Abstract
Yeast transcription factor IIIC (TFIIIC) plays a key role in assembling the transcription initiation factor TFIIIB on class III genes after TFIIIC-DNA binding. The second largest subunit of TFIIIC, τ131, is thought to initiate TFIIIB assembly by interacting with Brf1/TFIIIB70. In this work, we have analyzed a TFIIIC mutant (τ131-ΔTPR2) harboring a deletion in τ131 removing the second of its 11 tetratricopeptide repeats. Remarkably, this thermosensitive mutation was selectively suppressed in vivo by overexpression of B”/TFIIIB90, but not Brf1 or TATA-binding protein. In vitro, the mutant factor preincubated at restrictive temperature bound DNA efficiently but lost transcription factor activity. The in vitro transcription defect was abolished at high concentrations of B” but not Brf1. Copurification experiments of baculovirus-expressed proteins confirmed a direct physical interaction between τ131 and B”. τ131, therefore, appears to be involved in the recruitment of both Brf1 and B”.
Transcription by RNA polymerase (Pol) III requires the multistep assembly of transcription factor IIIC (TFIIIC) and TFIIIB into a preinitiation complex which is able to direct accurate initiation and multicycle transcription by Pol III (59). The assembly factor TFIIIC plays a primary role in preinitiation complex formation by recognizing the internal promoter elements (the A and B blocks) in tRNA genes (tDNA) or the TFIIIA-5S RNA gene complex and by facilitating the assembly of the initiation factor TFIIIB upstream of the transcription start site. TFIIIB is considered the initiation factor because the TFIIIB-DNA complex by itself is able to recruit Pol III productively in vitro (26).
Saccharomyces cerevisiae TFIIIB comprises three components: the TATA binding protein (TBP), Brf1/TFIIIB70, which is evolutionarily related to the class II factor TFIIB (9, 14, 27, 37), and B”/TFIIIB90 (28, 50, 51). The Pol III transcription system can use alternative pathways to build the TFIIIB-DNA complex. TFIIIB needs TFIIIC to bind to TATA-less Pol III genes like most tDNAs, but it can assemble by itself, at least in vitro, onto the TATA element of the SNR6 gene (24, 40, 41), although TFIIIC enhances the transcription efficiency (18). In vivo, the transcription of the SNR6 snRNA gene requires TFIIIC to direct TFIIIB binding and probably to overcome the repressive effect of chromatin (10, 20). Some tRNA genes that harbor a TATA-like element can also be transcribed, in vitro, without TFIIIC (17).
Yeast TFIIIC factor, also called τ, is a multiprotein complex comprising six subunits, τ138, τ131, τ95, τ91, τ60, and τ55 (5, 19, 46), each of which is essential for cell viability. Mutagenesis (3, 34, 45, 56) and protein-DNA and protein-protein interaction studies (12, 16, 30, 38) have provided a global view of TFIIIC-DNA complex formation and shed light on the role of TFIIIC subunits. Five subunits of TFIIIC could be photocross-linked to chemical probes specifically located within or immediately flanking the SUP4 tRNA and the 5S RNA genes (5, 8). This elegant approach has allowed their positioning within the factor-DNA complex. Subunits τ138 and τ91 interact around the B block and ensure the primary binding of TFIIIC to the B block of tDNA (3, 34). τ95 and τ55 interact physically with each other (38) and bind on the vicinity of the A block on opposite sides of the DNA helix (5). τ60 could not be cross-linked to DNA, but this polypeptide is located in τB, at least in part, together with τ138 and τ91 (16). Finally, in the TFIIIC-tDNA complex, τ131 is the only subunit cross-linked upstream of the transcription start site, in a region occupied by TFIIIB, and it also extends downstream between the A and B blocks (5, 6).
The mechanism by which TFIIIC recruits TFIIIB onto TATA-less class III genes appears to involve a stepwise series of intricate protein-protein interactions and conformational changes the submolecular details of which are poorly understood. In vitro experiments have shown that TFIIIB assembly starts with the recruitment of Brf1 by the TFIIIC-tDNA complex, through its interaction with τ131 (6, 12, 27, 30, 42, 57). A region of τ131 responsible for this interaction has been mapped in two-hybrid experiments to the first 168 residues (12). Entry of TBP in the complex, mediated by Brf1 (27, 36) and by τ60 (16), stabilizes the binding of TFIIIC to the DNA and dramatically increases the cross-linking of τ131 to upstream DNA (6). Addition of B" leads to further drastic changes in complex conformation and stability: (i) the TFIIIB-DNA complex becomes resistant to high salt or polyanions concentrations (29), and (ii) cross-linking of τ131 to upstream DNA is greatly reduced (6, 31). The change in cross-linking efficiency of τ131 during the TFIIIB assembly underscores the flexibility of this polypeptide within the complex. Upon binding to tDNA genes harboring long introns or long extra arms (or both as in some Leu or Ser tRNA genes), TFIIIC is able to stretch out between the A and B blocks, as seen by electron microscopy (54). The fact that TFIIIB could be assembled by TFIIIC at various distances from the A block further suggested that τ131 itself is able to stretch out along DNA, possibly due to its peculiar structure rich in tetratricopeptide repeat (TPR) motifs (23, 39, 49). Although the in vitro experiments are strongly suggestive of a multistep assembly pathway, it is not clear whether the in vivo recruitment of Brf1 and B” components of TFIIIB involves a one-step or two-step mechanism.
In this work, we have pursued the characterization of the τ131 subunit by investigating the effect of the deletion of its second TPR motif (ΔTPR2). We present biochemical and genetic evidence that this mutation causes a defective recruitment of the B” subunit. The transcription defect can be rescued by overdosage of B” both in vivo and in vitro. A direct interaction between τ131 and B” could be demonstrated. The results suggest that τ131 directs the recruitment of TFIIIB by interacting with both Brf1 and B”.
MATERIALS AND METHODS
Yeast strains, media, and genetic methods.
The yeast strains used in this study were constructed by genetic techniques based on transformation of lithium acetate-treated cells, sexual mating, and tetrad analysis with standard media and growth conditions (52).
Yeast strains are as follows: SC55 (a/α ura3 to 52/ura3-52 trp1-Δ1/trp1-Δ1 his3-Δ200/his3-Δ200 ade2-101/ade2-101 lys2-801/lys2-801 leu2-Δ1/+ cans/canR) (13), YCK107 (a ura3-52 trp1-Δ1 his3-Δ200 ade2-101 lys2-801 tfc4::HIS3 +pCK17) (39), YHD3 (a ura3-52 trp1-Δ1 his3-Δ200 ade2-101 lys2-801 tfc4-Δ::HIS3 +pCK17) (this work), YHD7 (a ura3-52 trp1-Δ1 his3-Δ200 ade2-101 lys2-801 tfc4-Δ::HIS3 +pUN45-τ131-ΔTPR2) (this work). All deletion mutants of τ131 used in this work have been described previously: τ131-ΔN1, τ131-ΔN2, τ131-ΔTPR1, τ131-ΔTPR2, τ131-ΔTPR3, τ131-ΔTPR4, τ131-ΔTPR5, τ131-ΔTPR6, τ131-ΔTPR7, τ131-ΔTPR8, τ131-ΔTPR9, τ131-Δbasic1, τ131-Δbasic2, τ131-ΔH1, τ131-Δloop, τ131-Δloop1, τ131-Δloop2, τ131-ΔH2, τ131-Δzipper, τ131-ΔTPR10, τ131-ΔTPR11 (12). The plasmids used for the in vivo suppression studies (pLR30, pL1, and pJR38, overexpressing Brf1, TBP, and B”, respectively) have been described by Lefebvre et al. (34) and Rüth et al. (51), except pFLΔTPR2, which was constructed by cloning the SalI-BamHI fragment of pNC14 (12) into pFL44L.
Deletion of TFC4 ORF.
The whole TFC4 open reading frame (ORF) (YGR047c) coding for τ131 was deleted using the direct deletion method (7). Two 40-mer oligonucleotides were used to amplify by PCR a DNA fragment containing the HIS3 gene flanked by the sequences upstream and downstream of TFC4. The PCR-amplified fragment was directly used to transform yeast strain SC55. The structure of the diploid His3+ transformants was checked by PCR analysis on genomic DNA, and these cells were transformed with plasmid pCK17 (39), which contains a wild-type copy of TFC4. After sporulation and dissection, a His3+ spore containing pCK17 was chosen to yield strain YHD3. Plasmids harboring mutant alleles of TFC4 were substituted to wild-type TFC4 in YHD3 by shuffling out the wild-type copy on plates containing 5-fluoro-orotic acid. Strain YHD7, which harbored a mutant TFC4 gene lacking the second TPR unit, was built in this way. Viable strains isolated at 30°C were also tested for growth at 37 and 16°C.
