Abstract
We demonstrate here that the E2F1 induced by DNA damage can bind to and promote the apoptotic function of p53 via the cyclin A binding site of E2F1. This function of E2F1 does not require its DP-1 binding, DNA binding, or transcriptional activity and is independent of mdm2. All the cyclin A binding E2F family members can interact and cooperate with p53 to induce apoptosis. This suggests a novel role for E2F in regulating apoptosis in response to DNA damage. Cyclin A, but not cyclin E, prevents E2F1 from interacting and cooperating with p53 to induce apoptosis. However, in response to DNA damage, cyclin A levels decrease, with a concomitant increase in E2F1-p53 complex formation. These results suggest that the binding of E2F1 to p53 can specifically stimulate the apoptotic function of p53 in response to DNA damage.
One of the most important functions of p53 in tumor suppression is its ability to induce apoptosis in response to stress signals (5, 25). In normal cells, the expression level of p53 is low; this is largely due to the short half-life of the protein. In response to stress signals such as DNA damage or hypoxia, p53 accumulates due to an increase in p53 protein stability (19). The induced p53 transactivates target genes such as p21waf1/cip1, which causes cell cycle arrest by inhibiting the phosphorylation of the retinoblastoma protein (Rb) (9). In addition, p53 is also able to induce apoptosis (5, 25). Interestingly, DNA damage induced p53 usually induces permanent G1 arrest or senescence in human primary fibroblast cells (23) but induces apoptosis in tumor cells (19). It is clear that there are many differences between a normal primary cell and a tumor cell, and the identification of the key factors responsible for the different cell fates would allow us to develop better strategies to treat cancer. Existing studies have shown that inactivation of both p53 and Rb tumor suppression pathways occur in most human cancers. Thus, one of the fundamental differences between normal and tumor cells is that in tumor cells, either the p53 or Rb pathway or both have been inactivated. Furthermore, there is growing evidence suggesting that the integrity of the Rb pathway could influence the activity of p53 and vice versa.
One of the best-known tumor suppression functions of Rb is its ability to negatively regulate the transcription factor E2F. To date, six different E2F-like proteins (E2F1, E2F2, E2F3, E2F4, E2F5, and E2F6) and two DP proteins (DP1 and DP2) have been isolated (8). E2F and DP proteins contain highly conserved DNA binding domains and dimerization domains. The C-terminal part of each of the E2F proteins (except E2F6) contains a potent transactivation domain. Dimerization with DP proteins enhances its DNA binding ability and increases E2F-dependent transactivation (1, 13). Formation of a complex between the E2F/DP and the Rb family protein regulates the ability of E2F to activate transcription. E2F1, E2F2, and E2F3 contain cyclin A binding domains and bind preferentially to Rb, whereas E2F4 and E2F5 interact mainly with p107 and p130 (41). Since the transactivation domain of E2F is embedded in the domain responsible for interaction with the RB family of (pocket) proteins, this may explain why in many different experimental systems, overexpressing Rb family proteins will inhibit E2F-dependent transcription. However, the phosphorylation status of Rb or Rb-related proteins plays an important role in regulating the protein complexes formed between E2F and Rb. It was found that only the hypophosphorylated species of the Rb protein family can interact with E2F during the cell cycle. In addition to Rb, cyclin A/cdk2 can bind to and phosphorylate E2F1, E2F2, and E2F3 and inhibit their DNA binding and transactivation functions (20, 21).
The specific properties of individual E2F proteins are not entirely clear, but it was suggested that different E2F/DP heterodimers can regulate different sets of E2F target genes (8). Furthermore, it was also demonstrated recently that only E2F1 can induce apoptosis in vitro (7). Predisposition to tumor development in E2F1-null mice but not in the knockout mice of other members of the E2F family indicated that the tumor suppression function is specific for E2F1 (10, 17, 24, 43). Studies carried out by us and others have demonstrated that overexpression of E2F1 can induce apoptosis independent of p53 (16, 29, 33). This ability of E2F1 requires its DNA binding function but not its transactivation function. There is also increasing evidence that E2F1 can cooperate with p53 to induce apoptosis (32, 35, 42), although less is known about the mechanisms through which this is achieved. One explanation is that E2F1 transactivates p14ARF (2) and stabilizes p53 by preventing mdm2-mediated degradation (34, 39, 44). Based on this hypothesis, the transactivation function of E2F1 would be essential for its ability to cooperate with p53. On the other hand, it is also possible that E2F1 could cooperate with p53 through a direct interaction since there have been reports that p53 can interact with E2F1/DP1 directly (11, 28, 30, 38). Furthermore, it has also been recently reported that DNA-damaging agents can induce the expression level of E2F1 with similar kinetics to those for p53 (3, 31). Nevertheless, nothing is known about the biological function of the induced E2F1.
To address these issues, we investigated whether there is an interaction between E2F1 and p53 in vivo since they both can be induced with similar kinetics by DNA-damaging agents such as UV. Here we show that the cyclin A interaction domain of E2F1 is required for binding to p53. This ability to bind p53 correlated with the capacity of E2F1 to enhance apoptosis induced by p53. Furthermore, cyclin A binding members of the E2F family can also bind and cooperate with p53. The binding of p53 and cyclin A to the same site on E2F1 hinted at possible competitive binding to E2F1. This was shown to be the case, since cyclin A inhibited E2F1 binding to p53 in vitro, and abrogated the ability of E2F1 to cooperate with p53 to induce apoptosis in vivo. Finally, we provide evidence that such an interaction and cooperation occurs in vivo, especially after DNA damage, and propose a model of how this might work.
MATERIALS AND METHODS
Cell culture, antibodies, and plasmids.
H1299, Saos-2, MCF-7, U20S, and RKO cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) in 10% CO2 unless otherwise stated. Anti-p53 antibodies DO-1 and DO-13 are mouse monoclonal antibodies. CM-1, CM5, and SK-79 are rabbit polyclonal antibodies specific to p53. The 9E10 epitope is recognized by the mouse monoclonal anti-Myc antibody, 9E10. The mouse monoclonal antibody PC-10 is specific to the proliferating-cell nuclear antigen (PCNA) protein. KH95, an anti-E2F1 monoclonal antibody used in the immunoprecipitation assays, was purchased from Santa Cruz Biotechnology, Inc. Anti-E2F1 (C-20), anti-E2F2 (C-20), anti-E2F3 (N-20), anti-E2F4 (C-108), anti-cyclin A (H-432), anti-cyclin E (M-20), and anti-bax (N-20) are rabbit polyclonal antibodies purchased from Santa Cruz Biotechnology, Inc. CD20Leu (Becton Dickinson) is a fluorescein isothiocyanate-(FITC)-conjugated monoclonal antibody specific for the cell surface marker CD20. All the expression plasmids used in this study were driven by the cytomegalovirus immediate-early promoter. The mdm2 binding-defective mutant p53, p53ΔI, was recloned into the pcDNA3 vector from the original plasmid provided by Karen Vousden. The E2F1 plasmids which are defective for cyclin A binding, E2F1(Δ79–103) and E2F1(Δ85–91), were recloned into the pcDNA3 vector in frame with a 9E10 epitope tag from the originals, referred to as Δ24 and Δ7, respectively in reference 20. These were gifts from William Krek. All the other E2F1 mutants were generated by PCR and cloned into the pcDNA-3 expression plasmid (Invitrogen) in frame with a 9E10 epitope tag at the N-terminal end. pRc/CMV cyclin A was a gift from Roger Watson, while pRc/CMV cyclin E was donated by Sybille Mittnacht. The Sf9 lysate containing cyclin A and the control lysate were kindly donated by Manuel Joaquin.
Immunoprecipitation and immunoblotting.
Cells were washed with phosphate-buffered saline (PBS) and then lysed on ice for 30 min with NET buffer containing 1% NP-40 (12) before being centrifuged at 20,800 × g at 4°C. The lysates were precleared with protein G beads and then incubated with the appropriate antibodies. The antibody complexes were isolated using protein G beads (if not previously cross-linked), washed three times with 1% NP-40/NET buffer and twice with NET buffer. The immunoprecipitate-protein G beads were boiled in sodium dodecyl sulfate (SDS) sample buffer, and the supernatant was loaded onto SDS-polyacrylamide gels.
For immunoblotting experiments, 20-μl portions of soluble cellular proteins (5 to 8 mg of total cellular protein per ml) were loaded on SDS-polyacrylamide gels in SDS sample buffer. After electrophoresis, the proteins were blotted onto nitrocellulose paper and then blocked with a 10% solution of reconstituted dried milk powder. Primary antibody was added to the blot and incubated for 2 to 3 h at room temperature or 4°C overnight. Unless otherwise stated, E2F1 and mutants were detected using a mouse monoclonal 9E10 antibody and PC-10 was used to probe for PCNA, which is used as a loading control because it is a very stable protein. Finally, a peroxidase-conjugated rabbit anti-mouse or goat anti-rabbit immunoglobin was incubated with the blot and bound immunocomplexes were detected by the enhanced chemiluminescence (ECL) method, as described by the manufacturer (Amersham).
In vitro translation and in vitro immunoprecipitation.
E2F1 (and its mutants), E2F2, E2F3, E2F4, and p53 were in vitro translated and labeled (when required) with [35S]methionine using the TNT T7 Quick-Coupled transcription-translation system (Promega). Samples (20 to 50 μl) of the lysates containing the appropriate in vitro-translated protein products were mixed and allowed to interact in PBS in a final volume of 200 μl at 30°C for 1 h and then at 4°C for a further 1 h on a rotating wheel. For competition assays, 5 to 10 μg of protein (from baculovirus-infected Sf9 cells expressing cyclin A or control lysate) was added to the reaction mixture together with the in vitro-translated protein products. The appropriate antibody (9E10, DO-1, DO-13, or KH95) immobilized on protein G-agarose beads was added to the binding-reaction mixtures and incubated with mixing at 4°C for 1 h. The beads were then washed with PBS or NET. The bound proteins were released in SDS gel sample buffer and analyzed by SDS-polyacrylamide gel electrophoresis (10% polyacrylomida). For detection, antibody from a different species from the immunoprecipitating antibody was used.
