Abstract
Transcriptional elongation by RNA polymerase II (RNAPII) is regulated by the positive transcription elongation factor b (P-TEFb). P-TEFb is composed of Cdk9 and C-type cyclin T1 (CycT1), CycT2a, CycT2b, or CycK. The role of the C-terminal region of CycT1 and CycT2 remains unknown. In this report, we demonstrate that these sequences are essential for the activation of transcription by P-TEFb via DNA, i.e., when CycT1 is tethered upstream or downstream of promoters and coding sequences. A histidine-rich stretch, which is conserved between CycT1 and CycT2 in this region, bound the C-terminal domain of RNAPII. This binding was required for the subsequent expression of full-length transcripts from target genes. Thus, P-TEFb could mediate effects of enhancers on the elongation of transcription.
Transcriptional elongation by RNA polymerase II (RNAPII) is a highly regulated process between negative transcription elongation factors (N-TEF), which pause RNAPII, and the positive transcription elongation factor b (P-TEFb), which facilitates its elongation (5, 45, 48). Transcription begins with the assembly of the preinitiation complex (PIC) on the promoter, which includes core elements (CP) and promoter-proximal regions (PPR) (4). CP consists of a TATA box, a TFIIB recognition element, an initiator (INR), and downstream promoter elements. PPR contain upstream sequences, where Sp1 and other activators bind (4). They recruit the PIC through general transcription factors (GTF; TFIIA to -J) and mediators (SRB, etc.), which interact with the nonphosphorylated C-terminal domain (CTD) of RNAPII and mediate the initiation of transcription (23, 31, 39, 47). TFIIH helps RNAPII to clear the promoter and partially phosphorylates the CTD (17, 56). Thus, transcription is initiated, but further elongation is blocked by N-TEF (11, 45), which includes at least two different complexes, the 5,6-dichloro-1β-d-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF) (19, 21, 57, 62). DSIF contains SPT4 (16 kDa) and SPT5 (160 kDa) (19, 21, 57). NELF is composed of four subunits, of which the smallest, NELF-E (or RD), contains an RNA recognition motif (62). At present, the mode of action of N-TEF is not well understood. Somehow it pauses RNAPII, resulting in arrested transcription (28, 44, 58, 61, 62). Finally, it is P-TEFb, consisting of Cdk9 and cyclin T1, T2a, T2b, or K (CycT1, differentially spliced CycT2a and CycT2b, and CycK), that overcomes effects of N-TEF and releases RNAPII from its arrest (8, 42, 45). P-TEFb phosphorylates the CTD, DSIF, and possibly other targets in these arrested complexes, thus catalyzing the transition from initiation to elongation of transcription (8, 11, 21, 27, 45, 59, 60).
Transcriptional enhancer sequences (enhancers) are defined as cis-acting DNA elements that increase levels of gene expression (4, 6, 36). Their effects are dependent on PPR and CP but independent of location, orientation, and distance with respect to the site of initiation of transcription (4, 6, 36). They can function independently of chromatin, i.e., on episomal plasmids and in in vitro transcription systems (49). Moreover, their effects are dependent on the CTD of RNAPII, which in primates contains 52 repeats of the heptapeptide YSPTSPS. Whereas sequences with 52 and 31 repeats support effects of different enhancers, those with 5 repeats have been found to be inactive in these assays (14). Thus, the CTD functions as a surface for the integration of different signals from activators and repressors on transcription. When unphosphorylated, the CTD is covered by mediators and recruits coactivators (50, 51). In sharp contrast, the phosphorylated CTD plays a central role in the cotranscriptional processing of mRNA by binding to the elongator complex (2). To date, an accepted model of how enhancers function is not available. The looping model suggests that proteins bound to enhancers interact directly with the PIC on promoters. Alternatively, the scanning model speculates that enhancer-binding proteins travel along the DNA until they meet their cognate promoter (4).
A well-defined system that incorporates important features of eukaryotic promoters and enhancers and reveals the central role that P-TEFb plays in the elongation of transcription by RNAPII is represented by the human immunodeficiency virus (HIV). On the HIV long terminal repeat (LTR) are found binding sites for TFE3, LEF-1, NF-AT, NF-κB, Sp1, TFIID, TFIII, and LBP-1 (24). Thus, there exists a classical arrangement of the enhancer, PPR, and CP (24). In the absence of enhancer binding proteins and Tat, the transcription of HIV is arrested at the transactivation response (TAR) element stem loop RNA, which is present at the 5′ ends of all viral transcripts (24). With the help of P-TEFb, Tat binds to TAR and promotes the elongation rather than the initiation of HIV transcription (3, 9, 13, 22, 24, 29, 59). NF-κB and other activators that bind to the HIV enhancer can substitute for the function of Tat (18) and/or act synergistically with it to increase levels of viral replication (40, 52). NF-κB also increases the expression of the immunoglobulin light chain kappa (Igκ) gene (43). As in HIV, RNAPII is engaged but does not elongate past this cellular promoter. The binding of NF-κB to Igκ enhancer then releases RNAPII from its pausing site (46). A similar regulation of transcription is found in the heat-shock genes from Drosophila melanogaster and the human proto-oncogene c-myc (32, 55). Tat transactivation also resembles enhancers in its requirement for the CTD of RNAPII for its effects (38). Thus, it is not surprising that the major histocompatibility complex class II (MHC II) transactivator (CIITA), which mediates effects of B-cell-specific and gamma interferon-inducible enhancers (54) and NF-κB (1), also binds P-TEFb.
Thus, we hypothesized that P-TEFb plays a general role in mediating effects of enhancers. Indeed, in this report, we demonstrate that P-TEFb activates transcription from proximal and distal sites upstream and downstream of the promoter and coding sequence of a reporter gene. PPR, CP, and the CTD of RNAPII were required for these effects of P-TEFb. A direct correlation between the binding of a histidine-rich stretch in the C-terminal region of CycT1 and the CTD could be made with this transcriptional activation, which also required the kinase activity of Cdk9. Importantly, adding these sequences of CycT1 to CycK, which lacks a long C-terminal extension, allowed this P-TEFb complex to activate transcription from the same distal sites. These effects were on the elongation rather than initiation of transcription, indicating that PPR and CP were sufficient to recruit and position RNAPII. We conclude that the interaction between C-terminal regions of RNAPII, CycT1, and distal sites enables P-TEFb to act as a transcriptional coactivator. Thus, P-TEFb can link activators at enhancers and PIC at promoters, which results in the productive elongation of transcription.
MATERIALS AND METHODS
Plasmid constructions.
The plasmid reporter pG6(5′Pro) was described previously (15). It contained six upstream activation sequence (UAS) repeats, which bind to residues 1 to 147 of the Gal4 DNA binding domain (DBD). UASs were located upstream of the three Sp1 sites, at position −119 of the HIV type 1 (HIV-1) LTR, relatively to the starting point of transcription. No TAR sequences were present in pG6(5′Pro). The HIV LTR was fused to the chloramphenicol acetyltransferase (CAT) reporter gene. pG6(3′Pro) contained the same sequences as pG6(5′Pro), the only difference being that UAS sites were placed 3′ to CAT and promoter sequences. pG5 reporter plasmid contained the EIb TATA box and was described previously (53).
