Abstract
Efficient transcription of the human T-cell leukemia virus type 1 (HTLV-1) genome requires Tax, a virally encoded oncogenic transcription factor, in complex with the cellular transcription factor CREB and the coactivators p300/CBP. To examine Tax transactivation in vitro, we used a chromatin assembly system that included recombinant core histones. The addition of Tax, CREB, and p300 to the HTLV-1 promoter assembled into chromatin activated transcription several hundredfold. Chromatin templates selectively lacking amino-terminal histone tails demonstrated enhanced transcriptional activation by Tax and CREB, with significantly reduced dependence on p300 and acetyl coenzyme A (acetyl-CoA). Interestingly, Tax/CREB activation from the tailless chromatin templates retained a substantial requirement for acetyl-CoA, indicating a role for acetyl-CoA beyond histone acetylation. These data indicate that during Tax transcriptional activation, the amino-terminal histone tails are the major targets of p300 and that tail deletion and acetylation are functionally equivalent.
Tax is a regulatory oncoprotein produced by the human T-cell leukemia virus type 1 (HTLV-1) that is required for high-level expression of the viral genome. The mechanism of Tax activation of HTLV-1 transcription has been intensely studied over the past several years, and many of the molecular events that lead to Tax transactivation have recently been characterized (see reference 50). The HTLV-1 promoter carries three highly conserved 21-bp enhancer elements, called viral CREs, which are critical to Tax-activated transcription. The cellular transcription factor CREB binds to the core viral CRE sequence, and Tax associates with CREB through protein-protein interactions (1, 16, 24, 83). Tax also forms protein-DNA interactions with GC-rich minor groove DNA sequences that immediately flank the CREB binding site, further stabilizing the nucleoprotein complex (39, 49, 55). The Tax-containing ternary complex serves as a high-affinity binding site for the recruitment of the multifunctional cellular coactivators p300/CBP (20, 44). Tax recruits the coactivators through multiple interactions with both the amino- and carboxy-terminal regions of p300/CBP (20, 28, 37, 44, 47, 70). The interaction between Tax and p300/CBP likely tethers the coactivator to the HTLV-1 promoter, leading to high-level transcription of the viral genome.
CBP and its paralog, p300, are very large, highly conserved coactivator proteins that serve as central mediators in the regulation of gene expression in metazoans (for reviews, see references 23 and 77). p300/CBP are conserved from Caenorhabditis elegans to humans and appear to be involved in essentially all known pathways regulating gene expression. These pathways include signal-dependent and -independent activation, cellular differentiation programs, and the execution of programmed cell death (for reviews, see references 21 and 23). p300/CBP are utilized by many structurally distinct transcription factors, such as c-fos, MyoD, and YY1, that represent nearly all known classes of DNA binding proteins (6, 18, 69).
The p300/CBP coactivators may function in a variety of ways to activate transcription. There is evidence that both p300 and CBP are intrinsic components of the RNA polymerase II holoenzyme complex and are therefore brought to target promoters simultaneous with polymerase recruitment (38, 61, 81). Perhaps consistent with these observations, several studies have demonstrated that p300 acts to enhance the formation of productive transcriptional initiation complexes (13, 41, 51). Recent studies have also suggested that p300/CBP may stabilize components of the general transcription machinery, including TFIIB and TBP (12, 45). Perhaps the best characterized coactivation property of p300 and CBP is their intrinsic histone acetyltransferase (HAT) activity (5, 62). Both p300 and CBP have been shown to acetylate lysine residues in the amino-terminal tails of all four core histones (H2A, H2B, H3, and H4), both free in solution and assembled into chromatin (42, 62). Following recruitment to a promoter, these coactivators have been shown to locally hyperacetylate the amino-terminal tails of nucleosomal histones assembled on the target promoter (43, 64). Although all four histones are acetylated by p300/CBP, studies have suggested that the H3 and H4 amino-terminal tails are the preferred substrates (5, 62).
Each of the four core histones consists of two structural and functional domains. The first is an α-helical histone fold domain that participates in formation of the histone octamer, the protein core of the nucleosome. The second domain is the apparently unstructured, lysine-rich amino-terminal tail. In native chromatin, these tails interact with DNA and α-helical regions of histones located outside their nucleosome of origin. Histone tails also participate in determining nucleosome structure and DNA sequence accessibility, but removal or acetylation of the N-terminal tails does not significantly affect histone octamer stability or nucleosome formation (3, 4, 52, 63, 73, 79). It appears that the primary structural role of the histone tails is the formation of higher-order folding of the chromatin fiber, which compacts and organizes the DNA in chromosomes (9, 19, 25, 58, 73, 75). It is through their role in chromatin compaction that the histone tails exert their greatest repressive effect on transcription factor access and transcriptional activation (25–27, 76). This repression is believed to be reversed through coactivator acetylation of histone tails. This nucleosomal modification is believed to increase the accessibility of the promoter DNA to binding by regulatory proteins and/or components of the basal transcription machinery, a critical and perhaps rate-limiting step in gene activation (22, 46, 78).
The HTLV-1 genome is naturally integrated into chromatin following retroviral infection, and thus, transcriptional activation by Tax requires interaction with the viral CRE enhancer elements as they exist in a chromatin environment. We designed this study to analyze the mechanistic role of p300 in Tax/CREB-mediated transcriptional activation in a chromatin context. We wished to distinguish between the functions of p300 as an acetyltransferase and as a mediator of basal factor recruitment. We also investigated the acetyl coenzyme A (acetyl-CoA) requirement in Tax transcriptional activation. Furthermore, we wanted to define the point in the HTLV-1 transcription process when histone tails exert their greatest regulatory effect. To address these questions, we utilized chromatin templates assembled with recombinant core histones. This system provides a powerful means to examine activator-coactivator interactions in a biochemically defined chromatin context. The recombinant histones in our system lack posttranslational modifications and can be manipulated to form chromatin templates lacking any or all of the histone amino-terminal tails.
We found that nucleosome assembly strongly repressed transcription from the HTLV-1 promoter, but the addition of Tax/CREB countered the repression in an acetyl-CoA-dependent fashion, leading to a ∼100-fold increase in RNA synthesis. The addition of exogenous p300 further increased transcriptional activation to a level greater than 300-fold over the basal level. Removal of the amino-terminal histone tails enhanced Tax/CREB transcriptional activation; however, the mutant chromatin templates exhibited a reduced dependence on p300 HAT activity. This suggests that histone tail deletion functionally mimics acetylation in the transactivation process. We also examined the role of acetyl-CoA in transcriptional activation from the wild-type and tail mutant chromatin templates. Although tailless templates displayed decreased sensitivity to acetyl-CoA than wild-type chromatin, transcriptional activation remained significantly dependent on the cofactor. Together, these results indicate that robust Tax transactivation is dependent upon the presence of chromatin and acetyl-CoA and is potentiated by the addition of the coactivator p300. These studies reveal the critical role of the histone tails in coactivator-mediated transcriptional activation and suggest that the primary role for p300 in Tax/CREB transcriptional activation is the acetylation of the core histone tails.
MATERIALS AND METHODS
Purification of recombinant proteins.