Antibody purification and cross-linking of monoclonal antibodies to protein G beads.
Antibody 9E10 was purified from the supernatant using ammonium sulfate precipitation and protein G beads. Purified monoclonal antibodies were cross-linked to protein G beads using a previously described method (12).
DNA transfection and flow cytometry.
For fluorescence-activated cell sorter (FACS) analysis, 106 Saos-2 cells in a 10-cm dish were transfected with 3 μg of p53 plasmid and 5 μg of plasmid expressing E2F1 or mutant E2F1 using the calcium phosphate methods as described previously (30). For competition assays, 5 to 10 μg of cyclin A plasmid or 15 to 20 μg of cyclin E plasmid was cotransfected. A plasmid expressing CD20 was used as a transfection marker. Sample collection, gating, data acquisition, and analysis using flow cytometry (FACS) is described elsewhere (15, 36).
In the figures, the cell cycle profiles are represented using histogram plots. The control plasmid-transfected cells are labeled as vector. In all experiments, transfected cells were gated based on the expression of CD20 (data not shown). Apoptosis was measured by the accumulation of cells with a sub-G1 DNA content in the region labeled M1 in the histogram plots. This may also be represented by a bar graph where the y axis corresponds to percentage of cells with sub-G1 DNA content, indicative of apoptosis (M1 in histogram plots).
UV irradiation.
Cells were plated at the required density on the day before irradiation. Prior to irradiation, the medium from the dishes was temporarily removed and stored under sterile conditions. The dishes were UV irradiated using 10 J m−2, the medium was replaced, and the dishes were returned to the incubator for the required length of time as indicated.
E2F peptide and its binding to p53.
The peptides were synthesized, purified by high-pressure liquid chromatography, and sequenced by the Advanced Biotechnology Centre (Imperial College School of Medicine, Charing Cross Hospital) by previously described methods (4). The E2F peptide, previously referred to as Tat-LDL, contains residues 86 to 93 of the cyclin A binding domain of E2F1 (PVKRRLDL). The control peptide, previously referred to as Tat-Umt, contains an unrelated sequence (ETDHQYLAESS). Both peptides are N-terminally linked to a sequence derived from human immunodeficiency virus Tat (YGRKKRRQRRRG), which directs the efficient uptake of heterologous peptides across cell membranes. A batch of fluorescein N-labeled peptides was also synthesized and shown to be efficiently taken up by almost all cells (data not shown).
35S-labeled p53 was in vitro translated as described above. This was incubated with 5 and 10 μl of 10 mM FITC-labeled E2F1 or control peptide for 30 min at 30°C and a further 2 h at 4°C. Then 5 μl of biotin-conjugated anti-FITC antibody was added to each binding-reaction mixture in an Eppendorf tube and allowed to rotate at 4°C for 2 h. The reaction products were washed with PBS-1% NP-40 to remove the excess antibody before 15 μl of strepavidin-conjugated Fe beads was added to each tube. The beads were then isolated using magnetic selection as described by the manufacturer (Dynabeads), washed twice with cold PBS-1% NP-40, and run on a 12% polyacrylamide gel.
p53-inducible cell line.
A system where p53 could be induced was constructed as specified by the manufacturer (Clontech Laboratories, Palo Alto, Calif.). Wild-type p53 was recloned into a Tet-responsive promoter, which was stably transfected in an inducible founder clone of H1299 cells (clone 7). The p53 protein was induced by 2 μg of doxycycline (an analogue of tetracycline) per ml, and its expression was tested by Western blotting and cell staining (data not shown). B225 is a clonal cell line that on treatment with doxycycline induces a low level of p53 which is unable to cause apoptosis after 24 to 48 h.
E2F peptide treatment.
H1299 B225 cells were split in six-well plates at a concentration of 105 cells/well. At 24 h later, they were induced with 2 μg of doxycycline per ml in RPMI 1640-10% FCS. The medium was removed 16 to 20 h later, the cells were irradiated with UV radiation at 10 J/m−2, and fresh medium (DMEM + 5% FCS with or without 2 μg of doxycycline per ml) was added. At 24 h after UV irradiation, the cells were washed once with Optimem (Gibco BRL) and treated with 100 μM respective peptide in Optimem. At 1 to 2 h later, the medium was supplemented with 5% FCS. Both floating and adherent cells were harvested 6 to 8 h later, stained with trypan blue, and scored for viability. For representation, data from a representative of two experiments, each performed using a different batch of peptides, was shown. The fold increase given is calculated by the following formula: fold increase of dead cells = percentage of dead cells after various treatment/percentage of dead cells when untreated/percentage of dead cells due to the respective treatments in Optimem alone.
RESULTS
p53 binds to the same site of E2F1 as cyclin A does.
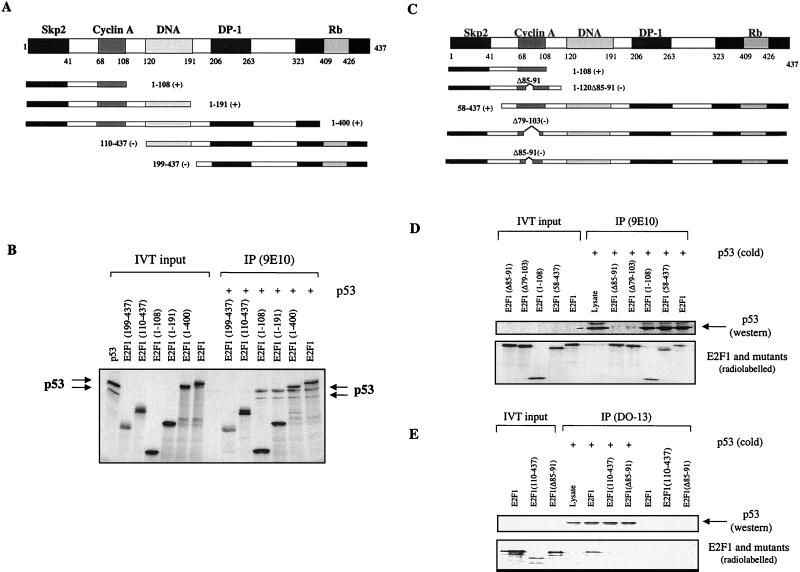
To better understand the biological implication of the physical interaction between p53 and E2F1, we used a series of truncation mutants of E2F1 which contained specific functional domains of E2F1 (diagrams in Fig. 1) to map the site on E2F1 responsible for its interaction with p53. Six major functional domains have been identified so far in E2F1; they are the p45SKP2 binding domain (1 to 41) (27), the cyclin A binding domain (amino acids 68 to 108), the DNA binding domain (120 to 191), the DP-1 binding domain (206 to 263), the transactivation domain (323 to 437) and the Rb binding domain (409 to 426). The first set of three mutants were truncated from the C terminus, and the remaining amino acids retained in the mutants are labeled E2F1(1–400), E2F1(1–191) and E2F1(1–108). The second set of two E2F1 mutants were truncated from the N terminus, and the remaining amino acids are labeled as E2F1(110–437) and E2F1(199–437) (Fig. 1A). All the mutants were tagged with a 9E10 antibody epitope at their N terminus to facilitate detection. Wild-type p53, E2F1, and E2F1 mutants were in vitro translated as indicated in Fig. 1B, and the lysates were incubated to allow binding to occur. The 9E10 antibody was used to capture tagged E2F1 and coprecipitate any bound complexes. As shown in Fig. 1B, p53 binds to all the C-terminally truncated E2F1 mutants, E2F1(1–400), E2F1(1–191), and E2F1(1–108), indicating that the first 108 amino acids of E2F1 were sufficient to bind p53. To corroborate this, the two N-terminally truncated E2F1 mutants, E2F1(110–437) and E2F1(199–437), failed to complex with p53 under the same conditions of this assay. Hence, p53 binds to the first 108 N-terminal amino acids of E2F1. Two functional domains have been mapped to this region of E2F1. The first 41 amino acids of E2F1 are involved in binding to p45SKP2 and regulating the stability of E2F1 (27), while residues 68 to 108 bind cyclin A, which is involved in the regulation of the DNA binding ability of E2F1 (20).
FIG. 1.
p53 binds to the cyclin A binding domain of the E2F1 protein. (A and C) Mapping was done using a series of E2F1 truncation mutants. The numbers refer to the residues the peptides contain, while the prefix Δ denotes the residues that are deleted. The E2F1 mutants that bind p53 are denoted by (+), while nonbinding mutants are indicated by (−). (B) Both in vitro-translated [35S]methionine-labeled E2F1 and p53 were used. The antibody 9E10 was used to immunoprecipitate 9E10-tagged E2F1 with the coimmunoprecipitation of radiolabeled p53. The presence of p53 and E2F1 mutants was detected by autoradiography. (D and E) Only in vitro-translated [35S]methionine-labeled E2F1 was used. The in vitro-translated p53 was unlabeled, and its presence was detected by Western blotting using rabbit polyclonal antibody SK-79 (D) or CM-1 (E). For panel E, an anti-p53 antibody (DO-13) was used to immunoprecipitate p53 and the corresponding binding of E2F1 was detected by autoradiography.