Cloning of the Kluyveromyces lactis τ131 ortholog.
The gene coding for the K. lactis ortholog of τ131 was cloned by genomic PCR using degenerate oligonucleotides targeted to sequences presumably conserved inside the TPR units. Two direct and two reverse oligonucleotides were devised, and one of the four PCR experiments yielded a PCR fragment of the predicted length. In the positive experiment, the sequence of the direct oligonucleotide was TGGGARTTYTGGAARATHGT, targeted at the start of TPR3 (peptide sequence WEFWKIV), and the sequence of the antiparallel oligonucleotide was GGNGTRAARAARTCDATNGC (antiparallel to peptide sequence AIDFFTP), targeted at position 20 of TPR7 (alanine at position 20 is the best-conserved residue in the TPR motif). A K. lactis genomic library in vector KEp6 (a gift from M. Weslowski-Louvel, Université Claude Bernard Lyon 1, Lyon, France) (58) was screened with this PCR fragment, and one clone was isolated that contained a whole ORF (3,087 bases). The resulting protein showed a strong homology to the τ131 sequence of S. cerevisiae. Sequencing on both strands by oligonucleotide walking was performed with an ABI 377 Sequencer.
Sequences of other τ131 orthologs.
The sequences of τ131 Schizosaccharomyces pombe ortholog (accession no. CAA20753), Caenorhabditis elegans (accession no. CAA94857, hypothetical protein ZK856.9), and Drosophila melanogaster (accession no. AAF57909) were identified with the NCBI Blast server (http://www.ncbi.nlm.nih.gov/blast/blast.cgi?Jform = 1) by running BlastP 2.1.2, using the S. cerevisiae sequence as entry. The sequence data from S. pombe were determined by the S. pombe Sequencing Group at the Sanger Centre and can be obtained from ftp://ftp.sanger.ac.uk/pub/yeast/sequences/pombe/. The three sequences have been already annotated as homolog to yeast TFC4 gene. Only amino acids 543 to 1491 of the C. elegans protein were retained, as it appeared that the CAA94857 protein was a fusion of two proteins ortholog to YHR040w and YGR047c (coding for τ131). The sequence of the τ131 Candida albicans ortholog was identified in the unpublished sequence named con6-2367 using the NCBI Blast server http://www.ncbi.nlm.nih.gov/Microb_blast/unfinishedgenome.html. Sequence data for C. albicans was obtained from the Stanford DNA Sequencing and Technology Center website at http://www-sequence.stanford.edu/group/candida. Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
Purification of wild-type and mutant TFIIIC.
TFIIIC was purified starting from about 14 g of S. cerevisiae cells using fast protein liquid chromatography grade resins. Cells were harvested in the exponential phase, and crude extracts were prepared as described by Huet et al. (22). The extracts were first diluted to 0.25 M ammonium sulfate (AS) with buffer I (20 mM Tris-HCl [pH 8.0], 0.5 mM EDTA, 10 mM β-mercaptoethanol, 10% [vol/vol] glycerol), then they were loaded at 2.5 ml/min onto a 25-ml heparin Hyper-D (BioSepra) column previously equilibrated with buffer I (0.25 M AS). The resin was then washed at 5 ml/min with 250 ml of buffer I (0.35 M AS). A linear gradient of AS from 0.35 to 0.70 M in 180 ml of buffer I was then applied at 2.5 ml/min. Fractions (2 ml) were collected and assayed for TFIIIC-DNA binding activity (see below). TFIIIC-containing fractions (0.45 to 0.55 M AS) were pooled and dialyzed against buffer I (0.07 M AS). Proteins were then loaded at 0.5 ml/min on a 1-ml Mono Q column (Amersham Pharmacia Biotech) previously equilibrated with buffer I (0.07 M AS). The column was then washed at 0.5 ml/min with 20 ml of buffer I (0.07 M AS). A linear gradient of AS from 0.07 to 0.4 M in 15 ml of buffer I was then applied at 0.5 ml/min. Fractions (200 μl) containing TFIIIC-DNA binding activity were eluted between 0.24 and 0.30 M AS. Based on Western blotting experiments using anti-τ55 and anti-τ60 polyclonal antibodies, the TFIIIC preparation from wild-type cells was found to contain fivefold more factor than the TFIIC-ΔTPR2 preparation.
DNA binding and in vitro transcription assays.
TFIIIC-tDNA interactions were monitored by gel retardation analysis as described previously (22, 34). Mono Q-purified TFIIIC fractions (100 ng of protein) were incubated with 32P-labeled DNA fragment (3 to 10 fmol; 4,000 to 10,000 cpm) carrying the tRNA3Leu (327 bp) or tRNA3Glu (198 bp) genes for 15 min at 25°C in a 15-μl reaction mixture containing 10 mM Tris-HCl (pH 8.0), 10% (vol/vol) glycerol, 180 mM monovalent cations (K+ and NH4+), 10 μg of bovine serum albumin, and 200 ng of competitor DNA (pBluescript-SK). Transcription mixtures (40 μl) contained standard transcription buffer (20 mM HEPES [pH 8.0], 5 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EDTA), 5% (vol/vol) glycerol, 8 units of RNasin (Amersham Pharmacia Biotech), 0.6 mM concentrations each of ATP, GTP, and CTP, 0.03 mM UTP and 10 μCi of [32P]UTP, recombinant TBP (250 ng), recombinant Brf1 (1.2 μg), and partially purified B” fraction (1.8 μg), RNA Pol III (50 ng), and TFIIIC (Mono Q fraction, 100 ng of protein). The concentration of monovalent cations was 110 mM. Protein fractions were prepared according to the method of Huet et al. (22). After 10 min of preincubation at 25°C, the transcription reaction was started by addition of 130 ng of plasmid pRS316-SUP4, containing the yeast SUP4 tRNATyr gene (a gift from B. D. Hall), and allowed to proceed for 45 min at 25°C. Transcripts were analyzed by electrophoresis on an 8 M urea gel (6% polyacrylamide) and revealed by autoradiography. Single-round transcription experiments were performed by incubating the reaction mixture in the absence of GTP (29). Addition of heparin and GTP (300-μg/ml and 0.6 mM final concentrations, respectively) allowed to pursue transcription for 10 min at 25°C. The artificially engineered tRNA Leu genes, displaying various A block-B block distances, have already been described (4).
Preparation of recombinant B” protein.
The vector expressing TFC5 gene (coding for B”) has been described previously (51). This construct was transformed into Escherichia coli strain BL21(LysS) for expression of recombinant B” (rB”) protein fused at its N terminus to six histidines and the T7-Tag (Novagen) epitope; cultures were grown at 37°C up to an optical density of 0.4 at 600 nm before adding isopropylthio-β-d-galactoside (IPTG) at 2 mM. After a further 2-h growth at 30°C, cells were harvested and proteins were purified by Ni2+ batch column, essentially as described by Chaussivert et al. (12). Western analysis was performed to monitor rB” purification with T7-Tag antibodies.
Production of recombinant baculoviruses.
PCR-mediated mutagenesis was used to insert unique restriction sites at the boundaries of the ORFs coding for τ131, B”, and TBP. The NheI/τ131/SpeI or NheI/Gea1/NotI (48) restriction endonuclease fragments were subcloned into the XbaI site of the PVLGST vector (1). The NheI/B”/NotI restriction endonuclease fragment was subcloned into the NheI/NotI sites of a modified PVL1393 (PharMingen) containing an octa-histidine tag. The NheI/TBP/NotI restriction endonuclease fragment was subcloned into the NheI/NotI sites of a modified PVL1393 (PharMingen) containing a calmodulin binding peptide (PCR amplified from vector pCAL-n; Stratagene, La Jolla, Calif.). The resulting transfer vectors were recombined with baculovirus DNA (Bac3000; Novagen) in High Five cells (Invitrogen). The High Five cells were grown in the express five medium complemented with l-glutamine (Life Technologies) at 28°C. The recombinant viruses were plaque purified; stocks were prepared by three-step growth amplification as described by O’Reilly et al. (44).
Coexpression and copurification of τ131 and B”.