The four core histones, both as wild-type and deletion mutant proteins, were individually expressed in Escherichia coli and purified to homogeneity as previously described (53). Histone octamers were prepared by denaturation, then renaturation with high levels of salt, followed by purification of the octamers by gel filtration and ion exchange chromatography. Drosophila NAP-1 (dNAP-1) (His6 tagged) was expressed from recombinant baculovirus (31) in Sf9 cells cultured in spinner flasks. The protein was purified by Ni2+-agarose batch binding and elution followed by Source 15Q column chromatography (33). We coexpressed FLAG-tagged ISWI and Acf1 from baculovirus in Sf9 cells and purified the complex by anti-FLAG affinity batch binding and elution as previously described (34). Full-length, recombinant CREB (16) was expressed and purified as previously described (20). Tax was expressed from the pTaxH6 expression plasmid (84), and purified as previously described (20). His6-tagged p300 wild-type and p300MutAT2 proteins were expressed from recombinant baculoviruses in Sf9 cells and purified as previously described (42).
Chromatin assembly and topological assays.
Chromatin templates were assembled as previously described (33). Briefly, histone octamers were incubated with an eightfold (wt/wt) excess of dNAP-1 on ice for 30 min. The core histone-dNAP-1 mixture was then added together with ACF (Acf1/ISWI; 0.87 fmol per ng of core histone octamer) and DNA to provide the empirically determined optimal core histone to DNA ratio. The p4TxRE/G-less plasmid DNA used in the assembly reactions carried four tandem copies of the HTLV-1 promoter-proximal viral CRE, cloned upstream of the HTLV-1 core promoter (2). Following the addition of the DNA, ATP (3 mM), creatine phosphokinase (1 μg/ml), and phosphocreatine (30 mM) were added to a 70-μl reaction mixture containing 10 mM HEPES (K+) (pH 7.6), 50 mM KCl, 5 mM MgCl2, 5% (vol/vol) glycerol, and 1% (wt/vol) polyethylene glycol. Assembly reaction mixtures were incubated for 4 to 18 h at 27°C. For topological assays, the p4TxRE/G-less plasmid DNA template was relaxed prior to assembly with 0.05 U of topoisomerase I (purified recombinant Drosophila topoisomerase I) per μg of DNA for 60 min at 37°C in a mixture of 50 mM Tris-HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.5 mM EDTA, and 30 μg of bovine serum albumin per ml. Immediately prior to assembly into chromatin, an additional 0.29 U of topoisomerase I per μg of DNA was added to the DNA template. Assembly reactions were stopped by the addition of 2 mM Na-EDTA (pH 8), 100 mM Tris-HCl (pH 7.5), 1% sodium dodecyl sulfate (SDS), 2 mM CaCl2, and 16 μg of proteinase K, and the samples were incubated at 45°C for 30 min. The samples were extracted, precipitated, and analyzed on a 1% agarose gel, and the degree of supercoiling was visualized by ethidium bromide staining.
In vitro transcription.
The supercoiled p4TxRE/G-less plasmid template was assembled into chromatin using NAP-1, ACF, and recombinant Xenopus histones, at a histone/DNA ratio that had been empirically determined to give full chromatin assembly. Following assembly, preinitiation complexes were formed on 75 fmol of the plasmid DNA in the absence or presence of Tax (8 nM), CREB (8 nM), p300 (3 nM), and p300 MutAT2 (3 nM). All reaction mixtures contained 100 μM acetyl-CoA unless indicated otherwise. CEM (HTLV-1-negative human T-lymphocyte) nuclear extract (70 μg) (14) was added immediately after the transcriptional activators. Following a 60-min preincubation reaction, RNA synthesis was initiated by the addition of 250 μM ATP, 250 μM CTP, 12 μM UTP, and 0.8 μM [α-32P]UTP (3,000 Ci/mmol). Transcription from unassembled templates was performed as described above but without the chromatin assembly step. Transcription reactions were processed and analyzed as previously described (48). Molecular weight markers (radiolabeled HpaII-digested pBR322) were used to estimate the size of the RNA products.
In vitro acetylation assay.
The p4TxRE/G-less plasmid template was assembled into chromatin using recombinant Xenopus histones (2 μg), NAP-1, and ACF as described above. Chromatin assembly was performed at empirically determined histone/DNA mass ratios. p300 (12 nM) and 14C-labeled acetyl-CoA (0.9 mM; 57 mCi/mmol) were added following chromatin assembly, along with 400 nM Tax and CREB where applicable, in a solution containing 50 mM Tris (pH 8), 10% glycerol, 10 mM sodium butyrate, 1 mM dithiothreitol, and 1 mM phenylmethylsulfonyl fluoride. Samples were incubated at 30°C for 60 min, and protein was precipitated by methanol-chloroform extraction and analyzed by SDS-18% polyacrylamide gel electrophoresis (PAGE).
RESULTS
Transcriptional activation by Tax/CREB and p300 on unassembled templates in vitro.
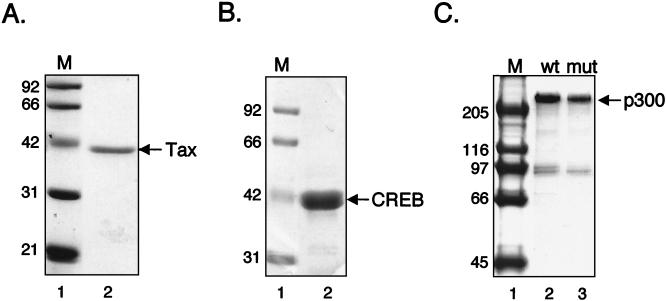
Several previous studies have suggested that the HTLV-1 Tax protein utilizes the coactivators p300/CBP to mediate transcriptional activation from the viral promoter (20, 28, 37, 44, 47, 70). We were interested in determining the mechanistic role of p300 in Tax/CREB-mediated transcriptional activation. Figure 1 shows the purified recombinant Tax, CREB, and p300 proteins used in this study (panels A to C). We tested the transcriptional activity of these proteins on unassembled DNA templates. We used the p4TxRE/G-less circular plasmid DNA template (Fig. 1D, schematic), which carries four copies of the Tax-responsive viral CRE driving synthesis of a 380-nucleotide (nt) guanine-less transcript (2). Transcription was examined in the presence of nuclear extracts prepared from the CEM cell line (HTLV-1-negative human T-lymphocyte). Figure 1D shows that the addition of Tax and CREB stimulated transcription from the p4TxRE/G-less template 4.5-fold. We analyzed p300 activity on a wide range of Tax/CREB levels and found that the coactivator produced only modest effects on activated transcription (an average of 1.6-fold above that of Tax/CREB alone) (Fig. 1D, lane 4; also data not shown). The addition of exogenous p300 had no effect on basal transcription (data not shown). Consistent with other studies, these results suggest that p300 may not function in the absence of chromatin (41, 43).
FIG. 1.
Analysis of the purified proteins used in the in vitro transcription assays. Purified recombinant Tax (A), purified recombinant CREB (B), and purified p300 and p300MutAT2 (C) were analyzed by SDS-PAGE and staining with Coomassie brilliant blue. The positions of molecular mass markers (lanes M) (in kilodaltons) are indicated to the left of the gels. In panel C, purified wild-type p300 (wt) and p300MutAT2 (mut) are shown. (D) Tax transactivation on unassembled DNA templates. The in vitro transcription assay was performed on a p4TxRE/G-less cassette template (2) shown schematically above the gel. Transcription reaction mixtures contained the p4TxRE/G-less template and CEM nuclear extract in the absence (−) or presence (+) of purified, recombinant Tax/CREB and p300, as indicated. All reactions were performed in the presence of acetyl-CoA. The positions of full-length 380-nt RNA transcript, labeled DNA recovery standard, and labeled DNA molecular size markers (in nucleotides) are indicated. This experiment is representative of at least five independent experiments.
Recombinant chromatin assembly system.
