Abstract
CAK1 encodes a protein kinase in Saccharomyces cerevisiae whose sole essential mitotic role is to activate the Cdc28p cyclin-dependent kinase by phosphorylation of threonine-169 in its activation loop. SMK1 encodes a sporulation-specific mitogen-activated protein (MAP) kinase homolog that is required to regulate the postmeiotic events of spore wall assembly. CAK1 was previously identified as a multicopy suppressor of a weakened smk1 mutant and shown to be required for spore wall assembly. Here we show that Smk1p, like other MAP kinases, is phosphorylated in its activation loop and that Smk1p is not activated in a cak1 missense mutant. Strains harboring a hyperactivated allele of CDC28 that is CAK1 independent and that lacks threonine-169 still require CAK1 to activate Smk1p. The data indicate that Cak1p functions upstream of Smk1p by activating a protein kinase other than Cdc28p. We also found that mutants lacking CAK1 are blocked early in meiotic development, as they show substantial delays in premeiotic DNA synthesis and defects in the expression of sporulation-specific genes, including IME1. The early meiotic role of Cak1p, like the postmeiotic role in the Smk1p pathway, is CDC28 independent. The data indicate that Cak1p activates multiple steps in meiotic development through multiple protein kinase targets.
Meiotic development is the program that generates haploid gametes in diploid organisms. Following induction, cells undergo a single round of DNA replication, which is followed by an elongated prophase, when synapsis and genetic recombination occur. Subsequently, two rounds of chromosome segregation take place without an intervening S-phase. The second meiotic division is coupled to differentiation programs that generate haploid gametes. In the yeast Saccharomyces cerevisiae, meiosis is coupled to spore formation (20). Our previous work has shown that the Smk1p mitogen-activated protein kinase (MAPK) homolog is a key regulator of spore formation in yeast cells (19, 41). We have also shown that the cyclin-dependent kinase-activating kinase (Cak1p) functions in the SMK1 pathway (42). This report focuses on the role of Cak1p and Smk1p in meiotic development.
MAPKs are a family of signaling enzymes that catalyze the transfer of phosphate from ATP to regulatory domains of target proteins (43). MAPKs are activated by the phosphorylation of a conserved threonine (T) and tyrosine (Y) in their activation loops by dual-specificity MAPK kinases (MEKs), which are in turn activated by upstream MEK kinases. MAPK cascades have been found in all eukaryotes examined to date and are required for cellular responses to a variety of adaptive, proliferative, and developmental signals. MAPK pathways are required at multiple steps during embryological development in multicellular organisms. MAPK pathways are also required to execute developmental programs in unicellular eukaryotes.
SMK1 encodes a MAPK homolog that is expressed specifically in sporulating cells (19). Diploid cells lacking Smk1p complete early meiotic events and the nuclear divisions but are defective in spore wall morphogenesis (19). smk1-Δ diploids exhibit a variety of abnormal spore wall structures. In contrast to the apparently random spore wall assembly phenotype seen in the null mutant, certain missense smk1 mutations appear to block spore morphogenesis at intermediate steps in the pathway (41). Distinct blocks in the pathway are seen in strains carrying different hypomorphic smk1 alleles. These intermediate blocks in the pathway are dependent on smk1 allelic dosage. These results suggest that the activation state of Smk1p is tightly regulated during sporulation and that quantitative changes in MAPK signaling are important to the mechanism by which this MAPK homolog controls the morphogenetic program.
Sporulation-specific genes can be broadly grouped based on their expression patterns into early, middle, and late temporal classes (1, 31). The expression of early genes peaks as premeiotic DNA synthesis and genetic recombination are occurring. Early gene promoters are regulated through DNA elements termed URS1s (upstream repression sequences), which specifically bind to Ume6p. Ume6p targets transcriptional repression complexes to early promoters in mitotic cells which are converted to activation complexes during early meiosis through a regulated pathway that requires association with the Ime1p transcriptional activator (20, 27, 39). Key regulated steps in this pathway include the transcription of IME1 itself, which is induced in response to nutrient signals (17, 32). Ime1p activity, binding of Ime1p to Ume6p, and activity of the Ume6p/Ime1p complex are also regulated by phosphorylation in response to nutritional and checkpoint signals that regulate meiotic progression (21, 25, 26, 40, 44).
Middle sporulation genes are expressed as the nuclear divisions and spore wall assembly are occurring. Middle gene promoters are regulated through a DNA element termed an MSE (middle sporulation element). MSEs activate expression during middle sporulation by binding the Ndt80p transcription factor, which is itself expressed as a middle gene (2, 11). Expression of active Ndt80p requires the completion of key steps in the pathway (genetic recombination) and is Ime1p dependent, thus coupling early events to the middle phases of the transcriptional cascade (2, 11, 37). A subclass of MSEs also repress gene expression during mitosis by binding the Sum1p transcriptional repressor (29, 45), which is degraded as cells enter middle sporulation (23). SMK1 is expressed as a tightly regulated middle gene through a single MSE (29) that binds Ndt80p during middle sporulation and Sum1p during mitosis (23, 45). Other Ndt80p-activated middle genes include key regulators of meiotic chromosome segregation, such as the B-type cyclins (CLBs) (2, 11). Thus, one mechanism that coordinates chromosome segregation with spore wall morphogenesis is the transcriptional cascade of sporulation which ensures that gene products required for these events are present at the same time. Late genes are expressed as spore formation is being completed and during spore maturation.
The expression pattern of SMK1 and the phenotypes of smk1 mutant strains suggest that the signal that activates this pathway is generated internally, since external signals do not regulate sporulation after meiotic chromosome segregation has occurred (20). Thus, in contrast to most characterized MAPKs, which couple to extracellular signals that induce downstream responses, Smk1p is required to complete a developmental program that has already been induced. One potential class of activating signal that could control Smk1p’s activity would be the completion of upstream events in the sporulation program, such as steps in the nuclear division pathway.
Both mitosis and meiosis require regulated changes in cyclin-dependent kinases (cdks) that are sequentially activated and inactivated by cyclin and inhibitor binding. In addition to binding of protein subunits, cdks are regulated by inhibitory and activating phosphorylations. The activating phosphorylation occurs on a conserved threonine in the activation loops of cdks. In Saccharomyces cerevisiae, the sole essential cdk, Cdc28p, is activated by a monomeric enzyme, named Cak1p, that phosphorylates threonine-169 (5, 16, 36).
We have previously shown that CAK1 functions positively in the spore morphogenesis pathway (42). First, multiple copies of CAK1 (2μm-CAK1 plasmids) were shown to suppress the spore morphogenesis phenotype of smk1 hypomorphs but not of an smk1 null mutant. Second, conditional alleles of CAK1 were isolated that caused specific defects in spore morphogenesis. Third, CAK1 mRNA levels increase as middle sporulation-specific genes such as SMK1 are expressed. These studies do not address whether Cak1p functions upstream or downstream of Smk1p. Furthermore, it is not known whether Cak1p activates Smk1p through Cdc28p or through a Cdc28p-independent pathway. Part of the uncertainty surrounding Cak1p’s role in spore formation stems from a lack of biochemical assays with which to study Smk1p.
In this report we show that Smk1p is modified by phosphorylation in its activation loop and describe an assay for this modification. We also show that Cak1p activates Smk1p through a Cdc28p-independent pathway. Cak1p is also required for at least one additional step in sporulation early in the program. This early step, like the Smk1p activation step, is Cdc28p independent. These results suggest that Cak1p phosphorylates multiple targets during sporulation to regulate multiple steps in the program. These regulatory interactions are relevant in considering how meiosis differs from mitosis and how meiotic events are coordinated with gametogenesis.
MATERIALS AND METHODS
Yeast strains.
The yeast strains used in this study are shown in Table 1. The coding region for three repeats of the hemagglutinin (HA) epitope was fused in-frame to the carboxy terminus of SMK1 in the chromosome of SK1 cells by the method of Longtine et al. (24) using kanamycin (G418) resistance as the selectable marker to generate DBY3. The CDC28-43244 allele is identical to CDC28-4324 described by Cross and Levine (3) except that it also contains a T18S substitution (substitutions are T18S, L61I, H78R, K83R, K96E, A125E, T169E, and A234V). CDC28-43244 was introduced into an SK1 haploid strain using the Kluyveromyces lactis URA3 adaptamer strategy described by Erdeniz et al. (4). The presence of all of the 43244 substitutions in CDC28 was confirmed by DNA sequence analysis of PCR fragments of the coding region generated from genomic DNA. This strain was crossed to MWY1079, a cak1::TRP1 SK1 derivative harboring the CAK1/URA3/CEN plasmid pMWY105 (42). Diploids lacking the CAK1 plasmid were identified after growth on 5-fluoroorotic acid medium, cells were sporulated, and cak1::TRP1 CDC28-43244 and CAK1 CDC2843244 MATa and MATα meiotic segregants were isolated and mated to generate homozygous diploids.