To map the precise binding site of p53 on E2F1, more mutants of E2F1 were employed (Fig. 1C): an N-terminal truncation mutant of E2F1, E2F1(58–437), without its first 58 amino acids and hence defective in binding to p45SKP2 (27), and three deletion mutants of E2F1, E2F1(1–120Δ85–91), E2F1(Δ85–91), and E2F1(Δ79–103), which were shown to be defective in binding to cyclin A (20). All the 9E10 epitope-tagged E2F1 mutants were efficiently immunoprecipitated with the 9E10 antibody (Fig. 1D, lower panel). However, p53 coimmunoprecipitated only with the wild-type E2F1, E2F1(1–108), and E2F1(58–437) but not with the two cyclin A binding-defective mutants of E2F1, E2F1(Δ79–103) and E2F1(Δ85–91) (Fig. 1D, upper panel). These results strongly suggest that p53 binds to the same site on E2F1 as cyclin A does. This was further confirmed by the observation that the anti-p53 antibody DO-13 immunoprecipitated p53 (Fig. 1E, upper panel), which specifically coimmunoprecipitated wild-type E2F1 but not the cyclin A binding-defective mutants E2F1(Δ85–91) and E2F1(110–437) (Fig. 1E, lower panel).
Binding to p53 is required for E2F1 to specifically enhance p53-induced apoptosis.
It has been shown previously that overexpression of E2F1 can cooperate with p53 to induce apoptosis. Having determined the region of E2F1 responsible for the interaction with p53, we tested whether this interaction between p53 and E2F1 would correlate with the ability of E2F1 to cooperate with p53 in the induction of apoptosis. Using Saos-2 cells, which are null for p53 and lack functional Rb, we titrated the expression of p53 or E2F1 alone to induce detectable but little apoptosis so that cooperation may be observed. Coexpression of p53 and E2F1 cooperated to induce 60.8% ± 7.1% of the transfected cells to die of apoptosis. This experimental system thus allowed us to test the ability of various E2F1 mutants to cooperate with p53 to induce apoptosis. We used the smallest mutant of E2F1 which could interact with p53 (the fragment containing only the first 108 amino acids from the N terminus) as well as two deletion mutants of E2F1, E2F1(Δ85–91) and E2F1(Δ79–103), which are specifically defective in binding to cyclin A and p53 (as shown previously).
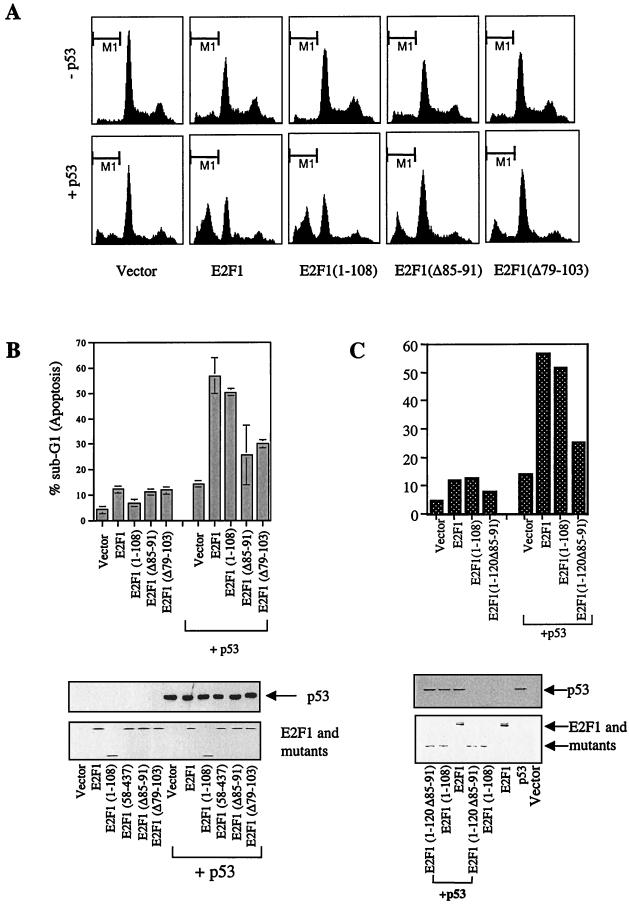
As shown in Fig. 2A and B, E2F1 and mutants, except E2F1(1–108), were able to induce p53-independent apoptosis in about 12% of transfected Saos-2 cells. E2F1(1–108), the only E2F1 mutant in this assay to lack the DNA binding domain, did not induce apoptosis when expressed on its own (6.7% ± 0.9%) but cooperated with p53 to induce apoptosis almost as effectively as did the full-length E2F1 (50.5% ± 0.7% and 56.8% ± 7.1% respectively). However, the two cyclin A and p53 binding-defective mutants of E2F1, E2F1(Δ85–91) and E2F1(Δ79–103), failed to cooperate with p53 to induce apoptosis (Fig. 2A and B) even though the mutants were expressed to similar levels to those of E2F1 (Fig. 2B, lower panel). The percentage of apoptosis detected in the cells when these two p53 binding-defective E2F1 mutants were cotransfected with p53 was higher (25 to 30%) than that detected with either p53 (14.4% ± 1.4%) or the E2F1 mutants (12 to 14%) alone. This was simply the sum of the apoptosis caused by the p53 and E2F mutants alone without any cooperation. These data show that an intact cyclin A binding domain of E2F1 and hence p53 binding is required for its ability to cooperate with p53 to induce apoptosis. This was further supported by the observation that the p53 and cyclin A binding-defective E2F1 mutant E2F1(1–120Δ85–91) failed to bind and cooperate with p53 to induce apoptosis (Fig. 2C). By testing all the mutants of E2F1 shown in Fig. 1, we found that the ability to cooperate with p53 to induce apoptosis perfectly correlated with their ability to bind p53 (Fig. 2 and data not shown).
FIG. 2.
Only E2F1 mutants that bind to p53 can cooperate with p53 to induce apoptosis. (A to C) FACS analysis showing identical-scale histogram plots (A) or bar graphs (B and C) of transiently transfected Saos-2 cells in the absence or presence of p53 (3 μg/10-cm dish) with E2F1 and its mutants (5 μg/10-cm dish) as indicated. The lower portions of panels B and C show the expression level of p53, E2F1, and mutants. The transfected p53 protein was detected with DO-1. (D) Results from a binding assay using E2F1 and E2F1(Δ85–91) and in vitro-translated p53 and p53 mutant 175his. (E) Cells undergoing apoptosis when p53 (3 μg) or the p53 mutants 175his and 175pro (5 μg/dish) were transfected with E2F1 (3 μg) or E2F1(Δ85–91) (3 μg) in Saos-2 cells. (F) Expression of p53, E2F1, and mutant. PCNA was used as a loading control.
Binding of p53 does not have a significant effect on the apoptotic function of E2F1.
It has been shown previously that E2F1 possesses an apoptotic function independent of p53. Therefore, we investigated the influence of the binding of p53 on the apoptotic function of E2F1. We used two tumor-derived p53 point mutations, 175pro and 175his, which are defective in inducing apoptosis. Although the mutation at residue 175 significantly alters the activity of p53, 175his, for example, still retained its ability to interact with the cyclin A binding domain of E2F1 as effectively as the wild-type p53 did (Fig. 2D). If the binding of p53 to E2F1 can affect the apoptotic function of E2F1, both mutants should alter the apoptotic function of E2F1 as effectively as the wild-type p53 did. As a control, a p53 binding-defective mutant of E2F1, E2F1(Δ85–91), was used. When expressed in Saos-2 cells, in agreement with the previous report, the apoptotic function of 175pro was reduced compared to that of the wild-type p53 (Fig. 2E) (36). Under the same conditions, the expression of 175His failed to induce significant apoptosis. The coexpression of E2F1 with 175pro or 175his failed to show the significant enhancement of apoptosis seen with wild-type p53. In E2F1- and wild-type p53-expressing cells, 50% of the cells die of apoptosis. This is significantly different from the percentage seen in cells expressing E2F1 alone (12%) or together with mutant p53 (18% for each mutant coexpressed with E2F1). As a control, we showed that the apoptosis induced by non-p53 binding E2F1(Δ85–91) was not significantly affected by the p53 mutants (Fig. 2E).
There was a small but detectable increase in apoptosis when apoptosis-deficient p53 mutants were cotransfected with E2F1. However, the increase was minimal in comparison to the effect of wild-type p53 together with E2F1, suggesting that the major effect of the E2F1-p53 complex is the enhancement of p53-induced apoptosis. The failure to show a significant enhancement of the apoptotic function of E2F1 was not due to the lack of protein expression of the two p53 mutants (Fig. 2F).
E2F1 can specifically cooperate with p53 to induce apoptosis, and this is independent of mdm2.
