Abstract
During culture, a chicken B cell line DT40 spontaneously mutates immunoglobulin (Ig) genes by gene conversion, which involves activation-induced cytidine deaminase (AID)-dependent homologous recombination of the variable (V) region gene with upstream pseudo-V genes. To explore whether this mutation mechanism can target exogenous non-Ig genes, we generated DT40 lines that bears a gene conversion substrate comprising the green fluorescent protein (GFP) gene as a donor and the blue fluorescent protein (BFP) gene as an acceptor. A few percent of the initially BFP-expressing cells converted their fluorescence from blue to green after culture for 2–3 weeks when the substrate construct was integrated in the Ig light chain locus, but not in the ovalbumin locus. This was the result of AID-dependent and the GFP gene-templated gene conversion of the BFP gene, thereby leading to the introduction of various sizes of GFP-derived gene segment into the BFP gene. Thus, G/B construct may be used to visualize gene conversion events. After switching off AID expression in DT40 cells, the mutant clones were isolated stably and maintained with their mutations being fixed. Thus, the gene conversion machinery in DT40 cells will be a useful means to engineer non-Ig proteins by a type of DNA shuffling.
INTRODUCTION
The immune system generates high-affinity antibodies (Abs) by the combination of two consecutive processes, hypermutation of immunoglobulin (Ig) variable region (V) genes followed by positive selection of B cells that acquired mutated high-affinity Abs (1). Thus, an Ab response is regarded as a highly efficient molecular evolution system. Distinct from human and mouse B cells, chicken B cells diversify Ig V genes not only by point mutation but also by a process termed gene conversion, which involves iterative homologous recombination of a rearranged V gene with a group of non-productive pseudo-V genes (2,3). Both point mutation and gene conversion of Ig genes have been shown to be mediated by a key enzyme, activation-induced cytidine deaminase (AID) (4–7). If the chicken gene conversion system is able to target exogenous genes other than Ig genes, it will provide a unique means for the mutational diversification of target proteins, which is generally a critical step in protein engineering. In addition, if an appropriate gene conversion substrate is designed to visualize gene conversion events in DT40 cells, it will be useful to analyze the molecular mechanism of AID-dependent gene conversion.
For exploring this possibility, a chicken B cell line DT40 has several advantages as follows: (i) DT40 cells constitutively express AID, and spontaneously mutate Ig genes by gene conversion during culture (6–9); (ii) An engineered DT40 line whose AID expression can be reversibly switched on or off by tamoxifen-regulated Cre recombinase has been established in our laboratory, thus enabling to fix desired mutations by stopping gene conversion (10); and (iii) Targeted integration of an exogenous gene can be conducted with high frequency, thereby making the cells genetically tractable (11). To test whether the gene conversion machinery in DT40 can target exogenous non-Ig protein genes, we designed a gene conversion substrate that contains the green fluorescence protein (GFP) gene as a donor and the blue fluorescence protein (BFP) gene as an acceptor. Here, we show that this gene construct worked as a substrate of chicken gene conversion system in the same fashion as Ig genes when it was integrated in the Ig light chain (IgL) locus.
MATERIALS AND METHODS
Cell culture and transfection
The DT40 cell line was obtained from RIKEN Cell Bank (Tsukuba, Japan). An engineered DT40 line, DT40-SW whose AID expression can be switched on or off by Cre-mediated recombination, was established as reported previously (10). DT40 cell lines were cultured in RPMI 1640 medium (ICN Pharmaceuticals) supplemented with 10% fetal bovine serum (Life Technologies), 1% chicken serum (Sigma), 50 µM 2-mercaptethanol, 2 mM glutamine, 1 mM pyruvic acid, 100 µg/ml penicillin G and 50 µg/ml streptomycin at 37°C in 5% CO2 and 95% air.
In transfection experiments, DT40 or DT40-SW cells were suspended in 250 µl of phosphate-buffered saline (PBS) at 2 × 107 cells/ml and transfected with 15 µg of a linearized vector by electroporation using Gene Pulser Xcell (Bio-Rad) at 550 V and 25 µF in 4 mm cuvette. Transfected DT40 cells were selected in a medium containing 50 µg/ml of blasticidin S (Invitrogen) or 0.5 µg/ml of puromycin (Sigma). Switching of AID expression by 4-OHT (Sigma) in DT40-SW cells was carried out as described previously (10).
Plasmid construction
Primers used in this study are listed as follows: CVLF6, CCMUCLAR, CVLR3 (12), SV-1, cCL3 (10), CPVL2 5′-CAGACATTATGTTTATGCTTTACAGCAGCA-3′, CVL3FR 5′-CTTCGGATCCAAGTCTGAACATGCC-3′, CVL5FR 5′-GAGCGGCCTGGATCCTGCGGAACCAA-3′, OVA1 5′-GATCCTCGAGGAACACTGCAAGTTCATATC-3′, OVA2 5′-TCAGATCTTGGTCGACAGGATAAGATGTTCTGA-3′, OVA3 5′-TTAGATCTCGTCGACCTGAAACTCTACAGTCTTC-3′, OVA4 5′-GAGCTCGAGCCACTACAGTAACAGCTGC-3′, OVAR 5′-GAAGCTCCTCAGTGCACAGGAATGGAGATA-3′, PCIR1 5′-CAGAGCAGATTGTACTGAGAGTGC-3′, SVR 5′-CTGCATTCTAGTTGTGGTTTGTCC-3′ and PCIF1 5′-GCACCTATTGGTCTTACTGACATC-3′. KOD Plus DNA polymerase (TOYOBO) was used for PCR amplification as instructed by the manufacturer. PCR products were confirmed by sequencing.
