Abstract
Assembly of antigen receptor genes by V(D)J recombination requires the site-specific recognition of two distinct DNA elements differing in the length of the spacer DNA that separates two conserved recognition motifs. Under appropriate conditions, V(D)J cleavage by the purified RAG1/RAG2 recombinase is similarly restricted. Double-strand breakage occurs only when these proteins are bound to a pair of complementary signals in a synaptic complex. We examine here the binding of the RAG proteins to signal sequences and find that the full complement of proteins required for synapsis of two signals and coupled cleavage can assemble on a single signal. This complex, composed of a dimer of RAG2 and at least a trimer of RAG1, remains inactive for double-strand break formation until a second complementary signal is provided. Thus, binding of the second signal activates the complex, possibly by inducing a conformational change. If synaptic complexes are formed similarly in vivo, one signal of a recombining pair may be the preferred site for RAG1/RAG2 assembly.
Mature immunoglobulin and T-cell receptor genes are assembled during lymphoid development through a series of site-specific DNA rearrangements collectively termed V(D)J recombination. Functional immunoglobulin and T-cell receptor molecules are constructed from three classes of gene segments: variable (V), diversity (D), and joining (J). The loci containing these gene segments may contain hundreds of copies of a given class of coding segment distributed over several megabases of DNA. Thus, an important requirement of V(D)J recombination is the targeting and uniting of two gene segments which may be separated by extensive portions of the chromosome (17).
Recombination signal sequences (RSS) flank V, D, and J coding segments and serve as the sites for recognition and cleavage by the recombinase. Each RSS consists of two conserved elements, the heptamer and the nonamer, and a spacer element of either 12 or 23 bases of fixed length but variable nucleotide composition. Recombination events are limited by the “12/23 rule” to those in which a pair of RSS participate, one 12-spacer signal and one 23-spacer signal. All coding segments of a given class (V, D, or J) have the same arrangement of spacer lengths, with the arrangement of signals limiting recombination events to those that could potentially encode a functional antigen receptor (17).
RAG1 and RAG2 carry out the initial stages of V(D)J recombination during which signal sequences are recognized and bound, and double-strand breaks are introduced at the border of the signal sequence and the coding segment (22). Studies with purified proteins have shown that double-strand break formation occurs in two steps (18). First, the RAG proteins introduce a single-strand nick at the 5′ end of the heptamer, adjacent to the coding DNA. A direct transesterification reaction follows, in which the free hydroxyl at the 3′ end of the coding sequence attacks the phosphodiester bond between the coding sequence and the RSS of the opposite strand, resulting in a blunt 5′ phosphorylated signal end and a covalently sealed hairpin coding end (18, 31).
Stable, site-specific binding and the two cleavage steps require both RAG1 and RAG2. In addition, a divalent metal ion is required for binding and cleavage. The identity of the metal ion profoundly influences the behavior of the recombinase (9, 32). Interdependent, or coupled, cleavage occurs with purified proteins when Mg2+ is the divalent metal ion (32). In this context, hairpin formation requires the presence of both a 12-signal and a 23-signal (9, 10, 14, 32). Restriction of hairpin formation by the 12/23 rule is further enhanced by DNA-bending proteins (HMG1 or HMG2) and nonspecific DNA (10, 14, 29). In contrast, hairpin formation can occur at an isolated 12- or 23RSS when the divalent metal ion is Mn2+ (18). Ca2+ can support RSS recognition but not nick or hairpin formation (11).
While the consequences of the 12/23 rule are evident at the level of hairpin formation, enforcement of this rule begins at the level of RSS recognition by RAG1 and RAG2. Individual RSS can be recognized by the RAG proteins, but they must be brought together in a synaptic complex to allow cleavage to occur. This synaptic complex forms when both a 12- and a 23-signal are present (10). The synaptic complex has been observed in mobility shift assays with purified proteins (RAG1, RAG2, and HMG1), Ca2+, and a pair of double-stranded oligonucleotides, one containing a 12RSS and one containing a 23RSS. When only one signal is present, RAG1 and RAG2 form a stable complex on the RSS, termed the single complex (SC). When a second oligonucleotide corresponding to the complementary signal is added, a slower-migrating band is observed, termed the paired complex (PC). This complex has been shown to be the functional complex for coupled cleavage (10). While it is clear that the RAG proteins and HMG are sufficient to form a 12/23 PC, little is known about the architecture or assembly of this complex.
Here we explore the precursor-product relationship between single-RSS complexes and the PC. We explore the protein content and intrinsic activities of each of these complexes. Two distinct complexes can form on a single site, one of which comprises the same complement of RAG1 and RAG2 proteins that is required to form the PC. However, this species remains unable to catalyze hairpin formation under coupled cleavage conditions. Only when a second complementary signal is bound does the complex become competent to form hairpins. These results suggest that binding of the second signal may induce a conformational change in the RAG proteins required for their catalytic activity.
MATERIALS AND METHODS
DNA manipulations.
Oligonucleotide substrates VDJ100/101 (12RSS) and VDJ132/133 (23RSS) were gel purified, annealed, and 5′-end labeled with 32P as described previously (7). Nonspecific competitor DNA was generated by annealing VDJ175 (5′-GCACTTCCAGACCACGCTACAAGCGATTAAACCACAGGGGCGACTTCGTGGA-3′) and VDJ176 (5′-TCCACGAAGTCGCCCCTGTGGTTTAATCGCTTGTAGCGTGGTCTGGAAGTGC-3′).
The 150-mer substrates were derived from plasmids 12RSS (+6) and 23RSS (+6) and prepared as described previously (15). The plasmid pNP107, expressing MBP-RAG1 core fusion protein (RAG1, amino acids 352 to 1040) in Escherichia coli from a pET11a (Novagen) cassette, was constructed in a pACYC184 backbone (New England Biolabs) that carries a chloramphenicol resistance marker. The resulting protein, used for copurification studies, does not contain a polyhistidine tag. (Construction details are available on request.)
Proteins.