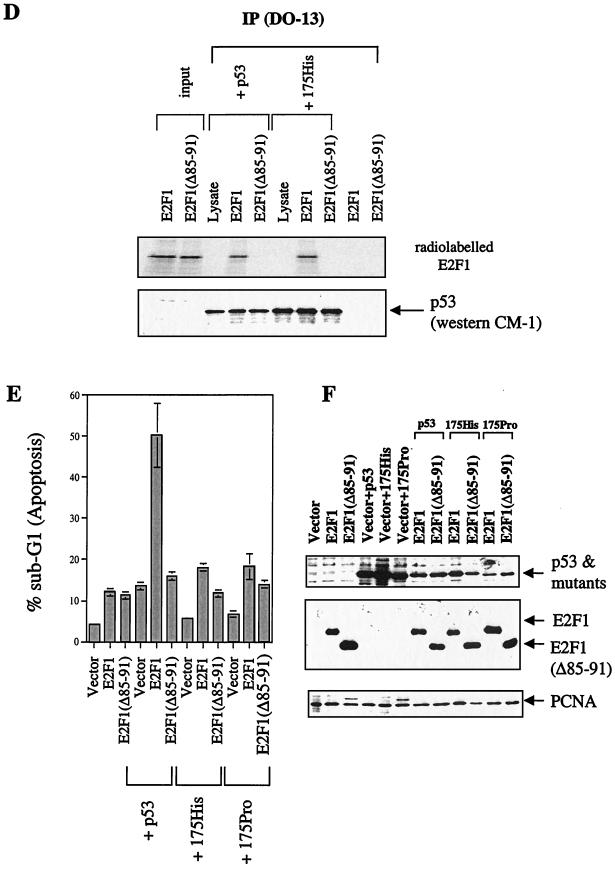
All the C-terminal truncation mutants of E2F1 that lacked their transactivation domains cooperated with p53 in these assays to induce apoptosis if they bound p53. Furthermore, these mutants failed to activate transcription from a promoter containing 3×E2F responsive elements, 3xwt linked to a luciferase reporter (data not shown), suggesting that the cooperation of E2F1 with p53 in the induction of apoptosis is independent of the transcription of E2F1. This is inconsistent with the proposal that E2F1 cooperated with p53 by up-regulating p14ARF (2), which can then stabilize p53 by preventing mdm2-mediated protein degradation of p53 (34, 44). In that hypothesis, the transactivation function is essential for the ability of E2F1 to enhance p53-induced apoptosis. The mechanism of cooperation between p53 and E2F1 observed here is independent of the transactivation function of E2F1 and hence cannot occur via p14ARF. To provide direct evidence for our hypothesis, we studied E2F1 and the E2F1(1–108) mutant for their ability to enhance the apoptotic function of Bax or a p53 mutant, p53ΔI. Bax-induced apoptosis was shown to be independent of p53 in Saos-2 cells (6). p53ΔI, deleted in the conserved box I(13–19), was shown to be active in transactivation assays and found to suppress transformation as well as wild-type p53 did but was unable to bind mdm2 (6).
As above, coexpression of E2F1 and E2F1(1–108) together with p53 enhanced the apoptotic function of p53 significantly. Interestingly, under these conditions, coexpression of E2F1 or E2F1(1–108) also stimulated the apoptotic function of the p53 mutant p53ΔI, arguing that E2F1 can cooperate with p53 to induce apoptosis independent of mdm2 (Fig. 3A), since p53ΔI is defective in mdm2 binding. This occurred even when all proteins were expressed to comparable levels (Fig. 3B). In contrast to p53, coexpression of E2F1 with Bax failed to result in a significant stimulation of the apoptotic function of Bax (Fig. 3C) like that observed with p53. Although the apoptosis induced by Bax and E2F1 is greater than with each plasmid alone, as argued above, it is only slightly greater than the sum of the apoptosis induced by the two expressed proteins alone. Hence, E2F1 cannot cooperate to enhance p53-independent apoptosis induced by Bax. This inability was not due to lack of E2F1 or Bax expression (Fig. 3D). These results indicate that E2F1 specifically cooperates with p53 to induce apoptosis and that this activity of E2F1 is independent of mdm2.
FIG. 3.
E2F1 can specifically cooperate with p53 to induce apoptosis independent of mdm2. (A) Cells undergoing apoptosis as determined by FACS analysis of Saos-2 cells were transfected with 3 μg of p53 or p53ΔI or 5 μg of E2F1 or E2F1(1–108). The numbers at the top right of each panel refer to the percentage in sub-G1 (labeled M1; indicative of apoptosis). (C) Bar graph showing the amount of apoptosis as determined by FACS analysis of Saos-2 cells transfected with 5 μg of E2F1 or E2F1(1–108) in the presence of 3 μg of p53 or 1 μg of Bax. (B and D) Immunoblotting to show the expression level of p53, E2F1, and mutants, of p53ΔI (B) and p53, and of E2F1 and Bax (D). The transfected p53 (and p53ΔI in panel B) were detected with the anti-p53 antibody DO-13, and the Bax protein (D) was detected using N-20.
All cyclin A binding E2F members can bind and cooperate with p53 to induce apoptosis.
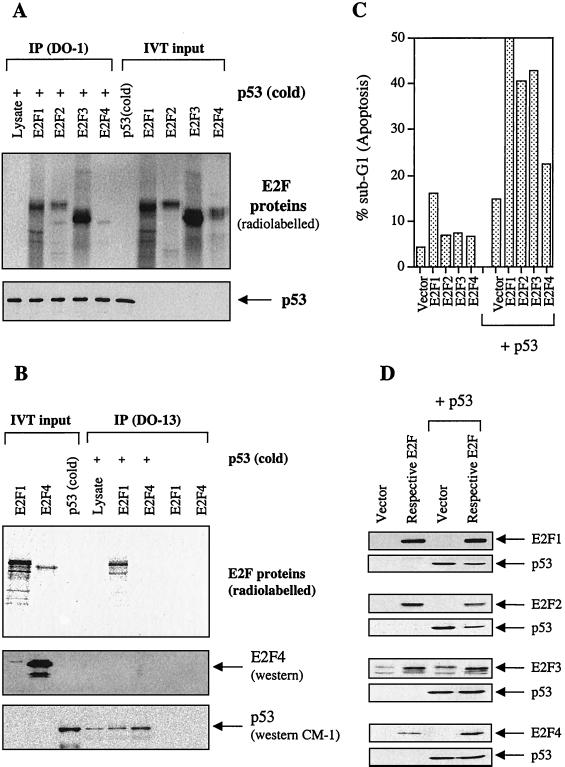
The cyclin A binding domain is conserved in E2F family members E2F1, E2F2, and E2F3 but not E2F4. We studied the ability of p53 to bind to E2F2 and E2F3. Since E2F4 does not contain the cyclin A binding region, we reasoned that if the cyclin A binding region was mediating the interaction between p53 and E2F2 and E2F3, p53 would not be able to interact with E2F4. In vitro-translated and [35S]methionine-labeled E2F1, E2F2, E2F3, and E2F4 were tested for coimmunoprecipitation with p53 by using the anti-p53 antibody DO-1. As shown in Fig. 4A, E2F1, E2F2, and E2F3 were specifically immunoprecipitated with p53. Under the same conditions, E2F4, which lacks the cyclin A binding domain, failed to coimmunoprecipitate with p53. The failure of E2F4 to interact with p53 was further supported by the observation that another anti-p53 antibody, DO-13, was able to immunoprecipitate E2F1 but not E2F4 with p53 under the same condition (Fig. 4B). To show that E2F4 was indeed expressed but that its radio signal was weak, we reloaded the lysate on an SDS-polyacrylamide gel and blotted it using an anti-E2F4 antibody to show that no E2F4 was immunoprecipitated with p53. These results suggested that, like E2F1, the expression of E2F2 and E2F3 might be able to influence the activity of p53 via protein-protein interactions.
FIG. 4.
Only E2F members containing the cyclin A binding domain (E2F1, E2F2, and E2F3) bind to and activate p53 to induce apoptosis. (A) In vitro binding assay showing the results of radiolabeled E2F1, E2F2, E2F3, and E2F4 and unlabeled p53 [referred to as p53 (cold)] immunoprecipitated with anti-p53 mouse monoclonal antibody DO-1. The presence of [35S]methionine-labeled E2Fs was detected by autoradiography, while the unlabeled p53 protein was detected using rabbit polyclonal antibody SK-79. (B) Immunoprecipitation using radiolabeled E2F1 and E2F4 with unlabeled p53 and immunoprecipitated by a anti-p53 antibody (DO-13). Since the radioactive E2F4 signal is rather weak, a blot showing that E2F4 is expressed but not immunoprecipitated under these conditions is included. (C) The percentage of apoptotic cells as determined by FACS of Saos-2 cells transfected with E2F1-4 (5 μg each/10-cm dish) in the absence or presence of p53 (3 μg/10-dish). (D) Immunoblotting showing the expression of the various E2F proteins using the respective antibodies (9E10 epitope-tagged E2F1 [9E10], E2F2 [C-20], E2F3 [N-20], and E2F4 [C-108]) and p53 using the antibody DO-1.
To further test our hypothesis that binding to p53 is required and sufficient for E2F1 to stimulate the apoptotic function of p53, we decided to investigate whether E2F2 and E2F3 could also cooperate with p53 to induce apoptosis. E2F4 was used as a control. If the binding were essential for the E2Fs to cooperate with p53, one would predict that E2F2 and E2F3 would augment p53-induced apoptosis since these E2F proteins have cyclin A binding domains which bind p53. Saos-2 cells were cotransfected with E2F1, E2F2, E2F3, or E2F4 together with p53. As shown in Fig. 4C, consistent with the notion that binding is required to activate p53 to induce apoptosis, coexpression of E2F1, E2F2, and E2F3 also produced a significant enhancement of the apoptotic function of p53. In contrast, coexpression of E2F4 failed to have such a significant effect on the apoptotic function of p53. These results demonstrated a functional difference between the E2F proteins containing a cyclin A binding domain and the rest of the family members in regulating the apoptotic function of p53. E2F1, E2F2, and E2F3 bind and activate p53 to induce apoptosis; E2F4 does not bind or cooperate with p53 to induce apoptosis. This was not because of a lack of protein expression (Fig. 4D).
Cyclin A can prevent E2F1 from cooperating with p53 to induce apoptosis.