To construct gene conversion substrates, the BFP gene in the NheI–NotI fragment of the pEBFP-N1 plasmid (Clontech) was modified with removal of the BglII–BamHI stretch, and inserted into the plasmid pCI-puro (13) to yield 0/B construct, pCIB1. The GFP gene in the SmaI–HpaI fragment derived from pEGFP-N1 (Clontech) was ligated into the blunt-ended BamHI site upstream of the BFP expression unit in the pCIB1 to yield a G/B construct, pCIGB1.
Targeting vectors of 0/B and G/B constructs for the IgL or OVA locus were constructed as follows. The 5′- and 3′-flanking regions of the rearranged IgL genomic DNA were amplified by PCR with primers CPVL2 and CVLR3, and CVLF6 and CCMUCLAR, respectively, and cloned into pCR-Blunt (Invitrogen). The EcoRI fragment of the cloned 3′-region was subcloned with a HindIII linker into the HindIII–EcoRI region of pEGFP-N1, and the BamHI fragment of the cloned 5′-region was inserted into the BglII site of the pEGFP-N1 in the same orientation with the 3′-region. A fragment containing the subcloned 5′- and 3′-franking regions was amplified with primers CVL5FR and CVL3FR, digested by BamHI, and inserted into the BamHI site of pCIB1 and pCIGB1 to yield targeting vectors of 0/B and G/B constructs for the IgL locus, pVLB1 and pVLGB1, respectively. Similarly, two genomic DNA fragments that contain the 5′-noncoding region and exons 4–6 of the OVA gene were amplified by PCR with primers OVA1 and OVA2, and OVA3 and OVA4, respectively, and cloned into pCR-Blunt. The XhoI fragment containing the 5′-noncoding region of OVA gene was ligated into the XhoI site of the plasmid containing the exons 4–6, and then the BglII fragment containing these two regions was inserted into the BamHI site of pCIB1 and pCIGB1 to yield targeting vectors of 0/B and G/B constructs for the OVA locus, pOVB1 and pOVGB1, respectively.
To apply G/B construct for DT40-SW cells where puror had been integrated as an element of the switching device of AID gene expression, the puror of pVLGB1 was replaced with the blasticidin S resistance gene (Bsr). The BamHI fragment of pLoxBsr (14) was modified by deleting the XhoI site, and inserted with a PvuII linker in the PvuII–BamHI stretch of pCIGB1. IgL gene fragments were inserted in the same manner with pVLGB1 to yield pVLGB2. To estimate the segment size of gene conversion of G/B construct, six silent mutations, C66T, C132A, C276T, C354T, C525A and C621T, were introduced into the GFP gene of pEGFP-N1 by QuickChange II site-directed mutagenesis kit (Stratagene) as instructed by the vender. The mutated GFP gene was used for constructing another G/B construct for targeting to the IgL locus, pVLMGB2, from pVLGB2.
An AID-disrupting vector has been described previously (10). For disruption of the second allele of the AID gene, the puror was replaced with the Bsr derived from pLoxBsr. Reporter constructs and AID-disrupting vectors were linearized by XhoI and KpnI, respectively, before transfection.
The targeted integration to the IgL and OVA loci was confirmed by PCR using primers cCL3 and PCIR1 for IgL loci (40 cycles of 15 s at 94°C, 30 s at 64°C and 3.5 min at 68°C), and OVAF and PCIR1 for OVA loci (35 cycles of 15 s at 94°C, 30 s at 64°C and 3.5 min at 68°C), respectively. The β-actin gene was used as an internal control (10). Targeting of 0/B or G/B construct to the rearranged IgL locus was confirmed by PCR with primers CVLF6 and CVLR3. Disruption and transcription of the AID gene was assessed by PCR and RT–PCR, respectively, as described previously (6,10).
Analysis of gene conversion products of G/B construct
The expression of fluorescent proteins was analyzed with FACS Calibur and FACS Aria (BD Biosciences). Green cells were sorted by using FACS Aria equipped with Auto Cell Deposit Unit for single cell isolation (BD Biosciences). The purity of the sorted cells was >95%. Cells were also analyzed with a confocal laser-scanning microscope MRC-1024 (Bio-Rad). To analyze sequences of the GFP and BFP genes of G/B constructs targeted in the IgL and OVA loci, genomic DNA was extracted from sorted green cells, and the GFP and BFP genes were amplified by PCR using primers cCL3 and PCIR1, or OVAR and PCIR1 for the GFP gene on the IgL or OVA locus, respectively (40 cycles of 15 s at 94°C, 30 s at 64°C and 2 min at 68°C), and PCIF1 and SVR for the BFP gene (40 cycles of 15 s at 94°C, 30 s at 58°C and 1 min at 68°C). In order to specifically amplify the GFP and BFP genes, primers were designed to be specific to the franking sequences of target genes. PCR products were cloned into pCR-Blunt, and sequenced with ABI PRISM 310 Genetic Analyzer (Applied Biosystems).