Recombinant RAG2 from vaccinia virus infection of HeLa cells was expressed and purified as previously described (18). Maltose-binding protein (MBP)-RAG2, from baculoviral infection of Sf9 cells with AcD25 was produced as described previously (18), except that the cells were infected in the absence of RAG1. Recombinant RAG1 and MBP-RAG1 were expressed and purified from E. coli containing pDRK534 or pDRK576 as previously described (13) or from Sf9 cells infected with the viruses AcD23 (30) and AcD26 (18). The two methods of expressing and purifying RAG1 and RAG2 produce proteins with the same properties in binding and cleavage assays (13, 25; C. Mundy, unpublished results). The experiments shown in Fig. 2B and 3 contain RAG1 isolated from Sf9 cells; all others used protein isolated from E. coli. To copurify mixed multimers of RAG1 containing RAG1 and MBP-RAG1, overnight cultures of DH5α containing both pDRK534 and pNP107 were diluted 40-fold into 1 liter of Luria broth containing 100 mg of ampicillin and 80 mg of chloramphenicol/ml. Cultures were grown to an optical density at 600 nm of 0.7 to 0.8, induced with IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM and incubated at 28°C with vigorous shaking for 90′ with the addition of 50 mg of ampicillin and 40 mg of chloramphenicol/ml. Harvesting, lysis, and Ni2+ purification were carried out as described previously (13). Human HMG1 (amino acids 1 to 162) was purified from E. coli containing the plasmid pDVG83 with sequential 2 and 10% tricholoroacetic acid extractions (23). The resulting pellet was washed with acetone, dried, and resuspended in buffer (25 mM Tris [pH 7.9], 1 mM EDTA, 1 mM dithiothreitol [DTT], 150 mM KCl, 10% glycerol).
FIG. 2.
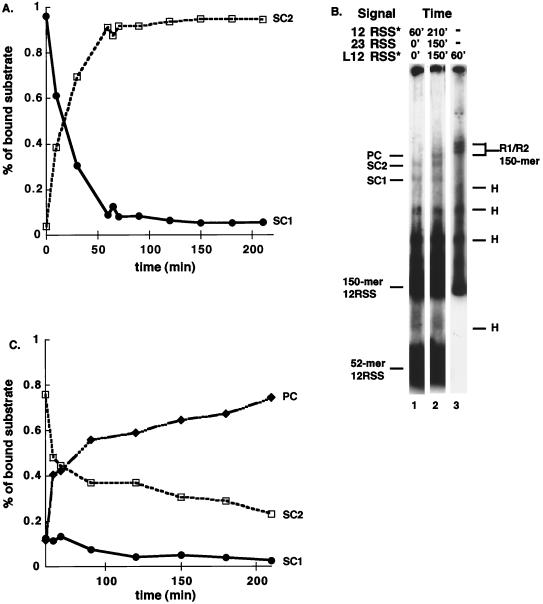
Time course of SC and PC assembly. (A) Time course for the assembly of SC1 and SC2 on a 12RSS. RAG1, RAG2, and HMG1 were incubated with a 32P labeled 12RSS for the indicated times, and the complexes were separated on a 4.5% polyacrylamide gel. The percentage of each complex formed at a given time relative to the total amount of bound substrate as determined by PhosphorImager analysis of the gel is shown. (B) RAG/RSS complexes do not continue to form after 1 h of incubation. All reactions contain RAG1, RAG2, and HMG1. The presence of radiolabeled (✽) or unlabeled (no asterisk) substrates is indicated. The length of time the substrate was present in the binding reaction is indicated. The total length of incubation was 60 min (lanes 1 and 3) or 210 min (lane 2). For complete details, see Results. L12RSS = 150-mer substrate. (C) Time course for the assembly of PC. RAG1, RAG2, and HMG1 were incubated with a 12-signal for 60 min. At 60 min, an unlabeled 23-signal was added to the reaction mixture, and binding was allowed to proceed for the times indicated prior to analysis. The gel was analyzed on a PhosphorImager as for panel A, and a graphic representation of the data is shown. Labeling is as described in Fig. 1 and 2A.
FIG. 3.
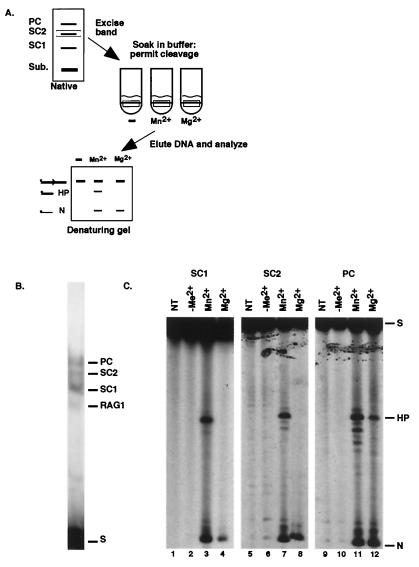
Functional analysis of individual RAG-RSS complexes. (A) Diagram of the assay. A representative analysis of the SC2 complex is shown. (B) Representative native gel lane from which individual complexes were excised. (C) Analyses of cleavage products formed by SC1 (left), SC2 (middle), and PC (right) are shown. Each slice was subject either to no additional treatment after gel excision (NT) or to incubation in reaction buffer lacking metal ion (−Me2+), or to incubation with Mn2+ or Mg2+ as indicated. N, nick; H, hairpin; S, substrate. Other labeling is as described in Fig. 1.
DNA binding assays.