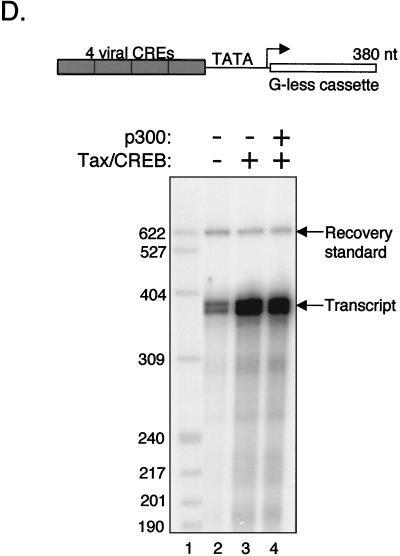
To test whether p300/CBP utilization by Tax might require a chromatin context, we developed a biochemically defined system for assembling chromatin templates from purified, recombinant proteins. The recombinant Drosophila chromatin assembly proteins NAP-1 and ACF (34), together with recombinant Xenopus core histones (54), provide the foundation for this system. NAP-1 is an H2A-H2B histone chaperone (31), and ACF is a two-subunit complex containing Acf1 and ISWI. The ACF complex acts catalytically in chromatin assembly and in the nucleosome remodeling that accompanies transcriptional activation (32). These assembly proteins are sufficient for the ATP-dependent formation of evenly spaced nucleosomal arrays (32, 34). Although these histone assembly and remodeling proteins are of Drosophila origin, they can be used with histones from several sources due to the evolutionary conservation of the histone proteins (P. J. Laybourn, unpublished observations). In addition, the Drosophila assembly system is compatible with transcriptional activators and nuclear extracts derived from human cells (41, 42, 57, 60, 65, 72). We have chosen to use recombinant Xenopus core histones as a source of nucleosomes in the assembly reaction. These histones are expressed in E. coli and are therefore unmodified and better suited than native histones for studies examining the HAT activity of p300 in Tax-activated transcription. Furthermore, a recombinant chromatin assembly system enables characterization of the roles of the core histone tails in transcriptional repression, transcriptional activation, and coactivator function through selective histone tail deletions. Finally, the templates are free of histone variants that are known to be present in native core histone preparations (see reference 80). The purified chromatin assembly factors Acf1, ISWI, and NAP-1, and the recombinant Xenopus core histones are shown in Fig. 2A to C.
FIG. 2.
Analysis of the purified proteins used in the chromatin assembly assay. Purified recombinant ACF (consisting of Acf1 and ISWI) (A), purified recombinant dNAP-1 (B), and purified recombinant Xenopus core histones were analyzed by SDS-PAGE and stained with Coomassie brilliant blue. The positions of molecular mass markers (lanes M) (in kilodaltons) are indicated to the left of the gels. (D) One-dimensional DNA topological assays comparing chromatin templates assembled with native Drosophila and recombinant Xenopus (rXenopus) core histones in the presence of ACF and NAP-1. The DNA topoisomers were resolved on an agarose gel, and the DNA was stained with ethidium bromide. The supercoiled (S), relaxed (R), and nicked DNA populations and the histone/DNA ratio are indicated over the gel. Relaxed DNA samples normally consist of multiple topoisomers (+2 to −2 supercoils) that resolve as multiple bands. A negative image of the ethidium-stained gel is shown to enhance visibility of the bands. Topoisomerase I-treated p4TxRE/G-less plasmid DNA was assembled with increasing amounts of purified, native Drosophila core histones (lanes 3 to 6) or recombinant Xenopus core histones (lanes 7 to 10). (E) Basal transcription from the p4TxRE/G-less template was assayed in the presence of increasing amounts of recombinant Xenopus core histones. Naked (unassembled) DNA was assayed as a positive control for basal transcription (lane 2). The positions of labeled DNA size markers (in nucleotides), recovery standard, and full-length G-less transcripts are indicated.
We used DNA topological analysis to determine whether our recombinant assembly factors efficiently deposit recombinant Xenopus histone octamers onto the HTLV-1 transcription template. Figure 2D compares the ability of native Drosophila core histones versus recombinant Xenopus core histones to assemble chromatin onto the p4TxRE/G-less template (lanes 3 to 10). In the presence of the assembly factors, increasing ratios (wt/wt) of the core histones to the DNA produced a concomitant increase in DNA supercoiling, indicating that nucleosomes were deposited onto the template. Figure 2D shows that at a recombinant Xenopus histone/DNA ratio of 1.2:1 (wt/wt), the DNA template was fully assembled into chromatin (lane 9). The highest recombinant Xenopus histone/DNA ratio, 1.6:1, produces an inhibition of the apparent supercoiling by interfering with DNA topoisomerase I activity (Fig. 2D, lane 10).
We were next interested in determining whether increasing nucleosome density produced an associated increase in repression of basal transcription. An in vitro transcription assay was performed following chromatin assembly with the recombinant Xenopus core histones (Fig. 2E). The core histone/DNA ratio that gave complete nucleosome assembly in the supercoiling assay (1.2:1) (Fig. 2D, lane 9) produced strong repression of basal HTLV-1 transcription (Fig. 2E, lane 5). These results indicate that our recombinant system efficiently forms chromatin and is functional for repression of basal transcription in vitro.
Transcriptional activation by Tax and p300 in a chromatin context.
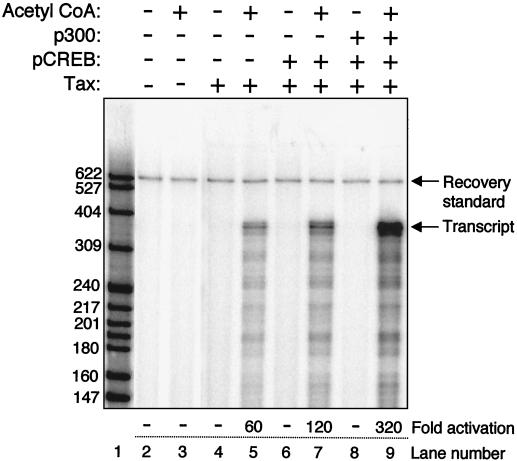
We next tested whether the p4TxRE/G-less template, assembled into chromatin with the unmodified recombinant Xenopus histones, could support Tax/CREB- and p300-mediated transcriptional activation. Figure 3 shows that the addition of Tax and Ser133-phosphorylated CREB (pCREB) together strongly activated transcription ∼100-fold (lane 7). Transcription was also stimulated by Tax and acetyl-CoA alone, presumably through interactions with endogenous CREB present in the nuclear extract (Fig. 3, lane 5). Exogenous p300 addition enhanced activated transcription by Tax/pCREB an additional threefold, leading to an overall activation level of ∼300-fold over basal transcription (Fig. 3, lane 9). This fold activation by p300 is consistent with the results of other studies (13, 41,42,43). We did not detect p300 stimulation of transcription in the absence of added Tax/pCREB (data not shown). Comparable Tax/pCREB and p300 stimulatory effects were observed on transcription templates assembled with native Drosophila core histones (data not shown). Transcriptional stimulation in the presence of the activators and coactivator was absolutely dependent upon the presence of acetyl-CoA in the reaction mixtures (Fig. 3, lanes 4 to 9); however, acetyl-CoA had no detectable effect on basal transcription (Fig. 3, lanes 2 and 3). These results are in agreement with several other studies showing that acetyl-CoA is required for transcriptional activation within a chromatin context (13, 36, 43). The overall degree of Tax/pCREB/p300 activation is far greater than that observed on unassembled templates. This result in a recombinant chromatin environment is similar to the strong Tax/CREB-dependent activation of HTLV-1 transcription and p300-mediated coactivation that is seen in vivo (8, 15, 40, 71, 74).