RESULTS
Construction of a gene conversion substrate and its integration in the IgL locus in DT40 cells
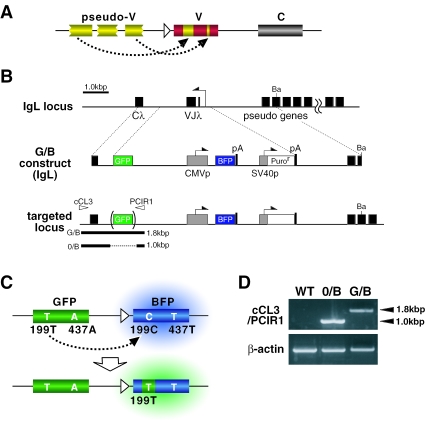
Since the gene conversion event introduces a sequence copied from an upstream pseudo-V gene (a donor) into a homologous site in the expressed V gene (an acceptor) (Figure 1A), we designed a gene conversion substrate named G/B construct, in which the GFP gene without promoter as a donor, the cytomegalovirus (CMV) promoter-driven BFP gene as an acceptor, and the SV40 promoter-driven puromycin-resistant gene (puror) were arranged in tandem in this order (Figure 1B). 0/B construct, the GFP-depleted version of G/B construct, was also prepared as the donor-negative control. BFP is a blue-shifted derivative of GFP. The humanized versions of BFP and GFP genes (714 bp) have the same nucleotide sequence except two nucleotides at positions 199 and 437, 199C and 437T for BFP and 199T and 437A for GFP, respectively (Figure 1C) (15). The codon containing the 199th nucleotide encodes the 67th amino acid, His for BFP and Tyr for GFP, which have been shown to be critical for their characteristic fluorescence colors (16).
Figure 1.
A gene conversion substrate using a non-Ig exogenous gene. (A) Gene conversion events in the Ig loci result in the introduction of pseudo-V gene segments into the expressed V genes. (B) The structure of G/B construct, a gene conversion substrate, and the strategy for its targeting to the IgL locus. C λ, the constant region of λL chain; VJ λ, the rearranged V region of λ L chain; Ba, BamHI; pA, the polyadenylation signal; CMVp, the cytomegalovirus promoter; SV40p, the simian virus 40 promoter. (C) Color shift (from blue to green) of BFP by C199T mutation caused by the gene conversion of G/B construct. (D) Confirmation of targeted integration of G/B or 0/B construct in the IgL locus. PCR using a primer pair [see arrowheads in (B)] gave the expected length of PCR products as illustrated in (B).
The G/B construct-transfected DT40 cells initially express BFP under the influence of the CMV promoter, and emit blue fluorescence. If the GFP gene-templated gene conversion modifies the BFP gene, some of the conversion events will result in the replacement of a segment covering 199C in the BFP gene with a corresponding sequence present in GFP, thus leading to C199T replacement. This may be monitored by the shift of the fluorescence color from blue to green (Figure 1C).
As chicken gene conversion machinery selectively targets Ig loci, we first examined the efficiency of the gene conversion of G/B construct that was integrated into the rearranged IgL locus. G/B or 0/B construct was inserted between the constant region (Cλ) gene and the proximal pseudo-V gene in the reverse direction against the IgL promoter using a targeting vector shown in Figure 1B. PCR analysis with a primer pair complementary for the indicated regions (Figure 1B) gave expected bands, thus confirming correct insertion of these construct (Figure 1D).
Efficient gene conversion of G/B construct in the IgL locus by the AID-dependent gene conversion
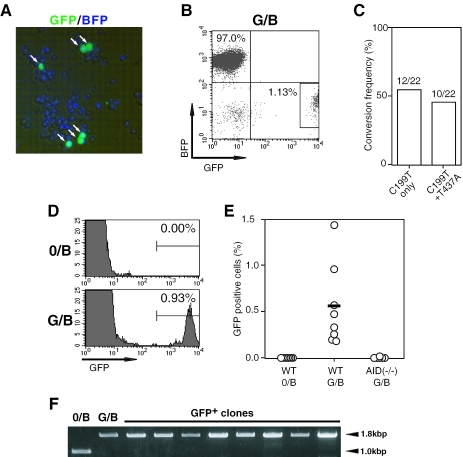
To examine whether the G/B construct could be a target of the gene conversion machinery, several BFP+ clones were selected from each transfected cell population, and further cultured for 3 weeks. Expanded cells from each clone were assessed for the appearance of green fluorescence-emitting cells (designated as green cells) by flow cytometry and fluorescence microscopy. In a typical G/B construct-bearing clone, 0.93%of originally BFP+ cells were found to emit green fluorescence after culture for 3 weeks (Figure 2A and B). Because vast majority of green cells that appeared after culture of G/B construct-bearing cells lost the BFP fluorescence (Figure 2B), the generation of green cells is considered to be due to the change of their fluorescence from blue to green. This was confirmed by sequencing the BFP gene in these green cells. All of the analyzed clones had a common mutation, C199T (Figure 2C), which has been reported to be critical for emitting green fluorescence (16). Of 22 clones analyzed, 12 clones had only C199T nucleotide replacement alone, and the rest 10 clones had an additional conversion, T437A (Figure 2C). No other mutations were found in all clones examined. Thus, the green cells were considered to be generated as the result of C199T mutation in the BFP gene. However, the C199T nucleotide replacement may not be due to point mutation, because analysis in multiple G/B or 0/B construct-bearing clones showed that the appearance of green cells (i.e. C199T mutants) required the donor GFP gene (Figure 2D and E). In addition, there was no structural change in the GFP gene in these green cells with respect to the nucleotide sequence and the configuration in the G/B construct (Figure 2F). These findings strongly support that the shift of fluorescence from blue to green in G/B construct-bearing DT40 cells was the result of the donor gene-templated gene conversion that is analogous to that occurring in Ig genes (2,3,8,9).
