Abstract
Free β-tubulin not in heterodimers with α-tubulin can be toxic, disrupting microtubule assembly and function. We are interested in the mechanisms by which cells protect themselves from free β-tubulin. This study focused specifically on the function of Rbl2p, which, like α-tubulin, can rescue cells from free β-tubulin. In vitro studies of the mammalian homolog of Rbl2p, cofactor A, have suggested that Rbl2p/cofactor A may be involved in tubulin folding. Here we show that Rbl2p becomes essential in cells containing a modest excess of β-tubulin relative to α-tubulin. However, this essential activity of Rbl2p/cofactorA does not depend upon the reactions described by the in vitro assay. Rescue of β-tubulin toxicity requires a minimal but substoichiometric ratio of Rbl2p to β-tubulin. The data suggest that Rbl2p binds transiently to free β-tubulin, which then passes into an aggregated form that is not toxic.
Studies of cellular control of microtubule assembly have focused primarily on the assembly reaction from α/β-tubulin heterodimers to microtubule polymers and on the identification of protein cofactors and structures that modulate this polymerization (8–10). Results obtained by several approaches suggest that cells may also regulate microtubule morphogenesis at stages preceding the polymerization reaction. Of particular interest are proteins that appear to interact with the α- xand β-tubulin polypeptides and modulate their activities. We are studying these proteins in the yeast Saccharomyces cerevisiae in order to understand their in vivo functions.
One of these yeast proteins is Rbl2p. Identified in a search for proteins that, when overexpressed, rescue cells from the toxicity of free β-tubulin (5), Rbl2p binds monomeric β-tubulin to form a heterodimer that excludes α-tubulin, both in vivo and in vitro (5). Pulse-labeling experiments demonstrate that Rbl2p can bind both newly synthesized β-tubulin before it is incorporated into α/β-tubulin heterodimers and β-tubulin released by dissociation of heterodimers (4). However, the precise function of Rbl2p in vivo is not known.
Biochemical experiments with the vertebrate homolog of Rbl2p, cofactor A, suggest one possible function. Cofactor A was purified from extracts based on its activity in an in vitro tubulin-folding assay that monitors the exchange of tubulin polypeptides released from the cytosolic chaperonin Tri-C into preexisting α/β-tubulin heterodimers (14, 30). Five cofactors facilitate this reaction. Three of them—cofactors C, D, and E—are necessary for the reaction. The functions of the other two—cofactors A and B—are a subset of the functions of cofactors D and E, respectively, and are not essential in the assay. However, their presence substantially stimulates the reaction (approximately fourfold for cofactor A [21]).
These experiments also suggest a pathway for the exchange reaction between unfolded tubulin polypeptides and heterodimers. When β-tubulin polypeptides are released from the cytosolic chaperonin, they are able initially to bind either cofactor A or cofactor D but all of the β-tubulin must subsequently be transferred to cofactor D in order to become competent to participate in heterodimer formation. In a parallel pathway, α-tubulin polypeptides released from the cytosolic chaperonin bind to either cofactor B or cofactor E. Those polypeptides that bind cofactor B are then transferred to cofactor E. The cofactor E/α-tubulin complex associates with the cofactor D/β-tubulin complex to generate α/β-tubulin polypeptides that are competent to exchange with exogenous, preexisting heterodimer.
Independently, the S. cerevisiae genes encoding homologs of four of these cofactors were identified in screens for a wide range of microtubule functions: sensitivity to microtubule-depolymerizing drugs (28), chromosome instability (18), sensitivity to undimerized β-tubulin (5), and functions of mitotic motors (15). Sequence homology identified the remaining cofactor homolog (11). The mutant phenotypes produced by deletion of these genes argue against a role for them in the primary pathway for tubulin heterodimer formation, as suggested by the in vitro results, because S. cerevisiae cells from which the cofactor homolog genes have been deleted, either singly or in combinations, are all viable (5, 11, 12, 18, 28, 31). Therefore, these cofactors are not required for the formation of tubulin heterodimers in the cell. There may be other genes, as yet undiscovered, that fulfill this function or are redundant with respect to the genes encoding the cofactor homologs. It is also possible that the catalysis of tubulin folding is not required in vivo. In this paper, we address specifically the in vivo role of Rbl2p/cofactor A. Rbl2p expression becomes essential in cells containing an excess of β-tubulin relative to α-tubulin. The data demonstrate that this essential activity of Rbl2p/cofactorA is not displayed by and does not depend upon Cin1p/cofactor D. Therefore, Rbl2p does not rescue cells from β-tubulin lethality by the pathway proposed for the in vitro exchange reaction described above. The results also demonstrate that increasing but substoichiometric levels of Rbl2p are necessary to rescue cells from increasing levels of free β-tubulin. Although Rbl2p is required for this activity, almost all of the cells’ free β-tubulin incorporates into an aggregate that is itself neither detrimental nor sufficient to suppress the toxicity of free β-tubulin. The results suggest that Rbl2p/cofactor A functions to protect cells from free β-tubulin by binding transiently to a subset of the free β-tubulin until it associates with an aggregate.
MATERIALS AND METHODS
Strains and media.
All of the yeast strains (Table 1) used in this study are derivatives of FSY182, -183, or -185 (35). We employed standard methods for yeast manipulations (17, 27).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| Strains | ||
| ASY560 | MATaura3-52 leu2-3,112 his3Δ200 lys2-801 ADE2 tub3Δ::hisG-URA3-hisG | This study |
| ASY561 | MATα ura3-52 leu2-3,112 his3Δ200 lys2-801 ade2 tub3Δ::hisG-URA3-hisG | This study |
| ASY566 | MATaura3-52 leu2-3,112 his3Δ200 lys2-801 ADE2 tub3Δ::hisG | This study |
| ASY568 | MATα ura3-52 leu2-3,112 his3Δ200 lys2-801 ade2 tub3Δ::hisG | This study |
| ASY587 | MATaura3-52 leu2-3,112 his3Δ200 lys2-801 ADE2 Δtub3::hisG Δpac10::HIS3 plus pJS30 | This study |
| ASY613 | ASY614 plus pJA33 | This study |
| ASY614 | MATaura3-52 leu2-3,112 his3Δ200 lys2-801 ADE2 rbl2Δ::hisG tub3Δ::hisG plus pJS30 | This study |
| FSY157 | MATaura3-52 leu2-3,112 his3Δ200 lys2-801 tub1Δ::HIS3 tub3Δ::TRP1 plus pRB634 | 24 |
| FSY183 | MATaura3-52 leu2-3,112 his3Δ200 lys2-801 ADE2 | 35 |
| FSY184 | MATα ura3-52 leu2-3,112 his3Δ200 lys2-801 ade2 | 35 |
| FSY185 | a/α ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 lys2-801/lys2-801 ade2/ADE2 | 35 |
| FSY534 | JAY47 plus pJA3 | 5 |