High Five cells (typically 2 × 108 cells) were coinfected with pairs of the appropriate baculovirus (B” + GST-τ131 or TBP + GST-τ131). Multiplicities of infection (between 2 and 10 PFU/cell) were adjusted so as to balance the amount of recombinant proteins simultaneously expressed from each virus. The cells were collected 72 h postinfection, washed in phosphate-buffered saline, and lysed by three cycles of freeze-thawing followed by sonication in buffer A (50 mM Tris [pH 7.5], 100 mM NaCl, 20% glycerol, 1% NP-40, 5 mM β-mercaptoethanol, and 1× protease inhibitor cocktail). The extract was then clarified by ultracentrifugation (30,000 × g, 1 h) at 4°C and loaded continuously overnight onto 2 ml of GSH-Fast Flow resin (Pharmacia) at 0.4 ml/min. The column was washed at 1 ml/min with 20 column volumes of buffer A and then washed with buffer B (50 mM Tris [pH 7.5], 100 mM NaCl, 20% glycerol, 5 mM β-mercaptoethanol). Elution was performed at 0.5 ml/min using five column volumes of buffer C (buffer B + 20 mM glutathione with pH adjusted to 8). The protein fractions were concentrated with StrataClean resin (Stratagene) and eluted in electrophoresis sample buffer (100 μl). Proteins retained were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (8% acrylamide) followed by either Coomassie blue staining or Western blotting using anti-histidine (QIAGEN) antibody.
Nucleotide sequence accession number.
The sequence determined in this study has been deposited in GenBank under accession no. AF229182.
RESULTS.
The number and characteristic grouping of TPR motifs in τ131 are conserved from yeasts to higher eukaryotes.
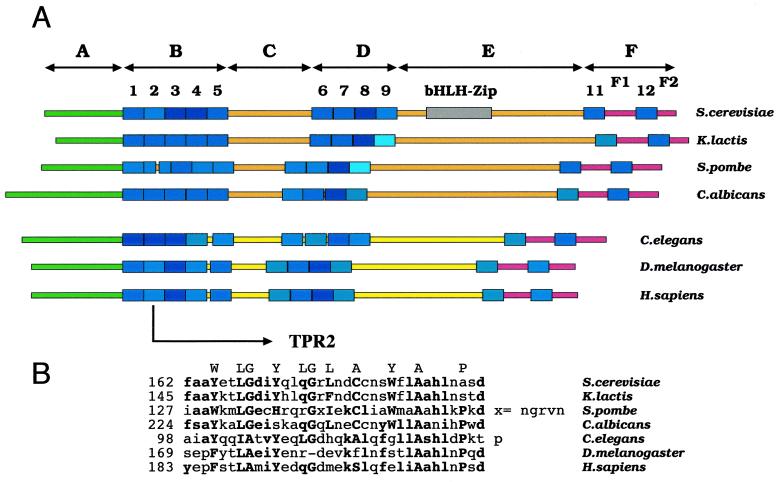
The sequence of the τ131 subunit is characterized by the presence of 11 TPR motifs and a bHLH-zipper motif (39). On the basis of sequence conservation within the TPR we have cloned the ortholog gene from K. lactis (GenBank accession no. AF229182). With the complete sequencing of several eukaryotic genomes, the sequences of four other orthologs have been found in S. pombe, C. albicans, C. elegans, and D. melanogaster. The human ortholog has been isolated through cDNA cloning by Roeder and coworkers (21). TPR motifs were identified in these ortholog sequences using a TPR consensus sequence derived from 200 TPR motifs of S. cerevisiae (H. Dumay-Odelot and C. Marck, unpublished data). All seven sequences retained the same typical 5-4-1-1 grouping of the 11 TPR motifs (Fig. 1A). This feature was used to sector the sequence of τ131 and its orthologs into six regions termed A to F. Outside the TPR regions, the sequences clearly segregated into two phylogenetic groups, the yeasts and higher eukaryotes. Regions C and E were the most divergent between the two groups, while regions F1 and F2, which flanked the last TPR unit, were more highly conserved. The conservation of this global structural organization, with an alternation of high- and low-homology regions, suggested that these proteins were built on a common TPR scaffold. Figure 1B presents the sequence of TPR2 in the seven orthologs: this TPR departed from the usual 34-residue consensus in S. pombe (addition of 4 residues) and D. melanogaster (deletion of 1 residue).
FIG. 1.
Conservation of the structure of seven orthologs of τ131 in four yeasts and three higher eukaryotes. (A) The sequence of S. cerevisiae (TFC4/YGR047c) (39) and the ortholog sequences of K. lactis (this work, GenBank accession number AF229182), S. pombe, and C. albicans (see Materials and Methods), C. elegans (11), D. melanogaster (2), and H. sapiens (21) are schematically presented. TPR motifs were located in the sequences and rated by using sequence information extracted from 200 TPR motifs of S. cerevisiae (Dumay-Odelot and Marck, unpublished). The light- to dark-blue color gradient indicates the fit to the TPR consensus (low to high). The bHLH-Zip motif, previously postulated in the S. cerevisiae sequence (39) is considered fortuitous, as it is not discernible in the six other sequences. Regions outside TPR units are color coded as follows: green, highly variable regions; orange, regions of good conservation between yeasts only; yellow, region of faint conservation in higher eukaryotes only; red, region of high conservation between all seven sequences. (B) Sequences of the second TPR unit in the seven sequences. Amino acids above sequences indicate the 11 best-conserved positions of the TPR motif as derived from 200 S. cerevisiae TPR motifs (Dumay-Odelot and Marck, unpublished). At these positions, conserved residues are indicated by boldface uppercase letters. Conserved residues at other positions are indicated by boldface lowercase letters.
Originally, it had been observed that τ131 contained a bHLH-zipper motif (39). From the analysis of S. cerevisiae τ131 and six of its orthologs, it appeared that most of the residues that defined the bHLH-zipper motif were not conserved, even in the closest ortholog, K. lactis (data not shown). For this reason, we have decided to consider the bHLH-zipper motif of τ131 as spurious. However, for the sake of clarity, we have continued using in this work the bHLH-zipper-related names already given to some mutants (e.g., ΔLoop, ΔZipper, etc.) (12).
Not all TPR motifs are essential for growth.
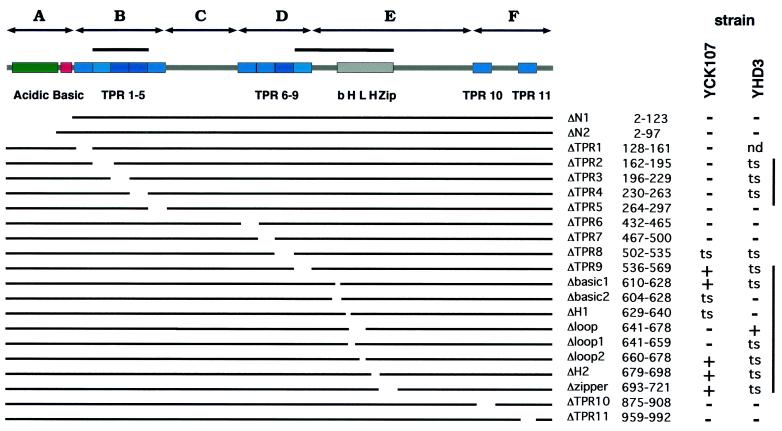
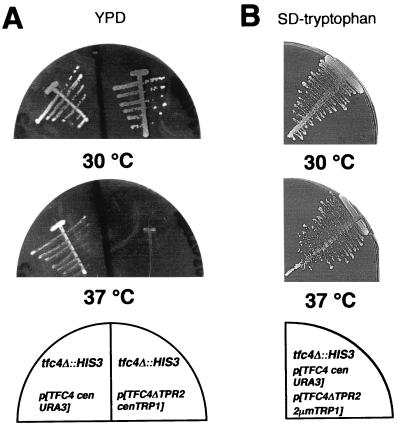
In a previous deletion analysis of τ131, most of the TPR motifs, when deleted individually, were found to be essential (12). However, in the strain YCK107 used for these experiments, the chromosomal copy of the wild-type TFC4 gene had not been fully deleted but simply inactivated by replacing the central part of the gene (residues 169 to 840, representing 65% of the protein) with a HIS3 cassette (39). This gene disruption perhaps resulted in the expression of a truncated form of τ131 containing the 168 first amino acids of the protein. Later, it was found that τ131 interacted, in the two-hybrid system, with Brf1 and that this interaction could be restricted to τ131 (residues 1 to 168) (12). Therefore, there was the possibility that the phenotype of the deletion mutants was altered by the presence of this putative truncated form of the wild-type protein, inasmuch as the N-terminal half of τ131 (residues 1 to 580) was found to inhibit, in vitro, the formation of Brf1-TFIIIC-tDNA complex (42). We thus performed the complete deletion of TFC4 (in strain SC55, yielding strain YHD3) and reexamined the phenotype of the τ131 deletion mutants (Fig. 2). Indeed, the phenotypes of deletions in the first TPR block (region B in Fig. 1A) changed from lethal to thermosensitive (deletions ΔTPR2, ΔTPR3, or ΔTPR4). On the other hand, the phenotypes of TPR deletions within region D (except for ΔTPR9) or region F remained unchanged. Deletions within regions B and E induced strain-dependent growth phenotypes. The thermosensitive and recessive phenotype of the τ131-ΔTPR2 mutation in strain YHD3 (previously determined as lethal in YCK107) is illustrated in Fig. 3A. The mutant cells grew slower than the wild type at permissive temperature (generation time, 2.5 instead of 2 h at 30°C) and stopped growing at 37°C. Overexpressing τ131-ΔTPR2 in a wild-type context showed that the phenotype of this mutation was not dominant negative (Fig. 3B). The loss of the wild-type copy of TFC4 was lethal at 30°C, showing that the wild-type copy was responsible for cell viability at permissive temperature (not shown). The conditional phenotype of the ΔTPR2 mutation prompted us to examine the properties of the mutant factor.