Figure 2.
Gene conversion machinery in DT40 cells can target a non-Ig exogenous gene. (A) Fluorescence microscopic observation of green cells (indicated by arrows) that appeared after 3 weeks culture of G/B construct-bearing DT40 cells, which had initially emitted blue fluorescence exclusively. (B) Loss of the BFP fluorescence in green cells (surrounded by a rectangle) derived from G/B construct-bearing DT40 cells that were initially BFP+GFP− exclusively. Flow cytometric analysis of G/B construct-bearing DT40 cells was carried out after culture for 3 weeks. (C) Mutation profile at the 199th and 437th nucleotides on the BFP gene in G/B construct that was integrated in the IgL locus of DT40 cells. A total of 22 DNA clones of the BFP gene obtained from green cells [shown in (B)] were analyzed. (D) Dependence of green cell generation on the donor GFP gene. Flow cytometric analysis was conducted after culture of 0/B or G/B construct-bearing DT40 cells for 3 weeks. (E) Green cell generation by AID-dependent gene conversion. Wild-type (WT) or AID-deficient DT40 cells were integrated with 0/B or G/B constructs in the IgL locus as indicated. BFP+ clones from each group (n = 8) were analyzed for the appearance of green cells after 3 weeks culture. Data indicate the percentage of GFP+-gated cells [see (D)] in each cultured clone. (F) Retention of structural configuration of the GFP gene in the G/B construct in green cells. The sequence covering the integrated-GFP gene in 9 clones was amplified by PCR using the same primer pair described in Figure 1D. Amplified bands of 1.8 kb were found in all clones tested, suggesting the retention of the GFP genes in the integrated construct. No mutations were found in the GFP gene
Because Ig gene conversion in chicken B cells as well as hypermutation and class switching in mouse and human B cells is an AID-dependent process (4–7), we examined whether the C199T replacement in the G/B construct-transfected cells required AID expression. In contrast to the wild-type cells, AID-deficient DT40 clones that were integrated with G/B construct did not generate green cells (Figure 2E), confirming that the fluorescence change was the result of AID-dependent gene conversion of G/B construct. Thus, we conclude that the chicken gene conversion machinery can target exogenous non-Ig genes.
Locus specificity in the efficiency of the gene conversion in G/B construct
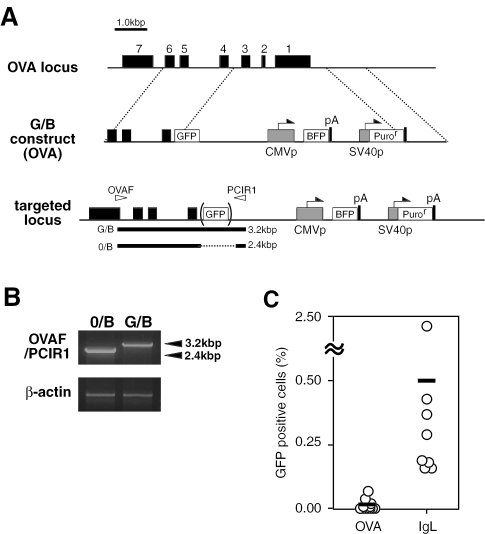
Next, we compared the efficiency of the gene conversion of G/B construct between two DT40 lines that were integrated with the construct in a site-directed fashion into the IgL locus and the OVA locus, respectively (Figures 1 and 3). Clones bearing G/B construct in the IgL locus developed green fluorescence with the frequency of approximately 0.2–2.4% after 3 weeks culture, while the conversion frequency did not exceed 0.05% under the same conditions when G/B construct was targeted to the OVA locus (Figure 3C). Similarly, only a low level of green cells (<0.1%) appeared in DT40 lines that were transfected with G/B construct in a non-targeted fashion (data not shown). In DT40 cells, one IgL allele has been rearranged, whereas the other remains in the germline configuration (8). The G/B construct inserted in the non-rearranged allele was converted in a frequency comparable to that in the rearranged counterpart (data not shown). Thus, the DT40 gene conversion machinery may target exogenous genes more efficiently when they were placed in the IgL locus to which this mutation mechanism is primarily directed.
Figure 3.