| FSY536 | JAY47 plus YCpGAL | 5 |
| FSY547 | JAY47 plus pA5 | 5 |
| FSY583 | JAY47 plus pA21A | 3 |
| FSY932 | FSY157 plus pGHR | 33 |
| JAY47 | a/α ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 lys2-801/lys2-801 ade2/ADE2 RBL2/RBL2 TUB2/TUB2-LEU2-GAL-TUB2 | 5 |
| JFY207 | MATα ura3-52 leu2-3,112 his3Δ200 lys2-801 ade2 cin1::hisG::URA3::hisG | This study |
| KCY46 | JAY47 plus pKC3 | This study |
| KCY48 | JAY47 plus pKC4 | This study |
| KCY74 | JAY47 plus pKC3 and pJA33 | This study |
| KCY76 | JAY47 plus pKC4 and pJA33 | This study |
| KCY110 | JAY47 plus pKC8 | This study |
| KCY162 | a/α, ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 lys2-801/lys2-801 ade2/ADE2 rbl2Δ::hisG/rbl2Δ::hisG TUB2/TUB2-LEU2-GAL-TUB2 | This study |
| KCY164 | KCY162 plus pA5 | This study |
| KCY166 | KCY162 plus pA21A | This study |
| KCY170 | KCY162 plus YCpGAL | This study |
| KCY1230 | ASY587 plus pRS317 | This study |
| KCY1234 | ASY 587 plus pKC45 | This study |
| KCY1681 | MATaura3-52 leu2-3,112 his3Δ200 lys2-801 ADE2 tub3::HIS5 | This study |
| KCY1755 | a/α ura3-52/ura3-52 leu2-3,112/leu2-3,112 his3Δ200/his3Δ200 lys2801/lys2-801 ade2/ADE2 cin1::hisG-URA3-hisG/CIN1 tub3::HIS5/TUB3 | This study |
| KCY1892 | MATaura3-52 leu2-3,112 his3Δ200 lys2-801 ADE2 tub3::HIS5 cin1::hisG pTUB3-URA3-CEN | This study |
| Plasmids | ||
| pA5 | GAL1-10-RBL2-URA3-CEN | 5 |
| pA21A | RBL2-URA3-CEN | 5 |
| pAS53 | tub3Δ-hisG knockout construct | This study |
| pFA6a-HIS3MX6 | S. pombe HIS5 | 34 |
| pGHR | GAL1-10-HIS6-RBL2-URA3-CEN | 5 |
| pJA33 | RBL2-HIS3-CEN | 3 |
| pJS30 | TUB3-URA3-CEN | 25 |
| pKC3 | GAL-TUB2-3′3/4-URA3-CEN | This study |
| pKC4 | GAL-TUB2-STOP-URA3-CEN | This study |
| pKC8 | MET25-RBL2-URA3-CEN | This study |
| pKC45 | RBL2-LYS2-CEN | This study |
| pPA10 | pGEM pac10::HIS3 | 2 |
| pRB624 | tub1-724-LEU2-CEN | 24 |
| pRS317 | LYS2-CEN | 26 |
| YcpGal | GAL1-10-URA3-CEN | 5 |
Plasmid and strain construction.
To construct a CEN plasmid overexpressing the C-terminal 75% of TUB2 under control of the GAL promoter, pWK67 (35) was digested with NotI and SacI to liberate a fragment containing the C-terminal coding region of TUB2. This fragment was ligated into pRS316-GAL (20) to produce pKC3. To construct a CEN plasmid overexpressing a tub2 cDNA that does not produce β-tubulin protein, pWK71 (35) was digested with NotI and SacI to release a fragment containing the TUB2 cDNA with three consecutive stop codons all before codon 5 Ile. This fragment was ligated into pRS316-GAL to generate pKC4. To construct the MET25-RBL2-URA3-CEN plasmid, pA5 (Table 1) was digested with BamHI and NgoMI to liberate a fragment containing the RBL2 open reading frame. This fragment was ligated into pB556 (22) to generate pKC8. To construct RBL2-LYS2-CEN, pA21A (Table 1) was digested with NotI and SalI and the resulting fragment was ligated into pRS317 to create pKC45.
To disrupt the TUB3 open reading frame with URA3, we used PCR to flank the hisG-URA3-hisG construct contained in pNKY51 (1) with 5′- and 3′-flanking regions of TUB3. The PCR primers used to generate the 1.3-kb 5′-flanking region from template genomic DNA were 5′-GGGAATATTCTCCTAGATATCAATTGG-3′ and 5′-GGGGACGTCTGTCTCAAGTCGCTTGC-3′. The resulting PCR fragment was cloned into pNKY51 with SspI and AatII to produce pAS52. The 1.3-kb 3′-flanking region was amplified from genomic DNA with PCR primers 5′-GGGGATCCGTGGCTCTGATCCCGATACCC-3′ and 5′GGGGTCGACCGTCGCCAGAACTTCTCGTTG-3′ and then cloned into pAS52 with BamHI and SalI to create pAS53. The tub3::hisG-URA3-hisG fragment was released from the plasmid with HpaI and MluI and transformed into wild-type haploids FSY183 and -184 to generate ASY560 and ASY561, respectively.
To delete the TUB3 open reading frame with Schizosaccharomyces pombe HIS5, we used the PCR-based-knockout strategy described by A. Wach and colleagues (34). By using primers containing both 40 bp of TUB3 flanking DNA and 20 bp of DNA found in the pFA6a-HIS3MX6 plasmid (a gift of P. Philippsen), we amplified a fragment containing the HIS5 gene surrounded by TUB3 flanking DNA. We purified this fragment and transformed it into wild-type haploid FSY183 to generate KCY1681. Proper integration was verified by PCR.
To construct ASY614 (rbl2::hisG tub3::hisG pTUB3-URA3-CEN), we transformed an rbl2::hisG strain containing pJA33 (RBL2-HIS3-CEN) with the tub3::hisG-URA3-hisG fragment of pAS53. Proper integration of the knockout construct at the TUB3 locus was verified by PCR. The rbl2Δ tub3Δ transformants containing pJA33 were streaked to medium containing 5-fluoroorotic acid (5-FOA) to select for loss of the URA3 gene. The resulting strain was subsequently transformed with pJS30 (pTUB3-URA3-CEN). Transformants were grown nonselectively in His+ synthetic complete medium to permit loss of the HIS3-marked pJA33 plasmid.
To construct KCY1892 (tub3::HIS5 cin1::hisG pTUB3-URA3), we crossed a cin1::hisG::URA3::hisG strain (JFY207) with a tub3::HIS5 strain (KCY1681) to generate a heterozygous diploid (KCY1755). After transformation with pDK44 (pTUB1-LYS2-CEN), we sporulated this diploid. We isolated a double-mutant haploid strain containing pDK44 and streaked it to medium containing 5-FOA to select for loss of the URA3 gene. The resulting strain was transformed with pJS30 (pTUB3-URA3-CEN). Transformants were grown in medium containing lysine to enable loss of the LYS2-marked TUB1 plasmid to generate KCY1892.
To construct ASY587 (tub3::hisG pac10::HIS3 pTUB3-URA3-CEN), we transformed a tub3::hisG strain containing pJS30 (ASY568) with a fragment created by BamHI digestion of pac10::HIS3 knockout plasmid pPA10. Integration of this disruption construct at the PAC10 locus was verified by PCR.
Affinity purification and gel filtration chromatography.
Affinity purification of Rbl2p/β-tubulin from cells overexpressing both Rbl2p and β-tubulin was performed as described in reference 4. To examine the state of β-tubulin by gel filtration chromatography, FSY185 and ASY566 cells were grown in YPD (17) until they reached mid-log phase (2 × 107 to 3 × 107 cells/ml). JAY47, FSY534, FSY547, and FSY932 cells were grown overnight in selective raffinose medium until they reached early log phase (107 cells/ml) and then induced with 2% galactose for 3 h. Approximately 1010 cells were prepared for cell lysis as previously described (4), except that 10% glycerol was included in the lysis buffer. The cells were lysed with a French press. A 0.5-ml volume of the cell lysate was injected into a Sephacryl S-300 HR column (Pharmacia) packed in MES buffer (80 mM morpholineethanesulfonic acid, 1 mM MgCl2, 1 mM EGTA, 0.2 mM GTP [pH 6.85]). The column was run with MES buffer at 0.5 ml/min, and 1-ml fractions were collected with a Bio-Rad Econo Pump and fraction collector. The column was calibrated with blue dextran, RNase A, chymotrypsinogen A, ovalbumin, and bovine serum albumin as sizing standards (Pharmacia Biotech).