TABLE 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| LNY3 | MATa/MAT αura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 | Lenore Neigeborne |
| JLY3 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 CDC28-43244/CDC28-43244 | This study |
| JLY4 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 CDC28-43244/CDC28-43244 cak1-Δ::TRP1/cak1-Δ::TRP1 | This study |
| DBY3 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 SMK1-HA/smk1-Δ::LEU2 | This study |
| MDPY10 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG lys2/lys2 ho::LYS2/ho::LYS2 smk1-Δ::LEU2/smk1-Δ::LEU2 | This study |
| MDPY6 | MATa/MATα ura3/ura3 leu2::hisG/leu2::hisG trp1::hisG/trp1::hisG lys2/lys2 ho::LYS2/ho::LYS2 his4-N/his4-G sps1-Δ::TRP1/sps1-Δ::TRP1 | This study |
Plasmids and site-directed mutagenesis.
The collection of plasmids used in this study is outlined in Table 2. The tagged SMK1-HA construct was generated from DNA amplified by PCR from genomic DNA isolated from strain DBY3. This PCR fragment was cloned into plasmid YEp352 using the endogenous KpnI site found 219 bp upstream of the initiator ATG of SMK1 and the PstI site that was introduced by the HA genomic construction 577 bp downstream of the SMK1 termination codon. The nucleotide sequence of the entire insert of this plasmid (named pSL-1) was determined and found not to contain substitutions that alter the coding information.
TABLE 2.
Plasmids
| Plasmid | Markers | Reference |
|---|---|---|
| YEp351 | 2μm URA3 | 12 |
| YEp352 | 2μm URA3 | 12 |
| pLac33 | CEN URA3 | 10 |
| pSL-1 | YEp352 + SMK1-HA | This study |
| pSL-2 | YEp352 + SMK1-HA D186R | This study |
| pSL-3 | YEp352 + SMK1-HA T207E Y209E | This study |
| pSL-5 | YEp352 + SMK1-HA T207A Y207F | This study |
| pSL-6 | YEp352 + SMK1-HA ΔMSE | This study |
| pSL-8 | YEp352 + SMK1-HA Y209F | This study |
| pSL-9 | YEp352 + SMK1-HA T207A | This study |
| YCpSL-1 | pLac33 + SMK1-HA | This study |
| YCpSL-2 | pLac33 + SMK1-HA D186R | This study |
| YCpSL-3 | pLac33 + SMK1-HA T207E Y209E | This study |
| YCpSL-5 | pLac33 + SMK1-HA T207A Y209F | This study |
| YCpSL-8 | pLac33 + SMK1-HA Y209F | This study |
| YCpSL-9 | pLac33 + SMK1-HA T207A | This study |
| pMWB106 | YEp352 + CAK1 | 42 |
| pMWY105 | CEN URA3 + CAK1 | 42 |
| PS303 | YEp351 + IME1 | 13 |
Site-directed mutagenesis of pSL1 was done by cloning PCR fragments generated with primers containing the desired mutations or using the Quik-Change (Stratagene) system according to the manufacturer’s recommendations. The entire coding region of all mutated derivatives was determined to ensure the presence of the desired substitutions and the absence of artifactual mutations. The following codon substitutions in SMK1 were generated: T207A, ACC to GCC; Y209F, TAC to TTC; T207E, ACC to GAA; Y209E, TAC to GAA; and D184R, GAT to AGA. For IME2, the T242A mutation was ACG to GCG.
To construct the ΔMSE SMK1 promoter driving the expression of HA-tagged Smk1p, the KpnI/BglII restriction endonuclease fragment containing the SMK1 promoter with a 4-bp substitution in the MSE core consensus from pMDP174 (29) was exchanged with the wild-type promoter in pSL-1 to construct pSL-6. The IME1 high-copy plasmid pS303 contains the 2.65-kbp BglII/BamHI IME1 fragment subcloned into Yep351 (Table 2).
Growth and sporulation of cells.
Vegetative cultures were grown in YEPD (1% yeast extract, 2% peptone, 2% glucose), SD (0.67% yeast nitrogen base without amino acids, 2% glucose plus nutrients essential for auxotrophic strains), or YEPA (1% yeast extract, 2% peptone, 2% potassium acetate). For synchronous sporulation, cells were grown for at least six doublings in YEPA to a density of 107 cells/ml (optical density at 600 nm [OD600] = 0.5), washed once with SM (2% potassium acetate plus 10 μg of adenine, 5 μg of histidine, 30 μg of leucine, 7.5 μg of lysine, 10 μg of tryptophan, and 5 μg of uracil per ml), and resuspended in SM at a final density of 4 × 107 cells/ml (OD600 = 2.0). Sporulating cultures were maintained with vigorous agitation at 30°C.
Immunological analysis of Smk1p.
Preparation of spore lysates for Western immunoblot analysis was done by a modification of the method of Yaffe and Schatz (46). Briefly, 10 OD600 units of cells at the indicated time points were harvested by centrifugation at 4°C. The pellet was then washed in 1 ml of ice-cold H2O and resuspended in 1 ml of H2O plus complete protease inhibitor cocktail mix (Boehringer Mannheim; 1 mM phenylmethylsulfonyl fluoride [PMSF], 25 mM glycerol phosphate, and 1 mM Na3VO4), and 150 μl of freshly prepared 2 N NaOH containing 8% β-mercaptoethanol was added prior to quick-freezing and storage at −80°C. Subsequently, samples were thawed on ice, and 150 μl of ice-cold 50% trichloroacetic acid was added to each tube. The tubes were incubated for 15 min at 4°C on a rocking platform and then centrifuged at 10,000 × g for 10 min at 4°C. The supernatants were aspirated, and the pellets were resuspended in 1 ml of ice-cold acetone. Samples were centrifuged at 10,000 × g for 2 min at 4°C. The acetone was aspirated, and the pellets were dried briefly under vacuum prior to resuspension in sample buffer.
Samples were run through 13-cm 8% polyacrylamide (29:1, acrylamide-bisacrylamide) resolving gels at 25 mA of constant current. Following electrophoresis, the proteins were blotted to Immobilon-P (Millipore). Filters were blocked with 1% nonfat dry milk in phosphate-buffered saline (PBS; 9.1 mM dibasic sodium phosphate, 1.7 mM monobasic sodium phosphate, 150 mM NaCl, pH 7.4) plus 0.1% Tween 20, then reacted with a 1:1,000 dilution of monoclonal antibody HA.11 (BAbCO clone 16B12 raw ascites fluid) for between 4 and 16 h on a rocking platform at 4°C. After thorough washing in PBS and 0.1% Tween 20, blots were incubated on a rocker for 1 to 4 h at room temperature with a 1:5,000 dilution of alkaline phosphatase-conjugated goat anti-mouse immunoglobulin antibody (Tropix) and developed as per the manufacturer’s instructions. In all experiments the recovery of total protein was monitored by Coomassie staining of samples analyzed in parallel.
For the preparative immunopurification of Smk1-HAp, lysates were prepared as follows. The cell pellets from 100 OD600 units of sporulating cells were resuspended in 10 ml of ice cold PBS-RIPA buffer (PBS plus 1% Nonidet P40, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate [SDS]) containing complete protease inhibitor cocktail mix (Boehringer Mannheim) and 1 mM PMSF and divided into six 2-ml screw-cap tubes. Glass beads (0.5-mm diameter zirconia-silica beads; Biospec Products) were added to fill each tube, and the cells were lysed with four 1-min bursts at 4°C in the Mini Bead Beater-8 cell disrupter (Biospec Products). Unlysed cells and debris were removed by centrifugation at 10,000 × g for 10 min at 4°C. Fifty micrograms of affinity-purified HA probe Y-11 (sc-805 rabbit anti-HA antibody; Santa Cruz Biotechnology) was added to each cleared lysate, and following 1 h on ice, a 100-μl bead volume of protein G-Sepharose (Pharmacia) equilibrated in PBS-RIPA buffer was added. Samples were incubated on a rocking platform at 4°C for 1 h, and then beads were harvested and washed thoroughly with PBS-RIPA, buffer followed by PBS.