Gene conversion machinery preferentially targets the G/B construct inserted in the IgL locus. (A) Targeting integration of G/B construct into the OVA locus of DT40 cells. (B) Confirmation of the targeting integration of G/B construct into the OVA locus by PCR. A primer pair used is indicated as arrowheads in (A). (C) Comparison of the gene conversion efficiency of G/B construct between two DT40 lines that were integrated with the construct in the IgL locus and the OVA locus, respectively. After 3 weeks culture, BFP+ clones from each group (n = 8) were analyzed for the appearance of GFP+-gated cells as described in Figure 2D and E.
Analysis of gene conversion events in G/B construct
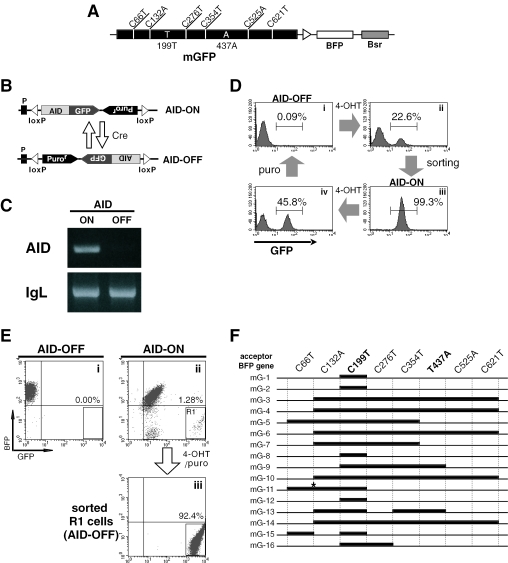
As described above, two types of green cell were generated from G/B construct-integrated DT40 cells with respect to the mutation profile in the BFP gene: one had a mutation C199T alone, and the other had A437T in addition to C199T (Figure 2C). Because BFP and GFP genes have an identical sequence except positions 199 and 437 (Figure 1C), it is impossible to determine the size of converted segments in the gene conversion products. To resolve this problem, the donor GFP gene was marked with silent mutations at six positions bearing C (Figure 4A). These included C66T, C132A, C276T, C354T, C525A and C621T. Thus, the modified GFP (mGFP) gene can be distinguished from the BFP gene with these six mutations in addition positions 199 and 437. We prepared a new construct named mG/B construct by replacing the GFP gene in G/B construct with the mGFP. It was confirmed that the mGFP gene expressed green fluorescence as efficiently as the original GFP gene when introduced to DT40 cells (data no shown).
Figure 4.
Analysis of the diversity of mutations in G/B construct: use of DT40-SW, an engineered DT40 line whose AID expression can be switched reversibly. (A) Structure of mG/B construct for estimation of the segment size that was replaced during gene conversion. mGFP was prepared by introducing six silent mutations into the GFP gene. (B) The device for switching AID expression by the use of tamoxifen-regulated Cre-loxP system [For details, see ref. (10)]. (C) Switching of AID expression in DT40-SW cells. Transcription of the AID gene in ‘AID-ON’ or ‘AID-OFF’ DT40-SW cells was examined by RT–PCR. Transcription of the IgL gene was used as a control. (D) Repeated switching of AID expression in DT40-SW cells by 4-OHT treatment. ‘AID-ON’ (GFP+) cells were purified by cell sorter (panel ii and iii), and treated with 4-OHT to switch to ‘AID-OFF’ (GFP−) cells, which was then isolated by passage through puromycin-containing medium (panel i and iv). (E) AID-dependent appearance of green cells in DT40-SW cells bearing mG/B construct in the IgL locus. First, an ‘AID-OFF’ DT40-SW clone bearing mG/B construct was generated. A single ‘AID-ON’ cell was isolated from the culture of this clone after 4-OHT treatment. After 3 weeks culture of each ‘AID-ON’ and ‘AID-OFF’ clones, generation of green cells (as gated in R1 in panel ii) was compared between them (panel i and ii). The R1-gated cells in panel ii were isolated as ‘AID-OFF’ cells after consecutive treatment with 4-OHT and puromycin (panel iii). (F) Sequence analysis of converted BFP genes from the sorted ‘AID-OFF’ green cells shown in panel iii of Figure 4E. Thick lines indicate putative gene conversion tracts. An asterisk indicates a point mutation.
In experiments analyzing gene conversion of mG/B construct, we took advantage of an engineered DT40 line, DT40-SW, in which gene conversion machinery can be reversibly switched on or off by controlling AID expression (10). Using DT40-SW bearing mG/B construct in the IgL locus, gene conversion that occurred in the BFP gene can be fixed at an appropriate time point by switching off AID expression. In DT40-SW, one AID allele was disrupted, and the other allele was replaced by the loxP-flanked AID construct (Figure 4B). The loxP-flanked AID gene is inverted by tamoxifen-regulated Cre recombinase that is expressed as a fusion protein with the estrogen receptor (17), thereby enabling to switch AID expression (10) (Figure 4C). The ‘AID-ON’ and the ‘AID-OFF’ states are discriminated by the expression of GFP and puromycin-resistance, respectively (Figure 4B). In this system, AID expression can be switched on or off repeatedly (Figure 4D). For instance, the ‘AID-ON’ cells can be isolated as cells expressing relatively low level of GFP (MFI = 5 × 101) by cell sorting (panels ii and iii in Figure 4D), and are subsequently converted to the ‘AID-OFF’ (GFP−) state by treating with 4-hydroxy-tamoxifen (4-OHT) followed by selection in the puromycin-containing medium (panels i and iv in Figure 4D). Note that the GFP fluorescence coupled with AID expression was totally abolished after puromycin-treatment.