The different plasmid effectors of Gal-CycT1 code for the chimeras of the full-length CycT1 [Gal-CycT(1–726)] and truncated forms Gal-CycT(1–590), Gal-CycT(1–420), and Gal-CycT(1–300) (see Fig. 1A), all fused at their N termini to the DBD of Gal4(1–147) and inserted at the BamHI/XbaI sites of pEF-Bos-myc(EF). pSG424-Gal-CycK(1–357) and pSG424-Gal-CycK(1–357)T(300–726) code for the chimeras of residues 1 to 357 CycK and of the same portion of CycK fused to CycT1 residues 300 to 726, respectively, linked to the Gal4 DBD protein at their N termini, and inserted at the EcoRI/XbaI sites of pEF-Bos-myc(EF). The plasmid effectors Gal-Cdk9 and Gal-Cdk9(D167N) expressed the chimeras of Cdk9 and its kinase-deficient mutant Cdk9(D167N) fused at their N termini to the DBD of Gal4 and inserted at the BamHI/XbaI sites of pEF-Bos-myc(EF). pSG424-Gal-VP16 codes for the activation domain of VP16 fused to Gal4 DBD at its N terminus. For in vitro transcription and translation, the different CycT1 truncation forms were subcloned into pEF-Bos-T7 in the BamHI/EcoRI sites. For in vitro glutathione S-transferase (GST) pull-down assays, CycT1 and its C-terminal truncation forms were cloned into pGEX-2TK (Amersham Pharmacia Biotech, Piscataway, N. J., Madison, Wis.) in the BamHI/EcoRI sites to express GST fusion proteins of CycT1 as described elsewhere (10).
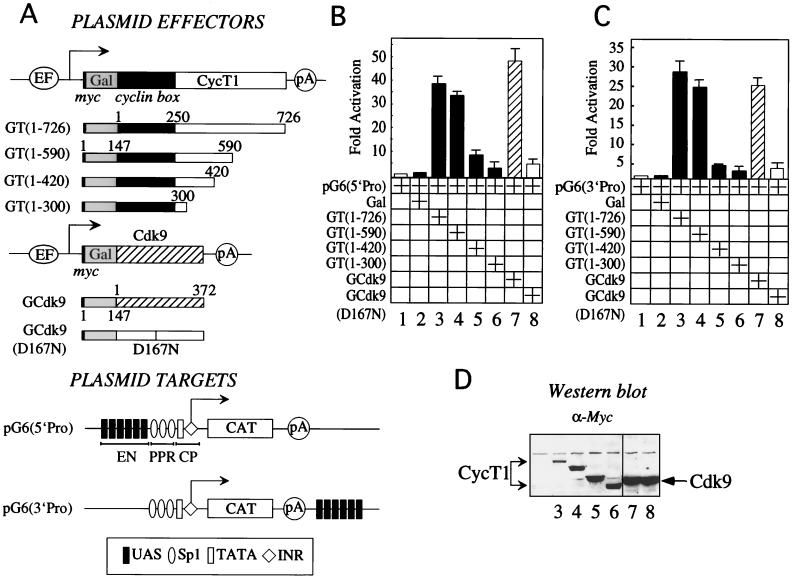
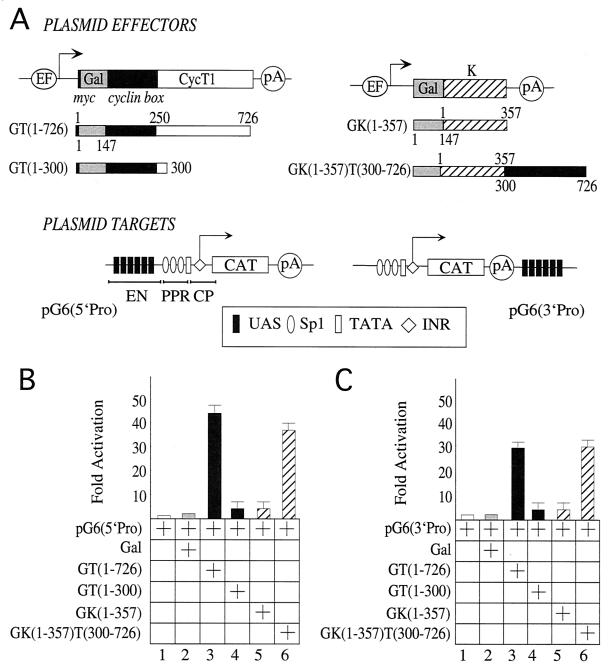
FIG. 1.
The C-terminal region of CycT1 is required for P-TEFb to activate transcription via DNA. (A) Plasmid effectors and targets used in this study. The plasmid effector GT(1–726) directed the expression of the full-length CycT1 protein from the EF-Bos promoter. CycT1 was fused at its N terminus to the Gal4 DBD (Gal, positions 1 to 147) and the c-Myc epitope tag (myc, black bar). Below the full-length chimera are Gal fusion proteins with the C-terminal truncations of CycT1. Also presented are the plasmid effectors GCdk9 and GCdk9(D167N), which expressed wild-type Cdk9 (striped bar) and the kinase-deficient mutant Cdk9(D167N) (white bar) fused at their N termini to Gal (gray bar). Also depicted are the EF-Bos promoter (EF), two cyclin boxes (positions 1 to 250, black box) of CycT1, and the polyadenylation site (pA) from simian virus 40. The plasmid target pG6(5′Pro) contained the HIV LTR linked to the CAT reporter gene. Six repeats of the synthetic Gal4-binding sites (UAS) were introduced upstream of the three Sp1 sites. These UAS repeats replaced the enhancer (EN) in the HIV LTR. Also presented is the promoter, which contained the CP, i.e., the TATA box and INR sequences, and the PPR, i.e., Sp1 sites, all placed 5′ to the CAT reporter gene (white bar). The arrow indicates the site of initiation of transcription. The plasmid target pG6(3′Pro) was identical to pG6(5′Pro) except that the UAS repeats were placed 3′ to CAT and promoter sequences. (B) P-TEFb activates transcription when presented upstream of the promoter via DNA. HeLa cells coexpressed pG6(5′Pro) and the indicated hybrid Gal-CycT1 (GT), Gal-Cdk9 (GCdk9), and dominant-negative Gal-Cdk9(D167N) proteins. The Gal4 DBD (Gal) was also expressed alone. CAT activities were measured as described in Materials and Methods. Data are relative to basal levels of the reporter gene alone (lane 1). They represent two independent experiments, and standard errors of the means are given. (C) P-TEFb activates transcription when presented via DNA downstream of the promoter and coding sequence. HeLa cells coexpressed pG6(3′Pro) with the indicated hybrid Gal-CycT1 proteins, wild-type Gal-Cdk9, and kinase-deficient Cdk9(D167N). CAT activities are relative to basal levels of the reporter gene alone (lane 1). They represent two independent experiments, and standard errors of the means are given. (D) Western blotting analysis with anti-Myc antibody revealed expression levels of hybrid Gal-CycT1 (lanes 3 to 6), wild-type Gal-Cdk9 (lane 7), and dominant-negative Gal-Cdk9(D167N) proteins (lane 8).
Binding assays.
Assays for the direct binding between the hybrid GST-CTD and different truncation proteins of CycT1 were performed as described previously (10). In brief, 5 μg of the hybrid GST-CTD was incubated at 4°C for 3 h in the presence of 35S-labeled in vitro transcribed-translated CycT1 or its C-terminal truncation forms (Promega) in 300 μl of binding buffer (buffer A) consisting of 20 mM HEPES-KOH (pH-7.9), 0.1% Triton X-100, 0.05% NP-40, 0.3% bovine serum albumin, 5 mM dithiothreitol, 5 mM EDTA, and 200 mM KCl. Following incubation, binding reaction products were washed extensively with buffer A containing 500 mM KCl. Washed beads were subjected to sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) and analyzed by autoradiography.