Binding reactions to detect single RSS complexes (SC1 and SC2) were carried out in 2.5 mM CaCl2, 25 mM K-morpholinepropanesulfonic acid (K-MOPS; pH 7.0), 60 mM potassium glutamate (K-Glu), 2 mM DTT, and 100 mg of bovine serum albumin/ml. Each 10-μl reaction contained 50 fmol of 32P-labeled duplex oligonucleotide substrate, VDJ100/101 (12RSS) or VDJ132/133 (23RSS) (7), 100 ng of RAG1 protein (1 pmol), 50 ng of RAG2 protein (1 pmol), and 2 pmol of unlabeled, duplex nonspecific oligonucleotide competitor DNA. To visualize the PC, 200 fmol of unlabeled VDJ100/101 or unlabeled VDJ132/133 and 10 ng of HMG1 protein were added to each reaction. All binding reactions were incubated at 37°C for 30′, unless otherwise noted, and were stopped with the addition of 2 μl of 50% (vol/vol) glycerol-0.1% bromophenol blue loading dye. Reaction products were separated through 4% polyacrylamide-TBE gels (19:1 acrylamide-bisacrylamide, 22.5 mM Tris, 22.5 mM boric acid, 0.5 mM EDTA) containing 6% glycerol, unless otherwise noted. Gels were run at 450 V at 4°C. For time course assays, binding reactions were performed as described above. Reactions were stopped by placement on ice in the presence of loading dye, conditions under which no further binding can occur (Y. Akamatsu and M. Oettinger, unpublished observations). Gels were visualized by autoradiography or by using a PhosphorImager (Molecular Dynamics). Phosphorimager data was quantified with ImageQuant software. Four independent time course experiments were performed with similar results; a representative analysis is shown.
Cleavage assays on preformed complexes.
To analyze theability of individual complexes to cleave DNA, binding reactions were carried out as described above, but with 200 fmol of labeled substrate and 200 fmol of the unlabeled complementary substrate. Complexes were separated on a 4% TBE gel (as described above), and the wet gel was exposed to film for 1 h. Slices containing individual complexes were cut from the gel and immersed in buffer (25 mM K-MOPS, 60 mM K-Glu, 2 mM DTT) containing either no additional divalent metal ion or 2 mM MgCl2 or 2 mM MnCl2. The gel slices were incubated at 37°C for 15 min, and then the reaction buffer was removed and replaced with 0.3 M sodium acetate (pH 5.2), 2 mM EDTA, and 0.2% sodium dodecyl sulfate. Tubes were heated to 68°C for 5′ to inactivate the RAG proteins and prevent further cleavage. The substrate DNA was eluted overnight and ethanol precipitated. Reaction products were analyzed on a 12.5% denaturing polyacrylamide TBE gel containing 7 M urea and 30% formamide.
RESULTS
RAG1 and RAG2 form multiple complexes on RSS.
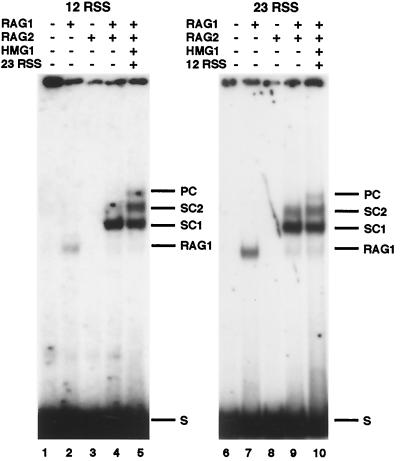
RAG-RSS interactions were examined in electrophoretic mobility shift assays in the presence of Ca2+. Binding reactions containing only RAG1 generated a single band (Fig. 1, lanes 2 and 7), a finding consistent with earlier work demonstrating that RAG1 can bind DNA in the absence of RAG2 (1, 4, 8, 21, 27, 28). Binding reactions containing RAG2 but not RAG1 did not show any DNA binding activity (Fig. 1, lanes 3 and 8). In the presence of RAG1/RAG2, a second complex band in addition to that seen before (1, 4, 10, 11, 21, 28) was seen under the high-resolution gel conditions used here. The faster-migrating complex is referred to here as SC1, and the slower-migrating species is referred to as SC2. In binding reactions where a second, unlabeled substrate with the complementary signal was added, the PC was evident as a third, distinct RAG1- and RAG2-dependent band of slower mobility than SC2. Consistent with prior studies (10), the PC was a distinct species, dependent on the simultaneous presence of both RAG proteins, HMG1, and a signal of complementary spacer length (see Fig. 2B and 6A). SC1, SC2, and PC all formed in the presence of Ca2+, Mn2+, or Mg2+, although in different amounts depending on the metal ion (data not shown).
FIG. 1.
Detection of multiple complexes of RAG1 and RAG2 bound to recombination signal sequences. Complexes formed on a labeled 12RSS (left panel) or labeled 23RSS (right panel) substrate are shown. The presence (+) or absence (−) of RAG1, RAG2, HMG1, and a complementary unlabeled RSS is indicated. The positions of the two SCs, SC1 and SC2, and the PC are as indicated. A complex containing RAG1 only (RAG1), which appears to a variable extent in these assays, is also indicated at right. S, substrate.
FIG. 6.
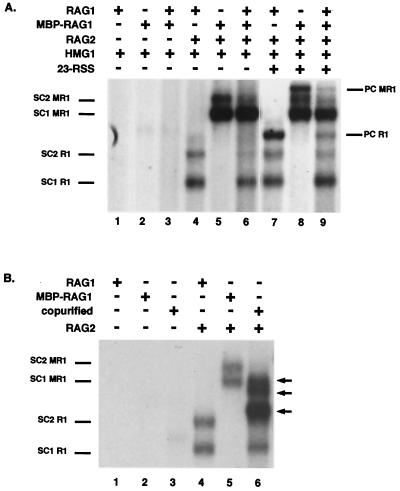
Purified RAG1 does not reassort during formation of SC1, SC2, or PC. (A) Gel shift analysis of complexes formed after incubation with core and MBP-tagged RAG1. All reactions contain a labeled 52-mer-12RSS. The presence (+) or absence (−) of RAG1, MBP-RAG1, RAG2, HMG1, and an unlabeled 23-signal is indicated. The positions of SC1 and SC2 derived from either RAG1 (R1) or MBP-RAG1 (MR1) in conjunction with RAG2 are indicated on the left. The positions of the PCs are indicated on the right. Lanes 1 to 6 and lanes 7 to 9 are derived from the same exposure of the same gel. (B) Core RAG1 and MBP-RAG1 are capable of forming heteromeric complexes when coexpressed. The presence (+) or absence (−) of RAG1, MBP-RAG1, coexpressed and copurified RAG1/MBP-RAG1 (copurified), and RAG2 is indicated. The positions of SC1 and SC2 derived from RAG1 or MBP-RAG1 in conjunction with RAG2 are indicated on the left. The arrows on the right indicate the positions of heteromeric forms isolated from copurification of RAG1 and MBP-RAG1.