mG/B construct was first integrated into the IgL locus in ‘AID-OFF’ DT40-SW cells. An isolated BFP+ clone was divided into two parts. One half was cultured persistently for 3 weeks without any treatment. The other half was first treated with 4-OHT for 48 h to switch on AID expression. A single ‘AID-ON’ cell sorted from the 4-OHT-treated population was expanded in the culture for 3 weeks. Green cells gated in R1 (panel ii in Figure 4E) that were similar to those observed in the gene conversion of G/B construct (Figure 2B) appeared in the culture of ‘AID-ON’ cells, but not that of ‘AID-OFF’ cells (panels i and ii in Figure 4E). The R1-containing ‘AID-ON’ cells were then treated with 4-OHT to terminate further gene conversion. The ‘AID-OFF’ green cells were isolated by passage through puromycin-containing medium, followed by cell sorting (panel iii in Figure 4E). As puromycin-treatment abolishes the expression of the AID-linked GFP gene (Figure 4D), the intense GFP fluorescence in puromycin-resistant green cells is considered to be derived from the gene conversion products of mG/B construct.
The DNA clones of the BFP gene amplified from the genomic DNA of the ‘AID-OFF’ green cells were analyzed for the occurrence of gene conversion. Of 16 clones examined, all clones were found to introduce various length of mGFP-derived segments resulting in C199T conversion (Figure 3F). Four clones (mG-1, -2, -8 and -12) underwent shorter segmental gene conversion that was limited to the proximal region of position 199. In other 5 clones (mG-3, -4, -6, -10 and -14), the longest region ranging from C132A to C621T were converted. However, it remains unclear whether the long-range conversion was the result of a single event or the consequence of more than two events. At least two consecutive events of gene conversion may have occurred in two clones, mG-13 and mG-15. In addition, a point mutation, C113G that occurred within a hot spot was observed in mG-11. In the ‘AID-OFF’ state, these mutant clones were cultured persistently without introducing further mutation. Collectively, at least nine different types of gene conversion were found in 16 DNA clones (Figure 4F), thus suggesting that the DT40 gene conversion machinery is highly potent in diversifying target genes other than Ig genes.
DISCUSSION
In the present paper, we showed that G/B construct as well as Ig genes worked as the substrate of the chicken gene conversion machinery in an AID-dependent and the donor gene-templated mechanism. The efficiency of the gene conversion in G/B construct is considered to be comparable to that of IgL genes (approximately one event/forty generations) (8). Our results are considered to be of significance in the following two respects: (i) Gene conversion machinery in DT40 can be used to engineer non-Ig proteins by a type of DNA shuffling; (ii) using G/B construct, the gene conversion event can be monitored visibly, thus contributing to, for instance, the dissection of putative factors involved in AID-dependent gene conversion and of functional domains in the AID molecule that may be responsible for either hypermutation or Ig isotype switching (18–20).
For efficient gene conversion to occur, it is likely that G/B construct must be placed in the IgL locus, because much lower efficiency was observed in cells that were integrated with the construct in the OVA locus or in a non-targeted fashion. Somatic hypermutation and gene conversion of Ig genes have been shown to be strictly dependent on AID expression (4–7). Although AID must be recruited specifically to Ig loci, the mechanism controlling this event remains to be elucidated. It has been suggested that the transcription of Ig loci that is coupled with histone modifications is involved in recruiting the mutation machinery to the target loci (21–25). Recently, phosphorylation of histone H2B has been found to be correlated with somatic hypermutation (26). However, the transcription alone may be not sufficient for the AID recruitment because in the OVA locus the BFP gene whose expression was equivalent with that in the IgL locus was not subjected to gene conversion (data not shown and Figure 2). Specific cis elements in the IgL locus may have a critical role in the targeted recruitment of the gene conversion machinery, for the reason that, when the BFP gene expression was driven by the CMV promoter in the active rearranged allele or the non-rearranged IgL allele where histones are originally less acetylated (27), G/B construct in either allele was comparably converted (data not shown and Figure 2). The Ig enhancer and matrix attachment region are known to be required for inducing hypermutations on IgV genes in mice (28–30). In the chicken Ig L locus, an enhancer has been found in the 3′ flanking region (31), and very recently the Ig L 3′ enhancer has been shown to be essential for gene expression and regulate gene conversion via the transcription factor E2A (32). However, it remains unclear how the 3′ enhancer is involved in recruiting the gene conversion machinery to the target locus.