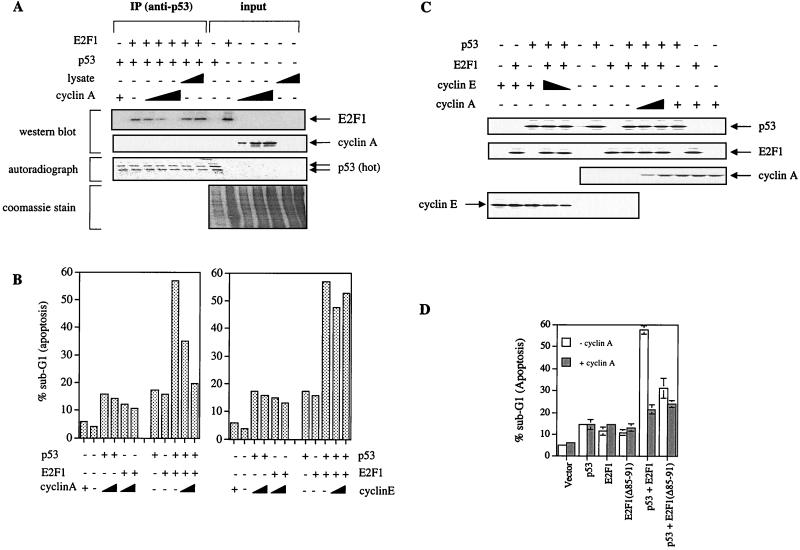
Our data show that p53 and cyclin A can bind to a similar site on E2F1. Therefore, we wondered if the binding of cyclin A and of p53 to E2F1 are mutually exclusive. An in vitro competition assay was performed to test if cyclin A could prevent E2F1 from binding to p53. As shown in Fig. 5A, immunoprecipitation of p53 pulled down E2F1. Addition of increasing concentrations of baculovirus-produced cyclin A reduced the amount of E2F1 immunoprecipitated with p53. Increasing concentrations of the same amount of baculovirus lysate (Fig. 5A, Coomassie stain of input) did not inhibit E2F1 binding to p53. This shows that cyclin A can inhibit E2F1 binding to p53 in a dose-dependent manner, suggesting that the binding of p53 and of cyclin A to E2F1 are mutually exclusive.
FIG. 5.
Cyclin A, but not cyclin E, competitively inhibits the cooperation of E2F1- and p53-induced apoptosis. (A) In vitro competition assay using baculovirus-expressed cyclin A (2, 5, or 10 μg) to compete out the binding of E2F1 to p53. Vector-infected baculovirus lysate was used as a control. The polyclonal anti-p53 (DO-13) was used to immunoprecipitate radiolabeled p53. (B) Apoptosis induced in Saos-2 cells transfected with E2F1 (5 μg/10-cm dish), p53 (3 μg/10-cm dish), and increasing concentrations of cyclin A or cyclin E (0, 5, and 10 μg/10-cm dish) as indicated. (C) Immunoblotting was used to show the expression of cyclin A using rabbit polyclonal antibody (H-432), cyclin E using rabbit polyclonal antibody (M-20), E2F1 using C-20 (anti-E2F1 polyclonal antibody), and p53 using DO-1. (D) Percentage of apoptotic cells as determined by FACS analysis of Saos-2 cells transfected with E2F1 and mutants (5 μg/10-cm dish) and p53 (3 μg/10-cm dish) in the absence or presence of cyclin A (10 μg/10-cm dish).
We then investigated the effect of the expression of cyclin A and cyclin E on the ability of E2F1 and p53 to cooperate to induce apoptosis. Expression of cyclin A or cyclin E alone with p53 or E2F1 did not induce a significant change in the amount of apoptosis over the respective levels in the transfected Saos-2 cells (Fig. 5B). Interestingly, coexpression of increasing concentrations of cyclin A (0, 5, and 10 μg) with p53 and E2F1 together exhibited a dose-dependent inhibition of the ability of E2F1 to augment p53 induced apoptosis (Fig. 5B, left graph, last three lanes). In the same experiment, corresponding amounts of cyclin E, which is known not to bind E2F1, did not reduce apoptosis resulting from the cooperation between E2F1 and p53 (Fig. 5B, right graph, last three lanes). Such effects were seen in the absence of perturbation of the protein levels of p53 and E2F1 by cyclin A or cyclin E (Fig. 5C). The specific effect of cyclin A in preventing E2F1 from stimulating the apoptotic function of p53 was further substantiated by the observation shown in Fig. 5D. The coexpression of cyclin A (10 μg) together with p53 and a cyclin A binding-defective mutant of E2F1, E2F1(Δ85–91), did not show a significant effect on p53-induced apoptosis. However, under the same conditions, cyclin A was able to effectively inhibit E2F1 from enhancing p53-induced apoptosis. Our results show that cyclin A competitively inhibited the binding of E2F1 to p53 in vitro. In vivo, this correlated with a dose-dependent inhibition by cyclin A, but not cyclin E, of the capacity of E2F1 to stimulate p53-induced apoptosis.
DNA damage induced E2F1 can complex with p53 in vivo and this correlates with decreased cyclin A levels.
One of the most important features of p53 is its accumulation in response to DNA damage (18, 26). Previous studies have demonstrated that almost all the p53-inducing agents can also induce E2F1 with similar kinetics (3, 31) and that the induced E2F1 is transcriptionally inactive (31). These observations suggested that p53 and E2F1 may cooperate in response to stress signals such as DNA damage.
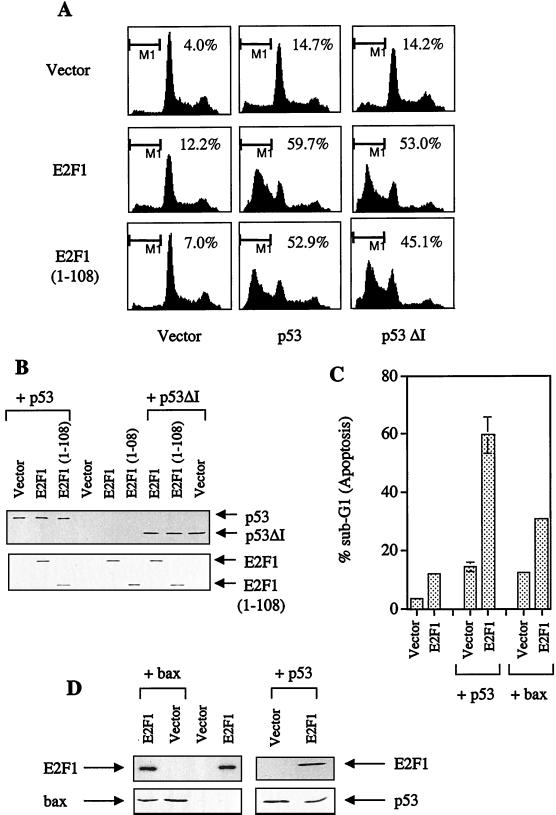
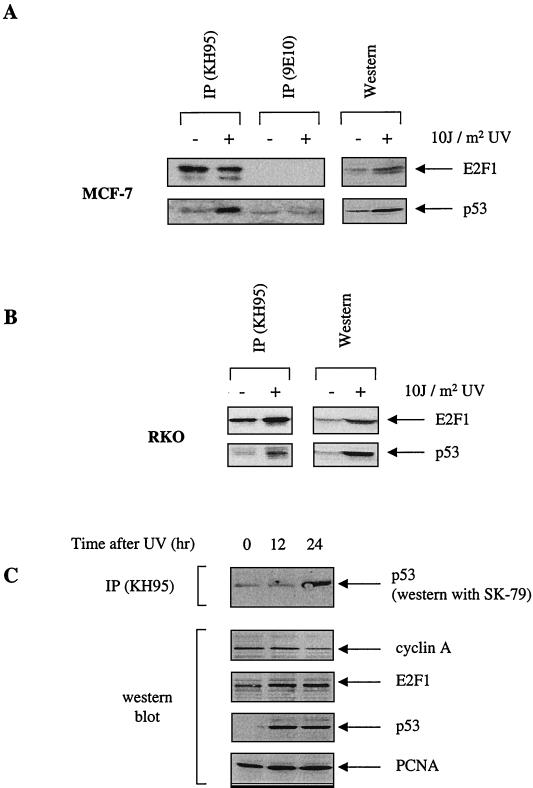
To determine whether an E2F1-p53 complex occurs under physiological conditions, two wild-type p53-expressing cell lines, RKO and MCF-7, were irradiated with UV radiation to cause DNA damage. As expected, an increase in the expression levels of p53 and E2F1 were detected (right panels of Fig. 6A and B). Using the anti-E2F1 antibody KH95, we found that UV-induced E2F1 can coimmunoprecipitate p53 in RKO and MCF-7 cells (left panels of Fig. 6A and B). This interaction is specific because a control antibody, 9E10, did not immunoprecipitate E2F1 or pull down p53 (Fig. 6A). This shows that E2F1 and p53 can interact in vivo and that this occurs under physiological conditions, in response to DNA damage by UV.
FIG. 6.
UV-induced endogenous E2F1 and p53 can complex in vivo. (A and B) MCF-7 cells (A) and RKO cells (B) were UV irradiated with 10 J m−2 (+) or left untreated (−) and then incubated for 16 h. The left parts of each panel show the results of the immunoprecipitation using the respective lysates with anti-E2F1 (KH95) and control antibody (9E10) (panel A only). The right parts of each panel show the corresponding Western blot of the cell lysate. In both cases, the rabbit polyclonal antibodies used were C-20 (for E2F1) and SK-79 (for p53). (C) Time course for U20S cells treated with UV. Immunoprecipitation using anti-E2F1 (KH95) and probed for p53 using the polyclonal antibody indicated is shown in the top part. The lower four parts show the results of a Western blot analysis using the corresponding lysate with endogenous levels of E2F1 (detected by C-20), p53 (detected by DO-1), and cyclin A (detected by H-432) shown at the indicated times after UV. PCNA (detected by PC-10) is used to show equal loading.
We next examined the possibility that cyclin A regulates the interaction between E2F1 and p53 in vivo. Interestingly, there is a concomitant decrease in the levels of cyclin A after UV irradiation in U20S cells (Fig. 6C, second panel from top). It is particularly interesting that at the 24-h time point, when cyclin A levels are lowest, complex formation between E2F1 and p53 appears to be at its highest (top panel). This reciprocal relationship between cyclin A levels and E2F1-p53 complex formation is consistent with the model that cyclin A prevents E2F1 from binding and cooperating with p53 to induce apoptosis.