FIG. 3.
Transcriptional activation from recombinant chromatin templates is dependent on acetyl-CoA. Transcriptional activation on p4TxRE/G-less chromatin templates was analyzed in the presence (+) of Ser133-phosphorylated CREB (pCREB), Tax, and p300 in the presence (+) and absence (−) of acetyl-CoA. The positions of molecular weight size markers, recovery standard, and full-length G-less transcripts are indicated. Basal transcription levels, although not visible in this exposure, were measurable and upon subtraction of the background signal generated a nonzero number that was set equal to 1 in terms of relative transcription. In this experiment, protein kinase A-phosphorylated CREB was used. However, we have found essentially no difference in the transcriptional activity of Tax in the presence of CREB versus pCREB. We have therefore used only unmodified CREB in subsequent experiments.
Deletion of histone tails abolishes p300 function.
The amino-terminal histone tails are thought to play a prominent role in the formation of higher-order chromatin structure. Posttranslational modification of the histone tails has been hypothesized to participate in the conversion of transcriptionally silent, highly compacted chromatin to a more relaxed chromatin that is accessible to the transcriptional machinery. We investigated whether the histone tails would be required for transcriptional repression as well as for the high levels of transcriptional activation mediated by Tax/CREB and p300. We expressed and purified each of the four recombinant Xenopus histones carrying amino-terminal deletions. The sites of proteolytic cleavage on nucleosomal histones have been previously characterized (7); we used these sites to define our deletion mutants. We then combined our histone tail mutants to generate the two types of histone octamers used in this study. The first, which we call ΔH3/H4, contains wild-type H2A and H2B assembled together with the H3 and H4 tail deletion mutants (H3Δ1-27 and H4Δ1-19) (Fig. 2C). The second, which we call tailless, contains deletion mutations in all four core histones (H2AΔ1-12,119-128, H2BΔ1-23, H3Δ1-27, and H4Δ1-19) (Fig. 2C).
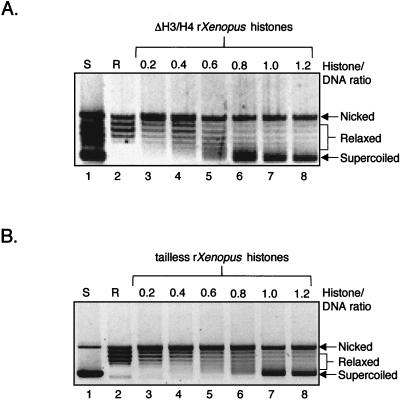
DNA topological analysis demonstrated that both mutant octamers assembled onto the p4TxRE/G-less plasmid template, with a degree of assembly comparable to that observed with wild-type octamer (Fig. 4). This is consistent with previous studies using both native, proteolyzed and recombinant histone tail deletion mutants that have demonstrated that removing the histone tails does not adversely affect nucleosome assembly, structure, or stability (3, 4, 11, 29, 52, 59).
FIG. 4.
One-dimensional DNA topological analysis of histone mutants. Topoisomerase I-treated p4TxRE/G-less cassette templates were assembled with ΔH3/H4 (A) and fully tailless (B) recombinant Xenopus (rXenopus) core histones in the presence of ACF and NAP-1. The DNA topoisomers were resolved on an agarose gel, and the DNA was stained with ethidium bromide. The supercoiled (S), relaxed (R), and nicked DNA populations and the histone/DNA ratio, are indicated over the gels. A negative image of the ethidium-stained gel is shown to enhance visibility of the bands.
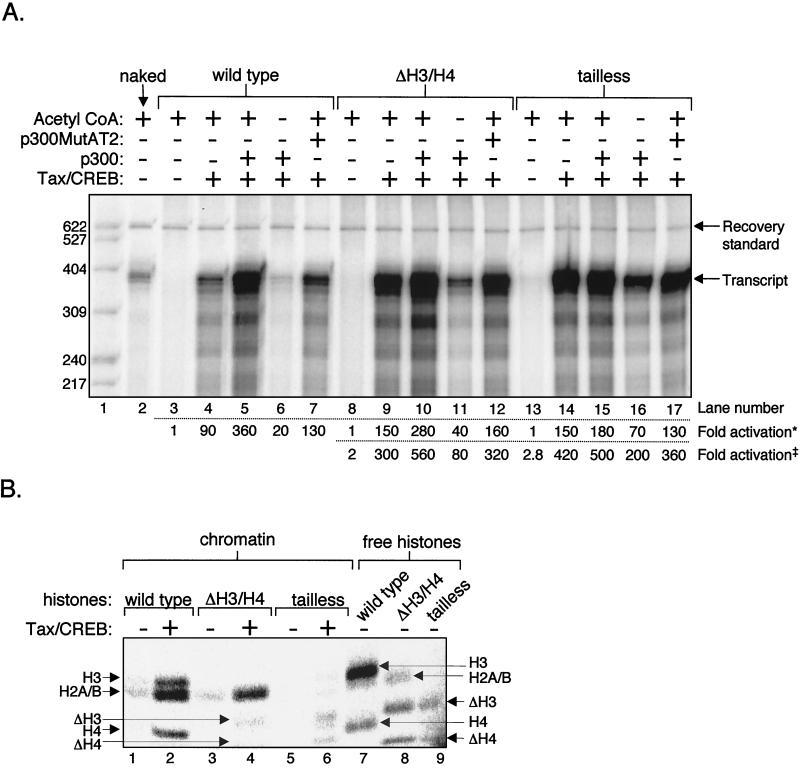
We tested whether the mutant histones affected Tax/CREB- and p300-mediated transcriptional activation. Nucleosomal templates formed with wild-type, ΔH3/H4, and tailless histone octamers each strongly repress basal transcription relative to transcription from naked DNA templates (Fig. 5A, lanes 2, 3, 8, and 13). Thus, the levels of transcriptional repression were not significantly affected by the deletion of the H3/H4 tails or of all four histone tails (Fig. 5A, compare lanes 3, 8, and 13). The addition of Tax/CREB produced robust transcriptional activation from both chromatin templates carrying the histone tail deletions, with a 1.7-fold increase in activation over that of wild-type chromatin (Fig. 5A, lanes 4, 9, and 14). The level of Tax/CREB activation from fully tailless templates was still 30-fold greater than that on free DNA templates (Fig. 1). We then compared the role of p300 in Tax/CREB-activated transcription using these mutant chromatin templates. Exogenous p300 activated Tax/CREB transcription approximately fourfold from the wild-type templates (Fig. 5A, lanes 4 and 5). In contrast, chromatin formed with histones carrying the H3/H4 tail deletions supported less than a twofold enhancement by p300, and the fully tailless histones were essentially unaffected by p300 addition (Fig. 5A, lanes 9 and 10 and lanes 14 and 15). The approximately twofold p300 stimulation from the ΔH3/H4 chromatin may be mediated through p300 acetylation of the intact H2A and H2B tails on these templates (Fig. 5B, lane 4).
FIG. 5.