Immunoblots.
Immunoblots against α-tubulin and β-tubulin were performed as previously described (5, 33). Immunoblots against Rbl2p were blocked for >12 h in 5% nonfat dry milk in TBST (0.5 M Tris, 0.5 M NaCl, 0.1% Tween 20) and incubated with affinity-purified anti-Rbl2p rabbit serum (no. 250) (5) for 2 h at a 1:100 dilution in 5% milk in TBST. The blots were washed with TBST once for 15 min and three times for 5 min. Blots were incubated with horseradish peroxidase-conjugated anti-rabbit secondary antibodies (Jackson ImmunoResearch) for 1 h at a 1:3,000 dilution and then washed in TBST as described above, rinsed for >30 min in phosphate-buffered saline, and developed with a chemiluminescence reagent kit (NEN Life Science Products). Immunoblot signals were scanned and quantitated with the IS-1000 digital imaging system (Alpha Innotech Corporation), phosphorimaging, or blue fluorescence imaging (Storm System; Molecular Dynamics).
Quantitation of cellular levels of Rbl2p and β-tubulin.
To quantitate protein levels in wild-type cells (see Fig. 2), cells were grown in the appropriate selective medium containing glucose. To examine protein levels in cells overexpressing RBL2 and β-tubulin (data not shown), cells were grown overnight to early log phase (107/ml) in selective raffinose medium and then induced with 2% galactose for 3 h. A total of 2 × 109 cells in mid-log phase (2 × 107 to 3 × 107 cells/ml) were harvested in all cases, and cell lysates were prepared by using a glass bead smash protocol as previously described (4). Serial dilutions of both the cell lysates and the standard (see below) were resolved on sodium dodecyl sulfate-14% (Rbl2p) or -7.5% (β-tubulin) polyacrylamide gels. β-Tubulin immunoblots were probed with an I125-protein A secondary antibody (NEN Dupont) as previously described (5). Due to the small amount of Rbl2p in wild-type cells, Rbl2p levels were quantified by chemiluminescence assay as described above.
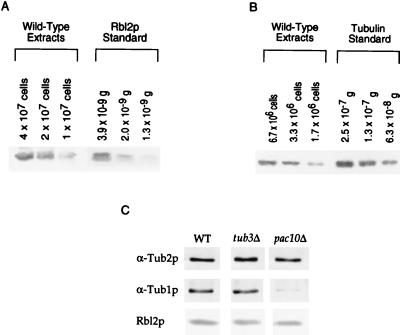
FIG. 2.
Quantitation of Rbl2p and β-tubulin in wild-type cells. (A) Immunoblots with anti-Rbl2p antibody of extracts of a known number of wild-type cells and of purified Rbl2p standards. (B) Immunoblots with anti-β-tubulin antibody of extracts of a known number of wild-type cells and of purified β-tubulin standards. (C) Immunoblots with anti-β-tubulin, anti-α-tubulin, and anti-Rbl2p antibodies of extracts from a known number of wild-type (WT), tub3Δ, and pac10Δ cells. The data shown are representative of at least three independent experiments.
Recombinant Rbl2p was generated as previously described (4). The concentration of the recombinant Rbl2p standard (1.3 μg/μl) was determined with reference to a bovine serum albumin standard (1 mg/ml) after staining with Ponceau Red. To calculate the number of Rbl2p molecules per cell, we compared the intensity of immunoblot signals generated by serial dilutions of cell lysates prepared from a known number of cells to that of a known amount of Rbl2p standard. For example, for the experiment shown in Fig. 2A, the signal generated by 4 × 107 cells is comparable to the signal generated by 3.9 × 10−9 g of purified Rbl2p. From this comparison, we determined the number of Rbl2p molecules per cell by dividing by the molecular mass of Rbl2p (12,373 Da) and multiplying the result by Avogadro’s number. All calculations are based on at least three independent experiments.
Purified, His6-tagged yeast tubulin was kindly provided by M. Gupta and R. Himes (University of Kansas). To remove any aggregated protein, the sample was spun for 20 min at 70,000 × g in a Beckman Airfuge Rotor A-100/18. The concentration of the resulting sample was determined to be 1.5 mg/ml with the Bio-Rad protein assay. To verify the purity of this sample, we separated 2 μg of the sample by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver stained the resulting gel. Only one band running at the size expected for α- and β-tubulin was observed. Since purified tubulin contains both α- and β-tubulin, we divided by two to obtain the concentration of β-tubulin in the sample. (The molecular weights of α- and β-tubulin are approximately equal.) To determine the number of β-tubulin molecules per cell, we compared the intensities of the immunoblot signals generated by serial dilutions of purified tubulin to that of the β-tubulin immunoblot signal generated by lysates of a known number of cells. For example, in the experiment shown in Fig. 2B, the signal from 6.7 × 106 cells was comparable to the signal generated by 1.3 × 107 g of tubulin standard. From this comparison, we determined the number of molecules per cell by dividing by the molecular mass of His6-tagged β-tubulin (51,821 Da) and multiplying the result by Avogadro’s number. The values discussed later in the text represent the means of three independent experiments.
Southern blot.
Yeast genomic DNA was digested with NsiI overnight. The digested DNA was resolved by electrophoresis in a 0.8% agarose gel and transferred to a Hybond-N membrane (Amersham Life Sciences) with a PosiBlot Pressure Blotter (Stratagene). The blots were probed with an 800-bp fragment produced by SspI digestion of a plasmid containing genomic RBL2 (pA21A) and radiolabeled with a Random Primed DNA Labeling Kit (Boehringer Mannheim). The blocking and hybridization procedures were performed as described in the PosiBlot Pressure Blotter manual.
RESULTS
Rbl2p/cofactor A does not require Cin1p/cofactorD to rescue cells from free β-tubulin.
Free β-tubulin (defined as β-tubulin that is not bound to α-tubulin) can be toxic to yeast cells. Rbl2p/cofactor A is a β-tubulin binding protein that protects cells from the toxicity of free β-tubulin. To alleviate the detrimental effects of free β-tubulin, Rbl2p could function as part of the tubulin folding pathway. For example, if it is the unfolded free β-tubulin that causes the toxicity, Rbl2p/cofactorA could neutralize this toxicity by catalyzing β-tubulin folding as part of its characterized effects in the in vitro β-tubulin-folding pathway. To test this model, we examined the genetic interactions among Rbl2p/cofactor A, Cin1p/cofactor D, and cells with the minor α-tubulin gene, TUB3, deleted. If Rbl2p protects cells from the toxicity of free β-tubulin via the β-tubulin-folding pathway, Rbl2p should require Cin1p to protect cells from the toxicity of free β-tubulin and all of the activities of Rbl2p should also be performed by Cin1p/cofactorD.