SC2 is a precursor to PC formation.
The identification of two distinct single signal complexes raised questions regarding their relationship to the PC and about whether each had the same catalytic activity. Incubations were carried out with a single signal to look at formation of SC1 and SC2. RAG1, RAG2, and HMG1 were incubated at 37°C with a labeled 12RSS for 3.5 h to allow SC1 and SC2 to form. Both SCs appear early in the time course, but after 1 h the majority of product is SC2, even though some SC1 persists (Fig. 2A).
We next sought to determine whether new binding of RAG1/RAG2 to substrate could occur throughout the duration of these experiments. After RAG1 and RAG2 were incubated with a labeled 52-mer substrate containing a 12RSS for 1 h, a labeled 150-mer substrate containing a 12RSS was added to the reaction mixture. While binding to the 52-mer substrate was readily detected, no binding to the 150-mer substrate was observed (Fig. 2B, lanes 1 and 2), although both RAG1 and RAG2 are present in molar excess. Independent binding assays demonstrated that the longer substrate can be bound and that SC1, SC2 and PC, as well as a complex with RAG1 alone, can all form (Fig. 2B, lane 3, and data not shown); therefore, after 1 h, the unbound RAG proteins are no longer active and are unable to assemble on an RSS. (Because independent activity of RAG2 cannot be measured, it is possible that RAG2 remains active.) Thus, the SC1 and SC2 observed at later time points does not arise from new DNA binding but rather reflects the persistence of stable complexes.
The relationship between SC1/SC2 and PC was explored by using time course experiments, taking advantage of the inactivation of RAG proteins under reaction conditions as discussed above. RAG1, RAG2, HMG1, and a labeled 12-signal were incubated at 37°C for 1 h to allow for both assembly of SC1 and SC2 and inactivation of unbound protein. After 1 h, an unlabeled 23-signal was added to the reaction, and binding was allowed to proceed for an additional 2.5 h. As shown in Fig. 2C, 5 min after the addition of the 23-signal, PC began to accumulate, and it continued to accumulate for the next 145 min of the experiment. While PC accumulated, SC2 decreased in abundance, strongly suggesting that SC2 is a functional precursor of PC. The PC observed here cannot arise from novel formation of SC1 or SC2 on the 23-signal and the subsequent synapsis of a 23-SC with a 12-SC, since RAG proteins not already complexed with DNA are no longer able to bind DNA at the time the 23RSS was added. Instead, PC must arise from the conversion of SC2 to PC solely via the addition of the second, complementary signal. Thus, the protein content of SC2 and PC must be the same, with the complexes differing only in their DNA content. SC1 cannot be the direct precursor of PC in these experiments since the amount of SC1 is too low to account for the total accumulation of PC and because, as discussed below, the protein content of SC1 is distinct from SC2 and PC.
Both SC1 and SC2 are active for hairpin formation in Mn2+ but not Mg2+.
While the two SCs were clearly structurally distinct from the PC, the functional capabilities of each were not known. Therefore, as outlined in Fig. 3A, we separated the individual complexes and analyzed their ability to cleave DNA. RAG-RSS complexes were formed in Ca2+ to permit complex formation in the absence of DNA cleavage. The complexes were then separated by native gel electrophoresis, and the wet gel was exposed to film. Slices containing the individual RAG-RSS complexes were cut from the gel (a representative portion of which is shown in Fig. 3B), and each slice was subjected to one of four subsequent treatments: direct extraction of the substrate DNA from the gel slice with no further treatment or immersion in reaction buffer containing either Mn2+, Mg2+, or no divalent metal ion. After 30 min, the reaction was stopped, and the DNA was eluted from the gel slices and analyzed by denaturing gel electrophoresis.
Analysis of SC1 showed that nicked product was generated when either Mn2+ or Mg2+ was present in the cleavage buffer (Fig. 3C, lanes 3 and 4). In the presence of Mn2+, SC1 was also able to form hairpins efficiently (Fig. 3C, lane 3), but no hairpins were formed in the presence of Mg2+ (Fig. 3C, lane 4). SC2 behaved identically to SC1 in these assays (Fig. 3C, lanes 7 and 8). Therefore, a full complement of RAG proteins by itself cannot bypass the requirement for a second signal in the presence of Mg2+. Conversely, the binding of a second signal is not required for nick and hairpin formation in Mn2+, in keeping with the findings of other studies (9, 10, 18, 31, 35). Only in the context of the PC did hairpin formation proceed in the presence of Mg2+ (as well as Mn2+) (Fig. 3C, lanes 11 and 12). Control experiments in which the slices received no further treatment or in which no additional divalent metal ion was present confirmed that no cleavage had taken place in the binding reaction or in the process of separating the complexes (Fig. 3C, lanes 1, 2, 5, 6, 9, and 10).
Stoichiometry of RAG-RSS complexes.
While multimeric forms of RAG1/RAG2 bound to a single RSS have been reported, questions about the stoichiometry of functional RAG/RSS complexes remain. In particular, the stoichiometry of RAG1 and RAG2 in the PC has not been explored. Thus, nothing is directly known about the protein content of a 12/23 restricted complex, the complex that represents the active form under the more physiological conditions where Mg2+ is the cation.
An approach similar to that described by Hope and Struhl (12) was used to determine the number of RAG2 proteins in SC1, SC2, and PC. Binding assays using mixtures of two forms of the same protein with different electrophoretic mobilities can reveal whether a protein binds to a DNA molecule as a dimer or other multimer. If, for example, the protein binds as a dimer, mixed reactions should show three bands: one for each of the homodimers and a third of intermediate size, resulting from the formation of a heterodimer of the larger and smaller proteins on the DNA substrate. It should be noted that the number of molecules of protein in a DNA-binding complex as determined by this assay may reflect the actual number of protein subunits, or it may represent mixtures of stable multimers that do not reassort freely. Therefore, these experiments determine the number of reassorting equivalents, or “N” number, which may or may not represent an absolute number of molecules.