It was demonstrated that the gene conversion of G/B construct in DT40 cells resulted in the generation of highly diversified BFP genes. As shown in Figure 3, of 16 mutated BFP clones examined, at least 9 clones had different conversion patterns. As this analysis was limited to clones bearing C199T mutation, more variable gene conversion products may have been generated in the DT40 cell culture. In usual protein engineering, mutation of the target gene, cloning of mutants, their expression in an appropriate host and functional evaluation of the gene products must be carried out step by step. In contrast, the use of DT40 cells makes it possible to conduct all these steps serially in a single cell. Thus, it is likely that the gene conversion machinery in DT40 can be used as an excellent tool to mutate non-Ig proteins towards those with improved functions.
Generally, mutational diversification of a target gene is a critical step in the methodology of protein engineering. Here, we showed that the DT40 gene conversion system enabled to manipulate non-Ig genes by a type of DNA shuffling, which have been regarded as a useful method to create novel proteins with improved physicochemical properties by exchanging functional motifs between two or more proteins (33). It might be argued that the high frequency of gene conversion of G/B construct is due to high sequence identity between the GFP and BFP genes (>98%). However, gene conversion events occur between the Vλ1 genes and the pseudogenes that display 4–20% divergence with the Vλ1 sequence (2). In addition, it is known that although the 5′ ends of the gene conversion tracts always begin in homologous regions between the pseudogene donor and acceptor V segment, the 3′ ends can occur in regions of no-homology and often encompass nucleotide insertions or deletions (34). Therefore, even in the case that homology between donor and acceptor genes is relatively low, short homologous patches in target genes could initiate gene conversion. Experiments are in progress to examine the efficiency of gene conversion between donor/acceptor pairs of lower sequence homology.
DT40 cells that spontaneously mutate their Ig genes during culture (8,9) are considered to be an excellent source for screening desired monoclonal antibodies. Recently, it has been reported that several monoclonal antibody-producing clones were isolated from a population of cultured DT40 cells (27,35). However, AID expression must be switched off to avoid further mutation in the isolated DT40 clones. The AID-switching device that we have developed will be a key technique to isolate and maintain desired DT40 clones producing either monoclonal Abs or engineered non-Ig proteins in a genetically fixed form (10). Cultured cell-based diversification of target genes by gene conversion coupled with the ON/OFF control of the mutation machinery will provide a novel means to engineer Ig and non-Ig proteins.
Acknowledgments
The authors thank Dr Michael Reth (Max-Planck-Institute for Immunology) for kindly providing the tamoxifen-regulated Cre recombinase system and Dr Hiroshi Arakawa (GSF National Research Center for Environment and Health, Neuherverg-Munich, Germany) for pLoxBsr. This work was supported by Grants-in-Aid from The Ministry of Education, Science, Sports and Culture of Japan to H.O. and N.K. K.T. was supported by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists. Funding to pay the Open Access publication charges for this article was provided by Grants-in-Aid from The Ministry of Education, Science, Sports and Culture of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 2.Reynaud C.A., Anquez V., Grimal H., Weill J.C. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell. 1987;48:379–388. doi: 10.1016/0092-8674(87)90189-9. [DOI] [PubMed] [Google Scholar]
- 3.Thompson C.B., Neiman P.E. Somatic diversification of the chicken immunoglobulin light chain gene is limited to the rearranged variable gene segment. Cell. 1987;48:369–378. doi: 10.1016/0092-8674(87)90188-7. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 5.Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 6.Arakawa H., Hauschild J., Buerstedde J.M. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 2002;295:1301–1306. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 7.Harris R.S., Sale J.E., Petersen-Mahrt S.K., Neuberger M.S. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 2002;12:435–438. doi: 10.1016/s0960-9822(02)00717-0. [DOI] [PubMed] [Google Scholar]
- 8.Buerstedde J.M., Reynaud C.A., Humphries E.H., Olson W., Ewert D.L., Weill J.C. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 1990;9:921–927. doi: 10.1002/j.1460-2075.1990.tb08190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S., Humphries E.H., Tjoelker L., Carlson L., Thompson C.B. Ongoing diversification of the rearranged immunoglobulin light-chain gene in a bursal lymphoma cell line. Mol. Cell. Biol. 1990;10:3224–3231. doi: 10.1128/mcb.10.6.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanayama N., Todo K., Reth M., Ohmori H. Reversible switching of immunoglobulin hypermutation machinery in a chicken B cell line. Biochem. Biophys. Res. Commun. 2005;327:70–75. doi: 10.1016/j.bbrc.2004.11.143. [DOI] [PubMed] [Google Scholar]
- 11.Buerstedde J.M., Takeda S. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 1991;67:179–188. doi: 10.1016/0092-8674(91)90581-i. [DOI] [PubMed] [Google Scholar]