FIG. 2.
Deletion analysis of τ131. The motifs noted in the τ131 protein (39) have been deleted one by one or in combination as described by Chaussivert et al. (12). Centromeric plasmids harboring various mutant copies of TFC4, expressed from its own wild-type promoter, were tested for their ability to confer viability, at 30 or 37°C, after the shuffling of the URA3 plasmid-borne wild-type copy, in the context of a chromosomal copy of TFC4 partially (65%) disrupted (strain YCK107) (12, 39) or of a totally deleted TFC4 gene (strain YHD3, this work). Phenotypes are indicated as lethal (−), wild type (+), temperature sensitive (ts), or not determined (nd).
FIG. 3.
Phenotype of TFIIIC mutant factor with TFC4-TPR2 mutant gene borne on centromeric or multicopy plasmids. (A) The ΔTPR2 mutation was constructed as described by Chaussivert et al. (12). A centromeric plasmid harboring the ΔTPR2 deletion mutant copy of TFC4, expressed from its own wild-type promoter, was tested at 30 or 37°C in selective medium for its ability to confer viability in the absence of TFC4 chromosomal copy. The ΔTPR2 mutation, when borne on a centromeric plasmid, confers a thermosensitive phenotype to yeast cells. (B) The phenotype of cells transformed by a multicopy plasmid harboring the ΔTPR2 mutant copy of TFC4 was tested in a wild-type TFC4 context, and when overexpressed, TFC4-ΔTPR2 was found not to have a dominant negative phenotype, even at 37°C. The loss of the wild-type copy of TFC4 on 5-FOA plates was lethal at 30°C, showing that TFC4 was responsible for the cell viability at this temperature (results not shown).
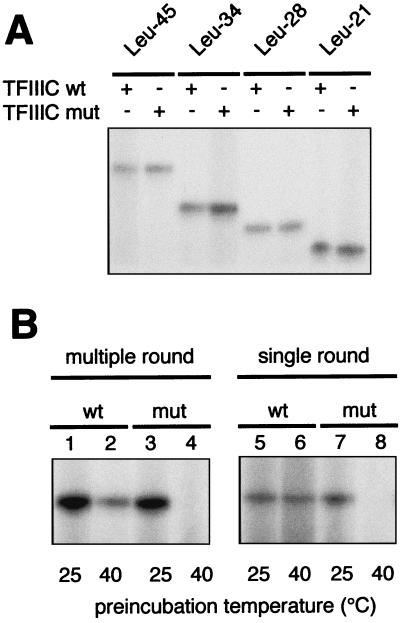
Mutant TFIIIC harboring the ΔTPR2 mutation (TFIIIC-ΔTPR2) was purified from YHD7 strain (tfc4-Δ;::HIS3 + pUN-τ131-ΔTPR2) and from a strain expressing a hemagglutinin-tagged version of τ138, the largest subunit of TFIIIC, to provide a control (33). Mutant and wild-type TFIIIC factors were partially purified, based on a tDNA binding assay, and compared for their ability to direct specific transcription of various tRNA genes in vitro. The same and limiting amount of both TFIIIC preparations, calibrated by Western blot analysis with anti-τ55 and anti-τ60 polyclonal antibodies, was used in transcription assays in the presence of reconstituted TFIIIB and purified Pol III. As shown in Fig. 4A, the transcriptional activity of the mutant TFIIIC was similar to that of the wild-type factor with several versions of the tRNA3Leu gene harboring different deletions in the region separating the A and B blocks (4). Remarkably, the deletion of TPR2 did not detectably affect the start site position, whatever the distance and the relative helical phasing of the A and B blocks. However, the mutant factor exhibited a marked thermosensitivity: in contrast to the wild-type factor, the mutant factor lost all its transcriptional activity in single-round or multiple-round transcription of the SUP4 tRNA gene, after 10 min of preincubation at 40°C (Fig. 4B). These results suggested that the heat treatment caused or enhanced a conformational change in the mutant factor, incompatible with proper initiation complex formation.
FIG. 4.
In vitro transcriptional activity of mutant TFIIIC-ΔTPR2. (A) Deletion of TPR2 does not affect the transcriptional start site. The plasmids used as templates (Leu-45, Leu-34, Leu-28, and Leu-21) harbor various versions of tRNA3Leu gene with different A block-to-B block distances of 45, 34, 28, and 21 bp, respectively (4). Transcriptions were performed in the presence of Mono Q-purified fractions of wild-type or mutant TFIIIC (100 and 500 ng, respectively), rTBP, rBrf1, partially purified B” fraction, and RNA polymerase III (see Materials and Methods). (B) Mutant TFIIIC is defective for the SUP4 tRNATyr in vitro synthesis. For each experiment, 100 ng of wild-type TFIIIC and 500 ng of mutant TFIIIC Mono Q fraction were preincubated for 10 min at 25 or 40°C (similar amounts of wild-type and mutant TFIIIC were used, based on Western blot experiments). Single-round and multiple transcriptions were performed at 25°C as described in Materials and Methods. The products were separated on a 7 M urea, 6% polyacrylamide gel and revealed by autoradiography.
DNA binding and TFIIIB recruitment by mutant TFIIIC-ΔTPR2 factor.
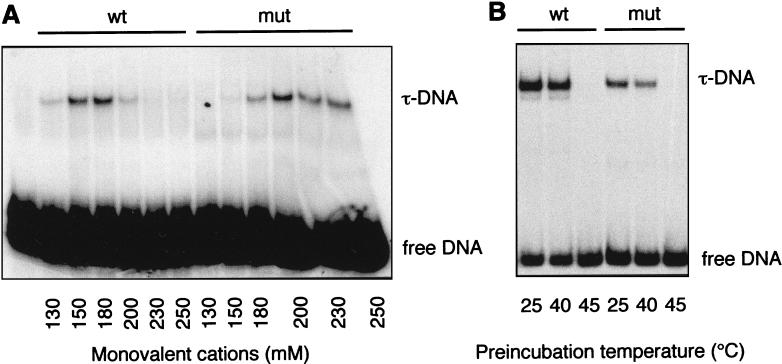
We first investigated whether the ΔTPR2 mutation affected the binding of mutant TFIIIC to tDNA using tRNA3Leu and tRNA3Glu genes as a probe. When the same amount of wild-type and mutant TFIIIC factors were used in gel-shift assays, the TFIIIC-ΔTPR2-DNA complex was formed as efficiently, but it displayed a sensitivity to salt concentration different from that of the wild-type factor (Fig. 5A). The optimum monovalent cation concentration for complex formation was 150 to 180 mM for the wild type and 200 to 230 mM for the mutant factor. This difference in salt concentration optima between wild-type and mutant factors remained unchanged at a twofold higher or lower concentration of competitor DNA (results not shown). In the following experiment, we selected the average monovalent cation concentration of 180 mM to examine the temperature sensitivity of factor-tDNA binding. Incubation at various temperatures, ranging from 25 to 45°C, was done prior to the formation of factor-tDNA complexes, under the same conditions as for transcription assays (Fig. 4). The mutant and wild-type factors showed the same thermosensitivity: DNA binding activities of the mutant and wild-type factors were only moderately affected after a 10-min preincubation at 40°C (Fig. 5B), in sharp contrast with the total loss of transcription activity (Fig. 4B). This observation suggested that the main transcriptional defect occurred after DNA binding, during or after TFIIIB assembly.
FIG. 5.