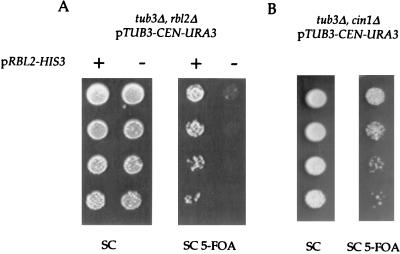
We created strains with TUB3 and either RBL2 or CIN1 deleted to assay the activity of the latter two genes in the presence of excess β-tubulin. TUB3 encodes the minor α-tubulin gene in yeast, accounting for approximately 15% of the cellular α-tubulin (23). Deletion of TUB3 does not lead to upregulation of TUB1 gene expression; tub3Δ cells have wild-type levels of Tub1p (see Fig. 2C). Therefore, approximately 15% of the β-tubulin in tub3Δ cells is free β-tubulin. Although tub3Δ cells are viable, they are supersensitive to the microtubule-depolymerizing drug benomyl (25). As the only defect in tub3Δ cells is deletion of the structural gene for an α-tubulin polypeptide, the folding pathway for β-tubulin is intact. Since cells with either RBL2 or CIN1 deleted have wild-type levels of α- and β-tubulin polypeptides (data not shown), the genetic interactions among TUB3, RBL2, and CIN1 can be attributed to the ability of Rbl2p and Cin1p to protect cells from free β-tubulin. The data demonstrate that Rbl2p is essential in tub3Δ cells. First, we recovered no viable rbl2Δ tub3Δ cells from sporulated rbl2Δ/RBL2 tub3Δ/TUB3 diploids; spores of all other genotypes were obtained (data not shown). Second, to demonstrate that the inviability of the double-mutant spores is not attributable to a germination defect, we created rbl2Δ tub3Δ double mutants carrying a low-copy plasmid expressing RBL2. These rbl2Δ tub3Δ double-mutant cells are inviable without plasmids that express either Rbl2p or Tub3p (Fig. 1A). This result demonstrates that wild-type levels of Rbl2p are essential to protect cells from the modest levels of free β-tubulin in tub3Δ cells.
FIG. 1.
Cells lacking the minor α-tubulin gene, TUB3, require RBL2 but not CIN1 for viability. (A) Serial dilutions of cells with both RBL2 and TUB3 deleted and carrying a low-copy TUB3 plasmid marked by URA3 were plated to synthetic complete medium (SC) and to synthetic complete medium containing 5-FOA (SC 5-FOA), which selects against retention of plasmids expressing URA3. Cells that contain pRBL2-HIS3-CEN (+) can grow on both media, but cells without that plasmid (−) cannot grow on 5-FOA. (B) Serial dilutions of cells with both CIN1 and TUB3 deleted and carrying a low-copy TUB3 plasmid marked by URA3 were plated to synthetic complete medium (SC) and to synthetic complete medium containing 5-FOA (SC 5-FOA). The cin1Δ tub3Δ double mutants can survive in the absence of an extra plasmid copy of TUB3.
To test whether Cin1p/cofactorD is required in cells with TUB3 deleted, we sporulated a CIN1/cin1Δ TUB3/tub3Δ diploid carrying a low-copy plasmid expressing TUB1 marked with LYS2. We isolated viable cin1Δ tub3Δ double mutants carrying the low-copy TUB1 plasmid and used plasmid shuffling to replace the plasmid expressing TUB1 with a URA3-marked plasmid expressing TUB3. By plating these cells (cin1Δ tub3Δ pTUB3-URA3) to 5-FOA, we showed that they can survive without the plasmid expressing α-tubulin (Fig. 1B; see Materials and Methods). This result demonstrates that Cin1p is not necessary for the protection of cells from free β-tubulin in tub3Δ cells. These results demonstrate that the ability of Rbl2p/cofactor A to protect cells from free β-tubulin is not shared by and does not depend upon Cin1p/cofactor D as the in vitro assay would predict. We cannot rule out the possibility that another yeast protein is the true functional homolog of cofactor D.
Levels of Rbl2p and tubulin in wild-type and mutant cells.
Rbl2p could protect tub3Δ cells from free β-tubulin—the amount of β-tubulin in excess of α-tubulin—by sequestering it in a complex. Biochemical and structural data demonstrate that the Rbl2p/β-tubulin complex is a 1:1 heterodimer (4, 21, 29). To determine whether the Rbl2p levels are sufficient for such a mechanism, we quantitated the amount of Rbl2p in wild-type cells and compared it to the level of free β-tubulin in mutant cells that require RBL2 (see above). We used immunoblots to compare the levels of Rbl2p and β-tubulin in extracts from a known number of wild-type cells to purified recombinant Rbl2p (see Materials and Methods) or purified yeast tubulin (a gift from M. Gupta and R. Himes) as a standard (Fig. 2A and B). The results of three independent trials demonstrate that wild-type cells contain 2.8 × 105 ± 0.9 × 105 molecules of β-tubulin per cell but only 5.2 × 103 ± 1.3 × 103 molecules of Rbl2p per cell or ∼2% of the level of β-tubulin. Cells with TUB3 deleted have wild-type levels of Rbl2p, β-tubulin, and TUB1-encoded α-tubulin (Fig. 2C); however, they have a 15% reduction of the total level of α-tubulin due to the deletion of the minor α-tubulin gene, TUB3 (25). As a result, approximately 15% of the β-tubulin in these cells is free (see definition above). Therefore, in tub3Δ cells, the level of free β-tubulin is about eightfold higher than the Rbl2p level.
Similarly, cells with the prefoldin component, PAC10, deleted require RBL2 (2). Quantitative immunoblots of pac10Δ extracts show that the levels of β-tubulin and α-tubulin polypeptides in pac10Δ cells are ∼40% and ∼60%, respectively, lower than wild-type levels. However, these cells contain wild-type levels of Rbl2p (Fig. 2C). Therefore, pac10Δ cells contain ∼8.4 × 104 molecules of undimerized β-tubulin or ∼16 times the wild-type level of Rbl2p. Based on the relative stoichiometries of Rbl2p and β-tubulin, Rbl2p cannot rescue pac10Δ or tub3Δ cells simply by sequestering the free β-tubulin in a stable heterodimer.
A threshold level of Rbl2p is required to rescue cells from high levels of free β-tubulin.
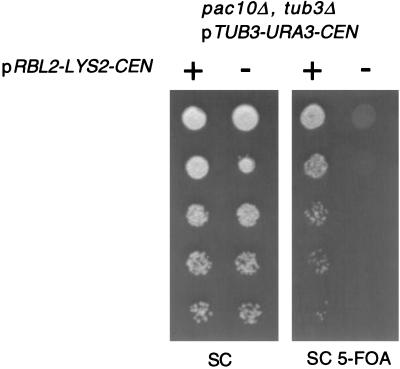
The measurement of β-tubulin and Rbl2p levels demonstrates that substoichiometric levels of Rbl2p can protect cells from free β-tubulin. However, when the amount of free β-tubulin increases, wild-type levels of Rbl2p are not sufficient to protect cells. For example, pac10Δ tub3Δ double mutants are inviable even in the presence of chromosomal RBL2. Introduction of a low-copy plasmid expressing RBL2 under its own promoter rescues these cells (Fig. 3).
FIG. 3.
An extra genomic copy of RBL2 is required to rescue cells from the synthetic lethality of tub3Δ pac10Δ. The inviability of cells with PAC10 and TUB3 deleted is suppressed by a plasmid expressing RBL2 (Fig. 1). SC, synthetic complete medium.