Proteins in which the core RAG1 or RAG2 sequences were fused with MBP were used to analyze the protein content of the three RAG1/RAG2 complexes. These functional fusion proteins (18, 30) generate distinct bands of slower mobility than the core proteins in gel shift assays. RAG1 was allowed to bind to a 12RSS in conjunction with either core RAG2, MBP-RAG2, or a mixture of both proteins. When the mixture of RAG2 proteins was used, a single novel band was observed with a mobility intermediate to that of the two forms of SC2 formed when RAG1 was incubated with either core or MBP-RAG2 (Fig. 4, lanes 4 to 6), indicating that two RAG2 moieties were present in SC2. No bands of intermediate mobility could be observed in the case of SC1, indicating that this complex contained a single RAG2 moiety.
FIG. 4.
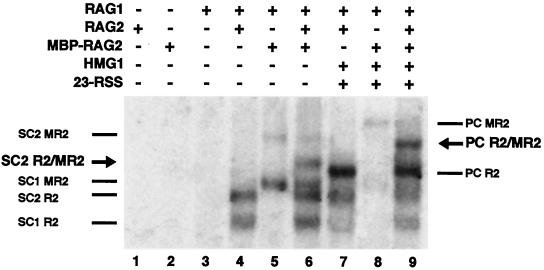
Stoichiometry of RAG2 in RAG-RSS complexes. The presence (+) or absence (−) of RAG1, RAG2, MBP-RAG2, HMG1, and an unlabeled 23-signal is indicated. Positions of SC1 and SC2 derived from either RAG2 (R2), MBP-RAG2 (MR2), or both (R2/MR2, see arrow) in conjunction with RAG1 are indicated on the left. Positions of each form of PC are indicated on the right, with the R2/MR2 marked by an arrow.
The same approach was used to demonstrate the RAG2 content of the PC. Binding reactions were carried out with an unlabeled 23RSS oligonucleotide present in addition to the labeled 12RSS oligonucleotide. These reactions also contained HMG1 to promote PC formation. A single band whose mobility was distinct and intermediate between PC formed by either core RAG2 or by MBP-RAG2 alone was generated when both proteins were present, indicating that PC contains two RAG2 moieties (Fig. 4, lanes 7 to 9). Reactions in which the 23-signal was the labeled partner produced the same result (data not shown).
Analysis of the RAG1 content of the complexes posed greater difficulties. Previous studies had demonstrated stable multimerization of RAG1 prior to DNA binding, since individually expressed forms of RAG1 fused to either one or two MBP moieties did not contribute to complexes of intermediate mobility when incubated with DNA. Only when the two were simultaneously expressed could mixed species of the two differently sized forms of RAG1 be observed (28). Therefore, two alternative approaches for assessing the RAG1 content of RAG-RSS complexes were pursued here.
The first approach determined the number of RAG1 molecules in each complex relative to the number of RAG2 molecules. We sought to determine whether SC1, SC2, and PC contained equal numbers of RAG1 and RAG2 moieties or whether one protein was present in molar excess. The logic of the experiment, diagrammed in Fig. 5A, is as follows. Binding reactions are set up with two combinations of proteins which differ in the placement of the MBP fusion partner: RAG1 plus MBP-RAG2 or MBP-RAG1 plus RAG2. Because the proteins expressed are identical except for the addition of the MBP moieties, any complex containing the same number of RAG1 and RAG2 molecules will have the same total molecular mass and should migrate similarly in mobility shift assays (Fig. 5A, left panel). Conversely, if one protein is present in molar excess in a given complex, it will bring along additional molecular mass from the presence of the MBP tags when it is present as the fusion protein but not when it is present as the unfused core (Fig. 5A, middle and right panels). If, for example, the numbers of RAG1 are greater than RAG2 in SC2, then the SC2 arising from MBP-RAG1/RAG2 should migrate with a slower mobility than SC2 formed from RAG1/MBP-RAG2 (see Fig. 5A, middle panel).
FIG. 5.
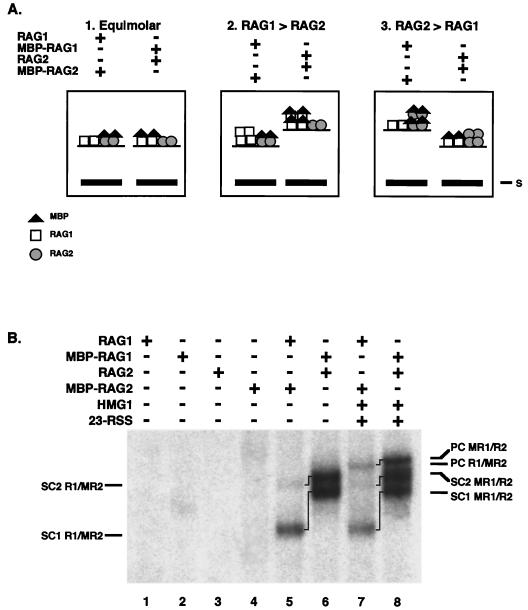
Relative content of RAG1 and RAG2 in SCs and PC. (A) Predicted relative mobilities of MBP-RAG1/RAG2 and RAG1/MBP-RAG2 complexes. The predicted relative mobilities of the RAG1/RAG2 complexes formed when the RAG proteins are present in equal numbers (left panel), when RAG1 is present in molar excess (middle), or when RAG2 is present in molar excess (right panel) are shown. (B) Actual relative mobilities of RAG1/RAG2 complexes. The presence (+) or absence (−) of RAG1, MBP-RAG1, RAG2, MBP-RAG-2, HMG1, and an unlabeled 23-signal is indicated. The positions of SC1 and SC2 and PC formed from the different combinations are indicated. Lines between bands mark the same complex formed from different combinations of protein.