For interaction between CycT1 and the endogenous RNAPII, HeLa nuclear extracts were incubated with the different GST-CycT1 fusion proteins at 4°C for 3 h in ELB binding buffer (50 mM HEPES-KOH [pH 7.9], 0.1% Triton X-100, 5 mM dithiothreitol, 5 mM EDTA, 150 mM NaCl). Following incubation, reaction products were washed extensively with ELB binding buffer containing 500 mM NaCl. Beads were subjected to 5% SDS-PAGE, followed by Western blotting with anti-RNAPII antibody (8WG16; BAbCO, Richmond, Calif.).
For interaction between CycT1 and the CTD of RNAPII, Raji cells (35) (kind gift of D. Eick) expressing hemagglutnin (HA) epitope-tagged RNAPII, where the CTD contains 52 or 5 repeats of the heptapeptide sequence, were transfected with 30 μg of effector plasmids. Lysates were prepared from these cells with ELB buffer and were immunoprecipitated with anti-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.). Proteins were subjected to SDS-PAGE, transferred to a nitrocellulose membrane, and probed with anti-HA antibody (Santa Cruz Biotechnology).
Transient-transfection assays.
HeLa cells were transfected with target plasmids (0.2 μg) with or without the different effector plasmids (1 μg) with Lipofectamine as recommended by the manufacturer (GIBCO/BRL, Gaithersburg, Md.). All transfections were balanced to total of 1.2 μg of DNA with pEF-Bos-myc plasmid DNA. At 48 h after transfection, cells were lysed in lysis buffer (250 mM Tris-HCl [pH 7.5] and 0.1% Triton X-100), and CAT activities were measured by a liquid scintillation assay as described previously (9). Western blot analysis was performed to evaluate expression levels of the proteins (see Fig. 1D). For the inhibition of kinase activity of Cdk9, cells were treated with DRB (Sigma, St. Louis, Mo.) (diluted in dimethyl sulfoxide) at various concentrations, 24 h before and 48 h after transfection. CAT activities were measured from cell lysates as described previously (9).
Raji cells expressing the different RNAPII CTD were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 100 μg of streptomycin per ml, 2 mM l-glutamine, 1 mg of G418 per ml, and 0.1 μg of tetracycline per ml. Before transfection, cells were washed three times with medium containing only 1% FCS and were grown for an additional 1 day with medium containing 10% FCS but no tetracycline. This enabled the expression of the different RNAPII-CTD forms. The next day, the different CycT1 plasmid effectors were transfected by electroporation (250V; 950 μF), and cells were grown for an additional 2 days. Cells were lysed with ELB buffer, and the lysates were subjected to immunoprecipitation with the anti-Myc antibody followed by Western blotting with anti-HA antibody (35).
Immunoprecipitation and Western blotting.
For determining the expression levels of proteins, 293T cells were transfected with various plasmids (2 μg). At 24 h after transfection, cells were lysed with ELB buffer. Proteins were subjected to SDS-PAGE, transferred onto Immobilon-NC membranes (Millipore, Bedford, Mass.), and reacted with the appropriate antibodies. Proteins were visualized by enhanced chemiluminescence (Amersham Inc., Evanston, Ill.).
For immunoprecipitation experiments, cells were lysed with ELB buffer and immunoprecipitated with the appropriate antibody. Following binding to the antibody, reaction mixtures were incubated with protein A-Sepharose beads at 4°C. Immunoprecipitate reaction products were washed extensively with ELB buffer and subjected to SDS-PAGE followed by Western blotting.
RPA.
COS cells were transfected with pG6TAR(5′Pro) (10 μg) and with Gal-CycT1 fusion proteins and C-terminal-truncation forms (10 μg). At 24 h after transfection, cells were harvested and RNA was purified. A 30-μg portion of RNA was used for RNase protection assays (RPA). Antisense probes MTX-147 and MTX-89 (kind gift of J. Karn) were linearized with XbaI and BamHI, respectively. Probes were transcribed in the presence of [α-32P]UTP by using MAXIscript (Ambion, Austin, Tex.) with T3 RNA polymerase to produce labeled RNA probe. Following synthesis, probes were subjected to polyacrylamide-urea gel electrophoresis and purified before use. RPA were performed using the RPAIII kit as recommended by the manufacturer (Ambion). Protected RNA fragments were separated on 11% polyacrylamide–urea sequencing gels. Gels were dried and subjected to autoradiography (38, 60).
RESULTS
P-TEFb can activate transcription from sites upstream and downstream of the promoter and coding sequence of a target gene.
To evaluate the hypothesis that P-TEFb can mediate effects of enhancers on the elongation of transcription, we fused CycT1 and Cdk9 to the DBD of Gal4 (Fig. 1A). These chimeras allowed us to present P-TEFb via UAS that bind Gal. This artificial DNA tethering reproduced the recruitment of P-TEFb by CIITA and RelA (1, 25) and should mimic the scenario with many as-yet-uncharacterized activators. Full-length CycT1 and its C-terminally truncated derivatives, fused to Gal, were then examined for their ability to activate transcription. We used a reporter gene which contained six UAS repeats located upstream of the HIV promoter, where TAR was deleted, and the CAT reporter gene [Fig. 1A, pG6(5′Pro)]. Importantly, these UAS repeats replaced the NF-κB binding sites, which represent the enhancer in the HIV LTR. Gal-CycT1 chimeras containing the wild-type 726 (Fig. 1B, lane 3) or N-terminal 590 (lane 4) residues activated transcription from pG6(5′Pro) 35- to 40-fold above basal levels. Further C-terminal truncations of CycT1 reduced the ability of the hybrid Gal-CycT1 proteins to activate transcription from pG6(5′Pro). Thus, Gal-CycT1 fusion proteins containing the N-terminal 420 (Fig. 1B, lane 5) or 300 (lane 6) residues displayed little or no activity on pG6(5′pro). Importantly, levels of expression of our mutant chimeras were even higher than that of the wild-type fusion protein (Fig. 1D, compare lanes 4, 5, and 6 to lane 3). We conclude that the C-terminal region of CycT1 is essential for the ability of CycT1 to activate transcription via DNA. Furthermore, the hybrid Gal-Cdk9 protein (Fig. 1B, lane 7) but not a kinase-deficient hybrid Gal-Cdk9 protein (lane 8) activated transcription 50-fold from the same reporter gene. Levels of activation by the Gal-Cdk9 chimera were similar to levels of activation by the Gal-VP16 fusion protein (data not presented). These results demonstrate that the direct recruitment of an active Cdk9 to a site upstream of a promoter activates transcription. We conclude that long CycT1 as well as Cdk9 can function when presented via DNA, indicating that P-TEFb mediates transcriptional effects via the C-terminal region of CycT1.