Analysis of transcriptional activity and acetylation states of chromatin templates assembled with recombinant Xenopus histone tail deletion mutants. (A) In vitro transcription assay of tail mutant chromatin templates. The p4TxRE/G-less template was assembled into chromatin using wild-type, ΔH3/H4, or tailless recombinant Xenopus histone octamers in the presence of NAP-1 and ACF. Transcriptional repression and activation on the assembled templates were analyzed in the presence (+) of CREB, Tax, wild-type p300, and the HAT-defective p300 mutant p300MutAT2, as indicated over the gel. Transcription was assayed in the presence (+) and absence (−) of acetyl-CoA. The positions of labeled DNA size markers (in nucleotides), recovery standard, and full-length G-less transcripts are indicated over the gel. Fold activation was calculated within each chromatin type (Fold activation*) and calculated relative to basal transcription from wild-type chromatin (Fold activation‡). (B) In vitro acetylation assay of recombinant histones. Wild-type (lanes 1 and 2), ΔH3/H4 (lanes 3 and 4), or tailless histones (lanes 5 and 6) were assembled into chromatin on the p4TxRE/G-less template in the presence of NAP-1 and ACF. Both assembled chromatin templates and free core histones (lanes 7 to 9) were acetylated in vitro in the presence of p300 and 14C-labeled acetyl-CoA. Tax and CREB were added to the acetylation reaction mixtures, as indicated over the gel.
To distinguish the role of acetylation from the role of basal factor recruitment by p300 in transcriptional activation, we tested a p300 mutant with compromised acetyltransferase function. This protein, called p300MutAT2, carries six point mutations located within the acetyltransferase domain and has been found to have <1% of the acetyltransferase activity of the wild-type p300 (42). SDS-PAGE was performed on purified p300MutAT2, and the result is shown in Fig. 1C (lane 3). We tested the transcriptional activation by p300MutAT2 in the presence of Tax and CREB on wild-type, ΔH3/H4, and tailless chromatin templates. Relative to wild-type p300, transcriptional coactivation by p300MutAT2 is reduced on all templates (Fig. 5A, compare lanes 5, 10, and 15 with lanes 7, 12, and 17, respectively). RNA synthesis levels produced by Tax/CREB/p300MutAT2 are approximately equal to that observed with Tax/CREB in the absence of added coactivator (Fig. 5A, compare lanes 4, 9, and 14 with lanes 7, 12, and 17, respectively). These data further support the idea that the transcriptional stimulation by p300 is largely due to the intrinsic histone acetyltransferase activity of the coactivator.
An acetyl-CoA requirement beyond histone tail acetylation.
We have already established a strong dependence on acetyl-CoA for Tax/CREB- and p300-mediated transcriptional activation from wild-type chromatin templates (Fig. 3). Since we have observed that p300 functions primarily through its intrinsic HAT activity in Tax transactivation, we were interested in assessing the requirement for acetyl-CoA in transcription from the mutant chromatin templates. As shown in Fig. 5A, we find a strong, but not complete, dependence on the addition of acetyl-CoA for Tax/CREB and p300 transcriptional activation on the wild-type, ΔH3/H4, and tailless templates. However, when the histone tails are progressively deleted, there is a concomitant decrease in the dependence on acetyl-CoA for transcriptional activation on these templates (Fig. 5A, compare lanes 6, 11, and 16). For example, on wild-type templates, transcription appears to be 95% dependent on acetyl-CoA (Fig. 5A, compare lanes 5 and 6), whereas on tailless templates, transcription is 60% dependent on acetyl-CoA (Fig. 5A, compare lanes 15 and 16). This observation demonstrates that histone tail acetylation promotes transcriptional activation and that acetylation becomes less important as the histone tails are removed. It is noteworthy that there is a significant acetyl-CoA requirement for transcriptional activation even in the absence of tails.
Tax/CREB stimulation of p300 HAT activity.
Recent studies have demonstrated that histone acetylation by p300 requires targeted recruitment by activators at promoter elements (43, 64). To directly measure the level and specificity of p300-dependent acetylation of the histones assembled into the chromatin templates and to determine the dependence upon Tax and CREB, we performed p300 acetylation assays on chromatin templates using 14C-labeled acetyl-CoA. Figure 5B shows that the addition of Tax/CREB strongly enhanced acetylation of all four core histones by p300, as assembled into chromatin (lanes 1 and 2). However, Tax and CREB do not stimulate p300-mediated acetylation of free histones (data not shown). As expected, acetylation of H3 and H4 was significantly reduced on chromatin formed with the ΔH3/H4 histones, without affecting H2A and H2B acetylation (Fig. 5B, lanes 3 and 4). We did not detect significant acetylation of chromatin formed with the fully tailless histones (Fig. 5B, lanes 5 and 6). We also assayed free wild-type, ΔH3/H4, and tailless histones in the absence of Tax and CREB as a positive control for p300 activity and histone acetylation (Fig. 5B, lanes 7 to 9). Our results confirm that p300 acetylates the amino-terminal tails of the recombinant Xenopus core histones assembled into chromatin. Additionally, p300 acetylation of histone tails on chromatin is strongly stimulated by Tax and CREB. This result, together with Fig. 5A, directly correlates core histone acetylation with p300 function in Tax/CREB-mediated activation of the HTLV-1 promoter.
DISCUSSION
The HTLV-1 genome is naturally integrated into chromatin following retroviral infection, and thus, transcriptional activation by Tax requires interaction with the viral CREs and stimulation of preinitiation complex formation in a chromatin environment. In this study, we have developed a biochemically defined chromatin template to examine the functional role of p300 in Tax transcriptional activation. To generate this template, we assembled recombinant core histones into nucleosomes on a Tax-responsive HTLV-1 template DNA. We selected recombinant Xenopus core histones as our source of nucleosomes, as these histones are expressed in E. coli, and therefore do not carry posttranslational modifications that might obscure or alter coactivator acetyltransferase function in the transcription assay. In addition, we can readily examine the role of histone tails in transcriptional regulation by forming octamers that selectively lack any or all of the tails. Finally, since the template is assembled in a defined system, it is free of histone variants that are known to be present in native core histone preparations (see reference 80). Our results indicate that the primary function of p300 in Tax/CREB-activated transcription from the HTLV-1 promoter is to counteract repression by the core histone tails through acetylation. By examining the activity of chromatin templates selectively lacking core histone tails, we observe that tail deletion functionally mimics acetylation and in fact abrogates the requirement for p300 for maximal transcriptional activation.
We found that the recombinant histones efficiently assemble into nucleosomes on the template DNA, resulting in strong repression of basal transcription. This is consistent with numerous studies demonstrating that nucleosomes are repressive to basal transcription in vitro (see reference 80). The addition of Tax and CREB reversed nucleosomal repression in an acetyl-CoA-dependent fashion and strongly stimulated RNA synthesis. This derepression and activation combined to increase RNA synthesis nearly 100-fold, which was further augmented three- to fourfold by the addition of the coactivator p300. Together, the activator/coactivator complex gave >300-fold activation relative to transcription observed in the absence of activators from the chromatin assembled HTLV-1 promoter. This degree of activation is far greater than that observed on unassembled templates and is comparable to the level of HTLV-1 Tax transactivation observed in vivo (8, 15, 40, 71, 74), suggesting that our chromatin-based transcription system accurately recapitulates critical aspects of viral gene regulation within the environment of an infected T cell. Using a p300 mutant, we demonstrate that p300 primarily functions as a HAT in HTLV-1 transcription, as mutations in the HAT domain abrogate the ability of p300 to augment Tax transactivation. This result is in agreement with previous work showing that p300 HAT mutants fail to activate Tax-dependent transcription from the HTLV-1 promoter in vivo (35). Consistent with this observation, wild-type p300-mediated activation is strongly dependent on the presence of acetyl-CoA in the transcription reaction mixtures. Finally, our in vitro HAT assay on the assembled templates reveals a strong dependence on Tax and CREB for efficient p300 acetylation of the four core histone tails. This effect is likely due to the targeted recruitment of p300 to the chromatin template by the Tax/CREB complex. Overall, we observe that maximal transcriptional activity (360-fold activation) from the wild-type chromatin template is dependent on p300 and acetyl-CoA. Furthermore, Tax and CREB strongly stimulate the HAT activity of p300. Together, these data provide direct functional evidence supporting a role for p300 HAT activity in mediating Tax transactivation.