Using this approach, it was clear that RAG1 is present in molar excess over RAG2 in all three complexes. Binding reactions, set up as outlined above, were analyzed in adjacent gel lanes to determine the relative sizes of the resulting RAG-RSS complexes. When a single substrate containing a 12RSS was present, both SC1 and SC2 formed from MBP-RAG1/RAG2 migrated more slowly than their counterparts formed from RAG1/MBP-RAG2 (Fig. 5B, lanes 5 and 6). Therefore, more MBP moieties must be present when they are attached to RAG1, rather than to RAG2, demonstrating that RAG1 is present in greater numbers in both complexes. Thus, SC1 must contain at least two molecules of RAG1, since it must contain more than the single molecule of RAG2, and SC2, which contains a dimer of RAG2, must contain more than a dimer of RAG1. While this experiment leaves open the formal possibility that the numbers of RAG1 differ in SC1 and SC2, additional studies (see below) strongly support the conclusion that SC1 and SC2 are comprised of equivalent numbers of RAG1.
The addition of an unlabeled 23-signal and HMG confirmed that the PC, like SC2, contains more RAG1 than RAG2 molecules. The PC generated by the MBP-RAG1/RAG2 mixture migrated more slowly than the PC generated by RAG1/MBP-RAG2 (Fig. 5B, lanes 7 and 8), again indicating that a greater number of MBP moieties and therefore a greater number of RAG1 molecules was present. Therefore, we can conclude that SC1 contains at least a dimer of RAG1 and that SC2 and PC contain a RAG1 multimer of three or more.
While these experiments indicated that SC1 contained at least a dimer of RAG1, it was unclear whether its RAG1 content was the same as SC2 and PC or whether it contained fewer molecules of RAG1 than either of the slower-migrating complexes. Because prior studies had suggested that RAG1 purified as stable dimers, the possibility that these dimers reassorted into the larger multimers observed in SC2 and PC remained open. Therefore, an experiment similar to those that had revealed the RAG2 content of the single and paired complexes was performed. Binding reactions were carried out in which core RAG1, MBP-RAG1, or both were incubated with RAG2 and a recombination signal sequence. When the two proteins were mixed, no complexes of intermediate mobility indicative of freely reassorting subunits were observed (Fig. 6A, lanes 4, 5, and 6), indicating that both SC1 and SC2 contain RAG1 that has been previously assembled into a stable multimer.
To determine whether reassortment of RAG1 could be observed as a result of synaptic complex formation, the PC was similarly examined. However, no PCs of intermediate mobility were observed from binding assays with mixtures of RAG1 and MBP-RAG1 (Fig. 6A, compare lanes 7 and 8 with lane 9). To avoid the potential problem of comigration of PCs formed from mixtures of RAG1 and MBP-RAG1 with SC1 or SC2 from homomultimers of MBP-RAG1, the experiments were repeated by using a 150-bp substrate containing a 23-signal as the binding partner. When this unlabeled substrate is partnered with a labeled 52-mer 12RSS, PCs formed with either RAG1 or MBP-RAG1 migrate more slowly than SC1 or SC2 arising from MBP-RAG1, precluding comigration. Again, no PCs of intermediate mobility were observed (data not shown).
In order to test the unlikely possibility that the mixed species were somehow comigrating with the homomeric forms of RAG1 or that MBP-RAG1 and RAG1 could not form complexes together, the two RAG1 derivatives were coexpressed, copurified, and subjected to gel shift analysis. As expected, a number of bands of intermediate mobilities were now observed, confirming that such intermediates could form and be detected (Fig. 6B, lane 6). (SC1 and SC2 from the homomer of MBP-RAG1 were not present, since the purification was achieved by using a nickel column specific for the histidine epitope tag present only on the core RAG1 form.) This method does not permit an accurate count of the total number of intermediate species, which could indicate the multimeric state of RAG1, since several bands appear as doublets. The presence of at least three novel bands (Fig. 6, arrows) again indicates that the RAG1 multimer is greater than a dimer.
Taken together, these results show that the functional PC observed here is not formed from the assembly of smaller RAG1 species into higher-order multimers. In other words, the functional multimeric form of RAG1 that is capable of binding to DNA to form SC1 and SC2 is the same form used to assemble PC. This result is consistent with the formation of PC arising simply from the addition of the partner signal DNA to SC2 rather than the binding of additional RAG1. The mixing experiments discussed above also strongly support the conclusion that SC1 contains the same number of RAG1 units as SC2 (and PC). If SC1 contained fewer RAG1 units than SC2 or PC, then mixing should have been possible and an SC2 or PC of intermediate mobility should have been observed. Since this was not the case, the RAG1 content of SC1 must be equal to that of SC2 and PC or else SC1 must be a dead-end product, unable to contribute to the formation of SC2 or PC. The early disappearance of SC1 in time course experiments, concomitant with the accumulation of SC2, is most consistent with SC2 forming from SC1 upon the addition of RAG2.
DISCUSSION
A common feature among site-specific recombinases is the need to ensure that only properly coupled events take place, thus preventing nonproductive events or potentially lethal breakage at a single site. In an effort to understand the molecular basis for the restriction of V(D)J cleavage to those events involving a 12/23 signal pair, we have compared the protein composition of PCs with complexes assembled on only a single recombination signal, determining the RAG content of functional complexes in each context. We have also isolated individual complexes bound to DNA under noncleavage conditions and subjected them to analysis under either coupled-cleavage conditions (Mg2+) or single-cleavage conditions (Mn2+) to determine the functional capabilities and limitations of each complex. Finally, we have examined the kinetic relationship between these complexes and identified at least one pathway for the assembly of a functional synaptic complex.
Molecular composition of functional V(D)J cleavage complexes.
The results presented here show that three distinct complexes of RAG1 and RAG2 bound to DNA can be formed (Fig. 1). As discussed above, SC2 and PC have the same protein content (Fig. 2, 4, and 6): a dimer (or 2N) of RAG2 (Fig. 4) and at least three RAG1 monomers (Fig. 5). An expectation of symmetry in the final complex would suggest that RAG1 is more likely a tetramer and gel filtration experiments of the isolated RAG1 protein suggest that this is likely to be the case (W. Yang, unpublished data). SC1 appears to have the same RAG1 content as SC2 and PC but contains only one RAG2 moiety.
