;24:128–137. doi: 10.1038/sj.emboj.7600501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollander M.C., Alamo I., Jackman J., Wang M.G., McBride O.W., Fornace A.J., Jr Analysis of the mammalian gadd45 gene and its response to DNA damage. J. Biol. Chem. 1993;268:24385–24393. [PubMed] [Google Scholar]
- 19.Bush A., Mateyak M., Dugan K., Obaya A., Adachi S., Sedivy J., Cole M. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng L., Pan H., Li S., Flesken-Nikitin A., Chen P.L., Boyer T.G., Lee W.H. Sequence-specific transcriptional corepressor function for BRCA1 through a novel zinc finger protein, ZBRK1. Mol. Cell. 2000;6:757–768. doi: 10.1016/s1097-2765(00)00075-7. [DOI] [PubMed] [Google Scholar]
- 21.Tchounwou P.B., Patlolla A.K., Centeno J.A. Carcinogenic and systemic health effects associated with arsenic exposure—a critical review. Toxicol. Pathol. 2003;31:575–588. doi: 10.1080/01926230390242007. [DOI] [PubMed] [Google Scholar]
- 22.Kitchin K.T. Recent advances in arsenic carcinogenesis: modes of action, animal model systems, and methylated arsenic metabolites. Toxicol. Appl. Pharmacol. 2001;172:249–261. doi: 10.1006/taap.2001.9157. [DOI] [PubMed] [Google Scholar]
- 23.Barchowsky A., Roussel R.R., Klei L.R., James P.E., Ganju N., Smith K.R., Dudek E.J. Low levels of arsenic trioxide stimulate proliferative signals in primary vascular cells without activating stress effector pathways. Toxicol. Appl. Pharmacol. 1999;159:65–75. doi: 10.1006/taap.1999.8723. [DOI] [PubMed] [Google Scholar]
- 24.Waxman S., Anderson K.C. History of the development of arsenic derivatives in cancer therapy. Oncologist. 2001;6(Suppl. 2):3–10. doi: 10.1634/theoncologist.6-suppl_2-3. [DOI] [PubMed] [Google Scholar]
- 25.Chen F., Lu Y., Zhang Z., Vallyathan V., Ding M., Castranova V., Shi X. Opposite effect of NF-kappa B and c-Jun N-terminal kinase on p53-independent GADD45 induction by arsenite. J. Biol. Chem. 2001;276:11414–11419. doi: 10.1074/jbc.M011682200. [DOI] [PubMed] [Google Scholar]
- 26.Liu J., Kadiiska M.B., Liu Y., Lu T., Qu W., Waalkes M.P. Stress-related gene expression in mice treated with inorganic arsenicals. Toxicol. Sci. 2001;61:314–320. doi: 10.1093/toxsci/61.2.314. [DOI] [PubMed] [Google Scholar]
- 27.Chen F., Zhang Z., Leonard S.S., Shi X. Contrasting roles of NF-kappaB and JNK in arsenite-induced p53-independent expression of GADD45alpha. Oncogene. 2001;20:3585–3589. doi: 10.1038/sj.onc.1204442. [DOI] [PubMed] [Google Scholar]
- 28.Tong T., Fan W., Zhao H., Jin S., Fan F., Blanck P., Alomo I., Rajasekaran B., Liu Y., Holbrook N.J., et al. Involvement of the MAP kinase pathways in induction of GADD45 following UV radiation. Exp. Cell Res. 2001;269:64–72. doi: 10.1006/excr.2001.5312. [DOI] [PubMed] [Google Scholar]
- 29.Yin F., Bruemmer D., Blaschke F., Hsueh W.A., Law R.E., Herle A.J. Signaling pathways involved in induction of GADD45 gene expression and apoptosis by troglitazone in human MCF-7 breast carcinoma cells. Oncogene. 2004;23:4614–4623. doi: 10.1038/sj.onc.1207598. [DOI] [PubMed] [Google Scholar]
- 30.Chen F., Castranova V., Li Z., Karin M., Shi X. Inhibitor of nuclear factor kappaB kinase deficiency enhances oxidative stress and prolongs c-Jun NH2-terminal kinase activation induced by arsenic. Cancer Res. 2003;63:7689–7693. [PubMed] [Google Scholar]
- 31.Chen C.Y., Gherzi R., Andersen J.S., Gaietta G., Jurchott K., Royer H.D., Mann M., Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 32.Katsanou V., Papadaki O., Milatos S., Blackshear P.J., Anderson P., Kollias G., Kontoyiannis D.L. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y., Lu Y., Yuan B.Z., Castranova V., Shi X., Stauffer J.L., Demers L.M., Chen F. The Human mineral dust-induced gene, mdig, is a cell growth regulating gene associated with lung cancer. Oncogene. 2005;24:4873–4882. doi: 10.1038/sj.onc.1208668. [DOI] [PubMed] [Google Scholar]
- 35.Datta S.R., Brunet A., Greenberg M.E. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 36.Brazil D.P., Park J., Hemmings B.A. PKB binding proteins. Getting in on the Akt. Cell. 2002;111:293–303. doi: 10.1016/s0092-8674(02)01083-8. [DOI] [PubMed] [Google Scholar]