- 12.Sale J.E., Calandrini D.M., Takata M., Takeda S., Neuberger M.S. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 2001;412:921–926. doi: 10.1038/35091100. [DOI] [PubMed] [Google Scholar]
- 13.Kanayama N., Kimoto T., Todo K., Nishikawa Y., Hikida M., Magari M., Cascalho M., Ohmori H. B cell selection and affinity maturation during an antibody response in the mouse with limited B cell diversity. J. Immunol. 2002;169:6865–6874. doi: 10.4049/jimmunol.169.12.6865. [DOI] [PubMed] [Google Scholar]
- 14.Arakawa H., Lodygin D., Buerstedde J.M. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 2001;1:7. doi: 10.1186/1472-6750-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang T.T., Cheng L., Kain S.R. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res. 1996;24:4592–4593. doi: 10.1093/nar/24.22.4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heim R., Prasher D.C., Tsien R.Y. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc. Natl Acad. Sci. USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Riesterer C., Ayrall A.M., Sablitzky F., Littlewood T.D., Reth M. Inducible site-directed recombination in mouse embryonic stem cells. Nucleic Acids Res. 1996;24:543–548. doi: 10.1093/nar/24.4.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ta V.T., Nagaoka H., Catalan N., Durandy A., Fischer A., Imai K., Nonoyama S., Tashiro J., Ikegawa M., Ito S., et al. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nature Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 19.Barreto V., Reina-San-Martin B., Ramiro A.R., McBride K.M., Nussenzweig M.C. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell. 2003;12:501–508. doi: 10.1016/s1097-2765(03)00309-5. [DOI] [PubMed] [Google Scholar]
- 20.Shinkura R., Ito S., Begum N.A., Nagaoka H., Muramatsu M., Kinoshita K., Sakakibara Y., Hijikata H., Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nature Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 21.Tumas-Brundage K., Manser T. The transcriptional promoter regulates hypermutation of the antibody heavy chain locus. J. Exp. Med. 1997;185:239–250. doi: 10.1084/jem.185.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters A., Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 23.Fukita Y., Jacobs H., Rajewsky K. Somatic hypermutation in the heavy chain locus correlates with transcription. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- 24.Bachl J., Carlson C., Gray-Schopfer V., Dessing M., Olsson C. Increased transcription levels induce higher mutation rates in a hypermutating cell line. J. Immunol. 2001;166:5051–5057. doi: 10.4049/jimmunol.166.8.5051. [DOI] [PubMed] [Google Scholar]
- 25.Woo C.J., Martin A., Scharff M.D. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity. 2003;19:479–489. doi: 10.1016/s1074-7613(03)00261-9. [DOI] [PubMed] [Google Scholar]
- 26.Odegard V.H., Kim S.T., Anderson S.M., Shlomchik M.J., Schatz D.G. Histone modifications associated with somatic hypermutation. Immunity. 2005;23:101–110. doi: 10.1016/j.immuni.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Seo H., Masuoka M., Murofushi H., Takeda S., Shibata T., Ohta K. Rapid generation of specific antibodies by enhanced homologous recombination. Nat. Biotechnol. 2005;23:731–735. doi: 10.1038/nbt1092. [DOI] [PubMed] [Google Scholar]
- 28.Yelamos J., Klix N., Goyenechea B., Lozano F., Chui Y.L., Gonzalez Fernandez A., Pannell R., Neuberger M.S., Milstein C. Targeting of non-Ig sequences in place of the V segment by somatic hypermutation. Nature. 1995;376:225–229. doi: 10.1038/376225a0. [DOI] [PubMed] [Google Scholar]
- 29.Betz A.G., Milstein C., Gonzalez-Fernandez A., Pannell R., Larson T., Neuberger M.S. Elements regulating somatic hypermutation of an immunoglobulin kappa gene: critical role for the intron enhancer/matrix attachment region. Cell. 1994;77:239–248. doi: 10.1016/0092-8674(94)90316-6. [DOI] [PubMed] [Google Scholar]
- 30.Sharpe M.J., Milstein C., Jarvis J.M., Neuberger M.S. Somatic hypermutation of immunoglobulin kappa may depend on sequences 3′ of C kappa and occurs on passenger transgenes. EMBO J. 1991;10:2139–2145. doi: 10.1002/j.1460-2075.1991.tb07748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lauster R., Reynaud C.A., Martensson I.L., Peter A., Bucchini D., Jami J., Weill J.C. Promoter, enhancer and silencer elements regulate rearrangement of an immunoglobulin transgene. EMBO J. 1993;12:4615–4623. doi: 10.1002/j.1460-2075.1993.tb06150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conlon T.M., Meyer K.B. The chicken Ig light chain 3′-enhancer is essential for gene expression and regulates gene conversion via the transcription factor E2A. Eur. J. Immunol. 2006;36:139–148. doi: 10.1002/eji.200535219. [DOI] [PubMed] [Google Scholar]
- 33.Patten P.A., Howard R.J., Stemmer W.P. Applications of DNA shuffling to pharmaceuticals and vaccines. Curr. Opin. Biotechnol. 1997;8:724–733. doi: 10.1016/s0958-1669(97)80127-9. [DOI] [PubMed] [Google Scholar]
- 34.McCormack W.T., Thompson C.B. Chicken IgL variable region gene conversions display pseudogene donor preference and 5′ to 3′ polarity. Genes Dev. 1990;4:548–558. doi: 10.1101/gad.4.4.548. [DOI] [PubMed] [Google Scholar]
- 35.Cumbers S.J., Williams G.T., Davies S.L., Grenfell R.L., Takeda S., Batista F.D., Sale J.E., Neuberger M.S. Generation and iterative affinity maturation of antibodies in vitro using hypermutating B-cell lines. Nat. Biotechnol. 2002;20:1129–1134. doi: 10.1038/nbt752. [DOI] [PubMed] [Google Scholar]