E2F1 can only stimulate p53-mediated apoptosis in response to UV irradiation.
So far, the results suggested that the physiological role of the E2F1-p53 complex formed by DNA damage-induced E2F1 and p53 is to promote the apoptotic function of p53 in response to DNA damage such as UV. Thus, the E2F1 peptide (4) containing 8 amino acids of the cyclin A binding site [E2F1(86–93)] might activate the apoptotic function of p53 in response to DNA damage. Consistent with this hypothesis, it has been shown previously that only the E2F1 peptide but not the unrelated control peptide can induce apoptosis in some of the wild-type p53-expressing cell lines such as U20S (4). However, it is difficult to demonstrate whether the cell killing induced by the E2F1 peptide was specific to p53 since the peptide also caused p53-independent cell death by inhibiting the activity of cyclin A/cdk2 kinase.
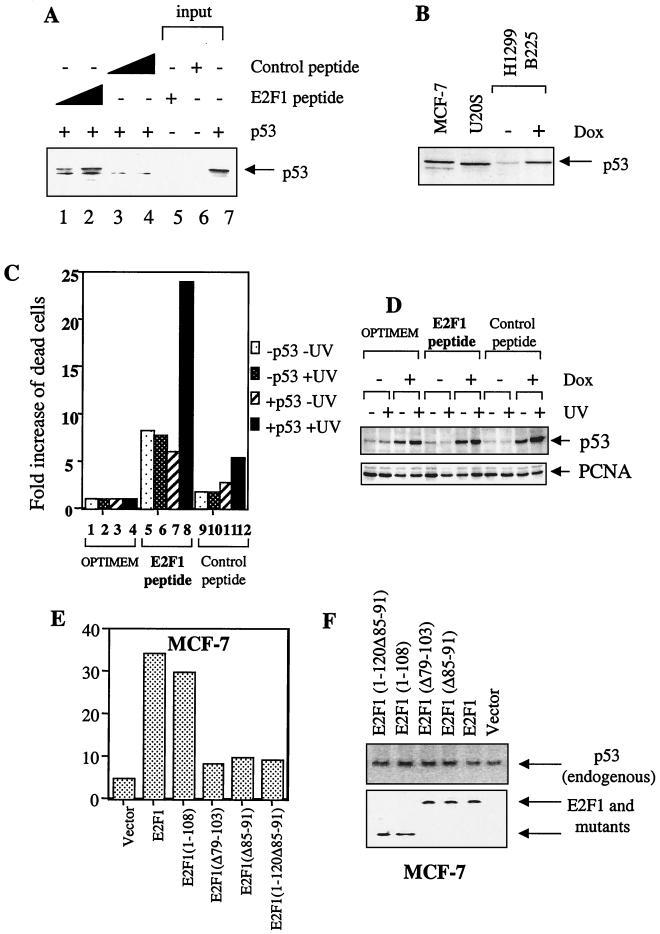
To test whether the cyclin A binding-competent E2F1 peptide can specifically promote the apoptotic function of p53 through direct binding, two peptides were synthesized as described in Materials and Methods and in previous papers (4). One batch was N labeled with FITC and used in the immunoprecipitation assays. The [35S]methionine-labeled p53 was coincubated with the control (unrelated sequence [4]; control peptide) or the cyclin A binding peptide of E2F1 (E2F1 peptide). As shown in Fig. 7A, the anti-FITC antibody was able to specifically coimmunoprecipitate p53 with the E2F1 peptide but not with the control peptide (Fig. 7A, compare lanes 1 and 2 and lanes 3 and 4). The amount of p53 coimmunoprecipitated by the anti-FITC antibody correlated with increasing amounts of the E2F1 peptide but not of the control peptide. This result clearly demonstrated that the 8 amino acids of the cyclin A binding peptide of E2F1 are sufficient to interact with p53 in vitro.
FIG. 7.
E2F1 can cooperate with endogenous p53 to induce apoptosis. p53 binding peptide of E2F1 can specifically enhance the apoptotic function of p53 in response to treatment with UV. (A) Immunoprecipitation data using biotin-conjugated anti-FITC to immunoprecipitate increasing concentrations of FITC-labeled E2F1 peptide and FITC-labeled control peptide. The presence of [35S]methionine-labeled p53 was detected using autoradiography. (B) Comparison of the levels of endogenous p53 in MCF7 and U20S and the p53 in the H1299 B225 p53-inducible cell line induced by 2 μg of doxycycline (Dox) per ml. The presence of p53 was detected using the mouse monoclonal antibody DO-1. (C) The p53-inducible cell line H1299 B225 was incubated in the absence (−p53) or presence (+p53) of 2 μg of doxycycline per ml untreated (−UV) or UV irradiated at 10 J m−2 (+UV) in medium only (Optimem) or 100 μM E2F1 peptide or the control peptide in Optimem as detailed in Materials and Methods. To take into account the p53-independent mechanism of cell killing exhibited by the peptide and the various effects of treatment, the relative fold increase in the number of dead cells is calculated and presented as described in Materials and Methods. (D) The corresponding Western blot where p53 was detected using DO-13 and the PC-10 supernatant used to detect PCNA. (E) Apoptosis was determined by FACS when E2F1 and mutants (10 μg) were transfected in wild-type expressing MCF7 cells. (F) Expression levels of transfected E2F1 and mutants and endogenous p53 detected using 9E10 and DO-1, respectively.
To further investigate the effect of the cyclin A binding peptide of E2F1 on the apoptotic function of p53, we constructed a wild-type p53-inducible H1299 human lung carcinoma cell line that is null for p53. The induced p53 is expressed in more than 80% of the cells (data not shown), and the expression level is lower than or comparable to the levels of p53 detected in two tumor cell lines, MCF-7 and U20S, expressing wild-type p53 (Fig. 7B). The expression of wild-type p53 is under the control of the tetracycline repressor (Tet-on system [Clontech]), and its expression can be induced in the presence of doxycyclin. Cyclin A can prevent E2F1 from cooperating with p53 to induce apoptosis, and the E2F1-p53 complex was detected mainly in DNA-damaged cells when the cyclin A level was at its lowest. Hence, the ability of the E2F1 peptide to stimulate the apoptotic function of p53 may best be observed in DNA-damaged cells. The p53-inducible cells were exposed to UV irradiation and low mitogenic stimuli (i.e., low serum [5% FCS]) to reduce the expression level of cyclin A. The induced p53 did not induce cell death (assessed by trypan blue uptake) in this cell line, even after UV irradiation (Fig. 7C). The presence of the E2F1 peptide induced cell death in the p53-inducible H1299 cell line (Fig. 7C, lanes 5 to 8). Interestingly, in the normally growing cells, the cell killing induced by the E2F1 peptide was independent of p53 since the induction of p53 did not influence the number of dead cells detected (lanes 5 and 7). These data are in agreement with previously published data (4). In addition, the number of dead cells detected in the H1299 cells in the absence of p53 was not influenced by UV treatment (compares lanes 5 and 6). However, on induction of p53 and on treatment with UV, the E2F1 peptide was able to cause a significant killing of the cells (lane 8). The effect of the E2F1 peptide is specific because the control peptide caused only a comparatively small degree of cell death under the same conditions (lanes 9 to 12). Furthermore, in agreement with the results obtained so far, the binding of E2F1 (mimicked by the E2F1 peptide in this case) to p53 (Fig. 6A) did not have a significant effect on the expression levels of p53 as shown in Fig. 7D. These results provided strong evidence that the binding of E2F1 to p53 was sufficient to promote the apoptotic function of p53 in response to DNA-damaging agents such as UV.
Finally, to show that binding to p53 is necessary and sufficient for E2F1 to enhance the apoptotic function of endogenous p53, we transfected p53 binding-competent and -defective E2F1 mutants in wild-type p53-expressing MCF-7 cells. E2F1(1–108) binds to p53, but E2F1(1–120Δ85–91) which lacks amino acids 85 to 91, like E2F1(Δ85–91), cannot bind p53. The expression of E2F1 or E2F1(1–108) induced a six- to sevenfold increase in apoptosis over vector control transfectants. In contrast, the p53 binding-defective E2F1 mutants, E2F1(Δ85–91), E2F1(Δ79–103), and E2F1(1–120Δ85–91), induced only about a twofold increase in apoptosis over vector control transfectants (Fig. 7E), even though all E2F1 and mutants were expressed to similar levels (Fig. 7F). Hence, we conclude that overexpression of E2F1 in these assays can cooperate with endogenous p53 only if it binds p53.
DISCUSSION
The E2F1-p53 complex specifically stimulate the apoptotic function of p53.
We provide evidence that there is a perfect correlation between the ability of E2F1 to bind p53 via its cyclin A binding domain and its ability to cooperate with p53 to induce apoptosis. Binding is absolutely required for E2F1 to cooperate with p53 to induce apoptosis. This differs from the ability of E2F1 to stabilize p53, since it did not correlate with E2F1 binding (28). In agreement with previous reports, the two cyclin A binding-defective mutants of E2F1, E2F1(Δ85–91) and E2F1(Δ79–103), actively induced apoptosis on their own (20). However, in comparison to E2F1, these two cyclin A and p53 binding-defective mutants failed to cooperate with p53 to induce apoptosis. Although we detected a modest increase in the number of apoptotic cells coexpressing p53 and the two cyclin A binding-defective mutants, they failed to stimulate the apoptotic function of p53 in the dramatic manner seen with the wild-type E2F1. These results are particularly important since both E2F1 mutants are active transcriptionally (20), even for the ARF promoter (28), yet they failed to promote the apoptotic function of p53 in Saos-2 cells, a cell line in which the overexpression of E2F1 was shown to induce p14ARF (2). In addition, E2F1(1–108), which even lacks binding domains for DNA and DP1, was able to bind p53 and cooperate with p53 to induce apoptosis. This is consistent with the hypothesis that the DP1 binding and transactivation functions of E2F1 are not essential for the ability of E2F1 to cooperate with p53.