For phosphatase treatment of immunopurified Smk1-HAp, protein G-anti-HA immunoprecipitates from sporulating cells were equilibrated in 20 mM Tris (pH 7.4), and treated in phosphatase buffer (50 mM Tris [pH 7.8], 5 mM dithiothreitol [DTT], 2 mM MnCl2, 100 μg of bovine serum albumin per ml) with 400 U of λ-protein phosphatase (Calbiochem) in the presence or absence of 20 mM Na3VO4. Following incubation at 37°C for 45 min, an equal volume of 2× Laemmli sample buffer was added, and products were analyzed by Western immunoblotting.
In vitro kinase assays.
The protein substrates for the Cak1p kinase assays were affinity- or immunopurified on beads and washed three times in 1× assay buffer (20 mM Tris [pH 7.4], 5 mM MgCl2, 1 mM DTT). Cak1p was purified as a glutathione S-transferase (GST) fusion from baculovirus-infected insect cells as previously described and added to the reactions at 4 ng/μl (14). Cdk2 epitope tagged with HA at 10 ng/μl was used as a positive control Cak1p substrate as previously described (15, 16). Smk1-HAp expressed from its own promoter during sporulation (8 h postinduction) or expressed from the GAL1 promoter in vegetative cells was immunopurified as described above. GST-Ime2 was expressed and purified as described by Zhu et al. (47) and added to reactions at 0.1 to 1.0 ng/μl. Reactions were initiated by addition of ATP (10 μM unlabeled ATP and 10 μCi of [γ-32P]ATP [NEN; 3,000 Ci/mmol]). Following incubation at 30°C for 30 min, an equal volume of 2× Laemmli sample buffer was added, and the samples were boiled for 5 min. Labeled proteins from the kinase reactions were analyzed by polyacrylamide gel electrophoresis and visualized by autoradiography.
RESULTS
Smk1p is expressed as a middle sporulation-specific protein.
We fused three repeats of HA epitope coding information to the last codon of the chromosomal SMK1 gene in the SK1 genetic background, which sporulates rapidly and synchronously. Diploid yeast cells harboring a single copy of the epitope-tagged SMK1 allele (SMK1-HA/smk1-Δ) sporulated indistinguishably from a congenic strain harboring the wild-type gene (SMK1/smk1-Δ) when viewed by phase-contrast microscopy.
The ability of the SMK1-HA and SMK1 pair of strains to form spore walls was also compared using the fluorescence assay, which measures incorporation of dityrosine into the outer layer of the spore wall, and using assays that measure spore-specific resistance to glusulase, ether, and heat shock treatments. Acquisition of these sporulation phenotypes requires different levels of SMK1 activity (29, 41), with heat shock sensitivity requiring the highest level (followed by ether, glusulase, and fluorescence, which have decreasing Smk1p threshold requirements). We have previously shown that promoter mutations that reduce SMK1 transcription by 40 to 60% in an SMK1/smk1-Δ heterozygote cause defects in the acquisition of heat shock resistance (29). Thus, if the HA epitope significantly decreased the activity of Smk1p, it is likely that phenotypic consequences would be detected in these assays. That a single copy of the SMK1-HA complements all of these phenotypes indistinguishably from the wild-type gene suggests that this C-terminal epitope does not significantly diminish the activity of Smk1p. These data indicate that the SMK1-HA allele is a good system with which to study Smk1p.
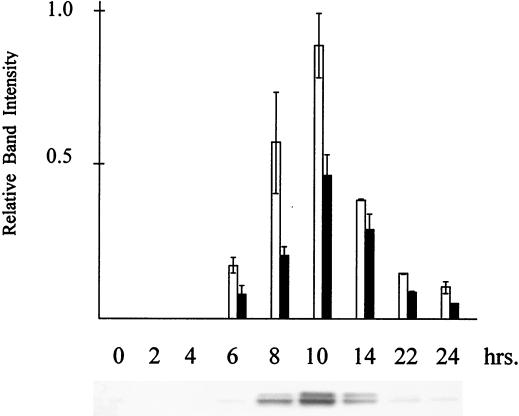
Smk1-HAp was followed in synchronously sporulating cells by immunoblot analysis (Fig. 1). Smk1-HAp is undetectable in vegetative cells, begins to accumulate approximately 6 h postinduction, reaches maximal levels at 10 h, and declines afterwards. The earliest appearance of Smk1-HAp in these experiments (6 h) corresponds to the time when middle sporulation genes begin to be expressed as cells exit pachytene and enter meiosis I. Smk1-HAp levels are highest when cells have completed meiosis II and when the major steps in spore morphogenesis are occurring (8 to 10 h). The kinetics of Smk1-HAp accumulation and its disappearance during sporulation are consistent with our previous analysis of SMK1 mRNA levels in the same SK1 genetic background (19, 29). These data show that Smk1p is expressed as a middle sporulation-specific protein when spore morphogenesis is occurring.
FIG. 1.
Smk1-HAp expression during sporulation. The SMK1-HA strain DBY3 (Table 2) was sporulated, aliquots were withdrawn at the indicated times, and total cellular protein was analyzed by immunoblot analysis with HA antibody. Quantitation of HA immunoreactivity in the lower band (open bars) and upper band (solid bars) is shown above. The closely spaced Smk1-HAp doublet migrates at around 45 kDa. In all of these experiments, meiosis I and II was first detected between 6 and 7 h and is mostly (>90%) complete by 9 h. The major steps in spore formation occur between 8 and 11 h. The data in the bar graph represent the averages of three independent experiments.
Assay for active Smk1p in sporulating cells.
A notable feature of Smk1-HAp immunoreactivity in sporulating cells is that it migrates as a closely spaced doublet, with the upper band accumulating more slowly than the lower band (Fig. 1). All MAPKs examined to date are phosphorylated on a conserved threonine (T) and tyrosine (Y) in their activating loops. In several cases these phosphorylations have been shown to retard the electrophoretic migration of the enzyme. SMK1 encodes a T and Y at positions 207 and 209, respectively, that align with the T’s and Y’s found in other MAPKs that are known to be phosphorylated by upstream MEKs. Furthermore, the sequence of amino acids surrounding these residues (presumed activation loop) in Smk1p is similar to the activation loops in other MAPKs. Treatment of Smk1-HAp that had been immunoprecipitated from extracts prepared from sporulated cells (9-h time point) with lambda phosphatase eliminated the upper form of Smk1-HAp in a vanadate-sensitive fashion, leaving an enzyme that migrated exclusively with the lower form (Fig. 2). These data indicate that the upper band of the Smk1-HAp doublet from sporulating cells corresponds to a phosphorylated form of the enzyme.
FIG. 2.
Smk1-HAp is phosphorylated. Smk1-HAp immunoprecipitates from DBY3 sporulated cell extracts (9-h time point) were treated with λ-phosphatase (λPPase) or λ-phosphatase and the inhibitor vanadate (VO4) as indicated.
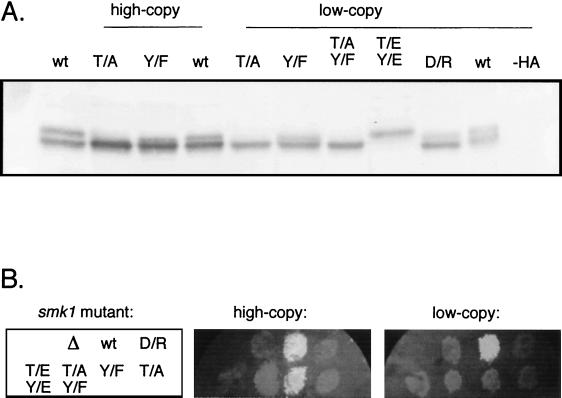
To address whether the upper Smk1p band may represent enzyme that is phosphorylated on the presumed activating T and Y, the Smk1-HAp T-207 and Y-209 were changed to the nonphosphorylatable alanine (A) and phenylalanine (F), respectively, both individually and in combination. When present as single-copy plasmids in an smk1-Δ/smk1-Δ strain, the T207A, Y209F, and double T207A/Y209F mutants all failed to form spores as assayed by phase-contrast microscopy and by the fluorescence plate assay for dityrosine (Fig. 3B). Thus, by functional assays, T207 and Y209 are required for Smk1p activity.
FIG. 3.
Site-directed mutagenesis of the Smk1p activating loop. (A) smk1-Δ/smk1-Δ strain MDPY10 harboring high-copy (2μm-based) or low-copy (CEN-based) SMK1-HA plasmids harboring the indicated codon substitutions were sporulated, and samples from the 9-h time point were analyzed by immunoblot analysis. −HA indicates a negative control strain containing an SMK1 plasmid lacking the HA epitope coding information. (B) Transformants harboring the mutant SMK1-HA derivative indicated by the key at left (smk1 mutant) were assayed for incorporation of dityrosine into spore walls by the fluorescence plate assay. This phenotype requires a relatively low level of SMK1 activity.