We also examined the roles of the core histone tails in Tax/CREB- and p300-mediated activation of the HTLV-1 promoter. We generated mutant chromatin templates lacking either H3/H4 or all four core histone tails. We found that progressive removal of the amino-terminal histone tails resulted in modest derepression of basal transcription and enhanced activation by the Tax/CREB complex. The level of Tax/CREB activation from fully tailless templates was 30-fold greater than that on free DNA templates. This effect is primarily due to the tailless nucleosomes still retaining significant ability to repress basal transcription while lacking any ability to inhibit Tax/CREB transcriptional activation. We found that Tax/CREB activation from tail deletion templates was refractory to stimulation by p300. Specifically, p300 addition modestly enhanced transcription from the ΔH3/H4 chromatin templates and had essentially no effect on the fully tailless chromatin templates. The approximately twofold p300 stimulation on the ΔH3/H4 chromatin was likely mediated through acetylation of the H2A and H2B tails, which were present (and acetylated) on the ΔH3/H4 chromatin templates. It is interesting to note that the absolute levels of Tax/CREB activation from the ΔH3/H4 and tailless chromatin templates were comparable to that observed from the wild-type templates in the presence of Tax/CREB and p300. This suggests that tail removal obviates the requirement for the coactivator. Together these observations suggest that all four histone tails serve as p300 acetylation targets in transcriptional activation.
Acetyl-CoA has previously been shown to be required for transcriptional activation from chromatin; however, its exact function in this process remains unknown (13, 36, 43). It has been widely speculated that acetyl-CoA is primarily utilized for histone tail acetylation during transcriptional activation. Yet, here we show that transcription in the absence of histone tails still exhibits a substantial requirement for acetyl-CoA. This result suggests the involvement of other acetyltransferases in HTLV-1 transcription, since addition of p300 had no effect on transcriptional activation from tailless templates. The basis for the acetyl-CoA requirement remains to be characterized but could reflect a number of recent observations concerning the role of acetyl-CoA during the transactivation process. Acetyl-CoA has been shown to enhance basal transcription on nonchromatin templates through a mechanism not involving acetyltransferase activity (17). Components of the RNA polymerase I and II general transcription machinery, such as UBF, TFIIE, and TFIIF, can also be acetylated, but the functional consequences of acetylation are unknown (10, 30, 66). Acetylation can increase the protein-DNA and protein-protein binding affinities of a variety of transcription factors, including EKLF, MyoD, and c-Myb (10, 67, 82). Unfortunately, we have found no evidence that Tax and CREB are acetylated. In summary, this is the first study to separate the requirement for acetyl-CoA in histone tail acetylation from other acetylation targets in Tax transcriptional activation. Our observations have significant implications for the role of acetyl-CoA as a critical cofactor in transcription beyond the process of histone tail acetylation.
The primary consequence of core histone tail deletion, which functionally mimics tail acetylation, during Tax/CREB transcriptional activation may be to reduce the chromatin barrier to activator function. It is possible that deletion of the histone tails increases the accessibility of the core promoter to preinitiation complex formation. It is also possible that deletion of histone tails serves to facilitate elongation by RNA polymerase II. For example, histone tail removal has been shown to enhance the elongation rate of T7 RNA polymerase through nucleosomes (68). However, the primary effect we observed is on activator function. The degree to which histone tail deletion contributes to enhanced regulatory protein binding, enhanced RNA polymerase recruitment, or enhanced rates of elongation, remains to be elucidated.
Although p300 functions primarily to acetylate core histone tails in Tax activated transcription, this does not appear to be generally true for all activators that utilize p300/CBP. For example, investigation of a panel of transcription factors demonstrated a differential requirement for p300 HAT activity during transcriptional activation (42). Because of the multifunctional nature of the p300/CBP coactivators, we have considered the possibility that p300 may have additional, HAT-independent functions in mediating Tax transcriptional activation that cannot be detected in our system. A recent study indicates that p300 forms a stable, template committed complex with chromatin, perhaps through interaction with histone H3 (56). This association of p300 with chromatin may serve to bring the coactivator into the vicinity of target genes, with promoter localization achieved following stable association with promoter-bound activators. Our ΔH3/H4 and tailless chromatin may be unable to carry out this first step in p300 recruitment, and therefore, the additional activation functions of p300, such as recruitment and/or stabilization of the general transcription machinery, would not be detected using the mutant chromatin templates. A second possibility is that p300 is present in the RNA polymerase II holoenzyme complex and that specific p300 activation functions distinct from histone acetylation accompany the RNA polymerase II holoenzyme that is supplied by the nuclear extract (38, 61, 81). The contribution of endogenous p300 in the nuclear extract used in these studies remains unknown.
In summary, these studies represent a major advance in our understanding of Tax- and p300-mediated transcriptional activation function and have the clear potential to extend our fundamental knowledge of coactivator-mediated transcription in a chromatin environment. The observation that progressive deletion of the histone tails is inversely proportional to coactivator utilization and acetyl-CoA dependence is a direct demonstration of the connection between p300 acetyltransferase activity, histone tail acetylation, and transcriptional activation. This supports the idea that histone tail deletion and acetylation are functionally equivalent during the transcriptional activation process. Finally, we uncover a prominent role for acetyl-CoA beyond histone acetylation during the transactivation process.
Acknowledgments
We thank James Kadonaga for the Acf1, ISWI, and dNAP-1 baculoviruses, and we thank Stephanie Price, Jes Kuruvilla, Sriwan (Apple) Wongwisansri, Holli A. Giebler, Raji Edayathumangalam, Stephanie Ebbesen, Robert Suto, and Jeanne Mick for significant contributions to this research.
This study was supported in part by grants from the National Institutes of Health (National Cancer Institute) (grant CA-87540 to J.K.N. and P.J.L. and grant CA-55035 to J.K.N.) and a grant from the Colorado Cancer League (P.J.L and J.K.N.). W.L.K. received a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and a grant from the National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases) (grant DK58110).