Compared DNA binding properties of mutant TFIIIC-ΔTPR2 and wild-type factors. (A) Differential salt sensitivity of mutant and wild-type TFIIIC-DNA complex formation. DNA binding reactions containing partially purified wild-type TFIIIC or TFIIIC-ΔTPR2 (in identical amounts) were carried out at 25°C for 10 min in the presence of various concentrations of KCl, 32P-labeled tRNA3Leu gene, and 200 ng of competitor DNA (pBluescript-SK). The concentrations of monovalent cations (K+ and NH4+) indicated take into account the ammonium sulfate brought by the protein fractions (25 mM final concentration). Analysis was done by gel retardation assay as described in Materials and Methods. (B) TFIIIC-DNA complex formation after preincubation of TFIIIC at different temperatures. Mutant or wild-type TFIIIC fractions were preincubated for 10 min at 25, 40, or 45°C as indicated in standard transcription buffer and further incubated in 180 mM monovalent cations with 32P-labeled tRNA3Glu gene and pBluescript-SK competitor DNA (200 ng) for 15 min at 25°C.
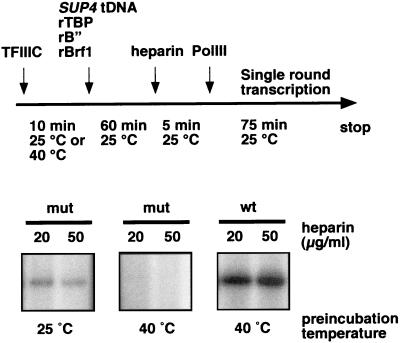
We next examined the ability of mutant TFIIIC to recruit TFIIIB and form heparin-resistant and transcriptionally competent preinitiation complexes onto the SUP4 tRNA gene. After formation of TFIIIB-TFIIIC-tDNA complexes using wild-type or mutant TFIIIC preincubated at 25 or 40°C, heparin was added to deplete them of TFIIIC (two concentrations of heparin were used to ensure a complete depletion of TFIIIC). We found that TFIIIB-TFIIIC-tDNA complexes stripped of mutant TFIIIC by heparin treatment retained transcriptional activity when the mutant factor was preincubated at 25°C, which was indicative of a stable and functional assembly of TFIIIB (Fig. 6). On the other hand, the complexes formed with the mutant TFIIIC preincubated at 40°C showed no detectable activity before or after stripping TFIIIC by heparin. These results suggested that ΔTPR2 mutation affected TFIIIB assembly rather than a later step of transcription like TFIIIC displacement from the transcriptional start site.
FIG. 6.
Heparin resistance of preinitiation complexes containing wild-type or mutant TFIIIC. Wild-type or mutant TFIIIC were preincubated for 10 min at 25 or 40°C as indicated. Biotin-coupled SUP4 tRNATyr gene was then reacted with streptavidin beads (Dynabeads), and preinitiation complexes were formed by addition of rBrf1, rTBP, and rB" onto immobilized DNA. After 60 min at 25°C, heparin (20 or 50 μg/ml) was added and incubation was pursued for 5 min. Beads were then washed to remove TFIIIC, and single-round transcription on SUP4 tDNATyr was performed as described in Materials and Methods. The products were run on a 7 M urea, 6% polyacrylamide gel and revealed by autoradiography. The signal obtained for the mutant factor (left) is weaker than that obtained with the wild-type factor (right), as less protein was used due to a salt concentration limitation.
The defect of mutant TFIIIC-ΔTPR2 is corrected by overdosage of the B” subunit of TFIIIB in vivo and in vitro.
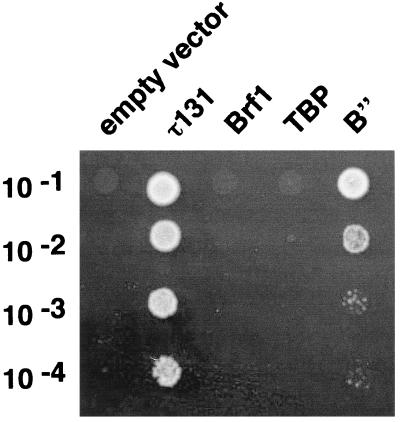
In order to determine the critical step of TFIIIB assembly that mutant TFIIIC-ΔTPR2 failed to correctly accomplish, we tested whether the overexpression of the components of TFIIIB, TBP, Brf1, or B” was able to suppress the τ131-ΔTPR2 mutation in vivo. TFIIIC-ΔTPR2 mutant cells were transformed with high-copy-number plasmids harboring the genes of TFIIIB components. A series of cell dilutions of YHD7 was plated on yeast extract-peptone-dextrose medium and incubated at permissive or nonpermissive temperature. Among the three genes coding for the components of TFIIIB, only TFC5 (encoding B”) was able to restore growth at 37°C (Fig. 7). This suppression had some allele specificity as other TPR deletions in τ131 that confer a thermosensitive phenotype could not be suppressed at nonpermissive temperature by overexpression of B” (ΔTPR3 and ΔTPR8) or were only partially suppressed (ΔTPR4) (data not shown).
FIG. 7.
In vivo suppression of the τ131-ΔTPR2 mutation and other ΔTPR mutations by overexpression of TFIIIB components. Multicopy suppression of τ131-ΔTPR2 by TFIIIB components. Stationary-phase cultures of mutant cells harboring the different multicopy plasmids indicated were diluted 10−1-, 10−2-, 10−3-, and 10−4-fold in water and spotted (5 μl) on solid yeast extract-peptone-dextrose medium and grown for three days. The different genes harbored on the high-copy-number vector pFL44L are as follows: pLR30, Brf1; pL1, TBP; pJR38, B”.
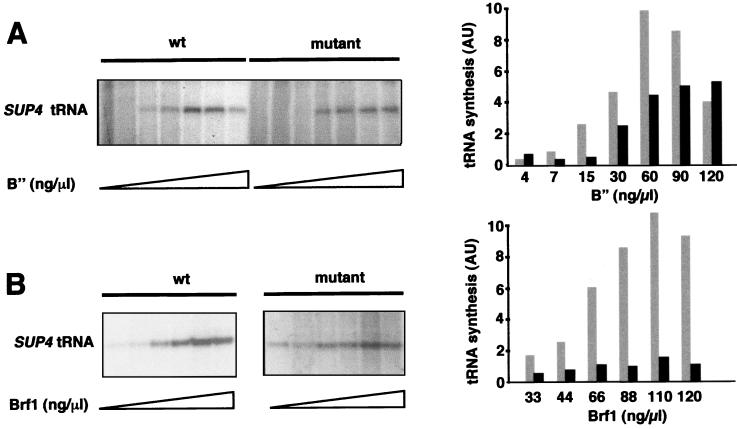
To confirm the involvement of τ131 in the recruitment of B” on a biochemical basis, we performed multiple-round transcription experiments with the SUP4 tRNA gene as a template at various concentrations of B”. Wild-type and mutant TFIIIC were preincubated at 40°C, and monovalent cation concentration was kept constant at 110 mM (Fig. 8A). The dosage-dependent effects of B” on transcriptional efficiency were clearly different for wild-type and mutant TFIIIC. Transcription with the wild-type factor displayed an optimum at B” concentrations around 60 ng/μl; an optimum concentration for B” has been previously observed (16). In contrast, with mutant TFIIIC, the transcription efficiency increased with B” concentration to reach the wild-type level at 120 ng/μl. On the other hand, increasing the concentration of rBrf1 did not correct the transcription defect of mutant TFIIIC (Fig. 8B). This result confirmed that the incorporation of B”, and not that of Brf1, into TFIIIB was the critical step of TFIIIB assembly directed by mutant TFIIIC on SUP4 tDNA. These results suggested that the assembly of B” by TFIIIC-ΔTPR2 factor was the main defective step of the preinitiation complex formation.
FIG. 8.
In vitro suppression of the τ131-ΔTPR2 mutation by overdosage of B” or Brf1. (A) In vitro suppression assay of the τ131-ΔTPR2 mutation by B”. Transcription mixtures contained identical amounts of wild-type and mutant TFIIIC (Mono Q fraction) preincubated for 10 min at 40°C; plasmid DNA harboring SUP4 tDNATyr, RNA Pol III fraction (50 ng), rTBP (250 ng), rBrf1 (1.2 μg, 30 ng/μl), and various amounts of B” (partially purified B” fraction), as indicated. The final concentration of monovalent cations (K+ and NH4+) was kept constant (at 110 mM) by taking into account the ammonium sulfate brought by the protein fractions. Transcripts were analyzed by electrophoresis and autoradiography (left panel). Transcripts were quantified using a PhosphorImager device and ImageQuant software (Molecular Dynamics). Arbitrary units are used to quantify transcription efficiency. Gray bars, wild-type TFIIIC; solid bars, mutant TFIIIC (right panel). (B) In vitro suppression assay of the τ131-ΔTPR2 mutation by Brf1. The same transcription mixtures as for panel A were used, with B” concentration set at 45 ng/μl and various amounts of rBrf1. The two autoradiograms for wild-type and mutant TFIIIC have been obtained from the same gel with two different exposures (the fraction of the gel showing mutant-factor-directed transcription is overexposed). Arbitrary units are used to quantify transcription efficiency. Gray bars, wild-type TFIIIC; solid bars, mutant TFIIIC (right panel).