Immunoblot analysis revealed that the T207A and T207A/Y209F mutant proteins migrate as single bands with the presumed unmodified form of Smk1-HAp (Fig. 3A). These data indicate that T207 is a site of activating phosphorylation. In contrast, the Y209F single mutant migrated as a compact doublet. Thus, these experiments do not address whether Y209 is phosphorylated (see Discussion). In addition, the Smk1-HAp T207 and Y209 were both changed to glutamic or aspartic acidic residues (E or D). The double Smk1-HAp T207E Y209E and T207D Y209D acidic substitutions resulted in proteins that migrated with the upper member of the Smk1-HAp doublet. However, these double acidic mutants did not complement an smk1-Δ diploid for the fluorescence defect (Fig. 3B) or when assayed by phase-contrast microscopy for spore formation (data not shown).
D186 in Smk1p aligns with the key aspartate that has been shown to be required for catalytic activity in other protein kinases. A D186R mutant behaved indistinguishably from the smk1-Δ mutant in the fluorescence assay (Fig. 3B) and in heat shock, ether, and glusulase resistance assays (data not shown). The Smk1-HAp D186R mutant protein is still phosphorylated, as shown by its migration as a doublet. This result indicates that Smk1p activity is not required for the activating phosphorylation, suggesting that there is a protein kinase other than Smk1p that phosphorylates the activating residue(s).
Interestingly, the Y/F single mutant but not the T/A single mutant or the T/A Y/F double mutant partially complemented the smk1-Δ sporulation defect when expressed from a high-copy (2μm) plasmid. We have previously shown that accumulation of dityrosine into insoluble material as measured using the fluorescence assay requires low levels of Smk1p activity, while the formation of morphologically normal spore walls as assayed by phase-contrast microscopy requires intermediate levels of Smk1p activity (41). The establishment of resistance to glusulase, ether, and heat shock treatment requires high levels. The Smk1-Y209Fp mutant expressed from a 2μm vector is indistinguishable from the wild type in the fluorescence assay (Fig. 3B) but does not form morphologically wild-type spore walls, as assayed by phase-contrast microscopy, and displays low resistance to glusulase, ether, and heat shock treatments (data not shown). This spectrum of phenotypes is similar to that seen in a weakened (hypomorphic) smk1-4 strain (41). These data suggest that the Smk1-Y209Fp mutant is able to accept phosphate on the activating threonine but that it becomes only partially activated during sporulation.
Smk1p activation requires Cak1p.
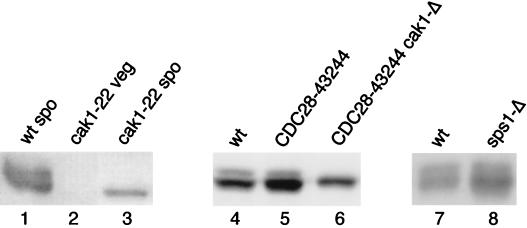
We previously identified the cdk-activating kinase CAK1 as a dosage (2μm) suppressor of a hypomorphic SMK1 mutant (42). Furthermore, cak1 mutants were identified that progress through the nuclear divisions but block with a smk1-Δ-like phenotype. These results indicate that Cak1p functions in the SMK1 pathway. To determine whether Smk1p activation requires Cak1p activity, we monitored the ratio of the phosphorylated and nonphosphorylated forms of Smk1-HAp in a cak1-22 diploid. Although, as shown below, CAK1 is required early in sporulation, the cak1-22 diploid progresses through these early events (albeit more slowly) to express middle genes (including SMK1). As shown in Fig. 4, the fraction of Smk1-HAp that is phosphorylated (upper band) is dramatically reduced in cak1-22 sporulated cells.
FIG. 4.
Smk1p activation requires Cak1p but does not require Sps1p. Lanes 1 to 3, Smk1p activation is defective in a cak1 missense mutant during sporulation. Extracts were assayed for the Smk1-HAp doublet by immunoblot analysis in DBY3 cells sporulated for 10 h (lane 1), homozygous cak1-22 diploid cells grown in rich medium (lane 2), and homozygous cak1-22 diploid cells sporulated for 16 h (lane 3). All growth and sporulation was at 30°C (semipermissive for cak1-22). The longer time point for the cak1-22 strain was chosen because it requires several hours longer to accumulate comparable levels of Smk1p. Lanes 4 to 6, Smk1-HAp ectopically expressed in vegetative cells is weakly activated and this activation requires CAK1. Smk1-HAp was ectopically expressed during vegetative growth using pSL-6 (Table 2). Extracts were assayed for the Smk1-HAp doublet by immunoblot analysis in a wild-type strain (lane 4), a strain whose sole source of CDC28 was from the CAK1-independent CDC28-43244 allele (lane 5), and in a CDC28-43244 strain lacking CAK1 (lane 6). Lanes 7 and 8, Smk1p activation does not require Sps1p. Extracts were assayed in wild-type (DBY3) cells sporulated for 9 h (lane 7) and in sps1-Δ/sps1-Δ (MDPY6) cells sporulated for 9 h (lane 8). The total amount of Smk1-HAp was found to be reproducibly higher in the sps1-Δ mutant (three independent clones tested).
In the experiments described below, strains were generated that completely lacked CAK1. We expressed Smk1-HAp using mutant promoters that lack the cis-acting MSE that normally represses expression of SMK1 in vegetative cells (see Materials and Methods). This approach allows a moderate level of Smk1-HAp expression in mitosis (15 to 25% of the maximally induced meiotic level). Smk1-HAp from these derepressed SMK1 promoters in mitotic cells was expressed and modified in both haploids and diploids. However, the ratio of modified to unmodified Smk1-HAp was substantially lower in mitosis than in meiosis. In cells lacking Cak1p (see below for the genetic approach used to construct these strains), the modified form of Smk1p was undetectable. These results show that Cak1p can promote Smk1p activation not only during meiosis but also during mitosis. Nonetheless, it should be pointed out that when Smk1p is expressed at high levels in mitotic cells using the strong galactose-inducible GAL1 promoter, the overwhelming majority of Smk1-HAp is in the unmodified form (data not shown).
These results suggest that certain SMK1 pathway signaling components are limiting for Smk1p activation in mitosis. However, Cak1p does not appear to be limiting for Smk1p activation in mitotic cells, since overexpression of Cak1p using 2μm vectors does not increase the proportion of Smk1-HAp that is in the upper member of the doublet (comparison of three independent 2μm-CAK1 and 2μm control isolates showed no significant difference; data not shown). The major conclusion of these experiments is that Cak1p functions upstream of Smk1p in the spore morphogenesis pathway.
SPS1 encodes a homolog of the Ste20p kinase, which is required in the mating pheromone MAPK pathway (9). SPS1 is transcribed as a middle sporulation-specific gene around the time when SMK1 is expressed, and sps1-Δ/sps1-Δ diploids are blocked in sporulation after meiosis II but before spore formation. Based on these observations, it has been suggested that Sps1p functions upstream in the Smk1p pathway (9, 19). We measured the ratio of the phosphorylated and nonphosphorylated forms of Smk1-HAp during sporulation in a strain lacking Sps1p. The activation state of Smk1-HAp was identical in a wild-type strain and an sps1-Δ/sps1-Δ mutant (Fig. 4). These data show that Sps1p is not required for Smk1p activation (see Discussion).
Cak1p functions in the spore morphogenesis pathway by a Cdc28p-independent mechanism.
Since Cak1p functions in the SMK1 pathway and also activates Cdc28p, it is possible that the role of Cak1p in activating Smk1p is Cdc28p dependent. For example, Cdc28p might phosphorylate Smk1p or an upstream component in the pathway. Alternatively, Cak1p might activate Smk1p in a Cdc28p-independent fashion. In this scenario, Cak1p might activate an upstream component of the pathway other than Cdc28p, or even Smk1p itself. Studying the role of Cak1p in sporulation is complicated because it is required to activate Cdc28p and is thus required for viability. Cross and Levine have described a series of CDC28 mutants that are able to support mitotic growth in the complete absence of CAK1 (3). These CAK1-independent alleles of CDC28 contain a mutation that changes the threonine that is normally phosphorylated by Cak1p (T-169) to an acidic residue as well as several additional hyperactivating substitutions that were identified in multiple rounds of sequential PCR-mediated mutagenesis and selection. CDC28-43244 is one of the CAK1-independent alleles identified in this study.