Given these results and since there is no directionality to UAS repeats, we hypothesized that P-TEFb should also function when placed at some distance downstream of a target gene. To examine this notion, six UAS repeats were positioned 3′ of the CAT gene, distant from the promoter and coding sequence [Fig. 1A, pG6(3′Pro)]. In this system, only the Gal-CycT(1–726) (Fig. 1C, lane 3) and Gal-CycT(1–590) (lane 4) chimeras activated transcription via pG6(3′Pro). Levels of activation were 25- to 30-fold above basal levels. C-terminal deletions of CycT1, i.e., the hybrids Gal-CycT(1–420) (Fig. 1C, lane 5) and Gal-CycT(1–300) (lane 6), had lower or no activity on this plasmid target. Moreover, the Gal-Cdk9 chimera (Fig. 1C, lane 7) but not the kinase-deficient Gal-Cdk9 protein (lane 8) also supported transcriptional activation from this distal site. Similar to the results with pG6(5′Pro), the C-terminal region of CycT1 was required for effects of our chimeras on pG6(3′Pro). We conclude that P-TEFb can function via sites which are located upstream or downstream of and at a distance from the promoter and coding sequence.
Sp1 is required for the activation of transcription by P-TEFb when presented via DNA.
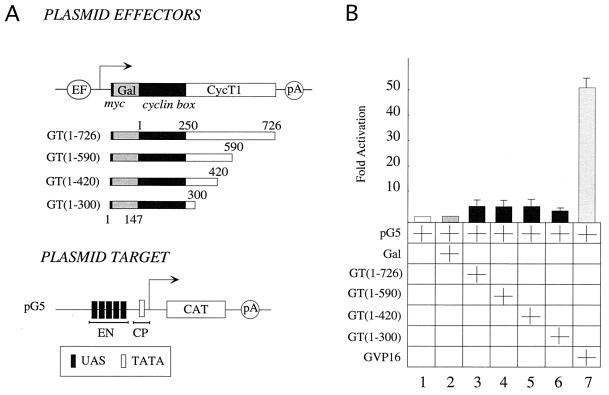
As P-TEFb could replace NF-κB when UAS were placed just upstream of the three Sp1 sites on the HIV LTR, we examined whether PPR is required for P-TEFb to mediate its effects. It should be noted that Sp1 sites are required for NF-κB to activate HIV transcription (24) and for the function of the 72-bp repeats of simian virus 40 (20). To this end, five UAS repeats were placed next to the CP (E1b TATA box) and upstream of the CAT reporter gene (Fig. 2A, pG5). Importantly, no Sp1 sites were present in pG5. None of our Gal-CycT1 fusion proteins activated transcription from pG5 (Fig. 2B, lanes 1 to 6). However, the Gal-VP16 chimera activated transcription via the same plasmid target (Fig. 2B, lane 7). Moreover, deletions of Sp1 sites in pG6(5′Pro) gave the same result, with three sites functioning optimally and their progressive removal resulting in no activity (data not presented). This suggests that Sp1 and CP recruit the PIC to initiate transcription. We conclude that PPR and CP are essential for the ability of P-TEFb to activate transcription by RNAPII via DNA.
FIG. 2.
Proximal promoter elements are required for the activation of transcription by P-TEFb via DNA. (A) Depicted are the hybrid GT(1–726) protein, C-terminal truncations of CycT1, and the plasmid target, pG5, which contained the E1b TATA box placed upstream of the CAT reporter gene. (B) HeLa cells coexpressed pG5 and our Gal-CycT1 (GT) chimeras (Fig. 1A). The Gal-VP16 fusion protein (GVP16) was used as a positive control. Results are relative to the basal level (lane 1) and represent two independent experiments, where standard errors of the means are given.
RNAPII interacts with the C-terminal region of CycT1 in vivo.
These results indicated that C-terminal residues of CycT1 promote transcriptional activation via distal sites. These interactions could occur by the looping of DNA. Indeed, some activators that bind to enhancers and coactivators that interact with activators and PIC also facilitate the bending of DNA (4, 36). The CTD of RNAPII is also required for these effects (14, 50). Accordingly, by interacting with the CTD, P-TEFb could function as a coactivator that links enhancers to the PIC. This interaction could lead to the phosphorylation of the CTD by Cdk9 and the activation of transcription.
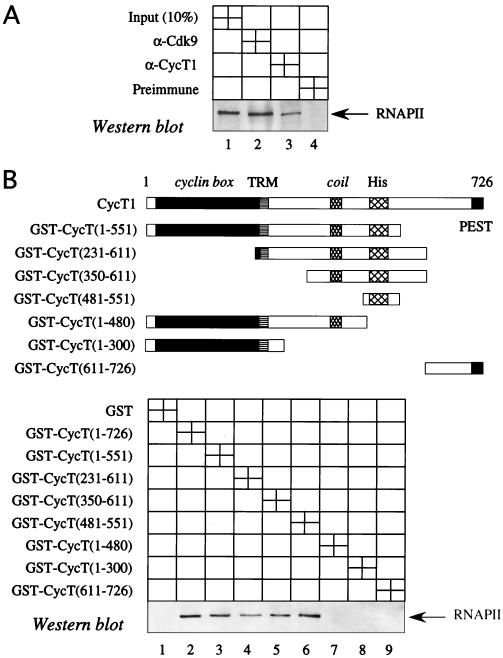
To evaluate this hypothesis, we examined whether P-TEFb interacts with RNAPII. Indeed, both CycT1 and Cdk9 coimmunoprecipitated this polymerase in HeLa cells (Fig. 3A, lanes 2 and 3). Moreover, we mapped the region of CycT1 that binds RNAPII (Fig. 3B). In vitro binding reactions using nuclear extracts and GST-CycT1 fusion proteins demonstrated that neither GST alone (Fig. 3B, lane 1) nor the fusion protein containing the N-terminal 300 residues (lane 8) interacted with RNAPII. The same was true for GST-CycT1 chimeras containing the first 480 residues (Fig. 3B, lane 7) and residues from positions 611 to 726 (lane 9). In contrast, full-length CycT1 (Fig. 3B, lane 2) and GST-CycT1 fusion proteins containing the 551 N-terminal residues (lane 3) or only sequences from positions 231 to 611 (lane 4) and 350 to 611 (lane 5) bound RNAPII. Moreover, a region of the hybrid GST-CycT1 protein from positions 481 to 551 (Fig. 3B, lane 6) still interacted with RNAPII. This region contains a histidine-rich stretch that could mediate the binding between CycT1 and RNAPII. Other GST-CycT1 chimeras that contained these 70 residues also binds RNAPII (Fig. 3B, lanes 4 and 5). We conclude that the C-terminal region of CycT1 between positions 481 and 551 interacts with RNAPII in vivo and with nuclear extracts in vitro.
FIG. 3.
P-TEFb interacts with RNAPII via the C-terminal region of CycT1 in vivo and in vitro. (A) P-TEFb interacts with RNAPII in vivo. Total nuclear extracts from HeLa cells were immunoprecipitated with anti-Cdk9 (lane 2) or anti-CycT1 (lane 3) antibodies. One tenth of the nuclear extract was used as an input control (lane 1). Preimmune serum was used as a control for immunoprecipitations (lane 4). Samples were separated by SDS-PAGE, transferred to a membrane, and probed with the antibody 8WG16, which recognizes RNAPII. (B) CycT1 interacts with RNAPII in vitro. GST-CycT1 chimeras that were examined for their interactions with the endogenous RNAPII are depicted. Also presented are important elements in CycT1: the cyclin box, the Tat-TAR recognition motif (TRM), the coil domain, the histidine-rich stretch (His), and PEST sequences. Equal amounts of the indicated GST-CycT1 fusion proteins, which were bound to glutathione-Sepharose beads (lanes 2 to 9), and the control GST alone were incubated with HeLa nuclear extracts as described in Materials and Methods. After a GST pull-down assay, Western blotting was performed with the 8WG16 antibody against RNAPII.