REFERENCES
- 1.Adya, N., and C. Z. Giam. 1995. Distinct regions in human T-cell lymphotropic virus type 1 Tax mediate interactions with activator protein CREB and basal transcription factors. J. Virol. 69:1834–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, M. G., K. E. Scoggin, C. M. Simbulan-Rosenthal, and J. A. Steadman. 2000. Identification of poly(ADP-ribose) polymerase as a transcriptional coactivator of the human T-cell leukemia virus type 1 Tax protein. J. Virol. 74:2169–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausio, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 206:451–463. [DOI] [PubMed] [Google Scholar]
- 4.Ausio, J., and K. E. van Holde. 1986. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry 25:1421–1428. [DOI] [PubMed] [Google Scholar]
- 5.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641–643. [DOI] [PubMed] [Google Scholar]
- 6.Bannister, A. J., and T. Kouzarides. 1995. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 14:4758–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohm, L., and C. Crane-Robinson. 1984. Proteases as structural probes for chromatin: the domain structure of histones. Biosci. Rep. 4:365–386. [DOI] [PubMed] [Google Scholar]
- 8.Cann, A. J., J. D. Rosenblatt, W. Wachsman, N. P. Shah, and I. S. Chen. 1985. Identification of the gene responsible for human T-cell leukaemia virus transcriptional regulation. Nature 318:571–574. [DOI] [PubMed] [Google Scholar]
- 9.Carruthers, L. M., and J. C. Hansen. 2000. The core histone N termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 275:37285–37290. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., M. Tini, and R. M. Evans. 2001. HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13:218–224. [DOI] [PubMed] [Google Scholar]
- 11.Clapier, C. R., G. Langst, D. F. Corona, P. B. Becker, and K. P. Nightingale. 2001. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol. Cell. Biol. 21:875–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallas, P. B., P. Yaciuk, and E. Moran. 1997. Characterization of monoclonal antibodies raised against p300: both p300 and CBP are present in intracellular TBP complexes. J. Virol. 71:1726–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dilworth, F. J., C. Fromental-Ramain, K. Yamamoto, and P. Chambon. 2000. ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol. Cell 6:1049–1058. [DOI] [PubMed] [Google Scholar]
- 14.Dynan, W. S. 1987. DNase I footprinting as an assay for mammalian gene regulatory proteins. Genet. Eng. 9:75–87. [Google Scholar]
- 15.Felber, B. K., H. Paskalis, C. Kleinman-Ewing, F. Wong-Staal, and G. N. Pavlakis. 1985. The pX protein of HTLV-I is a transcriptional activator of its long terminal repeats. Science 229:675–679. [DOI] [PubMed] [Google Scholar]
- 16.Franklin, A. A., M. F. Kubik, M. N. Uittenbogaard, A. Brauweiler, P. Utaisincharoen, M. A. Matthews, W. S. Dynan, J. P. Hoeffler, and J. K. Nyborg. 1993. Transactivation by the human T-cell leukemia virus Tax protein is mediated through enhanced binding of activating transcription factor-2 (ATF-2) response and cAMP element-binding protein (CREB). J. Biol. Chem. 268:21225–21231. [PubMed] [Google Scholar]
- 17.Galasinski, S. K., T. N. Lively, A. Grebe De Barron, and J. A. Goodrich. 2000. Acetyl coenzyme A stimulates RNA polymerase II transcription and promoter binding by transcription factor IID in the absence of histones. Mol. Cell. Biol. 20:1923–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galvin, K. M., and Y. Shi. 1997. Multiple mechanisms of transcriptional repression by YY1. Mol. Cell. Biol. 17:3723–3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Ramirez, M., C. Rocchini, and J. Ausio. 1995. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 270:17923–17928. [DOI] [PubMed] [Google Scholar]
- 20.Giebler, H. A., J. E. Loring, K. Van Orden, M. A. Colgin, J. E. Garrus, K. W. Escudero, A. Brauweiler, and J. K. Nyborg. 1997. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol. Cell. Biol. 17:5156–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giordano, A., and M. L. Avantaggiati. 1999. p300 and CBP: partners for life and death. J. Cell. Physiol. 181:218–230. [DOI] [PubMed] [Google Scholar]
- 22.Godde, J. S., Y. Nakatani, and A. P. Wolffe. 1995. The amino-terminal tails of the core histones and the translational position of the TATA box determine TBP/TFIIA association with nucleosomal DNA. Nucleic Acids Res. 23:4557–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553–1577. [PubMed] [Google Scholar]
- 24.Goren, I., O. J. Semmes, K. T. Jeang, and K. Moelling. 1995. The amino terminus of Tax is required for interaction with the cyclic AMP response element binding protein. J. Virol. 69:5806–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen, J. C., C. Tse, and A. P. Wolffe. 1998. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry 37:17637–17641. [DOI] [PubMed] [Google Scholar]
- 26.Hansen, J. C., and A. P. Wolffe. 1992. Influence of chromatin folding on transcription initiation and elongation by RNA polymerase III. Biochemistry 31:7977–7988. [DOI] [PubMed] [Google Scholar]
- 27.Hansen, J. C., and A. P. Wolffe. 1994. A role for histones H2A/H2B in chromatin folding and transcriptional repression. Proc. Natl. Acad. Sci. USA 91:2339–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harrod, R., Y. Tang, C. Nicot, H. S. Lu, A. Vassilev, Y. Nakatani, and C. Z. Giam. 1998. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol. Cell. Biol. 18:5052–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes, J. J., D. J. Clark, and A. P. Wolffe. 1991. Histone contributions to the structure of DNA in the nucleosome. Proc. Natl. Acad. Sci. USA 88:6829–6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imhof, A., X. J. Yang, V. V. Ogryzko, Y. Nakatani, A. P. Wolffe, and H. Ge. 1997. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 7:689–692. [DOI] [PubMed] [Google Scholar]
- 31.Ito, T., M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16:3112–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ito, T., M. Bulger, M. J. Pazin, R. Kobayashi, and J. T. Kadonaga. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90:145–155. [DOI] [PubMed] [Google Scholar]
- 33.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 14:1899–1907. [PMC free article] [PubMed] [Google Scholar]
- 34.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang, H., H. Lu, R. L. Schiltz, C. A. Pise-Masison, V. V. Ogryzko, Y. Nakatani, and J. N. Brady. 1999. PCAF interacts with Tax and stimulates Tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 19:8136–8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang, W., S. K. Nordeen, and J. T. Kadonaga. 2000. Transcriptional analysis of chromatin assembled with purified ACF and dNAP1 reveals that acetyl-CoA is required for preinitiation complex assembly. J. Biol. Chem. 275:39819–39822. [DOI] [PubMed] [Google Scholar]
- 37.Kashanchi, F., J. F. Duvall, R. P. Kwok, J. R. Lundblad, R. H. Goodman, and J. N. Brady. 1998. The coactivator CBP stimulates human T-cell lymphotrophic virus type I Tax transactivation in vitro. J. Biol. Chem. 273:34646–34652. [DOI] [PubMed] [Google Scholar]
- 38.Kim, T. K., T. H. Kim, and T. Maniatis. 1998. Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-beta enhanceosome in vitro. Proc. Natl. Acad. Sci. USA 95:12191–12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimzey, A. L., and W. S. Dynan. 1998. Specific regions of contact between human T-cell leukemia virus type I Tax protein and DNA identified by photocross-linking. J. Biol. Chem. 273:13768–13775. [DOI] [PubMed] [Google Scholar]
- 40.Kiyokawa, T., M. Seiki, S. Iwashita, K. Imagawa, F. Shimizu, and M. Yoshida. 1985. p27x-III and p21x-III, proteins encoded by the pX sequence of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. USA 82:8359–8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kraus, W. L., and J. T. Kadonaga. 1998. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 12:331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kraus, W. L., E. T. Manning, and J. T. Kadonaga. 1999. Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol. 19:8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kundu, T. K., V. B. Palhan, Z. Wang, W. An, P. A. Cole, and R. G. Roeder. 2000. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell 6:551–561. [DOI] [PubMed] [Google Scholar]