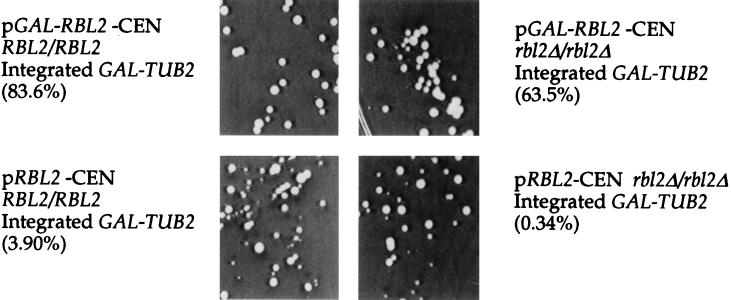
Induction of an extra copy of TUB2 under the control of the galactose-inducible promoter significantly increases β-tubulin expression. Colony formation by these cells requires still higher levels of Rbl2p (Table 2). For example, addition of a low-copy plasmid expressing RBL2 increases colony formation of a diploid containing integrated GAL-TUB2 approximately 200-fold (Table 2; compare FSY583 to FSY536). Surprisingly, the size of the colonies formed by cells that survive β-tubulin overexpression is unrelated to the efficiency of colony formation, even when that efficiency differs by orders of magnitude (Table 2 and Fig. 4). The relatively uniform colony size suggests that, in pCEN-RBL2 GAL-TUB2 cells, for example, the ∼5% plating efficiency does not correspond to the viability of the mitotic progeny. Instead, the data suggest that the progeny of cells that survive β-tubulin overexpression grow at an essentially wild-type rate. To test this hypothesis, pCEN-RBL2 GAL-TUB2 cells that survived β-tubulin overexpression were resuspended and replated to galactose medium. Ninety-eight percent of these cells survived β-tubulin overexpression and formed colonies on galactose (data not shown). This result demonstrates that pCEN-RBL2 GAL-TUB2 cells that survive β-tubulin overexpression acquire resistance to the toxicity of free β-tubulin.
TABLE 2.
Colony formation by β-tubulin-overexpressing strains containing various RBL2 dosagesa
| Strain | RBL2 locus | RBL2 plasmid | Mean % of colonies Gal/Glu ± SD |
|---|---|---|---|
| KCY170 | rbl2Δ/rbl2Δ | 0.002 ± 0.002 | |
| FSY536 | RBL2/RBL2 | 0.02 ± 0.01 | |
| KCY166 | rbl2Δ/rbl2Δ | pRBL2 | 0.34 ± 0.14 |
| FSY 583 | RBL2/RBL2 | pRBL2 | 3.90 ± 1.91 |
| KCY164 | rbl2Δ/rbl2Δ | pGAL-RBL2 | 63.5 ± 36.0 |
| FSY547 | RBL2/RBL2 | pGAL-RBL2 | 83.6 ± 11.8 |
All strains contain an extra integrated copy of TUB2 under the control of the galactose-inducible promoter. Cells were grown to saturation in selective glucose medium, counted, and plated quantitatively to both glucose and galactose-containing media. This experiment was performed at least three times with each strain.
FIG. 4.
Colonies similar in size formed by cells that survive β-tubulin overexpression. Cells containing an integrated copy of TUB2 under the control of the galactose promoter and a range of RBL2 gene dosages were plated to both glucose and galactose plates. The percentage of the cells able to form colonies on galactose is indicated in parentheses (Table 2). The colonies formed by the four strains are essentially identical in size, although there is some size variability within all four strains.
This resistance is explained by the accumulation of plasmids to upregulate RBL2 expression. For example, in pCEN-RBL2 GAL-TUB2 cells surviving β-tubulin overexpression, the RBL2 plasmid copy number increases four- to sevenfold relative to that in the same cells grown in noninducing medium (Fig. 5A). The increase in plasmid copy number accounts fully for the about four- to sevenfold increase in Rbl2p levels in these cells (Fig. 5B). β-Tubulin levels are still high in these cells; therefore, their increased Rbl2p levels likely indicate the minimum amount needed for cells to survive β-tubulin overexpression. By using quantitative immunoblots, as shown in Fig. 2, we determined that these β-tubulin-resistant pCEN-RBL2 GAL-TUB2 cells contain ∼2.5 × 105 molecules of β-tubulin in excess of α-tubulin and ∼5.7 × 104 molecules of Rbl2p. Approximately the same levels of Rbl2p and β-tubulin are found in pGAL-RBL2 GAL-TUB2 cells (FSY547). This ratio of about one molecule of Rbl2p for every four molecules of undimerized β-tubulin is about two- to fourfold higher than the ratio found in tub3Δ and pac10Δ cells (see above and Discussion).
FIG. 5.
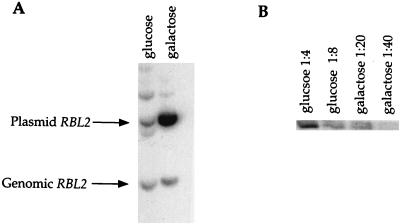
Levels of pRBL2-CEN and Rbl2p expression are upregulated in cells surviving β-tubulin overexpression. FSY583 cells containing a low-copy plasmid expressing RBL2 and an integrated copy of GAL-TUB2 were grown on glucose and galactose. DNA preparations from these cells were analyzed by Southern blotting to determine the level of the RBL2 sequence (A), and cell extracts were analyzed by immunoblotting to determine the level of Rbl2p (B) (see Materials and Methods).
Form of β-tubulin in rescued cells.
The stoichiometry of Rbl2p to free β-tubulin suggests that there is not enough Rbl2p in the cells to rescue them from the lethality of free β-tubulin by binding to the population of free β-tubulin in a 1:1 manner. To measure Rbl2p/β-tubulin heterodimer levels directly, we used affinity purification to quantitate the amount of the Rbl2p-His6/β-tubulin complex in cells overexpressing both Rbl2p-His6 and β-tubulin. This method was previously used to identify Rbl2p/β-tubulin complexes in pGAL-RBL2 tub1-724 cells (33). The mutant α-tubulin Tub1-724p binds β-tubulin with relatively low affinity. Rbl2p/β-tubulin complexes are readily formed when RBL2 is overexpressed in tub1-724 cells. As the stoichiometry of Rbl2p and free β-tubulin predicts, less than 5% of the total β-tubulin was bound to Rbl2p in cells overexpressing Rbl2p and β-tubulin (data not shown).
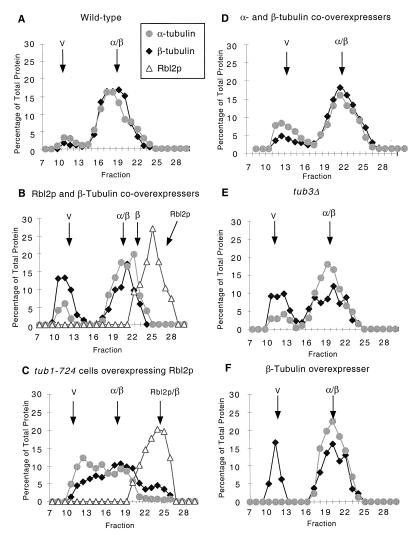
To determine the form of the β-tubulin in cells with free β-tubulin, we analyzed extracts of cells overexpressing both Rbl2p and β-tubulin by gel filtration (Sephacryl S-300 HR), followed by immunoblotting for Rbl2p and β- and α-tubulin. Figure 6B shows the elution profile of cells overexpressing both RBL2 and β-tubulin. As expected, all of the α-tubulin in the cell is bound to β-tubulin and elutes at the position expected for α/β-tubulin heterodimers, ∼100 kDa. There is no peak of β-tubulin eluting in the position expected for free monomeric β-tubulin; instead, β-tubulin is found in the void volume of the column (>1,500 kDa), behaving as aggregated β-tubulin. Rbl2p elutes as a single peak at ∼15 kDa; the position expected for monomeric Rbl2p. We did not observe any detectable peaks of Rbl2p/β-tubulin. This method can detect Rbl2p/β-tubulin complexes; in an analysis of extracts of tub1-724 cells overexpressing Rbl2p (see above), almost all of the cellular Rbl2p coeluted with β-tubulin at the position expected for Rbl2p/β-tubulin heterodimers, ∼70 kDa (Fig. 6C). (The Rbl2p peak appears much larger than the β-tubulin peak because the data for each protein is plotted as the percentage of the total protein.)