The C-terminal region of CycT1 binds the CTD of RNAPII in vitro and in vivo.
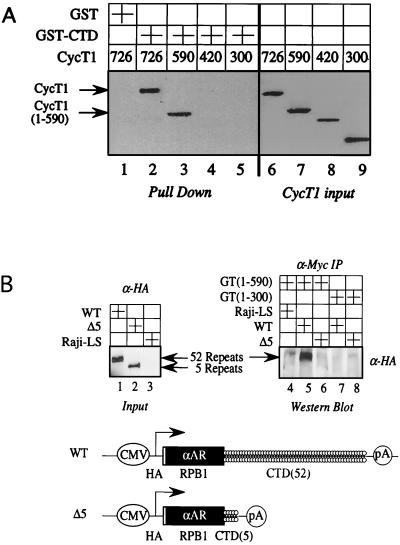
Next, we examined the binding between a GST-CTD fusion protein and 35S-labeled CycT1 protein, which was transcribed and translated with rabbit reticulocyte lysate in vitro. Only full-length CycT1 (Fig. 4A, lane 2) and CycT1 containing the N-terminal 590 residues (lane 3) bound the GST-CTD chimera. Since the wild-type CycT(1–726) did not bind GST alone, this interaction was specific (Fig. 4A, lane 1). Moreover, mutant CycT1 proteins containing the N-terminal 420 (Fig. 4A, lane 4) and 300 (lane 5) residues also did not bind the GST-CTD fusion protein. We conclude that the CTD of RNAPII binds the C-terminal residues of CycT1 in vitro.
FIG. 4.
The C-terminal region of CycT1 binds the CTD of RNAPII in vitro and in vivo. (A) The C-terminal region of CycT1 binds the CTD in vitro. The indicated 35S-labeled CycT1 proteins, which were transcribed and translated with rabbit reticulocyte lysate in vitro, were incubated with the GST-CTD fusion proteins bound to glutathione-Sepharose beads (lanes 2 to 5). As a control, GST alone was incubated with CycT(1–726). Binding reactions and steps leading to autoradiography were performed as described in Materials and Methods. CycT1 inputs represent 10% of amounts that were used for the binding reactions. (B) The C-terminal region of CycT1 interacts with the CTD in vivo. Raji cells stably expressing an HA epitope-tagged and α-amanitin-resistant RNAPII, which contained 52 or 5 repeats of the heptapeptide YSPTSPS of the CTD, coexpressed Gal-CycT(1–590) or Gal-CycT(1–300). Lysates prepared from these cells were immunoprecipitated with the anti-Myc antibody, subjected to SDS-PAGE, transferred to membranes, and probed with the anti-HA antibody. As a control, the Gal-CycT(1–590) chimera was expressed in parental Raji cells that do not express the HA epitope-tagged RNAPII molecules (Raji-LS). Lanes 1 to 3 represent the inputs of the different HA epitope-tagged RNAPII molecules, expressing 52 (wild type [WT]) or 5 (truncated [Δ5]) repeats of the CTD. Arrows indicate the wild-type and truncated large subunit of RNAPII (RPB1). Below the Western blots are the wild-type HA epitope-tagged RPB1 and Δ5 RPB1. The synthesis of the HA epitope-tagged and α-amanitin resistant (αAR) RNAPII was directed by the cytomegalovirus (CMV) promoter.
To correlate our binding studies in vitro with functional results in cells, we used Raji B cells that stably express HA epitope-tagged RNAPII with different lengths of the CTD, i.e., 52 or 5 repeats of the heptapeptide YSPTSPS (35). Previous studies demonstrated that only RNAPII containing 52 or 31 repeats of the CTD supported Tat transactivation and effects of enhancers (14, 38). Therefore, we performed immunoprecipitations using the hybrid Gal-CycT(1–590) and Gal-CycT(1–300) proteins, respectively. Only the N-terminal 590 residues of CycT1 were included because this fusion protein did not contain the C-terminal PEST region and was expressed at higher levels than the wild-type protein (Fig. 1D, compare lanes 3 and 4). Gal-CycT(1–590) but not Gal-CycT(1–300) interacted with the full-length CTD (Fig. 4B, compare lanes 5 and 7). Moreover, neither protein bound the RNAPII with only 5 repeats (Fig. 4B, compare lanes 6 and 8). Coexpression studies also revealed that only the full-length CTD supported the function of our Gal-CycT1 and Gal-Cdk9 chimeras on pG6(5′Pro) and pG6(3′Pro) in these Raji B cells (data not presented). We conclude that the C-terminal region of CycT1 binds the CTD of RNAPII in vitro and in vivo.
C-terminal region of CycT1 rescues the ability of CycK to mediate transcriptional activation.
Based on these results, the CTD appeared to be the key target for the C-terminal region of CycT1. The binding of these two domains was documented in several different ways and mapped to the histidine-rich stretch in CycT1. To prove the functional significance of this interaction, we examined the ability of a Gal-CycK chimera to activate transcription from distal sites via DNA. The same plasmid targets were used (Fig. 5A). By binding Cdk9, CycK forms an alternative P-TEFb complex (8). However, it contains only 357 residues, which include the N-terminal cyclin boxes but not the long C-terminal region of CycT1 and CycT2. Neither Gal-CycT(1–300) (Fig. 5B and C, lanes 4) nor Gal-CycK (lanes 5) activated transcription. In contrast, a Gal-CycK chimera which was fused to the C-terminal region of CycT1 (Fig. 5B and C, lanes 6) activated transcription when presented via DNA to levels of the Gal-CycT(1–726) fusion protein (lanes 3). Thus, the C-terminal residues of CycT1 rescued the ability of Gal-CycK fusion protein to support transcriptional activation from distal sites upstream and downstream of the promoter and coding sequence.
FIG. 5.
Addition of the C-terminal region of CycT1 to CycK rescues the activity of the Gal-CycK chimera via DNA. (A) Plasmid effector GT(1–726), which directed the expression of the Gal-CycT(1–726) fusion protein. Also illustrated are the hybrid proteins in which the N termini of CycK or of the hybrid CycK-CycT(300–726) protein were fused to the Gal4 DBD. Plasmid targets pG6(5′Pro) and pG6(3′Pro) (Fig. 1A) are also shown. (B) Addition of the C-terminal region of CycT1 to Cyc K rescues the activity of the Gal-CycK chimera from sites upstream of the promoter. HeLa cells coexpressed pG6(5′Pro) and Gal-CycT(1–726), truncated Gal-CycT(1–300), Gal-CycK(1–357), and Gal-CycK(1–357)-CycT(300–726) chimeras. Data are relative to basal levels of the reporter gene alone and represent two independent experiments; standard errors of the means are given. (C) Addition of the C-terminal region of CycT1 to CycK rescues the activity of the Gal-CycK chimera from sites downstream of the promoter and coding sequence. HeLa cells coexpressed pG6(3′Pro) and Gal-CycT(1–726), truncated Gal-CycT(1–300), Gal-CycK(1–357), and mixed Gal-CycK(1–357)-CycT(300–726) chimeras. Data are relative to basal levels of the reporter gene alone and represent two independent experiments, where standard errors of the means are given.
The kinase activity of Cdk9 is required for effects mediated by CycT1.