- 44.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380:642–646. [DOI] [PubMed] [Google Scholar]
- 45.Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger, R. G. Brennan, S. G. Roberts, M. R. Green, and R. H. Goodman. 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223–226. [DOI] [PubMed] [Google Scholar]
- 46.Lee, D. Y., J. J. Hayes, D. Pruss, and A. P. Wolffe. 1993. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 72:73–84. [DOI] [PubMed] [Google Scholar]
- 47.Lemasson, I., and J. K. Nyborg. 2001. Human T-cell leukemia virus type I Tax repression of p73β is mediated through competition for the C/H1 domain of CBP. J. Biol. Chem. 276:15720–15727. [DOI] [PubMed] [Google Scholar]
- 48.Lenzmeier, B. A., E. E. Baird, P. B. Dervan, and J. K. Nyborg. 1999. The tax protein-DNA interaction is essential for HTLV-I transactivation in vitro. J. Mol. Biol. 291:731–744. [DOI] [PubMed] [Google Scholar]
- 49.Lenzmeier, B. A., H. A. Giebler, and J. K. Nyborg. 1998. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol. Cell. Biol. 18:721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lenzmeier, B. A., and J. K. Nyborg. 1999. Molecular mechanisms of viral transcription and cellular deregulation associated with the HTLV-I Tax protein. Gene Ther. Mol. Biol. 3:327–345. [Google Scholar]
- 51.Li, Q., A. Imhof, T. N. Collingwood, F. D. Urnov, and A. P. Wolffe. 1999. p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J. 18:5634–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Libertini, L. J., J. Ausio, K. E. van Holde, and E. W. Small. 1988. Histone hyperacetylation. Its effects on nucleosome core particle transitions. Biophys. J. 53:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luger, K., T. J. Rechsteiner, A. J. Flaus, M. M. Waye, and T. J. Richmond. 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272:301–311. [DOI] [PubMed] [Google Scholar]
- 54.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304:3–19. [DOI] [PubMed] [Google Scholar]
- 55.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. S. Huang, J. P. Richards, R. G. Brennan, and R. H. Goodman. 1998. The human T-cell leukemia virus-1 transcriptional activator Tax enhances cAMP-responsive element-binding protein (CREB) binding activity through interactions with the DNA minor groove. J. Biol. Chem. 273:19251–19259. [DOI] [PubMed] [Google Scholar]
- 56.Manning, E. T., T. Ikehara, T. Ito, J. T. Kadonaga, and W. L. Kraus. 2001. p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21:3876–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayall, T. P., P. L. Sheridan, M. R. Montminy, and K. A. Jones. 1997. Distinct roles for P-CREB and LEF-1 in TCR alpha enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 11:887–899. [DOI] [PubMed] [Google Scholar]
- 58.McGhee, J. D., J. M. Nickol, G. Felsenfeld, and D. C. Rau. 1983. Histone hyperacetylation has little effect on the higher order folding of chromatin. Nucleic Acids Res. 11:4065–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morales, V., and H. Richard-Foy. 2000. Role of histone N-terminal tails and their acetylation in nucleosome dynamics. Mol. Cell. Biol. 20:7230–7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Naar, A. M., P. A. Beaurang, K. M. Robinson, J. D. Oliner, D. Avizonis, S. Scheek, J. Zwicker, J. T. Kadonaga, and R. Tjian. 1998. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 12:3020–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neish, A. S., S. F. Anderson, B. P. Schlegel, W. Wei, and J. D. Parvin. 1998. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 26:847–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953–959. [DOI] [PubMed] [Google Scholar]
- 63.Oliva, R., D. P. Bazett-Jones, L. Locklear, and G. H. Dixon. 1990. Histone hyperacetylation can induce unfolding of the nucleosome core particle. Nucleic Acids Res. 18:2739–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parekh, B. S., and T. Maniatis. 1999. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol. Cell 3:125–129. [DOI] [PubMed] [Google Scholar]
- 65.Pazin, M. J., P. L. Sheridan, K. Cannon, Z. Cao, J. G. Keck, J. T. Kadonaga, and K. A. Jones. 1996. NF-κB-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 10:37–49. [DOI] [PubMed] [Google Scholar]
- 66.Pelletier, G., V. Y. Stefanovsky, M. Faubladier, I. I. Hirschler-Laszkiewicz, J. Savard, L. I. Rothblum, J. Cote, and T. Moss. 2000. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol. Cell 6:1059–1066. [DOI] [PubMed] [Google Scholar]
- 67.Polesskaya, A., I. Naguibneva, A. Duquet, E. Bengal, P. Robin, and A. Harel-Bellan. 2001. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol. Cell. Biol. 21:5312–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Protacio, R. U., G. Li, P. T. Lowary, and J. Widom. 2000. Effects of histone tail domains on the rate of transcriptional elongation through a nucleosome. Mol. Cell. Biol. 20:8866–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Puri, P. L., M. L. Avantaggiati, C. Balsano, N. Sang, A. Graessmann, A. Giordano, and M. Levrero. 1997. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 16:369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scoggin, K. E., A. Ulloa, and J. K. Nyborg. 2001. The oncoprotein Tax binds the SRC-1-interacting domain of CBP/p300 to mediate transcriptional activation. Mol. Cell. Biol. 21:5520–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seiki, M., J. Inoue, T. Takeda, and M. Yoshida. 1986. Direct evidence that p40x of human T-cell leukemia virus type I is a trans-acting transcriptional activator. EMBO J. 5:561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheridan, P. L., C. T. Sheline, K. Cannon, M. L. Voz, M. J. Pazin, J. T. Kadonaga, and K. A. Jones. 1995. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 9:2090–2104. [DOI] [PubMed] [Google Scholar]
- 73.Simpson, R. T., J. P. Whitlock, Jr., M. Bina-Stein, and A. Stein. 1978. Histone-DNA interactions in chromatin core particles. Cold Spring Harbor Symp. Quant. Biol. 42:127–136. [DOI] [PubMed] [Google Scholar]
- 74.Sodroski, J., C. Rosen, W. C. Goh, and W. Haseltine. 1985. A transcriptional activator protein encoded by the x-lor region of the human T-cell leukemia virus. Science 228:1430–1434. [DOI] [PubMed] [Google Scholar]
- 75.Tse, C., and J. C. Hansen. 1997. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry 36:11381–11388. [DOI] [PubMed] [Google Scholar]
- 76.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Orden, K., and J. K. Nyborg. 2000. Insight into the tumor suppressor function of CBP through the viral oncoprotein Tax. Gene Expr. 9:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vettese-Dadey, M., P. A. Grant, T. R. Hebbes, C. Crane-Robinson, C. D. Allis, and J. L. Workman. 1996. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 79.Whitlock, J. P., Jr., and A. Stein. 1978. Folding of DNA by histones which lack their NH2-terminal regions. J. Biol. Chem. 253:3857–3861. [PubMed] [Google Scholar]
- 80.Wolffe, A. P. 1998. Chromatin: structure and function, 3rd ed. Academic Press, San Diego, Calif.
- 81.Yie, J., K. Senger, and D. Thanos. 1999. Mechanism by which the IFN-β enhanceosome activates transcription. Proc. Natl. Acad. Sci. USA 96:13108–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, W., S. Kadam, B. M. Emerson, and J. J. Bieker. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao, L. J., and C. Z. Giam. 1992. Human T-cell lymphotropic virus type I (HTLV-I) transcriptional activator, Tax, enhances CREB binding to HTLV-I 21-base-pair repeats by protein-protein interaction. Proc. Natl. Acad. Sci. USA 89:7070–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao, L. J., and C. Z. Giam. 1991. Interaction of the human T-cell lymphotrophic virus type I (HTLV-I) transcriptional activator Tax with cellular factors that bind specifically to the 21-base-pair repeats in the HTLV-I enhancer. Proc. Natl. Acad. Sci. USA 88:11445–11449. [DOI] [PMC free article] [PubMed] [Google Scholar]