Recombinant τ131 and B” proteins interact physically.
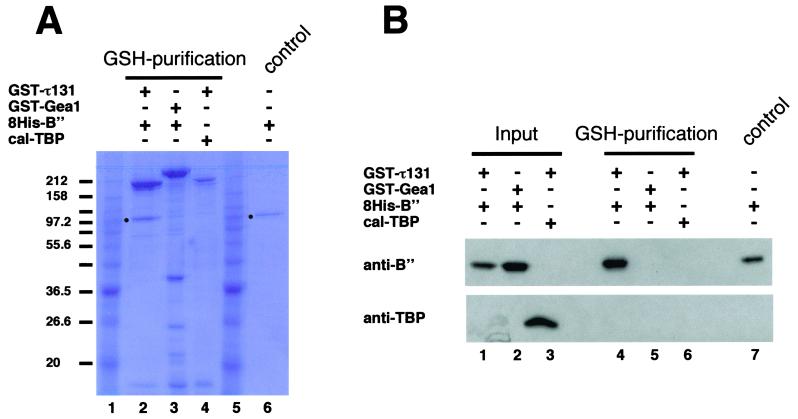
A direct interaction between τ131 (TFIIIC’s subunit) and B” (TFIIIB’s subunit) was suggested by previous two-hybrid experiments (51) and by the above multicopy suppression experiment. We therefore set out to determine whether τ131 could interact directly with B” in the absence of all the other components of TFIIIC and TFIIIB and DNA. To obtain full-length recombinant proteins, we used the baculovirus expression system. To facilitate the identification of the proteins and the purification of the protein complexes, tagged proteins were expressed (GST-τ131 and 8His-B”). As revealed by immunoblot analysis, cell crude extracts contained a majority of full-length recombinant proteins (data not shown). High Five insect cells were coinfected with three different pairs of recombinant baculoviruses, GST-τ131 and 8His-B”, GST-τ131 and calmodulin-TBP, or GST-Gea1 and 8His-B”. GST-τ131 or the GST-Gea1 control was purified from crude extracts on GSH-columns, and glutathione-eluted protein fractions were analyzed by SDS-PAGE followed by Coomassie blue staining and immunoblot analysis. As shown in Fig. 9A, a polypeptide of about 90 kDa specifically copurified with GST-τ131 from extracts of cells coexpressing GST-τ131 and 8His-B” but not from controls coexpressing GST-Gea1 and 8His-B” or GST-τ131 and TBP (Fig. 9A, compare lane 2 to lanes 3 and 4). On the other hand, in the same type of experiment, there was no detectable interaction between the tagged versions of τ131 and TBP. Immunoblotting experiments using anti-B” antibodies confirmed the specific copurification of B” with GST-τ131 (Fig. 9B, lane 4). No trace of B” was detected in the control experiment using GST-Gea1 instead of GST-τ131 (lane 5). Calmodulin-tagged TPB did not copurify with GST-τ131 (lane 6). These results showed that τ131 interacted directly and specifically with B”. A reverse experiment gave the same result: the GST-τ131 subunit was found to copurify with 8His-B” after immobilized metal ion affinity chromatography (IMAC) purification performed on extracts coexpressing GST-τ131 + 8His-B” (data not shown).
FIG. 9.
Physical interaction between τ131 and B”. Interaction between coexpressed τ131 and B” was revealed by copurification assay. High Five cells were coinfected with three different combinations of two recombinant baculoviruses, one expressing a GST-fused subunit (GST-τ131 or GST-Gea1) and the other expressing 8His-B” or calmodulin-TBP. Extracts were prepared as described in Materials and Methods. A glutathione purification was performed, and 50 μl of eluted fractions obtained were loaded onto SDS-PAGE gels. (A) Coomassie blue staining of an 8% polyacrylamide gel. Lanes 1 and 5 are molecular mass markers; some masses are indicated on the left (in kilodaltons). Combinations of recombinant baculoviruses are indicated at the tops of lanes 2, 3, and 4. Lane 6 is a control showing 8His-B” purified to homogeneity (heparine hyper-D and IMAC resin). The positions of B” are indicated with dots in lanes 2 and 6. (B) Input (10 mg of crude extracts) and bound proteins after glutathione purification (50 μl of eluted fractions) were analyzed by Western blotting. The membrane was probed with polyclonal antibodies directed against 8His-B” and TBP as indicated on the left. Lane 7 is the same control as that shown in lane 6 of panel A.
DISCUSSION
We present genetic and biochemical evidence that τ131, the TPR-containing and most upstream subunit of yeast TFIIIC, participates in the recruitment of B” on class III genes. The τ131 subunit of TFIIIC and the B” component of TFIIIB are two major actors of preinitiation complex assembly. τ131 extends from downstream of block A to within the TFIIIB binding site (5, 6), interacts with Brf1 (12, 30), and undergoes profound and successive conformational changes upon TFIIIB assembly onto DNA as evidenced by the altered pattern of τ131-DNA cross-linking during complex assembly and by the variable placement of TFIIIB on templates with altered 5′ sequences (23, 25, 27). While the interaction between Brf1 and τ131 is the limiting step for transcription complex assembly in vitro (27, 42, 43, 55), it is the recruitment of B” that confers the characteristic of fully assembled TFIIIB-DNA complex: much increased resistance to heparin and high-salt treatments, further DNA bending, accessibility of the transcription start site to DNase I, and rearrangement of τ131, Brf1, and TBP in the complex (26, 27, 31, 47). Of prime interest, therefore, is the mechanism of B” recruitment by TFIIIC. Deletion studies have shown that B” can be extensively truncated on either end and still retain TFIIIC-independent transcription activity on the TATA-containing SNR6 gene, but not TFIIIC-dependent activity on a tRNA gene (31). This striking observation suggested complex interactions of B” with TFIIIC components, likely the τ131 subunit (31).
We focused on a specific deletion mutant of τ131 removing precisely the TPR2 for several reasons. First, an attractive model of TFIIIB assembly suggested that τ131, with its many TPR motifs, provides the elasticity that allows variable placement of TFIIIB by the TFIIIC-DNA complex (23, 39). The deletion of the TPR motif vicinal to the N-terminal part of τ131 involved in Brf1 recruitment was expected to alter the geometric requisites of this interaction. However, we noted no qualitative defect in transcription accuracy of different templates (Fig. 4A and data not shown). Second, previous genetic studies had pointed to TPR2 as the central target of mutations enhancing transcription of tRNA gene bearing a defective A block by facilitating Brf1 recruitment (43, 49). Unexpectedly, however, the thermosensitive phenotype of the ΔTPR2 mutant was selectively suppressed by overexpression of B” and not by that of Brf1 or TBP. The TFIIIC-ΔTPR2 transcriptional defect was also suppressed in vitro by increasing the concentration of B”, not that of Brf1 (Fig. 9). These results suggested that TFIIIC-ΔTPR2 was primarily affected at the level of B” recruitment. Two-hybrid experiments between each of the six subunits of TFIIIC and B” gave a positive interaction signal with the τ131-B” pair only, and τ131-ΔTPR2 was even more efficient than wild-type τ131 (51). While quantitative two-hybrid responses are difficult to interpret in the absence of protein expression data, these observations suggested that τ131 is likely involved in B” binding and that the interaction does not require the TPR2 motif. The τ131-B” interaction domain remains to be identified. In vitro competition experiments did not disclose any interaction between B” and the N-terminal half of τ131 (τ131 residues 1 to 580) encompassing TPR 1 to 9, which, however, interacted detectably with Brf1 in solution (42). Two-hybrid experiments using τ131 (residues 1 to 579) were similarly negative with B” (51) but positive with Brf1 (12).
To investigate a possible direct interaction between τ131 and B”, we therefore turned to the baculovirus and insect cell system to coexpress the full-length proteins. Under these conditions, the formation of a τ131-B” complex was demonstrated by the selective copurification of the two proteins (Fig. 9). τ131 appears, therefore, to play a central role in TFIIIB assembly since it is the only TFIIIC subunit to detectably interact with both Brf1 and B”. Remarkably, no interaction between τ131 and TBP could be detected by the coexpression-copurification assay. As previously reported, the recruitment of TBP is directed by Brf1 (27, 30, 35) probably together with τ60 (16). It is notable that the mutant factor has modified DNA binding properties, which suggests an additional role for τ131 in TFIIIC-DNA binding. However, no evidence for DNA binding by recombinant τ131 was found (J. Acker, unpublished results); these results are much like those of Moir et al., who used recombinant τ131 (residues 1 to 580) (42).