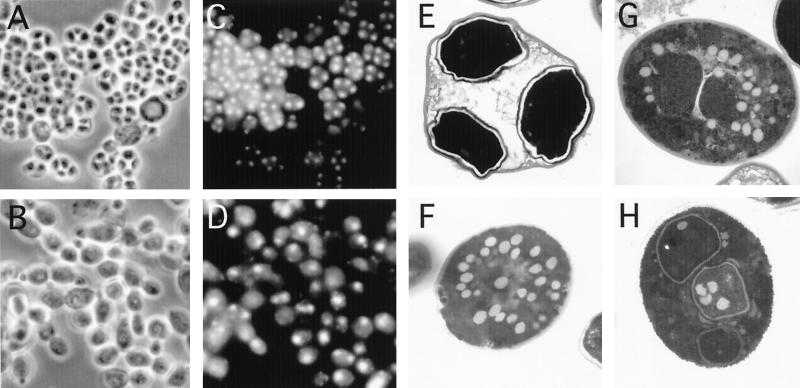
We generated a series of coisogenic CDC28-43244 SK1 diploid strains containing and lacking Cak1p. The mitotic growth rate of the cak1-Δ CDC28-43244 strain is only slightly slower than that of a CAK1 CDC28-43244 or a wild-type strain. The CAK1 CDC28-43244 strain completed the nuclear divisions (MI and MII) and formed spores with comparable kinetics and to the same extent as a wild-type control strain. Thus, at 24 h postinduction, 87% ± 6% of the CAK1 CDC28-43244 diploid cells had completed meiosis, as assayed using DAPI (4′,6′-diamidino-2-phenylindole) staining of DNA, compared with 91% ± 4% of the wild-type cells. In both strains, all of the cells that had completed meiosis formed spores, and only 6 to 9% in each strain completed only one of the meiotic divisions (>100 cells counted for three isolates of each genotype). In contrast, the cak1-Δ CDC28-43244 diploid showed substantial defects in the completion of MI and MII. Only 13% ± 2% of cells completed both divisions, while another 18% ± 2% of the cells completed only a single division (two DAPI foci). The numbers of meiotic cells did not increase even after 48 h. Moreover, none of the cells formed spores, as assayed by phase-contrast microscopy (Fig. 5A to D).
FIG. 5.
Phase-contrast and electron microscopy of cells with and without CAK1. CDC28-43244/CDC28-43244 strains containing CAK1 (A, C, and E) and lacking CAK1 (B, D, F, G, and H) were incubated in sporulation medium for 36 h. (A and B) Phase-contrast microscopy of CAK1+ and cak1 strains, respectively; (C and D) corresponding DAPI fluorescence images; (E) thin-section electron microscopic ultrastructure of a CAK1+ ascus (indistinguishable from wild type; data not shown); (F) ultrastructure of the predominant class of cak1 cell (blocked prior to the meiotic divisions); (G) a cak1 ascus in which only one meiotic division is evident (note the coalescence of vesicles around the edge of the nuclear compartment); (H) a cak1 ascus in which two meiotic divisions have occurred (three haploid products observed in the plane; note the abnormal spore wall structures).
Wild-type spore walls examined by thin-section electron microscopy consist of multiple layers which include two inner electron-lucent layers, a more diffuse chitin/chitosan layer, and the closely juxtaposed outer electron-dense dityrosine coat. The spore walls in the CDC28-43244 CAK1+ strain were indistinguishable from those seen in a wild-type control strain (Fig. 5E and data not shown). In contrast, no normal spore walls were observed in over 400 CDC28-43244 cak1-Δ cells examined. Most of these cak1-Δ cells showed no evidence of the meiotic divisions and contained large numbers of 0.2- to 0.4-μm-diameter vesicles (Fig. 5F). These ultrastructural observations are consistent with the notion that the majority of these cells are blocked early in the sporulation program.
In the cells that completed one or two meiotic divisions (Fig. 5G and H, respectively), a variety of different spore wall phenotypes were observed. Some cells appear to have been blocked early in spore wall morphogenesis, with vesicular structures surrounding the meiotic compartments (Fig. 5G), while others assembled layers around the haploid nuclei but the layers are abnormal or appear to resemble intermediates in the spore morphogenesis pathway. These experiments show that cells lacking Cak1p but with active Cdc28p appear to have defects in executing both meiosis and spore formation.
We tested whether a catalytically inactive mutant form of Cak1p (D169R) was able to function in the CDC28-43244 mutant background. The catalytically inactive cak1-D169R mutant was unable to promote meiosis or spore formation when present on a single (CEN) or multiple-copy (2μm) vector. Furthermore, a cak1 hypomorph (cak1-K31R) promoted meiosis and sporulation in the CDC28-43244 background when expressed from a 2μm but not from a CEN vector (data not shown). These results indicate that the catalytic activity of Cak1p is required for its functions in meiosis and spore formation.
The requirement for CAK1 in spore formation in the CDC28-43244 genetic background is consistent with our biochemical demonstration that Cak1p is required to activate Smk1p and demonstrates that Cak1p activates Smk1p by a Cdc28p-independent mechanism. The MI and MII defect of the cak1-Δ CDC28-43244 strains was unexpected. Temperature-sensitive cak1 mutations (cak1-17and cak1-22) in an otherwise wild-type background do cause blocks prior to MI and MII when cells are shifted to a temperature that is nonpermissive for vegetative growth (34°C or higher) (42). However, prior to this study, the block prior to the nuclear divisions in the cak1 strain at the nonpermissive temperature could have been explained solely by a decrease in the pool of Cdc28p that is phosphorylated on T-169. The Cdc28-43244p mutant protein lacks the T that is phosphorylated by Cak1p. That a CAK1/CAK1 CDC28-43244/CDC28-43244 strain completes meiosis normally while a cak1-Δ/cak1-Δ CDC28-43244/CDC28-43244 strain does not indicates that Cak1p promotes events prior to or at the meiotic divisions by phosphorylating a target other than Cdc28p.
Cak1p is required for timely premeiotic DNA synthesis and expression of sporulation genes.
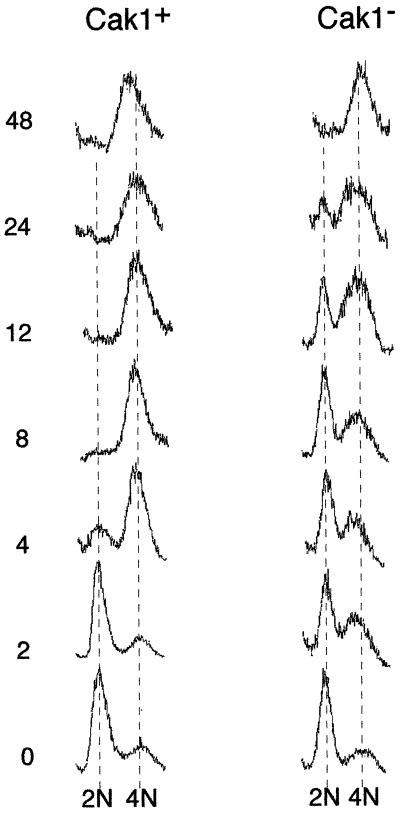
To further characterize Cak1p’s role in meiotic development, we monitored premeiotic DNA replication using fluorescence-activated cell sorting (FACS) (Fig. 6). The kinetics of premeiotic S-phase in a CDC28-43244/CDC28-43244 and wild-type strain was indistinguishable (data not shown). These results are consistent with the similar rates of MI, MII, and spore formation in these genetic backgrounds and show that the CDC28-43244 mutation does not affect the timing of S-phase in meiosis. However, the cak1-Δ/cak1-Δ CDC28-43244/CDC28-43244 strain showed a substantial delay in completing premeiotic S-phase. While the vast majority of CAK1/CAK1 CDC28-43244/CDC28-43244 cells duplicated their genomes by 4 h, genome replication in the cak1-Δ/cak1-Δ CDC28-43244/CDC28-43244 cells was undetectable until around 12 h postinduction. Even at 24 h, a significant percentage of the cak1-Δ cells had still not replicated their genomes. Complete replication was not observed until 48 h postinduction. These data suggest that Cak1p promotes sporulation early in the program, prior to or at S-phase.
FIG. 6.
Cells lacking Cak1p are delayed for premeiotic DNA synthesis. CDC28-43244/CDC28-43244 cells (Cak1+) and CDC28-43244/CDC28-43244 cak1-Δ/cak1-Δ cells (Cak1−) were sporulated using synchronous sporulation conditions, and cells were withdrawn at the indicated times and analyzed by FACS for genome replication.
We also monitored a CDC28-43244/CDC28-43244 diploid harboring a catalytically inactive cak1-D169R mutation by FACS analysis. The cak1-D169R mutant and the cak1-Δ null mutant were indistinguishable in the FACS time course, demonstrating that Cak1p’s catalytic activity is required for efficient progression through premeiotic S-phase (data not shown).