- 37.Chen F., Zhang Z., Bower J., Lu Y., Leonard S.S., Ding M., Castranova V., Piwnica-Worms H., Shi X. Arsenite-induced Cdc25C degradation is through the KEN-box and ubiquitin-proteasome pathway. Proc. Natl Acad. Sci. USA. 2002;99:1990–1995. doi: 10.1073/pnas.032428899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ricupero D.A., Poliks C.F., Rishikof D.C., Cuttle K.A., Kuang P.P., Goldstein R.H. Phosphatidylinositol 3-kinase-dependent stabilization of alpha1(I) collagen mRNA in human lung fibroblasts. Am. J. Physiol. Cell Physiol. 2001;281:C99–C105. doi: 10.1152/ajpcell.2001.281.1.C99. [DOI] [PubMed] [Google Scholar]
- 39.Barel M., Balbo M., Le Romancer M., Frade R. Activation of Epstein–Barr virus/C3d receptor (gp140, CR2, CD21) on human cell surface triggers pp60src and Akt-GSK3 activities upstream and downstream to PI 3-kinase, respectively. Eur. J. Immunol. 2003;33:2557–2566. doi: 10.1002/eji.200324059. [DOI] [PubMed] [Google Scholar]
- 40.Borgatti P., Martelli A.M., Tabellini G., Bellacosa A., Capitani S., Neri L.M. Threonine 308 phosphorylated form of Akt translocates to the nucleus of PC12 cells under nerve growth factor stimulation and associates with the nuclear matrix protein nucleolin. J. Cell Physiol. 2003;196:79–88. doi: 10.1002/jcp.10279. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Chen F. Reactive oxygen species (ROS), troublemakers between nuclear factor-kappaB (NF-kappaB) and c-Jun NH(2)-terminal kinase (JNK) Cancer Res. 2004;64:1902–1905. doi: 10.1158/0008-5472.can-03-3361. [DOI] [PubMed] [Google Scholar]
- 42.Fan J., Yang X., Wang W., Wood W.H., III, Becker K.G., Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl Acad. Sci. USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackman J., Alamo I., Jr, Fornace A.J., Jr Genotoxic stress confers preferential and coordinate messenger RNA stability on the five gadd genes. Cancer Res. 1994;54:5656–5662. [PubMed] [Google Scholar]
- 44.Abcouwer S.F., Schwarz C., Meguid R.A. Glutamine deprivation induces the expression of GADD45 and GADD153 primarily by mRNA stabilization. J. Biol. Chem. 1999;274:28645–28651. doi: 10.1074/jbc.274.40.28645. [DOI] [PubMed] [Google Scholar]
- 45.Rishi A.K., Sun R.J., Gao Y., Hsu C.K., Gerald T.M., Sheikh M.S., Dawson M.I., Reichert U., Shroot B., Fornace A.J., Jr, et al. Post-transcriptional regulation of the DNA damage-inducible gadd45 gene in human breast carcinoma cells exposed to a novel retinoid CD437. Nucleic Acids Res. 1999;27:3111–3119. doi: 10.1093/nar/27.15.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ginisty H., Sicard H., Roger B., Bouvet P. Structure and functions of nucleolin. J. Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 47.Srivastava M., Pollard H.B. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- 48.Zaidi S.H., Malter J.S. Nucleolin and heterogeneous nuclear ribonucleoprotein C proteins specifically interact with the 3′-untranslated region of amyloid protein precursor mRNA. J. Biol. Chem. 1995;270:17292–17298. doi: 10.1074/jbc.270.29.17292. [DOI] [PubMed] [Google Scholar]
- 49.Sengupta T.K., Bandyopadhyay S., Fernandes D.J., Spicer E.K. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J. Biol. Chem. 2004;279:10855–10863. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- 50.Skalweit A., Doller A., Huth A., Kahne T., Persson P.B., Thiele B.J. Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3′-untranslated region. Circ. Res. 2003;92:419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- 51.Singh K., Laughlin J., Kosinski P.A., Covey L.R. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J. Immunol. 2004;173:976–985. doi: 10.4049/jimmunol.173.2.976. [DOI] [PubMed] [Google Scholar]
- 52.Zheng X., Zhang Y., Chen Y., Castranova V., Shi X., Chen F. Inhibition of NF-kappaB stabilizes Gadd45alpha mRNA. Biochem. Biophys. Res. Commun. 2005;329:95–99. doi: 10.1016/j.bbrc.2005.01.105. [DOI] [PubMed] [Google Scholar]
- 53.Yang C., Maiguel D.A., Carrier F. Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res. 2002;30:2251–2260. doi: 10.1093/nar/30.10.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghisolfi-Nieto L., Joseph G., Puvion-Dutilleul F., Amalric F., Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J. Mol. Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- 55.Lu H., Li W., Noble W.S., Payan D., Anderson D.C. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J. Proteome Res. 2004;3:949–957. doi: 10.1021/pr0499592. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.