FIG. 6.
Rbl2p does not sequester free β-tubulin into an Rbl2p/β-tubulin complex. Total protein was prepared from four different strains and analyzed by Sephacryl S-300 HR gel filtration chromatography. Each fraction was assayed for α-tubulin, β-tubulin, and Rbl2p by immunoblotting, followed by scanning densitometry. The graphs plot the percentage of the total amount of each protein in each fraction and illustrate the elution positions of β-tubulin (diamonds), α-tubulin (circles), and Rbl2p (triangles). All of the experiments were repeated at least three times. Panels: A, wild-type cells; B, cells overexpressing both β-tubulin and Rbl2p; C, tub1-724 cells overexpressing Rbl2p; D, cells overexpressing β- and α-tubulin; E, tub3Δ cells; E, cells overexpressing β-tubulin. V, void volume; β, expected position of a peak of monomeric β-tubulin.
In strains in which the expression of Rbl2p is essential for survival—such as cells containing GAL-TUB2 and grown on galactose (Fig. 6B) or tub3Δ cells (Fig. 6E)—there is neither a detectable peak of β-tubulin comigrating with Rbl2p nor a peak of β-tubulin migrating as monomer. Aggregates of β-tubulin thus comprise the major portion of β-tubulin not in a heterodimer in these cells. Aggregates of β-tubulin form only when there is an excess of β-tubulin relative to α-tubulin. For example, we observed very little aggregated β-tubulin in either wild-type extracts (Fig. 6A) or extracts overexpressing both α- and β-tubulin (Fig. 6D). In addition, aggregates of β-tubulin form not only when there are high levels of free β-tubulin (GAL-TUB2 cells) (Fig. 6B and F) but also when only ∼15% of the β-tubulin in tub3Δ cells is not bound to α-tubulin (Fig. 6E). The aggregates are themselves not toxic, as both tub3Δ and pGAL-RBL2 GAL-TUB2 cells are viable. Furthermore, the formation of aggregates of β-tubulin does not require Rbl2p, as aggregates formed in rbl2Δ cells overexpressing β-tubulin (data not shown; the column profile looks identical to that in Fig. 6F). We concluded that the aggregates of β-tubulin serve as a nontoxic reservoir for free β-tubulin in cells with both high and low levels of free β-tubulin.
Role of aggregated β-tubulin in protecting cells from free β-tubulin.
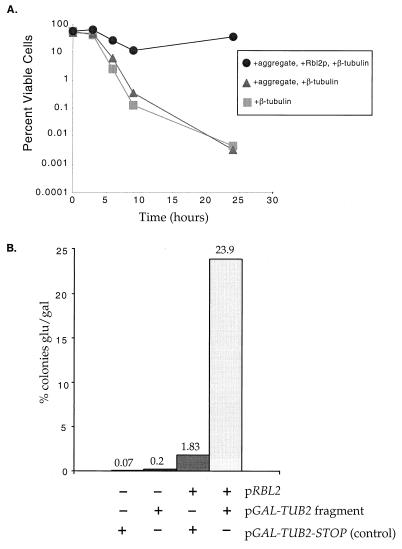
The finding that most free β-tubulin is sequestered in aggregates suggested the possibility that Rbl2p is not required to rescue cells from β-tubulin overexpression after aggregates form. To test this hypothesis, we induced simultaneous overexpression of β-tubulin under control of the GAL promoter and Rbl2p under control of the methionine-repressible promoter (MET25). After 3 h, significant aggregates containing β-tubulin had formed and the cells were completely viable. We then repressed overexpression of Rbl2p by adding methionine to the medium but continued to allow overexpression of β-tubulin. The cells containing the aggregate lost viability with the same rapid kinetics as cells in which only β-tubulin was overexpressed (Fig. 7A). Thus, Rbl2p is required to protect cells from the toxicity of free β-tubulin even in the presence of aggregates of β-tubulin.
FIG. 7.
Ability of aggregates to protect cells from β-tubulin overexpression. (A) Cells containing a single integrated copy of GAL-TUB2 and a plasmid copy of RBL2 under the control of the MET25 promoter were grown under inducing conditions for 3 h to allow an aggregate to form. The cells were then shifted to medium containing methionine to repress Rbl2p expression. Cells containing an aggregate preformed in the presence of Rbl2p (triangles) died from β-tubulin overexpression with the same kinetics as cells only overexpressing β-tubulin (squares). As a positive control, we included cells in which both Rbl2p and β-tubulin were overexpressed for the duration of the experiment (circles). (B) Cells containing an integrated copy of GAL-TUB2 with or without an extra genomic copy of RBL2 were transformed with either control plasmid pGAL-TUB2-STOP or plasmid pGAL-TUB2-3′3/4 overexpressing the C-terminal three-fourths of TUB2. The C-terminal fragment forms β-tubulin aggregates when overexpressed (data not shown). The pGAL-TUB2-STOP control plasmid does not affect the ability of cells to survive β-tubulin overexpression; pRBL2 alone is sufficient to allow >1% of the cells to survive β-tubulin overexpression (see Table 2). The pGAL-TUB2-STOP plasmid serves as a control for the reduction of β-tubulin overexpression that can occur when a second galactose-inducible promoter is introduced into cells. Although the aggregate formed by the β-tubulin fragment is a weak suppressor of β-tubulin overexpression, it enhances the suppression by RBL2. The data shown are representative of three independent experiments.
However, the formation of aggregates of β-tubulin can enhance the ability of Rbl2p to protect cells from the toxicity of free β-tubulin. Previous findings have shown that overexpression of the carboxy-terminal 75% of β-tubulin is not toxic and that this fragment of β-tubulin is not functional in vivo (35). By using gel filtration chromatography, we showed that when overexpressed, this β-tubulin fragment forms aggregates containing both the carboxy-terminal tubulin peptide and endogenous β-tubulin (data not shown). Like aggregates of full-length β-tubulin (see above), aggregates of the carboxy-terminal polypeptide are not sufficient to rescue cells from β-tubulin lethality but they do significantly enhance the rescue by pCEN-RBL2 (Fig. 7B). Overexpression of two other β-tubulin fragments that form aggregates (the amino-terminal half and the carboxy-terminal half of β-tubulin) has the same effect. The data suggest that aggregated tubulin helps protect cells from the toxicity of free β-tubulin only in the presence of excess Rbl2p.
DISCUSSION
Regulation of microtubule function is central to essential cellular functions such as chromosome segregation. Although the assembly of microtubules from heterodimeric subunits is a conspicuous context for such regulation, there is evidence that earlier steps in the heterodimerization pathway are also under cellular control. A specific example of this phenomenon is the GimC/prefoldin complex that mediates the transport of newly synthesized tubulin polypeptides to the cytoplasmic chaperonin (16, 32). Defects in prefoldin components lead to microtubule phenotypes and to downregulation of tubulin polypeptide levels (2, 16). This result suggests a relationship between efficient folding and the expression level of tubulin.