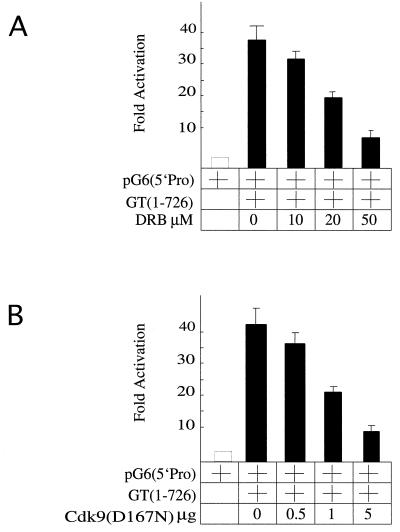
The interaction between the C-terminal region of CycT1 and the CTD of RNAPII was required for the activation of transcription. When presented via DNA, Cdk9 had the same activity (Fig. 1B and C). To demonstrate that the kinase activity of Cdk9 is also required for effects of Gal-CycT(1–726), we used DRB, a known inhibitor of Cdk9, or a kinase-deficient Cdk9 mutant [Cdk9(D167N)]. As shown in Fig. 6B, DRB inhibited the ability of the hybrid Gal-CycT(1–726) protein to activate transcription via pG6(5′Pro). These levels of inhibition were similar to the inhibition of Tat transactivation by DRB (data not presented). It is also known that at these levels, DRB does not inhibit other cyclin-dependent kinases, such as Cdk7 (43). Furthermore, the kinase-deficient Cdk9 protein [Cdk9(D167N)] also inhibited the activation of transcription by the Gal-CycT(1–726) fusion protein (Fig. 6C). Together, these results demonstrate that the kinase activity of Cdk9 is essential for the activation of transcription of target genes by P-TEFb from distal sites.
FIG. 6.
The kinase activity of Cdk9 is required for the activation of transcription by P-TEFb via DNA. (A) DRB inhibits the activation of transcription by the Gal-CycT(1–726) fusion protein. The Gal-CycT(1–726) hybrid was coexpressed with pG6(5′Pro) in HeLa cells. Cells were incubated for 24 h before and 48 h after the transfection with increasing concentrations of DRB. (B) A kinase-deficient Cdk9 protein inhibits the activation of transcription by the Gal-CycT(1–726) chimera. Increasing concentrations of a plasmid expressing the dominant-negative Cdk9(D167N) protein were cotransfected with plasmids expressing Gal-CycT(1–726) and pG6(5′Pro) into HeLa cells. CAT activities are relative to basal levels of the reporter gene alone. They represent two independent experiments, and standard errors of the means are given.
CycT1 and Cdk9 increase rates of elongation of transcription via DNA.
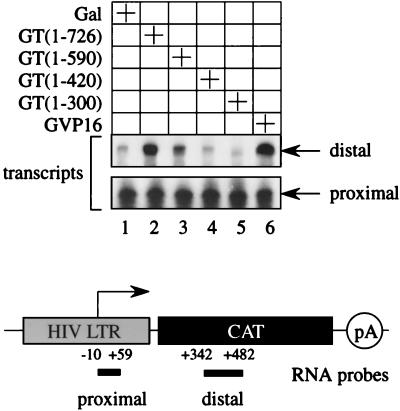
Our results demonstrated that the C-terminal region of CycT1 is required for transcriptional effects when P-TEFb is presented via DNA. This activation of transcription could have represented increased rates of initiation or elongation of transcription or both. It is established that on the HIV LTR, Tat and P-TEFb regulate the step of elongation of transcription. Nuclear run-on and steady-state RNA analyses give identical results in this system (1, 26). Because of the stem-loop structure, short transcripts, which correspond to TAR, have stabilities identical to that of the full-length mRNA species. Thus, to verify that effects of the Gal-CycT(1–726) chimera via DNA were also on this step of transcription, we performed RPA. RNA was purified from cells expressing pG6TAR(5′Pro) along with different Gal-CycT1 fusion proteins. The TAR sequence had no effect on the ability of the Gal-CycT1 hybrid to activate transcription via DNA (data not presented) (15). We used two different antisense RNA probes (60). A proximal probe hybridized to the TAR sequence and measured levels of initiated transcripts. A distal probe hybridized to the CAT sequence and measured levels of elongated transcripts (Fig. 7). Each probe was annealed to the purified RNA, digested with RNase T1, and separated by PAGE, and the protected species were detected by autoradiography.
FIG. 7.
Effects of the C-terminal region of CycT1 are on the elongation rather than initiation of transcription. RNA was purified from COS cells expressing the indicated Gal-CycT1 chimeras and the plasmid target pG6TAR(5′Pro) (lanes 1 to 5). Cells expressing Gal4-VP16 (GVP16) and Gal4 DBD (Gal) were used as the positive and negative controls, respectively. RPA were performed as described in Materials and Methods with two different RNA probes. A proximal probe corresponded to the HIV LTR from positions −10 to +59 and measured rates of initiation of transcription. A distal probe corresponded to the CAT sequence from positions 342 to 482 and measured rates of elongation.
As shown in Fig. 7, all Gal-CycT1 fusion proteins supported similar levels of promoter-proximal transcription. Levels of initiated transcripts were similar to those obtained with the Gal-VP16 fusion protein, which was used as a positive control (Fig. 7, lane 6, lower panel). However, differences between Gal-CycT1 fusion proteins were found when elongated transcripts were compared. Similar to the Gal-VP16 chimera, only Gal-CycT(1–726) (Fig. 7, lane 2) and Gal-CycT(1–590) (lane 3) increased levels of elongated transcripts (lanes 2 and 3, upper panel). In contrast, Gal-CycT(1–420) (Fig. 7, lane 4) and Gal-CycT(1–300) (lane 5) did not support these effects. These results confirm that the effects of P-TEFb via distal sites are also on the elongation of transcription.
DISCUSSION
In this report, we present evidence that the elongation of transcription can be regulated from sites upstream and downstream of target genes. P-TEFb, in this case composed of CycT1 and Cdk9, activated transcription when presented via DNA. CycT1 bound the CTD of RNAPII in vitro and in vivo, enabling P-TEFb to mediate its effects. This binding required more than five heptapeptide repeats in the CTD and the histidine-rich stretch, which is conserved in the C-terminal region of CycT1 and CycT2. The kinase activity of Cdk9 was required for P-TEFb to mediate its effects. Importantly, placing just this C-terminal region of CycT1 onto another cyclin that binds Cdk9 conferred upon CycK the ability to activate transcription via DNA. We conclude that the C-terminal region of CycT1 is required for the transcriptional effects of P-TEFb via DNA. In keeping with the enhancer hypothesis, this activation depended on promoter elements, such as CP and PPR, in this case Sp1, and the CTD of RNAPII. Thus, P-TEFb could mediate effects of enhancers on the elongation of transcription.
Our experiments confirm and extend previous studies on P-TEFb, where Gal-P-TEFb chimeras could activate transcription via UAS (34, 37). Distinct from promoter effects, our study revealed that P-TEFb can also function as a coactivator from sites distal to the target gene. Additionally, we documented that the CTD is the target of P-TEFb. Most importantly, the ability of P-TEFb to bind the CTD could be correlated directly with its effects on the elongation of transcription. Our study also extends data from D. melanogaster, where the CTD is essential for effects of P-TEFb (41). In this system, following heat shock, P-TEFb is recruited to regions upstream of promoters of over 200 transcriptional units, implying its requirement for the activated transcription of genes, where RNAPII is paused (33).