While there has been no other analysis of the protein content of the PC or the relationship between single and paired complexes, other work has considered the molecular makeup of single site complexes, as well as the multimeric state of isolated RAG protein. One study found that a RAG1/RAG2 complex bound to an individual 12RSS contained a monomer of RAG2 (28). A second study concluded that RAG2 is present as a dimer in solution and, by inference, that a dimer of RAG2 is also present when bound with RAG1 to a single RSS (4). Our work provides a possible explanation for this apparent discrepancy: the SC considered by Swanson and colleagues may have been composed primarily of SC1, while that analyzed by Bailin et al. may have been composed primarily of SC2. Variations in the amount of RAG2 present during binding can easily alter the relative representation of these two forms (unpublished observation).
Our conclusion that RAG1 is present in the RAG1/RAG2-DNA complex as a multimer containing at least three RAG1 monomers differs from previous reports which generally concluded that RAG1 binds as a dimer. However, these conclusions are generally based on less-direct methodologies than those employed here, and some reflect analysis of RAG1 in solution or bound to DNA in the absence of RAG2. For example, in one study at least three oligomeric complexes of RAG1 alone on a single RSS were identified, but it was unclear which of these forms are capable of assembling with RAG2 on DNA (25). Another study concluded that RAG1 is present as a dimer when complexed with RAG2 on DNA, but the Ferguson plot analysis employed is not very precise when the molecular mass is large and the complex may deviate from the assumed spherical shape (4, 20). Only one other study directly considered the number of RAG1 units bound in conjunction with RAG2 by using two differently sized RAG1 derivatives. The results indicated that at least a dimer of RAG1 was present but, as noted by the authors, it was difficult to rule out the presence of additional oligomers (28). The greater separation achieved in our gels makes it easier to discern the presence or absence of intermediate species. In addition, our comparison of mobilities of complexes formed from proteins carrying the MBP fusion partner on either RAG1 or RAG2 (Fig. 5B) clearly shows that RAG1 and RAG2 are not equimolar in any of the bound complexes and that at least a trimer of RAG1 is present in SC2 and PC.
Assembling a functional synaptic complex.
RAG-mediated cleavage displays different properties depending on the metal ion present in the reaction, with hairpin formation in Mg2+ restricted to 12/23 PCs, while occurring promiscuously on single signals in Mn2+. The molecular basis for these distinct behaviors has not been fully determined, although various possibilities have been suggested. Prior to the present study, it seemed likely that a single-site complex could not perform cleavage in Mg2+ because it lacked part of the full synaptic complement of RAG proteins in addition to the second RSS. Surprisingly, our results show that the RAG1 and RAG2 content of SC2 is the same as for PC. A full complement of RAG1/RAG2 protein can assemble on a single RSS, and a functional PC can then be formed by the addition of a second signal, but no more protein, to this complex. Thus, in contrast to expectation, the PC does not contain twice the RAG1/RAG2 protein found on a single RSS. Instead, the assembled RAG proteins require only the addition of a second, complementary signal to become catalytically competent for the strand-transfer step of V(D)J cleavage.
The observation that the addition of the second complementary signal is required to render the complex active for hairpin formation in Mg2+ suggests that this binding induces a conformational change that converts the recombinase from an inactive to an active form. The ability of SC1 and SC2 to form hairpins in Mn2+ may also be a reflection of such a requirement for a conformational change in the RAG proteins. It has been suggested that retroviral integrases may adopt different conformations in the presence of different divalent metal ions such as Mn2+ or Mg2+ (3). The same may be true for the RAG proteins where the binding of Mn2+, instead of Mg2+, at the active site could potentially mimic the conformational change normally promoted by binding the complementary DNA, allowing single site hairpinning in Mn2+.
Coordination of cleavage.
The need to coordinate cleavage at two participating DNA elements is a requirement faced by many site-specific recombinases. Studies of other cleavage reactions have led to the recognition of several strategies that are used, with more than one approach often being employed, ensuring even tighter control. One method, used by the MuA transposase to ensure coordinated cleavage in transposition reactions reconstituted in vitro, is to impose a requirement that both DNA ends be present before the necessary proteins for catalysis can be assembled. That is, the functional MuA tetramer assembles stably only in the presence of both transposon ends (5, 19). Our experiments suggest that the RAG proteins differ from Mu in this regard, since all of the catalytically required elements of the RAG1/RAG2 heteromultimer can assemble on a single end and remain stably associated with DNA for several hours.
While the RAG system does not avoid deleterious breakage by preventing protein assembly on a single site, control is exerted by maintaining the proteins in a catalytically inactive state (with respect to hairpin formation) until a proper second signal is bound. Such a strategy may also be employed by Mu, since studies with MuA have also suggested that a conformational adjustment is employed as an additional level of control (24, 33)
Other levels of control are also possible. MuA subunits of the active tetramer carry out cleavage and strand transfer not on the DNA molecule to which they are bound, but rather in trans, reaching across to DNA molecules bound by other MuA subunits within the synaptic complex (2, 26, 34). The Flp recombinase also requires that an appropriate multiprotein complex assemble for recombination to occur (see references 6 and 16 and references therein). Monomers of the recombinase are inactive; in the active complex, subunits bound on one DNA end donate catalytic residues so as to cleave the other DNA end. Whether similar architectural constraints are also used by the RAG proteins in the synaptic complex is currently unknown.
Physiological implications.
Our results suggest that synaptic complex formation in vitro does not occur via two half-sites (a 12- or 23RSS bound by just one-half of the protein found in the synaptic complex) coming together. SC1 is not a half-site that can pair with a similar complex on the complementary signal. If this is also the case in vivo, it may have interesting implications for the assembly of active cleavage complexes and the regulation of V(D)J cleavage. If the RAG proteins first assemble on one site and then search for a second naked signal to form a PC, the system may have an intrinsic asymmetry which could be exploited to effect regulation of site accessibility. For example, different chromatin modifications might be required to open a site for the initial binding of the RAG proteins than to bring in the complementary site. It is possible that an antigen receptor locus has an obligate order for RAG recognition of signal sequences, one RSS type being the preferred site of RAG binding and the other RSS type attaching to this complex.
Acknowledgments
We thank Tania Baker, Katrina Morshead, and Anne Clatworthy for helpful discussions and Wei Yang (NIH) for sharing unpublished information.