An in vitro assay suggested the possibility that tubulin polypeptides folded by the chaperonin are not competent to form heterodimers. Under the conditions of that assay, five proteins (cofactors A to E) are important to bring the tubulins released by the chaperonin to a state in which they can be incorporated into a heterodimer (30). The identification of yeast homologs of these factors, primarily by screens for genes affecting microtubule-dependent processes, enables the testing of their functions in vivo.
The above-described experiments focus on one of these proteins in budding yeast, Rbl2p/cofactor A. Overexpression of either RBL2 or murine cofactor A suppresses β-tubulin lethality in yeast (5). The in vitro assay that identified cofactor A suggests that if unfolded β-tubulin is toxic, Rbl2p/cofactor A could protect cells from excess β-tubulin by affecting tubulin folding. If Rbl2p/cofactor A rescues cells by this mechanism, the ability of Rbl2p to protect cells from free β-tubulin should be shared by and dependent upon Cin1p/cofactor D. We illustrate that Rbl2p/cofactor A, but not Cin1p/cofactor D, is required in cells that contain a modest excess of β-tubulin due to the deletion of the minor α-tubulin gene, TUB3. (This result contradicts a previous report that it was not possible to isolate tub3Δ cin1Δ double mutants by sporulating a heterozygous diploid [19]. This discrepancy may be explained by a germination defect in tub3Δ cin1Δ double mutants that we did not observe since we sporulated tub3Δ cin1Δ heterozygotes that were carrying a plasmid expressing TUB1.) This result shows that the ability of Rbl2p to suppress cells with free β-tubulin is distinct from and unrelated to the activities of Cin1p. Therefore, it is unlikely that Rbl2p rescues cells from free β-tubulin by acting in the pathway predicted by the in vitro tubulin-folding assay.
Substoichiometric levels of Rbl2p can rescue cells from free β-tubulin.
When cells contain high levels of free β-tubulin (defined as β-tubulin in excess of α-tubulin), overexpression of either Rbl2p or α-tubulin suppresses the lethality with comparable efficiency. Overexpression of α-tubulin rescues cells from the lethality of β-tubulin by forming a heterodimer with the excess β-tubulin (Fig. 6D). Although Rbl2p/cofactor A also forms a heterodimer with β-tubulin (4, 21), the suppression by Rbl2p does not involve sequestration of free β-tubulin into a stable heterodimer complex. Even in situations in which Rbl2p levels are sufficient to rescue cells fully from β-tubulin overexpression, Rbl2p levels are substantially substoichiometric with respect to β-tubulin. Furthermore, in extracts of such cells, very little Rbl2p/β-tubulin complex is detected by either affinity purification (data not shown) or gel filtration (Fig. 6B) experiments. Instead, the free β-tubulin is in an aggregate with an apparent molecular mass of >1,500 kDa.
Although substoichiometric levels of Rbl2p rescue cells from the toxicity of free β-tubulin, the efficiency of suppression depends upon the level of Rbl2p. For example, when overexpression generates high levels of free β-tubulin, cells containing a plasmid copy of RBL2 accumulate four to seven RBL2 plasmids in order to survive. Although CEN plasmids are normally present at a relatively low copy number (1 to 2 copies per cell), selective pressure can increase the copy number of CEN plasmids to ∼12 copies per cell (6, 13). The increase in Rbl2p levels in pCEN-RBL2 GAL-TUB2 cells due to the accumulation of multiple RBL2 plasmids defines the minimum amount of Rbl2p needed for cells to survive β-tubulin overexpression. In those cells, there is approximately one molecule of Rbl2p for every four molecules of free β-tubulin. This ratio of Rbl2p to β-tubulin is slightly higher than the calculated ratio of Rbl2p to free β-tubulin in either pac10Δ or tub3Δ cells, i.e., 1 to 16 or 1 to 8, respectively. This difference is discussed below in terms of a model to explain Rbl2p activity.
A mechanism for suppression of β-tubulin lethality.
Like other cells with impaired spindle function, cells overexpressing β-tubulin arrest in mitosis as large-budded cells (7, 35). However, unlike cells arrested with microtubule-depolymerizing drugs, for example, cells overexpressing β-tubulin do not recover when overexpression is repressed, suggesting that β-tubulin binds irreversibly to a target that normally binds α/β-tubulin heterodimers (35). In so doing, free β-tubulin could inhibit microtubule function by preventing microtubule nucleation or stabilization. One likely target for free β-tubulin is the spindle pole body; after long periods of β-tubulin overexpression, β-tubulin, but not α-tubulin, colocalizes with the spindle pole body (35).
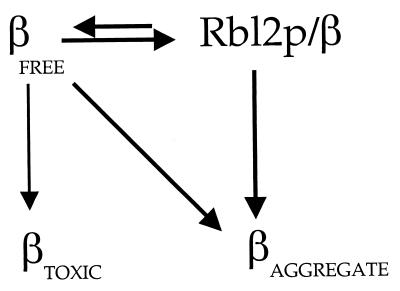
Although Rbl2p does not act by sequestering all of the free β-tubulin, the quantitative data presented above suggest that complex formation is important for rescue. We propose that Rbl2p interacts transiently with free β-tubulin, releasing it to three possible fates (Fig. 8). First, it can interact with some essential component or components of the cell (the target of β-tubulin toxicity) and so cause microtubule disassembly and cell death. Second, it can associate with other β-tubulin molecules—and possibly other proteins as well—to form an aggregate that is not toxic and that serves as a relatively stable reservoir for β-tubulin not associated with α-tubulin. Third, it can reassociate with Rbl2p. We suggest that the threshold level of Rbl2p required to rescue cells from high levels of free β-tubulin is the concentration of Rbl2p that can effectively compete with the essential target for the binding of free β-tubulin. This explains why the level of Rbl2p required for effective suppression increases with the level of excess β-tubulin, even though the two proteins are not stoichiometric.
FIG. 8.
A model for Rbl2p function in vivo. Rbl2p protects cells by binding transiently to free β-tubulin. Once released from Rbl2p, free β-tubulin has three possible fates: it can bind to the target of β-tubulin toxicity, be sequestered in the aggregate, or reassociate with Rbl2p. To protect cells from free β-tubulin, the level of Rbl2p in the cells must reach a threshold level so that Rbl2p can effectively compete with the target of β-tubulin toxicity for β-tubulin binding. Associations of β-tubulin with both the target of β-tubulin toxicity and the aggregate of β-tubulin appear to be irreversible. Rbl2p may facilitate aggregate formation by converting β-tubulin into a form that aggregates more readily.
The data show that the threshold level of Rbl2p needed to protect cells from free β-tubulin is higher in cells overexpressing β-tubulin than in cells with either TUB3 or PAC10 deleted (1:4 versus 1:8 and 1:16, respectively). This difference can be accounted for by the increased rate at which β-tubulin polypeptides are produced when β-tubulin is under the control of the galactose promoter. Under these circumstances, free β-tubulin polypeptides might be translated more quickly than they could be sequestered into an aggregate. As a result, a greater amount of free β-tubulin would be available to bind the toxic target and more Rbl2p would be needed to effectively compete for free β-tubulin.
Our data also suggest that the aggregates—although not sufficient to protect cells from free β-tubulin—can enhance rescue by low levels of Rbl2p (Fig. 7B). The aggregates provide a stable storage pool for free β-tubulin. However, the bimolecular rate at which free β-tubulin associates with the aggregate may be very slow, because there are likely to be relatively few aggregates of β-tubulin in the cell, thus limiting their capacity to rescue on their own.