Although CycT1 and Cdk9 were tethered artificially to DNA, this situation only mimicked several strong activators that can function on enhancers and which bind and recruit P-TEFb to these sequences. Previously, we made this connection with CIITA, which is the coactivator of MHC II genes (25). It functions from conserved upstream sequences in these promoters or from distal enhancers of these genes (54). Additionally, we found that NF-κB also recruits P-TEFb to the interleukin 8 gene (1). A recent report made the same connection with the androgen receptor (30). Thus, the recruitment of P-TEFb represents an essential step for transcriptional effects of these activators. Although the direct tethering to DNA made our analyses simpler, additional insights into complex DNA-protein assembly, looping, and transcriptional activation will come from more detailed studies with these individual cellular and viral activators.
This study also addresses two important issues regarding HIV replication. First, it supports the notion that DNA-bound activators, such as NF-κB, recruit P-TEFb (1). This interaction explains the ability of HIV to start transcribing its genome in the absence of Tat. Thus, cellular activation releases NF-κB to the nucleus, where it binds to two repeated sequences upstream of the three Sp1 sites. The recruitment of P-TEFb then leads to the synthesis of Tat. Second, P-TEFb on NF-κB could promote the autophosphorylation of Cdk9, which is necessary for productive interactions between Tat, TAR, and P-TEFb. This posttranslational modification is required for the creation of the optimal RNA-binding surface of the complex between Tat and P-TEFb (7, 12). Finally, this tripartite complex leads to sustained high levels of viral replication (59).
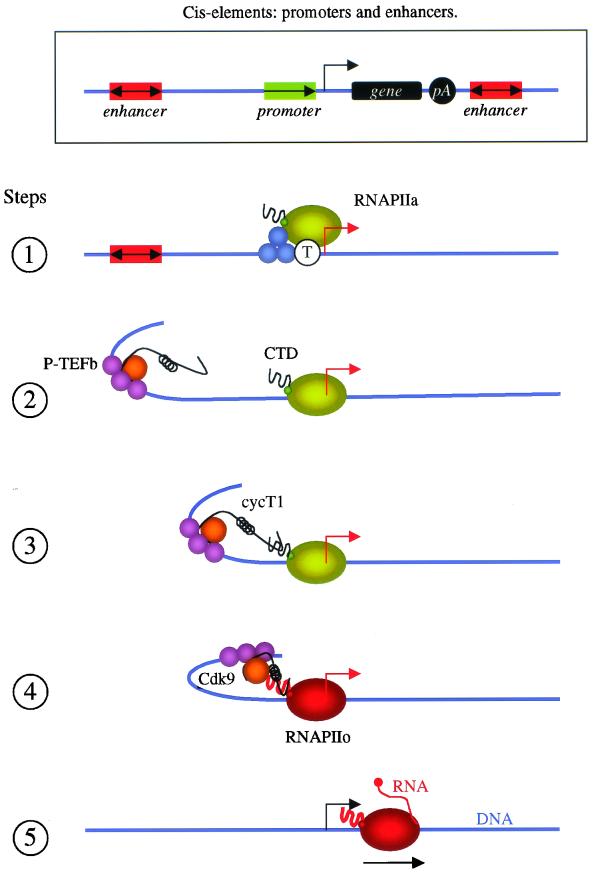
In fulfilling the criteria of enhancers, this study advances the concept that P-TEFb is an important coactivator for distal activators, i.e., that its interaction with the CTD of RNAPII can convey signals over long distances. Thus, our study suggests a model of how some enhancers could function (Fig. 8). First, the PIC is assembled and positioned on promoters. Proteins bound to TATA box, INR, and Sp1 sequences play important roles in this process (step 1). Other activators that bind to promoter and enhancer elements also remodel chromatin and recruit P-TEFb (step 2). Some proteins also bend DNA, enabling the C-terminal region of CycT1 to interact with the CTD of RNAPII (step 3). Cdk9 then phosphorylates the CTD (step 4), and possibly other proteins that interact with the paused RNAPII, resulting in the elongation of transcription (step 5). In summary, the interaction between the C-terminal region of CycT1 and the CTD of RNAPII allows P-TEFb to act as a coactivator that connects enhancer-binding proteins with the paused RNAPII at promoters, which leads to the elongation of transcription. This model brings closer the worlds of prokaryotic and eukaryotic transcription, where transcriptional antitermination, i.e., elongation, regulates among others the expression of bacteriophage λ genes (16).
FIG. 8.
Model of P-TEFb mediation of transcriptional effects from distal sites. The rectangle at the top shows cis elements of proximal (promoters) and distal (enhancers) transcriptional regulatory elements. The promoter consists of the CP (TATA box [T] and INR), downstream promoter element, and PPR, where activators like Sp1 (blue circles) bind. PIC is assembled and positioned at the promoter (step 1, green circles). Activators, such as NF-κB (purple circles), bind to the enhancer, positioned upstream (or downstream, not pictured) and in any orientation distant from the promoter and coding sequence. They affect chromatin remodeling and also recruit P-TEFb (step 2). Their ability to bend DNA enables the C-terminal region of CycT1 to bind the CTD of RNAPII (step 3). Cdk9 (orange circle) then phosphorylates the CTD of RNAPII (red circle) (step 4), resulting in the elongation of transcription (step 5).
Acknowledgments
We thank Paula Zapanc-Ecimovic for secretarial assistance, Dirk Eick and Jonathan Karn for reagents, Robert Tjian and Keith Yamamoto for advice and comments, and members of our laboratory for useful discussions.
Koh Fujinaga was supported by a fellowship from the Universitywide AIDS Research Program (F99-SF-118). This work was supported by the Howard Hughes Medical Institute.