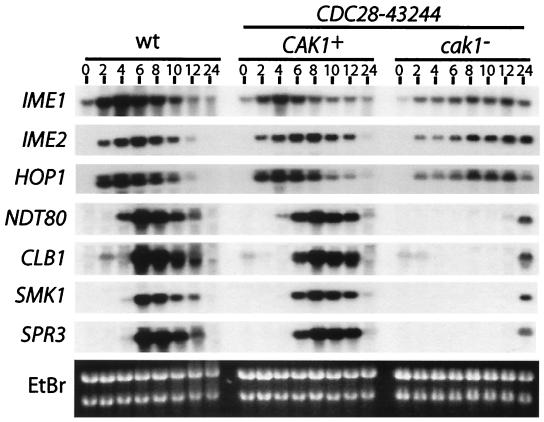
To further define the role of Cak1p in sporulation, the expression of early and middle sporulation genes was compared between the strains containing and lacking Cak1p by Northern blot hybridization analysis. As shown in Fig. 7, the CAK1/CAK1 CDC28-43244/CDC28-43244 and the wild-type control strain expressed the early sporulation-specific IME1, HOP1, and IME2 genes as well as the middle sporulation-specific NDT80, CLB1, SMK1, and SPR3 genes to comparable levels and with similar kinetics. Thus, the hyperactivated CDC28-43244 allele does not appear to substantially affect the transcriptional program under optimal sporulation conditions. In contrast, there are defects in the expression of both early and middle sporulation genes in the cak1-Δ/cak1-Δ CDC28-43244/CDC28-43244 mutant strain. The most striking quantitative defect is the almost complete absence of middle gene transcripts. Thus, induction of the middle SMK1, CLB1, NDT80, and SPR3 genes is undetectable in the strain lacking Cak1p at the 0- to 12-h time points. Low levels of these transcripts are observed at the 24-h time point.
FIG. 7.
Cells lacking Cak1p show defects in sporulation-specific gene expression. Wild-type (wt), CDC28-43244/CDC28-43244 (CAK1+), and CDC28-43244/CDC28-43244 cak1-Δ/cak1-Δ (cak1−) cells were sporulated in synchronous sporulation conditions. Cells were withdrawn at the indicated times, and total RNA was prepared and analyzed by Northern blot analysis with the indicated gene-specific probes. IME1, HOP1 and IME2 are classified as early sporulation-specific genes; NDT80, SMK1, and SPR3 are classified as middle sporulation-specific genes. CLB1 is expressed in mitosis but is strongly induced when middle genes are expressed.
Of particular interest is the expression of NDT80 and SMK1. Cells blocked in pachytene express both of these genes at moderate levels (2, 11, 23). The complete absence of these transcripts at hours 0 to 12 of the time course suggests that the cells lacking Cak1p stall early in the program prior to pachytene. This observation is consistent with the delay in premeiotic DNA synthesis. The absence of NDT80 mRNA can explain the defect in middle gene induction, as Ndt80p is required for the induction of middle promoters. NDT80 is induced as part of the transcriptional cascade, and active Ndt80p requires the expression of early genes as well as the completion of early events in the program (2, 11, 37). Thus, the middle gene expression defect in sporulating cells lacking Cak1p may be a consequence of a primary defect that occurs earlier in the program.
There are also defects in the expression of the early HOP1 and IME2 transcripts in the strain lacking Cak1p. While HOP1 and IME2 transcripts begin to increase at about the same time in all of the strains (2 h postinduction), the level of expression of these genes in the strain lacking Cak1p is less than that seen in the strains with Cak1p. Furthermore, while these early gene transcripts peak at 4 to 6 h and subsequently decline in the wild-type and CAK1/CAK1 CDC28-43244/CDC28-43244 cells, the maximally induced level is lower and the peaks are not reached until 8 to 12 h in the cak1-Δ/cak1-Δ CDC28-43244/CDC28-43244 cells.
IME1 encodes the key regulator of early gene expression in diploid cells. IME1 expression can be rate limiting for the induction of early promoters and meiotic induction in a/α cells (20). IME1 was induced slowly and to a low level in the strain lacking Cak1p (Fig. 7). These results raise the possibility that all of the defects in the transcriptional program can be explained by the deficit in IME1 transcript levels. The deficit in IME1 transcript levels in cells lacking Cak1p might also explain the DNA replication defect, as IME1 promotes premeiotic DNA synthesis (17).
We tested the effect of increasing IME1 transcript levels in cells lacking Cak1p using the high-copy IME1 plasmid pS303 (Table 2). This plasmid leads to substantial increases in IME1 mRNA in vegetative cells and can increase IME1 mRNA during sporulation to wild-type levels in mutants that are defective in IME1 induction (17, 22). pS303 and a control plasmid lacking IME1 (Yep351; see Table 2) were introduced into the cak1-Δ/cak1-Δ CDC28-43244/CDC28-43244 diploid. Total RNA was prepared from vegetative and sporulating cells (6 h postinduction), and the expression of IME1 and HOP1 (a representative IME1-responsive early gene) was tested by Northern-blot hybridization analysis. The expression of these genes was also tested in the CAK1/CAK1 CDC28-43244/CDC28-43244 strain in these experiments.
The level of IME1 in the cak1-Δ/cak1-Δ CDC28-43244/CDC28-43244 sporulating strain containing pS303 was almost twice that seen in this strain containing the empty control vector (1.9-fold ± 0.2-fold, n = 3). The expression of HOP1 was also substantially increased in the pS303 transformants at the 6-h time point (1.7-fold ± 0.2-fold). These levels of IME1 and HOP1 transcripts were indistinguishable from the levels seen in the CAK1/CAK1 CDC28-43244/CDC28-43244 strain (0.98-fold ± 0.3-fold and 0.95-fold ± 0.14-fold, respectively). Despite the increase in IME1 mRNA and in the expression of a representative IME1-responsive gene (HOP1), only 15% ± 7% of the CDC28-43244/CDC28-43244 cak1-Δ/cak1-Δ cells harboring pS303 completed MI and/or MII, as judged by DAPI staining. This compares with 12% ± 8% for cells harboring the Yep351 control vector lacking the IME1 insert (n = 200 for three independently derived transformants with each plasmid). Furthermore, the cak1-Δ/cak1-Δ CDC28-43244/CDC28-43244 cells harboring pS303 still had a substantial delay in premeiotic DNA replication, as monitored by FACS analysis (data not shown). These results demonstrate that the deficiency in IME1 transcript level is not sufficient to explain the early meiotic defect of cells lacking Cak1p. However, it should be noted that this does not necessarily imply that Cak1p does not function in the IME1 transcriptional activation pathway (see Discussion).
Cak1p does not directly phosphorylate Smk1p or Ime2p in vitro.
The results described above indicate that Cak1p regulates sporulation at multiple steps. The earliest Cdc28p-independent step takes place between induction and premeiotic DNA synthesis. A later Cdc28p-independent step occurs after the meiotic divisions and leads to activation of Smk1p. Two candidate kinase targets are suggested by these results. The first candidate is Ime2p. Ime2p contains a threonine at residue 242 in a region of the enzyme that shows similarity to the activating loops of other protein kinases. Furthermore, ime2-Δ/ime2-Δ strains have defects in early and middle gene expression and also exhibit a delay in premeiotic DNA synthesis (8, 28, 34). These phenotypes partially resemble those seen in the cak1-Δ/cak1-Δ time courses. The second candidate substrate of Cak1p is Smk1p itself. We expressed wild-type Ime2p and a mutant form of Ime2p in which the presumed activating threonine had been mutated to a nonphosphorylatable alanine (T242A) as GST fusion proteins and purified them using glutathione-agarose. We also prepared immunoprecipitates of Smk1-HAp and Smk1-T207A/Y209F-HAp either from vegetative cells in which these proteins were expressed using the GAL1 promoter (where more than 95% of Smk1-HAp is unphosphorylated) or from the endogenous SMK1 promoter from 8-h sporulation time points (where about 40% of Smk1-HAp is phosphorylated). The purified Ime2-GSTp and Smk1-HAp proteins were subsequently tested in vitro as Cak1p substrates using purified CDK2 (the human cdk that can function for Cdc28p in yeast in a CAK1-dependent fashion) as a positive control (16).