To further corroborate this and rule out the ARF pathway, a mutant p53, p53ΔI, which is able to induce apoptosis and growth suppression like the wild type but is unable to bind mdm2, still cooperates with E2F1 to induce apoptosis. This provides further evidence that this cooperation is independent of mdm2. Such a finding is entirely in agreement with existing literature showing that ARF expression does not respond to DNA damage (39) and that ARF-null cells exhibit an intact p53 response following UV irradiation (37). This would imply that promotion of p53-mediated apoptosis by E2F1 could occur even in tumour cells with defective p14ARF.
Finally, the p53-E2F1 complex does not have a significant effect on the apoptotic function of E2F1; p53 mutants such as 175his could not cooperate to significantly increase E2F1-induced apoptosis (Fig. 2E). These observations provided a mechanism by which E2F1 induced by DNA damage could be very important in regulating the apoptotic function of p53.
Cyclin A is an important inhibitor of E2F1-induced apoptosis.
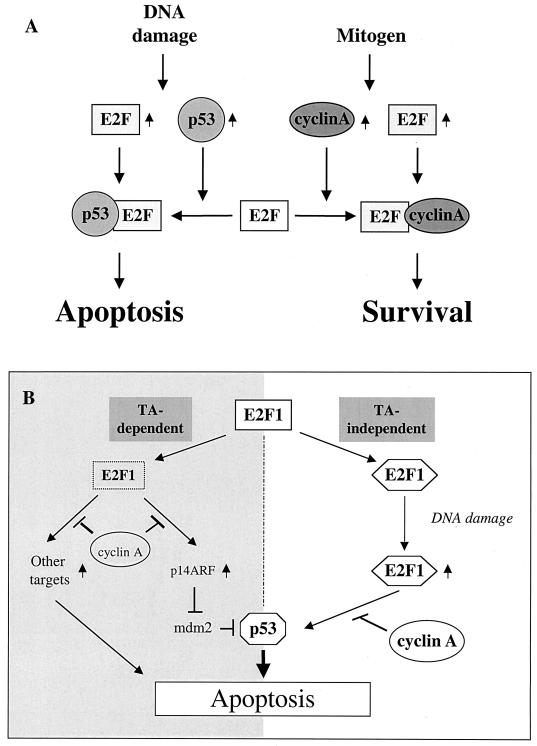
Cyclin A binding-defective E2F1 mutants were shown to induce apoptosis more effectively than did the wild-type E2F1 in NIH 3T3 cells in the presence of high serum levels (21). We do not think the results contradict our model. It is important to note that only in the presence of high serum levels or cotransfection with cyclin A does E2F1Δ24 [E2F1(Δ79–103), in our paper] show enhanced transcriptional or apoptotic activity with respect to wild-type E2F1 (20, 21). Our model is consistent with the notion that cyclin A is an important inhibitor of E2F1-mediated apoptosis. In the high-serum condition, cyclin A can inhibit the DNA binding activity of E2F1. Consequently, high levels of cyclin A (induced by high serum or exogenous expression) inhibit E2F1-induced apoptosis through pathways independent of p53 or dependent on ARF and p73 (Fig. 8B, left half). Our data shown here provide an additional mechanism through which cyclin A can inhibit the proapoptotic function of E2F1: augmentation of p53-induced apoptosis (Fig. 8B, right half). The level of cyclin A determines whether E2F1 can interact with p53 and cooperate with p53 to induce apoptosis in response to DNA damage (Fig. 8A).
FIG. 8.
(A) Method by which cyclin A may influence the apoptotic function of p53 via E2F1. (B) Summary of the various mechanisms by which E2F1 and p53 can cooperate to induce apoptosis under different conditions. The left half shows transactivation-dependent mechanisms, while the right half depicts the transactivation-independent mechanism we propose here.
Novel function for the E2F proteins containing a cyclin A binding domain.
The ability to bind p53 via its cyclin A binding domain enabled this group of E2F proteins (E2F1, E2F2, and E2F3) to participate in regulation of the apoptotic function of p53. This is in agreement with a recent report that all cyclin A binding E2F proteins can induce apoptosis in Rat-1 cells which express wild-type p53 (40). The phenotypes of E2F1 and E2F3 knockout mice would argue that E2F3 is more involved in mouse development than is E2F1 (10, 17, 43). Thus, it is possible that different cyclin A binding E2F proteins may function in different tissues. It is also important to note that all the cyclin A binding E2F proteins favor interaction with Rb rather than with pocket proteins p107 or p130. In human tumors, mutations were found mainly in the Rb gene but not in the p107 or p130 genes (41). Therefore, it is reasonable to suppose that in human tumor cells where there is a mutation or inactivation of Rb, there would be an up-regulated expression of E2F protein. This E2F can have either one of two possible fates depending on the other signals (Fig. 8A). One of these fates is that E2F proteins may complex with p53 to induce apoptosis in response to DNA damage, when cyclin A levels are reduced. This might explain partly why tumor cells are more sensitive to p53-induced apoptosis and to DNA damage than are normal primary cells.
DNA damage-induced E2F1 and p53 cooperate to induce apoptosis independent of the transactivation function of E2F1.
The studies done by us and by others demonstrate that many p53-inducing agents can also induce E2F1 with similar kinetics (3, 31). Even though the induced E2F1 was shown to bind DNA in vitro (14), it is unable to transactivate a consensus promoter (31); therefore it must function through some other mechanism. Interestingly, it has been shown previously that E2F1 can induce apoptosis independently of its transactivation function (16, 33). Furthermore, it is also well established that E2F1 can cooperate with p53 to induce apoptosis both in vitro and in vivo (32, 35, 42).
What is the biological function of the transcriptionally inactive E2F1 induced by DNA damage? The fact that the induction of E2F1 and of p53 expression by DNA damage have similar kinetics and the two induced proteins can form a complex in vivo (30) also requires an explanation. The observation that the E2F1-p53 complex was best detected in UV-irradiated cells, where cyclin A expression was the lowest (Fig. 6C), support the in vitro data that the interactions between E2F1-cyclin A and E2F1-p53 are mutually exclusive. The timing of the detection of the E2F1-p53 complex is particularly important because it may suggest the molecular mechanisms behind the timing at which p53 induces apoptosis in response to DNA damage. It is well documented that UV irradiation can induce the accumulation of E2F1 and p53 as early as 2 to 4 h and the expression level of both proteins peaks at around 8 to 12 h after the irradiation (3, 26, 31). However, even at 12 h after UV irradiation, we did not detect any U20S cells dying of apoptosis (data not shown). This would argue that initially, the DNA damage-induced p53 is latent to induce apoptosis. Apoptotic cells can be detected in UV-irradiated cells only 24 h or later after irradiation, suggesting that later p53 might be activated to induce apoptosis. Thus, the binding of E2F1 to p53 could be a factor in determining the switch from a latent p53 into an active form of p53. This interaction is determined by the levels of cellular cyclin A.
Consistent with this, treating the p53-inducible cell line with a p53 binding-competent peptide of E2F1 allowed us to demonstrate that the binding of E2F1 can indeed specifically promote the apoptotic function of p53 in response to DNA-damaging agents such as UV irradiation. Addition of the 8-amino-acid E2F1 peptide which bound p53 in vitro induces a seven- to ninefold increase in apoptosis which is independent of p53 induction. This agrees with previously published data, since this peptide also binds cyclin A. Significantly, the induction of p53 together with UV irradiation resulted in a 24-fold increase in apoptosis (Fig. 7C). This is best explained by the fact that the UV-induced p53 (mimicked by Dox-induced p53 in H1299 after UV irradiation) can complex with UV-induced E2F1 (mimicked in this case by the 8-amino-acid E2F1 peptide) in vivo. The observation that a Dox-induced p53 (without UV treatment) and E2F1 peptide together still do not result in such cooperation indicates that a DNA damage signal triggered by UV irradiation on p53 is important for cooperation. Such a signal is likely to involve posttranslational modification(s) of p53. Hence, the results shown in Fig. 7 together support a model in which DNA damage-induced E2F1 may dramatically enhance the apoptotic function of p53 after UV irradiation by forming an in vivo complex.
In normal cells, the activity of E2F is tightly regulated by the tumor suppressor Rb. Therefore, p53 induced by DNA damage cannot be activated to induce apoptosis by proteins like E2F. This could explain why a p53-dependent G1 arrest of DNA damage occurs in primary cells (23). However, in most tumor cells, inactivation of the Rb pathway results in deregulated E2F activity (41). We propose that the deregulated E2F could then complex with and promote p53 to induce apoptosis. This may partly explain the sensitivity of tumor cells to p53-dependent apoptosis after DNA damage (19). Cyclin A binding E2F would cooperate with p53 to induce apoptosis only in cells where the expression level of cyclin A were sufficiently low. Interestingly, reduced expression of cyclin A is often seen in cells in response to stress signals that also induce p53 and E2F1 (31). On the other hand, mitogen-induced E2F would transactivate genes like cyclin A (22), which complexes (20) and prevents E2F binding to p53, thereby keeping cells alive (Fig. 8A). It is already known that cyclin A can abolish the E2F1-dependent apoptosis by inhibition of the DNA binding ability of E2F1, which is required for its apoptotic capacity (Fig. 8A, left half). The results presented here illustrate a novel role for cyclin A in protecting cells from dying of apoptosis in response to DNA damage (Fig. 8A, right half).
Acknowledgments
J.-K. Hsieh and D. Yap contributed equally to this work.
We thank Paul Farrell and Elizabeth Slee for reading the manuscript.
This work was supported by Ludwig Institute for Cancer Research.