In summary, our data suggest that Rbl2p acts as molecular chaperone to protects cells from the toxicity of excess β-tubulin. Once it reaches a threshold level relative to the amount of free β-tubulin in the cell, Rbl2p occupies free β-tubulin so that it cannot bind the toxic target. We believe that it is the β-tubulin aggregate that provides a stable, nontoxic reservoir for free β-tubulin in the cell. The mechanisms by which Rbl2p and other yeast proteins contribute to aggregate formation are under study.
Acknowledgments
We thank R. Himes and M. Gupta at the University of Kansas for the gift of purified yeast tubulin. We thank the Philippsen (Universität Basel, Basel, Switzerland), Young (Massachusetts Institute of Technology [MIT]), and Fink (MIT) laboratories for plasmids. We thank A. Rushforth and other members of our laboratory, members of MIT M&M, A. Grossman (MIT), S. Bell (MIT), and S. Sanders (MIT) for valuable discussions.
K.C.A. and A.S. were supported in part by a training grant from the National Institute of General Medical Sciences to the Department of Biology, MIT. W.C. was supported in part by an HHMI Undergraduate Biological Sciences Education Program grant to the Department of Biology, MIT. This work was supported by a grant to F.S. from the National Institute of General Medical Sciences.
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez, P., A. Smith, J. Fleming, and F. Solomon. 1998. Modulation of tubulin polypeptide ratios by the yeast protein Pac10p. Genetics 149:857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Archer, J. E. 1996. PhD. thesis. Massachusetts Institute of Technology, Cambridge.
- 4.Archer, J. E., L. R. Vega, M. Magendantz, and F. Solomon. 1998. Formation and function of the Rbl2p–β-tubulin complex. Mol. Cell. Biol. 18:1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Archer, J. E., L. R. Vega, and F. Solomon. 1995. Rbl2p, a yeast protein that binds to β-tubulin and participates in microtubule function in vivo. Cell 82:425–434. [DOI] [PubMed] [Google Scholar]
- 6.Bitoun, R., and A. Zamir. 1986. Spontaneous amplification of yeast CEN ARS plasmids. Mol. Gen. Genet. 204:98–102. [DOI] [PubMed] [Google Scholar]
- 7.Burke, D., P. Gasdaska, and L. Hartwell. 1989. Dominant effects of tubulin overexpression in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassimeris, L. 1999. Accessory protein regulation of microtubule dynamics throughout the cell cycle. Curr. Opin. Cell Biol. 11:134–141. [DOI] [PubMed] [Google Scholar]
- 9.Cleveland, D. W. 1990. Microtubule MAPping. Cell 60:701–702. [DOI] [PubMed] [Google Scholar]
- 10.Desai, A. 1997. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 13:83–117. [DOI] [PubMed] [Google Scholar]
- 11.Feierbach, B., E. Nogales, K. Downing, and T. Stearns. 1999. Alf1p, a CLIP-170 domain containing protein, is functionally and physically associated with α-tubulin. J. Cell Biol. 144:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming, J. A., L. R. Vega, and F. Solomon. 2000. Function of tubulin binding proteins in vivo. Genetics 156:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Futcher, B., and J. Carbon. 1986. Toxic effects of excess cloned centromeres. Mol. Cell. Biol. 6:2213–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, Y., I. Vainberg, R. Chow, and N. Cowan. 1993. Two cofactors and cytoplasmic chaperonin are required for the folding of α- and β-tubulin. Mol. Cell. Biol. 13:2478–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiser, J. R., E. J. Schott, T. J. Kingsbury, N. B. Cole, L. J. Totis, G. Bhattacharyya, L. He, and M. A. Hoyt. 1997. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol. Biol. Cell 8:1035–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissler, S., K. Siegers, and E. Schiebel. 1998. A novel protein complex promoting formation of functional alpha- and gamma-tubulin. EMBO J. 17:952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guthrie, C., and G. Fink. 1991. Guide to yeast genetics and molecular biology, vol. 194. Academic Press, Inc., New York, N.Y.
- 18.Hoyt, M., T. Stearns, and D. Botstein. 1990. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol. Cell. Biol. 10:223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoyt, M. A., J. P. Macke, B. T. Roberts, and J. R. Geiser. 1997. Saccharomyces cerevisiae PAC2 functions with CIN1, 2 and 4 in a pathway leading to normal microtubule stability. Genetics 146:849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, H., J. Krizek, and A. Bretscher. 1992. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics 132:665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melki, R., H. Rommelaere, R. Leguy, J. Vandekerckhove, and C. Ampe. 1996. Cofactor A is a molecular chaperone required for beta-tubulin folding: functional and structural characterization. Biochemistry 35:10422–10435. [DOI] [PubMed] [Google Scholar]
- 22.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schatz, P. J., L. Pillus, F. Grisafi, F. Solomon, and D. Botstein. 1986. Two functional α-tubulin genes of the yeast Saccharomyces cerevisiae encode divergent proteins. Mol. Cell. Biol. 6:3711–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schatz, P., F. Solomon, and D. Botstein. 1988. Isolation and characterization of conditional-lethal mutation in the TUB1 alpha-tubulin gene of the yeast Saccharomyces cerevisiae. Genetics 120:681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schatz, P. J., F. Solomon, and D. Botstein. 1986. Genetically essential and nonessential α-tubulin genes specify functionally interchangeable proteins. Mol. Cell. Biol. 6:3722–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Solomon, F., L. Connell, D. Kirkpatrick, V. Praitis, and B. Weinstein. 1992. Methods for studying the yeast cytoskeleton. Oxford University Press, Oxford, England.
- 28.Stearns, T., M. Hoyt, and D. Botstein. 1990. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics 124:251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinbacher. 1999. Crystal structure of the post-chaperonin beta-tubulin binding cofactor Rbl2p. Nat. Struct. Biol. 6:1029–1032. [DOI] [PubMed] [Google Scholar]
- 30.Tian, G., Y. Huang, H. Rommelaere, J. Vandekerckhove, C. Ampe, and N. Cowan. 1996. Pathway leading to correctly folded β-tubulin. Cell 86:287–296. [DOI] [PubMed] [Google Scholar]
- 31.Tian, G., S. Lewis, B. Feierbach, T. Stearns, H. Rommelaere, C. Ampe, and N. Cowan. 1997. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J. Cell Biol. 138:821–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vainberg, I., S. Lewis, H. Rommelaere, C. Ampe, J. Vandekerckhove, H. Klein, and N. Cowan. 1998. Prefoldin, a chaperone that delivers unfolded proteins to the cytosolic chaperonin. Cell 93:863–873. [DOI] [PubMed] [Google Scholar]
- 33.Vega, L. R., J. Fleming, and F. Solomon. 1998. An alpha-tubulin mutant destabilizes the heterodimer: phenotypic consequences and interactions with tubulin-binding proteins. Mol. Biol. Cell 9:2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wach, A., A. Brachat, C. Alberti-Segui, C. Rebischung, and P. Philippsen. 1997. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in S. cerevisiae. Yeast 13:1065–1075. [DOI] [PubMed] [Google Scholar]
- 35.Weinstein, B., and F. Solomon. 1990. Phenotypic consequences of tubulin overproduction in Saccharomyces cerevisiae: differences between alpha-tubulin and beta-tubulin. Mol. Cell. Biol. 10:5295–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]