This work was supported by National Institutes of Health grant RO1 GM48026 (M.A.O.) and the Leukemia and Lymphoma Scholars Program (M.A.O.). A. G. W. Matthews is a Howard Hughes Medical Institute Predoctoral Fellow.
REFERENCES
- 1.Akamatsu, Y., and M. A. Oettinger. 1998. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol. 18:4670–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldaz, H., E. Schuster, and T. A. Baker. 1996. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell 85:257–269. [DOI] [PubMed] [Google Scholar]
- 3.Asante-Appiah, E., and A. M. Skalka. 1997. A metal-induced conformational change and activation of HIV-1 integrase. J. Biol. Chem. 272:16196–16205. [DOI] [PubMed] [Google Scholar]
- 4.Bailin, T., X. Mo, and M. J. Sadofsky. 1999. A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol. Cell. Biol. 19:4664–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, T. A., and K. Mizuuchi. 1992. DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev. 6:2221–2232 [DOI] [PubMed] [Google Scholar]
- 6.Chen, Y., U. Narendra, E. L. Iype, M. M. Cox, and A. P. Rice. 2000. Crystal structure of a Flp recombinase-Holliday junction complex: assembly of an active oligomer by helix swapping. Mol. Cell 6:885–897 [PubMed] [Google Scholar]
- 7.Cuomo, C. A., C. L. Mundy, and M. A. Oettinger. 1996. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 16:5683–5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Difilippantonio, M. J., C. J. McMahan, Q. M. Eastman, E. Spanopoulou, and D. G. Schatz. 1996. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell 87:253–262 [DOI] [PubMed] [Google Scholar]
- 9.Eastman, Q. M., T. M. Leu, and D. G. Schatz. 1996. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature 380:85–88 [DOI] [PubMed] [Google Scholar]
- 10.Hiom, K., and M. Gellert. 1998. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell 1:1011–1019 [DOI] [PubMed] [Google Scholar]
- 11.Hiom, K., and M. Gellert. 1997. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage.Cell 88:65–72 [DOI] [PubMed] [Google Scholar]
- 12.Hope, I. A., and K. Struhl. 1987. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA.EMBO J. 6:2781–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, D. R., Y. Dai, C. L. Mundy, W. Yang, and M. A. Oettinger. 1999. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase.Genes Dev. 13:3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, D. R., and M. A. Oettinger. 1998. Functional analysis of coordinated cleavage in V(D)J recombination. Mol. Cell. Biol. 18:4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon, J., A. N. Imbalzano, A. Matthews, and M. A. Oettinger. 1998. Accessibility of nucleosomal DNA to V(D)J cleavage of nucleosomal DNA is modulated by RSS positioning and HMG1. Mol. Cell 2:829–839 [DOI] [PubMed] [Google Scholar]
- 16.Lee, J., M. Jayaram, and I. Grainge. 1999. Wild-type Flp recombinase cleaves DNA in trans.EMBO J. 18:784–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis, S. M. 1994. The mechanism of V(D)J joining: Lessons from molecular, immunological, and comparative analyses. Adv. Immunol. 56:27–150 [DOI] [PubMed] [Google Scholar]
- 18.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387–395 [DOI] [PubMed] [Google Scholar]
- 19.Mizuuchi, K. 1992. Transpositional recombination: mechanistic insights from sturdies of Mu and other elements. Annu. Rev.Biochem. 61:1011–1051 [DOI] [PubMed] [Google Scholar]
- 20.Mo, X., T. Bailin, S. Noggle, and M. J. Sadofsky. 2000. A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res. 28:1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagawa, F., K. Ishiguro, A. Tsuboi, T. Yoshida, A. Ishikawa, T. Takemori, A. J. Otsuka, and H. Sakano. 1998. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol. Cell. Biol. 18:655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oettinger, M. A. 1999. V(D)J recombination: on the cutting edge. Curr. Opin. Cell Biol. 11:325–329 [DOI] [PubMed] [Google Scholar]
- 23.Paull, T. T., and R. C. Johnson. 1995. DNA looping by Saccharomyces cerevisiae high mobility group proteins NHP6A/B. Consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem. 270:8744–8754 [DOI] [PubMed] [Google Scholar]
- 24.Rice, P., and K. Mizuuchi. 1995. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell 82:209–220 [DOI] [PubMed] [Google Scholar]
- 25.Rodgers, K. K., I. J. Villey, L. Ptaszek, E. Corbett, D. G. Schatz, and J. E. Coleman. 1999. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 27:2938–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Savilahti, H., and K. Mizuuchi. 1996. Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell 85:271–280 [DOI] [PubMed] [Google Scholar]
- 27.Spanopoulou, E., F. Zaitseva, F. H. Wang, S. Santagata, D. Baltimore, and G. Panayotou. 1996. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell 87:263–276 [DOI] [PubMed] [Google Scholar]
- 28.Swanson, P. C., and S. Desiderio. 1999. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 19:3674–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Gent, D. C., K. Hiom, T. T. Paull, and M. Gellert. 1997. Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J. 16:2665–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Gent, D. C., J. F. McBlane, D. A. Ramsden, M. J. Sadofsky, J. E. Hesse, and M. Gellert. 1995. Initiation of V(D)J recombination in a cell-free system. Cell 81:925–934 [DOI] [PubMed] [Google Scholar]
- 31.van Gent, D. C., K. Mizuuchi, and M. Gellert. 1996. Similarities between initiation of V(D)J recombination and retroviral integration. Science 271:1592–1594 [DOI] [PubMed] [Google Scholar]
- 32.van Gent, D. C., D. A. Ramsden, and M. Gellert. 1996. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell 85:107–113 [DOI] [PubMed] [Google Scholar]
- 33.Williams, T. L., E. L. Jackson, A. Carritte, and T. A. Baker. 1999. Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev. 13:2725–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, J. Y., M. Jayaram, and R. M. Harshey. 1996. Positional information within the Mu transposase tetramer:catalytic contributions of the individual monomers. Cell 85:447–455 [DOI] [PubMed] [Google Scholar]
- 35.Yu, K., and M. R. Lieber. 2000. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 20:7914–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]