How to rationalize the effect of the ΔTPR2 mutation on the function of τ131 within TFIIIC? TPR2 does not provide a critical interaction site for TFIIIB or TFIIIC components, since the mutant cells are viable at permissive temperature. However, TPR2 could be involved in the conformation changes that occur during the TFIIIB assembly. Hence, the mutations in or in close vicinity of TPR2 could facilitate the Brf1 recruitment by favoring the conformation of τ131 required for that specific step (42). In the process of Brf1 loading, τ131 appears to undergo structural changes important for allowing the entry of B” into the preinitiation complex (27, 29, 31). The deletion of TPR2, in changing the geometry of τ131, could interfere with this dynamic process, for instance by stabilizing a transient conformation state favorable for Brf1 recruitment but unfavorable for B" assembly. This interpretation is in favor of a two-step assembly pathway for Brf1 and B”. The fact that multicopy expression of B” suppresses the ΔTPR2 phenotype also suggests the existence of a specific incorporation step for B”. If there are two predominant steps for TFIIIB assembly in vivo (B′ and then B”), then τ131 does get involved in both steps.
Based on the structures of two TPR proteins (15, 53), adjacent TPR motifs are tightly packed to form a superhelical structure that forms a concave surface used, in some case, in ligand binding (53). The deletion of TPR2 could mimic a reordering of the interactions within the TPR1-to-TPR5 block, with TPR1 and TPR3 interacting directly and TPR2 being flipped out of the TPR superhelix. The structure of the TPR motif calls for the presence of small hydrophobic residues at positions 8, 20, and 27 (32, 53), and we have noted that, in TPR2, residue 20 is not A as is usually found in the other TPR of τ131 (C in the four yeasts or F in D. melanogaster [Fig. 1B]). Four intervening residues are also found in the S. pombe ortholog, and one deletion occurs in the D. melanogaster ortholog. These deviations in the sequence of TPR2 suggest a special role for this motif. We were initially surprised to find that the deletion of TPR2 did not affect the transcription accuracy (i.e., TFIIIB placement). In fact, the N terminus of τ131 would be shortened by only 8.5 Å due to the tilting of the TPR α-helices with respect to the axis of the TPR superhelix (15). Further insights into such a complex trancription factor will require a structural analysis of τ131, isolated or reassembled into the τA complex.
Acknowledgments
We thank Emmanuel Favry for preparation of recombinant TBP, Brf1, B" fraction, and RNA polymerase III and Cécile Ducrot for technical assistance. We thank Micheline Wesolowski-Louvel (Université Claude Bernard Lyon 1) for the gift of the K. lactis genomic library. We thank Christine Conesa and Pierre Thuriaux for helpful discussions. Sequence data for C. albicans were obtained from the Stanford DNA Sequencing and Technology Center website at http://www-sequence.stanford.edu/group/candida. Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. The sequence data from S. pombe were produced by the S. pombe Sequencing Group at the Sanger Centre.
H.D.-O. acknowledges fellowships from the French Ministère de l’Éducation Nationale, de la Recherche et de la Technologie, and from the Association pour la Recherche contre le Cancer.
REFERENCES
- 1.Acker, J., M. de Graaff, I. Cheynel, V. Khazak, C. Kedinger, and M. Vigneron. 1997. Interactions between the human RNA polymerase II subunits. J. Biol. Chem. 272:16815–16821. [DOI] [PubMed] [Google Scholar]
- 2.Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, G. G. Sutton, J. R. Wortman, M. D. Yandell, Q. Zhang, L. X. Chen, R. C. Brandon, Y. H. Rogers, R. G. Blazej, M. Champe, B. D. Pfeiffer, K. H. Wan, C. Doyle, E. G. Baxter, G. Helt, C. R. Nelson, G. L. Gabor, J. F. Abril, A. Agbayani, H. J. An, C. Andrews-Pfannkoch, D. Baldwin, R. M. Ballew, A. Basu, J. Baxendale, L. Bayraktaroglu, E. M. Beasley, K. Y. Beeson, P. V. Benos, B. P. Berman, D. Bhandari, S. Bolshakov, D. Borkova, M. R. Botchan, J. Bouck, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- 3.Arrebola, R., N. Manaud, S. Rozenfeld, M. C. Marsolier, O. Lefebvre, C. Carles, P. Thuriaux, C. Conesa, and A. Sentenac. 1998. Tau91, an essential subunit of yeast transcription factor IIIC cooperates with tau138 in DNA binding. Mol. Cell. Biol. 18:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, R. E., S. Camier, A. Sentenac, and B. D. Hall. 1987. Gene size differentially affects the binding of yeast transcription factor τ to two intragenic regions. Proc. Natl. Acad. Sci. USA 84:8768–8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartholomew, B., G. A. Kassavetis, B. B. Braun, and E. P. Geiduschek. 1990. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 9:2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartholomew, B., G. A. Kassavetis, and E. P. Geiduschek. 1991. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol. Cell. Biol. 11:5181–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, B. R., B. Bartholomew, G. A. Kassavetis, and E. P. Geiduschek. 1992. Topography of transcription factor complexes on the Saccharomyces cerevisiae 5 S RNA gene. J. Mol. Biol. 228:1063–1077. [DOI] [PubMed] [Google Scholar]
- 9.Buratowski, S., and H. Zhou. 1992. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell 71:221–230. [DOI] [PubMed] [Google Scholar]
- 10.Burnol, A.-F., F. Margottin, J. Huet, G. Almouzni, M.-N. Prioleau, M. Mechali, and A. Sentenac. 1993. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature 362:475–477. [DOI] [PubMed] [Google Scholar]
- 11.C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282:2012–2018. [DOI] [PubMed] [Google Scholar]
- 12.Chaussivert, N., C. Conesa, S. Shaaban, and A. Sentenac. 1995. Complex interactions between yeast TFIIIB and TFIIIC. J. Biol. Chem. 270:15353–15358. [DOI] [PubMed] [Google Scholar]
- 13.Chiannilkulchai, N., R. Stalder, M. Riva, C. Carles, M. Werner, and A. Sentenac. 1992. RPC82 encodes the highly conserved, third-largest subunit of RNA polymerase C (III) from Saccharomyces cerevisiae. Mol. Cell. Biol. 12:4433–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colbert, T., and S. Hahn. 1992. A yeast TFIIB-related factor involved in RNA polymerase III transcription. Genes Dev. 6:1940–1949. [DOI] [PubMed] [Google Scholar]
- 15.Das, A. K., P. W. Cohen, and D. Barford. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 17:1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deprez, E., R. Arrebola, C. Conesa, and A. Sentenac. 1999. A subunit of yeast TFIIIC participates in the recruitment of TATA-binding protein. Mol. Cell. Biol. 19:8042–8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dieci, G., R. Percudani, S. Giuliodori, L. Bottarelli, and S. Ottonello. 2000. TFIIIC-independent in vitro transcription of yeast tRNA genes. J. Mol. Biol. 299:601–613. [DOI] [PubMed] [Google Scholar]
- 18.Eschenlauer, J. B., M. W. Kaiser, V. L. Gerlach, and D. A. Brow. 1993. Architecture of a yeast U6 RNA gene promoter. Mol. Cell. Biol. 13:3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gabrielsen, O. S., N. Marzouki, A. Ruet, A. Sentenac, and P. Fromageot. 1989. Two polypeptide chains in yeast transcription factor τ interact with DNA. J. Biol. Chem. 264:7505–7511. [PubMed] [Google Scholar]
- 20.Gerlach, V. L., S. K. Whitehall, E. P. Geiduschek, and D. A. Brow. 1995. TFIIIB placement on a yeast U6 RNA gene in vivo is directed primarily by TFIIIC rather than by sequence-specific DNA contacts. Mol. Cell. Biol. 15:1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh, Y. J., Z. Wang, R. Kovelman, and R. G. Roeder. 1999. Cloning and characterization of two evolutionarily conserved subunits (TFIIIC102 and TFIIIC63) of human TFIIIC and their involvement in functional interactions with TFIIIB and RNA polymerase III. Mol. Cell. Biol. 19:4944–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huet, J., N. Manaud, G. Dieci, G. Peyroche, C. Conesa, O. Lefebvre, A. Ruet, M. Riva, and A. Sentenac. 1996. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 273:249–267. [DOI] [PubMed] [Google Scholar]
- 23.Joazeiro, C. A., G. A. Kassavetis, and E. P. Geiduschek. 1996. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 10:725–739. [DOI] [PubMed] [Google Scholar]