Neither Ime2-GSTp or Smk1-HAp was phosphorylated under conditions in which CDK2 was phosphorylated in a robust fashion (data not shown). Moreover, in the case of Ime2p, a mutant form of Ime2p in which the putative activating T was changed to a nonphosphorylatable A complemented the sporulation defect of an ime2-Δ/ime2-Δ mutant strain. In addition, overexpression of IME2 or of a hyperactive mutant that lacks the C-terminal inhibitory domain (IME2-472) (35) did not increase the percentage of CDC28-43244/CDC28-43244 cak1-Δ/cak1-Δ cells that completed the meiotic divisions. The results do not provide any evidence to support the idea that Cak1p directly phosphorylates Ime2p or Smk1p.
DISCUSSION
In this work we have demonstrated that Smk1p is modified by phosphorylation in its activating loop. We have also shown that Cak1p is required to activate Smk1p and that it does so through a Cdc28p-independent pathway. In the course of these studies we discovered that Cak1p also functions early in meiotic development, prior to or at premeiotic DNA synthesis, and that this early Cak1p function, like the Smk1p activation function, is Cdc28p independent. Other studies have shown that Cak1p’s sole essential role in mitosis is to phosphorylate T-169 in Cdc28p (3, 5, 16, 36). We presume that Cak1p is also required to activate Cdc28p during meiosis, although this has not been tested here. Our results show that Cak1p’s role in meiosis differs fundamentally from its role in mitosis and that Cak1p plays a key role in regulating meiotic progression at multiple steps by activating multiple downstream targets.
Activation of Smk1p by phosphorylation.
Prior to this study, the notion that Smk1p is activated by phosphorylation was inferential and based on Smk1p’s sequence similarity to other MAPKs. Here we have described an assay for Smk1p based on its electrophoretic mobility as a doublet. This assay has been used to show that Smk1p is phosphorylated. Mutation of the T207 but not the Y209 eliminates the modified form of Smk1p. This result indicates that the T is a site of phosphorylation but does not address whether the Y is similarly modified. In this regard it is worth noting that the activated (T and Y dually phosphorylated) form of the mammalian Erk2 MAPK migrates as an upper member of a doublet in electrophoretic gels comparable to those used in this study. In Erk2, neither a T/A nor a Y/F single substitution mutation eliminates the doublet, while the T/A Y/F double mutation does (30). Phosphorylation of the activating T and Y in other MAPKs appears to occur via a two-step mechanism, with transient dissociation of the singly phosphorylated intermediate (7). With some MAPKs it appears that both residues need to be phosphorylated to generate an active enzyme and that this can be involved in establishing an all-or-none switch-like MAPK activation response. Smk1p appears to regulate multiple downstream steps in spore morphogenesis that require increasing levels of catalytic activity (41). The singly T-phosphorylated Smk1p enzyme appears to be partially active, since the smk1-Y209F mutant can activate the low-threshold event of dityrosine incorporation into insoluble material (as measured by the fluorescence assay) when it is overexpressed (Fig. 3B). It is possible that a singly T-phosphorylated Smk1p represents an intermediate activation state that exists in vivo during sporulation and that the Y may be phosphorylated only when Smk1p activity is at its highest. Thus, phosphorylation of one or the other of the activating residues in a kinase may provide a mechanism to control the output of a protein kinase in a variable fashion. Further studies are required to address these issues and to determine whether Smk1p is phosphorylated only on T207 or on both T207 and Y209.
Cak1p functions upstream of Smk1p.
CAK1 has previously been shown to function in the SMK1 pathway using genetic approaches. Here we have shown that Cak1p functions upstream in the Smk1p pathway by a Cdc28p-independent mechanism that requires catalytically active Cak1p. Cak1p has been shown by others to interact with Smk1p in a two-hybrid assay in mitotic cells (38). Although these collective observations suggest that Cak1p might directly phosphorylate Smk1p, we have found no evidence that purified Cak1p can directly phosphorylate Smk1p in vitro. Taken together, these results suggest a model in which Cak1p functions in the Smk1p pathway by phosphorylating an intermediary kinase that in turn leads to Smk1p activation. However, it is still conceivable that Cak1p activates Smk1p directly in vivo but that this requires specificity or cyclin-like factors that were missing from the in vitro reaction. Cak1p also phosphorylates and activates Cdc28p, and the activating phosphorylation is presumably required for Cdc28p’s role in the MI and MII divisions. The activation of the Cdc28p cdk and the SMK1 pathway by a single enzyme provides a potential regulatory mechanism for meiotic cells to couple the nuclear divisions to gametogenesis (spore morphogenesis).
While the identity of the putative protein kinase(s) that functions between Cak1p and Smk1p is unknown, several candidates can be ruled out. Diploid strains lacking the Ste7p, Pbs2p, Mkk1p, or Mkk2p MEK or the Ste11p MEKK form spores (unpublished results). Thus, none of these enzymes is uniquely required to activate Smk1p. Furthermore, while Sps1p (a member of the STE20 family of protein kinases) is required for spore wall assembly and is expressed specifically during middle sporulation, it is not required for Smk1p activation in the doublet assay (Fig. 4). These data suggest that Sps1p and Smk1p may function in different signaling pathways that control spore morphogenesis.
The only known in vivo substrate of Cak1p other than Cdc28p is Kin28p, an essential protein kinase that regulates transcriptional processes (6, 18). Kin28p phosphorylation by Cak1p is not required for sporulation, as a kin28 mutant lacking the threonine that is phosphorylated by Cak1p (kin28-T162A) is viable and homozygous diploids sporulate indistinguishably from wild-type cells (18; Jonathan Kimmelman and Mark Solomon, personal communication; our unpublished results). Thus, at this time, we do not know the identity of the putative kinase(s) that functions between Cak1p and Smk1p.
Cak1p functions early in meiosis.
During the course of this study, we made the surprising discovery that Cak1p is required not only to activate the SMK1 pathway but also to activate a CDC28-independent pathway early during the program. Thus, diploid CDC28-43244 cak1-Δ cells do not complete premeiotic DNA synthesis on schedule and show defects in the expression of early sporulation-specific genes. These results show that Cak1p functions early to promote meiotic development prior to or at premeiotic S-phase. An interesting feature of the S-phase delay is that it appears that cells can complete DNA synthesis once it has been initiated. We have previously shown that cak1 mutants can be isolated that complete early events, express middle genes (implying they have progressed through pachytene), and are blocked later in the program, after MI and MII but before spore morphogenesis (42). These data show that the early Cak1p functions can be genetically separated from the later functions.
One early defect in the cak1-Δ/cak1-Δ CDC28-43244/CDC28-43244 strain is a reduced and delayed level of IME1 transcript accumulation (Fig. 7). Overexpression of IME1 leads to increased expression of the HOP1 early gene during sporulation in a CDC28-43244/CDC28-43244 cak1-Δ/cak1-Δ background. However, these cells still show substantial delays in premeiotic DNA synthesis and in the completion of the nuclear divisions. Together, these observations suggest that the early meiotic defect in the strain lacking Cak1p cannot be explained exclusively by a defect in the Ime1 pathway. One class of model to explain these results is that Cak1p may positively regulate a step early in the program, such as DNA replication, whose completion is required to allow full expression of IME1 and early genes. The requirements for Cdc28p in premeiotic DNA replication are different from those in mitosis (33). It is possible that Cdc28p’s functions in regulating key events in DNA replication are replaced during meiosis by another Cak1p-activatable kinase or even by Cak1p itself.
The induction of early genes is inhibited by a checkpoint pathway when DNA replication is blocked by hydroxyurea (HU) (21). Induction of IME1 is weakly inhibited in HU-treated cells. Thus, the reduced expression of IME1 and early genes in the strains lacking Cak1p could be related to activation of a checkpoint pathway that couples meiotic gene expression to events that occur early in the program. That said, it should also be pointed out that multiple protein kinases regulate the Ime1 pathway and that overexpression of IME1 cannot suppress defects in all of them. For example, the Rim15p kinase regulates association of Ime1p with the Ume6p DNA-binding protein. A rim15-Δ/rim15-Δ strain, like a strain lacking Cak1p, expresses IME1 at a reduced level and with delayed kinetics, and the sporulation phenotypes of both of these strains cannot be suppressed by IME1 overexpression (40). These observations leave open the possibility that Cak1p might directly promote expression of certain key early genes by phosphorylating a kinase that functions in the Ime1 pathway, such as a positive regulator of Rim15p. Determining whether the defect in expression of IME1 and early genes seen in the cak1-Δ strain reflects a direct role for Cak1p in the IME1 pathway, a secondary consequence of Cak1p’s role in other early meiotic events, or both will require further experimentation.