REFERENCES
- 1.Barboric, M., R. M. Nissen, S. Kanazawa, N. Jabrane-Ferrat, and B. M. Peterlin. 2001. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327–337. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, D. 1999. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 11:347–351. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwood, E. M., and J. T. Kadonaga. 1998. Going the distance: a current view of enhancer action. Science 281:61–63. [DOI] [PubMed] [Google Scholar]
- 5.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375–380. [DOI] [PubMed] [Google Scholar]
- 6.Dynan, W. S. 1989. Modularity in promoters and enhancers. Cell 58:1–4. [DOI] [PubMed] [Google Scholar]
- 7.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20:5897–5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu, T. J., J. Peng, G. Lee, D. H. Price, and O. Flores. 1999. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J. Biol. Chem. 274:34527–34530. [DOI] [PubMed] [Google Scholar]
- 9.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garber, M. E., and K. A. Jones. 1999. HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11:460–465. [DOI] [PubMed] [Google Scholar]
- 12.Garber, M. E., T. P. Mayall, E. M. Suess, J. Meisenhelder, N. E. Thompson, and K. A. Jones. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20:6958–6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber, H. P., M. Hagmann, K. Seipel, O. Georgiev, M. A. West, Y. Litingtung, W. Schaffner, and J. L. Corden. 1995. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature 374:660–662. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh, S., M. J. Selby, and B. M. Peterlin. 1993. Synergism between Tat and VP16 in trans-activation of HIV-1 LTR. J. Mol. Biol. 23:610–619. [DOI] [PubMed] [Google Scholar]
- 16.Greenblatt, J., J. R. Nodwell, and S. W. Mason. 1993. Transcriptional antitermination. Nature 364:401–406. [DOI] [PubMed] [Google Scholar]
- 17.Hampsey, M., and D. Reinberg. 1999. RNA polymerase II as a control panel for multiple coactivator complexes. Curr. Opin. Gene Dev. 9:132–139. [DOI] [PubMed] [Google Scholar]
- 18.Harrich, D., J. Garcia, R. Mitsuyasu, and R. Gaynor. 1990. TAR independent activation of the human immunodeficiency virus in phorbol ester stimulated T lymphocytes EMBO J. 9:4417–4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNAPII. Genes Dev. 12:357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herr, W. 1993. The SV-40 enhancer: transcriptional regulation through a hierarchy of combinatorial interactions. Semin. Virol. 4:3–15. [Google Scholar]
- 21.Ivanov, D., Y. T. Kwak, J. Guo, and R. B. Gaynor. 2000. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 20:2970–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov, D., Y. T. Kwak, E. Nee, J. Guo, L. F. Garcia-Martinez, and R. B. Gaynor. 1999. Cyclin T1 domains involved in complex formation with Tat and TAR RNA. J. Mol. Biol. 288:41–56. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, P. F., and S. L. McKnight. 1989. Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem. 58:799–839. [DOI] [PubMed] [Google Scholar]
- 24.Jones, K. A., and B. M. Peterlin. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63:717–743. [DOI] [PubMed] [Google Scholar]
- 25.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 12:61–70. [DOI] [PubMed] [Google Scholar]
- 26.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489–493. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J. B., and P. A. Sharp. 2001. P-TEFb phosphorylates hSPT5 and RNA polymerase II CTD independently of CAK. J. Biol. Chem. 276:12317–12323. [DOI] [PubMed] [Google Scholar]
- 28.Kim, J. B., Y. Yamaguchi, T. Wada, H. Handa, and P. A. Sharp. 1999. Tat-SF1 protein associates with RAP30 and human SPT5 proteins. Mol. Cell. Biol. 19:5960–5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laspia, M. F., P. Wendel, and M. B. Mathews. 1993. HIV-1 Tat overcomes inefficient transcriptional elongation in vitro. J. Mol. Biol. 232:732–746. [DOI] [PubMed] [Google Scholar]
- 30.Lee, D. K., D. O. Duan, and C. Chang. 2001. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 276:9978–9984. [DOI] [PubMed] [Google Scholar]
- 31.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551–2569. [DOI] [PubMed] [Google Scholar]
- 32.Lis, J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63:347–356. [DOI] [PubMed] [Google Scholar]
- 33.Lis, J. T., P. Mason, J. Peng, D. H. Price, and J. Werner. 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 14:792–803. [PMC free article] [PubMed] [Google Scholar]
- 34.Majello, B., G. Napolitano, A. Giordano, and L. Lania. 1999. Transcriptional regulation by targeted recruitment of cyclin-dependent CDK9 kinase in vivo. Oncogene 18:4598–4605. [DOI] [PubMed] [Google Scholar]
- 35.Meininghaus, M., R. D. Chapman, M. Horndasch, and D. Eick. 2000. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J. Biol. Chem. 275:24375–24382. [DOI] [PubMed] [Google Scholar]
- 36.Muller, M. M., T. Gerster, and W. Schaffner. 1988. Enhancer sequences and the regulation of gene transcription. Eur. J. Biochem. 176:485–495. [DOI] [PubMed] [Google Scholar]
- 37.Napolitano, G., B. Majello, P. Licciardo, A. Giordano, and L. Lania. 2000. Transcriptional activity of positive transcription elongation factor b kinase in vivo requires the C-terminal domain of RNA polymerase II. Gene 254:139–145. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto, H., C. T. Sheline, J. L. Corden, K. A. Jones, and B. M. Peterlin. 1996. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc. Natl. Acad. Sci. USA 93:11575–11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 10:2657–2683. [DOI] [PubMed] [Google Scholar]
- 40.Pazin, M. J., P. L. Sheridan, K. Cannon, Z. Cao, J. G. Keck, J. T. Kadonaga, and K. A. Jones. 1996. NF-kappa B-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 10:37–49. [DOI] [PubMed] [Google Scholar]
- 41.Peng, J., N. F. Marshall, and D. H. Price. 1998. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 273:13855–13860. [DOI] [PubMed] [Google Scholar]
- 42.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pierce, J. W., M. Lenardo, and D. Baltimore. 1988. Oligonucleotide that binds nuclear factor NF-κB acts as a lymphoid-specific and inducible enhancer element. Proc. Natl. Acad. Sci. USA 85:1482–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ping, Y. H., and T. M. Rana. 2000. DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J. Biol. Chem. 276:12951–12958. [DOI] [PubMed] [Google Scholar]
- 45.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raschke, E. E., T. Albert, and D. Eick. 1999. Transcriptional regulation of the Ig kappa gene by promoter-proximal pausing of RNA polymerase II. J. Immunol. 163:4375–4382. [PubMed] [Google Scholar]
- 47.Reinberg, D., G. Orphanides, R. Ebright, S. Akoulitchev, J. Carcamo, H. Cho, P. Cortes, R. Drapkin, and O. Flores. 1998. The RNA polymerase II general transcription factors: past, present, and future. Cold Spring Harbor Symp. Quant. Biol. 63:83–103. [DOI] [PubMed] [Google Scholar]
- 48.Reines, D., R. C. Conaway, and J. W. Conaway. 1999. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr. Opin. Cell Biol. 11:342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seipel, K., O. Georgiev, H. P. Gerber, and W. Schaffner. 1993. C-terminal domain (CTD) of RNA-polymerase II and N-terminal segment of the human TATA binding protein (TBP) can mediate remote and proximal transcriptional activation, respectively. Nucleic Acids Res. 21:5609–5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seipel, K., O. Georgiev, H. P. Gerber, and W. Schaffner. 1994. Basal components of the transcription apparatus (RNA polymerase II, TATA-binding protein) contain activation domains: is the repetitive C-terminal domain (CTD) of RNA polymerase II a “portable enhancer domain”? Mol. Reprod. Dev. 39:215–225. [DOI] [PubMed] [Google Scholar]
- 51.Seipel, K., O. Georgiev, and W. Schaffner. 1992. Different activation domains stimulate transcription from remote (‘enhancer’) and proximal (‘promoter’) positions. EMBO J. 11:4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheridan, P. L., C. T. Sheline, K. Cannon, M. L. Voz, M. J. Pazin, J. T. Kadonaga, and K. A. Jones. 1995. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 9:2090–2104. [DOI] [PubMed] [Google Scholar]
- 53.Southgate, C. D., and M. R. Green. 1991. The HIV-1 Tat protein activates transcription from an upstream DNA-binding site: implications for Tat function. Genes Dev. 5:2496–2507. [DOI] [PubMed] [Google Scholar]
- 54.Steimle, V., L. A. Otten, M. Zufferey, and B. Mach. 1993. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 75:135–146. [PubMed] [Google Scholar]
- 55.Strobl, L. J., and D. Eick. 1992. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. EMBO J. 11:3307–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245–253. [DOI] [PubMed] [Google Scholar]
- 57.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395–7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451–462. [DOI] [PubMed] [Google Scholar]
- 60.West, M. J., and J. Karn. 1999. Stimulation of Tat-associated kinase-independent transcriptional elongation from the human immunodeficiency virus type-1 long terminal repeat by a cellular enhancer, EMBO J. 18:1378–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu-Baer, F., W. S. Lane, and R. B. Gaynor. 1998. Role of the human homolog of the yeast transcription factor SPT5 in HIV-1 Tat-activation. J. Mol. Biol. 277:179–197. [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41–51. [DOI] [PubMed] [Google Scholar]