REFERENCES
- 1.Bandara, L. R., V. M. Buck, M. Zamanian, L. H. Johnston, and N. La Thangue. 1993. Functional synergy between DP1 and E2F1 in the cell cycle-regulating transcription factor DRTF/E2F. EMBO J. 12:4317–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates, S., A. Phillips, P. Clark, F. Stott, G. Peters, R. Ludwig, and K. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124–125. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, C., A. Sparks, and D. Lane. 1999. Transcription factor E2F-1 is upregulated in response to DNA damage in a manner analogous to that of p53. Mol. Cell. Biol. 19:3704–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Y. N., S. K. Sharma, T. M. Ramsey, L. Jiang, M. S. Martin, K. Baker, P. D. B. Adams, and W. G. J. Kaelin. 1999. Selective killing of transformed cells by cyclin/cyclin-dependent kinase 2 antagonists. Proc. Natl. Acad. Sci. U-S-A 96:4325–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke, A. R., C. A. Purdie, D. J. Harrison, R. G. Morris, C. C. Bird, M. L. Hooper, and A. H. Wyllie. 1993. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362:849–852. [DOI] [PubMed] [Google Scholar]
- 6.Crook, T., N. J. Marston, E. A. Sara, and K. H. Vousden. 1994. Transcription activation by p53 correlates with suppression of growth but not transformation. Cell 79:817–827. [DOI] [PubMed] [Google Scholar]
- 7.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245–2262. [DOI] [PubMed] [Google Scholar]
- 9.El-Deiry, W., T. Tokino, V. E. Velculescu, D. B. Levy, V. E. Parson, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumour suppression. Cell 75:817–825. [DOI] [PubMed] [Google Scholar]
- 10.Field, S. J., F. Y. Tsai, F. Kuo, Z. A. M., J. W. G. Kaelin, D. M. Livingston, S. H. Orkin, and M. E. Greenberg. 1996. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85:549–561. [DOI] [PubMed] [Google Scholar]
- 11.Frank, D. K., T. J. Liu, M. J. Frederick, and G. L. Clayman. 1998. Combination E2F-1 and p53 gene transfer does not enhance growth inhibition in human squamous cell carcinoma of the head and neck. Clin. Cancer Res. 4:2265–2272. [PubMed] [Google Scholar]
- 12.Harlow, E. E., and D. P. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 13.Helin, K., C. L. Wu, A. R. Fattaey, J. A. Lees, B. D. Dynlacht, C. Ngwu, and E. Harlow. 1993. Heterodimerization of the transcription factors E2F1 and DP1 leads to cooperative trans-activation. Genes Dev. 7:1850–1861. [DOI] [PubMed] [Google Scholar]
- 14.Hofferer, M., C. Wirbelauer, B. Humar, and W. Krek. 1999. Increased levels of E2F-1-dependent DNA binding activity after UV- or γ-irradiation. Nucleic Acids Res. 27:491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsieh, J.-K., F. S. G. Chan, D. J. O’Connor, S. Mittnacht, S. Zhong, and X. Lu. 1999. Rb regulates the stability and the apoptotic function of p53 via mdm2. Mol. Cell 3:181–193. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh, J.-K., S. Fredersdorf, T. Kauzarides, K. Martin, and X. Lu. 1997. E2F1 induced apoptosis requires DNA binding but not transcriptional activity and is inhibited by the retinoblastoma protein through direct interaction. Genes Dev. 11:1840–1852. [DOI] [PubMed] [Google Scholar]
- 17.Humbert, P. O., R. Verona, J. M. Trimarchi, C. Rogers, S. Dandapani, and J. A. Lees. 2000. E2F3 is critical for normal cellular proliferation. Genes Dev. 14:690–703. [PMC free article] [PubMed] [Google Scholar]
- 18.Kastan, M. B., O. Onyekwere, D. Sidransky, B. Vogelstein, and R. W. Craig. 1991. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51:6304–6311. [PubMed] [Google Scholar]
- 19.Ko, L. J., and C. Prives. 1996. p53: puzzle and paradigm. Genes Dev. 10:1054–1072. [DOI] [PubMed] [Google Scholar]
- 20.Krek, W., M. E. Ewen, S. Shirodkar, Z. Arany, W. G. J. Kaelin, and D. M. Livingston. 1994. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell 78:161–172. [DOI] [PubMed] [Google Scholar]
- 21.Krek, W., G.-F. Xu, and D. M. Livingston. 1995. Cyclin A-kinase regulation of E2F-1 DNA binding function underlies suppression of an S phase checkpoint. Cell 83:1149–1158. [DOI] [PubMed] [Google Scholar]
- 22.Lam, E. W.-F., and N. B. La Thangue. 1994. DP and E2F proteins: coordinating transcription with cell cycle progression. Curr. Opin. Cell Biol. 6:859–866. [DOI] [PubMed] [Google Scholar]
- 23.Leonardo, A. D., S. P. Linke, K. Clarkin, and G. M. Wahl. 1994. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 8:2540–2551. [DOI] [PubMed] [Google Scholar]
- 24.Linderman, G. J., L. Dagnino, S. Gaubatz, Y. Xu, R. Bronson, H. B. Warren, and D. M. Livingston. 1998. A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. Genes Dev. 12:1092–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe, S. W., E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847–852. [DOI] [PubMed] [Google Scholar]
- 26.Lu, X., and D. P. Lane. 1993. Different induction of transcriptionally active p53 following UV or ionizing radiation: defects in chromosome instability syndromes? Cell 75:765–778. [DOI] [PubMed] [Google Scholar]
- 27.Marti, A., C. Wirbelauer, M. Scheffner, and W. Krek. 1999. Interaction between ubiquitin-protein ligase SCF SKP2 and E2F-1 underlies the regulation of E2F-1 degradation. Nat. Cell Biol. 1:14–19. [DOI] [PubMed] [Google Scholar]
- 28.Nip, J., D. K. Strom, C. M. Eischen, J. L. Cleveland, G. P. Zambetti, and S. W. Hiebert. 2001. E2F-1 induces the stabilization of p53 but blocks p53-mediated transactivation. Oncogene 20:910–920. [DOI] [PubMed] [Google Scholar]
- 29.Nip, J. S., D. K., B. E. Fee, G. Zambetti, J. L. Cleveland, and S. W. Hiebert. 1997. E2F1 cooperates with topoisomerase II inhibitor and DNA damage to selectively agument p53-independent apoptosis. Mol. Cell. Biol. 17:1049–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connor, D. J., E. W.-F. Lam, S. Griffin, S. Zhong, L. C. Leighton, S. A. Burbidge, and X. Lu. 1995. Physical and functional interactions between p53 and cell cycle co-operating transcription factors, E2F1 and DP1. EMBO J. 14:6184–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connor, D. J., and X. Lu. 2000. Stress signals induce transcriptionally inactive E2F1. Oncogene 19:2369–2376. [DOI] [PubMed] [Google Scholar]
- 32.Pan, H., C. Yin, N. J. Dyson, E. Harlow, L. Yamasaki, and T. Van Dyke. 1998. Key roles for E2F1 in signaling p53-dependent apoptosis and cell division within developing tumours. Mol. Cell 2:283–292. [DOI] [PubMed] [Google Scholar]
- 33.Phillips, A. C., S. Bates, K. M. Ryan, K. Helin, and K. H. Vousden. 1997. Induction of DNA synthesis and apoptosis are separable functions of E2F-1. Genes Dev. 11:1853–1863. [DOI] [PubMed] [Google Scholar]
- 34.Pomerantz, J., N. Schreiber-Agus, N. J. Liegeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H.-W. Lee, C. Cordon-Cardo, and R. A. DePinho. 1998. The ink4a tumour suppressor gene product, p19Arf, interacts with mdm2 and neutralizes mdm2’s inhibition of p53. Cell 92:713–723. [DOI] [PubMed] [Google Scholar]
- 35.Qin, X. Q., D. M. Livingston, W. G. J. Kaelin, and P. D. Adams. 1994. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc. Natl. Acad. Sci. USA 91:10918–10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowan, S., R. L. Ludwig, Y. Haupt, S. Bates, X. Lu, M. Oren, and K. H. Vousden. 1996. Specific loss of apoptotic but not cell-cycle arrest function in a human tumour derived p53 mutant. EMBO J. 15:827–838. [PMC free article] [PubMed] [Google Scholar]
- 37.Sherr, C. J. 1998. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 12:2984–2991. [DOI] [PubMed] [Google Scholar]
- 38.Sorensen, T. S., R. Girling, C.-W. Lee, J. Gannon, R. Bandara, and N. B. L. Thangue. 1996. Functional interaction between DP-1 and p53. Mol. Cell. Biol. 16:5888–5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stott, F., S. Bates, M. James, I. B. McConne, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. Vousden, and G. Peters. 1998. The alternative product from the human CDKN2A locus, p14 ARF participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vigo, E., H. Muller, E. Prosperini, G. Hateboer, P. Cartwright, M. C. Moroni, and K. Helin. 1999. CDC25A phosphatase is a target of E2F and is required for effecient E2F-induced S-phase. Mol. Cell. Biol. 19:6379–6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323–330. [DOI] [PubMed] [Google Scholar]
- 42.Wu, X., and A. J. Levine. 1994. p53 and E2F-1 cooperate to mediate apoptosis. Proc. Natl. Acad. Sci. USA 91:3602–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamasaki, L., T. Jacks, R. Bronson, E. Goillot, E. Harlow, and N. J. Dyson. 1996. Tumour induction and tissue atrophy in mice lacking E2F-1. Cell 85:537–548. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. Arf promotes mdm2 degradation and stablilizes p53: Arf-ink4a locus deletion impairs both the Rb and p53 tumour suppression pathways. Cell 92:725–734. [DOI] [PubMed] [Google Scholar]