In summary, the results described here show that Cak1p plays a key role in regulating meiotic development at multiple steps. The earliest requirement for Cak1p that we have detected occurs between induction of the program and premeiotic DNA synthesis. The latest requirement for Cak1p that we have detected is in the Smk1p signaling pathway that controls the postmeiotic steps in spore formation. Both the earliest and latest Cak1p steps are Cdc28p independent. Cak1p is also required to execute Cdc28p-dependent steps such as the nuclear divisions. Thus, in contrast to the mitotic cell cycle, Cak1p controls the meiotic program at multiple steps. Further studies of the role of Cak1p in meiosis should provide additional insights into how the regulation of meiosis differs from the regulation of mitosis.
Acknowledgments
We thank Fred Cross for the CDC28 plasmids used in these studies and Randy Strich and Erica Johnson for comments on the manuscript and helpful discussions. We also thank Allison Martin for carrying out and analyzing the control Northern blot experiments of IME1-overexpressing strains.
This work was supported by grants RPG-98-071-01-DDC from the American Cancer Society and RO1 GM61817 from the National Institutes of Health.
REFERENCES
- 1.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699–705. [DOI] [PubMed] [Google Scholar]
- 2.Chu, S., and I. Herskowitz. 1998. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell 1:685–696. [DOI] [PubMed] [Google Scholar]
- 3.Cross, F. R., and K. Levine. 1998. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol. Cell. Biol. 18:2923–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erdeniz, N., U. H. Mortensen, and R. Rothstein. 1997. Cloning-free PCR-based allele replacement methods. Genome Res. 7:1174–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinoza, F. H., A. Farrell, H. Erdjumentbromage, P. Tempst, and D. O. Morgan. 1996. A cyclin-dependent kinase-activating kinase (Cak) in budding yeast unrelated to vertebrate Cak. Science 273:1714–1717. [DOI] [PubMed] [Google Scholar]
- 6.Espinoza, F. H., A. Farrell, J. L. Nourse, H. M. Chamberlin, O. Gileadi, and D. O. Morgan. 1998. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol. 18:6365–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrell, J. E., Jr., and R. R. Bhatt. 1997. Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J. Biol. Chem. 272:19008–19016. [DOI] [PubMed] [Google Scholar]
- 8.Foiani, M., E. Nadjar-Boger, R. Capone, S. Sagee, T. Hashimshoni, and Y. Kassir. 1996. A meiosis-specific protein kinase, Ime2, is required for the correct timing of DNA replication and for spore formation in yeast meiosis. Mol. Gen. Genet. 253:278–288. [DOI] [PubMed] [Google Scholar]
- 9.Friesen, H., R. Lunz, S. Doyle, and J. Segall. 1994. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 8:2162–2175. [DOI] [PubMed] [Google Scholar]
- 10.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534. [DOI] [PubMed] [Google Scholar]
- 11.Hepworth, S. R., H. Friesen, and J. Segall. 1998. NDT80 and the meiotic recombination checkpoint regulate expression of middle sporulation-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5750–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill, J. E., A. M. Myers, T. J. Koerner, and A. Tzagoloff. 1986. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2:163–167. [DOI] [PubMed] [Google Scholar]
- 13.Honigberg, S. M., and R. H. Lee. 1998. Snf1 kinase connects nutritional pathways controlling meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:4548–4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaldis, P., A. Cheng, and M. J. Solomon. 2000. The effects of changing the site of activating phosphorylation in CDK2 from threonine to serine. J. Biol. Chem. 275:32578–32584. [DOI] [PubMed] [Google Scholar]
- 15.Kaldis, P., Z. W. Pitluk, I. A. Bany, D. A. Enke, M. Wagner, E. Winter, and M. J. Solomon. 1998. Localization and regulation of the cdk-activating kinase (Cak1p) from budding yeast. J. Cell Sci. 111:3585–3596. [DOI] [PubMed] [Google Scholar]
- 16.Kaldis, P., A. Sutton, and M. J. Solomon. 1996. The cdk-activating kinase (CAK) from budding yeast. Cell 86:553–564. [DOI] [PubMed] [Google Scholar]
- 17.Kassir, Y., D. Granot, and G. Simchen. 1988. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 52:853–862. [DOI] [PubMed] [Google Scholar]
- 18.Kimmelman, J., P. Kaldis, C. J. Hengartner, G. M. Laff, S. S. Koh, R. A. Young, and M. J. Solomon. 1999. Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol. 19:4774–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krisak, L., R. Strich, R. S. Winters, J. P. Hall, M. J. Mallory, D. Kreitzer, R. S. Tuan, and E. Winter. 1994. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8:2151–2161. [DOI] [PubMed] [Google Scholar]
- 20.Kupiec, M., B. Byers, R. Esposito, and A. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889–1036. In J. Pringle, J. Broach, and E. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Lamb, T. M., and A. P. Mitchell. 2001. Coupling of Saccharomyces cerevisiae early meiotic gene expression to DNA replication depends upon RPD3 and SIN3. Genetics 157:545–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee, R. H., and S. M. Honigberg. 1996. Nutritional regulation of late meiotic events in Saccharomyces cerevisiae through a pathway distinct from initiation. Mol. Cell. Biol. 16:3222–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindgren, A., D. Bungard, M. Pierce, J. Xie, A. Vershon, and E. Winter. 2000. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 19:6489–6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. [DOI] [PubMed] [Google Scholar]
- 25.Malathi, K., Y. Xiao, and A. P. Mitchell. 1999. Catalytic roles of yeast GSK3beta/shaggy homolog Rim11p in meiotic activation. Genetics 153:1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malathi, K., Y. Xiao, and A. P. Mitchell. 1997. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol. Cell. Biol. 17:7230–7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell, A. P. 1994. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol. Rev. 58:56–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell, A. P., S. E. Driscoll, and H. E. Smith. 1990. Positive control of sporulation-specific genes by the IME1 and IME2 products in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2104–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierce, M., M. Wagner, J. Xie, V. Gailus-Durner, J. Six, A. K. Vershon, and E. Winter. 1998. Transcriptional regulation of the SMK1 mitogen-activated protein kinase gene during meiotic development in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5970–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posada, J., and J. A. Cooper. 1992. Requirements for phosphorylation of MAP kinase during meiosis in Xenopus oocytes. Science 255:212–215. [DOI] [PubMed] [Google Scholar]
- 31.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415–423. [DOI] [PubMed] [Google Scholar]
- 32.Sagee, S., A. Sherman, G. Shenhar, K. Robzyk, N. Ben-Doy, G. Simchen, and Y. Kassir. 1998. Multiple and distinct activation and repression sequences mediate the regulated transcription of IME1, a transcriptional activator of meiosis-specific genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1985–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuster, E. O., and B. Byers. 1989. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics 123:29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith, H. E., and A. P. Mitchell. 1989. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:2142–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soushko, M., and A. P. Mitchell. 2000. An RNA-binding protein homologue that promotes sporulation-specific gene expression in Saccharomyces cerevisiae. Yeast 16:631–639. [DOI] [PubMed] [Google Scholar]
- 36.Thuret, J.-V., J.-G. Valay, G. Faye, and C. Mann. 1996. Civ1 (CAK in vivo), a novel cdk-activating kinase. Cell 86:565–576. [DOI] [PubMed] [Google Scholar]
- 37.Tung, K. S., E. J. Hong, and G. S. Roeder. 2000. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. USA 97:12187–12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623–627. [DOI] [PubMed] [Google Scholar]
- 39.Vershon, A. K., and M. Pierce. 2000. Transcriptional regulation of meiosis in yeast. Curr. Opin. Cell. Biol. 12:334–339. [DOI] [PubMed] [Google Scholar]
- 40.Vidan, S., and A. P. Mitchell. 1997. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol. Cell. Biol. 17:2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner, M., P. Briza, M. Pierce, and E. Winter. 1999. Distinct steps in yeast spore morphogenesis require distinct SMK1 MAP kinase thresholds. Genetics 151:1327–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner, M., M. Pierce, and E. Winter. 1997. The CDK-activating kinase CAK1 can dosage suppress sporulation defects of smk1 MAP kinase mutants and is required for spore wall morphogenesis in Saccharomyces cerevisiae. EMBO J. 16:1305–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79:143–180. [DOI] [PubMed] [Google Scholar]
- 44.Xiao, Y., and A. P. Mitchell. 2000. Shared roles of yeast glycogen synthase kinase 3 family members in nitrogen-responsive phosphorylation of meiotic regulator Ume6p. Mol. Cell. Biol. 20:5447–5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaffe, M. P., and G. Schatz. 1984. Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. USA 81:4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, H., J. F. Klemic, S. Chang, P. Bertone, A. Casamayor, K. G. Klemic, D. Smith, M. Gerstein, M. A. Reed, and M. Snyder. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26:283–289. [DOI] [PubMed] [Google Scholar]