- 24.Joazeiro, C. A. P., G. A. Kassavetis, and E. P. Geiduschek. 1994. Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol. Cell. Biol. 14:2798–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kassavetis, G. A., B. Bartholomew, J. A. Blanco, T. E. Johnson, and E. P. Geiduschek. 1991. Two essential components of the Saccharomyces cerevisiae transcription factor TFIIIB: transcription and DNA-binding properties. Proc. Natl. Acad. Sci. USA 88:7308–7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kassavetis, G. A., B. R. Braun, L. H. Nguyen, and E. P. Geiduschek. 1990. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 60:235–245. [DOI] [PubMed] [Google Scholar]
- 27.Kassavetis, G. A., C. A. P. Joazeiro, M. Pisano, E. P. Geiduschek, T. Colbert, S. Hahn, and J. A. Blanco. 1992. The role of the TATA-binding protein in the assembly and function of the multisubunit yeast RNA polymerase III transcription factor, TFIIIB. Cell 71:1055–1064. [DOI] [PubMed] [Google Scholar]
- 28.Kassavetis, G. A., S. T. Nguyen, R. Kobayashi, A. Kumar, C. A. P. Joazeiro, E. P. Geiduschek, and M. Pisano. 1995. Cloning, expression, and function of TFC5, the gene encoding the B” component of Saccharomyces cerevisiae RNA polymerase III transcription factor TFIIIB. Proc. Natl. Acad. Sci. USA 92:9786–9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kassavetis, G. A., D. L. Riggs, R. Negri, L. H. Nguyen, and E. P. Geiduschek. 1989. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol. Cell. Biol. 9:2551–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khoo, B., B. Brophy, and S. P. Jackson. 1994. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 8:2879–2890. [DOI] [PubMed] [Google Scholar]
- 31.Kumar, A., G. A. Kassavetis, E. P. Geiduschek, M. Hambalko, and C. J. Brent. 1997. Functional dissection of B” component of RNA Polymerase III transcription factor TFIIIB: a scaffolding protein with multiple roles in assembly and initiation of transcription. Mol. Cell. Biol. 17:1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamb, J. R., S. Tugendreich, and P. Hieter. 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem. Sci. 20:257–259. [DOI] [PubMed] [Google Scholar]
- 33.Lefebvre, O., C. Carles, C. Conesa, R. N. Swanson, F. Bouet, M. Riva, and A. Sentenac. 1992. TFC3: gene encoding the B-block binding subunit of the yeast transcription factor TFIIIC. Proc. Natl. Acad. Sci. USA 89:10512–10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefebvre, O., J. Ruth, and A. Sentenac. 1994. A mutation in the largest subunit of yeast TFIIIC affects tRNA and 5S RNA synthesis. Identification of two classes of suppressors. J. Biol. Chem. 269:23374–23381. [PubMed] [Google Scholar]
- 35.Librizzi, M. D., M. Brenowitz, and I. M. Willis. 1998. The TATA element and its context affect the cooperative interaction of TATA-binding protein with the TFIIB-related factor, TFIIIB70. J. Biol. Chem. 273:4563–4568. [DOI] [PubMed] [Google Scholar]
- 36.Librizzi, M. D., R. D. Moir, M. Brenowitz, and I. M. Willis. 1996. Expression and purification of the RNA polymerase III transcription specificity factor IIIB70 from Saccharomyces cerevisiae and its cooperative binding with TATA-binding protein. J. Biol. Chem. 271:32695–32701. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-De-Leon, A., M. Librizzi, K. Puglia, and I. Willis. 1992. PCF4 encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell 71:211–220. [DOI] [PubMed] [Google Scholar]
- 38.Manaud, N., R. Arrebola, B. Buffin-Meyer, O. Lefebvre, H. Voss, M. Riva, C. Conesa, and A. Sentenac. 1998. A chimeric subunit of yeast transcription factor IIIC forms a subcomplex with tau95. Mol. Cell. Biol. 18:3191–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marck, C., O. Lefebvre, C. Carles, M. Riva, N. Chaussivert, A. Ruet, and A. Sentenac. 1993. The TFIIIB-assembling subunit of yeast transcription factor TFIIIC has both tetratricopeptide repeat and basic-helix-loop-helix motifs. Proc. Natl. Acad. Sci. USA 90:4027–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margottin, F., G. Dujardin, M. Gerard, J. M. Egly, J. Huet, and A. Sentenac. 1991. Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science 251:424–426. [DOI] [PubMed] [Google Scholar]
- 41.Moenne, A., S. Camier, G. Anderson, F. Margottin, J. Beggs, and A. Sentenac. 1990. The U6 gene of Saccharomyces cerevisiae is transcribed by RNA polymerase C (III) in vivo and in vitro. EMBO J. 9:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moir, R. D., K. V. Puglia, and I. M. Willis. 2000. Interactions between the tetratricopeptide repeat-containing transcription factor TFIIIC131 and its ligand, TFIIIB70. Evidence for a conformational change in the complex. J. Biol. Chem. 275:26591–26598. [DOI] [PubMed] [Google Scholar]
- 43.Moir, R. D., I. Sethy-Coraci, K. Puglia, M. D. Librizzi, and I. M. Willis. 1997. A tetratricopeptide repeat mutation in yeast transcription factor III131 (TFIIIC131) facilitates recruitment of TFIIB-related factor TFIIIB70. Mol. Cell. Biol. 17:7119–7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculoviruses expression vectors. A laboratory manual. W. H. Freeman and Company, Oxford, England.
- 45.Parsons, M. C., and P. A. Weil. 1992. Cloning of TFC1, the Saccharomyces cerevisiae gene encoding the 95-kDa subunit of transcription factor TFIIIC. J. Biol. Chem. 267:2894–2901. [PubMed] [Google Scholar]
- 46.Parsons, M. C., and P. A. Weil. 1990. Purification and characterization of Saccharomyces cerevisiae transcription factor TFIIIC. Polypeptide composition defined with polyclonal antibodies. J. Biol. Chem. 265:5095–5103. [PubMed] [Google Scholar]
- 47.Persinger, J., S. M. Sengupta, and B. Bartholomew. 1999. Spatial organization of the core region of yeast TFIIIB-DNA complexes. Mol. Cell. Biol. 19:5218–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peyroche, A., S. Paris, and C. L. Jackson. 1996. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384:479–481. [DOI] [PubMed] [Google Scholar]
- 49.Rameau, R., K. Puglia, A. Crowe, I. Sethy, and I. Willis. 1994. A mutation in the second largest subunit of TFIIIC increases a rate-limiting step in transcription by RNA polymerase III. Mol. Cell. Biol. 14:822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts, S., S. J. Miller, W. S. Lane, S. Lee, and S. Hahn. 1996. Cloning and functional characterization of the gene encoding the TFIIIB90 subunit of RNA polymerase III transcription factor TFIIIB. J. Biol. Chem. 271:14903–14909. [DOI] [PubMed] [Google Scholar]
- 51.Rüth, J., C. Conesa, G. Dieci, O. Lefebvre, A. Düsterhöft, S. Ottonello, and A. Sentenac. 1996. A suppressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 52.Scherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3–20. [DOI] [PubMed] [Google Scholar]
- 53.Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F. U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199–210. [DOI] [PubMed] [Google Scholar]
- 54.Schultz, P., N. Marzouki, C. Marck, A. Ruet, P. Oudet, and A. Sentenac. 1989. The two DNA-binding domains of yeast transcription factor tau as observed by scanning transmission electron microscopy. EMBO J. 8:3815–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sethy, I., R. D. Moir, M. Librizzi, and I. M. Willis. 1995. In vitro evidence for growth regulation of tRNA gene transcription in yeast. A role for transcription factor (TF) IIIB70 and TFIIIC. J. Biol. Chem. 270:28463–28470. [DOI] [PubMed] [Google Scholar]
- 56.Sethy, L., and I. M. Willis. 1995. Recessive mutations in the second largest subunit of TFIIIC suggest a new step in RNA polymerase III transcription. Gene Exp. 5:35–47. [PMC free article] [PubMed] [Google Scholar]
- 57.Sethy-Coraci, I., R. D. Moir, A. Lopez-de-Leon, and I. M. Willis. 1998. A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res. 26:2344–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wesolowski-Louvel, M., C. Tanguy-Rougeau, and H. Fukuhara. 1988. A nuclear gene required for the expression of the linear DNA-associated killer system in the yeast Kluyveromyces lactis. Yeast 4:71–81. [DOI] [PubMed] [Google Scholar]
- 59.White, R. J. 1998. RNA polymerase III transcription, 2nd ed. Springer-Verlag/Landes Bioscience, New York, N.Y.